- 1Immunology and Molecular Oncology, Veneto Institute of Oncology IOV-IRCCS, Padova, Italy
- 2Eurac Research, Institute of Mountain Emergency Medicine, Bolzano, Italy
- 3Endoscopy Unit, Veneto Institute of Oncology IOV-IRCCS, Padova, Italy
- 4Department of Surgical Sciences, Oncology and Gastroenterology, University of Padova, Padova, Italy
Esophageal cancer (EC) is a very aggressive tumor, and no reliable prognostic markers exist especially for resectable advanced neoplasia. The principal aim of this study was to investigate the association of germline polymorphisms in nucleotide excision repair (NER) pathway genes with the overall survival (OS) of patients with advanced EC. As a second aim, we also studied the association of NER gene variants with response to cisplatin-based chemotherapy. Among the EC patients referred to our Institution between 2004 and 2012, we selected a cohort of 180 patients diagnosed with a clinical tumor stage ranging from IIB and IVA. Patients were genotyped for four NER variants, two in the ERCC1 (rs11615 and rs3212986) and two in the ERCC2/XPD (rs1799793 and rs13181) genes. Kaplan–Meier analyses and Cox proportional hazards model were used to evaluate the associations of the selected variants with OS; association with response to neoadjuvant therapy was investigated using logistic regression. Results showed that the ERCC1 rs3212986 and the ERCC2/XPD rs1799793 were significantly associated with shorter OS. On the contrary, response association analysis displayed that, while rs11615 and rs3212986 in ERCC1 were associated with response, both ERCC2/XPD variants were not. By creating survival prediction models, we showed that the rs3212986 and the rs1799793 have a better predictability of the tumor stage alone. Furthermore, they were able to improve the power of the clinical model (AUC = 0.660 vs. AUC = 0.548, p = 0.004). In conclusion, our results indicate that the ERCC1 rs3212986 and the ERCC2/XPD rs1799793 could be used as surrogate markers for a better stratification of EC patients with advanced resectable tumor.
Introduction
Esophageal cancer (EC) is a highly lethal malignancy, usually diagnosed at an advanced stage (1). Surgery is the standard of care for potentially resectable neoplasia, and very often is preceded by a cisplatin-based neoadjuvant chemotherapy. The two predominant histologic subtypes are adenocarcinoma (EADC) and squamous cell carcinoma (ESCC). EADC is thought to arise from an acquired precursor condition known as Barrett's esophagus caused by chronic gastro-esophageal reflux (2, 3). Other EADC risk factors, also shared by ESCC, include tobacco and alcohol consumption, habits that lead to a chronic inflammation status (4).
Epidemiological studies have shown that chronic inflammation predisposes to different types of cancer and, for this reason, inflammation has been proposed as the seventh hallmark of cancer (5). One of the mechanism involved in cancer-related inflammation is the induction of genetic instability resulting in random DNA alterations (5, 6). DNA repair genes are crucial for the maintenance of the integrity of DNA damaged by both endogenous and exogenous hazardous agents. Thus, DNA repair genes and their constitutive variants have been indicated as a possible cause of the inter-individual variability to chemotherapy and patient outcome or as a factor that could modify the risk of tumor development (7–14).
Among the different DNA repair mechanisms, nucleotide excision repair (NER) is one of the most relevant pathway involved in the repair of DNA damaged by tobacco, radiation, free radicals, and chemotherapeutic agents (15). Excision repair cross-complementing 1 (ERCC1) and excision repair cross-complementing 2/xeroderma pigmentosum group D (ERCC2/XPD) are considered two key rate-limiting enzymes in the multistep NER process, because of their pivotal role in the recognition and removal of damaged nucleotides. Both ERCC1 and ERCC2/XPD have single nucleotide polymorphisms (SNPs) that modulate their DNA repair capability (16–19). So far, numerous studies have investigated the potential predictive/prognostic power of constitutive genetic variants of these repair genes in a wide range of neoplasia, including EC, with contradictory results (20–24).
The first aim of this study was to define a possible association between variants in ERCC1 (rs11615 and rs3212986), and ERCC2/XPD (rs13181 and rs1799793) genes and OS of patients with advanced resectable EC; as a second aim, we also evaluated the association of the same genetic variants with response to cisplatin-based neoadjuvant chemotherapy. To restrain possible bias, among all the EC patients referred to our Institute between 2004 and 2012, we selected a cohort of patients, whose OS was not significantly affected by their clinico-pathological characteristics. The rs11615 (Asn118Asn) and rs3212986 (C8092A) in ERCC1, and the rs13181 (Lys751Gln) and rs1799793 (Asp312Asn) in ERCC2/XPD were selected based on their putative association with altered DNA repair capability (16–19), and their high frequency in the Caucasian population (minor allele frequency ranging from 28 to 42%).
