- Rapo Yerapeh Ltd., Jerusalem, Israel
With the recent success of oncolytic viruses in clinical trials, efforts toward improved monitoring of the viruses and their mechanism have intensified. Four main gene expression strategies have been employed to date including: analyzing overall gene expression in tumor cells, looking at gene expression of a few specific genes in the tumor cells, focusing on gene expression of specific transgenes introduced into the virus, and following gene expression of certain viral genes. Each strategy presents certain advantages and disadvantages over the others. Various methods to organize the dysregulated genes into clusters have provided a window into the mechanism of action for these viruses. Methodologically, the combined approach of looking at both overall gene expression, the tumor cells and gene expression of viral genes, enables researchers to assess correlation between the introduction of the virus and the changes in the tumor. This would seem to be the most productive approach for future studies, providing much information on mechanism and timing.
Introduction
With the development of recombinant deoxyribonucleic acid (DNA) technology in the early 1990s, the possibility of genetically engineering oncolytic viruses to improve virotherapy became a reality (1). Capitalizing on what was known about the mechanisms for oncolytic viruses, research focused on improving the safety profile, attenuating direct tumor lysis, and modulating the immune response. Beyond its potential therapeutic benefits, genetic engineering has generated new ways of monitoring and predicting sensitivity or resistance to oncolytic virotherapy (2). Various strategies, including animal models, cell lines, and even human in vivo gene expression studies, have been implemented to assess the effects of the oncolytic virus in a personalized way.
In 2015, the Food and Drug Administration approved the first oncolytic virus based on the results of a phase 3 trial for melanoma (3–5). Many other viruses are now being tested in clinical trials for various indications. This necessitates careful consideration of endpoints and creative new ways of monitoring therapeutic success.
