- Institute of Pathology, Faculty of Medicine, University of Ljubljana, Ljubljana, Slovenia
There is emerging evidence suggesting that epithelial-mesenchymal transition (EMT) and mesenchymal-epithelial transition (MET) play an important role in colorectal carcinoma (CRC), but their exact role remains controversial. Our aim was to analyze the miR-200 family as EMT markers and their target genes expression at invasive tumor front and in nodal and liver metastases. Sixty-three formalin-fixed paraffin-embedded tissue samples from 19 patients with CRC were included. Using a micropuncture technique, tissue was obtained from central part and invasive front of the primary tumor, and nodal and liver metastases. Expression of the miR-200 family and their target genes CDKN1B, ONECUT2, PTPN13, RND3, SOX2, TGFB2 and ZEB2 was analyzed using real-time PCR. We found miR-200 family down-regulation at invasive front compared to central part, and up-regulation of miRNA-200a/b/c and miR-429 in metastases compared to invasive front. At invasive front, TGFB2 was the only gene with inverse expression to the miR-200 family, whereas in metastases inverse expression was found for ONECUT2 and SOX2. CDKN1B, PTPN13 and ZEB2 were down-regulated at invasive front and up-regulated in metastases. Our results suggest the involvement of partial EMT at invasive tumor front, and partial MET in metastases in CRC, based on miR-200 family and its target genes expression.
Introduction
Significant progress has been made in recent decades in our understanding of cancer development, progression and metastasizing. Among pathogenetic mechanisms of cancerogenesis, epithelial-mesenchymal transition (EMT) has gained much attention. There is emerging evidence suggesting that EMT plays an important role in human cancer, but its role remains controversial (1–3).
EMT is a fundamental process during embryogenesis and is later physiologically silenced. In adult life, EMT can be reactivated in various conditions, such as wound healing, fibrosis and cancer (4, 5). During EMT, epithelial cells change from stationary polarized cells to mobile spindle cells. In cancer, this transition is assumed to be the basis for progression and metastasis, enabling cells to migrate from the primary tumor (6, 7). Once disseminated to a distant site, cancer cells are believed to regain epithelial properties in a reverse process, referred to as mesenchymal-epithelial transition (MET) (8, 9). Complete EMT includes a switch in intermediate filament expression, an alteration in intercellular junction composition and a concurrent change of cell morphology (10, 11). It appears that full EMT rarely occurs in human cancer though it does exist, e.g., in spindle cell carcinoma, which can be regarded as a “positive” control when studying the markers of EMT (12). Markers of EMT include classical and desmosomal cadherins, intermediate filaments and transcription factors, but their expression patterns can be variable in diseases in which EMT presumably plays an important role (13, 14).
In the majority of human cancer types, however, the morphologic criterion of a complete shift from an epithelial to a spindle phenotype during EMT is seldom met. Instead, cancer progression for the majority of cancers might only depend on partial EMT (15, 16). Cells undergoing partial EMT are difficult to identify due to their phenotypic heterogeneity, the transient and reversible nature of EMT and variable expression of EMT markers (15). Evidence of partial EMT often relies on activation of EMT transcription factors and their regulatory molecules, such as microRNAs (3, 17, 18). Previous studies suggest that miR-200 family is among the most sensitive markers of EMT and can be used to study full and partial EMT (12, 19–23).
A few studies have suggested that partial MET might occur at the metastatic site (24, 25) but the dynamics of EMT and MET in cancerogenesis is still poorly understood. The aim of this study was to analyze the expression of the miR-200 family and their target genes as markers of EMT in locations crucial for the progression of colorectal carcinoma (CRC), i.e., the central part of the tumor and invasive tumor front, as well as lymph node and liver metastases. The dynamics of EMT markers expression profiles might provide insight into the possible role of EMT/MET in cancer progression and metastasis formation.
Materials and methods
Patients and Tissue Samples
This retrospective study was approved by the National Medical Ethics Committee, Republic of Slovenia, Ministry of Health (reference number 0120-88/2020/3) and carried out following the rules of the Declaration of Helsinki. All authors had access to the study data and had reviewed and approved the final manuscript.
The study included patients with CRC who had been treated surgically, including resection of the primary tumor and regional lymph nodes, as well as liver metastases if present. Patients were divided into three groups: patients with CRC and lymph node metastases (N+ M0), patients with CRC and liver metastases but without lymph node metastases (N0 M+) and patients with CRC and both lymph node and liver metastases (N+ M+). Patients with neoadjuvant radiochemotherapy were excluded from the study. None of the patients had microsatellite unstable cancer.
After surgery, resection specimens were dealt with according to standard procedures. After 24-hour fixation in formalin, representative samples from the tumor, all lymph nodes and resection margins were taken, 3-4 µm-thick sections were cut and stained with eosin and hematoxylin. pTNM stage was determined after the primary resection (26).
For the purposes of this study, all slides were re-examined. Tumor budding was determined according to recommendations as Bd1-Bd3 (Bd1 0-4 buds/0.785 mm2, Bd2 5-9 buds/0.785 mm2, Bd3 ≥10 buds/0.785 mm2) (27). Poorly differentiated (PDC) were analyzed and graded as G1-G3 (G1: <5; G2: 5-9; G3: ≥ 10 PDC in a 20x objective lens field) (28, 29). Representative slides of the tumor and metastases were selected and corresponding paraffin blocks were retrieved from the archives of the Institute of Pathology, Faculty of Medicine, University of Ljubljana. Tissue was obtained from four locations using micropuncture technique, using a 0.6 mm needle: central part of the primary tumor, invasive tumor front and lymph node and liver metastases if present.
RNA Isolation
RNA Isolation From Formalin-Fixed Paraffin Embedded Tissue Samples
For the isolation procedure, 3 punches were made at each location. Total RNA isolation was performed using a MagMax FFPE DNA/RNA Ultra kit (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s protocol with a modification. Protease digestion was performed overnight at 56°C with shaking for 15 s at 300 rpm every 4 min. The concentration and quality of the isolates were assessed with a spectrophotometer ND-1000 or ND-One (Nanodrop, Thermo Fisher Scientific, Waltham, MA, USA) at the wavelengths 260, 280 and 230 nm.
RNA Quality Assessment
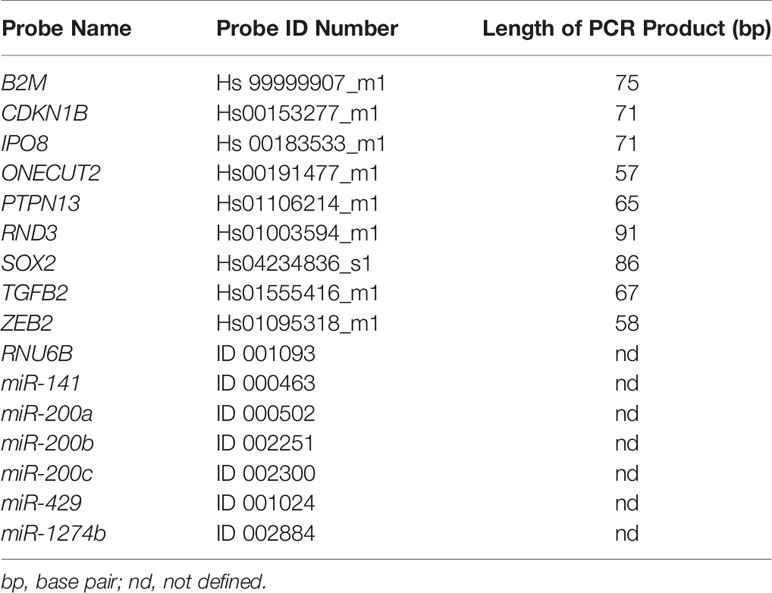
As quality control, reverse transcription (RT) of RNU6B, a housekeeping small nuclear RNA gene, was used followed by amplification using quantitative real-time PCR (qPCR) and TaqMan methodology (Thermo Fisher Scientific, Waltham, MA, USA). All of the samples included in the study had passed this initial quality control and those that did not amplify were not included in the study. Positive and negative amplification of RNU6B was in 100% correlation with positive and negative amplification of GAPDH (100 bp) used as initially quality control in previous research (data not shown) (19, 30). For selected genes, we chose TaqMan primers and probes that amplify and detect PCR products less than 100 bp long (Table 1).
Efficiency Testing
A pre-designed mixture of probes and primers specific for miRNAs or target gene (mRNAs) expression was used. Prior to qPCR, three pools of RNA samples were created, obtained from primary tumor, lymph node and liver metastases. After RT, the cDNA of miRNAs and pre-amplified cDNA of mRNAs was diluted in four steps, ranging from 5-point dilution to 625-point dilution, and the probes were tested for qPCR efficiency. All the qPCR efficiency reactions were performed on a RotorGene Q (Qiagen, Hilden, Germany) in triplicate.
