- 1Department of Clinical Epidemiology, Shengjing Hospital of China Medical University, Shenyang, China
- 2Clinical Research Center, Shengjing Hospital of China Medical University, Shenyang, China
- 3Department of Oncology, Shengjing Hospital of China Medical University, Shenyang, China
- 4Department of Pediatric, Shengjing Hospital of China Medical University, Shenyang, China
- 5Department of Endocrinology, Shanghai Fifth People’s Hospital, Fudan University, Shanghai, China
Globally, lung cancer is the most commonly diagnosed cancer and carries with it the greatest mortality rate, with 5-year survival rates varying from 4–17% depending on stage and geographical differences. For decades, researchers have studied disease mechanisms, occurrence rates and disease development, however, the mechanisms underlying disease progression are not yet fully elucidated, thus an increased understanding of disease pathogenesis is key to developing new strategies towards specific disease diagnoses and targeted treatments. Circular RNAs (circRNAs) are a class of non-coding RNA widely expressed in eukaryotic cells, and participate in various biological processes implicated in human disease. Recent studies have indicated that circRNAs both positively and negatively regulate lung cancer cell proliferation, migration, invasion and apoptosis. Additionally, circRNAs could be promising biomarkers and targets for lung cancer therapies. This review systematically highlights recent advances in circRNA regulatory roles in lung cancer, and sheds light on their use as potential biomarkers and treatment targets for this disease.
Introduction
Worldwide, lung cancer is the most commonly diagnosed cancer with the highest mortality (1). Every year, approximately 1.82 million people are diagnosed, and 1.6 million die from the disease (2). The 5-year survival rate varies from 4–17% depending on stage and geographical differences (3). Approximately 85% of lung cancers are non-small cell lung cancers (NSCLCs), mainly including lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC) (4, 5). For decades, researchers have studied disease mechanisms, occurrence rates and disease development, however, the mechanisms underlying disease progression are not yet fully elucidated, thus an increased understanding of disease pathogenesis is key to developing new strategies towards specific disease diagnoses and targeted treatments. In recent years, increased evidence has suggested that abnormal circular RNA (circRNA) expression is associated with several diseases, e.g., cardiovascular disease (6), osteoarthritis (7) and different cancers, including prostate (8), glioma (9) and breast cancer (10), etc However, the precise functions of circRNAs in lung cancer remain unclear.
CircRNAs were first discovered in the 1970s, and are a class of non-coding RNAs widely expressed in eukaryotic cells (11–15). Originally, they were believed to be rare products arising from mRNA splicing errors (15), however the first observations using electron microscopy in 1979 suggested these RNAs existed as circular forms (16). In the 21st century, ubiquitous circRNA expression detection has been greatly facilitated by RNA sequencing (RNA-seq) technologies and bioinformatics, which have allowed researchers to investigate millions of short sequencing reads representing all RNA isoforms. CircRNAs are believed to be differentially expressed, and appear to play opposing roles in lung cancer etiology. For instance, circFGFR1 (17) is upregulated in NSCLC and promotes lung cancer progression, while circPTK2 (18) and circNOL10 (19) are downregulated and inhibit lung cancer development. Hence, this review comprehensively encompasses the latest contemporary research, and focuses on the emerging, divergent roles of circRNAs in lung cancer. A basic overview of circRNAs, their role in lung cancer, and the limitations of current research are explored.
CircRNA Overview
CircRNAs are covalently-closed molecules lacking 5’ to 3’ ends, polarity and a polyadenylated tail, with single-stranded transcripts derived from pre-mRNA (20). These characteristics provide them a higher tolerance to RNA exonucleases. The molecular structure is highly stable, they are highly abundant and incredibly diverse, they exhibit tissue specific expression patterns, and they are known to be involved in cellular differentiation and pluripotency (21–23). The majority of reported circRNAs are non-coding, while some have been reported to encode polypeptides or proteins (24). CircRNAs are divided into four categories, based on their origin (Figure 1) (25); 1) circRNAs derived from exons, exonic circRNAs (ecircRNAs), 2) circular intronic RNAs (ciRNAs) derived from introns, 3) exon-intron circRNAs (ElciRNAs) derived from a combination of exons and introns, and 4) intergenic circRNAs (26). Several hypotheses have been proposed for circRNA biogenesis (27), amongst which alternative splicing, reverse complementary intronic sequence paring, or RNA binding protein regulation are the most accepted.
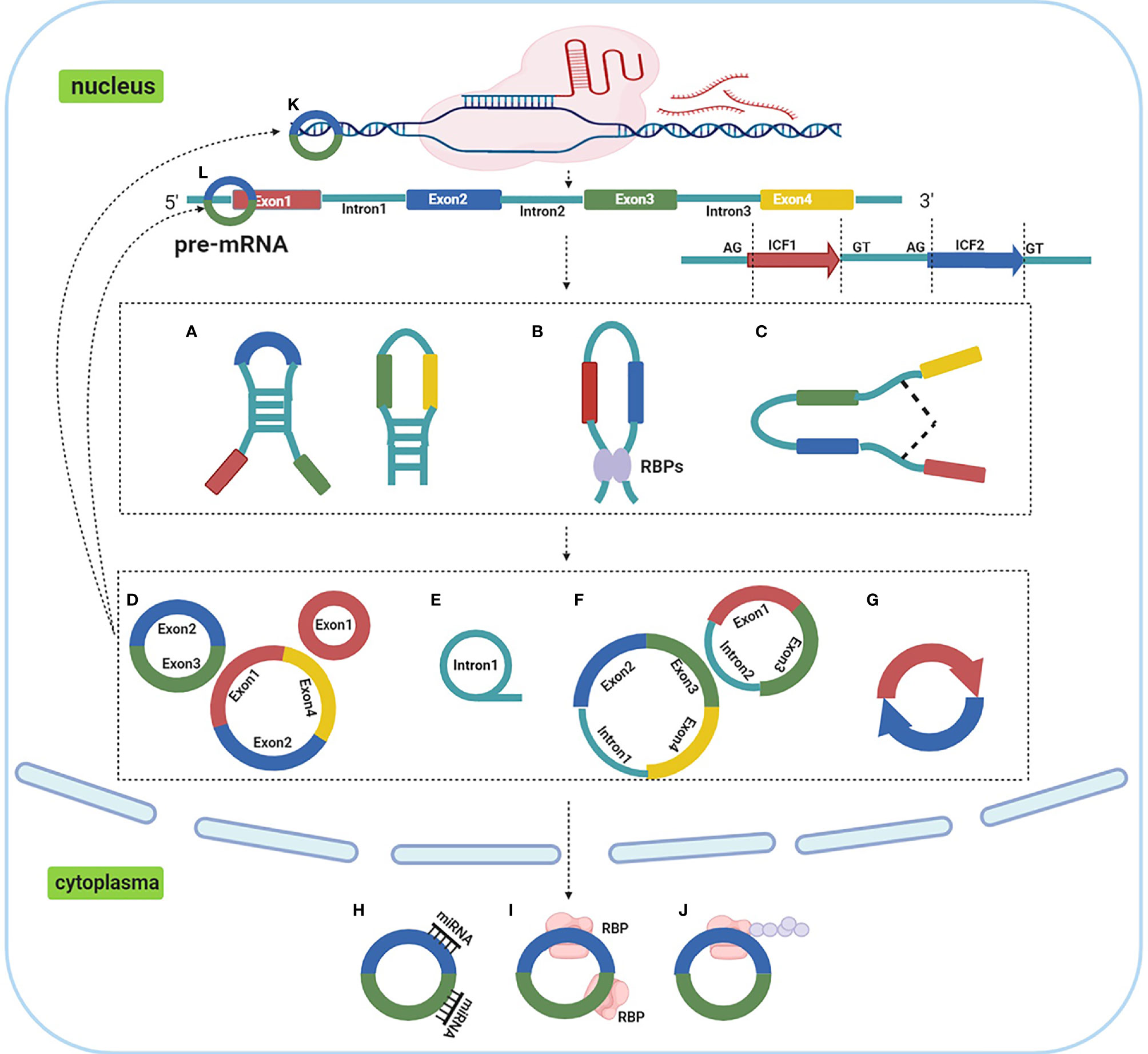
Figure 1 Biogenesis, categories and functions of circRNAs. (A). reverse complementary intronic sequence paring; (B) RNA binding protein regulation; (C) alternative splicing; (D). ecircRNAs; (E) ciRNAs; (F) ElciRNAs; (G) intergenic circRNAs; (H). act as ceRNA or miRNA molecular sponges; (I) interacting with RNA binding proteins; (J) peptide translation; (K) transcriptional regulation; (L) modulating miRNA stability.
Emerging evidence has suggested that circRNAs participate in various cellular biological processes including cell proliferation, differentiation, metastasis, senescence, apoptosis, etc (28), and are associated with lung cancer occurrence and development (29). CircRNAs functions primarily through the following five mechanisms; 1) act as competing endogenous RNA (ceRNA) or microRNA (miRNA) molecular sponges, 2) transcriptional regulation, 3) interacting with RNA binding proteins, 4) modulating miRNA stability and, 5) peptide translation (25, 30). Of these mechanisms, it is accepted that circRNAs act as miRNA sponges to regulate gene expression, and similarly, may have diagnostic or therapeutic potential for lung cancer treatment (31).
The Role of CircRNAs in Lung Cancer
CircRNAs Regulate Lung Cancer Cell Proliferation, Migration and Invasion
Cancer Associated Signaling Pathways
The Wnt/β-Catenin Pathway
Wnt activation has been observed in colorectal (32), breast (33) and lung cancer (34) and contributes to tumor onset and progression (35). CircRNAs are believed to promote or inhibit lung cancer cell proliferation, migration and invasion by regulating activation of the Wnt/β-catenin dependent or canonical pathway. CircRNAs regulating this pathway are summarized (Table 1).
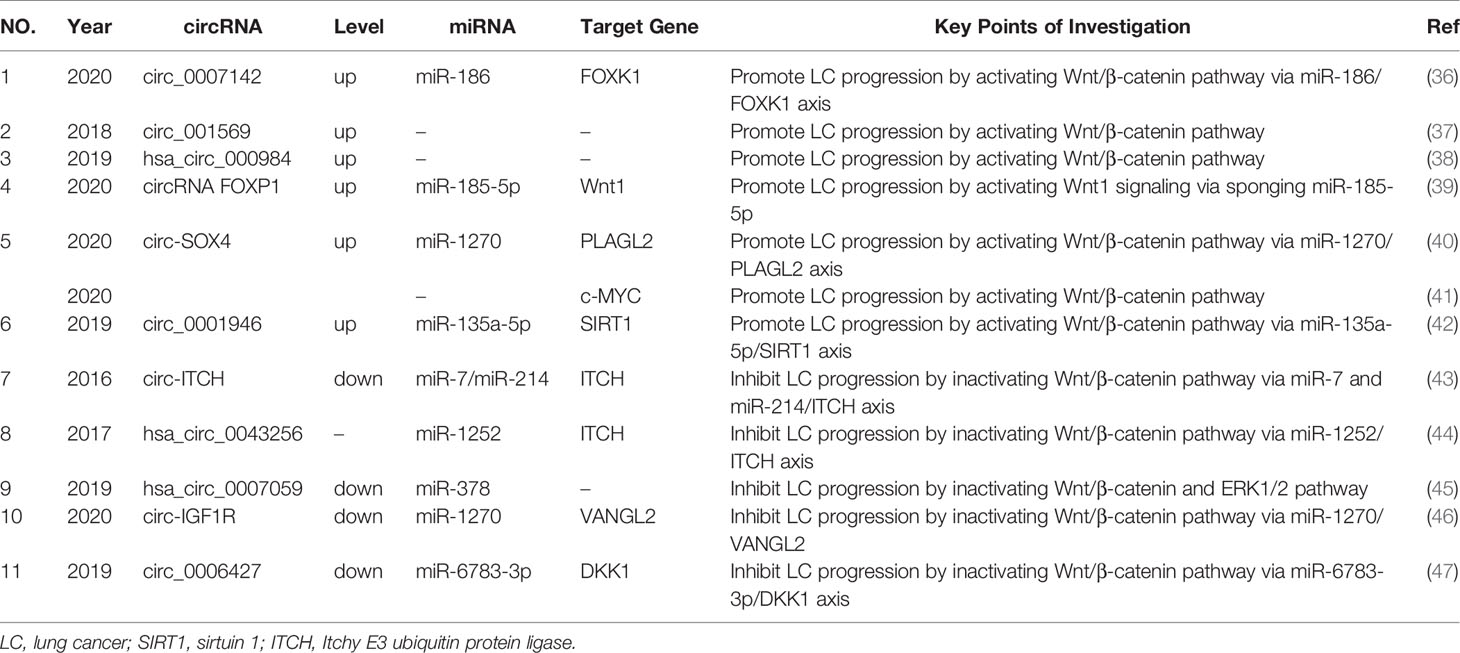
Table 1 The circRNAs regulate lung cancer proliferation, migration and invasion via Wnt/β-catenin pathway.
CircRNAs overexpressed in lung cancer cells and tissue drive in vitro cancer progression. For example, Liu et al. indicated that the circ_0007142/miR-186/FOXK1 axis played an important role in LUAD cell progression by activating the Wnt/β-catenin signaling pathway (36). Ding et al. observed that circ_001569 promoted cell proliferation by regulating the Wnt/β-catenin pathway in LUAD cells, whereas elevated circ_001569 expression demonstrated a poorer survival outcome (37). Li et al. also observed that hsa_circ_000984 exerted oncogenic functions by modulating Wnt/β-catenin pathway activation in LUAD cells (38). In other research, circRNA FOXP1 promoted LUAD cell proliferation by regulating the miR-185-5p/Wnt1 signaling pathway (39).
In addition, the role of several circRNAs with oncogenic functions in lung cancer mediated by the Wnt canonical pathway, were also demonstrated in in vivo studies. For instance, circ-SOX4 had oncogenic roles through the miR-1270/PLAGL2 axis, and subsequently activated the WNT signaling pathway in LUAD cells (40). In vivo data from this study showed that tumor volume and weight of xenograft nude mice were smaller in the circ-SOX4 silenced LUAD cell group when compared with the negative control (NC) group. Another study revealed that circ-SOX4 interacted with c-MYC by activating the Wnt/β-catenin pathway in LUAD cells, with circ-SOX4 down-regulating suppressed lung tumor-initiating cell proliferation, self-renewal, migration and invasion (41). Moreover, Yao et al. observed that circ_0001946 promoted LUAD cell growth by sponging miR-135a-5p to upregulate sirtuin 1 (SIRT1) expression (42). Moreover, in vivo experiments demonstrated that knockdown of circ_0001946 markedly suppressed tumor growth in nude mice. SIRT1 is a positive regulator of the Wnt/β-catenin signaling pathway (48, 49).
Several circRNAs have been downregulated in NSCLC, and appear to play an opposing role in lung cancer progression via inactivation of the Wnt canonical pathway. Itchy E3 ubiquitin protein ligase (ITCH) is a vital negative regulator of canonical Wnt signaling (44). Wan et al. found that circ-ITCH acted as sponge for oncogenic miR-7 and miR-214 to enhance ITCH expression, and thus suppressed activation of Wnt/β-catenin signaling (43). Tian et al. observed that hsa_circ_0043256 was upregulated in cinnamaldehyde treated NSCLC cells, and inhibited LUAD cell proliferation and induced apoptosis through ITCH in LUAD cell lines (44). Mechanistically, hsa_circ_0043256 functions as an miR-1252 sponge to directly target ITCH and inhibit Wnt/β-catenin pathway (44). Additionally, overexpressed hsa_circ_0007059 inhibited epithelial-to-mesenchymal transition (EMT) expression of the EMT proteins; vimentin, twist1 and zeb1, and elevated E-cadherin expression by suppressing miR-378 (45). Western blot assays revealed that overexpressed hsa_circ_0007059 inhibited Wnt3a, β-catenin and p-ERK1/2 expression in LUAD cell lines. Xu et al. observed that circ-IGF1R overexpression inhibited LUAD migration and invasion via the miR-1270/VANGL2 axis (46). VANGL2 is a Wnt signaling pathway related gene, and circ-IGF1R overexpression downregulated the Wnt pathway associated proteins, β‐catenin1 and vimentin (46). Yao et al. indicated that circ_0006427 overexpression effectively suppressed LUAD cell proliferation, migration and invasion (47) by inactivating the Wnt/β-catenin signaling pathway via miR-6783-3p sponging and DKK1 upregulation.
