- 1Department of Molecular Medicine, Sapienza University of Rome, Rome, Italy
- 2Genome Diagnostics Program, IFOM - The FIRC Institute of Molecular Oncology, Milan, Italy
- 3Cancer Risk Factors and Lifestyle Epidemiology Unit, Institute for Cancer Research, Prevention and Clinical Network (ISPRO), Florence, Italy
- 4Division of Pathological Anatomy, Department of Surgery and Translational Medicine, University of Florence, Florence, Italy
- 5Institute for Maternal and Child Health IRCCS Burlo Garofolo, Trieste, Italy
- 6Molecular Genetics Laboratory, Istituto Tumori Giovanni Paolo II, Bari, Italy
- 7Dipartimento di Patologia, ASST Settelaghi and Centro di Ricerca per lo studio dei tumori eredo-familiari, Università dell'Insubria, Varese, Italy
- 8Section of Medical Oncology, Department of Surgical and Oncological Sciences, University of Palermo, Palermo, Italy
- 9IRCCS Ospedale Policlinico San Martino, Genoa, Italy
- 10Department of Experimental Medicine, Sapienza University of Rome, Rome, Italy
- 11Istituto Scientifico Romagnolo per lo Studio e la Cura dei Tumori (IRST), Meldola, Italy
- 12Department of Oncology and Haematology, University of Modena and Reggio Emilia, Modena, Italy
- 13Unità di Oncogenetica e Oncogenomica Funzionale, Centro di Riferimento Oncologico di Aviano (CRO), IRCCS, Aviano, Italy
- 14Division of Cancer Prevention and Genetics, European Institute of Oncology (IEO), IRCCS, Milan, Italy
- 15Unità di Genetica Medica, Dipartimento di Oncologia Medica ed Ematologia, Fondazione IRCCS Istituto Nazionale dei Tumori (INT), Milan, Italy
- 16Immunology and Molecular Oncology Unit, Veneto Institute of Oncology IOV - IRCCS, Padua, Italy
- 17Unità di Ricerca Medicina Predittiva: Basi molecolari Rischio genetico e Test genetici, Dipartimento di Ricerca, Fondazione IRCCS Istituto Nazionale Tumori (INT), Milan, Italy
Inherited mutations in BRCA1, and, mainly, BRCA2 genes are associated with increased risk of male breast cancer (MBC). Mutations in PALB2 and CHEK2 genes may also increase MBC risk. Overall, these genes are functionally linked to DNA repair pathways, highlighting the central role of genome maintenance in MBC genetic predisposition. MUTYH is a DNA repair gene whose biallelic germline variants cause MUTYH-associated polyposis (MAP) syndrome. Monoallelic MUTYH variants have been reported in families with both colorectal and breast cancer and there is some evidence on increased breast cancer risk in women with monoallelic variants. In this study, we aimed to investigate whether MUTYH germline variants may contribute to MBC susceptibility. To this aim, we screened the entire coding region of MUTYH in 503 BRCA1/2 mutation negative MBC cases by multigene panel analysis. Moreover, we genotyped selected variants, including p.Tyr179Cys, p.Gly396Asp, p.Arg245His, p.Gly264Trpfs*7, and p.Gln338His, in a total of 560 MBC cases and 1,540 male controls. Biallelic MUTYH pathogenic variants (p.Tyr179Cys/p.Arg241Trp) were identified in one MBC patient with phenotypic manifestation of adenomatous polyposis. Monoallelic pathogenic variants were identified in 14 (2.5%) MBC patients, in particular, p.Tyr179Cys was detected in seven cases, p.Gly396Asp in five cases, p.Arg245His and p.Gly264Trpfs*7 in one case each. The majority of MBC cases with MUTYH pathogenic variants had family history of cancer including breast, colorectal, and gastric cancers. In the case-control study, an association between the variant p.Tyr179Cys and increased MBC risk emerged by multivariate analysis [odds ratio (OR) = 4.54; 95% confidence interval (CI): 1.17–17.58; p = 0.028]. Overall, our study suggests that MUTYH pathogenic variants may have a role in MBC and, in particular, the p.Tyr179Cys variant may be a low/moderate penetrance risk allele for MBC. Moreover, our results suggest that MBC may be part of the tumor spectrum associated with MAP syndrome, with implication in the clinical management of patients and their relatives. Large-scale collaborative studies are needed to validate these findings.
Introduction
Male Breast Cancer (MBC) is a rare disease whose etiology appears to be associated with genetic factors. Inherited mutations in BRCA1 and, mainly, BRCA2, predispose to MBC and account for up to 13% of all cases in the Italian population (1). Even though there is evidence supporting an association between increased MBC risk and pathogenic variants in PALB2 and CHEK2 (2–4), these two genes are unlikely to account for a substantial fraction of MBC cases. Thus, additional genes that may contribute to MBC genetic susceptibility need to be investigated.
BRCA1, BRCA2, PALB2, and CHEK2 belong to or are functionally linked to the Homologous Recombination (HR) mechanism, one of the most important DNA Double-Strand Break (DSB) repair pathways, highlighting the central role of genome maintenance in MBC predisposition (5). Overall, the maintenance of genomic integrity is achieved by a coordinated interplay of different mechanisms of DNA repair, including Mismatch Repair (MMR), Nucleotide Excision Repair (NER) and Base Excision Repair (BER), in addition to DSB repair (6, 7). While dysregulation of DSB repair is known to play a relevant role in breast cancer (BC) pathogenesis, the involvement of other DNA repair pathways in BC is much less established.
MUTYH encodes a DNA glycosylase involved in BER, preventing 8-oxo-G:A mispairs generated by oxidative damage (8). Oxidative DNA damage, including 8-oxoG, may be due to hormonal metabolism and may contribute to BC susceptibility (9, 10). In this context, it is noteworthy that BRCA1 and BRCA2 are also involved in 8-oxoG repair (11), thus further supporting a possible role of BER and, more specifically, MUTYH in BC pathogenesis.