Analysis of Expression Of mir-200 Family
Reverse Transcription (RT)
Looped primers for specific reverse transcription (RT) of miRNAs and a MicroRNA TaqMan RT kit (Applied Biosystems, Foster City, CA, USA) were utilized following the manufacturer’s protocol. RNU6B and miR-1247b were used as reference genes (RGs). MicroRNAs, miR-141, miR-200a, miR-200b, miR-200c and miR-429 were tested relative to the geometric mean of expression of RNU6B and miR-1247b (Table 1). Briefly, a 10 μL RT reaction master mix was performed with 10 ng of total RNA sample, 1.0 μL of MultiScribe Reverse Transcriptase (50 U/μL), 1.0 μL of Reverse Transcription Buffer (10×), 0.1 μL of dNTP (100 mM), 0.19 μL RNAase inhibitor (20 U/μL), and 2.0 μL of RT primer (5×). The reaction conditions were: 16°C for 30 min, 42°C for 30 min, 85°C for 5 min.
Quantitative Real-Time PCR (qPCR)
qPCR for miRNAs was carried out in a 10 μL PCR master mix containing 5.0 μL TaqMan 2× FastStart Essential DNA Probe Master (Roche, Basel, Switzerland), 0.5 μL TaqMan assay 20x, and 4.5 μL RT products diluted 100-fold. The qPCR reactions were performed on a RotorGene Q (Qiagen, Hilden, Germany) in duplicate, as follows: initial denaturation at 95°C for 10 min, 40 cycles for 15 s at 95°C (denaturation) and for 60 s at 60°C (primers annealing and elongation). The signal was collected at the endpoint of every cycle.
Analysis of Expression of miR-200 Family Target Genes
Reverse Transcription (RT)
Target mRNAs of the miR-200 family, CDKN1B, ONECUT2, PTPN13, RND3, SOX2, TGFB2, and ZEB2 (Table 1), were analyzed relatively to the geometric mean of RGs, IPO8 and B2M. mRNAs were reverse transcribed using a OneTaq RT-PCR Kit (New England Biolabs, Ipswich, MA, USA) using random primers according to the manufacturer’s instructions. Reverse transcription reactions were started with 3.0 µL (14-60 ng) of total RNA and 1.0 µL of Random Primer Mix incubated at 70°C for 5 min. The 10 μL RT master mix included 5.0 μL of M-MuLV Reaction Mix, 1.0 μL of M-MuLV reverse transcriptase and 4.0 μL of reaction mix after random priming. The reaction conditions were: 25°C for 5 min, 42°C for 60 min and 80°C for 4 min.
Pre-Amplification and Quantitative Real-Time PCR (qPCR)
Following RT, pre-amplification was performed using a TaqMan PreAmp Master Mix (Applied Biosystems, Foster City, CA, USA) in 10 µL according to the manufacturer’s protocol. The resulting PreAmp reaction was diluted 5-fold in all cases, except when investigating lymph node metastases, where it was diluted 25-fold. In the qPCR reaction 4.5 μL of the diluted sample was used in a 10 μL reaction volume with a 5.0 μL of 2x FastStart Essential DNA Probe Master Mix (Roche, Basel, Switzerland) and 0.5 μL of TaqMan 20X probe. Thermal conditions were applied as follows: 50°C for 2 min, initial denaturation at 95°C for 10 min and 40 cycles of denaturation at 95°C for 15 s and annealing at 60°C for 1 min. All qPCR analyses were performed on a Rotor Gene Q (Qiagen, Hilden, Germany) in duplicate. The signal was collected at the endpoint of each cycle.
Statistical Analysis of Experimental Data
The results were presented as relative gene expression. All Cqs were corrected for PCR efficiencies and the expression of the gene of interest (GOI, CqGOI) was calculated relative to a geometric mean of RGs (CqRG), named ΔCq. In CRC samples, mRNAs and miRNAs expression differences were compared between the central part of the tumor and the invasive front, invasive front and lymph node metastases and/or invasive front and liver metastases groups using ΔCq and the Wilcoxon Rank test. Lymph node and liver metastases groups were compared using ΔCq data and the Mann-Whitney U test. For all correlations/associations, Spearman rank-order correlation was used. Statistical analysis of data was performed using SPSS version 24 (SPSS Inc., Chicago, IL, USA). Differences were considered significant at p ≤ 0.05.
Results
Patients and Tissue Samples
In total, we analyzed 63 tissue samples from 19 patients (13 males and 6 females) with CRC with lymph node and/or liver metastases. Nine patients had liver metastases at the time of the initial presentation and resection of the primary tumor, whereas 3 patients developed liver metastases later in the disease course and underwent additional surgical resection of the liver metastases (5, 11 and 44 months after resection of the primary tumor). pTNM stage was determined after the primary resection according to the international guidelines (26). We divided the patients in 3 groups, based on the presence of lymph node and/or liver metastasis: the N+ M0 group included 7 patients (mean age 76.0 ± 13.5 years; male:female 6:1), the N0 M+ group included 3 patients (mean age 72.0 ± 6.1 years, male:female 2:1) and the group N+ M+ group included 9 patients (mean age 69.3 years ± 16.5; male:female 5:4). Tumor budding was determined as grade Bd1, Bd2 and Bd3 in 9, 8 and 2 patients respectively. PDC were graded as G1, G2 and G3 in 6, 8 and 5 patients respectively. Demographic and clinicopathologic features and information about the analyzed tissue samples are presented in Table 2.
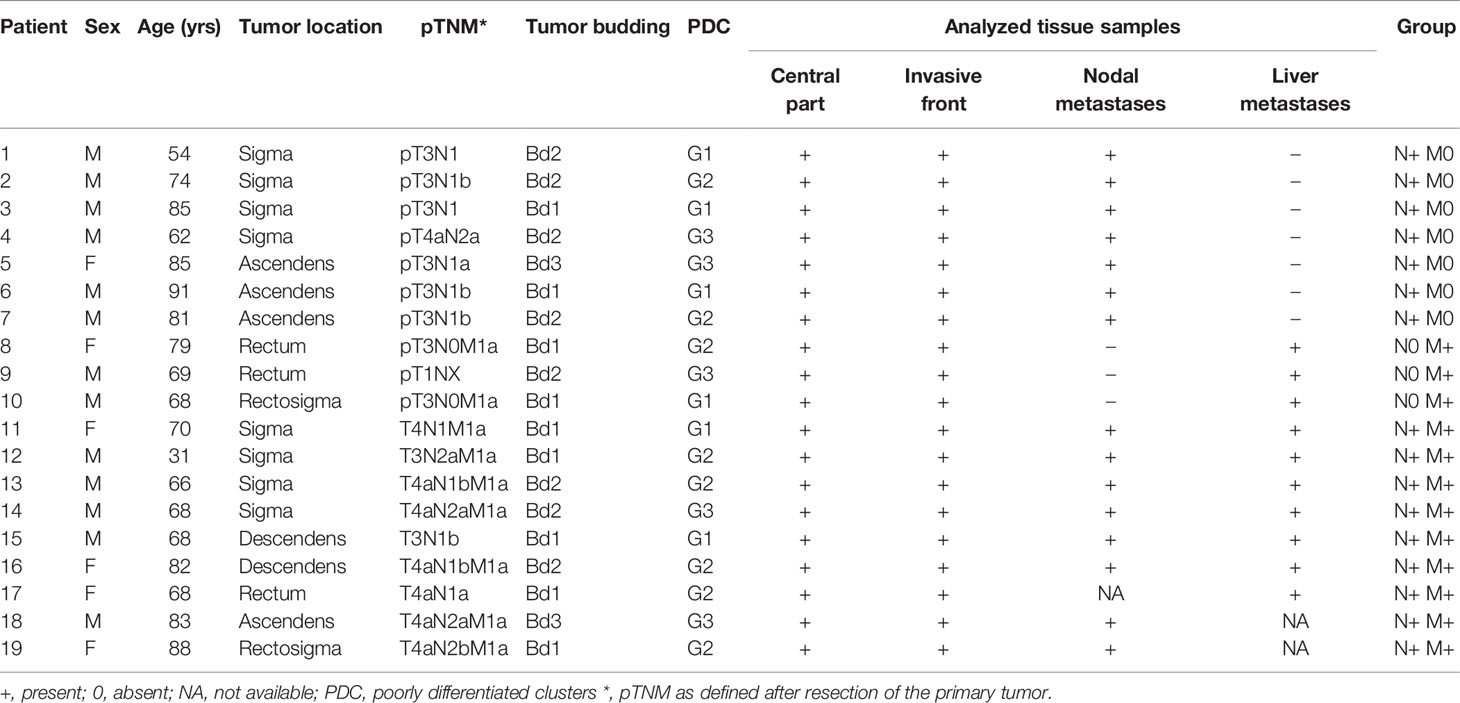
Table 2 Demographic and clinicopathologic features and information about the analyzed tissue samples.
Expression of the miR-200 Family
When comparing the invasive front to the central part of the tumor, all investigated miRNAs were down-regulated in all groups, as shown in Figure 1A.
