- 1Department of Pathology, Peking Union Medical College Hospital, Chinese Academy of Medical Sciences, Peking Union Medical College, Beijing, China
- 2Department of Pathology, Beijing Hospital of Traditional Chinese Medicine, Capital Medical University, Beijing, China
- 3Department of Oncology, Peking Union Medical College Hospital, Chinese Academy of Medical Sciences, Peking Union Medical College, Beijing, China
Objective: The identification of DNA polymerase epsilon (POLE) mutation subtypes in endometrial cancer is critical for molecular classification. The mutation of the POLE gene could only be detected by sequencing until now. We propose to validate and develop the feasibility of using BaseScope, an in situ hybridization (ISH) assay, for the detection of POLE mutations in high-grade endometrioid carcinomas (EC).
Methods: Among 51 paraffin-embedded samples of high-grade EC, BaseScope-ISH assays were used to detect the RNA mutation status of the POLE gene, mainly focusing on two hotspot mutations of P286R and V411L. The number of positive signals in the cytoplasm was counted, setting the positive threshold and determining the in situ hybridization results. The sensitivity and specificity of BaseScope-ISH assay were compared with that of the Sanger sequencing results.
Results: Based on the BaseScope assay, there were 19 positive samples and 32 negative samples in a total of 51 samples. Of the 19 positive samples, 10 samples showed P286R site mutations in the POLE gene, while the other nine samples were V411L site mutations. Only one sample with the V411L site mutation identified by Sanger sequencing showed negative signal value. The remaining 31 cases without the P286R site mutation or V411L site mutations all showed negative signal. This analysis result showed the sensitivity was 95% and the specificity was 100% for the BaseScope assay detecting POLE mutants in high-grade EC.
Conclusion: In the case of high-grade EC, combined with morphological characteristics, the BaseScope assay can effectively and specifically identify POLE mutation cases, providing a reliable foundation for the application of clinical diagnosis and molecular classification.
Introduction
Endometrial cancer is one of the most common gynecologic malignanciesin developed countries (1). In China, it ranks second in female reproductive system malignant tumors. According to the National Cancer Center, the incidence rate in 2015 was 0.643‰, and the mortality rate was 0.218‰ in China. Recently, the incidence of endometrial cancer has increased (2). Some gene alterations facilitating endometrial tumorigenesis have been identified (3–6). The Cancer Genome Atlas Research Network (TCGA) has divided endometrial carcinomas into four categories according to various genetic and epigenetic features: an ultramutator phenotype caused by DNA polymerase epsilon (POLE) mutations, a hypermutator phenotype caused by DNA mismatch repair deficiency (dMMR) leading to microsatellite instability (MSI), a copy number low phenotype, and a copy number high phenotype (3).
POLE, the catalytic subunit of DNA polymerase ε, is implicated in nuclear DNA replication and repair (7). Most studies show that the most common mutations of POLE were located in the exonuclease domain (exons 9–14), which are involved in proofreading activity during DNA replication. These mutations caused an unusually high mutational burden, leading to a hypermutated phenotype in colorectal and endometrial carcinomas (8). Endometrioid carcinoma (EC) is the primary subtype of endometrial carcinoma, accounting for 70–80% of cases. In EC patients, the most common mutations in POLE were in exon 9 (P286R and S297F) and exon 13 (V411L, L424V, and L424I), in which two mutations (P286R and V411L) accounted for 63–76% of all POLE point mutations. It has been established that POLE mutations like these contribute to a better prognosis and longer progression-free survival (3, 4, 9–11).
Meanwhile, POLE mutant cases have received much more attention because their morphological feature is inconsistent with the prognostic outcome. Currently, for EC patients, most tailored treatments recommended are based on the histological type, stage, and grade (12–15). However, it has been recently found that the efficacy of molecular typing in judging prognosis exceeds the histomorphological diagnosis to a certain extent, especially for high-grade endometrial carcinomas, which have significant overlaps in histopathologic and immunohistochemical characteristics (9, 16–19).
Molecular typing of endometrial cancer is of great significance for guiding clinical treatment and judging the prognosis of patients. Currently, this subtype is mainly recognized by conventional Sanger sequencing, and there are no other methods or alternatives for molecule detection used in clinical diagnosis. In our study, we will use specific BaseScope mutation probes to identify POLE mutations, and analyze the sensitivity and specificity of the BaseScope-ISH assay in ECs.
Materials and Methods
Human Samples
The information from “high-grade endometrioid carcinoma” cases was recorded in the archives of the Beijing Union Medical College Hospital from June 1, 2010, to November 30, 2017. A total of 202 cases were retrieved, and there were 161 cases with paraffin-embedded tumor tissues available. Fifty-one cases were selected for our studies, including 39 POLE gene (exon 9–14) mutation samples and 12 samples containing no POLE exon 9–14 mutations. All cases were under ethical approval (PUMCH committee, No. S-K688), and both informed and written consents were obtained from all patients of this study when they were admitted to the hospital (the written consents were kept in archives with every patient's files). The average time since the fixation of tissue blocks was 4 years (range 0–8 years). At least three expert histopathologists examined samples.
