- 1Department of Lung Cancer Surgery, Tianjin Medical University General Hospital, Tianjin, China
- 2Tianjin Key Laboratory of Lung Cancer Metastasis and Tumor Microenvironment, Lung Cancer Institute, Tianjin Medical University General Hospital, Tianjin, China
- 3Department of Respiratory and Critical Care, Tianjin Medical University General Hospital, Tianjin, China
- 4Department of Respiratory Medicine, Second Affiliated Hospital of Tianjin University of Traditional Chinese Medicine, Tianjin, China
Circular RNAs (circRNAs) are a class of endogenous non-coding RNAs (ncRNAs) with a structure of covalently closed continuous loops, which can regulate gene expression by acting as a microRNA sponge or through other mechanisms. Recent studies have identified that the expression of candidate circRNAs are dysregulated in various tumors and hence are considered as promising diagnostic or therapeutic targets across cancer types. However, the expression and function of circRNAs in lung adenocarcinoma (LUAD) remains unclear. In this article, we investigated the expression of circRNAs in LUAD via MiOncoCirc, which is the first and comprehensive database characterizing circRNAs across >2,000 cancer samples using an exome capture RNA sequencing. We identified seven abnormally expressed circRNAs in LUAD, including circCDR1-AS, circHIPK3, circFNDC3B, circPCMTD1, circRHOBTB3, circFAM13B, and circMAN1A2, as well as conducted a literature review about the function and features of these circRNAs. Previous studies have demonstrated that circCDR1-AS, circMAN1A2, and circHIPK3 were upregulated and significantly correlated with a poor survival, or promoted the tumor progression in lung cancer, whereas other circRNAs have not been fully explored. Besides, we reviewed all the publications regarding circRNAs and LUAD, and noticed that the dysregulation of these circRNAs impacts the development of LUAD through a variety of regulatory mechanisms. In conclusion, the underlying mechanisms of aberrant expression and functions of circRNAs in LUAD are worthy of being further investigated.
Introduction
Lung cancer, as the most common cancer, is the leading cause of cancer worldwide. Despite the improved prognosis of lung cancer patients resulting from surgery, chemotherapy, radiotherapy, tyrosine kinase inhibitor therapy, and immunotherapy, the 5-year survival rate is still low (1). Early detection with the use of low-dose CT decreases mortality in lung cancer (2). However, it is still necessary to develop more effective non-invasive biomarkers for the detection of early-stage lung cancers. As one of the main types of lung cancer, lung adenocarcinoma (LUAD) rates continue to increase in Eastern and Western populations, whereas the number of lung squamous cell carcinomas (LUSQ) gradually decreases (3). Because physicians always have to clarify diagnosis by tissue biopsy, a convenient, accurate, and non-invasive biomarker that has the ability to differentiate subtypes, and predict and monitor the progression of lung cancer is also significant.
Circular RNAs (circRNAs), first introduced by Sanger et al. in 1976 (4), are a kind of covalently closed circular non-coding RNAs (ncRNAs) without 5′ to 3′ polarity or polyadenylated tail (5). With the development of high-throughput RNA sequencing and bioinformatics tools, more and more novel circRNAs have been identified. After being reported in atherosclerotic vascular disease risk, neurological disorders, and prion diseases (6), circRNAs are also detected to play important roles in the progression of cancer (7). Owing to their unique structure, circRNAs are difficult to degrade and could be used as promising molecular biomarkers. Previous studies have confirmed that circRNAs have the potential to be diagnostic, prognostic, and predictive biomarkers (8–11). However, the biofunctions of circRNAs are still obscure. One of the major functions of circRNAs is regards miRNA sponges (12). For example, circ-AKT3 was reported to serve as a novel therapeutic to target clear cell renal cell carcinoma metastasis (13). CircFOXP1 was identified to predict and regulate the progression of gallbladder cancer (14). CircPLEKHM3 acted as a tumor suppressor in ovarian cancer (15). All these circRNAs play their role in the circRNA–miRNA–mRNA axis, although further studies still are needed to explore the function of circRNAs in lung cancer.
