- Oncology Department, Vall d’Hebron University Hospital, Barcelona, Spain
The phosphatidylinositol 3-kinases (PI3Ks) are members of a family of intracellular lipid kinases that phosphorylate the 3′-hydroxyl group of phosphatidylinositol and phosphoinositides. PI3K regulate signaling pathways for neoplasia, including cell proliferation, adhesion, survival, and motility. Different classes of PI3K have distinct roles in cellular signal transduction. PI3K pathway is activated by several different mechanisms in cancers, including, somatic mutation and gene amplification. In this review, we examine the literature addressing PI3K mutation status and gene amplification, with an emphasis on non-small cell lung cancer.
PI3K Classification
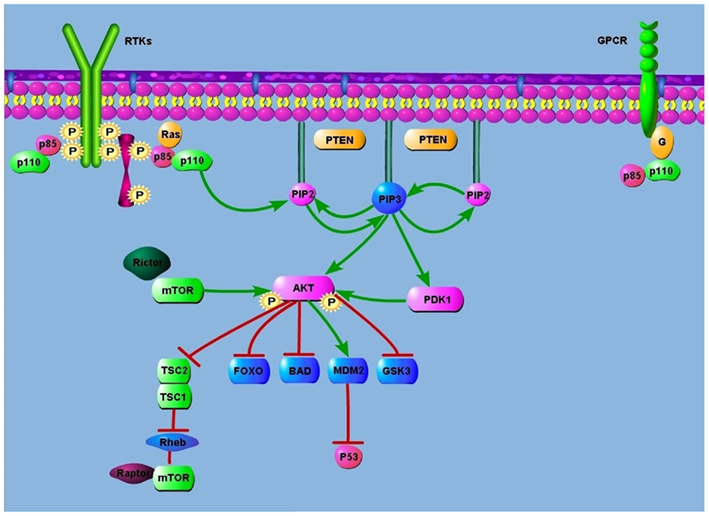
The phosphatidylinositol 3-kinases (PI3Ks) are members of a family of intracellular lipid kinases that phosphorylate the 3′-hydroxyl group of phosphatidylinositol and phosphoinositides. This reaction leads to the activation of many intracellular signaling pathways that regulate functions such as cell proliferation, adhesion, survival, and motility (Figure 1).
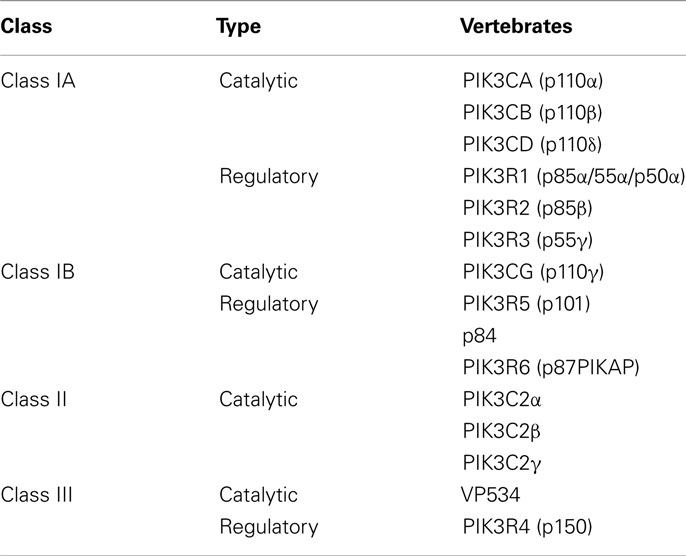
Phosphatidylinositol 3-kinases are grouped into three classes (I, II, III) according to their substrate preference and sequence homology. Different classes of PI3K have distinct roles in cellular signal transduction, as do the different isoforms that can exist within each class (Engelman et al., 2006).
In mammals, numerous genes encode different isoforms of PI3Ks (Fruman et al., 1998).
