- 1Cancer Institute (Key Laboratory of Cancer Prevention and Intervention, China National Ministry of Education), School of Medicine, The Second Affiliated Hospital, Zhejiang University, Hangzhou, China
- 2Department of Neurosurgery, School of Medicine, Second Affiliated Hospital, Zhejiang University, Hangzhou, China
- 3Interdisciplinary Institute of Neuroscience and Technology, Qiushi Academy for Advanced Studies, Zhejiang University, Hangzhou, China
- 4Department of Surgical Oncology, School of Medicine, The Second Affiliated Hospital, Zhejiang University, Hangzhou, China
- 5Department of Orthopedics, School of Medicine, The Second Affiliated Hospital, Zhejiang University, Hangzhou, China
Breast cancer (BC) remains the most frequently diagnosed cancer worldwide. Among breast cancer patients, distant metastasis and invasion is the leading cause of BC related death. Recently, long non-coding RNAs (lncRNAs), which used to be considered a genetic byproduct (owing to their unknown biological function), have been reported to be highly implicated in the development and progression of BC. In this review, we produce a summary of the functions and mechanisms of lncRNAs implicated in the different distant metastases of BC. The functions of lncRNAs have been divided into two types: oncogenic type and tumor suppressor. Furthermore, the majority of them exert their roles through the regulation of invasion, migration, epithelial—mesenchymal transition (EMT), and the metastasis process. In the final part, we briefly addressed future research prospects of lncRNAs, especially the testing methods through which to detect lncRNAs in the clinical work, and introduced several different tools with which to detect lncRNAs more conveniently. Although lncRNA research is still in the initial stages, it is a promising prognosticator and a novel therapeutic target for BC metastasis, which requires more research in the future.
Introduction
In past years, breast cancer (BC) has always been the most frequently diagnosed cancer worldwide, and is also the major reason for cancer-related mortality in women. It is estimated that there will be 266,120 new cases of female breast cancer and an estimated 40,920 people will die of this disease in the United States in 2018 (https://www.cancer.gov/types/common-cancers). Among these patients, distant metastatic invasion has been proposed as the leading cause of death (1–4). According to an estimation, 20–30% of patients with breast cancer will develop metastasis following diagnosis (5).
Long non-coding RNAs (lncRNAs), which are longer than 200 nucleotides in length, are one subtype of RNA transcripts which used to be considered a genetic byproduct (owing to their unknown biological function) (6). In recent years, the human genome has been gradually transcribed based on the development of DNA sequencing technologies, and it is unbelievable that more than 98% of genes of the whole human genome are non-encoding, receiving a considerable amount of concern in recent years. Thanks to biological techniques and high-throughput sequencing, such as SHAPE probing (7), ChIRP (8), and CHART (9), which can be used to reveal RNA structures and RNA–RNA, RNA–DNA, and RNA–protein interactions, researchers have uncovered a great amount of lncRNAs in past years, which have played significant roles in various biological processes, especially in the progression of malignant tumors (10, 11). LncRNAs can be classified into five categories—(1) sense, (2) antisense, (3) bidirectional, (4) intronic, or (5) intergenic—with respect to the nearest protein-coding transcripts (12). A diversity of malignant tumors has been reported to be involved with lncRNAs. In these studies, lncRNAs seem to play an indispensable role in the progression of cancer, both in the development of the primary tumor and in the metastatic procedure (13, 14).
