- 1Department of Dermatology, West China Hospital, Sichuan University, Chengdu, China
- 2State Key Laboratory of Biotherapy, West China Hospital, Sichuan University, Chengdu, China
Cutaneous squamous cell carcinoma derives from keratinocytes and is the second most common cause of non-melanoma skin cancer. Cutaneous squamous cell carcinoma (cSCC) develops rapidly and is also the leading cause of death in non-melanoma cancers. Lymph node metastasis occurs in 5% of cSCC patients, and some patients may even metastasize to the viscera. Patients with regional lymphatic metastasis or distant metastases have a <20% 10-year survival rate, indicating the substantial challenge in treating advanced and metastatic cSCC. Some lncRNAs have been found to be abnormally overexpressed in many tumor tissues, so that they can be considered as potential new biomarkers or targets that can be used in the diagnosis and treatment of cSCC in the future. In this review, we summarize the role of lncRNA in cutaneous squamous cell carcinoma to make a better understanding of mutations in cSCC and lay the foundation for effective target therapy of cSCC.
Introduction
Skin is the largest organ of the human body composed of epidermis, dermis, and subcutaneous tissue. The epidermis belongs to the multilayer epidermal epithelium, which is mainly composed of keratinocytes. The outermost cuticle of the epidermis can protect against physical damage, chemical stimulation, and microbial invasion. However, a variety of risk factors such as chemicals, ultraviolet radiation, smoke, and pollutants, can cause skin damage and even skin cancer (1, 2). Cutaneous squamous cell carcinoma is derived from keratinocytes and is the second most common cause of non-melanoma skin cancer (3). The occurrence of cutaneous squamous cell carcinoma is an adverse outcome caused by the interaction of multiple factors, including environmental factors and self-factors. The most important risk factors for cSCC included skin color and age reversal of tumor suppressor genes such as RAS, MYC, p53, and RUX3 (4). The adverse stimulation of the external environment is closely related to the occurrence of cSCC, such as sunlight exposure, chemical exposure, and viral infection. The occurrence of cSCC is generally believed that it is related to excessive ultraviolet irradiation (5). UV mainly causes DNA damage in cells, such as transformation of C-T and CC-TT pyrimidine dimer, activation of p53, and loss of function of regulating cell proliferation. In addition, mutations of p53 and RAS have been found in patients with actinic keratosis induced by UV. It can be inferred that mutation of p53 and RAS may be early changes of UV damage, which laid the foundation for the development of cSCC (6). Cutaneous squamous cell carcinoma develops rapidly, which is also the leading cause of death in non-melanoma cancers. Lymph node metastasis occurs in 5% of cSCC patients, and some patients may even metastasize to the viscera (7). Patients with regional lymphatic metastasis or distant metastases have a <20% 10-year survival rate, indicating the substantial challenge in treating advanced and metastatic cSCC (8). It has been reported that the down-regulation of p21 mediated by melanoma-associated antigen A12 (MAGEA12) may be involved in the pathogenesis of cSCC, which indicates that MAGEA12 might be a molecular biomarker of cSCC (9). And telomerase reverse transcriptase gene promoter (TERTp) mutation may be a molecular biomarker with prognostic significance for invasive cSCC, but further study is still needed (10). Meanwhile, Cortactin (CTTN) phosphorylation is closely related to the pathogenesis of cSCC and can be used as a molecular biomarker of cSCC (11). Positive pS6 seems to be the predictor of aggressiveness of cSCC combined with the operation history of cSCC, lesion-positive margin, degree of differentiation, and lesion size (12). The expression of cell division cycle 20 (CDC20) in cSCC tissues and cell lines increased significantly, which was related to the pathological differentiation of cSCC. It suggested that CDC20 may be a new biomarker for the prevention diagnosis and treatment of cSCC (13). LncRNA is a transcript with a length of more than 200 nucleotides without open reading frame and does not encode proteins. It is capped at the 5′ end and polyadenylated at the 3′ end, and transcribed by RNA polymerase II. Compared to mRNA, the expression of lncRNA is not abundant, and it has poor conservativeness among species (14). Although most lncRNAs do not encode proteins, it has been reported that about 8% of lncRNAs in humans can encode short peptides; these results demonstrated that lncRNA regulates biological processes by encoding short peptides (15). Some lncRNAs have been found to have an abnormal overexpression in many tumor tissues, which can be considered as new potential biomarkers that can predict the canceration of tissues.
