- 1Institute of Medicine, Chung Shan Medical University, Taichung, Taiwan
- 2Cancer Research Center, Changhua Christian Hospital, Changhua, Taiwan
- 3Department of Otorhinolaryngology-Head and Neck Surgery, Changhua Christian Hospital, Changhua, Taiwan
- 4Institute of Oral Sciences, Chung Shan Medical University, Taichung, Taiwan
- 5Department of Dentistry, Chung Shan Medical University Hospital, Taichung, Taiwan
- 6School of Medicine, Chung Shan Medical University, Taichung, Taiwan
- 7Department of Otolaryngology, Chung Shan Medical University Hospital, Taichung, Taiwan
- 8Department of Biomedical Sciences, College of Medicine Sciences and Technology, Chung Shan Medical University, Taichung, Taiwan
- 9Department of Medical Research, Chung Shan Medical University Hospital, Taichung, Taiwan
Long pentraxin 3 (PTX3) is produced by various cell types and is correlated with tumor progression in various tumor types. However, the clinical significance of PTX3 polymorphisms in oral cancer and their correlation with the risk of cancer are still unclear. In this study, we assessed the influence of PTX3 gene polymorphisms and environmental factors on susceptibility to oral tumorigenesis. We recruited 865 cases with oral cancer and 1,189 controls. Four single-nucleotide variations of the PTX3 gene (rs1840680, rs2305619, rs3816527, and rs2120243) were verified using a real-time polymerase chain reaction in control participants and cases with oral cancer. We found that rs3816527 in smokers was correlated with the development of late-stage cancer (odds ratio [OR], 2.328; 95% confidence interval [CI], 1.078–5.027) and increased lymph node metastasis (OR, 2.152; 95% CI, 1.047–4.422). Moreover, additional bioinformatics analysis results showed that the rs3816527 C allele variant to the A allele exhibited the strongest exonic splicing enhancer activity. In conclusion, our results suggested that PTX3 rs3816527 plays a role in oral cancer development.
Introduction
The incidence of oral cancer in Asia and more particularly Taiwan increases annually. In addition to tobacco smoking and alcohol consumption, betel quid chewing is a major cause of oral cancer (1–4). Moreover, the annual number of deaths due to oral cancer among men is increasing rapidly (5). The global 5 year mortality rate of oral cancer is ~50%. Although considerable progress has been made in surgery, chemotherapy, and radiotherapy, no considerable improvements have been made in the preceding 50 years (6).
Pentraxins (PTXs) are a superfamily of conserved proteins that contain the pentraxin domain. Proteins of the pentraxin family are multifunctional and participate in acute immunological responses (7). PTX3, also known as TGF14, is a transmembrane molecule expressed in a variety of human tissues (8, 9). PTX3 is secreted by natural immune cells in response to inflammatory cytokines, such as interleukin-1 β (IL-1β) and tumor necrosis factor α (TNFα), or selected pathogen-associated molecular patterns (9–11). Therefore, PTX3 may play a vital role at the crossroads of inflammation increase (12, 13), innate immunity (14–16), tissue repair stimulation (17, 18), and cancer (19–21). PTX3 promotes cell migration and invasion in several cancers, and the expression of PTX3 correlates with tumor progression in various human tumor types (19, 22–24).
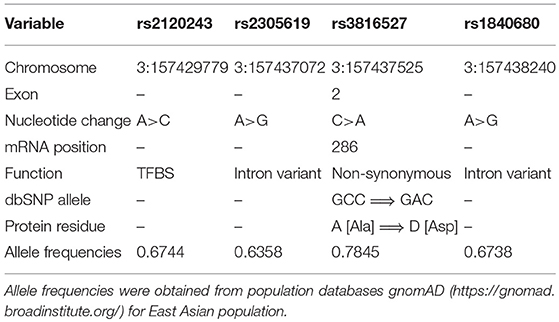
Genetic variations contribute to susceptibility to common diseases such as cardiovascular disease, diabetes, inflammatory disease, and cancer (25–29). Single-nucleotide variations (SNVs) may be a causative genetic variant that can affect the expression and structure of proteins, thereby directly contributing to disease (30). The PTX3 gene is located on chromosome 3 and contains three exons and two introns. Previous studies have described that SNVs in PTX3 (rs2305619 and rs1840680) have functional significance. Results indicated that the A allele of PTX3 SNVs (rs2305619 and rs1840680) is associated with higher plasma levels of PTX3 (31, 32). In addition, the A alleles of rs2305619 and rs1840680 are associated with susceptibility to Pseudomonas aeruginosa and Mycobacterium tuberculosis infections (33, 34). Carmo et al. also revealed that genetic variations in PTX3 (rs2305619 and rs1840680) and plasma levels were associated with hepatocellular carcinoma (35). Moreover, Hakelius et al. indicated that PTX3 played a major role in non-malignant and oral cancer malignant disease processes (36). However, few studies have investigated the association of PTX3 polymorphisms in oral cancer. Therefore, we investigated the relationship between four PTX3 gene polymorphisms (rs1840680, rs2305619, rs3816527, and rs2120243; Table 1) and clinicopathological characteristics of patients with oral cancer to identify those with an increased risk of oral cancer.