Numerous lncRNAs have been reported to be highly implicated in the development of BC (15–18), which could be categorized into two types: oncogenic type and tumor suppressor. Regardless of whether they act as a promoter or inhibitor during the progression of BC, the mechanism generally covers the following aspects: influence proliferation, invasion, apoptosis and drug resistance of BC cells. However, the molecular mechanisms in respect of how lncRNAs come into play in the distant metastasis of breast cancer remain insufficient. The functions and mechanisms of these lncRNAs—such as MALAT1 (19), HOTAIR (20) and NEAT1 (21)—have already been widely accepted regarding their interaction in the metastasis of BC. On account of the significant role that lncRNAs have played in BC metastasis, it is an urgent requirement to explore them as predictors for prognosis and as novel targets for therapy in BC patients.
With the inactivation of embryo dedifferentiation, epithelial–mesenchymal transition (EMT) increases the movement and proliferation of cancer cells (22). Transient EMT is thought to be the first necessary step in metastatic processes (23), which contributes to the migration and invasion of cancer cells, so that cancer cells leave the primary focus and form distant metastasis (24). In recent years, some researchers have indicated that EMT plays an indispensable role in the potential mechanism leading to the distant metastasis of BC (25).
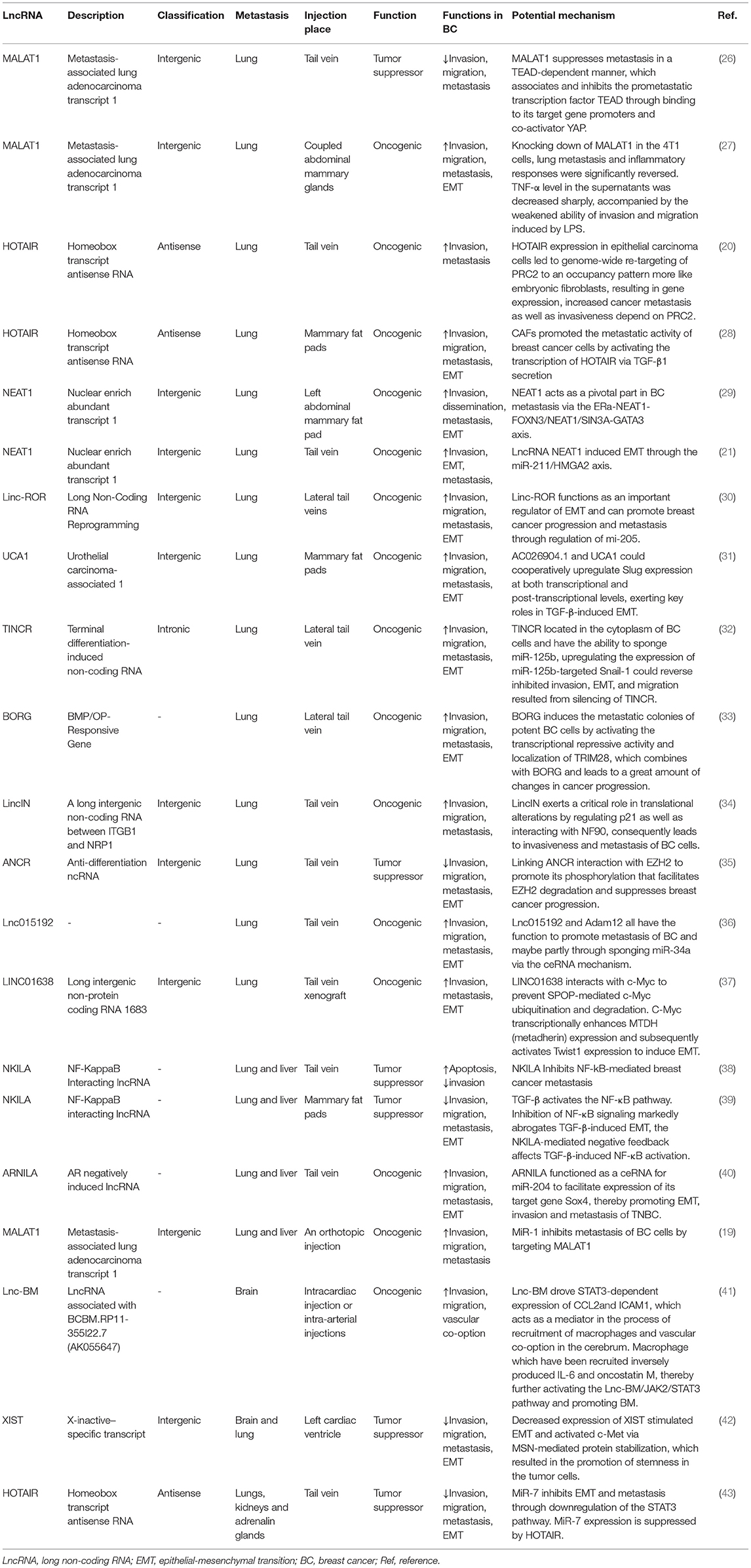
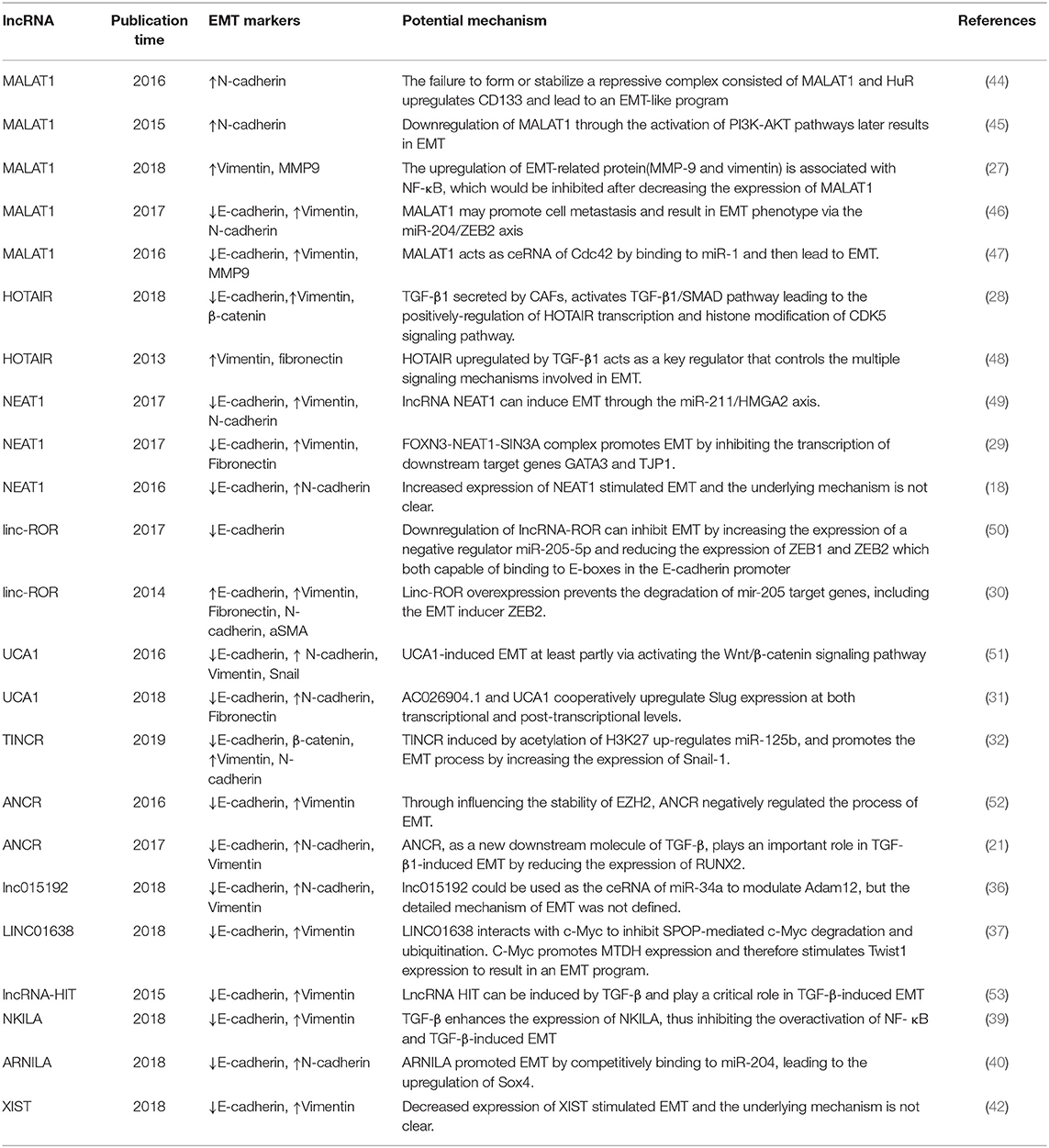
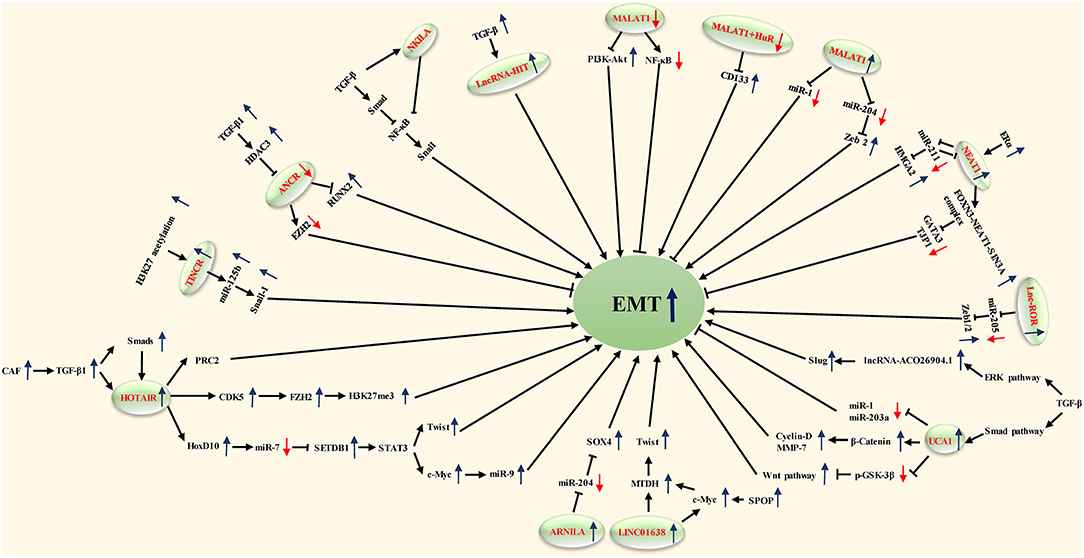
In this review, we produce a summary of the functions and mechanisms of lncRNAs implicated in the different distant metastases of BC (Table 1), especially via the EMT program (Table 2 and Figure 1).
LncRNAs in the Distant Metastasis of Breast Cancer
LncRNAs Implicated in the Lung Metastasis of BC
MALAT1
Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) was first found in non-small cell lung cancer (NSCLC) patients. Functioning as a poor prognosticator, its overexpression could be predictive of a higher stake related to distant metastasis in a series of cancers such as lung cancer, osteosarcoma, colorectal cancer, and especially in non-small cell lung cancer patients (54, 55). Zhang et al. have concluded that MALAT1 is actively involved in multiple physiological processes, such as myogenesis, alternative splicing, synapse formation, and epigenetic modification of gene expression, as well as in multiple pathological conditions (56).
In the past few decades, many in vitro and xenograft researches have reported the underlying functions of MALAT1 in regulating invasion, migration, EMT and the metastasis procedure of a diversity of malignancies, and have indicated inconsistent functions of MALAT1 with respect to the growth and progression of tumor cells (57–59). Experiments both in vitro and in vivo have revealed that MALAT1 is a proliferation promoter, as well as accelerating tumor development and metastasis in triple-negative breast cancer (TNBC). Moreover, it is even negatively correlated with the prognosis of breast cancer patients with an HER-2 positive ER-negative subtype or triple-negative subtype (60). Another study has demonstrated that 17b-Estradiol (E2) with a high level of concentration may inhibit cell growth, invasion and metastasis; in the meantime, the level of MALAT1 is reduced as well either in MCF7 cell lines (Luminal A) or in MDA-MB-231 cell lines (TNBC). Similar effects could be achieved through downregulated MALAT1, so E2 may influence tumor cells through regulating the lncRNA MALAT1 (61).
Xu et al. found that MALAT1 was downregulated in breast tumor cell lines and cancer tissue, and downregulation of MALAT1 in breast cancer cell lines through the activation of phosphatidylinositide-3 kinase-AKT (PI3K-AKT) pathways later results in EMT (58). Recently, MALAT1 was also considered a proinflammatory factor which regulated the lipopolysaccharide (LPS)-induced inflammatory response (62) and EMT process of breast cancer cells (63, 64). The upregulation of EMT-related protein (MMP-9 and vimentin) is associated with NF-κB, which would be inhibited after decreasing the expression of MALAT1 (27). CD133 (PROMININ1), which is one of the general cancer stem cell (CSC) markers, has been reported to possess the ability to facilitate EMT in breast cancer and other malignant tumors (65), and Latorre et al. recently showed that the failure to form or stabilize a repressive complex consisted of MALAT1 and the RNA-binding protein HuR in breast cancer upregulates CD133 and leads to an EMT-like program with the increase of N-cadherin (44).
