- 1Skull Base Research Center, Loghman Hakim Hospital, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- 2Department of Medical Genetics, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- 3Urogenital Stem Cell Research Center, Shahid Beheshti University of Medical Sciences, Tehran, Iran
Glioblastoma is the utmost aggressive diffuse kind of glioma which is originated from astrocytes, neural stem cells or progenitors. This malignant tumor has a poor survival rate. A number of genetic aberrations and somatic mutations have been associated with this kind of cancer. In recent times, the impact of long non-coding RNAs (lncRNAs) in glioblastoma has been underscored by several investigations. Up-regulation of a number of oncogenic lncRNAs such as H19, MALAT1, SNHGs, MIAT, UCA, HIF1A-AS2 and XIST in addition to down-regulation of other tumor suppressor lncRNAs namely GAS5, RNCR3 and NBAT1 indicate the role of these lncRNAs in the pathogenesis of glioblastoma. Several in vitro and a number of in vivo studies have demonstrated the contribution of these transcripts in the regulation of cell proliferation and apoptosis, cell survival, invasion and metastasis of glioblastoma cells. Moreover, some lncRNAs such as SBF2-AS1 are involved in conferring resistance to temozolomide. Finally, few circularRNAs have been identified that influence the evolution of glioblastoma. In this paper, we discuss the impacts of lncRNAs in the pathogenesis of glioblastoma, their applications as markers and their implications in the therapeutic responses in this kind of cancer.
Introduction
Being considered as grade IV glioma tumors, glioblastomas are the utmost aggressive diffuse kind of glioma originating from the astrocytes, neural stem cells or progenitors (1). This type of brain tumor includes about half of all glioma tumors and less than 20% of all primary brain tumors (2). Although being a rare tumor, the poor prognosis and low survival rate of glioblastoma have made it an important public health problem (3). It is more frequent in men compared with females, in Western countries compared with developing world and in some ethnicities such as Asians, Latinos and Whites (3). The etiology of this kind of tumor is largely unclarified, as no causal carcinogen has been linked with it. High dose ionizing radiation is the solitary environmental element that is highly associated with risk of glioblastoma (4). A number of genetic aberrations such as activation of growth factor cascade through amplification and mutations in receptor tyrosine kinase genes, induction of the PI3K proteins and loss of the p53 and Rb tumor suppressor genes have been identified in glioblastoma (5). Genome-wide and direct sequencing techniques have also detected recurrent disease-causing mutations in glioblastoma samples in a number of genes such as IDH1 (6) and TERT promoter (7). Moreover, contemporary studies have conveyed anomalous expression of long non-coding RNAs (lncRNAs) in glioblastoma samples indicating the impact of these transcripts in the pathobiology of this kind of cancer (8). These transcripts are larger than 200 nucleotides and regulate expression of numerous genes at transcriptional, post-transcriptional, and epigenetic phases (9). In the current paper, we discuss the impact of lncRNAs in the pathobiology of glioblastoma and their effects on the regulation of cell proliferation and apoptosis, cell survival, invasion and metastatic aptitude of glioblastoma cells.
Oncogenic lncRNAs in Glioblastoma
Several oncogenic lncRNAs have been up-regulated in glioblastoma samples. For instance, MIR22HG is an oncogenic lncRNA which has been shown to be highly dysregulated in glioblastoma via assessment of accessible datasets. This lncRNA hosts miR-22-3p and miR-22-5p. Further studies have unraveled over-expression of the MIR22HG/miR-22 route in glioblastoma and glioma stem-like cells. Over-expression of MIR22HG in glioblastoma samples has been related with poor patients’ outcome. Knock down of this lncRNA has led to inactivation of the Wnt/β-catenin route via modulating miR-22-3p and miR-22-5p expressions. Functionally, MIR22HG silencing has diminished cell proliferation, invasion and tumor growth in xenograft models. The mentioned miRNAs have been shown to target SFRP2 and PCDH15. Taken together, MIR22HG has been acknowledged as an important activator of the Wnt/β-catenin signaling pathway, and its silencing has been proposed as a therapeutic modality in this kind of cancer (10). The small nucleolar RNA host gene 5 (SNHG5) is another up-regulated lncRNA in glioblastoma which enhances cell proliferation and suppresses cell apoptosis in these cells. Expression of this lncRNA is activated by the Yin Yang 1 (YY1) transcription factor. This lncRNA exerts its oncogenic role via stimulation of the p38/MAPK axis (11). SNHG9 has also been demonstrated to be over-expressed in glioblastoma samples in association with poor survival of patients. SNHG9 has a role in suppression of miR-199a-5p expression and enhancement of Wnt2 expression in glioblastoma cells. This lncRNA has been revealed to enhance aerobic glycolysis and cell proliferation (12). Expression of SAMMSON has been increased in the plasma of patients with glioblastoma but not in those with diffuse neurosarcoidosis, a disorder that shares MRI signs with glioblastoma. This lncRNA has been displayed to suppress expression of miR-622 in glioblastoma cells and subsequently enhance cell (13). MIAT is another up-regulated lncRNA in glioblastoma. Bountali et al. have knocked down this lncRNA in glioblastoma cell lines and analyzed RNA profile of these cells via RNA sequencing method. They reported differential expression of several genes including those participating in cancer-associated functions, namely cell growth and viability, apoptotic features, reactive oxygen species creation and migration. Functionally, MIAT silencing abolishes long-term viability and migration and enhances apoptosis in these cells (14). A genome-wide expression profiling in glioblastoma cells has identified MALAT1 as one of the most remarkably over-expressed genes following treatment with temozolomide (TMZ). Expression of this lncRNA has been co-regulated by p50 and p53 through κB- and p53-binding sites which are located in coding sequence of this lncRNA. MALAT1 silencing has increased sensitivity of patient-originated glioblastoma cells to TMZ and improved the effects of this drug in xenograft mice models (15). UCA1 is another oncogenic lncRNA which enhances cell proliferation and migration, while suppressing cell apoptosis. Figure 1 depicts the molecular mechanisms through which UCA1 participates in the pathogenesis of glioblastoma.
