- 1Department of Neuroradiology, University of Alabama, Birmingham, AL, United States
- 2Russell H. Morgan Department of Radiology and Radiological Science, Johns Hopkins University School of Medicine, Baltimore, MD, United States
- 3Department of Neurosurgery, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, United States
- 4Department of Neurosurgery, University of Alabama, Birmingham, AL, United States
- 5Medical Scientist Training Program, University of California, Los Angeles, Los Angeles, CA, United States
- 6Senior Research Scientist, Uber AI Labs, San Francisco, CA, United States
- 7Department of Neurosurgery, Goethe University, Frankfurt, Germany
- 8Department of Surgery, Iranian Hospital, Dubai, United Arab Emirates
- 9Department of Mechanical Engineering, Arak University of Technology, Arak, Iran
- 10Division of Pediatric Hematology and Oncology, Department of Pediatrics, University of Alabama, Birmingham, AL, United States
Purpose: Artificial intelligence (AI) has accelerated novel discoveries across multiple disciplines including medicine. Clinical medicine suffers from a lack of AI-based applications, potentially due to lack of awareness of AI methodology. Future collaboration between computer scientists and clinicians is critical to maximize the benefits of transformative technology in this field for patients. To illustrate, we describe AI-based advances in the diagnosis and management of gliomas, the most common primary central nervous system (CNS) malignancy.
Methods: Presented is a succinct description of foundational concepts of AI approaches and their relevance to clinical medicine, geared toward clinicians without computer science backgrounds. We also review novel AI approaches in the diagnosis and management of glioma.
Results: Novel AI approaches in gliomas have been developed to predict the grading and genomics from imaging, automate the diagnosis from histopathology, and provide insight into prognosis.
Conclusion: Novel AI approaches offer acceptable performance in gliomas. Further investigation is necessary to improve the methodology and determine the full clinical utility of these novel approaches.
Introduction
Gliomas are the most common primary intracranial neoplasm (1), and comprise 1.8% of human malignancies and 2.3% of cancer deaths (2). World Health Organization (WHO) histopathological classification includes four grades: grades I and II are considered low-grade gliomas (LGG) and grades III and IV (glioblastoma-GBM) are considered high-grade gliomas (HGG) (3, 4). The resilience of gliomas derives from their infiltrative behavior, aggressive biology, genomics, and presence of the blood-brain barrier which reduces the effect of systemic chemotherapy (5). Treatment planning entails initial diagnosis, degree of infiltration, localization and segmentation, genomics, cell biology and clinical/imaging data. Post-treatment evaluation focuses on tumor progression/recurrence. Traditionally, these data are compiled manually by skilled physicians. In the future, artificial intelligence (AI) will augment clinical decision making in the management of oncologic patients, and usher in an era of personalized medicine. An example today is IBM Watson for oncology, a prototypic cloud-based AI, which helps physicians with treatment planning by analyzing extensive clinical, genetic and imaging databases (6–8).
Novel AI approaches such as deep learning (DL), neural networks (NN) and convolutional neural networks (CNN), have facilitated automated extraction of salient clinical data for treatment planning and post-treatment monitoring. Herein, we present a succinct description of foundational concepts of AI approaches and their relevance to clinical medicine. This is geared toward clinicians without computer science backgrounds. Also, we review novel AI approaches in the diagnosis and management of glioma. Finally, we discuss the future impact of AI on glioma diagnosis, genomics, perioperative planning, prognosis and post-treatment surveillance and its future challenges.
Artificial Intelligence
Succinctly, AI aims to create processes that analyze their environment and perform actions to optimize success toward a pre-determined goal. Machine learning is a sub-type of artificial intelligence focused on developing algorithms that can identify patterns within data without explicit specification. These algorithms can be classified into supervised and unsupervised learning (9). In supervised machine learning, the algorithm is trained on a human-labeled dataset, then the algorithm provides classification or regression on unlabeled data (e.g., for prediction of clinical outcomes). The rate-limiting step is a large human labeled dataset. The most common supervised machine learning techniques are linear and logistic regression, support vector machines (SVMs), naive Bayes, decision trees, and random forest methods. In unsupervised machine learning, algorithms identify hidden patterns for unlabeled datasets that are unknown to humans. The most common unsupervised machine learning methods include K-means, mean shift, affinity propagation, hierarchical clustering, Gaussian mixture modeling, and self-organizing maps. In machine learning, the input data for the above-mentioned algorithms are called features, which can be numerical or nominal values.
As an example, common features of neuroimaging data are location, size, shape, and signal intensity. In addition to these features, machine-learning algorithms are able to develop new inputs which are not readily visible to human eyes, including texture information, signal intensity gradient and skewness. Two main machine-learning algorithms, the SVM and the artificial neural network (ANN), have been applied to analysis of imaging data (Figure 1).
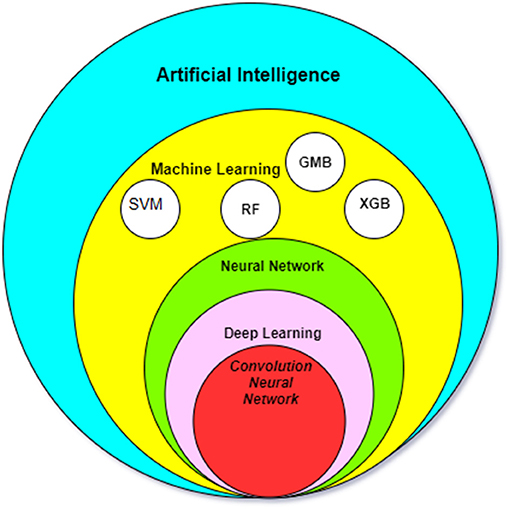
Figure 1. The relationship between the most common AI methods in medicine. SMV, Support Vector Machine; RF, Random forest algorithm; GBM, Gradient Boosting Machines; XGB, XGBoost.
SVM and random forest are relatively simple supervised machine learning algorithms that are useful for classifying an object into different categories. The SVM algorithm designs a decision surface in a high-dimensional space (hyper-plane) (Figure 2). SVM works well when the margin of separation between classes is maximized (10). Random forest algorithms develop multiple decision trees and merge them together to get a more accurate prediction. It is one of the most common AI algorithms and can be used for both classification (dividing pieces of data into different categories) and regression (predicting a quantitative response from a predictor variable) (Figure 3).
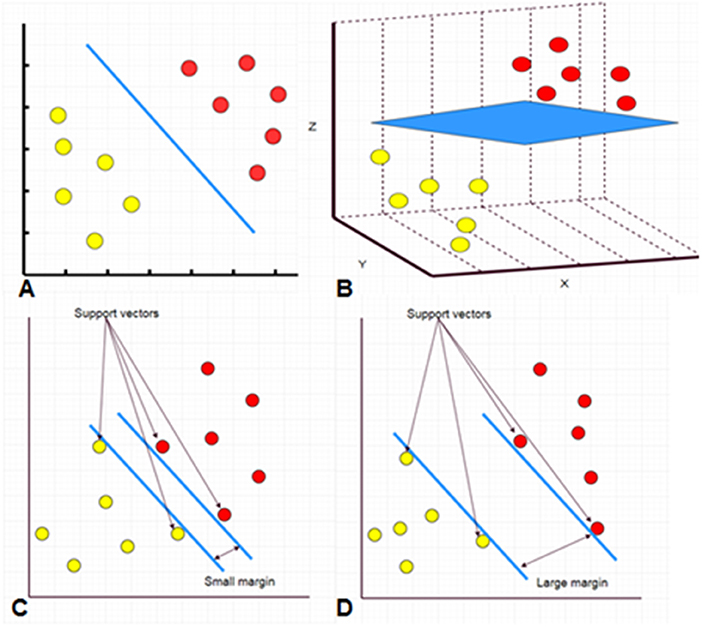
Figure 2. A hyperplane separating two classes of data points in 2D space (A, blue line). A hyperplane separating two classes of data points in 3D space (B, blue sheet). Separation in more dimensions is also performed but it is difficult to be presented on a 2D manuscript. The support vectors are data that are closer to the hyperplane. The larger the margin between the hyperplanes, the better the classification (C,D).