Four main strategies for monitoring oncolytic viruses were surveyed: overall gene expression in tumor cells, gene expression of a few specific genes in the tumor cells, gene expression of specific transgenes introduced into the virus, and gene expression of certain viral genes.
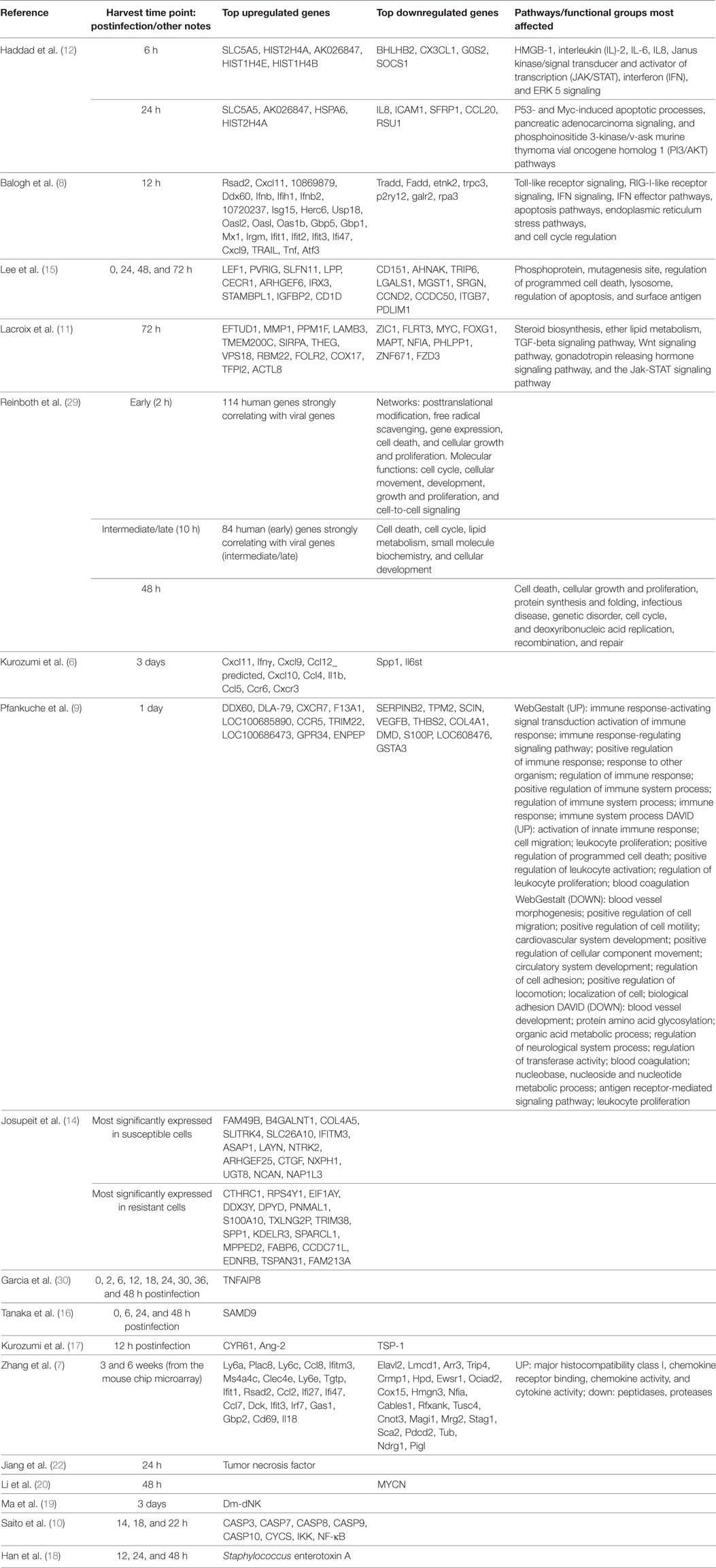
Monitoring Overall Gene Expression of Tumor Cells (See Tables 1–3)
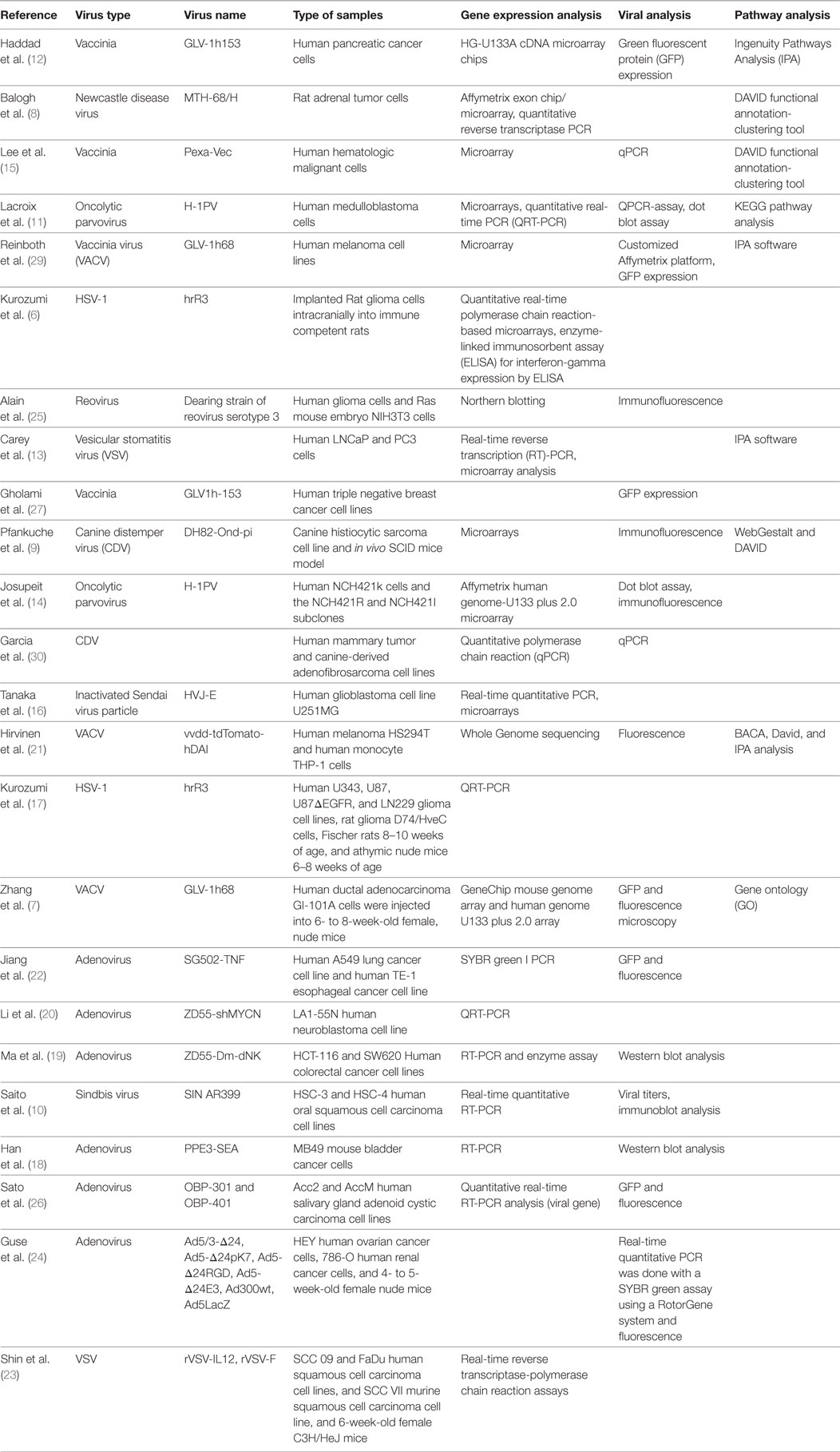
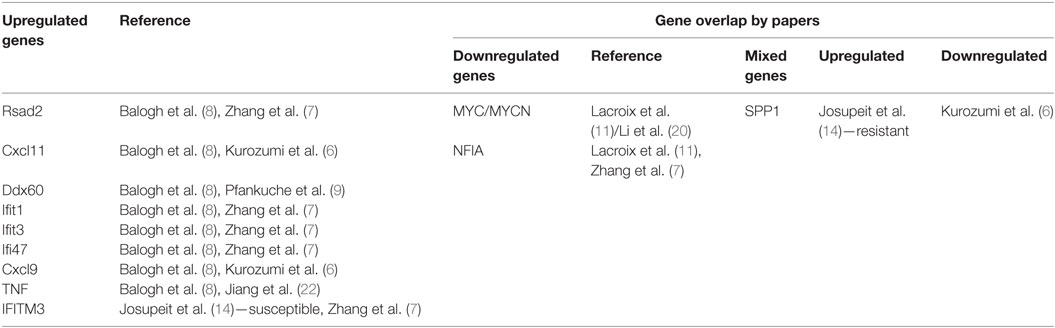
The first method of assessing the impact of oncolytic virotherapy involves monitoring overall gene expression of tumor cells before and after virotherapy. To identify genes whose expression was altered in tumor cells during infection with an oncolytic virus, genome-wide expression profiling needs to be performed. The studies surveyed included both animal studies and studies performed on human cell lines. Kurozumi et al. used a rat intracranial glioma model in immune competent rats (6). They introduced a type one herpes simplex virus (HSV-1) and used quantitative real-time polymerase chain reaction (PCR)-based microarrays to monitor tumor gene expression. They found that oncolytic virus treatment induced at least a twofold increase or decrease in the expression of 50 genes when compared with the control. More specifically, they found numerous genes were upregulated from the chemokine family (see Table 1). Unfortunately, Kurozumi et al. did not analyze gene clusters for these genes. Similarly, Zhang et al. also looked at tumor gene expression using an animal model. In this case, they used nude mice injected with metastatic human breast adenocarcinoma GI-101A cells (7). They monitored tumor gene expression after treatment with a vaccinia virus (VACV) using a mouse genome array and a human genome array. They found 681 genes differentially expressed when compared with controls. As opposed to the Kurozumi group, Zhang et al. used gene ontology to evaluate clustering and found upregulated genes related to the immune system, and downregulated genes related to enzymatic function (see Table 1).