Materials and Methods
Study Population
The participants of this case–control study were 865 male cases with oral cancer of squamous cell carcinoma recruited from Changhua Christian Hospital in Changhua and Chung Shan Medical University Hospital in Taichung, Taiwan, between 2007 and 2017, and 1,189 cancer-free male controls selected from the Taiwan Biobank. The oral cancers of squamous cell carcinoma in this study were pre-specified to include any cancers that originated from buccal mucosa (n = 314; 36.3%), tongue (n = 268; 31.0%), gingiva (n = 78; 9.0%), palate (n = 40; 4.6%), floor of the mouth (n = 25; 2.9%), and other areas (n = 140; 16.2%). The Institutional Review Board of Chung Shan Medical University approved this study (CSMUH No: CS13214-1). All participants provided written informed consent. Personal characteristics and information, including demographic characteristics; tobacco smoking, betel quid chewing, and alcohol consumption habits; and the medical histories of the participants, were investigated using interviewer-administered questionnaires.
Determination of Genotypes
Genomic DNA from peripheral blood leukocytes was extracted using a QIAamp DNA Blood Mini Kit (Qiagen, Valencia, CA, USA) following the manufacturer's protocol (37). DNA was dissolved in ethylenediaminetetraacetic acid (EDTA) buffer (10 mM Tris, 1 mM EDTA; pH 7.8) and then quantified by measuring absorbance at 260 nm. Finally, the preparation was stored in a −20°C refrigerator and later analyzed using a real-time polymerase chain reaction (PCR) system. Allelic discrimination for the PTX3 SNV was performed using a TaqMan assay (ID C_12069244_10 for rs1840680, C_22275654_10 for rs2305619, C_3035766_30 for rs3816527, and C_11796613_20 for rs2120243) with an ABI StepOne Real-Time PCR System (Applied Biosystems). The genotypic frequencies of PTX3 were further evaluated using the SDS v3.0 software program.
Bioinformatics Analysis
Several bioinformatics tools were used to assess the putative functional relevance of the rs3816527 PTX3 polymorphism. SNPinfo was used to predict the function of the rs351855 polymorphism. Splicing enhancement activity was analyzed using ESEfinder. We used the National Center for Biotechnology Information (NCBI) database to determine splicing types of PTX3. Furthermore, protein structure homology modeling of PTX3 was performed using SWISS-MODEL.
Statistical Analysis
The Mann–Whitney U-test was used to compare differences in demographic characteristics between healthy controls and cases with oral cancer. Multiple logistic regression models were used to determine the association between genotypic frequencies and oral cancer risk after adjustment for age, betel quid chewing, cigarette smoking, and alcohol consumption. Data were analyzed using SAS 9.1 statistical software. A p <0.05 was considered significant.
Results
Demographic Characteristics of the Participants
Table 2 displays the results of the statistical analysis of the participants' demographic characteristics. A total of 2,054 participants were enrolled, namely 865 male cases with oral cancer and 1,189 male controls. Results revealed significant differences in cigarette smoking (p < 0.001), betel quid chewing (p < 0.001), and alcohol consumption (p < 0.001) between the cases with oral cancer and the controls.
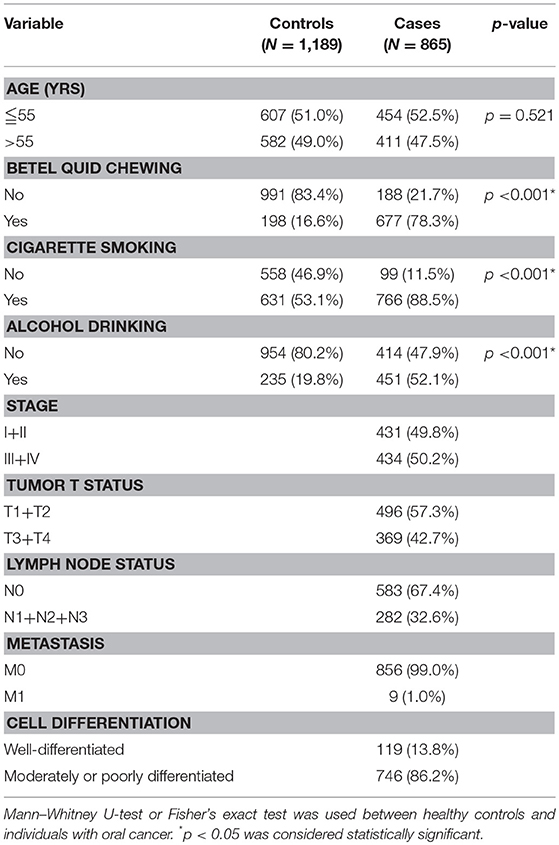
Table 2. Distribution of demographic characteristics in 1,189 controls and 865 male individuals with oral cancer.
PTX3 Gene Polymorphisms in Cases With Oral Cancer and Controls
To investigate the association between PTX3 gene polymorphisms and oral cancer risk, the genotypic, and allelic frequencies of PTX3 in the individuals with oral cancer and the controls were established in this investigation (Table 3). After adjustment for betel quid chewing, cigarette smoking, and alcohol consumption, no significant difference was observed between the participants with oral cancer who had rs1840680, rs2305619, rs3816527, and rs2120243 polymorphisms of the PTX3 gene and those with wild-type (WT) genes.