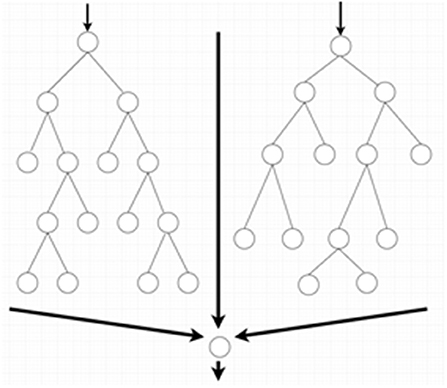
Figure 3. Random forest algorithms by developing multiple decision trees and merging them get more accurate predictions.
The ANN is a more complex machine learning algorithm with many variations that in rough terms attempt to mimic the functionality of biological neural networks. This algorithm is consistent with different nodes of input, hidden and output layers, each layer with a more abstract level of processing compared to the one prior. In the classical variant of ANN known as the multilayer perceptron, the nodes (“neurons”) within each successive layer are all connected to each other between the layers. Each neuron takes the inputs (weighted individually) from the previous layer and performs relatively simple mathematical operations to produce an output. The network can be trained (e.g., to use different weighting schemes) to fit a particular dataset by the use of learning algorithms. The ANN is very flexible for handling different types of data, but it is prone to data-overfitting and requires vast computational resources (Figure 4) (10). Another important disadvantage to neural network approaches is the lack of transparency in the “hidden layers” of neural networks, and the logic behind the mathematical transformations of each layer may not be readily understood. This is in contrast to other AI methods such as random forests, which allow full transparency due to access to the whole “tree” of information (although the decision trees can be rather complex). The lack of transparency inherent to some neural networks has been likened to a “black box”: the clinician cannot fully appreciate how exactly the AI arrives at a solution.
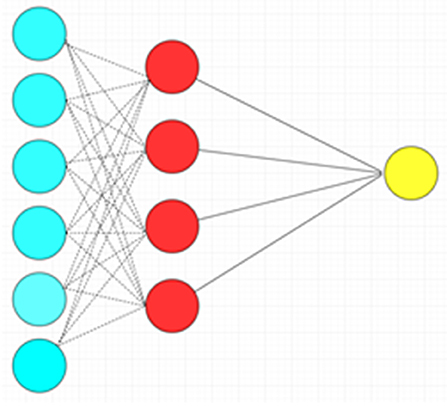
Figure 4. Artificial neural network with a single hidden layer. There is complete connection between layers. Blue circles: Input layer. Red circles: Hidden layer. Yellow circle: Output layer.
Deep learning is a variant of ANN that adds complexity by using multiple (“deep”) layers of an artificial neural network. The capability to implement deep learning methods using graphics processing units (GPU) and theoretical work over the past decade revived interest in neural networks and demonstrated the sophisticated capabilities of machine learning across many applications. The architecture of a neural network can vary in its complexity, resulting in a wide assortment of ANNs that can be used in deep learning. The standard neural network is termed “feedforward” as the hierarchy of data flows in only the forward direction (Figure 5).
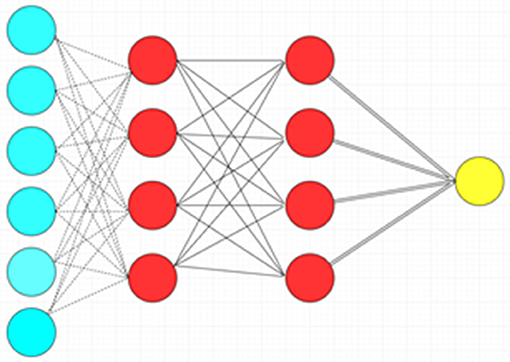
Figure 5. Deep feedforward neural network with two hidden layers. Blue circles: Input layer. Red circles: Hidden layer. Yellow circle: Output layer.
The convolution neural network (CNN) is another type of ANN in which imaging data is processed into reduced representations of features using mathematical transformations (most fundamentally, kernel convolution, followed by additional methods for dimensionality reduction), thereby enabling efficient deep learning. These and other innovations have improved the abilities of machine learning in the field of image processing (Figure 6) (10).
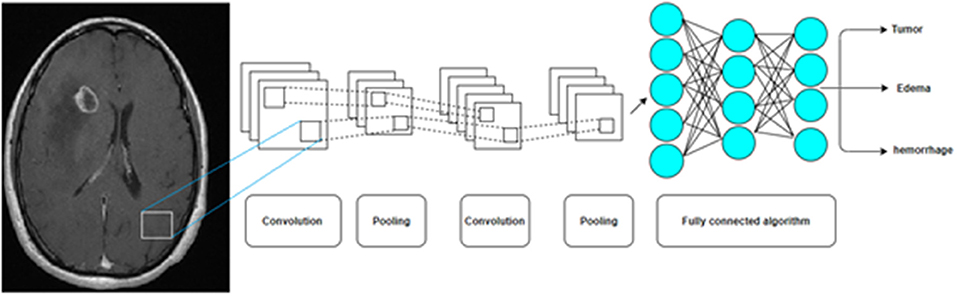
Figure 6. A Convolution Neural Network (CNN) by multiple pooling and convolution steps before a deep neural network is now the most common AI algorithm for image analysis.
Transfer Learning
Transfer learning is a machine learning method in which an already developed model for a task is reused as the starting point for a model on a second task. The advantage of using these pre-trained models is reducing the time of training. Currently, many of these pre-trained models are provided by major AI companies (e.g., GoogLeNet and AlexNet) which can be used to develop new AI applications.
Performance Analysis
As with other medical tests, AI algorithms can be evaluated by common biostatistical parameters, including sensitivity, specificity, positive/negative predictive values, and accuracy. Besides these parameters, performance can be evaluated with more dedicated tests. One of the most commonly used statistical tests for analyzing the performance of “classification modules” is the Receiver Operating Characteristic (ROC) Curve which shows the performance of a classification model at all classification thresholds. This model mainly depends on the true positive and false positive rates. In this test, the Area Under the ROC Curve (AUC) is the main indicator of the classification algorithm, represented as a value between 0 and 1. The closer this number to one, the better the performance of the classification algorithm. The “Dice score” is another common test used to evaluate the performance of image segmentation algorithms. This test compares a human-segmented image with the AI-segmented one and measures how similar they are. It is the size of the overlap of the two segmentations divided by the total size of the two objects. More sophisticated algorithms will require increasingly intricate metrics for evaluation of their performance and assessment of the suitability for clinical application.
Application of AI in the Management of Gliomas
Grading Prediction by Imaging
Predicting tumor grade on imaging using AI is approaching and may play an important role in future practice. The differentiation of LGG and HGG is crucial for treatment planning and prognosis. Traditionally, this task has been accomplished using anatomic images. LGGs (WHO grade I & II) usually do not show contrast enhancement and are without increased perfusion on MR perfusion sequences. Grade III gliomas may show punctate nodular enhancement and mildly increased perfusion. GBM (WHO grade IV) shows avid rim enhancement, marked hyper-perfusion and central necrosis (11–13). While these coarse imaging features that are appreciated by clinicians have reasonable predictive value, AI algorithms that can assess features not readily observed by humans may improve the differentiation of LGG vs. HGG.