Materials and Methods
Study Endpoints and Patient Selection
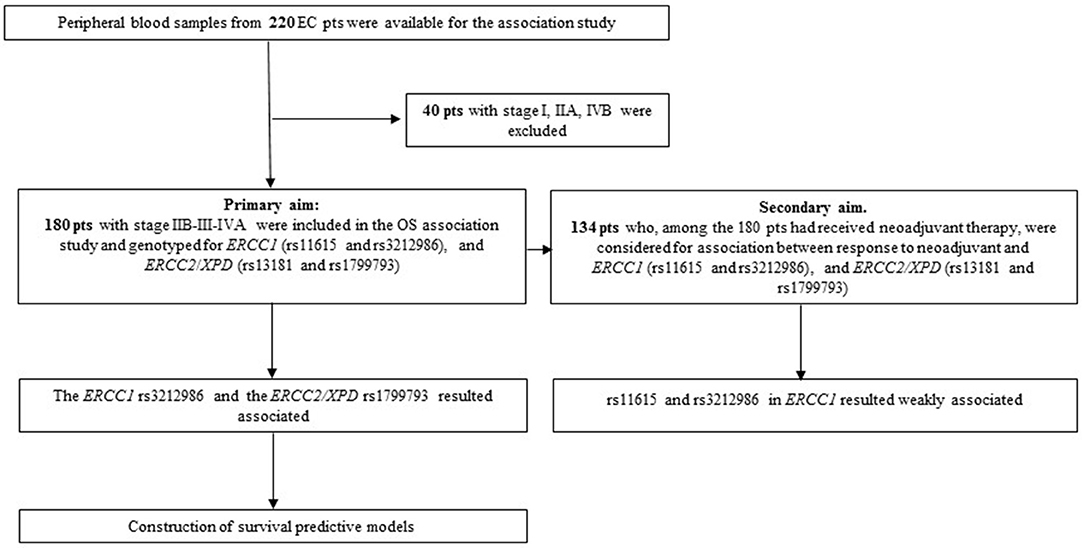
Blood samples from EC patients were collected between 2004 and 2012. Inclusion criteria for the overall survival (OS) association study were: a diagnosis of EC, availability of complete clinical data, clinical stage ranging from IIB and IVA, survival >3 months after diagnosis, follow-up >24 months for living patients. Differences in patient treatment (neo-adjuvant vs. up-front surgery) was not a discriminant. Patients who died from complication after surgical intervention were excluded. According to these criteria, 180 patients were included in the analysis (Figure 1). Outcome data were collected using clinical or anagraphic records; follow-up was stopped at 108 months; OS was defined as the interval between the date of diagnosis and the date of death from any cause. The 134 patients who had received neoadjuvant treatment, among the 180 patient cohort, were selected for the response association study; all of them completed the therapy cycles and had tumor restaging. Neoadjuvant chemotherapy mainly consisted in treatment with platinum in association with 5-Fluorouracil and concomitant radiation; patients were treated and followed-up exclusively at our Institution. Response criteria referred to RECIST version 1.1. Patients were divided into responder (R: complete and partial response) or non-responder (NR: stable disease and progressive disease). Tumor were staged according to the International Union Against Cancer tumor-node-metastasis (TNM) classification system (7th ed.) and those that were classified with the old edition were re-classified accordingly.
Genotyping
Genomic DNA was isolated from peripheral blood samples using Flexigene DNA kit (Qiagen Italia, Milano, Italy) following the manufacturer's instructions. DNA concentration and quality were assessed with the NanoDrop 1000 spectrophotometer instrument (Agilent Technologies, Santa Clara, CA, USA). Detection of polymorphism was performed using primers and conditions previously described (25), and applying the Restriction Fragment Length Polymorphism (RFLP)-PCR or the Amplification Refractory Mutation System (ARMS)-PCR method. Positive and negative controls were included in each analysis; re-genotyping of randomly selected samples was 100% concordant.
Statistical Analysis
Overall survival (OS) curves were estimated using the Kaplan-Meier methods and compared using the log-rank test. Hazard ratios (HRs) and 95% confidence interval were calculated with Cox proportional hazards univariate and multivariate regression models (MedCalc software, v.17.1). Adjustment was made for the following variables: histotype, stage, therapy and age at diagnosis. Association with response to neoadjuvant therapy was calculated by logistic regression, using the SNPStats software (https://www.snpstats.net/start.htm), and adjusted for histotype, stage and age at diagnosis. The receiver operating characteristic (ROC) curves method was used to create and discriminate the predictability of both genetic and clinical models. Comparison of the area under the curve (AUC) values of different ROC curves was performed by the DeLong method (MedCalc software, v.17.1).
Preliminary power estimation to detect association between SNPs and OS was performed using the power of genetic analysis package (26). The minimum detectable effect with odds ratio (OR) was calculated at various allele frequencies of SNP allele under a codominant model (alpha = 0.01; disease prevalence = 1/10,000). With this sample size, we have 80% power to detect an OR of 2.5 if the SNP allele frequency is 40% for single SNP analysis. To account for multiple comparisons (8 tests), the Benjamini and Hochberg false discovery rate (FDR) was used to correct the association with OS, which was our first aim; correction was not applied in the response association tests. P-values < 0.05 after correction were considered significant.
Results
Patient Cohort for Overall Survival (OS) Association Study
Tumor stage is, together with the response to therapy, one of the more reliable prognostic clinical features; however, a high level of uncertainty and inaccuracy exists for intermediate EC stages. To avoid the confounding effect of clinical stage that could hide the effects of constitutive genetic variants, we excluded from our cohort patients with stage I, IIA and IVB, and restricted the OS association study to 180 patients diagnosed with a clinical stage ranging from IIB to IVA. Table 1 reports the clinico-pathological characteristics of the patients. The median age at diagnosis was 63 years (range 25–86), 147 (82%) were males, adenocarcinoma and squamous carcinoma histotypes were equally represented, stage III was prevalent (73%), 74% received a cisplatin-based neoadjuvant treatment while 26% underwent an up-front surgery. The median survival of the entire cohort was 29.5 months (range 4–108) and the median follow-up for living patients was 66 months (range 28–108). None of the clinical features was statistically associated with the OS.
