- Virology Group, International Centre for Genetic Engineering and Biotechnology (ICGEB), New Delhi, India
Hepatotropic viruses such as hepatitis B virus (HBV) and hepatitis C virus (HCV) are the major etiological agents associated with development of hepatocellular carcinoma (HCC). Progression of HCC is a multistep process that requires sequential or parallel deregulation of oncogenic and tumor suppressive pathways leading to chromosomal instability and neoplastic phenotype. In the recent years, microRNAs (miRNAs) have carved their own niche alongside oncogenes and tumor suppressors, owing to their innate ability to receive and relay multiple signals. Not surprisingly, miRNAs are fast emerging as central player in myriads of malignancies including HCC. miRNAs are reported to participate in initiation and progression of HCC, and have also been clinically correlated with risk assessment, disease grade, aggressiveness, and prognosis. Despite extensive data available on the role of miRNAs in HCC, there is a pressing need to integrate and evaluate these datasets to find its correlation, if any, with causal agents in order to devise novel interventional modalities. Through this review, we attempt to bridge the gap by consolidating the current knowledge and concepts in the field of HCC-related miRNAs with special emphasis on HBV and HCV. Further, we assess the potential of common as well as unique signatures that may be useful in developing novel biomarkers and therapeutics.
Introduction
According to a WHO estimate, hepatitis B virus (HBV) and hepatitis C virus (HCV) together account for ~78% of hepatocellular carcinoma (HCC) incidence worldwide and are the second leading cause of cancer mortality (1). High morbidity observed in HCC is majorly attributed to lack of early detection markers and poor prognosis, which limit the options for chemotherapy, adjuvant therapies, or surgical procedures (2). Hence, exploration of novel frontiers in HCC diagnosis and therapeutics remain high priority research areas.
Recent studies suggest an indispensable role played by microRNAs (miRNAs) in tumor growth, and immune evasion. miRNAs constitute a major class of well-conserved, small non-coding RNAs that can up- or down-regulate gene expression (2, 3). Accordingly, miRNAs can function both as tumor suppressor (TS-miR) and oncogene (onco-miR) impacting both pro- and anti-proliferative cascades (4). To date, nearly 2000 different miRNAs have been identified in humans. Stability of miRNAs in blood circulation makes them ideal candidate for use in diagnosis and treatment of cancers. Further, tumor miRNA profiles can be helpful in defining relevant subtypes, patient survival, treatment response, and risk prediction (5). Given the vast repertoire of cancer-related pathways regulated by miRNAs, this class of biomolecules appears to be a driving force for oncogenesis as well as “Achilles’ heel” for therapeutic targeting. The intricate processes of miRNA biogenesis and maturation are easy targets of stealth hepatitis viruses (6). In the next section, we overview how these processes are modulated in HCC.
Transcriptional Regulation of miRNA
The transcription of miRNA is guided by RNA polymerase (pol II) regulators, which are often deregulated in case of liver pathologies. In HBV- and HCV-associated pathogenesis, c-Myc along with viral oncoproteins modulates miRNA expression, to create an oncogenic milieu (7) facilitating the binding of transcriptional repressor complexes to miRNA promoters to allow its sustained expression (8, 9). For example, c-Myc binding to miR-122 promoter (a liver-specific TS-miR) prevents RNA pol II recruitment and H3K9 acetylation (10). c-Myc also down-regulates HNF3β, a liver-specific transcription factor, involved in transcription of miR-122. Since c-Myc itself is a target of certain miRNAs, the transcriptional control of its regulators sets on a positive feedback loop for c-Myc expression in HCC (8, 11).
c-Myc is also reported to stimulate DROSHA promoter and thus, indirectly regulate the stability of DiGeorge syndrome critical region gene 8 mRNA (12). Although c-Myc normally functions as a negative regulator of miRNAs, the levels of onco-miRs like miR-21 and miR-17-92 polycistron are elevated in c-Myc microenvironment, which down-regulate tumor suppressors (13). HCV infection per se can cause down-regulation of TS-miR-181c by promoting the recruitment of CCAAT/enhancer binding protein β (C/EBP-β) (14). In contrast, HBV positively regulates miR-181a transcription – an index of poor survival whereas it negatively regulates tumor suppressors, WIF1 and DKK3 (controllers of Wnt signaling pathway and miR181a targets) possibly via miR181a (15–17). Inactivation of p53 in HCC leads to down-regulation of its transcriptional targets such as miR-34, miR-200, and miR-15/16, which allows cell proliferation and metastasis (18). p53 inactivation also alters miRNA biogenesis either by directly binding to DROSHA or via down-regulation of DICER-1 (19), which may define miRNA target genes in HCC.
Post-Transcriptional Regulation of miRNA
Hepatitis viruses often deregulate pro-proliferative pathways in HCC affecting phase-specific control of cyclins by miRNAs (6). Impairment of DICER-1 is also frequently observed in HCC combined with tumor stemness (20). Post-transcriptional regulators of Argonaute (Ago-1 and Ago-2) such as lin-41 are over-expressed in HCC in a c-Myc dependent fashion, which down-regulates Ago protein (21). Whereas low levels of miR-99 and miR-199a induce Ago-2 expression with consequent increase in miR-21 (22, 23). Modulation of TGFβ signaling in HCC also involves miRNAs to promote tumorigenesis (24, 25). Interestingly, TGFβ effector SMAD1/5 along with RNA helicase p68 increases the maturation rate of miR-21 and miR-199 contributing to vascularization (25).
Epigenetic Alterations of miRNA
Concordant hyper methylation of miRNA genes is frequently seen in HCC (26, 27). The master controllers of proliferation, like c-Met are epigenetically silenced by TS-miRs (28). An auto-feedback loop of hyper-methylation and gene suppression is suggested for miR-148a and miR-152 in HCC (29, 30). Acetylation status at the pri-miRNA promoters can also suppress miRNA and help relieve the suppressive effect of TS-miRs (31). Besides, sulfated glycolipids can facilitate intrahepatic metastasis (32).
HBV and HCV oncoproteins may also regulate miRNA expression (33) by engaging DNA methyltransferases (DNMT), which cause global hyper-methylation (34, 35). The HBx oncoprotein is known to sequester the epigenetic modifier PPARγ in order to down-regulate miR-122 in HCC (36). The HBx-mediated epigenetic silencing of miR-205 stabilizes HBx mRNA, and aggravates disease (37). Similarly, HCV core protein can down-regulate miR-124 and miR-345 levels by inducing DNMT expression and abrogating apoptosis (38).