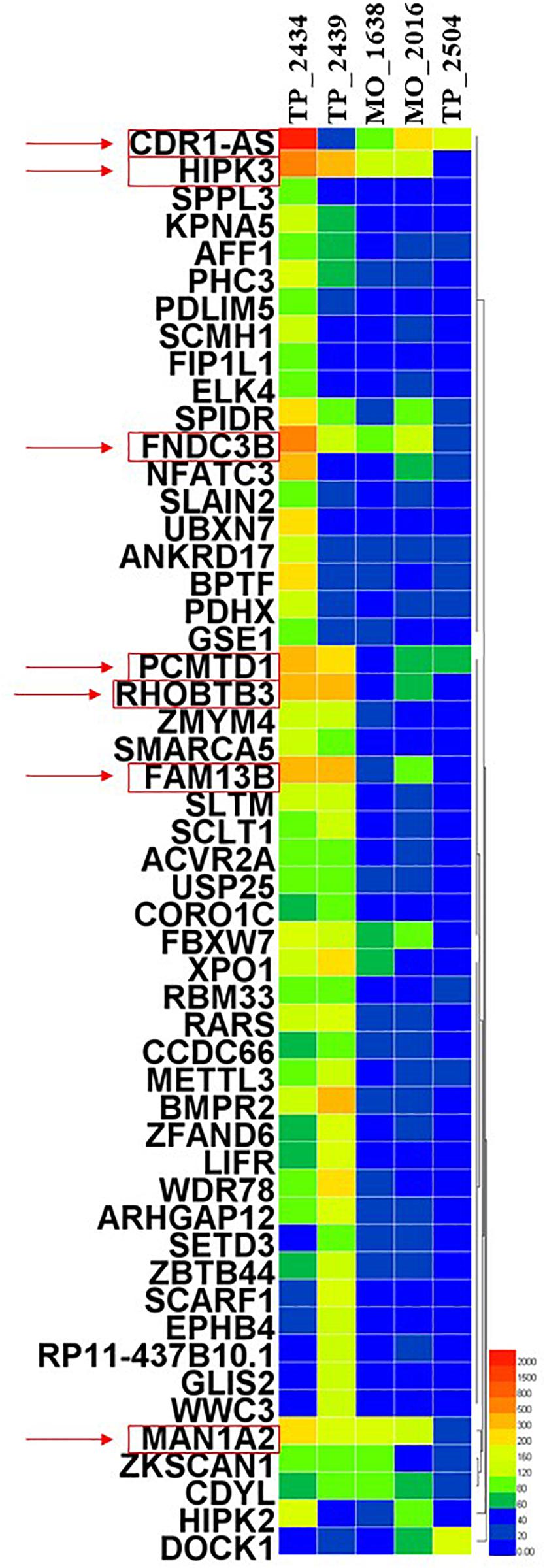
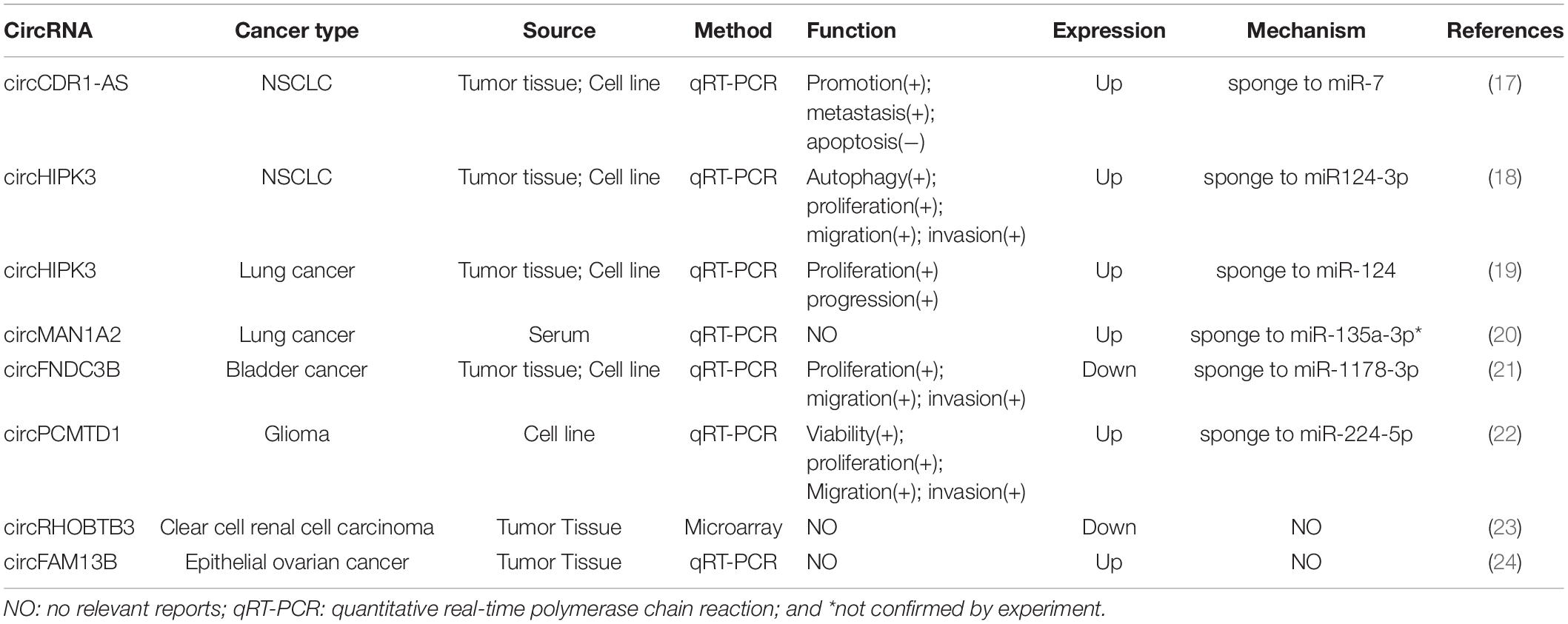
In this review, we aim to investigate the potential roles of these circRNAs in LUAD. By detecting more than 2,000 tumor samples and cell lines by exome capture RNA sequencing, researchers have collected all the data in MiOncoCirc database (16). Five patients with LUAD were selected in this database, including three female and two male patients. However, the detailed clinical information (e.g., staging and survival) for these patients was not provided in the database. To ensure the specificity of the selected circRNAs, the differentially expressed circRNAs were identified to be significant when fold-change (log2) was above 2. Relative expression heatmap based on expression of 52 different circRNAs revealed that seven candidate circRNAs have an abnormal expression profile (log2) that was above 6 (Figure 1). The detailed information of these circRNAs are summarized by searching the PubMed database. Baseline characteristics of these seven circRNAs are listed in Table 1. We further reviewed all published articles on circRNAs and LUAD and summarized their regulatory mechanisms (Table 2).