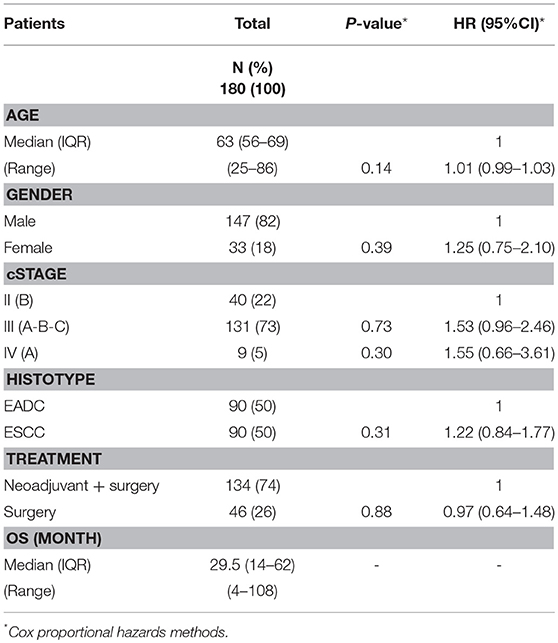
Table 1. Clinico-pathological characteristics of EC patients analyzed for association with overall survival.
Genotype Frequencies and Association With OS
All patients were successfully genotyped for rs11615 (Asn118Asn) and rs3212986 (C8092) in the ERCC1, and rs13181 (Lys751Gln) and rs1799793 (Asp312Asn) in the ERCC2/XPD. Genotype distribution respected the Hardy-Weinberg equilibrium (HWE) for rs11615, rs13181, and rs3212986 with p-values ranging from 0.76 to 0.12. On the contrary, rs1799793 in the ERCC2/XPD exhibited a p = 0.04, suggesting a possible role in EC onset for this genetic variant.
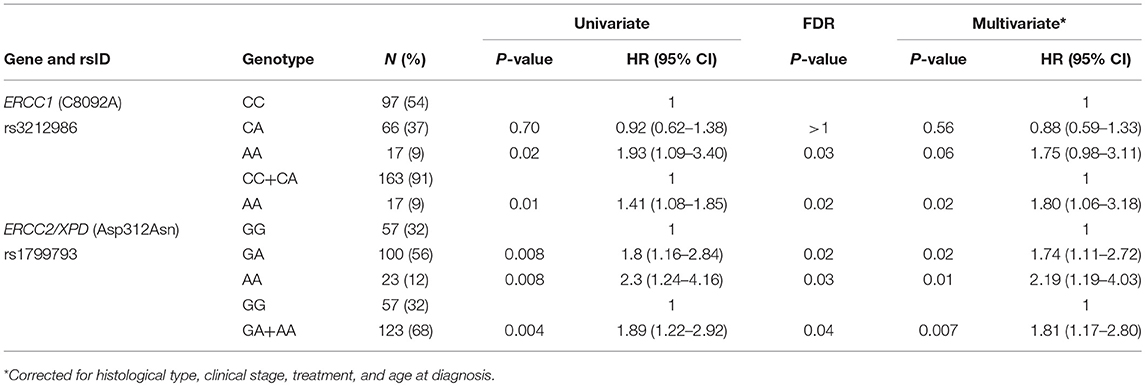
At first, association between OS and the variants was investigated using the Kaplan-Meier method under codominant genetic model (general model). As reported in Figure 2, among the analyzed variables, only the ERCC1 rs3212986 and the ERCC2/XPD rs1799793 resulted associated with OS (log-rank: p = 0.03 and p = 0.004, respectively). The median survival of patients carrying the A allele of ERCC1 rs3212986 in homozygosity was 16 months vs. > 32 months of patients with CC or CA genotypes. Concerning ERCC2/XPD rs1799793, the GA and AA genotypes exhibited a median survival of 26 and 19 months, respectively, vs. 47 months of the patients carrying the GG genotype. Comparable results were obtained when survival functions of the ERCC1 rs3212986 and the ERCC2/XPD rs1799793 were analyzed with the univariate Cox proportional method using the codominant model and, based on the Kaplan-Meier plots, the recessive (rs3212986) or the dominant (rs1799793) genetic model (Table 2). Both genetic variants remain statistically associated with survival even after the Benjamini and Hochberg multiple testing correction (Table 2). After adjustment for clinical variables, such as age at diagnosis, histotype, clinical stage, and therapeutic strategy, the association of the ERCC1 rs3212986 and the ERCC2/XPD rs1799793 with OS remained statistically significant (Table 2).