Single Nucleotide Polymorphism and Genetic Alterations
Single nucleotide polymorphism (SNP) in the regulatory or coding regions of miRNA is essential for onco-miR expression or silencing of TS-miRs in HCC. TS-miRs such as miR-34b/c are poorly expressed in HCC owing to SNP RS4938723, which inhibits the recruitment of transcription factor GATA (39). Alternately, SNPs in the coding region of miR-196a2 and promoter of miR-106b-25 up-regulate their expression and contribute toward HCC (40, 41). SNP in the stem–loop of pre-miR-146a affects its processing efficiency and increases the risk of HCC (42). As miRNA genes are often located close to fragile sites (43), their translocation is frequently observed in many cancers. The translocation of 5′ end of the hcr gene (encoding miR-122) to c-myc locus can cause a massive 50-fold increase in c-Myc expression and a consequent down-regulation of miR-122 in woodchuck hepatitis virus-related HCC (24, 44).
Thus, miRNAs have emerged as crucial players in virus–host interactions, where hepatitis viruses can alter miRNA biogenesis, which in turn control key cellular pathways to establish a successful infection as discussed in the next section.
Deregulation of miRNA in HCC
Deregulated miRNAs play a pivotal role in supporting viral replication and perturbing key cellular processes in HBV- and HCV-associated HCC as depicted in Figure 1.
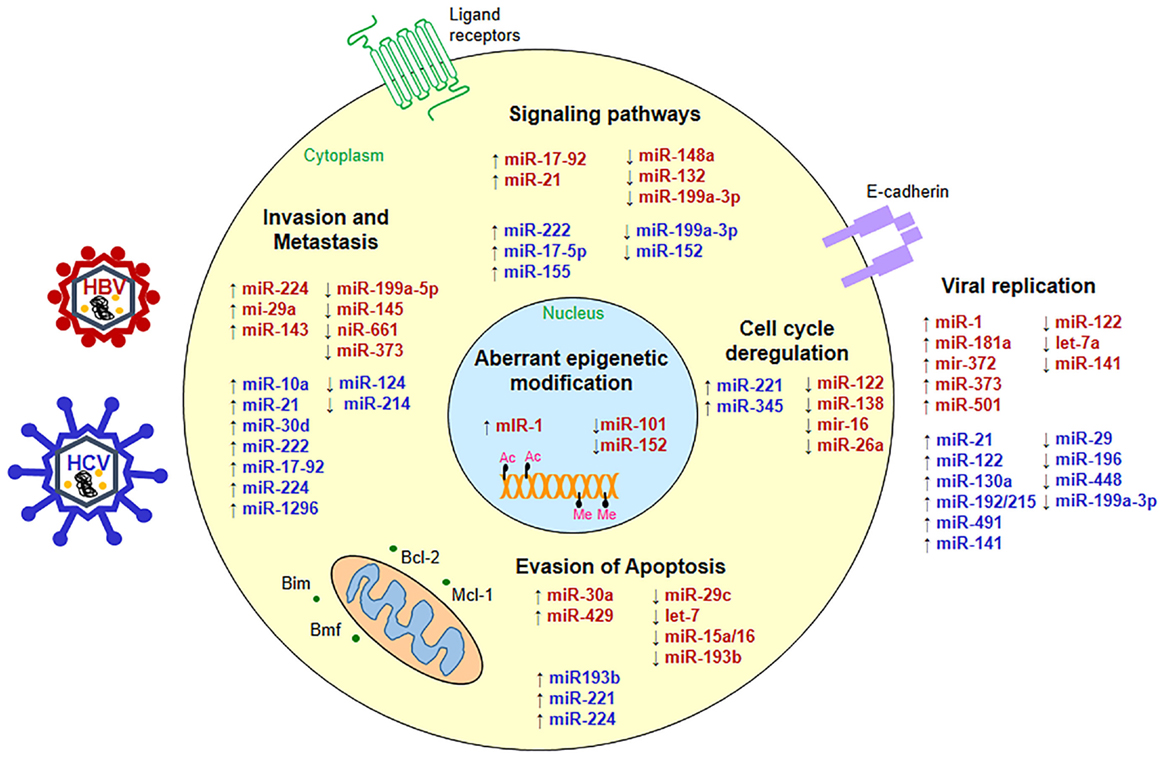
Figure 1. Status of micro-RNAs during hepatitis virus infection. miRNAs are up- (↑) or down-regulated (↓) in HBV- (red) or HCV-associated HCC (blue) and interfere with key cellular pathways such as cell signaling, apoptosis, cell cycle, metastasis as well as viral replication to promote hepatocarcinogenesis.
miRNA in the Regulation of Viral Replication
HBV and HCV engage host miRNAs to complete virus life cycle and establish chronic infection. For example, miR-122, which is often down-regulated in many liver pathologies, shows opposing effects on HBV and HCV replication (45). miR-122 restricts HBV replication by targeting cyclin G1 and allowing p53-mediated suppression of viral genes (46). Paradoxically, miR-122 may support HBV replication by down-regulating heme oxygenase-1 (HO-1), a negative regulator of HBV transcription (47). Further, miR-1 and miR-372 along with miR-373 can enhance HBV replication by activating farnesoid X receptor-alpha (FXRA) and nuclear factor I/B, respectively (48, 49). Interestingly, miR-15b can also promote viral replication by binding to hepatocyte nuclear factor 1α and inducing HBV enhancer 1, while down-regulated miR-15b levels lower virus load and allow persistent HBV infection (50).
In case of HCV, however, binding of miR-122 to the internal ribosome entry site (IRES) in the 5′-UTR of viral RNA augments viral replication and promotes HCV translation in an Ago-2-dependent manner (45, 51). miR-122 and miR-196 also target HO-1 by differentially regulating its repressor Bach-1 to complement or inhibit HCV replication, respectively (52, 53). Akin to miR-122, miR-199a-3p also binds to the IRES element and inhibits HCV replication (53). Interestingly, miR-27a is over-expressed in HCV cases which lowers the viral load to escape immune surveillance through a negative feedback loop (54).
Role of Deregulated miRNAs in Key Cellular Processes
The down-regulation of TS-miRs and concomitant elevation of onco-miRs also affect some key cellular processes, which support HCC.
Cell Cycle and Proliferation
microRNAs can target cell cycle regulators to override some checkpoints and support uncontrolled proliferation. For example, TS-miR-26a targets cyclin D2/E2 that control G1/S boundary of the cell cycle since its re-expression prevented disease progression in a pre-clinical HCC model (55). Further, down-regulated levels of miR-195 and miR-138 act as tumor suppressor by targeting cyclin D1 and cyclin D3 (56, 57), while onco-miRs miR-221 and miR-222 promote tumor growth by down-regulating p27, p57, and DNA damage inducible transcript 4 (58). The let-7 family of miRNAs, which is down-regulated in HCC, allows cell proliferation by negatively regulating STAT3 and c-Myc levels and positively regulating p16INK4A (59, 60). MiR-99a suppresses G1 arrest of cancer cells by targeting cyclin D1 via mTOR (61).