CircCDR1as
Zhang and colleagues have found that circCDR1as exhibited a much higher expression in non-small cell lung cancer (NSCLC) compared with normal lung tissue (17). The clinicopathological characteristics of NSCLC patients revealed that circCDR1as was positively correlated with the advanced stage and poor outcome of NSCLC patients. It was also found that circCDR1as exhibited a higher expression in LUAD patients compared with lung squamous cell carcinoma (sqCC; 79.2% vs. 61.1%). Moreover, in vitro gain and loss of function studies using A549 and H460 cell lines identified that circCDR1as acted as a powerful miR-7 sponge/inhibitor, which could regulate epidermal growth factor receptor (EGFR), cyclin E1 (CCNE1), and phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform (PIK3CD). Knockdown of circCDR1as or overexpression of miR-7 could suppress both LUAD and lung sqCC tumor cell proliferation via inducing apoptosis and G1/S arrest of NSCLC cells.
CircHIPK3
A previous study has demonstrated that the expression of circHIPK3 was much higher in several lung cancer cell lines, which is associated with proliferation, migration, and invasion of cancer cells. In serine/threonine kinase 11 (STK11) mutant lung cancer, circHIPK3 could modulate autography through miR-124-3p-STAT3-PRKAA/AMPKα signaling. Antagonistic regulation on autophagy has also been demonstrated between circHIPK3 and linear HIPK3. Moreover, it was found that the ratio between circHIPK3 and linHIPK3, detected by RT-PCR, was correlated with poor survival, especially in advanced-stage (II, III, and IV) LUAD patients, whereas circHIPK3 or linHIPK3 alone as a biomarker is meaningless. All these results suggest that circHIPK3 is a key autophagy regulator and could be used as a potential target in STK11 mutant lung cancer (18). In addition, another study also showed that the expression of circHIPK3 was upregulated in A549 cell line and in human lung cancer tissue. Further studies indicate that overexpression of circHIPK3 is associated with high proliferation and long survival of lung cancer cells. Suppressing the expression of circHIPK3 could inhibit the survival and proliferation of lung cancer cells, but lead to a remarkable cell apoptosis. Researchers also found that circHIPK3 could act as miR-124 sponges and regulate sphingosine kinase 1 (SphK1), cyclin-dependent kinase 4 (CDK4) as well as signal transducer and activator of transcription 3 (STAT3), all of which are the targets of miR-124. In conclusion, circHIPK3 may promote the growth of lung cancer cells by sponging miR-124, and STAT3 is generally accepted as a target of miR-124 (19).
CircMAN1A2
By comparing the serum from healthy donors and lung cancer patients, Fan et al. have found that lung cancer patients show a high level of circMAN1A2, which indicated that circMAN1A2 might be used as a potentially diagnostic biomarker in lung cancer patients. Bioinformatic analysis suggested that circMAN1A2 was most likely combined with has-miR-135a-3p, which was previously reported to be downregulated in ovarian cancer and inhibit cancer cell growth. Thus, circMAN1A2 seemed to exert its biofunction by acting as miR-135a-3p sponges. Further bioinformatic analysis indicated that solute carrier family 4 member 8 (SLC4A8), IKAROS family zinc finger 4 (IKZF4), small cell adhesion glycoprotein (SMAGP), Sp1 transcription factor (SP1), erb-b2 receptor tyrosine kinase 3 (ERBB3), and chromobox 5 (CBX5) were mostly probable binding partners for hsa-miR-135a-3p (20). However, there is no histology-based subgroup analysis in this study, and hence further research is necessary to explore its regulatory mechanism in LUAD and lung sqCC, respectively.
Other CircRNAs
The expression and function of circRNAs, circFNDC3B, circPCMTD1, circRHOBTB3, and circFAM13B, which express high in LUAD in our database analysis, have not been reported in lung cancer in previous studies. However, some of these circRNAs expressed with an abnormal level and participated in oncogenesis in other cancers. It was demonstrated that the expression of circFNDC3B was downregulated in bladder cancer tissue and cell lines, and interfered on tumor growth via miR-1178-3p/G3BP2/SRC/FAK axis (21). In glioma patients, circPCMTD1, as a newly discovered circRNA, was able to promote the progression of glioma by sponging to miR-224-5p (22). By using microarray analysis and statistical methods, Franz et al. have demonstrated that circRHOBTB3 could act as a diagnostic and prognostic biomarker of clear cell renal cell carcinoma (23). Besides, it was reported that circFAM13B was decreased in the epithelial ovarian cancer specimens, whereas the specific regulatory mechanism remained unclear (24).
