- 1Department of Neurosurgery, Zhujiang Hospital, Southern Medical University, Guangzhou, China
- 2Guangdong Provincial Key Laboratory on Brain Function Repair and Regeneration, The National Key Clinic Specialty, The Engineering Technology Research Center of Education Ministry of China, The Neurosurgery Institute of Guangdong Province, Southern Medical University, Guangzhou, China
- 3Department of Neurosurgery, Renmin Hospital of Wuhan University, Wuhan, China
- 4Department of Neurosurgery, Huzhou Central Hospital, Zhejiang University School of Medicine, Huzhou, China
- 5Department of Rheumatology, The Third Affiliated Hospital of Sun Yat-sen University, Guangzhou, China
Glioblastoma (GBM) is the most malignant tumor in the central nervous system and the treatment is still unsatisfactory because the mechanism of the disease remains unclear. The abnormal expression of miRNAs and its target proteins play a crucial role in the development of glioblastoma. In this study, we demonstrated that high expression of miR-9-5p and low expression of forkhead box P2 (FOXP2) were related with better outcome in patients with GBM, and down regulated FOXP2 expression was able to inhibit glioma cells proliferation by cell cycle arrest. Furthermore, we found that FOXP2 was the target protein of miR-9-5p in luciferase assay. The results of this study suggest a novel regulatory mechanism that miR-9-5p can inhibit glioma cells proliferation by downregulating FOXP2.
Introduction
Glioblastoma (GBM) is one of the most malignant tumors in adult central nervous system. With the development of modern medical technology, it is possible to remove the tumor through surgical resection followed by chemoradiation. However, it is still a challenge to completely cure due to the strong invasive and proliferative nature of GBM. In the past decade, the prognosis and treatment for GBM have not improved significantly, but there have been important gains in our understanding on genetic alterations associated with gliomagenesis, and the gene targeting therapy might be promising for glioblastoma (1, 2).
MicroRNAs (miRNA) are a class of small non-coding RNAs and usually regulate the expression of target functional proteins through the interaction with 3′ untranslated regions (3′-UTRs) of mRNA, thus regulating the physiological and pathological processes. In the past few decades, miRNAs have been widely studied as important molecules in tumor progression, among which, the abnormal expression of miR-9 can be found in many malignancies. Interests of miR-9 have been grown with the aim of using it as a diagnostic and prognostic marker for tumors (1–3).
MiR-9 as a tumor promoter promotes the metastasis and invasion of non-small cell lung cancer (NSCLC) cells by inhibiting the expression of E-cadherin (4, 5). On the other hand, miR-9 functions as a tumor inhibitor suppressing cell proliferation and invasive ability through the SDF-1/CXCR4 pathway in epithelial ovarian cancer (6). The miR-9 can also induce cell arrest and apoptosis of oral squamous cell carcinoma via CDK 4/6 pathway (7), or down-regulate TNFAIP8 to inhibit the gastric cancer cell proliferation (8). In glioma, miR-9 inhibits glioma cells growth through various signals and promote apoptosis (9, 10). In the EGFRvIII pathway, miR-9 plays a negative role on tumorigenic capacity (11). Low expression of miR-9-3p results in a high level of Herpud1, which may protect against apoptosis in glioma (12). Over expressed miR-9 in U87 and U251 cells increases apoptosis by structural maintenance of chromosomes 1A (SMC1A) (13). It is also reported that miR-9 can reduce cell migration and the invasion of Glioblastoma cell lines through MAPK14 pathway (14). However, other researches showed different results. For example, Wu et al. found increased expression of microRNA-9 predicts an unfavorable prognosis in human Glioma (15). However, its important to note that the researches made no distinguishing between miR-9, miR-9-3p, or miR-9-5p in various tumors reported functions of miR-9.
Forkhead box P2 (FOXP2) was first found as a transcription factor involved in speech and language acquisition (16). Recently, abnormal expression of FOXP2 was found in kinds of tumors. However, the results are still controversial. Studies showed downregulation of FOXP2 in breast cancer, hepatocellular carcinoma and gastric cancer biopsies (16–18), while overexpression of FPXP2 was found in multiple myelomas, MGUS (Monoclonal Gammopathy of Undetermined Significance), several subtypes of lymphomas, osteosarcoma, neuroblastomas, and ERG fusion-negative prostate cancers (19–21).
Our previous work demonstrated miR-9-5p was a positive marker for prognosis in GBM, while FOXP2 a negative marker. FOXP2 is predicted as a direct target of miR-9-5p. In this study, miR-9-5p inhibited cell proliferation in 3 GMB cell lines in vitro. Reverse test was used to prove FOXP2 as a key point in miR-9-5p inhibiting GBM cells proliferation. Finally, we identified miR-9-5p inhibit tumor growth in mouse model by reducing FOXP2 expressing.
Combining these results, we conclude that miR-9-5p, through directly downregulating FOXP2 and inhibiting proliferation, is a tumor suppressor in GBM.
Materials and Methods
GBM Tissue Collection
All the 110 samples were collected from March 2015 to April 2017 in the Department of Neurosurgery of Renmin Hospital of Wuhan University (Wuhan, China) and Department of neurosurgery, Zhujiang Hospital of Southern Medical University (Guangzhou, China). All the tumor samples were pathologically diagnosed as glioblastoma. The study was approved by local Ethics Committee of Wuhan University and Zhujiang hospital of Southern Medical University.
Cells and Cell Culture
Three human glioblastoma-derived cancer cell lines, U251, A118MG, and U87MG, were used in the study. The cells were purchased from the Cell Bank Type Culture Collection of Chinese Academy of Sciences, and were cultured in DMEM (Gibco) containing 10% fetal bovine serum (Gibco) and 1% penicillin-streptomycin (Sigma-Aldrich) at a temperature of 37°C and a humidified atmosphere of 5% CO2.
