- 1Department of Immunology, Jiangsu Key Laboratory of Laboratory Medicine, School of Medicine, Jiangsu University, Zhenjiang, China
- 2Zhenjiang Hospital of Chinese Traditional and Western Medicine, Zhenjiang, China
Metabolism is a complex network of regulatory system. Cells often alter their metabolism in response to the changes in their environment. These adaptive changes are particularly pronounced in tumor cells, known as metabolic reprogramming. Metabolic reprogramming is considered to be one of the top 10 characteristics of tumor cells. Glucose and lipid metabolism are important components of metabolic reprogramming. A large number of experimental studies have shown that long non-coding RNAs (lncRNAs) play an important role in glucose and lipid metabolism. The current review briefly introduces the regulatory effect of lncRNAs on glucose and lipid metabolism of tumor cells, and the significance of lncRNA-mediated metabolism in tumor therapy, hoping to provide new strategies for clinical targeting tumor therapy.
Introduction
Long non-coding RNAs (lncRNAs) are a class of RNA molecules <200 bp in length. LncRNAs are normally transcribed by RNA polymerase II and undergoes 5′ end capping, RNA splicing, and polyadenylation procedures (1, 2). LncRNAs do not code or have very low protein-coding capacity (3), and they have been regarded as “transcriptional noise” without any biological effects in the past (4). However, increasing evidence shows that lncRNAs are involved in gene regulation (5), cell cycle regulation (6), cell differentiation (7), immune response (8), tumor metabolism (9), and other processes (10, 11). LncRNAs often perform different functions in the cytoplasm and the nucleus (2). Interestingly, environmental transitions or infections can also induce changes of the lncRNA localization in the nucleus and the cytoplasm. For example, the nuclear export and mitochondrial localization of lncRNA RMRP is regulated by G-rich RNA sequence-binding factor 1 and human antigen R in the environments (12). In general, the molecular mechanisms of lncRNAs mainly include signals, decoys, guides, and scaffolds (13). As signal molecules, lncRNAs could mark the regulation of space, time, developmental stage, and gene expression. A signal molecule, linc-p21 as an example, is found to play an important role in apoptotic response after DNA damage, and this depends on the p53 pathway (14). In addition, lnc-c/EBPβ and lnc-chop are found to regulate the immunosuppressive function of myeloid-derived suppressor cells (MDSCs) in tumor and inflammatory environments, which as scaffolds bring together multiple proteins to form complexes (15, 16).
In the tumor microenvironment, lncRNAs may show oncogenic or tumor-suppressive functions due to the changes in its expression (17). The hypoxia, low pH, and energy stress of the tumor microenvironment are all factors that cause alteration in lncRNAs. It is reported that energy stress-induced lncRNA FILNC1 and lncRNA HAND2-AS1 could repress tumor development by regulating energy metabolism (18, 19). Hypoxia is also one of the microenvironmental cues responsible for lncRNAs changes, for example, the overexpression of lncRNA EIF3J-AS1 in atocellular carcinoma and lncRNA-HAL in breast cancer (20, 21). HIF-1α and c-Myc are often involved in the regulation of lncRNAs under hypoxia conditions. The potential mechanisms of dysregulation of lncRNAs will not be described in detail; here, we will focus on the regulatory effects of lncRNAs on glucose and lipid metabolism.
It is commonly believed that the balance of glucose, fatty acid, and protein metabolism plays an essential role in mammals. Once this metabolic balance is broken, it may cause various diseases, even tumor (22). Abundant evidence reveals that during the process of various diseases, especially cell carcinogenesis (23), the metabolic pattern changes significantly, involving glycolysis, mitochondrial oxidative phosphorylation, fatty acid oxidation, and other aspects. The researchers called this phenomenon the metabolic reprogramming of tumor cells (24–27). Glucose metabolism and lipid metabolism are the main energy metabolism modes of organisms, and they are closely related. Moon et al. reported that the products of glucose metabolism are substrates of lipid synthesis. Androgen can stimulate the conversion between metabolisms, and it increases the utilization of glucose by activating HK2 and PFKFB2 to provide a sufficient carbon source for fatty acid synthesis (28). Researchers also detected that the lactic acid from active TCA cycle is the primary raw material for fatty acid synthesis in glioblastoma cells by using 13C NMR spectroscopy (29). It has been reported that in the tumor microenvironment, the majority of tumor cells choose to increase glycolysis to meet their own energy and material needs, but some prefer to fatty acids as an energy source. In adipocytes and breast cancer cell co-culture models, breast cancer cells prefer to use β-oxidation of fatty acids (FAO) to supply the energies for proliferation and migration, which may be related to the tissue specificity of the breast (30). There are also some reports that suggest that under metabolic pressure, tumor cells ingest fatty acids in the surroundings and perform FAO to provide nutrients when glucose levels are low (31). Also, the amount of ATP produced by the full oxidation of each fatty acid molecule is more than twice that of glucose (32).
In conclusion, the metabolic mode of tumor cells is complex and changeable, and under pathological conditions, tumor cells will choose the optimal metabolic mode for their survival according to the different environments in which they are located. Recently, with the development of metabolic reprogramming of tumor cells, the roles of lncRNAs in glucose and lipid metabolism become a research hotspot, especially in glucose metabolism. Numerous evidence shows that lncRNAs can reprogram glucose and lipid metabolism in tumor cells, which influence tumor initiation, development, and progression, and may serve as a promising novel target for diagnosis and treatment of tumor.
LncRNAs Participate in Glucose Metabolism in Tumor Cells
As the most important energy source of tumor cells, glucose metabolism mainly includes glucose uptake (primary entrance), entry into mitochondrial oxidative phosphorylation, and excretion of lactate (two primary exits) (33). Also, a little bit goes into pentose phosphate pathway (PPP). Glucose metabolic reprogramming is an important feature of tumor cells, providing sufficient ATP and NADPH for tumor cells to adapt to the changes in survival conditions and rapid proliferation. Under normal physiological conditions, mitochondrial oxidative phosphorylation is the main source of ATP, while tumor cells tend to have enhanced glycolysis even under sufficient oxygen conditions (34–36). Compared to oxidative phosphorylation, which produces 36 molecules of ATP, glycolysis only produces two molecules of ATP. However, ATP is formed faster than oxidative phosphorylation and it provides abundant macromolecular precursors to meet the demands for rapid growth and differentiation of tumor cells to the maximum extent. This is known as Warburg effect (37). In addition, the PPP is also an important process for the synthesis of biomacromolecules and reduction of equivalent (38). In the process of metabolic reprogramming, glucose transporters and many key enzymes determine the metabolic rate of glucose in most cases. The expression levels of these dominating enzymes usually markedly change during cell carcinogenesis (27). It is reported that lncRNAs regulate glucose metabolism primarily by regulating key enzymes in the glucose metabolism pathway.
LncRNAs Participate in Glucose Uptake
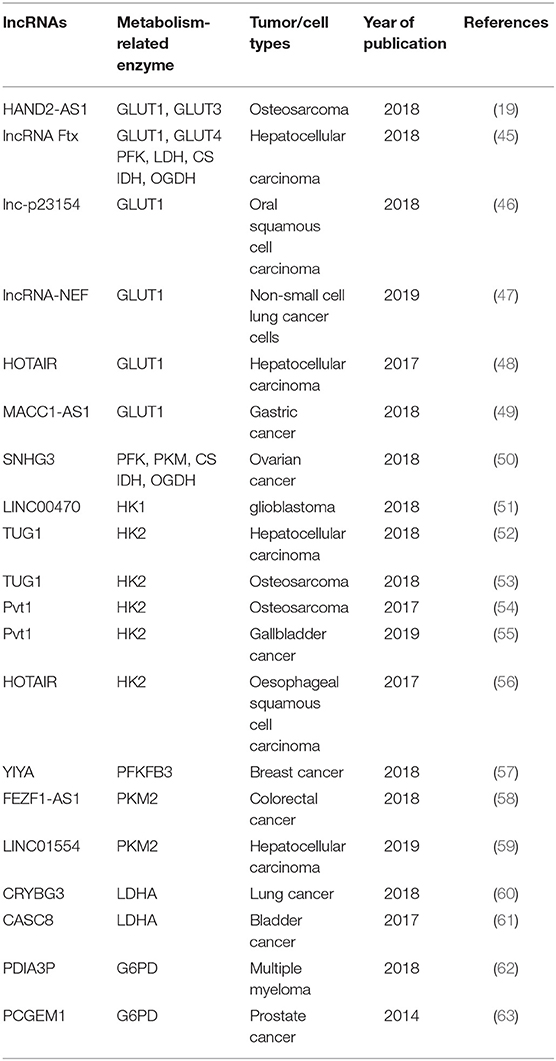
Glucose transporters (GLUTs) are a family of transmembrane protein that regulate the entry of extracellular glucose into cells during the processes of glucose metabolism (39). There are many subtypes of GLUTs, of which GLUT4, GLUT3, GLUT2, and GLUT1 are mostly relevant to the glucose metabolism. Insulin-regulated GLUT4 membrane transport is the most important glucose transporter in adipocytes and skeletal muscle cells (40). GLUT3 is a kind of glucose transporter mainly distributed in nerve tissue (41). GLUT2 is critical for the maintenance of hepatocytes' glucose balance and meanwhile plays a key role in the glucose perception of the nervous system (42). GLUT1 is a kind of glucose transporter that is widely distributed in many tissues and organs (43). It regulates basal glucose uptake during maintenance of normal physiological functions of most cells. This may be closely related to the rapid growth of tumor cells requiring a large amount of glucose. The expression and dysfunction of GLUTs are related to many diseases, especially in the metabolism of tumor cells (44). For example, the deficiency of lncRNA HAND2-AS1 is identified to up-regulate GLUT1 and GLUT3 in the process of promoting glucose uptake in osteosarcoma (19) (Figure 1 and Table 1).