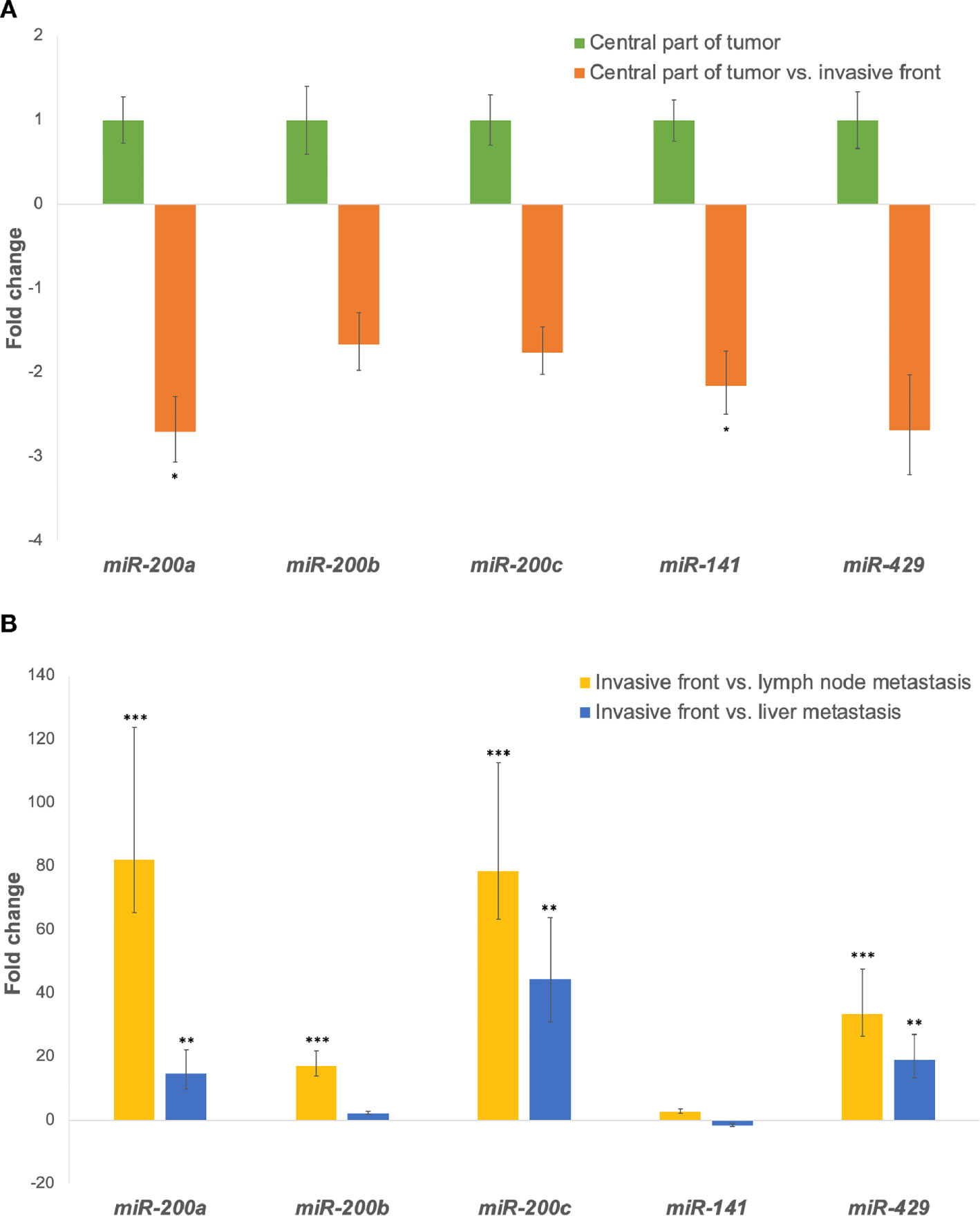
Figure 1 (A) Expression of the miR-200 family at the invasive front in comparison to the central part of the tumor. *p ≤ 0.05. (B) Expression of the miR-200 family in lymph node and/or liver metastases in comparison to the invasive tumor front. **p ≤ 0.01; ***p ≤ 0.001.
All investigated miRNAs were up-regulated in lymph node metastases compared to the invasive front. Additionally, expression of all investigated miRNAs, except miR-141, were up-regulated in liver metastases when compared to the invasive front, as shown in Figure 1B. Fold changes and p-values for all investigated comparisons are presented in Table 3.
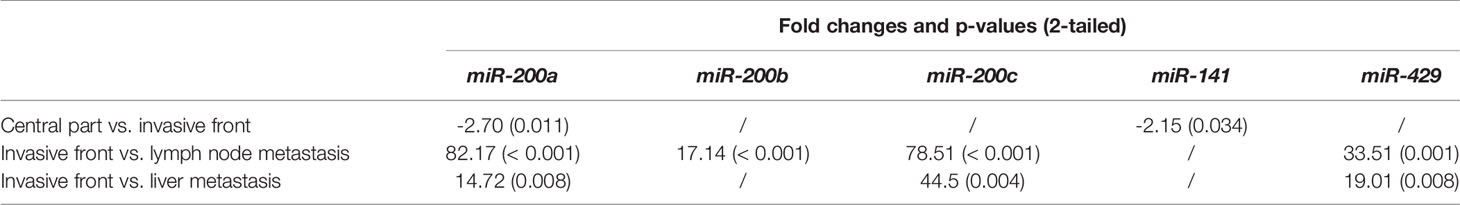
Table 3 Fold changes and p-values for all investigated comparisons for the miR-200 family with Wilcoxon signed ranks test.
When investigating the differences between lymph node and liver metastases, all miRNAs were down-regulated in liver metastases when comparing them independently to lymph node metastases. Statistically significant differences were found for miR-200a (p = 0.003), miR-200b (p = 0.001), miR-141 (p = 0.005) and miR-429 (p = 0.016), as shown in Figure 2.
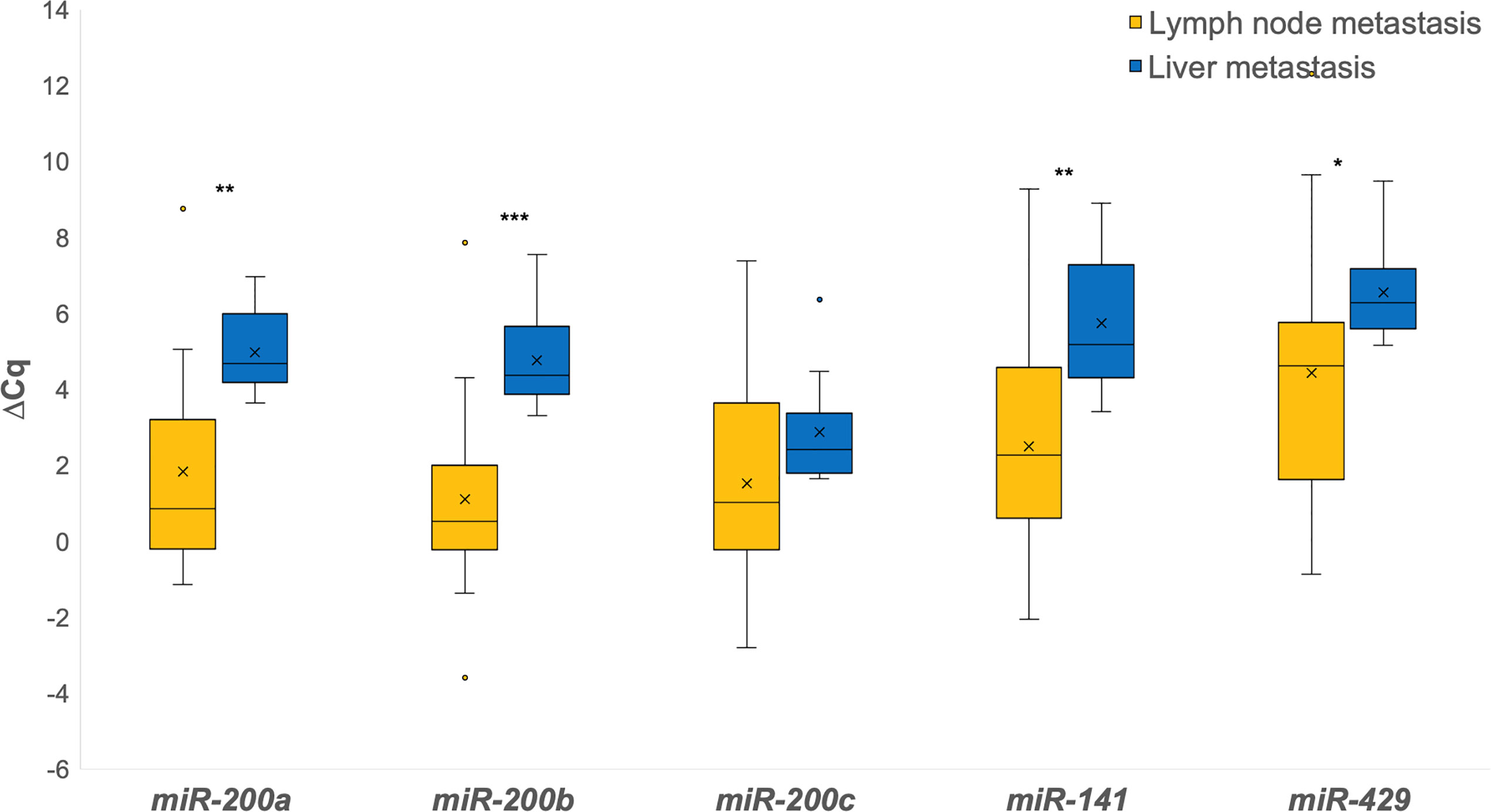
Figure 2 Expression of the miR-200 family in lymph node metastases in comparison to liver metastases. *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001.