DNA and RNA Transfection
Human FOXP2 (GenBank ID: NM_148898) full length cDNA was subcloned into a pcDNA3.1 vector to generate expression construct. All cell lines in this study were transfected with plasmids by using Lipofectamine 3000 (Thermo Fisher, USA) according to the manufacturers' protocols. FOXP2 small interfering ribonucleic acid (siRNA) (5′GACAGGCAGTTAACACTTAAT3′) and a non-specific si-RNA (as a negative control) transfections were conducted in non-serum-containing conditions using Lipofectamine 3000. All si-RNAs were used at a final concentration of 20 nM. Has-miR-9-5p mimics (5′UCUUUGGUUAUCUAGCUGUAUGA3′), negative control miRNA mimics (miR-ctrl mimics), and inhibitor (5′AGAAACCAAUAGAUCGACAUACU′3) were constructed by RiboBio (Guangzhou, China). Transfections were conducted in non-serum-containing conditions using Lipofectamine 3000 according to the manufacturer's instructions.
Western Blot
Cell lysates were prepared by sonicating cells briefly in a modified RIPA buffer (0.1% SDS, 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.1% sodium deoxycholate, 1% Nonidet P-40) with proteinase and phosphatase inhibitors. BCA protein assay (TaKaRa) was used for protein quantifying. The antibody used were as follows: anti-FOXP2 (Abcam, ab16046, USA), anti-p21 (Abcam, ab109520, USA), Goat anti-Rabbit, and Goat anti-Mouse infrared dye secondary antibodies (800 CW), which were purchased from LI-COR Biosciences (Lincoln, NE, USA). Proteins were visualized with Odyssey Bioanalyzer (LI-COR).
Real Time PCR
Total RNAs of human tissues and cells were extracted using TRIzol reagent (Invitrogen). Quantitative real-time PCR technology was used to measure the miR-9-5p expression by using the All in-One miRNA qRT-PCR Detection Kit (GeneCopoeia, Rockville,MD, USA). hsa-miR-9-5p forward primer: 5′TGCGCTCTTTGGTTATCTAGCTG3′; reverse primer: 5′CCAGTGCAGGGTCCGAGGTATT3′; U6 forward primer: 5′CGCTTCGGCAGCACATATAC 3′; reverse primer: 5′AAATATGGAACGCTTCACGA3′. All qRT-PCR processes and analyses were carried out using Applied Biosystems 7500 Fast Real-Time PCR system (Life Technologies). Relative expression of miRNA and mRNA was calculated using the 2−ΔΔCT method.
Luciferase Reporter Assay
Candidate targets and its putative binding site of miR-9-5p were predicted by miRNA database (http://www.microrna.org/microrna/home.do). The 3′UTR of FOXP2, containing the wild-type or mutant miR-9-5p binding sequence, was cloned into the pMIRREPORT vector (Ambion, USA). U251 cells were cultured in 24-well plates and transfected with 0.1 μg of luciferase reporter vectors with miR-9-5p mimics or miR-ctrl mimics. The pRL-TK vector (Promega, USA) containing Renilla luciferase was also co-transfected for normalization in all experiments. Cells were harvested 48 h after transfection, and Firefly and Renilla luciferase activities were measured using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer's protocol.
Cell Proliferation Assay
U251, A118MG, and U87MG cell growth was measured 24, 48, and 72 h after transfection with FTL si-RNA by using the Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc., Rockville, MD, USA), according to the manufacturer's protocol. On average, six replicates for each time point were statistically analyzed. EdU assay was also used to measure the cell growth, Cell-Light EdU Apollo488 in vitro Flow Cytometry Kit (20T) (RiboBio, China) was used according to the manufacturer's instructions.
Flow Cytometry
Transfected glioma cells were trypsinized and fixed in 70% icecold ethanol at 20°C overnight. After centrifugation and wash with phosphate-buffered saline (PBS), the cells were suspended in propidium iodide (PI) working solution (50 mg/ml PI, 0.2 mg/ml RNase A, and 0.1% Triton X-100) for 30 min at 37°C. Twenty thousand cells were harvested and analyzed by FACS Calibur flow cytometry (BD Biosciences, USA).
Tumor Formation Assay in a Nude Mouse Model
U251 cells were collected at a concentration of 2 × 107 cells/mL and 0.1 ml was subcutaneously injected into either side of the armpit of male BALB/c nude mice (4–5 weeks old) the next day. Mice were purchased from Shanghai Experimental Animal Center of the Chinese Academy of Sciences (Shanghai, China). AgomiR-9-5p [micrON hsa-miR-9-5p agomiR was purchased from RiboBio (GuangZhou, China)] or agomiR control were injected into tumor at 1 nmol every 4 days for 4 times after transplanted. Tumor volumes and weights were measured every 4 days and tumor volumes were calculated using the following equation: V = 0.5 × D × d2 (V, volume; D, longest diameter; d, diameter perpendicular to the longest diameter). On the 20th day after injection, mice were killed, and the subcutaneous growth of each tumor was examined. Primary tumors were excised and tumor tissues were used to perform qPCR analysis of miR-9-5p levels. This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the Committee on the Ethics of Animal Experiments of the Southern Medical University.
Statistical Analysis
Data were expressed as mean ± standard error. Statistical analysis was performed with SPSS 20.0 software. Differences between means were assessed by student's t-test for normal distribution data, or Mann–Whitney U-test for non-normal distribution data. In multiple comparisons, one-way analysis of variance (ANOVA) was used. Pearson's test was used to detect the correction of two groups and compare quantitative values of expression. Survival curves were plotted by the Kaplan-Meier method and compared by log-rank test. A value of P <0.05 was considered statistically significant.