Class IA PI3K
This is the most clearly implicated class in human cancers (Yuan and Cantley, 2008). Class IA PI3K is a heterodimer that consists of a p85 regulatory subunit and a p110 catalytic subunit. Three genes, PIK3R1, PIK3R2, and PIK3R3 encode the p85α, p85β, and p55γ isoforms of the p85 regulatory subunit, respectively. The PIK3R1 gene also gives rise to two shorter isoforms, p55α and p50α. The Class IA p85 regulatory isoforms have a common core structure consisting of a p110-binding domain [also called the inter-Src-homology 2 (inter-SH2) domain] flanked by two SH2 domains (Huang et al., 2007). The p85 regulatory subunit is crucial in mediating the activation of class IA PI3K by receptor tyrosine kinases (RTKs).
Three genes – PIK3CA, PIK3CB, and PIK3CD – encode the highly homologous p110 catalytic subunit isoforms p110α, p110β, and p110δ, respectively (Cantley, 2002). The p110α subunit of PI3Kα has five domains: an N-terminal domain called adaptor binding domain (ABD), that binds to p85α, a Ras-binding domain (RBD), a domain called C2 that has been proposed to bind to cellular membranes, a helical domain, and a kinase catalytic domain (Vanhaesebroeck and Waterfield, 1999; Cantley, 2002; Huang et al., 2007).
In mammals, class I PI3Ks are involved in cellular growing, proliferation, motility, cell survival, and intracellular traffic. Class I PI3K also regulates glucose homeostasis: insulin receptor substrate.
Class IB PI3K
Class IB PI3K is a heterodimer consisting of a p101 regulatory subunit and a p110γ catalytic subunit. In 2005 two other regulatory subunits, p84 and p87 PIKAP (phosphoinositide 3-kinase gamma adapter protein of 87 kDa) were described.
Class II PI3K
Class II PI3K consists of only a p110-like catalytic subunit. The three isoforms of class II-PIK3C2α, PIK3C2β, and PIK3C2γ are encoded by distinct genes. Relatively little is known about the function of class II.
Class III PI3K
Class III PI3Ks consist of a single member, Vps34 (vacuolar sorting protein 34) which has been implicated in phagocytosis in the system immune and in traffic proteins and vesicles.
PI3K Signaling Cascade
All major elements of the PI3K pathway have been found mutated or amplified in a broad range of cancers. The axis of PI3K signaling in cancer begins with engagement of growth factors by receptor tyrosine kinases (RTKs). These RTKs are often mutated, amplified, or overexpressed, causing aberrant PI3K activation. For example, PI3K is activated by epithelial growth factor receptor (EGFR) in lung cancers harboring somatic activating mutations in EGFR (Skolnik et al., 1991; Engelman and Settleman, 2008). In this cancer, EGFR directly binds and activates PI3K. The regulatory subunit, p85, directly binds to phosphotyrosine residues on RTKs and/or adaptors (Carpenter et al., 1993). This binding relieves the intermolecular inhibition of the p110 catalytic subunit by p85 and localizes PI3K to the plasma membrane where its substrate, phosphatidylinositol 4,5-bisphosphate (PIP2), resides (Zhao and Vogt, 2008). PI3K can also be stimulated by activated Ras, which directly binds p110 (Shaw and Cantley, 2006). Additionally, the p110β catalytic subunit can be activated by G-protein coupled receptors (Katso et al., 2001).
Phosphatidylinositol 3-kinases is then recruited to plasma membrane-anchored receptors and is activated and phosphorylates PIP2 on the 3′OH position to produce phosphatidylinositol 3,4,5-trisphosphate (PIP3). The tumor suppressor phosphatase and tensin homolog deleted on chromosome 10 (PTEN) negatively regulates PI3K; dephosphorylates PIP3 to PIP2, thereby terminating PI3K-dependent signaling (Courtney et al., 2010). PIP3 propagates intracellular signaling by directly binding pleckstrin homology (PH) domains of various signaling proteins (Cantley, 2002). Phosphatidylinositol-3,4,5-trisphosphate (PIP3) propagates intracellular signaling as a second messenger activating many downstream molecules. The protein serine/threonine kinase AKT (also known as PKB) is a principal target of PIP3 (Franke et al., 1997). Binding of PIP3 to AKT leads to the membrane recruitment of AKT and subsequent phosphorylation by the mammalian target of rapamycin (mTOR)-rictor kinase complex and by 3-phosphoinositide-dependent kinase (PDK1; Sarbassou et al., 2005). The full activation of AKT phosphorylates many target proteins, such as forkhead (FOXO) family of transcription factors.