Expression of the Target Genes of the miR-200 Family
Expression of all investigated genes between the central part of the tumor and the invasive front as fold changes is shown in Figure 3A. All investigated genes in this comparison were down-regulated, except TGFB2, which was up-regulated.
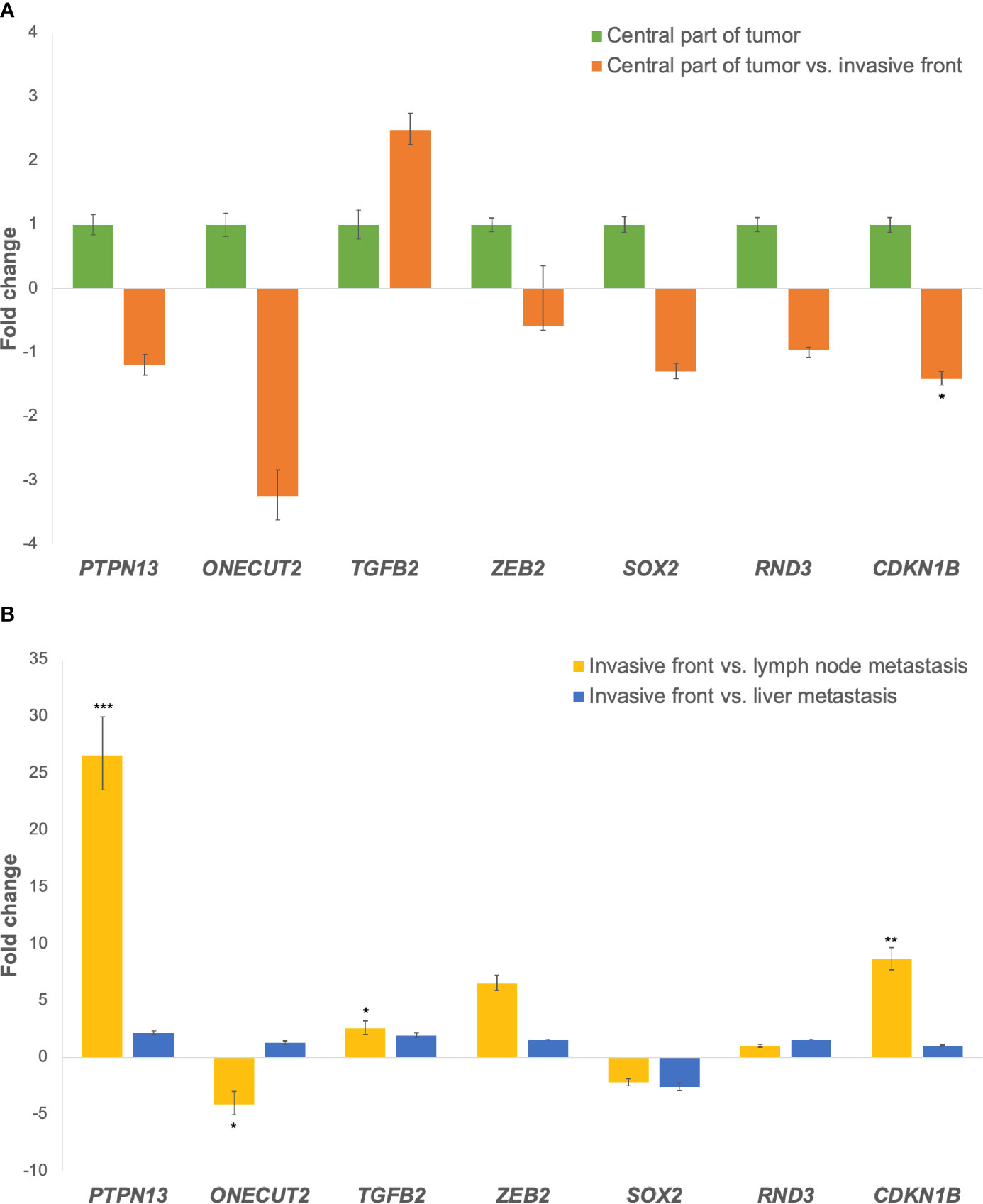
Figure 3 (A) Expression of the miR-200 family target genes at the invasive tumor front in comparison to the central part of the tumor. Legend: *p ≤ 0.05. (B) Expression of the miR-200 family target genes in lymph node and liver metastases in comparison to the invasive tumor front. Legend: *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001.
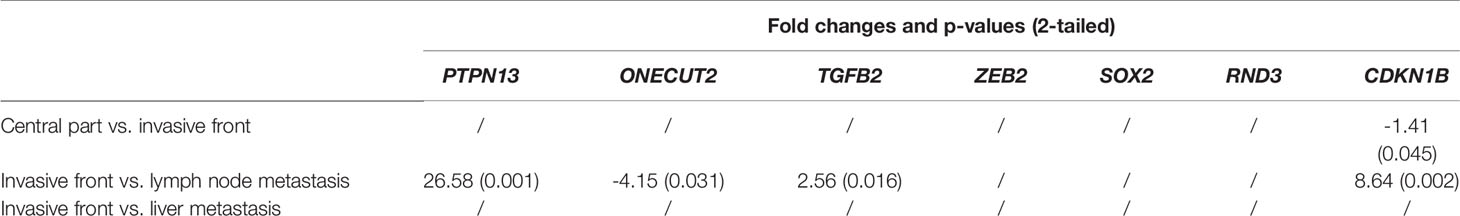
In lymph node metastases compared to the invasive front of the tumor, ONECUT2 and SOX2 were down-regulated, while PTPN13, TGFB2, ZEB2, RND3 and CDKN1B were up-regulated. When comparing liver metastases to the invasive tumor front, SOX2 was down-regulated, while PTPN13, ONECUT2, TGFB2, ZEB2, RND3 and CDKN1B were up-regulated, as shown in Figure 3B. Fold changes and p-values for all investigated comparisons among groups are shown in Table 4.
When comparing liver metastases to lymph node metastases for the investigated genes, PTPN13 (p = 0.012), ZEB2, RND3, CDKN1B (p = 0.036) were down-regulated, while ONECUT2 (p = 0.046), TGFB2, SOX2 were up-regulated in the liver metastases as shown in Figure 4.