Some studies showed that MALAT1 inhibited the expression of E-cadherin and induced the expression of vimentin at mRNA and protein levels, while miR-1 inhibited the expression of vimentin and MMP-9 while enhancing the expression of E-cadherin in Western blot results, which can be summarized as MALAT1 and miR-1 having opposite effects on the migration and invasion of breast cancer cells. In conclusion, MALAT1 acts as ceRNA of Cdc42 by binding to miR-1 and then leads to EMT in human breast cancer cell lines (47). miR-204 expression was downregulated by MALAT1 via acting as an endogenous sponge. MiR-204 inhibited the expression of ZEB2 by binding to the non-coding region of ZEB2 3-UTR. Therefore, MALAT1 regulated the miR-204/ZEB2 axis in breast cancer. In view of the fact that ZEB2 is a key factor in EMT, it was speculated that MALAT1 may promote cell metastasis and result in an EMT phenotype via the miR-204/ZEB2 axis (46).
As part of in vivo studies, Zhang et al. generated mice models with a deleted-3-kb genomic site encompassing the 5′end of lncRNA MALAT1 as well as its promoter region (66). Another study reported that the systemic knockdown or genetic loss of MALAT1 in the MMTV (mouse mammary tumor virus)-PyMT (polyomavirus middle T antigen) (67) rats model led to slower growth of tumor cells as well as a reduction in lung metastases (68). They found a diminishment of branching morphogenesis in Her2/neu-amplified and MMTV-PyMT tumor tissues and cell migration accompanied by an enhancement of cell adhesion in MALAT1-loss cells; however, the potential mechanism regarding the reduced metastasis remained unclear. It is considered that this MALAT1-loss model is usually accompanied by the upregulation of substantial adjacent genes of MALAT1, including Neat1, Tigd3, Frmd8, Ehbp1l1, and so on (66). In order to explore whether this high level of expression was caused by the deletion of MALAT1 or the loss of regulatory sequences for its adjacent genes, Kim et al. adopted a MALAT1-knockout animal model wherein a transcriptional terminator was inserted 69-bp downstream of the transcriptional start region of MALAT1. When the expression of MALAT1 was restored in breast tumor tissue, the distant metastasis in the lung was reduced. Besides, they proposed that MALAT1 suppresses metastasis in a TEAD-dependent manner, which associates and inhibits the prometastatic transcription factor TEAD through binding to its target gene promoters and coactivator YAP (26).
The opposite observation results from disparate studies suggested the complicacy of MALAT1 in BC, which may be according to the specific tumor subtypes or different cell types.
HOTAIR
HOTAIR, which is an lncRNA in the mammalian HOXC locus that targets and binds to polycomb repressive complex 2 (PRC2) to the HOXD locus, is located on a disparate chromosome (69).
HOTAIR expression is upregulated in breast cancer tissue and its metastasis, and could also predict the eventual metastasis and death according to different expression levels. LncRNA HOTAIR has been proven through interacting with PRC2, which can promote breast cancer progression (20). For example, the interaction of HOTAIR with PRC2 or LSD1 can be inhibited by HOTAIR targeting small molecular inhibitors, thereby reducing the metastasis of breast cancer (70).
Some findings suggest that HOTAIR is involved in the activation of the genetic process that promotes EMT. Padua Alves et al. have shown that HOTAIR is an important regulator of EMT-related genes in breast cancer cells, and transforming growth factor-β1 (TGF-β1) supports this role of HOTAIR (48). TGF-β-1 induced the upregulated expression of HOTAIR, which may be a central event in promoting PRC2 to inhibit E-cadherin gene expression. This hypothesis is supported by other extended analyses of gene expression profiles, PRC2 occupation, and H3K27me3 (20). In these analyses, they observed the activation of EMT in cells overexpressing HOTAIR. Other experiments generally support the view that TGF-β1 secreted by cancer-associated fibroblasts (CAFs) activates the TGF-β1/SMAD pathway in breast cancer cells, whereby leading to the positive regulation of HOTAIR transcription and histone modification of the CDK5 signaling pathway. Therefore, EMT can be induced to promote the progression and lung metastasis of breast cancer in vivo (28). Moreover, HOTAIR can inhibit the expression of miR-7. In turn, can significantly induce the expression of E-cadherin, and reverse EMT by downregulating the STAT3 pathway (43).
NEAT1
Nuclear-enriched abundant transcript 1 (NEAT1) is a sub-nuclear structure localized exclusively to paraspeckles (71). Its function is that of modulating RNA splicing as well as transcription. It has been proposed that NEAT1 functions as an oncogenic lncRNA in various kinds of malignancies, such as BC, and it is suggested to induce EMT in cancer progression (72).
Several studies have reported that lncRNA NEAT1 accelerates the proliferation and progression of malignancies. EMT is one of the essential contributors to invasion and metastasis in breast carcinoma (25). Zhang et al. found that the expression of EMT markers was altered according to the expression of lncRNA NEAT1. The expression of E-cadherin was increased and N-cadherin was decreased by inhibiting lncRNA NEAT1, suggesting that lncRNA NEAT1 might regulate EMT in BC; however, the underlying mechanism remains unclear (18).
HMGA2 has been reported to regulate EMT transcription factor (EMT-TF) networks such as Snail1, Snail2, ZEB1, ZEB2, and Twist1 (58). There was mutual inhibition between NEAT1 and miR-211. In addition, EMT inducer HMGA2 was identified as a downstream target of miR-211. This concluded that lncRNA NEAT1 can induce EMT through the miR-211/HMGA2 axis (21). As part of an in vivo experiment, cells transduced with sh-NEAT1 were inoculated into animal models through the tail vein, and it was found that compared with the control group, the experimental group generated smaller distant metastatic colonies in the lung, meaning that inhibiting the activity of lncRNA NEAT1 reduced the metastatic ability of cancer cells in vivo (21).
FOXN3 is a transcription inhibitor associated with SIN3A repressor complexes in estrogen receptor-positive (ER+) cells. It has been shown that an FOXN3–NEAT1–SIN3A complex promotes the invasion of breast cancer cells and EMT in vitro and the proliferation and metastasis of breast cancer in vivo by inhibiting the transcription of downstream target genes GATA3 and TJP1. After inoculating the engineered MCF-7 breast cancer cells into the left abdominal mammary fat pad, it was found that overexpression of either NEAT1 or FOXN3 efficiently promoted lung metastasis in animal models (29).
Linc-ROR
It has been reported for lncRNA-regulator of reprogramming (lincRNA-ROR) that the expression is upregulated in several different solid tumors, which suppresses the invasion of a tumor through reducing the EMT marker's expression (73). Additionally, lincRNA-ROR also potently promotes the invasion and distant metastasis ability of BC through the EMT program.
Hou et al. first identified the role of linc-ROR in the control of EMT and metastasis in breast cancer cells. Linc-ROR is associated with miRNPs and acts as an endogenous RNA competing with mi-205. Specifically, linc-ROR overexpression prevents the degradation of mir-205 target genes in breast cancer cells, including the EMT inducer ZEB2. Therefore, linc-ROR is an important regulator of EMT and promotes the progression and metastasis of breast cancer by regulating miRNAs (30). It has been verified that in MDA-MB-231 cells, the downregulated expression of lncRNA-ROR can inhibit the EMT of breast cancer by increasing the expression of negative regulator miR-205-5p and reducing the expression of ZEB1 and ZEB2, key EMT-TFs that, more than others, play a central role in controlling EMT activation (74), both of which include two zinc finger domains capable of binding to E-boxes in the gene promoter region, such as the E-cadherin promoter (50).
UCA1
Urothelial carcinoma-associated 1 (UCA1), which is an lncRNA that contains three exons and encodes two transcripts (75), was first detected overexpression in bladder cancer (76). Its expression level is related to cellular proliferation and migration (77). The expression levels of AC026904.1 and UCA1 in metastatic breast cancer were reported to be higher than those in non-metastatic breast cancer (31).
Huang et al. proved UCA1 to be an oncogenic lncRNA in BC either in vitro or in vivo. UCA1 bound to hnRNP I (heterogeneous nuclear ribonucleoprotein I) and formed a complex which made itself more stabilized. While promoting the translational ability of p27 through hnRNP I alone, the protein content of p27 was widely suppressed when it interacted with UCA1 (78). Current research has observed that TGF-β activates UCA1 and AC026904.1 through Smad and ERK pathways, respectively. AC026904.1 activates the cis-transcription of the Slug gene in the form of eRNA. UCA1 promotes the expression of Slug in breast cancer by directly titrating miR-1 and miR-203a at the post-transcriptional level. Both of them cooperated with each other to upregulate the expression of Slug at the transcriptional and post-transcriptional levels (31). The Wnt/ β-catenin signaling pathway is recognized as a common signaling pathway related to EMT and the metastasis of breast cancer (79, 80). After transfection of si-UCA1 into MDA-MB-231 cells, the expression of negative regulators p-gsk-3 β and gsk-3 β in the Wnt signaling pathway was significantly increased. At the same time, UCAl gene knockout inhibited the protein expression of β-catenin and its downstream genes (including cyclinD1 and MMP7) (51).