AKT promotes cell survival by inhibiting pro-apoptotic Bcl2 family members BAD and BAX (Cantley, 2002). AKT also can phosphorylate MDM2 leading to p53 degradation (Vivanco and Sawyers, 2002). AKT phosphorylates and inactivates the FOXO family of transcription factors. FOXO proteins promote the expression of pro-apoptotic genes, such as Bim and Fas and p27Kip and retinoblastoma-like2 (RBL2) to inhibit cell-cycle entry and cell survival.
AKT mediates cell metabolism by activating glycogen synthase through the inhibition of glycogen synthase kinase 3 (GSK3). AKT regulate protein synthesis by phosphorylating the tuberous sclerosis complex 2 (TSC2) protein tuberin, and therefore inhibits the GTPase-activating protein (GAP) activity of the TSC1–TSC2 complex toward Rheb. This allows GTP-bound Rheb to accumulate and activate the mTOR-raptor kinase complex, which in turn mediates phosphorylation of 4E-BP1 and p70, ultimately leading to increased protein synthesis (Richardson et al., 2004).
Implications for Diabetes
The p85 regulatory subunit is required for the stabilization of p110 and for the activation of PI3K by the insulin receptor. A partial reduction in p85 levels leads to improved PI3K signaling and increased insulin sensitivity in vivo. PI3K signaling mediates different cellular responses depending on the tissue context, and defective PI3K signaling in many tissues contributes collectively to the complex metabolic defects associated with type-2 diabetes (Hansen et al., 1997; Barroso et al., 2003). Elevated levels of p85 have been observed in women with pregnancy-induced insulin resistance (Kirwan et al., 2004). Similarly, elevated levels of p85, but not p110, were observed in muscles of type-2 diabetic individuals, indicating that increased levels of p85 might contribute to muscle insulin resistance in diabetes.
Frequency of Mutations and Amplifications in the PI3K Pathway in NSCLC
Receptor tyrosine kinases upstream of PI3K, the p110α catalytic subunit of PI3K, the downstream kinase, AKT, and the negative regulator, PTEN, are all frequently altered in cancer. The PIK3CA gene that encodes p110α is also amplified at high frequencies in squamous cell lung carcinoma (Engelman et al., 2006). PIK3CA and PIK3R1 are somatically mutated in cancers, and these mutations promote activation of the PI3K pathway (Ikenoue et al., 2005).
Huang et al. (2007) reported a 3.0-Å resolution structure of a complex between p110α and a polypeptide containing the p110α-binding domains of p85α, a protein required for its enzymatic activity (Vanhaesebroeck and Waterfield, 1999). The structure showed that many of the mutations occurred at residues lying at the interfaces between p110α and p85α or between the kinase domain of p110α and other domains within the catalytic subunit. The two most common genetic mutations that directly activate the PI3K signaling pathway are somatic activating mutations of p110α (PIK3CA) and loss of the tumor suppressor PTEN.
Additionally, amplification of PIK3CA and AKT are occasionally observed in epithelial cancers (Engelman et al., 2006). In non-small cell lung cancer (NSCLC), mutations in PIK3CA and PTEN are uncommon, although there are reports demonstrating evidence for loss of PTEN protein expression and PIK3CA amplification (Massion et al., 2002; Marsit et al., 2005). Somatic mutations in PIK3CA have been identified in a variety of human tumors, including NSCLC. Most of these mutations in p110α cluster to two hot spot regions in exons 9 and 20 (>75%). Exon 20 encodes the catalytic domain of p110α (H1047R); exon 9 encodes the helical domain of p110α (E545K; Samuels and Velculescu, 2004). A smaller cluster of mutations is also found in the N-terminal p85 interacting domain. While activating mutations in PIK3CA have been identified in NSCLC, no oncogenic mutations have been verified in p110β, p110δ, or the class IB catalytic isoform p110γ. The expression of these p110α mutants in cells confers AKT activation in the absence of growth factor stimulation (Samuels et al., 2005).