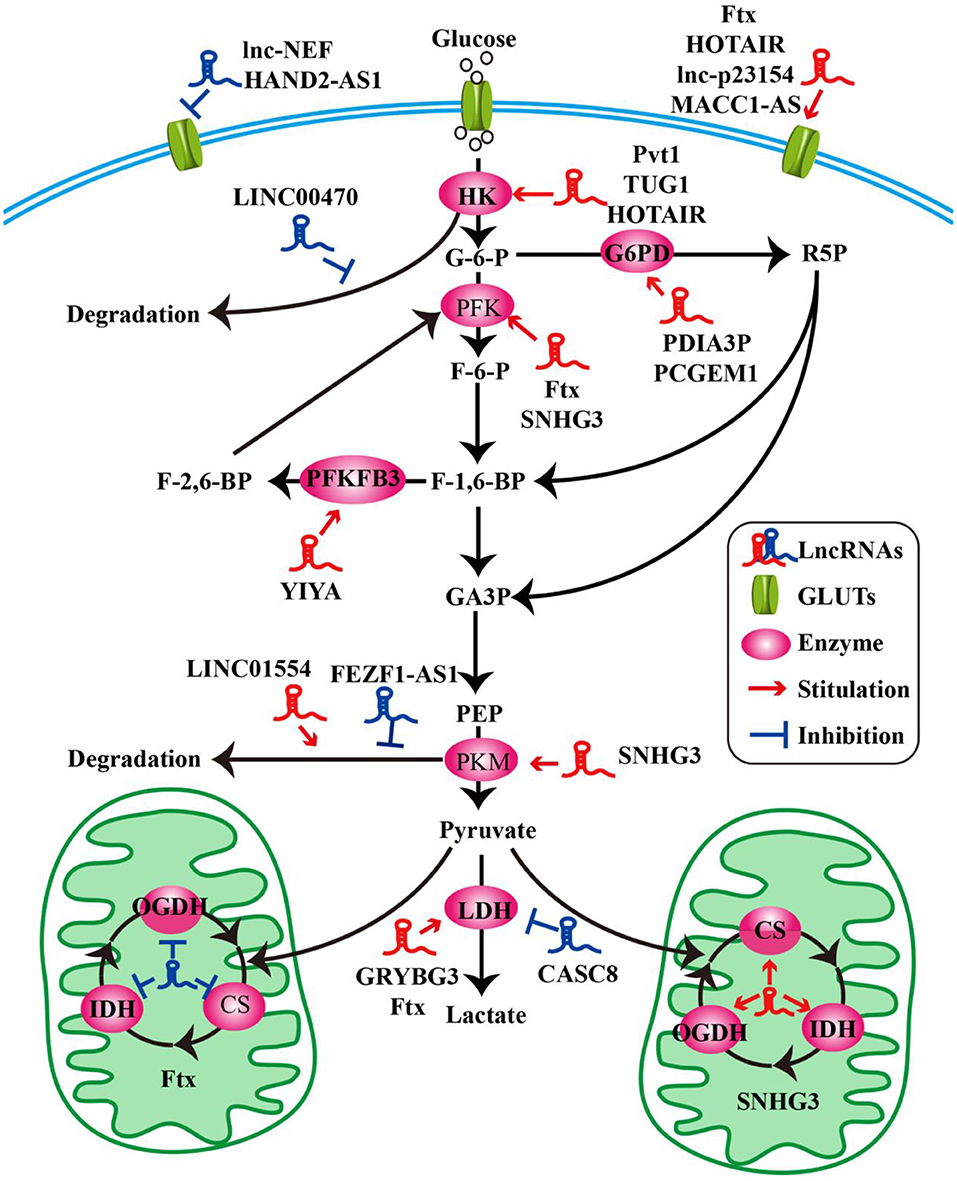
Figure 1. LncRNAs modulate the molecules of glucose metabolism in tumor cells. LncRNAs modulate glucose uptake by GLUTs. LncRNAs influence glycolysis, oxidative phosphorylation, and pentose phosphate pathway by key enzymes.
In pathological conditions, impaired membrane transport or functions of certain GLUTs are an important cause of disorders of glucose levels in various tumor cells, such as hepatocellular carcinoma (HCC) cells (45, 48), renal tumor cells (64), osteosarcoma cells (65), and so on. The high expression of GLUTs satisfies the energy needs of tumor cells. In a recent study, a new lncRNA named lnc-p23154 was found to promote glucose uptake and glycolysis by GLUT1, and it affected oral squamous cell carcinoma metastasis and invasion. A further study demonstrated that lnc-p23154 inhibited the transcription of miR-378a-3p by interacting with the promoter of miR-378a-3p. MiR-378a-3p could then bind to the 3′UTR of GLUT1 directly and repressed GLUT1 expression both at the mRNA and at the protein level (46). In non-small-cell lung cancer (NSCLC), the lncRNA-NEF is down-regulated, comparing adjacent healthy tissues and tumor tissues in patients with NSCLC. The overexpression of lncRNA-NEF inhibits NSCLC cell proliferation and glucose uptake and down-regulated GLUT1 expression. In short, lncRNA-NEF targets GLUT1 to influence the proliferation of NSCLC cells (47). The relationship between glucose uptake and HOTAIR has also been revealed in HCC cells. HOTAIR mediates the expression of GLUT1 via activating mammalian target of rapamycin (mTOR) signaling, which may provide a therapeutic strategy for HCC (48) (Figure 1 and Table 1).
Besides influencing the expression of GLUT1, lncRNAs are also likely to affect the distribution of GLUT1 in tumor cells to regulate the uptake of glucose. In gastric cancer cells, the expression of GLUT1 surrounding the cell membrane is induced by MACC1-AS1, which is a lncRNA, that is highly expressed under metabolic pressure. This is an indication that MACC1-AS1 is likely to promote glucose uptake and then promote glycolysis by increasing the distribution of GLUT1 in the vicinity of the cell membrane. However, the specific relationship between the MACC1-AS1 and GLUT1 remains to be further studied (49). The above results indicate that the role of lncRNAs in glycolysis-mediated proliferation or metastasis of tumor cells depends on GLUTs (Figure 1 and Table 1).
