- 1Molecular Pharmacology Program and Department of Medicine, Memorial Sloan Kettering Cancer Center, New York, NY, United States
- 2Department of Respiratory Medicine, Nagasaki University Graduate School of Biomedical Sciences, Nagasaki, Japan
- 3Department of Medicine, Weill Cornell Medical College, New York, NY, United States
Small cell lung cancer (SCLC) is an aggressive malignancy characterized by rapid growth, early metastasis, and acquired therapeutic resistance. A majority of patients with SCLC have extensive-stage (ES) disease, defined as the presence of metastatic disease outside the hemithorax at first diagnosis. SCLC has been considered “a graveyard for drug development,” with chemotherapy remaining the standard treatment for first- and second-line management until quite recently. In contrast to NSCLC, identifying therapeutic targets in SCLC has been challenging, partly because driver mutations are primarily loss of function, involving the tumor suppressor genes RB1 and TP53 or currently untargetable (e.g., amplification of MYC family members). Recent gene expression profiling of SCLC cells lines, patient samples and representative murine models, have led to a proposed delineation of four major subtypes for SCLC distinguished by differential expression of four key transcriptional regulators (ASCL1, NEUROD1, POU2F3, and YAP1). Our understanding of the biology of SCLC has indeed significantly improved recently due to the continued efforts of the dedicated investigators in this field, but the therapeutic options remain dismal. While recent results from immunotherapy trials are encouraging, most patients demonstrate either primary or rapid acquired resistance to current regimens, highlighting the clear need to improve the effectiveness and expand the scope of current therapeutic strategies. In this opinion article, we will discuss recent developments in the treatment of SCLC, focused on current understanding of the signaling pathways, the role of immunotherapy and targeted therapy, and emerging biomarkers of response to therapy in SCLC.
Introduction
Small cell lung cancer (SCLC) is an aggressive malignancy characterized by rapid growth, early metastasis, and acquired therapeutic resistance (1–3). A majority of patients with SCLC have extensive-stage (ES) disease, defined as the presence of metastatic disease outside the hemithorax at first diagnosis. Although the clinical treatment for non-small cell lung cancer (NSCLC) has changed dramatically and rapidly, SCLC has been considered “a graveyard for drug development,” with chemotherapy remaining the standard treatment for first- and second-line management for over four decades until quite recently. In contrast to NSCLC, identifying therapeutic targets in SCLC has been challenging, partly because driver mutations are primarily loss of function, involving the tumor suppressor genes RB1 and TP53 (4, 5) or currently untargetable (e.g., amplification of MYC family members). Recent gene expression profiling of SCLC cells lines, patient samples and representative murine models, have led to a proposed delineation of four major subtypes for SCLC distinguished by differential expression of four key transcriptional regulators (ASCL1, NEUROD1, POU2F3, and YAP1). Our understanding of the biology of SCLC has indeed significantly improved recently due to the continued efforts of the dedicated investigators in this field, but the therapeutic options remain dismal. While recent results from immunotherapy trials are encouraging, most patients demonstrate either primary or rapid acquired resistance to current regimens, highlighting the clear need to improve the effectiveness and expand the scope of current therapeutic strategies. In this opinion article, we will discuss recent developments in the treatment of SCLC, focused on current understanding of the signaling pathways, the role of immunotherapy and targeted therapy, and emerging biomarkers of response to therapy in SCLC (Figure 1).
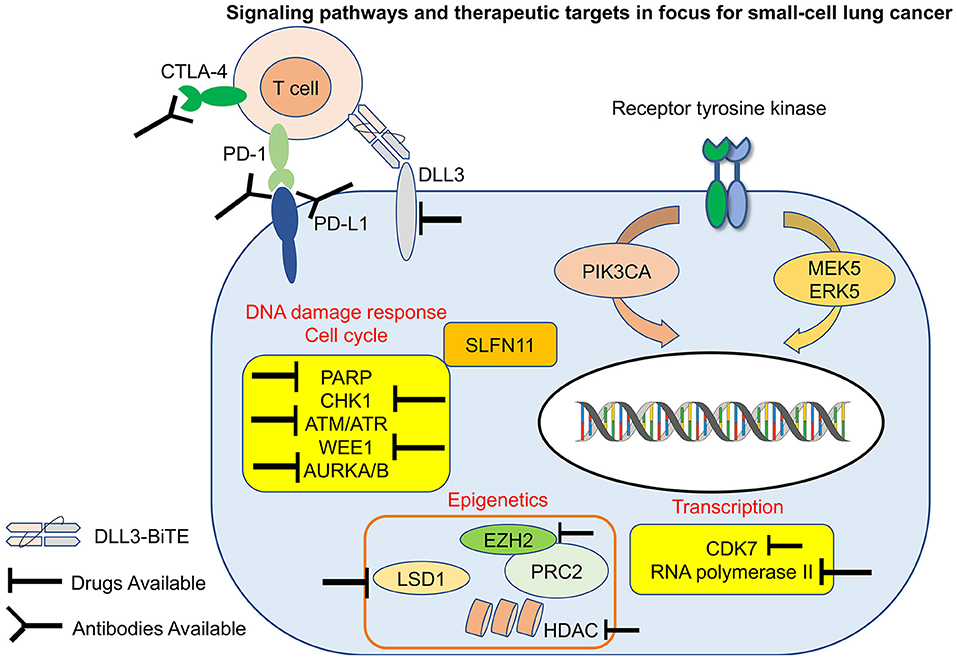
Figure 1. Signaling pathways and therapeutic targets in focus for small-cell lung cancer (SCLC). Notable targets and evolving treatment strategies in SCLC including immunotherapy, targeted therapy, antibody drug conjugates. PD-1, programmed death-1; PD-L1, programmed death ligand-1; CTLA-4, cytotoxic T lymphocyte associated protein 4; DLL3, delta-like 3; PIK3CA, phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha; AURKA/B, aurora kinase A/B; CHK1, checkpoint kinase 1; PARP1, poly-ADP ribose polymerase 1; EZH2, enhancer of zeste 2; LSD1, lysine-specific demethylase 1A; HDAC, histone deacetylase; ATR, ataxia telangiectasia and RAD3-related protein; ATM, ataxia telangiectasia mutated; PRC2, polycomb repressor complex 2; CDK7, cyclin-dependent kinase 7; SLFN11, schlafen11.