Based on these in vitro and in vivo studies, the Wnt pathway appears to be an important signaling pathway for circRNAs in regulating lung cancer progression, thus further exploration as a clinical therapy target is warranted. While these mechanisms are primarily via the canonical Wnt pathway, circRNA effects towards non-canonical pathways (Wnt/Ca2+ pathway and planar cell polarity pathway) are rarely studied, therefore these pathways could potentially open up new exploratory avenues for lung cancer research.
The Mitogen-Activated Protein Kinase (MAPK) Signaling Pathway
MAPK cascades are key signaling pathways which regulate a variety of cellular processes (e.g. proliferation, differentiation and apoptosis) (50, 51), and include four signaling families: the MAPK/extracellular signal-regulated kinase (ERK) family or the classical pathway, Big MAP kinase-1 (BMK-1), c-Jun N-terminal kinase (JNK) and p38 signaling families (52). Several circRNAs are upregulated in lung cancer and participate in tumorigenesis via MAPK signaling pathways. Chen et al. observed that hsa_circ_0007580 sponged miR-545-3p and subsequently inhibited protein kinase Ca (PKCA) to promote NSCLC cell proliferation and invasion by activating p38/MAPK signaling (53). In a xenograft tumor model, downregulated hsa_circ_0007580 inhibited NSCLC tumorigenesis by inactivating p38/MAPK signaling. Moreover, circ_0074027 exerted oncogenic properties in NSCLC cells via sponging miR‐185‐3p to upregulate bromodomain‐containing protein 4 (BRD4) and MAPK‐activating death domain containing protein (MADD) expression levels (54). Zhang et al. found that hsa_circRNA_101237 promoted MAPK1 expression via miRNA-490-3p sponging, thereby affecting NSCLC proliferation, migration and invasion, via its role as an important onco-circRNA (55).
Interestingly, Wang et al. found that circ-ZKSCAN1 (hsa_circ_0001727) acted as a sponge for carcinogenic miR-330-5p to increase the expression of family with sequence similarity 83(FAM83)-member A, inhibit the MAPK signal transduction pathway, and promote NSCLC progress (56). Overexpression of circZKSCAN1 significantly decreased JNK, p38 and ERK expression in NSCLC cells (56). Accordingly, these studies suggested that circRNAs promoted lung cancer progression by activating MAPK signaling, except for circ-ZKSCAN1, which exerted its oncogenic role by inhibiting MAPK signal transduction.
The Nuclear Factor-κB (NF-κB) Signaling Pathway
Early studies revealed that the NF- κB signaling pathway played important roles in cancer proliferation and apoptosis (57–59). Evidence showed that circRNAs appeared to interact with the NF- κB signaling pathway during lung cancer progression. Liu et al. observed that circ_cMras, alpha-beta hydrolase domain 5 (ABHD5) and adipose triglyceride lipase (ATGL) were downregulated in LUAD tissue and cells (60). Furthermore, the upregulation of circ_cMras inhibited LUAD cell proliferation, migration and invasion, and inhibited in vivo tumor growth via the ABHD5/ATGL axis, which targeted the NF-κB signaling pathway.
Proteins Related to DNA Repair and RNA Splicing or Translation
High Mobility Group (HMG) Proteins
The HMG of proteins, i.e., HMGA, HMGB and HMGN are non-histone nuclear proteins, which modulate DNA repair efficiency in all the major cellular pathways, i.e., nucleotide excision, base excision, double-stand break and mismatch repair (61), but also participate in cancer onset and progression (62–64). Studies have indicated that circRNAs upregulated in NSCLC promote lung cancer progression by positively influencing HMGA2 and HMGB3 expression. For example, Xu et al. identified that aspartate beta-hydroxylase (ASPH) RNA (circASPH/hsa_circ_0084606) was regulated by HMGA2 overexpression, by screening 6576 circRNAs using RNA-seq analysis (65). CircASPH promoted tumor growth in LUAD cells by sponging miR-370 and abrogating miR-370-mediated inhibition of HMGA2 (65). Li et al. also demonstrated that another circRNA, circ_100565 promoted proliferation, migration and invasion of NSCLC by upregulating HMGA2 via miR-506-3p sponging (66). Moreover, circEPSTI1 aggravated in vitro NSCLC progression by elevating HMGB3 expression via miR-145 sponging (67). Similarly, circEPSTI1 silencing restrained NSCLC tumor growth in vivo (67). Zhou et al. found that circRNA_102179 facilitated NSCLC proliferation, migration and invasion via the miR-330-5p/HMGB3 axis (68).
The Trinucleotide Repeat Containing 6 (TNRC6 or GW182) Family of Proteins
This family of proteins are core components of RNA interference (RNAi) molecules, and consist of three paralogs, i.e., TNRC6A, TNRC6B and TNRC6C (69). Argonaute and TNRC6 proteins, which form the RNA-induced silencing complex, mediate the fine-tuning of gene expression and are involved in several key biological processes (70), thereby greatly influencing disease development, especially cancer (71). The circRNAs, circABCC4 and circ0006916, were shown to interact with TNRC6 to positively and negatively regulate lung cancer progression, respectively. CircABCC4 was upregulated in LUAD and promoted cell proliferation, migration and invasion via miR-3186-3p sponging to upregulate TNRC6B (72). However, Circ0006910 was downregulated in lung cancer cells and tissue, and acted as a tumor suppressor in NSCLC via miR-522-3p sponging, and inhibiting pleckstrin homology domain and leucine rich repeat protein phosphatase 1 (PHLPP1) activity, thus inhibiting cell proliferation (73). Similarly, TNRC6A may also promote circ0006916 expression (73).
The MYC Family
The MYC oncogene family is dysregulated in > 50% of cancers, and is frequently associated with poor prognosis and unfavorable patient survival rates (74). The MYC oncogene encodes the transcription factor, MYC, which triggers selective gene expression amplification to promote cancer cell growth and proliferation (75). Two circRNAs were upregulated in lung cancer tissues and cell lines, with oncogenic roles that positively regulated MYC expression. Hsa_circRNA_103809 promoted lung cancer cell proliferation and invasion by promoting ZNF121 expression via miR-4302 sponging, and thus elevated ZNF121 expression levels, subsequently increasing MYC levels in lung cancer (76). Additionally, Zhang et al. indicated that circRNA_010763 promoted NSCLC proliferation, migration and invasion by sponging miR-715 to modulate its inhibitory effects on oncogenic c-MYC (77).
The Forkhead Box Protein M1 (FOXM1)
FOXM1 is a critical proliferation-associated transcription factor widely expressed during the cell cycle (78). In most cancers, including lung cancer, FOXM1 is oncogenic in nature thanks to its repeated upregulation, thereby generating a poor prognosis for patients (79, 80). Data from two studies showed that circRNAs upregulated in lung cancer tissue and cells, promoted lung cancer progression by regulating FOXM1 via miRNA sponging. Cheng et al. suggested that elevated circTP63 promoted LUSC cell proliferation both in vivo and in vitro, however, the ectopic expression of circTP63 exerted no significant effects on LUSC migration and invasion (81). Mechanistically, circTP63 overexpression promoted cell progression from the G1/S to the G2/M phase, suggesting increased cell cycle progression via sponging miR-873-3p to upregulate FOXM1. This subsequently regulated expression of the cell cycle related proteins, CENPA, histone H3 variant and CENPB heterochromatin (81). Additionally, Lu et al. observed that circHIPK3 overexpression promoted both cell proliferation and invasion in vitro, and tumorigenesis and metastasis in vivo, by sponging miR-149 and subsequently upregulating FOXM1 expression (82).
Moreover, a circRNA derived from FOXM1was closely associated with NSCLC progression. Yu et al. indicated that has_circ_0025039 was derived from exons 4 and 5 of FOXM1, labeling it circFOXM1 (83). This circRNA was upregulated in NSCLC tissue, potentially predicting an unfavorable overall survival (OS) for NSCLC patients (83). These findings suggested that circFOXM1 may have an oncogenic role in NSCLC progression by influencing the expression of cell cycle-related genes via miR-614 sponging, and FAM83D upregulation. Overall, these studies indicated that circRNAs promoting FOXM1 expression or derived from FOXM1 itself, could promote lung cancer progression, suggesting FOXM1 may function as a new molecular target for lung cancer treatment.
Cancer Associated Biological Processes
The EMT Process
EMT is a biological process where non-motile epithelial cell changes occur in a mesenchymal phenotype, but with invasive capacities (84). This phenomenon has been well documented in multiple biological processes, including embryogenesis, fibrosis, tumor progression and metastasis (84, 85). The EMT process plays a key role in the migration and invasion of malignant tumors, including NSCLC (86). Several circRNAs are upregulated in NSCLC, with oncogenic roles to positively regulate in vitro EMT processes. For instance, circ-LDLRAD3 promoted proliferation and EMT in NSCLC cells by downregulating miR-137, and subsequently upregulating glutamine transporter solute carrier family A1 member 5 (SLC1A5) (87). Notably, SLC1A5 was identified as participating in NSCLC progression and regulation, and similarly, its inactivation inhibited NSCLC cell viability (88). The circ_0012673 also facilitated LUAD proliferation and invasion (89). Loss-of-function studies indicated that circ_0012673 knockdown restricted proliferation, motility and EMT, but induced apoptosis by targeting miR-320a, and subsequently upregulating LIM domain kinase 1 in LUAD cell lines (89). Li et al. also noted that hsa_circ_0079530 upregulation promoted NSCLC cell migration and invasion by regulating EMT processes (90). Liu et al. found that hsa_circ_0023404 affected the expression of EMT related proteins, by regulating the miR-217/zinc finger E-box-binding homeobox 1 (ZEB1) axis, and contributing to in vitro NSCLC cell growth (91).
Moreover, several in vivo and in vitro studies have also indicated that circRNAs facilitate lung cancer progression by promoting EMT processes. Qu et al. observed that hsa_circ_0020123 enhanced proliferation, migration and invasion, while inhibiting in vitro NSCLC apoptosis, by suppressing miR-144 and upregulating ZEB1 and EZH2, respectively (92). Further studies also indicated that hsa_circ_ 0020123 knockdown inhibited NSCLC growth and metastasis both in vitro and in vivo (92). In their study, Chi et al. suggested that circPIP5K1A (Circ_0014130) potentially functioned as an miR-600 sponge to facilitate NSCLC proliferation and metastasis, by promoting hypoxia-inducible factor (HIF)-1α and reversing the inhibitory effects of miR-600 on EMT-related proteins (93). Furthermore, in vivo studies further illustrated that circPIP5K1A silencing suppressed tumor growth and pulmonary metastasis (93). Wang et al. found that circP4HB enhanced EMT processes in NSCLC via miR-133a-5p sequestration, leading to the in vitro upregulation of vimentin, and facilitating in vivo xenograft metastasis (94). Additionally, hsa_circ_0007534 positively regulated cell migration and invasion by affecting EMT in NSCLC cells, and promoting tumor growth in nude mice (95), however the underlying hsa_circ_0007534 mechanisms have yet to be elucidated.
However, several circRNAs are downregulated in NSCLC, and inhibit disease progression both in vitro and in vivo by negatively regulating EMT processes. Wang et al. found that hsa_circ_00008305 (circPTK2) acted as a sponge for miR-429/miR-200b-3p, and was positively correlated with transcriptional intermediary factor 1-γ (TIF1-γ) expression in human NSCLC tissue (18). CircPTK2 overexpression augmented TIF1-γ expression, and inhibited TGF-β-induced EMT and NSCLC cell invasion. In vivo studies showed that circPTK2 overexpression suppressed NSCLC cell metastasis (18). CircPTPRA also suppressed EMT processes in NSCLC cell lines and reduced in vivo metastasis in the murine xenograft model, by sequestering miR-96-5p and upregulating Ras association domain-containing protein 8 (RASSF8) (96). These findings have provided new EMT-mediated insights into the role of circRNAs in lung cancer.
Glycolysis
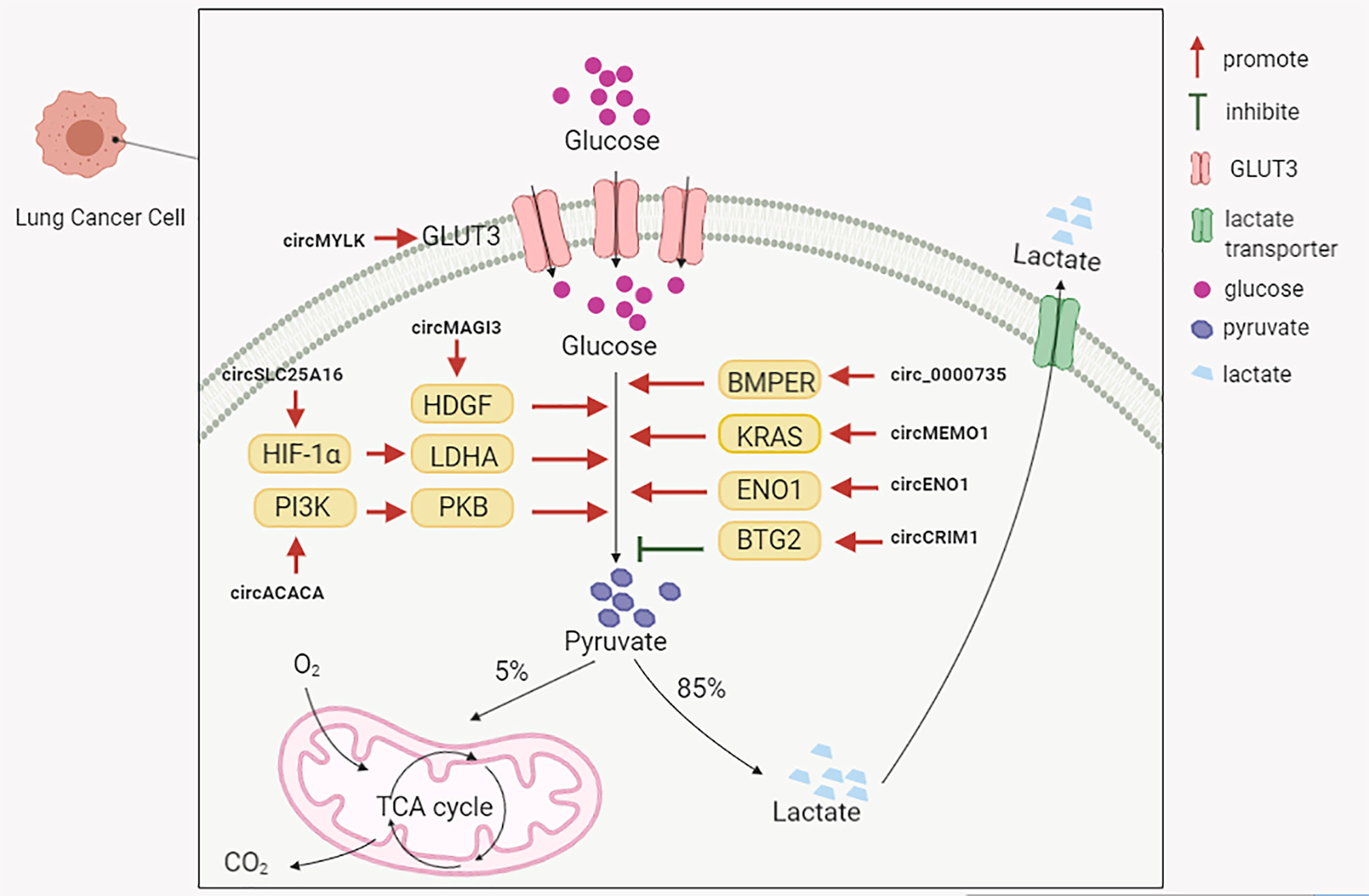
Glycolysis is the predominant energy producing pathway for cancer cells under both aerobic and hypoxic conditions, and is a biochemical fingerprint representing one of the “hallmarks of cancer” (97). The abnormal expression of circRNAs in lung cancer appears to influence glycolytic mechanisms (Figure 2). Several circRNAs upregulated in NSCLC could positively regulate glycolysis to promote disease progression. For instance, Guo et al. observed that circMAGI3 functioned as a sponge for miR-515-5p to promote its target gene, hepatoma-derived growth factor (HDGF) expression, thereby accelerating glycolytic mechanisms in NSCLC (98). Hong et al. found that circSLC25A16 accelerated glycolysis and promoted NSCLC proliferation by interacting with miR-488-3p and enhancing HIF-1α expression, which transcriptionally activated lactate dehydrogenase A (LDHA) (99). A study by Wu et al. indicated that circ-ACACA promoted in vitro NSCLC proliferation, migration and glycolysis by negatively regulating miR-1183 to activate the phosphoinositide 3-kinase/protein kinase B(PI3K/PKB) pathway (100). Huang et al. observed that circ_0000735 overexpression enhanced NSCLC proliferation, migration, invasion, and glycolysis by targeting the miR-940/bone morphogenetic protein binding endothelial cell precursor-derived regulator (BMPER, a secreted glycoprotein) axis (101). Ding et al. reported that circ-MEMO1 elevated expression in NSCLC patients was associated with a poor prognosis (102). Elevated circ-MEMO1 levels accelerated cell proliferation, cell cycle progression and aerobic glycolysis, and inhibited NSCLC apoptosis via the miR-101-3p/KRAS axis (102). Equally, the circRNA knockdown or silencing of these aforementioned molecules also suppressed tumor growth effects in in vivo xenograft assays.
