- 1Department of Oncology, The First Affiliated Hospital of Zhengzhou University, Zhengzhou, China
- 2The Academy of Medical Science, Zhengzhou University, Zhengzhou, China
Natural killer/T-cell lymphoma (NKTCL) is a rare and aggressive subtype of non-Hodgkin's lymphoma that is associated with a poor outcome. Non-coding RNAs (ncRNAs), which account for 98% of human RNAs, lack the function of encoding proteins but instead have the important function of regulating gene expression, including transcription, translation, RNA splicing, editing, and turnover. However, the roles and mechanisms of aberrantly expressed ncRNAs in NKTCL are not fully clear. Aberrant expressions of microRNA (miRNAs) affect the PI3K/AKT signaling pathways (miRNA-21, miRNA-155, miRNA-150, miRNA-142, miRNA-494), NF-κB (miRNA-146a, miRNA-155) and cell cycle signaling pathways to regulate cell function. Moreover, Epstein-Barr virus (EBV) encoded miRNAs and EBV oncoprotein LMP-1 regulated the expression of cellular genes that induce invasion, metastasis, cell cycle progression and cellular transformation. In addition, NKTCL-associated Long non-coding RNA (lncRNA) ZFAS1 regulated certain pathways and lncRNA MALAT1 acted as a predictive marker. This review article provides an overview of ncRNAs associated with NKTCL, summarizes the function of significantly differentially expressed hotspot non-coding RNAs that contribute to the pathogenesis, diagnoses, treatment and prognosis of NKTCL and discusses the relevance of these ncRNAs to clinical practice.
Introduction
Non-Hodgkin's lymphoma (NHL) originates from B-lymphocytes, T-lymphocytes and natural killer (NK) lymphocytes and ranges from the indolent to the very aggressive (1). Each subtype could be further classified according to its origin, genetic signature or clinical features (2). Natural killer/T-cell lymphoma (NKTCL) is a rare and aggressive subtype of NHL that has a high incidence in East Asia and Latin America and that is associated with a poor outcome (3, 4). Extranodal NKTCL can be further classified into nasal NKTCL, which primarily affects the nasal cavity, nasopharynx and the upper aerodigestive tract, and non-nasal NKTCL, which involves the outside of the nasopharyngeal region, such as the skin, gastrointestinal tract and testis (5).
Deletion of chromosome 6q and down-regulation of tumor suppressor genes located on 6q21 regions, including PRDM1 (6), FOXO3 (7), PTPRK, HACE1, ATG5, and AIM1 (8) were observed through oligo-array comparative genomic hybridization (CGH) and gene-expression profiling. High mutation frequencies of FAS (9), TP53 and DDX3X (10) had a trend toward advanced stage and poor prognosis in NKTCL. Recurrent gene mutations in the JAK-STAT pathway were demonstrated, including high expression of phosphorylated-JAK3 and phosphorylated-STAT3 (11). Moreover, the expression of Epstein-Barr virus (EBV)-encoded RNA (EBER) was specific for diagnosis in clinical practice (12).
Non-coding RNAs (ncRNAs), which account for 98% of all human RNAs, lack a protein-coding function, but rather, they have the important function of regulating gene expression, including transcription, translation, RNA splicing, editing, and turnover (13). NcRNAs include microRNAs (miRNAs), small nuclear RNAs, PIWI-interacting RNAs, long non-coding RNAs (lncRNA), and circular RNAs. With the development of next-generation sequencing and bioinformatics approaches, ncRNAs show great biological importance in cancers. However, the roles and mechanisms of aberrantly expressed non-coding RNAs in NKTCL have not been fully clarified. This review article provides an overview of the recent advancements of ncRNAs associated with NKTCL and discusses their relevance to clinical practice.
MiRNAs
MiRNAs are 18–24-nucleotide-long single-stranded ncRNAs that can regulate translation via binding to 3′-untranslated regions (3′-UTRs) of target mRNAs in order to affect cell function (14, 15).
MiRNA-21
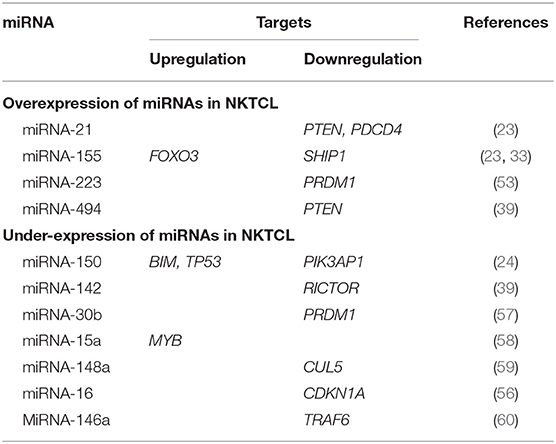
MiRNA-21 regulates various genes and signaling pathways involved in cancer pathogenesis, progression and metastasis (16). MiRNA-21 is overexpressed in various solid tumor types including breast, colon, lung, pancreas, prostate, and stomach (17) tumors and is also upregulated in hematological malignancies such as chronic lymphocytic leukemia (18), acute and chronic myeloid leukemia (19), diffuse large B-cell lymphoma (20), cutaneous T-cell lymphoma (21) and Hodgkin lymphoma (22). The expression of miRNA-21 was found to be higher in NK-cell lymphoma-derived cell lines and in samples of primary NKTCL compared with normal natural killer cells (23, 24). MiRNA-21 regulated apoptosis of NK-cell lymphoma cell lines via the PTEN/AKT signaling pathway, and the downregulation of miRNA-21 led to the upregulation of phosphatase and tensin homolog (PTEN), programmed cell death 4 (PDCD4) and the downregulation of pAKT. PTEN, served as a multi-functional tumor suppressor, commonly lost in human cancer and negatively regulating AKT/PKB signaling pathway (25, 26). PDCD4, a tumor suppresser, inhibited neoplastic transformation and invasion (27–29). In addition, the proapoptotic protein Bim was found to be increased (23, 30). The role of miRNA-21 in the pathogenesis of NKTCL suggests that miRNA-21 can serve as a new biomarker or target in the treatment of NKTCL.
MiRNA-155
MiRNA-155 is overexpressed in various hematological and solid malignancies (16). MiRNA-155 regulates inflammation, immune cells, and the differentiation and maturation of tumor cells (31). The expressions of miRNA-155 were higher in NK-cell lymphoma cell lines and primary NKTCL specimens than in normal NK cells (23, 24). Among various NK-cell lymphoma cell lines, the expression of miRNA-155 was reported to be highest in SNK-6 cells (32). MiRNA-155 regulated apoptosis via the PTEN/AKT and NF-κB signaling pathways in NK-cell lymphoma cell lines. MiRNA-155 directly down regulated Src homology-2 domain-containing inositol 5-phosphatase 1 (SHIP1), which inhibited signaling in the phosphoinositide 3-kinase (PI3K)-AKT pathway and further inhibited p21 and p27 (23). Moreover, the expression of miRNA-155 was related to several inflammatory factors, such as interleukin 6 (IL-6), interleukin 13 (IL-13), and tumor necrosis factor (TNF), in NKTCL (33). In addition, whole DNA hypomethylation was observed to occur with locus-specific hypermethylation, especially on promoter-associated CpG islands, which resulted in the silencing of downstream genes and ncRNAs (34, 35). In NKTCL patients, the prevalence of miRNA-155-3p methylation has been discovered to occur distinctively with the overexpression of LT-β (35). MiRNA-155 is a potential molecular marker of NKTCL (33).