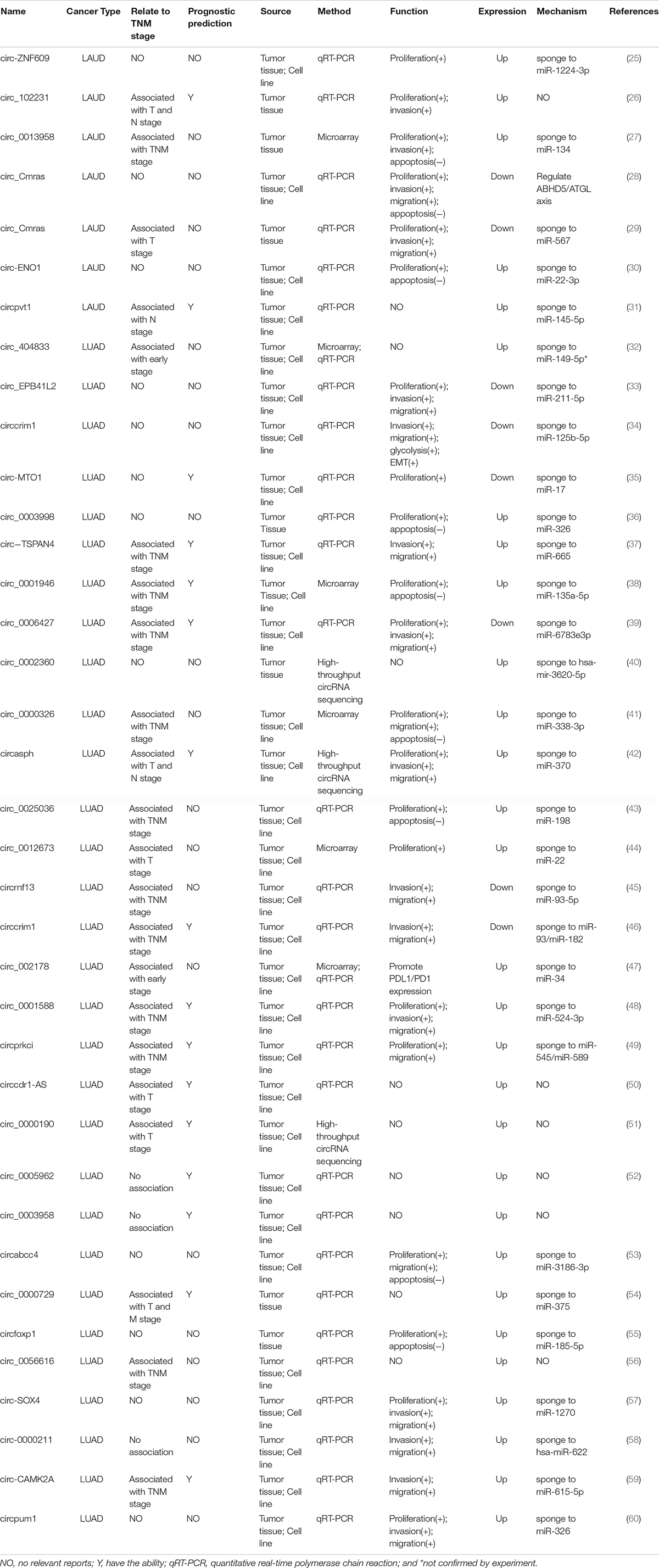
To more comprehensively explore the detailed function of circRNAs in LUAD, we reviewed all the publications regarding circRNAs and LUAD (Table 2). The most common regulatory mechanism of these circRNAs is acting as miRNA sponges, and one circRNA could sponge with more than one miRNA which impacts the tumor progression. Some studies also suggested that circRNAs could bind protein to regulate LUAD cell progression. In these studies, the abnormally expressed circRNAs were correlated with LUAD cell proliferation, invasion, migration, apoptosis, epithelial–mesenchymal transition, and drug resistance. Some of these circRNAs were also related to the TMN stage as well as prognosis in LUAD patients (25–60). Moreover, a previous study indicated that circRNA-002178 could enhance PD-L1 expression by absorbing miR-28-5p in CD8+ T cells (47).
Discussion
Circular RNAs, a large class of ncRNAs, have been demonstrated to dysregulate in LUAD patients. From the current studies, the abnormal expression of circRNAs is correlated with the poor prognosis, disease progression, metastasis, and recurrence. Most of these circRNAs are isolated from tissues and tumor cells. There are some reports suggesting that the abnormal expression of circRNAs could be found in plasma from cancer patients (61–63). Furthermore, Wang et al. demonstrated that circRNAs had the potential role to distinguish LUAD and LUSQ, indicating that circRNAs may play a different role in the initiation and development of LUAD and LUSQ (64). In recent years, owing to the popularization of CT as a diagnostic method of early lung cancer, the mortality rate of lung cancer patients has been significantly reduced (65). However, it is not easy to distinguish benign and malignant nodules in many cases, and it is also difficult to differentiate the types of lung cancers. Previous studies have reported that this special class of endogenous ncRNAs could be considered as reliable biomarkers for the diagnosis and evaluation in cancers because of their stable structure and convenient way to obtain.
It is well known that circRNAs exert their biofunctions by playing the role of miRNA sponges. In this review, we have noticed that most circRNAs work in this way. For example, circCDR1 maintains the expression of EGFR, CCNE1, and PIK3CD by acting as a sponge of miR-7. Besides, there are other reports suggesting that circRNAs are able to exert their biofunctions by other approaches, such as acting as sponges or decoys for proteins, directly interacting with specific proteins to enhance their function, functioning as protein scaffolds to promote their reaction kinetics, recruiting proteins, and combining with ribosome to play the role of templates for translation (66). However, most of these studies investigated more about the dysregulated circRNAs in tumor tissues or cell lines instead of peripheral blood. Thus, further studies are needed to explore the detailed functions of circRNAs in different resources.