Suppression of Apoptosis
Cancer cells evade apoptosis by perturbing the balance between pro and anti-apoptotic factors. In aggressive HCC cases, miR-25 and miR-221 are over-expressed, which target pro-apoptotic proteins Bim and Bmf, respectively (62, 63). Down-regulated miR-29, miR-101, and miR-122 relieve suppression of their anti-apoptotic targets Bcl-2, Mcl-1, and Bcl-w, respectively, leading to increased survival in HCC (64). HBx oncoprotein down-regulates let-7 and miR-15a/16 leading to increased anti-apoptotic activity (65). Similarly, low levels of TS-miR-29c in HCC target tumor necrosis factor alpha-induced protein 3 to prevent apoptosis (66).
Alteration of Signaling Pathways
Tumor suppressor PTEN, a negative regulator of PI3K/AKT signaling is frequently targeted by elevated levels of miR-21, miR-222, and miR-29a in HCC (59, 64). miR-199-3p targets mTOR and c-Met, a HGF receptor, which controls downstream PAK4/Raf/MEK/ERK pathway (67). Restoration of miR-199-3p levels in hepatoma cells leads to G1 arrest, enhanced susceptibility to hypoxia and drug (67). HBx suppresses the p53-dependent activation of miR-148a, resulting in upregulation of AKT, ERK, and activation of mTOR pathway, which promotes tumor growth and metastasis in a HCC mouse model (68). miR-17-5p, a member of onco-miR-17-92 cluster, activates p38-MAPK- and Hsp27 pathway to promote cell migration and proliferation (69). Further, miR-222 overexpression confers metastatic potential on cancer cells by activating Akt pathway (70). Besides its pro-inflammatory role, miR-155 upregulation by HCV results in increased nuclear accumulation of signal transducer β-catenin to promote HCC (71).
Epithelial–Mesenchymal Transition and Metastasis
Prior to metastasis, tumor cells undergo epithelial–mesenchymal transition (EMT) involving loss of E-cadherin, gain of vimentin, collagen I, and fibronectin. miR-224, a highly expressed miRNA in HCC, promotes tumor growth and metastasis by silencing its target genes Cdc42, CDH1, PAK2, and BCL-2 (72). Elevated levels of miR-29a and miR-148a, which target PTEN and miR-143 also down-regulate fibronectin type III domain-containing protein 3B to enhance hepatoma cell migration (73, 74). HBx engages miR-661 and miR-373 to stimulate metastasis associated-1 protein and suppress E-cadherin, respectively (75). TS-mir-34a and miR-125b that, respectively, target c-Met and LIN28B2 oncogenes are inversely correlated with metastasis in HCC (76, 77). In contrast, miR-200c suppresses EMT of liver cancer stem cells (CSCs) by upregulating E-cadherin, reducing vimentin, and inhibiting metastasis through repression of neurotrophic receptor tyrosine kinase 2 (78). Zeb1 and Zeb2 transcription factors are also targeted by miR-141/200c to alter E-cadherin and related gene expression involved in cell polarity (79). Not surprisingly, miR-141/200c cluster is often silenced in cancer by DNA methylation (80).
Thus, miRNAs can initiate multistep process of cancer by perturbing normal cell homeostasis and endowing cells with the ability to invade and metastasize. Further, the crosstalk between miRNAs could have a multiplier effect on their target genes as well as drug resistance cancer phenotype.
miRNAs as Diagnostic Markers in HCC
The lack of effective diagnostic methods for early HCC has rendered the overall survival rate to a low 0–14% from the time of clinical diagnosis (81). Modest precision of currently used diagnostic markers such as alpha-fetoprotein (AFP) calls for testing the prospect of miRNAs as HCC biomarkers. Circulating miRNAs are highly stable in serum owed to their resistance to RNAse, extreme pH, and temperature, hence are perfect as biomarkers for detecting early stage, presymptomatic diseases such as HCC as depicted in (Figure 2).
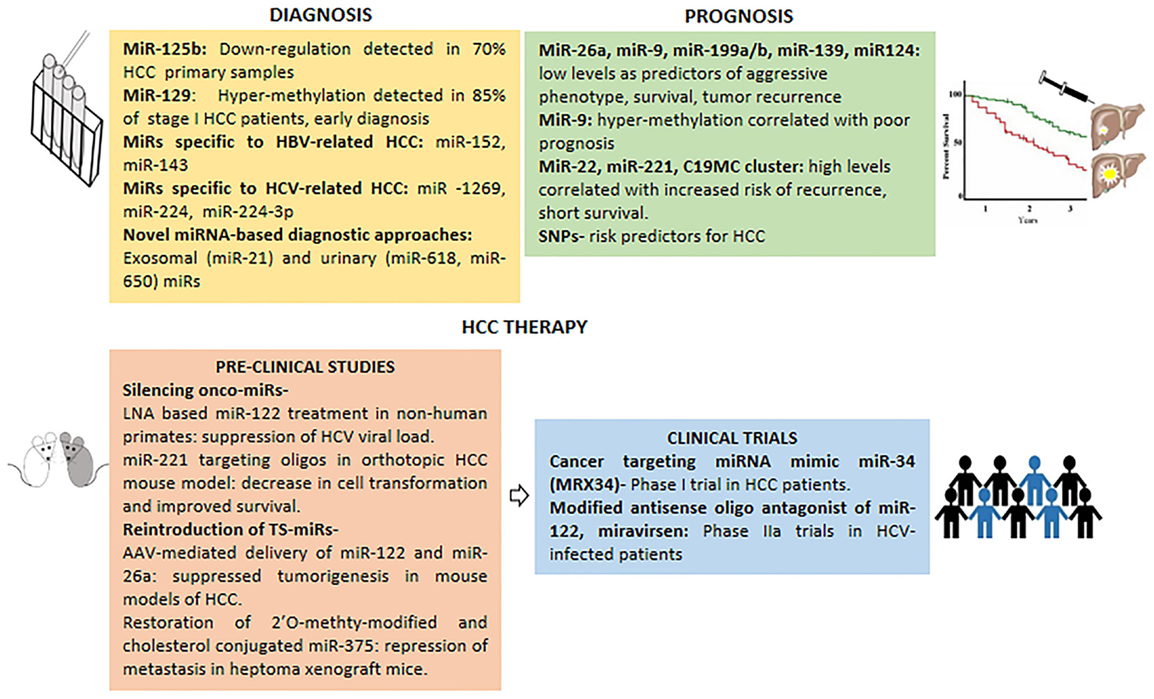
Figure 2. Micro-RNAs in clinical management of HCC. miRNAs with potential in HCC diagnosis, prognosis and therapy (pre-clinical studies and clinical trials).
The serum levels of miRNAs undergo alterations in HCC patients as evident from low levels of miR-16, miR-199a, and high levels of miR-21, miR-221, miR-222, miR-223, and miR-224 in serum samples (5). miR-125b is down-regulated in ~70% of primary HCC samples thus could be a good candidate for diagnosis (77). Further to expand the repertoire of prospective miRNAs in early diagnosis of HCC, down-regulated levels of TS-miR-129 in HCC can be detected in plasma samples from 85% of stage I HCC patients as compared to AFP in just 10% of stage I cases (82).
