- Department of Biochemistry and Molecular Biology, McGovern Medical School, UTHealth, Houston, TX, United States
Calcium is a critical regulator of cell death pathways. One of the most proximal events leading to cell death is activation of plasma membrane and endoplasmic reticulum-resident calcium channels. A large body of evidence indicates that defects in this pathway contribute to cancer development. Although we have a thorough understanding of how downstream elevations in cytosolic and mitochondrial calcium contribute to cell death, it is much less clear how calcium channels are activated upstream of the apoptotic stimulus. Recently, it has been shown that protein lipidation is a potent regulator of apoptotic signaling. Although classically thought of as a static modification, rapid and reversible protein acylation has emerged as a new signaling paradigm relevant to many pathways, including calcium release and cell death. In this review, we will discuss the role of protein lipidation in regulating apoptotic calcium signaling with direct therapeutic relevance to cancer.
Introduction
Apoptotic cell death is important for embryonic development and tissue homeostasis, and dysfunction of this pathway can contribute to various disease states, including cancer. The intrinsic apoptosis pathway is activated by cellular stress and leads to Bcl-2 protein activation, mitochondrial membrane permeabilization, and release of proapoptotic proteins (1, 2). In addition to their roles in mitochondrial membrane permeabilization, Bcl-2 family proteins are also essential and direct regulators of intracellular calcium during apoptosis by binding to a surprising variety of channels, pumps, and exchangers (3–11). Elevated cytosolic calcium then contributes to the apoptotic program in a multitude of ways including mitochondrial permeabilization and further activation of proapoptotic Bcl-2 proteins (12).
The extrinsic apoptosis pathway is activated by ligand binding to death receptors of the tumor necrosis factor-α (TNFα) superfamily. Ligand binding to the death receptor results in the activation of the initiator caspases 8 and 10 (13–15). Caspase 8/10 can directly cleave and activate effector caspases and/or cleave the proapoptotic Bcl-2 family protein Bid. Truncated Bid leads to mitochondrial permeabilization and release of proapoptotic proteins (16). Cell which do not engage the mitochondrial pathway are called “type I” cells, and those which lead to mitochondrial permeabilization are called “type II” cells (17). Thus, in type II cells the extrinsic pathway converges with the intrinsic pathway at the mitochondria. Calcium also contributes to the progression of the extrinsic pathway (18–21); however, this is less well understood.
Recently, it has been found that multiple proteins which regulate apoptotic calcium release in both the intrinsic and extrinsic pathways are subject to lipidation. Protein lipidation is the cotranslational or posttranslational covalent addition of a variety of lipids, including fatty acids, isoprenoids, and cholesterol, to target proteins. Such modifications regulate protein localization and function in many signaling processes. Recent advances in detecting lipidated proteins by proteomic and targeted approaches have revealed that lipidation of signaling proteins is essential for regulating a wide variety of signaling pathways. Stimulus-dependent lipidation of the apoptotic machinery is likely a central regulator of cell death, and defects in this pathway may be contributing factors in cancer development. In this review, we will discuss how protein lipidation plays an essential role in apoptotic signaling and the relevance to cancer therapeutics.
Types of Lipidation
Lipidation can be categorized into two types based on the location of the modified proteins: those that are modified in the ER lumen and secreted and those that are modified in the cytoplasm or on the cytoplasmic face of membrane (22). The former type includes glycosylphosphatidylinositol (GPI) anchor and cholesterylation, and the latter includes N-myristoylation, acylation, and prenylation.
Glycosylphosphatidylinositol anchor was first discovered in the parasite Trypanosoma brucei where the highly expressed variant surface glycoprotein is anchored to the cell surface via a glycolipid containing phosphatidylinositol (23–25). Since then, many proteins in mammals and lower eukaryotes such as protozoa have been shown to contain GPI anchors with enormous structural variety, most of which include an ethanolamine attached to the carboxyl terminus of the protein, a glycan core, inositol, and lipid moieties (26–28). GPI-anchored peptides often include a cleavable N-terminal signal sequence, which directs the peptide to ER lumen, and a hydrophobic C-terminal sequence that is cleaved at the time of GPI anchor addition (27, 29, 30). GPI anchors facilitate tethering of proteins to the extracellular face of the plasma membrane and are important for many cellular functions, including adhesion, membrane trafficking, and immune system signaling (31–33).
Cholesterylation is a characteristic of the mammalian Hedgehog family, which are secreted signaling proteins that regulate embryonic patterning of many tissues and structures (34, 35). The Hedgehog protein undergoes an autocatalytic processing that internally cleaves between the conserved Gly257 and Cys258 at the GCF motif and yields a ~20 kDa N-terminal signaling domain and a ~25 kDa C-terminal catalytic domain (36, 37). The N-terminal domain receives a cholesterol moiety and is active in signaling (35, 37, 38). Interestingly, multiple studies have detected other potentially cholesterylated proteins (35, 39). However, the identification of these potential cholesterylation targets remains to be elucidated.
N-myristoylation is the attachment of the 14-carbon myristic acid to an N-terminal Gly residue via an amide bond (40). It was first identified as a blocking group that prevents Edman degradation on the N-terminus of the catalytic subunit of cyclic AMP-dependent protein kinase and the calcium-binding β-subunit of calcineurin (41, 42). Many other proteins regulating key signaling pathways, including the Src family non-receptor protein tyrosine kinases (43, 44) and Gα proteins (45, 46) were shown to be myristoylated. These proteins contain the N-terminal sequence Met–Gly– and often have a Ser/Thr/Cys at position 6 (40, 47). Myristoylation can happen cotranslationally following the removal of the initiator methionine residue (48). Although myristoylation is required for membrane targeting of many proteins, it is not sufficient for stable membrane anchoring due to its weak hydrophobic nature and often needs subsequent lipid modifications (49–51). Additionally, myristoylation can also happen posttranslationally during apoptosis following the caspase cleavage of substrate proteins that exposes an internal glycine (52–55). Many apoptotic proteins, including Bid, gelsolin, and p21-activated kinase 2, require posttranslational myristoylation following caspase cleavage for proper subcellular localization and subsequent functions (52–54).
Acylation is the addition of various fatty acids, such as palmitic acid, oleic acid, and stearic acid, on different amino acid residues (56–58). One of the best studied types of acylation is S-palmitoylation, which is characterized by the reversible addition of the 16-carbon saturated palmitic acid to Cys residues via labile thioester bonds (58, 59). Despite the presence of multiple algorithms to predict palmitoylation sites, there is no validated consensus sequence for palmitoylation (60–62). One key aspect of palmitoylation is that its reversibility allows for a palmitoylation and depalmitoylation cycle that regulates the posttranslational trafficking and functions of target proteins, such as H- and N-Ras (63). Palmitoylation can also occur on large transmembrane proteins, including ion channels and G protein-coupled receptors (64–68). The roles of transmembrane protein palmitoylation include regulation of channel maturation/quality control and association with lipid rafts (69–72).
Prenylation is the addition of the 15-carbon farnesyl or the 20-carbon geranylgeranyl isoprenoid lipid on cysteine residues via stable thioether bonds (73, 74). It requires a C-terminal CAAX motif, where C is a cysteine, A is aliphatic amino acids, and X can be any amino acid. Prenylation at the CAAX motif is found in many proteins, including mammalian Ras proteins (75, 76). In addition to its role in membrane association, prenylation can also regulate protein–protein interaction and subcellular distribution of the modified targets (77, 78).
Enzymatic Regulators of Protein Lipidation
Protein lipidation is catalyzed by specific enzymatic regulators crucial for the addition (and removal in the case of S-acylation) of the lipid moieties. The GPI precursor, formed in ER lumen, is transferred to target proteins by GPI transamidase, a membrane-bound multi-subunit enzyme (79–82). GPI transamidase cleaves the C-terminal signal peptide of the target proteins, and forms an enzyme-substrate intermediate, allowing the nucleophilic attack by the terminal amino group of preformed GPI (83). On the other hand, cholesterylation of the N-terminal signaling domain of Hedgehog seems to be only dependent on the presence of the C-terminal catalytic domain, suggesting that this process is autocatalytic (35).
N-myristoylation is catalyzed by N-myristoyltransferases (NMTs) (84–86). NMTs bind first to myristoyl-CoA and then to the peptide, followed by a direct nucleophilic addition–elimination reaction and subsequent release of CoA and the myristoylated peptide (87). Studies in various tissues and cell types have shown that the enzymatic activity of NMTs is predominantly distributed in the cytosolic fraction (88–91). Some studies have shown that low levels of myristoyl-CoA may be rate limiting for NMT activity (92, 93). However, the transcriptional upregulation of NMT under pathological conditions suggests that this might not always be the case (94).
Protein S-acylation is regulated by palmitoyl acyltransferases (PATs) that catalyze lipid attachment to cysteine residues and acyl-protein thioesterases (APTs), which remove them. There are 23 PATs in mammals, all of which share a common DHHC (Asp–His–His–Cys) motif within a cysteine-rich domain (95, 96). PATs are polytopic membrane proteins that are localized to distinct subcellular compartments, primarily the Golgi apparatus and the plasma membrane (97). Some DHHC enzymes show preference for certain types of proteins (i.e., transmembrane proteins), and in some cases the same substrates can be palmitoylated by multiple DHHC enzymes (95, 98–100). Compared to the large amount of studies on PATs, thioesterases are relatively poorly characterized. Two protein palmitoyl thioesterases (PPT1/2) and two APTs (APT1/2) have been identified (101–104). PPTs predominantly localize to the lysosomal lumen and are involved in depalmitoylation during protein degradation, whereas APTs have cytosolic localization and are shown to depalmitoylate and recycle signaling proteins such as Ras and growth associate protein (GAP-43) from the plasma membrane back to Golgi (102–105). Very recently, two independent groups found that the α/β hydrolase fold (ABHD) family of serine hydrolases is potent depalmitoylating enzymes for select substrates, including PSD-95 and N-Ras (106, 107).