Histotype and Grade Assignment
We had original diagnoses from the archived data on both diagnostic (curettage specimens) and final hysterectomy specimens. Also, two gynecologic subspecialty pathologists (Zhaohui Lu and Shuangni Yu) reviewed hematoxylin and eosin stained slides from diagnostic and final hysterectomy specimens with the goal of assigning histotype and grade. Only those specimens of grade 3 or 2–3 and endometrioid carcinoma were kept in the study. These pathologists were blind to the original pathology reports and to each other's interpretations.
TMA Construction
For all diagnostic endometrioid carcinoma samples (endometrial curettage and hysterectomy specimens), two pieces of tissue microarrays were constructed using 2 mm cores per case in duplicate at random.
Sanger Sequencing
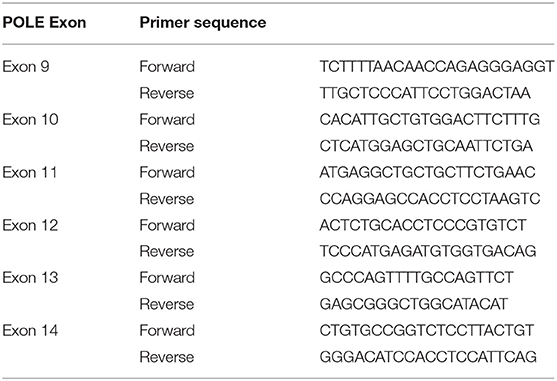
DNA extracted using the QiaAMP DNA micro kit (QIAGEN Ltd, Manchester, UK) was used for polymerase chain reaction (PCR) to amplify POLE exons 9–14. PCR primers were designed (Table 1), and PCR products (150–200 bp) were amplified with the input of 100 ng FFPE derived DNA. Sequencing was performed using BigDye v3.1 terminator cycle sequencing chemistry on the ABI 3730 DNA analyzer (Applied Biosystems Inc., Foster City, CA). All validated POLE mutations were bidirectionally sequenced twice using Sanger sequencing.
BaseScope Assay
BaseScope assays were performed and assisted following guidelines (BaseScope™ Detection Reagent Kit-RED User Manual) provided by the supplier (Advanced Cell Diagnostics, Newark, CA). Briefly, sections were cut at 4 μm thickness onto Superfrost plus slides (Thermo Scientific, New Hampshire, USA) and allowed to dry overnight at room temperature (RT). Sections were then baked at 60°C for one h before being deparaffinized in xylene (2 ×5 min) and ethanol (2 ×2 min), then dried by baking at 60°C for 2 min. Pretreat 1 (hydrogen peroxide) was applied for 10 min at RT, Pretreat 2 (target retrieval) for 15 min at 100°C and Pretreat 3 (protease IV) for 30 min (tissue sections) at 40°C, with two rinses in distilled water between pretreatments. BaseScope probes (four types of candidate probes were all purchased from Advanced Cell Diagnostics) (see Table 2) were then applied for 2 h at 40°C in a HybEZ oven before incubation with reagents AMP0 (30 min at 40°C), AMP1 (15 min at 40°C), AMP2 (30 min at 40°C), AMP3 (30 min at 40°C), AMP4 (15 min at 40°C), AMP5 (30 min at RT), and AMP6 (15 min at RT). Slides were rinsed with wash buffer (2 ×2 min) between AMP incubations. Finally, slides were incubated with Fast Red for 10 min at room temperature in the dark. Then, slides were counterstained with hematoxylin before drying for 15 min at 60°C.
Signal Quantification and Statistical Analysis
The signals were observed and counted by eye under the microscopy (at 400 × magnification) as a preliminary investigation and digital images of signals were photographed for the convenience of display and a second look. The hybridized images were scanned into digital data, the cytoplasm signals were recorded, and statistical analysis was made for each sample (400 × High magnification vision field). The average value was calculated, and the cutoff value was set at 31 red dots signal/3.14 mm2 (≈10/mm2), positive samples: ≥31 red dots signal/3.14 mm2; negative samples: <31 red dots signal/3.14 mm2. All data were represented in mean ± standard deviation (SD), and statistical significance was set at p <0.05 (two-tailed). We also calculated the sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV). Statistical analyses were performed using Student's t-test.
Results
Validation of BaseScope Probes in High-Grade EC
First, we used the positive control probe to detect RNA status in paraffin-embedded high-grade EC samples, and the results showed numerous strong positive hybridization signals (Figures 1A,D); then we used two types of BaseScope probes, one specific for the P286R point mutation (857C > G) and the other for the V411L point mutation (1231G > C/T), respectively, to test high-grade EC samples by ISH. The results showed positive staining signals (Figures 1C,F), but little to no signal in the negative samples (<31 positive signals/1 core of tissue array, or 10 positive signals/mm2) (Figures 1B,E).