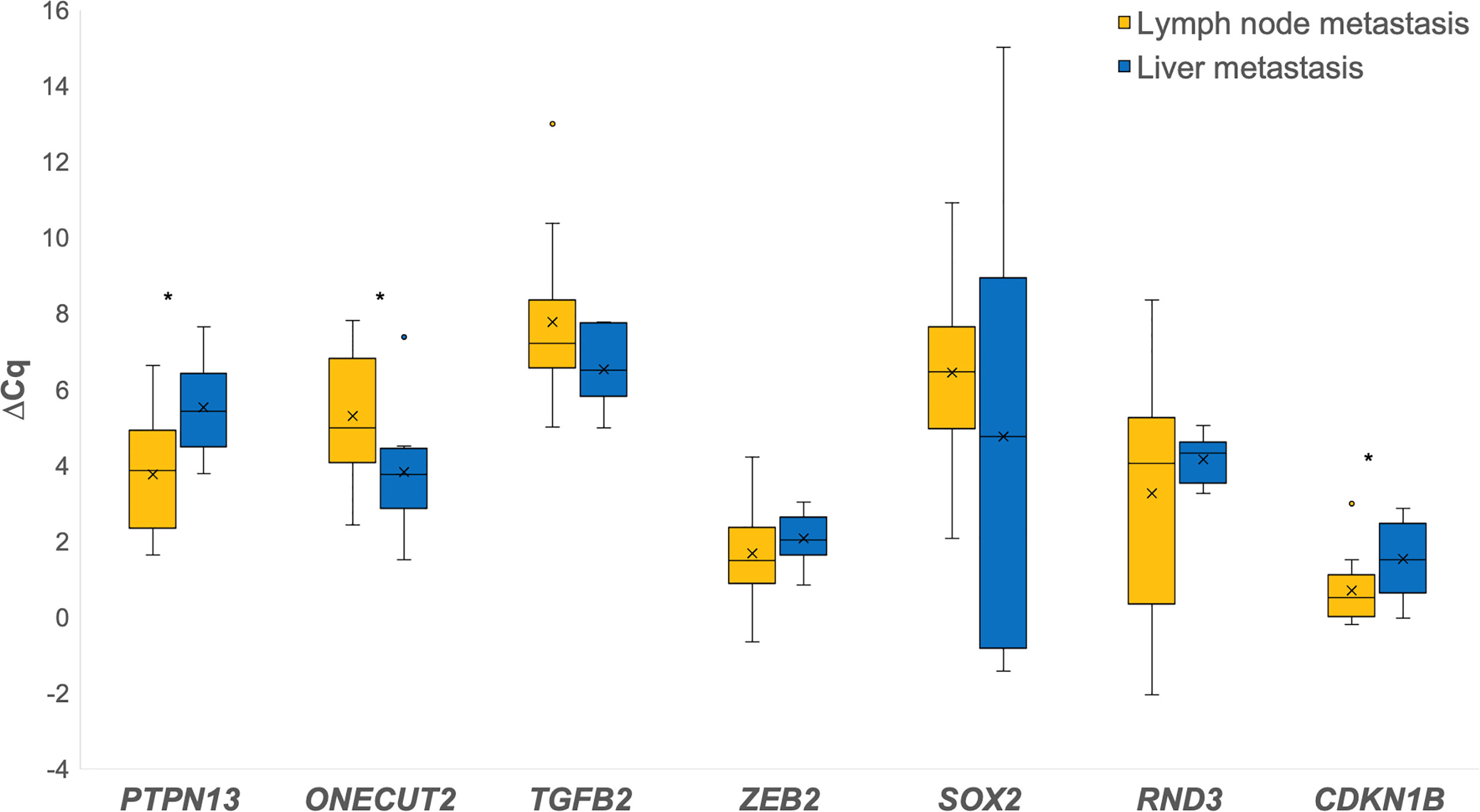
Figure 4 Expression of the investigated genes in lymph node metastases in comparison to liver metastases. *p ≤ 0.05.
Correlations Between miR-200 family and Their Target Genes
Spearman coefficient of correlation revealed moderate or weak positive correlation between miR-200a and PTPN13, ZEB2, and CDKN1B, respectively. miR-200b was in weak positive correlation with PTPN13 and ZEB2. miR-200c was in weak positive correlation with PTPN13, ZEB2 and CDKN1B. miR-141 did not correlate significantly with any of the investigated genes, while miR-429 was in weak positive correlation with ZEB2. Results are summarized in Table 5.
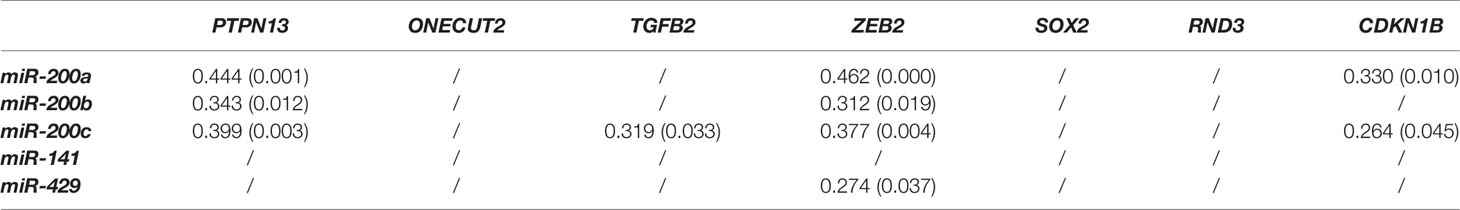
Table 5 Comparison between the miR-200 family and investigated genes expression (Spearman correlation coefficients and corresponding p-values in brackets).
Discussion
We analyzed the expression of the miR-200 family, known to be the master regulator of EMT, and their target genes, to investigate the role of EMT in CRC invasion and metastasizing. We obtained tumor tissue from locations essential for tumor growth and progression, i.e., the invasive tumor front and lymph node and/or liver metastases. The results of our study suggest the involvement of partial EMT at the invasive tumor front and the involvement of partial MET in both lymph node and liver metastases. This is based on the expression patterns of the miR-200 family in these critical locations, showing miR-200 family down-regulation at the invasive front compared to the central part of the tumor, suggesting the involvement of EMT in invasive tumor growth, and up-regulation in both lymph node and liver metastases compared to the invasive front, suggesting the involvement of MET in metastasis formation.
Similar to previous studies, we did not observe any morphologic evidence of full EMT, i.e., transition from an epithelioid to a spindle phenotype of carcinoma cells at the invasive front. Furthermore, no morphologic evidence of transition from a spindle to an epithelioid phenotype was noted in the metastases. These findings support the hypothesis that EMT and MET do play an important role in CRC growth and metastases, not as full EMT/MET but only as partial EMT/MET.
When we compared the invasive front to the central part of the tumor, all five investigated miRNAs (miR-200a/b/c, miR-141, miR-429) were down-regulated in all samples, the differences being significant for miR-200a and miR-141. Several previous studies have suggested that impaired miR-200 expression may lead to EMT initiation and eventually to cancer dissemination. Decreased expression of miR-200 has previously been reported at the invasive front of metastatic CRC (24, 25, 31–33). Specifically, Paterson et al. found miR-200a/b/c to be down-regulated at the invasive front with degraded basement membrane (25) and Knudsen et al. demonstrated miR-200b down-regulation in the tumor budding cells of CRC (31). Muto et al. also showed a statistically significant difference in miR-200c expression between the invasive front of metastatic and non-metastatic tumors (32), whereas no significant difference was observed by Jepsen et al. (33). It is believed that down-regulation of miR-200 and subsequent up-regulation of their target genes promotes a switch in cell phenotype, resulting in an increased invasiveness and metastatic potential of tumor cells.
Furthermore, we found strong up-regulation of miRNA-200a/b/c and miR-429 in both lymph node and liver metastases in comparison to the invasive tumor front. All differences were statistically significant except for miR-200b in liver metastases. Only a few previous studies have focused on the expression of EMT markers in locations critical for carcinoma progression and metastasizing. Similar to our study, Hur et al. used the miR-200 family as markers of EMT and demonstrated down-regulation of miR-200c at the CRC invasive front and up-regulation in matched liver metastases (24). They concluded that miR-200c probably plays a pivotal role in CRC metastases. Our results support their findings, since we also showed significant up-regulation of miR-200c in matched lymph node and liver metastases. In contrast, Li et al. demonstrated down-regulation of miR-200c in matched CRC liver metastases (34). Previous studies using miR-200a on matched metastases in CRC are very scarce. Cristobal et al. observed no significant difference in the expression of miR-200a between primary CRC and matched liver metastases (35). Interestingly, we found miR-200a to be a reliable marker, showing both significant down-regulation at the invasive front and up-regulation in the metastases. Paterson et al. immunostained tumors and local lymph node metastases with miR-200b and demonstrated weak staining of miR-200b at the invasive front and intense staining in metastases and vascular carcinoma deposits. Their results provide some evidence of cells undergoing EMT at the invasive front and recapitulating the phenotype of the primary tumor at metastatic sites (25).
To the best of our knowledge, this is the first study to compare the expression of the miR-200 family in CRC with matched lymph node and liver metastases. Our results suggest that partial EMT/MET is involved in metastasis formation regardless of hematogenous or lymphogenic tumor spread. Except for miR-141, all tested miRNAs from the miR-200 family showed up-regulation in both lymph node and liver metastases. miR-141 showed a completely different pattern to other miR-200, with a significant down-regulation in liver metastases and an insignificant up-regulation in the lymph node metastases. This result is different from previously reported studies. Hur et al. demonstrated a significant up-regulation of miR-141 in liver metastases (24), whereas no significant change between the two sites was observed by Cristobal et al. (35).
Interestingly, we observed some differences between lymph node and liver metastases. Significant up-regulation in both lymph node and liver metastases was observed for miR-200a, miR-200c and miR-429, whereas miR-200b was significantly up-regulated only in lymph node metastases. Furthermore, up-regulation was more pronounced in lymph node metastases than in liver metastases, which was an unexpected finding but is in accordance with a recent research suggesting that lymph node and distant metastases develop through fundamentally different evolutionary mechanisms (36).
We also investigated the expressions of genes PTPN13, ONECUT2, TGFB2, ZEB2, SOX2, RND3 and CDKN1B, previously validated as target genes of the miR-200 family in CRC (37). Among these genes, we focused our attention on those with inverse expression to their miRNA partners, either at the invasive front or in metastases.