Generally, according to its relative position with coding genes, lncRNA can be basically divided into intergenic lncRNA, intronic lncRNA, sense lncRNA, antisense lncRNA, and bidirectional lncRNA. The location of lncRNA transcription in genomes often determines its related function (14, 16). In addition, it can be classified into four categories according to the function of lncRNA: signal, decoy, guide, and scaffold. LncRNA participated in the transmission of some signaling pathways when they acted as signals; some lncRNAs can regulate downstream gene transcription. As decoys, lncRNA can bind and remove some transcription factors or proteins to regulate gene expression. As guides, lncRNA can recruit cis- or trans-acting target genes of chromatin modifying enzymes. As scaffolds, lncRNA can bind a variety of proteins to form complex and modify histone in chromatin (17). As for the regulation mechanism of lncRNA, it can interact with DNA, RNA, or protein to regulate gene expression via various pathways. Firstly, lncRNA can be scaffolds or guides to regulate the related chromatin modifying enzymes in transcriptional processes (18). Secondly, lncRNA affected gene expression by regulating epichromatin modification (19). Thirdly, lncRNA regulated gene expression after transcription and regulated the level of mRNA and miRNA by different mechanisms such as competing endogenous RNA (20). Consequently, we reviewed the role of lncRNA in cutaneous squamous cell carcinoma to make a better understanding of mutations in cSCC and lay the foundation for effective target therapy of cSCC. It has been reported that the expression of lncRNA MALAT1 was upregulated in tongue squamous cell carcinoma (TSCC) and was related to cervical lymph node metastasis. Further mechanism studies showed that MALAT1 can inhibit tumor cell apoptosis and induce cell migration and invasion by regulating the Wnt/beta-catenin signaling pathway, and overexpression of MALAT1 can induce epithelial mesenchymal transition (EMT) (21). Meanwhile, high MALAT1 level was found in 54 cases of oral squamous cell carcinoma (OSCC) with poor prognosis (22). Compared with normal tissues, the expression of HOTAIR in OSCC was increased, which negatively correlated with E-cadherin level (23). And HOTAIR is highly expressed in TSCC, which was involved in the regulation of proliferation and apoptosis of TSCC (24). It was found that the expression of GAS5 in OSCC was lower than that in normal tissues, suggesting that the overexpression of GAS5 inhibited the proliferation, migration, and invasion of tumors (25).
Role of lncRNA in Cutaneous Squamous Cell Carcinoma
PICSAR
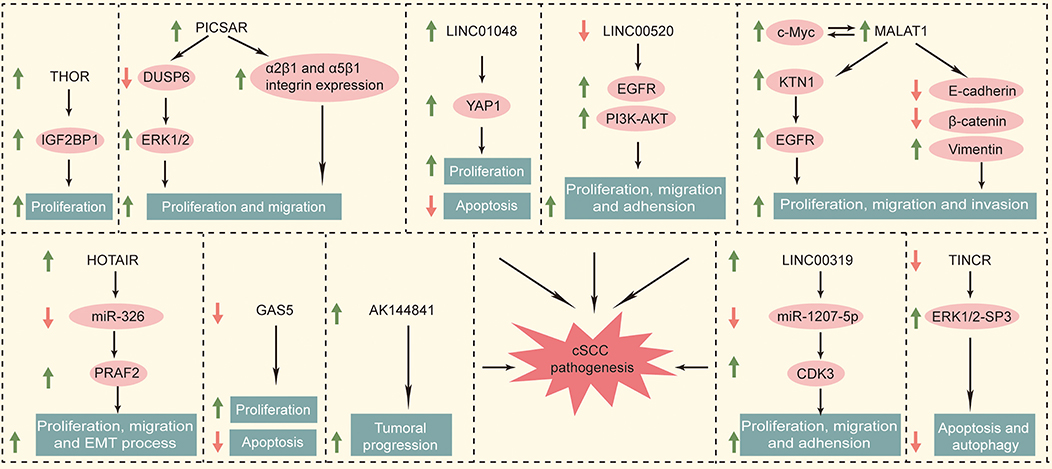
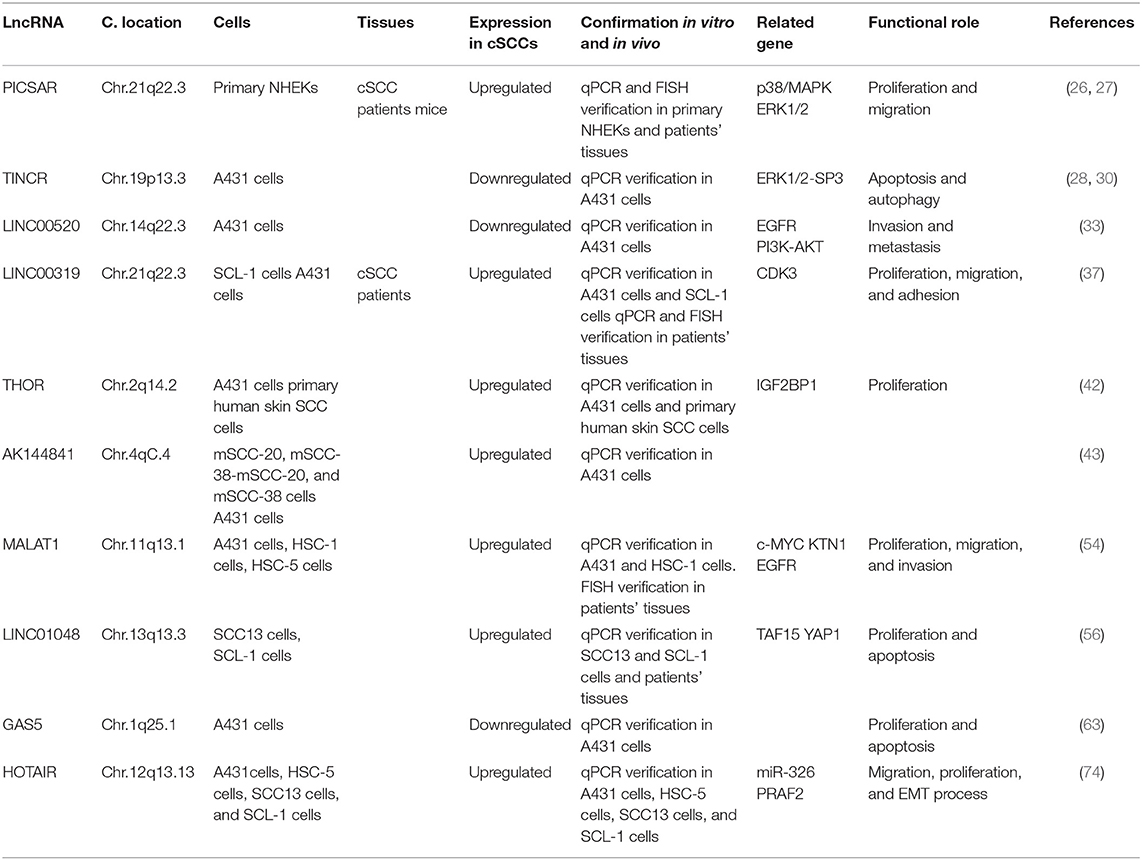
PICSAR, LINC00162, was a kind of p38 inhibited cutaneous squamous cell carcinoma associated with lincRNA, which was first reported in 2016. Piipponen et al. obtained cSCC cell lines from patients' skin tissue by surgical resection. By whole-transcriptome analyses, it has been found that several kinds of lncRNAs were differentially expressed in cSCC cell compared with primary NHEKs (normal human epidermal keratinocytes). Among them, long intergenic non-protein coding RNA 162 (LINC00162) was the most significantly up-regulated lncRNA. RNA in situ hybridization analysis showed that PICSAR was specifically expressed by tumor cells in cSCCs, but not by keratinocytes in normal skin in vivo. In addition, PICSAR played a carcinogenic role by regulating the mitogen-activated protein kinase/extracellular signal-regulated kinase (MAPK/ERK) signaling pathway, which is