MiRNA-142
MiRNA-142 has two different forms (miRNA-142-3p and miRNA-412-5p) that participate in the regulation of hematopoietic differentiation and immune response (36). MiRNA-142 upregulates various proteins such as the IL-6, interleukin 6 signal transducer (IL6ST), toll-like receptor 2 (TLR2), prostaglandin E receptor 2 (PGE2), and TNF (37). The miRNA-142-5p and miRNA-142-3p were under-expression in NKTCL compared with EBV-negative lymphomas (38). MiR-142-3p down regulated RICTOR, one of components of the mTOR complex, and further affected pAKT in YT cell line (39). In addition, the down-regulation of miR-142-3p led to the upregulation of IL1A in NKTCL (38). MiRNA-142-3p is a potential target of therapy (39).
MiRNA-494
As a tumor suppressor miRNA, miRNA-494 played a role in various tumors (40, 41). MiRNA-494 induced PTEN downregulation in cervical cancer cells (42) and myeloid cells (43). In addition, TGF-β1 was a tumor-derived factor that was associated with the upregulation of miRNA-494 in MDSCs and MMPs, which led to tumor cell invasion and metastasis (43). In an NK-cell lymphoma cell line NK92, miRNA-494-3p was also found to down-regulate PTEN, which activated AKT in accordance with previous reports (39). Moreover, miRNA-494-3p worked in coordination with the EBV-encoded miRNA-BART20-5p, which inhibited the T-bet-PTEN pathway, with subsequent upregulation of AKT and suppression of TP53 (39). Antagomir to miRNA-494-3p may serve as a potential target of therapy of NKTCL (39).
MiRNA-150
MiRNA-150 as a key regulator of the differentiation and activation (44) of immune cells, such as B-, T-, and NK-lymphocytes (45), abnormally expressed in solid (46) and hematological malignancies (44). MiRNA-150 was found to be apparently lower in lymphoma cell lines and primary lymphoma specimens compared with normal NK cells, while no significant difference was found between resting and activated NK cells (24). Furthermore, miRNA-150 down regulated PIK3AP1 and AKT2, which were part of the PI3K-AKT pathway and upregulated Bim and p53. MiRNA-150 led to cancer cell anti-apoptosis and immortality, as pAKTser473/4 acted on telomerase via phosphorylation of hTERT (24). In addition, miRNA-150 down regulated DKC1, which functioned in regulating pseudouridine in RNA and the telomerase RNA subunit hTR in NKTCL cells (24, 47). MiRNA-150 provides novel strategy upstream of AKT in the treatment of NKTCL (24).
MiRNA-223
MiRNA-223 is strongly expressed in the bone marrow and bone marrow cells but is absent in B- and T-lymphocytes (48). In resting NK cells, miRNA-223 downregulated in the case of cytokine activation and controls GzmB translation in resting NK cells (49). Overexpression of miRNA-223 can decrease cancer cell proliferation (50, 51). For instance, miRNA-223 expression was reported to be lower in CD19+ lymphocytes in patients with mantle cell lymphoma compared with healthy donors (50). In NKTCL cells, overexpression of miRNA-223 is associated with cell differentiation (52). Positive regulatory domain containing I (PRDM1), a tumor suppressor gene in NK cell, was directly downregulated by miRNA-223 in NKTCL patient samples and NKTCL cell lines (53). All miRNA-223-positive samples from patients with NKTCL showed EBV infection, which implied that EBV infection may be responsible for miRNA-223 overexpression (53).
MiRNA-16
Members of the miRNA-16 family function as tumor suppressors in a number of cancers via the regulation of the cell apoptosis pathway (54) and the cell cycle (55). In NK-cell lymphoma cell lines and primary tissue, miRNA-16 was found to be under expressed (56). MiRNA-16 and SAHA shared common therapeutic targets and induce senescence and apoptosis in NKTCL. However, in Kitadate's study, NK-cell lymphoma cell lines with non-functional p53 (KHYG1) did not show senescence caused by miRNA-16 or by SAHA. It has been confirmed that miRNA-16 or SAHA induces apoptosis, downregulates survivin and upregulates cleaved caspase-3 and CDKN1A (also known as p21) (56). This finding suggests that miRNA-16 has the potential to serve as a novel target in NKTCL treatment.
The miRNAs and their identified target genes are listed in Table 1.
EBV-Encoded MiRNAs
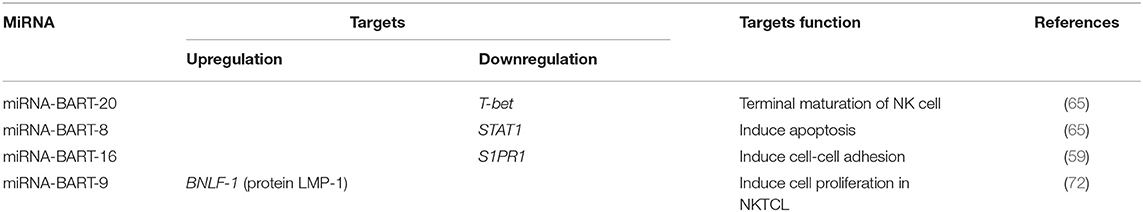
Since NKTCL is an EBV associated lymphoma, researchers illustrated that EBV infection promoted the progression from a lesion into NKTCL (33, 61, 62) via the regulation of 44 microRNAs (59). The most common outcome of EBV infection was viral latency, including type I, II and III (63). Latency I was reported to only express characterized EBNA-1, while latency II expressed EBNA-1 as well as LMP-1 and 2. Moreover, latency III with B cell infection expressed all EBNAs and LMPs (64). Furthermore, latency patterns were distinct in different malignant subtypes due to the expression of subsets of the latent genes (65, 66).
LMP-1, encoded by the BNLF-1 gene, is the principal EBV oncoprotein and regulates the expression of cellular genes that induce invasion, metastasis, cell cycle progression and cellular transformation (63, 67, 68). LMP-1 mediates NF-κB and PI3K/AKT activation in EBV-positive NKTCL cell lines and inhibits cell apoptosis by promoting survivin expression (69–71). In addition, LMP1 also regulates cell function through regulation of the expression of other miRNAs; for instance, LMP1 inhibits the cell cycle via the downregulation of miRNA-15a, which inhibits MYB and cyclin D1 in NKTCL cells (58). In addition, EBV-miRNA-BART9 upregulates LMP1 (72).