Different techniques of machine learning applied to various imaging modalities have been studied for glioma grading. On the simple end of the spectrum, logistic regression classifiers have been developed to predict grade based on texture features in 34 GBMs and 73 LGG. This model achieved accuracy of 93%, a sensitivity of 97%, a negative predictive value of 99%, and an AUC of 0.94 in differentiating LGG vs. GBM (12). In comparison to LGG, HGGs have more complex anatomical morphology and BOLD-fMRI features. SVM has been employed to predict glioma grading based on resting-state functional MRI images, although these are not typically acquired in clinical practice. This model achieved accuracy of 89% in grading prediction (14). In another study, the SVM classifiers were developed to diagnose low-grade vs. high-grade and grade III vs. IV gliomas. SVM models detected 30 and 28 optimal features for classifying LGGs from HGGs and grades III from IV, respectively. The AUC was 0.987 for classifying LGGs from HGGs, and 0.992 for classifying grades III from IV (15). In a study of 130 gliomas, an artificial neural network used 41 features based on 2D T1-weighted MRI images. The algorithm was able to differentiate LGG vs. HGG with an accuracy of 90.3%, mean sensitivity of 87.8% and mean specificity of 92.5%. The AUC was 0.9486 (16). Ranjith et al. (17) differentiated LGG (only WHO grade II) from high-HGG (WHO grade III and IV) using MRI spectroscopy and machine algorithms in 38 patients (including multilayer perceptron, support vector machine, random forest, and locally weighted learning). They reported a AUC of more than 0.8 in three algorithms; with the best AUC using the random forest algorithm (0.911) and the best sensitivity using locally weighted learning (86%).
By using deep learning algorithms and transfer learning, researchers were able to predict glioma grading from T1-weighted, contrast-enhanced images before surgery in 113 patients with the high performance of the GoogLeNet and AlexNet software. They showed that the performance of transferred learning algorithms outperformed the trained CNN algorithm. Overall, the performance of GoogLeNet was better than AlexNet. The mean value of validation accuracy, test accuracy and test AUC of GoogLeNet was 0.867, 0.909, and 0.939, respectively. For AlexNet, the mean value of validation accuracy, test accuracy and test AUC were 0.866, 0.855, and 0.895, respectively (18). In this study, grades II and III were considered as low-grade and grade IV as high-grade.
In summary, these studies suggest grade prediction based on imaging features is feasible by AI algorithms. In comparison to humans, the machine learning algorithms are capable of using large numbers of imaging features which may improve the grading prediction power. It is still unclear if automated grading can change clinical management. Comparison of the various studies is difficult since a universal definition of HGG and LGG by imaging characteristics was not used. In addition, it is unclear which machine learning algorithm works best. In one study, 25 common machine-learning algorithms were compared to predict glioma grading; the SVM exhibited superior performance to other classifiers. In this study, grade II, III and IV were considered as high-grade which makes comparison with other studies challenging (19).
Genetic Information
Clinicians have attempted to estimate histopathologic features and glioma genomics, which provide important classifications of the tumor that guide treatment and prognosis, from non-invasive imaging studies with the use of AI. Traditionally, the diagnosis of glioma depends on the histopathological examination, but this practice has changed in recent decades due to advancement in genetic studies. Current WHO classification of CNS malignancies is in part based on genetic mutations. For example, the classification of astrocytomas and oligodendrogliomas depends on the mutation status of IDH1/2, ATRX loss, and p53 mutations (in astrocytomas), and co-deletion of 1p and 19q (in oligodendrogliomas). In other tumors, such as midline gliomas, the presence of the H3K27M mutation can redefine the grade of a tumor from a low-grade (by histo-anatomic diagnosis) to a grade IV entity known as “diffuse midline glioma, H3K27M-mutant.” Through the advancement of imaging modalities, it is possible to predict the pattern of genetic mutation based on the evaluation of the radiologic features. AI has a major impact on this “radiomics” approach. Radiomics describes a broad set of computational methods that extract quantitative features from radiographic images that are often beyond the ability of human eyes to see (20, 21). Notably, most of the radiomics approaches depend on the hand-engineered features including size, shape, location, texture, intensity and peri-tumoral features; however, these approaches are limited in the information they can evaluate. In this context, several researchers have developed machine learning-based algorithms to predict genetic mutations from the imaging. So far, most of the research was dedicated to predict the isocitrate dehydrogenase (IDH) mutations, O (6)-methylguanine-DNA methyltransferase (MGMT) promoter methylation, and co-deletion of chromosome arms 1p/19q with better results in comparison to hand-engineered radiomics (22). These genetic features are often associated with better treatment response and survival rates (23–31).
Akkus et al. (32) trained a CNN algorithm based on T1 post contrast and T2 sequences and was able to predict the co-deletion of chromosome arms 1p/19q with a sensitivity of 93.3%, specificity of 82.2%, and accuracy of 87.7%. By using a residual convolutional neural network for each MR sequence including the FLAIR, T2, T1 pre-, and post-contrast, other researchers predicted the presence of IDH mutation with an accuracy of 85.7% without including the patients' age and 89.1% after including the age factor (33). Wu et al. (34) utilized 126 glioma patients, 704 radiomic features and eight classical machine learning methods in an effort to predict IDH genotype in diffuse gliomas before surgery. They reported high predictive performance with a random forest algorithm (accuracy 0.885 ± 0.041, AUC 0.931 ± 0.036) but low predictive performance with a neural network (accuracy 0.829 ± 0.064, AUC 0.878 ± 0.052). They concluded that a random forest algorithm is suitable for IDH genomic prediction before surgery of glioma. Random forest algorithms also have been tested on grade III and IV gliomas for prediction of IDH mutation with an accuracy of 89% and AUC of 0.9231 (35). Deep learning based radiomics algorithms has been used to extract data from imaging for prediction of IDH1 mutations in LGG with 6 convolutional layers of 4,096 neurons resulting in an AUC of 92% (36). Other researchers have developed a machine learning based algorithm to estimate the chance of IDH1 mutations in LGG with an AUC of 0.95 (34).
By developing deep learning models, researchers were able to predict the chance of MGMT methylation in GBM with accuracy as high as 95% (37, 38). In a study on archive images of patients with LGG and HGG, researchers developed a deep learning convolutional neural network and were able to estimate codeletion of chromosome arms 1p/19q, IDH1 mutation, and MGMT methylation with an accuracy of 92, 94, and 83%, respectively (39). Liu et al. (40) have used the combined model of CNN features and a support-vector-machine classifier to automatically predict genotypes of midline gliomas (H3 K27M mutation) with an accuracy of 94.8% to predict the mutation.
In summary, prediction of tumor genomics from imaging data is feasible by application of AI algorithm radiomics. The advantages of AI are its ability to detect multiple radiologic features which are too numerous and too subtle for the human eye. Most reported algorithms achieved high performance with accuracy above 80–90%. So far, the majority of genetic studies utilize imaging data to predict gene mutations. Machine learning algorithms have also been used to extract data from genetic databases. These algorithms were able to classify the patients with GBM into different clusters and predict the prognosis and treatment response (41). Accuracy of about 89% has been reported with the application of neural network-based classifiers to help differentiate the transcriptional subtypes of GBM (e.g., mesenchymal, classical, proneural, and neural subtypes) (42). AI algorithms are a promising approach to help analyze gene expression and predict the histopathology of GBM subtypes.