Biallelic (homozygous or compound heterozygous) MUTYH variants occur in 0.01–0.04% of European descent populations and cause MUTYH-associated polyposis syndrome (MAP), which predisposes patients to develop colorectal polyps and colorectal cancer (12–19). Monoallelic (heterozygous) MUTYH variants occur in 1–2% of European descent populations and are associated with an increased risk of colorectal cancer (14, 16–21). Several studies on extracolonic cancers in carriers of MUTYH variants have been performed (21–26). The association of MUTYH variants with malignancies other than colon cancer is less robust, especially when establishing cancer risks in heterozygous MUTYH individuals. Increased risks of bladder and ovarian cancers have been reported for biallelic mutation carriers, while slightly increased risks of gastric, hepatobiliary, endometrial, and breast cancer have been observed in monoallelic mutation carriers (27).
Overall, the association between MUTYH mutations and BC risk remains controversial, some studies have shown an increased BC risk among MUTYH mutation carriers, while others have not (22–26, 28–30). An increased risk of BC associated with biallelic and monoallelic variants of MUTYH has been reported in BRCA1/2 mutation negative individuals (21–23, 26). A higher frequency of monoallelic MUTYH mutations in families with both breast and colorectal cancer has been also reported compared to general population (21). Recently, an increased BC risk has been also reported for women with the common p.Gln338His variant (31).
To date the possible association between MUTYH variants and MBC risk has not been investigated. MBC is recognized as being primarily a hormone-dependent malignancy and is widely accepted as an estrogen-driven disease specifically related to hyperestrogenism (32) thus, oxidative DNA damage, due to hormonal metabolism, may particularly contribute to BC susceptibility in men. In this context, impairment of MUTYH activity due to inactivating/pathogenic variants may contribute to increase MBC risk.
To assess if MUTYH germline variants may contribute to MBC susceptibility, we screened a large series of BRCA1/2 mutation negative MBC patients by sequencing the entire MUTYH coding region. Furthermore, to explore whether MUTYH variants were significantly associated with MBC risk, we performed a case-control study of selected MUTYH variants.
Patients and Methods
Study Population
A total of 560 BRCA1/2 mutation negative MBC cases and 1,540 male controls, enrolled in the frame of the ongoing Italian Multicenter Study on MBC (33), were included in the present study. For each MBC case, information on the main clinical-pathologic characteristics were collected as previously described (33, 34). Controls were male individuals without personal history of cancer, enrolled under research or clinical protocols, or blood donors. All controls were recruited in the same geographical area of cases. For each study participant, samples of blood or DNA from peripheral blood leukocytes were collected. DNA from blood samples was extracted and quantified as previously described (35). The study was approved by Local Ethical Committee (Sapienza University of Rome, Prot. 669/17) and informed consent for using information and biological samples was obtained from all participants to the study.
MUTYH Gene Sequencing
A total of 503 MBC cases underwent next generation sequencing (NGS) of a custom panel of 50 cancer susceptibility genes including MUTYH. Briefly, paired-end libraries were prepared using the Nextera Rapid Capture Custom Enrichment kit (Illumina, San Diego, California, USA), pooled and loaded into a MiniSeq system (Illumina) for automated cluster generation, sequencing, and data analysis, including variant calling. Variant annotation and filtering was performed with Illumina Variant Studio Software version 2.2 against the human reference genome GRCh37. Variants were classified as pathogenic or likely pathogenic (collectively termed, pathogenic) according to the American College of Medical Genetics and Genomics (ACMG) recommendations (36). Briefly, variants were classified as pathogenic if they had a truncating, initiation codon or splice donor/acceptor effect or if pathogenicity was demonstrated by functional studies supportive of a damaging effect on the gene or gene product. All pathogenic variants were confirmed by double-stranded Sanger Sequencing (primer sequences are available upon request). Variants were named according to Human Genome Variation Society nomenclature (HGVS, http://www.hgvs.org).
Genotyping Analysis
Genotyping analysis of five MUTYH variants, rs34612342 (c.536A>G; p.Tyr179Cys), rs36053993 (c.1187G>A; p.Gly396Asp), rs140342925 (c.734G>A; p.Arg245His), rs587780751 (c.933+3A>C; p.Gly264Trpfs*7), and rs3219489 (c.1014G>C; p.Gln338His), identified by NGS and selected because previously proposed to be associated with increased risk of extracolonic cancer, including BC, was performed by allelic discrimination real-time PCR, in an ABI 7500 fast real-time PCR instrument (Life Technologies, Carlsbad, California, USA), using commercially available TaqMan SNP genotyping assays (Life Technologies) and according to the manufacturer's instructions. The specific assay IDs used are: C_32911941_10 (rs36512342), C_27860250_10 (rs36053993), C_166223223_10 (rs140342925), C_362043726_10 (rs587780751), and C_27504565_10 (rs3219489). In each experiment, positive (cases for which genotype was confirmed by Sanger Sequencing) and negative (water) controls were always included. A total of 560 MBC cases, including the 503 cases analyzed by NGS, and 1,540 male controls were genotyped.
Statistical Analysis
Chi-square test was performed in a case-case analysis in order to evaluate potential associations between pathogenic variants and specific clinical-pathologic characteristics.
The genotype frequency for each variant was evaluated in both series of cases and controls. The association between each variant and overall MBC risk was measured by the odds ratio (OR) and its corresponding 95% confidence interval (CI) by univariate logistic regression, and also by a multivariate analysis including adjustment for age, center and type of enrolment. A p-value < 0.05 was considered statistically significant. All the analyses were performed using STATA version 13.1 statistical program.
Results
Clinical-Pathologic Characteristics of MBC Cases
The study population consisted of 560 BRCA1/2 mutation negative MBC cases, enrolled in the frame of the ongoing Italian Multicenter Study on MBC. Overall, mean age at first BC diagnosis was 61.8 years (range 22–91 years); 91 cases (16.2%) reported first-degree family history of breast and/or ovarian cancer (BC/OC), 247 cases (44.1%) had first-degree family history of cancer and 101 cases (18%) had a personal history of cancer in addition to BC, mostly colorectal and prostate cancer. The majority of male breast tumors were invasive ductal carcinomas (85.9%), estrogen receptor positive (ER+, 94.2%), progesterone receptor positive (PR+ 88.4%), and HER2 negative (79.2%).