However, some reservations must be expressed regarding the use of Affymetrix mouse arrays. Since they are relatively species specific, genes that are identified as differentially expressed may primarily represent host cells infiltrating the tumor. When assessing the changes of potential cross hybridization of human genes to the mouse chip, less than 50% of the genes identified with the mouse arrays were differentially expressed according to the human chip. Furthermore, the immune-related genes that were differentially expressed displayed very low fluorescence intensities in human as compared with very high intensities displayed by the same genes in the mouse array. This would suggest that perhaps comparisons of gene expression between species should be evaluated with caution.
Balogh et al. used a rat adrenal tumor cell line, namely, pheochromocytoma cells, as the model, and looked at gene expression after infection with a recombinant oncolytic viral strain of Newcastle disease virus called MTH-68/H (8). They used a rat specific microarray chip to monitor gene expression and confirmed changes via quantitative reverse transcriptase PCR. They found that 729 genes were upregulated and 612 genes were downregulated with oncolytic viral treatment compared with controls. Balogh et al. relied on DAVID functional annotation-clustering tool to group the genes according to function. This clustering tool was far more elaborate than the method used by Zhang. They found pathways including receptor signaling, apoptosis, and cellular stress to be involved (see Table 1). The detailed report they provided was extremely helpful in beginning to understand the mechanisms involved in oncolytic viral therapy. In addition, Pfankuche et al. used a canine sarcoma cell line as the tumor model and injected a canine distemper virus (CDV) called DH82-Ond-pi (9). They visualized the cells using immunofluorescence and used a canine specific microarray chip to evaluate tumor gene expression. They identified 892 upregulated genes and 869 downregulated genes when compared with controls. They analyzed these results using WebGestalt and DAVID and found that upregulated genes were primarily related to immune processes, cell migration, apoptosis, and blood coagulation (see Table 1). Certainly, the role of oncolytic viruses in impairing angiogenesis demands further attention based on the mechanism related clues provided by this gene expression study.
Beyond the studies looking at animal cell lines, there are a few studies that looked at tumor gene expression in human cell lines. Saito et al. used two human oral squamous cell carcinoma cell lines called tHSC-4 and HSC-3 and injected the Sindbis SIN AR399 oncolytic virus (10). Using real-time quantitative reverse transcription (RT)-PCR to monitor the tumor gene expression after treatment with the virus, they found that Caspases 7, 8, and 10 were upregulated in both HSC-3 and HSC-4 cells, but that Caspases 3 and 9, cytochrome c, NF-κB, and IKK were only upregulated in HSC-3 cells. The likely interpretation of these data is that SIN induced oncolysis in HASC-3 cells by activating a few apoptotic pathways. Similarly, Lacroix et al. looked at six human medulloblastoma cell lines (MB) and how they are affected by treatment with the oncolytic parvovirus H-1 (H-1PV) (11). They used microarray and quantitative real-time PCR (QRT-PCR) to evaluate gene expression after oncolytic virus treatment. They focused on the 25 most significantly upregulated and the 25 most significantly downregulated genes. They used KEGG pathway analysis to identify clusters of genes and found that five pathways were particularly impacted by H-1PV infection. These included the pathways for steroid biosynthesis, ether lipid metabolism, TGF-beta signaling, Wnt signaling, gonadotropin releasing hormone signaling, and Jak-STAT signaling. One advantage in the methodology employed by the Lacroix group was focusing on the top 25. As opposed to the other groups that tried to find patterns from all the up and downregulated genes, Lacroix et al. potentially eliminated noise and were able to better identify the most important pathways. Finally, Haddad et al. looked at the PANC-1 human pancreatic cancer cell line infected with GLV-1h153, an oncolytic VACV (12). They used cDNA microarray chips to monitor gene expression after infection with the virus and used a cutoff of a twofold change to identify the most relevant genes. At 6 h postinfection, they found that 139 genes were up- or downregulated, but by 24 h after infection 5,698 genes were dysregulated. They analyzed the pathways using the Ingenuity Pathways Analysis (IPA) software and found that downregulated genes clustered around pathways associated with cell death, cell cycle, and DNA repair. Upregulated genes were associated with mechanisms related to infection. In contrast to the groups surveyed so far, Haddad et al. also labeled the virus with green fluorescent protein (GFP) and used this to determine if GFP-marker gene expression can be correlated with viral copy number. Analyzing GFP expression levels in the cells infected by the virus was shown to be both time and multiplicity of infection dependent. Considering that by 24 h postinfection almost 70% of live cells expressed GFP, and that the amount of dysregulated genes was significantly higher by 24 h, it seems reasonable to assume that the virus significantly affected the tumor. The pathway analysis enabled Haddad et al. to go a step further and begin to understand the method by which the viruses affect the tumor cells. More robust examples of combined testing of both tumor gene expression and viral levels will be reviewed in a later section.