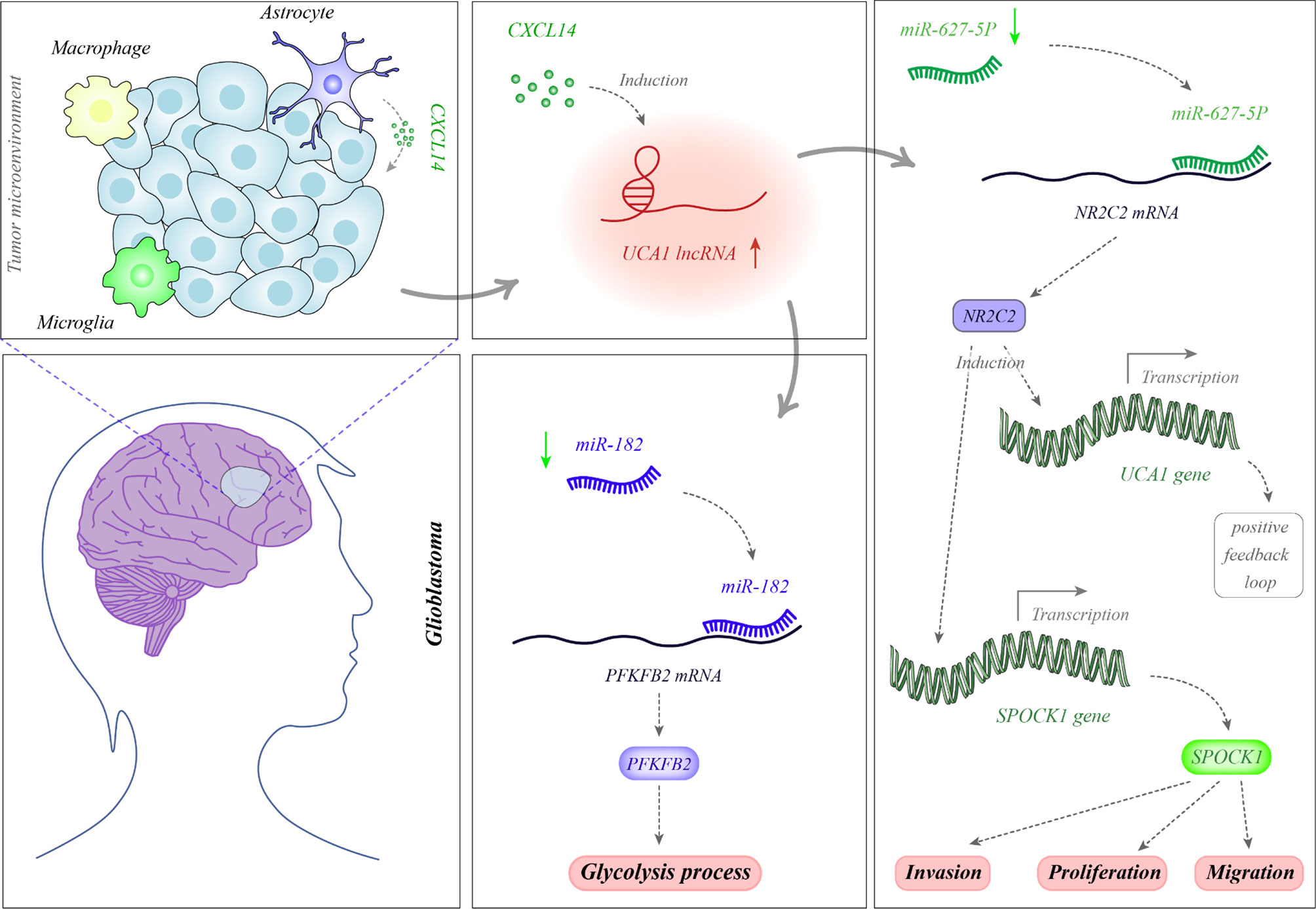
Figure 1 Glioblastoma-associated stromal cells (GASCs) are special cells in the tumor microenvironment which are phenotypically and functionally similar to the cancer-associated fibroblasts. These cells produce CXCL14 which functions as a paracrine factor to enhance expression of UCA1. UCA1 serves as a sponge for miR-182. Since miR-182 suppresses expression of PFKFB2, UCA1 up-regulation results in up-regulation of PFKFB2 through sequestering miR-182. PFKFB2 protein increases glycolysis in the tumor cells (16). In addition, UCA1 decreases miR-627-5p levels. As miR-627-5p inhibits NR2C2 expression, down-regulation of miR-627-5p by UCA1 enhances expression of NR2C2. NR2C2 binds with the promoter region of UCA1 and increase its expression through a positive feedback loop. Moreover, NR2C2 enhances expression of SPOCK1 increasing cell proliferation, migration and invasiveness of tumor cells (17).
Table 1 reviews the function of oncogenic lncRNAs in glioblastoma.
Tumor Suppressor lncRNAs In Glioblastoma
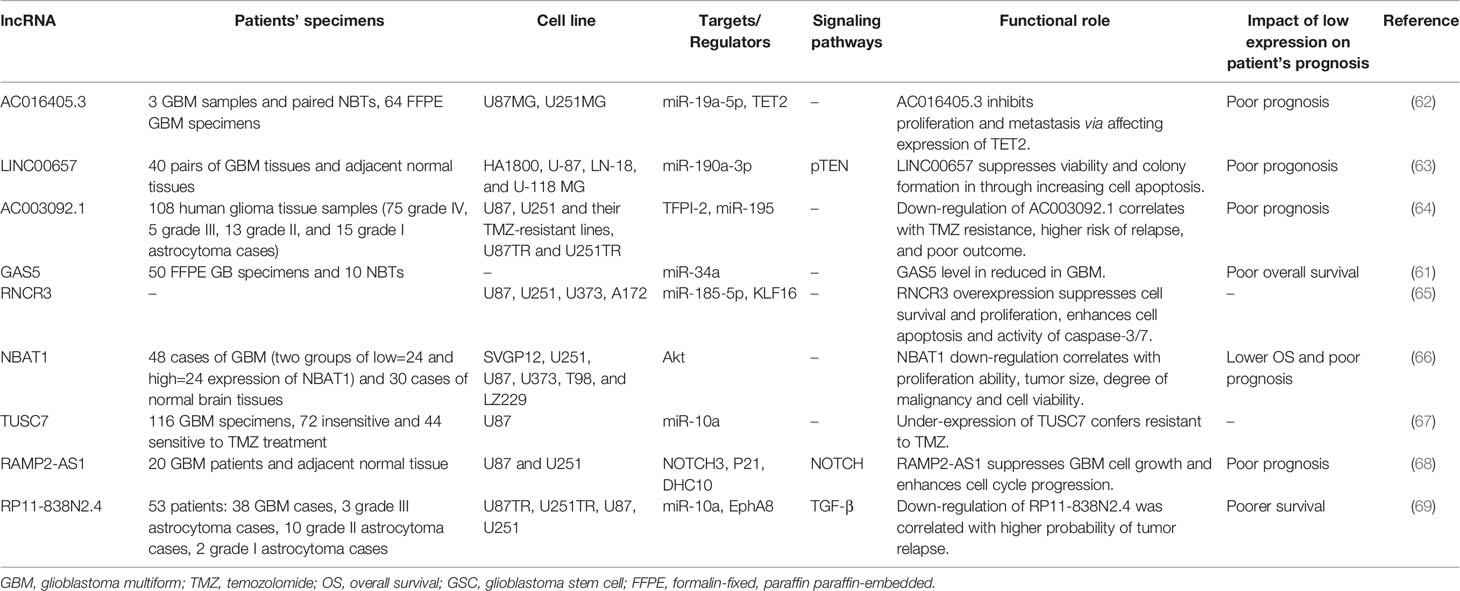
Expression of GAS5 has been decreased in glioblastoma and its levels have been negatively correlated with miR-34a levels (61). In addition, expression of AC016405.3 has been decreased in glioblastoma tissues in association with numerous aggressive characteristics of this type of cancer. Up-regulation of this lncRNA inhibits proliferation and metastatic ability of glioblastoma cells. The oncogenic miRNA, miR-19a-5p has been identified as a downstream miRNA of AC016405.3. AC016405.3 has been shown to be targeted by miR-19a-5p. Functionally, AC016405.3 inhibits cell proliferation and metastasis via regulation of TET2 by serving as a sponge for miR-19a-5p (62). LINC00657 is another tumor suppressor lncRNA whose expression has been decreased in glioblastoma sections compared with neighboring normal section. Up-regulation of this lncRNA has suppressed cell proliferation, colony formation, invasiveness and migratory potential of glioma cells through activating cell apoptosis. LINC00657 has been acknowledged as a direct target of miR-190a-3p, a miRNA that negatively regulates PTEN expression. The tumor suppressive role of LINC00657 has also been verified in xenograft models (63). The lncRNA AC003092.1 has been shown to be down-regulated in TMZ resistance cells compared with their original cells. Moreover, down-regulation of this