Detection of HBV- or HCV-positive HCC cases, especially those independent of cirrhosis etiology, poses a greater challenge due to lack of biomarkers. In light of this, unique expression profile of serum miRNAs in HBV- and HCV-positive HCC patients can serve as a fingerprint for distinguishing between HBV and HCV cases. Not surprisingly, miRNAs such as miR-1269, miR-224, and miR-224-3p are significantly altered specifically in HCV-associated HCCs (83). In contrast, miR-152 and miR-143 are aberrantly regulated in HBV-related HCC and hence constitute potential diagnostic markers for HBV-related HCC cases (84, 85).
Serum miRNAs enriched in exosomes can also serve as valuable non-invasive HCC biomarkers for both diagnostic and prognostic purposes. Indeed, recent findings have shown that serum exosomal miR-21 from HCC patients provides increased sensitivity of detection compared to whole serum (86). Interestingly, a report on urinary miRNAs such as miR-618 and miR-650 has opened the prospect to use them as biomarkers for early detection of HCV-induced HCC (87).
Predictive Prognostic Value of miRNAs in HCC
It is being increasingly realized that miRNAs may possess an edge over mRNA as prognostic indicators owed to their stability in clinical samples and robust expression patterns. In fact, expression profiles of a panel of 20 miRNAs can be used as metastatic predictor, correlating with survival as well as relapse rates in HCC (88). As indicated in Figure 2, hyper-methylation of miRNA promoters seems to correlate with poor prognosis, exemplified by aberrant methylation of miR-9 and corroborates with clinical outcomes (89). Further, miR-199a/b and miR-139, which are frequently down-regulated in most HCC patients, show a significant correlation with poor survival (89, 90). Likewise, low expression of miR-124 in HCC seems to be associated with more aggressive behavior and shorter survival (89). While, high levels of miR-22, miR-221, and C19MC miRNA correlate with increased risk of tumor recurrence and shorter survival (59).
Interestingly, the patterns of miRNA expression as well as SNPs in some miRNAs seem potent as predictors of patient response to various therapeutic strategies as well as disease risk (63). In Chinese and Turkish populations, a positive association has been noticed between rs11614913 (C →T) SNP in miR-196a-2 and HCC susceptibility (91, 92). Similarly, “TTCA” insertion (rs3783553) in 3′-UTR of IL-α gene, which ablates the binding site for miR-122 and miR-378 leading to upregulation of IL1-α, correlates well with HCC development (93).
miRNAs in HCC Therapy
Alterations in miRNA expression are frequently associated with HCC disease physiology; hence miRNAs could be used as potential druggable targets in HCC management. One of the approaches in miRNA-based HCC therapy involves using antagomirs against onco-miRs. Non-human primates chronically infected with HCV when treated with locked nucleic acid (LNA) specific for miR-122 (a positive regulator of HCV replication), exhibited long-term suppression of HCV viral load, supporting therapeutic use of miRNA in HCC (2). In another pre-clinical study, involving an orthotopic HCC mouse model, oligonucleotides targeting miR-221 inhibited cell transformation and improved survival, underscoring its potential in HCC therapeutics (5). Another potential therapeutic approach for HCC treatment entails restoration of TS-miRs, thus serving as anti-cancer agents. For instance, AAV-mediated delivery of miR-122 and miR-26a and systemic restoration of miR-124 can suppress tumorigenesis in animal models of HCC. Similarly, restoration of miR-375 (2′O-methyl-modified and cholesterol conjugated form) and miR-29 could inhibit tumorigenesis in pre-clinical HCC models (2, 5).
Delivery of miRNA mimics can also be used in HCC therapy. Indeed, a cancer-targeting miRNA mimic of miR-34 (MRX34) is in phase I clinical trial performed by Mirna Therapeutics Inc., in HCC patients (5, 94). Interestingly, the mimic miRNAs are delivered using Smarticles®, which are anionic at neural pH, but attain cationic charge in acidic tumor environment thus minimizing off-target effects (5). Importantly, the therapeutic potential of miR-122 antagonist, miravirsen was evident from a multi-centric phase IIA trial, which showed sequestration of mature miR-122 and reduction of viral load (95).
Though miRNAs possess tremendous therapeutic potential for HCC, a major concern remains their delivery system. miRNAs can be incorporated into PEG-gylated stable nucleic acid lipid particles (SNALPS), to extend the circulation time. Further, virus-like particle (VLP) dependent delivery gives the leverage of natural tissue tropism, albeit with risk of eliciting immune response (5).
The emergence of miRNAs as novel clinical biomarkers is set to change the face of HCC diagnosis and therapeutic procedures. The proposition of miRNA profiles serving as signatures to distinguish HCV from HBV cases, though awaiting clinical evaluation, offers the advantage of accurate diagnosis and appropriate therapeutic course.
Conclusion
The heterogeneity in individual cases of cancer including HCC demands development of personalized medicine to ensure most effective treatment with minimal side effects. In such a scenario, unique tumor specific miRNA signatures, as reviewed here, will help design accurate diagnostics and therapeutics tailored to individual needs. The ease of delivering oligonucleotides to liver and high tolerance of normal liver cells to supplements of deficient miRNAs make HCC an ideal model to test miRNA therapy (2). Preliminary evidence in non-human primates indicates that miRNA-based treatment of chronic HCV infection provides prolonged alleviation of virus-induced liver pathology with a high barrier to viral resistance suggesting miRNAs may function as better antivirals than conventional drugs (2). CSCs, resistant to conventional chemotherapy and cause of cancer recurrence, form the major obstacle in successful treatment of the disease. The discovery of stemness associated miRNAs in liver cancer such as miR-181, miR-150, and miR-548c-5p warrants further evaluation of their potential as druggable targets in therapies against liver CSCs (96). The gender specific discrepancy in HCC incidence can be in part explained by microRNA signatures such as higher levels of TS miR-26a and miR-26b in females that may provide them a protective edge over males (97). Elevated levels of miR-18a in HCC female patients, which suppresses protective effects of estrogen by targeting estrogen receptor-alpha, can be used as a risk predictor of HCC development in female population (98). Finally, the quest for yet elusive HBV and HCV encoded miRNAs poses a challenge, which when overcome may help in devising novel strategies to silence viral miRNA and cure liver cancer.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Stewart BW, Wild CP. World Cancer Report 2014. Lyon: International Agency for Research on Cancer, World Health Organization (2014).