Although targeted therapy and immunotherapy are widely used in LUAD, immunotherapy is only efficient in approximately 20% of all patients and targeted therapy could only benefit those patients with specific gene mutations. Also, all the patients would develop drug resistance inevitably. Thus, it is urgent to find novel treatment strategies. Considering the essential role of circRNAs, whether these endogenous ncRNA could be the potentially therapeutic target deserves further investigation. Moreover, owing to the difficulty of tissue biopsy in many cancer patients, plasma circRNAs may be a promising non-invasive biomarker in the diagnosis, individualized treatment evaluation, and dynamic follow-up. From the literature review (Table 2), we found that most of the listed circRNAs were correlated with the TNM stage and prognosis in LUAD patients, suggesting that a panel of circRNAs may have clinical value in the diagnosis and predicting prognosis in LUAD patients. However, the exact mechanisms of some circRNAs in LUAD are still not clear, which need further investigation.
In conclusion, circRNAs could be considered as not only important biomarkers for early diagnosis and prognosis of LUAD but also the potential targets for LUAD treatment.
Data Availability Statement
The datasets analyzed for this study can be found in the MiOncoCirc database (https://mioncocirc.github.io/).
Author Contributions
SKZ wrote the article. SL, WL, YYW, XFL, SZ, and XL completed the figures and tables. SX conceived, organized, and edited the text. All authors contributed to the article and approved the submitted version.
Funding
The present study was funded by the National Natural Science Foundation of China (No. 81772464) and the Tianjin Key Project of Natural Science Foundation (No. 17JCZDJC36200).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature. (2018) 553:446–54. doi: 10.1038/nature25183
2. National Lung Screening Trial Research Team, Aberle DR, Adams AM, Berg CD, Black WC, Clapp JD, et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med. (2011) 365:395–409. doi: 10.1056/NEJMoa1102873
3. Lortet-Tieulent J, Soerjomataram I, Ferlay J, Rutherford M, Weiderpass E, Bray F. International trends in lung cancer incidence by histological subtype: adenocarcinoma stabilizing in men but still increasing in women. Lung Cancer. (2014) 84:13–22. doi: 10.1016/j.lungcan.2014.01.009
4. Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci USA. (1976) 73:3852–6. doi: 10.1073/pnas.73.11.3852
5. Hu W, Bi ZY, Chen ZL, Liu C, Li LL, Zhang F, et al. Emerging landscape of circular RNAs in lung cancer. Cancer Lett. (2018) 427:18–27. doi: 10.1016/j.canlet.2018.04.006
6. Qu S, Yang X, Li X, Wang J, Gao Y, Shang R, et al. Circular RNA: a new star of noncoding RNAs. Cancer Lett. (2015) 365:141–8. doi: 10.1016/j.canlet.2015.06.003
7. Zhang HD, Jiang LH, Sun DW, Hou JC, Ji ZL. CircRNA: a novel type of biomarker for cancer. Breast Cancer. (2018) 25:1–7. doi: 10.1007/s12282-017-0793-9
8. Zhang X, Wang S, Wang H, Cao J, Huang X, Chen Z, et al. Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol Cancer. (2019) 18:20. doi: 10.1186/s12943-018-0935-5
9. Yu J, Xu QG, Wang ZG, Yang Y, Zhang L, Ma JZ, et al. Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J Hepatol. (2018) 68:1214–27. doi: 10.1016/j.jhep.2018.01.012
10. Zhang J, Liu H, Hou L, Wang G, Zhang R, Huang Y, et al. Circular RNA_LARP4 inhibits cell proliferation and invasion of gastric cancer by sponging miR-424-5p and regulating LATS1 expression. Mol Cancer. (2017) 16:151. doi: 10.1186/s12943-017-0719-3
11. Han D, Li J, Wang H, Su X, Hou J, Gu Y, et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. (2017) 66:1151–64. doi: 10.1002/hep.29270
12. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature. (2013) 495:384–8. doi: 10.1038/nature11993
13. Xue D, Wang H, Chen Y, Shen D, Lu J, Wang M, et al. Circ-AKT3 inhibits clear cell renal cell carcinoma metastasis via altering miR-296-3p/E-cadherin signals. Mol Cancer. (2019) 18:151. doi: 10.1186/s12943-019-1072-5