Prenylation is catalyzed by the enzymes farnesyltransferase (FTase), geranylgeranyltransferase I (GGTase 1), and Rab geranylgeranyltransferase (GGTase 2) (108–110). The prenylating enzymes localize to the cytosol and conjugate isoprenoids generated from mevalonate/HMG-CoA reductase pathway to target proteins. Specifically, the isoprenoids farnesyl and geranylgeranyl are transferred to a C-terminal CAAX motif on target proteins. Unlike FTase and GGTase 1, geranylgeranyl transfer by GGTase 2 requires the co-factor REP (Rab escort protein) (111). GGTase 1 and FTase generally have high specificity for the protein targets, depending on the X residue (112–114). However, they can act on each other’s substrates. One example is that K-Ras and N-Ras, usually targets of FTase, can be geranylgeranylated in Ras-mutant human cancer cells treated with FTase inhibitors (115, 116). Removal of the –AAX tripeptide and methylation of the prenyl-cysteine, catalyzed by the ER membrane proteins RCE1 and ICMT, respectively, are two post-prenylation steps required for maturation of prenylated proteins (117–119).
Lipidation and Apoptotic Calcium Release
As noted in the Section “Introduction,” calcium regulates many cellular processes and plays a prominent role in cell death signaling. Both intrinsic (12, 120–122) and extrinsic (18–21, 123) apoptotic stimuli lead to cytosolic, nuclear, and mitochondrial calcium elevations, which contribute to the execution of the apoptotic program. It is well known that many proteins that regulate cytosolic calcium and apoptotic calcium release are also subject to lipidation including pumps (124), exchangers (125), channels (126), and regulatory proteins (127). Perhaps best studied is protein palmitoylation due to the proliferation of proteomic studies using acyl-biotin exchange (ABE) to identify fatty-acylated proteins (128). Proteins are often assumed to be palmitoylated in ABE experiments, but clearly other lipids may also be conjugated with a thioester bond to target protein cysteine residues (129).
In order to understand how palmitoylation contributes to apoptotic calcium signaling, it is worthwhile to considering the kinetics of the enzymatic machinery. In many proteins which do not have a transmembrane domain, N-terminal myristoylation precedes palmitoylation (130). Over 15 years ago, it was shown that β-adrenergic stimulation resulted in rapid palmitoylation (130, 131) or depalmitoylation (132) of Gαs. The model was based upon availability of free Gαs: dissociation from the βγ subunits allowed putative palmitoylating and depalmitoylation enzymes access to the protein. Under this model, regulated palmitoylation and depalmitoylation cycles would be restricted to proteins which, under physiologic conditions, had regulated exposure of potential palmitoylation sites. Indeed, Gαs is one of only a very select few proteins in which direct palmitoylation within minutes of cellular stimulation has been conclusively determined [although other proteins such as PSD-95, eNOS, and Ras clearly have much higher turnover of palmitoyl groups in response to various stimuli, suggesting rapid cycling of lipid (133)].
Many proteins associated with cell death signaling are modified by lipids. Our group (18) and others (134–137) have investigated the role of palmitoylation in regulating death receptor signaling. We found that components of the T cell receptor (TCR) complex, such as Lck, Zap-70, PLC-γ1 and other TCR components were required for apoptotic calcium release in T cells after engagement of the Fas receptor with Fas ligand (19). The Src kinase Lck is myristoylated and doubly palmitoylated on the N-terminus, and this regulates plasma membrane localization and partitioning into lipid rafts. It is known that the Fas macromolecular complex assembles and signals in lipid rafts (138), so we asked whether Fas stimulation resulted in rapid palmitoylation of Lck. Fas stimulation resulted in a rapid increase in de novo palmitoylation of Lck detectable within minutes of Fas receptor engagement (18). Unexpectedly, the lipid moiety was removed from Lck almost as quickly, and Lck palmitoylation was almost undetectable by 30 min. These kinetics closely matched the phosphorylation and de-phosphorylation of canonical TCR components, such as Zap-70 and PLC-γ1 (18). These findings strongly suggest that the enzymatic mechanisms controlling stimulus-dependent protein palmitoylation and depalmitoylation likely are directly activated by components of the Fas signaling pathway. In the case of Fas signaling, the plasma membrane-localized DHHC21 protein is essential (18). Presumably Fas stimulation rapidly activates DHHC21 and a yet unidentified acyl-protein thioesterase to regulate Lck lipidation levels (Figure 1). How the activation of these enzymes occurs is unknown, but could possibly be regulated either directly or indirectly by calcium ions.
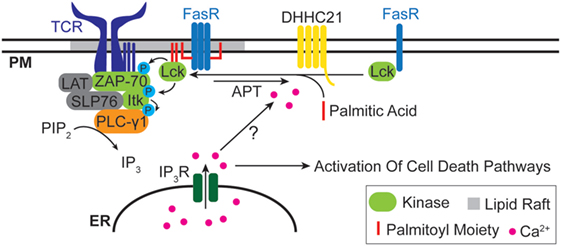
Figure 1. Lipidation and Fas death receptor signaling. The Src kinase Lck is rapidly palmitoylated upon Fas stimulation and partitioned into lipid rafts, where it interacts with the T cell receptor (TCR) complex and leads to downstream apoptotic calcium release. The plasma membrane-localized palmitoyl acyltransferases DHHC21 is essential for Lck palmitoylation. The identity of the depalmitoylating enzyme(s) for Lck is unclear.
In addition to palmitoylation, there has been intense interest in how other lipid modifications are coupled to the cell signaling/cell death machinery. As mentioned above myristoylation of Src kinases regulates their localization to the plasma membrane, which would have implications for coupling to the apoptotic calcium release machinery. The Src-related kinase c-Abl is recruited to membranes via a myristoyl/phosphotyrosine switch (139, 140). The c-Abl kinase sequesters the myristoyl group in a hydrophobic pocket, a conformation which is essential for autoinhibition of the enzyme (140). Binding of phosphotyrosine ligands to the SH2 domain causes a conformational change which results in displacement of the myristoyl group from the pocket, activation of the kinase, and membrane localization. Thus, even though myristoylation is a stable modification, the regulated exposure of the lipid group allows stimulus-dependent regulation of the kinase. During cell death induced by calcium store depletion and subsequent ER stress, c-Abl translocates to mitochondrial membranes to stimulate cytochrome c release (141). It is important to note that the myristoyl switch is not conserved in all Src-related kinases. For example, Lck has bulky residues which occlude the myristoyl binding pocket, and combined with biochemical evidence (142, 143), suggest that a myristoyl switch does not operate in Lck.
Prenylation is a prominent modification of many disease relevant proteins. Small GTPases, such as Rab, Rho, and Ras, are arguably the best studied prenylated proteins; however, as mentioned above, many other proteins have also been shown to be prenylated including chaperones, kinases, enzymes, and receptors (110). How prenylation is linked to apoptotic calcium release has mostly been elucidated by studying the effects of HMG-CoA reductase inhibitors (more commonly known as statins). Many studies have shown that statins induce cell death of multiple cell types, and it is generally assumed to be due to decreased prenylation of GTPases such as Ras and Rho (144–147). We showed that simvastatin was a potent inducer of calcium-dependent apoptosis in human leiomyoma cells (148). The mechanism included inhibition of ERK signaling downstream of Ras and activation of plasma membrane L-type calcium channels via a still unknown mechanism. We were able to extend these finding to xenograft models (149) and showed in a nested case–control study that statin users had a lower risk of uterine fibroid tumors and related symptoms (150). Statins may also be effective for inhibiting the proliferation of many other tumor types including breast (151), ovarian (152), colon (153), and other cancers.
Lipidation and Calcium-Dependent Tumor Progression
In addition to regulating cell death signaling, calcium is also a key factor in proliferative and pro-metastatic signaling. In many types of cancer, alterations in expression, localization, and functions of calcium pumps and channels are observed, resulting in ectopic calcium flux across the plasma membrane or intracellular organelles (154). Studies in prostate (155–157), colon (158), breast (159, 160), and ovarian (161) cancers demonstrated that multiple transient receptor potential (TRP) channels, a family of calcium permeable ion channels, are overexpressed and regulate proliferation in primary tumors. Additionally, TRP channels contribute to tumor cell migration by generating localized calcium signals that guide the direction of movement toward growth factors (162, 163). Another calcium-dependent pathway related to metastasis is the store-operated calcium entry (SOCE) mediated by ER calcium sensor stromal interaction molecule 1 and the plasma membrane calcium channel ORAI1. This pathway has been extensively studied in breast cancer, where it accelerates the turnover rate of focal adhesions by reorganizing the actin cytoskeleton in a Ras and Rac-dependent manner (164). The SOCE pathway is activated by G-protein coupled receptor-mediated activation of the phospholipase C-IP3R pathway, which results in calcium release from ER stores and contributes to metastasis by promoting actin assembly (165, 166).
Many proteins involved in the calcium-dependent proliferative and pro-metastatic pathways are regulated by lipidation (Figure 2). The Wnt signaling pathway is an extensively studied mediator of tumor progression. Immature Wnt proteins (with the exception of WntD) require N-glycosylation and lipidation, specifically palmitate/palmitoleic acylation on conserved C77/C93 and S209/S24 residues for proper secretion and subsequent recognition by Frizzled (Fzd) receptors (167–169). In addition to canonical β-catenin-dependent Wnt signaling, Wnt ligands such as Wnt5a bind Fzd receptors and activate PLC via G-proteins leading to IP3R-mediated increase in cytosolic calcium levels (170). Activation of the non-canonical Wnt/Ca2+ pathway has been implicated in multiple cancer types, including melanoma where it promotes invasion by initiating epithelial-to-mesenchymal transition (171, 172). Increasing in vitro data indicate that Wnt lipidation at the two sites is differentially regulated and activates distinct canonical versus non-canonical pathways (173), but their exact functions in different types of cancers remain unclear. Therefore, further characterization of Wnt lipidation and the mechanisms through which they regulate calcium-dependent proliferation and migration is necessary.