New Therapeutic Targets for SCLC
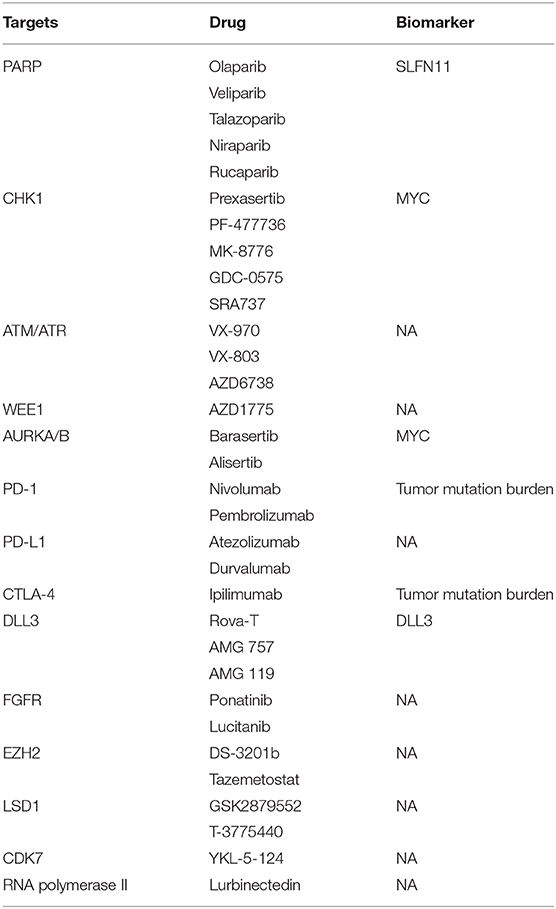
SCLC tumors typically carry a high mutation burden and have evident genomic instability manifest by aneuploidy and multiple intra- and inter-chromosomal rearrangements. Almost all SCLC tumors have functional inactivation of both TP53 and RB1; however, attempts to target these genomic alterations have resoundingly failed. Recent studies using comprehensive whole exome and whole genome sequencing in SCLC have revealed that SCLC tumors include other recurrent genomic alternations (4–6). Visualization of the SCLC genomic landscape has led to the identification of new targets such as PTEN loss (7), activating PI3K mutations (8, 9), and FGFR1 amplifications (10, 11). The novel therapeutic targets, corresponding drugs and the predictive biomarkers were summarized in Table 1.
DNA Damage Repair Pathway and Cell Cycle
The high mutation burden of SCLC is largely attributable to the strong association of this disease with heavy tobacco exposure, with only 2% of cases occurring in never smokers (12–14). The loss of cell cycle checkpoint controls due to inactivation of RB1 and TP53 may increase susceptibility of SCLC to DNA damage. Indeed, multiple reports in the past few years have convincingly pointed to DNA damage response (DDR) pathways as critical vulnerabilities in SCLC. Targeting central DDR mediators, such as poly ADP-ribose polymerase (PARP), checkpoint kinase 1 (CHK1), Ataxia telangiectasia and RAD3-related protein (ATR), Ataxia telangiectasia mutated (ATM), and WEE1, have demonstrated promising therapeutic opportunities in SCLC.
The anti-tumor activities of PARP inhibitors occur through multiple mechanisms, including (1) trapping the enzyme to the single-strand DNA breaks (SSBs) by preventing the utilization of nicotinamide adenine dinucleotide (NAD), and (2) inhibiting poly ADP-ribosylation (PARylation) and binding of PARP to DNA (15). The PARP inhibitor AZD2281 was found to have greater against SCLC cell lines than NSCLCs (16). PARP inhibitors with PARP trapping activity sensitized SCLC cell lines and patient-derived xenografts to ionizing radiation (17). A phase 1 trial demonstrated initial promising activity of the potent PARP trapping drug talazoparib, including in patients with SCLC (18).
SCLC cell lines have a higher median CHK1 protein and gene expression than NSCLC lines, and the CHK1 inhibitor prexasertib demonstrated strong anti-tumor activity in SCLC cell lines, SCLC syngeneic, genetically-engineered mouse (GEM) and chemo-resistant models (19). The effectiveness of targeting CHK1/ATR axis in SCLC was later confirmed in an independent preclinical study with ATR inhibitors in particular demonstrating activity against SCLC in both in vitro and in vivo models (20). Activation of ATR through DNA damage stimulates multiple downstream targets including CHK1, which halts cell cycle progression at the G2-M phase (21, 22). The G2/M checkpoint regulator WEE1 is also upregulated in SCLC cell lines relative to normal lung tissue or NSCLCs, and the WEE1 inhibitor AZD1775 showed activity in several SCLC cell lines (23).