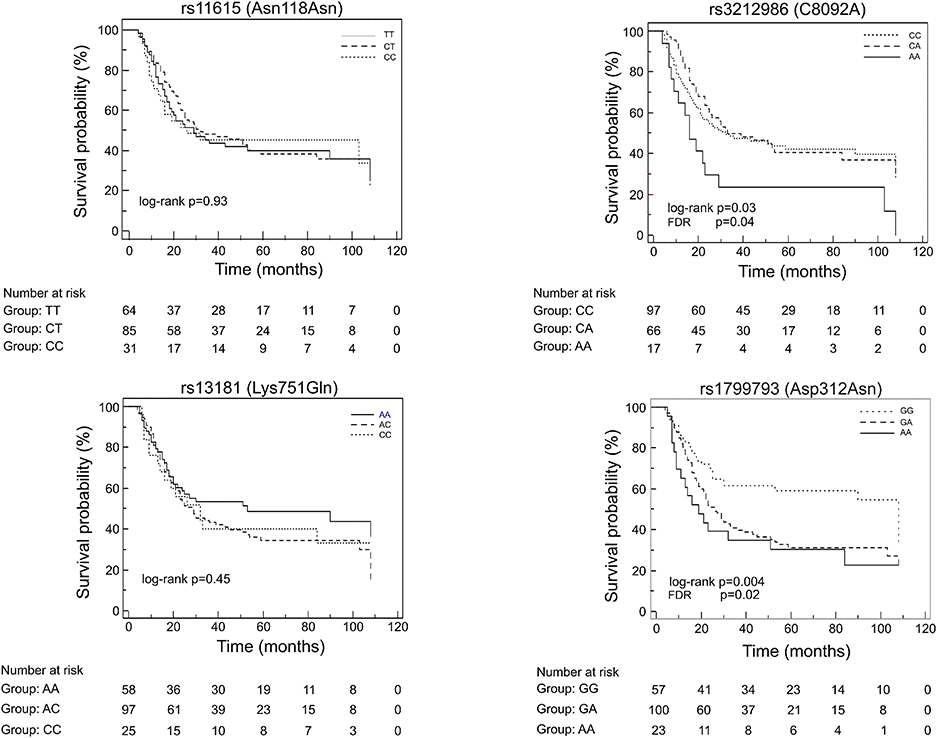
Figure 2. Kaplan-Meier plots of the association between the ERCC1 rs11615 and rs3212986, and the ERCC2/XPD rs13181 and 1799793 and OS.
Survival Predictive Model
We constructed survival prediction models to explore whether the survival-associated rs3212986 and rs1799793 could increase the predictability of the clinical features. We found that the genetic variants together had a better predictability (area under the curve (AUC) = 0.624) of the clinical stage alone (AUC = 0.548), and of the clinical characteristics (stage, histotype and therapy) together (AUC = 0.606) (Figures 3A,B). When the genetic variables were added to the clinical models, the predictability had a slight increase. This improvement was significant for the clinical stage model: AUC = 0.660, p = 0.004 (Figure 3A), but not for the stage, histotype and therapy model: AUC = 0.669, p = 0.09 (Figure 3B).
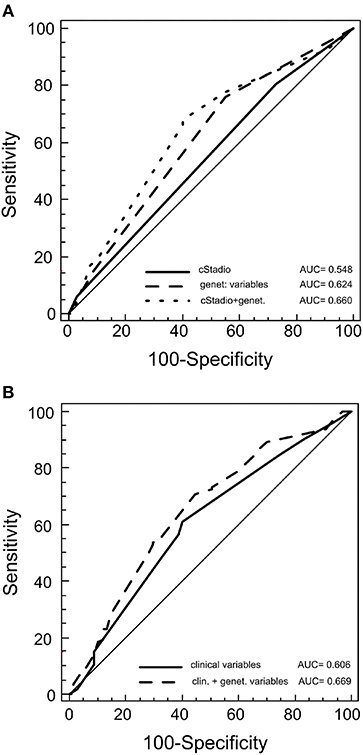
Figure 3. Receiver operating characteristic (ROC) for predicting patient outcome. The plots represent the curves and the area under the curve (AUC) values of the predictive model of germline variants with clinical stage (A) or with germline variants with other clinical features (histotype, therapy and stage) (B).
Patient Cohort for Response Association Study
The second objective of the study was to see whether the same NER variants were associated with response to neoadjuvant therapy. Thus, 134 patients who had had a cisplatin-based neoadjuvant treatment in the total EC cohort were analyzed using the SNPStats software (Figure 1). The clinical and pathological features of this subgroup are reported in Table 3. Median age at diagnosis was 61 years (range 25–80), 81% of patients were males, and the two histotypes were equally represented. All patients received cisplatin-based neoadjuvant therapy and 83% had concomitant radiotherapy. Fifty seven percent of patients were classified as responder (complete and partial response), while 43% were classified as non-responder (patients with stable disease or in progression) following the RECIST 1.1 criteria. No association was observed between response and the other clinical features (Table 3).
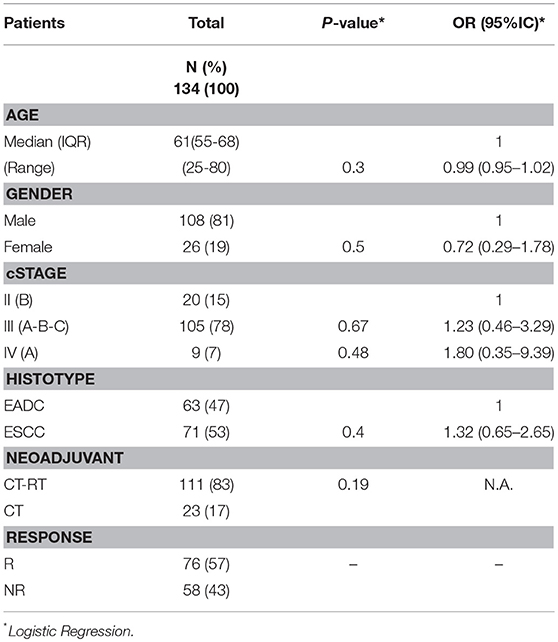
Table 3. Clinical characteristics of EC patients analyzed for the association with neoadjuvant treatment.