well-known to be dysregulated in cSCCs. It has been further determined that the carcinogenic mechanism of PICSAR was through inhibiting ERK2's negative regulator dual specificity phosphate 6 (DUSP6), while PICSAR can be inhibited by P38 MAPK. Besides, knockout of PICSAR suppressed the proliferation and migration of cSCC cells and inhibited the growth of human cSCC xenografts in vivo (26). Meanwhile, it has been mentioned that knockdown of PICSAR increased adhesion and decreased cell migration on collagen I and fibronectin by downregulating α2β1 and α5β1 integrin expression (27).
TINCR
The gene of TINCR is located between SAFB2 and ZNRF4 genes on chromosome 19, which can promote epidermal differentiation through post-transcriptional mechanism. Studies show that the number of layers in human epidermal tissue layered granules decreased by 81.4%. Caspase decreased 83.7% in the absence of TINCR, which hydrolyzed protein and promoted apoptosis and is needed to maintain the function of the epidermal barrier (28, 29). Cutaneous squamous cell carcinoma derives from keratinocytes in the epidermis, which is closely related to epidermal differentiation. In human squamous cell carcinoma specimens, the expression of TINCR was down-regulated, which is consistent with the decrease in the differentiation of squamous cell carcinoma (28). Other research suggested that TINCR is involved in ALA-PDT-induced (5-aminolevulinic acid- photodynamic therapy-induced) apoptosis and autophagy in A431 cells. ALA-PDT promoted the expression of TINCR in A431 cells, and then TINCR promoted ALA-PDT-induced apoptosis and autophagy via the ERK1/2-SP3 (specificity protein 3) pathway (30).
LINC00520
As a new type of lncRNA, LINC00520 has been reported only in a few tumors. The expression of LINC00520 increased in breast cancer. The oncogenes SRC, PIK3CA, and STAT3 can regulate the expression of LINC00520 and affect the progress of breast cancer (31). The expression of LINC00520 was upregulated in laryngeal squamous cell carcinoma, and it was associated with lymph node metastasis (32). In cutaneous squamous cell carcinoma, LINC00520 suppressed the invasion and metastasis of A431 cells via inhibiting EGFR and inactivating the PI3K-AKT signaling pathway (33).
LINC00319
LINC00319 was a newly discovered cancer-related lncRNA transcribed from the intergenic region of chromosome 21, which has been reported as a carcinogen in several human cancers, such as lung cancer (34), nasopharyngeal carcinoma (35), and ovarian cancer (36). It has been reported that LINC00319 was significantly upregulated in cSCC tissues and cell lines. Functional studies showed that LINC00319 promoted the proliferation of CSCC cells, accelerated the cell cycle process, promoted cell migration and invasion, and inhibited cell apoptosis. In mechanistic studies, LINC00319 promoted cell proliferation, migration, and invasion through the regulation of CDK3 (cyclin-dependent kinase 3) in A431 cells mediated by miR-1207-5p (37).
THOR
LncRNA THOR was a highly conserved long non-coding RNA mainly expressed in normal testis and tumors (38), which has also been shown to be closely related to the biological functions of tumors. For example, THOR can promote the proliferation of hepatocellular carcinoma cells and renal cancer cells, and even mediate cisplatin resistance in nasopharyngeal carcinoma (39–41). Knockdown of THOR in A431 cells downregulated IGF2BP1-dependent mRNAs, and then suppressed A431cell survival and proliferation. Targeting IGF2BP1 by Lnc-THOR silencing might be a novel strategy to inhibit cSCCs (42).