Moreover, circRNA-ENO1 promoted glycolysis so as to promote LUAD proliferation, migration and EMT processes via the miR-22-3p/enolase 1 (ENO1) axis (103). Additionally, also circ-ENO1 promoted tumor growth and metastasis in vivo. Xiong et al. observed that circMYLK overexpression was closely associated with a poor prognosis in NSCLC patients (104). At the molecular level, circMYLK promoted glycolysis and NSCLC proliferation by sponging miR-195-5p, and up-regulating the expression of glucose transporter member 3 (GLUT3) (104).
However, distinct to the functionalities of these aforementioned circRNAs, Zhang et al. observed that circCRIM1 and BTG anti-proliferation factor 2 (BTG2) were downregulated, and miR-125b-5p upregulated in LUAD tissue and cells (105). CircCRIM1 upregulation inhibited in vitro LUAD cell migration, invasion, EMT processes and glycolysis by sponging miR-125b-5p to promote BTG2 expression, and negatively affect in vitro tumor growth (105). These studies demonstrated that circRNAs regulated glucose metabolism during lung cancer, and are potential therapeutic targets for the disease.
Other Mechanisms
In addition to the circRNA mechanisms summarized above, circRNAs may also function via other distinct mechanisms (Table 2).
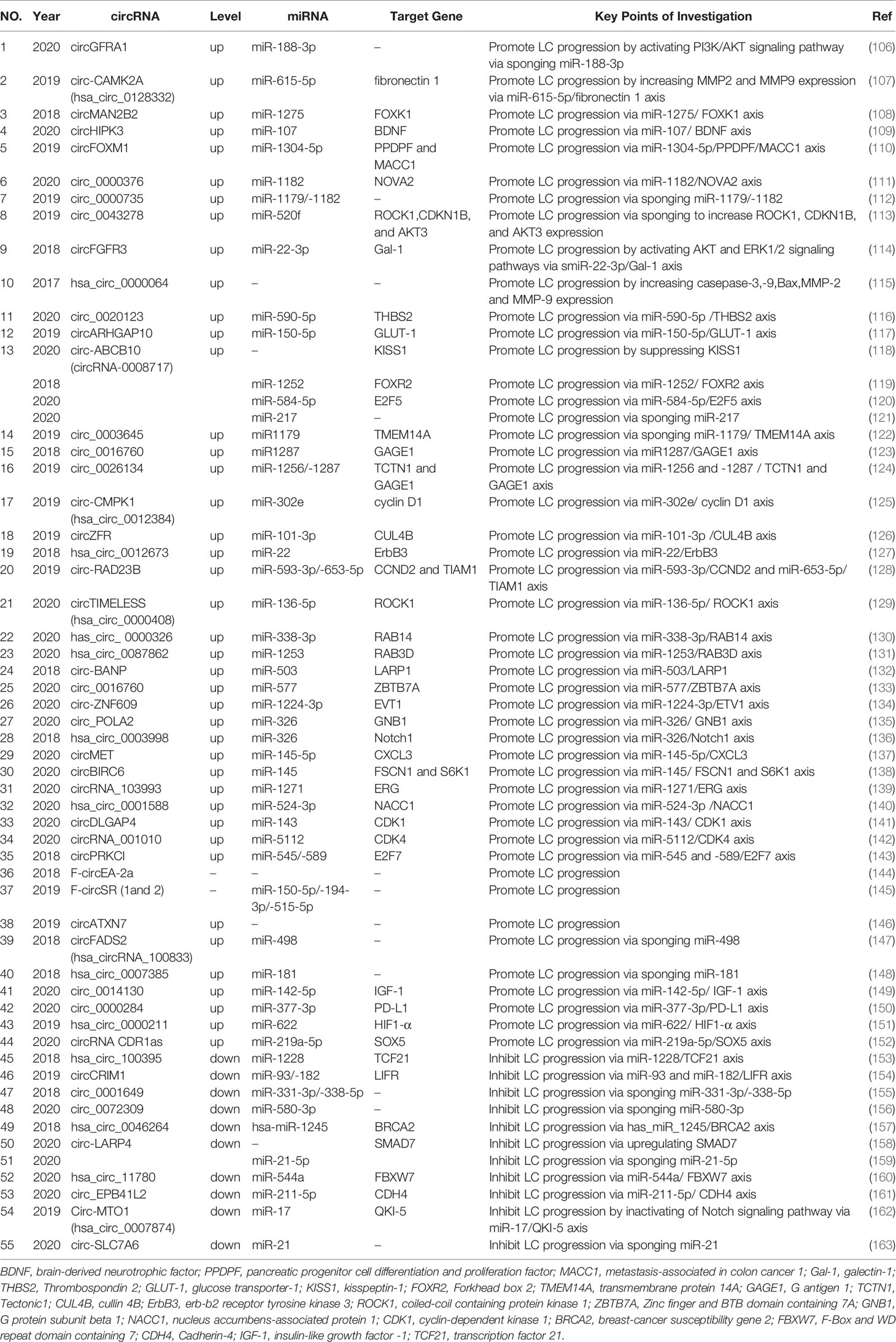
Table 2 The circRNAs regulate lung cancer proliferation, migration and invasion via other mechanisms.
Several circRNAs are upregulated in NSCLC, and exert oncogenic roles during lung cancer (108–152). For instance, the phosphatidylinositol 3-kinase-regulated protein kinase, Akt, plays an important role in cancer initiation and progression (164). Mammalian cells express three Akt isoforms (Akt1–3), which are encoded by three distinct genes (164). Yao et al. reported that circGFRA1 acted as an miR-188-3p sponge, to regulate NSCLC proliferation via the PI3K/Akt signaling pathway (106). Moreover, matrix metalloproteinases (MMPs) play vital roles in many biological processes (165). In particular, MMP-2 and MMP-9 are both implicated in NSCLC invasion and tumor metastasis (166). A recent study indicated that circ-CAMK2A (hsa_circ_0128332) up-regulated the expression levels of fibronectin 1 by sponging miR-615-5p, thereby increasing MMP-2 and MMP-9 expression levels to promote LUAD metastasis (107).
In contrast, some circRNAs are downregulated in NSCLC and have roles as cancer suppressors (154–163). For example, Chen et al. observed that hsa_circ_100395 overexpression dramatically inhibited in vitro NSCLC cell proliferation, arrested cell-cycle progression and reduced cell migration and invasion (153). Data from a flow cytometry (FACS) study indicated that hsa_circ_100395 overexpression increased cells arrested in G0/G1 phase, with fewer cells in S and G2/M phase (153). Mechanistically, hsa_circ_100395 promoted expression of transcription factor 21, also known as a suppressor, by sponging miR-1228 in lung cancer (153). These data exemplified the multi-mechanistic roles of circRNAs in lung cancer progression.
CircRNAs Regulate Lung Cancer Cell Death
Signal Transducer and Activator of Transcription (STAT) Proteins
These proteins are cytoplasmic transcription factors implicated in many cellular biological processes (167). Numerous studies have reported that STAT overexpression enhances carcinogenesis and affects prognosis in cancer patients (168), including NSCLC (169, 170). Chen et al. investigated circHIPK3 mechanisms in regulating lung cancer cell death (171). Their data indicated that autophagy was induced upon loss of circHIPK3, via the miR124-3p-STAT3-PRKAA/AMPKa axis in STK11 mutant lung cancer cell lines (171). At the molecular level, circHIPK3 negatively regulated miR124-3p expression and subsequently upregulated STAT3 expression (171). Further research has indicated that STAT3 silencing and inhibition induced autophagy via the PRKAA/AMPKa pathway in STK11 mutant cells. A recent study provided evidence that silencing circHIPK3 induced lung cancer cell death and apoptosis, by sponging miR-124 and regulating miR-124 targets, including SphK1, STAT3 and CDK4 proteins (172). Additionally, Wang et al. showed that STAT3 activated circCCDC66 transcription, and thereby promoted circCCDC66 expression in NSCLC cells, and similarly circCCDC66 inhibited cell apoptosis via the miR-33a-5p/KPNA4 axis in these cells (173). These studies highlighted the key mechanistic interactions between circRNAs and STAT3 in regulating lung cancer death.
B-Cell Lymphoma 2 (Bcl-2) and/or Bcl-2-Associated X (Bax)
Bcl-2 and related cytoplasmic proteins are key regulators of apoptosis, including apoptosis inhibitory mechanisms (174). Bcl-2 functions as an oncogene not only by blocking apoptosis, but also by blocking autophagy (175). Bax is a proapoptotic protein which is an essential component of the intrinsic apoptosis signaling pathway, thereby promoting apoptosis (176). Several studies have indicated that some circRNAs upregulated in lung cancer inhibit cell death, and promote disease progression via upregulating Bcl-2 and/or downregulating Bax expression. For instance, Wang et al. reported that circVANGL1 overexpression behaved as an oncogene by sponging miR-195, and subsequently activating Bcl-2 to inhibit in vitro NSCLC apoptosis (177). Western blot data confirmed that circVANGL1 knockdown increased expression of proapoptotic Bax, while it decreased the expression of antiapoptotic Bcl-2 in LUAD cells (177). Hsa_circ_0109320 inhibited apoptosis in NSCLC by sponging miR-595 to upregulate E2F transcription factor 7 expression, and subsequently upregulating Bcl-2, downregulating Bax and cleaving caspase 3 in vitro (178).
Furthermore, several researches also identified the effects of circRNAs on tumorigenesis in in vivo studies. For example, Geng et al. observed that hsa_circ_0014130 had oncogenic roles in NSCLC, and functioned as a ceRNA of miR-136-5p, to activate Bcl-2 which inhibited NSCLC apoptosis (179). Qin et al. showed that circPVT1 promoted NSCLC progression by regulating the miR-497/Bcl-2 axis, and inhibiting apoptosis (180). Chen et al. found that circPUM1 was significantly upregulated in both LUAD cell lines and tissue, whereas circPUM1 silencing impaired LUAD proliferation, migration and invasion abilities, and increased apoptosis (181). Mechanistically, circPUM1 could sponge miR-326 to promote expression of its downstream proteins, Bcl-2 and cyclin D1 (181). Furthermore, these studies also indicated that knockdown or silencing hsa_circ_0014130, circPVT1 and circPUM1 inhibited in vivo lung cancer tumorigenesis in subcutaneous xenograft mouse models (179–181).
In contrast, circNOL10 was downregulated in lung cancer, and inhibited lung cancer development both in vivo and in vitro by promoting apoptosis (19). At the molecular level, circNOL10 promoted expression of the transcription factor sex comb on midleg-like 1 (SCML1) by inhibiting transcription factor ubiquitination, and thereby affecting humanin polypeptide family regulation by SCML1. Ultimately, circNOL10 promoted lung cancer cell apoptosis by increasing Bax and caspase-9 expression, and in comparison, decreasing Bcl-2 expression (19). These data indicated that circRNAs regulated lung cancer apoptosis via members of the Bcl-2 family, which may be exploited as potential therapeutic targets.
The Caspase Family
Caspases are a family of endoproteases, broadly classified by their roles in apoptosis (caspase-3, -6, -7, -8, and -9 in mammals) and inflammation (caspase-1, -4, -5, -12 in humans, and caspase-1, -11, and -12 in mice) (182). Caspase dysregulation underlies several human diseases, including cancer and inflammatory disorders (182). CircRNAs upregulated in lung cancer have played oncogenic roles by regulating caspases expression to inhibit apoptosis. For instance, circRNA ecto-5’-nucleotidase (circNT5E) silencing induced in vitro cell apoptosis by increasing the activity of caspase-3 and the cleavages of poly (ADP-ribose) polymerase, via miR-134 sponging (183). Moreover, circRNA 100146 functioned as an oncogene in NSCLC cells by interacting with the splicing factor SF3 family (SF3B3, SF3B2 and SF3A1), and binding miR-361-3p and miR-615-5p to regulate multiple downstream mRNAs (i.e., NFAT5, COL1A1, TRAF3 and MEF2C) (184). In further xenograft nude mouse model studies, surgically removed tumors were analyzed and revealed decreased PCNA and p53 levels, and increased caspase-9 and E-cadherin levels in the circRNA 100146 silenced group (184). Yang et al. observed that circRNA TUBA1C accelerated NSCLC progression by sponging miR-143-3p (185). By conducting in vivo nude mice studies, these authors also observed that circTUBA1C silencing increased the protein expression of cleaved caspase-3 and Bax (185). These findings indicated that circRNAs are implicated in regulating caspase expression.
CircRNAs as Potential Biomarkers for Lung Cancer
A biomarker is defined as “a characteristic that is objectively measured and evaluated as an indicator of normal biological processes, pathogenic processes, or pharmacologic responses to a therapeutic intervention” (186). Traditional clinical blood biomarkers of lung cancer include; neuron-specific enolase (NSE), progastrin-releasing peptide (pro-GRP), carcinoembryonic antigen (CEA), CYFRA 21-1, cancer antigen 15 (CA125) and squamous cell carcinoma antigen (SCC). These biomarkers facilitate differential diagnoses, and determine prognostic and monitoring responses to systemic lung cancer therapies. However, these biomarkers still lack comprehensive sensitivity and/or specificity for early diagnoses. Study data from Tarro et al. demonstrated that for early stage I disease detection, NSCLC patients showed a sensitivity of 33.3% for CA19-9, 11.1% for CYFRA 21-1 and CA125, and 0% for CEA; whereas the specificity for all biomarkers was 100% (187). Additionally, Li et al. observed that serum tumor marker sensitivity was 28.46% for CEA, 19.51% for CA 125, 3.25% for NSE, 50.41% for CYFRA 21‐1, 26.02% for SCC and 11.38% for pro‐GRP in lung cancer patients (188).