Results
Expression of miR-9-5p and FOXP2 in GBM and Clinical Features
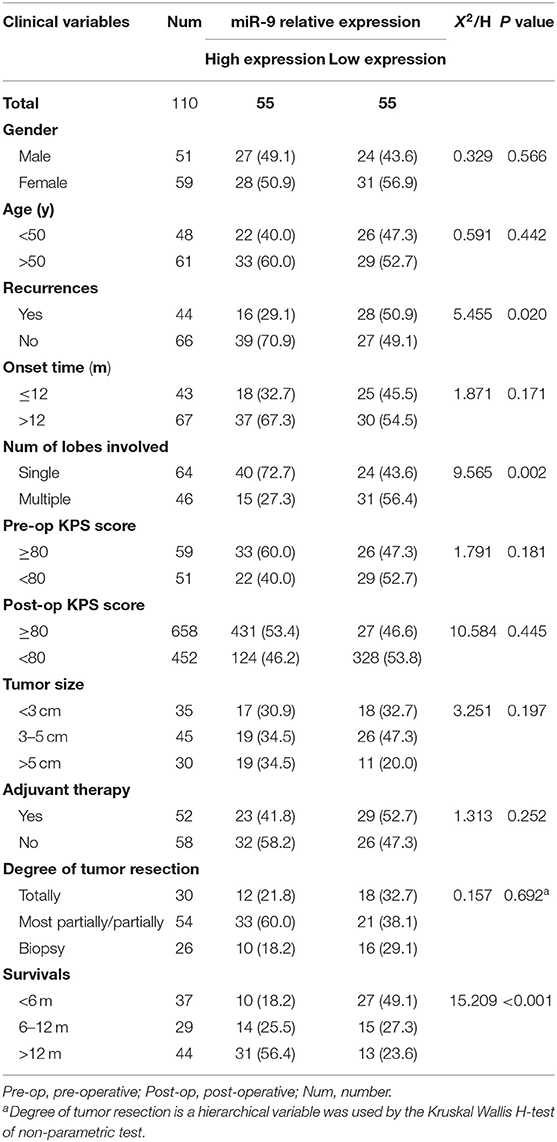
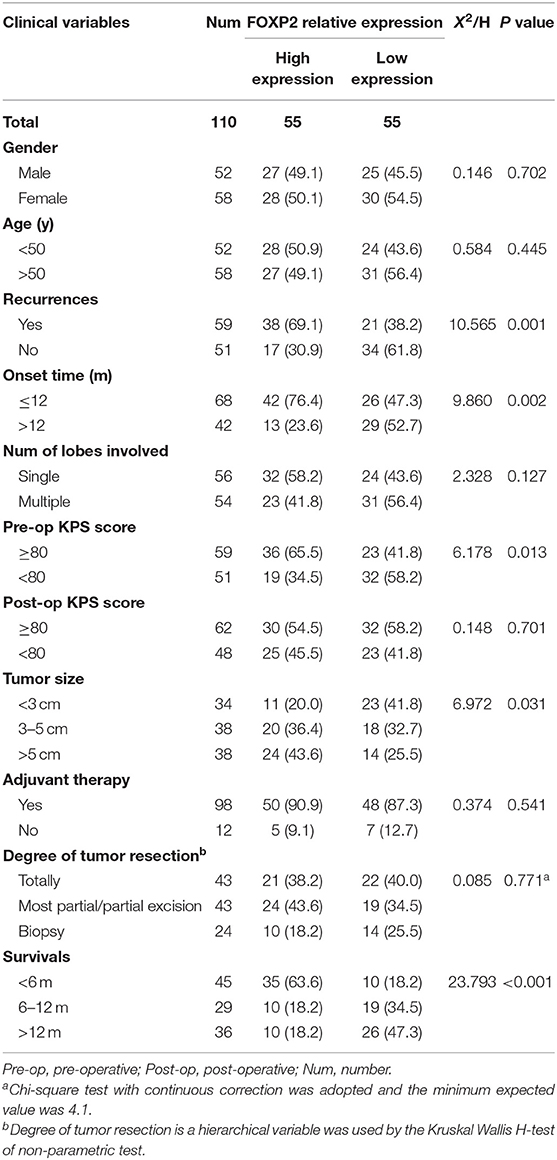
To detect the expression of miR-9-5p and FOXP2 in GBM, 110 GBM samples with complete clinical and follow-up survey data were collected for this study. According to the expression level of miR-9-5p or FOXP2, cases were divided into high expression group and low expression group (Figures 1A,B). The clinical features and relative expression of miR-9-5p and FOXP2 are presented in Tables 1, 2. The cases with high expression of miR-9-5p and low expression of FOXP2 showed higher overall survival rate (Figures 1C,D).
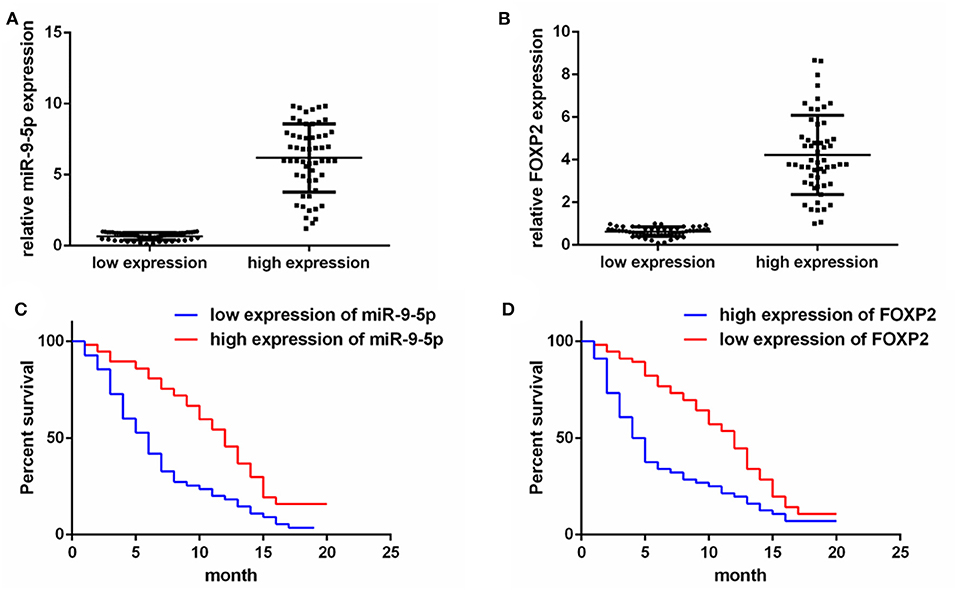
Figure 1. The expression of miR-9-5p and FOXP2 in glioblastoma and patients' survival. (A) Cases are divided into two groups according to the expression of miR-9-5p in GBM. (B) Cases are divided into two groups according to the expression of FOXP2 in GBM. (C) Kaplan-Meir survival curve analysis reveals that lower miR-9-5p predicts poorer survival (110 GBM patients). (D) Kaplan-Meir survival curve analysis reveals that higher FOXP2 predicts poorer survival (110 GBM patients).