14. Wang S, Zhang Y, Cai Q, Ma M, Jin LY, Weng M, et al. Circular RNA FOXP1 promotes tumor progression and Warburg effect in gallbladder cancer by regulating PKLR expression. Mol Cancer. (2019) 18:145. doi: 10.1186/s12943-019-1078-z
15. Zhang L, Zhou Q, Qiu Q, Hou L, Wu M, Li J, et al. CircPLEKHM3 acts as a tumor suppressor through regulation of the miR-9/BRCA1/DNAJB6/KLF4/AKT1 axis in ovarian cancer. Mol Cancer. (2019) 18:144. doi: 10.1186/s12943-019-1080-5
16. Vo JN, Cieslik M, Zhang Y, Shukla S, Xiao L, Zhang Y, et al. The landscape of circular RNA in cancer. Cell. (2019) 176:869–81.e13. doi: 10.1016/j.cell.2018.12.021
17. Zhang X, Yang D, Wei Y. Overexpressed CDR1as functions as an oncogene to promote the tumor progression via miR-7 in non-small-cell lung cancer. Onco Targets Ther. (2018) 11:3979–87. doi: 10.2147/OTT.S158316
18. Chen X, Mao R, Su W, Yang X, Geng Q, Guo C, et al. Circular RNA circHIPK3 modulates autophagy via MIR124-3p-STAT3-PRKAA/AMPKalpha signaling in STK11 mutant lung cancer. Autophagy. (2019) 16:659–71. doi: 10.1080/15548627.2019.1634945
19. Yu H, Chen Y, Jiang P. Circular RNA HIPK3 exerts oncogenic properties through suppression of miR-124 in lung cancer. Biochem Biophys Res Commun. (2018) 506:455–62. doi: 10.1016/j.bbrc.2018.10.087
20. Fan C-M, Wang J-P, Tang Y-Y, Zhao J, He S-Y, Xiong F, et al. circMAN1A2 could serve as a novel serum biomarker for malignant tumors. Cancer Sci. (2019) 110:2180–8. doi: 10.1111/cas.14034
21. Liu H, Bi J, Dong W, Yang M, Shi J, Jiang N, et al. Invasion-related circular RNA circFNDC3B inhibits bladder cancer progression through the miR-1178-3p/G3BP2/SRC/FAK axis. Mol Cancer. (2018) 17:161. doi: 10.1186/s12943-018-0908-8
22. Zheng S-Q, Qi Y, Wu J, Zhou F-L, Yu H, Li L, et al. CircPCMTD1 acts as the sponge of miR-224-5p to promote glioma progression. Front Oncol. (2019) 9:398. doi: 10.3389/fonc.2019.00398
23. Franz A, Ralla B, Weickmann S, Jung M, Rochow H, Stephan C, et al. Circular RNAs in clear cell renal cell carcinoma: their microarray-based identification, analytical validation, and potential use in a clinico-genomic model to improve prognostic accuracy. Cancers. (2019) 11:1473. doi: 10.3390/cancers11101473
24. Ning L, Long B, Zhang W, Yu M, Wang S, Cao D, et al. Circular RNA profiling reveals circEXOC6B and circN4BP2L2 as novel prognostic biomarkers in epithelial ovarian cancer. Int J Oncol. (2018) 53:2637–46. doi: 10.3892/ijo.2018.4566
25. Zuo Y, Shen W, Wang C, Niu N, Pu J. Circular RNA Circ-ZNF609 promotes lung adenocarcinoma proliferation by modulating miR-1224-3p/ETV1 signaling. Cancer Manag Res. (2020) 12:2471–9. doi: 10.2147/CMAR.S232260
26. Zong L, Sun Q, Zhang H, Chen Z, Deng Y, Li D, et al. Increased expression of circRNA_102231 in lung cancer and its clinical significance. Biomed Pharmacother. (2018) 102:639–44. doi: 10.1016/j.biopha.2018.03.084
27. Zhu X, Wang X, Wei S, Chen Y, Chen Y, Fan X, et al. hsa_circ_0013958: a circular RNA and potential novel biomarker for lung adenocarcinoma. FEBS J. (2017) 284:2170–82. doi: 10.1111/febs.14132
28. Zhou Q, Sun Y. Circular RNA cMras suppresses the progression of lung adenocarcinoma through ABHD5/ATGL axis using NF-κB signaling pathway. Cancer Biother Radiopharm. (2020). doi: 10.1089/cbr.2020.3709 [Epub ahead of print].
29. Yu C, Tian F, Liu J, Su M, Wu M, Zhu X, et al. Circular RNA cMras inhibits lung adenocarcinoma progression via modulating miR-567/PTPRG regulatory pathway. Cell Prolif. (2019) 52:e12610. doi: 10.1111/cpr.12610
30. Zhou J, Zhang S, Chen Z, He Z, Xu Y, Li Z. CircRNA-ENO1 promoted glycolysis and tumor progression in lung adenocarcinoma through upregulating its host gene ENO1. Cell Death Dis. (2019) 10:885. doi: 10.1038/s41419-019-2127-7
31. Zheng F, Xu R. CircPVT1 contributes to chemotherapy resistance of lung adenocarcinoma through miR-145-5p/ABCC1 axis. Biomed Pharmacother. (2020) 124:109828. doi: 10.1016/j.biopha.2020.109828
32. Zhao J, Li L, Wang Q, Han H, Zhan Q, Xu M. CircRNA expression profile in early-stage lung adenocarcinoma patients. Cell Physiol Biochem. (2017) 44:2138–46. doi: 10.1159/000485953