Pre-operative Planning
The 3D volumetric measurement of the viable/enhancing tumoral component and peripheral edema is essential for surgical planning and post-operative follow-up. Manual 3D segmentation methods are time-consuming (43). AI has been used for tumor segmentation (44, 45). To differentiate voxels representing viable neoplasm vs. edema vs. normal brain tissue, several machine-learning algorithms have been used. So far, the most promising techniques are SVM, random forest and CNNs. The CNN models have the best performance (22, 46–49). In one study a tumor localization network (a fully convolutional network in conjunction with transfer learning technology) was used to localize the tumor (50). This two-step protocol was faster than and at least as accurate as the prior reported methods in the literature (50). A 3D U-Net CNN has also been used for automated segmentation of gliomas on 18F-fluoroethyl-tyrosine (18F-FET) PET with 88% sensitivity, 99% specificity, a 78% positive predictive value, and a 99% negative predictive value (51). In another study, a multipathway convolutional neural network and fully connected conditional random field were implemented for 3D FLAIR images for segmentation of a LGG with a Dice similarity coefficient of 0.85 (52). Additionally, SMV has been used to differentiate viable HGG from peri-tumoral edema in a study containing 9 patients with HGG: the SMV was able to differentiate viable “non-enhancing” tumor from peripheral edema with a misclassification error of 8.4%. When SVM output was smoothed using a mean filter, the misclassification error was reduced to 2.4 % (53). Automated algorithms have been developed by using random forest models combined with voxel texture features on contrast-enhanced T1 and FLAIR MRI for glioma segmentation. This model had an overall moderate accuracy with better performance to segment high-grade enhancing neoplasm and edema in comparison to non-enhancing LGG or necrosis (54).
Gliomas, specifically GBM, are infiltrative neoplasms. They often invade the tissues beyond the enhancing area on MRI. Given the fact that surgical resection of glioma includes resection of mostly enhancing tissue, non-enhancing tumor can be left behind. AI can help physicians predict the location of subsequent recurrence. Currently, an AI algorithm has been developed to perform this task with AUC of 84% (55).
In summary, different AI techniques can segment tumor, edema and normal tissue. In this context, the CNNs, SVM, and random forest algorithms are promising with at least moderate accuracy.
Intra-Operative Treatment Planning
Gliomas display an infiltrative behavior. Differentiation of tumor vs. normal tissue is challenging, not only by imaging, but also during surgery. In this context, intraoperative MRI scanners have been used at some oncologic centers for this purpose, although their application is limited by their cost, availability and logistics. An interesting application of AI has been introduced to help neurosurgeons resect the maximum amount of tumor and minimum amount of normal tissue. In this technique, deep learning methods are used to analyze the images from hyperspectral imaging (a non-contact, non-ionizing, label-free and intraoperative imaging modality) during surgery with accuracy about 80% to differentiate neoplastic tissue from adjacent non-tumoral brain tissues (56).
Histopathologic Diagnosis
In classical medical practice, pathologists and clinicians usually analyze the histopathologic features of the disease. In some settings, such as during surgery, the amount of time from preparation of tissue to diagnosis may be a limiting factor. The diagnosis depends on pathologist's expertise and is critical for the management of the patient, both intraoperatively and for subsequent treatment (57–59). Considering these aforementioned restrictions, computational histopathology is becoming more popular (60, 61). The application of AI to pathology has been aided by the advent of slide scanners, which can convert microscopic slides to high-quality image files. Once digitized, the slides are amenable to computation. In the future, AI methods may assist neuropathologists with identifying characteristics of a tumor and allow for interpretation of subtle histological features that cannot be easily appreciated by humans.
Microscopically, glioma biopsy specimens have the appearance of a collection of variably pleomorphic neoplastic glial cells, sometimes mixed with normal brain tissue. Different tissue architecture (e.g., mixed mesenchymal and glial areas in gliosarcoma) may be present. Immune cells, blood vessels, and areas of necrosis may also be seen. The morphology of the neoplastic glial cells can assist in the classification of the tumor; in addition, the observation and frequency of mitotic figures are important for determination of HGG vs. LGG to patients. Additional pathological features, such as the presence of vascular endothelial hyperplasia or palisading necrosis, also play a role in defining HGG. The complex picture provided by sometimes limited specimens provides a rich substrate for AI approaches for analysis.
While usually readily identifiable, non-neoplastic cells may sometimes mimic glioma cells, confounding their differentiation. For example, reactive astrocytes may appear similar to neoplastic astrocytes, but are differentiated by the cellular spacing and nuclear atypia; likewise, prior radiation treatment may result in abnormal or bizarre-appearing cells that can be challenging to distinguish from GBM recurrence. Abas et al. (62) used multiple machine learning methods, such as the SVM and decision trees, to segment and classify these cells on smear preparations of glioma. SVM and random forest algorithms have also been used for nucleus segmentation on histopathologic images from a glioma database and were more than 98% accurate for classification of LGG or HGG (63). Beyond cytology, Yonekura et al. (64) reported that the CNNs could extract significant features from GBM histopathology slide images with an accuracy of about 98%. SVM algorithms have been used for diagnosis and glioma grading (grade II, III, and IV) on slide images with promising accuracy of about 90% (65). In a recent study, a machine learning technique (Google Inception V3 convolutional neural network) was used for diagnosis of glioma. The haemotoxylin and eosin (H&E) stain images from 50 normal brain, 45 LGG, and 59 HGG were analyzed. The accuracy of this algorithm was 100% for the diagnosis of HGG vs. normal brain and 98% for diagnosing glioma (LGG or HGG) vs. normal brain (66).
In addition to diagnosis and grading, AI can help to predict prognosis by combining features from paraffin-embedded tissue specimens and molecular pathology, such as mutations in IDH1/2 genes and codeletion of chromosomes 1p and 19q. In a study on 769 gliomas including LGGs (grade II and III) and HGGs (grade IV), the CNN was able to predict survival rate with accuracy equal to the manual histologic-grade baseline models (67). While the application of AI to the histopathology of gliomas is only in its infancy, such approaches promise to aid in the evaluation of large tissue specimens and improve diagnostic accuracy for patients in the near future. Pathologists may benefit from segmentation algorithms which rapidly highlight and classify cells (such as mitotic figures), or which can define regions of increased cellularity. In the future, it is not inconceivable that AI-assisted methods may define new pathologic subsets of gliomas with their own characteristic responsiveness to treatments and prognosis, and therefore open new avenues in the pathologic examination of gliomas.
Radiation
AI has the potential to positively impact the field of radiotherapy. Patient selection, simulation, treatment planning, quality assurance, and follow-up are the most important steps in radiotherapy. Simulation of glioma is less challenging than other visceral malignancies with respiratory motions. Treatment planning includes identification of glioma and adjacent organs which may be damaged by radiation, Organs At Risk (OAR). Segmentation of tumor and OARs are traditionally performed by automated software with atlas-based segmentation tools techniques. The atlas-based segmentation tools are accurate for segmentation of high contrast organs (e.g., lung and bone) but they are suboptimal for tissues with close densities (soft tissues). In this context, machine and deep learning techniques approaches may improve the segmentation and treatment planning (56).