Various studies have attempted to determine the clinical significance of miRNA-BARTs. In one study, the higher expression of miRNA-BART2-5p, miRNA-BART7-3p and miRNA-BART13-3p led to a poorer prognosis in patients with NKTCL (73). MiRNA-BART20-5p and miRNA-BART8 led to cell apoptosis via the inhibition of the IFN-γ-STAT1 pathway and the downregulation of miRNA-let7 in NKTCLs (65). Additionally, miRNA-BART20-5p inhibited TP53 via T-bet (74, 75), a member of the T-box family that involved in tumor development (76). MiR-BART16 down-regulated the sphingosin-1-phosphate receptor 1 (S1PR1), which expressed in cells of lymphoid origin and named as CD363 antigen (59).
These studies have implications in the mechanisms of lymphomagenesis, and future experiments should be directed at the investigation of the role of EBV miRNAs and their regulation of cellular targets. The EBV-encoded miRNAs and their identified target genes are listed in Table 2 and associations are shown in Figure 2.
Other miRNAs
In NK-cell lymphoma cell lines, various miRNAs function in the regulation of tumor development (77), including miRNA-101, miRNA-26a, miRNA-26b, miRNA-28-5, and miRNA-363 (57).
MiRNA-20, miRNA-26a, miRNA-92, miRNA-103, and miRNA-181 were shown to be overexpressed in patients of NKTCL (23). Moreover, miRNA-424 (38) and miRNA-16 (56) were shown to be under-expressed in NK-cell lymphoma cell lines and tumor tissue. The expression of miRNA-221 in the serum of NKTCL patients might be a prognostic factor since high expression leads to a poorer overall survival (OS) (78).
Furthermore, miRNAs regulate gene expression. For example, the expression of PRDM1 was directly downregulated by miRNA-30b in NKTCL (57). In addition, CUL5 is a target of deregulated miRNA-148a in NKTCL (59).
MiRNA-15a was reported to inhibit the cell cycle by blocking G1/S progression in NK-cell lymphoma cell lines (58). Specifically, miRNA-15a upregulated MYB and cyclin D1 which were essential for the proliferation of NK-cell lymphoma cells (58).
As a tumor suppressor, miRNA-34a (79) was found to be hypermethylated in both myeloma and lymphoma cell lines (80). Furthermore, in lymphoma primary patient samples, methylation of miRNA-34a was found to be more frequently in NKTCL than in B- or T-cell lymphoma (80).
MiRNA-146a also exhibited hypermethylation in NKTCL, and down-regulated its target gene TRAF6 and NF-κB signaling pathway (60). In clinical study, low miRNA-146a expression was an independent poor prognostic factor.
In summary, we found that dysregulation of miRNAs might be a key feature of the pathogenesis of NKTCL. Aberrant expression of miRNAs might affect the AKT, NF-κB and cell cycle signaling pathways to regulate cell function. The signaling pathway model has been integrated in Figure 1. Hypermethylation is another way by which cell function is regulated. These findings provide new thought about the pathogenesis of NKTCL.
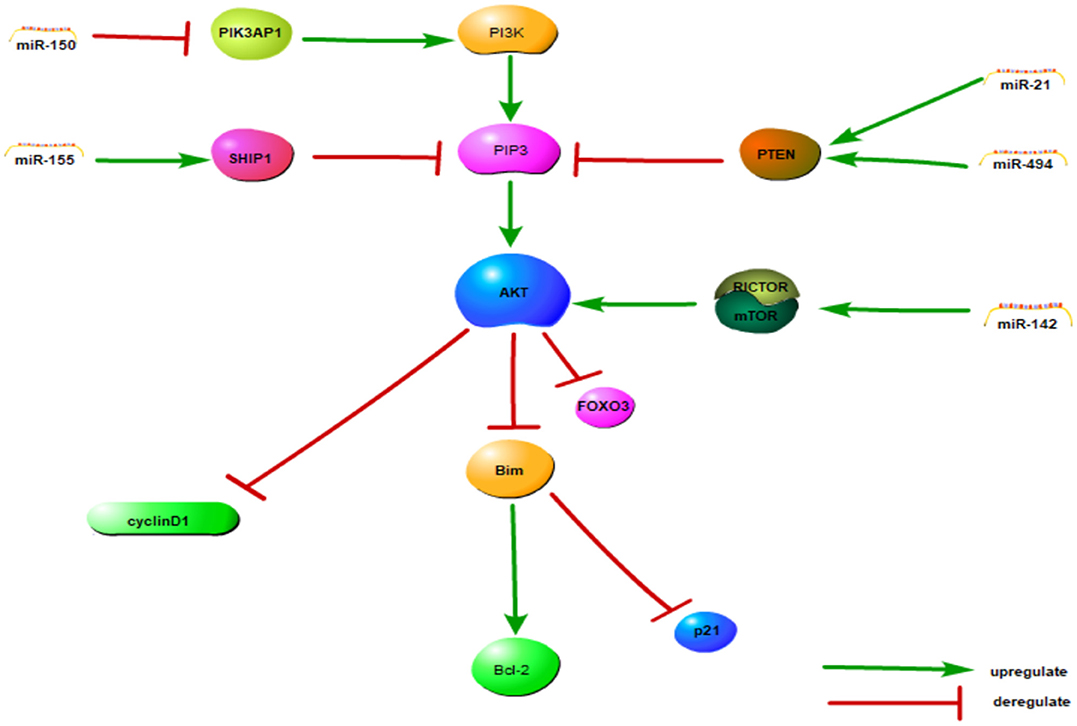
Figure 1. The PI3K/AKT and cell cycle signaling pathway model of NKTCL. MiRNAs can influence almost every cellular behavior from transcription to translation by diverse mechanisms including the cell cycle and the PI3K/AKT pathways, among others. All the miRNAs in this figure are upregulated in NKTCL sample, except miRNA-150. This figure adapted from Chen et al. (39) and the appropriate copyright permissions has been obtained from the copyright holder of this work.
LncRNAs
LncRNAs are a group of RNAs >200 nucleotides in length that regulate gene expression by transcriptional and posttranscriptional destabilization (81, 82). LncRNAs have great potential value in the pathogenesis, diagnosis, treatment and prognosis of malignant tumors (83, 84).
Baytak et al. (85) conducted whole transcriptome sequencing (WTS) analysis on NKTCL cases, normal NK-cells and NK-cell lymphoma cell lines. They revealed 166 lncRNAs with more than 1.5-fold overexpression, such as RAB30-AS1, ARAP-AS1 and PRMT5-AS1 which may have biological function on cell growth.