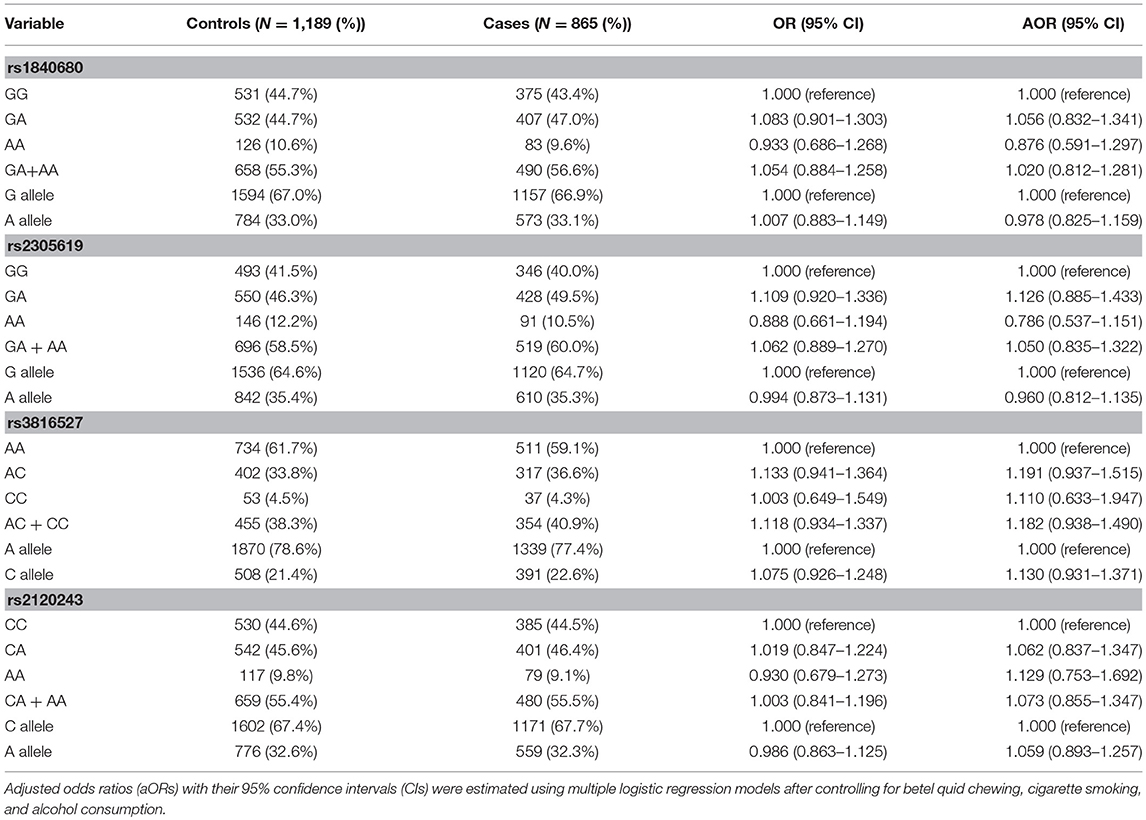
Table 3. Genotyping and allelic frequency of PTX3 single-nucleotide variations (SNVs) in individuals with oral cancer and controls.
Combined Effects of Environmental Factors and PTX3 Gene Polymorphisms on Oral Cancer
To determine the combined effects of environmental factors and PTX3 gene SNVs on oral cancer susceptibility, we conducted further analysis on 1,397 smokers (Table 4). As presented in Table 4, participants with at least one A allele of rs1840680, one A allele of rs2305619, one C allele of rs3816527, or one A allele of rs2120243 exhibited 12.622-fold (95% CI: 8.725–18.259), 13.171-fold (95% CI: 8.944–19.395), 13.798-fold (95% CI: 9.538–19.960), and 12.794-fold (95% CI: 8.787–18.628) higher risks of oral cancer, respectively, compared with individuals with WT homozygotes who did not chew betel quid.
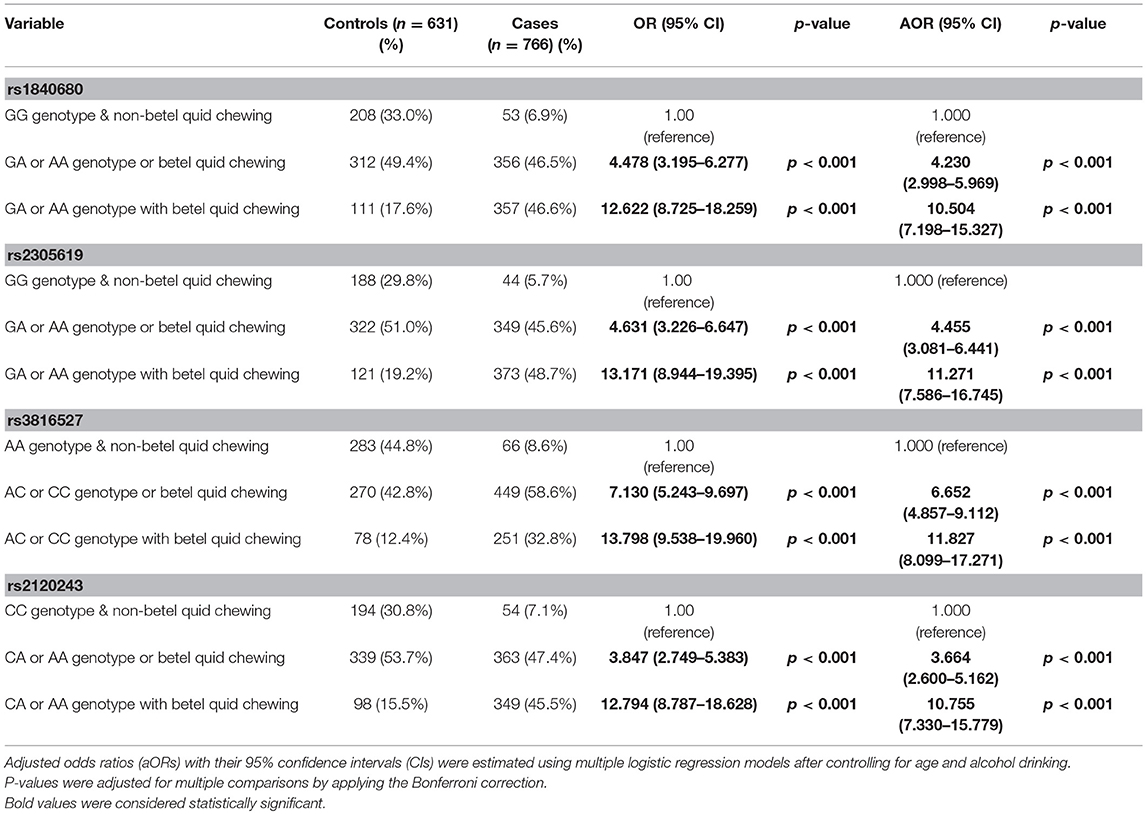
Table 4. Association of the combined effect of PTX3 gene polymorphisms and betel quid chewing with susceptibility to oral cancer among 1,397 smokers.