AK144841
AK144841 is a new long noncoding found by Gilles et al., the expression of which in cSCCs was 40 times higher than that in healthy skin. AK144841 was absent from normal keratinocytes, indicating that it may play a possible role in tumoral progression (43).
MALAT1
MALAT1, a bona fide lncRNA, was highly transformed in mammals and widely expressed in human tissues (44) and played an important role in angiogenesis (45). MALAT1 showed abnormally high expression in breast cancer (46), liver cancer (47), gastric cancer (48), and tongue squamous cell carcinoma (49), which promoted the migration and proliferation of cancer cells (50) and affected the drug resistance of cancer cells (51). In addition, it was related to the adverse prognosis of various solid tumors (52, 53). MALAT1 was characterized to be highly expressed in cSCC tissues and cell lines. Zhang et al. established a novel c-MYC-assisted MALAT1-KTN1-EGFR axis. MALAT1 regulated the protein expression of EGFR but did not affect the EGFR mRNA expression. Transcriptional sequencing identified KTN1 as the key mediator regulating EGFR. Mechanism studies have shown that MALAT1 interacted with c-MYC to form a complex, which bound directly to the promoter region of KTN1 gene to enhance its activation, thereby actively regulating the protein expression of EGFR (54). Meanwhile, knocking down MALAT1 significantly increased the protein expression of E-cadherin and β-catenin and decreased the protein expression of vimentin (55).
LINC01048
According to data from the TCGA database, upregulation of intergenic length non-protein coding RNA 1048 (LINC01048) was associated with a low overall survival rate in cSCCs. Knockout of LINC01048 inhibited cell proliferation and promoted cell apoptosis, suggesting the carcinogenic role of LINC01048 in cSCCs. Mechanism studies showed that LINC01048 increased the binding of TAF15 to the YAP1 promoter, thereby activating YAP1 in cSCC cells (56).
GAS5
GAS5, a tumor suppressor (57), was usually induced by stress such as serum deficiency and cell-to-cell contact inhibition (58). The expression level of GAS5 in renal, breast, lung, prostate, and bladder transitional cell carcinomas was significantly lower than that in normal tissues, which may lead to apoptosis avoidance and cell cycle disorder (57–62). Studies showed that GAS5 in normal tissues was significantly higher than that in cSCC tissues. Overexpression of GAS5 inhibited proliferation and promoted apoptosis in A431 cells (63).
HOTAIR
HOTAIR is the first lncRNA found to have a trans-acting effect (64). The HOTAIR gene sequence is less conserved except for specific regions, and evolved faster than the HOXC gene cluster located nearby (65). These two genes not only are closely related to the human embryonic development but also played a key regulatory role in the adult organ and tissue formation (66). HOTAIR is widely involved in the regulation of the malignant process such as proliferation, apoptosis, angiogenesis, invasion, and metastasis (67). HOTAIR monitored epigenetic modification by the histone H3K27me3 (68) and regulated WIF-1 and PTEN, thereby effecting the Wnt and Akt signaling pathways (69, 70). Another study showed that HOTAIR (HOX transcript antisense RNA) knockdown inhibited the motility and invasiveness of A375 cells and reduced the degradation of the extracellular matrix (71). Meanwhile, Liu et al. showed that overexpression of HOTAIR upregulated the PKR expression to activate PI3K/AKT and NF-κB pathways in keratinocytes (HACAT cells). This promoted the UVB-induced apoptosis and inflammatory injury (72). It has been shown that HOTAIR had higher expression levels in melanomas than in non-tumor tissues (73). It has been reported that the expression of HOTAIR in cSCC cells increased significantly. The overexpression of HOTAIR promoted the migration, proliferation, and epithelial–mesenchymal transitions by competitively combining with miR-326 to regulate the expression of PRAF2 (74). We summarized the functional roles of specific deregulated lncRNAs in cataneous squamous cell carcinoma in Figure 1.
Conclusion
Cutaneous squamous cell carcinoma derives from keratinocytes and is the second most common cause of non-melanoma skin cancer. Lymph node metastasis occurs in 5% of cSCC patients, and patients with regional lymphatic metastasis or distant metastases have a <20% 10-year survival rate, suggesting that it is necessary to further understand the pathogenesis of cSCC. In this review, we summarized the role of lncRNA in cutaneous squamous cell carcinoma to make a better understanding of mutations in cSCC and lay the foundation for effective target therapy of cSCC. The expression of PICSAR, LINC00319, THOR, AK144841, MALAT1, LINC10148, and HOTAIR was upregulated in cSCCs. However, that of TINCR, LINC00520, and GAS5 was downregulated in cSCCs. MALAT1 and HOTAIR were upregulated in TSCC, OSCC, and cSCC, which may play a role in SCC diagnosis. Meanwhile, GAS5 was downregulated in OSCC and cSCC. PICSAR, LINC00319, THOR, AK144841, LINC10148, TINCR, and LINC00520 were only found in cSCC, which have a potential to be specific markers of cSCC. LncRNAs played different roles in cSCC, and we summarized them in Table 1 for a clearer understanding. Among them, PICSAR and TINCR regulated cSCC via the ERK1/2 pathway. We believed that there will be more lncRNA-related findings in future studies to improve cSCC therapy.