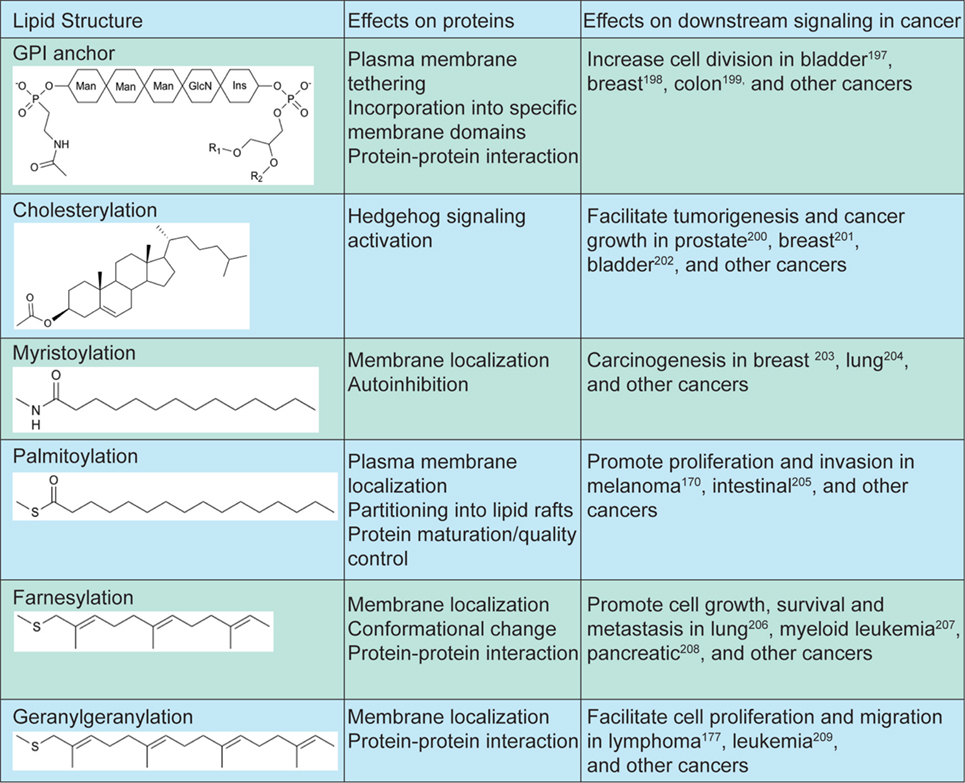
Figure 2. Lipidation structures and effects on downstream signaling in cancer. Increase GPI transamidase activity leads to increased cell proliferation in bladder (174), breast (175), and colon (176) cancers. Elevated hedgehog signaling is linked to sporadic tumorigenesis in prostate (177), breast (178), and bladder (179) cancers. Increased NMT activity and myristoylated Src kinases are linked to increased cell proliferation in breast (180), lung (181), and other cancers. Palmitoylation of signaling proteins in multiple pathways are linked to proliferation and invasion in melanoma (171), intestinal (182), and other cancers. Targeting farnesylation of Ras proteins slows down tumor progression in lung (183), leukemia (184), pancreatic (185), and other cancers. Geranylgeranylation of small GTPases facilitate cell proliferation and migration in lymphoma (186), leukemia (187), and other cancers.
Another class of lipidated proteins that are involved in calcium-mediated cancer progression is the small GTPases, including Ras, Rho and Rac, which are known regulators of the calcium-dependent cytoskeletal rearrangement (164, 188). All three major mammalian isoforms of Ras (H-Ras, N-Ras and K-Ras4B) are farnesylated at the C-terminus (189). Ras farnesylation is required for membrane localization and activation of downstream pathways to induce tumorigenesis (190). Additionally, H-Ras and N-Ras are palmitoylated in the Golgi and subsequently localized to the plasma membrane, where it can be depalmitoylated and cycled back to the Golgi, leading to spatial and temporal control of Ras signaling (191). Rho and Rac are geranylgeranylated near the C-terminus and blocking geranylgeranylation leads to reduced cancer cell proliferation and migration (186, 192).
Lipidation as a Drug Target in Cancer
Many oncogenic proteins require lipidation for proper function. Indeed, Ras is one of the most commonly mutated proteins in cancer (193). As such, the enzymes that mediate these modifications are excellent targets for drug development. Inhibitors of prenylation enzymes GGTase 1 and FTase are being developed to treat cancer. The GGTase 1 inhibitor PTX-200 (GGTI-2418) is being tested in clinical trials by Prescient Therapeutics for breast cancer and multiple myeloma; however, the Phase I trial data has not been published. Many more drugs have been developed which target FTases. There have been several clinical trials conducted with FTase inhibitors, such as lonafarnib, and tipifarnib (194); however, the results have been mostly disappointing (195). Interestingly, positive clinical responses were not correlated to Ras mutation status, suggesting that the drugs target other pathways or substrates (196). Drugs targeting the post-prenylation processing enzymes RCE1 and ICMT are in pre-clinical development (195, 197–199).
As mentioned above, many oncogenic proteins require palmitoylation for proper function, including Ras and Src proteins. There are very few pharmacological tools to target the DHHC enzymes; however, the irreversible lipid-based inhibitor 2-bromopalmitate (200) and several non-lipid reversible inhibitors (201) are widely used as research tools to probe the role of palmitoylation in biological processes. There are no drugs sufficiently well developed to initiate clinical trials, most likely due to the fact that selective DHHC enzyme inhibitors would likely need to be developed. As there are so many enzymes with potentially overlapping substrates, this seems to be a very daunting task. However, new targeted screening strategies for therapeutically relevant substrates, such as Ras, show great promise (202).
As mentioned above, statins are potent inhibitors of the mevalonate pathway and thus may be an attractive target for further development as anti-cancer agents. Indeed, several prospective and retrospective studies have shown that statins have activity against a wide variety of cancers (203–209). There are a multitude of prospective clinical trials currently underway to evaluate the potential of statins as anti-cancer therapeutics. Interestingly, the mechanism of action for inhibiting cancer progression by statins may reflect targeting of multiple pathways, including prenylation of oncogenic proteins and the production of hormones synthesized from cholesterol such as estrogen and testosterone which can drive tumor growth. Statins are one of the most widely prescribed drugs in the world with excellent safety and tolerability profiles (210). If statins prove to be efficacious as a cancer preventative or treatment, this would have the potential to revolutionize cancer care and survival.
Author Contributions
The content of this review was conceived by DB and written with equal effort by JC and DB.
Conflict of Interest Statement
The authors declare that this review was written in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding
This work is supported by National Institutes of Health (NIH) grant R01GM081685 (DB).
References
1. Schweizer M, Richter C. Peroxynitrite stimulates the pyridine nucleotide-linked Ca2+ release from intact rat liver mitochondria. Biochemistry (1996) 35(14):4524–8. doi:10.1021/bi952708+
2. Kidd JF, Pilkington MF, Schell MJ, Fogarty KE, Skepper JN, Taylor CW, et al. Paclitaxel affects cytosolic calcium signals by opening the mitochondrial permeability transition pore. J Biol Chem (2002) 277(8):6504–10. doi:10.1074/jbc.M106802200
3. Pinton P, Ferrari D, Magalhães P, Schulze-Osthoff K, Di Virgilio F, Pozzan T, et al. Reduced loading of intracellular Ca(2+) stores and downregulation of capacitative Ca(2+) influx in Bcl-2-overexpressing cells. J Cell Biol (2000) 148(5):857–62. doi:10.1083/jcb.148.5.857
4. Foyouzi-Youssefi R, Arnaudeau S, Borner C, Kelley WL, Tschopp J, Lew DP, et al. Bcl-2 decreases the free Ca2+ concentration within the endoplasmic reticulum. Proc Natl Acad Sci U S A (2000) 97(11):5723–8. doi:10.1073/pnas.97.11.5723
5. Chen R, Valencia I, Zhong F, McColl KS, Roderick HL, Bootman MD, et al. Bcl-2 functionally interacts with inositol 1,4,5-trisphosphate receptors to regulate calcium release from the ER in response to inositol 1,4,5-trisphosphate. J Cell Biol (2004) 166(2):193–203. doi:10.1083/jcb.200309146
6. Rong YP, Aromolaran AS, Bultynck G, Zhong F, Li X, McColl K, et al. Targeting Bcl-2-IP3 receptor interaction to reverse Bcl-2’s inhibition of apoptotic calcium signals. Mol Cell (2008) 31(2):255–65. doi:10.1016/j.molcel.2008.06.014
7. Scorrano L, Oakes SA, Opferman JT, Cheng EH, Sorcinelli MD, Pozzan T, et al. BAX and BAK regulation of endoplasmic reticulum Ca2+: a control point for apoptosis. Science (2003) 300(5616):135–9. doi:10.1126/science.1081208
8. Oakes SA, Scorrano L, Opferman JT, Bassik MC, Nishino M, Pozzan T, et al. Proapoptotic BAX and BAK regulate the type 1 inositol trisphosphate receptor and calcium leak from the endoplasmic reticulum. Proc Natl Acad Sci U S A (2005) 102(1):105–10. doi:10.1073/pnas.0408352102
9. Vervliet T, Parys JB, Bultynck G. Bcl-2 proteins and calcium signaling: complexity beneath the surface. Oncogene (2016) 35(39):5079–92. doi:10.1038/onc.2016.31
10. Dremina ES, Sharov VS, Kumar K, Zaidi A, Michaelis EK, Schöneich C. Anti-apoptotic protein Bcl-2 interacts with and destabilizes the sarcoplasmic/endoplasmic reticulum Ca2+-ATPase (SERCA). Biochem J (2004) 383(Pt 2):361–70. doi:10.1042/BJ20040187
11. Ferdek PE, Gerasimenko JV, Peng S, Tepikin AV, Petersen OH, Gerasimenko OV. A novel role for Bcl-2 in regulation of cellular calcium extrusion. Curr Biol (2012) 22(13):1241–6. doi:10.1016/j.cub.2012.05.002
12. Orrenius S, Zhivotovsky B, Nicotera P. Regulation of cell death: the calcium-apoptosis link. Nat Rev Mol Cell Biol (2003) 4(7):552–65. doi:10.1038/nrm1150
13. Boldin MP, Goncharov TM, Goltsev YV, Wallach D. Involvement of MACH, a novel MORT1/FADD-interacting protease, in Fas/APO-1- and TNF receptor-induced cell death. Cell (1996) 85(6):803–15. doi:10.1016/S0092-8674(00)81265-9
14. Muzio M, Chinnaiyan AM, Kischkel FC, O’Reourke K, Shevchenko A, Ni J, et al. FLICE, a novel FADD-homologous ICE/CED-3-like protease, is recruited to the CD95 (Fas/APO-1) death-inducing signaling complex. Cell (1996) 85(6):817–27. doi:10.1016/S0092-8674(00)81266-0
15. Fernandes-Alnemri T, Armstrong RC, Krebs J, Srinivasula SM, Wang L, Bullrich F, et al. In vitro activation of CPP32 and Mch3 by Mch4, a novel human apoptotic cysteine protease containing two FADD-like domains. Proc Natl Acad Sci U S A (1996) 93(15):7464–9. doi:10.1073/pnas.93.15.7464
16. Scaffidi C, Fulda S, Srinivasan A, Friesen C, Li F, Tomaselli KJ, et al. Two CD95 (APO-1/Fas) signaling pathways. EMBO J (1998) 17(6):1675–87. doi:10.1093/emboj/17.6.1675
17. Barnhart BC, Alappat EC, Peter ME. The CD95 type I/type II model. Semin Immunol (2003) 15(3):185–93. doi:10.1016/S1044-5323(03)00031-9
18. Akimzhanov AM, Boehning D. Rapid and transient palmitoylation of the tyrosine kinase Lck mediates Fas signaling. Proc Natl Acad Sci U S A (2015) 112(38):11876–80. doi:10.1073/pnas.1509929112
19. Akimzhanov AM, Wang X, Sun J, Boehning D. T-cell receptor complex is essential for Fas signal transduction. Proc Natl Acad Sci U S A (2010) 107(34):15105–10. doi:10.1073/pnas.1005419107
20. Steinmann C, Landsverk ML, Barral JM, Boehning D. Requirement of inositol 1,4,5-trisphosphate receptors for tumor-mediated lymphocyte apoptosis. J Biol Chem (2008) 283(20):13506–9. doi:10.1074/jbc.C800029200
21. Wozniak AL, Wang X, Stieren ES, Scarbrough SG, Elferink CJ, Boehning D. Requirement of biphasic calcium release from the endoplasmic reticulum for Fas-mediated apoptosis. J Cell Biol (2006) 175(5):709–14. doi:10.1083/jcb.200608035
22. Nadolski MJ, Linder ME. Protein lipidation. FEBS J (2007) 274(20):5202–10. doi:10.1111/j.1742-4658.2007.06056.x
23. Ferguson MA, Low MG, Cross GA. Glycosyl-sn-1,2-dimyristylphosphatidylinositol is covalently linked to Trypanosoma brucei variant surface glycoprotein. J Biol Chem (1985) 260(27):14547–55.