Inhibition of Aurora kinase A or B inhibits the proliferation, growth of SCLC in vitro and in vivo (24, 25). A recently reported clinical trial demonstrated that the aurora kinase A inhibitor alisertib plus paclitaxel had significantly improved PFS vs. paclitaxel alone in patients with cMYC positive SCLC (26).
Finally, several preclinical and clinical trials have demonstrated that combining DDR inhibitors with chemotherapy or other targeted agents could be a promising strategy (16, 23, 27–31).
Targeting Epigenetic Modifiers in SCLC
Visualizing the human epigenome using next generation sequencing highlighted the role of epigenetic processes in cancer generally, and SCLC in particular (32–34). Here we focus on two of the most promising epigenetic regulatory proteins; enhancer of zeste homology 2 (EZH2) and lysine-specific demethylase 1A (LSD1), both of which are now being tested in current and upcoming SCLC clinical trials.
EZH2 is one of the enzymatic histone-lysine N-methyltransferase subunits of polycomb repressor complex 2 (PRC2), which primarily inhibits gene expression by promoting tri-methylation of Histone 3 on lysine at position 27. EZH2 expression is higher in SCLC than in any tumor type included in the Cancer Genome Atlas (34), and preclinical analysis showed that an EZH2 inhibitor augmented chemotherapeutic efficacy and could prevent emergence of acquired chemotherapy resistance in multiple in vivo SCLC patient-derived xenograft models (35). A phase I/II study to test this strategy in clinic has been launched, using the EZH1/2 inhibitor DS-3201b together with irinotecan in patients with recurrent SCLC (NCT03879798). Further raising interest in EZH2 as a target in SCLC, it has been recently demonstrated that PRC2 transcriptionally suppresses MHC class I expression in SCLC, suggesting that EZH2 inhibition may also augment SCLC response to immune check point inhibitors (36).
LSD1, a monoamine oxidase that demethylates mono- or di-methylated lysine 4 or lysine 9 of histone H3, has been implicated in oncogenesis and depending on context can either activate or repress gene transcription (37). LSD1 inhibitors has been shown to exert anticancer effects against SCLC in vitro and in vivo through inhibition of the interaction between LSD1 and SNAG domain proteins; insulinoma-associated protein 1 (INSM1) or Growth factor independence 1B (38, 39).
Immunotherapy Regimens and Combinations for SCLC
Immunotherapy using checkpoint inhibitory monoclonal antibodies blocking programmed cell death 1 (PD-1), programmed death-ligand 1 (PD-L1), and cytotoxic T-lymphocyte-associated protein 4 (CTLA-4), either as single agents or in combination, have led to a revolution in the treatment of several solid tumors, including NSCLC. Inhibition of these immune checkpoint molecules can prompt reactivation of cytotoxic T cell immunity that had been held in check, in some cases resulting in durable anticancer responses even in patients with advanced disease.
Recently, the United States Food and Drug Administration approved the PD-1 inhibitors, nivolumab and pembrolizumab, as the third line treatment options for SCLC patients. The phase 1/2 CheckMate 032 trial, which explored the efficacy of nivolumab alone or in combination with two different doses of ipilimumab, and the phase 1b KEYNOTE-028 and phase 2 KEYNOTE-158 trials, which examined the efficacy of pembrolizumab for pretreated patients with SCLC with PD-L1 positive tumors, demonstrated efficacy in previously treated SCLC patients (40–42). Unfortunately, while response rates on these trials ranged from 11 to 33%, most patients treated did not appear to benefit, as evidenced by median progression-free survivals of only 1.4–2 months. A small number of patients, <10%, demonstrate long-term responses on these trials, prompting intensive and ongoing investigation into biomarkers that might discriminate these patients, and exploration of combination therapies that might increase the fraction of patients with durable benefit.
A pair of recent landmark studies in SCLC have explored the efficacy of PD-L1 inhibitors combined with cytotoxic agents for newly diagnosed extensive stage patients. In the first line setting, the PD-L1 inhibitor atezolizumab combined with carboplatin plus etoposide was approved by the FDA based on the results of the IMPOWER133 study (43). Similarly, an initial report from the CASPIAN trial demonstrated that first-line durvalumab plus platinum–etoposide also significantly improved OS in patients with ES-SCLC (44). These studies established the benefit of adding a PD-L1 inhibitor to platinum plus etoposide and confirmed the promise of immune check point inhibitors for the treatment of patients with SCLC. Notably a third trial, KEYNOTE-604, assessing the addition of pembrolizumab to first line carboplatin and etoposide, demonstrated a similar improvement in PFS to the studies of the PD-L1 inhibitors above, but narrowly missed statistical significance for an OS benefit; detailed results of this study have not yet been presented or published.
Building on these initial important but limited successes, ongoing preclinical and clinical studies are now exploring combined therapies with PD-1 or PD-L1 inhibitors and compounds which target epigenetic modifiers, DDR proteins, or cell cycle regulators. Some of these have already shown promising results for SCLC in preclinical studies. As noted above, treatment with an EZH2 inhibitor can upregulate MHC class I expression, promoting better antigenic presentation by tumor cells and significant tumor suppression when combined with immune checkpoint inhibitors (36). Furthermore, the combination of a CHK1 inhibitor and low dose gemcitabine enhanced the effect of PD-L1 inhibition (45), and concomitant treatment with a DDR inhibitor remarkably potentiated the antitumor effects of PD-L1 inhibition in mouse models of SCLC (46). Treatment with a cyclin-dependent kinase 7 inhibitor, YKL-5-124, was also found to enhance anti-tumor efficacy of a PD-1 inhibitor in SCLC preclinical models (47). Several clinical trials using immune checkpoint inhibitor combinations are ongoing (48).
