Abstract
Retroviruses that have the ability to infect germ line cells can become an integral and inherited part of the host genome. About 8% of the human chromosomal DNA consists of sequences derived from infections by retroviruses that presumably circulated 2–40 millions of years ago, and some elements are actually much older. Post-insertional recombinations, deletions, and mutations have rendered all known human endogenous retroviruses (HERVs) non-infectious. However some, particularly the most recently acquired proviruses of the HERV-K(HML-2) family, can expresses viral proteins and produce viral particles. In this review we will first discuss the major aspects of the endogenization process and peculiarities of the different HERV-K families. We will then focus on the genes and proteins encoded by HERV-K(HML-2) as well as inactivation of these proviruses by postinsertional mutations and their inhibition by antiretroviral factors. After describing the evolutionary interplay between host and endogenous retrovirus we will delve deeper into the currently limited understanding of HERV-K and its possible association with disease, particularly tumorigenesis.
Introduction
Human chromosomes contain numerous retroviral sequences that form part of our genetic legacy. In total, human endogenous retroviruses (HERVs) sequences account for ∼8% of our genome. With more than 500,000 elements, there are over 20-fold more HERV sequences than there are human genes. These elements typically have their origin in ancient infections of germ cells by exogenous retroviruses that circulated ∼2–40 million years ago. The human genome and the genomes of other vertebrate species that contain endogenous retroviruses can therefore be regarded as museums of ancient retroviruses. Phylogenetic studies have allowed these elements to be sorted into distinct groups that are usually termed “families.” Similar to artifacts in an archeological museum, time has left its traces on these retroviral elements and very often only “shards” of the once complete sequence remain. All known proviruses in the human genome have suffered from postinsertional mutations, deletions, and recombinations that render them non-infectious. Most often, only a single long terminal repeat (LTR) remained. In contrast to humans, several other species including mice and sheep carry infectious endogenous retroviruses (1–3).
The most recently integrated and best preserved human proviruses belong to a subgroup of the HERV-K elements dubbed HERV-K(HML-2). The HERV-K group can be divided into 10 families (4). The designation “K” comes from their use of a lysine tRNA to prime reverse transcription and human mouse mammary tumor virus like-2 (HML-2) indicates their relationship to the murine betaretrovirus mouse mammary tumor virus (MMTV). More than 90 proviruses of the HML-2 family are exceptionally well preserved and maintain open reading frames encoding functional viral proteins (4). These proteins are expressed and non-infectious particles are released in various cells, particularly in tumors and cancer cell lines derived from teratocarcinomas and melanomas (5–7). Moreover, two research groups have demonstrated that consensus sequences of HERV-K(HML-2) elements are able to produce infectious particles (8, 9). The infection and replication of these particles is inhibited by a variety of restriction factors including members of the APOBEC family, tetherin and most likely other, yet to be identified antiviral factors that have evolved over millions of years of coevolution between this virus and its primate hosts.
HERV-K proviruses can be classified into two types. Only type II proviruses express the accessory Rec protein that functionally resembles the HIV Rev and HTLV Rex proteins (10, 11). Type I proviruses have a 292-bp deletion that prevents expression of the rec gene but can instead express an alternative protein called Np9. Both Rec and Np9 are associated with malignancies. In addition to the evidence for the protein-induced or promoted tumorigenesis of somatic cells, HERV-K elements are also implicated in cancer development at the DNA or RNA level. In this context, HERV-K proviruses have been shown to be involved in recombination events that in one setting left an LTR-promoter closely upstream of exons 5–12 of the ETV1 gene that encodes a transcription factor of the Ets family. Androgen-dependent expression of the protein fragment driven by the viral LTR was correlated with an increase in the invasiveness of prostate cancer (12). Promoters of other HERV elements and retrotransposons have also been shown to stimulate expression of nearby genes that promote malignancy (13). The number of known genes that are under definite transcriptional control by HERVs and our understanding of the mechanisms involved have dramatically increased in recent years (14).
In this regard it should be stressed that transcription of HERV-K and other HERV elements is usually suppressed by epigenetic factors such as DNA methylation and heterochromatin-silencing by histone modifications (15). Nevertheless, it is estimated that even in normal healthy cells, retroviral LTRs initiate 10 times as many transcripts as regular promoters (16–19). The transcripts induced include many RNAs that are complementary to regular gene transcripts and can downregulate the expression of cellular genes (17). During tumorigenesis (and also, presumably, in other poorly understood situations) this epigenetic control can be partially or selectively weakened and particular retroviral promoters become able to vigorously activate the transcription of cellular genes.
Endogenization of Retroviruses with a Focus on HERV-K(HML-2)
The prerequisite for acquisition of a new endogenous retrovirus is infection of a germ line cell that results in integration of the proviral DNA into the cell’s chromosomal DNA. If this integration and the possible replication of this founder virus do not prevent fertilization, a fetus can form that carries the retroviral element in every one of its somatic and germ line cells. From a genetic perspective, the provirus becomes a regular host gene that is passed down the generations, being inherited in a Mendelian fashion (20–22). Such endogenization and transfer to offspring has been demonstrated in the laboratory by infection of fertilized eggs with recombinant avian leucosis virus (23). The viremia in the carriers can be attributed to replication of either the exogenous or the endogenous form, which results in repeated infection of somatic cells. It can be assumed that frequent recombination events occur at that stage by copackaging of vRNAs, one from an inherited endogenous element and one from an infecting exogenous virus with template switching by the reverse transcriptase (RT) occurring during subsequent infection of a new host cell. To some degree, germ cells might also be repeatedly infected, leading to further colonization of the germ line and amplification of the number of viral elements belonging to the same family. Other modes of amplification are also conceivable and have been documented, including retrotransposition or duplication of chromosomal regions that contain HERV elements (24). However, in the case of HERV-K(HML-2) it appears that the predominant mode of amplification within the human genome has been reinfection by circulating infectious viruses over a period of at least 30–40 million years (25). Colonization of the human genome by this betaretrovirus did not occur at the same frequency all the time: there were bursts of integration, during which the replicative activity of the virus is assumed to have been high, interspaced with longer periods of low integration (25). In recent years the number of known human-specific HERV-K(HML-2) loci has risen (4, 10, 26) and the lack of these loci in chimpanzees, our closest primate relatives, proves that elements of this family continued to reinfect our ancestors after their split from the chimpanzee lineage about five to six million years ago. Although there is no evidence for it being ongoing in humans, retroviral integration is not something that only occurred in the distant past. In Australia we are currently witnessing a fulminant endogenization process in koala bears involving a retrovirus presumed to have been acquired from a species of Asian mouse only a hundred or so years ago (27–29).
The reproductive success of individuals carrying a germ line provirus at a particular chromosomal locus determines the frequency of this element in the population, i.e., it is subject to host selection (Figure 1). Selective forces and fluctuations in the population size (first and foremost bottlenecks that simply by chance increase the relative ratio of the carriers in the population) might eventually result in fixation of the element. This means it is present in all members of the population. It can be inferred that the likelihood of an element reaching fixation correlates negatively with its virulence if it compromises the reproductive success of the carrier. It is therefore likely that neutral elements or elements that even endow the carrier with a selective advantage are more likely to reach fixation whereas those with a negative impact will not and will eventually vanish. One piece of solid evidence in favor of this hypothesis is the orientation of the endogenous elements found in introns. Although integration has no preference with regard to direction, the majority of HERVs have an orientation opposite to that of the gene they have integrated next to, usually rendering the splice sites and other regulatory elements inactive. Elements that occurred in the same orientation as the gene vanished with higher frequency because they can modify expression of the encoded cellular protein to the detriment of the host. In contrast, there are only very few elements that are obviously under strong positive selective pressure, none of which belong to the HERV-K(HML-2) family (30).
Figure 1
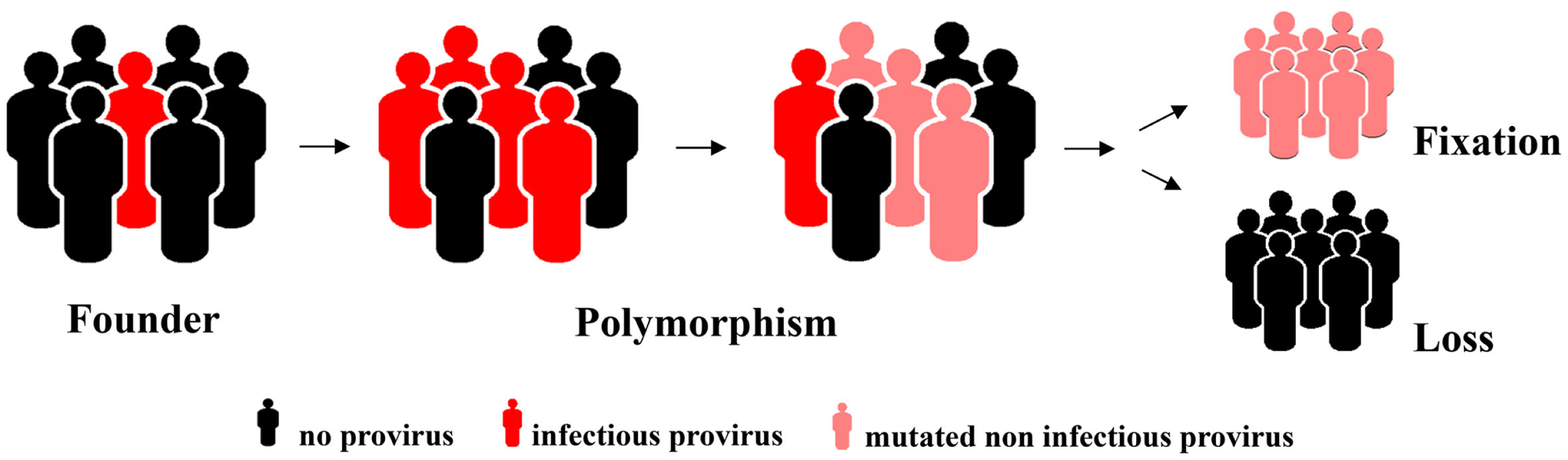
Hypothetical model for an increase in allelic frequency and eventual loss or fixation of an endogenized element integrated at a particular chromosomal locus.