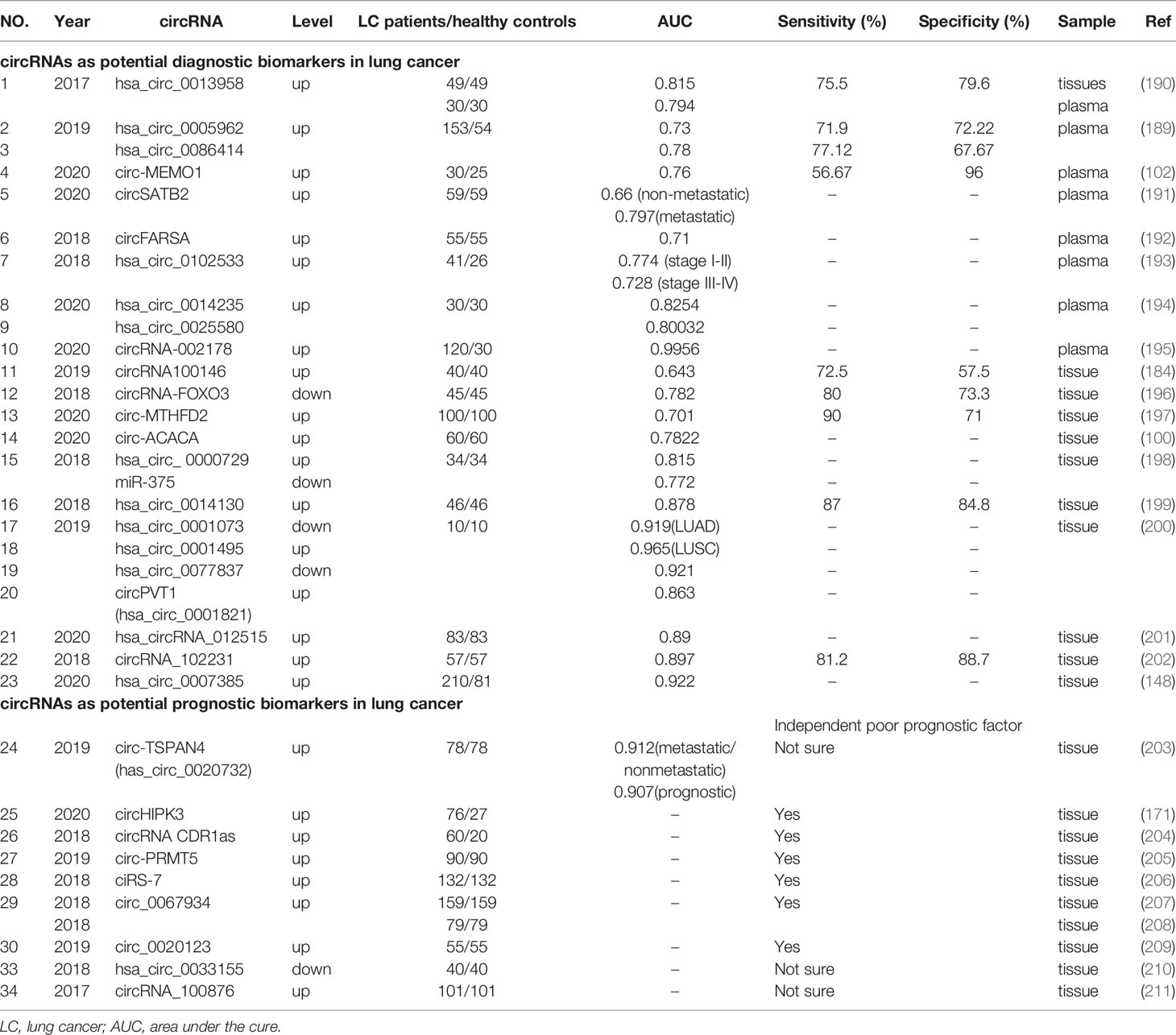
Therefore in the future, more biomarkers which influence clinical decision-making and improve patient care are required. High numbers of circRNAs have exhibited aberrant expression in human peripheral blood and tissue from lung cancer patients, which are easy to detect. Several studies have demonstrated differential expression patterns of circRNAs under different pathological conditions, thereby indicating the non-invasive biomarker potential of circRNAs in lung cancer (189, 190). These potential circRNA biomarkers are shown (Table 3).
Diagnostic circRNAs Biomarkers in Lung Cancer
Several circRNAs of potential diagnostic value have been detected in human plasma, and could be exploited as highly sensitive and specific non-invasive biomarkers for NSCLC. Zhu et al. reported that hsa_circ_0013958 was upregulated in all LUAD tissue, cells and even plasma (190). Additionally, these hsa_circ_0013958 levels were closely related to lymphatic metastasis. The area under the receiver operating characteristic (ROC) cure was 0.815, and sensitivity and specificity for diagnosis were 75.5% and 79.6%, respectively. In addition, plasma hsa_circ_0013958 levels distinguished LUAD cases from healthy controls, with an area under the cure (AUC) of 0.794 (190). These observations indicated that hsa_circ_0013958 could be exploited as a non-invasive biomarker, with high sensitivity and specificity for screening high-risk individuals and early-stage LUAD patients (190). After comparing 153 primary LUAD and 54 normal plasma samples, Liu et al. observed that has_circ_0005962 was upregulated in LUAD, whereas has_circ_0086414 was downregulated (189). The AUC for hsa_circ_0005962 was 0.73, and the optimal cut-off value was 1.21, with a sensitivity of 71.90% and specificity of 72.22%. For hsa_circ_0086414, the AUC was 0.78 and the cut-off value was 0.39, with a sensitivity and specificity of 77.12% and 66.67%, respectively. Additionally, plasma hsa_circ_0005962 showed differential expression patterns in LUAD patients before and after surgical resection, indicating expression may be closely related to patient tumor burden (189). Another study indicated that exosomal circ-MEMO1 levels were higher in NSCLC patient serum, when compared with healthy controls. The AUC was approximately 0.76, with a diagnostic sensitivity and specificity of 56.67% and 96%, respectively (102). Zhang et al. found that circSATB2 was implicated in NSCLC progression by positively regulating fascin homolog 1, actin-bundling protein 1 (FSCN1) expression via miR-326 (191). The AUC of exosomal circSATB2 was 0.660 in serum from lung cancer patients, and 0.797 in serum from metastatic lung cancer patients. These observations indicated that exosomal circSATB2 had the potential to provide blood detection indices for the diagnosis of lung and lung cancer metastasis, with high sensitivity and specificity (191). CircFARSA was upregulated in NSCLC patient plasma, and had an AUC diagnostic value of 0.71 (192). Hsa_circ_0102533 served as a blood-based biomarker for stage I-II cancer detection in NSCLC patients (AUC = 0.774), and stage III-IV detection in NSCLC patients (AUC = 0.728) (193). Another study revealed diagnostic indices for hsa_circ_0014235 (AUC = 0.8254) and hsa_circ_0025580 (AUC = 0.80032) in LUSC plasma samples, suggesting high diagnostic values for LUSC (194). Wang et al. compared serum samples from 30 healthy controls and 120 LUAD patients without treatment, and observed that circRNA-002178 was upregulated, with an AUC of 0.9956 (195).
In addition, circRNA expression indices, with potential diagnostic value, have been detected in cancer tissue only, but not plasma. Chen et al. demonstrated that the AUC for circRNA100146 was 0.643 in NSCLC tissue, and the sensitivity and specificity were 72.5% and 57.5%, respectively (184). Zhang et al. observed that the AUC for circRNA-FOXO3 was 0.782, and sensitivity and specificity indices were 80.0% and 73.3%, respectively (196). Geng et al. also reported that the AUC of circ-MTHFD2 was 0.701, with a cut-off value of 3.534, and 90% sensitivity and 71% specificity (197). The diagnostic accuracy of circ-ACACA was assessed using ROC curve analysis, and showed that the AUC was 0.7822 (100). Li et al. observed that hsa_circ_ 0000729 was upregulated in LUAD tissue and sponged miR‐375, both of which had significant diagnostic accuracy, with AUC values of 0.815 and 0.772, respectively (198). Hsa_circ_0014130 exhibited significant overexpression in NSCLC tissue, with an AUC of 0.878 and an optimal cutoff value 0.573, with sensitivity and specificity indices measured at 87.0% and 84.8%, respectively (199). Wang et al. in an RNA-Seq analysis observed that four circRNAs from 17,952 circRNAs, had diagnostic values for NSCLC (200). Specifically, the AUC for hsa_circ_0001073 was 0.919 in LUAD tissue, and 0.965 for hsa_circ_0001495 in LUSC tissue, indicating both molecules could serve as effective diagnostic biomarkers for LUAD and LUSC prediction, respectively. For the other circRNAs, the AUC for hsa_circ_0077837 was 0.921, and for hsa_circ_0001821 (circPVT1), 0.863. However, these circRNAs were unable to distinguish LUAD and LUSC subtypes (200).
Furthermore, several circRNAs have exhibited high diagnostic accuracy, while the aberrant expression of these circRNAs has been closely associated with cancer prognoses. Fu et al. observed that hsa_circRNA_012515 exhibited a high diagnostic accuracy for NSCLC, with an AUC of 0.89 (201). Kaplan-Meier (K-M) survival analyses also indicated that OS and disease free survival (DFS) indices were considerably shortened in patients expressing high levels of hsa_circRNA_012515, when compared to those patients with low expression (201). Zong et al. indicated that circRNA_102231 expression was significantly upregulated in LUAC tissue, and was associated with advanced TNM stage, lymph node metastasis and poor OS in lung cancer patients, with an AUC of 0.897 (202). CircRNA_102231 also exhibited a good sensitivity of 81.2%, and specificity of 88.7%. Hsa_circ_0007385 was upregulated in NSCLC tissue with an AUC of 0.922; multivariate regression analyses indicated that the high expression of this circRNA independently predicted a worse OS status (148). These studies showed that some circRNAs may be considered good (non-invasive) biomarkers for lung cancer diagnostics, and some are useful as prognostic biomarkers.
Prognostic circRNA Biomarkers in Lung Cancer
CircRNAs exhibited different expression patterns and levels in lung cancer tissue. These expression levels are closely related to the stage and prognosis of disease, potentially making them new prognostic biomarkers in clinical settings. For instance, Ying et al. observed that circ‐TSPAN4 (has_circ_0020732) expression levels were increased in LUAD tissue and cell lines, suggesting it as a promising prognostic biomarker for LUAD patients (203). The AUC for circ‐TSPAN4 expression levels for metastatic versus non-metastatic LUAD patients was 0.912. More importantly, the OS status for patients with high circ‐TSPAN4 expression levels was significantly shorter than patients with low levels, and the prognostic utility of circ‐TSPAN4 for LUAD patients was high, with an AUC of 0.907 (203). Moreover, Chen et al. observed that circHIPK3 was upregulated in NSCLC, while its linear counterpart, linear HIPK3 (linHIPK3) was significantly downregulated in diseased tissue, when compared to adjacent normal tissue (171). The expression ratio between circHIPK3 and linHIPK3 was significantly higher in tumors when compared to normal tissue, and indicated poor survival, especially for advanced-stage NSCLC patients.
In addition, several circRNAs showed higher expression in NSCLC samples than the normal counterparts that was highly associated with TNM stage and lymph node metastasis, but not associated with other factors such as, gender, age, smoking, histology, etc, which could function as an independent poor prognostic factor. For example, Zhang et al. showed that circRNA CDR1as was upregulated in NSCLC tissue (204). Further data indicated that patient’s expressing high circRNA CDR1as levels had shorter OS than those with low levels. Similar results were also generated for circRNA PRMT5 (circ-PRMT5); high expression levels indicated lung cancer patients were more likely to develop poor progression-free survival (PFS) and OS (205). In their investigation, Yan et al. indicated that high ciRS-7 expression was correlated with shorter DFS and OS status, and multivariate Cox’s proportional hazard regression analyses showed this high expression was an independent factor for predicting a poor prognosis (206). Both Wang et al. and Zou et al. reported that circ_0067934 expression was significantly increased in NSCLC tissue and cell lines (207, 208). K-M curves showed that high expression of this circRNA was an independent risk factor for OS status in patients with NSCLC in both studies. The K-M analyses by Wan et al. indicated that high circ_0020123 expression levels were associated with decreased OS in NSCLC patients (209).
Furthermore, several studies have demonstrated the biomarker potential of other circRNAs, however their role as independent prognostic factors requires further study. For instance, Gu et al. observed that the aberrant expression of hsa_circ_0033155 in NSCLC tissue was correlated with lymphatic metastasis, however no other significant indices were associated with other clinicopathological characteristics (210). Yao et al. proposed a close correlation between circRNA_100876 upregulated expression and lymph node metastasis and tumor staging (211). Moreover, K-M survival analyses demonstrated that the OS for NSCLC patients with high circRNA_100876 expression levels was significantly shorter for patients with low levels (211). Thus, these findings suggested that circRNAs could serve as prognostic markers for lung cancer, however they require further investigation to prove their utility as independent prognostic factors.
The Therapeutic Potential of circRNAs in Lung Cancer
The Role of circRNAs in Lung Cancer Drug Resistance
The greatest hurdle to targeted cancer therapy is the inevitable emergence of drug resistance (212); tumors can develop this resistance during the early or late phase of drug treatment. Resistance is typically classified into two therapeutic categories: (1) intrinsic or primary resistance and, (2) acquired or secondary resistance (212). In recent years, an increased volume of studies have focused on mechanisms of drug resistance in lung cancer (213, 214). Such mechanisms include; the Thr790Met mutation in the epidermal growth factor receptor (EGFR) which induces resistance to tyrosine kinase inhibitors (TKIs) (215), and tumor-reprogramming of the lung microenvironment which induces resistance to angiogenesis and immune checkpoint molecular target therapies in lung cancer (216).
However, in recent years, the role of circRNAs in drug resistance has gained considerable traction with different research groups. Here, we summarize the current data exemplifying the role of circRNAs in therapeutic resistance and related mechanisms of disease.
Platinum Cytotoxic Drugs: Cisplatin (CDDP)
Cisplatin mainly exerts its cytotoxic effects in tumor cells via the generation of DNA-platinum adducts, and subsequent DNA damage responses (212). Despite major clinical successes over recent decades, cisplatin-mediated drug resistance in tumor cells has hindered the clinical utility of this drug (217). Notwithstanding this, a number of mechanisms have emerged contributing to resistance onset. For instance, STAT3 may be attributable to CDDP resistance, and its inhibitors could reverse CDDP-resistance (218). Recently, circRNAs upregulated in NSCLC were found to promote CDDP resistance by activating STAT3 expression; Dong et al.reported that circ_0076305 positively regulated CDDP resistance by upregulating STAT3 by targeting miR-296-5p (219). Moreover, Xu et al. found that circAKT3 inhibited glycolysis and CDDP sensitivity in lung cancer cells by regulating the miR-516b-5p/STAT3 axis (220).
In addition, several other circRNAs were overexpressed in NSCLC, and positively regulated CDDP resistance via other mechanisms. Hong et al. observed that circ-CPA4 promoted cell invasion and EMT processes in NSCLC via the let-7 miRNA/PD-1 axis (221). In addition, NSCLC cell derived PD-L1 exosomes self-regulated cell stemness to increase NSCLC resistance to CDDP, blockading PD-L1 sensitized chemoresistant NSCLC cells to CDDP (221). CircRNACDR1as promoted LUSC metastasis by sponging miR-671-5p and regulating Golgi trafficking (222), thus, participating in CDDP-resistance in NSCLC. Moreover, Zhao et al. indicated that circRNACDR1as regulated stemness properties mediated by CDDP resistance in NSCLC cells by targeting the miR−641/HOXA9 axis (223). Huang et al. screened 31 × eukaryotic initiation factor 3 (EIF3)-derived circRNAs, and correlated two circEIF3 molecules (hsa_circ_0004350 and hsa_circ_0092857) with CDDP drug sensitivity in lung cancer (224); downregulation of these molecules reversed CDDP resistance in lung cancer cells. Pang et al. observed that levels of circ‐PRMT5 and EV3‐like DNA‐directed polymerase ζ catalytic subunit (REV3L) were markedly increased, while miR‐4458 was downregulated in CDDP-resistant NSCLC tissue and cells (225). Circ‐PRMT5 absence contributed to CDDP sensitivity via the miR‐4458/REV3L axis. Importantly, circ‐PRMT5 silencing affected CDDP treatment to expedite an in vivo decrease in tumor growth (225). Li et al. observed that circ_0072083 depletion contributed to CDDP-triggered inhibition of NSCLC tumors via the miR-545-3p/CBLL1 axis (226). Lu et al. proposed that hsa_circ_0096157 expression contributed to CDDP resistance in NSCLC cells by regulating the cell cycle signaling pathway, EMT processes and the expression of apoptosis associated proteins (227). Hsa_circ_0085131 enhanced CDDP-resistance in NSCLC cells by sponging miR-654-5p to upregulate ATG7, leading to cell autophagy (228). Circ_0000376 functioned with oncogenic roles in NSCLC, and enhanced in vitro NSCLC CDDP-resistance by repressing miR-384 (229). Xiao et al. reported that circRNA_103762 expression was upregulated in CDDP-resistant NSCLC cells, and enhanced multidrug resistance by inhibiting CHOP (DNA damage inducible transcript 3) expression in NSCLC cells (230).