Effects of Polymorphic Genotypes of PTX3 on Clinical Status of Oral Cancer
We analyzed the relationship between the combined effect of cigarette smoking and PTX3 variants on oral cancer development. As depicted in Table 5, individuals who possessed the CC allele of rs3816527 and smoked cigarettes were more prone to developing late-stage tumors (stage III/IV: OR, 2.328; 95% CI, 1.078–5.027; p = 0.0314) and lymph node metastasis (OR, 2.152; 95% CI, 1.047–4.422; p = 0.0371) compared with individuals who were homozygous for the WT allele of rs3816527 (Table 5).
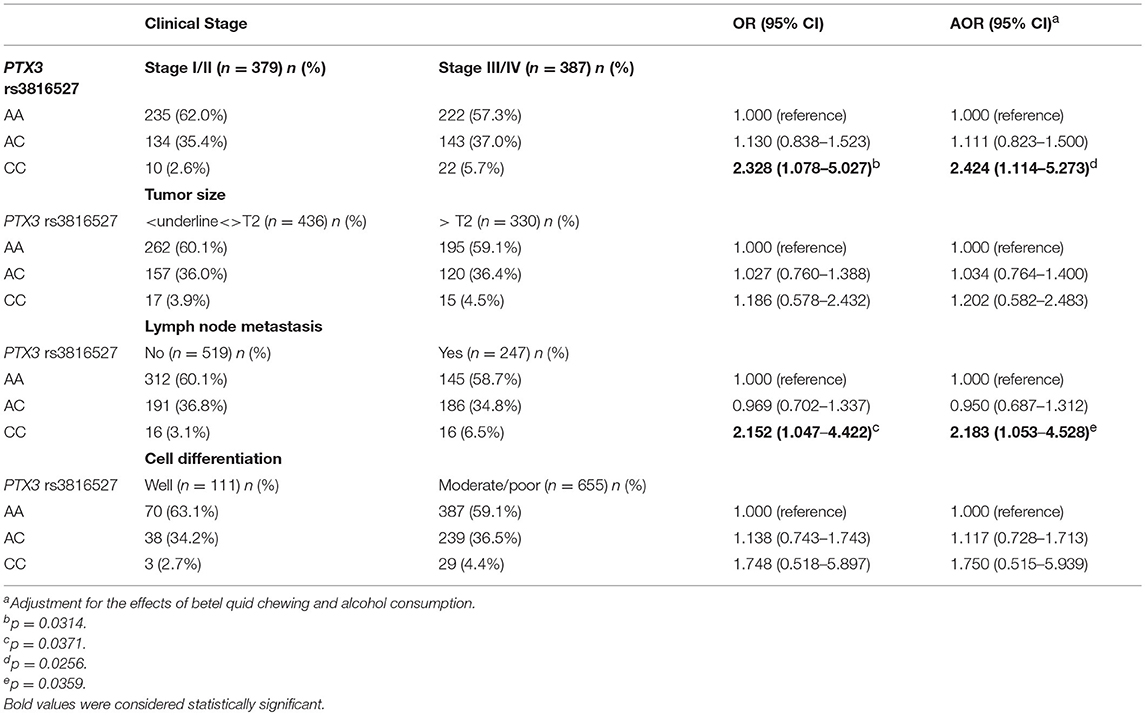
Table 5. Genotyping frequency of the PTX3 rs3816527 polymorphism on clinical status of oral cancer among 766 smokers.
Functional Connotation of the PTX3 rs3816527 Locus
We investigated the functional connotation of the rs3816527 SNV on the PTX3 gene. The function of PTX3 rs3816527 was performed using several bioinformatics tools, namely SNPinfo, the NCBI database, ESEfinder, and SWISS-MODEL. Data indicated that PTX3 rs3816527 was located on the exonic splice enhancer sequence of the PTX3 gene (Figure 1A). The C allele had stronger enhancement activity than did the A allele and may be more likely to promote the splicing modification of the PTX3 gene (Figure 1B). Two splicing forms of PTX3 existed in the NCBI database (Figure 1C).