HERV-K Families
To date, there is little known about members of the HERV-K superfamily other than HERV-K(HML-2). All HML families belong to the class II/betaretrovirus group, which also contain the endogenous and exogenous MMTV, Mason–Pfizer monkey virus (MPMV), and Jaagsiekte sheep retrovirus (JSRV) (31). Elements such as HML-4 and HML-6, which are related to but clearly distinct from the HERV-K(HML-2) prototype virus HERV-K10 (32), were identified more than 20 years ago by extensive lab work using low-stringency Southern blot analyses with MMTV-derived gag/pol probes (33). These data were later complemented by PCR and the sequences were grouped into the HERV-K families HML-1 to HML-6, based on the sequences of a conserved RT region (34). These results were subsequently compiled to yield our current understanding that there are up to 10 HERV-K(HML) families in addition to a related family HERV-K(14C) present in the human genome (35). As the complete human genome has now been sequenced, identification of HERV elements can now be performed with relative ease by database analyses. Searching for sequences derived from the cloned HERV-K (HML-1 to HML-10) elements reveals a probable total of about 550 HERV-K proviruses and 6400 solitary LTRs in the human genome (36), ranging from 150 HML-3 proviral elements to 5 HML-4 proviruses, for example (24, 37). Not all HML families are described in detail using in silico generated consensus sequences or by analysis of individual elements, a summary of the features of HERV-K families is given in Table 1, although these numbers may need to be revised as was recently done for HML-2 solo LTRs, being adjusted from ∼2500 to 944 (4, 38).
Table 1
| Name | Alternative names | Estimated time of integration (mya) | Estimated copy number in genome | Reference | |
|---|---|---|---|---|---|
| Proviruses | Solo LTRs* | ||||
| HML-1 | NMWV6, HERV-K14I | ∼39 | 68 | 350 | Franklin et al. (33), Medstrand and Blomberg (34), Flockerzi et al. (41) |
| HML-2 | NMWV1, HDTV | ∼35–2 | 91 | 944 | Subramanian et al. (4), Bannert and Kurth (24), Ono (42) |
| HML-3 | NMWV5, HERV-K9I | ∼36 | 140 | 700 | Franklin et al. (33), Medstrand and Blomberg (34), Mayer and Meese (43) |
| HML-4 | HERVK13I, HERV-K-T47D | ∼55–35 | 5 | 800 | Medstrand and Blomberg (34), Seifarth et al. (37) |
| HML-5 | NMWV2, HERV-K22I | ∼55 | 139 | 600 | Franklin et al. (33), Lavie et al. (40) |
| HML-6 | NMWV4, HERV-K3I | ∼30 | 30–40 | 50 | Franklin et al. (33), Medstrand et al. (44), Yin et al. (45) |
| HML-7 | NMWV7, HERV-K11D1 | n.d. | 20 | 140 | Franklin et al. (33), Andersson et al. (35) |
| HML-8 | NMWV3, HERV-K11I | n.d. | 60 | 600 | Franklin et al. (33), Andersson et al. (35) |
| HML-9 | NMWV9 | n.d. | 10 | 40 | Franklin et al. (33), Andersson et al. (35) |
| HML-10 | HERV-K(C4) | ∼30 | 10–50 | 100 | Jern et al. (31), Tassabehji et al. (46) |
Class II/HERV-K (human MMTV-like) families.
* Most copy numbers of solo LTRs given in Ref (24); mya, million years ago; n.d., not determined.
The oldest group within the HERV-K superfamily appears to be the HERV-K(HML-5) family. Based on the traditional criterion for naming the HERV elements, HML-5 proviruses could constitute the HERV-M family, as the primer binding site of the consensus sequence is identical to the 3′ end of methionine tRNA. These elements are not found in prosimians, and are the only HERV-K proviruses in both Old World and New World monkeys, suggesting that the integration event must have taken place 35–55 million years ago. An even more precise estimate for the time of integration can be made by examining the LTRs. At that point in time, both LTRs in the viral genome would have been identical, but in the host genome they would accumulate random mutations at a rate of 2.3–5 × 10−9 substitutions per site per year (39), i.e., one difference between the two LTRs would be generated every 200,000–450,000 years. Although this kind of estimation has several limitations, including the fact that there were certainly periods of varying mutational rates, the integration of HML-5, based on LTR–LTR divergence, is estimated to have occurred ∼55 million years ago. It is interesting to speculate whether the age of HML-5 makes it the ancestor of other HERV-K families. However, sequence comparisons show that HML-5 does not lie at the root of the HERV-K phylogenetic tree, suggesting that this is probably not the case (40).
Remarkably, within the HML-1 family seven proviruses were found in which gag, pro, and pol sequences have been replaced by similar regions from HERV-W, resulting in hybrid structures (41). Such recombination events can occur by template switching by RT or at the DNA level. However, the differing flanking cellular sequences indicate that at some point the initial hybrid provirus was able to form progeny proviruses in the genome through a transcript.
HERV Genes, Transcripts, Proteins, and Structure
To varying degrees, endogenous retroviruses are phylogenetically similar to exogenous retroviruses and it is therefore common to categorize them into three different classes. While class I HERV elements (e.g., HERV-W, HERV-FRD) have similarities to exogenous gammaretroviruses (e.g., MLV, GaLV) and class III HERV (e.g., HERV-L) are distantly related to lenti- or spuma-like retroviruses (e.g., HFV), the structure of a HERV-K(HML-1 to HML-10) element resembles a typical betaretrovirus (31). As HERV-K(HML-2) elements, in addition to the four major ORFs for gag, pro, pol, and env, also encode an accessory gene similar to those of delta-, lenti-, and spumaviruses they were defined as complex endogenous retroviruses. HML-2 type II proviruses encode an HIV-1 Rev- or HTLV-1 Rex-like protein termed Rec and only 26% of HML-2 elements are classified as type I expressing the protein Np9 (4, 47, 48). The genomic organization of these proviruses and their resulting RNA transcripts are shown in Figure 2.
Figure 2
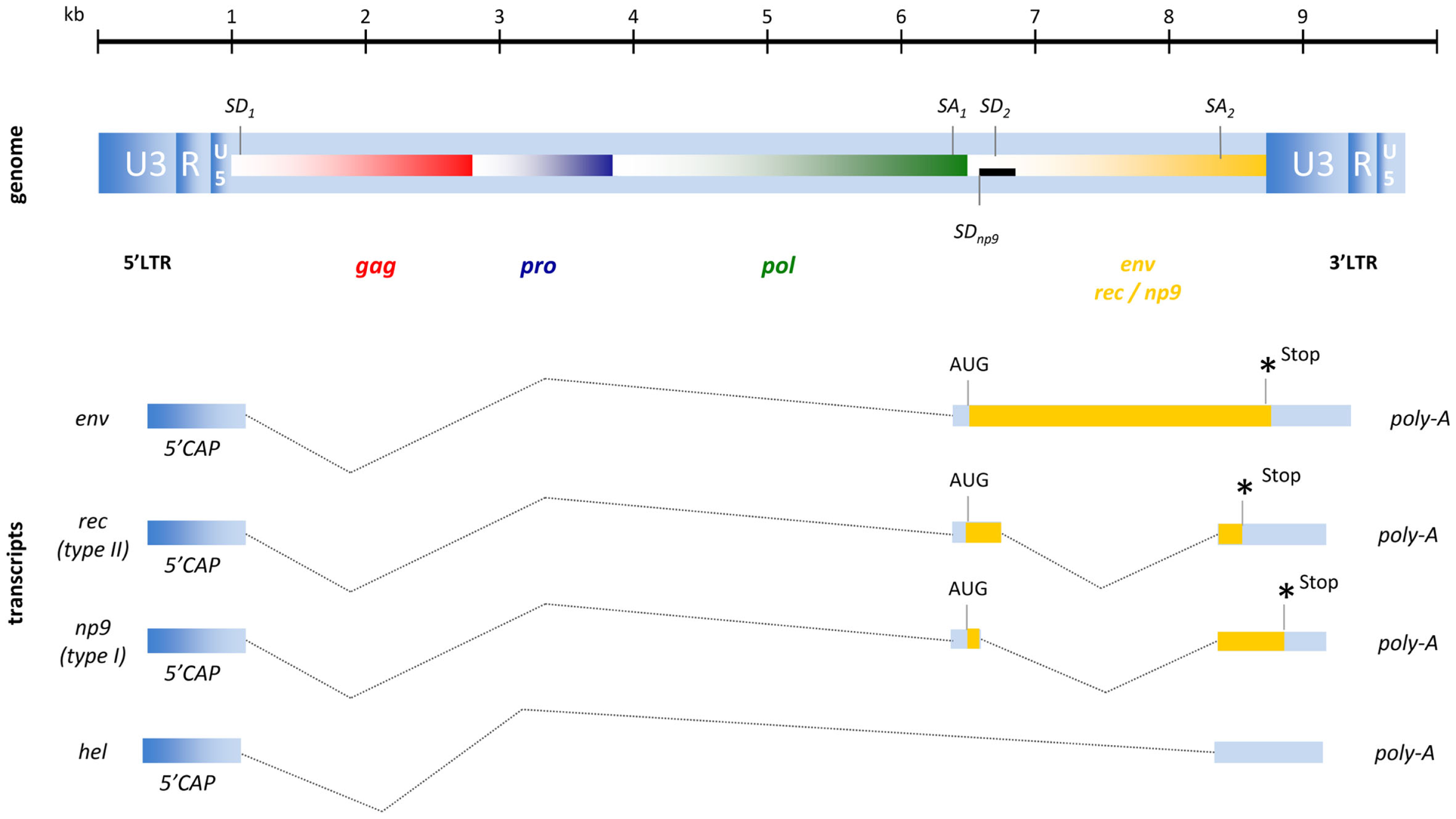
Proviral organization of HERV-K(HML-2) and RNA transcripts. The black bar represents a 292-bp deletion that results in the Np9 gene product for type I proviruses instead of the Rec (an HIV-1 Rev homolog) characteristic of the type II proviruses. Hel is a poorly described RNA without protein coding function.