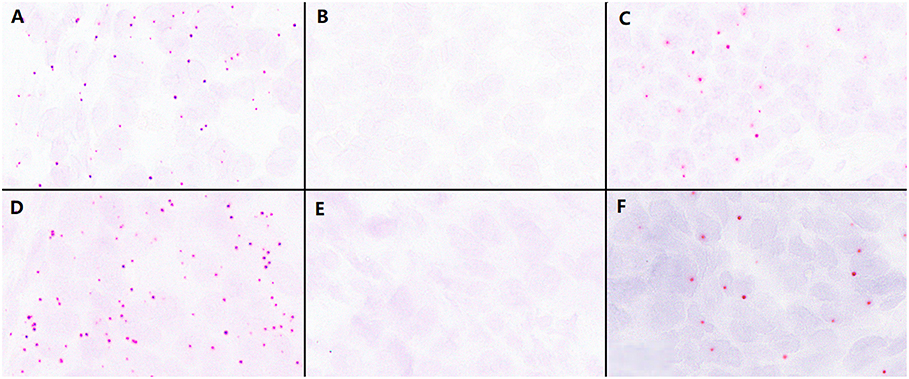
Figure 1. Validation of the effectiveness of the BaseScope probe in tumor tissues. (A) The analysis of RNA quality by the P286R positive control probes in P286R mutated high-grade endometrioid carcinoma (EC), showing a large number of positive signals. (B) Negative control probes showed no positive signal in P286R mutated EC. (C) The detection of P286R mutations in high-grade EC by BaseScope in situ hybridization, showing multiple strong red signals in the cytoplasm. (D) The analysis of RNA quality by V411L positive control probes in V411L mutated high-grade EC, showing a large number of positive signals. (E) Negative control probes showed no positive signal in V411L mutated EC. (F) The detection of V411L mutation in high-grade EC by BaseScope in situ hybridization, showing multiple strong red signals in the cytoplasm; Scale bars represent 60 microns.
POLE Mutation Sequencing and Sensitivity and Specificity of the BaseScope Assay
We sequenced POLE gene exons 9–14 in 161 high-grade EC samples from our center, and the results showed 39 patients possessed point mutations in their POLE gene exons 9–14. By reverse sequencing, the results confirmed that these 39 samples included mutations in the POLE exon region, 10 cases with the P286R site mutation, and 10 with the V411L site mutation, which comprised 51% of all the mutations.
We made tissue microarrays using 51 cases of high-grade EC, including the 10 cases of the P286R mutation and 10 cases of the V411L mutation. Twenty cases with other point mutations in POLE exons 9–14 (including one case with P286R mutation and another point mutation of the POLE exons 9–14), and 12 cases without POLE exon 9–14 mutations were used as controls.
The number of positive signal points of the BaseScope-ISH in each tissue microarray was collected; they are shown in Table 3, ranging between 0 and 640/3.14 mm2. The positive threshold was set to 31/3.14 mm2 (10/mm2). BaseScope-ISH results showed that 19 of 51 cases were positive based on the threshold (Figure 2). Ten of 19 positive samples were P286R point mutations, and nine cases were V411L site mutations, which is entirely consistent with sequencing results. In the BaseScope-ISH negative samples, only one case in which a V411L site mutation was identified by sequencing showed a few signal points (3/3.14 mm2), which was below the positive threshold and inconsistent with sequencing results. The remaining cases were all negative, the same as the sequencing results. There was a significant difference in the number of signal points between the P286R probe positive group (mean ± standard deviation: 427 ± 155) and the negative group (mean ± standard deviation: 3 ± 4) (P <0.001). The number of signal points in the V411L probe positive group (mean ± standard deviation: 339 ± 176) and the negative group (mean ± standard deviation: 6 ± 6) were also significantly different (P <0.001). For the sensitivity and specificity of BaseScope detection, the P286R probe was 100% (10 samples were all detected); the sensitivity of the V411L probe was 90% (only 1 of 10 samples was not detected), and the specificity was 100%. The positive and negative predictive value of the P286R site mutation in the POLE gene were both 100%; for the V411L site mutation, they were 90 and 97.6%, respectively.