The human studies reviewed until this point compared between tumor cells and normal cells. A slightly different methodology was employed in a few studies in which gene expression was compared between susceptible and resistant cancer cells. For example, Carey et al. capitalized on the fact that vesicular stomatitis virus (VSV) replicates selectively in cancer cells that have defects in the interferon (IFN)-I pathway (13). They looked at two different lines of prostate cancer cells. The first, human LNCaP cells, possess a defective IFN-I response, making them sensitive to VSV infection. The second, human PC3 prostate cancer cells, on the other hand, have functional IFN-I signaling, making them resistant to VSV infection. They employed real-time RT-PCR analysis and found that primary transcription, secondary transcription, and viral protein synthesis were delayed in PC3 cells compared with LNCaP cells. To look at gene expression, they used microarray and found that PC3 cells expressed many antiviral gene products compared with LNCaP cells. Furthermore, they looked at 80 different signaling pathways using IPA software and found specific pathways to be associated with a difference in gene expression between the two cell lines. Predictably, the IFN pathway had the highest percentage of differentially expressed genes. This research suggests the possibility of sensitivity markers for VSV treatment and hints at the mechanism of action of the virus. Similarly, Josupeit et al. looked at human NCH421k glioblastoma multiforme cells, which are susceptible to infection by parvovirus H-1 (H-1PV) and compared its response to H-1PV with NCH421R cells, which are a subclone resistant to H-1PV (14). They used “stem like” cell lines in NOD/SCID mice. They found a decrease in metabolic activity in the sensitive cell line compared with the resistant cell line when treated with H-1PV. When they analyzed gene expression using the Affymetrix Human Genome-U133 plus 2.0 microarray, they found 201 genes that were differentially expressed by at least threefold. They used unsupervised clustering to group the differentially expressed genes into three different categories. Some of these gene products are involved in regulating the antiviral immune response. A further example of comparing the response to oncolytic viruses in susceptible and non-susceptible cell lines is the study done by Lee et al. They compared the response of sensitive human myeloid leukemia lines and resistant human lymphoid leukemia cell lines to Pexa-Vec, a VACV engineered to express human granulocyte-macrophage colony-stimulating factor and β-galactosidase (15). Using quantitative PCR (qPCR) they found that 660 genes were upregulated at least twofold and 776 genes were downregulated at least twofold in the lymphoid cancer cell lines. In the case of the upregulated genes, changes were particularly remarkable, with more than 50 genes induced fivefold or higher, and 150 genes that were expressed three or fourfold higher, than the control. They used the DAVID functional annotation-clustering tool to classify the genes into 319 functional gene clusters. Some of the clusters included genes related to: viral replication and regulation of apoptosis.
Gene Expression of Specific Genes in the Tumor Cells
A second strategy currently employed to monitor the efficacy of oncolytic virotherapy is to monitor the expression of specific genes in the tumor cells. Interestingly, to the best of our knowledge, no such human studies have been published. However, a number of human cell-line studies have been done. For example, Tanaka et al. focused on the sterile alpha motif containing domain (SAMD9) gene in the human glioblastoma cell line called U251MG (16). They treated this line with Sendai virus particle (HVJ-E). Using real-time quantitative PCR and microarray analysis they found that SAMD9 gene was upregulated in tumor cells treated with the virus and the SAMD9 messenger ribonucleic acid (mRNA) was upregulated in a time and dose-dependent manner. In a study by Kurozumi et al., they also looked at a limited number of genes in the tumor cells, but they looked at both human and animal cell lines (17). They looked at 10 genes in glioma cell lines following treatment with HSV-1 virus hrR3 and compared the response to controls. When they used QRT-PCR, they found that three genes in particular were dysregulated. The first, the antiangiogenic factor TSP-1 was downregulated, and the other two, angiogenic factors CYR61 and Ang-2 were significantly upregulated when compared with controls. The advantage of this study over the previous one is that they were able to correlate the upregulation of CYR61 gene expression with the presence of the virus in the tumor tissue in vivo. CYR61 is a known protein that is involved in apoptosis, angiogenesis, the cell cycle, and extracellular matrix formation. By focusing on known genes but looking at the tumor cell expression in response to viral therapy and looking at the presence of the virus, researchers are able to better pinpoint the mechanisms at work when a virus infects a tumor cell.
Monitoring Transgene Expression
A third and particularly innovative strategy currently employed to monitor the efficacy of oncolytic virotherapy is the introduction of transgenes into the oncolytic virus and then analyzing gene expression of the transgene specifically. As we saw with the specific gene studies, there were no known published human studies. However, we will analyze one animal line study and a number of studies looking at human cell lines. Han et al. developed an oncolytic adenovirus PPE3-SEA that expressed the superantigen Staphylococcus enterotoxin A (SEA) and that has improved tumor specificity due to regulating the expression of E1A and E1B genes (18). They tested the PPE3-SEA virus against MB49 mouse bladder cancer cells in vitro and in vivo. They found that the mice treated with the virus had a significantly lower mean tumor volume than the control group. This seemed to correlate well with the increased expression of the virus mRNA in vitro.
In the human cell line studies, for example, Ma et al. constructed an adenoviral vector ZD55-Dm-dNK, containing the Drosophila melanogaster multisubstrate deoxyribonucleoside kinase—an important suicide gene (19). They looked at the expression of Dm-dNK human in colorectal cancer cells (HCT-116 and SW620) using RT-PCR. They found higher expression of the virus in the colorectal cancer cell lines, and lower levels of expression in the normal cell controls.