2. Callegari E, Gramantieri L, Domenicali M, D’Abundo L, Sabbioni S, Negrini M. microRNAs in liver cancer: a model for investigating pathogenesis and novel therapeutic approaches. Cell Death Differ (2015) 22:46–57. doi: 10.1038/cdd.2014.136
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
3. Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science (2007) 318:1931–4. doi:10.1126/science.1149460
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
4. Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, et al. microRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci U S A (2007) 104:15805–10. doi:10.1073/pnas.0707628104
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
5. Yang N, Ekanem NR, Sakyi CA, Ray SD. Hepatocellular carcinoma and microRNA: new perspectives on therapeutics and diagnostics. Adv Drug Deliv Rev (2014) 81C:62–74. doi:10.1016/j.addr.2014.10.029
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
6. Chu R, Mo G, Duan Z, Huang M, Chang J, Li X, et al. miRNAs affect the development of hepatocellular carcinoma via dysregulation of their biogenesis and expression. Cell Commun Signal (2014) 12:45. doi:10.1186/s12964-014-0045-y
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
7. Bui TV, Mendell JT. Myc: maestro of microRNAs. Genes Cancer (2010) 1:568–75. doi:10.1177/1947601910377491
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
8. Han H, Sun D, Li W, Shen H, Zhu Y, Li C, et al. A c-Myc-microRNA functional feedback loop affects hepatocarcinogenesis. Hepatology (2013) 57:2378–89. doi:10.1002/hep.26302
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
9. Wang L, Zhang X, Jia LT, Hu SJ, Zhao J, Yang JD, et al. c-Myc-mediated epigenetic silencing of microRNA-101 contributes to dysregulation of multiple pathways in hepatocellular carcinoma. Hepatology (2014) 59:1850–63. doi:10.1002/hep.26720
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
10. Wang B, Hsu SH, Wang X, Kutay H, Bid HK, Yu J, et al. Reciprocal regulation of microRNA-122 and c-Myc in hepatocellular cancer: role of E2F1 and transcription factor dimerization partner 2. Hepatology (2014) 59:555–66. doi:10.1002/hep.26712
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
11. O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature (2005) 435:839–43. doi:10.1038/nature03677
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
12. Wang X, Zhao X, Gao P, Wu M. c-Myc modulates microRNA processing via the transcriptional regulation of Drosha. Sci Rep (2013) 3:1942. doi:10.1038/srep01942
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
13. Connolly E, Melegari M, Landgraf P, Tchaikovskaya T, Tennant BC, Slagle BL, et al. Elevated expression of the miR-17-92 polycistron and miR-21 in hepadnavirus-associated hepatocellular carcinoma contributes to the malignant phenotype. Am J Pathol (2008) 173:856–64. doi:10.2353/ajpath.2008.080096
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
14. Mukherjee A, Shrivastava S, Bhanja Chowdhury J, Ray R, Ray RB. Transcriptional suppression of miR-181c by hepatitis C virus enhances homeobox A1 expression. J Virol (2014) 88:7929–40. doi:10.1128/JVI.00787-14
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
15. Zou C, Li Y, Cao Y, Zhang J, Jiang J, Sheng Y, et al. Up-regulated microRNA-181a induces carcinogenesis in hepatitis B virus-related hepatocellular carcinoma by targeting E2F5. BMC Cancer (2014) 17(14):97. doi:10.1186/1471-2407-14-97
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
16. Ji D, Chen Z, Li M, Zhan T, Yao Y, Zhang Z, et al. microRNA-181a promotes tumor growth and liver metastasis in colorectal cancer by targeting the tumor suppressor WIF-1. Mol Cancer (2014) 13:86. doi:10.1186/1476-4598-13-86
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
17. Ding Z, Qian YB, Zhu LX, Xiong QR. Promoter methylation and mRNA expression of DKK-3 and WIF-1 in hepatocellular carcinoma. World J. Gastroenterol (2009) 15:2595–601. doi:10.3748/wjg.15.2595
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
18. Feng Z, Zhang C, Wu R, Hu W. Tumor suppressor p53 meets microRNAs. J Mol Cell Biol (2011) 3:44–50. doi:10.1093/jmcb/mjq040
19. Hermeking H. microRNAs in the p53 network: micromanagement of tumour suppression. Nat Rev Cancer (2012) 12:613–26. doi:10.1038/nrc3318
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
20. Iliou MS, da Silva-Diz V, Carmona FJ, Ramalho-Carvalho J, Heyn H, Villanueva A, et al. Impaired DICER1 function promotes stemness and metastasis in colon cancer. Oncogene (2014) 33:4003–15. doi:10.1038/onc.2013.398
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
21. Chen YL, Yuan RH, Yang WC, Hsu HC, Jeng YM. The stem cell E3-ligase Lin-41 promotes liver cancer progression through inhibition of microRNA-mediated gene silencing. J Pathol (2013) 229:486–96. doi:10.1002/path.4130
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
22. Zhang J, Jin H, Liu H, Lv S, Wang B, Wang R, et al. MiRNA-99a directly regulates Ago2 through translational repression in hepatocellular carcinoma. Oncogenesis (2014) 3:e97. doi:10.1038/oncsis.2014.11
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
23. Hou J, Lin L, Zhou W, Wang Z, Ding G, Dong Q, et al. Identification of miRNomes in human liver and hepatocellular carcinoma reveals miR-199a/b-3p as therapeutic target for hepatocellular carcinoma. Cancer Cell (2011) 19:232–43. doi:10.1016/j.ccr.2011.01.001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
24. Mott JL. microRNAs involved in tumor suppressor and oncogene pathways: implications for hepatobiliary neoplasia. Hepatology (2009) 50:630–7. doi:10.1002/hep.23010
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
25. Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature (2008) 454:56–61. doi:10.1038/nature07086
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
26. Pogribny IP, Rusyn I. Role of epigenetic aberrations in the development and progression of human hepatocellular carcinoma. Cancer Lett (2014) 342:223–30. doi:10.1016/j.canlet.2012.01.038
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
27. Anwar SL, Albat C, Krech T, Hasemeier B, Schipper E, Schweitzer N, et al. Concordant hypermethylation of intergenic microRNA genes in human hepatocellular carcinoma as new diagnostic and prognostic marker. Int J Cancer (2013) 133:660–70. doi:10.1002/ijc.28068
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
28. Buurman R, Gürlevik E, Schäffer V, Eilers M, Sandbothe M, Kreipe H, et al. Histone deacetylases activate hepatocyte growth factor signaling by repressing microRNA-449 in hepatocellular carcinoma cells. Gastroenterology (2012) 143:811–20. doi:10.1053/j.gastro.2012.05.033