LncRNAs Participate in Glycolysis and Oxidative Phosphorylation
As we all know, the cells absorb glucose in the cytoplasm and catalyze the formation of pyruvate through a series of key enzymes of glucose metabolism. The process by which pyruvate enters the mitochondria to produce large amounts of energy is called mitochondrial oxidative phosphorylation, while the process in which pyruvate is oxidized directly in the cytoplasm to produce lactic acid that does not enter the mitochondria is called glycolysis. Glycolysis and mitochondrial oxidative phosphorylation are inseparable. The process of glycolysis and mitochondrial oxidative phosphorylation involve a large number of enzymes, of which the major ones include hexokinases (HKs) (66), pyruvate kinase enzyme M (PKM) (67), lactate dehydrogenase (LDH) (68), citrate synthase (CS), and so on. Alterations of lncRNAs drive tumor cells to aerobic glycolysis and mitochondrial oxidative phosphorylation through regulation of metabolic enzymes involving these pathways. In HCC progression, lncRNA Ftx affects glucose metabolism reprogramming through the PPARγ pathway. On the one hand, it promotes glycolysis by promoting the expression and activity of phosphofructokinase (PFK) and LDH, and at the same time weakens the activity and expression of tricarboxylic acid cycle key enzymes CS, isocitrate dehydrogenase (IDH), and α-ketoglutarate dehydrogenase (OGDH) (45). The lncRNA SNHG3 is likely to play a similar role. Li et al. found that SNHG3 could up-regulate the expression of the metabolic enzymes PFK, PKM, CS, IDH, and OGDH to regulate the energy metabolism of ovarian cancer through mitochondrial proteomics analysis (50) (Figure 1 and Table 1).
HKs catalyzes the first and irreversible step of glycolysis (69). Hexokinase 1 (HK1) is one of the subtypes of HK. A cytoplasmic lncRNA LINC00470 involving fused in sarcoma (Fus), AKT, and HK1 pathway promotes glycolysis in glioblastoma cells by repressing HK1 ubiquitination (51). In addition to HK1, there are three other enzymes of HK identified, out of which hexokinase 2 (HK2) is the major enzyme that is closely involved in tumor cell glycolysis (70). For example, lncRNA taurine up-regulated gene 1 (TUG1) is disclosed to be related to HK2 involved in glycolytic metabolism and cell metastasis in HCC. The process involves a series of cascades, including TUG1/p21/miR-455-3p/AMPKβ2/HK2/Snail (52). TUG1 has also been associated with the HK2-mediating glycolysis regulating the viability ability of osteosarcoma cells, although the exact mechanism is unknown (53). In addition, one of the main functions of lncRNAs is to interact with microRNAs acting as a molecular sponge. It has been reported that lncRNA Pvt1, which is usually regarded as an oncogene, influences the expression of HK2, promoting glycolysis, and tumor progression in osteosarcoma and gallbladder cancer through acting as a competitive endogenous RNA to directly bind to miR-497 or miR-143, respectively (54, 55). Similarly, HOTAIR binding to miR-125 and miR-143 directly promotes the expression HK2 in esophageal squamous cell carcinoma cells (56). The above research findings fully show that HK2 is implicated in the regulation of tumor progression and metabolic programs involving lncRNAs (Figure 1 and Table 1).
PFKFB3 and PKM2 are also key enzymes regulating glycolysis and oxidative phosphorylation. LncRNA YIYA regulates the PFKFB3 phosphorylation in cyclin-dependent kinase 6-dependent pathway, promoting glycolysis in breast cancer cell (57). The overexpression of lncRNA FEZF1-AS1 in colorectal cancer cells reduced the ubiquitination degradation of PKM2. As a result, the expression of cytoplasmic and nuclear PKM2 protein in glucose metabolism process increased (58). Contrary to FEZF1-AS1, LINC01554 shows a suppressive function because of the acceleration in PKM2 ubiquitination degradation in HCC cells (59) (Figure 1 and Table 1).
LDHA catalyzes the last step of aerobic glycolysis, which is highly critical to the glycolysis phenotype of tumor cells (71). Overexpression of LDHA promotes tumor cell malignant transformation and growth, indicating the important role of LDHA in tumor initiation or maintenance (68). In lung cancer cell lines, it is reported that lncRNA CRYBG3 regulates glycolysis rather than oxidative phosphorylation to increase lung cancer cell proliferation through interacting with LDHA. Knockdown of lncRNA CRYBG3 reduces the expression and activity of LDHA and then decreases the consumption of glucose and pyruvate; however, there are no notable changes in the level of oxidative phosphorylation including all kinds of tricarboxylic acid cycle intermediates regardless of overexpression or knockdown lncRNA CRYBG3 (60). In bladder cancer, the lncRNA cancer susceptibility candidate 8 (CASC8) is reported to function as a tumor suppressor and reduces glycolysis via inhibiting fibroblast growth factor receptor 1-mediated LDHA phosphorylation at Tyr10 (61) (Figure 1 and Table 1).
LncRNAs Participate in the PPP
The PPP is another way of catabolizing glucose, which can produce large amounts of NADPH, providing a reducing agent for various synthesis reactions of tumor cells. The ribose-5-phosphate (R5P) produced by this pathway can also provide raw materials for the synthesis of many substances (72). Glucose-6-phosphate dehydrogenase (G6PD) is the first key enzyme in the oxidation phase of the PPP, which transfers glucose-6-phosphate to 6-phosphogluconate. G6PD is up-regulated in many tumor cells and tumor tissues and generally its level correlates with the overall survival of patients. The lncRNA protein disulfide isomerase family A member 3 pseudogene 1 (PDIA3P) is a 2099-bp lncRNA located at chromosome 1q21.1. It has been reported that PDIA3P regulates multiple myeloma growth and drug resistance through up-regulating G6PD in the PPP. Researchers further discovered that PDIA3P increased the G6PD expression and the PPP flux through enhancing c-MYC transactivation activity bound to the G6PD promoter (62). Similar to PDIA3P, the lncRNA prostate cancer gene expression marker 1 (PCGEM1) also affects pentose phosphate shunt to facilitate biosynthesis of nucleotide and generates NADPH for redox homeostasis as a coactivator for c-MYC and androgen receptor (63). Although the PPP is not the main pathway of glucose metabolism and there are not many studies on lncRNAs and PPP, it is reasonable to believe that PPP is also an important link in the regulation of lncRNAs during tumor progression (Figure 1 and Table 1).
LncRNAs Participate in Lipid Metabolism in Tumor Cells
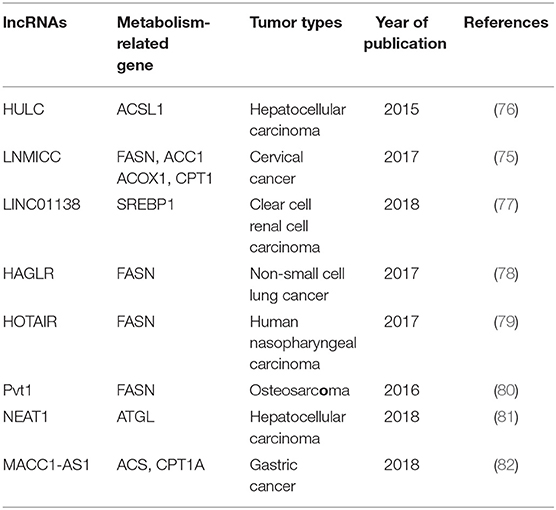
The metabolism of tumor cells is completely different from that of normal cells; in addition to glucose metabolism, lipid metabolism may also undergo adaptive changes (73, 74). Such changes in the overall metabolic pattern constitute the metabolic reprogramming of tumor cells (27). Lipid biosynthesis is not surprising as a part of metabolic abnormalities in tumor cells, which require a large amount of lipid to synthesize biofilms, organelles, and important signaling molecules during rapid proliferation (32). As such, FAO also could provide ATP for tumor cells (31), and one molecule of fatty acid produces much more ATP than glucose. Numerous key enzymes related to lipid synthesis and lipolysis are highly expressed in tumor cells. LncRNAs can be involved in the regulation of multiple lipid metabolism-related genes in tumor cells. It is reported that lncRNA associated with lymph node metastasis in cervical cancer (LNMICC) facilitates fatty acid metabolism reprogramming to promote lymph node metastasis of cervical cancer cells by regulating fatty acid-binding protein 5 (FABP5), which is a carrier of fatty acid uptake and transport. Many key genes involved in fatty acid metabolism, including fatty acid synthase (FASN), carnitine palmitoyl transferase 1 A (CPT1A), acyl-CoA oxidase 1 (ACOX1), and acetyl-CoA carboxylase 1 (ACC1), are all altered by the gain- and loss-of-function strategies of LNMICC, and this regulation of LNMICC on lipid metabolism depends on FABP5 (75) (Figure 2 and Table 2).