Author Contributions
YW, BS, GH, and XJ designed the study. YW, DH, DD, and XW performed the research. YW, BS, XW, and DH analyzed the data. YW, DD, GH, and XJ wrote the article.
Funding
This research was supported by grants from the National Natural Science Foundation (Nos. 21772131 and 81872535) and the Fundamental Research Funds of Science & Technology Department of Sichuan Province (Grant Nos. 2019YFSY0004 and 2017JY0075).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Luch A. Nature and nurture - lessons from chemical carcinogenesis. Nat Rev Cancer. (2005) 5:113–25. doi: 10.1038/nrc1546
2. Poirier MC. Chemical-induced DNA damage and human cancer risk. Discov Med. (2012) 14:283–8. doi: 10.1038/nrc1410
3. Motaparthi K, Kapil JP, Velazquez EF. Cutaneous squamous cell carcinoma: review of the eighth edition of the american joint committee on cancer staging guidelines, prognostic factors, and histopathologic variants. Adv Anatomic Pathol. (2017) 24:171–94. doi: 10.1097/PAP.0000000000000157
4. Wesers-Attemal A, Joosten VM, Roozeboom MH, Nelemans PJ, Loh-Man BG, Botterweck AA, et al. Correlation between histological findings on punch biopsy specimens and subsequent excision specimens in cutaneous squamous cell carcinoma. Acta Derm Venereol. (2015) 95:181–5. doi: 10.2340/00015555-1826
5. Thompson AK, Kelley BF, Prokop LJ, Murad MH, Baum CL. Risk factors for cutaneous squamous cell carcinoma recurrence, metastasis, and disease-specific death: a systematic review and meta-analysis. JAMA Dermatol. (2016) 152:419–28. doi: 10.1001/jamadermatol.2015.4994
6. Fernandez Figueras MT. From actinic keratosis to squamous cell carcinoma: pathophysiology revisited. J Eur Acad Dermatol Venereol. (2017) 31:5–7. doi: 10.1111/jdv.14151
7. Khullar G, Saikia UN, De D, Handa S, Das Radotra B. Predisposing factors and histopathological variants of cutaneous squamous cell carcinoma: experience from a North Indian teaching hospital. Indian J Dermatol Venereol Leprol. (2016) 82:273–8. doi: 10.4103/0378-6323.168936
8. Hillen U, Ulrich M, Alter M, Becker JC, Gutzmer R, Leiter U, et al. Cutaneous squamous cell carcinoma: a review with consideration of special patient groups. Hautarzt. (2014) 65:590–9. doi: 10.1007/s00105-013-2734-7
9. Zhao G, Bae JY, Zheng Z, Park HS, Chung KY, Roh MR, et al. Overexpression and implications of melanoma-associated antigen A12 in pathogenesis of human cutaneous squamous cell carcinoma. Anticancer Res. (2019) 39:1849–57. doi: 10.21873/anticanres.13292
10. Campos MA, Macedo S, Fernandes M, Pestana A, Pardal J, Batista R, et al. TERT promoter mutations are associated with poor prognosis in cutaneous squamous cell carcinoma. J Am Acad Dermatol. (2019) 80:660–9. e666. doi: 10.1016/j.jaad.2018.08.032
11. Zhu L, Cho E, Zhao G, Roh MR, Zheng Z. The pathogenic effect of cortactin tyrosine phosphorylation in cutaneous squamous cell carcinoma. In Vivo. (2019) 33:393–400. doi: 10.21873/invivo.11486
12. Khandelwal AR, Ma X, Egan P, Kaskas NM, Moore-Medlin T, Caldito G, et al. Biomarker and pathologic predictors of cutaneous squamous cell carcinoma aggressiveness. Otolaryngol Head Neck Surg. (2016) 155:281–8. doi: 10.1177/0194599816641913
13. Chu Z, Zhang X, Li Q, Hu G, Lian CG, Geng S. CDC20 contributes to the development of human cutaneous squamous cell carcinoma through the Wnt/β-catenin signaling pathway. Int J Oncol. (2019) 54:1534–44. doi: 10.3892/ijo.2019.4727
14. Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. (2009) 136:629–41. doi: 10.1016/j.cell.2009.02.006
15. Banfai B, Jia H, Khatun J, Wood E, Risk B, Gundling WE Jr, et al. Long noncoding RNAs are rarely translated in two human cell lines. Genome Res. (2012) 22:1646–57. doi: 10.1101/gr.134767.111
16. Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, et al. Landscape of transcription in human cells. Nature. (2012) 489:101–8. doi: 10.1038/nature11233
17. Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. (2011) 43:904–14. doi: 10.1016/j.molcel.2011.08.018
18. Castelo-Branco G, Amaral PP, Engstrom PG, Robson SC, Marques SC, Bertone P, et al. The non-coding snRNA 7SK controls transcriptional termination, poising, and bidirectionality in embryonic stem cells. Genome Biol. (2013) 14:R98. doi: 10.1186/gb-2013-14-9-r98