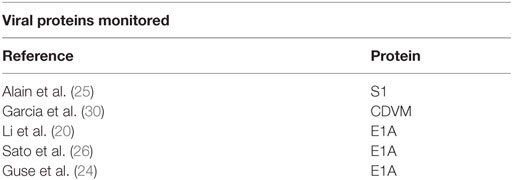
Similarly, Li et al. constructed ZD55-shMYCN, an oncolytic adenovirus ZD55 targeting the MYCN gene (20). They treated a p53-null and MYCN amplified human neuroblastoma cell line LA1-55N with the new virus. Using a two-step real-time reverse transcription (RT)-PCR procedure, they found that the virus selectively replicated and significantly downregulated the MYCN expression and that it was capable of effectively silencing the MYCN gene and inducing apoptosis in the tumor cells. Furthermore, they were able to demonstrate that the virus inhibited the growth of xenograft tumor in vivo.
In addition, Hirvinen et al. developed an oncolytic VACV that expressed intracellular pattern recognition receptor DNA-dependent activator of IFN− regulatory factors (DAI) to stimulate the innate immune system and to activate adaptive immune cells in the tumor (21). They tested this virus on two different human cell lines: human melanoma cells (HS294T) and human monocyte cells (THP-1). They used the BACA representing tools via the DAVID database to analyze the genes. They found that in the THP-1 cell line, there were a lot more upregulated genes than in the melanoma cell line. They used IPA and found that the most upregulated networks involved pathways connected to the activation of the immune system. More specifically, pathways related to dendritic cell growth, communication between innate and adaptive immune systems, and recognition of viruses were dysregulated. Significantly, they also found a sevenfold upregulation of DAI in both cell lines.
Finally, Jiang et al. developed a recombinant adenovirus using SG502 and tumor necrosis factor (TNF), yielding SG502-TNF (22). They looked at the effect of the virus on two different human cell lines: human A549 lung cancer cell line and human TE-1 esophageal cancer cell line. With the help of SYBR green I PCR, they found that the expression of TNF protein increased in both cell lines after infection with the virus. Furthermore, they found that the virus attacked the tumor cells specifically, and that they regulated the apoptotic-signaling pathway.
Shin et al. investigated human squamous cell carcinoma cell lines, murine squamous cell carcinoma cell lines, as well as a murine model (23). They used a VSV that was engineered to express the murine interleukin (IL) 12 gene called rVSV-IL12 and compared it to a non-cytokine carrying VSV virus called rVSV-F. They found that both viruses demonstrated similar infection efficiency. Real-time RT-PCR and enzyme-linked immunosorbent assay were used to look at viral replication and IL12 expression. They found that human squamous cell carcinoma cell lines infected with rVSV-IL12 had a high level of IL12 expression at 48 h postinfection. In the murine model, the animals treated with virus had a smaller tumor area than the control group. The mice treated with rVSV-IL12 had a much greater reduction of the tumor compared with the mice treated with rVSV-F. In vivo they showed that by day 30, none of the control mice survived, yet 3 animals injected with rVSV-F and 10 animals injected with rVSV-IL12, survived beyond day 30.
Monitoring Viral Gene Expression (See Table 4)
In assessing the impact of oncolytic virotherapy, a fourth method is to look at the gene expression of the virus itself inside of the tumor cells over time. By marking the virus with GFP and measuring the viral gene expression over time within the tumor cells, there is a clear indicator of viral growth, followed by a decrease in viral presence in the tumor cell as the tumor cell is destroyed. This method has been demonstrated in a number of studies including: animal studies (in vivo), and in vitro studies with cell lines, but no human studies have been published to date to the best of our knowledge.
Studies Where Human Cell Lines Implanted into Animals (In Vivo)
In conducting animal studies, human cell lines are implanted into animals (in vivo). For example, Guse et al. used two different murine xenograft models, one for renal cancer and a second for ovarian adenocarcinoma (24). They co-injected a luciferase-encoding virus with eight different adenoviruses. In the ovarian cancer cell model, they found using PCR that the mice infected with some of the adenoviruses had an over 3 log increase in luciferase gene expression, a luminescence gene, compared with mice infected with other adenoviruses. They also found that gene copies of luciferase genes were increased in some of the models and decreased in others. In the renal cancer model, they used qPCR to monitor gene copies and found that they increased by three orders of magnitude in some of the lines but not at all in others. Bioluminescence demonstrated photoemission for all of the tumors that were treated which implies that the virus entered the tumors.
Human and Mice Cell Line Studies (In Vitro)
Shifting to in vitro studies, Alain et al. looked at human glioma cells, Ras-transformed mouse embryo NIH3T3 cells, reovirus resistant human glioma cells, and untransformed NIH3t3 cells (25). They infected these four cell lines with mammalian orthoreoviruses. Using Northern blotting, they found that outer capsid protein sigma-1 reovirus transcripts were found only in the Ras-transformed cell line and the susceptible cell line. However, they found that when the cells were treated with an E64 protease inhibitor it successfully blocked the virus. Treating with an infectious subviral particle enabled the virus to be detected even in the resistant cell line and even in the presence of E64. Furthermore they found that the level of active cathepsin B and L was increased in tumors.
Sato et al. looked at human salivary gland adenoid cystic carcinoma cell lines (26). They infected the cells with a telomerase-specific replication-selective adenovirus (OBP-301) and OBP-401, a genetically engineered adenovirus with the GFP gene. Using quantitative real-time RT-PCR, they found that E1A expression increased in infected cells. When using the virus with GFP, the intensity of the fluorescence increased in a dose-dependent manner. Another in vitro study was conducted by Gholami et al. They used the human triple negative breast cancer cell lines HCC38 and MDA-MB-468 (27). They infected the cells with a VACV GLV-1h153 that was engineered to express the human sodium iodine symporter gene. Using GFP, they found that the virus infected the cell lines in a time-dependent way that was proportional to the concentration of the virus.