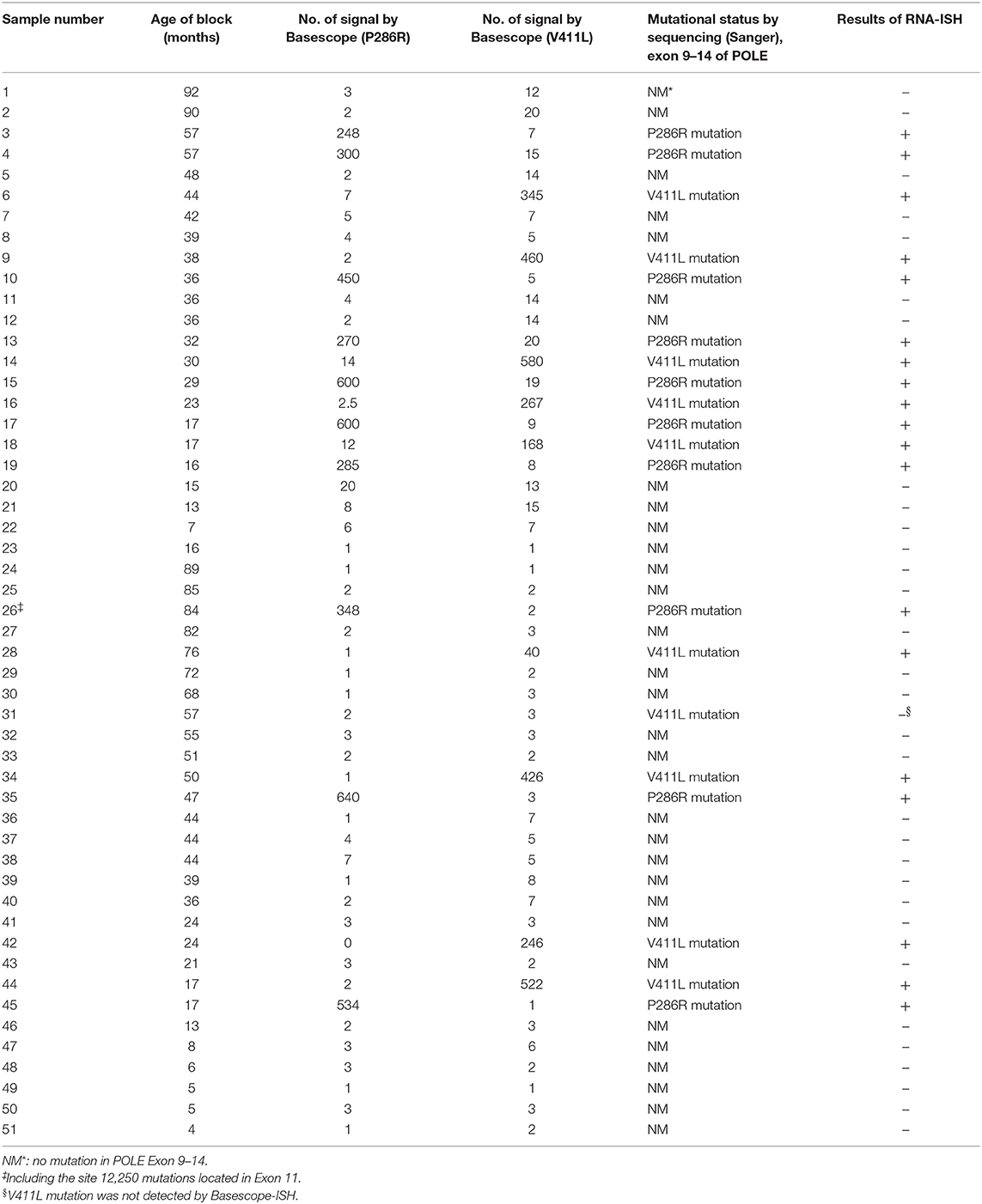
Table 3. Number of positive signals in high-grade endometrioid carcinoma tissues (mean value of two tissue microarray core per case, each tissue microarray core area is about 3.14 mm2).
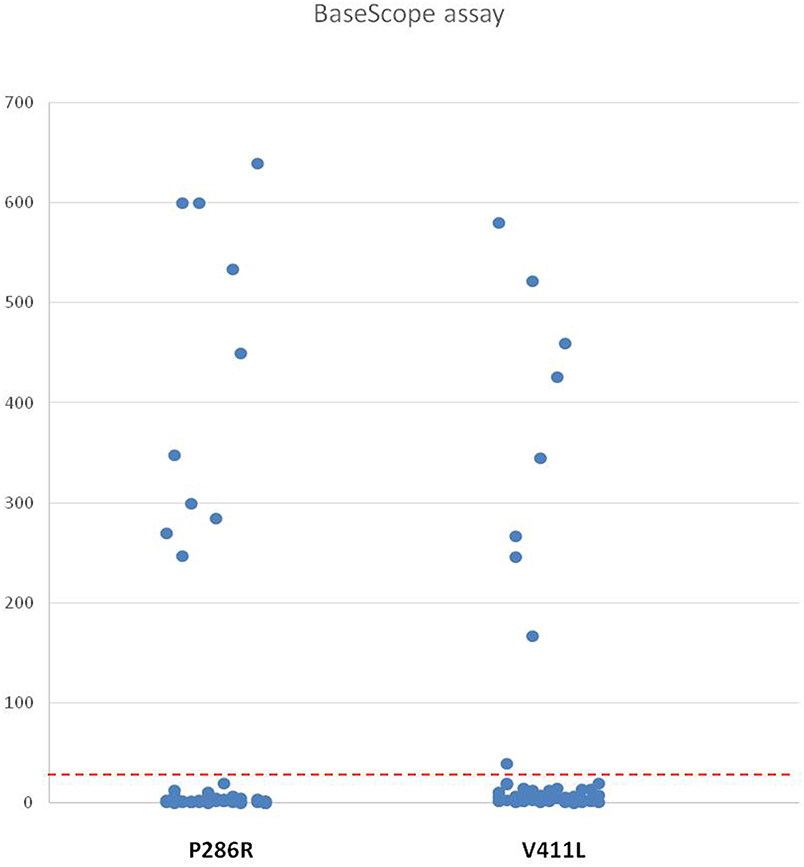
Figure 2. The Number of the Positive Signal of Two Probes in High-Grade EC. The ordinate shows the value of the positive signal of P286R and V411L probes in high-grade EC microarrays (51 cases), and the red dashed line represents the positive threshold (31/3.14 mm2).