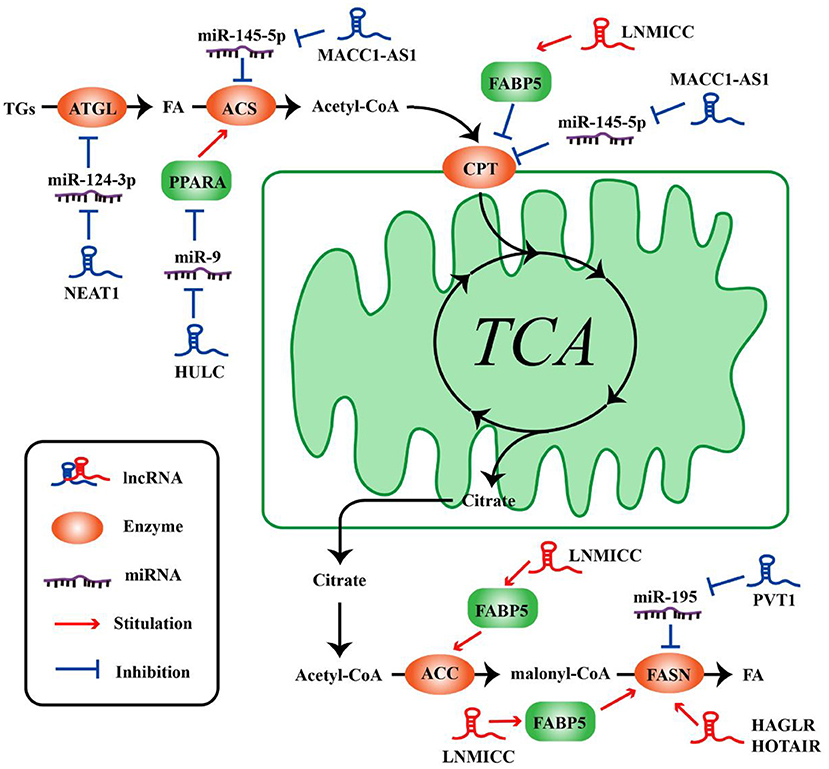
Figure 2. LncRNAs regulate the molecules of lipid metabolism in tumor cells. LncRNAs regulate lipid synthesis and decomposition by different ways and molecules.
Lipid metabolism balance includes lipid uptake, synthesis, catabolism, and secretion. Currently, abundant lncRNAs have been confirmed to be involved in the regulation of lipid metabolism (83, 84). However, lncRNAs involved in lipid metabolism is mostly associated with cardiovascular disease or hepatic disease. There are few reports on the lipid metabolism functions of lncRNAs in tumor cells. The following headlines below principally discusses the role of lncRNAs in lipid synthesis and lipolysis.
LncRNAs Participate in Lipid Synthesis
Lipid synthesis is a landmark change in tumor cells. Synthesis of lipid can not only provide a large number of membrane phospholipids as raw materials for the continuous division and proliferation of tumor cells, but also synthesize a series of cancer-promoting lipid signaling molecules, such as phosphatidylinositol and sphingomyelin, etc.
FASN is a key rate-limiting enzyme catalyzing de novo fatty acid biogenesis (85). Since FASN was discovered, its role in tumor growth and intracellular signal transduction has been widely studied. HAGLR, also called HOXD-AS1, is generally elevated in NSCLC and it is regarded as a predictor of poor patient survival. The proliferation and invasion ability of NSCLC cells decreased after HAGLR knockdown and cells mainly remained in the G1 phase. Furthermore, the expression of FASN and the amount of free fatty acids significantly reduced with the decrease of HAGLR expression in the non-small cells (78). In human nasopharyngeal carcinoma cells, high expression of HOTAIR is positively correlated with FASN expression. Adenovirus encoded short hairpin RNA knockdown of HOTAIR causes the decrease of free fatty acid and FASN at transcriptional and post-transcriptional levels in nasopharyngeal carcinoma cells (79). Although the specific mechanism, of how lncRNAs regulate lipid synthesis involving FASN is unclear, they at least partly demonstrate that lncRNAs play an important role in tumor cells by FASN-mediated lipid synthesis. In osteosarcoma cells, lncRNA Pvt1 was found to act as a sponge molecule to adsorb miR-195. As a result, the expression of FASN, B-cell lymphoma 2 protein, and Cyclin D1 increased. At the same time, the study also showed that silencing Pvt1 reduced cell invasion, and FASN could reverse this effect (80). Sterol regulatory element binding protein 1 (SREBP1) is a major transcription factor involved in lipid accumulation and desaturation. Zhang et al. in their study indicated that the lncRNA LINC01138 increased the arginine methylation of SREBP1 at the post-transcriptional level to regulate lipid desaturation and cells growth of clear cell renal cell carcinoma by interacting with protein arginine methyltransferase 5 (77) (Figure 2 and Table 2).
LncRNAs Participate in Lipolysis
Fat mobilization is the first step in lipid catabolism, and adipose triglyceride lipase (ATGL) is a first key lipase that has been discovered in recent years to initiate fat mobilization (86, 87). It is vital to maintain the balance of lipid storage mobilization (88). In fact, ATGL has been reported to be reduced in leiomyosarcoma, non-small cell lung cancer as well as pancreatic adenocarcinoma, and the levels of ATGL are associated with patients survival (89). In the diet-induced murine HCC model of steatohepatitis and human HCC, the expression of ATGL is also found to be decreased by Di Leo (90). Although it is frequently reported that ATGL shows anti-neoplastic effect, some studies have proposed its tumor-promoting function (91). Interestingly, Liu et al. found that the expression of ATGL at transcriptional level and post-transcriptional level is higher in human HCC tissues and orthotopic HCC mouse model, which is exactly the opposite of Di Leo. It is showed that high expression of ATGL promoted the growth of HCC cells by catalyzing the production of diacylglycerol (DAG) and free fatty acids (FFA). This opposite phenomenon may be due to differences in the clinicopathological features of the patients and the mouse model. Simultaneously, the study also indicated that the lncRNA NEAT1 could regulate the expression of ATGL and affect the abnormal lipidosis of HCC cells through ATGL. Further research confirmed miR-124-3p/ATGL/DAG+FFA to participate in the regulation effect of NEAT1 on the lipid decomposition in HCC and thus promoted the progress of HCC (92) (Figure 2 and Table 2).
It is well-known that the most important form of fatty acid decomposition is β-oxidation of fatty acids (FAO), and studies have shown that lncRNAs are also extremely important in the oxidation of fatty acids. Acyl-CoA synthetase (ACS) is the rate-limiting enzyme in the first step of the fatty acid oxidation reaction. HULC is the first lncRNA identified to be specifically elevated in HCC compared with normal liver tissues (93). Cui et al. revealed that, in hepatoma carcinoma cells, HULC could modulate lipogenesis by the pathway involving miR-9, peroxisome proliferator-activated receptor alpha (PPARA), and ACS long-chain family members 1 (ACSL1), and interestingly, ACSL1 also seems to have a positive feedback effect on HULC expression during this process (81). In gastric cancer cells, transforming growth factor β1 secreted by mesenchymal stem cells could promote the high expression of lncRNA MACC1-AS1. The high expression of MACC1-AS1 relieved the inhibitory effect of miR-145-5p on FAO by inhibiting miR-145-5p, ultimately promoting the stemness and chemoresistance depending on FAO of gastric cancer cells (94). Taken together, lncRNAs participate in lipid decomposition in different ways to affect tumor progress (Figure 2 and Table 2).
LncRNAs Participate in the Adjuvant Therapy of Tumor Possibly
Tumor is the leading killer of human health. Luo et al. demonstrated that lncRNA MALAT1 was crucial in arsenite-induced hepatotoxicity. In the process of cell carcinogenesis induced by arsenic, there is also a phenomenon of enhanced glycolysis, which is mainly manifested by the accumulation of lactate, the acceleration of glucose consumption, and the increased expression of a series of glycolytic related genes, such as HK2, Eno1, and GLUT4. MALAT1 further stabilizes HIF-1α, enhancing the arsenite-induced glycolysis process by disrupting the VHL–HIF-1α interaction (76). It provides new insight into the mechanism of arsenic toxicity on the human body. For a long time, people have been committed to exploring and studying the mechanisms of tumor occurrence and development, trying to find a more effective therapy for tumors. LncRNAs are abnormally expressed in many tumors, and lncRNA-mediated metabolic reprogramming plays a key role in promoting and maintaining tumor formation and progression. These have shown the clinical potential as a therapeutic target of lncRNAs.