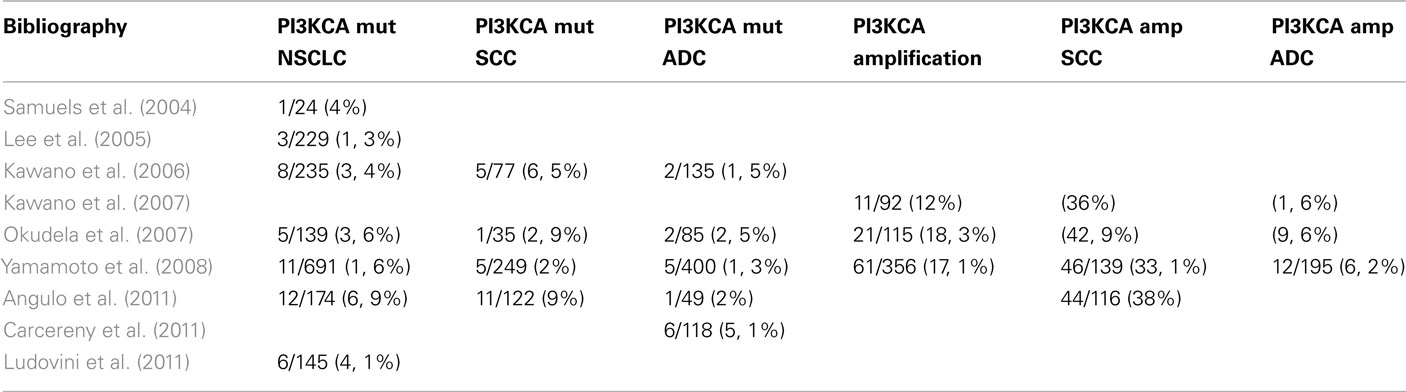
Samuels et al. (2004) sequenced PI3K genes in human cancers and corresponding normal tissue and identified 8 PI3K and 8 PI3K-like genes. Sequences containing the kinase domain of identified PI3Ks were extracted from the Celera or Public draft human genome sequences. Primers for PCR amplification and sequencing were designed using the primer 3 program. They examined the sequences in a total of 396 tumors. Mutations in PIK3CA were identified in 1 (E545K) of 24 lung cancers (4%); and >75% of alterations occurred in two small clusters in the helical and kinase domains. Data suggest that mutant PIK3CA was likely to function as an oncogene in human cancers.
Lee et al. (2005) analyzed somatic mutations of PIK3CA gene (exons 9, 20) in the 668 tissue samples from gastric, breast, and hepatocellular carcinomas, acute leukemia, and NSCLC. The mutational analysis based on PCR, single-strand conformation polymorphism (SSCP) analysis, and sequencing analysis ensures the specificity of the results. They analyzed 229 NSCLC: 111 squamous cell carcinomas, 108 adenocarcinomas, and 10 large-cell carcinomas and detected PIK3CA somatic mutations in 3 of 229 NSCLC (1.3%). No significant correlation was found between PIK3CA mutations and the histologic subtypes of NSCLC. PIK3CA mutation hot spots, E545K, and H1047R, were detected in 50% of samples.
Gymnopoulos et al. (2007) suggested three groups of PIK3CA mutants, defined by their location in distinct functional domains of the protein. They hypothesized that these three groups could induce a gain in PI3K function by a different molecular mechanism. Kawano et al. (2006) genotyped the PIK3CA gene in Japanese lung cancer patients. The study included 235 lung cancer specimens obtained at lung cancer surgery at Nogoya Hospital from 1997 to 2003. The two PIK3CA mutation hot spots (exon 9, 20) were analyzed by real-time PCR, and then confirmed by direct sequencing. In exon 9, somatic mutations were found in eight patients (3 E542K, 3 E545K, 1 E542Q, 1 Q546K); in exon 20 there were no mutations. Around the eight mutations that objectified (8/235, 3.4%), two were adenocarcinomas, five were squamous cell carcinomas, and one adenosquamous carcinoma. PIK3CA mutation incidence was significantly lower in adenocarcinoma (2/135; 1.5%) than in squamous cell carcinoma (5/79; 6.5%, p = 0.0495). Of the eight patients with PIK3CA mutation, three also harbored EGFR mutations. PIK3CA mutations did not correlate with gender, age, or smoking status. Overall, there was no significant difference in survival between patients with PIK3CA wild type (N = 227, 77 died) and patients with PIK3CA mutation (N = 8, 4 died).