24. Ferguson MA, Homans SW, Dwek RA, Rademacher TW. The glycosylphosphatidylinositol membrane anchor of Trypanosoma brucei variant surface glycoprotein. Biochem Soc Trans (1988) 16(3):265–8. doi:10.1042/bst0160265
25. Ferguson MA, Williams AF. Cell-surface anchoring of proteins via glycosyl-phosphatidylinositol structures. Annu Rev Biochem (1988) 57:285–320.
26. Thomas JR, Dwek RA, Rademacher TW. Structure, biosynthesis, and function of glycosylphosphatidylinositols. Biochemistry (1990) 29(23):5413–22. doi:10.1021/bi00475a001
27. Englund PT. The structure and biosynthesis of glycosyl phosphatidylinositol protein anchors. Annu Rev Biochem (1993) 62:121–38. doi:10.1146/annurev.bi.62.070193.001005
28. Paulick MG, Bertozzi CR. The glycosylphosphatidylinositol anchor: a complex membrane-anchoring structure for proteins. Biochemistry (2008) 47(27):6991–7000. doi:10.1021/bi8006324
29. Caras IW, Weddell GN, Davitz MA, Nussenzweig V, Martin DW Jr. Signal for attachment of a phospholipid membrane anchor in decay accelerating factor. Science (1987) 238(4831):1280–3. doi:10.1126/science.2446389
30. Takeda J, Kinoshita T. GPI-anchor biosynthesis. Trends Biochem Sci (1995) 20(9):367–71. doi:10.1016/S0968-0004(00)89078-7
31. Chatterjee S, Mayor S. The GPI-anchor and protein sorting. Cell Mol Life Sci (2001) 58(14):1969–87. doi:10.1007/PL00000831
32. Ferguson MAJ, Kinoshita T, Hart GW. Glycosylphosphatidylinositol anchors. In: Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, et al., editors. Essentials of Glycobiology. NY: Cold Spring Harbor (2009).
33. Fujita M, Kinoshita T. Structural remodeling of GPI anchors during biosynthesis and after attachment to proteins. FEBS Lett (2010) 584(9):1670–7. doi:10.1016/j.febslet.2009.10.079
34. Nusslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature (1980) 287(5785):795–801.
35. Porter JA, Young KE, Beachy PA. Cholesterol modification of hedgehog signaling proteins in animal development. Science (1996) 274(5285):255–9. doi:10.1126/science.274.5285.255
36. Lee JJ, Ekker SC, von Kessler DP, Porter JA, Sun BI, Beachy PA. Autoproteolysis in hedgehog protein biogenesis. Science (1994) 266(5190):1528–37. doi:10.1126/science.7985023
37. Porter JA, von Kessler DP, Ekker SC, Young KE, Lee JJ, Moses K, et al. The product of hedgehog autoproteolytic cleavage active in local and long-range signalling. Nature (1995) 374(6520):363–6. doi:10.1038/374363a0
38. Fan CM, Porter JA, Chiang C, Chang DT, Beachy PA, Tessier-Lavigne M. Long-range sclerotome induction by sonic hedgehog: direct role of the amino-terminal cleavage product and modulation by the cyclic AMP signaling pathway. Cell (1995) 81(3):457–65. doi:10.1016/0092-8674(95)90398-4
39. Heal WP, Jovanovic B, Bessin S, Wright MH, Magee AI, Tate EW. Bioorthogonal chemical tagging of protein cholesterylation in living cells. Chem Commun (Camb) (2011) 47(14):4081–3. doi:10.1039/c0cc04710d
40. Towler DA, Eubanks SR, Towery DS, Adams SP, Glaser L. Amino-terminal processing of proteins by N-myristoylation. Substrate specificity of N-myristoyl transferase. J Biol Chem (1987) 262(3):1030–6.
41. Carr SA, Biemann K, Shoji S, Parmelee DC, Titani K. n-Tetradecanoyl is the NH2-terminal blocking group of the catalytic subunit of cyclic AMP-dependent protein kinase from bovine cardiac muscle. Proc Natl Acad Sci U S A (1982) 79(20):6128–31. doi:10.1073/pnas.79.20.6128
42. Aitken A, Cohen P, Santikarn S, Williams DH, Calder AG, Smith A, et al. Identification of the NH2-terminal blocking group of calcineurin B as myristic acid. FEBS Lett (1982) 150(2):314–8. doi:10.1016/0014-5793(82)80759-X
43. Buss JE, Sefton BM. Myristic acid, a rare fatty acid, is the lipid attached to the transforming protein of Rous sarcoma virus and its cellular homolog. J Virol (1985) 53(1):7–12.
44. Linder ME, Burr JG. Nonmyristoylated p60v-src fails to phosphorylate proteins of 115–120 kDa in chicken embryo fibroblasts. Proc Natl Acad Sci U S A (1988) 85(8):2608–12. doi:10.1073/pnas.85.8.2608
45. Buss JE, Mumby SM, Casey PJ, Gilman AG, Sefton BM. Myristoylated alpha subunits of guanine nucleotide-binding regulatory proteins. Proc Natl Acad Sci U S A (1987) 84(21):7493–7. doi:10.1073/pnas.84.21.7493
46. Mumby SM, Heukeroth RO, Gordon JI, Gilman AG. G-protein alpha-subunit expression, myristoylation, and membrane association in COS cells. Proc Natl Acad Sci U S A (1990) 87(2):728–32. doi:10.1073/pnas.87.2.728
47. Maurer-Stroh S, Eisenhaber B, Eisenhaber F. N-terminal N-myristoylation of proteins: prediction of substrate proteins from amino acid sequence. J Mol Biol (2002) 317(4):541–57. doi:10.1006/jmbi.2002.5426
48. Wilcox C, Hu JS, Olson EN. Acylation of proteins with myristic acid occurs cotranslationally. Science (1987) 238(4831):1275–8. doi:10.1126/science.3685978
49. Peitzsch RM, McLaughlin S. Binding of acylated peptides and fatty acids to phospholipid vesicles: pertinence to myristoylated proteins. Biochemistry (1993) 32(39):10436–43. doi:10.1021/bi00090a020
50. Ames JB, Tanaka T, Stryer L, Ikura M. Portrait of a myristoyl switch protein. Curr Opin Struct Biol (1996) 6(4):432–8. doi:10.1016/S0959-440X(96)80106-0
51. Seykora JT, Myat MM, Allen LA, Ravetch JV, Aderem A. Molecular determinants of the myristoyl-electrostatic switch of MARCKS. J Biol Chem (1996) 271(31):18797–802. doi:10.1074/jbc.271.31.18797
52. Zha J, Weiler S, Oh KJ, Wei MC, Korsmeyer SJ. Posttranslational N-myristoylation of BID as a molecular switch for targeting mitochondria and apoptosis. Science (2000) 290(5497):1761–5. doi:10.1126/science.290.5497.1761
53. Utsumi T, Sakurai N, Nakano K, Ishisaka R. C-terminal 15 kDa fragment of cytoskeletal actin is posttranslationally N-myristoylated upon caspase-mediated cleavage and targeted to mitochondria. FEBS Lett (2003) 539(1–3):37–44. doi:10.1016/S0014-5793(03)00180-7
54. Sakurai N, Utsumi T. Posttranslational N-myristoylation is required for the anti-apoptotic activity of human tGelsolin, the C-terminal caspase cleavage product of human gelsolin. J Biol Chem (2006) 281(20):14288–95. doi:10.1074/jbc.M510338200
55. Martin DD, Vilas GL, Prescher JA, Rajaiah G, Falck JR, Bertozzi CR, et al. Rapid detection, discovery, and identification of post-translationally myristoylated proteins during apoptosis using a bio-orthogonal azidomyristate analog. FASEB J (2008) 22(3):797–806. doi:10.1096/fj.07-9198com
56. Liang X, Lu Y, Neubert TA, Resh MD. Mass spectrometric analysis of GAP-43/neuromodulin reveals the presence of a variety of fatty acylated species. J Biol Chem (2002) 277(36):33032–40. doi:10.1074/jbc.M204607200
57. Takada R, Satomi Y, Kurata T, Ueno N, Norioka S, Kondoh H, et al. Monounsaturated fatty acid modification of Wnt protein: its role in Wnt secretion. Dev Cell (2006) 11(6):791–801. doi:10.1016/j.devcel.2006.10.003
58. Magee AI, Koyama AH, Malfer C, Wen D, Schlesinger MJ. Release of fatty acids from virus glycoproteins by hydroxylamine. Biochim Biophys Acta (1984) 798(2):156–66. doi:10.1016/0304-4165(84)90298-8
59. Kaufman JF, Krangel MS, Strominger JL. Cysteines in the transmembrane region of major histocompatibility complex antigens are fatty acylated via thioester bonds. J Biol Chem (1984) 259(11):7230–8.