The full-length HML-2 mRNA encodes the structural proteins of Gag, termed matrix (MA, p15), capsid (CA, p27), nucleocapsid (NC, p10) plus three smaller peptides as well as the putative phosphoprotein (pp15) (49). Due to two slippery sites and the resulting −1 bp ribosomal frameshifts, the Gag-Pro and Gag-Pro-Pol precursor proteins are produced and processed into the protease (Pro) enzyme and into polymerase (Pol) that exhibits RT and integrase (IN) activity. The single spliced mRNA for Env encodes the typical retroviral Env protein consisting of a signal peptide, a surface unit (SU; ∼44 kDa), and a transmembrane unit (TM; ∼26k Da). A functional Env protein has only been described for the HERV-K108 provirus (50), although the infectivity of another HERV-K(HML-2) element (K113) could be restored by back mutation of putative post-insertional amino acid changes (51). In addition to another two mRNAs coding for the accessory proteins Rec or Np9 that will be addressed in detail later, a 1.5-kb transcript termed hel has been found in human teratocarcinoma cell lines (48) although, as this mRNA does not encode a protein, its function requires clarification.
Some exogenous and endogenous retroviruses code for a dUTPase in addition to the enzymes essential for the retroviral life cycle such as RT, RNase H, Pro, and IN. dUTPase catalyzes the degradation of dUTP in order to prevent the toxic misincorporation of uracil into the newly forming proviral DNA. Both betaretroviruses and non-primate lentiviruses [i.e., Maedi-Visna virus (MVV) and equine infectious anemia virus (EIAV)] feature a dUTPase, albeit located in different regions of the genome (31, 52). The dUTPase in betaretroviruses is found within the N-terminal region of pro whereas in non-primate lentiviruses it is in pol.
Postintegrational Mutations and Replication Potential of HERV-K
It can be assumed that unless antiretroviral cellular factors of the APOBEC-superfamily were deaminating their genomes during reverse transcription, the majority of HERV-K(HML-2) elements were replication competent for some period of time after integration into the germ line cell genome. HERV-K(HML-2) is particularly susceptible to APOBEC 3F and APOBEC 3G deamination as shown in infectivity assays using infectious viruses produced with HERV-K(HML-2) consensus sequences (9, 53). Moreover, 2 of 16 (53) most recently integrated elements known are substantially mutated by APOBEC deaminases, indicating that these antiviral factors were already active several millions of years ago. How long elements that escape restriction by deaminases are usually able to produce infectious viruses is an open question. It is known that some endogenous retroviruses of pigs, sheep, and other animals have resided in their host’s genomes for tens or even hundreds of thousands of years and yet retain open reading frames and remain both replication competent and infectious (21). Some replication competent porcine endogenous retroviruses in Old World pig species are presumably even older (3).
It is known that during their residency in the host cell genome, endogenous elements acquire postinsertional mutations and deletions that can inactivate the provirus. Recombination events frequently take place that delete most of the proviral sequence, leaving just an LTR (solo LTR). With HERV-K, solo LTRs outnumber elements still framed by both LTRs by a factor of at least 10. In contrast to these single LTR remnants, HERV-K(HML-2) proviruses also exist that possess open reading frames for all viral genes. Such elements include variants (haplotypes) of HERV-K113 and HERV-K108 that are thought to be two of the most recently acquired elements (4, 26). However, post-insertional mutations other than those that generate premature stop codons have since rendered these viruses replication incompetent (36, 54): infection could not be achieved using supernatants from germ line cells that release HERV-K(HML-2) particles. Indeed, none of the currently known HERV-K(HML-2) elements appear to have the capacity to produce infectious particles. It is not known when the most recent HERV-K(HML-2) endogenization occurred and when the last provirus lost its ability to replicate. In fact, although unlikely, the existence of an infectious HERV-K(HML-2) provirus with low prevalence in the current human population cannot be completely ruled out.
Even if no functional HERV-K(HML-2) element is currently extant in humans, an infectious virus might nevertheless be reborn as a chimera of partially inactivated parental elements. That infectious retroviruses can form by recombination between the RNA genomes of two inactivated parental elements is well documented, with the two most recent occasions involving murine gammaretroviruses, including the notorious XMRV (55, 56). Similar events could also result in the reconstitution of a replication competent HERV-K(HML-2) element. Indeed, Heidmann and Co-workers have demonstrated in vitro the reconstitution of an infectious HERV-K(HML-2) element through recombination of three HERV-K(HML-2) elements (8). Compared to exogenous retroviruses, the infectivity of such reconstituted particles is rather poor and productive replication in cell culture has yet to be demonstrated. It is therefore likely that despite the demonstration of in vitro infection and integration, restriction factors including APOBEC have evolved to prevent productive replication in cell culture and in vivo. However, HERV-K(HML-2) does not appear to be a target of human Trim5alpha, an antiretroviral factor attacking the capsids of infecting retroviruses (9).
Role of HERV-K in Human Evolution and Physiology
In addition to the examples described in more detail below, transposable elements, including HERVs, generally drive alterations of the human genome by providing RT, an enzyme that can facilitate gene duplication for example, and by the addition or modulation of promoter activities. Even before the human genome was sequenced, it was suggested that constant updating of the transcriptional pattern through evolution of cis-acting elements might contribute more to species diversity than do changes in coding regions themselves (57). The simple insertion of an LTR can replace the original promoter, alter tissue specificity, or add enhancer activity (16). Of course, the LTRs acting as mobile genomic elements originate from full-length, inserted retroviruses.
Hypothetically, there are three possible outcomes of retroviral host genome invasion, i.e., genetic modification by the foreign DNA and/or the impact at the specific point of integration can be beneficial, harmful, or neutral for the individual. Retroviral integrations that severely harm the host genome, for example by interfering with the regulation of gene expression through promoter activity or direct destruction of coding regions, cannot play a significant role in human evolution because affected individuals are subject to negative selection and are removed from the gene pool. On the other hand, positive effects of altered gene expression through retroviral integration are well known. For example, the amylase gene is expressed in the pancreas of all mammalian species. However, in primates, rodents, and lagomorphs (rabbits, hares, and pikas), amylase is also expressed in the salivary gland as a result of gene duplication and insertion of a retroviral sequence (58), an event that probably helped these species to include starch in their diets.
Perhaps the most striking example of mammalian exploitation of retroviral sequences is the co-option of an endogenous retrovirus envelope gene to form the syncytiotrophoblast during pregnancy. The main function of the retroviral envelope protein is to mediate fusion of virus and target cell membranes during infection, making it an ideal “tool” to facilitate cell–cell fusion. The HERV-W (59) and HERV-FRD (60) glycoproteins, expressed exclusively in the placenta, are therefore termed syncytin-1 and -2, respectively. Furthermore, the envelope proteins are involved in retrovirus-induced immunosuppression (61) and as the placenta, as far as the mother’s immune system is concerned, is an allogeneic organ, local immunological tolerance is necessary to prevent rejection of the fetus. This capacity for immunosuppression, shown for syncytin-2, but so far not for syncytin-1 (59), may therefore be another reason that the endogenous retroviral env is expressed in the human placenta.
Further evidence for the deliberate co-option of retroviral genes during host evolution includes the presence of syncytin-A and -B genes in the murine genome, genes that have the same functions as their human counterparts but are clearly distinct at the sequence level (62). With mice it is possible to directly study the impact of these genes and “knocking out” syncytin-A results in disrupted placental architecture and the in utero death of embryos (63). The later discovery of syncytin genes in other mammals demonstrates that the exploitation of retroviral env genes has occurred independently and on multiple occasions throughout mammalian evolution. It is exciting to speculate that the first retroviral env integration in an oviparous species marked the beginning of the change from egg-laying prototherians to placental mammals (64).
A more direct advantage for the host of endogenization may be protection from infection by related exogenous pathogenic retroviruses. One well-studied example of this is sheep that carry an endogenous form of JSRV and are therefore protected against infection by exogenous JSRV (65) through receptor interference by the endogenous retroviral envelope. Furthermore, the Gag protein of defective transdominant endogenous JSRV interferes at a late stage of replication with the exogenous form of the protein (66), a phenomenon also described in mice in which expression of an endogenous retroviral Gag protein with homology to the HERV-L family blocks various strains of exogenous MLV (67). To date, no comparable function for HERV-derived proteins is known, but it is very plausible that the open reading frames retained for some HERVs serve a similar purpose.
Expression and Reactivation of HERV-K Loci in Health and Disease
Although HERV-K(HML-2) gene expression is largely repressed in most cells by epigenetic silencing, low-level expression occurs in some tissues. Various groups have found that between 7 and 30% of all HERV sequences in the genome are transcriptionally active (18, 68, 69). In particular, the testes and placenta appear to be privileged tissues for HERV expression (69). There is an ongoing and active debate about HERV expression in healthy tissues, with some authors assuming a possible advantage for cells that express HERV-K proteins (70). Expression of the HERV-K envelope protein TM subunit in placental cytotrophoblast cells suggests its potential involvement in placentogenesis and pregnancy (71), as discussed above for syncytin-1 and -2. Moreover, Wang-Johanning and Colleagues have detected low-level expression of HERV-K env mRNAs and proteins in healthy ovarian tissues (72).
Andersson and Colleagues made the very interesting discovery of HERV-K rec transcripts in the placenta and in various other normal human fetal tissues, suggesting that this small RNA transport molecule may play a role in the development and differentiation of human tissues (73).
Although HERV-K is expressed at only very low levels (if at all) in healthy tissues, activation of transcription, and protein production is associated with diseases such as cancer and autoimmune disorders. Indeed, the possible role played by upregulation of HERV-K expression in autoimmune diseases such as multiple sclerosis (MS) and rheumatoid arthritis (RA) is being investigated.
Rheumatoid arthritis is a chronic, systemic inflammatory disorder that especially affects synovial joints. Serological and molecular assays show a significant increase of HERV-K gag expression in RA patients compared to inflammatory and healthy control groups (74). Furthermore, exogenous viral protein expression and proinflammatory cytokines enhance HERV-K gag transcription. Finally, anti-HERV-K antibodies with possible cross-reactivity to host proteins could cause autoimmune responses through molecular mimicry (74).