However, several circRNAs downregulated in lung cancer have different roles in CDDP-resistance in lung cancer, distinct to the aforementioned circRNAs. Huang et al. indicated that hsa_circ_0001946 exhibited tumor suppressive roles in lung cancer cells, and affected NSCLC cell sensitivity to CDDP via modulation of the Nucleotide excision repair signaling pathway (231). Additionally, a recent study showed that circRNA epithelial splicing regulatory protein-1 (ESRP1) was significantly downregulated in chemoresistant lung cancer cells (232). CircRNA cESRP1 was sensitized to small cell lung cancer (SCLC) cells upon chemotherapy (i.e., doxorubicin, CDDP and etoposide) by sponging miR-93-5p, and upregulating expression of the miR-93-5p downstream target, Smad7/p21(CDKN1A). This formed a negative feedback loop that regulated transforming growth factor-β (TGF-β) mediated EMT processes (232). These studies highlighted the crucial role of circRNAs in CDDP resistance, in both NSCLC and SCLC, and as such may be considered potential novel therapeutic targets for drug resistance in lung cancer.
Antimetabolite Drugs: Pemetrexed and Gemcitabine
Pemetrexed is a novel multitargeted antifolate that inhibits ≥ three enzymes involved in folate metabolism and purine and pyrimidine synthesis (233). Pemetrexed and CDDP combination chemotherapies are widely used to treat NSCLC (234). Mao et al. observed that circRNA CDR1-as was highly expressed in pemetrexed and CDDP resistant LUAD tissues and cell lines (235). CircRNA CDR1-as promoted pemetrexed and CDDP chemoresistance via the EGFR/PI3K signaling pathway in LUAD (235). Zheng et al. reported that circPVT1 overexpression was positively related to chemotherapy insensitivity in LUAD patients (236). Mechanistically, circPVT1 was shown to contribute to pemetrexed and CDDP chemotherapy resistance via the miR-145-5p/ABCC1 axis (236).
Gemcitabine (2’,2’-difluoro 2’-deoxycytidine, dFdC) is the most important cytidine analogue developed since cytosine arabinoside (Ara-C) (237). The cytotoxic activity of gemcitabine may result from several actions against DNA synthesis (237). The combination of gemcitabine and CDDP chemotherapy may be a viable treatment for advanced NSCLC (238). Lu et al. observed that circPVT1 expression was decreased after gemcitabine and CDDP combination treatment; circPVT1 expression in chemotherapy-resistant patients was higher than chemotherapy-sensitive patients (239). Thus, it may be feasible to determine therapy effects post-chemotherapy by detecting circPTV1 expression in patient serum. However, Tong et al. observed that circ-SMARCA5 overexpression enhanced chemosensitivity to gemcitabine and CDDP in treated cells, when compared to overexpression control cells (240). These findings indicated that circRNAs may facilitate rescue for pemetrexed/gemcitabine and CDDP combined chemotherapy resistance in patients.
Plant Derived Chemotherapy Drugs: Paclitaxel (taxol)
Paclitaxel (taxol) was the first member of the taxane family used for cancer chemotherapy; taxanes exert their cytotoxic effects by arresting mitosis via microtubule stabilization, resulting in cellular apoptosis (241). Paclitaxel use has become a broadly accepted option for the treatment of patients with NSCLC (241). A recent study observed that circ_0011292 facilitated tumorigenesis and paclitaxel resistance in NSCLC by regulating the miR-379-5p/TRIM65 axis, suggesting circ_0011292 was a promising therapeutic target for NSCLC chemotherapy (242). Circ_0011292 silencing also reduced paclitaxel resistance in vivo (242). However, Li et al. indicated that circ_0002483 overexpression significantly inhibited NSCLC cell proliferation and invasion in vitro and in vivo, and enhanced NSCLC sensitivity to taxol by sponging miR-182-5p to release the inhibition on GRB2, FOXO1 and FOXO3 mRNAs (243). These studies demonstrated the different roles circRNA exert towards paclitaxel resistance in lung cancer, suggesting their potential as therapeutic strategies in the future.
Targeted Drugs: Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitors (EGFR-TKIs)
These molecules, which include gefitinib, erlotinib and osimertinib (AZD9291), have become important treatment options for NSCLC patients with EGFR sensitive mutations (244). The acquired resistance mechanisms are currently unclear, except for the Thr790Met mutation (215). Recent studies have indicated that circRNAs may play important roles in EGFR-TKI resistance. For instance, Zhou et al. reported that hsa_circ_0004015 contributed to disease progression and gefitinib resistance in NSCLC patients; mechanistically, circ_0004015 sponged miR-1183 and subsequently targeted 3-phosphoinositide dependent protein kinase 1 (PDPK1) (245). PDPK1 is a classic effector of the EGF signaling pathway, which prevents apoptosis and mediates drug resistance in pancreatic cancer (246). Wen et al. found that hsa_circ_0000567 was upregulated and hsa_circ_0006867 downregulated in gefitinib-resistant NSCLC cell lines, when compared to sensitive cells, indicating these circRNAs may be implicated in acquired gefitinib resistance via a circRNA-miRNA-mRNA interactive network (247). Moreover, by analyzing 52 NSCLC patients treated with gefitinib, Liu et al. reported that elevated hsa_circ_0109320, identified from 1,377 circRNAs, was associated with longer PFS in gefitinib-treated NSCLC patients (248). Thus, this circRNA may be a potential biomarker for EGFR-TKI efficacy in these patients.
In addition, Joseph et al. observed that hepatocyte growth factor/c-Met regulated expression of the SUMO-activating enzyme subunit 2(SAE2) and circRNA CCDC66 to increase gefitinib and erlotinib resistance in LUAD cells (249). Mechanistically, SAE2 maintained protein stability, including AAA domain-containing 3A and the EMT markers, vimentin and paxillin, which were crucial for metastatic potential and drug resistance. Critically, these bio-parameters correlated with prognoses in LUAD patients (249).
Furthermore, Ma et al. observed that hsa_circRNA_0002130 was highly expressed in osimertinib-resistant NSCLC cells, and in serum exosomes derived from osimertinib-resistant NSCLC patients (250). Mechanistically, this circRNA facilitated osimertinib-resistance in NSCLC patients by sponging miR-498 to upregulate the GLUT1, hexokinase 2 and LDHA (250). Chen et al. identified 15,504 circRNAs that were differentially expressed in AZD9291-resistant NSCLC cell lines versus control cell lines, and revealed that circRNAs may have roles in NSCLC-AZD9291 resistance. These authors also suggested these circRNAs may be promising molecular candidates for gene therapy (251). These findings demonstrated the importance of circRNAs in TKI-resistance, and their potential as biomarkers for TKI treatment efficacy. However, further studies are required to explore circRNA molecular mechanisms so that target gene therapy studies may proceed to clinical trials.
Immunotherapeutic Drugs: Programmed Cell Death 1/Programmed Cell Death-Ligand 1 (PD-1/PDL-1)
Immune checkpoint inhibitors (ICIs) have dramatically changed the landscape of NSCLC treatment. The PD-1/PDL-1 inhibitors now forms part of first-line NSCLC mono-therapy treatments, or combined with chemotherapy or chemoradiotherapy in patients with stage III unresectable NSCLC (252). Anti-PD-1/PDL-1 ICIs have indicated promising efficacies (~30% response rates), and improved survival of patients with metastatic NSCLC (253). The mechanism of drug resistance during immunotherapy remains unclear, however circRNAs have been shown to exert important functions in NSCLC immunotherapy resistance. CircRNA fibroblast growth factor receptor 1 (circFGFR1/hsa_circ_0084003) promoted NSCLC progression and resistance to anti-PD-1-based therapy (17). CircFGFR1 directly interacted with miR-381-3p, then upregulated the expression of its target gene, C-X-C motif chemokine receptor 4 (CXCR4). Additionally, Luo et al. observed that lung cancer patients with positive PDL-1 expression (≥ 1%) expressed higher level of hsa_circ_0000190 (254). Moreover, long-term follow-up of immunotherapy treated cases indicated that upregulated plasma hsa_circ_0000190 levels correlated with poor responses to systemic therapy and immunotherapy (254). These findings demonstrated that circRNAs played key roles in ICI-resistance, providing potential therapeutic targets for these mechanisms. Similarly, these data also indicated that circRNAs are potential biomarkers for ICI treatment efficacy.
The Role of circRNAs in Radiotherapy Resistance of Lung Cancer
Radiotherapy can be applied in local and regional advanced NSCLC with no surgical chance, as neoadjuvant in the group that has the potential to have surgery and can be applied as adjuvant considering some risk factors after surgery (255). Similarly, radioresistance mechanisms in NSCLC are currently unclear. One potential radioresistance mechanism involves the generation of exosomes. Exosomes are 40–150 nm vesicles released by cancer cells and contain pathogenic components, such as proteins, mRNAs, DNA fragments, non-coding RNAs and lipids. It has been proposed that radiation-derived exosomes may promote radioresistance (256). For instance, Fan et al. observed that both intracellular and extracellular miR-1246 levels were upregulated after irradiation in a time-dependent pattern, resulting in increased NSCLC radioresistance via mTOR-inhibited autophagy activation (257). Moreover, circRNA CDR1as sequestered miR-1246 and antagonized its effects towards radioresistance mechanisms in NSCLC cell lines (257). Accordingly, it suggested that circRNAs could play vital roles in radioresistance mechanisms in lung cancer, however more studies are required to explore such mechanisms initiated by circRNAs in radioresistance.
Conclusion and Perspectives
RNA-seq technologies have provided unprecedented insights into the human genome; the clinical value of circRNAs has been unraveled for various diseases, including lung cancer. As outlined here, circRNAs participate in lung cancer proliferation, migration, invasion and apoptosis, and have the extraordinary potential to be recognized as biomarkers for molecular therapeutics. As good biomarkers, circRNAs have stable molecular structure and are more abundant than liner RNAs. However, research in this area is far from complete, therefore we propose three critical research areas which will define the success of these molecules as biomarkers; (1) The role of circRNAs in other lung cancer pathological processes is little known, therefore more studies are required to investigate tumor microenvironments, molecular heterogeneity and inflammation, etc. (2) More circRNA biomarker research must be conducted to distinguish different pathological types of lung cancer. (3) More comprehensive and in-depth clinical trials must be conducted to verify these biomarkers as potential therapeutics for lung cancer. (4) In-depth studies on cells, animals and population cohorts are needed to further explore the value of circRNAs in evaluating the prognosis of lung cancer.
By implementing these three key strategies, we will gain a better understanding of the regulatory roles of circRNAs in lung cancer, as they will provide new molecular insights into circRNA mechanisms behind the disease. Thus, this review has systematically and comprehensively highlighted the recent advances in circRNAs in lung cancer etiology, and has exemplified the diagnostic and therapeutic potential of circRNAs in this disease.