Other Emerging Targets in SCLC
Genomic profiling of SCLC has demonstrated common inactivating mutations in the primary Notch family members (4) and aberrant overexpression of a key negative regulator of Notch signaling, delta-like protein 3 (DLL3) was found in the majority of SCLC tumors (49). Initial clinical evaluation of an anti-DLL3 antibody-drug conjugate rovalpituzumab teserine (Rova-T) demonstrated promising activity, although the ultimate utility of this agent was compromised by toxicities (50, 51). DLL3 remains a target of substantial interest for drug development in SCLC, with active strategies including an anti-DLL3/anti-CD3 bispecific T-cell engager (NCT03319940).
The association of SCLC with selective activation of master transcriptional regulators has raised interest in strategies focused on modulation of transcriptional control. Lurbinectidin, a DNA binding agent that appears to function as a selective inhibitor of RNA polymerase II transcription, has demonstrated substantial activity against SCLC both as a single agent and in combination with doxorubicin (52–54).
Other targets of recently emerging interest include selective metabolomic dependencies of SCLC. MEK5 and ERK5 have been recently identified as critical regulators of lipid metabolism of SCLC cells, suggesting these kinases as possible therapeutic targets (55). MYC-driven SCLC cells have been reported to be highly dependent on arginine-regulated pathways including polyamine biosynthesis and mTOR pathway activation; selective arginine depletion appeared to be highly effective in MYC-driven SCLC preclinical models (56).
Biomarkers in SCLC
Biomarkers of Targeted Therapy
Biomarkers predictive of response to the therapy are urgently needed to guide treatment selection for patients with SCLC. Studies from multiple groups have suggested that Schlafen11 (SLFN11) expression is a potential biomarker of sensitivity of both DNA damaging chemotherapy and PARP inhibition (29, 35, 57, 58). Genetic alteration of MYC, most commonly gene amplification, was observed in approximately 20% of SCLC, placing it among the most common genetic abnormalities after TP53 and RB1. Higher expression or amplification of MYC predicted sensitivity to CHK1 inhibition in SCLC (19, 59, 60) and Aurora Kinase inhibition (61). High expression of DLL3 on cell surface of SCLC was associated with better response to Rova-T in both preclinical and clinical studies (49–51).
Biomarkers of Immunotherapy
Expression of PD-L1 on tumor cells has been shown to be an enrichment factor for efficacy of PD-1 inhibition in many solid tumors including NSCLC (62, 63). The predictive value of PD-L1 expression for the treatment of SCLC with PD-1 inhibitors is currently unknown. Challenging the utility of PD-L1 expression as a useful biomarker in SCLC is a lower prevalence, heterogeneous expression on SCLC tumors, and the lack of clear correlation between PD-L1 expression and the effect of immunotherapy (40, 41). In contrast to the expression of PD-L1, a detailed retrospective biomarker analysis of patients enrolled in the CheckMate 032 trial suggested improved ORR, OS, and PFS of nivolumab monotherapy or nivolumab plus ipilimumab combination therapy in patients with a high tumor mutation burden (TMB) relative to patients with a low/medium TMB (64).
The presence of a high number of tumor-infiltrating lymphocytes (TILs), mismatch repair (MMR) deficiency, or a high frequency of microsatellite instability (MSI) also predicted improved response to immune checkpoint inhibitors in other types of cancers (65–69). The recent phase II clinical trial, KEYNOTE-158, demonstrated the clinical benefit of therapy with pembrolizumab among patients with previously treated unresectable or metastatic, high MSI, DNA MMR deficient, non-colorectal cancer (70); with only four patients with SCLC enrolled in this study, further investigation will be required.
Future Strategies for SCLC
As in other solid tumors, therapies directed toward induction of anti-tumor immunity are likely to play an increasingly important role in the treatment of SCLC. Immune checkpoint inhibitors have shown modest yet promising effects when combined with platinum plus etoposide for patients with ES SCLC. However, continued efforts need to be made to achieve more durable and more broadly effective immunotherapy responses in SCLC. Future research efforts should seek to identify not only effective combinatorial regimens, but also predictive markers of immunotherapy in SCLC. Another area of current intensive investigation is the analysis of SCLC subtype-specific therapeutic vulnerabilities and predictive biomarkers associated with particular treatment outcomes for each of the four major subtypes. Recent insights into the biology of SCLC have promoted the development of molecular targeted and immunologic strategies for what has been a particularly refractory disease. Future research and improved clinical prospects for patients with SCLC will depend on continued focus on an integrated platform of basic discovery and clinical translational research, identifying novel biomarker-driven approaches to integrate immunotherapy and other targeted therapies.
Author Contributions
HT, TS, and CR contributed to write the manuscript.
Funding
This work was supported by Parker Institute of Cancer Immunotherapy grant (TS) and NCI U24 CA213274 and NCI R01 CA197936 (CR).