60. Xue Y, Chen H, Jin C, Sun Z, Yao X. NBA-Palm: prediction of palmitoylation site implemented in Naive Bayes algorithm. BMC Bioinformatics (2006) 7:458. doi:10.1186/1471-2105-7-458
61. Ren J, Wen L, Gao X, Jin C, Xue Y, Yao X. CSS-Palm 2.0: an updated software for palmitoylation sites prediction. Protein Eng Des Sel (2008) 21(11):639–44. doi:10.1093/protein/gzn039
62. Li YX, Shao YH, Deng NY. Improved prediction of palmitoylation sites using PWMs and SVM. Protein Pept Lett (2011) 18(2):186–93. doi:10.2174/092986611794475084
63. Rocks O, Gerauer M, Vartak N, Koch S, Huang ZP, Pechlivanis M, et al. The palmitoylation machinery is a spatially organizing system for peripheral membrane proteins. Cell (2010) 141(3):458–71. doi:10.1016/j.cell.2010.04.007
64. O’Brien PJ, Zatz M. Acylation of bovine rhodopsin by [3H]palmitic acid. J Biol Chem (1984) 259(8):5054–7.
65. Schmidt JW, Catterall WA. Palmitylation, sulfation, and glycosylation of the alpha subunit of the sodium channel. Role of post-translational modifications in channel assembly. J Biol Chem (1987) 262(28):13713–23.
66. O’Dowd BF, Hnatowich M, Caron MG, Lefkowitz RJ, Bouvier M. Palmitoylation of the human beta 2-adrenergic receptor. Mutation of Cys341 in the carboxyl tail leads to an uncoupled nonpalmitoylated form of the receptor. J Biol Chem (1989) 264(13):7564–9.
67. Hawtin SR, Tobin AB, Patel S, Wheatley M. Palmitoylation of the vasopressin V1a receptor reveals different conformational requirements for signaling, agonist-induced receptor phosphorylation, and sequestration. J Biol Chem (2001) 276(41):38139–46. doi:10.1074/jbc.M106142200
68. Zhang L, Foster K, Li Q, Martens JR. S-acylation regulates Kv1.5 channel surface expression. Am J Physiol Cell Physiol (2007) 293(1):C152–61. doi:10.1152/ajpheart.00268.2006
69. Hayashi T, Rumbaugh G, Huganir RL. Differential regulation of AMPA receptor subunit trafficking by palmitoylation of two distinct sites. Neuron (2005) 47(5):709–23. doi:10.1016/j.neuron.2005.06.035
70. Zheng H, Chu J, Qiu Y, Loh HH, Law PY. Agonist-selective signaling is determined by the receptor location within the membrane domains. Proc Natl Acad Sci U S A (2008) 105(27):9421–6. doi:10.1073/pnas.0802253105
71. Gonnord P, Delarasse C, Auger R, Benihoud K, Prigent M, Cuif MH, et al. Palmitoylation of the P2X7 receptor, an ATP-gated channel, controls its expression and association with lipid rafts. FASEB J (2009) 23(3):795–805. doi:10.1096/fj.08-114637
72. Hayashi T, Thomas GM, Huganir RL. Dual palmitoylation of NR2 subunits regulates NMDA receptor trafficking. Neuron (2009) 64(2):213–26. doi:10.1016/j.neuron.2009.08.017
73. Kamiya Y, Sakurai A, Tamura S, Takahashi N. Structure of rhodotorucine A, a novel lipopeptide, inducing mating tube formation in Rhodosporidium toruloides. Biochem Biophys Res Commun (1978) 83(3):1077–83. doi:10.1016/0006-291X(78)91505-X
74. Farnsworth CC, Gelb MH, Glomset JA. Identification of geranylgeranyl-modified proteins in HeLa cells. Science (1990) 247(4940):320–2. doi:10.1126/science.2296721
75. Hancock JF, Magee AI, Childs JE, Marshall CJ. All ras proteins are polyisoprenylated but only some are palmitoylated. Cell (1989) 57(7):1167–77. doi:10.1016/0092-8674(89)90054-8
76. Casey PJ, Solski PA, Der CJ, Buss JE. p21ras is modified by a farnesyl isoprenoid. Proc Natl Acad Sci U S A (1989) 86(21):8323–7. doi:10.1073/pnas.86.21.8323
77. Hoffman GR, Nassar N, Cerione RA. Structure of the Rho family GTP-binding protein Cdc42 in complex with the multifunctional regulator RhoGDI. Cell (2000) 100(3):345–56. doi:10.1016/S0092-8674(00)80670-4
78. Berg TJ, Gastonguay AJ, Lorimer EL, Kuhnmuench JR, Li R, Fields AP, et al. Splice variants of SmgGDS control small GTPase prenylation and membrane localization. J Biol Chem (2010) 285(46):35255–66. doi:10.1074/jbc.M110.129916
79. Benghezal M, Lipke PN, Conzelmann A. Identification of six complementation classes involved in the biosynthesis of glycosylphosphatidylinositol anchors in Saccharomyces cerevisiae. J Cell Biol (1995) 130(6):1333–44. doi:10.1083/jcb.130.6.1333
80. Hamburger D, Egerton M, Riezman H. Yeast Gaa1p is required for attachment of a completed GPI anchor onto proteins. J Cell Biol (1995) 129(3):629–39. doi:10.1083/jcb.129.3.629
81. Benghezal M, Benachour A, Rusconi S, Aebi M, Conzelmann A. Yeast Gpi8p is essential for GPI anchor attachment onto proteins. EMBO J (1996) 15(23):6575–83.
82. Fraering P, Imhof I, Meyer U, Strub JM, van Dorsselaer A, Vionnet C, et al. The GPI transamidase complex of Saccharomyces cerevisiae contains Gaa1p, Gpi8p, and Gpi16p. Mol Biol Cell (2001) 12(10):3295–306. doi:10.1091/mbc.12.10.3295
83. Chen R, Anderson V, Hiroi Y, Medof ME. Proprotein interaction with the GPI transamidase. J Cell Biochem (2003) 88(5):1025–37. doi:10.1002/jcb.10439
84. Duronio RJ, Towler DA, Heuckeroth RO, Gordon JI. Disruption of the yeast N-myristoyl transferase gene causes recessive lethality. Science (1989) 243(4892):796–800. doi:10.1126/science.2644694
85. Ntwasa M, Egerton M, Gay NJ. Sequence and expression of Drosophila myristoyl-CoA:protein N-myristoyl transferase: evidence for proteolytic processing and membrane localisation. J Cell Sci (1997) 110(Pt 2):149–56.
86. Giang DK, Cravatt BF. A second mammalian N-myristoyltransferase. J Biol Chem (1998) 273(12):6595–8. doi:10.1074/jbc.273.12.6595
87. Rudnick DA, McWherter CA, Rocque WJ, Lennon PJ, Getman DP, Gordon JI. Kinetic and structural evidence for a sequential ordered Bi Bi mechanism of catalysis by Saccharomyces cerevisiae myristoyl-CoA:protein N-myristoyltransferase. J Biol Chem (1991) 266(15):9732–9.
88. Boutin JA, Ferry G, Ernould A, Maes P, Remond G, Vincent M. Myristoyl-CoA: protein N-myristoyltransferase activity in cancer cells. Purification and characterization of a cytosolic isoform from the murine leukemia cell line L1210. Eur J Biochem (1993) 214(3):853–67.
89. McIlhinney RA, McGlone K, Willis AC. Purification and partial sequencing of myristoyl-CoA:protein N-myristoyltransferase from bovine brain. Biochem J (1993) 290(Pt 2):405–10. doi:10.1042/bj2900405
90. Raju RV, Kalra J, Sharma RK. Purification and properties of bovine spleen N-myristoyl-CoA protein:N-myristoyltransferase. J Biol Chem (1994) 269(16):12080–3.
91. McIlhinney RA, McGlone K. Immunocytochemical characterization and subcellular localization of human myristoyl-CoA: protein N-myristoyltransferase in HeLa cells. Exp Cell Res (1996) 223(2):348–56. doi:10.1006/excr.1996.0090
92. van der Vusse GJ, van Bilsen M, Glatz JF, Hasselbaink DM, Luiken JJ. Critical steps in cellular fatty acid uptake and utilization. Mol Cell Biochem (2002) 239(1–2):9–15. doi:10.1023/A:1020538119691
93. Colombo S, Longhi R, Alcaro S, Ortuso F, Sprocati T, Flora A, et al. N-myristoylation determines dual targeting of mammalian NADH-cytochrome b5 reductase to ER and mitochondrial outer membranes by a mechanism of kinetic partitioning. J Cell Biol (2005) 168(5):735–45. doi:10.1083/jcb.200407082
94. Selvakumar P, Lakshmikuttyamma A, Shrivastav A, Das SB, Dimmock JR, Sharma RK. Potential role of N-myristoyltransferase in cancer. Prog Lipid Res (2007) 46(1):1–36. doi:10.1016/j.plipres.2006.05.002
95. Fukata M, Fukata Y, Adesnik H, Nicoll RA, Bredt DS. Identification of PSD-95 palmitoylating enzymes. Neuron (2004) 44(6):987–96. doi:10.1016/j.neuron.2004.12.005
96. Mitchell DA, Vasudevan A, Linder ME, Deschenes RJ. Protein palmitoylation by a family of DHHC protein S-acyltransferases. J Lipid Res (2006) 47(6):1118–27. doi:10.1194/jlr.R600007-JLR200
97. Ohno Y, Kihara A, Sano T, Igarashi Y. Intracellular localization and tissue-specific distribution of human and yeast DHHC cysteine-rich domain-containing proteins. Biochim Biophys Acta (2006) 1761(4):474–83. doi:10.1016/j.bbalip.2006.03.010
98. Roth AF, Wan J, Bailey AO, Sun B, Kuchar JA, Green WN, et al. Global analysis of protein palmitoylation in yeast. Cell (2006) 125(5):1003–13. doi:10.1016/j.cell.2006.03.042
99. Fang C, Deng L, Keller CA, Fukata M, Fukata Y, Chen G, et al. GODZ-mediated palmitoylation of GABA(A) receptors is required for normal assembly and function of GABAergic inhibitory synapses. J Neurosci (2006) 26(49):12758–68. doi:10.1523/JNEUROSCI.4214-06.2006
100. Fernández-Hernando C, Fukata M, Bernatchez PN, Fukata Y, Lin MI, Bredt DS, et al. Identification of Golgi-localized acyl transferases that palmitoylate and regulate endothelial nitric oxide synthase. J Cell Biol (2006) 174(3):369–77. doi:10.1083/jcb.200601051
101. Camp LA, Hofmann SL. Purification and properties of a palmitoyl-protein thioesterase that cleaves palmitate from H-Ras. J Biol Chem (1993) 268(30):22566–74.