Post-treatment Follow-Up
Differentiation of post-treatment changes including radiation necrosis and pseudo-progression vs. true tumor progression/recurrence is a very common challenge in neuroradiology. This task is difficult for anatomic images; therefore, many advanced imaging techniques have been proposed (e.g., MR spectroscopy, MR perfusion and PET with difference tracers) with persistent uncertainty. Very few studies are available involving application of AI to differentiate post-treatment changes vs. CNS tumor progression. The SVM classifier has been trained to diagnose pseudo-progression vs. recurrence in patients with glioma treated with surgery and chemotherapy. In this study of 31 patients, the sensitivity and specificity of the classifier for pseudoprogression was 89.91 and 93.72%, respectively with AUC of 0.94; of note, the best predictor image sequences were DWI and MRP (68). A CNN has been developed to differentiate true vs. pseudo-progression in patients with GBM status post resection and chemo-radiation with acceptable performance and an AUC of 0.83 (69). In one study, AI was applied for this task with a high performance of SVM classifiers outperforming two expert neuroradiologists with an AUC for FLAIR sequence equal to 0.79. The main limitation in this study is the fact that researchers included the primary and metastatic CNS tumors in combination (70).
Outcome Prediction
There are many models to predict the overall survival rate in patients with glioma. Most of them are based on clinical data, genetic information, and imaging. Recently, many AI algorithms have been developed to predict survival rate in these patients, and potentially have superior accuracy compared to conventional methods (71). Nie et al. (72) have developed a multi-module and multi-channel deep survival prediction model for glioma using CNNs to analyze MRI images (including the T1 post contrast, DTI and resting state functional MRI) in association with a SVM (which contains information on tumor histology, tumor size and patients age) to predict the overall survival of patients with glioma with an accuracy of 90.66%. In another study, the Pathway-Associated Sparse Deep Neural Network (PASNet) outperformed other models to predict the outcome of GBM with an AUC of 0.66 (73). Machine learning by means of SVM in combination with whole-tumor rCBV histogram analysis has been used for outcome prediction of glioma with an AUC of 0.7 to 0.8 (74). Additionally, SVM algorithms predicted the outcome of gliomas from MR images with an 80% accuracy of 80% (61). SVM has been used to analyze the anatomic (DTI) and functional (rs-fMRI) data to predict good outcomes (>650-day survival) vs. poor outcomes (<650-day) in high-grade glioma with an accuracy of 75% (75). By developing a convolutional neural base network, Lao et al. (76) predicted the survival rate of GBM patients with a C-index of 0.71, which was subsequently improved after inclusion of the clinical data. C-index is equal to the AUC and ranges from 0.5 to 1. Values above 0.7 indicate a good test.
In summary, the overall accuracy of machine learning methods to predict glioma outcomes from imaging and clinical data approaches 80%, as evident on the meta-analysis done by Sarkiss and Germano (71) on 29 studies (including glioma and brain metastasis). In the future, AI will help to integrate data from disparate fields (clinical examination, imaging, and pathology) to guide treatment and prognosis. One major uncertainty about these prognostic algorithms is the heterogeneity of the patient population. One recent study of machine learning on a small heterogeneous population of glioma using 21 features (demographic, clinical, genetic, histopathologic data) achieved accuracy of <70% (77).
Future Challenges
AI may demonstrate a crucial role in patient selection for clinical trials, specifically with biomarkers and radiomics. Classically, in glioma clinical trials, the most common biomarkers are the status of MGMT promoter methylation and IDH mutation but on the horizon are radiomic markers which can predict the treatment response to a particular treatment. This would help improve clinical trial designs with more finely targeted inclusion and exclusion criteria and potentially improve trial outcome (6, 78–80). Additionally, there are commercially available AI techniques for extraction of patient data, to link them to large databases and to select the best trial and medication for those specific patients (81).
Several challenges must be addressed before the adaptation of AI in oncology and specifically, the management of glioma. Developing accurate AI needs large standardized and annotated data sets and high-quality ground truth data. The multi-institutional nature of most clinical trials for gliomas complicates the ability to acquire uniform data sets (82). Although AI algorithms use numerous features for decision making, their analysis processes are not always readily understood to humans, so many of the previously described AI algorithms are not amenable to be re-created by other investigators. There is ongoing research to solve this “black-box” nature of AI, and in the future, may help to follow otherwise impenetrable processes step-by-step in transparent algorithms (39).
Generalizability of the AI algorithms is one of the major challenges preventing their widespread clinical adaptation. So far most of the AI applications in oncology and gliomas have been trained on relatively small patient populations. Performance of an AI algorithm developed on a small population is not optimal, especially for the large and heterogeneous population of gliomas (83). In addition, the inclusion criteria in the aforementioned glioma studies were very heterogeneous. Before the clinical adaptation of an algorithm, it is critical to have a universal definition for LGG and HGG. Large, standardized datasets from multiple institutions with clinical, neuroimaging, and neuropathologic data that cover diverse patient populations are needed to fully realize the power of AI for the diagnosis and treatment of gliomas.
Given the inchoate stage of development for AI in gliomas, there are no comprehensive cost-benefit studies or prospective studies to confirm that AI can improve patient outcome. Another obstacle against universal adaptation of AI in oncology and gliomas is clinician-engineer interaction. Currently, physicians receive very little training in computer/data science and most of the computer scientists are not familiar with the complexity of clinical patient management (81).
Thus, the abovementioned challenges, as well as many unanswered legal and ethical questions, must be addressed before the adaptation of AI in the daily practice of oncologic centers (84).
Conclusion
Many AI approaches have been created to help with glioma management. AI techniques have been developed to predict grading from imaging data; survival rates from clinical, genetic and imaging data; and molecular genetics from imaging data. AI techniques have also been developed to automate diagnosis from histopathologic slides to segment tissues for surgical planning and to monitor the patients after treatment. Most of these techniques suggest acceptable performance, but the application of AI to glioma diagnosis and treatment has only now begun in earnest. The promise and performance of these techniques in daily clinical practice and their effect on patient outcomes warrant further development.
Author Contributions
HS, OS, ES, and MB contributed conception, design of the study, and wrote the first draft of the manuscript. HS organized the database. JB, GE, JC, PS, GC, FG, AS, and GF wrote sections of the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
Funding
This research was supported in part by grants from the Rally Foundation for Childhood Cancer Research, Hyundai Hope on Wheels, and the Kaul Pediatric Research Institute to GF.