Comparison of Mixed Probes to Single Probes
In order to detect the two mutation sites simultaneously, we used two probe mixtures to hybridize 51 high-grade EC samples and found 19 mutations samples, including 10 cases of P286R point mutations, and nine cases of V411L point mutations. The case involving the V411L mutation identified by sequencing but negative by the single probe still did not reach the positive threshold (the number of positive signals is 17/3.14 mm2)using mixed probes. The result of mixed probes was utterly consistent with the single probe detection with a sensitivity of 95% and a specificity of 100% (Figures 3A–D).
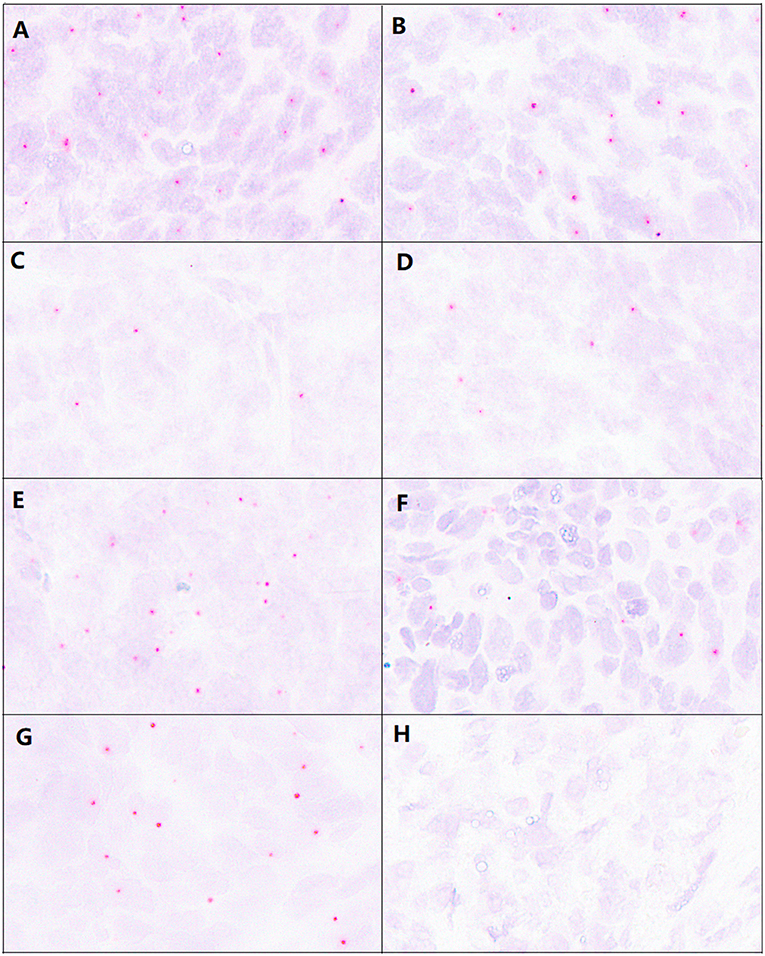
Figure 3. Comparison of detection results from mixed and single probes and paraffin section signal images before and after 5 months. (A) The signal of the P286R probe in P286R mutated high-grade EC. (B) The signal of mixed probes (P286R and V411L probe) in P286R mutated high-grade EC. (C) The signal of the V411L probe in V411L mutated high-grade EC. (D) The signal of mixed probes (P286R and V411L probe) in V411L mutated high-grade EC. (E,F) The detection using the P286R probe in P286R high-grade EC paraffin section before (E) and after (F) 5 months, the hybridization signal counts were significantly reduced 5 months after in paraffin sections. (G,H) The detection using the V411L probe in V411L high-grade EC paraffin section before (E) and after (F) 5 months, the hybridization signal counts were significantly reduced 5 months after in paraffin sections.
Comparison of Slide Detection at 5 Months and After
We prepared one tissue microarray with 22 tumor tissue samples, including seven cases of the P286R site mutation, five cases of the V411L site mutation, six cases of other mutations within POLE exons 9–14, and four cases without exon 9–14 mutations in the POLE gene. We placed these sample slices at room temperature for 5 months and then repeated BaseScope-ISH.
For the samples with P286R site mutations in their POLE gene, the number of positive signal points decreased (Figures 3E,F), but the number of positive samples was still 7, and the positive detection rate had no change (100%). In the POLE gene V411L point mutation samples, the number of positive signal points was decreased (Figures 3G,H), the number of signals in two samples did not reach the positive threshold, and the positive detection rate decreased by 40%. However, the specificity of these two probe detections was still perfect (100%) (Figure 4).