33. Zhang SJ, Ma J, Wu JC, Hao ZZ, Zhang YN, Zhang YJ. CircRNA EPB41L2 inhibits tumorigenicity of lung adenocarcinoma through regulating CDH4 by miR-211-5p. Eur Rev Med Pharmacol Sci. (2020) 24:3749–60. doi: 10.26355/eurrev_202004_20839
34. Zhang SJ, Ma J, Wu JC, Hao ZZ, Zhang YA, Zhang YJ. Circular RNA circCRIM1 suppresses lung adenocarcinoma cell migration, invasion, EMT, and glycolysis through regulating miR-125b-5p/BTG2 axis. Eur Rev Med Pharmacol Sci. (2020) 24:3761–74. doi: 10.26355/eurrev_202004_20841
35. Zhang B, Chen M, Jiang N, Shi K, Qian R. A regulatory circuit of circ-MTO1/miR-17/QKI-5 inhibits the proliferation of lung adenocarcinoma. Cancer Biol Ther. (2019) 20:1127–35. doi: 10.1080/15384047.2019.1598762
36. Yu W, Peng W, Sha H, Li J. Hsa_circ_0003998 promotes chemoresistance via modulation of miR-326 in lung adenocarcinoma cells. Oncol Res. (2019) 27:623–8. doi: 10.3727/096504018X15420734828058
37. Ying X, Zhu J, Zhang Y. Circular RNA circ-TSPAN4 promotes lung adenocarcinoma metastasis by upregulating ZEB1 via sponging miR-665. Mol Genet Genomic Med. (2019) 7:e991. doi: 10.1002/mgg3.991
38. Yao Y, Hua Q, Zhou Y, Shen H. CircRNA has_circ_0001946 promotes cell growth in lung adenocarcinoma by regulating miR-135a-5p/SIRT1 axis and activating Wnt/β-catenin signaling pathway. Biomed Pharmacother. (2019) 111:1367–75. doi: 10.1016/j.biopha.2018.12.120
39. Yao Y, Hua Q, Zhou Y. CircRNA has_circ_0006427 suppresses the progression of lung adenocarcinoma by regulating miR-6783-3p/DKK1 axis and inactivating Wnt/β-catenin signaling pathway. Biochem Biophys Res Commun. (2019) 508:37–45. doi: 10.1016/j.bbrc.2018.11.079
40. Yan Y, Zhang R, Zhang X, Zhang A, Zhang Y, Bu X. RNA-Seq profiling of circular RNAs and potential function of hsa_circ_0002360 in human lung adenocarcinom. Am J Transl Res. (2019) 11:160–75.
41. Xu Y, Yu J, Huang Z, Fu B, Tao Y, Qi X, et al. Circular RNA hsa_circ_0000326 acts as a miR-338-3p sponge to facilitate lung adenocarcinoma progression. J Exp Clin Cancer Res. (2020) 39:57. doi: 10.1186/s13046-020-01556-4
42. Xu L, Ma Y, Zhang H, Lu QJ, Yang L, Jiang GN, et al. HMGA2 regulates circular RNA ASPH to promote tumor growth in lung adenocarcinoma. Cell Death Dis. (2020) 11:593. doi: 10.1038/s41419-020-2726-3
43. Wu S, Li H, Lu C, Zhang F, Wang H, Lu X, et al. Aberrant expression of hsa_circ_0025036 in lung adenocarcinoma and its potential roles in regulating cell proliferation and apoptosis. Biol Chem. (2018) 399:1457–67. doi: 10.1515/hsz-2018-0303
44. Wang X, Zhu X, Zhang H, Wei S, Chen Y, Chen Y, et al. Increased circular RNA hsa_circ_0012673 acts as a sponge of miR-22 to promote lung adenocarcinoma proliferation. Biochem Biophys Res Commun. (2018) 496:1069–75. doi: 10.1016/j.bbrc.2018.01.126
45. Wang L, Liu S, Mao Y, Xu J, Yang S, Shen H, et al. CircRNF13 regulates the invasion and metastasis in lung adenocarcinoma by targeting miR-93-5p. Gene. (2018) 671:170–7. doi: 10.1016/j.gene.2018.04.069
46. Wang L, Liang Y, Mao Q, Xia W, Chen B, Shen H, et al. Circular RNA circCRIM1 inhibits invasion and metastasis in lung adenocarcinoma through the microRNA (miR)-182/miR-93-leukemia inhibitory factor receptor pathway. Cancer Sci. (2019) 110:2960–72. doi: 10.1111/cas.14131
47. Wang J, Zhao X, Wang Y, Ren F, Sun D, Yan Y, et al. circRNA-002178 act as a ceRNA to promote PDL1/PD1 expression in lung adenocarcinoma. Cell Death Dis. (2020) 11:32. doi: 10.1038/s41419-020-2230-9
48. Sun Z. Circular RNA hsa_circ_0001588 promotes the malignant progression of lung adenocarcinoma by modulating miR-524-3p/NACC1 signaling. Life Sci. (2020) 259:118157. doi: 10.1016/j.lfs.2020.118157
49. Qiu M, Xia W, Chen R, Wang S, Xu Y, Ma Z, et al. The circular RNA circPRKCI promotes tumor growth in lung adenocarcinoma. Cancer Res. (2018) 78:2839–51. doi: 10.1158/0008-5472.CAN-17-2808
50. Mao Y, Xu R. Circular RNA CDR1-AS contributes to pemetrexed and cisplatin chemoresistance through EGFR/PI3K signaling pathway in lung adenocarcinoma. Biomed Pharmacother. (2020) 123:109771. doi: 10.1016/j.biopha.2019.109771