LncRNA ZNFX1 antisense RNA 1 (ZFAS1) transcribed from the antisense gene ZNFX1 (86), was overexpressed in mammary gland while under-expressed in breast tumors (87). However, ZFAS1 functioned as oncogene in various tumors, such as glioma (88), colorectal cancer (89), Gastric cancer (90), hepatocellular carcinoma (91) and ovarian cancer (92) for the high expression in tumor tissues. Moreover, ZFAS1 also showed high expression in AML cell lines (93). LncRNA ZFAS1 was observed to be upregulated in NKTCL and further demonstrated 483 relevant genes (70 genes with strong positive correlation, while 413 genes with strong negative correlation). Overall, ZFAS1-correlated genes associated with the upregulation of certain pathways, including non-sense-mediated mRNA decay, NF-κB signaling, β-catenin independent WNT signaling and p53-dependent apoptosis and the cell cycle pathways (Figure 2) (85). In addition, lncRNA ZFAS1 regulates p53 via invasion and metastasis related genes MDM2, and p53 can further regulate the NF-κB, WNT, and NOTCH1 pathways (85).
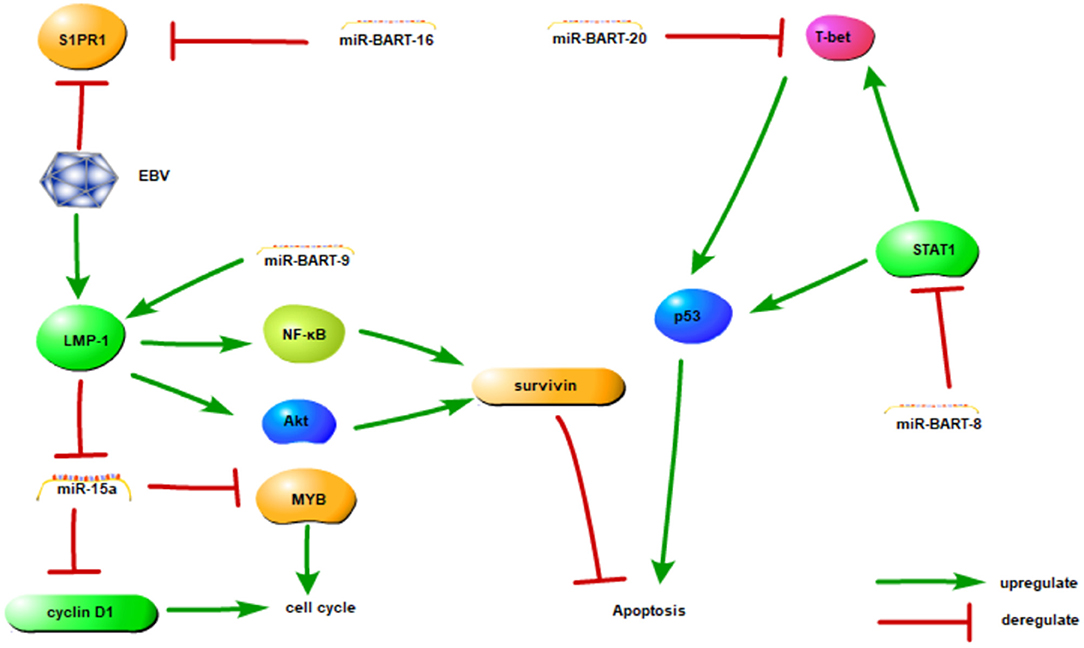
Figure 2. The association between EBV-miRNAs and targets in NKTCL. This figure adapted from Huang and Lin (65) and the appropriate copyright permission has been obtained.
LncRNA metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), also known as nuclear-enriched transcript 2 (NEAT2), is discovered as a predictive marker for metastasis and survival in early-stage, non-small cell lung cancer (94). The high expression of MALAT1 have been found in various cancer types (95, 96) and predicted metastasis or poor prognosis (97, 98). LncRNA MALAT1 was highly expressed in NKTCL, but in the absence of expression, inferior OS is observed. MALAT1 was important in sustaining PRC2-induced H3K27me3, which led to the subsequent activation of BMI1 which predicted the clinically aggressive behaviors in NKTCL (99, 100).
The functions and regulations of lncRNAs were not isolated. Positive regulatory domain containing I (PRDM1), which acted as a tumor suppressor gene, was silenced in NKTCL as previously mentioned (7). PRDM1α regulated 212 lncRNAs (169 upregulation and 103 downregulation). Among them, MIRNA-155HG and TERC may be an indirect target of PRDM1 in NK-cell lymphoma cells. However, biological functions of most lncRNAs regulated by PRDM1 in NKTCL remained unknown, and further studies should involve functional characterization (85).
Conclusion
With the development of next-generation sequencing and bioinformatics, non-coding RNAs have shown promising value in tumor research in recent years. However, the focus on B-cell and T-cell lymphoma encompasses much more than NKTCL. This article summarizes the function of significantly differentially expressed hotspot non-coding RNAs that contribute to the pathogenesis, diagnoses, treatment and prognosis of NKTCL. The detailed mechanisms and the function of the non-coding RNAs mentioned above still remain to be clarified. Further connections among these non-coding RNAs can be supplemented. Moreover, research on novel non-coding RNAs such as piwi RNAs, circular RNAs and tiRNAs that are associated with NKTCL is needed. Clinical research of thoroughly studied non-coding RNAs could be performed and lead to the early diagnoses of NKTCL and could be useful in drug resistance or targeted therapy.
Author Contributions
MZ and MM designed the study. MM collected data and wrote the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (No. 81570203).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
I would like to show gratitude to all members in oncology department of the First Affiliated Hospital of Zhengzhou University.
Abbreviations
NHL, Non-Hodgkin's lymphoma; EBER, EBV-encoded RNA; NKTCL, Natural killer/T-cell lymphoma; EBV, Epstein-Barr virus; ncRNAs, non-coding RNAs; CGH, comparative genomic hybridization; miRNAs, microRNAs; 3′-UTRs, 3′ - untranslated regions; IL-6, immune cytokines interleukin 6; IL6ST, interleukin 6 signal transducer; TLR2, toll-like receptor 2; PGE2, prostaglandin E receptor 2; TNF, tumor necrosis factor; OS, overall survival; WTS, whole transcriptome sequencing; PTEN, phosphatase and tensin homolog; PDCD4, programmed cell death 4; SHIP1, Src homology-2 domain-containing inositol 5-phosphatase 1; PI3K, phosphoinositide 3-kinase; PRDM1, Positive regulatory domain containing I; S1PR1, sphingosin-1-phosphate receptor 1; ZFAS1, ZNFX1 antisense RNA 1; MALAT1, metastasis-associated lung adenocarcinoma transcript 1; NEAT2, nuclear-enriched transcript 2.