TINCR
TINCR (terminal differentiation-induced non-coding RNA) is a spliced lncRNA which produces a 3.7-kb transcript. It was initially discovered from well-differentiated human somatic tissue and was proven to be an indispensable component of normal differentiation of the epidermis (81). Liu and his colleagues reported TINCR to be an oncogenic factor for BC (82); however, whether it is because of drug resistance or resistance-induced tumor metastasis remains unknown.
Compared with other sensitive cells, TINCR was critically upregulated in trastuzumab-resistant tumor cells. While the drug resistance and EMT program could be reversed through the knockdown of TINCR, Dong et al. demonstrated that TINCR induced by acetylation of H3K27 upregulates miR-125b, and promotes the EMT process by increasing the expression of Snail-1. Experimental distant metastasis in the lung was formed by injections into the lateral tail vein of mouse models. In vivo, compared to the sh-NC group, both the deep lung metastasis and the visible metastasis on the surface of the lung were distinctly lesser in the experimental group (32).
BORG
BMP/OP-responsive gene (BORG) in a C2C12 mouse myoblast cell line which trans-differentiates into osteoblastic cells reacts to bone morphogenetic proteins (BMPs) (83), particularly alters the proliferation of disseminated BC cells.
Alex et al. revealed that high expression of BORG is closely associated with recurrent and metastatic disease, making it a potential biomarker which is able to identify women who are at high risk of developing BC with high invasiveness or metastatic ability. In theory, BORG induces the metastatic colonies of potent BC cells by activating the transcriptional repressive activity and localization of TRIM28, which combines with BORG and leads to a great amount of changes in cancer progression. In addition, downregulated BORG in metastatic BC enhances their metastatic ability in animal models, indicating that BORG could promote alterations at both epigenetic and genetic levels, therefore underlying the development of recurrent and metastatic BC (33).
LincIN
LincIN is a long intergenic non-coding RNA between ITGB1 and NRP1 (LincIN), using a high-density SNP array-based gene expression approach to assess the lncRNA transcriptome in paired tumor tissue vs. normal samples.
Compared to adjacent normal tissue, it is frequently observed with highly expressed LincIN in tumors, and it is reported that LincIN is significantly correlated with aggressive BC. Moreover, analysis of TCGA data also supports that upregulation of LincIN is associated with poor prognosis in BC patients. Conversely, deletion of LincIN suppresses the invasion and migration of tumor cells in experimental studies, and transcriptome analysis of the LincIN-deletion tumor cells agrees with this result. Importantly, the loss of LincIN decreases the amount of lung metastasis in tail vein injected- mice models. A study suggested that LincIN exerts a critical role in translational alterations by cooperating with NF90 to suppress the translation of p21, and the upregulation of nuclear p21 by LincIN knockdown may be related to a less aggressive phenotype in a metastasis model (34).
ANCR
An lncRNA termed anti-differentiation ncRNA (ANCR or DANCR), an 855-nucleotide lncRNA, was first discovered during the differentiation process with its downregulated expression (84).
Li and his colleagues revealed that ANCR exerts a pivotal role in BC progression and metastasis, mostly through diminishing EZH2 (enhancer of zeste homolog 2) stability. EZH2, as a pivotal inducer and regulator of EMT at the epigenetic level, contributes to multiple cancer metastases. They initially found that in contrast to normal breast tissue, the expression level of ANCR was lower both in BC cell lines and in BC samples. ANCR-mediated EZH2 degradation may play a key role in weakening the ability of initiating EMT and metastasis programs in breast cancer cells. ANCR interacts with EZH2 to promote the binding of CDK1 to EZH2 and its phosphorylation at Thr-345 and Thr-487 sites, which leads to the degradation of EZH2. Through influencing the stability of EZH2, ANCR negatively regulated the process of EMT (35).
Another study showed that ANCR, as a new downstream molecule of TGF-β, plays an important role in TGF-β1-induced EMT by reducing the expression of RUNX2. TGF-β1 induces the increasing expression of HDAC3; HDAC3 then binds to the promoter region of ANCR which inhibited the transcription of ANCR. Besides, ANCR attenuates the TGF-βsignaling pathway at least in part by inhibiting the phosphorylation of Smad2/3. In an in vivo study, the expression level of RUNX2 in lung tissue sections was measured, uncovering a decreasing level of RUNX2 in the lung tissue of mice models which had been injected with MDA-MB-231-ANCR cells (52).
Lnc015192
In a recent study, Huang et al. found that Adam12 is overexpressed in breast cancer cell lines, and promotes mesenchymal-epithelial transition (MET) and inhibits migration and invasiveness. The knockout of Adam12 and lnc015192 inhibits migration, invasion and EMT in breast cancer cells. It has been shown that lnc015192 could be used as the ceRNA of miR-34a to modulate Adam12, but the detailed mechanism of EMT has not been defined. In order to explore the influence of lnc015192 and Adam12 on tumor metastasis in vivo, either lnc015192 or Adam12-deletion 4T1 cells were injected into mice models, and the results showed that a smaller number of lung metastatic focuses were induced in sh-lnc015192-injected or sh-Adam12-injected animal models than in the control group, suggesting that the loss of lnc015192 or Adam12 suppresses the invasion, migration and metastasis ability of BC cells (36).
LINC01638
A validated lncRNA, LINC01638 (long intergenic non-protein coding RNA 1683), was reported with low expression in various normal human tissues (85) and may be associated with the EMT program of HCC cells (86).
Current research has suggested that LINC01638 is expressed at a high level in TNBC cells and tissue. LINC01638 maintains the mesenchymal characteristics of TNBC cells, containing a cancer stem cell-like state and enriched EMT signature. The knockdown of LINC01638 inhibits tumor cell proliferation and invasiveness either in vivo or in vitro. Overexpression of LINC01638 is regarded as a predictor of poor prognosis of BC patients. In theory, LINC01638 interacts with c-Myc to inhibit SPOP-mediated c-Myc degradation and ubiquitination. C-Myc promotes the expression of MTDH (metadherin) at a transcriptional level and, therefore, stimulates Twist1 expression, resulting in an EMT program.
In addition, to assess the impact of LINC01638 on BC metastasis in vivo, LINC01638-deletion MDA-MB-231 cells were inoculated into animal models via the tail vein. A smaller number of lung metastatic nodules were revealed from the experimental group than from the control group after several days, but restoring the expression of c-Myc diminished the impact of LINC01638 deletion on tumor metastasis in vivo.
These findings verified LINC01638-mediated signal transduction and emphasized the key role that LINC01638 played in TNBC development and metastasis (37).
LncRNA-HIT
In addition, lncRNA-HIT (HOXA-associated transcript induced by TGF-β) also plays a key role in TGFβ-induced EMT. E-cadherin was identified as being one of the major targets of lncRNA-HIT. The effects of lncRNA-HIT on EMT, invasion and migration in a 4T1 orthotopic mouse xenograft model were rescued by the introduction of ectopic E-cadherin. These results suggest that lncRNA-HIT can be induced by TGF-β and play a critical role in TGF-β-induced EMT (53).
LncRNAs Implicated in the Liver Metastasis of BC
NKILA
Nuclear factor-κB interacting lncRNA (NKILA) is a main suppressing checkpoint for NF-κB activation in BC. A gene at chromosome 20q13 encoded NKILA, which first exerted the antimetastatic abilities of breast cancer cells. NKILA is a novel lncRNA which was detected recently by Liu et al. (38), who attempted and successfully proposed that lncRNA NKILA suppresses the activation of NF-κB signaling through combining with the NF-κB/IκB complex to cover up the phosphorylation regions of IκB; therefore, the NF-κB/IκB complex is stabilized. Additionally, both tumor samples and in vivo studies validated that the downregulation of NKILA in high-grade BC promotes distant metastasis (38).
A recent study found that NF-κB plays an important role in TGF-β-induced EMT. TGF-β enhanced the expression of NKILA, thus inhibiting the overactivation of NF-κB by interacting with I-κB. The expression of NKILA controls the switch “on and off,” negatively regulating NF-κB. Additionally, the upregulation of NKILA expression notably decreased liver metastasis induced by TGF-β of a malignant tumor in vivo. Similar to the outcomes of animal models, the expression of NKILA was negatively associated with different EMT phenotypes in BC tissue. In conclusion, it has been shown that lncRNA NKILA can modulate the TGF-β-induced EMT of breast cancer (39).