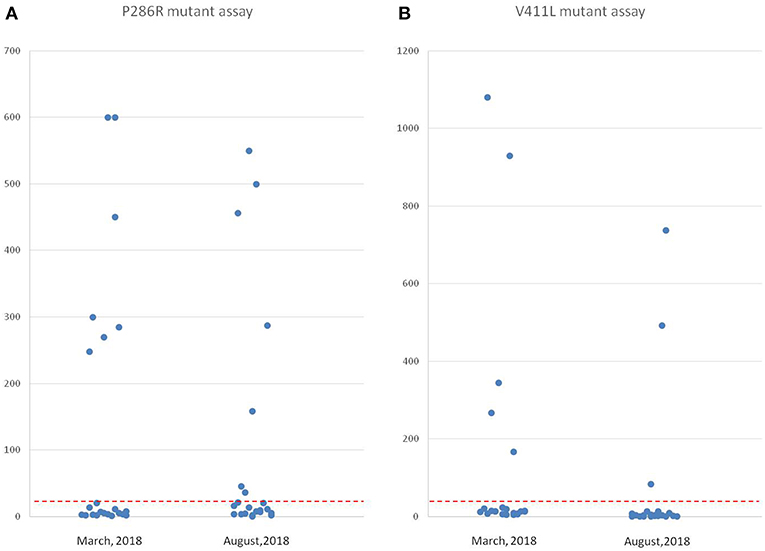
Figure 4. Comparison of point mutation signals detected by in situ hybridization 5 months before and after in paraffin sections using two probes. (A) The values of the positive signal of POLE P286R mutations from the same group of high-grade EC tissues at two time points (22 cases). The left side is the result from March 2018, and the right side is the result from August 2018 (the number of positive signals decreased). (B) The values of the positive signal of POLE V411L mutations from the same group of high-grade EC tissues at two-time points (22 cases). The left side is the result from March 2018, and the right side is the result from August 2018 (the number of positive signals decreased remarkably, and the signal values of two cases fell below the threshold).
Discussion
BaseScope-ISH is an effective, highly sensitive, and specific method for detecting point mutations in RNA level (20). It has been reported to be used in studying tumor heterogeneity in colon cancer tumor tissues (21). Some researchers have successfully detected the ALK and PD-L1 gene mutations in lung cancer (22, 23). In our study, we used the BaseScope probes of two hot POLE gene mutations to test the high-grade EC samples, which surmounted the disadvantages of screening mutant samples by sequencing. It is more useful for the samples with less tissue because it does not require additional tissue for DNA extraction. Meanwhile, we could observe and count the positive signals under microscopy by eye when tumor cell morphology was directly combined as reference. Assessment of signals by eye provided acceptably high sensitivity and specificity, which would allow this methodology to be more widely used in clinical practice. Equally important we performed signal interpretation by intuitively and closely combining tumor cell morphology. The positive signals were counted only when they were within the cytoplasm or nucleus of tumor cells. The bulky nuclei of tumor cells were outlined by hematoxylin. Signals presenting in normal endometrium, vascular wall, and fibrotic or necrotic tissues were interpreted as false negative signals and wouldn't be included. This combination of signal reading with cell morphology would bring us an accurate evaluation of assay.
The individualized treatment of tumors is closely related with the molecular typing of tumor genes (24–26), and tumor with the POLE gene mutation, representing a unique subtype of EC, owns a quite better prognosis. In particular, these tumors often appear to be high-grade. And in the absence of the knowledge of their POLE gene mutation status, they would be mistakenly included into a category of tumors with much worse prognosis than they actually have. So, the identification of this mutated subgroup is essential for better personalized treatment and prognosis analysis of endometrial cancer (3, 17), and our findings made a vital step toward this goal.
The POLE gene mutation rate in high-grade EC is 10–26% (9, 16, 17, 27), and the mutation rate in our center's samples was 24% (39/161 cases), which is consistent with the reported mutation rate. There are many different kinds of point mutations in the POLE gene, which are mainly distributed in the exon 9–14 region. The most common mutations are at site 857 (amino acid 286, P286R) and 1231 (amino acid 411, V411L), accounting for 63–76% of all mutation types (3, 27, 28). The mutation rate of these two hotspots in our samples was 51% of the entire exon 9–14 region mutations. Compared with the literature, the ratio was slightly low. We found that the mutation site in our samples distribution spectrum was wide—which might be a characteristic of disease-related mutations in our Chinese population.
Our study also showed that two mutation points could be detected at the same time if we use mixed probes to test samples, with over half of the mutations recognized in one test. This significantly improves the efficiency of mutation case screening, and has great significance for clinical diagnosis. Further studies would be carried out to look for the probes that are able to recognize the other identifiable mutations, as well as to answer the questions about how many such probes could be effectively combined.
There are two subtypes of mutated bases for the POLE gene V411L (1231G > C/T) site mutation (4, 9, 27): G/C and G/T. In 10 cases of the V411L mutation samples we tested, five cases were G-mutated to C, and five cases were G-mutated to T. We detected these two mutations simultaneously when using BaseScope-ISH, indicating that this probe can recognize two mutations at the same site. In practical applications, it will remarkably increase the detection types and detection efficiency. For POLE gene V411L mutation detection, the number of positive signals in most mutant samples was much higher than the threshold. However, in one sample, the number of positive signals was 40, slightly higher than the positive threshold (31 signals); and the number in another sample did not reach the positive threshold. This may be caused by the small proportion of mutant cells or sample degradation before fixation. The possibility of uneven distribution caused by tumor heterogeneity is less likely because all of the samples were randomly acquired, and signals were detected in all other mutant samples. So, for the majority of samples, the tissue microarray is considered to be a good representative.