miR-9-5p Was a Negative Regulator of GBM Cell Proliferation
We transfected the miR-9-5p mimics, inhibitor and mi-control into the U251 and U118MG cell, and q-PCR assay showed miR-9-5p expression were significantly different (Figure 2A). CCK-8 assay was used to observe the proliferation change. Comparing with the cells transfected with mi-control, the cellular viability of the groups transfected with mimics and inhibitor was significantly decreased and increased, respectively (Figure 2B). Flow cytometry was used in EdU assay, and the results showed that less marked cells were detected in the group transfected with mimics, while more in the group with inhibitor (Figure 2C). The expression level of miR-9-5p had a significant influence on G1 phase (Figure 2D). Further, we detected proteins functioning in the cell cycle process though Western blot assay, and the expression of p21 was significantly higher in miR-9-5p (Figure 2E).
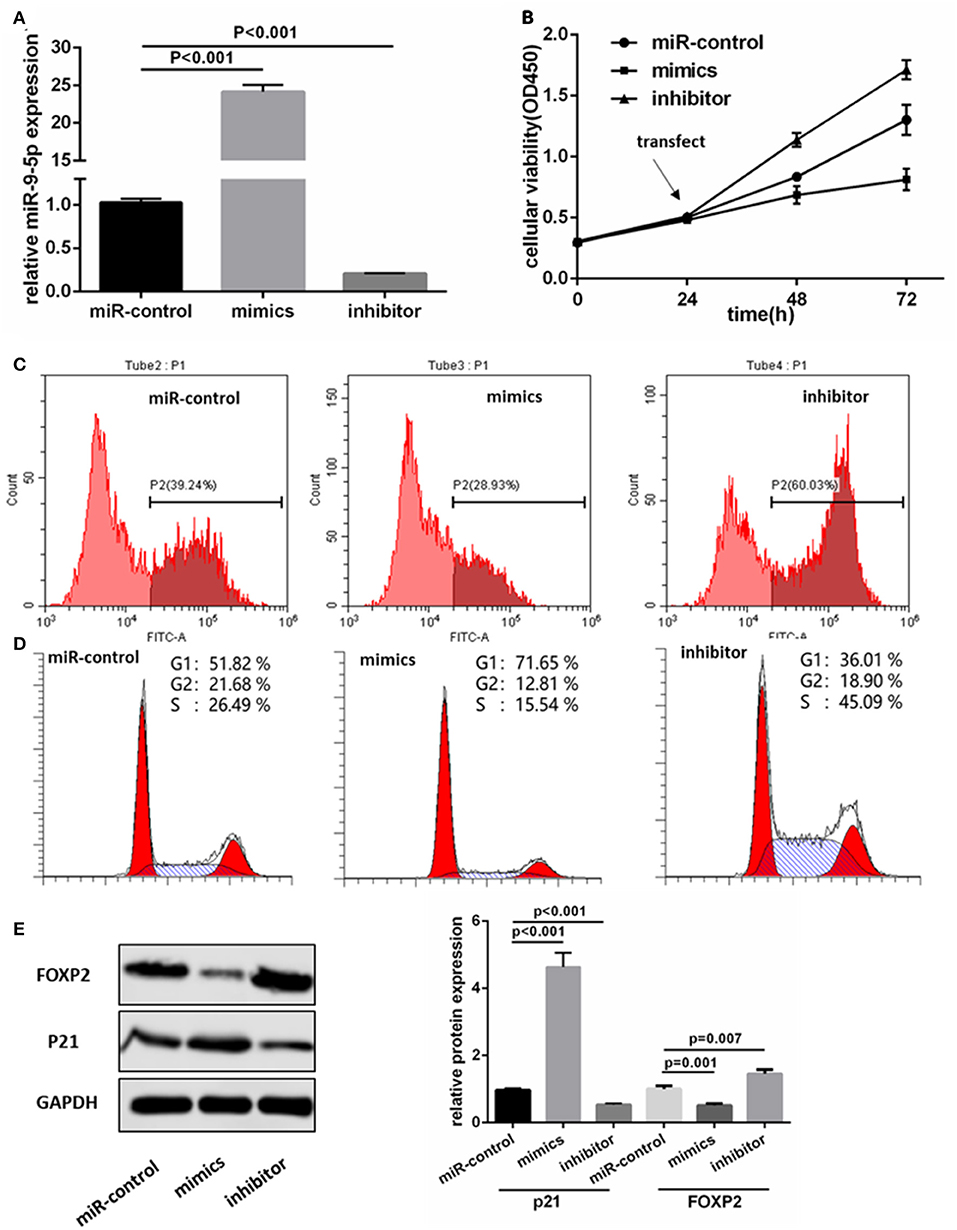
Figure 2. MiR-9-5p inhibits GBM cell growth through cell cycle arrest in vitro. (A) Expression of miR-9-5p in U251 cell transfected with miRNA. (B,C) CCK-8 (B) and EdU (C) show that high expression of miR-9-5p inhibits cell growth while low expression leads to the opposite effect. (D) Cell cycle is arrested in cells with high expression of miR-9-5p analyzed by Flow cytometry. (E) Over expression of miR-9-5p leads to down regulation of FOXP2 (p = 0.001) and up regulation of p21 (p = 0.001); while the inhibited miR-9-5p leads to up regulation of FOXP2 (p = 0.003) and down regulation of p21 (p < 0.001).
FOXP2 Was a Positive Regulator of GBM Cell Proliferation
To demonstrate that FOXP2 exerts positive effects on GBM cell proliferation, we intervened in the expression of FOXP2 (Figure 3A). Similar assays were used to analyze the cell proliferation, cell cycle and cell cycle associated proteins. Results showed that low expression of FOXP2 slowed down the cell proliferation (Figures 3B,C) and G1 arrested (Figure 3D) and inhibited p21 high expression (Figure 3A).