References
1. Swerdlow SH, Campo E, Pileri SA, Harris NL, Stein H, Siebert R, et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. (2016) 127:2375–90. doi: 10.1182/blood-2016-01-643569
2. Tagawa H. microRNA in malignant lymphoma. Adv Exp Med Biol. (2015) 889:41–50. doi: 10.1007/978-3-319-23730-5_3
3. Harabuchi Y, Takahara M, Kishibe K, Moriai S, Nagato T, Ishii H. Nasal natural killer (NK)/T-cell lymphoma: clinical, histological, virological, and genetic features. Int J Clin Oncol. (2009) 14:181–90. doi: 10.1007/s10147-009-0882-7
4. Li X, Cheng Y, Zhang M, Yan J, Li L, Fu X, et al. Activity of pembrolizumab in relapsed/refractory NK/T-cell lymphoma. J Hematol Oncol. (2018) 11:15. doi: 10.1186/s13045-018-0559-7
5. Tse E, Kwong YL. Practical management of natural killer/T-cell lymphoma. Cur Opin Oncol. (2012) 24:480–6. doi: 10.1097/CCO.0b013e3283556142
6. Kucuk C, Iqbal J, Hu X, Gaulard P, De Leval L, Srivastava G, et al. PRDM1 is a tumor suppressor gene in natural killer cell malignancies. Proc Nat Acad Sci USA. (2011) 108:20119–24. doi: 10.1073/pnas.1115128108
7. Karube K, Nakagawa M, Tsuzuki S, Takeuchi I, Honma K, Nakashima Y, et al. Identification of FOXO3 and PRDM1 as tumor-suppressor gene candidates in NK-cell neoplasms by genomic and functional analyses. Blood. (2011) 118:3195–204. doi: 10.1182/blood-2011-04-346890
8. Huang Y, de Reynies A, de Leval L, Ghazi B, Martin-Garcia N, Travert M, et al. Gene expression profiling identifies emerging oncogenic pathways operating in extranodal NK/T-cell lymphoma, nasal type. Blood. (2010) 115:1226–37. doi: 10.1182/blood-2009-05-221275
9. Takakuwa T, Dong Z, Nakatsuka S, Kojya S, Harabuchi Y, Yang WI, et al. Frequent mutations of Fas gene in nasal NK/T cell lymphoma. Oncogene. (2002) 21:4702–5. doi: 10.1038/sj.onc.1205571
10. Zhang Y, Li C, Xue W, Zhang M, Li Z. Frequent mutations in natural killer/T cell lymphoma. Cell Physiol Biochem. (2018) 49:1–16. doi: 10.1159/000492835
11. Chen YW, Guo T, Shen L, Wong KY, Tao Q, Choi WW, et al. Receptor-type tyrosine-protein phosphatase kappa directly targets STAT3 activation for tumor suppression in nasal NK/T-cell lymphoma. Blood. (2015) 125:1589–600. doi: 10.1182/blood-2014-07-588970
12. Yamaguchi M, Oguchi M, Suzuki R. Extranodal NK/T-cell lymphoma: Updates in biology and management strategies. Best Pract Res Clin Haematol. (2018) 31:315–21. doi: 10.1016/j.beha.2018.07.002
13. Mattick JS, Makunin IV. Non-coding RNA. Human Mol Genet. (2006) 15 R17-29. doi: 10.1093/hmg/ddl046
14. Kotaki R, Koyama-Nasu R, Yamakawa N, Kotani A. miRNAs in Normal and Malignant Hematopoiesis. Int J Mol Sci. (2017) 18:1495. doi: 10.3390/ijms18071495
15. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. (2009) 136:215–33. doi: 10.1016/j.cell.2009.01.002
16. Pan Y, Meng M, Zhang G, Han H, Zhou Q. Oncogenic microRNAs in the genesis of leukemia and lymphoma. Curr Pharmaceut Design. (2014) 20:5260–7. doi: 10.2174/1381612820666140128211724
17. Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Nat Acad Sci USA. (2006) 103:2257–61. doi: 10.1073/pnas.0510565103
18. Fulci V, Chiaretti S, Goldoni M, Azzalin G, Carucci N, Tavolaro S, et al. Quantitative technologies establish a novel microRNA profile of chronic lymphocytic leukemia. Blood. (2007) 109:4944–51. doi: 10.1182/blood-2006-12-062398
19. Espadinha AS, Prouzet-Mauleon V, Claverol S, Lagarde V, Bonneu M, Mahon FX, et al. A tyrosine kinase-STAT5-miR21-PDCD4 regulatory axis in chronic and acute myeloid leukemia cells. Oncotarget. (2017) 8:76174–88. doi: 10.18632/oncotarget.19192
20. Lawrie CH, Soneji S, Marafioti T, Cooper CD, Palazzo S, Paterson JC, et al. MicroRNA expression distinguishes between germinal center B cell-like and activated B cell-like subtypes of diffuse large B cell lymphoma. Int J Cancer. (2007) 121:1156–61. doi: 10.1002/ijc.22800
21. van der Fits L, van Kester MS, Qin Y, Out-Luiting JJ, Smit F, Zoutman WH, et al. MicroRNA-21 expression in CD4+ T cells is regulated by STAT3 and is pathologically involved in Sezary syndrome. J Invest Dermatol. (2011) 131:762–8. doi: 10.1038/jid.2010.349
22. Navarro A, Gaya A, Martinez A, Urbano-Ispizua A, Pons A, Balague O, et al. MicroRNA expression profiling in classic Hodgkin lymphoma. Blood. (2008) 111:2825–32. doi: 10.1182/blood-2007-06-096784
23. Yamanaka Y, Tagawa H, Takahashi N, Watanabe A, Guo YM, Iwamoto K, et al. Aberrant overexpression of microRNAs activate AKT signaling via down-regulation of tumor suppressors in natural killer-cell lymphoma/leukemia. Blood. (2009) 114:3265–75. doi: 10.1182/blood-2009-06-222794
24. Watanabe A, Tagawa H, Yamashita J, Teshima K, Nara M, Iwamoto K, et al. The role of microRNA-150 as a tumor suppressor in malignant lymphoma. Leukemia. (2011) 25:1324–34. doi: 10.1038/leu.2011.81
25. Chen CY, Chen J, He L, Stiles BL. PTEN: Tumor suppressor and metabolic regulator. Front Endocrinol. (2018) 9:338. doi: 10.3389/fendo.2018.00338
26. Pfeifer M, Grau M, Lenze D, Wenzel SS, Wolf A, Wollert-Wulf B, et al. PTEN loss defines a PI3K/AKT pathway-dependent germinal center subtype of diffuse large B-cell lymphoma. Proc Nat Acad Sci USA. (2013) 110:12420–5. doi: 10.1073/pnas.1305656110