Summary
The same group in 2007 investigated PIK3CA copy number in NSCLC (Kawano et al., 2007). They included 92 Japanese lung carcinoma patients who had undergone surgery at Nagoya Hospital. PIK3CA copy number was analyzed by quantitative real-time PCR; PIK3CA amplification (increased PIK3CA copy number) was defined as >3copies. Incidence of PIK3CA amplification was 12% (11/92). Among the 11 patients with PIK3CA amplification, 2 harbored PIK3CA mutations (E542Q, E545K). A correlation between PIK3CA amplification and PIK3CA mutation was not found (p = 0.7534). Among the 11 patients with PIK3CA amplification, no EGFR mutation was found. PIK3CA amplification status was significantly different in regard to: gender (male 11/63 versus female 0/29; p = 0.040), smoking history (current/former 11/59 versus never 0/33; p = 0.021), histology (squamous 10/28 versus adenocarcinoma 1/64; p = 0.0001). Overall survival of 92 patients in regard to PIK3CA amplification status showed a significant difference in survival between patients with PIK3CA normal copy number (N = 81, 37 died) versus patients with PIK3CA amplification (N = 11, 9 died), Log-rank test p = 0.0045. Using cox regression model, only pathologic stage (p = 0.0337) but not PIK3CA amplification (p = 0.2655) was a prognostic factor.
Okudela et al. (2007) analyzed samples from 148 Japanese patients with lung cancer who were surgically treated at Hamamatsu Hospital and Mikatahara Hospital from 1997 to 2006. Fragments of PI3K were analyzed by PCR; DNA sequence was analyzed from 139 of the 148 tissues. PIK3CA mutations were found in 5/139 patients (3.6%). Copy number gains of PIK3CA locus were found in 21/115 patients by FISH (18.3%). No patients were found to harbor both PI3KCA mutation and alteration in copy number.
Yamamoto et al. (2008) analyzed 691 tumor samples from patients from Japan (N = 323), Taiwan (N = 148), USA (N = 150), Australia (N = 70) who underwent surgical resection. They identified PIK3CA mutations in 11/691 (1.6%). Mutations occurred in the following histological subtypes: 5 of 249 (2%) squamous cell carcinoma, 5 of 400 (1.3%) adenocarcinoma, and 1 of 42 (2.4%) other NSCLC. Sufficient DNA was available from 356 of these tumors for PIK3CA gene copy number analysis by real-time quantitative PCR which was detected in 61/356 (17.1%): squamous cell carcinoma 46/139 (33.1%) and adenocarcinoma 12/195 (6.2%).
Angulo et al. (2011) analyzed PIK3CA gene mutations (exon 9, 20) in 178 NSCLC: 123 squamous cell carcinoma, 51 adenocarcinoma, and 4 large-cell carcinoma. Screening PIK3CA gene mutation by PCR and direct sequencing was performed in 174. They identified 12 PIK3CA mutations (6.9%); in squamous cell carcinoma 11/122 (9%), and in adenocarcinoma 1/49 (2%). The analyses of PIK3CA gene amplification by FISH was limited to squamous cell carcinoma and identified in 44 cases (38%). Tumors with PI3KCA mutation do not always display amplification of the gene; only 2.6% of the samples had both alterations concomitantly. These results would indicate a complementary relationship between PIK3CA amplification and mutations in NSCLC.
Carcereny et al. (2011) examined the presence and potential influence of PIK3CA mutations on outcome in 118 NSCLC patients with EGFR mutations treated with erlotinib. They detected six PIK3CA mutations (5.1%); 84% of patients had adenocarcinoma. The response rate was 50% for patients with PIK3CA mutation versus 70% for those with PIK3CA wild type (p = 0.03). A non-significant trend toward shorter progression-free survival was observed in the six patients with PIK3CA mutations (9 versus 16 months; p = 0.59).