Author Contributions
H-HC and T-NZ contributed equally to this work. H-HC and T-NZ wrote the manuscript. Q-JW and Y-HZ is responsible for research supervision. Y-HZ is responsible for funding acquisition. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Key R&D Program of China (No. 2017YFC0907401 to Y-HZ), the Natural Science Foundation of China (No. 82073647 to Q-JW), and LiaoNing Revitalization Talents Program (No. XLYC1907102 to Q-JW and No. XLYC1802095 to Y-HZ).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Nasim F, Sabath BF, Eapen GA. Lung Cancer. Med Clinics North Am (2019) 103:463–73. doi: 10.1016/j.mcna.2018.12.006
2. Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer Incidence and Mortality Worldwide: Sources, Methods and Major Patterns in GLOBOCAN 2012. Int J Cancer (2015) 136:E359–86. doi: 10.1002/ijc.29210
3. Hirsch FR, Scagliotti GV, Mulshine JL, Kwon R, Curran WJ, Wu Y-L, et al. Lung Cancer: Current Therapies and New Targeted Treatments. Lancet (Lond Eng) (2017) 389:299–311. doi: 10.1016/S0140-6736(16)30958-8
4. Smith RA, Manassaram-Baptiste D, Brooks D, Doroshenk M, Fedewa S, Saslow D, et al. Cancer Screening in the United States, 2015: A Review of Current American Cancer Society Guidelines and Current Issues in Cancer Screening. CA: Cancer J Clin (2015) 65:30–54. doi: 10.3322/caac.21261
5. Ettinger DS, Wood DE, Akerley W, Bazhenova LA, Borghaei H, Camidge DR, et al. Non-Small Cell Lung Cancer, Version 6.2015. J Natl Compr Cancer Network: JNCCN (2015) 13:515–24. doi: 10.6004/jnccn.2015.0071
6. Altesha MA, Ni T, Khan A, Liu K, Zheng X. Circular RNA in Cardiovascular Disease. J Cell Physiol (2019) 234:5588–600. doi: 10.1002/jcp.27384
7. Xiang S, Li Z, Bian Y, Weng X. RNA Sequencing Reveals the Circular RNA Expression Profiles of Osteoarthritic Synovium. J Cell Biochem (2019) 120:18031–40. doi: 10.1002/jcb.29106
8. Hua JT, Chen S, He HH. Landscape of Noncoding RNA in Prostate Cancer. Trends Genet: TIG (2019) 35:840–51. doi: 10.1016/j.tig.2019.08.004
9. Hua L, Huang L, Zhang X, Feng H, Shen B. Knockdown of Circular RNA CEP128 Suppresses Proliferation and Improves Cytotoxic Efficacy of Temozolomide in Glioma Cells by Regulating miR-145-5p. Neuroreport (2019) 30:1231–8. doi: 10.1097/WNR.0000000000001326
10. Liu Z, Zhou Y, Liang G, Ling Y, Tan W, Tan L, et al. Circular RNA Hsa_Circ_001783 Regulates Breast Cancer Progression via Sponging miR-200c-3p. Cell Death Dis (2019) 10:55. doi: 10.1038/s41419-018-1287-1
11. Arnberg AC, Van Ommen GJ, Grivell LA, Van Bruggen EF, Borst P. Some Yeast Mitochondrial RNAs Are Circular. Cell (1980) 19:313–9. doi: 10.1016/0092-8674(80)90505-X
12. Kos A, Dijkema R, Arnberg AC, van der Meide PH, Schellekens H. The Hepatitis Delta (Delta) Virus Possesses a Circular RNA. Nature (1986) 323:558–60. doi: 10.1038/323558a0
13. Chandler B, Hayashi M, Hayashi MN, Spiegelman S. Circularity of the Replicating Form of a Single-Stranded DNA Virus. Sci (New York NY) (1964) 143:47–9. doi: 10.1126/science.143.3601.47
14. Bretscher MS. Direct Translation of a Circular Messenger DNA. Nature (1968) 220:1088–91. doi: 10.1038/2201088a0
15. Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are Single-Stranded Covalently Closed Circular RNA Molecules Existing as Highly Base-Paired Rod-Like Structures. Proc Natl Acad Sci USA (1976) 73:3852–6. doi: 10.1073/pnas.73.11.3852
16. Hsu MT, Coca-Prados M. Electron Microscopic Evidence for the Circular Form of RNA in the Cytoplasm of Eukaryotic Cells. Nature (1979) 280:339–40. doi: 10.1038/280339a0
17. Zhang P-F, Pei X, Li K-S, Jin L-N, Wang F, Wu J, et al. Circular RNA Circfgfr1 Promotes Progression and Anti-PD-1 Resistance by Sponging miR-381-3p in Non-Small Cell Lung Cancer Cells. Mol Cancer (2019) 18:179. doi: 10.1186/s12943-019-1111-2
18. Wang L, Tong X, Zhou Z, Wang S, Lei Z, Zhang T, et al. Circular RNA Hsa_Circ_0008305 (Circptk2) Inhibits TGF-β-Induced Epithelial-Mesenchymal Transition and Metastasis by Controlling TIF1γ in Non-Small Cell Lung Cancer. Mol Cancer (2018) 17:140. doi: 10.1186/s12943-018-0889-7
19. Nan A, Chen L, Zhang N, Jia Y, Li X, Zhou H, et al. Circular RNA Circnol10 Inhibits Lung Cancer Development by Promoting SCLM1-Mediated Transcriptional Regulation of the Humanin Polypeptide Family. Adv Sci (2019) 6(2):1800654. doi: 10.1002/advs.201800654
20. Qu S, Yang X, Li X, Wang J, Gao Y, Shang R, et al. Circular RNA: A New Star of Noncoding RNAs. Cancer Lett (2015) 365:141–8. doi: 10.1016/j.canlet.2015.06.003
21. Maass PG, Glažar P, Memczak S, Dittmar G, Hollfinger I, Schreyer L, Sauer AV, et al. A Map of Human Circular RNAs in Clinically Relevant Tissues. J Mol Med (Berlin Germany) (2017) 95:1179–89. doi: 10.1007/s00109-017-1582-9
22. Kristensen LS, Okholm TLH, Venø MT, Kjems J. Circular RNAs are Abundantly Expressed and Upregulated During Human Epidermal Stem Cell Differentiation. RNA Biol (2018) 15:280–91. doi: 10.1080/15476286.2017.1409931
23. Yu C-Y, Li T-C, Wu Y-Y, Yeh C-H, Chiang W, Chuang C-Y, et al. The Circular RNA Circbirc6 Participates in the Molecular Circuitry Controlling Human Pluripotency. Nat Commun (2017) 8:1149. doi: 10.1038/s41467-017-01216-w
24. Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, et al. Translation of CircRNAs. Mol Cell (2017) 66(1):9–21.e7. doi: 10.1016/j.molcel.2017.02.021
25. Meng S, Zhou H, Feng Z, Xu Z, Tang Y, Li P, et al. CircRNA: Functions and Properties of a Novel Potential Biomarker for Cancer. Mol Cancer (2017) 16:94. doi: 10.1186/s12943-017-0663-2
26. Gao Y, Wang J, Zhao F. CIRI: An Efficient and Unbiased Algorithm for De Novo Circular RNA Identification. Genome Biol (2015) 16:4. doi: 10.1186/s13059-014-0571-3
27. Li X, Yang L, Chen L-L. The Biogenesis, Functions, and Challenges of Circular RNAs. Mol Cell (2018) 71:428–42. doi: 10.1016/j.molcel.2018.06.034
28. Wilusz JE. A 360° View of Circular RNAs: From Biogenesis to Functions. Wiley Interdiscip Rev RNA (2018) 9:e1478. doi: 10.1002/wrna.1478
29. Ng WL, Mohd Mohidin TB, Shukla K. Functional Role of Circular RNAs in Cancer Development and Progression. RNA Biol (2018) 15(8):995–1005. doi: 10.1080/15476286.2018.1486659
30. Kristensen LS, Hansen TB, Venø MT, Kjems J. Circular RNAs in Cancer: Opportunities and Challenges in the Field. Oncogene (2018) 37:555–65. doi: 10.1038/onc.2017.361
31. Shang Q, Yang Z, Jia R, Ge S. The Novel Roles of circRNAs in Human Cancer. Mol Cancer (2019) 18:6. doi: 10.1186/s12943-018-0934-6
32. Zhan T, Ambrosi G, Wandmacher AM, Rauscher B, Betge J, Rindtorff N, et al. MEK Inhibitors Activate Wnt Signalling and Induce Stem Cell Plasticity in Colorectal Cancer. Nat Commun (2019) 10:2197. doi: 10.1038/s41467-019-09898-0
33. Khramtsov AI, Khramtsova GF, Tretiakova M, Huo D, Olopade OI, Goss KH. Wnt/beta-Catenin Pathway Activation Is Enriched in Basal-Like Breast Cancers and Predicts Poor Outcome. Am J Pathol (2010) 176:2911–20. doi: 10.2353/ajpath.2010.091125
34. Krishnamurthy N, Kurzrock R. Targeting the Wnt/beta-Catenin Pathway in Cancer: Update on Effectors and Inhibitors. Cancer Treat Rev (2018) 62:50–60. doi: 10.1016/j.ctrv.2017.11.002
35. Li YF, Zhang J, Yu L. Circular RNAs Regulate Cancer Onset and Progression via Wnt/β-Catenin Signaling Pathway. Yonsei Med J (2019) 60:1117–28. doi: 10.3349/ymj.2019.60.12.1117
36. Ma D, Liu H, Qin Y, Li D, Cui Y, Li L, et al. Circ_0007142/miR-186/FOXK1 Axis Promoted Lung Adenocarcinoma Progression. Am J Trans Res (2020) 12:4728–38.
37. Ding L, Yao W, Lu J, Gong J, Zhang X. Upregulation of Circ_001569 Predicts Poor Prognosis and Promotes Cell Proliferation in Non-Small Cell Lung Cancer by Regulating the Wnt/β-Catenin Pathway. Oncol Lett (2018) 16:453–8. doi: 10.3892/ol.2018.8673
38. Li XY, Liu YR, Zhou JH, Li W, Guo HH, Ma HP. Enhanced Expression of Circular RNA Hsa_Circ_000984 Promotes Cells Proliferation and Metastasis in Non-Small Cell Lung Cancer by Modulating Wnt/β-Catenin Pathway. Eur Rev Med Pharmacol Sci (2019) 23:3366–74. doi: 10.26355/eurrev_201904_17700
39. Li O, Kang J, Zhang JJ, Wang J, Hu LW, Li L, et al. Circle RNA FOXP1 Promotes Cell Proliferation in Lung Cancer by Regulating miR-185-5p/Wnt1 Signaling Pathway. Eur Rev Med Pharmacol Sci (2020) 24:6767–78. doi: 10.26355/eurrev_202006_21665
40. Gao N, Ye B. Circ-SOX4 Drives the Tumorigenesis and Development of Lung Adenocarcinoma via Sponging miR-1270 and Modulating PLAGL2 to Activate WNT Signaling Pathway. Cancer Cell Int (2020) 20:2. doi: 10.1186/s12935-019-1065-x
41. Wang L, Zheng C, Wu X, Zhang Y, Yan S, Ruan L, et al. Circ-SOX4 Promotes Non-Small Cell Lung Cancer Progression by Activating the Wnt/β-Catenin Pathway. Mol Oncol (2020) 14(12):3253. doi: 10.1002/1878-0261.12656
42. Yao Y, Hua Q, Zhou Y, Shen H. CircRNA has_Circ_0001946 Promotes Cell Growth in Lung Adenocarcinoma by Regulating miR-135a-5p/SIRT1 Axis and Activating Wnt/β-Catenin Signaling Pathway. Biomed Pharmacother (2019) 111:1367–75. doi: 10.1016/j.biopha.2018.12.120
43. Wan L, Zhang L, Fan K, Cheng Z-X, Sun Q-C, Wang J-J. Circular RNA-ITCH Suppresses Lung Cancer Proliferation via Inhibiting the Wnt/β-Catenin Pathway. BioMed Res Int (2016) 2016:1579490. doi: 10.1155/2016/1579490
44. Tian F, Yu CT, Ye WD, Wang Q. Cinnamaldehyde Induces Cell Apoptosis Mediated by a Novel Circular RNA Hsa_Circ_0043256 in Non-Small Cell Lung Cancer. Biochem Biophys Res Commun (2017) 493:1260–6. doi: 10.1016/j.bbrc.2017.09.136
45. Gao S, Yu Y, Liu L, Meng J, Li G. Circular RNA Hsa_Circ_0007059 Restrains Proliferation and Epithelial-Mesenchymal Transition in Lung Cancer Cells via Inhibiting microRNA-378. Life Sci (2019) 233:116692. doi: 10.1016/j.lfs.2019.116692
46. Xu Z, Xiang W, Chen W, Sun Y, Qin F, Wei J, et al. Circ-IGF1R Inhibits Cell Invasion and Migration in Non-Small Cell Lung Cancer. Thoracic Cancer (2020) 11:875–87. doi: 10.1111/1759-7714.13329
47. Yao Y, Hua Q, Zhou Y. CircRNA has_Circ_0006427 Suppresses the Progression of Lung Adenocarcinoma by Regulating miR-6783-3p/DKK1 Axis and Inactivating Wnt/β-Catenin Signaling Pathway. Biochem Biophys Res Commun (2019) 508:37–45. doi: 10.1016/j.bbrc.2018.11.079
48. Luo Y, Chen J-J, Lv Q, Qin J, Huang Y-Z, Yu M-H, et al. Long non-Coding RNA NEAT1 Promotes Colorectal Cancer Progression by Competitively Binding miR-34a With SIRT1 and Enhancing the Wnt/β-Catenin Signaling Pathway. Cancer Lett (2019) 440-441:11–22. doi: 10.1016/j.canlet.2018.10.002
49. Liu S, Yang H, Hu B, Zhang M. Sirt1 Regulates Apoptosis and Extracellular Matrix Degradation in Resveratrol-Treated Osteoarthritis Chondrocytes via the Wnt/β-Catenin Signaling Pathways. Exp Ther Med (2017) 14:5057–62. doi: 10.3892/etm.2017.5165
50. Fang JY, Richardson BC. The MAPK Signalling Pathways and Colorectal Cancer. Lancet Oncol (2005) 6:322–7. doi: 10.1016/S1470-2045(05)70168-6
51. Guo Y-J, Pan W-W, Liu S-B, Shen Z-F, Xu Y, Hu L-L. ERK/MAPK Signalling Pathway and Tumorigenesis. Exp Ther Med (2020) 19:1997–2007. doi: 10.3892/etm.2020.8454
52. Sun Y, Liu W-Z, Liu T, Feng X, Yang N, Zhou H-F. Signaling Pathway of MAPK/ERK in Cell Proliferation, Differentiation, Migration, Senescence and Apoptosis. J Recept Signal Transduction Res (2015) 35:600–4. doi: 10.3109/10799893.2015.1030412
53. Chen S, Lu S, Yao Y, Chen J, Yang G, Tu L, et al. Downregulation of Hsa_Circ_0007580 Inhibits Non-Small Cell Lung Cancer Tumorigenesis by Reducing miR-545-3p Sponging. Aging (2020) 12:14329–40. doi: 10.18632/aging.103472
54. Gao P, Wang Z, Hu Z, Jiao X, Yao Y. Circular RNA Circ_0074027 Indicates a Poor Prognosis for NSCLC Patients and Modulates Cell Proliferation, Apoptosis, and Invasion via miR-185-3p Mediated BRD4/MADD Activation. J Cell Biochem (2019) 121:2632–42. doi: 10.1002/jcb.29484
55. Zhang Z-Y, Gao X-H, Ma M-Y, Zhao C-L, Zhang Y-L, Guo S-S. CircRNA_101237 Promotes NSCLC Progression via the miRNA-490-3p/MAPK1 Axis. Sci Rep (2020) 10:9024. doi: 10.1038/s41598-020-65920-2
56. Wang Y, Xu R, Zhang D, Lu T, Yu W, Wo Y, et al. Circ-ZKSCAN1 Regulates FAM83A Expression and Inactivates MAPK Signaling by Targeting miR-330-5p to Promote Non-Small Cell Lung Cancer Progression. Trans Lung Cancer Res (2019) 8:862–75. doi: 10.21037/tlcr.2019.11.04
57. Shen Y, Xue C, Li X, Ba L, Gu J, Sun Z, et al. Effects of Gastric Cancer Cell-Derived Exosomes on the Immune Regulation of Mesenchymal Stem Cells by the NF-kB Signaling Pathway. Stem Cells Dev (2019) 28:464–76. doi: 10.1089/scd.2018.0125
58. Dolcet X, Llobet D, Pallares J, Matias-Guiu X. NF-kB in Development and Progression of Human Cancer. Virchows Archiv: An Int J Pathol (2005) 446:475–82. doi: 10.1007/s00428-005-1264-9