MUTYH Gene Sequencing in MBC Cases
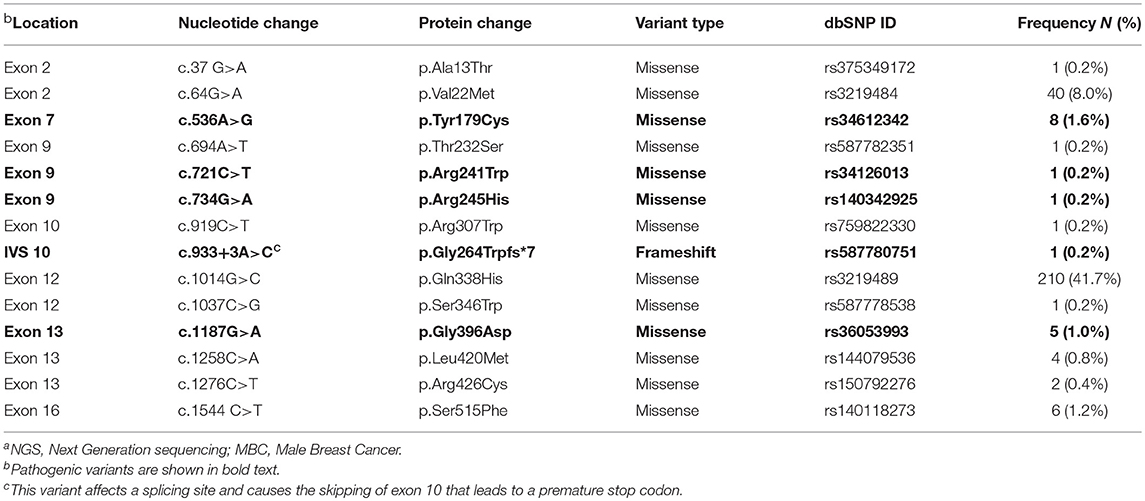
The entire coding region of MUTYH was screened in 503 BRCA1/2 mutation negative MBC cases, by a custom multigene panel using NGS technologies. MUTYH variants detected are shown in Table 1. p.Tyr179Cys and p.Gly396Asp variants were the most frequently detected pathogenic variants and were identified in 1.6 and 1.0% of the MBC cases, respectively. The common variant p.Gln338His was identified in 41.7% of the MBC cases (Table 1).
Overall, pathogenic variants were identified in 15 (3.0%) MBC cases (Table 2), 14 cases were carriers of monoallelic (heterozygous) pathogenic variants and one case was carrier of the biallelic p.Tyr179Cys/p.Arg241Trp (compound heterozygous) pathogenic variants. The majority of MBC cases with MUTYH pathogenic variants had family history of cancer including breast, colorectal, and gastric cancers (Table 2). In particular, the biallelic MUTYH pathogenic variant carrier was a man diagnosed with BC at 51 years of age who developed colon cancer, with phenotypic manifestation of adenomatous polyposis, at early age (41 years) and had a first-degree relative affected by melanoma at young age (26 years). With the exception of this case, clinical features of the other MBC patients with MUTYH pathogenic variants did not suggest a MAP phenotype.
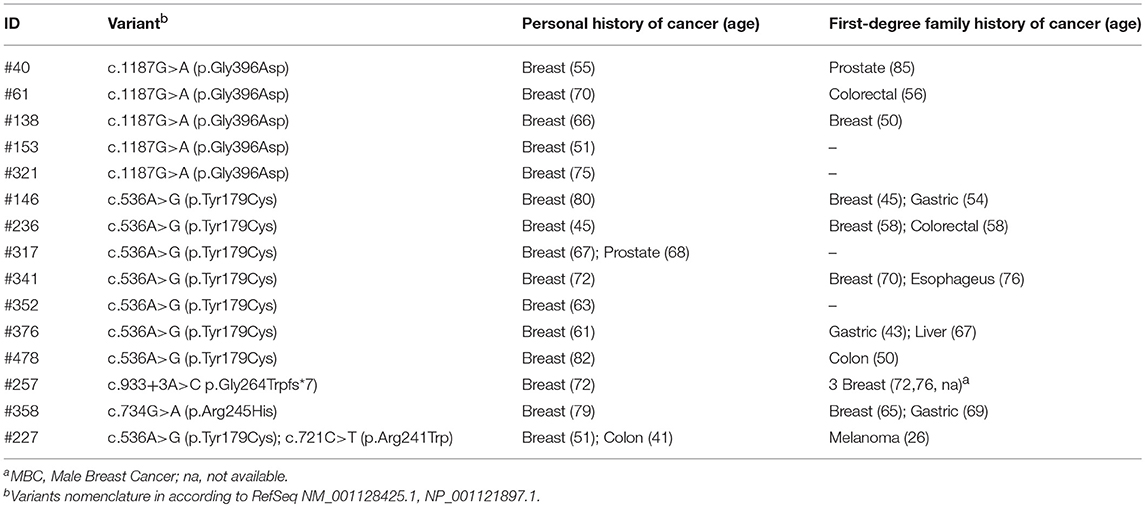
Table 2. Personal and family history of cancer in MBC cases with germline MUTYH pathogenic variantsa.