Figure 1. Functional prediction profiling of PTX3 SNV rs3816527. (A) The sequence of PTX3 is from the NCBI reference sequences (RefSeq) (NM_002852.3). The cis-elements (5′ splice site, 3′ splice site, and branch site) and exonic splice enhancer (ESE) are indicated by various color squares. PTX3 SNV rs3816527 is located on the ESE sequence. (B) The functional prediction of rs3816527 from SNPinfo web server is presented in this Figure. The relative frequency of the nucleotide for the ESE sequence is illustrated by the sequence logo. (C) PTX3 gene consists of three exons. The first exon encodes the leader peptide, and the second and third exons encode N-terminal and C-terminal domains, respectively. Two common splicing forms of PTX3 are accessed from the NCBI database.
Discussion
Head and neck cancers are common worldwide. Approximately 90% of head and neck cancers are squamous cell carcinomas, of which ~50% occur in the oral cavity (38, 39). In south Central Asia, oral cancer is the third most common type of cancer (40). Oral cancer is associated with the use of chronic stimuli such as tobacco smoking, alcohol consumption, and betel quid chewing in particular (3, 4, 41). PTX3 was recognized by proteomics as a critical candidate biomarker for liposarcoma (42), lung cancer (43), prostate cancer (44), and pancreatic cancer (22). For example, a high expression of PTX3 was found in pancreatic cancer cell lines and a direct relationship was found between tumor metastasis and PTX3 expression (22). Moreover, several studies have reported the role of PTX3 in epithelial cancer progression due to EMT induction (45–48). A low expression of PTX3 has been associated with increased susceptibility to epithelial carcinogenesis (46). Previous studies have demonstrated that PTX3 mediated the induction of the EMT by reducing the expression of E-cadherin, increasing the expression of N-cadherin and vimentin, and promoting the migration of HK-2 cells (45). Moreover, results indicated that the expression of pentraxin family members was significantly associated with the poor prognosis of patients with pancreatic cancer (22). Furthermore, overexpression of PTX3 could promote the proliferation and invasion of cervical cancer in vitro and in vivo (24). However, few studies have discussed the role of PTX3 in oral cancer. In the present study, the combined effect of environmental factors and PTX3 polymorphisms considerably increased the risk of oral cancer (Table 4). Moreover, patients with PTX3 SNV rs3816527 with CC had the highest risk of tumors (Table 5).
The occurrence of cancer stems from the genetic and epigenetic alterations of the basic mechanisms of the normal cell cycle, such as replication control and cell death (49, 50). Most genes in the human genome consist of multiple introns and exons that are modified through splicing to form mature messenger ribonucleic acid and protein products (51, 52). Although alternative splicing (AS) provides cells with protein diversity, studies have found that pathological changes in splicing can contribute to the development of cancers. For instance, mutations and gene expression changes affect the splicing regulatory sequences of key cancer-related genes (53) and the core or accessory components of spliceosome complexes (54–59).
PTX3 has two splice variants; one contains exon 1 or 2 (length of 453 bp) and the other contains exon 3 (length of 931 bp) (Figure 1C). ESEfinder analysis results revealed that the rs3816527 C allele had superior splicing enhancement activity for the A allele (Figure 1B). In addition, in the protein structure constructed by SWISS-MODEL, PTX3 was split into an N terminus and C terminus after splicing. Although the mechanism of action for this structure remains unclear, a molecular study indicated that the overexpression of PTX3 at the N terminus can considerably inhibit the oncogenic activity of transgenic adenocarcinoma mouse prostate-C2 transfectants, whereas C-terminal overexpression has only a minor effect on tumor growth (60).
We revealed an impact of PTX3 gene variations on the development of oral cancer; however, there are some limitations in the study. As this study only included a discovery population and not a second independent study to replicate the findings, the associations between PTX3 variations and oral cancer should be considered preliminary. The other concern is that we failed to exclude the possibility of potential selection bias and since the control group was enrolled among subjects without cancer on a hospital basis. In addition, determining the functional role of PTX3 in the development of oral cancer still requires further investigation.
In conclusion, our results suggest that the allelic effects of PTX3 SNVs (rs1840680, rs2305619, rs3816527, and rs2120243) enhance the risk and progression of oral cancer in the presence of environmental factors such as tobacco smoking and betel quid chewing. This genetic association was observed most markedly in smokers. These results expose a novel genetic–environmental predisposition to oral cancer carcinogenesis.
Data Availability
The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.
Ethics Statement
The Institutional Review Board of Chung Shan Medical University approved this study (CSMUH No: CS13214-1). All participants provided informed consent.
Author Contributions
C-MY, C-WL, S-FY, and M-KC contributed to conception, design and critically revised the manuscript. C-MY, S-FY, and M-KC contributed to conception and drafted the manuscript, C-YC, Y-FL, and C-HC contributed to performed experiments and analyzed data.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was supported by grants from Ministry of Science and Technology, Taiwan (MOST-106-2314-B-371-005-MY3).
Abbreviations
PTX3, long pentraxin 3; OSCC, oral squamous cell carcinoma; OR, odds ratio; SNV, single-nucleotide variation.