lncRNA has been correlated with resistance to TMZ, higher possibility of tumor relapse, and poor patients’ outcome. Cell line studies has shown improvement of TMZ sensitivity following up-regulation of AC003092.1. The effect of this lncRNA in the modulation of TMZ sensitivity is exerted via regulation of TFPI-2–associated cell apoptosis through sponging miR-195 (64). RNCR3 is another down-regulated lncRNA in glioblastoma. Over-expression of this lncRNA significantly suppresses cell survival and proliferation of glioblastoma cells, while enhancing cell apoptosis and activity caspase‐3/7. Besides, up-regulation of this lncRNA enhances expression of Krüppel‐like factor 16 (KLF16) via suppressing miR‐185‐5p (65). Table 2 gives an outline of studies which assessed function of tumor suppressor lncRNAs in glioblastoma.
Diagnostic and Prognostic Value of lncRNAs in Glioblastoma
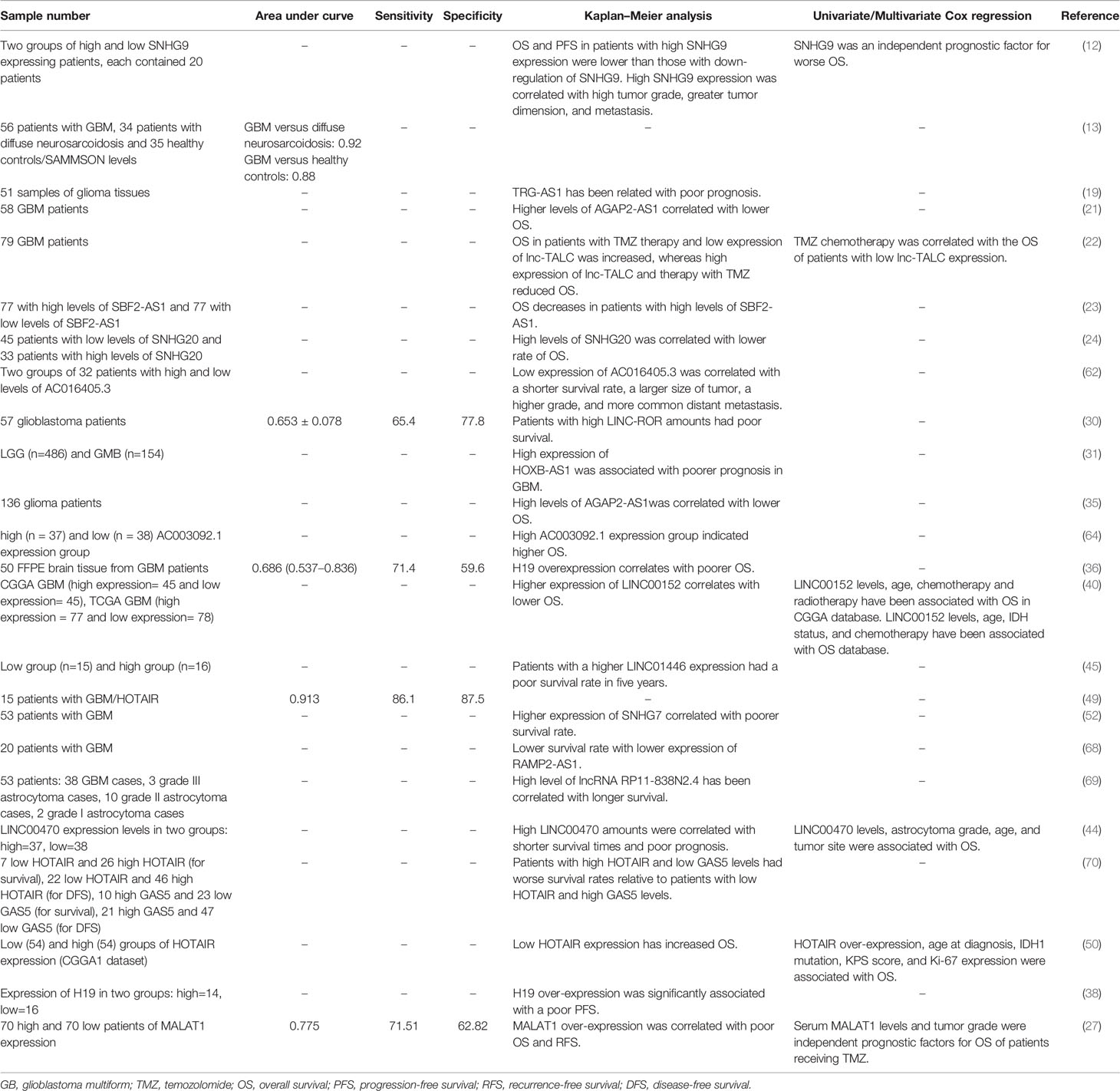
Expression levels of lncRNAs can distinguish patients with glioblastoma from cancer-free individuals. Moreover, these transcripts can possibly differentiate different brain tumors. For instance, plasma levels of SAMMSON can differentiate glioblastoma from both diffuse neurosarcoidosis and healthy controls (13). Among lncRNAs whose diagnostic power has been assessed in glioblastoma, HOTAIR has exhibited the most promising results. Tan et al. have demonstrated significant higher levels of this lncRNA in sera of glioblastoma patients compared with controls. The area under the receiver operating characteristic (ROC) curve was 0.913 indicating the ideal feature of HOTAIR for this purpose. Moreover, they reported significant correlation between its levels and high tumor grade. Notably, there was significant correlation between tumor and serum levels of this lncRNA. Finally, exosomes extracted from the serum samples have been shown to contain this lncRNA, further emphasizing the application of this lncRNA in the prognostic and diagnostic processes in glioblastoma (49). In addition, Kaplan-Meier analysis has indicated the correlation between expression levels of several lncRNAs such as SNHG9, TRG-AS1, AGAP2-AS1, lnc-TALC, SBF2-AS1, SNHG20, AC016405.3, LINC-ROR, HOXB-AS1, H19, LINC00152, RAMP2-AS1 and GAS5 and patients’ prognosis in the terms of overall survival, disease-free survival and progression free survival. Table 3 gives a summary of studies which assessed such aspect of lncRNAs in glioblastoma.