102. Soyombo AA, Hofmann SL. Molecular cloning and expression of palmitoyl-protein thioesterase 2 (PPT2), a homolog of lysosomal palmitoyl-protein thioesterase with a distinct substrate specificity. J Biol Chem (1997) 272(43):27456–63. doi:10.1074/jbc.272.43.27456
103. Duncan JA, Gilman AG. A cytoplasmic acyl-protein thioesterase that removes palmitate from G protein alpha subunits and p21(RAS). J Biol Chem (1998) 273(25):15830–7. doi:10.1074/jbc.273.25.15830
104. Tomatis VM, Trenchi A, Gomez GA, Daniotti JL. Acyl-protein thioesterase 2 catalyzes the deacylation of peripheral membrane-associated GAP-43. PLoS One (2010) 5(11):e15045. doi:10.1371/journal.pone.0015045
105. Camp LA, Verkruyse LA, Afendis SJ, Slaughter CA, Hofmann SL. Molecular cloning and expression of palmitoyl-protein thioesterase. J Biol Chem (1994) 269(37):23212–9.
106. Yokoi N, Fukata Y, Sekiya A, Murakami T, Kobayashi K, Fukata M. Identification of PSD-95 depalmitoylating enzymes. J Neurosci (2016) 36(24):6431–44. doi:10.1523/JNEUROSCI.0419-16.2016
107. Lin DT, Conibear E. ABHD17 proteins are novel protein depalmitoylases that regulate N-Ras palmitate turnover and subcellular localization. Elife (2015) 4:e11306. doi:10.7554/eLife.11306
108. Gibbs JB. Ras C-terminal processing enzymes – new drug targets? Cell (1991) 65(1):1–4. doi:10.1016/0092-8674(91)90352-Y
109. Yoshida Y, Kawata M, Katayama M, Horiuchi H, Kita Y, Takai Y. A geranylgeranyltransferase for rhoA p21 distinct from the farnesyltransferase for ras p21S. Biochem Biophys Res Commun (1991) 175(2):720–8. doi:10.1016/0006-291X(91)91625-M
110. Maurer-Stroh S, Koranda M, Benetka W, Schneider G, Sirota FL, Eisenhaber F. Towards complete sets of farnesylated and geranylgeranylated proteins. PLoS Comput Biol (2007) 3(4):e66. doi:10.1371/journal.pcbi.0030066
111. Andres DA, Seabra MC, Brown MS, Armstrong SA, Smeland TE, Cremers FP, et al. cDNA cloning of component A of Rab geranylgeranyl transferase and demonstration of its role as a Rab escort protein. Cell (1993) 73(6):1091–9. doi:10.1016/0092-8674(93)90639-8
112. Taylor JS, Reid TS, Terry KL, Casey PJ, Beese LS. Structure of mammalian protein geranylgeranyltransferase type-I. EMBO J (2003) 22(22):5963–74. doi:10.1093/emboj/cdg571
113. Hartman HL, Hicks KA, Fierke CA. Peptide specificity of protein prenyltransferases is determined mainly by reactivity rather than binding affinity. Biochemistry (2005) 44(46):15314–24. doi:10.1021/bi0509503
114. Zverina EA, Lamphear CL, Wright EN, Fierke CA. Recent advances in protein prenyltransferases: substrate identification, regulation, and disease interventions. Curr Opin Chem Biol (2012) 16(5–6):544–52. doi:10.1016/j.cbpa.2012.10.015
115. Ahearn IM, Haigis K, Bar-Sagi D, Philips MR. Regulating the regulator: post-translational modification of RAS. Nat Rev Mol Cell Biol (2011) 13(1):39–51. doi:10.1038/nrm3255
116. Rao S, Cunningham D, de Gramont A, Scheithauer W, Smakal M, Humblet Y, et al. Phase III double-blind placebo-controlled study of farnesyl transferase inhibitor R115777 in patients with refractory advanced colorectal cancer. J Clin Oncol (2004) 22(19):3950–7. doi:10.1200/JCO.2004.10.037
117. Hrycyna CA, Sapperstein SK, Clarke S, Michaelis S. The Saccharomyces cerevisiae STE14 gene encodes a methyltransferase that mediates C-terminal methylation of a-factor and RAS proteins. EMBO J (1991) 10(7):1699–709.
118. Boyartchuk VL, Ashby MN, Rine J. Modulation of Ras and a-factor function by carboxyl-terminal proteolysis. Science (1997) 275(5307):1796–800. doi:10.1126/science.275.5307.1796
119. Dai Q, Choy E, Chiu V, Romano J, Slivka SR, Steitz SA, et al. Mammalian prenylcysteine carboxyl methyltransferase is in the endoplasmic reticulum. J Biol Chem (1998) 273(24):15030–4. doi:10.1074/jbc.273.24.15030
120. Orrenius S, Gogvadze V, Zhivotovsky B. Calcium and mitochondria in the regulation of cell death. Biochem Biophys Res Commun (2015) 460(1):72–81. doi:10.1016/j.bbrc.2015.01.137
121. Boehning D, Patterson RL, Sedaghat L, Glebova NO, Kurosaki T, Snyder SH. Cytochrome c binds to inositol (1,4,5) trisphosphate receptors, amplifying calcium-dependent apoptosis. Nat Cell Biol (2003) 5(12):1051–61. doi:10.1038/ncb1063
122. Garcia MI, Chen JJ, Boehning D. Genetically encoded calcium indicators for studying long-term calcium dynamics during apoptosis. Cell Calcium (2017) 61:44–9. doi:10.1016/j.ceca.2016.12.010
123. Boehning D, van Rossum DB, Patterson RL, Snyder SH. A peptide inhibitor of cytochrome c/inositol 1,4,5-trisphosphate receptor binding blocks intrinsic and extrinsic cell death pathways. Proc Natl Acad Sci U S A (2005) 102(5):1466–71. doi:10.1073/pnas.0409650102
124. Hernandez JL, Davda D, Majmudar JD, Won SJ, Prakash A, Choi AI, et al. Correlated S-palmitoylation profiling of snail-induced epithelial to mesenchymal transition. Mol Biosyst (2016) 12(6):1799–808. doi:10.1039/c6mb00019c
125. Wan J, Savas JN, Roth AF, Sanders SS, Singaraja RR, Hayden MR, et al. Tracking brain palmitoylation change: predominance of glial change in a mouse model of Huntington’s disease. Chem Biol (2013) 20(11):1421–34. doi:10.1016/j.chembiol.2013.09.018
126. Fredericks GJ, Hoffmann FW, Rose AH, Osterheld HJ, Hess FM, Mercier F, et al. Stable expression and function of the inositol 1,4,5-triphosphate receptor requires palmitoylation by a DHHC6/selenoprotein K complex. Proc Natl Acad Sci U S A (2014) 111(46):16478–83. doi:10.1073/pnas.1417176111
127. Lynes EM, Raturi A, Shenkman M, Ortiz Sandoval C, Yap MC, Wu J, et al. Palmitoylation is the switch that assigns calnexin to quality control or ER Ca2+ signaling. J Cell Sci (2013) 126(Pt 17):3893–903. doi:10.1242/jcs.125856
128. Hannoush RN, Sun J. The chemical toolbox for monitoring protein fatty acylation and prenylation. Nat Chem Biol (2010) 6(7):498–506. doi:10.1038/nchembio.388
129. Greaves J, Munro KR, Davidson SC, Riviere M, Wojno J, Smith TK, et al. Molecular basis of fatty acid selectivity in the zDHHC family of S-acyltransferases revealed by click chemistry. Proc Natl Acad Sci U S A (2017) 114(8):E1365–74. doi:10.1073/pnas.1612254114
130. Mumby SM, Kleuss C, Gilman AG. Receptor regulation of G-protein palmitoylation. Proc Natl Acad Sci U S A (1994) 91(7):2800–4. doi:10.1073/pnas.91.7.2800
131. Degtyarev MY, Spiegel AM, Jones TL. Increased palmitoylation of the Gs protein alpha subunit after activation by the beta-adrenergic receptor or cholera toxin. J Biol Chem (1993) 268(32):23769–72.