References
1. Chiba I. Prevention of Betel Quid Chewers' Oral Cancer in the Asian-Pacific Area. Asian Pac J Cancer Prev. (2001) 2:263−9.
2. Jeng JH, Chang MC, Hahn LJ. Role of areca nut in betel quid-associated chemical carcinogenesis: current awareness and future perspectives. Oral Oncol. (2001) 37:477–92. doi: 10.1016/S1368-8375(01)00003-3
3. Su SC, Lin CW, Liu YF, Fan WL, Chen MK, Yu CP, et al. Exome sequencing of oral squamous cell carcinoma reveals molecular subgroups and novel therapeutic opportunities. Theranostics. (2017) 7:1088–99. doi: 10.7150/thno.18551
4. Yang SF, Huang HD, Fan WL, Jong YJ, Chen MK, Huang CN, et al. Compositional and functional variations of oral microbiota associated with the mutational changes in oral cancer. Oral Oncol. (2018) 77:1–8. doi: 10.1016/j.oraloncology.2017.12.005
5. Petti S, Scully C. How many individuals must be screened to reduce oral cancer mortality rate in the Western context? Chall Oral Dis. (2015) 21:949–54. doi: 10.1111/odi.12372
6. Miller C, Shay A, Tajudeen B, Sen N, Fidler M, Stenson K, et al. Clinical features and outcomes in young adults with oral tongue cancer. Am J Otolaryngol. (2019) 40:93–6. doi: 10.1016/j.amjoto.2018.09.022
7. Gewurz H, Zhang XH, Lint TF. Structure and function of the pentraxins. Curr Opin Immunol. (1995) 7:54–64. doi: 10.1016/0952-7915(95)80029-8
8. Garlanda C, Bottazzi B, Bastone A, Mantovani A. Pentraxins at the crossroads between innate immunity, inflammation, matrix deposition, and female fertility. Ann Rev Immunol. (2005) 23:337–66. doi: 10.1146/annurev.immunol.23.021704.115756
9. Lee GW, Lee TH, Vilcek J. TSG-14, a tumor necrosis factor- and IL-1-inducible protein, is a novel member of the pentaxin family of acute phase proteins. J Immunol. (1993) 150:1804–12.
10. Breviario F, d'Aniello EM, Golay J, Peri G, Bottazzi B, Bairoch A, et al. Interleukin-1-inducible genes in endothelial cells. Cloning of a new gene related to C-reactive protein and serum amyloid P component. J Biol Chem. (1992) 267:22190–7.
11. Vouret-Craviari V, Matteucci C, Peri G, Poli G, Introna M, Mantovani A. Expression of a long pentraxin, PTX3, by monocytes exposed to the mycobacterial cell wall component lipoarabinomannan. Infect Immun. (1997) 65:1345–50. doi: 10.1006/cyto.1997.0242
12. Bonavita E, Gentile S, Rubino M, Maina V, Papait R, Kunderfranco P, et al. PTX3 is an extrinsic oncosuppressor regulating complement-dependent inflammation in cancer. Cell. (2015) 160:700–14. doi: 10.1016/j.cell.2015.01.004
13. Daigo K, Mantovani A, Bottazzi B. The yin-yang of long pentraxin PTX3 in inflammation and immunity. Immunol Lett. (2014) 161:38–43. doi: 10.1016/j.imlet.2014.04.012
14. Jaillon S, Moalli F, Ragnarsdottir B, Bonavita E, Puthia M, Riva F, et al. The humoral pattern recognition molecule PTX3 is a key component of innate immunity against urinary tract infection. Immunity. (2014) 40:621–32. doi: 10.1016/j.immuni.2014.02.015
15. Kabbani D, Bhaskaran A, Singer LG, Bhimji A, Rotstein C, Keshavjee S, et al. Pentraxin 3 levels in bronchoalveolar lavage fluid of lung transplant recipients with invasive aspergillosis. J Heart Lung Transpl. (2017) 36:973–9. doi: 10.1016/j.healun.2017.04.007
16. Zhang J, Zhao GQ, Qu J, Lin J, Che CY, Yang XJ. Early expression of PTX3 in Aspergillus fumigatus infected rat cornea. Inter J Ophthalmol. (2018) 11:1084–9. doi: 10.18240/ijo.2018.07.02
17. Doni A, Garlanda C, Mantovani A. PTX3 orchestrates tissue repair. Oncotarget. (2015) 6:30435–6. doi: 10.18632/oncotarget.5453
18. Doni A, Musso T, Morone D, Bastone A, Zambelli V, Sironi M, et al. An acidic microenvironment sets the humoral pattern recognition molecule PTX3 in a tissue repair mode. J Exp Med. (2015) 212:905–25. doi: 10.1084/jem.20141268
19. Chang WC, Wu SL, Huang WC, Hsu JY, Chan SH, Wang JM, et al. PTX3 gene activation in EGF-induced head and neck cancer cell metastasis. Oncotarget. (2015) 6:7741–57. doi: 10.18632/oncotarget.3482