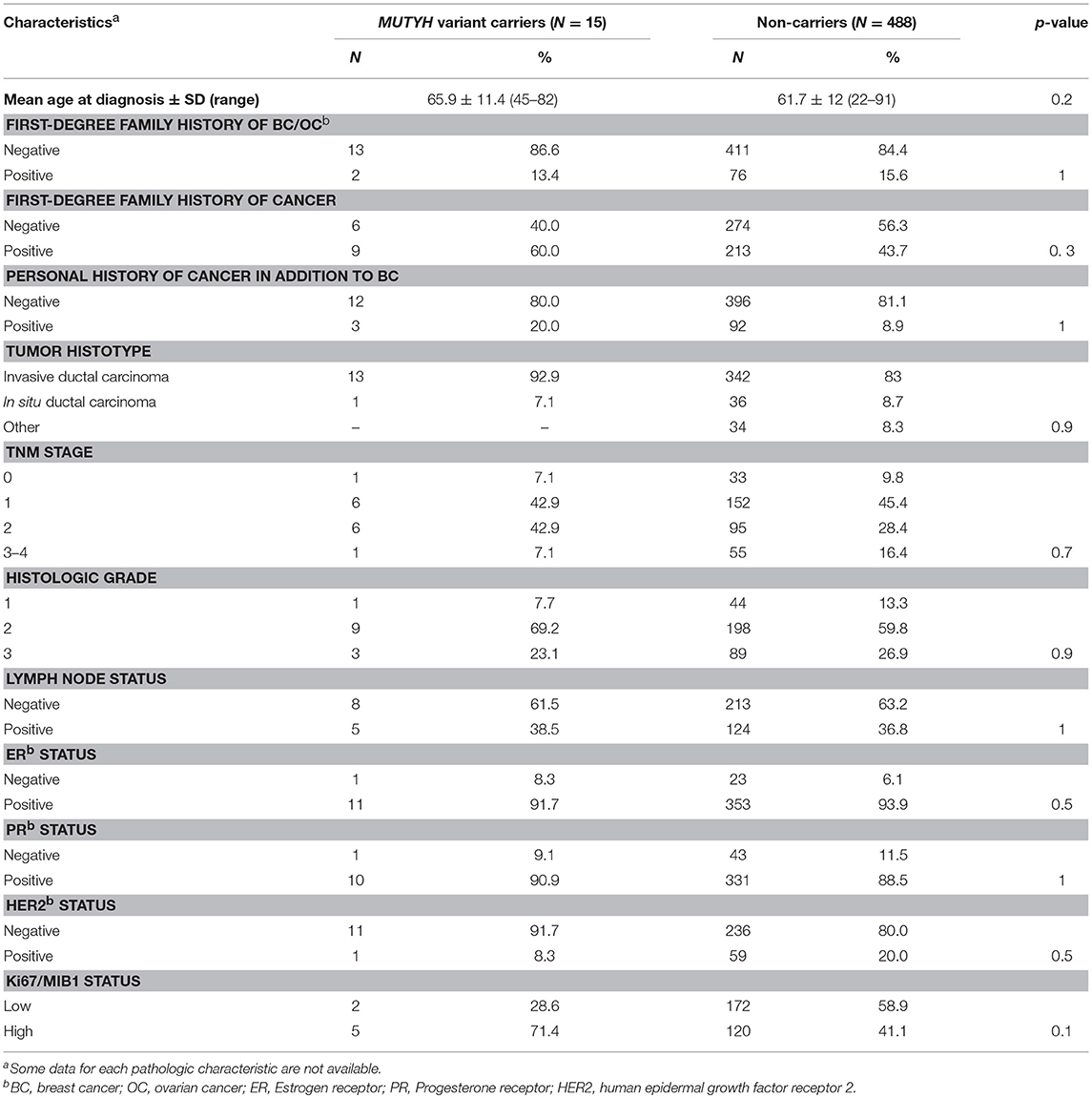
Overall, comparison of the clinical-pathologic characteristics between MUTYH pathogenic variant carriers and non-carriers did not show any statistically significant differences (Table 3).
Genotyping Analysis of Selected MUTYH Variants in MBC Cases and Controls
MUTYH pathogenic variants, including p.Tyr179Cys (rs34612342), p.Gly396Asp (rs36053993), p.Arg245His (rs140342925), p.Gly264Trpfs*7 (rs587780751), and the common variant p.Gln338His (rs3219489), were genotyped in 560 cases and 1,540 male controls. Overall, pathogenic variants were detected at significantly higher frequency (p = 0.04) in MBC cases (15/560 2.7%) than in controls (21/1540, 1.3%).
The distribution of genotype frequencies and the estimates for the association between each genotyped variant and overall MBC risk are summarized in Table 4. Significant differences in the distribution of genotypes between MBC cases and controls emerged for p.Tyr179Cys (rs34612342) variant. The analysis of the genotype-specific risks showed that men with heterozygous genotype for MUTYH p.Tyr179Cys variant were at increased BC risk both in the univariate (OR = 5.56; 95%CI:1.67–18.55; p = 0.005) and in the multivariate analysis (OR = 4.54; 95%CI:1.17–17.58; p = 0.028). No statistically significant differences in genotype distribution between case and controls emerged for the other variants analyzed.
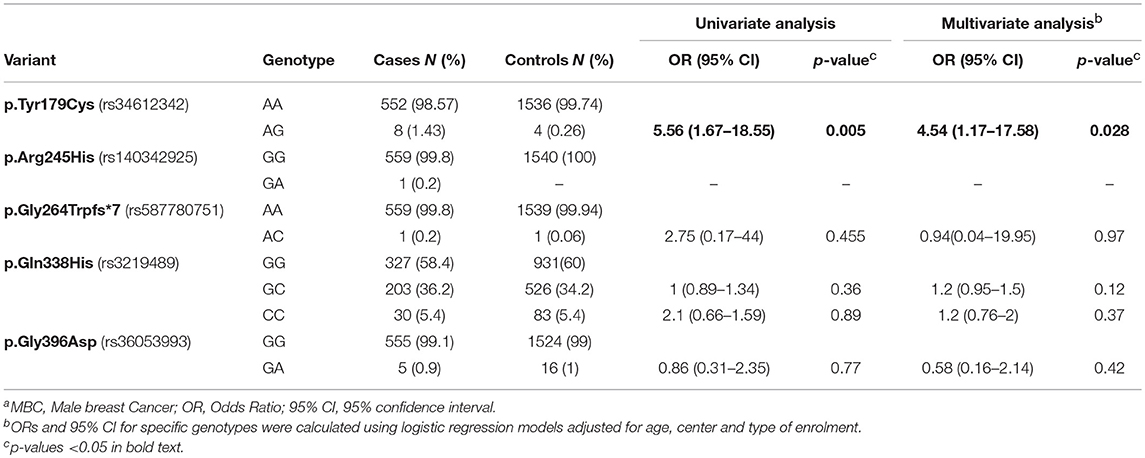
Table 4. Distribution of 560 BRCA1/2 negative MBC cases and 1,540 controls according to genotype frequencies and MBC risk estimates for selected MUTYH variantsa.