Genotype Frequencies and Association With Response
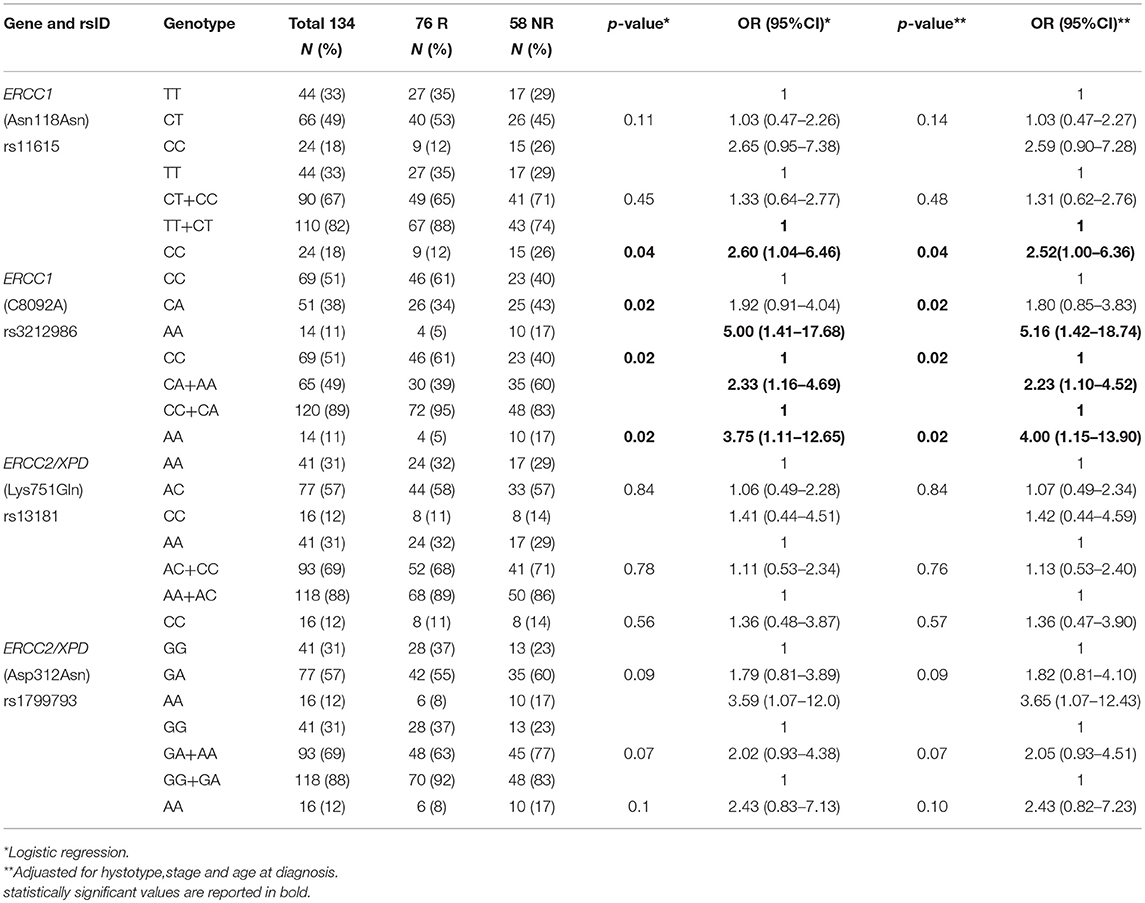
By analyzing the 134 EC patients treated with neoadjuvant cisplatin-based chemotherapy, no association with response was found for the ERCC2/XPD rs13181 and the ERCC2/XPD rs1799793. On the contrary, the minor A allele of ERCC1 rs3212986 both in homozygosity and heterozygosity exhibited a weak association (p = 0.02) as well as the CC genotype of the ERCC1 rs11615 (p = 0.04) (Table 4). Interestingly, the ERCC2/XPD rs1799793 though not associated with response (see Table 4), was still associated with OS (p = 0.009, under the dominant genetic model) (Figure 4).
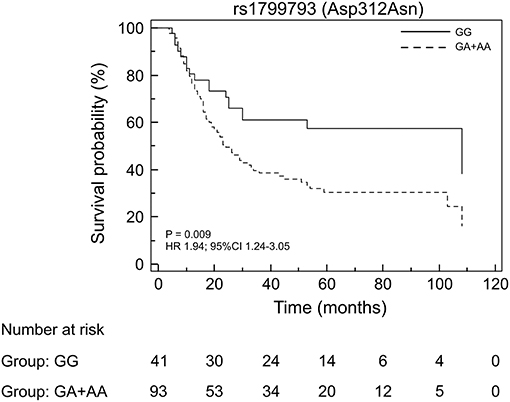
Figure 4. Kaplan-Meier plots of the association of the ERCC2/XPD rs1799793 with OS in the EC patients that underwent to neoadjuvant treatment.
Discussion
Although tumor stage and resection status are the main prognostic factors in EC, other reliable biomarkers that provide a better prediction of outcome, especially for the advanced stages, are still needed. Germline variants in DNA repair pathway genes have been extensively studied to check their influence in tumor onset, response to chemotherapy and OS. However, inconsistent results were obtained, with some studies highlighting the relevance of some genetic variants and others emphasizing opposite results (20, 21, 24, 27).