Ludovini et al. (2011) realized a retrospective analysis to investigate the role of PIK3CA, EGFR, and KRAS gene mutations in predicting response and survival in 166 NSCLC patients treated with EGFR-TKIs. PIK3CA (exons 9, 20), EGFR, and KRAS mutations were analyzed using PCR. Of 166 patients, PIK3CA mutations were evaluated in 145 with 6 found to have PIK3CA mutations (4.1%). One adenocarcinoma patient with PIK3CA mutation (M1043I) had EGFR mutation (selection 19). PIK3CA mutation correlated with shorter median time-to-progression (2.3 months in mutated versus 6 months in wild type, HR 2.9, CI 0.66–12.8; p = 0.01), and worse overall survival (9.9 months in mutated versus 30.2 months in wild type, HR 4.76, CI 0.85–26.57; p < 0.001). The authors suggested that PIK3CA seems to be an indicator of poor survival in patients with NSCLC treated with EGFR-TKIs.
In conclusion, several studies have analyzed the PI3K pathway in NSCLC and reported frequent alterations. At present ongoing studies are addressing the role of PI3K inhibitors in NSCLC in the hope that they may lead to targeted therapies in the not too distant future.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Angulo, B., Conde, E., Suarez-Gauthier, A., Garcia-Lujan, R., Sanchez-Cespedes, M., Lopez-Encuentra, A., Paz-Ares, L., and Lopez-Rios, F. (2011). PIK3CA alterations in lung squamous cell carcinomas: role for mutations and amplification. IASLC. abstr. 037.06, Amsterdam.
Barroso, I., Luan, J., Middelberg, R. P. S., Harding, A. H., Franks, P. W., Jakes, R. W., Clayton, D., Schafer, A. J., O’Rahilly, S., and Wareham, N. J. (2003). Candidate gene association study in type 2 diabetes indicates a role for genes involved in β-cell function as well as insulin action. PLoS Biol. 1, e20. doi:10.1371/journal.pbio.0000020
Carcereny, E., Molina, M. A., Sanchez, J. J., Bertran-Alamillo, J., Mayo, C., Aldeguer, E., Gimenez-Capotan, A., Yeste, Z., Costa, C., Benlloch, S., Martinez, A., Buges, C., Bosch, J., Isla, D., Domine, M., Provencio, M., Sanchez, J. M., Camps, C., Taron, M., and Rosell, R. (2011). Mutations of the catalytic subunit α of PI3K (PIK3CA) in erlotinib-treated non-samll-cell lung cancer (NSCLC) patients (p) with epidermal growth factor receptor (EGFR) mutations. J. Clin. Oncol. 29(Suppl.) abstr. 7588.
Carpenter, C. L., Auger, K. R., Chanudhuri, M., Yoakim, M., Schaffhausen, B., Shoelson, S., and Cantley, L. C. (1993). Phosphoinositide 3-kinase is activated by phosphopeptides that bind to the SH2 domains of the 85-KDa subunit. J. Biol. Chem. 268, 9478–9483.
Courtney, K. D., Corcoran, R. B., and Engelman, J. A. (2010). The PI3K pathway as drug target in human cancer. J. Clin. Oncol. 28, 1075–1083.
Engelman, J. A., Luo, J., and Cantley, L. C. (2006). The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat. Rev. Genet. 7, 606–619.
Engelman, J. A., and Settleman, J. (2008). Acquired resistance to tyrosine kinase inhibitors during cancer therapy. Curr. Opin. Genet. Dev. 18, 73–79.
Franke, T. F., Kaplan, D. R., Cantley, L. C., and Toker, A. (1997). Direct regulation of the AKT proto-oncogene product by phosphatidylinositol-3,4-bisphosphate. Science 275, 665–668.
Fruman, D. A., Meyers, R. E., and Cantley, L. C. (1998). Phosphoinositide kinases. Annu. Rev. Biochem. 67, 481–507.