27. Cmarik JL, Min H, Hegamyer G, Zhan S, Kulesz-Martin M, Yoshinaga H, et al. Differentially expressed protein Pdcd4 inhibits tumor promoter-induced neoplastic transformation. Proc Nat Acad Sci USA. (1999) 96:14037–42. doi: 10.1073/pnas.96.24.14037
28. Jansen AP, Camalier CE, Colburn NH. Epidermal expression of the translation inhibitor programmed cell death 4 suppresses tumorigenesis. Cancer Res. (2005) 65:6034–41. doi: 10.1158/0008-5472.CAN-04-2119
29. Leupold JH, Yang HS, Colburn NH, Asangani I, Post S, Allgayer H. Tumor suppressor Pdcd4 inhibits invasion/intravasation and regulates urokinase receptor (u-PAR) gene expression via Sp-transcription factors. Oncogene. (2007) 26:4550–62. doi: 10.1038/sj.onc.1210234
30. O'Connor L, Strasser A, O'Reilly LA, Hausmann G, Adams JM, Cory S, et al. Bim: a novel member of the Bcl-2 family that promotes apoptosis. EMBO J. (1998) 17:384–95. doi: 10.1093/emboj/17.2.384
31. Higgs G, Slack F. The multiple roles of microRNA-155 in oncogenesis. J Clin Bioinformat. (2013) 3:17. doi: 10.1186/2043-9113-3-17
32. Ji WG, Zhang XD, Sun XD, Wang XQ, Chang BP, Zhang MZ. miRNA-155 modulates the malignant biological characteristics of NK/T-cell lymphoma cells by targeting FOXO3a gene. J Huazhong Univ Sci Technol Med Sci. (2014) 34:882–8. doi: 10.1007/s11596-014-1368-z
33. Zhang X, Ji W, Huang R, Li L, Wang X, Li L, et al. MicroRNA-155 is a potential molecular marker of natural killer/T-cell lymphoma. Oncotarget. (2016) 7:53808–19. doi: 10.18632/oncotarget.10780
34. Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer. (2011) 11:726–34. doi: 10.1038/nrc3130
35. Yim RL, Wong KY, Kwong YL, Loong F, Leung CY, Chu R, et al. Methylation of miR-155-3p in mantle cell lymphoma and other non-Hodgkin's lymphomas. Oncotarget. (2014) 5:9770–82. doi: 10.18632/oncotarget.2390
36. Sharma S. Immunomodulation: A definitive role of microRNA-142. Dev Comp Immunol. (2017) 77:150–6. doi: 10.1016/j.dci.2017.08.001
37. Wong JJ, Au AY, Gao D, Pinello N, Kwok CT, Thoeng A, et al. RBM3 regulates temperature sensitive miR-142-5p and miR-143 (thermomiRs), which target immune genes and control fever. Nucleic Acids Res. (2016) 44:2888–97. doi: 10.1093/nar/gkw041
38. Motsch N, Alles J, Imig J, Zhu J, Barth S, Reineke T, et al. MicroRNA profiling of Epstein-Barr virus-associated NK/T-cell lymphomas by deep sequencing. PLoS ONE. (2012) 7:e42193. doi: 10.1371/journal.pone.0042193
39. Chen HH, Huang WT, Yang LW, Lin CW. The PTEN-AKT-mTOR/RICTOR pathway in nasal natural killer cell lymphoma is activated by miR-494-3p via PTEN but inhibited by miR-142-3p via RICTOR. Am J Pathol. (2015) 185:1487–99. doi: 10.1016/j.ajpath.2015.01.025
40. Yuan J, Wang K, Xi M. MiR-494 inhibits epithelial ovarian cancer growth by targeting c-Myc. Med Sci Monitor. (2016) 22:617–24. doi: 10.12659/MSM.897288
41. Cheng L, Kong B, Zhao Y, Jiang J. miR-494 inhibits cervical cancer cell proliferation through upregulation of SOCS6 expression. Oncol Lett. (2018) 15:3075–80. doi: 10.3892/ol.2017.7651
42. Yang YK, Xi WY, Xi RX, Li JY, Li Q, Gao YE. MicroRNA-494 promotes cervical cancer proliferation through the regulation of PTEN. Oncol Rep. (2015) 33:2393–401. doi: 10.3892/or.2015.3821
43. Liu Y, Lai L, Chen Q, Song Y, Xu S, Ma F, et al. MicroRNA-494 is required for the accumulation and functions of tumor-expanded myeloid-derived suppressor cells via targeting of PTEN. J Immunol. (2012) 188:5500–10. doi: 10.4049/jimmunol.1103505
44. He Y, Jiang X, Chen J. The role of miR-150 in normal and malignant hematopoiesis. Oncogene. (2014) 33:3887–93. doi: 10.1038/onc.2013.346
45. Bezman NA, Chakraborty T, Bender T, Lanier LL. miR-150 regulates the development of NK and iNKT cells. J Exp Med. (2011) 208:2717–31. doi: 10.1084/jem.20111386
46. Wang F, Ren X, Zhang X. Role of microRNA-150 in solid tumors. Oncol. Lett. (2015) 10:11–6. doi: 10.3892/ol.2015.3170
47. Mitchell JR, Wood E, Collins K. A telomerase component is defective in the human disease dyskeratosis congenita. Nature. (1999) 402:551-5. doi: 10.1038/990141
48. Li T, Niu L, Wu L, Gao X, Li M, Liu W, et al. A functional polymorphism in microRNA-196a2 is associated with increased susceptibility to non-Hodgkin lymphoma. Tumour Biol. (2015) 36:3279–84. doi: 10.1007/s13277-014-2957-y
49. Fehniger TA, Wylie T, Germino E, Leong JW, Magrini VJ, Koul S, et al. Next-generation sequencing identifies the natural killer cell microRNA transcriptome. Genome Res. (2010) 20:1590–604. doi: 10.1101/gr.107995.110
50. Zhou K, Feng X, Wang Y, Liu Y, Tian L, Zuo W, et al. miR-223 is repressed and correlates with inferior clinical features in mantle cell lymphoma through targeting SOX11. Exp Hematol. (2018) 58:27–34.e1. doi: 10.1016/j.exphem.2017.10.005
51. Wang H, Chao K, Ng SC, Bai AH, Yu Q, Yu J, et al. Pro-inflammatory miR-223 mediates the cross-talk between the IL23 pathway and the intestinal barrier in inflammatory bowel disease. Genome Biol. (2016) 17:58. doi: 10.1186/s13059-016-0901-8
52. Ti HJ, Nong L, Wang W, Zhang S, Li T. [Expression of microRNA in extranodal NK/T cell lymphoma, nasal type]. Chin J Pathol. (2011) 40:610–5.