Circular RNAs and Glioblastoma
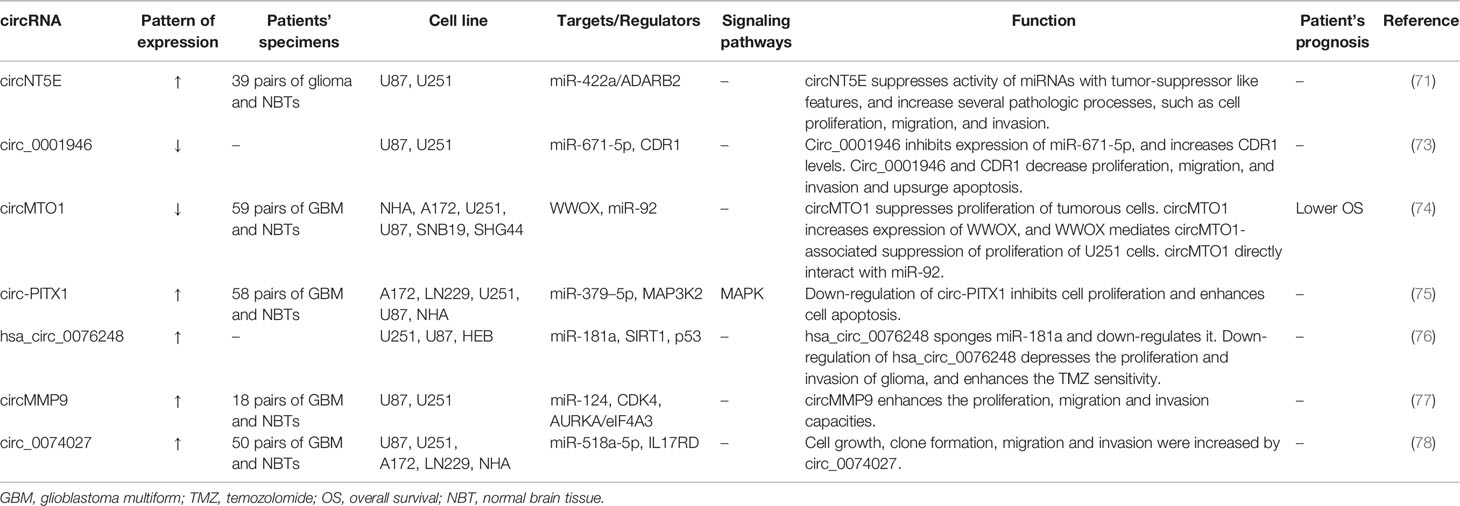
In addition to lncRNAs, Circular RNAs (circRNAs) can act as miRNA sponges to modulate expression of their target genes. Numerous studies have assessed expression and function of circRNAs in glioblastoma. For instance, Wang et al. have reported over-expression of some circRNAs and lncRNAs in miR-422a–downregulated glioblastoma samples. They have also recognized a new circRNA originated from NT5E, termed circNT5E. Expression of this circRNA is modulated by ADARB2 through binding to sites neighboring circRNA-creating introns. circNT5E has been shown to regulate cell proliferation, migration, and invasion of glioblastoma cells through binding with miR-422a and suppressing its activity (71). Li et al. have demonstrated down-regulation of circ_0001946 and CDR1, while up-regulation of miR‐671‐5p in glioblastoma cells. Circ_0001946 has been shown to inhibit expression of miR‐671‐5p, therefore enhancing CDR1 expression. Circ_0001946 and CDR1 decrease cell proliferation, migration, and invasion and induce apoptosis in glioblastoma cells as verified by both in vitro and in vivo assays (72). Table 4 summarizes the expression and function of circRNAs in glioblastoma.
Discussion
Both candidate gene and high throughput expression studies have reported anomalous expression of several lncRNAs in glioblastoma samples indicating the oncogenic roles for some lncRNAs and tumor suppressor roles for a number of other lncRNAs. Yet, the function of the former group of lncRNAs has been more assessed in this kind of cancer. Like other cancers, the role of lncRNAs in the pathogenesis of glioblastoma can be exerted through their effects on the expression of miRNAs. Accordingly, several lncRNA/miRNA/mRNA axes have been identified in this context among them are SNHG9/miR-199a-5p/Wnt2, MIR155HG/miR-185/ANXA2, TRG-AS1/miR-877-5p/SUZ12, LINC01579/miR‐139‐5p/EIF4G2, AC016405.3/miR-19a-5p/TET2, AC003092.1/miR-195/TFPI-2, LINC00657/miR-190a-3p/PTEN, RNCR3/miR‐185‐5p/KLF16, and MALAT1/miR-203/thymidylate synthase axes. Thus, comprehensive assessment of these three types of transcripts would facilitate identification of the molecular pathways underlying the pathogenesis of this type of cancer. Moreover, a number of recent studies revealed the role of circRNAs in regulation of expression of miRNAs, thus adding an extra level of complexity in gene regulation networks. An example of the circRNA/miRNA/mRNA functional axis in glioblastoma is represented by circ_0001946/miR‐671‐5p/CDR1.