ARNILA
Androgen receptor negatively induced lncRNA (ARNILA) is located in the cytoplasm of TNBC tissues and cells, which have been demonstrated to promote invasion, EMT and metastasis in in vivo and in vitro experiments. In addition, ARNILA was reported to function as a ceRNA for miR-204 to promote its target gene Sox4's expression, which subsequently contributed to the EMT program and induced the progression of breast cancer, thereby facilitating the invasion, EMT and metastasis of TNBC (40).
MALAT1
In a recent study, which identified MALAT1 as a target of miR-1, it was found that the expression level of miR-1 was negatively correlated with the expression of MALAT1 in BC samples. MiR-1 suppressed the development of BC, decreased cell motility and activated apoptosis by regulating MALAT1. The knockdown of MALAT1 could partly imitate the tumor-inhibitive impact of miR-1. As part of in vivo metastatic experiments, MCF7 cells transduced stably with miR-1 formed fewer and smaller lung and liver metastases than the control group. Such outcomes indicated that miR-1 could suppress the metastasis of BC cells by regulating MALAT1 in vivo (19).
lncRNAs Implicated in the Brain Metastasis of BC
lnc-BM
LncRNA associated with brain metastasis (Lnc-BM) has been demonstrated to be a prognostic factor of the progression of brain metastasis in BC patients. In preclinical experiments, upregulated expression of Lnc-BM promoted BCBM, while the knockdown of Lnc-BM with nanoparticle-encapsulated siRNAs effectively improved BCBM in murine models. In BC cells, Lnc-BM drove the STAT3-dependent expression of CCL2 and ICAM1, which acts as a mediator in the process of recruitment of macrophages and vascular co-option in the cerebrum. Macrophages which were recruited inversely produced IL-6 and oncostatin M, thereby further activating the Lnc-BM/JAK2/STAT3 pathway and promoting BCBM by mediating interaction between BC cells and the brain microenvironment (41).
XIST
X-inactive-specific transcript (XIST) is a lncRNA that participates at the beginning of X chromosome inactivation during early embryogenesis. Numerous human malignant tumors are accompanied by the deficiency of inactivated X chromosomes (Xi) as well as the duplication of active X chromosomes (87). Such a phenomenon is more common in several cancers such as breast cancer.
Here, Xing et al. found that XIST was notably decreased in BM tissue from patients with BC. The expression level of XIST was inversely associated with BM in BC patients. A loss of XIST preferentially facilitated the BM growth of XIST-high cells in xenograft animal models. Additionally, deletion of XIST in the breast of mice models enhanced the growth of BC and BM. Downregulation of XIST activated EMT and stimulated c-Met through stabilizing MSN-mediated protein, which contributed to the activation of tumor cells' stemness. Deletion of XIST also drove the secretion of exosomal miRNA-503, which consequently triggered M1–M2 polarization of microglia. However, the underlying mechanism of EMT remains unclear (42).
Others
In addition, there are some other lncRNAs which have been reported to be associated with the brain metastasis of breast cancer patients, such as NCT02915744 III, which is related to metastatic BC with brain metastases, and NCT02000882 II, which is found in TNBC patients with brain metastases (88).
LncRNAs Implicated in Other Metastasis of BC
HOTAIR
MiR-7, which was initially detected from human MDA-MB-231 and MCF-7 cell lines, was lowly expressed in breast cancer stem cells (BCSCs). Zhang and his colleagues found that MiR-7 suppressed the metastasis of BCSCs in many different organs, such as kidneys, lungs as well as adrenal glands, in NOD/SCID mice. Moreover, downregulated miR-7 may be indirectly owing to HOTAIR through regulating the expression level of HoxD10, which enhances miR-7 expression in BCSCs (43).
Conclusion
In the past few decades, lncRNAs have experienced a long history, from being regarded as non-functional (6) to being confirmed as an indispensable contributor to the development and progression of a variety of malignant tumors, attracting a considerable amount of attention from multiple investigators who have devoted themselves to research on lncRNAs and malignant tumors, especially tumor metastasis (89), which would directly result in cancer-related death.
In this review, we briefly illustrated those lncRNAs involved in regulating the metastatic ability of breast cancer and tried to describe the potential mechanisms, especially through the EMT process, as well as attempting to explore some novel strategies for targeting highly invasive BC. Depending on the screening results of lncRNAs, a large number of them are demonstrated to function as a promoter in the process of invasion, migration, EMT or metastasis of BC, while a small number have contradictory effects to inhibit the metastasis of BC. They regulated the distant metastatic process at different levels such as transcription levels or post-transcription levels, and it seems that there are several signaling pathways and transcription factors that are similar among different lncRNA-involved pathways, which may be novel therapeutic targets for cancer patients in the future. However, the underlying mechanisms regarding how most of the lncRNAs regulate breast cancer remain under-defined (90).
Some studies verified whether different lncRNAs affect the distant metastasis of breast cancer through injecting the breast cancer cells with or without lncRNAs into the animal models, and observe the size and the quantity of the distant metastatic colonizations to discover the underlying mechanisms among the process of distant metastasis (28, 35, 37, 46). It has been reported that EMT mediates the dissemination of tumor cells from primary lesions, while MET is essential for the seeding of circulating tumor cells to distal organs (91–93). We intended to summarize the metastatic mechanisms to different organs accordingly, however, most of them focused on the initiation mechanism of metastasis, such as EMT, there are less studies tried to find the mechanism associated with the different metastatic organs. Huang et al. reported that lnc015192 knockdown enhanced the MET process in the lung metastasis while the underlying mechanism remains unclear (36). Another study showed that ANCR, as a new downstream molecule of TGF-β, plays an important role in TGF- β1-induced EMT by reducing the expression of RUNX2. They uncovered a reduced RUNX2 level in nude mice lung tissues injected with MDA-MB-231-ANCR cells (52). Brain metastasis has been reported with several organ-specific lncRNAs in our review, XIST expression was found significantly downregulated in brain metastatic lesions compared with other metastatic tumors based on an organ-specific cohort analysis, deletion of XIST also drove the secretion of exosomal miRNA-503, which consequently triggered M1–M2 polarization of microglia in the brain (42). Lnc-BM drove STAT3-dependent expression of CCL2and ICAM1, acting as a mediator in the process of recruitment of macrophages and vascular co-option in the cerebrum. And then promoted BCBM via activating the Lnc-BM/JAK2/STAT3 pathway (41). It seems that some lncRNAs are still related with organ-specific metastasis in breast cancer patients, but without enough studies to support an exact classification, further studies should pay more attention on this direction.
The aforementioned studies demonstrated that the lncRNAs upregulated or downregulated in BC cells or tissue are promising targets for therapy and predictors of prognosis. Nevertheless, the current problem with their clinical application is how to find a way in which to detect these lncRNAs conveniently. As we all know, lncRNAs that are usually located in the cytoplasm or nucleus of tumor cells may interact with DNA, mRNA, miRNA or protein to perform their various functions (94, 95). Nowadays, several scientists are exploring the detection of lncRNAs in the circulatory system. LncRNA HOTAIR has been detected in patients' serum with various malignant tumors and might be a potential biomarker for diagnosis and prognosis prediction of BC (96). Extracellular lncRNA-SNHG14 was capable of being equipped in exosomes and then transmitted to sensitive cells, consequently diminishing drug resistance (97). In addition, circulating PVT1 DNA increases notably in the serum of BC patients to be detected. Compared with PVT1 RNA, DNA is a more common form of the PVT1-derived segment (49). These findings totally indicate that lncRNAs-relevant factors play a critical role in BC and are possible targets for examination and therapy, which might be a more convenient method to act as a prognosticator for BC patients in the future.
Author Contributions
All authors participated in designing the concept of this manuscript. YW, LW, CY, and KH reviewed the literature and drafted the article. CP, SZ, and AS finalized the paper and provided suggestions to improve it.