Conflict of Interest Statement
JB has positions/equity in CITC Ltd and Avidea Technologies and is a member of the Scientific Board of Advisors for POCKiT Diagnostics. PS was employed by Uber AI Lab.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Dolecek TA, Propp JM, Stroup NE, Kruchko C. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2005–2009. Neuro Oncol. (2012) 14 (Suppl. 5):v1–49. doi: 10.1093/neuonc/nos218
2. Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. (2015) 136:E359–386. doi: 10.1002/ijc.29210
3. Ahmed R, Oborski MJ, Hwang M, Lieberman FS, Mountz JM. Malignant gliomas: current perspectives in diagnosis, treatment, and early response assessment using advanced quantitative imaging methods. Cancer Manag Res. (2014) 6:149–70. doi: 10.2147/CMAR.S54726
4. Wesseling P, Capper D. WHO 2016 Classification of gliomas. Neuropathol Appl Neurobiol. (2018) 44:139–50. doi: 10.1111/nan.12432
5. Aldape K, Brindle KM, Chesler L, Chopra R, Gajjar A, Gilbert MR, et al. Challenges to curing primary brain tumours. Nat Rev Clin Oncol. (2019) 16:509–20. doi: 10.1038/s41571-019-0177-5
6. Lee WS, Ahn SM, Chung JW, Kim KO, Kwon KA, Kim Y, et al. Assessing concordance with watson for oncology, a cognitive computing decision support system for colon cancer treatment in Korea. JCO Clin Cancer Inform. (2018) 2:1–8. doi: 10.1200/CCI.17.00109
7. Choi YI, Chung JW, Kim KO, Kwon KA, Kim YJ, Park DK, et al. Concordance rate between clinicians and watson for oncology among patients with advanced gastric cancer: early, real-world experience in Korea. Can J Gastroenterol Hepatol. (2019) 2019:8072928. doi: 10.1155/2019/8072928
8. Hamilton JG, Genoff Garzon M, Westerman JS, Shuk E, Hay JL, Walters C, et al. “A tool, not a crutch”: patient perspectives about IBM Watson for oncology trained by memorial sloan kettering. J Oncol Pract. (2019) 15:e277–e288. doi: 10.1200/JOP.18.00417
9. McBee MP, Awan OA, Colucci AT, Ghobadi CW, Kadom N, Kansagra AP, et al. Deep learning in radiology. Acad Radiol. (2018) 25:1472–80. doi: 10.1016/j.acra.2018.02.018
10. Lee EJ, Kim YH, Kim N, Kang DW. Deep into the brain: artificial intelligence in stroke imaging. J Stroke. (2017) 19:277–85. doi: 10.5853/jos.2017.02054
11. Wu CC, Guo WY, Chen MH, Ho DM, Hung AS, Chung HW. Direct measurement of the signal intensity of diffusion-weighted magnetic resonance imaging for preoperative grading and treatment guidance for brain gliomas. J Chin Med Assoc. (2012) 75:581–8. doi: 10.1016/j.jcma.2012.08.019
12. Li-Chun Hsieh K, Chen CY, Lo CM. Quantitative glioma grading using transformed gray-scale invariant textures of MRI. Comput Biol Med. (2017) 83:102–8. doi: 10.1016/j.compbiomed.2017.02.012
13. Wen PY, Huse JT. 2016 World Health Organization classification of central nervous system tumors. Continuum. (2017) 23:1531–47. doi: 10.1212/CON.0000000000000536
14. Wu J, Qian Z, Tao L, Yin J, Ding S, Zhang Y, et al. Resting state fMRI feature-based cerebral glioma grading by support vector machine. Int J Comput Assist Radiol Surg. (2015) 10:1167–74. doi: 10.1007/s11548-014-1111-z
15. Tian Q, Yan LF, Zhang X, Zhang X, Hu YC, Han Y, et al. Radiomics strategy for glioma grading using texture features from multiparametric MRI. J Magn Reson Imaging. (2018) 48:1518–28. doi: 10.1002/jmri.26010
16. Mao Y, Liao W, Cao D, Zhao L, Wu X, Kong L, et al. [An artificial neural network model for glioma grading using image information]. Zhong Nan Da Xue Xue Bao Yi Xue Ban. (2018) 43:1315–22. doi: 10.11817/j.issn.1672-7347.2018.12.006
17. Ranjith G, Parvathy R, Vikas V, Chandrasekharan K, Nair S. Machine learning methods for the classification of gliomas: initial results using features extracted from MR spectroscopy. Neuroradiol J. (2015) 28:106–11. doi: 10.1177/1971400915576637
18. Yang Y, Yan LF, Zhang X, Han Y, Nan HY, Hu YC, et al. Glioma grading on conventional MR images: a deep learning study with transfer learning. Front Neurosci. (2018) 12:804. doi: 10.3389/fnins.2018.00804
19. Zhang X, Yan LF, Hu YC, Li G, Yang Y, Han Y, et al. Optimizing a machine learning based glioma grading system using multi-parametric MRI histogram and texture features. Oncotarget. (2017) 8:47816–30. doi: 10.18632/oncotarget.18001
20. Sonoda Y, Shibahara I, Kawaguchi T, Saito R, Kanamori M, Watanabe M, et al. Association between molecular alterations and tumor location and MRI characteristics in anaplastic gliomas. Brain Tumor Pathol. (2015) 32:99–104. doi: 10.1007/s10014-014-0211-3
21. Zhou M, Scott J, Chaudhury B, Hall L, Goldgof D, Yeom KW, et al. Radiomics in brain tumor: image assessment, quantitative feature descriptors, and machine-learning approaches. Am J Neuroradiol. (2018) 39:208–16. doi: 10.3174/ajnr.A5391
22. Lotan E, Jain R, Razavian N, Fatterpekar GM, Lui YW. State of the art: machine learning applications in glioma imaging. AJR Am J Roentgenol. (2019) 212:26–37. doi: 10.2214/AJR.18.20218
23. Scheie D, Andresen PA, Cvancarova M, Bo AS, Helseth E, Skullerud K, et al. Fluorescence in situ hybridization (FISH) on touch preparations: a reliable method for detecting loss of heterozygosity at 1p and 19q in oligodendroglial tumors. Am J Surg Pathol. (2006) 30:828–37. doi: 10.1097/01.pas.0000213250.44822.2e
24. Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. (2008) 321:1807–12. doi: 10.1126/science.1164382
25. Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, et al. IDH1 and IDH2 Mutations in Gliomas. N Engl J Med. (2009) 360:765–73. doi: 10.1056/NEJMoa0808710
26. Hartmann C, Hentschel B, Wick W, Capper D, Felsberg J, Simon M, et al. Patients with IDH1 wild type anaplastic astrocytomas exhibit worse prognosis than IDH1-mutated glioblastomas, and IDH1 mutation status accounts for the unfavorable prognostic effect of higher age: implications for classification of gliomas. Acta Neuropathol. (2010) 120:707–18. doi: 10.1007/s00401-010-0781-z
27. Houillier C, Wang X, Kaloshi G, Mokhtari K, Guillevin R, Laffaire J, et al. IDH1 or IDH2 mutations predict longer survival and response to temozolomide in low-grade gliomas. Neurology. (2010) 75:1560–6. doi: 10.1212/WNL.0b013e3181f96282
28. Jansen NL, Schwartz C, Graute V, Eigenbrod S, Lutz J, Egensperger R, et al. Prediction of oligodendroglial histology and LOH 1p/19q using dynamic [18F]FET-PET imaging in intracranial WHO grade II and III gliomas. Neuro Oncol. (2012) 14:1473–80. doi: 10.1093/neuonc/nos259
29. Fellah S, Caudal D, De Paula AM, Dory-Lautrec P, Figarella-Branger D, Chinot O, et al. Multimodal MR imaging (diffusion, perfusion, and spectroscopy): is it possible to distinguish oligodendroglial tumor grade and 1p/19q codeletion in the pretherapeutic diagnosis? Am J Neuroradiol. (2013) 34:1326–33. doi: 10.3174/ajnr.A3352
30. Bourdillon P, Hlaihel C, Guyotat J, Guillotton L, Honnorat J, Ducray F, et al. Prediction of anaplastic transformation in low-grade oligodendrogliomas based on magnetic resonance spectroscopy and 1p/19q codeletion status. J Neurooncol. (2015) 122:529–37. doi: 10.1007/s11060-015-1737-x