Association between lncRNA expression levels and resistance to TMZ has been assessed in several studies. Notably, expressions of oncogenic lncRNAs lnc-TALC, LncSBF2-AS1, MALAT1, TP73-AS1, and H19 as well as expression of tumor suppressor lncRNAs AC003092.1, TUSC7, and RP11-838N2.4 have been shown to alter this phenotype in glioblastoma cells. Therefore, a panel of these lncRNAs might be applied to predict response of pateints to this chemotherapeutic agent and establish a personalized strategy for these patients.
Finally, several oncogenic and tumor suppressor lncRNAs have been identified as modulators of glioblastoma patients’ survival indicating the appropriateness of these transcripts as prognostic biomarkers. The diagnostic power of lncRNAs SAMMSON, HOTAIR, MALAT1, H19, and LINR-ROR has been assessed in serum or tissue samples of pateints with glioblastoma revealing the best results for the first two mentioned lncRNAs based on the high values of the area under the reciver operating characteristic curves. Considering the unavialbility of tissue samples for the purpose of early diagnosis and ambiguity of imaging techniques in early stages of the disease, assessment of expression of lncRNAs in serum samples provides a non-invasive method for early detection of this kind of malignant tumor.
In brief, dysregulation of several lncRNAs has been deteceted in glioblastoma cells leading to abnormal regualtion of cancer-associated pathways and cellular processes namely apoptosis, proliferation and survival. These transcripts provide promising tools for early detection of glioblastoma and prediction of patients’ prognosis and response to therapeutic choices particularly TMZ. However, a limitation of in vitro studies in this regard is that most of them has been executed using traditional serum-grown cell lines such as U87 or U251. Furhther functional in vitro and in vivo investigations are required to verify the obtained data.
Author Contributions
MT and SG-F wrote the draft and revised it. KHT, GS, and OR performed the data collection and designed the tables. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol (2016) 131(6):803–20. doi: 10.1007/s00401-016-1545-1
2. Ostrom QT, Gittleman H, Farah P, Ondracek A, Chen Y, Wolinsky Y, et al. CBTRUS statistical report: Primary brain and central nervous system tumors diagnosed in the United States in 2006-2010. Neuro-oncology (2013) 15 Suppl 2(Suppl 2):ii1–56. doi: 10.1093/neuonc/not151
3. Hanif F, Muzaffar K, Perveen K, Malhi SM, Simjee SU. Glioblastoma Multiforme: A Review of its Epidemiology and Pathogenesis through Clinical Presentation and Treatment. Asian Pac J Cancer Prev (2017) 18(1):3–9. doi: 10.22034/APJCP.2017.18.1.3
4. Braganza MZ, Kitahara CM, Berrington de González A, Inskip PD, Johnson KJ, Rajaraman P. Ionizing radiation and the risk of brain and central nervous system tumors: a systematic review. Neuro-oncology (2012) 14(11):1316–24. doi: 10.1093/neuonc/nos208
5. Network CGAR. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature (2008) 455(7216):1061. doi: 10.1038/nature07385
6. Parsons DW, Jones S, Zhang X, Lin JC-H, Leary RJ, Angenendt P, et al. An integrated genomic analysis of human glioblastoma multiforme. Science (2008) 321(5897):1807–12. doi: 10.1126/science.1164382
7. Nonoguchi N, Ohta T, Oh J-E, Kim Y-H, Kleihues P, Ohgaki H. TERT promoter mutations in primary and secondary glioblastomas. Acta Neuropathol (2013) 126(6):931–7. doi: 10.1007/s00401-013-1163-0
8. Li J, Zhu Y, Wang H, Ji X. Targeting Long Noncoding RNA in Glioma: A Pathway Perspective. Mol Ther Nucleic Acids (2018) 13:431–41. doi: 10.1016/j.omtn.2018.09.023
9. Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet (2009) 10(3):155–9. doi: 10.1038/nrg2521
10. Han M, Wang S, Fritah S, Wang X, Zhou W, Yang N, et al. Interfering with long non-coding RNA MIR22HG processing inhibits glioblastoma progression through suppression of Wnt/β-catenin signalling. Brain (2020) 143(2):512–30. doi: 10.1093/brain/awz406
11. Chen L, Gong X, Huang M. YY1-activated long noncoding RNA SNHG5 promotes Glioblastoma cell proliferation through p38/MAPK signaling pathway. Cancer Biother Radiopharm (2019) 34(9):589–96. doi: 10.1089/cbr.2019.2779
12. Zhang H, Qin D, Jiang Z, Zhang J. SNHG9/miR-199a-5p/Wnt2 axis regulates cell growth and aerobic glycolysis in glioblastoma. J Neuropathol Exp Neurol (2019) 78(10):939–48. doi: 10.1093/jnen/nlz078
13. Xie J, Wang X, Liu S, Chen C, Jiang F, Mao K, et al. LncRNA SAMMSON overexpression distinguished glioblastoma patients from patients with diffuse neurosarcoidosis. NeuroReport (2019) 30(12):817–21. doi: 10.1097/WNR.0000000000001278
14. Bountali A, Tonge DP, Mourtada-Maarabouni M. RNA sequencing reveals a key role for the long non-coding RNA MIAT in regulating neuroblastoma and glioblastoma cell fate. Int J Biol Macromol (2019) 130:878–91. doi: 10.1016/j.ijbiomac.2019.03.005
15. Voce DJ, Bernal GM, Wu L, Crawley CD, Zhang W, Mansour NM, et al. Temozolomide treatment induces lncRNA MALAT1 in an NF-κB and p53 codependent manner in glioblastoma. Cancer Res (2019) 79(10):2536–48. doi: 10.1158/0008-5472.CAN-18-2170
16. He Z, You C, Zhao D. Long non-coding RNA UCA1/miR-182/PFKFB2 axis modulates glioblastoma-associated stromal cells-mediated glycolysis and invasion of glioma cells. Biochem Biophys Res Commun (2018) 500(3):569–76. doi: 10.1016/j.bbrc.2018.04.091
17. Fan Z, Zheng J, Xue Y, Liu X, Wang D, Yang C, et al. NR2C2-uORF targeting UCA1-miR-627-5p-NR2C2 feedback loop to regulate the malignant behaviors of glioma cells. Cell Death Dis (2018) 9(12):1–18. doi: 10.1038/s41419-018-1149-x