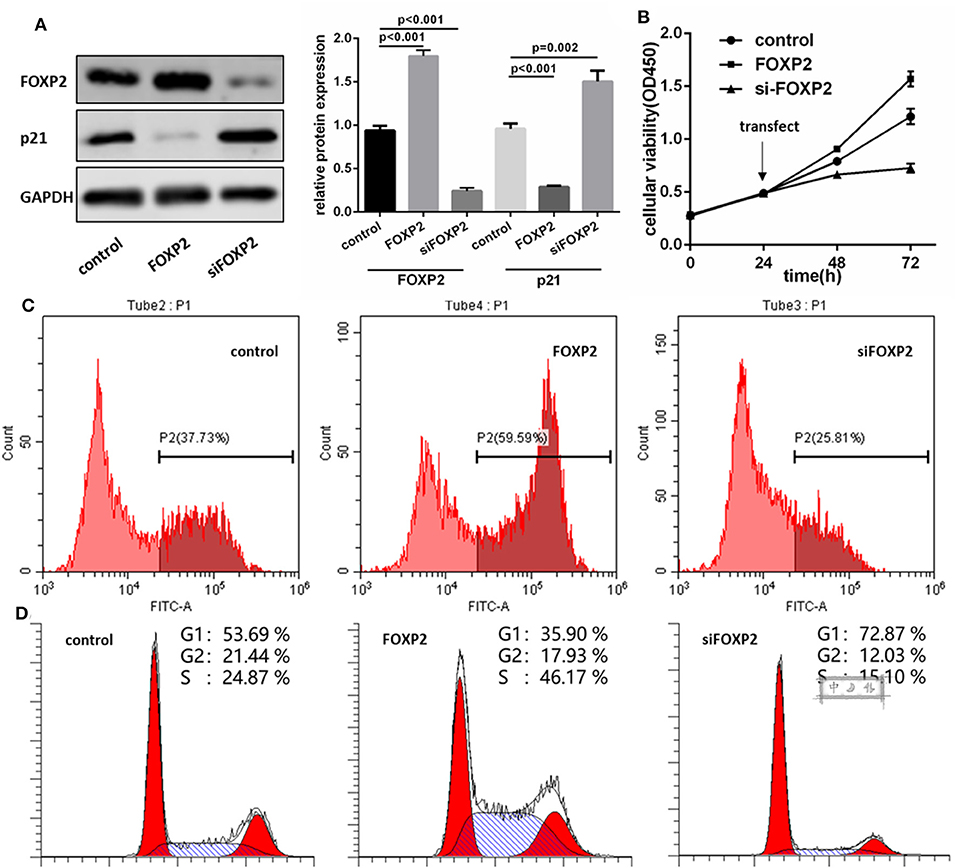
Figure 3. FOXP2 is a positive regulator of U87MG cell proliferation. (A) pcDNA3.1-FOXP2 and siRNA transfection is used to regulate the FOXP2 expression in U251 cell. Western blot shows that FOXP2 expresses as expected (p < 0.001). Upregulated FOXP2 leads to low expression of p21 (p < 0.001) and down regulated FOXP2 leads to high expression of p21 (p = 0.002). (B,C) CCK-8 (B) and EdU (C) shows high expression of FOXP2 contributes to cell proliferation while low expression leads to the opposite effect. (D) Cell cycle is arrested in cells with low expression of miR-9-5p analyzed by Flow cytometry.
miR-9-5p Directly Regulated FOXP2 Expression
Figure 3A shows a significant, negative relationship between the expression of FOXP2 and miR-9-5p. In GBM cells, miR-9-5p mimics resulted in FOXP2 down regulation, while the inhibitor exert a opposite effect (Figure 2E). Luciferase reporter assay suggested that FOXP2 was the direct target of miR-9-5p (Figure 4A).
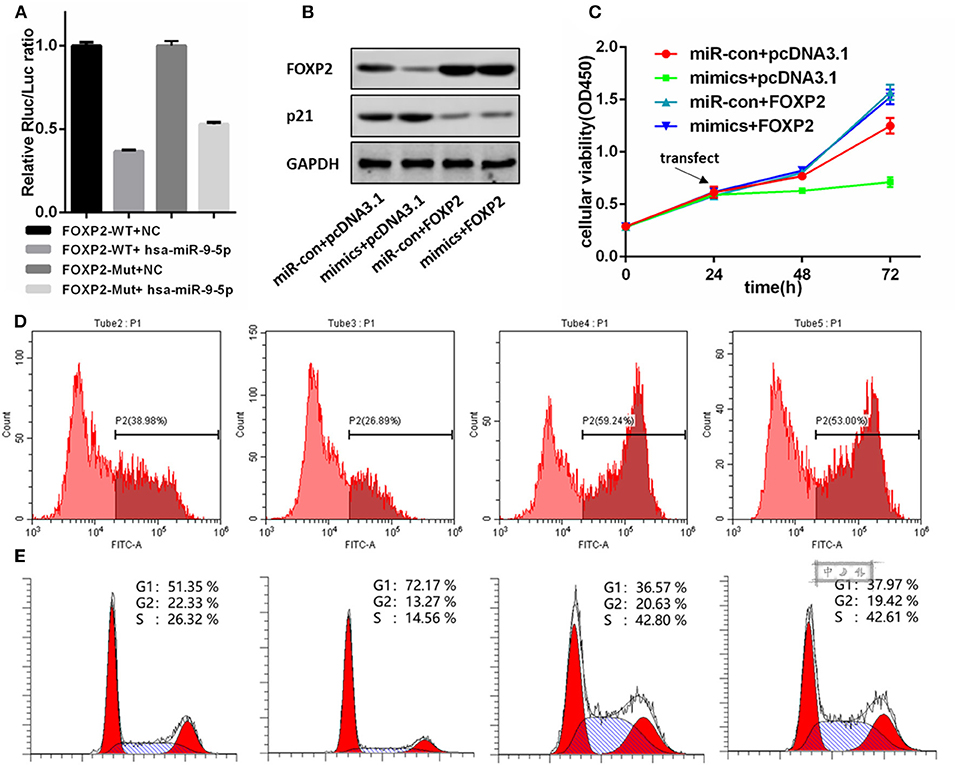
Figure 4. FOXP2 is indispensable for miR-9-5p suppressing U87MG cell proliferation. (A) Luciferase reporter assay suggests FOXP2 is the direct target of miR-9-5p. (B) Western bolt shows that the DNA transfection hold back the down regulation of FOXP2. Further, the up regulation of p21 caused by miR-9-5p is also restrained. (C,D) CCK-8 (C) and EdU (D) show that cell proliferation inhibition caused by miR-9-5p is restrained by stable expression of FOXP2. (E) Cell cycle arrest is blocked by the stable FOXP2.