Conflict of Interest
CR has previously consulted for AbbVie, Amgen, Ascentage, Astra Zeneca, Celgene, Daiichi Sankyo, Genentech/Roche, Ipsen, Loxo, PharmaMar, Harpoon Therapeutics, and Bridge Medicines.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Rudin CM, Poirier JT. Small-cell lung cancer in 2016: shining light on novel targets and therapies. Nat Rev Clin Oncol. (2017) 14:75–6. doi: 10.1038/nrclinonc.2016.203
2. Sen T, Gay CM, Byers LA. Targeting DNA damage repair in small cell lung cancer and the biomarker landscape. Transl Lung Cancer Res. (2018) 7:50–68. doi: 10.21037/tlcr.2018.02.03
3. Gazdar AF, Bunn PA, Minna JD. Small-cell lung cancer: what we know, what we need to know and the path forward. Nat Rev Cancer. (2017) 17:725–37. doi: 10.1038/nrc.2017.87
4. George J, Lim JS, Jang SJ, Cun Y, Ozretic L, Kong G, et al. Comprehensive genomic profiles of small cell lung cancer. Nature. (2015) 524:47–53. doi: 10.1038/nature14664
5. Rudin CM, Durinck S, Stawiski EW, Poirier JT, Modrusan Z, Shames DS, et al. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat Genet. (2012) 44:1111–6. doi: 10.1038/ng.2405
6. Peifer M, Fernandez-Cuesta L, Sos ML, George J, Seidel D, Kasper LH, et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet. (2012) 44:1104–10. doi: 10.1038/ng.2396
7. Yokomizo A, Tindall DJ, Drabkin H, Gemmill R, Franklin W, Yang P, et al. PTEN/MMAC1 mutations identified in small cell, but not in non-small cell lung cancers. Oncogene. (1998) 17:475–9. doi: 10.1038/sj.onc.1201956
8. Shibata T, Kokubu A, Tsuta K, Hirohashi S. Oncogenic mutation of PIK3CA in small cell lung carcinoma: a potential therapeutic target pathway for chemotherapy-resistant lung cancer. Cancer Lett. (2009) 283:203–11. doi: 10.1016/j.canlet.2009.03.038
9. Umemura S, Mimaki S, Makinoshima H, Tada S, Ishii G, Ohmatsu H, et al. Therapeutic priority of the PI3K/AKT/mTOR pathway in small cell lung cancers as revealed by a comprehensive genomic analysis. J Thorac Oncol. (2014) 9:1324–31. doi: 10.1097/jto.0000000000000250
10. Schultheis AM, Bos M, Schmitz K, Wilsberg L, Binot E, Wolf J, et al. Fibroblast growth factor receptor 1 (FGFR1) amplification is a potential therapeutic target in small-cell lung cancer. Mod Pathol. (2014) 27:214–21. doi: 10.1038/modpathol.2013.141
11. Park JS, Lee JS, Kim EY, Jung JY, Kim SK, Chang J, et al. The frequency and impact of FGFR1 amplification on clinical outcomes in Korean patients with small cell lung cancer. Lung Cancer. (2015) 88:325–31. doi: 10.1016/j.lungcan.2015.03.002
12. Varghese AM, Zakowski MF, Yu HA, Won HH, Riely GJ, Krug LM, et al. Small-cell lung cancers in patients who never smoked cigarettes. J Thorac Oncol. (2014) 9:892–6. doi: 10.1097/JTO.0000000000000142
13. Pesch B, Kendzia B, Gustavsson P, Jockel KH, Johnen G, Pohlabeln H, et al. Cigarette smoking and lung cancer–relative risk estimates for the major histological types from a pooled analysis of case-control studies. Int J Cancer. (2012) 131:1210–9. doi: 10.1002/ijc.27339
14. Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature. (2013) 500:415–21. doi: 10.1038/nature12477
15. Murai J, Huang SY, Das BB, Renaud A, Zhang Y, Doroshow JH, et al. Trapping of PARP1 and PARP2 by clinical PARP inhibitors. Cancer Res. (2012) 72:5588–99. doi: 10.1158/0008-5472.Can-12-2753
16. Byers LA, Wang J, Nilsson MB, Fujimoto J, Saintigny P, Yordy J, et al. Proteomic profiling identifies dysregulated pathways in small cell lung cancer and novel therapeutic targets including PARP1. Cancer Discov. (2012) 2:798–811. doi: 10.1158/2159-8290.Cd-12-0112
17. Laird JH, Lok BH, Ma J, Bell A, de Stanchina E, Poirier JT, et al. Talazoparib is a potent radiosensitizer in small cell lung cancer cell lines and xenografts. Clin Cancer Res. (2018) 24:5143–52. doi: 10.1158/1078-0432.Ccr-18-0401
18. de Bono J, Ramanathan RK, Mina L, Chugh R, Glaspy J, Rafii S, et al. Phase I, dose-escalation, two-part trial of the PARP inhibitor talazoparib in patients with advanced germline BRCA1/2 mutations and selected sporadic cancers. Cancer Discov. (2017) 7:620–9. doi: 10.1158/2159-8290.Cd-16-1250
19. Sen T, Tong P, Stewart CA, Cristea S, Valliani A, Shames DS, et al. CHK1 inhibition in small-cell lung cancer produces single-agent activity in biomarker-defined disease subsets and combination activity with cisplatin or olaparib. Cancer Res. (2017) 77:3870–84. doi: 10.1158/0008-5472.Can-16-3409