Funding
This work was funded by China Postdoctoral Science Foundation (2017M612010), National Natural Science Foundation of China (81701144), National Natural Science Foundation of China (81802628), and Zhejiang Provincial Natural Science Foundation (LQ17H160012).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Gupta GP, Massague J. Cancer metastasis: building a framework. Cell. (2006) 127:679–95. doi: 10.1016/j.cell.2006.11.001
2. Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. (2010) 127:2893–917. doi: 10.1002/ijc.25516
3. Forouzanfar MH, Foreman KJ, Delossantos AM, Lozano R, Lopez AD, Murray CJ, et al. Breast and cervical cancer in 187 countries between 1980 and 2010: a systematic analysis. Lancet. (2011) 378:1461–84. doi: 10.1016/s0140-6736(11)61351-2
4. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. (2011) 144:646–74. doi: 10.1016/j.cell.2011.02.013
5. Fitzmaurice C, Akinyemiju TF, Al Lami FH, Alam T, Alizadeh-Navaei R, Allen C, et al. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 29 cancer groups, 1990 to 2016: a systematic analysis for the global burden of disease study. JAMA Oncol. (2018) 4:1553–68. doi: 10.1001/jamaoncol.2018.2706
6. Struhl K. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat Struct Mol Biol. (2007) 14:103–5. doi: 10.1038/nsmb0207-103
7. Chillon I, Pyle AM. Inverted repeat Alu elements in the human lincRNA-p21 adopt a conserved secondary structure that regulates RNA function. Nucleic Acids Res. (2016) 44:9462–71. doi: 10.1093/nar/gkw599
8. Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell. (2011) 44:667–78. doi: 10.1016/j.molcel.2011.08.027
9. West JA, Davis CP, Sunwoo H, Simon MD, Sadreyev RI, Wang PI, et al. The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol Cell. (2014) 55:791–802. doi: 10.1016/j.molcel.2014.07.012
10. Yang F, Huo XS, Yuan SX, Zhang L, Zhou WP, Wang F, et al. Repression of the long noncoding RNA-LET by histone deacetylase 3 contributes to hypoxia-mediated metastasis. Mol Cell. (2013) 49:1083–96. doi: 10.1016/j.molcel.2013.01.010
11. Zhang EB, Yin DD, Sun M, Kong R, Liu XH, You LH, et al. P53-regulated long non-coding RNA TUG1 affects cell proliferation in human non-small cell lung cancer, partly through epigenetically regulating HOXB7 expression. Cell Death Dis. (2014) 5:e1243. doi: 10.1038/cddis.2014.201
12. Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. (2008) 133:704–15. doi: 10.1016/j.cell.2008.03.027
13. He Z, Wang Y, Huang G, Wang Q, Zhao D, Chen L. The lncRNA UCA1 interacts with miR-182 to modulate glioma proliferation and migration by targeting iASPP. Arch Biochem Biophys. (2017) 623–24:1–8. doi: 10.1016/j.abb.2017.01.013
14. Jin Y, Cui Z, Li X, Jin X, Peng J. Upregulation of long non-coding RNA PlncRNA-1 promotes proliferation and induces epithelial-mesenchymal transition in prostate cancer. Oncotarget. (2017) 8:26090–9. doi: 10.18632/oncotarget.15318
15. Conte F, Fiscon G, Chiara M, Colombo T, Farina L, Paci P. Role of the long non-coding RNA PVT1 in the dysregulation of the ceRNA-ceRNA network in human breast cancer. PLoS ONE. (2017) 12:e0171661. doi: 10.1371/journal.pone.0171661
16. Wu ZJ, Li Y, Wu YZ, Wang Y, Nian WQ, Wang LL, et al. Long non-coding RNA CCAT2 promotes the breast cancer growth and metastasis by regulating TGF-beta signaling pathway. Eur Rev Med Pharmacol Sci. (2017) 21:706–14.
17. Xi J, Feng J, Li Q, Li X, Zeng S. The long non-coding RNA lncFOXO1 suppresses growth of human breast cancer cells through association with BAP1. Int J Oncol. (2017) 50:1663–70. doi: 10.3892/ijo.2017.3933
18. Zhang M, Wu WB, Wang ZW, Wang XH. lncRNA NEAT1 is closely related with progression of breast cancer via promoting proliferation and EMT. Eur Rev Med Pharmacol Sci. (2017) 21:1020–6.
19. Liu R, Li J, Lai Y, Liao Y, Liu R, Qiu W. Hsa-miR-1 suppresses breast cancer development by down-regulating K-ras and long non-coding RNA MALAT1. Int J Biol Macromol. (2015) 81:491–7. doi: 10.1016/j.ijbiomac.2015.08.016
20. Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. (2010) 464:1071–6. doi: 10.1038/nature08975
21. Li X, Wang S, Li Z, Long X, Guo Z, Zhang G, et al. The lncRNA NEAT1 facilitates cell growth and invasion via the miR-211/HMGA2 axis in breast cancer. Int J Biol Macromol. (2017) 105(Pt 1):346–53. doi: 10.1016/j.ijbiomac.2017.07.053
22. Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. (2009) 139:871–90. doi: 10.1016/j.cell.2009.11.007
23. Brabletz T. EMT and MET in metastasis: where are the cancer stem cells? Cancer Cell. (2012) 22:699–701. doi: 10.1016/j.ccr.2012.11.009
24. Bill R, Christofori G. The relevance of EMT in breast cancer metastasis: correlation or causality? FEBS Lett. (2015) 589:1577–87. doi: 10.1016/j.febslet.2015.05.002
25. Zeng J, Wei M, Shi R, Cai C, Liu X, Li T, et al. MiR-204-5p/Six1 feedback loop promotes epithelial-mesenchymal transition in breast cancer. Tumour Biol. (2016) 37:2729–35. doi: 10.1007/s13277-015-4039-1
26. Kim J, Piao HL, Kim BJ, Yao F, Han Z, Wang Y, et al. Long noncoding RNA MALAT1 suppresses breast cancer metastasis. Nat Genet. (2018) 50:1705–15. doi: 10.1038/s41588-018-0252-3
27. Li Z, Xu L, Liu Y, Fu S, Tu J, Hu Y, et al. LncRNA MALAT1 promotes relapse of breast cancer patients with postoperative fever. Am J Transl Res. (2018) 10:3186–97.
28. Ren Y, Jia HH, Xu YQ, Zhou X, Zhao XH, Wang YF, et al. Paracrine and epigenetic control of CAF-induced metastasis: the role of HOTAIR stimulated by TGF-ss1 secretion. Mol Cancer. (2018) 17:5. doi: 10.1186/s12943-018-0758-4
29. Li W, Zhang Z, Liu X, Cheng X, Zhang Y, Han X, et al. The FOXN3-NEAT1-SIN3A repressor complex promotes progression of hormonally responsive breast cancer. J Clin Invest. (2017) 127:3421–40. doi: 10.1172/JCI94233
30. Hou P, Zhao Y, Li Z, Yao R, Ma M, Gao Y, et al. LincRNA-ROR induces epithelial-to-mesenchymal transition and contributes to breast cancer tumorigenesis and metastasis. Cell Death Dis. (2014) 5:e1287. doi: 10.1038/cddis.2014.249
31. Li GY, Wang W, Sun JY, Xin B, Zhang X, Wang T, et al. Long non-coding RNAs AC026904.1 and UCA1: a “one-two punch” for TGF-beta-induced SNAI2 activation and epithelial-mesenchymal transition in breast cancer. Theranostics. (2018) 8:2846–61. doi: 10.7150/thno.23463
32. Dong H, Hu J, Zou K, Ye M, Chen Y, Wu C, et al. Activation of LncRNA TINCR by H3K27 acetylation promotes Trastuzumab resistance and epithelial-mesenchymal transition by targeting MicroRNA-125b in breast cancer. Mol Cancer. (2019) 18:3. doi: 10.1186/s12943-018-0931-9
33. Gooding AJ, Zhang B, Jahanbani FK, Gilmore HL, Chang JC, Valadkhan S, et al. The lncRNA BORG drives breast cancer metastasis and disease recurrence. Sci Rep. (2017) 7:12698. doi: 10.1038/s41598-017-12716-6
34. Jiang Z, Slater CM, Zhou Y, Devarajan K, Ruth KJ, Li Y, et al. LincIN, a novel NF90-binding long non-coding RNA, is overexpressed in advanced breast tumors and involved in metastasis. Breast Cancer Res. (2017) 19:62. doi: 10.1186/s13058-017-0853-2