20. Garlanda C, Bottazzi B, Magrini E, Inforzato A, Mantovani A. PTX3, a humoral pattern recognition molecule in innate immunity, tissue repair, and cancer. Physiol Rev. (2018) 98:623–39. doi: 10.1152/physrev.00016.2017
21. Stallone G, Cormio L, Netti GS, Infante B, Selvaggio O, Fino GD, et al. Pentraxin 3: a novel biomarker for predicting progression from prostatic inflammation to prostate cancer. Cancer Res. (2014) 74:4230–8. doi: 10.1158/0008-5472.CAN-14-0369
22. Kondo S, Ueno H, Hosoi H, Hashimoto J, Morizane C, Koizumi F, et al. Clinical impact of pentraxin family expression on prognosis of pancreatic carcinoma. Br J Cancer. (2013) 109:739–46. doi: 10.1038/bjc.2013.348
23. Tung JN, Ko CP, Yang SF, Cheng CW, Chen PN, Chang CY, et al. Inhibition of pentraxin 3 in glioma cells impairs proliferation and invasion in vitro and in vivo. J Neuro Oncol. (2016) 129:201–9. doi: 10.1007/s11060-016-2168-z
24. Ying TH, Lee CH, Chiou HL, Yang SF, Lin CL, Hung CH, et al. Knockdown of Pentraxin 3 suppresses tumorigenicity and metastasis of human cervical cancer cells. Sci Rep. (2016) 6:29385. doi: 10.1038/srep29385
25. Eccles D, Tapper W. The influence of common polymorphisms on breast cancer. Cancer Treat Res. (2010) 155:15–32. doi: 10.1007/978-1-4419-6033-7_2
26. Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. (2000) 100:57–70. doi: 10.1016/S0092-8674(00)81683-9
27. Hua KT, Liu YF, Hsu CL, Cheng TY, Yang CY, Chang JS, et al. 3'UTR polymorphisms of carbonic anhydrase IX determine the miR-34a targeting efficiency and prognosis of hepatocellular carcinoma. Sci Rep. (2017) 7:4466. doi: 10.1038/s41598-017-04732-3
28. Schmith VD, Campbell DA, Sehgal S, Anderson WH, Burns DK, Middleton LT, et al. Pharmacogenetics and disease genetics of complex diseases. Cell Mol Life Sci. (2003) 60:1636–46. doi: 10.1007/s00018-003-2369-4
29. Su SC, Hsieh MJ, Lin CW, Chuang CY, Liu YF, Yeh CM, et al. Impact of HOTAIR gene polymorphism and environmental risk on oral cancer. J Dent Res. (2018) 97:717–24. doi: 10.1177/0022034517749451
30. Sunyaev S, Ramensky V, Koch I, Lathe W 3rd, Kondrashov AS, Bork P. Prediction of deleterious human alleles. Hum Mol Genet. (2001) 10:591–7. doi: 10.1093/hmg/10.6.591
31. Barbati E, Specchia C, Villella M, Rossi ML, Barlera S, Bottazzi B, et al. Influence of pentraxin 3 (PTX3) genetic variants on myocardial infarction risk and PTX3 plasma levels. PLoS ONE. (2012) 7:e53030. doi: 10.1371/journal.pone.0053030
32. Diamond JM, Meyer NJ, Feng R, Rushefski M, Lederer DJ, Kawut SM, et al. Variation in PTX3 is associated with primary graft dysfunction after lung transplantation. Am J Resp Crit Care Med. (2012) 186:546–52. doi: 10.1164/rccm.201204-0692OC
33. Chiarini M, Sabelli C, Melotti P, Garlanda C, Savoldi G, Mazza C, et al. PTX3 genetic variations affect the risk of Pseudomonas aeruginosa airway colonization in cystic fibrosis patients. Genes Immun. (2010) 11:665–70. doi: 10.1038/gene.2010.41
34. Olesen R, Wejse C, Velez DR, Bisseye C, Sodemann M, Aaby P, et al. DC-SIGN (CD209), pentraxin 3 and vitamin D receptor gene variants associate with pulmonary tuberculosis risk in West Africans. Genes Immun. (2007) 8:456–67. doi: 10.1038/sj.gene.6364410
35. Carmo RF, Aroucha D, Vasconcelos LR, Pereira LM, Moura P, Cavalcanti MS. Genetic variation in PTX3 and plasma levels associated with hepatocellular carcinoma in patients with HCV. J Viral Hepatit. (2016) 23:116–22. doi: 10.1111/jvh.12472
36. Hakelius M, Reyhani V, Rubin K, Gerdin B, Nowinski D. Normal oral keratinocytes and head and neck squamous carcinoma cells induce an innate response of fibroblasts. Anticanc Res. (2016) 36:2131–7.
37. Su SC, Hsieh MJ, Liu YF, Chou YE, Lin CW, Yang SF. ADAMTS14 gene polymorphism and environmental risk in the development of oral cancer. PLoS ONE. (2016) 11:e0159585. doi: 10.1371/journal.pone.0159585