59. Oeckinghaus A, Hayden MS, Ghosh S. Crosstalk in NF-κb Signaling Pathways. Nat Immunol (2011) 12:695–708. doi: 10.1038/ni.2065
60. Zhou Q, Sun Y. Circular RNA Cmras Suppresses the Progression of Lung Adenocarcinoma Through ABHD5/ATGL Axis Using NF-kappaB Signaling Pathway. Cancer Biother Radiopharm (2020). doi: 10.1089/cbr.2020.3709
61. Reeves R. High Mobility Group (HMG) Proteins: Modulators of Chromatin Structure and DNA Repair in Mammalian Cells. DNA Repair (2015) 36:122–36. doi: 10.1016/j.dnarep.2015.09.015
62. Sgarra R, Pegoraro S, Ros G, Penzo C, Chiefari E, Foti D, et al. (HMGA) Proteins: Molecular Instigators of Breast Cancer Onset and Progression. Biochim Biophys Acta Rev Cancer (2018) 1869:216–29. doi: 10.1016/j.bbcan.2018.03.001
63. Hawsawi O, Henderson V, Burton LJ, Dougan J, Nagappan P, Odero-Marah V. High Mobility Group A2 (HMGA2) Promotes EMT via MAPK Pathway in Prostate Cancer. Biochem Biophys Res Commun (2018) 504:196–202. doi: 10.1016/j.bbrc.2018.08.155
64. Wu X-J, Chen Y-Y, Gong C-C, Pei D-S. The Role of High-Mobility Group Protein Box 1 in Lung Cancer. J Cell Biochem (2018) 119:6354–65. doi: 10.1002/jcb.26837
65. Xu L, Ma Y, Zhang H, Lu Q-J, Yang L, Jiang G-N, et al. HMGA2 Regulates Circular RNA ASPH to Promote Tumor Growth in Lung Adenocarcinoma. Cell Death Dis (2020) 11:593. doi: 10.1038/s41419-020-2726-3
66. Li L, Wei H, Zhang H, Xu F, Che G. Circ_100565 Promotes Proliferation, Migration and Invasion in Non-Small Cell Lung Cancer Through Upregulating HMGA2 via Sponging miR-506-3p. Cancer Cell Int (2020) 20:160. doi: 10.1186/s12935-020-01241-8
67. Xie Y, Wang L, Yang D. CircEPSTI1 Promotes the Progression of Non-Small Cell Lung Cancer Through miR-145/HMGB3 Axis. Cancer Manage Res (2020) 12:6827–36. doi: 10.2147/CMAR.S252893
68. Zhou Z-F, Wei Z, Yao J-C, Liu S-Y, Wang F, Wang Z, et al. CircRNA_102179 Promotes the Proliferation, Migration and Invasion in Non-Small Cell Lung Cancer Cells by Regulating miR-330-5p/HMGB3 Axis. Pathol Res Pract (2020) 216:153144. doi: 10.1016/j.prp.2020.153144
69. Rehwinkel J, Behm-Ansmant I, Gatfield D, Izaurralde E. A Crucial Role for GW182 and the DCP1:DCP2 Decapping Complex in miRNA-Mediated Gene Silencing. RNA (New York NY) (2005) 11:1640–7. doi: 10.1261/rna.2191905
70. Riedmann LT, Schwentner R. miRNA, siRNA, piRNA and Argonautes: News in Small Matters. RNA Biol (2010) 7:133–9. doi: 10.4161/rna.7.2.11288
71. Wu J, Yang J, Cho WC, Zheng Y. Argonaute Proteins: Structural Features, Functions and Emerging Roles. J Advanced Res (2020) 24:317–24. doi: 10.1016/j.jare.2020.04.017
72. Liu M, Wang P, Sui X, Ding F, Liu L, Gao Z, et al. Circular RNA Circabcc4 Regulates Lung Adenocarcinoma Progression via miR-3186-3p/TNRC6B Axis. J Cell Biochem (2020) 121:4226–38. doi: 10.1002/jcb.29627
73. Dai X, Zhang N, Cheng Y, Yang T, Chen Y, Liu Z, et al. RNA-Binding Protein Trinucleotide Repeat-Containing 6A Regulates the Formation of Circular RNA Circ0006916, With Important Functions in Lung Cancer Cells. Carcinogenesis (2018) 39:981–92. doi: 10.1093/carcin/bgy061
74. Chen H, Liu H, Qing G. Targeting Oncogenic Myc as a Strategy for Cancer Treatment. Signal transduction Targeted Ther (2018) 3:5. doi: 10.1038/s41392-018-0008-7
75. Stine ZE, Walton ZE, Altman BJ, Hsieh AL, Dang CV. MYC, Metabolism, and Cancer. Cancer Discovery (2015) 5:1024–39. doi: 10.1158/2159-8290.CD-15-0507
76. Liu W, Ma W, Yuan Y, Zhang Y, Sun S. Circular RNA Hsa_circRNA_103809 Promotes Lung Cancer Progression via Facilitating ZNF121-Dependent MYC Expression by Sequestering miR-4302. Biochem Biophys Res Commun (2018) 500:846–51. doi: 10.1016/j.bbrc.2018.04.172
77. Zhang P, Xue XF, Ling XY, Yang Q, Yu Y, Xiao J, et al. CircRNA_010763 Promotes Growth and Invasion of Lung Cancer Through Serving as a Molecular Sponge of miR-715 to Induce C-Myc Expression. Eur Rev Med Pharmacol Sci (2020) 24:7310–9. doi: 10.26355/eurrev_202007_21892
78. Liao G-B, Li X-Z, Zeng S, Liu C, Yang S-M, Yang L, et al. Regulation of the Master Regulator FOXM1 in Cancer. Cell Commun Signaling: CCS (2018) 16:57. doi: 10.1186/s12964-018-0266-6
79. Nandi D, Cheema PS, Jaiswal N, Nag A. FoxM1: Repurposing an Oncogene as a Biomarker. Semin Cancer Biol (2018) 52:74–84. doi: 10.1016/j.semcancer.2017.08.009
80. Gartel AL. FOXM1 in Cancer: Interactions and Vulnerabilities. Cancer Res (2017) 77:3135–9. doi: 10.1158/0008-5472.CAN-16-3566
81. Cheng Z, Yu C, Cui S, Wang H, Jin H, Wang C, et al. Circtp63 Functions as a ceRNA to Promote Lung Squamous Cell Carcinoma Progression by Upregulating FOXM1. Nat Commun (2019) 10:3200. doi: 10.1038/s41467-019-11162-4
82. Lu H, Han X, Ren J, Ren K, Li Z, Sun Z. Circular RNA HIPK3 Induces Cell Proliferation and Inhibits Apoptosis in Non-Small Cell Lung Cancer Through Sponging miR-149. Cancer Biol Ther (2020) 21:113–21. doi: 10.1080/15384047.2019.1669995
83. Yu C, Cheng Z, Cui S, Mao X, Li B, Fu Y, et al. Circfoxm1 Promotes Proliferation of Non-Small Cell Lung Carcinoma Cells by Acting as a ceRNA to Upregulate FAM83D. J Exp Clin Cancer Res: CR (2020) 39:55. doi: 10.1186/s13046-020-01555-5
84. Serrano-Gomez SJ, Maziveyi M, Alahari SK. Regulation of Epithelial-Mesenchymal Transition Through Epigenetic and Post-Translational Modifications. Mol Cancer (2016) 15:18. doi: 10.1186/s12943-016-0502-x
85. Yilmaz M, Christofori G. EMT, the Cytoskeleton, and Cancer Cell Invasion. Cancer Metastasis Rev (2009) 28:15–33. doi: 10.1007/s10555-008-9169-0
86. Mittal V. Epithelial Mesenchymal Transition in Tumor Metastasis. Annu Rev Pathol (2018) 13:395–412. doi: 10.1146/annurev-pathol-020117-043854
87. Xue M, Hong W, Jiang J, Zhao F, Gao X. Circular RNA Circ-LDLRAD3 Serves as an Oncogene to Promote Non-Small Cell Lung Cancer Progression by Upregulating SLC1A5 Through Sponging miR-137. RNA Biol (2020) 17(12):1811–22. doi: 10.1080/15476286.2020.1789819
88. van Geldermalsen M, Wang Q, Nagarajah R, Marshall AD, Thoeng A, Gao D, et al. ASCT2/SLC1A5 Controls Glutamine Uptake and Tumour Growth in Triple-Negative Basal-Like Breast Cancer. Oncogene (2016) 35:3201–8. doi: 10.1038/onc.2015.381
89. Qin H, Liu J, Du ZH, Hu R, Yu YK, Wang QA. Circular RNA Hsa_Circ_0012673 Facilitates Lung Cancer Cell Proliferation and Invasion via miR-320a/LIMK18521 Axis. Eur Rev Med Pharmacol Sci (2020) 24:1841–52. doi: 10.26355/eurrev_202002_20362
90. Li J, Wang J, Chen Z, Chen Y, Jin M. Hsa_circ_0079530 Promotes Cell Proliferation and Invasion in Non-Small Cell Lung Cancer. Gene (2018) 665:1–5. doi: 10.1016/j.gene.2018.04.059
91. Liu C, Zhang Z, Qi D. Circular RNA Hsa_Circ_0023404 Promotes Proliferation, Migration and Invasion in Non-Small Cell Lung Cancer by Regulating miR-217/ZEB1 Axis. OncoTargets Ther (2019) 12:6181–9. doi: 10.2147/OTT.S201834
92. Qu D, Yan B, Xin R, Ma T. A Novel Circular RNA Hsa_Circ_0020123 Exerts Oncogenic Properties Through Suppression of miR-144 in Non-Small Cell Lung Cancer. Am J Cancer Res (2018) 8:1387–402.
93. Chi Y, Luo Q, Song Y, Yang F, Wang Y, Jin M, et al. Circular RNA Circpip5k1a Promotes Non-Small Cell Lung Cancer Proliferation and Metastasis Through miR-600/HIF-1α Regulation. J Cell Biochem (2019) 120:19019–30. doi: 10.1002/jcb.29225
94. Wang T, Wang X, Du Q, Wu N, Liu X, Chen Y, et al. The circRNA Circp4hb Promotes NSCLC Aggressiveness and Metastasis by Sponging miR-133a-5p. Biochem Biophys Res Commun (2019) 513:904–11. doi: 10.1016/j.bbrc.2019.04.108
95. Qi Y, Zhang B, Wang J, Yao M. Upregulation of Circular RNA Hsa_Circ_0007534 Predicts Unfavorable Prognosis for NSCLC and Exerts Oncogenic Properties In Vitro and In Vivo. Gene (2018) 676:79–85. doi: 10.1016/j.gene.2018.07.028
96. Wei S, Zheng Y, Jiang Y, Li X, Geng J, Shen Y, et al. The circRNA circPTPRA Suppresses Epithelial-Mesenchymal Transitioning and Metastasis of NSCLC Cells by Sponging miR-96-5p. EBioMedicine (2019) 44:182–93. doi: 10.1016/j.ebiom.2019.05.032
97. Ganapathy-Kanniappan S, Geschwind J-FH. Tumor Glycolysis as a Target for Cancer Therapy: Progress and Prospects. Mol Cancer (2013) 12:152. doi: 10.1186/1476-4598-12-152
98. Guo F, Li S, Guo C, Xu X, Zhou X, Ma D, et al. Circular RNA Circmagi3 Accelerates the Glycolysis of Non-Small Cell Lung Cancer Through miR-515-5p/HDGF. Am J Trans Res (2020) 12:3953–63.
99. Shangguan H, Feng H, Lv D, Wang J, Tian T, Wang X. Circular RNA Circslc25a16 Contributes to the Glycolysis of Non-Small-Cell Lung Cancer Through Epigenetic Modification. Cell Death Dis (2020) 11:437. doi: 10.1038/s41419-020-2635-5
100. Wu W, Xi W, Li H, Yang M, Yao X. Circular RNA Circ−ACACA Regulates Proliferation, Migration and Glycolysis in Non−Small−Cell Lung Carcinoma via miR−1183 and PI3K/PKB Pathway. Int J Mol Med (2020) 45:1814–24. doi: 10.3892/ijmm.2020.4549
101. Huang W, Xu X, Liu M, Cui W, Peng G. Downregulation of Hsa_circ_0000735 Inhibits the Proliferation, Migration, Invasion, and Glycolysis in Non-Small-Cell Lung Cancer by Targeting miR-940/BMPER Axis. OncoTargets Ther (2020) 13:8427–39. doi: 10.2147/OTT.S253474
102. Ding C, Xi G, Wang G, Cui D, Zhang B, Wang H, et al. Exosomal Circ-MEMO1 Promotes the Progression and Aerobic Glycolysis of Non-Small Cell Lung Cancer Through Targeting MiR-101-3p/KRAS Axis. Front Genet (2020) 11:962. doi: 10.3389/fgene.2020.00962
103. Zhou J, Zhang S, Chen Z, He Z, Xu Y, Li Z. CircRNA-ENO1 Promoted Glycolysis and Tumor Progression in Lung Adenocarcinoma Through Upregulating its Host Gene ENO1. Cell Death Dis (2019) 10(12):885. doi: 10.1038/s41419-019-2127-7
104. Xiong S, Li D, Wang D, Huang L, Liang G, Wu Z, et al. Circular RNA MYLK Promotes Glycolysis and Proliferation of Non-Small Cell Lung Cancer Cells by Sponging miR-195-5p and Increasing Glucose Transporter Member 3 Expression. Cancer Manage Res (2020) 12:5469–78. doi: 10.2147/CMAR.S257386
105. Zhang SJ, Ma J, Wu JC, Hao ZZ, Zhang YA, Zhang YJ. Circular RNA Circcrim1 Suppresses Lung Adenocarcinoma Cell Migration, Invasion, EMT, and Glycolysis Through Regulating miR-125b-5p/BTG2 Axis. Eur Rev Med Pharmacol Sci (2020) 24:3761–74. doi: 10.26355/eurrev_202004_20841
106. Yao J, Xu G, Zhu L, Zheng H. Circgfra1 Enhances NSCLC Progression by Sponging miR-188-3p. OncoTargets Ther (2020) 13:549–58. doi: 10.2147/OTT.S230795
107. Du J, Zhang G, Qiu H, Yu H, Yuan W. The Novel Circular RNA Circ-CAMK2A Enhances Lung Adenocarcinoma Metastasis by Regulating the miR-615-5p/Fibronectin 1 Pathway. Cell Mol Biol Lett (2019) 24:72. doi: 10.1186/s11658-019-0198-1
108. Ma X, Yang X, Bao W, Li S, Liang S, Sun Y, et al. Circular RNA Circman2b2 Facilitates Lung Cancer Cell Proliferation and Invasion via miR-1275/FOXK1 Axis. Biochem Biophys Res Commun (2018) 498:1009–15. doi: 10.1016/j.bbrc.2018.03.105
109. Hong W, Zhang Y, Ding J, Yang Q, Xie H, Gao X. Circhipk3 Acts as Competing Endogenous RNA and Promotes Non-Small-Cell Lung Cancer Progression Through the miR-107/BDNF Signaling Pathway. BioMed Res Int (2020) 2020:6075902. doi: 10.1155/2020/6075902
110. Liu G, Shi H, Deng L, Zheng H, Kong W, Wen X, et al. Circular RNA Circ-FOXM1 Facilitates Cell Progression as ceRNA to Target PPDPF and MACC1 by Sponging miR-1304-5p in Non-Small Cell Lung Cancer. Biochem Biophys Res Commun (2019) 513:207–12. doi: 10.1016/j.bbrc.2019.03.213
111. Li C, Liu H, Niu Q, Gao J. Circ_0000376, a Novel circRNA, Promotes the Progression of Non-Small Cell Lung Cancer Through Regulating the miR-1182/NOVA2 Network. Cancer Manage Res (2020) 12:7635–47. doi: 10.2147/CMAR.S258340
112. Li W, Jiang W, Liu T, Lv J, Guan J. Enhanced Expression of Circ_0000735 Forecasts Clinical Severity in NSCLC and Promotes Cell Progression via Sponging miR-1179 and miR-1182. Biochem Biophys Res Commun (2019) 510:467–71. doi: 10.1016/j.bbrc.2019.01.134
113. Cui J, Li W, Liu G, Chen X, Gao X, Lu H, et al. A Novel Circular RNA, Hsa_Circ_0043278, Acts as a Potential Biomarker and Promotes Non-Small Cell Lung Cancer Cell Proliferation and Migration by Regulating miR-520f. Artif Cells Nanomed Biotechnol (2019) 47:810–21. doi: 10.1080/21691401.2019.1575847
114. Qiu B-Q, Zhang P-F, Xiong D, Xu J-J, Long X, Zhu S-Q, et al. CircRNA Fibroblast Growth Factor Receptor 3 Promotes Tumor Progression in Non-Small Cell Lung Cancer by Regulating Galectin-1-AKT/ERK1/2 Signaling. J Cell Physiol (2019) 234:11256–64. doi: 10.1002/jcp.27783
115. Luo YH, Zhu XZ, Huang KW, Zhang Q, Fan YX, Yan PW, et al. Emerging Roles of Circular RNA Hsa_Circ_0000064 in the Proliferation and Metastasis of Lung Cancer. BioMed Pharmacother (2017) 96:892–8. doi: 10.1016/j.biopha.2017.12.015