The POLE mutations can be detected in archived paraffin-embedded samples(The earliest sample used in this study was collected in 2010). However, for paraffin tissue slices placed at room temperature for 5 months, some RNA degradation may occur, resulting in a significantly reduced detection rate, and the omission of some positive cases. Therefore, it is no longer suitable for RNA in situ hybridization detection to be used on long-term, preserved slices stored at room temperature. In daily work, we should pay attention to sample preservation, and use freshly prepared paraffin slices for ISH detection of RNA point mutations when possible.
Conclusion
BaseScope-ISH is a significant advancement and improvement in screening paraffin-embedded EC tissue samples for POLE mutations, which can enhance the identification of POLE mutation subtypes and provide a powerful strategy for the molecular classification and detection of other tumors involving known gene mutations.
Data Availability
The datasets analyzed in this manuscript are not publicly available. Requests to access the datasets should be directed to bHV6aGhAcHVtY2guY24=.
Author Contributions
ZL: experimental design, supervision, and results analysis. SY and HS: experiments, manuscript writing, and data analysis. XB: sample collection and data analysis. HKZ and JC: critical reading and revision of the manuscript. YY and XM: evaluation of positive signals. NZ: figure and table construction. HZ: preparation of tissue slice and technical supports.
Funding
This work was supported by the CAMS Science and Technology Innovation Program Fund for Medical Sciences and Health (Grant No. 2016-I2M-1-001), the National Science Foundations of China (Grant Nos. 81672648, 81472326, 81400664, and 81341070), and special funds from the CAMS Molecular Pathology Center and the Central Public Welfare Institutions of CAMS (Grant Nos. 2016ZX310176-4 and 2017PT31008).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We sincerely thank Yuhan Zhang for technical support with tissue microarray preparation.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. (2018) 68:7–30. doi: 10.3322/caac.21442
2. Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, et al. Cancer statistics in China, 2015. CA Cancer J Clin. (2016) 66:115–32. doi: 10.3322/caac.21338
3. Kandoth C, Schultz N, Cherniack AD, Akbani R, Liu Y, Shen H, et al. Integrated genomic characterization of endometrial carcinoma. Nature. (2013) 497:67–73. doi: 10.1038/nature12113
4. Stelloo E, Bosse T, Nout RA, MacKay HJ, Church DN, Nijman HW, et al. Refining prognosis and identifying targetable pathways for high-risk endometrial cancer; a TransPORTEC initiative. Mod Pathol. (2015) 28:836–44. doi: 10.1038/modpathol.2015.43
5. McMeekin DS, Tritchler DL, Cohn DE, Mutch DG, Lankes HA, Geller MA, et al. Clinicopathologic significance of mismatch repair defects in endometrial cancer: an NRG Oncology/Gynecologic Oncology Group Study. J Clin Oncol. (2016) 34:3062–8. doi: 10.1200/JCO.2016.67.8722
6. Alkushi A, Kobel M, Kalloger SE, Gilks CB. High-grade endometrial carcinoma: serous and grade 3 endometrioid carcinomas have different immunophenotypes and outcomes. Int J Gynecol Pathol. (2010) 29:343–50. doi: 10.1097/PGP.0b013e3181cd6552
7. Rayner E, van Gool IC, Palles C, Kearsey SE, Bosse T, Tomlinson I, et al. A panoply of errors: polymerase proofreading domain mutations in cancer. Nat Rev Cancer. (2016) 16:71–81. doi: 10.1038/nrc.2015.12
8. Shinbrot E, Henninger EE, Weinhold N, Covington KR, Goksenin AY, Schultz N, et al. Exonuclease mutations in DNA polymerase epsilon reveal replication strand specific mutation patterns and human origins of replication. Genome Res. (2014) 24:1740–50. doi: 10.1101/gr.174789.114
9. Church DN, Stelloo E, Nout RA, Valtcheva N, Depreeuw J, ter Haar N, et al. Prognostic significance of POLE proofreading mutations in endometrial cancer. J Natl Cancer Inst. (2015) 107:402. doi: 10.1093/jnci/dju402
10. Lancaster JM, Powell CB, Chen LM, Richardson DL. Society of Gynecologic Oncology statement on risk assessment for inherited gynecologic cancer predispositions. Gynecol Oncol. (2015) 136:3–7. doi: 10.1016/j.ygyno.2014.09.009
11. Stelloo E, Nout RA, Osse EM, Jurgenliemk-Schulz IJ, Jobsen JJ, Lutgens LC, et al. Improved risk assessment by integrating molecular and clinicopathological factors in early-stage endometrial cancer-combined analysis of the PORTEC cohorts. Clin Cancer Res. (2016) 22:4215–24. doi: 10.1158/1078-0432.CCR-15-2878