Multiple sclerosis is more controversial, and although the etiology of this disease remains unknown it is likely that the trigger for MS is multifactorial. One interesting hypotheses involves a neurotropic infectious agent, possibly a virus, and HERV-K(HML-2) is one candidate in this context (75). Indeed, Tai and Colleagues found HERV-K18 expressing a superantigen in its env gene to be a risk factor for MS (76) and although this observation was made in a very small cohort, it is exciting because expression of HERV-K18 superantigen is also elevated in juvenile RA (77). There is also some evidence that, depending on the population, the LTR sequences of HERVs such as HRES-1 show polymorphisms within MS patient haplotypes while those from non-MS patients are identical without geographic restrictions (78). However, Moyes and Colleagues have published data showing that HERV-K113 and HERV-K115 are not significantly increased in patients with MS compared to the combined parent group (79, 80) and their results do not therefore support an association between these particular endogenous retroviruses and MS (79, 81). Finally, various groups have reported an association between HERV-K18 polymorphisms and type 1 diabetes (82, 83). In summary, a correlation between HERV-K expression and autoimmune disease is neither refuted nor proven. A definite conclusion will need to be based on a great deal more data.
In contrast to the situation with autoimmune diseases, the picture concerning HERV-K expression in malignant diseases is much clearer. As already mentioned, the expression of HERV-K(HML-2) in malignant tissues such as germ cell tumors (GCTs), melanomas, or ovarian cancers is considerably higher than in healthy tissues (5, 6, 70, 72, 84–86). A number of malignant diseases suggested to be associated with HERV-K expression are listed in Table 2. HERV-K expression was first observed in GCTs (87–89) and is often associated with the release of HERV-K(HML-2) particles. The frequent induction of an immune response to HERV-K(HML-2) Gag and Env proteins (90, 91) is well documented in patients with GCT disease. The presence of antibodies specific for HERV-K(HML-2) proteins correlates strongly with the clinical manifestation of GCTs and may therefore be used as a diagnostic and prognostic marker for these types of cancer (92). Interestingly, HERV-K expression in teratocarcinoma cell lines can be further enhanced by androgens (93), although, it is still unknown whether the androgen receptor (AR) binds directly to HERV-K LTRs or acts via other host proteins. Direct binding of other cellular proteins and transcription factors to the HERV-K(HML-2) LTR have already been demonstrated (94, 95).
Table 2
| Tissue | Cancer | HERV-K(HML-2) activity | Reference |
|---|---|---|---|
| Skin | Melanoma | Retroviral particles | Buscher et al. (6), Muster et al. (86), Hirschl et al. (124) |
| Enhanced transcription | |||
| RT activity | |||
| Expression of Env, Rec, Np9 | |||
| Testes | Germ cell tumors, gonadoblastoma, seminoma | Anti-Gag/Env-Ab | Boller et al. (87), Kleiman et al. (92), Boller et al. (125) |
| Expression of Rec, Np9 | |||
| Ovary | Ovarian clear cell carcinoma; ovarian epithelial tumors | Expression of Gag and Env | Gotzinger et al. (15), Wang-Johanning et al. (72), Iramaneerat et al. (126) |
| Breast | Breast cancer | Free viral RNA | Wang-Johanning et al. (112), Contreras-Galindo et al. (127), Wang-Johanning et al. (128) |
| RT activity | |||
| Virus particles | |||
| Specific CTLs | |||
| Prostate | Prostate cancer | Enhanced Gag-production due to fusion to androgen-dependent ETV1 and ETS genes | Tomlins et al. (12), Lamprecht et al. (13), Ishida et al. (111) |
| Blood | Lymphoma | Free RNA | Contreras-Galindo et al. (127) |
| RT activity | |||
| Virus-like particles |
List of malignant diseases that are associated with HERV-K(HML-2) activity.
Beside teratocarcinomas HERV-K proteins are expressed in 90% of epithelial ovarian tumors but at only very low levels, if at all, in healthy tissues or benign tumors (72). Activation of HERV-K expression in ovarian cancer might occur in response to stimulating transcriptional factors that are expressed in malignant ovarian cells (96). Such mechanisms, which lead to hypomethylation and subsequent enhancement of HERV-K transcription, have been already described for HERV-H and HERV-W elements in the ovaries (97–100) and for HERV-K in urothelial cancer and GCTs (101, 102).
Expression of HERV-K in melanomas has been the subject of intense study by many groups (6, 86). Both mRNA and Gag, Env, Rec, Np9, and RT proteins have been detected in specimens from primary and metastatic melanoma biopsies and melanoma cell lines but not in melanocytes or normal lymph nodes (6, 86). Muster and colleagues even observed retrovirus-like particles consisting of mature forms of Gag and Env and having RT activity (86). About 85% of malignant melanocytes express HERV-K MEL that is produced by a pseudo-gene incorporated into the HERV-K env gene and which has already been defined as a marker for melanoma risk (103–105). In accordance with the high levels of HERV-K expression, antibodies specific for Env and Gag were detectable in the sera of 16–22% of melanoma patients but not in those of healthy controls (6, 106). Hahn et al. showed in 2008 that the immune response of melanoma patients against HERV-K gene products correlates negatively with their survival (106). In line with existing knowledge about melanoma development, HERV-K expression can be triggered by UV radiation and may therefore be able to accelerate carcinogenesis (107, 108).
In addition to teratocarcinomas, ovary tumors, and melanomas, elevated levels of HERV-K(HML-2) env RNA were also detected in breast cancer tissues and gag RNA was found in the peripheral blood cells of leukemia patients as well as in breast and prostate cancers (109–112). The possible influence of steroid hormones on HERV-K activity in both breast cancer and prostate cancer is being discussed, with various groups reporting a stimulating effect of estroprogestin on HERV-K env transcription in breast cancer cells (110, 112, 113).
It was reported in early 2006 that, in contrast to patients undergoing successful HAART or to seronegative controls, HERV-K(HML-2) RNA can be found in the plasma of HIV-1 infected individuals whose antiretroviral therapy fails to control virus replication (114, 115). Subsequently, it was even possible to observe virus-like particles by electron microscopy and by immunostaining using HERV-K Env and Gag specific antibodies (116). A possible mechanism for preferential HERV-K expression in HIV-1 infected cells could be the interaction between HIV-1 Rev and the Rec response region in HERV-K(HML-2) RNA (11, 117) as well as the recently demonstrated susceptibility of the HERV-K (HML-2) transcriptional promoter to HIV-1 Tat (118).
The putative association between HERV-K(HML-2) expression and HIV-1 infection is strengthened by the detection of HERV-K specific cytotoxic T-cells in HIV-1 infected individuals (119). Interestingly, such HERV-specific T-cells are not found in HTLV-1 infected individuals (120), although this latter study cannot completely rule out an involvement of HERV activation in HTLV-1 pathogenesis. HERV-K specific CD8+ T-cells that can recognize and kill cells presenting the cognate peptides as a result of HIV-induced expression of HERV proteins (119) may play an important part in the natural immune response to HIV (121). In contrast to HIV specific CD8+ T-cells that are compromised in their ability to cope with the profound diversity of HIV-1 and subsequent appearance of escape mutants, the HERV proteins remain unchanged throughout the course of infection. Having an unchanging target allows HERV-specific T-cell clones from HIV-1 infected individuals to kill, in vitro at least, human CD4+ cells infected with viruses as diverse as HIV-1, HIV-2, and SIV (122). The stimulation of HERV-specific T-cells may therefore offer a new approach to inducing a broadly specific anti-HIV immune response. Of course, this approach is currently hampered by our limited understanding of HERV antigen expression in different individuals, tissues, and diseases (123).
Implications for Cancer Development
Whether HERV-K expression is an initiator of cancerous processes or is simply a consequence of malignant degeneration remains a topic of discussion. Many groups have found evidence for the involvement of HERV-K in tumor processes and various hypotheses concerning its potential contribution to oncogenesis have been postulated. These possible mechanisms include the misregulation of oncogenes, protooncogenes, or growth factors by HERV-K LTRs or HERV loci. Several examples of transcriptionally active HERV loci that influence neighboring genes by providing alternative promotors are known (13, 129). Hypomethylation often affects HERV-derived solo LTRs that function as alternate promoters, for example the HERV-derived LTR is hypomethylated in B cell-derived Hodgkin’s lymphoma and is responsible for deregulated expression of the colony-stimulating factor 1 receptor (CSF1R), a protooncogene (13). Another possible mechanism for HERV loci to affect cellular gene transcripts is by the provision of alternative splice and polyadenylation sites [reviewed in Ref. (16)].
A second mechanism of tumor induction/promotion by HERV-K is the inactivation of tumor suppressor genes by de novo insertion or translocation of retroelements within the genome (70, 130). Recombination involving HERV-K sequences might also lead to chromosomal instability and large-scale chromosomal anomalies such as those typically observed in aberrant cells (131, 132).
A third mechanism is the expression of HERV proteins such as Rec and Np9 that function as oncogenes or of proteins that induce cell–cell fusion such as Env. It is possible that cell fusions initiated by Env contribute to tumor progression or might even play an important role in metastasis (133). Furthermore, Env is considered to have immunosuppressive properties and may therefore play a role in tumor escape processes (133–135). HERV-K may trigger the pathological processes that lead to melanoma development and contribute to the cellular modifications implicated in melanoma maintenance and progression (96, 136). UV radiation, hormone action, or mutating viruses lead to enhanced expression of HERV-K proteins such as Env or the HERV-K MEL-antigen that is particularly expressed in melanomas (96, 107, 137). The HERV-K Env protein is a molecular mimic of the oxygen responsive element binding protein (OREBP), a human nuclear factor that controls the expression of glutathione peroxidase, resulting in reduced production of the enzyme (137). This further increases the toxicity of free radicals that are no longer under proper control and enhances the risk of cancer.
Potential Oncogenes: Rec and Np9
In recent years evidence has accumulated to indicate a role for the accessory HERV-K(HML-2) protein Rec and for Np9 in tumor development. Rec is a 14.5-kDa protein, translated from a doubly spliced transcript (Figure 2), that mediates the transport of unspliced or partially spliced HERV-K(HML-2) mRNAs from the nucleus into the cytoplasm (11). In 2005, Galli and Co-workers demonstrated that transgenic mice expressing Rec show disturbances in germ cell development and that some animals develop lesions similar to the predecessor lesions of classical seminomas in humans (138). This indicates that HERV-K(HML-2) Rec expression might play a direct role in cell transformation, creating a pre-cancerous state, although the molecular mechanisms and pathways involved remain elusive.