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
29. Long XR, He Y, Huang C, Li J. microRNA-148a is silenced by hypermethylation and interacts with DNA methyltransferase 1 in hepatocellular carcinogenesis. Int J Oncol (2014) 44:1915–22. doi:10.3892/ijo.2014.2373
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
30. Huang J, Wang Y, Guo Y, Sun S. Down-regulated microRNA-152 induces aberrant DNA methylation in hepatitis B virus-related hepatocellular carcinoma by targeting DNA methyltransferase 1. Hepatology (2010) 52:60–70. doi:10.1002/hep.23660
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
31. Yuan JH, Yang F, Chen BF, Lu Z, Huo XS, Zhou WP, et al. The histone deacetylase 4/SP1/microrna-200a regulatory network contributes to aberrant histone acetylation in hepatocellular carcinoma. Hepatology (2011) 54:2025–35. doi:10.1002/hep.24606
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
32. Dong YW, Wang R, Cai QQ, Qi B, Wu W, Zhang YH, et al. Sulfatide epigenetically regulates miR-223 and promotes the migration of human hepatocellular carcinoma cells. J Hepatol (2013) 60:792–801. doi:10.1016/j.jhep.2013.12.004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
33. Okamoto Y, Shinjo K, Shimizu Y, Sano T, Yamao K, Gao W, et al. Hepatitis virus infection affects DNA methylation in mice with humanized livers. Gastroenterology (2013) 146:562–72. doi:10.1053/j.gastro.2013.10.056
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
34. Wei X, Tan C, Tang C, Ren G, Xiang T, Qiu Z, et al. Epigenetic repression of miR-132 expression by the hepatitis B virus x protein in hepatitis B virus-related hepatocellular carcinoma. Cell Signal (2013) 25:1037–43. doi:10.1016/j.cellsig.2013.01.019
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
35. Wei X, Xiang T, Ren G, Tan C, Liu R, Xu X, et al. miR-101 is down-regulated by the hepatitis B virus x protein and induces aberrant DNA methylation by targeting DNA methyltransferase 3A. Cell Signal (2013) 25:439–46. doi:10.1016/j.cellsig.2012.10.013
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
36. Song K, Han C, Zhang J, Lu D, Dash S, Feitelson M, et al. Epigenetic regulation of microRNA-122 by peroxisome proliferator activated receptor-gamma and hepatitis b virus X protein in hepatocellular carcinoma cells. Hepatology (2013) 58:1681–92. doi:10.1002/hep.26514
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
37. Zhang T, Zhang J, Cui M, Liu F, You X, Du Y, et al. Hepatitis B virus X protein inhibits tumor suppressor miR-205 through inducing hypermethylation of miR-205 promoter to enhance carcinogenesis. Neoplasia (2013) 15:1282–91.
38. Shiu TY, Huang SM, Shih YL, Chu HC, Chang WK, Hsieh TY. Hepatitis C virus core protein down-regulates p21(Waf1/Cip1) and inhibits curcumin-induced apoptosis through microRNA-345 targeting in human hepatoma cells. PLoS One (2013) 8:e61089. doi:10.1371/journal.pone.0061089
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
39. Xu Y, Liu L, Liu J, Zhang Y, Zhu J, Chen J, et al. A potentially functional polymorphism in the promoter region of miR-34b/c is associated with an increased risk for primary hepatocellular carcinoma. Int J Cancer (2011) 128:412–7. doi:10.1002/ijc.25342
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
40. Li XD, Li ZG, Song XX, Liu CF. A variant in microRNA-196a2 is associated with susceptibility to hepatocellular carcinoma in Chinese patients with cirrhosis. Pathology (2010) 42:669–73. doi:10.3109/00313025.2010.522175
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
41. Liu Y, Zhang Y, Wen J, Liu L, Zhai X, Liu J, et al. A genetic variant in the promoter region of miR-106b-25 cluster and risk of HBV infection and hepatocellular carcinoma. PLoS One (2012) 7:e32230. doi:10.1371/journal.pone.0032230
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
42. Xu T, Zhu Y, Wei QK, Yuan Y, Zhou F, Ge YY, et al. A functional polymorphism in the miR-146a gene is associated with the risk for hepatocellular carcinoma. Carcinogenesis (2008) 29:2126–31. doi:10.1093/carcin/bgn195
43. Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A (2004) 101:2999–3004. doi:10.1073/pnas.0307323101
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
44. Möröy T, Marchio A, Etiemble J, Trépo C, Tiollais P, Buendia MA. Rearrangement and enhanced expression of c-myc in hepatocellular carcinoma of hepatitis virus infected woodchucks. Nature (1986) 324:276–9. doi:10.1038/324276a0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
45. Bandiera S, Pfeffer S, Baumert TF, Zeisel MB. MiR-122 – a key factor and therapeutic target in liver disease. J Hepatol (2014) 62:448–57. doi:10.1016/j.jhep.2014.10.004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
46. Wang S, Qiu L, Yan X, Jin W, Wang Y, Chen L, et al. Loss of microRNA 122 expression in patients with hepatitis B enhances hepatitis B virus replication through cyclin G(1)-modulated P53 activity. Hepatology (2012) 55:730–41. doi:10.1002/hep.24809
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
47. Qiu L, Fan H, Jin W, Zhao B, Wang Y, Ju Y, et al. miR-122-induced down-regulation of HO-1 negatively affects miR-122-mediated suppression of HBV. Biochem Biophys Res Commun (2010) 398:771–7. doi:10.1016/j.bbrc.2010.07.021
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
48. Zhang X, Zhang E, Ma Z, Pei R, Jiang M, Schlaak JF, et al. Modulation of hepatitis B virus replication and hepatocyte differentiation by microRNA-1. Hepatology (2011) 53:1476–85. doi:10.1002/hep.24195
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
49. Guo H, Liu H, Mitchelson K, Rao H, Luo M, Xie L, et al. microRNAs-372/373 promote the expression of hepatitis B virus through the targeting of nuclear factor I/B. Hepatology (2011) 54:808–19. doi:10.1002/hep.24441
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
50. Dai X, Zhang W, Zhang H, Sun S, Yu H, Guo Y, et al. Modulation of HBV replication by microRNA-15b through targeting hepatocyte nuclear factor 1α. Nucleic Acids Res (2014) 42:6578–90. doi:10.1093/nar/gku260
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
51. Luna JM, Scheel TKH, Danino T, Shaw KS, Mele A, Fak JJ, et al. Hepatitis C virus RNA functionally sequesters miR-122. Cell (2015) 160:1099–110. doi:10.1016/j.cell.2015.02.025
52. Shan Y, Zheng J, Lambrecht RW, Bonkovsky HL. Reciprocal effects of micro-RNA-122 on expression of heme oxygenase-1 and hepatitis C virus genes in human hepatocytes. Gastroenterology (2007) 133:1166–74. doi:10.1053/j.gastro.2007.08.002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
53. Gupta P, Cairns MJ, Saksena NK. Regulation of gene expression by microRNA in HCV infection and HCV-mediated hepatocellular carcinoma. Virol J (2014) 11:64. doi:10.1186/1743-422X-11-64