At the invasive tumor front, TGFB2 was the only gene with inverse expression to the miR-200 family. This is in accordance with our previous study demonstrating a negative correlation between TGFB2 and three of five members of the miR-200 family, namely miR-200a, miR-200c and miR-141, in CRC with lymph node metastases (19). Increased expression of TGFB2 mRNAs and protein has also been reported in CRC progression (38, 39).
In the metastases, we observed inverse expression of ONECUT2 and SOX2 in comparison to the miR-200 family. Interestingly, ONECUT2, a target of miR-429, was down-regulated only in lymph node but not in liver metastases. An emerging role of ONECUT2 in tumor biology has recently been recognized (40, 41). It has been shown that miR-429 inhibits the initiation of EMT by targeting ONECUT2 (42). It has also been suggested that miR-429 could reverse TGF-β induced EMT by interfering with ONECUT2 in CRC (42). However, neither miR-429 nor ONECUT2 expression have been previously described in CRC metastases. Our results suggest that miR-429 may contribute to lymph node metastasis formation by reversing EMT related changes in CRC. SOX2, a target of both miR-200c and miR-429, is a stemness transcription factor, promoting proliferation, migration and invasion in CRC (43, 44). We observed down-regulation of SOX2 in both lymph node and liver metastases in comparison to the invasive front. It has been previously suggested that reduced expression of SOX2 causes the restoration of growth and metastasis in vivo and in vitro (43). Expression of SOX2 has been previously investigated in CRC liver metastases (45, 46). However, there is limited data about the expression of SOX2 in lymph node metastases, and on comparison between its expression in metastases and primary CRC. Our results suggest that down-regulation of SOX2 in distant metastases may play a role in metastases formation. This is in accordance with previous research showing that down-regulation of SOX2 by miR-429 decreases cell apoptosis (44).
Furthermore, we were interested in genes that showed inverse expression in metastases compared to the invasive tumor front, irrespective of their regulatory miRNAs expression. We found CDKN1B, PTPN13 and ZEB2 to be down-regulated at the invasive front and up-regulated in the metastases. CDKN1B is a target of miR-200b, which is believed to have a tumor-promoting role in CRC (47). A previous study showed that in CRC, loss of p27 protein, encoded by CDKN1B, probably promotes lymph node metastases and is correlated with a poor prognosis (48). Prior research on an mRNA level found only discrete changes in protein expression between liver metastases and the primary tumor (48). In contrast, our findings showed CDKN1B gene down-regulation in lymph node metastases, suggesting an important role of post-transcriptional regulators, such as miRNAs. While there are no available data about the expression of PTPN13 (miR-200c target) in metastases, a number of its potential interacting partners point to its role in tumor progression, including modification of cell shape, motility and cell signaling (49, 50). PTPN13 has been reported to be an anti-apoptotic factor in CRC (51). There is also little information about the RND3 gene. In our study, it showed only a slight increase in expression between the invasive front and lymph node and liver metastases. This result is consistent with a previously described elevated expression of the RND3 protein in lymph node metastases compared to the primary tumor (52). ZEB2, a target of all microRNAs of the miR-200 family, is an EMT-inducing transcription factor, previously shown to be associated with progression of CRC and a risk of distal but not local recurrence (53, 54). It has been shown that it is overexpressed at the invasion front of liver metastases in comparison to the center of metastases (55).
There are several advantages and limitations of our study. The main advantage is the use of the micropuncture technique, which enabled us to obtain tissue from the locations of interest, determined by microscopic analysis. Importantly, direct comparison of distinct locations within the tumor and matched metastases from the same patient enabled the avoidance of inappropriate comparison of miRNA expression between different tumors/patients. This is important, since miRNAs might be tissue specific, and depend on age and gender (56). However, using this technique, both tumor and stromal cells were present in tissue samples, not allowing a comparison between the tumor and stromal cells. Another limitation of our study is a small number of patients with heterogenous tumors regarding the stage, tumor budding and poorly differentiated clusters. Correlation between these features and the expression of EMT markers was therefore not possible. Furthermore, patients’ selection was biased because only those with lymph node and/liver metastases were included. The study groups were also uneven, since a sufficient number of patients matching the designed groups could not be obtained.
In conclusion, our results suggest the involvement of partial EMT at the invasive tumor front, and a reverse process - partial MET in both lymph node and liver metastases in CRC, based on analysis of the miR-200 family expression (Figure 5). The expression patterns of some of the postulated target genes further support these results. Interestingly, some differences were observed between lymph node and liver metastases, encouraging further studies, which will hopefully result in the development of new treatment modalities.
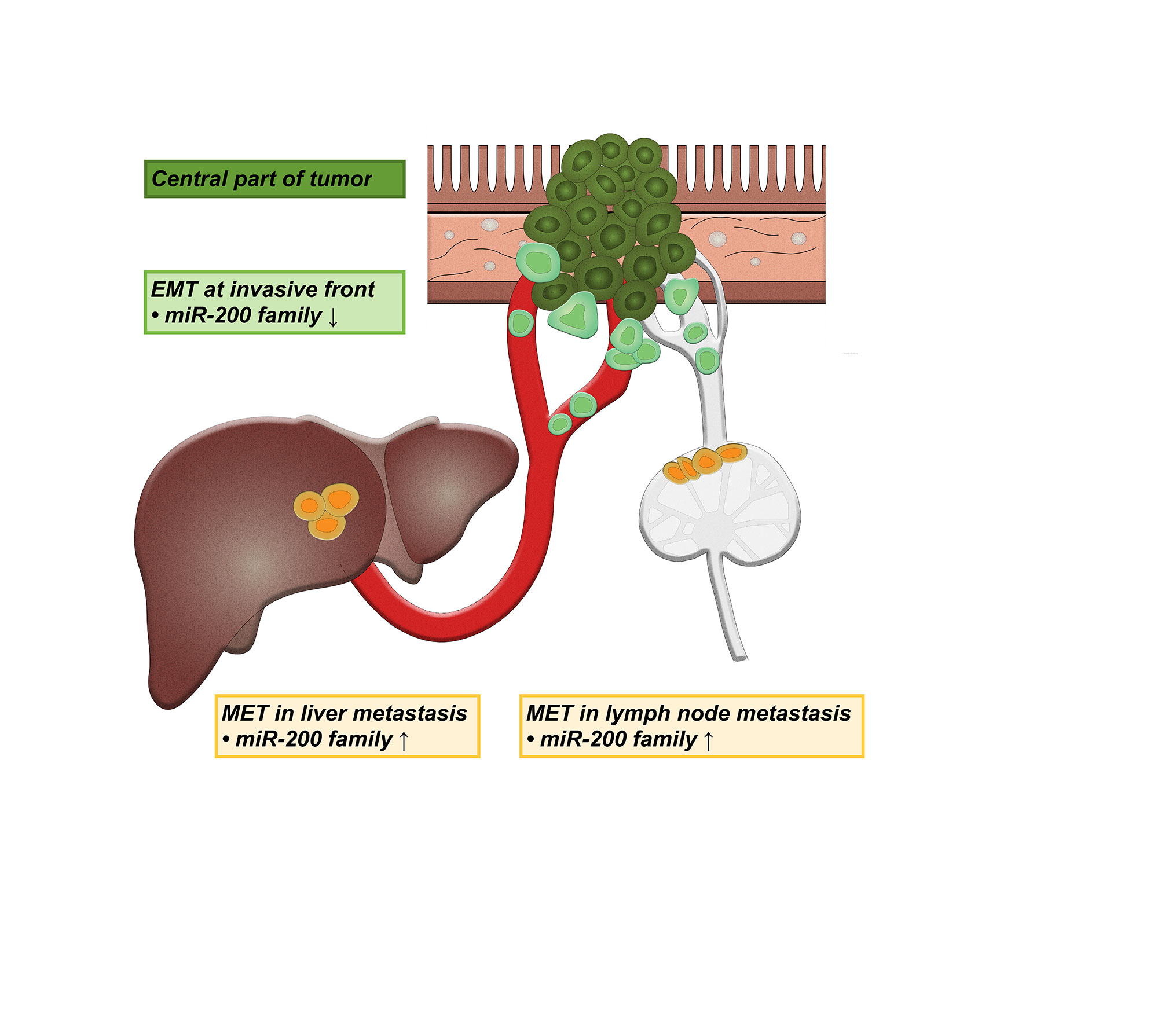
Figure 5 Schematic representation of the involvement of partial epithelial-mesenchymal transition (EMT) and partial mesenchymal-epithelial transition (MET) in lymph node and liver metastases. ↑ up-regulation, ↓ down-regulation.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
Patient consent was waived due to the following reason. As stated in the approval document, the study is retrospective, observational, performed on tissue samples that were obtained during routine diagnostic/therapeutic procedures, consisted of either excision or resection. Therefore, enough tissue was available for routine analysis and research. Moreover, tissue is still available for any additional analysis in the future. Our State Ethical Committee does not require informed consent from patients in such studies. However, the informed consent was obtained before the routine procedure.