The cellular interaction partners of Rec are also poorly defined, although some have recently been identified. Rec interacts with the promyelocytic leukemia zinc-finger protein (PLZF) that is essential for the self-renewal of spermatogonial stem cells in mice and which may also be involved in certain human leukemias (139–142). Transient co-expression of Rec with PLZF interferes with its function as a negative regulator of the c-myc protooncogene and cells stably transfected with PLZF and Rec show increased cell proliferation (142). Recently, Kaufmann et al. reported that Rec forms trimeric complexes with the testicular zinc-finger protein (TZFP) and the AR (143). These data support the findings of other groups, including our own. Rec interacts directly with the human small glutamine-rich tetratricopeptide repeat-containing protein (hSGT), a co-chaperone and negative regulator of the AR (93, 144). Binding of Rec to the co-chaperone abrogates its ability to bind to the AR and to maintain it in an inactive state (93), which in turn leads to enhanced levels of AR activity. It is interesting that HERV-K transcription (and subsequently Rec expression) can be stimulated by hormones, including androgens, and various steroid hormone-binding sites are predicted to be located within the HERV-K LTR (89, 93, 95, 113).
In contrast to Rec, Np9 has no known physiological function in HERV-K replication. This 9 kDa protein is produced by viruses lacking a 292-bp sequence within the pol-env boundary. Np9, whose first 15 amino acids are identical to Env and Rec, is expressed exclusively in various tumor tissues such as breast cancer, GCTs, ovary cancer, and leukemia (47, 145). It is therefore important to identify host proteins potentially interacting with Np9.
Np9, like Rec, binds to the PLZF protein, albeit in a different region. Also like Rec, Np9 is able to abrogate the regulator of the c-myc protooncogene, leading to increased c-myc transcription that in turn leads to enhanced cell growth and reduced apoptosis (142).
Np9 has also been shown to interact with the ligand of Numb protein X (LNX), a RING-type E3 ubiquitin ligase that ubiquitylates Numb and marks it for degradation by 26S proteasomes (146). Numb is an antagonist of the transcription factor Notch and both proteins regulate the pro-proliferative Ras signaling cascade. Misregulation of this system has been linked to breast cancer, leukemias, and GCTs (147–151). Furthermore, Chen and Co-workers recently reported that Np9 is a potent viral oncogene in human leukemia (145). This small protein promotes growth of myeloid and lymphoblastic leukemic cells by activation of ERK, AKT, and Notch1 pathways and by upregulation of β-catenin. The latter is essential for survival of leukemia stem/progenitor cells.
Concluding Remarks and Outlook
The examples of HERV activity described in this review make it clear that these elements are not simply pieces of “junk DNA” that litter the genome because a mechanism to completely remove an integrated retrovirus from the germ line does not exist. Despite having been discovered in the late 1960s, it appears that many of the intricate evolutionary and physiological roles in health and disease played by the retroviral part of our own genome still remain to be revealed. In addition, many questions concerning the endogenization process, the interplay of coexisting endogenous and exogenous forms, population dynamics, and the fixation of ERVs remain open. The regulation of human genes by HERV-K(HML-2) and other HERV families and the silencing and restriction mechanisms by which the host controls their activities are currently of particular interest.
From a virological point of view, the reconstitution of consensus sequences and expression of original HERV-K(HML-2) proteins have greatly facilitated studies into the functional role of viral proteins in the replication cycle and in cellular processes. This, as well as the increasing availability of whole genome sequences, significantly enriches the exciting and novel field of “paleoretrovirology.”
Statements
Acknowledgments
The authors are indebted to Steve Norley for critical reading of the manuscript and inspiring discussions.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1
BentvelzenPDaamsJHHagemanPCalafatJ. Genetic transmission of viruses that incite mammary tumor in mice. Proc Natl Acad Sci U S A (1970) 67(1):377–84.10.1073/pnas.67.1.377
2
PalmariniMMuraMSpencerTE. Endogenous betaretroviruses of sheep: teaching new lessons in retroviral interference and adaptation. J Gen Virol (2004) 85(Pt 1):1–13.10.1099/vir.0.19547-0
3
PatienceCTakeuchiYWeissRA. Infection of human cells by an endogenous retrovirus of pigs. Nat Med (1997) 3(3):282–6.10.1038/nm0397-282
4
SubramanianRPWildschutteJHRussoCCoffinJM. Identification, characterization, and comparative genomic distribution of the HERV-K (HML-2) group of human endogenous retroviruses. Retrovirology (2011) 8:90.10.1186/1742-4690-8-90
5
LowerRLowerJFrankHHarzmannRKurthR. Human teratocarcinomas cultured in vitro produce unique retrovirus-like viruses. J Gen Virol (1984) 65(Pt 5):887–98.10.1099/0022-1317-65-5-887
6
BuscherKTrefzerUHofmannMSterryWKurthRDennerJ. Expression of human endogenous retrovirus K in melanomas and melanoma cell lines. Cancer Res (2005) 65(10):4172–80.10.1158/0008-5472.CAN-04-2983
7
SchmittKReichrathJRoeschAMeeseEMayerJ. Transcriptional profiling of human endogenous retrovirus group HERV-K(HML-2) loci in melanoma. Genome Biol Evol (2013) 5(2):307–28.10.1093/gbe/evt010
8
DewannieuxMHarperFRichaudALetzelterCRibetDPierronGet alIdentification of an infectious progenitor for the multiple-copy HERV-K human endogenous retroelements. Genome Res (2006) 16(12):1548–56.10.1101/gr.5565706
9
LeeYNBieniaszPD. Reconstitution of an infectious human endogenous retrovirus. PLoS Pathog (2007) 3(1):e10.10.1371/journal.ppat.0030010
10
MacfarlaneCSimmondsP. Allelic variation of HERV-K(HML-2) endogenous retroviral elements in human populations. J Mol Evol (2004) 59(5):642–56.10.1007/s00239-004-2656-1
11
MaginCLowerRLowerJ. cORF and RcRE, the Rev/Rex and RRE/RxRE homologues of the human endogenous retrovirus family HTDV/HERV-K. J Virol (1999) 73(11):9496–507.
12
TomlinsSALaxmanBDhanasekaranSMHelgesonBECaoXMorrisDSet alDistinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature (2007) 448(7153):595–9.10.1038/nature06024
13
LamprechtBWalterKKreherSKumarRHummelMLenzeDet alDerepression of an endogenous long terminal repeat activates the CSF1R proto-oncogene in human lymphoma. Nat Med (2010) 16(5):571–9. 1p following 579,10.1038/nm.2129
14
BuzdinAKovalskaya-AlexandrovaEGogvadzeESverdlovE. At least 50% of human-specific HERV-K (HML-2) long terminal repeats serve in vivo as active promoters for host nonrepetitive DNA transcription. J Virol (2006) 80(21):10752–62.10.1128/JVI.00871-06
15
GotzingerNSauterMRoemerKMueller-LantzschN. Regulation of human endogenous retrovirus-K Gag expression in teratocarcinoma cell lines and human tumours. J Gen Virol (1996) 77(Pt 12):2983–90.10.1099/0022-1317-77-12-2983
16
CohenCJLockWMMagerDL. Endogenous retroviral LTRs as promoters for human genes: a critical assessment. Gene (2009) 448(2):105–14.10.1016/j.gene.2009.06.020
17
ConleyABMillerWJJordanIK. Human cis natural antisense transcripts initiated by transposable elements. Trends Genet (2008) 24(2):53–6.10.1016/j.tig.2007.11.008
18
ConleyABPiriyapongsaJJordanIK. Retroviral promoters in the human genome. Bioinformatics (2008) 24(14):1563–7.10.1093/bioinformatics/btn243
19
GogvadzeEStukachevaEBuzdinASverdlovE. Human-specific modulation of transcriptional activity provided by endogenous retroviral insertions. J Virol (2009) 83(12):6098–105.10.1128/JVI.00123-09
20
VogtPKFriisRR. An avian leukosis virus related to RSV(O): properties and evidence for helper activity. Virology (1971) 43(1):223–34.10.1016/0042-6822(71)90240-6
21
WeissRA. The discovery of endogenous retroviruses. Retrovirology (2006) 3:67.10.1186/1742-4690-3-S1-S67
22
WeissRAPayneLN. The heritable nature of the factor in chicken cells which acts as a helper virus for Rous sarcoma virus. Virology (1971) 45(2):508–15.10.1016/0042-6822(71)90351-5
23
SalterDWSmithEJHughesSHWrightSECrittendenLB. Transgenic chickens: insertion of retroviral genes into the chicken germ line. Virology (1987) 157(1):236–40.
24
BannertNKurthR. The evolutionary dynamics of human endogenous retroviral families. Annu Rev Genomics Hum Genet (2006) 7:149–73.10.1146/annurev.genom.7.080505.115700
25
BelshawRDawsonALWoolven-AllenJReddingJBurtATristemM. Genomewide screening reveals high levels of insertional polymorphism in the human endogenous retrovirus family HERV-K(HML2): implications for present-day activity. J Virol (2005) 79(19):12507–14.10.1128/JVI.79.19.12507-12514.2005
26
TurnerGBarbulescuMSuMJensen-SeamanMIKiddKKLenzJ. Insertional polymorphisms of full-length endogenous retroviruses in humans. Curr Biol (2001) 11(19):1531–5.10.1016/S0960-9822(01)00455-9
27
FiebigUHartmannMGBannertNKurthRDennerJ. Transspecies transmission of the endogenous koala retrovirus. J Virol (2006) 80(11):5651–4.10.1128/JVI.02597-05
28
TarlintonREMeersJYoungPR. Retroviral invasion of the koala genome. Nature (2006) 442(7098):79–81.10.1038/nature04841
29
ShojimaTHoshinoSAbeMYasudaJShogenHKobayashiTet alConstruction and characterization of an infectious molecular clone of Koala retrovirus. J Virol (2013) 87(9):5081–8.10.1128/JVI.01584-12
30
BonnaudBBoutonOOriolGCheynetVDuretLMalletF. Evidence of selection on the domesticated ERVWE1 env retroviral element involved in placentation. Mol Biol Evol (2004) 21(10):1895–901.10.1093/molbev/msh206
31
JernPSperberGOBlombergJ. Use of endogenous retroviral sequences (ERVs) and structural markers for retroviral phylogenetic inference and taxonomy. Retrovirology (2005) 2:50.10.1186/1742-4690-2-S1-S50
32
OnoMYasunagaTMiyataTUshikuboH. Nucleotide sequence of human endogenous retrovirus genome related to the mouse mammary tumor virus genome. J Virol (1986) 60(2):589–98.