Radiotherapy is a common method of tumor treatment. Li et al. unveiled the role of UCA1 in the radiotherapy of cervical cancer from the perspective of glucose metabolism and proposed UCA1 as a potential target for improving the efficacy of radiotherapy for cervical cancer. The radiosensitivity of cervical cancer cells was enhanced after treating with glycolysis inhibitor 2-DG. Knockdown of UCA1 contributed to reduce glucose consumption and lactic acid production by down-regulation of HK2 expression rather than GLUTs or PKM. The discovery of the UCA1/HK2/glycolysis pathway in the radiotherapy of cervical cancer provides a new method to improve the radiotherapy effect (82). Abnormal energy metabolism is a characteristic of tumor cells, and biguanides are often used as potential tumor therapy drugs because of their role in inhibiting the mitochondrial respiratory chain (95). Phenformin is a type of biguanides used in the treatment of tumor. In this treatment with phenformin, lncRNA NBR2 has been discovered to have a potential role in the adaptive response of tumor cells. Elevated NBR2 expression reduced the apoptosis of tumor cells induced by phenformin. As a result, the number of tumor cells increased (96). Thus, the role of lncRNA-mediated metabolic reprogramming in tumor therapy cannot be ignored. They can, therefore, be used as an adjunct to other therapies to treat tumors.
Summary and Prospect
In recent years, the studies of lncRNAs on tumor cells' metabolic reprogramming have developed rapidly. Numerous reports have been published on the different lncRNAs that affect the proliferation, migration, invasion, and other pathological processes of tumor cells by regulating the energy metabolism. As an important part of tumor microenvironments, similar to tumor cells, there is also a phenomenon of metabolic reprogramming in immune cells (97). Immune cells of different states and different stages of differentiation show different metabolic phenotypes (98, 99). For example, naive T cells are primarily dependent on FAO and oxidative phosphorylation to maintain their quiescence (100), while activated T cells and effector T cells, including Th1, Th2, and Th17, have a similar metabolic pattern to tumor cells and mainly obtain energy by aerobic glycolysis (98). A recent study found that the expression of lncRNA Malat1 was significantly different in macrophages treated with LPS and IL-4, respectively. Knockdown of Malat1 promoted the M2 macrophage polarization induced by IL-4. After the use of mitochondrial pyruvate carriers inhibitor UK-5099 or mitochondrial oxidative phosphorylation complex 1 inhibitor, the oxidative phosphorylation of macrophages was prevented, and the effect of Malat1 knockdown on M2 macrophage polarization was also eliminated (101), thus suggesting that Malat1-mediated alternative activation of macrophages is dependent on the elevated mitochondrial oxidative phosphorylation.
Notwithstanding, the regulation of lncRNAs on immune cell metabolism has rarely been reported, but the regulation of lncRNAs on immune cells has been extensively studied. MDSCs are a group of heterogeneous cells with significant immunosuppressive activity derived from bone marrow. It abnormally expands during inflammation, infection, and tumor microenvironments (102). Previous experiments in our laboratory showed that lncRNA RUNXOR accelerates MDSC-mediated immunosuppression via targeting RUNX1 in lung cancer (103). Pvt1 regulates the immunosuppression activity of granulocytic myeloid-derived suppressor cells (G-MDSCs), which is a subgroup of MDSCs, and Pvt1 is up-regulated by HIF-1α under hypoxia conditions in tumor-bearing mice (104). In turn, different living environments and metabolic state will also affect the phenotype and functions of immune cells, making immune cells more suitable for their functional needs (105–107). Also, Hossain et al. revealed that the oxidation of fatty acids could induce the immunosuppressive function of MDSCs and ultimately promoted the progression of the tumor (108). In nasopharyngeal carcinoma cells, the promotion of latent membrane proteins 1 on MDSC expansion also depends on GLUT1-mediated glycolysis (109).
Therefore, it is reasonable to believe that lncRNAs is also likely to regulate the function of immune cells by regulating the metabolic reprogramming. Of course, this conclusion remains to be confirmed by more studies. In-depth discussion of the effect of lncRNAs on the metabolic function of tumor cells and immune cells will help to further understand the mechanism of tumor occurrence, development, and the function of immune cells in tumor microenvironments, and could provide new ideas and strategies for clinical diagnosis and anti-tumor therapy.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
This work was supported by the China Postdoctoral Science Foundation (Grant No. 2016 M600382) and the Jiangsu Postdoctoral Science Foundation (Grant No. 1601082B). Clinical Medical Science and Technology Development Foundation of Jiangsu University (Grant No. JLY20160102).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
lncRNAs, long non-coding RNAs; MDSCs, myeloid derived suppressor cells; FAO, β-oxidation of fatty acids; GLUTs, glucose transporters; HCC, hepatocellular carcinoma; NSCLC, non-small-cell lung cancer; HK, hexokinase; PK, pyruvate kinase isozyme; LDH, lactate dehydrogenase; CS, citrate synthase; PFK, phosphofructokinase; IDH, isocitrate dehydrogenase; OGDH, α-ketoglutarate dehydrogenase; TUG1, lncRNA taurine up-regulated gene 1; CASC8, the lncRNA cancer susceptibility candidate 8; PPP, pentose phosphate pathway; G6PD, glucose-6-phosphate dehydrogenase; PDIA3P, protein disulfide isomerase family A member 3 pseudogene 1; PCGEM1, long non-coding RNA prostate cancer gene expression marker 1; LNMICC, lncRNA associated with lymph node metastasis in cervical cancer; FABP5, fatty acid-binding protein 5; FASN, fatty acid synthase; CPT1A, carnitine palmitoyl transferase 1 A; ACOX1, acyl-CoA oxidase 1; ACC1, acetyl-CoA carboxylase 1; ACSL1, Acyl-CoA synthetase long-chain family members 1; SREBP1, sterol regulatory element binding protein 1; ATGL, adipose triacylglyceride lipase.
References
1. Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. (2016) 17:47–62. doi: 10.1038/nrg.2015.10
2. Chen LL. Linking long noncoding RNA localization and function. Trends Biochem Sci. (2016) 41:761–72. doi: 10.1016/j.tibs.2016.07.003
3. Okazaki Y, Furuno M, Kasukawa T, Adachi J, Bono H, Kondo S, et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. (2002) 420:563–73. doi: 10.1038/nature01266
4. Struhl K. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat Struct Mol Biol. (2007) 14:103–5. doi: 10.1038/nsmb0207-103
5. Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. (2013) 152:1298–307. doi: 10.1016/j.cell.2013.02.012
6. Kitagawa M, Kitagawa K, Kotake Y, Niida H, Ohhata T. Cell cycle regulation by long non-coding RNAs. Cell Mol Life Sci. (2013) 70:4785–94. doi: 10.1007/s00018-013-1423-0
7. Fatica, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. (2014) 15:7–21. doi: 10.1038/nrg3606
8. Chen YG, Satpathy AT, Chang HY. Gene regulation in the immune system by long noncoding RNAs. Nat Immunol. (2017) 18:962–72. doi: 10.1038/ni.3771
9. Fan C, Tang Y, Wang J, Xiong F, Guo C, Wang Y, et al. Role of long non-coding RNAs in glucose metabolism in cancer. Mol Cancer. (2017) 16:130. doi: 10.1186/s12943-017-0699-3
10. Su YJ, Yu J, Huang YQ, Yang J. Circulating long noncoding RNA as a potential target for prostate cancer. Int J Mol Sci. (2015) 16:13322–38. doi: 10.3390/ijms160613322
11. Jarroux J, Morillon A, Pinskaya M. History, discovery, and classification of lncRNAs. Adv Exp Med Biol. (2017) 1008:1–46. doi: 10.1007/978-981-10-5203-3_1
12. Noh JH, Kim KM, Abdelmohsen K, Yoon JH, Panda AC, Munk R, et al. HuR and GRSF1 modulate the nuclear export and mitochondrial localization of the lncRNA RMRP. Genes Dev. (2016) 30:1224–39. doi: 10.1101/gad.276022.115
13. Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. (2011) 43:904–14. doi: 10.1016/j.molcel.2011.08.018
14. Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. (2010) 142:409–19. doi: 10.1016/j.cell.2010.06.040
15. Gao Y, Sun W, Shang W, Li Y, Zhang D, Wang T, et al. Lnc-C/EBPbeta negatively regulates the suppressive function of myeloid-derived suppressor cells. Cancer Immunol Res. (2018) 6:1352–63. doi: 10.1158/2326-6066.CIR-18-0108
16. Gao Y, Wang T, Li Y, Zhang Y, Yang R. Lnc-chop promotes immunosuppressive function of myeloid-derived suppressor cells in tumor and inflammatory environments. J Immunol. (2018) 200:2603–14. doi: 10.4049/jimmunol.1701721
17. Bhan, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. (2017) 77:3965–81. doi: 10.1158/0008-5472.CAN-16-2634
18. Xiao ZD, Han L, Lee H, Zhuang L, Zhang Y, Baddour J, et al. Energy stress-induced lncRNA FILNC1 represses c-Myc-mediated energy metabolism and inhibits renal tumor development. Nat Commun. (2017) 8:783. doi: 10.1038/s41467-017-00902-z
19. Kang Y, Zhu X, Xu Y, Tang Q, Huang Z, Zhao Z, et al. Energy stress-induced lncRNA HAND2-AS1 represses HIF1alpha-mediated energy metabolism and inhibits osteosarcoma progression. Am J Cancer Res. (2018) 8:526–537.