132. Wedegaertner PB, Bourne HR. Activation and depalmitoylation of Gs alpha. Cell (1994) 77(7):1063–70. doi:10.1016/0092-8674(94)90445-6
133. Tsutsumi R, Fukata Y, Fukata M. Discovery of protein-palmitoylating enzymes. Pflugers Arch (2008) 456(6):1199–206. doi:10.1007/s00424-008-0465-x
134. Guardiola-Serrano F, Rossin A, Cahuzac N, Lückerath K, Melzer I, Mailfert S, et al. Palmitoylation of human FasL modulates its cell death-inducing function. Cell Death Dis (2010) 1:e88. doi:10.1038/cddis.2010.62
135. Rossin A, Durivault J, Chakhtoura-Feghali T, Lounnas N, Gagnoux-Palacios L, Hueber AO. Fas palmitoylation by the palmitoyl acyltransferase DHHC7 regulates Fas stability. Cell Death Differ (2015) 22(4):643–53. doi:10.1038/cdd.2014.153
136. Rossin A, Derouet M, Abdel-Sater F, Hueber AO. Palmitoylation of the TRAIL receptor DR4 confers an efficient TRAIL-induced cell death signalling. Biochem J (2009) 419(1):185–92, 2 p following 192. doi:10.1042/BJ20081212
137. Berg V, Rusch M, Vartak N, Jüngst C, Schauss A, Waldmann H, et al. miRs-138 and -424 control palmitoylation-dependent CD95-mediated cell death by targeting acyl protein thioesterases 1 and 2 in CLL. Blood (2015) 125(19):2948–57. doi:10.1182/blood-2014-07-586511
138. Gajate C, Mollinedo F. Lipid rafts and raft-mediated supramolecular entities in the regulation of CD95 death receptor apoptotic signaling. Apoptosis (2015) 20(5):584–606. doi:10.1007/s10495-015-1104-6
139. Hantschel O, Nagar B, Guettler S, Kretzschmar J, Dorey K, Kuriyan J, et al. A myristoyl/phosphotyrosine switch regulates c-Abl. Cell (2003) 112(6):845–57. doi:10.1016/S0092-8674(03)00191-0
140. Nagar B, Hantschel O, Young MA, Scheffzek K, Veach D, Bornmann W, et al. Structural basis for the autoinhibition of c-Abl tyrosine kinase. Cell (2003) 112(6):859–71. doi:10.1016/S0092-8674(03)00194-6
141. Ito Y, Pandey P, Mishra N, Kumar S, Narula N, Kharbanda S, et al. Targeting of the c-Abl tyrosine kinase to mitochondria in endoplasmic reticulum stress-induced apoptosis. Mol Cell Biol (2001) 21(18):6233–42. doi:10.1128/MCB.21.18.6233-6242.2001
142. Abraham N, Veillette A. Activation of p56lck through mutation of a regulatory carboxy-terminal tyrosine residue requires intact sites of autophosphorylation and myristylation. Mol Cell Biol (1990) 10(10):5197–206. doi:10.1128/MCB.10.10.5197
143. Bijlmakers MJ, Isobe-Nakamura M, Ruddock LJ, Marsh M. Intrinsic signals in the unique domain target p56(lck) to the plasma membrane independently of CD4. J Cell Biol (1997) 137(5):1029–40. doi:10.1083/jcb.137.5.1029
144. Blanco-Colio LM, Villa A, Ortego M, Hernández-Presa MA, Pascual A, Plaza JJ, et al. 3-Hydroxy-3-methyl-glutaryl coenzyme A reductase inhibitors, atorvastatin and simvastatin, induce apoptosis of vascular smooth muscle cells by downregulation of Bcl-2 expression and Rho A prenylation. Atherosclerosis (2002) 161(1):17–26. doi:10.1016/S0021-9150(01)00613-X
145. Matzno S, Yasuda S, Juman S, Yamamoto Y, Nagareya-Ishida N, Tazuya-Murayama K, et al. Statin-induced apoptosis linked with membrane farnesylated Ras small G protein depletion, rather than geranylated Rho protein. J Pharm Pharmacol (2005) 57(11):1475–84. doi:10.1211/jpp.57.11.0014
146. Brinkkoetter PT, Gottmann U, Schulte J, van der Woude FJ, Braun C, Yard BA. Atorvastatin interferes with activation of human CD4(+) T cells via inhibition of small guanosine triphosphatase (GTPase) activity and caspase-independent apoptosis. Clin Exp Immunol (2006) 146(3):524–32. doi:10.1111/j.1365-2249.2006.03217.x
147. Kah J, Wüstenberg A, Keller AD, Sirma H, Montalbano R, Ocker M, et al. Selective induction of apoptosis by HMG-CoA reductase inhibitors in hepatoma cells and dependence on p53 expression. Oncol Rep (2012) 28(3):1077–83. doi:10.3892/or.2012.1860
148. Borahay MA, Kilic GS, Yallampalli C, Snyder RR, Hankins GD, Al-Hendy A, et al. Simvastatin potently induces calcium-dependent apoptosis of human leiomyoma cells. J Biol Chem (2014) 289(51):35075–86. doi:10.1074/jbc.M114.583575
149. Borahay MA, Vincent K, Motamedi M, Sbrana E, Kilic GS, Al-Hendy A, et al. Novel effects of simvastatin on uterine fibroid tumors: in vitro and patient-derived xenograft mouse model study. Am J Obstet Gynecol (2015) 213(2):196.e1–8. doi:10.1016/j.ajog.2015.03.055
150. Borahay MA, Fang X, Baillargeon JG, Kilic GS, Boehning DF, Kuo YF. Statin use and uterine fibroid risk in hyperlipidemia patients: a nested case-control study. Am J Obstet Gynecol (2016) 215(6):750.e1–8. doi:10.1016/j.ajog.2016.06.036
151. Muck AO, Seeger H, Wallwiener D. Inhibitory effect of statins on the proliferation of human breast cancer cells. Int J Clin Pharmacol Ther (2004) 42(12):695–700. doi:10.5414/CPP42695
152. Martirosyan A, Clendening JW, Goard CA, Penn LZ. Lovastatin induces apoptosis of ovarian cancer cells and synergizes with doxorubicin: potential therapeutic relevance. BMC Cancer (2010) 10:103. doi:10.1186/1471-2407-10-103
153. Cho SJ, Kim JS, Kim JM, Lee JY, Jung HC, Song IS. Simvastatin induces apoptosis in human colon cancer cells and in tumor xenografts, and attenuates colitis-associated colon cancer in mice. Int J Cancer (2008) 123(4):951–7. doi:10.1002/ijc.23593
154. Monteith GR, Davis FM, Roberts-Thomson SJ. Calcium channels and pumps in cancer: changes and consequences. J Biol Chem (2012) 287(38):31666–73. doi:10.1074/jbc.R112.343061
155. Tsavaler L, Shapero MH, Morkowski S, Laus R. Trp-p8, a novel prostate-specific gene, is up-regulated in prostate cancer and other malignancies and shares high homology with transient receptor potential calcium channel proteins. Cancer Res (2001) 61(9):3760–9. Available from: http://cancerres.aacrjournals.org/content/61/9/3760.long
156. Prevarskaya N, Flourakis M, Bidaux G, Thebault S, Skryma R. Differential role of TRP channels in prostate cancer. Biochem Soc Trans (2007) 35(Pt 1):133–5. doi:10.1042/BST0350133
157. Czifra G, Varga A, Nyeste K, Marincsák R, Tóth BI, Kovács I, et al. Increased expressions of cannabinoid receptor-1 and transient receptor potential vanilloid-1 in human prostate carcinoma. J Cancer Res Clin Oncol (2009) 135(4):507–14. doi:10.1007/s00432-008-0482-3
158. Zhuang L, Peng JB, Tou L, Takanaga H, Adam RM, Hediger MA, et al. Calcium-selective ion channel, CaT1, is apically localized in gastrointestinal tract epithelia and is aberrantly expressed in human malignancies. Lab Invest (2002) 82(12):1755–64. doi:10.1097/01.LAB.0000043910.41414.E7
159. Aydar E, Yeo S, Djamgoz M, Palmer C. Abnormal expression, localization and interaction of canonical transient receptor potential ion channels in human breast cancer cell lines and tissues: a potential target for breast cancer diagnosis and therapy. Cancer Cell Int (2009) 9:23. doi:10.1186/1475-2867-9-23
160. Dhennin-Duthille I, Gautier M, Faouzi M, Guilbert A, Brevet M, Vaudry D, et al. High expression of transient receptor potential channels in human breast cancer epithelial cells and tissues: correlation with pathological parameters. Cell Physiol Biochem (2011) 28(5):813–22. doi:10.1159/000335795
161. Yang SL, Cao Q, Zhou KC, Feng YJ, Wang YZ. Transient receptor potential channel C3 contributes to the progression of human ovarian cancer. Oncogene (2009) 28(10):1320–8. doi:10.1038/onc.2008.475
162. Wei C, Wang X, Chen M, Ouyang K, Song LS, Cheng H. Calcium flickers steer cell migration. Nature (2009) 457(7231):901–5. doi:10.1038/nature07577
163. Fiorio Pla A, Ong HL, Cheng KT, Brossa A, Bussolati B, Lockwich T, et al. TRPV4 mediates tumor-derived endothelial cell migration via arachidonic acid-activated actin remodeling. Oncogene (2012) 31(2):200–12. doi:10.1038/onc.2011.231
164. Yang S, Zhang JJ, Huang XY. Orai1 and STIM1 are critical for breast tumor cell migration and metastasis. Cancer Cell (2009) 15(2):124–34. doi:10.1016/j.ccr.2008.12.019
165. Peng HH, Hodgson L, Henderson AJ, Dong C. Involvement of phospholipase C signaling in melanoma cell-induced endothelial junction disassembly. Front Biosci (2005) 10:1597–606. doi:10.2741/1643
166. Baljinnyam E, De Lorenzo MS, Xie LH, Iwatsubo M, Chen S, Goydos JS, et al. Exchange protein directly activated by cyclic AMP increases melanoma cell migration by a Ca2+-dependent mechanism. Cancer Res (2010) 70(13):5607–17. doi:10.1158/0008-5472.CAN-10-0056
167. Burrus LW, McMahon AP. Biochemical analysis of murine Wnt proteins reveals both shared and distinct properties. Exp Cell Res (1995) 220(2):363–73. doi:10.1006/excr.1995.1327
168. Willert K, Brown JD, Danenberg E, Duncan AW, Weissman IL, Reya T, et al. Wnt proteins are lipid-modified and can act as stem cell growth factors. Nature (2003) 423(6938):448–52. doi:10.1038/nature01611
169. Kurayoshi M, Yamamoto H, Izumi S, Kikuchi A. Post-translational palmitoylation and glycosylation of Wnt-5a are necessary for its signalling. Biochem J (2007) 402(3):515–23. doi:10.1042/BJ20061476
170. Slusarski DC, Yang-Snyder J, Busa WB, Moon RT. Modulation of embryonic intracellular Ca2+ signaling by Wnt-5A. Dev Biol (1997) 182(1):114–20. doi:10.1006/dbio.1996.8463
171. Medrano EE. Wnt5a and PKC, a deadly partnership involved in melanoma invasion. Pigment Cell Res (2007) 20(4):258–9. doi:10.1111/j.1600-0749.2007.00383.x
172. Dissanayake SK, Weeraratna AT. Detecting PKC phosphorylation as part of the Wnt/calcium pathway in cutaneous melanoma. Methods Mol Biol (2008) 468:157–72. doi:10.1007/978-1-59745-249-6_12