Gymnopoulos, M., Elsliger, M. A., and Vogt, P. K. (2007). Rare cancer-specific mutations in PIK3CA show gain of function. Proc. Natl. Acad. Sci U.S.A. 104, 5569–5574.
Hansen, T., Andersen, C. B., Echwald, S. M., Urhammer, S. A., Clausen, J. O., Vestergaard, H., Owens, D., Hansen, L., and Pedersen, O. (1997). Identification of a common amino acid polymorphism in the p85α regulatory subunit of phosphatidylinositol 3-kinase: effects on glucose disappearance constant, glucose effectiveness, and the insulin sensitivity index. Diabetes 46, 494–501.
Huang, C. H., Mandelker, D., Schmidt-Kittler, O., Samuels, Y., Velculescu, V. E., Kinzler, K. W., Vogelstein, B., Gabelli, S. B., and Amzel, M. (2007). The structure of a human p110α/p85α complex elucidates the effects of oncogenic PI3K mutations. Science 318, 1744–1748.
Ikenoue, T., Kanai, F., and Hikiba, Y. (2005). Functional analysis of PIK3CA gene mutations in human colorectal cancer. Cancer Res. 65, 4562–4567.
Katso, R., Okkenhaug, K., Ahmadi, K., White, S., Timms, J., and Waterfield, M. D. (2001). Cellular function of phosphoinositide 3-kinases: implications for development, homeostasis, and cancer. Annu. Rev. Cell Dev. Biol. 17, 615–675.
Kawano, O., Sasaki, H., Endo, K., Suzuki, E., Haneda, H., Yukiue, H., Kobayashi, Y., Yano, M., and Fujii, Y. (2006). PIK3CA mutation status in Japanese lung cancer patients. Lung Cancer 54, 209–215.
Kawano, O., Sasaki, H., Okuda, K., Yukiue, H., Yokoyama, T., Yano, M., and Fujii, Y. (2007). PIK3CA gene amplification in Japanese non-small cell lung cancer. Lung Cancer 58, 159–160.
Kirwan, J., Varastehpour, A., Jing, M., Presley, L., Shao, J., Friedman, J. E., and Catalano, P. M. (2004). Reversal of insulin resistance postpartum in linked to enhanced skeletal muscle insulin signalling. J. Clin. Endocrinol. Metab. 89, 4678–4684.
Lee, J. W., Soung, Y. H., Kim, S. Y., Lee, H. W., Park, W. S., Nam, S. W., Kim, S. H., Lee, J. Y., Yoo, N. J., and Lee, S. H. (2005). PIK3CA gene is frequently mutated in breast carcinomas and hepatocellular carcinomas. Oncogene 24, 1477–1480.
Ludovini, V., Bianconi, F., Pistola, L., Chiari, R., Minotti, V., Colella, R., Giuffrida, D., Tofanetti, F. R., Siggillino, A., Flacco, A., Baldelli, E., Iacono, D., Mameli, M. G., Cavaliere, A., and Crino, L. (2011). Phosphoinositide-3-kinase catalytic alpha and KRAS mutations are important predictors of resistance to therapy with epidermal growth factor receptor tyrosine kinase inhibitors in patients with advanced non-small cell lung cancer. J. Thorac. Oncol. 6, 707–715.
Marsit, C. J., Zheng, S., Aldape, K., Hinds, P. W., Nelson, H. H., Wiencke, J. K., and Kelsey, K. T. (2005). PTEN expression in non-small-cell lung cancer: evaluating its relation to tumor characteristics, allelic loss, and epigenetic alteration. Hum. Pathol. 36, 768–776.
Massion, P. P., Kuo, W. L., Stokoe, D., Olshen, A. B., Treseler, P. A., Chin, K., Chen, C., Polikoff, D., Jain, A. N., Pinkel, D., Albertson, D. G., Jablons, D. M., and Gray, J. W. (2002). Genomic copy number analysis of non-small cell lung cancer using array comparative genomic hybridization: implications of the phosphatidylinositol 3-kinase pathway. Cancer Res. 62, 3636–3640.