20. Yang X, Yao B, Niu Y, Chen T, Mo H, Wang L, et al. Hypoxia-induced lncRNA EIF3J-AS1 accelerates hepatocellular carcinoma progression via targeting miR-122-5p/CTNND2 axis. Biochem Biophys Res Commun. (2019) 518:239–45. doi: 10.1016/j.bbrc.2019.08.039
21. Garcia-Venzor A, Mandujano-Tinoco EA, Lizarraga F, Zampedri C, Krotzsch E, Salgado RM, et al. Microenvironment-regulated lncRNA-HAL is able to promote stemness in breast cancer cells. Biochim Biophys Acta Mol Cell Res. (2019) 1866:118523. doi: 10.1016/j.bbamcr.2019.118523
22. Heindel JJ, Blumberg B, Cave M, Machtinger R, Mantovani A, Mendez MA, et al. Metabolism disrupting chemicals and metabolic disorders. Reprod Toxicol. (2017) 68:3–33. doi: 10.1016/j.reprotox.2016.10.001
23. Muschen M. Metabolic gatekeepers to safeguard against autoimmunity and oncogenic B cell transformation. Nat Rev Immunol. (2019) 19:337–48. doi: 10.1038/s41577-019-0154-3
24. Xie H, Simon MC. Oxygen availability and metabolic reprogramming in cancer. J Biol Chem. (2017) 292:16825–32. doi: 10.1074/jbc.R117.799973
25. Nilsson A, Nielsen J. Genome scale metabolic modeling of cancer. Metab Eng. (2017) 43:103–12. doi: 10.1016/j.ymben.2016.10.022
26. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. (2011) 144:646–74. doi: 10.1016/j.cell.2011.02.013
27. Li Z, Zhang H. Reprogramming of glucose, fatty acid and amino acid metabolism for cancer progression. Cell Mol Life Sci. (2016) 73:377–92. doi: 10.1007/s00018-015-2070-4
28. Moon JS, Jin WJ, Kwak JH, Kim HJ, Yun MJ, Kim JW, et al. Androgen stimulates glycolysis for de novo lipid synthesis by increasing the activities of hexokinase 2 and 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 2 in prostate cancer cells. Biochem J. (2011) 433:225–33. doi: 10.1042/BJ20101104
29. DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, et al. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci USA. (2007) 104:19345–50. doi: 10.1073/pnas.0709747104
30. Nieman KM, Kenny HA, Penicka CV, Ladanyi A, Buell-Gutbrod R, Zillhardt MR, et al. Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth. Nat Med. (2011) 17:1498–503. doi: 10.1038/nm.2492
31. Jeon SM, Chandel NS, Hay N. AMPK regulates NADPH homeostasis to promote tumour cell survival during energy stress. Nature. (2012) 485:661–5. doi: 10.1038/nature11066
32. Boroughs LK, DeBerardinis RJ. Metabolic pathways promoting cancer cell survival and growth. Nat Cell Biol. (2015) 17:351–9. doi: 10.1038/ncb3124
33. Mulukutla BC, Yongky A, Le T, Mashek DG, Hu WS. Regulation of glucose metabolism - a perspective from cell bioprocessing. Trends Biotechnol. (2016) 34:638–51. doi: 10.1016/j.tibtech.2016.04.012
34. Koppenol WH, Bounds PL, Dang CV. Otto Warburg's contributions to current concepts of cancer metabolism. Nat Rev Cancer. (2011) 11:325–37. doi: 10.1038/nrc3038
35. Lu J, Tan M, Cai Q. The Warburg effect in tumor progression: mitochondrial oxidative metabolism as an anti-metastasis mechanism. Cancer Lett. (2015) 356:156–64. doi: 10.1016/j.canlet.2014.04.001
36. Pavlova NN, Thompson CB. The emerging hallmarks of cancer metabolism. Cell Metab. (2016) 23:27–47. doi: 10.1016/j.cmet.2015.12.006
37. Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. (2009) 324:1029–33. doi: 10.1126/science.1160809
38. Stincone A, Prigione A, Cramer T, Wamelink MM, Campbell K, Cheung E, et al. The return of metabolism: biochemistry and physiology of the pentose phosphate pathway. Biol Rev Cambridge Philos Soc. (2015) 90:927–63. doi: 10.1111/brv.12140
39. Mueckler M, Thorens B. The SLC2 (GLUT) family of membrane transporters. Mol Aspects Med. (2013) 34:121–38. doi: 10.1016/j.mam.2012.07.001
40. Lauritzen HP. Insulin- and contraction-induced glucose transporter 4 traffic in muscle: insights from a novel imaging approach. Exerc Sport Sci Rev. (2013) 41:77–86. doi: 10.1097/JES.0b013e318275574c
41. Simpson IA, Dwyer D, Malide D, Moley KH, Travis A, Vannucci SJ. The facilitative glucose transporter GLUT3: 20 years of distinction. Am J Physiol Endocrinol Metab. (2008) 295:E242–53. doi: 10.1152/ajpendo.90388.2008
42. Thorens B. GLUT2, glucose sensing and glucose homeostasis. Diabetologia. (2015) 58:221–32. doi: 10.1007/s00125-014-3451-1
43. Deng D, Xu C, Sun P, Wu J, Yan C, Hu M, et al. Crystal structure of the human glucose transporter GLUT1. Nature. (2014) 510:121–5. doi: 10.1038/nature13306
44. Ancey PB, Contat C, Meylan E. Glucose transporters in cancer - from tumor cells to the tumor microenvironment. FEBS J. (2018) 285:2926–43. doi: 10.1111/febs.14577
45. Li X, Zhao Q, Qi J, Wang W, Zhang D, Li Z, et al. lncRNA Ftx promotes aerobic glycolysis and tumor progression through the PPARgamma pathway in hepatocellular carcinoma. Int J Oncol. (2018) 53:551–66. doi: 10.3892/ijo.2018.4418
46. Wang Y, Zhang X, Wang Z, Hu Q, Wu J, Li Y, et al. LncRNA-p23154 promotes the invasion-metastasis potential of oral squamous cell carcinoma by regulating Glut1-mediated glycolysis. Cancer Lett. (2018) 434:172–83. doi: 10.1016/j.canlet.2018.07.016
47. Chang L, Xu W, Zhang Y, Gong F. Long non-coding RNA-NEF targets glucose transportation to inhibit the proliferation of non-small-cell lung cancer cells. Oncol Lett. (2019) 17:2795–801. doi: 10.3892/ol.2019.9919
48. Wei S, Fan Q, Yang L, Zhang X, Ma Y, Zong Z, et al. Promotion of glycolysis by HOTAIR through GLUT1 upregulation via mTOR signaling. Oncol Rep. (2017) 38:1902–8. doi: 10.3892/or.2017.5840
49. Zhao Y, Liu Y, Lin L, Huang Q, He W, Zhang S, et al. The lncRNA MACC1-AS1 promotes gastric cancer cell metabolic plasticity via AMPK/Lin28 mediated mRNA stability of MACC1. Mol Cancer. (2018) 17:69. doi: 10.1186/s12943-018-0820-2
50. Li N, Zhan X, Zhan X. The lncRNA SNHG3 regulates energy metabolism of ovarian cancer by an analysis of mitochondrial proteomes. Gynecol Oncol. (2018) 150:343–54. doi: 10.1016/j.ygyno.2018.06.013
51. Liu C, Zhang Y, She X, Fan L, Li P, Feng J, et al. A cytoplasmic long noncoding RNA LINC00470 as a new AKT activator to mediate glioblastoma cell autophagy. J Hematol Oncol. (2018) 11:77. doi: 10.1186/s13045-018-0619-z