12. Colombo N, Preti E, Landoni F, Carinelli S, Colombo A, Marini C, et al. Endometrial cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. (2013) 24 Suppl. 6:vi33–8. doi: 10.1093/annonc/mdt353
13. Kong TW, Chang SJ, Paek J, Lee Y, Chun M, Ryu HS. Risk group criteria for tailoring adjuvant treatment in patients with endometrial cancer: a validation study of the Gynecologic Oncology Group criteria. J Gynecol Oncol. (2015) 26:32–9. doi: 10.3802/jgo.2015.26.1.32
14. Kwon JS, Qiu F, Saskin R, Carey MS. Are uterine risk factors more important than nodal status in predicting survival in endometrial cancer? Obstet Gynecol. (2009) 114:736–43. doi: 10.1097/AOG.0b013e3181b96ec6
15. Soslow RA. High-grade endometrial carcinomas–strategies for typing. Histopathology. (2013) 62:89–110. doi: 10.1111/his.12029
16. Billingsley CC, Cohn DE, Mutch DG, Hade EM, Goodfellow PJ. Prognostic significance of POLE exonuclease domain mutations in high-grade endometrioid endometrial cancer on survival and recurrence: a subanalysis. Int J Gynecol Cancer. (2016) 26:933–8. doi: 10.1097/IGC.0000000000000681
17. Meng B, Hoang LN, McIntyre JB, Duggan MA, Nelson GS, Lee CH, et al. POLE exonuclease domain mutation predicts long progression-free survival in grade 3 endometrioid carcinoma of the endometrium. Gynecol Oncol. (2014) 134:15–9. doi: 10.1016/j.ygyno.2014.05.006
18. Bosse T, Nout RA, McAlpine JN, McConechy MK, Britton H, Hussein YR, et al. Molecular classification of grade 3 endometrioid endometrial cancers identifies distinct prognostic subgroups. Am J Surg Pathol. (2018) 42:561–8. doi: 10.1097/PAS.0000000000001020
19. Britton H, Huang L, Lum A, Leung S, Shum K, Kale M, et al. Molecular classification defines outcomes and opportunities in young women with endometrial carcinoma. Gynecol Oncol. (2019) 153:487–95. doi: 10.1016/j.ygyno.2019.03.098
20. Erben L, He M-X, Laeremans A, Park E, Buonanno A. A novel ultrasensitive in situ hybridization approach to detect short sequences and splice variants with cellular resolution. Mol Neurobiol. (2018) 55:6169–81. doi: 10.1007/s12035-017-0834-6
21. Baker AM, Huang WN, Wang XMM, Jansen M, Ma XJ, Kim J, et al. Robust RNA-based in situ mutation detection delineates colorectal cancer subclonal evolution. Nat Commun. (2017) 8:8. doi: 10.1038/s41467-017-02295-5
22. Nakajima N, Yoshizawa A, Kondo K, Rokutan-Kurata M, Hirata M, Furuhata A, et al. Evaluating the effectiveness of RNA in-situ hybridization for detecting lung adenocarcinoma with anaplastic lymphoma kinase rearrangement. Histopathology. (2017) 71:143–9. doi: 10.1111/his.13198
23. Yu H, Batenchuk C, Badzio A, Boyle TA, Czapiewski P, Chan DC, et al. PD-L1 Expression by two complementary diagnostic assays and mRNA in situ hybridization in small cell lung cancer. J Thorac Oncol. (2017) 12:110–20. doi: 10.1016/j.jtho.2016.09.002
24. Hussein YR, Soslow RA. Molecular insights into the classification of high-grade endometrial carcinoma. Pathology. (2018) 50:151–61. doi: 10.1016/j.pathol.2017.09.010
25. Talhouk A, Derocher H, Schmidt P, Leung S, Milne K, Gilks B, et al. Molecular subtype not immune response drives outcomes in endometrial carcinoma. Clin Cancer Res. (2019) 25:2537–48. doi: 10.1158/1078-0432.CCR-18-3241
26. Bell DW, Ellenson LH. Molecular genetics of endometrial carcinoma. Annu Rev Pathol. (2019) 14:339–67. doi: 10.1146/annurev-pathol-020117-043609
27. McConechy MK, Talhouk A, Leung S, Chiu D, Yang W, Senz J, et al. Endometrial carcinomas with POLE exonuclease domain mutations have a favorable prognosis. Clin Cancer Res. (2016) 22:2865–73. doi: 10.1158/1078-0432.CCR-15-2233
Keywords: POLE, endometrioid carcinoma, detection, BaseScope-ISH, P286R, V411L
Citation: Yu S, Shao H, Ban X, Zhang H, You Y, Zhou N, Mao X, Zhao H, Chen J and Lu Z (2019) Detection of POLE Subtypes in High-Grade Endometrioid Carcinoma by BaseScope-ISH Assay. Front. Oncol. 9:831. doi: 10.3389/fonc.2019.00831
Received: 06 June 2019; Accepted: 13 August 2019;
Published: 04 September 2019.
Edited by:
Jian-Jun Wei, Northwestern University, United StatesReviewed by:
Kruti P. Maniar, Northwestern University, United StatesUmberto Malapelle, University of Naples Federico II, Italy
Copyright © 2019 Yu, Shao, Ban, Zhang, You, Zhou, Mao, Zhao, Chen and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhaohui Lu, bHV6aGhAcHVtY2guY24=
†These authors have contributed equally to this work
 Shuangni Yu
Shuangni Yu Huilin Shao
Huilin Shao Xinchao Ban
Xinchao Ban Hongkai Zhang
Hongkai Zhang Yan You
Yan You Na Zhou
Na Zhou Xinxin Mao
Xinxin Mao He Zhao1
He Zhao1 Jie Chen
Jie Chen Zhaohui Lu
Zhaohui Lu