19. Di Ruscio A, Ebralidze AK, Benoukraf T, Amabile G, Goff LA, Terragni J, et al. DNMT1-interacting RNAs block gene-specific DNA methylation. Nature. (2013) 503:371. doi: 10.1038/nature12598
20. Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. (2014) 505:344–52. doi: 10.1038/nature12986
21. Liang J, Liang L, Ouyang K, Li Z, Yi X. MALAT1 induces tongue cancer cells' EMT and inhibits apoptosis through Wnt/β-catenin signaling pathway. J Oral Pathol Med. (2017) 46:98–105. doi: 10.1111/jop.12466
22. Zhou X, Liu S, Cai G, Kong L, Zhang T, Ren Y, et al. Long non coding RNA MALAT1 promotes tumor growth and metastasis by inducing epithelial-mesenchymal transition in oral squamous cell carcinoma. Sci Rep. (2015) 5:15972. doi: 10.1038/srep15972
23. Wu Y, Zhang L, Zhang L, Wang Y, Li H, Ren X, et al. Long non-coding RNA HOTAIR promotes tumor cell invasion and metastasis by recruiting EZH2 and repressing E-cadherin in oral squamous cell carcinoma. Int J Oncol. (2015) 46:2586–94. doi: 10.3892/ijo.2015.2976
24. Guo W, Kong L, Sun S, Wang Y, Zhao M, Zhou X, et al. Effects of long non-coding RNA HOTAIR on proliferation and apoptosis of human tongue squamous cell carcinoma in vitro and in vivo. Tianjin Med J. (2016) 44:1185–9. doi: 10.11958/20150403
25. Yang M, Xiong X, Chen L, Yang L, Li X. Identification and validation long non-coding RNAs of oral squamous cell carcinoma by bioinformatics method. Oncotarget. (2017) 8:107469–76. doi: 10.18632/oncotarget.18178
26. Piipponen M, Nissinen L, Farshchian M, Riihila P, Kivisaari A, Kallajoki M, et al. Long noncoding RNA PICSAR promotes growth of cutaneous squamous cell carcinoma by regulating ERK1/2 activity. J Invest Dermatol. (2016) 136:1701–10. doi: 10.1016/j.jid.2016.03.028
27. Piipponen M, Heino J, Kahari V-M, Nissinen L. Long non-coding RNA PICSAR decreases adhesion and promotes migration of squamous carcinoma cells by downregulating alpha 2 beta 1 and alpha 5 beta 1 integrin expression. Biol Open. (2018) 7:bio037044. doi: 10.1242/bio.037044
28. Kretz M, Siprashvili Z, Chu C, Webster DE, Zehnder A, Qu K, et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. (2013) 493:231–45. doi: 10.1038/nature11661
29. Chen Z, Liu Y, He A, Li J, Chen M, Zhan Y, et al. Theophylline controllable RNAi-based genetic switches regulate expression of lncRNA TINCR and malignant phenotypes in bladder cancer cells. Sci Rep. (2016) 6:30798. doi: 10.1038/srep30798
30. Zhou W, Zhang S, Li J, Li Z, Wang Y, Li X. lncRNA TINCR participates in ALA-PDT-induced apoptosis and autophagy in cutaneous squamous cell carcinoma. J Cell Biochem. (2019) 120:13893–902. doi: 10.1002/jcb.28662
31. Henry WS, Hendrickson DG, Beca F, Glass B, Lindahl-Allen M, He L, et al. LINC00520 is induced by Src, STAT3, and PI3K and plays a functional role in breast cancer. Oncotarget. (2016) 7:81981–94. doi: 10.18632/oncotarget.11962
32. Wu YY, Gao W, Zhang YL, Niu M, Cui JJ, Xiang CX, et al. Expression and clinical significance of long non-coding RNA LINC00520 in laryngeal squamous cell carcinoma. Lin Chung Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. (2018) 32:91–5. doi: 10.13201/j.issn.1001-1781.2018.02.003
33. Mei X-L, Zhong S. Long noncoding RNA LINC00520 prevents the progression of cutaneous squamous cell carcinoma through the inactivation of the PI3K/Akt signaling pathway by downregulating EGFR. Chin Med J. (2019) 132:454–65. doi: 10.1097/CM9.0000000000000070
34. Zhou B, Yuan W, Li X. Long intergenic noncoding RNA 319 (linc00319) promotes cell proliferation and invasion in lung cancer cells by directly downregulating the tumor suppressor MiR-32. Oncol Res. (2017). doi: 10.3727/096504017X15016337254650
35. Song P, Yin S-C. Long non-coding RNA 319 facilitates nasopharyngeal carcinoma carcinogenesis through regulation of miR-1207-5p/KLF12 axis. Gene. (2019) 680:51–8. doi: 10.1016/j.gene.2018.09.032
36. Du W, Feng Z, Sun Q. LncRNA LINC00319 accelerates ovarian cancer progression through miR-423-5p/NACC1 pathway. Biochem Biophys Res Commun. (2018) 507:198–202. doi: 10.1016/j.bbrc.2018.11.006
37. Li F, Liao J, Duan X, He Y, Liao Y. Upregulation of LINC00319 indicates a poor prognosis and promotes cell proliferation and invasion in cutaneous squamous cell carcinoma. J Cell Biochem. (2018) 119:10393–405. doi: 10.1002/jcb.27388