In this study, we investigated the association between four germline variants in DNA repair genes, two in the ERCC1 (rs11615 and rs3212986), and two in the ERCC2/XPD (rs13181 and rs1799793) and the outcome of patients with advanced EC. ERCC1 and ERCC2/XPD are considered two pivotal proteins of the NER pathway. The ERCC1 protein is responsible for the recognition and excision of the damaged DNA while the ERCC2/XPD codes for a helicase that, besides being a major player in DNA repair, is also a component of transcription factor II H (TFIIH) complex (18, 28, 29).
We observed that, among the analyzed variants, the ERCC1 rs3212986 and the ERCC2/XPD rs1799793 remained statistically associated with a poor OS after correction for multiple tests. Patients carrying the minor A allele of the rs3212986 in homozygosity had a median survival of 16 months vs. > 32 months in patients homozygote or heterozygote for the major C allele. Concerning the rs1799793, patients carrying the variant A allele both in homozygosity or heterozygosity exhibited a significant shorter survival compared to patients with the GG genotype (median survival 23 months vs. 47 months). Adjustment with clinical features did not change the significance suggesting that they are independent prognostic biomarkers. When, as a second aim, we analyzed the association of the same NER variants with the response to neoadjuvant therapy, only the rs3212986 and rs11615 in ERCC1 exhibited an association. Interestingly, both ERCC2/XPD variants were not associated. This last finding strengthens the hypothesis that ERCC2/XPD rs1799793 may only have a prognostic value.
Association of ERCC1 rs3212986 and ERCC2/XPD rs1799793 with survival has been previously investigated in a few EC Caucasian patient cohorts and results are in contrast with those reported in this study (24). In particular, Bradbury and co-workers reported that the presence of the CA and AA genotypes of the rs3212986, and the GA and AA genotypes of the rs1799793 correlated with a better outcome (30). Similarly, in a previous study conducted in a small cohort of Caucasian EADC and ESCC patients, we also found a positive association between OS and the minor allele of ERCC1 rs3212986 (25). In the same cohort, no association between survival and rs1799793 was found. We believe that the inclusion of a broad range of different tumor stages (from early to metastatic) in the work by Bradbury et al, and the small sample size analyzed in our previous study are the major cause of these discrepancies. Indeed, owing to the relatively low hazard risk of each single germline variant, the samples homogeneity and size are the most critical points in pharmacogenetic studies.
The observed deviation from HWE of the ERCC2/XPD rs1799793 in our cohort might reflect its involvement in development of the EC (31, 32). The contribution of rs1799793 to EC onset might also explain its impact in patient survival by promoting tumor progression and aggressiveness, rather than by influencing the response to therapeutic treatment.
So far, the altered DNA repair activity of the ERCC1 and ERCC2/XPD variants has been considered central for its association with tumor risk and cancer patient outcome (17, 19). However, ERCC2/XPD is a helicase also endowed with transcriptional activity, and the codon Asp312Asn is located in the Arch transcriptional domain (that encompasses codons S246-D439) of the protein. Thus, it is conceivable that this germline amino acid substitution might principally affect its transcriptional activity, rather than DNA repair, as reported for mutations located in this domain (33, 34). Furthermore, this possibility might also explain why the rs1799793 influences patient survival but not the efficacy of chemotherapy, which is considered more related to the removal of DNA adducts.
Although our data clearly indicate that the ERCC1 rs3212986 and the ERCC2/XPD rs1799793 affect OS in advanced EC, the study presents some limitations. Besides its retrospective nature, other constraints are the limited number of SNPs analyzed and the candidate gene approach. This can explain the poor clinical impact of the rs3212986 and the rs1799793 (AUC 0.669) as prognostic biomarkers, despite their statistical significant association with OS. Indeed, although there is no doubt about the relevance of constitutive variants on cancer onset and outcome, their limited power is still the greatest obstacle to their clinical use. It has been suggested that the restricted influence of germline SNPs can be overcome by enlarging the pharmacogenetic analyses to additional variants and by calculating the overall risk (i.e., polymorphic risk score) (35–37). Thus, further studies are needed to find other germline variants that, together with the rs3212986 and the rs1799793, could generate a prognostic panel with increased clinical impact. Nevertheless, until the discovery of a more powerful prognostic signature, we believe that both the ERCC1 rs3212986 and the ERCC2/XPD rs1799793 could contribute to the better stratification of patients with advanced resectable EC.
Ethics Statement
The study had the approval of the Comitato Etico per la Sperimentazione Clinica (CESC) of the Veneto Institute of Oncology IOV-IRCCS (cod. number CE IOV 2012/65), and was carried out according to the Code of Ethics of the World Medical Association (Declaration of Helsinki). All patients gave written informed consent in accordance with the declaration of Helsinki.
Author Contributions
EB, SM, ER, and DS provided the concept and design of the study. SM, ER, GB, and AR participated to the acquisition of data, analysis, and interpretation. EB, SM, and DS analyzed and interpretated the data, drafted the article. AA and AR critically revised the manuscript.
Funding
Institutional funding of the Veneto Institute of Oncology IOV-IRCCS and Associazione Italiana per la Ricerca sul Cancro IG2013-14032 supported this work.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We express our profound gratitude to all patients that participated to this study.