20. Doerr F, George J, Schmitt A, Beleggia F, Rehkamper T, Hermann S, et al. Targeting a non-oncogene addiction to the ATR/CHK1 axis for the treatment of small cell lung cancer. Sci Rep. (2017) 7:15511. doi: 10.1038/s41598-017-15840-5
21. Rundle S, Bradbury A, Drew Y, Curtin NJ. Targeting the ATR-CHK1 axis in cancer therapy. Cancers. (2017) 9:E41. doi: 10.3390/cancers9050041
22. Smith J, Tho LM, Xu N, Gillespie DA. The ATM-Chk2 and ATR-Chk1 pathways in DNA damage signaling and cancer. Adv Cancer Res. (2010) 108:73–112. doi: 10.1016/b978-0-12-380888-2.00003-0
23. Sen T, Tong P, Diao L, Li L, Fan Y, Hoff J, et al. Targeting AXL and mTOR pathway overcomes primary and acquired resistance to WEE1 inhibition in small-cell lung cancer. Clin Cancer Res. (2017) 23:6239–53. doi: 10.1158/1078-0432.Ccr-17-1284
24. Lu Y, Liu Y, Jiang J, Xi Z, Zhong N, Shi S, et al. Knocking down the expression of Aurora-A gene inhibits cell proliferation and induces G2/M phase arrest in human small cell lung cancer cells. Oncol Rep. (2014) 32:243–9. doi: 10.3892/or.2014.3194
25. Helfrich BA, Kim J, Gao D, Chan DC, Zhang Z, Tan AC, et al. Barasertib (AZD1152), a small molecule aurora B inhibitor, inhibits the growth of SCLC cell lines in vitro and in vivo. Mol Cancer Ther. (2016) 15:2314–22. doi: 10.1158/1535-7163.Mct-16-0298
26. Owonikoko TK, Niu H, Nackaerts K, Csoszi T, Ostoros G, Mark Z, et al. Randomized phase II study of paclitaxel plus alisertib versus paclitaxel plus placebo as second-line therapy for small-cell lung cancer: primary and correlative biomarker analyses. J Thorac Oncol. (2019) 15:274–87. doi: 10.1016/j.jtho.2019.10.013
27. Pietanza MC, Kadota K, Huberman K, Sima CS, Fiore JJ, Sumner DK, et al. Phase II trial of temozolomide in patients with relapsed sensitive or refractory small cell lung cancer, with assessment of methylguanine-DNA methyltransferase as a potential biomarker. Clin Cancer Res. (2012) 18:1138–45. doi: 10.1158/1078-0432.Ccr-11-2059
28. Owonikoko TK, Zhang G, Deng X, Rossi MR, Switchenko JM, Doho GH, et al. Poly (ADP) ribose polymerase enzyme inhibitor, veliparib, potentiates chemotherapy and radiation in vitro and in vivo in small cell lung cancer. Cancer Med. (2014) 3:1579–94. doi: 10.1002/cam4.317
29. Lok BH, Gardner EE, Schneeberger VE, Ni A, Desmeules P, Rekhtman N, et al. PARP Inhibitor activity correlates with SLFN11 expression and demonstrates synergy with temozolomide in small cell lung cancer. Clin Cancer Res. (2017) 23:523–35. doi: 10.1158/1078-0432.Ccr-16-1040
30. Owonikoko TK, Dahlberg SE, Sica GL, Wagner LI, Wade JL III, Srkalovic G, et al. Randomized phase II trial of cisplatin and etoposide in combination with veliparib or placebo for extensive-stage small-cell lung cancer: ECOG-ACRIN 2511 study. J Clin Oncol. (2019) 37:222–9. doi: 10.1200/jco.18.00264
31. Cardnell RJ, Feng Y, Mukherjee S, Diao L, Tong P, Stewart CA, et al. Activation of the PI3K/mTOR pathway following PARP inhibition in small cell lung cancer. PLoS ONE. (2016) 11:e0152584. doi: 10.1371/journal.pone.0152584
32. Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer. (2011) 11:726–34. doi: 10.1038/nrc3130
33. Kalari S, Jung M, Kernstine KH, Takahashi T, Pfeifer GP. The DNA methylation landscape of small cell lung cancer suggests a differentiation defect of neuroendocrine cells. Oncogene. (2013) 32:3559–68. doi: 10.1038/onc.2012.362
34. Poirier JT, Gardner EE, Connis N, Moreira AL, de Stanchina E, Hann CL, et al. DNA methylation in small cell lung cancer defines distinct disease subtypes and correlates with high expression of EZH2. Oncogene. (2015) 34:5869–78. doi: 10.1038/onc.2015.38
35. Gardner EE, Lok BH, Schneeberger VE, Desmeules P, Miles LA, Arnold PK, et al. Chemosensitive relapse in small cell lung cancer proceeds through an EZH2-SLFN11 axis. Cancer Cell. (2017) 31:286–99. doi: 10.1016/j.ccell.2017.01.006
36. Burr ML, Sparbier CE, Chan KL, Chan YC, Kersbergen A, Lam EYN, et al. An Evolutionarily conserved function of polycomb silences the mhc class i antigen presentation pathway and enables immune evasion in cancer. Cancer Cell. (2019) 36:385–401.e8. doi: 10.1016/j.ccell.2019.08.008
37. Klose RJ, Zhang Y. Regulation of histone methylation by demethylimination and demethylation. Nat Rev Mol Cell Biol. (2007) 8:307–18. doi: 10.1038/nrm2143
38. Mohammad HP, Smitheman KN, Kamat CD, Soong D, Federowicz KE, Van Aller GS, et al. A DNA hypomethylation signature predicts antitumor activity of LSD1 inhibitors in SCLC. Cancer Cell. (2015) 28:57–69. doi: 10.1016/j.ccell.2015.06.002