Using Marker Peptides to Monitor Viruses Instead of PCR
Peng et al. developed oncolytic viruses that could be tracked via marker peptides (28). They used the Edmonston vaccine strain of measles virus (MV-Edm) to express either human carcinoembryonic antigen (CEA) or beta-human chorionic gonadotropin (βhCG). They injected MV-shCEA or MV-βhCG into two groups of transgenic mice. They compared detection of CEA in serum to RT-PCR for nucleocapsid RNA and found that serum CEA was a more sensitive method than PCR.
Combination Studies (Viral and Host Gene Expression)
Until now we have reviewed studies that primarily looked either at tumor gene expression or viral gene expression. Perhaps the most interesting studies are the studies that combine both. Combination studies can potentially assess the correlation between the viral replication and the host response. Reinboth et al. looked at two different human melanoma cell lines (888-MEL and 1936-MEL) (29). They infected the cell lines with an attenuated VACV GLV-1h68. They used a platform called 36K to monitor human gene expression and they used a customized platform to monitor viral expression. To monitor viral expression, various markers were used including RUC-GFP, gusA, and the viral IFN-α/β-receptor-like secreted glycoprotein. The levels of the first two markers increased after infection, and the glycoprotein was expressed exclusively by GLV-1h68. To analyze the relationship between host cell transcription and viral replication, they assessed the correlation between viral and human gene expression at 2 h postinfection (early) and at 10 h postinfection (intermediate/late). At 2 h postinfection, they found 7 VACV genes and which were correlated to 114 human genes. Analysis with IPA demonstrated that these genes were related to the following pathways: apoptosis and the cell cycle, posttranslational modification, cellular growth and signaling, and other networks (Table 1). They then assessed whether human early gene transcription was predictive of VACV intermediate/late transcription and found 84 human early genes that correlated. These genes were important in processes such as cellular development and death, and lipid metabolism amongst others. Upon looking at expression 48 h after infection (late), there was a significant change in the expression of genes related to cellular growth, cell death, protein synthesis and folding, DNA replication, and DNA repair.
Similarly, Garcia et al. used three human mammary tumor cell lines along with a cell line for adenofibrosarcoma of canine origin, and infected them with CDV (30). They were testing the sensitivity to CDV infection, cell proliferation, apoptosis, mitochondrial membrane potential and expression of tumor necrosis factor-alpha-induced protein 8 (TNFAIP8). Using qPCR they were able to quantify both TNFAIP8 gene, and the virus CDVM gene expression; they found that both TNFAIP8 and CDVM gene expression were positively correlated in all cell lines.
Critical Analysis: Advantages/Disadvantages of Four Strategies
In critically assessing the advantages and limitations of each of the four strategies described earlier, various conclusions became clear. For example, when looking at overall gene expression changes of tumor cells, the advantages include that it generates the broadest view of what is happening inside the tumor cells after infection with the virus without the initial bias of inserting a gene and looking specifically at that gene. Of course, it is also provides a much wider scope which is more advantageous than looking at only one or two genes. The disadvantages of this method, on the other hand, is that they provide less specific information about specific pathways or mechanisms through which the tumor increases or decreases due to the wider lens used. In addition, this method provides no information on the viral gene expression or the correlation between the virus and the decrease in tumor size.
Upon analyzing the second method, namely, gene expression of specific genes in the tumor cells, we saw various strengths and weaknesses as well. For example, this method could potentially provide more information on the mechanism for tumor shrinkage or growth. In addition, since the method is more specific it makes it slightly easier to execute. Furthermore, once the specific genes are chosen, it enables a more in depth analysis of how these genes are important and how gene expression changes in the tumor cells after viral infection can have an impact on the cells.
On the other hand, looking at specific genes in the tumor cells might not reflect the overall tumor status since the broad picture of what is going on in the tumor cells is missing. Similar to the limitation of the first method, this method also does not provide any information on the viral gene expression or the correlation between the virus and the decrease in the tumor.
The third method of looking at transgenes is advantageous because it is a relatively simple study to do, allowing for effective monitoring of a specific transgene, and it is not “shooting in the dark” and looking for a wide variety of genes. In this way, researchers can test how effective a specific transgene performs. However, disadvantages include that it lacks the broader perspective of a study looking at overall gene expression and does not provide any indication of how the expression of genes in the tumor cells change after viral infection.
Monitoring viral gene expression is helpful in terms of providing information about how effectively the virus is infecting and replicating in the host cell, but provides no information about gene expression changes in the tumor cell or the mechanisms for tumor growth or shrinkage. Compared with analyzing marker peptides to monitor viral growth and replication, gene expression might not be as sensitive, but this requires further studies to confirm this finding.
Overall, in terms of future directions, it would seem that combination studies are the optimal method for studying gene expression changes. They potentially allow both a broad picture of gene expression changes in tumor cells, and the ability to correlate the tumor shrinkage or growth with viral replication. Furthermore, much information about the precise mechanisms for how the virus attacks the tumor cells can be culled from this type of study. As more combination studies are performed, patterns in the clusters of genes involved will become apparent enabling researchers to pinpoint exactly how various viruses attack tumors and providing fruitful ideas for developing a new generation of recombinant viruses that are more effective.
Author Contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication. AA drafted the article, reviewed the relevant literature, made substantial contributions to conception and design, interpreted the data and approved final version. JR drafted the article, reviewed the relevant literature, made substantial contributions to conception and design, interpreted the data, revised the article critically and approved final version. PZ made substantial contributions to conception and design, participated in revising it critically, interpreted the data, and approved final version. BG participated in revising it critically, interpreted the data, and approved final version.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The authors are all employees of Rapo Yerapeh Ltd.