18. Wang J, Quan X, Peng D, Hu G. Long non−coding RNA DLEU1 promotes cell proliferation of glioblastoma multiforme. Mol Med Rep (2019) 20(2):1873–82. doi: 10.3892/mmr.2019.10428
19. Xie H, Shi S, Chen Q, Chen Z. LncRNA TRG-AS1 promotes glioblastoma cell proliferation by competitively binding with miR-877-5p to regulate SUZ12 expression. Pathol-Res Pract (2019) 215(8):152476. doi: 10.1016/j.prp.2019.152476
20. Chai Y, Xie M. LINC01579 promotes cell proliferation by acting as a ceRNA of miR-139-5p to upregulate EIF4G2 expression in glioblastoma. J Cell Physiol (2019) 234(12):23658–66. doi: 10.1002/jcp.28933
21. Luo W, Li X, Song Z, Zhu X, Zhao S. Long non-coding RNA AGAP2-AS1 exerts oncogenic properties in glioblastoma by epigenetically silencing TFPI2 through EZH2 and LSD1. Aging (Albany NY) (2019) 11(11):3811. doi: 10.18632/aging.102018
22. Wu P, Cai J, Chen Q, Han B, Meng X, Li Y, et al. Lnc-TALC promotes O 6-methylguanine-DNA methyltransferase expression via regulating the c-Met pathway by competitively binding with miR-20b-3p. Nat Commun (2019) 10(1):1–15. doi: 10.1038/s41467-019-10025-2
23. Zhang SY, Huang SH, Gao SX, Wang YB, Jin P, Lu FJ. Upregulation of lncRNA RMRP promotes the activation of cardiac fibroblasts by regulating miR−613. Mol Med Rep (2019) 20(4):3849–57. doi: 10.3892/mmr.2019.10634
24. Gao X, He H, Zhu X, Xie S, Cao Y. LncRNA SNHG20 promotes tumorigenesis and cancer stemness in glioblastoma via activating PI3K/Akt/mTOR signaling pathway. Neoplasma (2019) 2019:532–42. doi: 10.4149/neo_2018_180829N656
25. Li H, Yuan X, Yan D, Li D, Guan F, Dong Y, et al. Long non-coding RNA MALAT1 decreases the sensitivity of resistant glioblastoma cell lines to temozolomide. Cell Physiol Biochem (2017) 42(3):1192–201. doi: 10.1159/000478917
26. Cai T, Liu Y, Xiao J. Long noncoding RNA MALAT 1 knockdown reverses chemoresistance to temozolomide via promoting micro RNA-101 in glioblastoma. Cancer Med (2018) 7(4):1404–15. doi: 10.1002/cam4.1384
27. Chen W, Xu X-K, Li J-L, Kong K-K, Li H, Chen C, et al. MALAT1 is a prognostic factor in glioblastoma multiforme and induces chemoresistance to temozolomide through suppressing miR-203 and promoting thymidylate synthase expression. Oncotarget (2017) 8(14):22783. doi: 10.18632/oncotarget.15199
28. Wu W, Yu T, Wu Y, Tian W, Zhang J, Wang Y. The miR155HG/miR-185/ANXA2 loop contributes to glioblastoma growth and progression. J Exp Clin Cancer Res (2019) 38(1):1–14. doi: 10.1186/s13046-019-1132-0
29. Mazor G, Levin L, Picard D, Ahmadov U, Carén H, Borkhardt A, et al. The lncRNA TP73-AS1 is linked to aggressiveness in glioblastoma and promotes temozolomide resistance in glioblastoma cancer stem cells. Cell Death Dis (2019) 10(3):1–14. doi: 10.1038/s41419-019-1477-5
30. Toraih EA, El-Wazir A, Hussein MH, Khashana MS, Matter A, Fawzy MS, et al. Expression of long intergenic non-coding RNA, regulator of reprogramming, and its prognostic value in patients with glioblastoma. Int J Biol Markers (2019) 34(1):69–79. doi: 10.1177/1724600818814459
31. Chen X, Li L, Qiu X, Wu H. Long non-coding RNA HOXB-AS1 promotes proliferation, migration and invasion of glioblastoma cells via HOXB-AS1/miR-885-3p/HOXB2 axis. Neoplasma (2019) 2019. doi: 10.4149/neo_2018_180606N377
32. Chen H, Zong J, Wang S. LncRNA GAPLINC promotes the growth and metastasis of glioblastoma by sponging miR-331-3p. Eur Rev Med Pharmacol Sci (2019) 23(1):262–70. doi: 10.26355/eurrev_201901_16772
33. Liao K, Ma X, Chen B, Lu X, Hu Y, Lin Y, et al. Upregulated AHIF-mediated radioresistance in glioblastoma. Biochem Biophys Res Commun (2019) 509(2):617–23. doi: 10.1016/j.bbrc.2018.12.136
34. Dai X, Liao K, Zhuang Z, Chen B, Zhou Z, Zhou S, et al. AHIF promotes glioblastoma progression and radioresistance via exosomes. Int J Oncol (2019) 54(1):261–70. doi: 10.3892/ijo.2018.4621
35. Tian Y, Zheng Y, Dong X. AGAP2-AS1 serves as an oncogenic lncRNA and prognostic biomarker in glioblastoma multiforme. J Cell Biochem (2019) 120(6):9056–62. doi: 10.1002/jcb.28180
36. Fawzy MS, Ellawindy A, Hussein MH, Khashana MS, Darwish MK, Abdel-Daim MM, et al. Long noncoding RNA H19, and not microRNA miR-326, is over-expressed and predicts survival in glioblastoma. Biochem Cell Biol (2018) 96(6):832–9. doi: 10.1139/bcb-2018-0122
37. Wu W, Hu Q, Nie E, Yu T, Wu Y, Zhi T, et al. Hypoxia induces H19 expression through direct and indirect Hif-1α activity, promoting oncogenic effects in glioblastoma. Sci Rep (2017) 7:45029. doi: 10.1038/srep45029
38. Jiang X, Yan Y, Hu M, Chen X, Wang Y, Dai Y, et al. Increased level of H19 long noncoding RNA promotes invasion, angiogenesis, and stemness of glioblastoma cells. J Neurosurg (2016) 124(1):129–36. doi: 10.3171/2014.12.JNS1426
39. Li W, Jiang P, Sun X, Xu S, Ma X, Zhan R. Suppressing H19 modulates tumorigenicity and stemness in U251 and U87MG glioma cells. Cell Mol Neurobiol (2016) 36(8):1219–27. doi: 10.1007/s10571-015-0320-5
40. Cai J, Zhang J, Wu P, Yang W, Ye Q, Chen Q, et al. Blocking LINC00152 suppresses glioblastoma malignancy by impairing mesenchymal phenotype through the miR-612/AKT2/NF-κB pathway. J Neuro-Oncol (2018) 140(2):225–36. doi: 10.1007/s11060-018-2951-0
41. Reon BJ, Karia BTR, Kiran M, Dutta A. LINC00152 promotes invasion through a 3′-hairpin structure and associates with prognosis in glioblastoma. Mol Cancer Res (2018) 16(10):1470–82. doi: 10.1158/1541-7786.MCR-18-0322