39. Takagi S, Ishikawa Y, Mizutani A, Iwasaki S, Matsumoto S, Kamada Y, et al. LSD1 Inhibitor T-3775440 inhibits SCLC cell proliferation by disrupting LSD1 interactions with SNAG domain proteins INSM1 and GFI1B. Cancer Res. (2017) 77:4652–62. doi: 10.1158/0008-5472.CAN-16-3502
40. Antonia SJ, Lopez-Martin JA, Bendell J, Ott PA, Taylor M, Eder JP, et al. Nivolumab alone and nivolumab plus ipilimumab in recurrent small-cell lung cancer (CheckMate 032): a multicentre, open-label, phase 1/2 trial. Lancet Oncol. (2016) 17:883–95. doi: 10.1016/s1470-2045(16)30098-5
41. Ott PA, Elez E, Hiret S, Kim DW, Morosky A, Saraf S, et al. Pembrolizumab in patients with extensive-stage small-cell lung cancer: results from the phase Ib KEYNOTE-028 study. J Clin Oncol. (2017) 35:3823–9. doi: 10.1200/jco.2017.72.5069
42. Chung HC, Lopez-Martin JA, Kao SC-H, Miller WH, Ros W, Gao B, et al. Phase 2 study of pembrolizumab in advanced small-cell lung cancer (SCLC): KEYNOTE-158. J Clin Oncol. (2018) 36:8506. doi: 10.1200/JCO.2018.36.15_suppl.8506
43. Horn L, Mansfield AS, Szczesna A, Havel L, Krzakowski M, Hochmair MJ, et al. First-line atezolizumab plus chemotherapy in extensive-stage small-cell lung cancer. N Engl J Med. (2018) 379:2220–9. doi: 10.1056/NEJMoa1809064
44. Paz-Ares L, Dvorkin M, Chen Y, Reinmuth N, Hotta K, Trukhin D, et al. Durvalumab plus platinum–etoposide versus platinum–etoposide in first-line treatment of extensive-stage small-cell lung cancer (CASPIAN): a randomised, controlled, open-label, phase 3 trial. Lancet. (2019) 394:1929–39. doi: 10.1016/s0140-6736(19)32222-6
45. Sen T, Della Corte CM, Milutinovic S, Cardnell RJ, Diao L, Ramkumar K, et al. Combination treatment of the oral CHK1 inhibitor, SRA737, and low-dose gemcitabine enhances the effect of programmed death ligand 1 blockade by modulating the immune microenvironment in SCLC. J Thorac Oncol. (2019) 14:2152–63. doi: 10.1016/j.jtho.2019.08.009
46. Sen T, Rodriguez BL, Chen L, Corte CMD, Morikawa N, Fujimoto J, et al. Targeting DNA damage response promotes antitumor immunity through sting-mediated T-cell activation in small cell lung cancer. Cancer Discov. (2019) 9:646–61. doi: 10.1158/2159-8290.CD-18-1020
47. Zhang H, Christensen CL, Dries R, Oser MG, Deng J, Diskin B, et al. CDK7 inhibition potentiates genome instability triggering anti-tumor immunity in small cell lung cancer. Cancer Cell. (2020) 37:37–54.e9. doi: 10.1016/j.ccell.2019.11.003
48. Yang S, Zhang Z, Wang Q. Emerging therapies for small cell lung cancer. J Hematol Oncol. (2019) 12:47. doi: 10.1186/s13045-019-0736-3
49. Saunders LR, Bankovich AJ, Anderson WC, Aujay MA, Bheddah S, Black K, et al. A DLL3-targeted antibody-drug conjugate eradicates high-grade pulmonary neuroendocrine tumor-initiating cells in vivo. Sci Transl Med. (2015) 7:302ra136. doi: 10.1126/scitranslmed.aac9459
50. Rudin CM, Pietanza MC, Bauer TM, Ready N, Morgensztern D, Glisson BS, et al. Rovalpituzumab tesirine, a DLL3-targeted antibody-drug conjugate, in recurrent small-cell lung cancer: a first-in-human, first-in-class, open-label, phase 1 study. Lancet Oncol. (2017) 18:42–51. doi: 10.1016/s1470-2045(16)30565-4
51. Morgensztern D, Besse B, Greillier L, Santana-Davila R, Ready N, Hann CL, et al. Efficacy and safety of rovalpituzumab tesirine in third-line and beyond patients with DLL3-expressing, relapsed/refractory small-cell lung cancer: results from the phase II TRINITY study. Clin Cancer Res. (2019) 25:6958–66. doi: 10.1158/1078-0432.Ccr-19-1133
52. Santamaria Nunez G, Robles CM, Giraudon C, Martinez-Leal JF, Compe E, Coin F, et al. Lurbinectedin specifically triggers the degradation of phosphorylated RNA polymerase II and the formation of DNA breaks in cancer cells. Mol Cancer Ther. (2016) 15:2399–412. doi: 10.1158/1535-7163.Mct-16-0172
53. Calvo E, Moreno V, Flynn M, Holgado E, Olmedo ME, Lopez Criado MP, et al. Antitumor activity of lurbinectedin (PM01183) and doxorubicin in relapsed small-cell lung cancer: results from a phase I study. Ann Oncol. (2017) 28:2559–66. doi: 10.1093/annonc/mdx357
54. Farago AF, Drapkin BJ, Lopez-Vilarino de Ramos JA, Galmarini CM, Nunez R, Kahatt C, et al. ATLANTIS: a Phase III study of lurbinectedin/doxorubicin versus topotecan or cyclophosphamide/doxorubicin/vincristine in patients with small-cell lung cancer who have failed one prior platinum-containing line. Future Oncol. (2019) 15:231–9. doi: 10.2217/fon-2018-0597
55. Cristea S, Coles GL, Hornburg D, Gershkovitz M, Arand J, Cao S, et al. The MEK5-ERK5 kinase axis controls lipid metabolism in small cell lung cancer. Cancer Res. (2020) 80:1293–303. doi: 10.1158/0008-5472.CAN-19-1027