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
54. Shirasaki T, Honda M, Shimakami T, Horii R, Yamashita T, Sakai Y, et al. microRNA-27a regulates lipid metabolism and inhibits hepatitis C virus replication in human hepatoma cells. J Virol (2013) 87:5270–86. doi:10.1128/JVI.03022-12
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
55. Chen L, Zheng J, Zhang Y, Yang L, Wang J, Ni J, et al. Tumor-specific expression of microRNA-26a suppresses human hepatocellular carcinoma growth via cyclin-dependent and -independent pathways. Mol Ther (2011) 19:1521–8. doi:10.1038/mt.2011.64
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
56. Xu T, Zhu Y, Xiong Y, Ge YY, Yun JP, Zhuang SM. microRNA-195 suppresses tumorigenicity and regulates G1/S transition of human hepatocellular carcinoma cells. Hepatology (2009) 50:113–21. doi:10.1002/hep.22919
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
57. Wang W, Zhao LJ, Tan YX, Ren H, Qi ZT. MiR-138 induces cell cycle arrest by targeting cyclin D3 in hepatocellular carcinoma. Carcinogenesis (2012) 33:1113–20. doi:10.1093/carcin/bgs113
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
58. Pineau P, Volinia S, McJunkin K, Marchio A, Battiston C, Terris B, et al. miR-221 overexpression contributes to liver tumorigenesis. Proc Natl Acad Sci USA (2010) 107:264–9. doi:10.1073/pnas.0907904107
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
59. Sun J, Lu H, Wang X, Jin H. microRNAs in hepatocellular carcinoma: regulation, function, and clinical implications. ScientificWorldJournal (2013) 2013:924206. doi:10.1155/2013/924206
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
60. Lan FF, Wang H, Chen YC, Chan CY, Ng SS, Li K, et al. Hsa-let-7g inhibits proliferation of hepatocellular carcinoma cells by downregulation of c-Myc and upregulation of p16(INK4A). Int J Cancer (2011) 128:319–31. doi:10.1002/ijc.25336
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
61. Li D, Liu X, Lin L, Hou J, Li N, Wang C, et al. microRNA-99a inhibits hepatocellular carcinoma growth and correlates with prognosis of patients with hepatocellular carcinoma. J Biol Chem (2011) 286:36677–85. doi:10.1074/jbc.M111.270561
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
62. Li Y, Tan W, Neo TW, Aung MO, Wasser S, Lim SG, et al. Role of the miR-106b-25 microRNA cluster in hepatocellular carcinoma. Cancer Sci (2009) 100:1234–42. doi:10.1111/j.1349-7006.2009.01164.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
63. Gramantieri L, Fornari F, Ferracin M, Veronese A, Sabbioni S, Calin GA, et al. microRNA-221 targets Bmf in hepatocellular carcinoma and correlates with tumor multifocality. Clin Cancer Res (2009) 15:5073–81. doi:10.1158/1078-0432.CCR-09-0092
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
64. Law PT, Wong N. Emerging roles of microRNA in the intracellular signaling networks of hepatocellular carcinoma. J Gastroenterol Hepatol (2011) 26:437–49. doi:10.1111/j.1440-1746.2010.06512.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
65. Liu N, Zhang J, Jiao T, Li Z, Peng J, Cui Z, et al. Hepatitis B virus inhibits apoptosis of hepatoma cells by sponging the microRNA 15a/16 cluster. J Virol (2013) 87:13370–8. doi:10.1128/JVI.02130-13
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
66. Wang CM, Wang Y, Fan CG, Xu FF, Sun WS, Liu YG, et al. MiR-29c targets TNFAIP3, inhibits cell proliferation and induces apoptosis in hepatitis B virus-related hepatocellular carcinoma. Biochem Biophys Res Commun (2011) 411:586–92. doi:10.1016/j.bbrc.2011.06.191
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
67. Fornari F, Milazzo M, Chieco P, Negrini M, Calin GA, Grazi GL, et al. MiR-199a-3p regulates mTOR and c-Met to influence the doxorubicin sensitivity of human hepatocarcinoma cells. Cancer Res (2010) 70:5184–93. doi:10.1158/0008-5472.CAN-10-0145
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
68. Xu X, Fan Z, Kang L, Han J, Jiang C, Zheng X, et al. Hepatitis B virus X protein represses miRNA-148a to enhance tumorigenesis. J Clin Invest (2013) 123:630–45. doi:10.1172/JCI64265
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
69. Yang F, Yin Y, Wang F, Wang Y, Zhang L, Tang Y, et al. miR-17-5p Promotes migration of human hepatocellular carcinoma cells through the p38 mitogen-activated protein kinase-heat shock protein 27 pathway. Hepatology (2010) 51:1614–23. doi:10.1002/hep.23566
70. Wong QW, Ching AK, Chan AW, Choy KW, To KF, Lai PB, et al. MiR-222 overexpression confers cell migratory advantages in hepatocellular carcinoma through enhancing AKT signaling. Clin Cancer Res (2010) 16:867–75. doi:10.1158/1078-0432.CCR-09-1840
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
71. Zhang Y, Wei W, Cheng N, Wang K, Li B, Jiang X, et al. Hepatitis C virus-induced up-regulation of microRNA-155 promotes hepatocarcinogenesis by activating Wnt signaling. Hepatology (2012) 56:1631–40. doi:10.1002/hep.25849
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
72. Zhang Y, Takahashi S, Tasaka A, Yoshima T, Ochi H, Chayama K. Involvement of microRNA-224 in cell proliferation, migration, invasion, and anti-apoptosis in hepatocellular carcinoma. J Gastroenterol Hepatol (2013) 28:565–75. doi:10.1111/j.1440-1746.2012.07271.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
73. Kong G, Zhang J, Zhang S, Shan C, Ye L, Zhang X. Upregulated microRNA-29a by hepatitis B virus X protein enhances hepatoma cell migration by targeting PTEN in cell culture model. PLoS One (2011) 6:e19518. doi:10.1371/journal.pone.0019518
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
74. Yuan K, Lian Z, Sun B, Clayton MM, Ng IO, Feitelson MA. Role of miR-148a in hepatitis B associated hepatocellular carcinoma. PLoS One (2012) 7:e35331. doi:10.1371/journal.pone.0035331
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
75. Tian Y, Yang W, Song J, Wu Y, Ni B. Hepatitis B virus X protein-induced aberrant epigenetic modifications contributing to human hepatocellular carcinoma pathogenesis. Mol Cell Biol (2013) 33:2810–6. doi:10.1128/MCB.00205-13
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
76. Li N, Fu H, Tie Y, Hu Z, Kong W, Wu Y, et al. miR-34a inhibits migration and invasion by down-regulation of c-Met expression in human hepatocellular carcinoma cells. Cancer Lett (2009) 275:44–53. doi:10.1016/j.canlet.2008.09.035
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
77. Liang L, Wong CM, Ying Q, Fan DN, Huang S, Ding J, et al. microRNA-125b suppressed human liver cancer cell proliferation and metastasis by directly targeting oncogene LIN28B2. Hepatology (2010) 52:1731–40. doi:10.1002/hep.23904