35. Li Z, Hou P, Fan D, Dong M, Ma M, Li H, et al. The degradation of EZH2 mediated by lncRNA ANCR attenuated the invasion and metastasis of breast cancer. Cell Death Differ. (2017) 24:59–71. doi: 10.1038/cdd.2016.95
36. Huang X, Xie X, Liu P, Yang L, Chen B, Song C, et al. Adam12 and lnc015192 act as ceRNAs in breast cancer by regulating miR-34a. Oncogene. (2018) 37:6316–26. doi: 10.1038/s41388-018-0410-1
37. Luo L, Tang H, Ling L, Li N, Jia X, Zhang Z, et al. LINC01638 lncRNA activates MTDH-Twist1 signaling by preventing SPOP-mediated c-Myc degradation in triple-negative breast cancer. Oncogene. (2018) 37:6166–79. doi: 10.1038/s41388-018-0396-8
38. Liu B, Sun L, Liu Q, Gong C, Yao Y, Lv X, et al. A cytoplasmic NF-kappaB interacting long noncoding RNA blocks IkappaB phosphorylation and suppresses breast cancer metastasis. Cancer Cell. (2015) 27:370–81. doi: 10.1016/j.ccell.2015.02.004
39. Wu W, Chen F, Cui X, Yang L, Chen J, Zhao J, et al. LncRNA NKILA suppresses TGF-beta-induced epithelial-mesenchymal transition by blocking NF-kappaB signaling in breast cancer. Int J Cancer. (2018) 143:2213–24. doi: 10.1002/ijc.31605
40. Yang F, Shen Y, Zhang W, Jin J, Huang D, Fang H, et al. An androgen receptor negatively induced long non-coding RNA ARNILA binding to miR-204 promotes the invasion and metastasis of triple-negative breast cancer. Cell Death Differ. (2018) 25:2209–20. doi: 10.1038/s41418-018-0123-6
41. Wang S, Liang K, Hu Q, Li P, Song J, Yang Y, et al. JAK2-binding long noncoding RNA promotes breast cancer brain metastasis. J Clin Invest. (2017) 127:4498–515. doi: 10.1172/jci91553
42. Xing F, Liu Y, Wu SY, Wu K, Sharma S, Mo YY, et al. Loss of XIST in breast cancer activates MSN-c-Met and reprograms microglia via exosomal miRNA to promote brain metastasis. Cancer Res. (2018) 78:4316–30. doi: 10.1158/0008-5472.CAN-18-1102
43. Zhang H, Cai K, Wang J, Wang X, Cheng K, Shi F, et al. MiR-7, inhibited indirectly by lincRNA HOTAIR, directly inhibits SETDB1 and reverses the EMT of breast cancer stem cells by downregulating the STAT3 pathway. Stem Cells. (2014) 32:2858–68. doi: 10.1002/stem.1795
44. Latorre E, Carelli S, Raimondi I, D'Agostino V, Castiglioni I, Zucal C, et al. The ribonucleic complex HuR-MALAT1 represses CD133 expression and suppresses epithelial-mesenchymal transition in breast cancer. Cancer Res. (2016) 76:2626–36. doi: 10.1158/0008-5472.CAN-15-2018
45. Xu S, Sui S, Zhang J, Bai N, Shi Q, Zhang G, et al. Downregulation of long noncoding RNA MALAT1 induces epithelial-to-mesenchymal transition via the PI3K-AKT pathway in breast cancer. Int J Clin Exp Pathol. (2015) 8:4881–91.
46. Wang Y, Zhou Y, Yang Z, Chen B, Huang W, Liu Y, et al. MiR-204/ZEB2 axis functions as key mediator for MALAT1-induced epithelial-mesenchymal transition in breast cancer. Tumour Biol. (2017) 39:1010428317690998. doi: 10.1177/1010428317690998
47. Chou J, Wang B, Zheng T, Li X, Zheng L, Hu J, et al. MALAT1 induced migration and invasion of human breast cancer cells by competitively binding miR-1 with cdc42. Biochem Biophys Res Commun. (2016) 472:262–9. doi: 10.1016/j.bbrc.2016.02.102
48. Padua Alves C, Fonseca AS, Muys BR, de Barros ELBR, Burger MC, de Souza JE, et al. Brief report: the lincRNA hotair is required for epithelial-to-mesenchymal transition and stemness maintenance of cancer cell lines. Stem Cells. (2013) 31:2827–32. doi: 10.1002/stem.1547
49. Li X, Chen W, Wang H, Wei Q, Ding X, Li W. Amplification and the clinical significance of circulating cell-free DNA of PVT1 in breast cancer. Oncol Rep. (2017) 38:465–71. doi: 10.3892/or.2017.5650
50. Zhang HY, Liang F, Zhang JW, Wang F, Wang L, Kang XG. Effects of long noncoding RNA-ROR on tamoxifen resistance of breast cancer cells by regulating microRNA-205. Cancer Chemother Pharmacol. (2017) 79:327–37. doi: 10.1007/s00280-016-3208-2
51. Xiao C, Wu CH, Hu HZ. LncRNA UCA1 promotes epithelial-mesenchymal transition (EMT) of breast cancer cells via enhancing Wnt/beta-catenin signaling pathway. Eur Rev Med Pharmacol Sci. (2016) 20:2819–24.
52. Li Z, Dong M, Fan D, Hou P, Li H, Liu L, et al. LncRNA ANCR down-regulation promotes TGF-beta-induced EMT and metastasis in breast cancer. Oncotarget. (2017) 8:67329–43. doi: 10.18632/oncotarget.18622
53. Richards EJ, Zhang G, Li ZP, Permuth-Wey J, Challa S, Li Y, et al. Long non-coding RNAs (LncRNA) regulated by transforming growth factor (TGF) beta: LncRNA-hit-mediated TGFbeta-induced epithelial to mesenchymal transition in mammary epithelia. J Biol Chem. (2015) 290:6857–67. doi: 10.1074/jbc.M114.610915
54. Ji P, Diederichs S, Wang W, Boing S, Metzger R, Schneider PM, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. (2003) 22:8031–41. doi: 10.1038/sj.onc.1206928
55. Wu Y, Huang C, Meng X, Li J. Long noncoding RNA MALAT1: insights into its biogenesis and implications in human disease. Curr Pharm Des. (2015) 21:5017–28. doi: 10.2174/1381612821666150724115625
56. Zhang X, Hamblin MH, Yin KJ. The long noncoding RNA Malat1: its physiological and pathophysiological functions. RNA Biol. (2017) 14:1705–14. doi: 10.1080/15476286.2017.1358347
57. Xu C, Yang M, Tian J, Wang X, Li Z. MALAT-1: a long non-coding RNA and its important 3' end functional motif in colorectal cancer metastasis. Int J Oncol. (2011) 39:169–75. doi: 10.3892/ijo.2011.1007
58. Dong Y, Liang G, Yuan B, Yang C, Gao R, Zhou X. MALAT1 promotes the proliferation and metastasis of osteosarcoma cells by activating the PI3K/Akt pathway. Tumour Biol. (2015) 36:1477–86. doi: 10.1007/s13277-014-2631-4
59. Hu L, Wu Y, Tan D, Meng H, Wang K, Bai Y, et al. Up-regulation of long noncoding RNA MALAT1 contributes to proliferation and metastasis in esophageal squamous cell carcinoma. J Exp Clin Cancer Res. (2015) 34:7. doi: 10.1186/s13046-015-0123-z
60. Jadaliha M, Zong X, Malakar P, Ray T, Singh DK, Freier SM, et al. Functional and prognostic significance of long non-coding RNA MALAT1 as a metastasis driver in ER negative lymph node negative breast cancer. Oncotarget. (2016) 7:40418–36. doi: 10.18632/oncotarget.9622
61. Zhao Z, Chen C, Liu Y, Wu C. 17beta-Estradiol treatment inhibits breast cell proliferation, migration and invasion by decreasing MALAT-1 RNA level. Biochem Biophys Res Commun. (2014) 445:388–93. doi: 10.1016/j.bbrc.2014.02.006
62. Zhao Y, Kong X, Li X, Yan S, Yuan C, Hu W, et al. Metadherin mediates lipopolysaccharide-induced migration and invasion of breast cancer cells. PLoS ONE. (2011) 6:e29363. doi: 10.1371/journal.pone.0029363
63. Huang NS, Chi YY, Xue JY, Liu MY, Huang S, Mo M, et al. Long non-coding RNA metastasis associated in lung adenocarcinoma transcript 1 (MALAT1) interacts with estrogen receptor and predicted poor survival in breast cancer. Oncotarget. (2016) 7:37957–65. doi: 10.18632/oncotarget.9364
64. Miao Y, Fan R, Chen L, Qian H. Clinical significance of long non-coding RNA MALAT1 expression in tissue and serum of breast cancer. Ann Clin Lab Sci. (2016) 46:418–24.