38. Hosono Y, Niknafs YS, Prensner JR, Iyer MK, Dhanasekaran SM, Mehra R, et al. Oncogenic role of THOR, a conserved cancer/testis long non-coding RNA. Cell. (2017) 171:1559. doi: 10.1016/j.cell.2017.11.040
39. Cheng Z, Lei Z, Yang P, Si A, Xiang D, Zhou J, et al. Long non-coding RNA THOR promotes cell proliferation and metastasis in hepatocellular carcinoma. Gene. (2018) 678:129–36. doi: 10.1016/j.gene.2018.08.035
40. Gao L, Cheng X-L, Cao H. LncRNA THOR attenuates cisplatin sensitivity of nasopharyngeal carcinoma cells via enhancing cells stemness. Biochimie. (2018) 152:63–72. doi: 10.1016/j.biochi.2018.06.015
41. Ye X-T, Huang H, Huang W-P, Hu W-L. LncRNA THOR promotes human renal cell carcinoma cell growth. Biochem Biophys Res Commun. (2018) 501:661–7. doi: 10.1016/j.bbrc.2018.05.040
42. Liu Z, Wu G, Lin C, Guo H, Xu J, Zhao T. IGF2BP1 over-expression in skin squamous cell carcinoma cells is essential for cell growth. Biochem Biophys Res Commun. (2018) 501:731–8. doi: 10.1016/j.bbrc.2018.05.057
43. Ponzio G, Rezzonico R, Bourget I, Allan R, Nottet N, Popa A, et al. A new long noncoding RNA (lncRNA) is induced in cutaneous squamous cell carcinoma and down-regulates several anticancer and cell differentiation genes in mouse. J Biol Chem. (2017) 292:12483–95. doi: 10.1074/jbc.M117.776260
44. Gutschner T, Haemmerle M, Diederichs S. MALAT1- a paradigm for long noncoding RNA function in cancer. J Mol Med. (2013) 91:791–801. doi: 10.1007/s00109-013-1028-y
45. Michalik KM, You X, Manavski Y, Doddaballapur A, Zoernig M, Braun T, et al. Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. Circul Res. (2014) 114:1389–97. doi: 10.1161/CIRCRESAHA.114.303265
46. Arun G, Spector DL. MALAT1 long non-coding RNA and breast cancer. RNA Biol. (2019) 16:860–3. doi: 10.1080/15476286.2019.1592072
47. Abbastabar M, Sarfi M, Golestani A, Khalili E. Lncrna involvement in hepatocellular carcinoma metastasis and prognosis. EXCLI J. (2018) 17:900–13. doi: 10.17179/excli2018-1541
48. Wang J, Su L, Chen X, Li P, Cai Q, Yu B, et al. MALAT1 promotes cell proliferation in gastric cancer by recruiting SF2/ASF. Biomed Pharmacother. (2014) 68:557–64. doi: 10.1016/j.biopha.2014.04.007
49. Zhu M, Zhang C, Chen D, Chen S, Zheng H. lncRNA MALAT1 potentiates the progression of tongue squamous cell carcinoma through regulating miR-140-5p-PAK1 pathway. Oncotargets Ther. (2019) 12:1365–77. doi: 10.2147/OTT.S192069
50. Zhuang M, Zhao S, Jiang Z, Wang S, Sun P, Quan J, et al. MALAT1 sponges miR-106b-5p to promote the invasion and metastasis of colorectal cancer via SLAIN2 enhanced microtubules mobility. EBiomedicine. (2019) 41:286–98. doi: 10.1016/j.ebiom.2018.12.049
51. Xi Z, Si J, Nan J. LncRNA MALAT1 potentiatesautophagy-associated cisplatin resistance by regulating the microRNA-30b/autophagy-related gene 5 axis in gastric cancer. Int J Oncol. (2019) 54:239–48. doi: 10.3892/ijo.2018.4609
52. Pang E-J, Yang R, Fu X-B, Liu Y-F. Overexpression of long non-coding RNA MALAT1 is correlated with clinical progression and unfavorable prognosis in pancreatic cancer. Tumor Biol. (2015) 36:2403–7. doi: 10.1007/s13277-014-2850-8
53. Chen Y, Huang W, Sun W, Zheng B, Wang C, Luo Z, et al. LncRNA MALAT1 promotes cancer metastasis in osteosarcoma via activation of the PI3K-Akt signaling pathway. Cell Physiol Biochem. (2018) 51:1313–26. doi: 10.1159/000495550
54. Zhang Y, Gao L, Ma S, Ma J, Wang Y, Li S, et al. MALAT1-KTN1-EGFR regulatory axis promotes the development of cutaneous squamous cell carcinoma. Cell Death Different. (2019) 26:2061–73. doi: 10.1038/s41418-019-0288-7
55. Li SS, Zhou L, Gao L, Wang YH, Ding ZH. Role of long noncoding RNA MALAT1 promotes the occurrence and progression of cutaneous squamous cell carcinoma. Nan Fang Yi Ke Da Xue Xue Bao. (2018) 38:421–7. doi: 10.3969/j.issn.1673-4254.2018.04.09
56. Chen L, Chen Q, Kuang S, Zhao C, Yang L, Zhang Y, et al. USF1-induced upregulation of LINC01048 promotes cell proliferation and apoptosis in cutaneous squamous cell carcinoma by binding to TAF15 to transcriptionally activate YAP1. Cell Death Dis. (2019) 10:296. doi: 10.1038/s41419-019-1516-2