52. Lin YH, Wu MH, Huang YH, Yeh CT, Cheng ML, Chi HC, et al. Taurine up-regulated gene 1 functions as a master regulator to coordinate glycolysis and metastasis in hepatocellular carcinoma. Hepatology. (2018) 67:188–203. doi: 10.1002/hep.29462
53. Han X, Yang Y, Sun Y, Qin L, Yang Y. LncRNA TUG1 affects cell viability by regulating glycolysis in osteosarcoma cells. Gene. (2018) 674:87–92. doi: 10.1016/j.gene.2018.06.085
54. Song J, Wu X, Liu F, Li M, Sun Y, Wang Y, et al. Long non-coding RNA PVT1 promotes glycolysis and tumor progression by regulating miR-497/HK2 axis in osteosarcoma. Biochem Biophys Res Commun. (2017) 490:217–24. doi: 10.1016/j.bbrc.2017.06.024
55. Chen J, Yu Y, Li H, Hu Q, Chen X, He Y, et al. Long non-coding RNA PVT1 promotes tumor progression by regulating the miR-143/HK2 axis in gallbladder cancer. Mol Cancer. (2019) 18:33. doi: 10.1186/s12943-019-0947-9
56. Ma J, Fan Y, Feng T, Chen F, Xu Z, Li S, et al. HOTAIR regulates HK2 expression by binding endogenous miR-125 and miR-143 in oesophageal squamous cell carcinoma progression. Oncotarget. (2017) 8:86410–22. doi: 10.18632/oncotarget.21195
57. Xing Z, Zhang Y, Liang K, Yan L, Xiang Y, Li C, et al. Expression of long noncoding RNA YIYA promotes glycolysis in breast cancer. Cancer Res. (2018) 78:4524–32. doi: 10.1158/0008-5472.CAN-17-0385
58. Bian Z, Zhang J, Li M, Feng Y, Wang X, Zhang J, et al. LncRNA-FEZF1-AS1 promotes tumor proliferation and metastasis in colorectal cancer by regulating PKM2 signaling. Clin Cancer Res. (2018) 24:4808–19. doi: 10.1158/1078-0432.CCR-17-2967
59. Zheng YL, Li L, Jia YX, Zhang BZ, Li JC, Zhu YH, et al. LINC01554-mediated glucose metabolism reprogramming suppresses tumorigenicity in hepatocellular carcinoma via downregulating PKM2 expression and inhibiting Akt/mTOR signaling pathway. Theranostics. (2019) 9:796–810. doi: 10.7150/thno.28992
60. Chen H, Pei H, Hu W, Ma J, Zhang J, Mao W, et al. Long non-coding RNA CRYBG3 regulates glycolysis of lung cancer cells by interacting with lactate dehydrogenase A. J Cancer. (2018) 9:2580–8. doi: 10.7150/jca.24896
61. Hu R, Zhong P, Xiong L, Duan L. Long noncoding rna cancer susceptibility candidate 8 suppresses the proliferation of bladder cancer cells via regulating glycolysis. DNA Cell Biol. (2017) 36:767–74. doi: 10.1089/dna.2017.3785
62. Yang X, Ye H, He M, Zhou X, Sun N, Guo W, et al. LncRNA PDIA3P interacts with c-Myc to regulate cell proliferation via induction of pentose phosphate pathway in multiple myeloma. Biochem Biophys Res Commun. (2018) 498:207–13. doi: 10.1016/j.bbrc.2018.02.211
63. Hung CL, Wang LY, Yu YL, Chen HW, Srivastava S, Petrovics G, et al. A long noncoding RNA connects c-Myc to tumor metabolism. Proc Natl Acad Sci USA. (2014) 111:18697–702. doi: 10.1073/pnas.1415669112
64. Almeida L, Silva R, Cavadas B, Lima J, Pereira L, Soares P, et al. GLUT1, MCT1/4 and CD147 overexpression supports the metabolic reprogramming in papillary renal cell carcinoma. Histol Histopathol. (2017) 32:1029–40. doi: 10.14670/hh-11-863
65. Yuan G, Zhao Y, Wu D, Gao C. Mir-150 up-regulates Glut1 and increases glycolysis in osteosarcoma cells. Asian Pacif J Cancer Preven. (2017) 18:1127–31. doi: 10.22034/apjcp.2017.18.4.1127
66. Tan VP, Miyamoto S. HK2/hexokinase-II integrates glycolysis and autophagy to confer cellular protection. Autophagy. (2015) 11:963–4. doi: 10.1080/15548627.2015.1042195
67. Dayton TL, Jacks T, Vander Heiden MG. PKM2, cancer metabolism, and the road ahead. EMBO Rep. (2016) 17:1721–30. doi: 10.15252/embr.201643300
68. Feng Y, Xiong Y, Qiao T, Li X, Jia L, Han Y. Lactate dehydrogenase A: a key player in carcinogenesis and potential target in cancer therapy. Cancer Med. (2018) 7:6124–36. doi: 10.1002/cam4.1820
69. Martinez-Outschoorn UE, Peiris-Pages M, Pestell RG, Sotgia F, Lisanti MP. Cancer metabolism: a therapeutic perspective. Nat Rev Clin Oncol. (2017) 14:11–31. doi: 10.1038/nrclinonc.2016.60
70. Roberts DJ, Miyamoto S. Hexokinase II integrates energy metabolism and cellular protection: akting on mitochondria and TORCing to autophagy. Cell Death Differ. (2015) 22:248–57. doi: 10.1038/cdd.2014.173
71. Graziano F, Ruzzo A, Giacomini E, Ricciardi T, Aprile G, Loupakis F, et al. Glycolysis gene expression analysis and selective metabolic advantage in the clinical progression of colorectal cancer. Pharmacogenom J. (2017) 17:258–64. doi: 10.1038/tpj.2016.13
72. Jiang P, Du W, Wu M. Regulation of the pentose phosphate pathway in cancer. Protein Cell. (2014) 5:592–602. doi: 10.1007/s13238-014-0082-8
73. Corbet C, Feron O. Emerging roles of lipid metabolism in cancer progression. Curr Opin Clin Nutri Metab Care. (2017) 20:254–60. doi: 10.1097/MCO.0000000000000381
74. Corbet C, Pinto A, Martherus R, Santiago de Jesus JP, Polet F, Feron O. Acidosis drives the reprogramming of fatty acid metabolism in cancer cells through changes in mitochondrial and histone acetylation. Cell Metab. (2016) 24:311–23. doi: 10.1016/j.cmet.2016.07.003
75. Shang C, Wang W, Liao Y, Chen Y, Liu T, Du Q, et al. LNMICC promotes nodal metastasis of cervical cancer by reprogramming fatty acid metabolism. Cancer Res. (2018) 78:877–90. doi: 10.1158/0008-5472.CAN-17-2356
76. Luo F, Liu X, Ling M, Lu L, Shi L, Lu X, et al. The lncRNA MALAT1, acting through HIF-1alpha stabilization, enhances arsenite-induced glycolysis in human hepatic L-02 cells. Biochim Biophys Acta. (2016) 1862:1685–95. doi: 10.1016/j.bbadis.2016.06.004
77. Zhang X, Wu J, Wu C, Chen W, Lin R, Zhou Y, et al. The LINC01138 interacts with PRMT5 to promote SREBP1-mediated lipid desaturation and cell growth in clear cell renal cell carcinoma. Biochem Biophys Res Commun. (2018) 507:337–42. doi: 10.1016/j.bbrc.2018.11.036
78. Lu C, Ma J, Cai D. Increased HAGLR expression promotes non-small cell lung cancer proliferation and invasion via enhanced de novo lipogenesis. Tumour Biol. (2017) 39:1010428317697574. doi: 10.1177/1010428317697574
79. Ma DD, Yuan LL, Lin LQ. LncRNA HOTAIR contributes to the tumorigenesis of nasopharyngeal carcinoma via up-regulating FASN. Eur Rev Med Pharmacol Sci. (2017) 21:5143–52.