173. Galli LM, Burrus LW. Differential palmit(e)oylation of Wnt1 on C93 and S224 residues has overlapping and distinct consequences. PLoS One (2011) 6(10):e26636. doi:10.1371/journal.pone.0026636
174. Guo Z, Linn JF, Wu G, Anzick SL, Eisenberger CF, Halachmi S, et al. CDC91L1 (PIG-U) is a newly discovered oncogene in human bladder cancer. Nat Med (2004) 10(4):374–81. doi:10.1038/nm1004-1139b
175. Wu G, Guo Z, Chatterjee A, Huang X, Rubin E, Wu F, et al. Overexpression of glycosylphosphatidylinositol (GPI) transamidase subunits phosphatidylinositol glycan class T and/or GPI anchor attachment 1 induces tumorigenesis and contributes to invasion in human breast cancer. Cancer Res (2006) 66(20):9829–36. doi:10.1158/0008-5472.CAN-06-0506
176. Nagpal JK, Dasgupta S, Jadallah S, Chae YK, Ratovitski EA, Toubaji A, et al. Profiling the expression pattern of GPI transamidase complex subunits in human cancer. Mod Pathol (2008) 21(8):979–91. doi:10.1038/modpathol.2008.76
177. Zunich SM, Douglas T, Valdovinos M, Chang T, Bushman W, Walterhouse D, et al. Paracrine sonic hedgehog signalling by prostate cancer cells induces osteoblast differentiation. Mol Cancer (2009) 8:12. doi:10.1186/1476-4598-8-12
178. Sims-Mourtada J, Yang D, Tworowska I, Larson R, Smith D, Tsao N, et al. Detection of canonical hedgehog signaling in breast cancer by 131-iodine-labeled derivatives of the sonic hedgehog protein. J Biomed Biotechnol (2012) 2012:639562. doi:10.1155/2012/639562
179. Fei DL, Sanchez-Mejias A, Wang Z, Flaveny C, Long J, Singh S, et al. Hedgehog signaling regulates bladder cancer growth and tumorigenicity. Cancer Res (2012) 72(17):4449–58. doi:10.1158/0008-5472.CAN-11-4123
180. Hitosugi T, Sato M, Sasaki K, Umezawa Y. Lipid raft specific knockdown of SRC family kinase activity inhibits cell adhesion and cell cycle progression of breast cancer cells. Cancer Res (2007) 67(17):8139–48. doi:10.1158/0008-5472.CAN-06-4539
181. Uno F, Sasaki J, Nishizaki M, Carboni G, Xu K, Atkinson EN, et al. Myristoylation of the fus1 protein is required for tumor suppression in human lung cancer cells. Cancer Res (2004) 64(9):2969–76. doi:10.1158/0008-5472.CAN-03-3702
182. Kaemmerer E, Gassler N. Wnt lipidation and modifiers in intestinal carcinogenesis and cancer. Cancers (Basel) (2016) 8(7):E69. doi:10.3390/cancers8070069
183. Liu M, Sjogren AK, Karlsson C, Ibrahim MX, Andersson KM, Olofsson FJ, et al. Targeting the protein prenyltransferases efficiently reduces tumor development in mice with K-RAS-induced lung cancer. Proc Natl Acad Sci U S A (2010) 107(14):6471–6. doi:10.1073/pnas.0908396107
184. Raponi M, Lancet JE, Fan H, Dossey L, Lee G, Gojo I, et al. A 2-gene classifier for predicting response to the farnesyltransferase inhibitor tipifarnib in acute myeloid leukemia. Blood (2008) 111(5):2589–96. doi:10.1182/blood-2007-09-112730
185. Baines AT, Xu D, Der CJ. Inhibition of Ras for cancer treatment: the search continues. Future Med Chem (2011) 3(14):1787–808. doi:10.4155/fmc.11.121
186. van de Donk NW, Schotte D, Kamphuis MM, van Marion AM, van Kessel B, Bloem AC, et al. Protein geranylgeranylation is critical for the regulation of survival and proliferation of lymphoma tumor cells. Clin Cancer Res (2003) 9(15):5735–48. Available from: http://clincancerres.aacrjournals.org/content/9/15/5735.long
187. Sjogren AK, Andersson KM, Khan O, Olofsson FJ, Karlsson C, Bergo MO. Inactivating GGTase-I reduces disease phenotypes in a mouse model of K-RAS-induced myeloproliferative disease. Leukemia (2011) 25(1):186–9. doi:10.1038/leu.2010.242
188. Wojciak-Stothard B, Potempa S, Eichholtz T, Ridley AJ. Rho and Rac but not Cdc42 regulate endothelial cell permeability. J Cell Sci (2001) 114(Pt 7):1343–55. Available from: http://jcs.biologists.org/content/114/7/1343.long
189. Gelb MH. Protein prenylation, et cetera: signal transduction in two dimensions. Science (1997) 275(5307):1750–1. doi:10.1126/science.275.5307.1750
190. Sun J, Qian Y, Hamilton AD, Sebti SM. Both farnesyltransferase and geranylgeranyltransferase I inhibitors are required for inhibition of oncogenic K-Ras prenylation but each alone is sufficient to suppress human tumor growth in nude mouse xenografts. Oncogene (1998) 16(11):1467–73. doi:10.1038/sj.onc.1201656
191. Rocks O, Peyker A, Kahms M, Verveer PJ, Koerner C, Lumbierres M, et al. An acylation cycle regulates localization and activity of palmitoylated Ras isoforms. Science (2005) 307(5716):1746–52. doi:10.1126/science.1105654
192. Xiao H, Qin X, Ping D, Zuo K. Inhibition of Rho and Rac geranylgeranylation by atorvastatin is critical for preservation of endothelial junction integrity. PLoS One (2013) 8(3):e59233. doi:10.1371/journal.pone.0059233
193. Montalvo SK, Li L, Westover KD. Rationale for RAS mutation-tailored therapies. Future Oncol (2017) 13(3):263–71. doi:10.2217/fon-2016-0363
194. Moorthy NS, Sousa SF, Ramos MJ, Fernandes PA. Farnesyltransferase inhibitors: a comprehensive review based on quantitative structural analysis. Curr Med Chem (2013) 20(38):4888–923. doi:10.2174/09298673113206660262
195. Berndt N, Hamilton AD, Sebti SM. Targeting protein prenylation for cancer therapy. Nat Rev Cancer (2011) 11(11):775–91. doi:10.1038/nrc3151
196. Braun T, Fenaux P. Farnesyltransferase inhibitors and their potential role in therapy for myelodysplastic syndromes and acute myeloid leukaemia. Br J Haematol (2008) 141(5):576–86. doi:10.1111/j.1365-2141.2008.07099.x
197. Winter-Vann AM, Casey PJ. Post-prenylation-processing enzymes as new targets in oncogenesis. Nat Rev Cancer (2005) 5(5):405–12. doi:10.1038/nrc1612
198. Mohammed I, Hampton SE, Ashall L, Hildebrandt ER, Kutlik RA, Manandhar SP, et al. 8-Hydroxyquinoline-based inhibitors of the Rce1 protease disrupt Ras membrane localization in human cells. Bioorg Med Chem (2016) 24(2):160–78. doi:10.1016/j.bmc.2015.11.043
199. Lau HY, Ramanujulu PM, Guo D, Yang T, Wirawan M, Casey PJ, et al. An improved isoprenylcysteine carboxylmethyltransferase inhibitor induces cancer cell death and attenuates tumor growth in vivo. Cancer Biol Ther (2014) 15(9):1280–91. doi:10.4161/cbt.29692
200. Davda D, El Azzouny MA, Tom CT, Hernandez JL, Majmudar JD, Kennedy RT, et al. Profiling targets of the irreversible palmitoylation inhibitor 2-bromopalmitate. ACS Chem Biol (2013) 8(9):1912–7. doi:10.1021/cb400380s
201. Ducker CE, Griffel LK, Smith RA, Keller SN, Zhuang Y, Xia Z, et al. Discovery and characterization of inhibitors of human palmitoyl acyltransferases. Mol Cancer Ther (2006) 5(7):1647–59. doi:10.1158/1535-7163.MCT-06-0114
202. Ganesan L, Shieh P, Bertozzi CR, Levental I. Click-chemistry based high throughput screening platform for modulators of Ras palmitoylation. Sci Rep (2017) 7:41147. doi:10.1038/srep41147
203. Hung MS, Chen IC, Lee CP, Huang RJ, Chen PC, Tsai YH, et al. Statin improves survival in patients with EGFR-TKI lung cancer: a nationwide population-based study. PLoS One (2017) 12(2):e0171137. doi:10.1371/journal.pone.0171137
204. Wang A, Aragaki AK, Tang JY, Kurian AW, Manson JE, Chlebowski RT, et al. Statin use and all-cancer survival: prospective results from the Women’s Health Initiative. Br J Cancer (2016) 115(1):129–35. doi:10.1038/bjc.2016.149
205. Habis M, Wroblewski K, Bradaric M, Ismail N, Yamada SD, Litchfield L, et al. Statin therapy is associated with improved survival in patients with non-serous-papillary epithelial ovarian cancer: a retrospective cohort analysis. PLoS One (2014) 9(8):e104521. doi:10.1371/journal.pone.0104521
206. Nayan M, Punjani N, Juurlink DN, Finelli A, Austin PC, Kulkarni GS, et al. Statin use and kidney cancer survival outcomes: a systematic review and meta-analysis. Cancer Treat Rev (2017) 52:105–16. doi:10.1016/j.ctrv.2016.11.009
207. Nevadunsky NS, Van Arsdale A, Strickler HD, Spoozak LA, Moadel A, Kaur G, et al. Association between statin use and endometrial cancer survival. Obstet Gynecol (2015) 126(1):144–50. doi:10.1097/AOG.0000000000000926
208. Kozak MM, Anderson EM, von Eyben R, Pai JS, Poultsides GA, Visser BC, et al. Statin and metformin use prolongs survival in patients with resectable pancreatic cancer. Pancreas (2016) 45(1):64–70. doi:10.1097/MPA.0000000000000470
209. Lee HS, Lee SH, Lee HJ, Chung MJ, Park JY, Park SW, et al. Statin use and its impact on survival in pancreatic cancer patients. Medicine (Baltimore) (2016) 95(19):e3607. doi:10.1097/MD.0000000000003607
Keywords: calcium, apoptosis, lipidation, Fas, cancer, inositol phosphates, kinases, statins
Citation: Chen JJ and Boehning D (2017) Protein Lipidation As a Regulator of Apoptotic Calcium Release: Relevance to Cancer. Front. Oncol. 7:138. doi: 10.3389/fonc.2017.00138
Received: 12 April 2017; Accepted: 16 June 2017;
Published: 29 June 2017
Edited by:
Cesar Cardenas, Universidad de Chile, ChileReviewed by:
Olivier Micheau, Institut national de la santé et de la recherche médicale (INSERM), FranceCatherine Brenner Jan, Institut national de la santé et de la recherche médicale (INSERM), France
Copyright: © 2017 Chen and Boehning. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Darren Boehning, ZGFycmVuLmYuYm9laG5pbmdAdXRoLnRtYy5lZHU=
 Jessica J. Chen
Jessica J. Chen Darren Boehning
Darren Boehning