FOXP2 Was Indispensable for miR-9-5p Suppressing Tumor Growth
We re-expressed FOXP2 in the U251 cell with high level miR-9-5p by DNA transfection, and the result showed that the original cell proliferation inhibition and the cell cycle arrest caused by miR-9-5p were changed (Figures 4C–E). Further, western blot showed no difference in p21 between cells with high level miR-9-5p and the control group (Figure 4B).
miR-9-5p Up-Regulation Inhibited GBM Growth in vivo
To further investigate whether the level of miR-9-5p expression could inhibit GBM growth in vivo, we inoculated U251 cell into male nude mice, and injected miR-9-5p mimics or mi-control (as control) into their tail vein. Twenty-one days after injection, all mice developed xenograft tumors at the injection site and the tumor size of the mimics group was significantly smaller compared with that of the control group (Figure 5A). Moreover, the growth of tumors was significantly slower in the mimics group than that in the mi-control group (Figure 5A). The injected group showed a higher expression of miR-9-5p (Figure 5B), a lower expression of FOXP2 expression and higher p21 (Figure 5C).
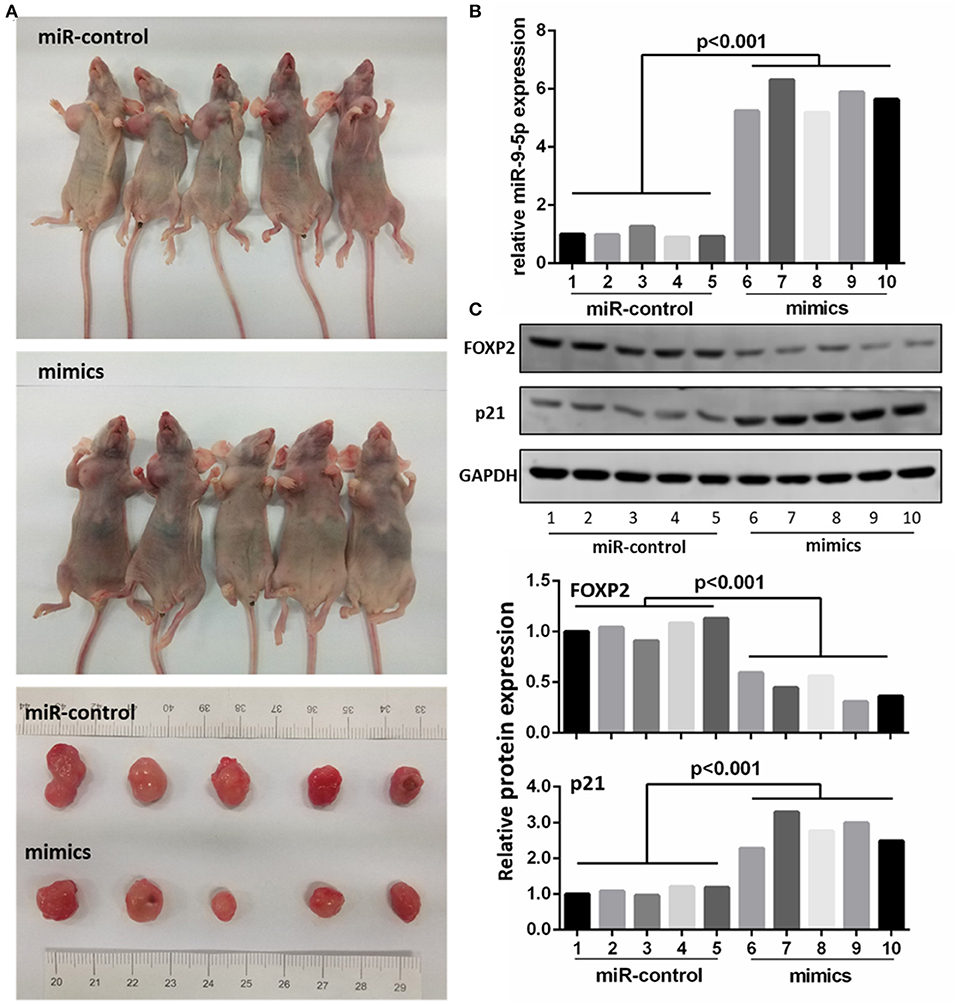
Figure 5. MiR-9-5p inhibits tumor growth in vivo. (A) Mice injected with miR-9-5p mimics have smaller tumors compared with control group. (B) q-PCR shows that miR-9-5p expression of mimics group is extremely higher than that of control group (p < 0.001). (C) Western blot shows that in mimics group, FOXP2 expression is lower (p < 0.001) and p21 expression is higher (p < 0.001).
Discussion
A better understanding of key pathways driving glioma growth and development has the potential to improve the treatment of GBM. Previous studies have demonstrated that high expression of miR-9 led to lower survival rate in patients with high grade (WHO III IV) glioma (15). However, opposite results were presented in other studies. Participating in mutant EGFR signaling, the most common abnormal signaling in GBM, suppressed miR-9 expression increased GBM cells proliferation (11, 22). On the basis of the malignant behavior, Glioma is mainly divided into two types according to WHO I–IV classification (23, 24). In this study, we found that low expression of miR-9-5p led to poor prognosis while high miR-9-5p expression inhibited tumor growth by cell cycle arrest. Similar results were found in neck cancer cells (25).