33
FranklinGCChretienSHansonIMRochefortHMayFEWestleyBR. Expression of human sequences related to those of mouse mammary tumor virus. J Virol (1988) 62(4):1203–10.
34
MedstrandPBlombergJ. Characterization of novel reverse transcriptase encoding human endogenous retroviral sequences similar to type A and type B retroviruses: differential transcription in normal human tissues. J Virol (1993) 67(11):6778–87.
35
AnderssonMLLindeskogMMedstrandPWestleyBMayFBlombergJ. Diversity of human endogenous retrovirus class II-like sequences. J Gen Virol (1999) 80(Pt 1):255–60.
36
MayerJMeeseE. Human endogenous retroviruses in the primate lineage and their influence on host genomes. Cytogenet Genome Res (2005) 110(1–4):448–56.10.1159/000084977
37
SeifarthWBaustCMurrASkladnyHKrieg-SchneiderFBluschJet alProviral structure, chromosomal location, and expression of HERV-K-T47D, a novel human endogenous retrovirus derived from T47D particles. J Virol (1998) 72(10):8384–91.
38
MagerDLMedstrandP. Retroviral repeat sequences. In: CooperD, editor. Nature Encyclopedia of the Human Genome. London: Nature Publishing Group (2003). p. 57–63.
39
JohnsonWECoffinJM. Constructing primate phylogenies from ancient retrovirus sequences. Proc Natl Acad Sci U S A (1999) 96(18):10254–60.10.1073/pnas.96.18.10254
40
LavieLMaldenerEBrouhaBMeeseEUMayerJ. The human L1 promoter: variable transcription initiation sites and a major impact of upstream flanking sequence on promoter activity. Genome Res (2004) 14(11):2253–60.10.1101/gr.2745804
41
FlockerziABurkhardtSSchemppWMeeseEMayerJ. Human endogenous retrovirus HERV-K14 families: status, variants, evolution, and mobilization of other cellular sequences. J Virol (2005) 79(5):2941–9.10.1128/JVI.79.5.2941-2949.2005
42
OnoM. Molecular cloning and long terminal repeat sequences of human endogenous retrovirus genes related to types A and B retrovirus genes. J Virol (1986) 58(3):937–44.
43
MayerJMeeseEU. The human endogenous retrovirus family HERV-K(HML-3). Genomics (2002) 80(3):331–43.10.1006/geno.2002.6839
44
MedstrandPMagerDLYinHDietrichUBlombergJ. Structure and genomic organization of a novel human endogenous retrovirus family: HERV-K (HML-6). J Gen Virol (1997) 78(Pt 7):1731–44.
45
YinHMedstrandPKristoffersonADietrichUAmanPBlombergJ. Characterization of human MMTV-like (HML) elements similar to a sequence that was highly expressed in a human breast cancer: further definition of the HML-6 group. Virology (1999) 256(1):22–35.10.1006/viro.1998.9587
46
TassabehjiMStrachanTAndersonMCampbellRDCollierSLakoM. Identification of a novel family of human endogenous retroviruses and characterization of one family member, HERV-K(C4), located in the complement C4 gene cluster. Nucleic Acids Res (1994) 22(24):5211–7.10.1093/nar/22.24.5211
47
ArmbruesterVSauterMKrautkraemerEMeeseEKleimanABestBet alA novel gene from the human endogenous retrovirus K expressed in transformed cells. Clin Cancer Res (2002) 8(6):1800–7.
48
LowerRTonjesRRKorbmacherCKurthRLowerJ. Identification of a Rev-related protein by analysis of spliced transcripts of the human endogenous retroviruses HTDV/HERV-K. J Virol (1995) 69(1):141–9.
49
GeorgeMSchweckeTBeimfordeNHohnOChudakCZimmermannAet alIdentification of the protease cleavage sites in a reconstituted Gag polyprotein of an HERV-K(HML-2) element. Retrovirology (2011) 8:30.10.1186/1742-4690-8-30
50
DewannieuxMBlaiseSHeidmannT. Identification of a functional envelope protein from the HERV-K family of human endogenous retroviruses. J Virol (2005) 79(24):15573–7.10.1128/JVI.79.24.15573-15577.2005
51
HankeKKramerPSeeherSBeimfordeNKurthRBannertN. Reconstitution of the ancestral glycoprotein of human endogenous retrovirus k and modulation of its functional activity by truncation of the cytoplasmic domain. J Virol (2009) 83(24):12790–800.10.1128/JVI.01368-09
52
MayerJMeeseEU. Presence of dUTPase in the various human endogenous retrovirus K (HERV-K) families. J Mol Evol (2003) 57(6):642–9.10.1007/s00239-003-2514-6
53
LeeYNMalimMHBieniaszPD. Hypermutation of an ancient human retrovirus by APOBEC3G. J Virol (2008) 82(17):8762–70.10.1128/JVI.00751-08
54
BeimfordeNHankeKAmmarIKurthRBannertN. Molecular cloning and functional characterization of the human endogenous retrovirus K113. Virology (2008) 371(1):216–25.10.1016/j.virol.2007.09.036
55
PaprotkaTDelviks-FrankenberryKACingozOMartinezAKungHJTepperCGet alRecombinant origin of the retrovirus XMRV. Science (2011) 333(6038):97–101.10.1126/science.1205292
56
YoungGREksmondUSalcedoRAlexopoulouLStoyeJPKassiotisG. Resurrection of endogenous retroviruses in antibody-deficient mice. Nature (2012) 491(7426):774–8.10.1038/nature11599
57
KingMCWilsonAC. Evolution at two levels in humans and chimpanzees. Science (1975) 188(4184):107–16.10.1126/science.1090005
58
TingCNRosenbergMPSnowCMSamuelsonLCMeislerMH. Endogenous retroviral sequences are required for tissue-specific expression of a human salivary amylase gene. Genes Dev (1992) 6(8):1457–65.
59
MiSLeeXLiXVeldmanGMFinnertyHRacieLet alSyncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature (2000) 403(6771):785–9.10.1038/35001608
60
BlaiseSde ParsevalNBenitLHeidmannT. Genomewide screening for fusogenic human endogenous retrovirus envelopes identifies syncytin 2, a gene conserved on primate evolution. Proc Natl Acad Sci U S A (2003) 100(22):13013–8.10.1073/pnas.2132646100
61
Schlecht-LoufGRenardMMangeneyMLetzelterCRichaudADucosBet alRetroviral infection in vivo requires an immune escape virulence factor encrypted in the envelope protein of oncoretroviruses. Proc Natl Acad Sci U S A (2010) 107(8):3782–7.10.1073/pnas.0913122107
62
DupressoirAMarceauGVernochetCBenitLKanellopoulosCSapinVet alSyncytin-A and syncytin-B, two fusogenic placenta-specific murine envelope genes of retroviral origin conserved in Muridae. Proc Natl Acad Sci U S A (2005) 102(3):725–30.10.1073/pnas.0406509102
63
DupressoirAVernochetCBawaOHarperFPierronGOpolonPet alSyncytin-A knockout mice demonstrate the critical role in placentation of a fusogenic, endogenous retrovirus-derived, envelope gene. Proc Natl Acad Sci U S A (2009) 106(29):12127–32.10.1073/pnas.0902925106
64
DupressoirALavialleCHeidmannT. From ancestral infectious retroviruses to bona fide cellular genes: role of the captured syncytins in placentation. Placenta (2012) 33(9):663–71.10.1016/j.placenta.2012.05.005
65
SpencerTEMuraMGrayCAGriebelPJPalmariniM. Receptor usage and fetal expression of ovine endogenous betaretroviruses: implications for coevolution of endogenous and exogenous retroviruses. J Virol (2003) 77(1):749–53.10.1128/JVI.77.1.749-753.2003
66
ArnaudFCaporaleMVarelaMBiekRChessaBAlbertiAet alA paradigm for virus-host coevolution: sequential counter-adaptations between endogenous and exogenous retroviruses. PLoS Pathog (2007) 3(11):e170.10.1371/journal.ppat.0030170
67
BestSLe TissierPTowersGStoyeJP. Positional cloning of the mouse retrovirus restriction gene Fv1. Nature (1996) 382(6594):826–9.10.1038/382826a0
68
OjaMPeltonenJBlombergJKaskiS. Methods for estimating human endogenous retrovirus activities from EST databases. BMC Bioinformatics (2007) 8(Suppl 2):S11.10.1186/1471-2105-8-S2-S11
69
PerotPMugnierNMontgiraudCGimenezJJaillardMBonnaudBet alMicroarray-based sketches of the HERV transcriptome landscape. PLoS One (2012) 7(6):e40194.10.1371/journal.pone.0040194
70
KurthRBannertN. Beneficial and detrimental effects of human endogenous retroviruses. Int J Cancer (2010) 126(2):306–14.10.1002/ijc.24902
71
KammererUGermeyerAStengelSKappMDennerJ. Human endogenous retrovirus K (HERV-K) is expressed in villous and extravillous cytotrophoblast cells of the human placenta. J Reprod Immunol (2011) 91(1–2):1–8.10.1016/j.jri.2011.06.102
72
Wang-JohanningFLiuJRycajKHuangMTsaiKRosenDGet alExpression of multiple human endogenous retrovirus surface envelope proteins in ovarian cancer. Int J Cancer (2007) 120(1):81–90.10.1002/ijc.22256
73
AnderssonACVenablesPJTonjesRRSchererJErikssonLLarssonE. Developmental expression of HERV-R (ERV3) and HERV-K in human tissue. Virology (2002) 297(2):220–5.10.1006/viro.2002.1428
74
FreimanisGHooleyPEjtehadiHDAliHAVeitchARylancePBet alA role for human endogenous retrovirus-K (HML-2) in rheumatoid arthritis: investigating mechanisms of pathogenesis. Clin Exp Immunol (2010) 160(3):340–7.10.1111/j.1365-2249.2010.04110.x
75
TselisA. Evidence for viral etiology of multiple sclerosis. Semin Neurol (2011) 31(3):307–16.10.1055/s-0031-1287656
76
TaiAKO’ReillyEJAlroyKASimonKCMungerKLHuberBTet alHuman endogenous retrovirus-K18 Env as a risk factor in multiple sclerosis. Mult Scler (2008) 14(9):1175–80.10.1177/1352458508094641
77
SicatJSutkowskiNHuberBT. Expression of human endogenous retrovirus HERV-K18 superantigen is elevated in juvenile rheumatoid arthritis. J Rheumatol (2005) 32(9):1821–31.