Discussion
In this study, we aimed to evaluate the contribution of MUTYH variants in MBC susceptibility. To this purpose, we obtained NGS data of the entire coding region of MUTYH from a large series of BRCA1/2 mutation negative MBC cases, from the ongoing Italian Multicenter Study on MBC, and further genotyped selected variants in a case-control study. To date, there is contrasting evidence on the impact of MUTYH pathogenic variants on risk of BC in women and, to the best of our knowledge, no study has been performed in MBC.
By NGS, we identified 15 MBC patients (3.0%) with germline MUTYH pathogenic variants, including one biallelic and 14 monoallelic variant carriers. The MBC patient with biallelic MUTYH pathogenic variants was affected by colorectal cancer at early age with phenotypic manifestation of adenomatous polyposis. Thus, our results allowed a molecular diagnosis of MAP. To the best of our knowledge, to date, only another MBC case has been reported with MAP syndrome (23). Taking into account the rarity of both MBC and MAP, the occurrence of MBC in MAP patients may underline a possible common genetic pathway and suggest that MBC could be considered a MAP-related malignancy.
Overall, MUTYH monoallelic pathogenic variants, including p.Tyr179Cys, p.Gly396Asp, p.Arg245His, and p.Gly264Trpfs*7, were found with a frequency of 2.8% in our MBC series. p.Tyr179Cys and p.Gly396Asp were the most frequently variants detected and were identified in 2.4% of the cases. Published data showed that these two variants are the most frequent pathogenic variants in populations of European origin and account for 50 to 90% of MUTYH pathogenic variants identified in MAP patients (13, 14, 37, 38). The p.Arg245His variant was identified in a MBC patient with family history of breast and gastric cancers. This variant has been reported strongly associated with familial colorectal cancer (23, 39), and has also been identified in patients with suspected Lynch Syndrome and in a patient with gastric cancer (23, 40). The p.Gly264Trpfs*7 variant was identified in a MBC patient, from North-East of Italy, where it occurs as a founder mutation accounting for about 15.0% of the MUTYH pathogenic variants identified in MAP patients (41). By contrast, this variant has been reported with lower frequency, ranging from 1.0 to 8.0%, in MAP patients from other populations of Caucasian ethnicity (23, 41–47).
To investigate whether MBC arising in MUTYH pathogenic variant carriers may be characterized by specific features, we compared clinical-pathologic characteristics between carriers and non-carriers. No statistically significant association emerged for any of the clinical features tested. However, the great majority of MBC patients with MUTYH pathogenic variants had family history of cancer, including, breast, colorectal, and gastric cancers. These findings, if confirmed by additional data, may be useful in decisions concerning clinical management of patients and their families.
To further investigate the role of MUTYH in MBC, we evaluated the risk of MBC associated with selected MUTYH variants previously proposed to be associated with increased cancer risk, including BC risk (21, 27, 31), by performing a case-control study. Among the pathogenic variants examined, the p.Tyr179Cys variant was associated with an increased MBC risk (OR = 4.54, 95%CI = 1.17–17.58). A higher frequency of p.Tyr179Cys has been reported in families with both breast and colorectal cancer compared to the general population (21), but an association between p.Tyr179Cys variant and increased BC risk has not been observed (25, 26, 28, 30). Our results, suggest that p.Tyr179Cys variant may be a low/moderate penetrance risk allele for BC in men. This variant, located at 8-oxo-G binding site, causes major structural protein changes and a reduction in functionality (48, 49). Thus, oxidative DNA damage due to hormonal metabolism, like estrogen-induced 8-oxo-dG generation, may particularly contribute to MBC susceptibility, as BC in men is primarily a hormone-dependent tumor, specifically related to hyperestrogenism. Furthermore, it can be hypothesized that MBC, unencumbered by the many confounding factors that exist in female BC (i.e., reproductive factors and high frequency) might facilitate the identification of genetic factors and molecular mechanisms that may influence BC risk in general (50).
We also assessed whether the common p.Gln338His variant, reported to increase BC risk in women (31), was associated with MBC risk. We did not observe any significant differences in p.Gln338His genotypes distribution between MBC cases and controls inconsistent with a possible role of this variant in MBC risk. The other common variant, p.Val22Met, has not been reported to be associated with cancer risk (51–53) and was not examined in this study.
Overall, we observed that the majority of MBC patients with pathogenic MUTYH variants have first-degree family history of cancers. This raises the question of whether MUTYH variants, especially the Tyr179Cys variant, may be associated with MBC risk only, or with the risk of familial or multi-syndromic diseases, including MBC. Further clinical/phenotype assessments and detailed statistical analyses would be useful in future studies to answer this question.
In conclusion, our study suggests that MUTYH pathogenic variants may have a role in MBC, in particular, p.Tyr179Cys variant may be a low/moderate penetrance risk allele for MBC. Our findings also suggest that MBC may be part of the tumor spectrum associated with MAP syndrome, with implications in the clinical management of the patients and their relatives.
Although we have a large series of MBC cases, this study may be underpowered to detect smaller risk effects and large-scale collaborative studies are needed to investigate any possible association with rarer variants and to have a more comprehensive examination and characterization of the link between MUTYH variants and MBC risk.
Data Availability Statement
Datasets are available on request. The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.