31. Iwadate Y, Shinozaki N, Matsutani T, Uchino Y, Saeki N. Molecular imaging of 1p/19q deletion in oligodendroglial tumours with 11C-methionine positron emission tomography. J Neurol Neurosurg Psychiatry. (2016) 87:1016–21. doi: 10.1136/jnnp-2015-311516
32. Akkus Z, Ali I, Sedlar J, Agrawal JP, Parney IF, Giannini C, et al. Predicting deletion of chromosomal Arms 1p/19q in Low-grade gliomas from MR images using machine intelligence. J Digit Imaging. (2017) 30:469–76. doi: 10.1007/s10278-017-9984-3
33. Chang K, Bai HX, Zhou H, Su C, Bi WL, Agbodza E, et al. Residual convolutional neural network for the determination of IDH status in low- and high-grade gliomas from MR imaging. Clin Cancer Res. (2018) 24:1073–81. doi: 10.1158/1078-0432.CCR-17-2236
34. Wu S, Meng J, Yu Q, Li P, Fu S. Radiomics-based machine learning methods for isocitrate dehydrogenase genotype prediction of diffuse gliomas. J Cancer Res Clin Oncol. (2019) 145:543–50. doi: 10.1007/s00432-018-2787-1
35. Zhang B, Chang K, Ramkissoon S, Tanguturi S, Bi WL, Reardon DA, et al. Multimodal MRI features predict isocitrate dehydrogenase genotype in high-grade gliomas. Neuro Oncol. (2017) 19:109–17. doi: 10.1093/neuonc/now121
36. Li Z, Wang Y, Yu J, Guo Y, Cao W. Deep Learning based Radiomics (DLR) and its usage in noninvasive IDH1 prediction for low grade glioma. Sci Rep. (2017) 7:5467. doi: 10.1038/s41598-017-05848-2
37. Korfiatis P, Kline TL, Lachance DH, Parney IF, Buckner JC, Erickson BJ. Residual deep convolutional neural network predicts MGMT methylation status. J Digit Imaging. (2017) 30:622–8. doi: 10.1007/s10278-017-0009-z
38. Han L, Kamdar MR. MRI to MGMT: predicting methylation status in glioblastoma patients using convolutional recurrent neural networks. Pac Symp Biocomput. (2018) 23:331–42. doi: 10.1142/9789813235533_0031
39. Chang P, Grinband J, Weinberg BD, Bardis M, Khy M, Cadena G, et al. Deep-learning convolutional neural networks accurately classify genetic mutations in gliomas. Am J Neuroradiol. (2018) 39:1201–7. doi: 10.3174/ajnr.A5667
40. Liu J, Chen F, Pan C, Zhu M, Zhang X, Zhang L, et al. A cascaded deep convolutional neural network for joint segmentation and genotype prediction of brainstem gliomas. IEEE Trans Biomed Eng. (2018) 65:1943–52. doi: 10.1109/TBME.2018.2845706
41. Young JD, Cai C, Lu X. Unsupervised deep learning reveals prognostically relevant subtypes of glioblastoma. BMC Bioinformatics. (2017) 18 (Suppl. 11):381. doi: 10.1186/s12859-017-1798-2
42. Vasudevan P, Murugesan T. Cancer subtype discovery using prognosis-enhanced neural network classifier in multigenomic data. Technol Cancer Res Treat. (2018) 17:1533033818790509. doi: 10.1177/1533033818790509
43. Gordillo N, Montseny E, Sobrevilla P. State of the art survey on MRI brain tumor segmentation. Magn Reson Imaging. (2013) 31:1426–38. doi: 10.1016/j.mri.2013.05.002
45. Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, et al. A survey on deep learning in medical image analysis. Med Image Anal. (2017) 42:60–88. doi: 10.1016/j.media.2017.07.005
46. Tustison NJ, Shrinidhi KL, Wintermark M, Durst CR, Kandel BM, Gee JC, et al. Optimal symmetric multimodal templates and concatenated random forests for supervised brain tumor segmentation (simplified) with ANTsR. Neuroinformatics. (2015) 13:209–25. doi: 10.1007/s12021-014-9245-2
47. Pereira S, Pinto A, Alves V, Silva CA. Brain tumor segmentation using convolutional neural networks in MRI images. IEEE Trans Med Imaging. (2016) 35:1240–51. doi: 10.1109/TMI.2016.2538465
48. Havaei M, Davy A, Warde-Farley D, Biard A, Courville A, Bengio Y, et al. Brain tumor segmentation with deep neural networks. Med Image Anal. (2017) 35:18–31. doi: 10.1016/j.media.2016.05.004
49. Kamnitsas K, Ledig C, Newcombe VFJ, Simpson JP, Kane AD, Menon DK, et al. Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med Image Anal. (2017) 36:61–78. doi: 10.1016/j.media.2016.10.004
50. Cui S, Mao L, Jiang J, Liu C, Xiong S. Automatic semantic segmentation of brain gliomas from MRI images using a deep cascaded neural network. J Healthc Eng. (2018) 2018:4940593. doi: 10.1155/2018/4940593
51. Blanc-Durand P, Van Der Gucht A, Schaefer N, Itti E, Prior JO. Automatic lesion detection and segmentation of 18F-FET PET in gliomas: a full 3D U-Net convolutional neural network study. PLoS ONE. (2018) 13:e0195798. doi: 10.1371/journal.pone.0195798
52. Li Z, Wang Y, Yu J, Shi Z, Guo Y, Chen L, et al. Low-grade glioma segmentation based on CNN with fully connected CRF. J Healthc Eng. (2017) 2017:9283480. doi: 10.1155/2017/9283480
53. Sengupta A, Agarwal S, Gupta PK, Ahlawat S, Patir R, Gupta RK, et al. On differentiation between vasogenic edema and non-enhancing tumor in high-grade glioma patients using a support vector machine classifier based upon pre and post-surgery MRI images. Eur J Radiol. (2018) 106:199–208. doi: 10.1016/j.ejrad.2018.07.018
54. Bonte S, Goethals I, Van Holen R. Machine learning based brain tumour segmentation on limited data using local texture and abnormality. Comput Biol Med. (2018) 98:39–47. doi: 10.1016/j.compbiomed.2018.05.005
55. Akbari H, Macyszyn L, Da X, Bilello M, Wolf RL, Martinez-Lage M, et al. Imaging surrogates of infiltration obtained via multiparametric imaging pattern analysis predict subsequent location of recurrence of glioblastoma. Neurosurgery. (2016) 78:572–80. doi: 10.1227/NEU.0000000000001202
56. Fabelo H, Halicek M, Ortega S, Shahedi M, Szolna A, Pineiro JF, et al. Deep learning-based framework for in vivo identification of glioblastoma tumor using hyperspectral images of human brain. Sensors. (2019) 19:E920. doi: 10.3390/s19040920
57. Bilgin C, Demir C, Nagi C, Yener B. Cell-graph mining for breast tissue modeling and classification. Conf Proc IEEE Eng Med Biol Soc. (2007) 2007:5311–4. doi: 10.1109/IEMBS.2007.4353540
58. Sertel O, Kong J, Catalyurek UV, Lozanski G, Saltz JH, Gurcan MN. Histopathological image analysis using model-based intermediate representations and color texture: follicular lymphoma grading. J Signal Process Syst. (2008) 55:169. doi: 10.1007/s11265-008-0201-y
59. Gurcan MN, Boucheron LE, Can A, Madabhushi A, Rajpoot NM, Yener B. Histopathological image analysis: a review. IEEE Rev Biomed Eng. (2009) 2:147–71. doi: 10.1109/RBME.2009.2034865
60. Rodenacker K, Bengtsson E. A feature set for cytometry on digitized microscopic images. Anal Cell Pathol. (2003) 25:1–36. doi: 10.1155/2003/548678
61. Marchevsky AM, Wick MR. Evidence-based medicine, medical decision analysis, and pathology. Hum Pathol. (2004) 35:1179–88. doi: 10.1016/j.humpath.2004.06.004
62. Abas FS, Gokozan HN, Goksel B, Otero JJ, Gurcan MN. Intraoperative Neuropathology of Glioma Recurrence: Cell Detection and Classification. San Diego, CA: SPIE (2016).