Funding
The research was funded by Rapo Yerapeh Ltd.
Abbreviations
ACC, adenoid cystic carcinoma; βhCG, beta-human chorionic gonadotropin; CEA, carcinoembryonic antigen; CDV, canine distemper virus; DAI, DNA-dependent activator of IFN− regulatory factors; DNA, deoxyribonucleic acid; ELISA, enzyme-linked immunosorbent assay; FDA, Food and Drug Administration; GBM, glioblastoma multiforme; GM-CSF, granulocyte-macrophage colony-stimulating factor; GnRH, gonadotropin releasing hormone; HSV, herpes simplex virus; IL, interleukin; IFN, interferon; IPA, Ingenuity Pathways Analysis; ISVP, infectious subviral particle; mRNA, messenger ribonucleic acid; MOI, multiplicity of infection; MV, measles virus; NDV, Newcastle disease virus; NOS/SCID, non-obese diabetic/severe combined immunodeficient; PCR, polymerase chain reaction; qPCR, quantitative polymerase chain reaction; RT-PCR, reverse transcription-polymerase chain reaction; ruc-GFP, Renilla luciferase-Aequorea-green fluorescent protein; SEA, superantigen Staphylococcus enterotoxin A; TNBC, triple negative breast cancer; TNF, tumor necrosis factor; VACV, vaccinia virus; VSV, vesicular stomatitis virus.
References
1. Kelly E, Russell SJ. History of oncolytic viruses: genesis to genetic engineering. Mol Ther (2007) 15(4):651–9. doi:10.1038/sj.mt.6300108
2. Liu TC, Kirn D. Gene therapy progress and prospects cancer: oncolytic viruses. Gene Ther (2008) 15(12):877–84. doi:10.1038/gt.2008.72
3. Kaufman HL, Bines SD. OPTIM trial: a phase III trial of an oncolytic herpes virus encoding GM-CSF for unresectable stage III or IV melanoma. Future Oncol (2010) 6(6):941–9. doi:10.2217/fon.10.66
4. Andtbacka RH, Kaufman HL, Collichio F, Amatruda T, Senzer N, Chesney J, et al. Talimogene laherparepvec improves durable response rate in patients with advanced melanoma. J Clin Oncol (2015) 33(25):2780–8. doi:10.1200/JCO.2014.58.3377
5. Pol J, Kroemer G, Galluzzi L. First oncolytic virus approved for melanoma immunotherapy. Oncoimmunology (2015) 5(1):e1115641. doi:10.1080/2162402X.2015.1115641
6. Kurozumi K, Hardcastle J, Thakur R, Yang M, Christoforidis G, Fulci G, et al. Effect of tumor microenvironment modulation on the efficacy of oncolytic virus therapy. J Natl Cancer Inst (2007) 99(23):1768–81. doi:10.1093/jnci/djm229
7. Zhang Q, Yu YA, Wang E, Chen N, Danner RL, Munson PJ, et al. Eradication of solid human breast tumors in nude mice with an intravenously injected light-emitting oncolytic vaccinia virus. Cancer Res (2007) 67(20):10038–46. doi:10.1158/0008-5472.CAN-07-0146
8. Balogh A, Bator J, Marko L, Nemeth M, Pap M, Setalo G Jr, et al. Gene expression profiling in PC12 cells infected with an oncolytic Newcastle disease virus strain. Virus Res (2014) 185:10–22. doi:10.1016/j.virusres.2014.03.003
9. Pfankuche VM, Spitzbarth I, Lapp S, Ulrich R, Deschl U, Kalkuhl A, et al. Reduced angiogenic gene expression in morbillivirus-triggered oncolysis in a translational model for histiocytic sarcoma. J Cell Mol Med (2017) 21(4):816–30. doi:10.1111/jcmm.13023
10. Saito K, Uzawa K, Kasamatsu A, Shinozuka K, Sakuma K, Yamatoji M, et al. Oncolytic activity of Sindbis virus in human oral squamous carcinoma cells. Br J Cancer (2009) 101(4):684–90. doi:10.1038/sj.bjc.6605209
11. Lacroix J, Schlund F, Leuchs B, Adolph K, Sturm D, Bender S, et al. Oncolytic effects of parvovirus H-1 in medulloblastoma are associated with repression of master regulators of early neurogenesis. Int J Cancer (2014) 134(3):703–16. doi:10.1002/ijc.28386
12. Haddad D, Socci N, Chen CH, Chen NG, Zhang Q, Carpenter SG, et al. Molecular network, pathway, and functional analysis of time-dependent gene changes associated with pancreatic cancer susceptibility to oncolytic vaccinia virotherapy. Mol Ther Oncolytics (2016) 3:16008. doi:10.1038/mto.2016.8
13. Carey BL, Ahmed M, Puckett S, Lyles DS. Early steps of the virus replication cycle are inhibited in prostate cancer cells resistant to oncolytic vesicular stomatitis virus. J Virol (2008) 82(24):12104–15. doi:10.1128/JVI.01508-08
14. Josupeit R, Bender S, Kern S, Leuchs B, Hielscher T, Herold-Mende C, et al. Pediatric and adult high-grade glioma stem cell culture models are permissive to lytic infection with parvovirus H-1. Viruses (2016) 8(5):138. doi:10.3390/v8050138
15. Lee NH, Kim M, Oh SY, Kim SG, Kwon HC, Hwang TH. Gene expression profiling of hematologic malignant cell lines resistant to oncolytic virus treatment. Oncotarget (2017) 8(1):1213–25. doi:10.18632/oncotarget.13598