57. Qiao H-P, Gao W-S, Huo J-X, Yang Z-S. Long non-coding RNA GAS5 functions as a tumor suppressor in renal cell carcinoma. Asian Pacific J Cancer Prev. (2013) 14:1077–82. doi: 10.7314/APJCP.2013.14.2.1077
58. Zhang Z, Zhu Z, Watabe K, Zhang X, Bai C, Xu M, et al. Negative regulation of lncRNA GAS5 by miR-21. Cell Death Different. (2013) 20:1558–68. doi: 10.1038/cdd.2013.110
59. Pickard MR, Mourtada-Maarabouni M, Williams GT. Long non-coding RNA GAS5 regulates apoptosis in prostate cancer cell lines. Biochim Biophys Acta Mol Basis Dis. (2013) 1832:1613–23. doi: 10.1016/j.bbadis.2013.05.005
60. Pickard MR, Williams GT. Regulation of apoptosis by long non-coding RNA GAS5 in breast cancer cells: implications for chemotherapy. Breast Cancer Res Treat. (2014) 145:359–70. doi: 10.1007/s10549-014-2974-y
61. Shi X, Sun M, Liu H, Yao Y, Kong R, Chen F, et al. A critical role for the long non-coding RNA GAS5 in proliferation and apoptosis in non-small-cell lung cancer. Mol Carcinog. (2015) 54:E1–12. doi: 10.1002/mc.22120
62. Cao Q, Wang N, Qi J, Gu Z, Shen H. Long non-coding RNA-GAS5 acts as a tumor suppressor in bladder transitional cell carcinoma via regulation of chemokine (C-C motif) ligand 1 expression. Mol Med Rep. (2016) 13:27–34. doi: 10.3892/mmr.2015.4503
63. Wang T-H, Chan C-W, Fang J-Y, Shih Y-M, Liu Y-W, Wang T-CV, et al. 2-O-Methylmagnolol upregulates the long non-coding RNA, GAS5, and enhances apoptosis in skin cancer cells. Cell Death Dis. (2017) 8: E1–12. doi: 10.1038/cddis.2017.66
64. Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xiao X, Brugmann SA, et al. Functional Demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. (2007) 129:1311–23. doi: 10.1016/j.cell.2007.05.022
65. Oulion S, Debiais-Thibaud M, D'Aubenton-Carafa Y, Thermes C, Silva CD, Bernard-Samain S, et al. Evolution of hox gene clusters in gnathostomes: insights from a survey of a shark (Scyliorhinus canicula) transcriptome. Mol Biol Evolut. (2010) 27:2829–38. doi: 10.1093/molbev/msq172
66. He S, Liu S, Zhu H. The sequence, structure and evolutionary features of HOTAIR in mammals. BMC Evolut Biol. (2011) 11:102. doi: 10.1186/1471-2148-11-102
67. Sun G, Wang Y, Zhang J, Lin N, You Y. MiR-15b/HOTAIR/p53 form a regulatory loop that affects the growth of glioma cells. J Cell Biochem. (2018) 119:4540–7. doi: 10.1002/jcb.26591
68. Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. (2010) 464:1071–6. doi: 10.1038/nature08975
69. Kai W. Epigenetic disruption of the WNT/β-catenin signaling pathway in human cancers. Epigenetics. (2009) 4:307–12. doi: 10.4161/epi.4.5.9371
70. Dandan L, Jiapeng F, Tianyi W, Yandong W, Yanan S, Jingyuan R, et al. Long intergenic noncoding RNA HOTAIR is overexpressed and regulates PTEN methylation in laryngeal squamous cell carcinoma. Am J Pathol. (2013) 182:64–70. doi: 10.1016/j.ajpath.2012.08.042
71. Tang L, Zhang W, Su B, Yu B. Long noncoding RNA HOTAIR is associated with motility, invasion, and metastatic potential of metastatic melanoma. BioMed Res Int. (2013) 2013:251098. doi: 10.1155/2013/251098
72. Liu G, Zhang W, Liu G, Zhang W. Long non-coding RNA HOTAIR promotes UVB-induced apoptosis and inflammatory injury by up-regulation of PKR in keratinocytes. Brazil J Med Biol Res. (2018) 51:e6896. doi: 10.1590/1414-431x20186896
73. Tang H, Wu Z, Zhang J, Su B. Salivary lncRNA as a potential marker for oral squamous cell carcinoma diagnosis. Mol Med Rep. (2013) 7:761–6. doi: 10.3892/mmr.2012.1254
Keywords: cutaneous squamous cell carcinoma, lncRNA, ERK1/2, skin, cancer
Citation: Wang Y, Sun B, Wen X, Hao D, Du D, He G and Jiang X (2020) The Roles of lncRNA in Cutaneous Squamous Cell Carcinoma. Front. Oncol. 10:158. doi: 10.3389/fonc.2020.00158
Received: 16 October 2019; Accepted: 29 January 2020;
Published: 28 February 2020.
Edited by:
Alfons Navarro, University of Barcelona, SpainReviewed by:
Fengbiao Mao, University of Michigan, United StatesAbdullah Al Emran, Centenary Institute Australia, Australia
Copyright © 2020 Wang, Sun, Wen, Hao, Du, He and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gu He, aGVndUBzY3UuZWR1LmNu; Xian Jiang, amVubnl4aWFuakAxNjMuY29t
†These authors have contributed equally to this work and share first authorship
 Yujia Wang
Yujia Wang Bensen Sun
Bensen Sun Xiang Wen
Xiang Wen Dan Hao
Dan Hao Dan Du
Dan Du Gu He
Gu He Xian Jiang
Xian Jiang