56. Chalishazar MD, Wait SJ, Huang F, Ireland AS, Mukhopadhyay A, Lee Y, et al. MYC-driven small-cell lung cancer is metabolically distinct and vulnerable to arginine depletion. Clin Cancer Res. (2019) 25:5107–21. doi: 10.1158/1078-0432.Ccr-18-4140
57. Allison Stewart C, Tong P, Cardnell RJ, Sen T, Li L, Gay CM, et al. Dynamic variations in epithelial-to-mesenchymal transition (EMT), ATM, and SLFN11 govern response to PARP inhibitors and cisplatin in small cell lung cancer. Oncotarget. (2017) 8:28575–87. doi: 10.18632/oncotarget.15338
58. Murai J, Feng Y, Yu GK, Ru Y, Tang SW, Shen Y, et al. Resistance to PARP inhibitors by SLFN11 inactivation can be overcome by ATR inhibition. Oncotarget. (2016) 7:76534–50. doi: 10.18632/oncotarget.12266
59. Hook KE, Garza SJ, Lira ME, Ching KA, Lee NV, Cao J, et al. An integrated genomic approach to identify predictive biomarkers of response to the aurora kinase inhibitor PF-03814735. Mol Cancer Ther. (2012) 11:710–9. doi: 10.1158/1535-7163.Mct-11-0184
60. Brockmann M, Poon E, Berry T, Carstensen A, Deubzer HE, Rycak L, et al. Small molecule inhibitors of aurora-a induce proteasomal degradation of N-myc in childhood neuroblastoma. Cancer Cell. (2013) 24:75–89. doi: 10.1016/j.ccr.2013.05.005
61. Mollaoglu G, Guthrie MR, Bohm S, Bragelmann J, Can I, Ballieu PM, et al. MYC drives progression of small cell lung cancer to a variant neuroendocrine subtype with vulnerability to aurora kinase inhibition. Cancer Cell. (2017) 31:270–85. doi: 10.1016/j.ccell.2016.12.005
62. Reck M, Rodriguez-Abreu D, Robinson AG, Hui R, Csoszi T, Fulop A, et al. Pembrolizumab versus chemotherapy for PD-L1-positive non-small-cell lung cancer. N Engl J Med. (2016) 375:1823–33. doi: 10.1056/NEJMoa1606774
63. Mok TSK, Wu YL, Kudaba I, Kowalski DM, Cho BC, Turna HZ, et al. Pembrolizumab versus chemotherapy for previously untreated, PD-L1-expressing, locally advanced or metastatic non-small-cell lung cancer (KEYNOTE-042): a randomised, open-label, controlled, phase 3 trial. Lancet. (2019) 393:1819–30. doi: 10.1016/s0140-6736(18)32409-7
64. Hellmann MD, Callahan MK, Awad MM, Calvo E, Ascierto PA, Atmaca A, et al. Tumor mutational burden and efficacy of nivolumab monotherapy and in combination with ipilimumab in small-cell lung cancer. Cancer Cell. (2018) 33:853–61.e4. doi: 10.1016/j.ccell.2018.04.001
65. Teng MW, Ngiow SF, Ribas A, Smyth MJ. Classifying cancers based on T-cell infiltration and PD-L1. Cancer Res. (2015) 75:2139–45. doi: 10.1158/0008-5472.Can-15-0255
66. Solomon B, Young RJ, Bressel M, Urban D, Hendry S, Thai A, et al. Prognostic Significance of PD-L1(+) and CD8(+) immune cells in HPV(+) oropharyngeal squamous cell carcinoma. Cancer Immunol Res. (2018) 6:295–304. doi: 10.1158/2326-6066.Cir-17-0299
67. Taube JM, Anders RA, Young GD, Xu H, Sharma R, McMiller TL, et al. Colocalization of inflammatory response with B7-h1 expression in human melanocytic lesions supports an adaptive resistance mechanism of immune escape. Sci Transl Med. (2012) 4:127ra137. doi: 10.1126/scitranslmed.3003689
68. Dudley JC, Lin MT, Le DT, Eshleman JR. Microsatellite instability as a biomarker for PD-1 blockade. Clin Cancer Res. (2016) 22:813–20. doi: 10.1158/1078-0432.Ccr-15-1678
69. Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. (2015) 372:2509–20. doi: 10.1056/NEJMoa1500596
70. Marabelle A, Le DT, Ascierto PA, Di Giacomo AM, De Jesus-Acosta A, Delord JP, et al. Efficacy of pembrolizumab in patients with noncolorectal high microsatellite instability/mismatch repair-deficient cancer: results from the phase II KEYNOTE-158 study. J Clin Oncol. (2020) 38:1–10. doi: 10.1200/jco.19.02105
Keywords: SCLC, targeted therapy, immune therapy, DNA damage repair pathway, biomarker
Citation: Taniguchi H, Sen T and Rudin CM (2020) Targeted Therapies and Biomarkers in Small Cell Lung Cancer. Front. Oncol. 10:741. doi: 10.3389/fonc.2020.00741
Received: 17 February 2020; Accepted: 20 April 2020;
Published: 20 May 2020.
Edited by:
Sonam Puri, University of Utah, United StatesReviewed by:
Antonio Passaro, European Institute of Oncology (IEO), ItalyK. Shilo, The Ohio State University, United States
Copyright © 2020 Taniguchi, Sen and Rudin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Triparna Sen, c2VudEBtc2tjYy5vcmc=; Charles M. Rudin, cnVkaW5jQG1za2NjLm9yZw==
 Hirokazu Taniguchi
Hirokazu Taniguchi Triparna Sen
Triparna Sen Charles M. Rudin
Charles M. Rudin