42. Liu X, Zhao H, Luo Y, Ma X, Xu M. LncRNA LINC00152 promoted glioblastoma progression through targeting the miR-107 expression. Environ Sci Pollut Res (2018) 25(18):17674–81. doi: 10.1007/s11356-018-1784-x
43. Liu C, Fu H, Liu X, Lei Q, Zhang Y, She X, et al. LINC00470 Coordinates the Epigenetic Regulation of ELFN2 to Distract GBM Cell Autophagy. Mol Ther (2018) 26(9):2267–81. doi: 10.1016/j.ymthe.2018.06.019
44. Liu C, Zhang Y, She X, Fan L, Li P, Feng J, et al. A cytoplasmic long noncoding RNA LINC00470 as a new AKT activator to mediate glioblastoma cell autophagy. J Hematol Oncol (2018) 11(1):77. doi: 10.1186/s13045-018-0619-z
45. Zhang L, Wang Q, Wang F, Zhang X, Tang Y, Wang S. LncRNA LINC01446 promotes glioblastoma progression by modulating miR-489-3p/TPT1 axis. Biochem Biophys Res Commun (2018) 503(3):1484–90. doi: 10.1016/j.bbrc.2018.07.067
46. Zhou Y, Dai W, Wang H, Pan H, Wang Q. Long non-coding RNA CASP5 promotes the malignant phenotypes of human glioblastoma multiforme. Biochem Biophys Res Commun (2018) 500(4):966–72. doi: 10.1016/j.bbrc.2018.04.217
47. Wang H, Li L, Yin L. Silencing LncRNA LOXL1-AS1 attenuates mesenchymal characteristics of glioblastoma via NF-κB pathway. Biochem Biophys Res Commun (2018) 500(2):518–24. doi: 10.1016/j.bbrc.2018.04.133
48. Gao Y, Xu Y, Wang J, Yang X, Wen L, Feng J. LncRNA MNX1-AS1 promotes glioblastoma progression through inhibition of miR-4443. Oncol Res Featuring Preclinical Clin Cancer Ther (2019) 27(3):341–7. doi: 10.3727/096504018X15228909735079
49. Tan SK, Pastori C, Penas C, Komotar RJ, Ivan ME, Wahlestedt C, et al. Serum long noncoding RNA HOTAIR as a novel diagnostic and prognostic biomarker in glioblastoma multiforme. Mol Cancer (2018) 17(1):74. doi: 10.1186/s12943-018-0822-0
50. Zhou X, Ren Y, Zhang J, Zhang C, Zhang K, Han L, et al. HOTAIR is a therapeutic target in glioblastoma. Oncotarget (2015) 6(10):8353. doi: 10.18632/oncotarget.3229
51. Zhang K, Sun X, Zhou X, Han L, Chen L, Shi Z, et al. Long non-coding RNA HOTAIR promotes glioblastoma cell cycle progression in an EZH2 dependent manner. Oncotarget (2015) 6(1):537. doi: 10.18632/oncotarget.2681
52. Ren J, Yang Y, Xue J, Xi Z, Hu L, Pan S-J, et al. Long noncoding RNA SNHG7 promotes the progression and growth of glioblastoma via inhibition of miR-5095. Biochem Biophys Res Commun (2018) 496(2):712–8. doi: 10.1016/j.bbrc.2018.01.109
53. Chen Q, Cai J, Wang Q, Wang Y, Liu M, Yang J, et al. Long noncoding RNA NEAT1, regulated by the EGFR pathway, contributes to glioblastoma progression through the WNT/β-catenin pathway by scaffolding EZH2. Clin Cancer Res (2018) 24(3):684–95. doi: 10.1158/1078-0432.CCR-17-0605
54. Gong W, Zheng J, Liu X, Ma J, Liu Y, Xue Y. Knockdown of NEAT1 restrained the malignant progression of glioma stem cells by activating microRNA let-7e. Oncotarget (2016) 7(38):62208. doi: 10.18632/oncotarget.11403
55. Su R, Cao S, Ma J, Liu Y, Liu X, Zheng J, et al. Knockdown of SOX2OT inhibits the malignant biological behaviors of glioblastoma stem cells via up-regulating the expression of miR-194-5p and miR-122. Mol Cancer (2017) 16(1):1–22. doi: 10.1186/s12943-017-0737-1
56. Cai H, Liu X, Zheng J, Xue Y, Ma J, Li Z, et al. Long non-coding RNA taurine upregulated 1 enhances tumor-induced angiogenesis through inhibiting microRNA-299 in human glioblastoma. Oncogene (2017) 36(3):318–31. doi: 10.1038/onc.2016.212
57. Mineo M, Ricklefs F, Rooj AK, Lyons SM, Ivanov P, Ansari KI, et al. The long non-coding RNA HIF1A-AS2 facilitates the maintenance of mesenchymal glioblastoma stem-like cells in hypoxic niches. Cell Rep (2016) 15(11):2500–9. doi: 10.1016/j.celrep.2016.05.018
58. Yao Y, Ma J, Xue Y, Wang P, Li Z, Liu J, et al. Knockdown of long non-coding RNA XIST exerts tumor-suppressive functions in human glioblastoma stem cells by up-regulating miR-152. Cancer Lett (2015) 359(1):75–86. doi: 10.1016/j.canlet.2014.12.051
59. Cao Y, Wang P, Ning S, Xiao W, Xiao B, Li X. Identification of prognostic biomarkers in glioblastoma using a long non-coding RNA-mediated, competitive endogenous RNA network. Oncotarget (2016) 7(27):41737. doi: 10.18632/oncotarget.9569
60. Tang G, Luo L, Zhang J, Zhai D, Huang D, Yin J, et al. lncRNA LINC01057 promotes mesenchymal differentiation by activating NF-κB signaling in glioblastoma. Cancer Lett (2020) 498:152–64. doi: 10.1016/j.canlet.2020.10.047
61. Toraih EA, Alghamdi SA, El-Wazir A, Hosny MM, Hussein MH, Khashana MS, et al. Dual biomarkers long non-coding RNA GAS5 and microRNA-34a co-expression signature in common solid tumors. PLoS One (2018) 13(10):e0198231. doi: 10.1371/journal.pone.0198231
62. Ren S, Xu Y. AC016405. 3, a novel long noncoding RNA, acts as a tumor suppressor through modulation of TET2 by microRNA-19a-5p sponging in glioblastoma. Cancer Sci (2019) 110(5):1621. doi: 10.1111/cas.14002
63. Chu L, Yu L, Liu J, Song S, Yang H, Han F, et al. Long intergenic non-coding LINC00657 regulates tumorigenesis of glioblastoma by acting as a molecular sponge of miR-190a-3p. Aging (Albany NY) (2019) 11(5):1456. doi: 10.18632/aging.101845
64. Xu N, Liu B, Lian C, Doycheva DM, Fu Z, Liu Y, et al. Long noncoding RNA AC003092. 1 promotes temozolomide chemosensitivity through miR-195/TFPI-2 signaling modulation in glioblastoma. Cell Death Dis (2018) 9(12):1–16. doi: 10.1038/s41419-018-1183-8
65. Zhang L, Cao Y, Wei M, Jiang X, Jia D. Long noncoding RNA-RNCR3 overexpression deleteriously affects the growth of glioblastoma cells through miR-185-5p/Krüppel-like factor 16 axis. J Cell Biochem (2018) 119(11):9081–9.