References
1. Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. (2017) 67:7–30. doi: 10.3322/caac.21387
2. Saraggi D, Fassan M, Bornschein J, Farinati F, Realdon S, Valeri N, et al. From Barrett metaplasia to esophageal adenocarcinoma: the molecular background. Histol Histopathol. (2016) 31:25–32. doi: 10.14670/HH-11-659
3. Rugge M, Fassan M, Cavallin F, Cavallini F, Zaninotto G. Re: risk of malignant progression in Barrett's esophagus patients: results from a large population-based study. J Natl Cancer Inst. (2012) 104:1771–2. doi: 10.1093/jnci/djs426
4. Domper Arnal MJ, Ferrández Arenas Á, Lanas Arbeloa Á. Esophageal cancer: risk factors, screening and endoscopic treatment in Western and Eastern countries. World J Gastroenterol. (2015) 21:7933–43. doi: 10.3748/wjg.v21.i26.7933
5. Colotta F, Allavena P, Sica A, Garlanda C, Mantovani A. Cancer-related inflammation, the seventh hallmark of cancer: links to genetic instability. Carcinogenesis (2009) 30:1073–81. doi: 10.1093/carcin/bgp127
6. Lin R, Zhang C, Zheng J, Tian D, Lei Z, Chen D, et al. Chronic inflammation-associated genomic instability paves the way for human esophageal carcinogenesis. Oncotarget (2016) 7:24564–71. doi: 10.18632/oncotarget.8356
7. Pan J, Lin J, Izzo JG, Liu Y, Xing J, Huang M, et al. Genetic susceptibility to esophageal cancer: the role of the nucleotide excision repair pathway. Carcinogenesis (2009) 30:785–92. doi: 10.1093/carcin/bgp058
8. Goode EL, Ulrich CM, Potter JD. Polymorphisms in DNA repair genes and associations with cancer risk. Cancer Epidemiol Biomarkers Prev. (2002) 11:1513–30.
9. Ming-Zhong S, Hui-Xiang J, Zhong-Wei Z, Hao J, Rong Z. Genetic variants of the DNA damage repair genes XRCC4 and RAD51 are associated with susceptibility to esophageal cancer. Clin Res Hepatol Gastroenterol. (2015) 39:379–83. doi: 10.1016/j.clinre.2014.12.002
10. Rafiq R, Bhat GA, Lone MM, Masood A, Dar NA. Potential risk of esophageal squamous cell carcinoma due to nucleotide excision repair XPA and XPC gene variants and their interaction among themselves and with environmental factors. Tumour Biol. (2016) 37:10193–207. doi: 10.1007/s13277-016-4895-3
11. Tudek B. Base excision repair modulation as a risk factor for human cancers. Mol Aspects Med. (2007) 28:258–75. doi: 10.1016/j.mam.2007.05.003
12. Paszkowska-Szczur K, Scott RJ, Serrano-Fernandez P, Mirecka A, Gapska P, Górski B, et al. Xeroderma pigmentosum genes and melanoma risk. Int J Cancer (2013) 133:1094–100. doi: 10.1002/ijc.28123
13. McCullough LE, Santella RM, Cleveland RJ, Millikan RC, Olshan AF, North KE, et al. Polymorphisms in DNA repair genes, recreational physical activity and breast cancer risk. Int J Cancer (2014) 134:654–63. doi: 10.1002/ijc.28383
14. Dylawerska A, Barczak W, Wegner A, Golusinski W, Suchorska WM. Association of DNA repair genes polymorphisms and mutations with increased risk of head and neck cancer: a review. Med Oncol. (2017) 34:197. doi: 10.1007/s12032-017-1057-4
15. Neumann AS, Sturgis EM, Wei Q. Nucleotide excision repair as a marker for susceptibility to tobacco-related cancers: a review of molecular epidemiological studies. Mol Carcinog. (2005) 42:65–92. doi: 10.1002/mc.20069
16. Seker H, Butkiewicz D, Bowman ED, Rusin M, Hedayati M, Grossman L, et al. Functional significance of XPD polymorphic variants: attenuated apoptosis in human lymphoblastoid cells with the XPD 312 Asp/Asp genotype. Cancer Res. (2001) 61:7430–4.
17. Wolfe KJ, Wickliffe JK, Hill CE, Paolini M, Ammenheuser MM, Abdel-Rahman SZ. Single nucleotide polymorphisms of the DNA repair gene XPD/ERCC2 alter mRNA expression. Pharmacogenet Genomics (2007) 17:897–905. doi: 10.1097/FPC.0b013e3280115e63
18. Chen P, Wiencke J, Aldape K, Kesler-Diaz A, Miike R, Kelsey K, et al. Association of an ERCC1 polymorphism with adult-onset glioma. Cancer Epidemiol Biomarkers Prev. (2000) 9:843–7.