16. Tanaka M, Shimbo T, Kikuchi Y, Matsuda M, Kaneda Y. Sterile alpha motif containing domain 9 is involved in death signaling of malignant glioma treated with inactivated Sendai virus particle (HVJ-E) or type I interferon. Int J Cancer (2010) 126(8):1982–91. doi:10.1002/ijc.24965
17. Kurozumi K, Hardcastle J, Thakur R, Shroll J, Nowicki M, Otsuki A, et al. Oncolytic HSV-1 infection of tumors induces angiogenesis and upregulates CYR61. Mol Ther (2008) 16(8):1382–91. doi:10.1038/mt.2008.112
18. Han C, Hao L, Chen M, Hu J, Shi Z, Zhang Z, et al. Target expression of Staphylococcus enterotoxin A from an oncolytic adenovirus suppresses mouse bladder tumor growth and recruits CD3+ T cell. Tumor Biol (2013) 34:2863–9. doi:10.1007/s13277-013-0847-3
19. Ma S, Qu W, Mao L, Zhu Z, Jia L, Zhao L, et al. Antitumor effects of oncolytic adenovirus armed with Drosophila melanogaster deoxyribonucleoside kinase in colorectal cancer. Oncol Rep (2012) 27(5):1443–50. doi:10.3892/or.2012.1665
20. Li Y, Zhang B, Zhang H, Zhu X, Feng D, Zhang D, et al. Oncolytic adenovirus armed with shRNA targeting MYCN gene inhibits neuroblastoma cell proliferation and in vivo xenograft tumor growth. J Cancer Res Clin Oncol (2013) 139(6):933–41. doi:10.1007/s00432-013-1406-4
21. Hirvinen M, Capasso C, Guse K, Garofalo M, Vitale A, Ahonen M, et al. Expression of DAI by an oncolytic vaccinia virus boosts the immunogenicity of the virus and enhances antitumor immunity. Mol Ther Oncolytics (2016) 3:16002. doi:10.1038/mto.2016.2
22. Jiang YQ, Zhang Z, Cai HR, Zhou H. Killing effect of TNF-mediated by conditionally replicating adenovirus on esophageal cancer and lung cancer cell lines. Int J Clin Exp Pathol (2015) 8(11):13785–94.
23. Shin EJ, Wanna GB, Choi B, Aguila D III, Ebert O, Genden EM, et al. Interleukin-12 expression enhances vesicular stomatitis virus oncolytic therapy in murine squamous cell carcinoma. Laryngoscope (2007) 117(2):210–4. doi:10.1097/01.mlg.0000246194.66295.d8
24. Guse K, Dias JD, Bauerschmitz GJ, Hakkarainen T, Aavik E, Ranki T, et al. Luciferase imaging for evaluation of oncolytic adenovirus replication in vivo. Gene Ther (2007) 14(11):902–11. doi:10.1038/sj.gt.3302949
25. Alain T, Kim TS, Lun X, Liacini A, Schiff LA, Senger DL, et al. Proteolytic disassembly is a critical determinant for reovirus oncolysis. Mol Ther (2007) 15(8):1512–21. doi:10.1038/sj.mt.6300207
26. Sato D, Kurihara Y, Kondo S, Shirota T, Urata Y, Fujiwara T, et al. Antitumor effects of telomerase-specific replication-selective oncolytic viruses for adenoid cystic carcinoma cell lines. Oncol Rep (2013) 30(6):2659–64. doi:10.3892/or.2013.2738
27. Gholami S, Chen CH, Belin LJ, Lou E, Fujisawa S, Antonacci C, et al. Vaccinia virus GLV-1h153 is a novel agent for detection and effective local control of positive surgical margins for breast cancer. Breast Cancer Res (2013) 15(2):R26. doi:10.1186/bcr3404
28. Peng KW, Facteau S, Wegman T, O’Kane D, Russell SJ. Non-invasive in vivo monitoring of trackable viruses expressing soluble marker peptides. Nat Med (2002) 8(5):527–31. doi:10.1038/nm0502-527
29. Reinboth J, Ascierto ML, Chen NG, Zhang Q, Yu YA, Aguilar RJ, et al. Correlates between host and viral transcriptional program associated with different oncolytic vaccinia virus isolates. Hum Gene Ther Methods (2012) 23(5):285–96. doi:10.1089/hgtb.2012.057
30. Garcia JA, Ferreira HL, Vieira FV, Gameiro R, Andrade AL, Eugenio FR, et al. Tumour necrosis factor-alpha-induced protein 8 (TNFAIP8) expression associated with cell survival and death in cancer cell lines infected with canine distemper virus. Vet Comp Oncol (2017) 15(2):336–44. doi:10.1111/vco.12168
Keywords: oncolytic viruses, gene expression, oncolytic virotherapy, microarray analysis, transgenes
Citation: Ansel A, Rosenzweig JP, Zisman PD and Gesundheit B (2017) Monitoring the Efficacy of Oncolytic Viruses via Gene Expression. Front. Oncol. 7:264. doi: 10.3389/fonc.2017.00264
Received: 24 August 2017; Accepted: 19 October 2017;
Published: 06 November 2017
Edited by:
Anahid Jewett, University of California, Los Angeles, United StatesReviewed by:
Zong Sheng Guo, Harvard University, United StatesViktor Umansky, Deutsches Krebsforschungszentrum (DKFZ), Germany
Copyright: © 2017 Ansel, Rosenzweig, Zisman and Gesundheit. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Beni Gesundheit, Yi5nZXN1bmRAZ21haWwuY29t
 Ashley Ansel
Ashley Ansel Joshua P. Rosenzweig
Joshua P. Rosenzweig Philip D. Zisman
Philip D. Zisman Beni Gesundheit
Beni Gesundheit