66. Liu J, Wang W, Zhang X, Du Q, Li H, Zhang Y. Effect of downregulated lncRNA NBAT1 on the biological behavior of glioblastoma cells. Eur Rev Med Pharmacol Sci (2018) 22(9):2715–22. doi: 10.26355/eurrev_201805_14968
67. Shang C, Tang W, Pan C, Hu X, Hong Y. Long non-coding RNA TUSC7 inhibits temozolomide resistance by targeting miR-10a in glioblastoma. Cancer Chemother Pharmacol (2018) 81(4):671–8. doi: 10.1007/s00280-018-3522-y
68. Liu S, Mitra R, Zhao M-M, Fan W, Eischen CM, Yin F, et al. The potential roles of long noncoding RNAs (lncRNA) in glioblastoma development. Mol Cancer Ther (2016) 15(12):2977–86. doi: 10.1158/1535-7163.MCT-16-0320
69. Liu Y, Xu N, Liu B, Huang Y, Zeng H, Yang Z, et al. Long noncoding RNA RP11-838N2. 4 enhances the cytotoxic effects of temozolomide by inhibiting the functions of miR-10a in glioblastoma cell lines. Oncotarget (2016) 7(28):43835. doi: 10.18632/oncotarget.9699
70. Shen J, Hodges TR, Song R, Gong Y, Calin GA, Heimberger AB, et al. Serum HOTAIR and GAS5 levels as predictors of survival in patients with glioblastoma. Mol Carcinog (2018) 57(1):137–41. doi: 10.1002/mc.22739
71. Wang R, Zhang S, Chen X, Li N, Li J, Jia R, et al. CircNT5E acts as a sponge of miR-422a to promote glioblastoma tumorigenesis. Cancer Res (2018) 78(17):4812–25. doi: 10.1158/0008-5472.CAN-18-0532
72. Li X, Diao H. Circular RNA circ_0001946 acts as a competing endogenous RNA to inhibit glioblastoma progression by modulating miR-671-5p and CDR1. J Cell Physiol (2019) 234(8):13807–19. doi: 10.1002/jcp.28061
73. Qi X, Yu XJ, Wang XM, Song TN, Zhang J, Guo XZ, et al. Knockdown of KCNQ1OT1 Suppresses Cell Invasion and Sensitizes Osteosarcoma Cells to CDDP by Upregulating DNMT1-Mediated Kcnq1 Expression. Mol Ther Nucleic Acids (2019) 17:804–18. doi: 10.1016/j.omtn.2019.06.010
74. Zhang X, Zhong B, Zhang W, Wu J, Wang Y. Circular RNA CircMTO1 inhibits proliferation of Glioblastoma cells via miR-92/WWOX signaling pathway. Med Sci Monit: Int Med J Exp Clin Res (2019) 25:6454. doi: 10.12659/MSM.918676
75. Lv X, Wang M, Qiang J, Guo S. Circular RNA circ-PITX1 promotes the progression of glioblastoma by acting as a competing endogenous RNA to regulate miR-379–5p/MAP3K2 axis. Eur J Pharmacol (2019) 863:172643. doi: 10.1016/j.ejphar.2019.172643
76. Lei B, Huang Y, Zhou Z, Zhao Y, Thapa AJ, Li W, et al. Circular RNA hsa_circ_0076248 promotes oncogenesis of glioma by sponging miR-181a to modulate SIRT1 expression. J Cell Biochem (2019) 120(4):6698–708. doi: 10.1002/jcb.27966
77. Wang R, Zhang S, Chen X, Li N, Li J, Jia R, et al. EIF4A3-induced circular RNA MMP9 (circMMP9) acts as a sponge of miR-124 and promotes glioblastoma multiforme cell tumorigenesis. Mol Cancer (2018) 17(1):1–12. doi: 10.1186/s12943-018-0911-0
Keywords: lncRNA, circRNA, glioblastoma, expression, polymorphism
Citation: Rezaei O, Tamizkar KH, Sharifi G, Taheri M and Ghafouri-Fard S (2021) Emerging Role of Long Non-Coding RNAs in the Pathobiology of Glioblastoma. Front. Oncol. 10:625884. doi: 10.3389/fonc.2020.625884
Received: 04 November 2020; Accepted: 22 December 2020;
Published: 03 February 2021.
Edited by:
Yaohua Liu, Shanghai First People’s Hospital, ChinaReviewed by:
Maite Verreault, INSERM U1127 Institut du Cerveau et de la Moelle épinière (ICM), FranceKristin Huntoon, University of Texas MD Anderson Cancer Center, United States
Copyright © 2021 Rezaei, Tamizkar, Sharifi, Taheri and Ghafouri-Fard. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mohammad Taheri, bW9oYW1tYWRfODIzQHlhaG9vLmNvbQ==; Soudeh Ghafouri-Fard, cy5naGFmb3VyaWZhcmRAc2JtdS5hYy5pcg==
 Omidvar Rezaei1
Omidvar Rezaei1 Kasra Honarmand Tamizkar
Kasra Honarmand Tamizkar Guive Sharifi
Guive Sharifi Mohammad Taheri
Mohammad Taheri