53. Liang L, Nong L, Zhang S, Zhao J, Ti H, Dong Y, et al. The downregulation of PRDM1/Blimp-1 is associated with aberrant expression of miR-223 in extranodal NK/T-cell lymphoma, nasal type. J Exp Clin Cancer Res. (2014) 33:7. doi: 10.1186/1756-9966-33-7
54. Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Nat Acad Sci USA. (2005) 102:13944–9. doi: 10.1073/pnas.0506654102
55. Liu Q, Fu H, Sun F, Zhang H, Tie Y, Zhu J, et al. miR-16 family induces cell cycle arrest by regulating multiple cell cycle genes. Nucleic Acids Res. (2008) 36:5391–404. doi: 10.1093/nar/gkn522
56. Kitadate A, Ikeda S, Teshima K, Ito M, Toyota I, Hasunuma N, et al. MicroRNA-16 mediates the regulation of a senescence-apoptosis switch in cutaneous T-cell and other non-Hodgkin lymphomas. Oncogene. (2016) 35:3692–704. doi: 10.1038/onc.2015.435
57. Ng SB, Yan J, Huang G, Selvarajan V, Tay JL, Lin B, et al. Dysregulated microRNAs affect pathways and targets of biologic relevance in nasal-type natural killer/T-cell lymphoma. Blood. (2011) 118:4919–29. doi: 10.1182/blood-2011-07-364224
58. Komabayashi Y, Kishibe K, Nagato T, Ueda S, Takahara M, Harabuchi Y. Downregulation of miR-15a due to LMP1 promotes cell proliferation and predicts poor prognosis in nasal NK/T-cell lymphoma. Am J Hematol. (2014) 89:25–33. doi: 10.1002/ajh.23570
59. Alles J, Menegatti J, Motsch N, Hart M, Eichner N, Reinhardt R, et al. miRNA expression profiling of Epstein-Barr virus-associated NKTL cell lines by Illumina deep sequencing. FEBS Open Bio. (2016) 6:251–63. doi: 10.1002/2211-5463.12027
60. Paik JH, Jang JY, Jeon YK, Kim WY, Kim TM, Heo DS, et al. MicroRNA-146a downregulates NFkappaB activity via targeting TRAF6 and functions as a tumor suppressor having strong prognostic implications in NK/T cell lymphoma. Clin Cancer Res. (2011) 17:4761–71. doi: 10.1158/1078-0432.CCR-11-0494
61. Zhou Z, Li Z, Sun Z, Zhang X, Lu L, Wang Y, et al. S100A9 and ORM1 serve as predictors of therapeutic response and prognostic factors in advanced extranodal NK/T cell lymphoma patients treated with pegaspargase/gemcitabine. Sci Rep. (2016) 6:23695. doi: 10.1038/srep23695
62. Kanavaros P, Lescs MC, Brière J, Divine M, Galateau F, Joab I, et al. Nasal T-cell lymphoma: a clinicopathologic entity associated with peculiar phenotype and with Epstein-Barr virus. Blood. (1993) 81:2688–95.
63. Zhang L, Hong K, Zhang J, Pagano JS. Multiple signal transducers and activators of transcription are induced by EBV LMP-1. Virology. (2004) 323:141–52. doi: 10.1016/j.virol.2004.03.007
64. Imig J, Motsch N, Zhu JY, Barth S, Okoniewski M, Reineke T, et al. microRNA profiling in Epstein-Barr virus-associated B-cell lymphoma. Nucleic Acids Res. (2011) 39:1880–93. doi: 10.1093/nar/gkq1043
65. Huang WT, Lin CW. EBV-encoded miR-BART20-5p and miR-BART8 inhibit the IFN-gamma-STAT1 pathway associated with disease progression in nasal NK-cell lymphoma. Am J Pathol. (2014) 184:1185–97. doi: 10.1016/j.ajpath.2013.12.024
66. Pfeffer S, Zavolan M, Grasser FA, Chien M, Russo JJ, Ju J, et al. Identification of virus-encoded microRNAs. Science. (2004) 304:734–6. doi: 10.1126/science.1096781
67. Wakisaka N, Pagano JS. Epstein-Barr virus induces invasion and metastasis factors. Anticancer Res. (2003) 23:2133–8.
68. Eliopoulos AG, Dawson CW, Mosialos G, Floettmann JE, Rowe M, Armitage RJ, et al. CD40-induced growth inhibition in epithelial cells is mimicked by Epstein-Barr Virus-encoded LMP1: involvement of TRAF3 as a common mediator. Oncogene. (1996) 13:2243–54.
69. Takada H, Imadome KI, Shibayama H, Yoshimori M, Wang L, Saitoh Y, et al. EBV induces persistent NF-kappaB activation and contributes to survival of EBV-positive neoplastic T- or NK-cells. PLoS ONE. (2017) 12:e0174136. doi: 10.1371/journal.pone.0174136
70. Sun L, Zhao Y, Shi H, Ma C, Wei L. LMP1 promotes nasal NK/T-cell lymphoma cell function by eIF4E via NF-kappaB pathway. Oncol. Rep. (2015) 34:3264–71. doi: 10.3892/or.2015.4305
71. Sun L, Zhao Y, Shi H, Ma C, Wei L. LMP-1 induces survivin expression to inhibit cell apoptosis through the NF-kappaB and PI3K/Akt signaling pathways in nasal NK/T-cell lymphoma. Oncol Rep. (2015) 33:2253–60. doi: 10.3892/or.2015.3847
72. Ramakrishnan R, Donahue H, Garcia D, Tan J, Shimizu N, Rice AP, et al. Epstein-Barr virus BART9 miRNA modulates LMP1 levels and affects growth rate of nasal NK T cell lymphomas. PLoS ONE. (2011) 6:e27271. doi: 10.1371/journal.pone.0027271
73. Komabayashi Y, Kishibe K, Nagato T, Ueda S, Takahara M, Harabuchi Y. Circulating Epstein-Barr virus-encoded micro-RNAs as potential biomarkers for nasal natural killer/T-cell lymphoma. Hematol Oncol. (2017) 35:655–63. doi: 10.1002/hon.2360
74. Jenner RG, Townsend MJ, Jackson I, Sun K, Bouwman RD, Young RA, et al. The transcription factors T-bet and GATA-3 control alternative pathways of T-cell differentiation through a shared set of target genes. Proc Nat Acad Sci USA. (2009) 106:17876–81. doi: 10.1073/pnas.0909357106
75. Steiner DF, Thomas MF, Hu JK, Yang Z, Babiarz JE, Allen CD, et al. MicroRNA-29 regulates T-box transcription factors and interferon-gamma production in helper T cells. Immunity. (2011) 35:169–81. doi: 10.1016/j.immuni.2011.07.009
76. Naiche LA, Harrelson Z, Kelly RG, Papaioannou VE. T-box genes in vertebrate development. Ann Rev Genet. (2005) 39:219–39. doi: 10.1146/annurev.genet.39.073003.105925
77. Garzon R, Croce CM. MicroRNAs in normal and malignant hematopoiesis. Curr Opin Hematol. (2008) 15:352–8. doi: 10.1097/MOH.0b013e328303e15d
78. Guo HQ, Huang GL, Guo CC, Pu XX, Lin TY. Diagnostic and prognostic value of circulating miR-221 for extranodal natural killer/T-cell lymphoma. Disease Mark. (2010) 29:251–8. doi: 10.1155/2010/474692
79. Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. (2007) 26:745–52. doi: 10.1016/j.molcel.2007.05.010
80. Chim CS, Wong KY, Qi Y, Loong F, Lam WL, Wong LG, et al. Epigenetic inactivation of the miR-34a in hematological malignancies. Carcinogenesis. (2010) 31:745–50. doi: 10.1093/carcin/bgq033
81. Li C, Chen J, Zhang K, Feng B, Wang R, Chen L. Progress and prospects of long noncoding RNAs (lncRNAs) in hepatocellular carcinoma. Cell Physiol Biochem. (2015) 36:423–34. doi: 10.1159/000430109
82. Shi X, Wu Y, Ai Z, Liu X, Yang L, Du J, et al. AICAR sustains J1 mouse embryonic stem cell self-renewal and pluripotency by regulating transcription factor and epigenetic modulator expression. Cell Physiol Biochem. (2013) 32:459–75. doi: 10.1159/000354451
83. Li DX, Fei XR, Dong YF, Cheng CD, Yang Y, Deng XF, et al. The long non-coding RNA CRNDE acts as a ceRNA and promotes glioma malignancy by preventing miR-136-5p-mediated downregulation of Bcl-2 and Wnt2. Oncotarget. (2017) 8:88163–78. doi: 10.18632/oncotarget.21513
84. Zhang T, Wu DM, Deng SH, Han R, Liu T, Li J, et al. Integrated analysis reveals that long non-coding RNA TUBA4B can be used as a prognostic biomarker in various cancers. Cell Physiol Biochem. (2018) 49:530–44. doi: 10.1159/000492991
85. Baytak E, Gong Q, Akman B, Yuan H, Chan WC, Kucuk C. Whole transcriptome analysis reveals dysregulated oncogenic lncRNAs in natural killer/T-cell lymphoma and establishes MIR155HG as a target of PRDM1. Tumour Biol. (2017) 39:1010428317701648. doi: 10.1177/1010428317701648
86. Askarian-Amiri ME, Crawford J, French JD, Smart CE, Smith MA, Clark MB, et al. SNORD-host RNA Zfas1 is a regulator of mammary development and a potential marker for breast cancer. RNA. (2011) 17:878–91. doi: 10.1261/rna.2528811
87. Zhang Z, Weaver DL, Olsen D, deKay J, Peng Z, Ashikaga T, et al. Long non-coding RNA chromogenic in situ hybridisation signal pattern correlation with breast tumour pathology. J Clin Pathol. (2016) 69:76–81. doi: 10.1136/jclinpath-2015-203275
88. Gao K, Ji Z, She K, Yang Q, Shao L. Long non-coding RNA ZFAS1 is an unfavourable prognostic factor and promotes glioma cell progression by activation of the Notch signaling pathway. Biomed. Pharmacother. (2017) 87:555–60. doi: 10.1016/j.biopha.2017.01.014
89. Thorenoor N, Faltejskova-Vychytilova P, Hombach S, Mlcochova J, Kretz M, Svoboda M, et al. Long non-coding RNA ZFAS1 interacts with CDK1 and is involved in p53-dependent cell cycle control and apoptosis in colorectal cancer. Oncotarget. (2016) 7:622–37. doi: 10.18632/oncotarget.5807
90. Zhou H, Wang F, Chen H, Tan Q, Qiu S, Chen S, et al. Increased expression of long-noncoding RNA ZFAS1 is associated with epithelial-mesenchymal transition of gastric cancer. Aging. (2016) 8:2023–38. doi: 10.18632/aging.101048
91. Li T, Xie J, Shen C, Cheng D, Shi Y, Wu Z, et al. Amplification of long noncoding RNA ZFAS1 promotes metastasis in hepatocellular carcinoma. Cancer Res. (2015) 75:3181–91. doi: 10.1158/0008-5472.CAN-14-3721
92. Xia B, Hou Y, Chen H, Yang S, Liu T, Lin M, et al. Long non-coding RNA ZFAS1 interacts with miR-150-5p to regulate Sp1 expression and ovarian cancer cell malignancy. Oncotarget. (2017) 8:19534–46. doi: 10.18632/oncotarget.14663
93. Guo H, Wu L, Zhao P, Feng A. Overexpression of long non-coding RNA zinc finger antisense 1 in acute myeloid leukemia cell lines influences cell growth and apoptosis. Exp Therap Med. (2017) 14:647–51. doi: 10.3892/etm.2017.4535
94. Ji P, Diederichs S, Wang W, Boing S, Metzger R, Schneider PM, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. (2003) 22:8031–41. doi: 10.1038/sj.onc.1206928
95. Hu L, Wu Y, Tan D, Meng H, Wang K, Bai Y, et al. Up-regulation of long noncoding RNA MALAT1 contributes to proliferation and metastasis in esophageal squamous cell carcinoma. J Exp Clin Cancer Res. (2015) 34:7. doi: 10.1186/s13046-015-0123-z
96. Wu XS, Wang XA, Wu WG, Hu YP, Li ML, Ding Q, et al. MALAT1 promotes the proliferation and metastasis of gallbladder cancer cells by activating the ERK/MAPK pathway. Cancer Biol Therap. (2014) 15:806–14. doi: 10.4161/cbt.28584
97. Ji Q, Zhang L, Liu X, Zhou L, Wang W, Han Z, et al. Long non-coding RNA MALAT1 promotes tumour growth and metastasis in colorectal cancer through binding to SFPQ and releasing oncogene PTBP2 from SFPQ/PTBP2 complex. Br J Cancer. (2014) 111:736–48. doi: 10.1038/bjc.2014.383
98. Ma KX, Wang HJ, Li XR, Li T, Su G, Yang P, et al. Long noncoding RNA MALAT1 associates with the malignant status and poor prognosis in glioma. Tumour Biol. (2015) 36:3355–9. doi: 10.1007/s13277-014-2969-7
99. Kim SH, Kim SH, Yang WI, Kim SJ, Yoon SO. Association of the long non-coding RNA MALAT1 with the polycomb repressive complex pathway in T and NK cell lymphoma. Oncotarget. (2017) 8:31305–17. doi: 10.18632/oncotarget.15453
Keywords: non-coding RNAs, microRNAs, EBV-encoded miRNAs, lncRNAs, natural killer/T-cell lymphoma (NKTCL)
Citation: Mei M and Zhang M (2019) Non-coding RNAs in Natural Killer/T-Cell Lymphoma. Front. Oncol. 9:515. doi: 10.3389/fonc.2019.00515
Received: 23 February 2019; Accepted: 29 May 2019;
Published: 13 June 2019.
Edited by:
Anjali Mishra, Sidney Kimmel Cancer Center, United StatesReviewed by:
Jun-ichi Kawada, Nagoya University Hospital, JapanVemika Chandra, Children's Hospital of Philadelphia, United States
Copyright © 2019 Mei and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mingzhi Zhang, bWluZ3poaV96aGFuZzFAMTYzLmNvbQ==
 Mei Mei
Mei Mei Mingzhi Zhang
Mingzhi Zhang