63. Fukuma K, Prasath VBS, Kawanaka H, Aronow BJ, Takase H. A study on nuclei segmentation, feature extraction and disease stage classification for human brain histopathological images. Procedia Computer Sci. (2016) 96:1202–10. doi: 10.1016/j.procs.2016.08.164
64. Yonekura A, Kawanaka H, Prasath VBS, Aronow BJ, Takase H. Automatic disease stage classification of glioblastoma multiforme histopathological images using deep convolutional neural network. Biomed Eng Lett. (2018) 8:321–7. doi: 10.1007/s13534-018-0077-0
65. Wang X, Wang D, Yao Z, Xin B, Wang B, Lan C, et al. Machine Learning Models for Multiparametric Glioma Grading With Quantitative Result Interpretations. Front Neurosci. (2018) 12:1046. doi: 10.3389/fnins.2018.01046
66. Ker J, Bai Y, Lee HY, Rao J, Wang L. Automated brain histology classification using machine learning. J Clin Neurosci. (2019) 66:239–45. doi: 10.1016/j.jocn.2019.05.019
67. Mobadersany P, Yousefi S, Amgad M, Gutman DA, Barnholtz-Sloan JS, Velazquez Vega JE, et al. Predicting cancer outcomes from histology and genomics using convolutional networks. Proc Natl Acad Sci USA. (2018) 115:E2970–E2979. doi: 10.1073/pnas.1717139115
68. Hu X, Wong KK, Young GS, Guo L, Wong ST. Support vector machine multiparametric MRI identification of pseudoprogression from tumor recurrence in patients with resected glioblastoma. J Magn Reson Imaging. (2011) 33:296–305. doi: 10.1002/jmri.22432
69. Jang BS, Jeon SH, Kim IH, Kim IA. Prediction of pseudoprogression versus progression using machine learning algorithm in glioblastoma. Sci Rep. (2018) 8:12516. doi: 10.1038/s41598-018-31007-2
70. Tiwari P, Prasanna P, Wolansky L, Pinho M, Cohen M, Nayate AP, et al. Computer-extracted texture features to distinguish cerebral radionecrosis from recurrent brain tumors on multiparametric mri: a feasibility study. AJNR Am J Neuroradiol. (2016) 37:2231–6. doi: 10.3174/ajnr.A4931
71. Sarkiss CA, Germano IM. Machine learning in neuro-oncology: can data analysis from 5,346 patients change decision making paradigms? World Neurosurg. (2019) 124:287–94. doi: 10.1016/j.wneu.2019.01.046
72. Nie D, Lu J, Zhang H, Adeli E, Wang J, Yu Z, et al. Multi-channel 3D deep feature learning for survival time prediction of brain tumor patients using multi-modal neuroimages. Sci Rep. (2019) 9:1103. doi: 10.1038/s41598-018-37387-9
73. Hao J, Kim Y, Kim TK, Kang M. PASNet: pathway-associated sparse deep neural network for prognosis prediction from high-throughput data. BMC Bioinformatics. (2018) 19:510. doi: 10.1186/s12859-018-2500-z
74. Emblem KE, Pinho MC, Zollner FG, Due-Tonnessen P, Hald JK, Schad LR, et al. A generic support vector machine model for preoperative glioma survival associations. Radiology. (2015) 275:228–34. doi: 10.1148/radiol.14140770
75. Liu L, Zhang H, Rekik I, Chen X, Wang Q, Shen D. Outcome prediction for patient with high-grade gliomas from brain functional and structural networks. Med Image Comput Comput Assist Interv. (2016) 9901:26–34. doi: 10.1007/978-3-319-46723-8_4
76. Lao J, Chen Y, Li ZC, Li Q, Zhang J, Liu J, et al. A Deep Learning-Based Radiomics Model for Prediction of Survival in Glioblastoma Multiforme. Sci Rep. (2017) 7:10353. doi: 10.1038/s41598-017-10649-8
77. Panesar SS, D'Souza RN, Yeh FC, Fernandez-Miranda JC. Machine learning versus logistic regression methods for 2-year mortality prognostication in a small, heterogeneous glioma database. World Neurosurg. (2019) 2:100012. doi: 10.1016/j.wnsx.2019.100012
78. Chang K, Zhang B, Guo X, Zong M, Rahman R, Sanchez D, et al. Multimodal imaging patterns predict survival in recurrent glioblastoma patients treated with bevacizumab. Neuro Oncol. (2016) 18:1680–7. doi: 10.1093/neuonc/now086
79. Grossmann P, Narayan V, Chang K, Rahman R, Abrey L, Reardon DA, et al. Quantitative imaging biomarkers for risk stratification of patients with recurrent glioblastoma treated with bevacizumab. Neuro Oncol. (2017) 19:1688–97. doi: 10.1093/neuonc/nox092
80. Bi WL, Hosny A, Schabath MB, Giger ML, Birkbak NJ, Mehrtash A, et al. Artificial intelligence in cancer imaging: clinical challenges and applications. CA Cancer J Clin. (2019) 69:127–57. doi: 10.3322/caac.21552
81. Kann BH, Thompson R, Thomas CRJr, Dicker A, Aneja S. Artificial intelligence in oncology: current applications and future directions. Oncology. (2019) 33:46–53.
82. Choy G, Khalilzadeh O, Michalski M, Do S, Samir AE, Pianykh OS, et al. Current applications and future impact of machine learning in radiology. Radiology. (2018) 288:318–28. doi: 10.1148/radiol.2018171820
83. Rudie JD, Rauschecker AM, Bryan RN, Davatzikos C, Mohan S. Emerging applications of artificial intelligence in neuro-oncology. Radiology. (2019) 290:607–18. doi: 10.1148/radiol.2018181928
Keywords: glioma, artificial intelligence, neural network, deep neural network, convolution neural network, support vector machines
Citation: Sotoudeh H, Shafaat O, Bernstock JD, Brooks MD, Elsayed GA, Chen JA, Szerip P, Chagoya G, Gessler F, Sotoudeh E, Shafaat A and Friedman GK (2019) Artificial Intelligence in the Management of Glioma: Era of Personalized Medicine. Front. Oncol. 9:768. doi: 10.3389/fonc.2019.00768
Received: 31 May 2019; Accepted: 30 July 2019;
Published: 14 August 2019.
Edited by:
Sandro M. Krieg, Technical University of Munich, GermanyReviewed by:
Saul Oswaldo Lugo Reyes, National Institute of Pediatrics, MexicoSeunggu Jude Han, Oregon Health & Science University, United States
Copyright © 2019 Sotoudeh, Shafaat, Bernstock, Brooks, Elsayed, Chen, Szerip, Chagoya, Gessler, Sotoudeh, Shafaat and Friedman. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Houman Sotoudeh, aHNvdG91ZGVoQHVhYm1jLmVkdQ==
 Houman Sotoudeh
Houman Sotoudeh Omid Shafaat
Omid Shafaat Joshua D. Bernstock
Joshua D. Bernstock Michael David Brooks1
Michael David Brooks1 Jason A. Chen
Jason A. Chen Gustavo Chagoya
Gustavo Chagoya Ehsan Sotoudeh
Ehsan Sotoudeh Amir Shafaat
Amir Shafaat Gregory K. Friedman
Gregory K. Friedman