For the past few years, researches have demonstrated the involvement of FOXP2 in oncogenesis (26), but it exerts opposite function in different cancers. In neuroblastomas, multiple myelomas and several subtypes of lymphomas, FOXP2 is overexpressed (19, 20, 27). In the present study, we demonstrated that FOXP2 was a tumor promoter, and it was able to accelerate the cell cycle and increase the proliferation of GBM cells. It has been reported that FOXP2 functions in neurogenesis in embryonic development (28). FOXP2 null mutant mice developed cerebellar hypoplasia (29). Consider the important effects of FOXP2 on central nervous system, we presume that once unbalanced, it may lead to GBM oncogenesis. A previous study has found that the downregulation of FOXP2 in TP53 associated glioma cell apoptosis (30), which probably indicates that FOXP2 acts as a cancer-promoting gene in GBM.
In the clinical data, FOXP2 was relatively high expressed in recurrent GBM. It is widely considered that chemotherapy resistance is a vital factor leading to GBM recurrence. FOXP2 was reported to directly target the adenosine triphosphate (ATP)-binding cassette (ABC) family proteins such as ABCA6 and ABCG2, which were transporters expelling chemical compounds (31–33). This might be a mechanism in chemotherapy resistance of GBM and a reason for why relatively high expression of FOXP2 was found in recurrent GBM.
Conclusion
In this study, we confirmed that the high expression of miR-9-5p, which down regulated FOXP2, was able to suppress the proliferation of GBM via p21-dependent cell cycle arrest both in vivo and in vitro. It suggests a potential pathway of miR-9-5p-FOXP2 signal which may be applied to GBM therapy in the future.
Data Availability Statement
The datasets during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics Statement
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the Committee on the Ethics of Animal Experiments of the Southern Medical University.
Author Contributions
HZ and YL planned and designed the experimental scheme and the initial draft of the paper, tumor sample collection, and performed experiment. YT, QL, SJ, and DL participated in the cell culture, microRNA extraction experiments, and participated in the collation of experimental data, statistics, and results analysis. QC and SZ were fully responsible for the implementation and supervision of the subject.
Funding
This study was supported by China postdoctoral science foundation (2018M640802 to HZ), and the National Natural Science Foundation of China (81671240 to SZ), and the Guangdong Provincial Clinical Medical Center for Neurosurgery (2013B020400005 to SZ).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Tang J, Li Y, Liu K, Zhu Q, Yang WH, Xiong LK, et al. Exosomal miR-9-3p suppresses HBGF-5 expression and is a functional biomarker in hepatocellular carcinoma. Minerva Med. (2018) 109:15–23. doi: 10.23736/S0026-4806.17.05167-9
2. Moridikia A, Mirzaei H, Sahebkar A, Salimian J. MicroRNAs: potential candidates for diagnosis and treatment of colorectal cancer. J Cell Physiol. (2018) 233:901–13. doi: 10.1002/jcp.25801
3. Cui Y, Xue Y, Dong S, Zhang P. Plasma microRNA-9 as a diagnostic and prognostic biomarker in patients with esophageal squamous cell carcinoma. J Int Med Res. (2017) 45:1310–7. doi: 10.1177/0300060517709370
4. Zhou B, Xu H, Xia M, Sun C, Li N, Guo E, et al. Overexpressed miR-9 promotes tumor metastasis via targeting E-cadherin in serous ovarian cancer. Front Med. (2017) 11:214–22. doi: 10.1007/s11684-017-0518-7
5. Wang H, Wu Q, Zhang Y, Zhang HN, Wang YB, Wang W. TGF-beta1-induced epithelial-mesenchymal transition in lung cancer cells involves upregulation of miR-9 and downregulation of its target, E-cadherin. Cell Mol Biol Lett. (2017) 22:22. doi: 10.1186/s11658-017-0053-1
6. He L, Zhang L, Wang M, Wang W. miR-9 functions as a tumor inhibitor of cell proliferation in epithelial ovarian cancer through targeting the SDF-1/CXCR4 pathway. Exp Ther Med. (2017) 13:1203–8. doi: 10.3892/etm.2017.4118
7. Shang A, Lu WY, Yang M, Zhou C, Zhang H, Cai ZX, et al. miR-9 induces cell arrest and apoptosis of oral squamous cell carcinoma via CDK 4/6 pathway. Artif Cells Nanomed Biotechnol. (2018) 46:1754–62. doi: 10.1080/21691401.2017.1391825
8. Gao HY, Huo FC, Wang HY, Pei DS. MicroRNA-9 inhibits the gastric cancer cell proliferation by targeting TNFAIP8. Cell Prolif. (2017) 50:e12331. doi: 10.1111/cpr.12331
9. Peng Z, Ying L. Effects of TNFalpha on cell viability, proliferation and apoptosis of glioma cells U251. J Buon. (2014) 19:733–41.