78
ClausenJ. Endogenous retroviruses and MS: using ERVs as disease markers. Int MS J (2003) 10(1):22–8.
79
MoyesDLGorisABanMCompstonAGriffithsDJSawcerSet alHERV-K113 is not associated with multiple sclerosis in a large family-based study. AIDS Res Hum Retroviruses (2008) 24(3):363–5.10.1089/aid.2007.0196
80
MoyesDLMartinASawcerSTempertonNWorthingtonJGriffithsDJet alThe distribution of the endogenous retroviruses HERV-K113 and HERV-K115 in health and disease. Genomics (2005) 86(3):337–41.10.1016/j.ygeno.2005.06.004
81
AntonyJMDeslauriersAMBhatRKEllestadKKPowerC. Human endogenous retroviruses and multiple sclerosis: innocent bystanders or disease determinants?Biochim Biophys Acta (2011) 1812(2):162–76.10.1016/j.bbadis.2010.07.016
82
ConradBWeissmahrRNBoniJArcariRSchupbachJMachB. A human endogenous retroviral superantigen as candidate autoimmune gene in type I diabetes. Cell (1997) 90(2):303–13.10.1016/S0092-8674(00)80338-4
83
MargueratSWangWYToddJAConradB. Association of human endogenous retrovirus K-18 polymorphisms with type 1 diabetes. Diabetes (2004) 53(3):852–4.10.2337/diabetes.53.3.852
84
BuscherKHahnSHofmannMTrefzerUOzelMSterryWet alExpression of the human endogenous retrovirus-K transmembrane envelope, Rec and Np9 proteins in melanomas and melanoma cell lines. Melanoma Res (2006) 16(3):223–34.10.1097/01.cmr.0000215031.07941.ca
85
HerbstHSauterMMueller-LantzschN. Expression of human endogenous retrovirus K elements in germ cell and trophoblastic tumors. Am J Pathol (1996) 149(5):1727–35.
86
MusterTWaltenbergerAGrassauerAHirschlSCaucigPRomirerIet alAn endogenous retrovirus derived from human melanoma cells. Cancer Res (2003) 63(24):8735–41.
87
BollerKKonigHSauterMMueller-LantzschNLowerRLowerJet alEvidence that HERV-K is the endogenous retrovirus sequence that codes for the human teratocarcinoma-derived retrovirus HTDV. Virology (1993) 196(1):349–53.10.1006/viro.1993.1487
88
KurthRLöwerRLöwerJHarzmannRPfeifferRSchmidtGCet alOncornavirus synthesis in human teratocarcinoma cultures and an increased antiviral immune reactivity in corresponding patients. In: EssexMTodaroGJzur HausenH, editors. Viruses in Naturally Occurring Cancers. New York: Cold Spring Harbor Laboratory (1980). p. 835–46.
89
OnoMKawakamiMUshikuboH. Stimulation of expression of the human endogenous retrovirus genome by female steroid hormones in human breast cancer cell line T47D. J Virol (1987) 61(6):2059–62.
90
GoedertJJSauterMEJacobsonLPVessellaRLHilgartnerMWLeitmanSFet alHigh prevalence of antibodies against HERV-K10 in patients with testicular cancer but not with AIDS. Cancer Epidemiol Biomarkers Prev (1999) 8(4 Pt 1):293–6.
91
SauterMSchommerSKremmerERembergerKDolkenGLemmIet alHuman endogenous retrovirus K10: expression of Gag protein and detection of antibodies in patients with seminomas. J Virol (1995) 69(1):414–21.
92
KleimanASenyutaNTryakinASauterMKarseladzeATjulandinSet alHERV-K(HML-2) GAG/ENV antibodies as indicator for therapy effect in patients with germ cell tumors. Int J Cancer (2004) 110(3):459–61.10.1002/ijc.11649
93
HankeKChudakCKurthRBannertN. The Rec protein of HERV-K(HML-2) upregulates androgen receptor activity by binding to the human small glutamine-rich tetratricopeptide repeat protein (hSGT). Int J Cancer (2013) 132(3):556–67.10.1002/ijc.27693
94
KnosslMLowerRLowerJ. Expression of the human endogenous retrovirus HTDV/HERV-K is enhanced by cellular transcription factor YY1. J Virol (1999) 73(2):1254–61.
95
MangheraMDouvilleRN. Endogenous retrovirus-K promoter: a landing strip for inflammatory transcription factors?Retrovirology (2013) 10:16.10.1186/1742-4690-10-16
96
CegolonLSalataCWeiderpassEVineisPPaluGMastrangeloG. Human endogenous retroviruses and cancer prevention: evidence and prospects. BMC Cancer (2013) 13:4.10.1186/1471-2407-13-4
97
de ParsevalNAlkabbaniHHeidmannT. The long terminal repeats of the HERV-H human endogenous retrovirus contain binding sites for transcriptional regulation by the Myb protein. J Gen Virol (1999) 80(Pt 4):841–5.
98
LeeWJKwunHJKimHSJangKL. Activation of the human endogenous retrovirus W long terminal repeat by herpes simplex virus type 1 immediate early protein 1. Mol Cells (2003) 15(1):75–80.
99
MenendezLBenignoBBMcDonaldJF. L1 and HERV-W retrotransposons are hypomethylated in human ovarian carcinomas. Mol Cancer (2004) 3:12.10.1186/1476-4598-3-12
100
SjottemEAnderssenSJohansenT. The promoter activity of long terminal repeats of the HERV-H family of human retrovirus-like elements is critically dependent on Sp1 family proteins interacting with a GC/GT box located immediately 3’ to the TATA box. J Virol (1996) 70(1):188–98.
101
FlorlARLowerRSchmitz-DragerBJSchulzWA. DNA methylation and expression of LINE-1 and HERV-K provirus sequences in urothelial and renal cell carcinomas. Br J Cancer (1999) 80(9):1312–21.10.1038/sj.bjc.6690524
102
LavieLKitovaMMaldenerEMeeseEMayerJ. CpG methylation directly regulates transcriptional activity of the human endogenous retrovirus family HERV-K(HML-2). J Virol (2005) 79(2):876–83.10.1128/JVI.79.2.876-883.2005
103
GrangeJMKroneBKolmelKFMastrangeloG. Can prior vaccinations against certain infections confer protection against developing melanoma?Med J Aust (2009) 191(9):478–9.
104
KolmelKFGefellerOHaferkampB. Febrile infections and malignant melanoma: results of a case-control study. Melanoma Res (1992) 2(3):207–11.10.1097/00008390-199209000-00009
105
SchiavettiFThonnardJColauDBoonTCouliePG. A human endogenous retroviral sequence encoding an antigen recognized on melanoma by cytolytic T lymphocytes. Cancer Res (2002) 62(19):5510–6.
106
HahnSUgurelSHanschmannKMStrobelHTonderaCSchadendorfDet alSerological response to human endogenous retrovirus K in melanoma patients correlates with survival probability. AIDS Res Hum Retroviruses (2008) 24(5):717–23.10.1089/aid.2007.0286
107
ReicheJPauliGEllerbrokH. Differential expression of human endogenous retrovirus K transcripts in primary human melanocytes and melanoma cell lines after UV irradiation. Melanoma Res (2010) 20(5):435–40.10.1097/CMR.0b013e32833c1b5d
108
SchanabOHumerJGleissAMikulaMSturlanSGruntSet alExpression of human endogenous retrovirus K is stimulated by ultraviolet radiation in melanoma. Pigment Cell Melanoma Res (2011) 24(4):656–65.
109
DepilSRocheCDussartPPrinL. Expression of a human endogenous retrovirus, HERV-K, in the blood cells of leukemia patients. Leukemia (2002) 16(2):254–9.10.1038/sj.leu.2402355
110
EjthadiHDMartinJHJunyingJRodenDALahiriMWarrenPet alA novel multiplex RT-PCR system detects human endogenous retrovirus-K in breast cancer. Arch Virol (2005) 150(1):177–84.10.1007/s00705-004-0378-8
111
IshidaTObataYOharaNMatsushitaHSatoSUenakaAet alIdentification of the HERV-K gag antigen in prostate cancer by SEREX using autologous patient serum and its immunogenicity. Cancer Immun (2008) 8:15.
112
Wang-JohanningFFrostARJianBEppLLuDWJohanningGL. Quantitation of HERV-K env gene expression and splicing in human breast cancer. Oncogene (2003) 22(10):1528–35.10.1038/sj.onc.1206241
113
GolanMHiziAResauJHYaal-HahoshenNReichmanHKeydarIet alHuman endogenous retrovirus (HERV-K) reverse transcriptase as a breast cancer prognostic marker. Neoplasia (2008) 10(6):521–33.