Okudela, K., Sukuki, M., Kageyama, S., Bunai, T., Nagura, K., Igarashi, H., Takamochi, K., Suzuki, K., Yamada, T., Niwa, H., Ohashi, R., Ogawa, H., Mori, H., Kitamura, H., Kaneko, T., Tsuneyoshi, T., and Sugimura, H. (2007). PIK3CA mutation and amplification in human lung cancer. Pathol. Int. 57, 664–671.
Richardson, C. J., Schalm, S. S., and Blenis, J. (2004). PI3-kinase and TOR: PIKTORing cell growth. Semin. Cell Dev. Biol. 15, 147–159.
Samuels, Y., Diaz, L. A., Schmidt-Kittler, O., Cummins, J. M., Delong, L., Cheong, I., Rago, C., Huso, D. L., Lengauer, C., Kinzler, K. W., Vogelstein, B., and Velculescu, V. E. (2005). Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer Cell 7, 561–573.
Samuels, Y., and Velculescu, V. E. (2004). Oncogenic mutations of PIK3CA in human cancers. Cell Cycle 3, 1221–1224.
Samuels, Y., Wang, Z., Bardelli, A., Silliman, N., Ptak, J., Szabo, S., Yan, H., Gazdar, A., Powell, S. M., Riggins, G. J., Willson, J. K., Markowitz, S., Kinzler, K. W., Vogelstein, B., and Velculescu, V. E. (2004). High frequency of mutations of the PIK3CA gene in human cancer. Science 304, 554.
Sarbassou, D., Guertin, D. D., Ali, S. M., and Sabatini, D. M. (2005). Phosphorylation and regulation of AKT/PKB by the rector-mTOR complex. Science 307, 1098–1101.
Shaw, R. J., and Cantley, L. C. (2006). Ras, PI(3)K and mTOR signalling controls tumour cell growth. Nature 441, 424–430.
Skolnik, E. Y., Margolis, B., Mohammadi, M., Lowenstein, E., Fischer, R., Drepps, A., Ullrich, A., and Schlessinger, J. (1991). Cloning of PI3Kinase-associated p85 utilizing a novel method for expression/cloning of target proteins for receptor tyrosine kinases. Cell 65, 83–90.
Vanhaesebroeck, B., and Waterfield, M. D. (1999). Signaling by distinct classes of phosphoinositide 3-kinases. Exp. Cell Res. 253, 239.
Vivanco, I., and Sawyers, C. L. (2002). The phosphatidylinositol 3-kinase AKT pathway in human cancer. Nat. Rev. Cancer 2, 489–501.
Yamamoto, H., Shigematsu, H., Nomura, M., Lockwood, W. W., Sato, M., Okumura, N., Soh, J., Suzuki, M., Wistuba, I. I., Fong, K. M., Lee, H., Toyooka, S., Date, H., Lam, W. L., Minna, J. D., and Gazdar, A. F. (2008). PIK3CA mutations and copy number gains in human lung cancers. Cancer Res. 68, 6913–6921.
Yuan, T. L., and Cantley, L. C. (2008). PI3K pathway alterations in cancer. Variations on a theme. Oncogene 27, 5497–5510.
Keywords: PI3KCA, NSCLC, mutations, gene amplification, copy number, p110α, exon 9, exon 20
Citation: Martinez-Martí A and Felip E (2012) PI3K Pathway in NSCLC. Front. Oncol. 1:55. doi: 10.3389/fonc.2011.00055
Received: 29 September 2011;
Accepted: 12 December 2011;
Published online: 06 January 2012.
Edited by:
Oliver Gautschi, Luzerner Kantonsspital, SwitzerlandReviewed by:
Solange Peters, Centre Hospitalier Universitaire Vaudois, SwitzerlandSacha I. Rothschild, University Hospital Basel, Switzerland
Copyright: © 2012 Martinez-Martí and Felip. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Enriqueta Felip, Department of Medical Oncology Service, Vall d’Hebron University Hospital, Passeig Vall d’Hebron119-129, Barcelona 08035, Spain. e-mail:ZWZlbGlwQHZoZWJyb24ubmV0