10. Song Y, Mu L, Han X, Li Q, Dong B, Li H, et al. MicroRNA-9 inhibits vasculogenic mimicry of glioma cell lines by suppressing Stathmin expression. J Neurooncol. (2013) 115:381–90. doi: 10.1007/s11060-013-1245-9
11. Gomez GG, Volinia S, Croce CM, Zanca C, Li M, Emnett R, et al. Suppression of microRNA-9 by mutant EGFR signaling upregulates FOXP1 to enhance glioblastoma tumorigenicity. Cancer Res. (2014) 74:1429–39. doi: 10.1158/0008-5472.CAN-13-2117
12. Yang L, Mu Y, Cui H, Liang Y, Su X. MiR-9-3p augments apoptosis induced by H2O2 through down regulation of Herpud1 in glioma. PLoS ONE. (2017) 12:e174839. doi: 10.1371/journal.pone.0174839
13. Zu Y, Zhu Z, Lin M, Xu D, Liang Y, Wang Y, et al. MiR-9 promotes apoptosis via suppressing SMC1A expression in GBM cell lines. Curr Chem Genom Transl Med. (2017) 11:31–40. doi: 10.2174/2213988501711010031
14. Ben-Hamo R, Zilberberg A, Cohen H, Efroni S. hsa-miR-9 controls the mobility behavior of glioblastoma cells via regulation of MAPK14 signaling elements. Oncotarget. (2016) 7:23170–81. doi: 10.18632/oncotarget.6687
15. Wu Z, Wang L, Li G, Liu H, Fan F, Li Z, et al. Increased expression of microRNA-9 predicts an unfavorable prognosis in human glioma. Mol Cell Biochem. (2013) 384:263–8. doi: 10.1007/s11010-013-1805-5
16. Yan X, Zhou H, Zhang T, Xu P, Zhang S, Huang W, et al. Downregulation of FOXP2 promoter human hepatocellular carcinoma cell invasion. Tumour Biol. (2015) 36:9611–9. doi: 10.1007/s13277-015-3701-y
17. Cuiffo BG, Campagne A, Bell GW, Lembo A, Orso F, Lien EC, et al. MSC-regulated microRNAs converge on the transcription factor FOXP2 and promote breast cancer metastasis. Cell Stem Cell. (2014) 15:762–74. doi: 10.1016/j.stem.2014.10.001
18. Jia WZ, Yu T, An Q, Yang H, Zhang Z, Liu X, et al. MicroRNA-190 regulates FOXP2 genes in human gastric cancer. Onco Targets Ther. (2016) 9:3643–51. doi: 10.2147/OTT.S103682
19. Wong KK, Gascoyne DM, Soilleux EJ, Lyne L, Spearman H, Roncador G, et al. FOXP2-positive diffuse large B-cell lymphomas exhibit a poor response to R-CHOP therapy and distinct biological signatures. Oncotarget. (2016) 7:52940–56. doi: 10.18632/oncotarget.9507
20. Khan FH, Pandian V, Ramraj S, Natarajan M, Aravindan S, Herman TS, et al. Acquired genetic alterations in tumor cells dictate the development of high-risk neuroblastoma and clinical outcomes. BMC Cancer. (2015) 15:514. doi: 10.1186/s12885-015-1463-y
21. Gascoyne DM, Spearman H, Lyne L, Puliyadi R, Perez-Alcantara M, Coulton L, et al. The Forkhead transcription factor FOXP2 is required for regulation of p21WAF1/CIP1 in 143B osteosarcoma cell growth arrest. PLoS ONE. (2015) 10:e128513. doi: 10.1371/journal.pone.0128513
22. Frederick L, Wang XY, Eley G, James CD. Diversity and frequency of epidermal growth factor receptor mutations in human glioblastomas. Cancer Res. (2000) 60:1383–7.
23. Morgan LL. The epidemiology of glioma in adults: a “state of the science” review. Neuro Oncol. (2015) 17:623–4. doi: 10.1093/neuonc/nou358
24. Olar A, Aldape KD. Using the molecular classification of glioblastoma to inform personalized treatment. J Pathol. (2014) 232:165–77. doi: 10.1002/path.4282
25. Hersi HM, Raulf N, Gaken J, Folarin N, Tavassoli M. MicroRNA-9 inhibits growth and invasion of head and neck cancer cells and is a predictive biomarker of response to plerixafor, an inhibitor of its target CXCR4. Mol Oncol. (2018) 12:2023–41. doi: 10.1002/1878-0261.12352
26. Herrero MJ, Gitton Y. The untold stories of the speech gene, the FOXP2 cancer gene. Genes Cancer. (2018) 9:11–38. doi: 10.18632/genesandcancer.169
27. Campbell AJ, Lyne L, Brown PJ, Launchbury RJ, Bignone P, Chi J, et al. Aberrant expression of the neuronal transcription factor FOXP2 in neoplastic plasma cells. Br J Haematol. (2010) 149:221–30. doi: 10.1111/j.1365-2141.2009.08070.x
28. Tsui D, Vessey JP, Tomita H, Kaplan DR, Miller FD. FoxP2 regulates neurogenesis during embryonic cortical development. J Neurosci. (2013) 33:244–58. doi: 10.1523/JNEUROSCI.1665-12.2013
29. Shu W, Cho JY, Jiang Y, Zhang M, Weisz D, Elder GA, et al. Altered ultrasonic vocalization in mice with a disruption in the Foxp2 gene. Proc Natl Acad Sci USA. (2005) 102:9643–8. doi: 10.1073/pnas.0503739102
30. Seznec J, Weit S, Naumann U. Gene expression profile in a glioma cell line resistant to cell death induced by the chimeric tumor suppressor-1 (CTS-1), a dominant-positive variant of p53–the role of NFkappaB. Carcinogenesis. (2010) 31:411–8. doi: 10.1093/carcin/bgp319
31. Hedditch EL, Gao B, Russell AJ, Lu Y, Emmanuel C, Beesley J, et al. ABCA transporter gene expression and poor outcome in epithelial ovarian cancer. J Natl Cancer Inst. (2014) 106:dju149. doi: 10.1093/jnci/dju149
32. Dlugosz A, Janecka A. ABC transporters in the development of multidrug resistance in cancer therapy. Curr Pharm Des. (2016) 22:4705–16. doi: 10.2174/1381612822666160302103646
Keywords: miR-9-5p, FOXP2, glioblastoma, proliferation, glioma
Citation: Zhang H, Li Y, Tan Y, Liu Q, Jiang S, Liu D, Chen Q and Zhang S (2019) MiR-9-5p Inhibits Glioblastoma Cells Proliferation Through Directly Targeting FOXP2 (Forkhead Box P2). Front. Oncol. 9:1176. doi: 10.3389/fonc.2019.01176
Received: 07 May 2019; Accepted: 18 October 2019;
Published: 19 November 2019.
Edited by:
Liam Chen, Johns Hopkins University, United StatesReviewed by:
Kamalakannan Palanichamy, The Ohio State University, United StatesAkira Asai, University of Shizuoka, Japan
Copyright © 2019 Zhang, Li, Tan, Liu, Jiang, Liu, Chen and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qianxue Chen, Y2hlbnF4NjY2QHdodS5lZHUuY24=; Shizhong Zhang, emhhbmdzaGl6aG9uZ0BzbXUuZWR1LmNu
†These authors have contributed equally to this work
 Hongbo Zhang
Hongbo Zhang Yuntao Li3,4†
Yuntao Li3,4† Yinqiu Tan
Yinqiu Tan Qianxue Chen
Qianxue Chen Shizhong Zhang
Shizhong Zhang