114
Contreras-GalindoRGonzalezMAlmodovar-CamachoSGonzalez-RamirezSLorenzoEYamamuraY. A new real-time-RT-PCR for quantitation of human endogenous retroviruses type K (HERV-K) RNA load in plasma samples: increased HERV-K RNA titers in HIV-1 patients with HAART non-suppressive regimens. J Virol Methods (2006) 136(1–2):51–7.10.1016/j.jviromet.2006.03.029
115
Contreras-GalindoRKaplanMHMarkovitzDMLorenzoEYamamuraY. Detection of HERV-K(HML-2) viral RNA in plasma of HIV type 1-infected individuals. AIDS Res Hum Retroviruses (2006) 22(10):979–84.10.1089/aid.2006.22.979
116
Contreras-GalindoRKaplanMHContreras-GalindoACGonzalez-HernandezMJFerlenghiIGiustiFet alCharacterization of human endogenous retroviral elements in the blood of HIV-1-infected individuals. J Virol (2012) 86(1):262–76.10.1128/JVI.00602-11
117
YangJBogerdHPPengSWiegandHTruantRCullenBR. An ancient family of human endogenous retroviruses encodes a functional homolog of the HIV-1 Rev protein. Proc Natl Acad Sci U S A (1999) 96(23):13404–8.10.1073/pnas.96.23.13404
118
Gonzalez-HernandezMJSwansonMDContreras-GalindoRCookinhamSKingSRNoelRJJret alExpression of human endogenous retrovirus type K (HML-2) is activated by the Tat protein of HIV-1. J Virol (2012) 86(15):7790–805.10.1128/JVI.07215-11
119
GarrisonKEJonesRBMeiklejohnDAAnwarNNdhlovuLCChapmanJMet alT cell responses to human endogenous retroviruses in HIV-1 infection. PLoS Pathog (2007) 3(11):e165.10.1371/journal.ppat.0030165
120
JonesRBLealFEHasenkrugAMSeguradoACNixonDFOstrowskiMAet alHuman endogenous retrovirus K(HML-2) Gag and Env specific T-cell responses are not detected in HTLV-I-infected subjects using standard peptide screening methods. J Negat Results Biomed (2013) 12:3.10.1186/1477-5751-12-3
121
SenGuptaDTandonRVieiraRGNdhlovuLCLown-HechtROrmsbyCEet alStrong human endogenous retrovirus-specific T cell responses are associated with control of HIV-1 in chronic infection. J Virol (2011) 85(14):6977–85.10.1128/JVI.00179-11
122
JonesRBGarrisonKEMujibSMihajlovicVAidarusNHunterDVet alHERV-K-specific T cells eliminate diverse HIV-1/2 and SIV primary isolates. J Clin Invest (2012) 122(12):4473–89.10.1172/JCI64560
123
van der KuylAC. HIV infection and HERV expression: a review. Retrovirology (2012) 9:6.10.1186/1742-4690-9-6
124
HirschlSSchanabOSeppeleHWaltenbergerAHumerJWolffKet alSequence variability of retroviral particles derived from human melanoma cells melanoma-associated retrovirus. Virus Res (2007) 123(2):211–5.10.1016/j.virusres.2006.08.010
125
BollerKSchonfeldKLischerSFischerNHoffmannAKurthRet alHuman endogenous retrovirus HERV-K113 is capable of producing intact viral particles. J Gen Virol (2008) 89(Pt 2):567–72.10.1099/vir.0.83534-0
126
IramaneeratKRattanatunyongPKhemapechNTriratanachatSMutiranguraA. HERV-K hypomethylation in ovarian clear cell carcinoma is associated with a poor prognosis and platinum resistance. Int J Gynecol Cancer (2011) 21(1):51–7.10.1097/IGC.0b013e3182021c1a
127
Contreras-GalindoRKaplanMHLeissnerPVerjatTFerlenghiIBagnoliFet alHuman endogenous retrovirus K (HML-2) elements in the plasma of people with lymphoma and breast cancer. J Virol (2008) 82(19):9329–36.10.1128/JVI.00646-08
128
Wang-JohanningFRadvanyiLRycajKPlummerJBYanPSastryKJet alHuman endogenous retrovirus K triggers an antigen-specific immune response in breast cancer patients. Cancer Res (2008) 68(14):5869–77.10.1158/0008-5472.CAN-07-6838
129
DunnCAMedstrandPMagerDL. An endogenous retroviral long terminal repeat is the dominant promoter for human beta1,3-galactosyltransferase 5 in the colon. Proc Natl Acad Sci U S A (2003) 100(22):12841–6.10.1073/pnas.2134464100
130
RuprechtKMayerJSauterMRoemerKMueller-LantzschN. Endogenous retroviruses and cancer. Cell Mol Life Sci (2008) 65(21):3366–82.10.1007/s00018-008-8496-1
131
MullinsCSLinnebacherM. Human endogenous retroviruses and cancer: causality and therapeutic possibilities. World J Gastroenterol (2012) 18(42):6027–35.10.3748/wjg.v18.i42.6027
132
RomanishMTCohenCJMagerDL. Potential mechanisms of endogenous retroviral-mediated genomic instability in human cancer. Semin Cancer Biol (2010) 20(4):246–53.10.1016/j.semcancer.2010.05.005
133
OricchioESciamannaIBeraldiRTolstonogGVSchumannGGSpadaforaC. Distinct roles for LINE-1 and HERV-K retroelements in cell proliferation, differentiation and tumor progression. Oncogene (2007) 26(29):4226–33.10.1038/sj.onc.1210214
134
MangeneyMPothlichetJRenardMDucosBHeidmannT. Endogenous retrovirus expression is required for murine melanoma tumor growth in vivo. Cancer Res (2005) 65(7):2588–91.10.1158/0008-5472.CAN-04-4231
135
SinghSKayeSGoreMEMcClureMOBunkerCB. The role of human endogenous retroviruses in melanoma. Br J Dermatol (2009) 161(6):1225–31.10.1111/j.1365-2133.2009.09415.x
136
SerafinoABalestrieriEPierimarchiPMatteucciCMoroniGOricchioEet alThe activation of human endogenous retrovirus K (HERV-K) is implicated in melanoma cell malignant transformation. Exp Cell Res (2009) 315(5):849–62.10.1016/j.yexcr.2008.12.023
137
KroneBKolmelKFHenzBMGrangeJM. Protection against melanoma by vaccination with Bacille Calmette-Guerin (BCG) and/or vaccinia: an epidemiology-based hypothesis on the nature of a melanoma risk factor and its immunological control. Eur J Cancer (2005) 41(1):104–17.10.1016/j.ejca.2004.08.010
138
GalliUMSauterMLecherBMaurerSHerbstHRoemerKet alHuman endogenous retrovirus rec interferes with germ cell development in mice and may cause carcinoma in situ, the predecessor lesion of germ cell tumors. Oncogene (2005) 24(19):3223–8.10.1038/sj.onc.1208543
139
BoeseASauterMGalliUBestBHerbstHMayerJet alHuman endogenous retrovirus protein cORF supports cell transformation and associates with the promyelocytic leukemia zinc finger protein. Oncogene (2000) 19(38):4328–36.10.1038/sj.onc.1203794
140
BuaasFWKirshALSharmaMMcLeanDJMorrisJLGriswoldMDet alPlzf is required in adult male germ cells for stem cell self-renewal. Nat Genet (2004) 36(6):647–52.10.1038/ng1366
141
CostoyaJAHobbsRMBarnaMCattorettiGManovaKSukhwaniMet alEssential role of Plzf in maintenance of spermatogonial stem cells. Nat Genet (2004) 36(6):653–9.10.1038/ng1367
142
DenneMSauterMArmbruesterVLichtJDRoemerKMueller-LantzschN. Physical and functional interactions of human endogenous retrovirus proteins Np9 and rec with the promyelocytic leukemia zinc finger protein. J Virol (2007) 81(11):5607–16.10.1128/JVI.02771-06
143
KaufmannSSauterMSchmittMBaumertBBestBBoeseAet alHuman endogenous retrovirus protein Rec interacts with the testicular zinc-finger protein and androgen receptor. J Gen Virol (2010) 91(Pt 6):1494–502.10.1099/vir.0.014241-0
144
BuchananGRicciardelliCHarrisJMPrescottJYuZCJiaLet alControl of androgen receptor signaling in prostate cancer by the cochaperone small glutamine rich tetratricopeptide repeat containing protein alpha. Cancer Res (2007) 67(20):10087–96.10.1158/0008-5472.CAN-07-1646
145
ChenTMengZGanYWangXXuFGuYet alThe viral oncogene Np9 acts as a critical molecular switch for co-activating beta-catenin, ERK, Akt and Notch1 and promoting the growth of human leukemia stem/progenitor cells. Leukemia (2013) 27(7):1469–78.10.1038/leu.2013.8
146
ArmbruesterVSauterMRoemerKBestBHahnSNtyAet alNp9 protein of human endogenous retrovirus K interacts with ligand of numb protein X. J Virol (2004) 78(19):10310–9.10.1128/JVI.78.19.10310-10319.2004
147
AdamahDJGokhalePJEastwoodDJRajpert De-MeytsEGoepelJWalshJRet alDysfunction of the mitotic:meiotic switch as a potential cause of neoplastic conversion of primordial germ cells. Int J Androl (2006) 29(1):219–27.10.1111/j.1365-2605.2005.00569.x
148
BeverlyLJCapobiancoAJ. Perturbation of Ikaros isoform selection by MLV integration is a cooperative event in Notch(IC)-induced T cell leukemogenesis. Cancer Cell (2003) 3(6):551–64.10.1016/S1535-6108(03)00137-5
149
CallahanRRaafatA. Notch signaling in mammary gland tumorigenesis. J Mammary Gland Biol Neoplasia (2001) 6(1):23–36.10.1023/A:1009512414430
150
HayashiTKageyamaYIshizakaKXiaGKiharaKOshimaH. Requirement of Notch 1 and its ligand jagged 2 expressions for spermatogenesis in rat and human testes. J Androl (2001) 22(6):999–1011.
151
HayashiTYamadaTKageyamaYKiharaK. Expression failure of the notch signaling system is associated with the pathogenesis of testicular germ cell tumor. Tumour Biol (2004) 25(3):99–105.10.1159/000079140
Summary
Keywords
human endogenous retrovirus, HERV-K, replication, Rec, cancer
Citation
Hohn O, Hanke K and Bannert N (2013) HERV-K(HML-2), the Best Preserved Family of HERVs: Endogenization, Expression, and Implications in Health and Disease. Front. Oncol. 3:246. doi: 10.3389/fonc.2013.00246
Received
19 July 2013
Accepted
05 September 2013
Published
20 September 2013
Volume
3 - 2013
Edited by
Iyoko Katoh, University of Yamanashi, Japan
Reviewed by
Holger Kalthoff, Institute for Experimental Cancer Research, Germany; Shashidhar Jatiani, Forest Research Institute, USA
Copyright
© 2013 Hohn, Hanke and Bannert.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Norbert Bannert, Division for HIV and Other Retroviruses, Robert Koch Institute, Nordufer 20, 13353 Berlin, Germany e-mail: bannertn@rki.de
This article was submitted to Molecular and Cellular Oncology, a section of the journal Frontiers in Oncology.
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.