38. Boyle P, Levin B. World Cancer Report 2008. Lyon: IARC Press, International Agency for Research on Cancer (2008).
39. Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. (2009) 59:225–49. doi: 10.3322/caac.20006
40. Petersen PE. The World Oral Health Report 2003: continuous improvement of oral health in the 21st century–the approach of the WHO Global Oral Health Programme. Commun Dentist Oral Epidemiol. (2003) 31(Suppl 1):3–23. doi: 10.1046/j.2003.com122.x
41. Su CW, Chien MH, Lin CW, Chen MK, Chow JM, Chuang CY, et al. Associations of genetic variations of the endothelial nitric oxide synthase gene and environmental carcinogens with oral cancer susceptibility and development. Nitric Oxide Biol Chem. (2018) 79:1–7. doi: 10.1016/j.niox.2018.06.005
42. Willeke F, Assad A, Findeisen P, Schromm E, Grobholz R, von Gerstenbergk B, et al. Overexpression of a member of the pentraxin family (PTX3) in human soft tissue liposarcoma. Euro J Cancer. (2006) 42:2639–46. doi: 10.1016/j.ejca.2006.05.035
43. Diamandis EP, Goodglick L, Planque C, Thornquist MD. Pentraxin-3 is a novel biomarker of lung carcinoma. Clin Cancer Res. (2011) 17:2395–9. doi: 10.1158/1078-0432.CCR-10-3024
44. Sardana G, Jung K, Stephan C, Diamandis EP. Proteomic analysis of conditioned media from the PC3, LNCaP, and 22Rv1 prostate cancer cell lines: discovery and validation of candidate prostate cancer biomarkers. J Proteome Res. (2008) 7:3329–38. doi: 10.1021/pr8003216
45. Hung TW, Tsai JP, Lin SH, Lee CH, Hsieh YH, Chang HR. Pentraxin 3 activates JNK signaling and regulates the epithelial-to-mesenchymal transition in renal fibrosis. Cell Physiol Biochem. (2016) 40:1029–38. doi: 10.1159/000453159
46. Ronca R, Di Salle E, Giacomini A, Leali D, Alessi P, Coltrini D, et al. Long pentraxin-3 inhibits epithelial-mesenchymal transition in melanoma cells. Mol Cancer Therap. (2013) 12:2760–71. doi: 10.1158/1535-7163.MCT-13-0487
47. Scimeca M, Antonacci C, Colombo D, Bonfiglio R, Buonomo OC, Bonanno E. Emerging prognostic markers related to mesenchymal characteristics of poorly differentiated breast cancers. Tumour Biol. (2016) 37:5427–35. doi: 10.1007/s13277-015-4361-7
48. Scimeca M, Antonacci C, Toschi N, Giannini E, Bonfiglio R, Buonomo CO, et al. Breast Osteoblast-like cells: a reliable early marker for bone metastases from breast cancer. Clin Breast Cancer. (2018) 18:e659–69. doi: 10.1016/j.clbc.2017.11.020
49. Feinberg AP, Koldobskiy MA, Gondor A. Epigenetic modulators, modifiers and mediators in cancer aetiology and progression. Nat Rev Genet. (2016) 17:284–99. doi: 10.1038/nrg.2016.13
50. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. (2011) 144:646–74. doi: 10.1016/j.cell.2011.02.013
51. Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. (2008) 40:1413–5. doi: 10.1038/ng.259
52. Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. (2008) 456:470–6. doi: 10.1038/nature07509
53. Jung H, Lee D, Lee J, Park D, Kim YJ, Park WY, et al. Intron retention is a widespread mechanism of tumor-suppressor inactivation. Nat Genet. (2015) 47:1242–8. doi: 10.1038/ng.3414
54. Anczukow O, Rosenberg AZ, Akerman M, Das S, Zhan L, Karni R, et al. The splicing factor SRSF1 regulates apoptosis and proliferation to promote mammary epithelial cell transformation. Nat Struct Mol Biol. (2012) 19:220–8. doi: 10.1038/nsmb.2207
55. Jensen MA, Wilkinson JE, Krainer AR. Splicing factor SRSF6 promotes hyperplasia of sensitized skin. Nat Struct Mol Biol. (2014) 21:189–97. doi: 10.1038/nsmb.2756
56. Karni R, de Stanchina E, Lowe SW, Sinha R, Mu D, Krainer AR. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat Struct Mol Biol. (2007) 14:185–93. doi: 10.1038/nsmb1209
57. Quesada V, Conde L, Villamor N, Ordonez GR, Jares P, Bassaganyas L, et al. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat Genet. (2011) 44:47–52. doi: 10.1038/ng.1032
58. Wang L, Lawrence MS, Wan Y, Stojanov P, Sougnez C, Stevenson K, et al. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N Engl J Med. (2011) 365:2497–506. doi: 10.1056/NEJMoa1109016
59. Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. (2011) 478:64–9. doi: 10.1038/nature10496
Keywords: long pentraxin 3, oral squamous cell carcinoma, single-nucleotide variation, metastasis, splicing, tumor progression
Citation: Yeh C-M, Lin C-W, Chuang C-Y, Liu Y-F, Chou C-H, Yang S-F and Chen M-K (2019) Functional Genetic Variant of Long Pentraxin 3 Gene Is Associated With Clinical Aspects of Oral Cancer in Male Patients. Front. Oncol. 9:581. doi: 10.3389/fonc.2019.00581
Received: 25 March 2019; Accepted: 14 June 2019;
Published: 03 July 2019.
Edited by:
Victor C. Kok, Asia University, TaiwanReviewed by:
Jiiang-Huei Jeng, National Taiwan University, TaiwanAmanda Ewart Toland, The Ohio State University, United States
Copyright © 2019 Yeh, Lin, Chuang, Liu, Chou, Yang and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shun-Fa Yang, eXNmQGNzbXUuZWR1LnR3; Mu-Kuan Chen, NTM3ODBAY2NoLm9yZy50dw==
 Chia-Ming Yeh1,2,3
Chia-Ming Yeh1,2,3 Yu-Fan Liu
Yu-Fan Liu Shun-Fa Yang
Shun-Fa Yang Mu-Kuan Chen
Mu-Kuan Chen