Authors Contributions
PiR drafted the manuscript, performed NGS and statistical analyses and interpreted the results. VS performed genotyping and statistical analyses, and interpreted the results. AB and IC performed genotyping analysis. VZ and VV performed NGS analysis. IZ, GM, SB AS, ST, MT, AR, LV, AC, DC, LC, AV, BB, JA, SM, MM, PaR, and DP recruited samples and collected clinicalpathologic data. PP contributed to study design, recruited samples and collected clinical pathologic data. LO conceived, designed and coordinated the study, and drafted the manuscript. All authors reviewed, edited, and approved the manuscript for publication.
Funding
This study was supported by Associazione Italiana per la Ricerca sul Cancro (AIRC IG 16933) to LO.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank all the participants in this study and the institutions and their staff who supported the recruitment of patients and the collection of samples and data.
Abbreviations
ACMG, American College of Medical Genetics and Genomics; BC, breast cancer; BER, base excision repair; DSB, double-strand break; ER, estrogen receptor; HER2, human epidermal growth factor receptor 2; HGVS, Human Genome Variation Society; HR, homologous recombination; MAP, MUTYH-associated polyposis; MBC, male breast cancer; MMR, mismatch repair; NER, nucleotide excision repair; NGS, next generation sequencing; OC, ovarian cancer; OR, Odds Ratio; PR, progesterone receptor; CI, confidence interval.
References
1. Rizzolo P, Silvestri V, Tommasi S, Pinto R, Danza K, Falchetti M, et al. Male breast cancer: genetics, epigenetics, and ethical aspects. Ann Oncol. (2013) 24(Suppl 8):viii75–82. doi: 10.1093/annonc/mdt316
2. Falchetti M, Lupi R, Rizzolo P, Ceccarelli K, Zanna I, Calò V, et al. BRCA1/BRCA2 rearrangements and CHEK2 common mutations are infrequent in Italian male breast cancer cases. Breast Cancer Res Treat. (2008) 110:161–7. doi: 10.1007/s10549-007-9689-2
3. Silvestri V, Zelli V, Valentini V, Rizzolo P, Navazio AS, Coppa A, et al. Whole-exome sequencing and targeted gene sequencing provide insights into the role of PALB2 as a male breast cancer susceptibility gene. Cancer (2017) 123:210–8. doi: 10.1002/cncr.30337
5. Nielsen FC, van Overeem Hansen T, Sørensen CS. Hereditary breast and ovarian cancer: new genes in confined pathways. Nat Rev Cancer (2016) 16:599–612. doi: 10.1038/nrc.2016.72
6. Tham KC, Kanaar R, Lebbink JH. Mismatch repair and homeologous recombination. DNA Repair (2016) 38:75–83. doi: 10.1016/j.dnarep.2015.11.010
7. Tubbs A, Nussenzweig A. Endogenous DNA Damage as a Source of Genomic Instability in Cancer. Cell (2017) 168:644–56. doi: 10.1016/j.cell.2017.01.002
8. Mazzei F, Viel A, Bignami M. Role of MUTYH in human cancer. Mutat Res. (2013) 743–744:33–43. doi: 10.1016/j.mrfmmm.2013.03.003
9. Li D, Zhang W, Zhu J, Chang P, Sahin A, Singletary E, et al. Oxidative DNA damage and 8-hydroxy-2-deoxyguanosine DNA glycosylase/apurinic lyase in human breast cancer. Mol Carcinog. (2001) 31:214–23. doi: 10.1002/mc.1056
10. Dziaman T, Huzarski T, Gackowski D, Rozalski R, Siomek A, Szpila A, et al. Elevated level of 8-oxo-7,8-dihydro-2'-deoxyguanosine in leukocytes of BRCA1 mutation carriers compared to healthy controls. Int J Cancer (2009) 125:2209–13. doi: 10.1002/ijc.24600
11. Le Page F, Randrianarison V, Marot D, Cabannes J, Perricaudet M, Feunteun J, et al. BRCA1 and BRCA2 are necessary for the transcription-coupled repair of the oxidative 8-oxoguanine lesion in human cells. Cancer Res. (2000) 60:5548–52.
12. Al-Tassan N, Eisen T, Maynard J, Bridle H, Shah B, Fleischmann C, et al. Inherited variants of MYH associated with somatic G:C–>T:A mutations in colorectal tumors. Nat Genet. (2002) 30:227. doi: 10.1038/ng828
13. Sieber OM, Lipton L, Crabtree M, Heinimann K, Fidalgo P, Phillips RK, et al. Multiple colorectal adenomas, classic adenomatous polyposis, and germ-line mutations in MYH. N Engl J Med. (2003) 348:791–9. doi: 10.1056/NEJMoa025283
14. Cleary SP, Cotterchio M, Jenkins MA, Kim H, Bristow R, Green R, et al. Germline MutY human homologue mutations and colorectal cancer: a multisite case–control study. Gastroenterology (2009) 136:1251–60. doi: 10.1053/j.gastro.2008.12.050
15. Lubbe SJ, Di Bernardo MC, Chandler IP, Houlston RS. Clinical implications of the colorectal cancer risk associated with MUTYH mutation. J Clin Oncol. (2009) 27:3975–80. doi: 10.1200/JCO.2008.21.6853
16. Theodoratou E, Campbell H, Tenesa A, Houlston R, Webb E, Lubbe S, et al. A large-scale meta-analysis to refine colorectal cancer risk estimates associated with MUTYH variants. Br J Cancer (2010) 103:1875–84. doi: 10.1038/sj.bjc.6605966
17. Win AK, Dowty JG, Cleary SP, Kim H, Buchanan DD, Young JP, et al. Risk of colorectal cancer for carriers of mutations in MUTYH, with and without a family history of cancer. Gastroenterology (2014) 146:1208–11. doi: 10.1053/j.gastro.2014.01.022