19. Formica V, Doldo E, Antonetti FR, Nardecchia A, Ferroni P, Riondino S, et al. Biological and predictive role of ERCC1 polymorphisms in cancer. Crit Rev Oncol Hematol. (2017) 111:133–43. doi: 10.1016/j.critrevonc.2017.01.016
20. Findlay JM, Middleton MR, Tomlinson I. A systematic review and meta-analysis of somatic and germline DNA sequence biomarkers of esophageal cancer survival, therapy response and stage. Ann Oncol. (2015) 26:624–44. doi: 10.1093/annonc/mdu449
21. Li Y, Liu Z, Liu H, Wang LE, Tan D, Ajani JA, et al. ERCC1 and ERCC2 variants predict survival in gastric cancer patients. PLoS ONE (2013) 8:e71994. doi: 10.1371/journal.pone.0071994
22. Qin Q, Zhang C, Yang X, Zhu H, Yang B, Cai J, et al. Polymorphisms in XPD gene could predict clinical outcome of platinum-based chemotherapy for non-small cell lung cancer patients: a meta-analysis of 24 studies. PLoS ONE (2013) 8:e79864. doi: 10.1371/journal.pone.0079864
23. Assis J, Pereira C, Nogueira A, Pereira D, Carreira R, Medeiros R. Genetic variants as ovarian cancer first-line treatment hallmarks: a systematic review and meta-analysis. Cancer Treat Rev. (2017) 61:35–52. doi: 10.1016/j.ctrv.2017.10.001
24. Rumiato E, Boldrin E, Amadori A, Saggioro D. Predictive role of host constitutive variants in neoadjuvant therapy of esophageal cancer. Pharmacogenomics (2016) 17:805–20. doi: 10.2217/pgs-2016-0009
25. Rumiato E, Cavallin F, Boldrin E, Cagol M, Alfieri R, Basso D, et al. ERCC1 C8092A (rs3212986) polymorphism as a predictive marker in esophageal cancer patients treated with cisplatin/5-FU-based neoadjuvant therapy. Pharmacogenet Genomics (2013) 23:597–604. doi: 10.1097/FPC.0b013e3283653afc
26. Menashe I, Rosenberg PS, Chen BE. PGA: power calculator for case-control genetic association analyses. BMC Genet. (2008) 9:36. doi: 10.1186/1471-2156-9-36
27. Pérez-Ramírez C, Cañadas-Garre M, Molina M, Robles AI, Faus-Dáder MJ, Calleja-Hernández M. Contribution of genetic factors to platinum-based chemotherapy sensitivity and prognosis of non-small cell lung cancer. Mutat Res. (2017) 771:32–58. doi: 10.1016/j.mrrev.2016.11.003
28. Spitz MR, Wu X, Wang Y, Wang LE, Shete S, Amos CI, et al. Modulation of nucleotide excision repair capacity by XPD polymorphisms in lung cancer patients. Cancer Res. (2001) 61:1354–7.
29. Lee MS, Liu CY, Su L, Christiani DC. Polymorphisms in ERCC1 and ERCC2/XPD genes and carcinogen DNA adducts in human lung. Lung Cancer (2015) 89:8–12. doi: 10.1016/j.lungcan.2015.05.001
30. Bradbury PA, Kulke MH, Heist RS, Zhou W, Ma C, Xu W, et al. Cisplatin pharmacogenetics, DNA repair polymorphisms, and esophageal cancer outcomes. Pharmacogenet Genomics (2009) 19:613–25. doi: 10.1097/FPC.0b013e32832f3010
31. Duan XL, Gong H, Zeng XT, Ni XB, Yan Y, Chen W, et al. Association between XPD Asp312Asn polymorphism and esophageal cancer susceptibility: a meta-analysis. Asian Pac J Cancer Prev. (2012) 13:3299–303. doi: 10.7314/APJCP.2012.13.7.3299
32. Boldrin E, Rumiato E, Fassan M, Rugge M, Cagol M, Marino D, et al. Genetic risk of subsequent esophageal cancer in lymphoma and breast cancer long-term survival patients: a pilot study. Pharmacogenomics J. (2016) 16:266–71. doi: 10.1038/tpj.2015.41
33. Abdulrahman W, Iltis I, Radu L, Braun C, Maglott-Roth A, Giraudon C, et al. ARCH domain of XPD, an anchoring platform for CAK that conditions TFIIH DNA repair and transcription activities. Proc Natl Acad Sci USA. (2013) 110:E633–42. doi: 10.1073/pnas.1213981110
34. Rump A, Benet-Pages A, Schubert S, Kuhlmann JD, Janavičius R, Macháčková E, et al. Identification and functional testing of ERCC2 mutations in a multi-national cohort of patients with familial breast- and ovarian cancer. PLoS Genet. (2016) 12:e1006248. doi: 10.1371/journal.pgen.1006248
35. Hertz DL, McLeod HL. Using pharmacogene polymorphism panels to detect germline pharmacodynamic markers in oncology. Clin Cancer Res. (2014) 20:2530–40. doi: 10.1158/1078-0432.CCR-13-2780
36. Wheeler HE, Maitland ML, Dolan ME, Cox NJ, Ratain MJ. Cancer pharmacogenomics: strategies and challenges. Nat Rev Genet. (2013) 14:23–34. doi: 10.1038/nrg3352
Keywords: esophageal cancer, overall survival, germline variants, ERCC2/XPD rs1799793, XPD Asp312Asn, ERCC1 rs3212986, ERCC1 C8092A
Citation: Boldrin E, Malacrida S, Rumiato E, Battaglia G, Ruol A, Amadori A and Saggioro D (2019) Association Between ERCC1 rs3212986 and ERCC2/XPD rs1799793 and OS in Patients With Advanced Esophageal Cancer. Front. Oncol. 9:85. doi: 10.3389/fonc.2019.00085
Received: 03 December 2018; Accepted: 29 January 2019;
Published: 21 February 2019.
Edited by:
Jianguang Ji, Lund University, SwedenCopyright © 2019 Boldrin, Malacrida, Rumiato, Battaglia, Ruol, Amadori and Saggioro. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daniela Saggioro, ZC5zYWdnaW9yb0B1bmlwZC5pdA==; ZGFuaWVsYS5zYWdnaW9yb0Bpb3YudmVuZXRvLml0
†These authors have contributed equally to this work and are co-first authors
 Elisa Boldrin
Elisa Boldrin Sandro Malacrida
Sandro Malacrida Enrica Rumiato1
Enrica Rumiato1 Alberto Amadori
Alberto Amadori Daniela Saggioro
Daniela Saggioro