Author Contributions
Conceptualization, NZ, AP and EB. Methodology, AP, KU, KŠ, EB and NZ. Software, KU and EB; validation, AP, KU and EB. Formal analysis, AP, KU, KŠ, EB and NZ. Investigation, AP, KŠ, KU and EB. Resources, NZ. Data curation, AP, KU, KŠ, EB and NZ. Writing—original draft preparation, AP. Writing—review and editing, AP, KU, EB and NZ. Visualization, AP and NZ. Supervision, EB and NZ. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Slovenian Research Agency (ARRS) under research core funding number P3-0054.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Eva Pavlič for Figure 5 creation.
References
1. Cao H, Xu E, Liu H, Wan L, Lai M. Epithelial-Mesenchymal Transition in Colorectal Cancer Metastasis: A System Review. Pathol Res Pract (2015) 211(8):557–69. doi: 10.1016/j.prp.2015.05.010
2. Bakir B, Chiarella AM, Pitarresi JR, Rustgi AK. Emt, MET, Plasticity, and Tumor Metastasis. Trends Cell Biol (2020) 30(10):764–76. doi: 10.1016/j.tcb.2020.07.003
3. Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 Family Determines the Epithelial Phenotype of Cancer Cells by Targeting the E-cadherin Repressors ZEB1 and ZEB2. Genes Dev (2008) 22(7):894–907. doi: 10.1101/gad.1640608
4. Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-Mesenchymal Transitions in Development and Disease. Cell (2009) 139(5):871–90. doi: 10.1016/j.cell.2009.11.007
5. Nieto MA, Huang RY, Jackson RA, Thiery JP. Emt: 2016. Cell (2016) 166(1):21–45. doi: 10.1016/j.cell.2016.06.028
6. Kalluri R, Weinberg RA. The Basics of Epithelial-Mesenchymal Transition. J Clin Invest (2009) 119(6):1420–8. doi: 10.1172/JCI39104
7. Pearson GW. Control of Invasion by Epithelial-to-Mesenchymal Transition Programs During Metastasis. J Clin Med (2019) 8(5):646–60. doi: 10.3390/jcm8050646
8. Brabletz T. EMT and MET in Metastasis: Where are the Cancer Stem Cells? Cancer Cell (2012) 22(6):699–701. doi: 10.1016/j.ccr.2012.11.009
9. Yao D, Dai C, Peng S. Mechanism of the Mesenchymal-Epithelial Transition and its Relationship With Metastatic Tumor Formation. Mol Cancer Res (2011) 9(12):1608–20. doi: 10.1158/1541-7786.MCR-10-0568
10. Tarin D, Thompson EW, Newgreen DF. The Fallacy of Epithelial Mesenchymal Transition in Neoplasia. Cancer Res (2005) 65(14):5996–6000; discussion -1. doi: 10.1158/0008-5472.CAN-05-0699
11. Voulgari A, Pintzas A. Epithelial-Mesenchymal Transition in Cancer Metastasis: Mechanisms, Markers and Strategies to Overcome Drug Resistance in the Clinic. Biochim Biophys Acta (2009) 1796(2):75–90. doi: 10.1016/j.bbcan.2009.03.002
12. Zidar N, Boštjančič E, Malgaj M, Gale N, Dovšak T, Didanovič V. The Role of Epithelial-Mesenchymal Transition in Squamous Cell Carcinoma of the Oral Cavity. Virchows Arch (2018) 472(2):237–45. doi: 10.1007/s00428-017-2192-1
13. Gurzu S, Silveanu C, Fetyko A, Butiurca V, Kovacs Z, Jung I. Systematic Review of the Old and New Concepts in the Epithelial-Mesenchymal Transition of Colorectal Cancer. World J Gastroenterol (2016) 22(30):6764–75. doi: 10.3748/wjg.v22.i30.6764
14. Zidar N, Boštjančič E, Gale N, Kojc N, Poljak M, Glavač D, et al. Down-Regulation of microRNAs of the miR-200 Family and miR-205, and an Altered Expression of Classic and Desmosomal Cadherins in Spindle Cell Carcinoma of the Head and Neck–Hallmark of Epithelial-Mesenchymal Transition. Hum Pathol (2011) 42(4):482–8. doi: 10.1016/j.humpath.2010.07.020
15. Teeuwssen M, Fodde R. Cell Heterogeneity and Phenotypic Plasticity in Metastasis Formation: The Case of Colon Cancer. Cancers (Basel) (2019) 11(9):1368–87. doi: 10.3390/cancers11091368
16. Pastushenko I, Blanpain C. EMT Transition States During Tumor Progression and Metastasis. Trends Cell Biol (2019) 29(3):212–26. doi: 10.1016/j.tcb.2018.12.001
17. Kaller M, Hermeking H. Interplay Between Transcription Factors and microRNAs Regulating Epithelial-Mesenchymal Transitions in Colorectal Cancer. Adv Exp Med Biol (2016) 937:71–92. doi: 10.1007/978-3-319-42059-2_4
18. Alidadiani N, Ghaderi S, Dilaver N, Bakhshamin S, Bayat M. Epithelial Mesenchymal Transition Transcription Factor (Tf): The Structure, Function and microRNA Feedback Loop. Gene (2018) 674:115–20. doi: 10.1016/j.gene.2018.06.049
19. Ranković B, Zidar N, Žlajpah M, Boštjančič E. Epithelial-Mesenchymal Transition-Related microRNAs and Their Target Genes in Colorectal Cancerogenesis. J Clin Med (2019) 8(10):1603–24. doi: 10.3390/jcm8101603
20. Vu T, Datta PK. Regulation of EMT in Colorectal Cancer: A Culprit in Metastasis. Cancers (Basel) (2017) 9(12):171–93. doi: 10.3390/cancers9120171
21. O’Brien SJ, Carter JV, Burton JF, Oxford BG, Schmidt MN, Hallion JC, et al. The Role of the miR-200 Family in Epithelial-Mesenchymal Transition in Colorectal Cancer: A Systematic Review. Int J Cancer (2018) 142(12):2501–11. doi: 10.1002/ijc.31282
22. Pichler M, Ress AL, Winter E, Stiegelbauer V, Karbiener M, Schwarzenbacher D, et al. MiR-200a Regulates Epithelial to Mesenchymal Transition-Related Gene Expression and Determines Prognosis in Colorectal Cancer Patients. Br J Cancer (2014) 110(6):1614–21. doi: 10.1038/bjc.2014.51
23. Hur K, Toiyama Y, Okugawa Y, Ide S, Imaoka H, Boland CR, et al. Circulating microRNA-203 Predicts Prognosis and Metastasis in Human Colorectal Cancer. Gut (2017) 66(4):654–65. doi: 10.1136/gutjnl-2014-308737
24. Hur K, Toiyama Y, Takahashi M, Balaguer F, Nagasaka T, Koike J, et al. MicroRNA-200c Modulates Epithelial-to-Mesenchymal Transition (EMT) in Human Colorectal Cancer Metastasis. Gut (2013) 62(9):1315–26. doi: 10.1136/gutjnl-2011-301846
25. Paterson EL, Kazenwadel J, Bert AG, Khew-Goodall Y, Ruszkiewicz A, Goodall GJ. Down-Regulation of the miRNA-200 Family At the Invasive Front of Colorectal Cancers With Degraded Basement Membrane Indicates EMT is Involved in Cancer Progression. Neoplasia (2013) 15(2):180–91. doi: 10.1593/neo.121828
26. Brierley J, Gospodarowicz MK, Wittekind C. TNM Classification of Malignant Tumours. Eighth edition. Chichester, West Sussex, UK; Hoboken, NJ: John Wiley & Sons, Inc (2017).