18. Peterlongo P, Mitra N, Sanchez de Abajo A, de la Hoya M, Bassi C, Bertario L, et al. Increased frequency of disease-causing MYH mutations in colon cancer families. Carcinogenesis (2006) 27:2243–9. doi: 10.1093/carcin/bgl093
19. Peterlongo P, Mitra N, Chuai S, Kirchhoff T, Palmer C, Huang H, et al. Colorectal cancer risk in individuals with biallelic or monoallelic mutations of MYH. Int J Cancer (2005) 114:505–7. doi: 10.1002/ijc.20767
20. Jones N, Vogt S, Nielsen M, Christian D, Wark PA, Eccles D, et al. Increased colorectal cancer incidence in obligate carriers of heterozygous mutations in MUTYH. Gastroenterology (2009) 137:489–94. doi: 10.1053/j.gastro.2009.04.047
21. Wasielewski M, Out AA, Vermeulen J, Nielsen M, van den Ouweland A, Tops CM, et al. Increased MUTYH mutation frequency among Dutch families with breast cancer and colorectal cancer. Breast Cancer Res Treat (2010) 124:635–41. doi: 10.1007/s10549-010-0801-7
22. Nielsen M, Franken PF, Reinards TH, Weiss MM, Wagner A, van der Klift H, et al. Multiplicity in polyp count and extracolonic manifestations in 40 Dutch patients with MYHassociated polyposis coli (MAP). J Med Genet. (2005) 42:e54. doi: 10.1136/jmg.2005.033217
23. Vogt S, Jones N, Christian D, Engel C, Nielsen M, Kaufmann A, et al. Expanded extracolonic tumor spectrum in MUTYH-associated polyposis. Gastroenterology (2009) 137:1976–85.e1-10. doi: 10.1053/j.gastro.2009.08.052
24. Win AK, Cleary SP, Dowty JG, Baron JA, Young JP, Buchanan DD, et al. Cancer risks for monoallelic MUTYH mutation carriers with a family history of colorectal cancer. Int J Cancer (2011) 129:2256–62. doi: 10.1002/ijc.25870
25. Out AA, Wasielewski M, Huijts PE, van Minderhout IJ, Houwing-Duistermaat JJ, Tops CM, et al. MUTYH gene variants and breast cancer in a Dutch case–control study. Breast Cancer Res Treat. (2012) 134:219–27. doi: 10.1007/s10549-012-1965-0
26. Rennert G, Lejbkowicz F, Cohen I, Pinchev M, Rennert HS, Barnett-Griness O. MutYH mutation carriers have increased breast cancer risk. Cancer (2012) 118:1989–93. doi: 10.1002/cncr.26506
27. Win AK, Reece JC, Dowty JG, Buchanan DD, Clendenning M, Rosty C, et al. Risk of extracolonic cancers for people with biallelic and monoallelic mutations in MUTYH. Int J Cancer (2016) 139:1557–63. doi: 10.1002/ijc.30197
28. Beiner M, Zhang W, Zhang S, Gallinger S, Sun P, Narod SA. Mutations of the MYH gene do not substantially contribute to the risk of breast cancer. Breast Cancer Res Treat. (2009) 114:575–8. doi: 10.1007/s10549-008-0042-1
29. Zhu M, Chen X, Zhang H, Xiao N, Zhu C, He Q, et al. AluYb8 insertion in the MUTYH gene and risk of early-onset breast and gastric cancers in the Chinese population. Asian Pac J Cancer Prev. (2011) 12:1451–5.
30. Boesaard EP, Vogelaar IP, Bult P, Wauters CA, van Krieken JH, Ligtenberg MJ. Germline MUTYH gene mutations are not frequently found in unselected patients with papillary breast carcinoma. Hered Cancer Clin Pract. (2014) 12:21. doi: 10.1186/1897-4287-12-21
31. Kappil M, Terry MB, Delgado-Cruzata L, Liao Y, Santella RM. Mismatch repair polymorphisms as markers of breast cancer prevalence in the breast cancer family registry. Anticancer Res. (2016) 36:4437–41. doi: 10.21873/anticanres.10987
32. Brinton LA, Cook MB, McCormack V, Johnson KC, Olsson H, Casagrande JT, et al. Anthropometric and hormonal risk factors for male breast cancer: male breast cancer pooling project results. J Natl Cancer Inst. (2014) 106:djt465. doi: 10.1093/jnci/djt465
33. Silvestri V, Rizzolo P, Zelli V, Valentini V, Zanna I, Bianchi S, et al. A possible role of FANCM mutations in male breast cancer susceptibility: results from a multicenter study in Italy. Breast (2018) 38:92–7. doi: 10.1016/j.breast.2017.12.013
34. Ottini L, Silvestri V, Rizzolo P, Falchetti M, Zanna I, Saieva C, et al. Clinical and pathologic characteristics of BRCA-positive and BRCA-negative male breast cancer patients: results from a collaborative multicenter study in Italy. Breast Cancer Res Treat. (2012) 134:411–8. doi: 10.1007/s10549-012-2062-0
35. Ottini L, Silvestri V, Saieva C, Rizzolo P, Zanna I, Falchetti M, et al. Association of low-penetrance alleles with male breast cancer risk and clinicopathological characteristics: results from a multicenter study in Italy. Breast Cancer Res Treat. (2013) 138:861–8. doi: 10.1007/s10549-013-2459-4
36. Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. (2015) 17(5):405–24. doi: 10.1038/gim.2015.30
37. Kanter-Smoler G, Björk J, Fritzell K, Engwall Y, Hallberg B, Karlsson G, et al. Novel findings in Swedish patients with MYH-associated polyposis: mutation detection and clinical characterization. Clin Gastroenterol Hepatol. (2006) 4:499–506. doi: 10.1016/j.cgh.2006.01.005
38. Aretz S, Tricarico R, Papi L, Spier I, Pin E, Horpaopan S, et al. MUTYH-associated polyposis (MAP): evidence for the origin of the common European mutations p.Tyr179Cys and p.Gly396Asp by founder events. Eur J Hum Genet. (2014) 22:923–9. doi: 10.1038/ejhg.2012.309