65. Grosse-Gehling P, Fargeas CA, Dittfeld C, Garbe Y, Alison MR, Corbeil D, et al. CD133 as a biomarker for putative cancer stem cells in solid tumours: limitations, problems and challenges. J Pathol. (2013) 229:355–78. doi: 10.1002/path.4086
66. Zhang B, Arun G, Mao YS, Lazar Z, Hung G, Bhattacharjee G, et al. The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep. (2012) 2:111–23. doi: 10.1016/j.celrep.2012.06.003
67. Guy CT, Cardiff RD, Muller WJ. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol Cell Biol. (1992) 12:954–61.
68. Arun G, Diermeier S, Akerman M, Chang KC, Wilkinson JE, Hearn S, et al. Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev. (2016) 30:34–51. doi: 10.1101/gad.270959.115
69. Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. (2007) 129:1311–23. doi: 10.1016/j.cell.2007.05.022
70. Tsai MC, Spitale RC, Chang HY. Long intergenic noncoding RNAs: new links in cancer progression. Cancer Res. (2011) 71:3–7. doi: 10.1158/0008-5472.Can-10-2483
71. Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, et al. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. (2009) 33:717–26. doi: 10.1016/j.molcel.2009.01.026
72. Lu Y, Li T, Wei G, Liu L, Chen Q, Xu L, et al. The long non-coding RNA NEAT1 regulates epithelial to mesenchymal transition and radioresistance in through miR-204/ZEB1 axis in nasopharyngeal carcinoma. Tumour Biol. (2016) 37:11733–41. doi: 10.1007/s13277-015-4773-4
73. Li L, Gu M, You B, Shi S, Shan Y, Bao L, et al. Long non-coding RNA ROR promotes proliferation, migration and chemoresistance of nasopharyngeal carcinoma. Cancer Sci. (2016) 107:1215–22. doi: 10.1111/cas.12989
74. Simeone P, Trerotola M, Franck J, Cardon T, Marchisio M, Fournier I, et al. The multiverse nature of epithelial to mesenchymal transition. Semin. Cancer Biol. (2018) doi: 10.1016/j.semcancer.2018.11.004 [Epub ahead of print].
75. Yang YT, Wang YF, Lai JY, Shen SY, Wang F, Kong J, et al. Long non-coding RNA UCA1 contributes to the progression of oral squamous cell carcinoma by regulating the WNT/beta-catenin signaling pathway. Cancer Sci. (2016) 107:1581–9. doi: 10.1111/cas.13058
76. Wang XS, Zhang Z, Wang HC, Cai JL, Xu QW, Li MQ, et al. Rapid identification of UCA1 as a very sensitive and specific unique marker for human bladder carcinoma. Clin Cancer Res. (2006) 12:4851–8. doi: 10.1158/1078-0432.Ccr-06-0134
77. Wang F, Li X, Xie X, Zhao L, Chen W. UCA1, a non-protein-coding RNA up-regulated in bladder carcinoma and embryo, influencing cell growth and promoting invasion. FEBS Lett. (2008) 582:1919–27. doi: 10.1016/j.febslet.2008.05.012
78. Huang J, Zhou N, Watabe K, Lu Z, Wu F, Xu M, et al. Long non-coding RNA UCA1 promotes breast tumor growth by suppression of p27 (Kip1). Cell Death Dis. (2014) 5:e1008. doi: 10.1038/cddis.2013.541
79. Cai J, Guan H, Fang L, Yang Y, Zhu X, Yuan J, et al. MicroRNA-374a activates Wnt/beta-catenin signaling to promote breast cancer metastasis. J Clin Invest. (2013) 123:566–79. doi: 10.1172/jci65871
80. Shan S, Lv Q, Zhao Y, Liu C, Sun Y, Xi K, et al. Wnt/beta-catenin pathway is required for epithelial to mesenchymal transition in CXCL12 over expressed breast cancer cells. Int J Clin Exp Pathol. (2015) 8:12357–67.
81. Kretz M. TINCR, staufen1, and cellular differentiation. RNA Biol. (2013) 10:1597–601. doi: 10.4161/rna.26249
82. Liu Y, Du Y, Hu X, Zhao L, Xia W. Up-regulation of ceRNA TINCR by SP1 contributes to tumorigenesis in breast cancer. BMC Cancer. (2018) 18:367. doi: 10.1186/s12885-018-4255-3
83. Takeda K, Ichijo H, Fujii M, Mochida Y, Saitoh M, Nishitoh H, et al. Identification of a novel bone morphogenetic protein-responsive gene that may function as a noncoding RNA. J Biol Chem. (1998) 273:17079–85.
84. Kretz M, Webster DE, Flockhart RJ, Lee CS, Zehnder A, Lopez-Pajares V, et al. Suppression of progenitor differentiation requires the long noncoding RNA ANCR. Genes Dev. (2012) 26:338–43. doi: 10.1101/gad.182121.111
85. Fagerberg L, Hallstrom BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J, et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics. (2014) 13:397–406. doi: 10.1074/mcp.M113.035600
86. Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF, et al. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. (2014) 25:666–81. doi: 10.1016/j.ccr.2014.03.010
87. Weakley SM, Wang H, Yao Q, Chen C. Expression and function of a large non-coding RNA gene XIST in human cancer. World J Surg. (2011) 35:1751–6. doi: 10.1007/s00268-010-0951-0
88. Rodriguez Bautista R, Ortega Gomez A, Hidalgo Miranda A, Zentella Dehesa A, Villarreal-Garza C, Avila-Moreno F, et al. Long non-coding RNAs: implications in targeted diagnoses, prognosis, and improved therapeutic strategies in human non- and triple-negative breast cancer. Clin Epigen. (2018) 10:88. doi: 10.1186/s13148-018-0514-z
89. Yang Q, Wang X, Tang C, Chen X, He J. H19 promotes the migration and invasion of colon cancer by sponging miR-138 to upregulate the expression of HMGA1. Int J Oncol. (2017) 50:1801–9. doi: 10.3892/ijo.2017.3941
90. Wang G, Liu C, Deng S, Zhao Q, Li T, Qiao S, et al. Long noncoding RNAs in regulation of human breast cancer. Brief Funct Genomics. (2016) 15:222–6. doi: 10.1093/bfgp/elv049
91. Gunasinghe NP, Wells A, Thompson EW, Hugo HJ. Mesenchymal-epithelial transition (MET) as a mechanism for metastatic colonisation in breast cancer. Cancer Metastasis Rev. (2012) 31:469–78. doi: 10.1007/s10555-012-9377-5
92. Aiello NM, Bajor DL, Norgard RJ, Sahmoud A, Bhagwat N, Pham MN, et al. Metastatic progression is associated with dynamic changes in the local microenvironment. Nat Commun. (2016) 7:12819. doi: 10.1038/ncomms12819
93. Pattabiraman DR, Bierie B, Kober KI, Thiru P, Krall JA, Zill C, et al. Activation of PKA leads to mesenchymal-to-epithelial transition and loss of tumor-initiating ability. Science. (2016) 351:aad3680. doi: 10.1126/science.aad3680
94. Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. (2011) 25:1915–27. doi: 10.1101/gad.17446611
95. Novikova IV, Hennelly SP, Sanbonmatsu KY. Sizing up long non-coding RNAs: do lncRNAs have secondary and tertiary structure? Bioarchitecture. (2012) 2:189–99. doi: 10.4161/bioa.22592
96. Lu R, Zhang J, Zhang W, Huang Y, Wang N, Zhang Q, et al. Circulating HOTAIR expression predicts the clinical response to neoadjuvant chemotherapy in patients with breast cancer. Cancer Biomark. (2018) 22:249–56. doi: 10.3233/cbm-170874
Keywords: long non-coding RNA, breast cancer, metastasis, invasion, mechanism
Citation: Wu Y, Shao A, Wang L, Hu K, Yu C, Pan C and Zhang S (2019) The Role of lncRNAs in the Distant Metastasis of Breast Cancer. Front. Oncol. 9:407. doi: 10.3389/fonc.2019.00407
Received: 28 January 2019; Accepted: 30 April 2019;
Published: 31 May 2019.
Edited by:
Cesar Wong, Hong Kong Polytechnic University, Hong KongReviewed by:
Allen Chi-Shing Yu, Codex Genetics Limited, Hong KongDaniele Vergara, University of Salento, Italy
Copyright © 2019 Wu, Shao, Wang, Hu, Yu, Pan and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anwen Shao, MjExMTgxMTZAemp1LmVkdS5jbg==; YW53ZW5zaGFvQHNpbmEuY29t; Suzhan Zhang, enVjaUB6anUuZWR1LmNu
†These authors have contributed equally to this work
 Yinan Wu1†
Yinan Wu1† Anwen Shao
Anwen Shao