51. Luo YH, Yang YP, Chien CS, Yarmishyn AA, Ishola AA, Chien Y, et al. Plasma level of circular RNA hsa_circ_0000190 correlates with tumor progression and poor treatment response in advanced lung cancers. Cancers. (2020) 12:1740. doi: 10.3390/cancers12071740
52. Liu XX, Yang YE, Liu X, Zhang MY, Li R, Yin YH, et al. A two-circular RNA signature as a non-invasive diagnostic biomarker for lung adenocarcinoma. J Transl Med. (2019) 17:50. doi: 10.1186/s12967-019-1800-z
53. Liu M, Wang P, Sui X, Ding F, Liu L, Gao Z, Cheng Z, et al. Circular RNA circABCC4 regulates lung adenocarcinoma progression via miR-3186-3p/TNRC6B axis. J Cell Biochem. (2020) 121:4226–4238. doi: 10.1002/jcb.29627
54. Li S, Sun X, Miao S, Lu T, Wang Y, Liu J, et al. hsa_circ_0000729, a potential prognostic biomarker in lung adenocarcinoma. Thorac Cancer. (2018) 9:924–30. doi: 10.1111/1759-7714.12761
55. Li O, Kang J, Zhang JJ, Wang J, Hu LW, Li L, et al. Circle RNA FOXP1 promotes cell proliferation in lung cancer by regulating miR-185-5p/Wnt1 signaling pathway. Eur Rev Med Pharmacol Sci. (2020) 24:6767–78. doi: 10.26355/eurrev_202006_21665
56. He F, Zhong X, Lin Z, Lin J, Qiu M, Li X, et al. Plasma exo-hsa_circRNA_0056616: a potential biomarker for lymph node metastasis in lung adenocarcinoma. J Cancer. (2020) 11:4037–46. doi: 10.7150/jca.30360
57. Gao N, Ye B. Circ-SOX4 drives the tumorigenesis and development of lung adenocarcinoma via sponging miR-1270 and modulating PLAGL2 to activate WNT signaling pathway. Cancer Cell Int. (2020) 20:2. doi: 10.1186/s12935-019-1065-x
58. Feng D, Xu Y, Hu J, Zhang S, Li M, Xu L. A novel circular RNA, hsa-circ-0000211, promotes lung adenocarcinoma migration and invasion through sponging of hsa-miR-622 and modulating HIF1-α expression. Biochem Biophys Res Commun. (2020) 521:395–401. doi: 10.1016/j.bbrc.2019.10.134
59. Du J, Zhang G, Qiu H, Yu H, Yuan W. The novel circular RNA circ-CAMK2A enhances lung adenocarcinoma metastasis by regulating the miR-615-5p/fibronectin 1 pathway. Cell Mol Biol Lett. (2019) 24:72. doi: 10.1186/s11658-019-0198-1
60. Chen J, Xu S, Chen S, Zong Z, Han X, Zhao Y, et al. CircPUM1 promotes the malignant behavior of lung adenocarcinoma by regulating miR-326. Biochem Biophys Res Commun. (2019) 508:844–9. doi: 10.1016/j.bbrc.2018.11.176
61. Rong D, Lu C, Zhang B, Fu K, Zhao S, Tang W, et al. CircPSMC3 suppresses the proliferation and metastasis of gastric cancer by acting as a competitive endogenous RNA through sponging miR-296-5p. Mol Cancer. (2019) 18:25. doi: 10.1186/s12943-019-0958-6
62. Li T, Shao Y, Fu L, Xie Y, Zhu L, Sun W, et al. Plasma circular RNA profiling of patients with gastric cancer and their droplet digital RT-PCR detection. J Mol Med. (2018) 96:85–96. doi: 10.1007/s00109-017-1600-y
63. Tang W, Fu K, Sun H, Rong D, Wang H, Cao H. CircRNA microarray profiling identifies a novel circulating biomarker for detection of gastric cancer. Mol Cancer. (2018) 17:137. doi: 10.1186/s12943-018-0888-8
64. Wang C, Tan S, Liu W-R, Lei Q, Qiao W, Wu Y, et al. RNA-Seq profiling of circular RNA in human lung adenocarcinoma and squamous cell carcinoma. Mol Cancer. (2019) 18:134. doi: 10.1186/s12943-019-1061-8
65. Boiselle PM. Computed tomography screening for lung cancer. JAMA. (2013) 309:1163–70. doi: 10.1001/jama.2012.216988
Keywords: circular RNAs, lung adenocarcinoma, biomarker, sponge, MiOncoCirc
Citation: Zhao S, Li S, Liu W, Wang Y, Li X, Zhu S, Lei X and Xu S (2020) Circular RNA Signature in Lung Adenocarcinoma: A MiOncoCirc Database-Based Study and Literature Review. Front. Oncol. 10:523342. doi: 10.3389/fonc.2020.523342
Received: 04 February 2020; Accepted: 14 September 2020;
Published: 09 October 2020.
Edited by:
Jian-Guo Zhou, University of Erlangen Nuremberg, GermanyReviewed by:
Huaxin Sheng, Duke University Medical Center, United StatesBei Liu, The Ohio State University, United States
Copyright © 2020 Zhao, Li, Liu, Wang, Li, Zhu, Lei and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Song Xu, eHVzb25nMTk4QGhvdG1haWwuY29t
†These authors have contributed equally to this work
 Shikang Zhao
Shikang Zhao Shuo Li3†
Shuo Li3† Xiongfei Li
Xiongfei Li Song Xu
Song Xu