39. Ali M, Kim H, Cleary S, Cupples C, Gallinger S, Bristow R. Characterization of mutant MUTYH proteins associated with familial colorectal cancer. Gastroenterology (2008) 135:499–507. doi: 10.1053/j.gastro.2008.04.035
40. Yurgelun MB, Allen B, Kaldate RR, Bowles KR, Judkins T, Kaushik P, et al. Identification of a variety of mutations in cancer predisposition genes in patients with suspected lynch syndrome. Gastroenterology (2015) 149(3):604–13.e20. doi: 10.1053/j.gastro.2015.05.006
41. Pin E, Pastrello C, Tricarico R, Papi L, Quaia M, Fornasarig M, et al. MUTYH c.933+3A>C, associated with a severely impaired gene expression, is the first Italian founder mutation in MUTYH-Associated Polyposis. Int J Cancer (2013) 132:1060–9. doi: 10.1002/ijc.27761
42. Sampson JR, Dolwani S, Jones S, Eccles D, Ellis Evans A, et al. Autosomal recessive colorectal adenomatous polyposis due to inherited mutations of MYH. Lancet (2003) 362:39–41. doi: 10.1016/S0140-6736(03)13805-6
43. Eliason K, Hendrickson BC, Judkins T, Norton M, Leclair B, Lyon E, et al. The potential for increased clinical sensitivity in genetic testing for polyposis colorectal cancer through the analysis of MYH mutations in North American patients. J Med Genet. (2005) 42:95–6. doi: 10.1136/jmg.2004.025973
44. Aretz S, Uhlhaas S, Goergens H, Siberg K, Vogel M, Pagenstecher C, et al. MUTYH-associated polyposis: 70 of 71 patients with biallelic mutations present with an attenuated or atypical phenotype. Int J Cancer (2006) 119:807–14. doi: 10.1002/ijc.21905
45. O'Shea AM, Cleary SP, Croitoru MA, Kim H, Berk T, Monga N, et al. Pathological features of colorectal carcinomas in MYH-associated polyposis. Histopathology (2008) 53:184–94. doi: 10.1111/j.1365-2559.2008.03071.x
46. Nielsen M, Joerink-van de Beld MC, Jones N, Vogt S, Tops CM, Vasen HF, et al. Analysis of MUTYH genotypes and colorectal phenotypes in patients With MUTYH - associated polyposis. Gastroenterology (2009) 136:471–6. doi: 10.1053/j.gastro.2008.10.056
47. Morak M, Laner A, Bacher U, Keiling C, Holinski-Feder E. MUTYH-associated polyposis - variability of the clinical phenotype in patients with biallelic and monoallelic MUTYH mutations and report on novel mutations. Clin Genet. (2010) 78:353–63. doi: 10.1111/j.1399-0004.2010.01478.x
48. Fromme JC, Banerjee A, Huang SJ, Verdine GL. Structural basis for removal of adenine mispaired with 8-oxoguanine by MutY adenine DNA glycosylase. Nature (2004) 427:652–6. doi: 10.1038/nature02306
49. D'Agostino VG, Minoprio A, Torreri P, Marinoni I, Bossa C, Petrucci TC, et al. Functional analysis of MUTYH mutated proteins associated with familial adenomatous polyposis. DNA Repair (2010) 9:700–7. doi: 10.1016/j.dnarep.2010.03.008
50. Ottini L. Male breast cancer: a rare disease that might uncover underlying pathways of breast cancer. Nat Rev Cancer (2014) 14:643. doi: 10.1038/nrc3806
51. Görgens H, Krüger S, Kuhlisch E, Pagenstecher C, Höhl R, Schackert HK, et al. Microsatellite stable colorectal cancers in clinically suspected hereditary nonpolyposis colorectal cancer patients without vertical transmission of disease are unlikely to be caused by biallelic germline mutations in MYH. J Mol Diagn. (2006) 8:178–82. doi: 10.2353/jmoldx.2006.050119
52. Shin EJ, Chappell E, Pethe V, Hersey K, van der Kwast T, Fleshner N, et al. MYH mutations are rare in prostate cancer. J Cancer Res Clin Oncol. (2007) 133:373–8. doi: 10.1007/s00432-006-0181-x
Keywords: male breast cancer, genetic susceptibility, BRCA1/2, MUTYH, NGS, MUTYH-associated polyposis (MAP) syndrome, breast cancer risk
Citation: Rizzolo P, Silvestri V, Bucalo A, Zelli V, Valentini V, Catucci I, Zanna I, Masala G, Bianchi S, Spinelli AM, Tommasi S, Tibiletti MG, Russo A, Varesco L, Coppa A, Calistri D, Cortesi L, Viel A, Bonanni B, Azzollini J, Manoukian S, Montagna M, Radice P, Palli D, Peterlongo P and Ottini L (2018) Contribution of MUTYH Variants to Male Breast Cancer Risk: Results From a Multicenter Study in Italy. Front. Oncol. 8:583. doi: 10.3389/fonc.2018.00583
Received: 24 July 2018; Accepted: 19 November 2018;
Published: 04 December 2018.
Edited by:
Heather Cunliffe, University of Otago, New ZealandReviewed by:
Sevtap Savas, Memorial University of Newfoundland, CanadaRengyun Liu, Johns Hopkins University, United States
Copyright © 2018 Rizzolo, Silvestri, Bucalo, Zelli, Valentini, Catucci, Zanna, Masala, Bianchi, Spinelli, Tommasi, Tibiletti, Russo, Varesco, Coppa, Calistri, Cortesi, Viel, Bonanni, Azzollini, Manoukian, Montagna, Radice, Palli, Peterlongo and Ottini. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Laura Ottini, bGF1cmEub3R0aW5pQHVuaXJvbWExLml0
 Piera Rizzolo
Piera Rizzolo Valentina Silvestri1
Valentina Silvestri1 Irene Catucci
Irene Catucci Alessandro Mauro Spinelli
Alessandro Mauro Spinelli Maria Grazia Tibiletti
Maria Grazia Tibiletti Antonio Russo
Antonio Russo Anna Coppa
Anna Coppa Alessandra Viel
Alessandra Viel Jacopo Azzollini
Jacopo Azzollini Siranoush Manoukian
Siranoush Manoukian Paolo Radice
Paolo Radice Paolo Peterlongo
Paolo Peterlongo Laura Ottini
Laura Ottini