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
78. Hung CH, Chiu YC, Chen CH, Hu TH. microRNAs in hepatocellular carcinoma: carcinogenesis, progression, and therapeutic target. Biomed Res Int (2014) 2014:486407. doi:10.1155/2014/486407
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
79. Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev (2008) 22:894–907 Erratum in: Genes Dev (2009) 23(11):1378. doi:10.1101/gad.1640608
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
80. Vrba L, Jensen TJ, Garbe JC, Heimark RL, Cress AE, Dickinson S, et al. Role for DNA methylation in the regulation of miR-200c and miR-141 expression in normal and cancer cells. PLoS One (2010) 5:e8697. doi:10.1371/journal.pone.0008697
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
81. D’Anzeo M, Faloppi L, Scartozzi M, Giampieri R, Bianconi M, Del Prete M, et al. The role of micro-RNAs in hepatocellular carcinoma: from molecular biology to treatment. Molecules (2014) 19(5):6393–406. doi:10.3390/molecules19056393
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
82. Lu CY, Lin KY, Tien MT, Wu CT, Uen YH, Tseng TL. Frequent DNA methylation of MiR-129-2 and its potential clinical implication in hepatocellular carcinoma. Genes Chromosomes Cancer (2013) 52(7):636–43. doi:10.1002/gcc.22059
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
83. Hou W, Bonkovsky HL. Non-coding RNAs in hepatitis C-induced hepatocellular carcinoma: dysregulation and implications for early detection, diagnosis and therapy. World J Gastroenterol (2013) 19(44):7836–45. doi:10.3748/wjg.v19.i44.7836
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
84. Li LM, Hu ZB, Zhou ZX, Chen X, Liu FY, Zhang JF, et al. Serum microRNA profiles serve as novel biomarkers for HBV infection and diagnosis of HBV-positive hepatocarcinoma. Cancer Res (2010) 70(23):9798–807. doi:10.1158/0008-5472.CAN-10-1001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
85. Wei YF, Cui GY, Ye P, Chen JN, Diao HY. microRNAs may solve the mystery of chronic hepatitis B virus infection. World J Gastroenterol (2013) 19(30):4867–76. doi:10.3748/wjg.v19.i30.4867
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
86. Wang H, Hou L, Li A, Duan Y, Gao H, Song X. Expression of serum exosomal microRNA-21 in human hepatocellular carcinoma. Biomed Res Int (2014) 2014:864894. doi:10.1155/2014/864894
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
87. Abdalla MA, Haj-Ahmad Y. Promising candidate urinary microRNA biomarkers for the early detection of hepatocellular carcinoma among high-risk hepatitis C virus egyptian patients. J Cancer (2011) 3:19–31. doi:10.7150/jca.3.19
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
88. Budhu A, Jia HL, Forgues M, Liu CG, Goldstein D, Lam A, et al. Identification of metastasis-related microRNAs in hepatocellular carcinoma. Hepatology (2008) 47(3):897–907. doi:10.1002/hep.22160
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
89. Anwar SL, Lehmann U. DNA methylation, microRNAs, and their crosstalk as potential biomarkers in hepatocellular carcinoma. World J Gastroenterol (2014) 20:7894–913. doi:10.3748/wjg.v20.i24.7894
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
90. Wang C, Song B, Song W, Liu J, Sun A, Wu D, et al. Underexpressed microRNA-199b-5p targets hypoxia-inducible factor-1α in hepatocellular carcinoma and predicts prognosis of hepatocellular carcinoma patients. J Gastroenterol Hepatol (2011) 26(11):1630–7. doi:10.1111/j.1440-1746.2011.06758.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
91. Guo J, Jin M, Zhang M, Chen K. A genetic variant in miR-196a2 increased digestive system cancer risks: a meta-analysis of 15 case-control studies. PLoS One (2012) 7(1):e30585. doi:10.1371/journal.pone.0030585
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
92. Akkız H, Bayram S, Bekar A, Akgöllü E, Ulger Y. A functional polymorphism in pre-microRNA-196a-2 contributes to the susceptibility of hepatocellular carcinoma in a Turkish population: a case-control study. J Viral Hepat (2011) 18(7):e399–407. doi:10.1111/j.1365-2893.2010.01414.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
93. Gao Y, He Y, Ding J, Wu K, Hu B, Liu Y, et al. An insertion/deletion polymorphism at miRNA-122-binding site in the interleukin-1alpha 3’ untranslated region confers risk for hepatocellular carcinoma. Carcinogenesis (2009) 30(12):2064–9. doi:10.1093/carcin/bgp283
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
94. Shaikh F, Goff LW. Decoding hepatocellular carcinoma: the promise of microRNAs. Hepatobiliary Surg Nutr (2014) 3(2):93–4. doi:10.3978/j.issn.2304-3881.2014.02.08
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
95. Janssen HL, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, et al. Treatment of HCV infection by targeting microRNA. N Engl J Med (2013) 368:1685–94. doi:10.1056/NEJMoa1209026
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
96. Ma L, Chua MS, Andrisani O, So S. Epigenetics in hepatocellular carcinoma: an update and future therapy perspectives. World J Gastroenterol (2014) 20(2):333–45. doi:10.3748/wjg.v20.i2.333
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
97. Gao J, Liu QG. The role of miR-26 in tumors and normal tissues (Review). Oncol Lett (2011) 2(6):1019–23.
98. Liu WH, Yeh SH, Lu CC, Yu SL, Chen HY, Lin CY, et al. microRNA-18a prevents estrogen receptor-alpha expression, promoting proliferation of hepatocellular carcinoma cells. Gastroenterology (2009) 136(2):683–93. doi:10.1053/j.gastro.2008.10.029
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: hepatitis B virus, hepatitis C virus, hepatocellular carcinoma, microRNA, onco-miR
Citation: Sidhu K, Kapoor NR, Pandey V and Kumar V (2015) The “macro” world of microRNAs in hepatocellular carcinoma. Front. Oncol. 5:68. doi: 10.3389/fonc.2015.00068
Received: 01 February 2015; Accepted: 08 March 2015;
Published online: 25 March 2015.
Edited by:
Rupert Langer, University of Bern, SwitzerlandReviewed by:
Francesca Fanini, Istituti di Ricovero e Cura a Carattere Scientifico, ItalyToru Furukawa, Tokyo Women’s Medical University, Japan
Copyright: © 2015 Sidhu, Kapoor, Pandey and Kumar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Vijay Kumar, J. C. Bose Fellow, Virology Group, International Centre for Genetic Engineering and Biotechnology (ICGEB), Aruna Asaf Ali Marg, New Delhi 110067, India e-mail:dmlqYXlAaWNnZWIucmVzLmlu
†Kaveri Sidhu, Neetu Rohit Kapoor and Vijaya Pandey have contributed equally to this work.
 Kaveri Sidhu
Kaveri Sidhu Neetu Rohit Kapoor
Neetu Rohit Kapoor Vijaya Pandey
Vijaya Pandey Vijay Kumar
Vijay Kumar