27. Lugli A, Kirsch R, Ajioka Y, Bosman F, Cathomas G, Dawson H, et al. Recommendations for Reporting Tumor Budding in Colorectal Cancer Based on the International Tumor Budding Consensus Conference (Itbcc) 2016. Mod Pathol (2017) 30(9):1299–311. doi: 10.1038/modpathol.2017.46
28. Ueno H, Kajiwara Y, Shimazaki H, Shinto E, Hashiguchi Y, Nakanishi K, et al. New Criteria for Histologic Grading of Colorectal Cancer. Am J Surg Pathol (2012) 36(2):193–201. doi: 10.1097/PAS.0b013e318235edee
29. Shivji S, Conner JR, Barresi V, Kirsch R. Poorly Differentiated Clusters in Colorectal Cancer: A Current Review and Implications for Future Practice. Histopathology (2020) 77(3):351–68. doi: 10.1111/his.14128
30. Žlajpah M, Hauptman N, Boštjančič E, Zidar N. Differential Expression of Extracellular Matrixrelated Genes DCN, Epha4, FN1, Sparc, SPON2 and SPP1 in Colorectal Carcinogenesis. Oncol Rep (2019) 42:1539–48. doi: 10.3892/or.2019.7274
31. Knudsen KN, Lindebjerg J, Nielsen BS, Hansen TF, Sorensen FB. MicroRNA-200b is Downregulated in Colon Cancer Budding Cells. PloS One (2017) 12(5):e0178564. doi: 10.1371/journal.pone.0178564
32. Muto Y, Suzuki K, Kato T, Tsujinaka S, Ichida K, Takayama Y, et al. Heterogeneous Expression of Zinc-Finger E-box-binding Homeobox 1 Plays a Pivotal Role in Metastasis Via Regulation of miR-200c in Epithelial-Mesenchymal Transition. Int J Oncol (2016) 49(3):1057–67. doi: 10.3892/ijo.2016.3583
33. Jepsen RK, Novotny GW, Klarskov LL, Christensen IJ, Hogdall E, Riis LB. Investigating Intra-Tumor Heterogeneity and Expression Gradients of miR-21, miR-92a and miR-200c and Their Potential of Predicting Lymph Node Metastases in Early Colorectal Cancer. Exp Mol Pathol (2016) 101(2):187–96. doi: 10.1016/j.yexmp.2016.08.003
34. Li W, Chang J, Tong D, Peng J, Huang D, Guo W, et al. Differential microRNA Expression Profiling in Primary Tumors and Matched Liver Metastasis of Patients With Colorectal Cancer. Oncotarget (2017) 8(22):35783–91. doi: 10.18632/oncotarget.16206
35. Cristobal I, Rincon R, Manso R, Carames C, Aguilera O, Madoz-Gurpide J, et al. Deregulation of miR-200b, miR-200c and miR-429 Indicates its Potential Relevant Role in Patients With Colorectal Cancer Liver Metastasis. J Surg Oncol (2014) 110(4):484–5. doi: 10.1002/jso.23661
36. Reiter JG, Hung WT, Lee IH, Nagpal S, Giunta P, Degner S, et al. Lymph Node Metastases Develop Through a Wider Evolutionary Bottleneck Than Distant Metastases. Nat Genet (2020) 52(7):692–700. doi: 10.1038/s41588-020-0633-2
37. Humphries B, Yang C. The microRNA-200 Family: Small Molecules With Novel Roles in Cancer Development, Progression and Therapy. Oncotarget (2015) 6(9):6472–98. doi: 10.18632/oncotarget.3052
38. Bellone G, Carbone A, Tibaudi D, Mauri F, Ferrero I, Smirne C, et al. Differential Expression of Transforming Growth factors-beta1, -Beta2 and -Beta3 in Human Colon Carcinoma. Eur J Cancer (2001) 37(2):224–33. doi: 10.1016/S0959-8049(00)00391-9
39. Kouvidou C, Latoufis C, Lianou E, Kouvatseas G, Kakouri E, Anagnostakis D, et al. Expression of Smad4 and TGF-beta2 in Colorectal Carcinoma. Anticancer Res (2006) 26(4B):2901–7.
40. Yu J, Li D, Jiang H. Emerging Role of ONECUT2 in Tumors. Oncol Lett (2020) 20(6):328. doi: 10.3892/ol.2020.12192
41. Rotinen M, You S, Yang J, Coetzee SG, Reis-Sobreiro M, Huang WC, et al. ONECUT2 is a Targetable Master Regulator of Lethal Prostate Cancer That Suppresses the Androgen Axis. Nat Med (2018) 24(12):1887–98. doi: 10.1038/s41591-018-0241-1
42. Sun Y, Shen S, Liu X, Tang H, Wang Z, Yu Z, et al. MiR-429 Inhibits Cells Growth and Invasion and Regulates EMT-related Marker Genes by Targeting Onecut2 in Colorectal Carcinoma. Mol Cell Biochem (2014) 390(1-2):19–30. doi: 10.1007/s11010-013-1950-x
43. Lu YX, Yuan L, Xue XL, Zhou M, Liu Y, Zhang C, et al. Regulation of Colorectal Carcinoma Stemness, Growth, and Metastasis by an miR-200c-Sox2-negative Feedback Loop Mechanism. Clin Cancer Res (2014) 20(10):2631–42. doi: 10.1158/1078-0432.CCR-13-2348
44. Li J, Du L, Yang Y, Wang C, Liu H, Wang L, et al. MiR-429 is an Independent Prognostic Factor in Colorectal Cancer and Exerts its Anti-Apoptotic Function by Targeting SOX2. Cancer Lett (2013) 329(1):84–90. doi: 10.1016/j.canlet.2012.10.019
45. Lundberg IV, Lofgren Burstrom A, Edin S, Eklof V, Oberg A, Stenling R, et al. SOX2 Expression is Regulated by BRAF and Contributes to Poor Patient Prognosis in Colorectal Cancer. PloS One (2014) 9(7):e101957. doi: 10.1371/journal.pone.0101957
46. Humphries HN, Wickremesekera SK, Marsh RW, Brasch HD, Mehrotra S, Tan ST, et al. Characterization of Cancer Stem Cells in Colon Adenocarcinoma Metastasis to the Liver. Front Surg (2017) 4:76. doi: 10.3389/fsurg.2017.00076
47. Fu Y, Liu X, Zhou N, Du L, Sun Y, Zhang X, et al. MicroRNA-200b Stimulates Tumour Growth in TGFBR2-null Colorectal Cancers by Negatively Regulating P27/Kip1. J Cell Physiol (2014) 229(6):772–82. doi: 10.1002/jcp.24497
48. Li JQ, Miki H, Wu F, Saoo K, Nishioka M, Ohmori M, et al. Cyclin A Correlates With Carcinogenesis and Metastasis, and p27(kip1) Correlates With Lymphatic Invasion, in Colorectal Neoplasms. Hum Pathol (2002) 33(10):1006–15. doi: 10.1053/hupa.2002.125774
49. Freiss G, Chalbos D. PTPN13/PTPL1: An Important Regulator of Tumor Aggressiveness. Anticancer Agents Med Chem (2011) 11(1):78–88. doi: 10.2174/187152011794941262
50. Schickel R, Park SM, Murmann AE, Peter ME. miR-200c Regulates Induction of Apoptosis Through CD95 by Targeting FAP-1. Mol Cell (2010) 38(6):908–15. doi: 10.1016/j.molcel.2010.05.018
51. Yao H, Song E, Chen J, Hamar P. Expression of FAP-1 by Human Colon Adenocarcinoma: Implication for Resistance Against Fas-mediated Apoptosis in Cancer. Br J Cancer (2004) 91(9):1718–25. doi: 10.1038/sj.bjc.6602136
52. Zhou J, Yang J, Li K, Mo P, Feng B, Wang X, et al. Rhoe is Associated With Relapse and Prognosis of Patients With Colorectal Cancer. Ann Surg Oncol (2013) 20(1):175–82. doi: 10.1245/s10434-012-2472-6
53. Sreekumar R, Harris S, Moutasim K, DeMateos R, Patel A, Emo K, et al. Assessment of Nuclear ZEB2 as a Biomarker for Colorectal Cancer Outcome and TNM Risk Stratification. JAMA Netw Open (2018) 1(6):e183115. doi: 10.1001/jamanetworkopen.2018.3115
54. Li MZ, Wang JJ, Yang SB, Li WF, Xiao LB, He YL, et al. ZEB2 Promotes Tumor Metastasis and Correlates With Poor Prognosis of Human Colorectal Cancer. Am J Transl Res (2017) 9(6):2838–51.
55. Kahlert C, Lahes S, Radhakrishnan P, Dutta S, Mogler C, Herpel E, et al. Overexpression of ZEB2 At the Invasion Front of Colorectal Cancer is an Independent Prognostic Marker and Regulates Tumor Invasion In Vitro. Clin Cancer Res (2011) 17(24):7654–63. doi: 10.1158/1078-0432.CCR-10-2816
Keywords: epithelial-mesenchymal transition, mesenchymal-epithelial transition, colon cancer, metastases, miRNA
Citation: Pavlič A, Urh K, Štajer K, Boštjančič E and Zidar N (2021) Epithelial-Mesenchymal Transition in Colorectal Carcinoma: Comparison Between Primary Tumor, Lymph Node and Liver Metastases. Front. Oncol. 11:662806. doi: 10.3389/fonc.2021.662806
Received: 01 February 2021; Accepted: 12 April 2021;
Published: 11 May 2021.
Edited by:
Ulrike Stein, Charité – Universitätsmedizin Berlin, GermanyReviewed by:
Ibrahim Halil Sahin, Moffitt Cancer Center, United StatesWilma Mesker, Leiden University Medical Center, Netherlands
Valeria Barresi, University of Verona, Italy
Copyright © 2021 Pavlič, Urh, Štajer, Boštjančič and Zidar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nina Zidar, bmluYS56aWRhckBtZi51bmktbGouc2k=
 Ana Pavlič
Ana Pavlič Kristian Urh
Kristian Urh Katarina Štajer
Katarina Štajer Emanuela Boštjančič
Emanuela Boštjančič Nina Zidar
Nina Zidar