80. Zhou Q, Chen F, Zhao J, Li B, Liang Y, Pan W, et al. Long non-coding RNA PVT1 promotes osteosarcoma development by acting as a molecular sponge to regulate miR-195. Oncotarget. (2016) 7:82620–33. doi: 10.18632/oncotarget.13012
81. Cui M, Xiao Z, Wang Y, Zheng M, Song T, Cai X, et al. Long noncoding RNA HULC modulates abnormal lipid metabolism in hepatoma cells through an miR-9-mediated RXRA signaling pathway. Cancer Res. (2015) 75:846–57. doi: 10.1158/0008-5472.CAN-14-1192
82. Fan L, Huang C, Li J, Gao T, Lin Z, Yao T. Long noncoding RNA urothelial cancer associated 1 regulates radioresistance via the hexokinase 2/glycolytic pathway in cervical cancer. Int J Mol Med. (2018) 42:2247–59. doi: 10.3892/ijmm.2018.3778
83. Zeng Y, Ren K, Zhu X, Zheng Z, Yi G. Long noncoding RNAs: advances in lipid metabolism. Adv Clin Chem. (2018) 87:1–36. doi: 10.1016/bs.acc.2018.07.001
84. van Solingen C, Scacalossi KR, Moore KJ. Long noncoding RNAs in lipid metabolism. Curr Opin Lipidol. (2018) 29:224–32. doi: 10.1097/MOL.0000000000000503
85. Menendez JA, Lupu R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat Rev Cancer. (2007) 7:763–77. doi: 10.1038/nrc2222
86. Haemmerle G, Lass A, Zimmermann R, Gorkiewicz G, Meyer C, Rozman J, et al. Defective lipolysis and altered energy metabolism in mice lacking adipose triglyceride lipase. Science. (2006) 312:734–7. doi: 10.1126/science.1123965
87. Zimmermann R, Strauss JG, Haemmerle G, Schoiswohl G, Birner-Gruenberger R, Riederer M, et al. Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science. (2004) 306:1383–6. doi: 10.1126/science.1100747
88. Cerk IK, Wechselberger L, Oberer M. Adipose triglyceride lipase regulation: an overview. Curr Protein Peptide Sci. (2018) 19:221–33. doi: 10.2174/1389203718666170918160110
89. Al-Zoughbi W, Pichler M, Gorkiewicz G, Guertl-Lackner B, Haybaeck J, Jahn SW, et al. Loss of adipose triglyceride lipase is associated with human cancer and induces mouse pulmonary neoplasia. Oncotarget. (2016) 7:33832–40. doi: 10.18632/oncotarget.9418
90. Di Leo L, Vegliante R, Ciccarone F, Salvatori I, Scimeca M, Bonanno E, et al. Forcing ATGL expression in hepatocarcinoma cells imposes glycolytic rewiring through PPAR-alpha/p300-mediated acetylation of p53. Oncogene. (2019) 38:1860–75. doi: 10.1038/s41388-018-0545-0
91. Vegliante R, Di Leo L, Ciccarone F, Ciriolo MR. Hints on ATGL implications in cancer: beyond bioenergetic clues. Cell Death Dis. (2018) 9:316. doi: 10.1038/s41419-018-0345-z
92. Liu X, Liang Y, Song R, Yang G, Han J, Lan Y, et al. Long non-coding RNA NEAT1-modulated abnormal lipolysis via ATGL drives hepatocellular carcinoma proliferation. Mol Cancer. (2018) 17:90. doi: 10.1186/s12943-018-0838-5
93. Xie H, Ma H, Zhou D. Plasma HULC as a promising novel biomarker for the detection of hepatocellular carcinoma. BioMed Res Int. (2013) 2013:136106. doi: 10.1155/2013/136106
94. He W, Liang B, Wang C, Li S, Zhao Y, Huang Q, et al. MSC-regulated lncRNA MACC1-AS1 promotes stemness and chemoresistance through fatty acid oxidation in gastric cancer. Oncogene. (2019) 38:4637–54. doi: 10.1038/s41388-019-0747-0
95. Pollak M. Potential applications for biguanides in oncology. J Clin Invest. (2013) 123:3693–700. doi: 10.1172/JCI67232
96. Liu X, Gan B. lncRNA NBR2 modulates cancer cell sensitivity to phenformin through GLUT1. Cell Cycle. (2016) 15:3471–81. doi: 10.1080/15384101.2016.1249545
97. Biswas SK. Metabolic reprogramming of immune cells in cancer progression. Immunity. (2015) 43:435–49. doi: 10.1016/j.immuni.2015.09.001
98. Ghesquiere B, Wong BW, Kuchnio A, Carmeliet P. Metabolism of stromal and immune cells in health and disease. Nature. (2014) 511:167–76. doi: 10.1038/nature13312
99. Pearce EL, Poffenberger MC, Chang CH, Jones RG. Fueling immunity: insights into metabolism and lymphocyte function. Science. (2013) 342:1242454. doi: 10.1126/science.1242454
100. MacIver NJ, Michalek RD, Rathmell JC. Metabolic regulation of T lymphocytes. Annu Rev Immunol. (2013) 31:259–83. doi: 10.1146/annurev-immunol-032712-095956
101. Cui H, Banerjee S, Guo S, Xie N, Ge J, Jiang D, et al. Long noncoding RNA Malat1 regulates differential activation of macrophages and response to lung injury. JCI Insight. (2019) 4:124522. doi: 10.1172/jci.insight.124522
102. Gabrilovich DI. Myeloid-derived suppressor cells. Cancer Immunol Res. (2017) 5:3–8. doi: 10.1158/2326-6066.CIR-16-0297
103. Tian X, Ma J, Wang T, Tian J, Zheng Y, Peng R, et al. Long non-coding RNA RUNXOR accelerates MDSC-mediated immunosuppression in lung cancer. BMC Cancer. (2018) 18:660. doi: 10.1186/s12885-018-4564-6
104. Zheng Y, Tian X, Wang T, Xia X, Cao F, Tian J, et al. Long noncoding RNA Pvt1 regulates the immunosuppression activity of granulocytic myeloid-derived suppressor cells in tumor-bearing mice. Mol Cancer. (2019) 18:61. doi: 10.1186/s12943-019-0978-2
105. Zeng H, Chi H. Metabolic control of regulatory T cell development and function. Trends Immunol. (2015) 36:3–12. doi: 10.1016/j.it.2014.08.003
106. Sica A, Strauss L. Energy metabolism drives myeloid-derived suppressor cell differentiation and functions in pathology. J Leukocyte Biol. (2017) 102:325–34. doi: 10.1189/jlb.4MR1116-476R
107. Reina-Campos M, Moscat J, Diaz-Meco M. Metabolism shapes the tumor microenvironment. Curr Opin Cell Biol. (2017) 48:47–53. doi: 10.1016/j.ceb.2017.05.006
108. Hossain F, Al-Khami AA, Wyczechowska D, Hernandez C, Zheng L, Reiss K, et al. Inhibition of fatty acid oxidation modulates immunosuppressive functions of myeloid-derived suppressor cells and enhances cancer therapies. Cancer Immunol Res. (2015) 3:1236–47. doi: 10.1158/2326-6066.CIR-15-0036
Keywords: lncRNAs, tumor, glucose, lipid, metabolism, therapy
Citation: Lu W, Cao F, Wang S, Sheng X and Ma J (2019) LncRNAs: The Regulator of Glucose and Lipid Metabolism in Tumor Cells. Front. Oncol. 9:1099. doi: 10.3389/fonc.2019.01099
Received: 19 July 2019; Accepted: 07 October 2019;
Published: 26 November 2019.
Edited by:
Alfons Navarro, University of Barcelona, SpainReviewed by:
Maria Rosa Ciriolo, University of Rome Tor Vergata, ItalyDohoon Kim, University of Massachusetts Medical School, United States
Copyright © 2019 Lu, Cao, Wang, Sheng and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiumei Sheng, c2hlbmd4aXVtZWlAdWpzLmVkdS5jbg==; Jie Ma, anNkeG1hamllQHVqcy5lZHUuY24=
 Wei Lu
Wei Lu Fenghua Cao2
Fenghua Cao2 Shengjun Wang
Shengjun Wang Xiumei Sheng
Xiumei Sheng Jie Ma
Jie Ma