116. Wang L, Zhao L, Wang Y. Circular RNA Circ_0020123 Promotes Non-Small Cell Lung Cancer Progression by Sponging miR-590-5p to Regulate THBS2. Cancer Cell Int (2020) 20:387. doi: 10.1186/s12935-020-01444-z
117. Jin M, Shi C, Yang C, Liu J, Huang G. Upregulated circRNA ARHGAP10 Predicts an Unfavorable Prognosis in NSCLC Through Regulation of the miR-150-5p/GLUT-1 Axis. Mol Ther Nucleic Acids (2019) 18:219–31. doi: 10.1016/j.omtn.2019.08.016
118. Zheng JP, Dai YM, Chen Z, Chen Q, Zheng Y, Lin X, et al. Circular RNA Circ-ABCB10 Promotes Non-Small Cell Lung Cancer Proliferation and Inhibits Cell Apoptosis Through Repressing KISS1. Eur Rev Med Pharmacol Sci (2020) 24:2518–24. doi: 10.26355/eurrev_202003_20519
119. Tian X, Zhang L, Jiao Y, Chen J, Shan Y, Yang W. CircABCB10 Promotes Nonsmall Cell Lung Cancer Cell Proliferation and Migration by Regulating the miR-1252/FOXR2 Axis. J Cell Biochem (2019) 120:3765–72. doi: 10.1002/jcb.27657
120. Ma D, Qin Y, Huang C, Chen Y, Han Z, Zhou X, et al. Circular RNA ABCB10 Promotes Non-Small Cell Lung Cancer Progression by Increasing E2F5 Expression Through Sponging miR-584-5p. Cell Cycle (Georgetown Tex) (2020) 19:1611–20. doi: 10.1080/15384101.2020.1761617
121. Hu TY, Zhu QX, Duan QY, Jin XY, Wu R. CircABCB10 Promotes the Proliferation and Migration of Lung Cancer Cells Through Down-Regulating microRNA-217 Expression. Eur Rev Med Pharmacol Sci (2020) 24:6157–65. doi: 10.26355/eurrev_202006_21511
122. An J, Shi H, Zhang N, Song S. Elevation of Circular RNA Circ_0003645 Forecasts Unfavorable Prognosis and Facilitates Cell Progression via miR-1179/TMEM14A Pathway in Non-Small Cell Lung Cancer. Biochem Biophys Res Commun (2019) 511:921–5. doi: 10.1016/j.bbrc.2019.03.011
123. Li Y, Hu J, Li L, Cai S, Zhang H, Zhu X, et al. Upregulated Circular RNA Circ_0016760 Indicates Unfavorable Prognosis in NSCLC and Promotes Cell Progression Through miR-1287/GAGE1 Axis. Biochem Biophys Res Commun (2018) 503:2089–94. doi: 10.1016/j.bbrc.2018.07.164
124. Chang H, Qu J, Wang J, Liang X, Sun W. Circular RNA Circ_0026134 Regulates Non-Small Cell Lung Cancer Cell Proliferation and Invasion via Sponging miR-1256 and miR-1287. Biomed Pharmacother (2019) 112:108743. doi: 10.1016/j.biopha.2019.108743
125. Cui D, Qian R, Li Y. Circular RNA Circ-CMPK1 Contributes to Cell Proliferation of Non-Small Cell Lung Cancer by Elevating Cyclin D1 via Sponging miR-302e. Mol Genet Genomic Med (2020) 8:e999. doi: 10.1002/mgg3.999
126. Zhang H, Wang X, Hu B, Zhang F, Wei H, Li L. Circular RNA ZFR Accelerates Non-Small Cell Lung Cancer Progression by Acting as a miR-101-3p Sponge to Enhance CUL4B Expression. Artif Cells Nanomed Biotechnol (2019) 47:3410–6. doi: 10.1080/21691401.2019.1652623
127. Wang X, Zhu X, Zhang H, Wei S, Chen Y, Chen Y, et al. Increased Circular RNA Hsa_Circ_0012673 Acts as a Sponge of miR-22 to Promote Lung Adenocarcinoma Proliferation. Biochem Biophys Res Commun (2018) 496:1069–75. doi: 10.1016/j.bbrc.2018.01.126
128. Han W, Wang L, Zhang L, Wang Y, Li Y. Circular RNA Circ-RAD23B Promotes Cell Growth and Invasion by miR-593-3p/CCND2 and miR-653-5p/TIAM1 Pathways in Non-Small Cell Lung Cancer. Biochem Biophys Res Commun (2019) 510:462–6. doi: 10.1016/j.bbrc.2019.01.131
129. Zhang W, Shi J, Cheng C, Wang H. CircTIMELESS Regulates the Proliferation and Invasion of Lung Squamous Cell Carcinoma Cells via the miR-136-5p/ROCK1 Axis. J Cell Physiol (2020) 235:5962–71. doi: 10.1002/jcp.29521
130. Xu Y, Yu J, Huang Z, Fu B, Tao Y, Qi X, et al. Circular RNA Hsa_Circ_0000326 Acts as a miR-338-3p Sponge to Facilitate Lung Adenocarcinoma Progression. J Exp Clin Cancer Res (2020) 39:57. doi: 10.1186/s13046-020-01556-4
131. Li L, Wan K, Xiong L, Liang S, Tou F, Guo S. CircRNA Hsa_Circ_0087862 Acts as an Oncogene in Non-Small Cell Lung Cancer by Targeting miR-1253/RAB3D Axis. OncoTargets Ther Volume (2020) 13:2873–86. doi: 10.2147/ott.S243533
132. Han J, Zhao G, Ma X, Dong Q, Zhang H, Wang Y, et al. CircRNA Circ-BANP-Mediated miR-503/LARP1 Signaling Contributes to Lung Cancer Progression. Biochem Biophys Res Commun (2018) 503:2429–35. doi: 10.1016/j.bbrc.2018.06.172
133. Hao Y, Xi J, Peng Y, Bian B, Hao G, Xi Y, et al. Circular RNA Circ_0016760 Modulates Non-Small-Cell Lung Cancer Growth Through the miR-577/ZBTB7A Axis. Cancer Manage Res (2020) 12:5561–74. doi: 10.2147/CMAR.S243675
134. Zuo Y, Shen W, Wang C, Niu N, Pu J. Circular RNA Circ-ZNF609 Promotes Lung Adenocarcinoma Proliferation by Modulating miR-1224-3p/ETV1 Signaling. Cancer Manage Res (2020) 12:2471–9. doi: 10.2147/CMAR.S232260
135. Fan Z, Bai Y, Zhang Q, Qian P. CircRNA Circ_POLA2 Promotes Lung Cancer Cell Stemness via Regulating the miR-326/GNB1 Axis. Environ Toxicol (2020) 35:1146–56. doi: 10.1002/tox.22980
136. Yu W, Jiang H, Zhang H, Li J. Hsa_circ_0003998 Promotes Cell Proliferation and Invasion by Targeting miR-326 in Non-Small Cell Lung Cancer. OncoTargets Ther (2018) 11:5569–77. doi: 10.2147/OTT.S174750
137. Pei X, Chen S-W, Long X, Zhu S-Q, Qiu B-Q, Lin K, et al. circMET Promotes NSCLC Cell Proliferation, Metastasis, and Immune Evasion by Regulating the miR-145-5p/CXCL3 Axis. Aging (2020) 12:13038–58. doi: 10.18632/aging.103392
138. Yang H, Zhao M, Zhao L, Li P, Duan Y, Li G. CircRNA BIRC6 Promotes Non-Small Cell Lung Cancer Cell Progression by Sponging microRNA-145. Cell Oncol (Dordrecht) (2020) 43:477–88. doi: 10.1007/s13402-020-00503-x
139. Lv YS, Wang C, Li LX, Han S, Li Y. Effects of circRNA_103993 on the Proliferation and Apoptosis of NSCLC Cells Through miR-1271/ERG Signaling Pathway. Eur Rev Med Pharmacol Sci (2020) 24:8384–93. doi: 10.26355/eurrev_202008_22635
140. Sun Z. Circular RNA Hsa_Circ_0001588 Promotes the Malignant Progression of Lung Adenocarcinoma by Modulating miR-524-3p/NACC1 Signaling. Life Sci (2020) 259:118157. doi: 10.1016/j.lfs.2020.118157
141. Wang B, Hua P, Zhao B, Li J, Zhang Y. Circular RNA Circdlgap4 Is Involved in Lung Cancer Development Through Modulating microRNA-143/CDK1 Axis. Cell Cycle (Georgetown Tex) (2020) 19:2007–17. doi: 10.1080/15384101.2020.1786649
142. Wang Q, Kang PM. CircRNA_001010 Adsorbs miR-5112 in a Sponge Form to Promote Proliferation and Metastasis of Non-Small Cell Lung Cancer (NSCLC). Eur Rev Med Pharmacol Sci (2020) 24:4271–80. doi: 10.26355/eurrev_202004_21007
143. Qiu M, Xia W, Chen R, Wang S, Xu Y, Ma Z, et al. The Circular RNA circPRKCI Promotes Tumor Growth in Lung Adenocarcinoma. Cancer Res (2018) 78:2839–51. doi: 10.1158/0008-5472.CAN-17-2808
144. Tan S, Sun D, Pu W, Gou Q, Guo C, Gong Y, et al. Circular RNA F-circEA-2a Derived From EML4-ALK Fusion Gene Promotes Cell Migration and Invasion in Non-Small Cell Lung Cancer. Mol Cancer (2018) 17:138. doi: 10.1186/s12943-018-0887-9
145. Wu K, Liao X, Gong Y, He J, Zhou J-K, Tan S, et al. Circular RNA F-circSR Derived From SLC34A2-ROS1 Fusion Gene Promotes Cell Migration in Non-Small Cell Lung Cancer. Mol Cancer (2019) 18:98. doi: 10.1186/s12943-019-1028-9
146. Huang Q, Wang S, Li X, Yang F, Feng C, Zhong K, et al. Circular RNA ATXN7 Is Upregulated in Non-Small Cell Lung Cancer and Promotes Disease Progression. Oncol Lett (2019) 17:4803–10. doi: 10.3892/ol.2019.10168
147. Zhao F, Han Y, Liu Z, Zhao Z, Li Z, Jia K. CircFADS2 Regulates Lung Cancer Cells Proliferation and Invasion via Acting as a Sponge of miR-498. Biosci Rep (2018) 38(4):BSR20180570. doi: 10.1042/BSR20180570
148. Lin Y, Su W, Lan G. Value of Circular RNA 0007385 in Disease Monitoring and Prognosis Estimation in Non-Small-Cell Lung Cancer Patients. J Clin Lab Anal (2020) 34:e23338. doi: 10.1002/jcla.23338
149. Wang M, Shi J, Jiang H, Xu K, Huang Z. Circ_0014130 Participates in the Proliferation and Apoptosis of Nonsmall Cell Lung Cancer Cells via the miR-142-5p/IGF-1 Axis. Cancer Biotherapy Radiopharm (2020) 35:233–40. doi: 10.1089/cbr.2019.2965
150. Li L, Zhang Q, Lian K. Circular RNA Circ_0000284 Plays an Oncogenic Role in the Progression of Non-Small Cell Lung Cancer Through the miR-377-3p-Mediated PD-L1 Promotion. Cancer Cell Int (2020) 20:247. doi: 10.1186/s12935-020-01310-y
151. Feng D, Xu Y, Hu J, Zhang S, Li M, Xu L. A Novel Circular RNA, Hsa-Circ-0000211, Promotes Lung Adenocarcinoma Migration and Invasion Through Sponging of hsa-miR-622 and Modulating HIF1-α Expression. Biochem Biophys Res Commun (2020) 521:395–401. doi: 10.1016/j.bbrc.2019.10.134
152. Li Y, Zhang J, Pan S, Zhou J, Diao X, Liu S. CircRNA CDR1as Knockdown Inhibits Progression of Non-Small-Cell Lung Cancer by Regulating miR-219a-5p/SOX5 Axis. Thorac Cancer (2020) 11:537–48. doi: 10.1111/1759-7714.13274
153. Chen D, Ma W, Ke Z, Xie F. CircRNA Hsa_Circ_100395 Regulates miR-1228/TCF21 Pathway to Inhibit Lung Cancer Progression. Cell Cycle (Georgetown Tex) (2018) 17:2080–90. doi: 10.1080/15384101.2018.1515553
154. Wang L, Liang Y, Mao Q, Xia W, Chen B, Shen H, et al. Circular RNA Circcrim1 Inhibits Invasion and Metastasis in Lung Adenocarcinoma Through the microRNA (Mir)-182/miR-93-Leukemia Inhibitory Factor Receptor Pathway. Cancer Sci (2019) 110:2960–72. doi: 10.1111/cas.14131
155. Liu T, Song Z, Gai Y. Circular RNA Circ_0001649 Acts as a Prognostic Biomarker and Inhibits NSCLC Progression via Sponging miR-331-3p and miR-338-5p. Biochem Biophys Res Commun (2018) 503:1503–9. doi: 10.1016/j.bbrc.2018.07.070
156. Pang W, Huang F, Zhang X, Ye M, Huang Y, Huang X, et al. Circular RNA Hsa_Circ_0072309 Inhibits Non-Small Cell Lung Cancer Progression by Sponging miR-580-3p. Biosci Rep (2020) 40(5):BSR20194237. doi: 10.1042/BSR20194237
157. Yang L, Wang J, Fan Y, Yu K, Jiao B, Su X. Hsa_circ_0046264 Up-Regulated BRCA2 to Suppress Lung Cancer Through Targeting hsa-miR-1245. Respir Res (2018) 19:115. doi: 10.1186/s12931-018-0819-7
158. Shi JQ, Wang B, Cao XQ, Wang YX, Cheng X, Jia CL, et al. Circular RNA_LARP4 Inhibits the Progression of Non-Small-Cell Lung Cancer by Regulating the Expression of SMAD7. Eur Rev Med Pharmacol Sci (2020) 24:1863–9. doi: 10.26355/eurrev_202002_20364
159. Yang Z, Xu X, Song C. Circular RNA La-Related Protein 4 Inhibits Nonsmall Cell Lung Cancer Cell Proliferation While Promotes Apoptosis Through Sponging microRNA-21-5p. Cancer Biotherapy Radiopharm (2020). doi: 10.1089/cbr.2020.3707
160. Liu Y, Yang C, Cao C, Li Q, Jin X, Shi H. Hsa_circ_RNA_0011780 Represses the Proliferation and Metastasis of Non-Small Cell Lung Cancer by Decreasing FBXW7 via Targeting miR-544a. Onco Targets Ther (2020) 13:745–55. doi: 10.2147/OTT.S236162
161. Zhang SJ, Ma J, Wu JC, Hao ZZ, Zhang YN, Zhang YJ. CircRNA EPB41L2 Inhibits Tumorigenicity of Lung Adenocarcinoma Through Regulating CDH4 by miR-211-5p. Eur Rev Med Pharmacol Sci (2020) 24:3749–60. doi: 10.26355/eurrev_202004_20839
162. Zhang B, Chen M, Jiang N, Shi K, Qian R. A Regulatory Circuit of Circ-MTO1/miR-17/QKI-5 Inhibits the Proliferation of Lung Adenocarcinoma. Cancer Biol Ther (2019) 20:1127–35. doi: 10.1080/15384047.2019.1598762
163. Wang Y, Zheng F, Wang Z, Lu J, Zhang H. Circular RNA Circ-SLC7A6 Acts as a Tumor Suppressor in Non-Small Cell Lung Cancer Through Abundantly Sponging miR-21. Cell Cycle (Georgetown Tex) (2020) 19:2235–46. doi: 10.1080/15384101.2020.1806449
164. Linnerth-Petrik NM, Santry LA, Petrik JJ, Wootton SK. Opposing Functions of Akt Isoforms in Lung Tumor Initiation and Progression. PloS One (2014) 9:e94595. doi: 10.1371/journal.pone.0094595
165. Nagase H, Visse R, Murphy G. Structure and Function of Matrix Metalloproteinases and TIMPs. Cardiovasc Res (2006) 69:562–73. doi: 10.1016/j.cardiores.2005.12.002
166. Wu Z, He D, Zhao S, Wang H. IL-17a/IL-17RA Promotes Invasion and Activates MMP-2 and MMP-9 Expression via P38 MAPK Signaling Pathway in Non-Small Cell Lung Cancer. Mol Cell Biochem (2019) 455:195–206. doi: 10.1007/s11010-018-3483-9
167. Berg T. Signal Transducers and Activators of Transcription as Targets for Small Organic Molecules. Chembiochem: Eur J Chem Biol (2008) 9:2039–44. doi: 10.1002/cbic.200800274
168. Heppler LN, Frank DA. Targeting Oncogenic Transcription Factors: Therapeutic Implications of Endogenous STAT Inhibitors. Trends Cancer (2017) 3:816–27. doi: 10.1016/j.trecan.2017.10.004
169. Yang M, Chen H, Zhou L, Chen K, Su F. Expression Profile and Prognostic Values of STAT Family Members in Non-Small Cell Lung Cancer. Am J Trans Res (2019) 11:4866–80.
170. Mehta HJ, Patel V, Sadikot RT. Curcumin and Lung Cancer–a Review. Targeted Oncol (2014) 9:295–310. doi: 10.1007/s11523-014-0321-1
171. Chen X, Mao R, Su W, Yang X, Geng Q, Guo C, et al. Circular RNA Modulates Autophagy via -STAT3-PRKAA/Ampkα Signaling in STK11 Mutant Lung Cancer. Autophagy (2020) 16:659–71. doi: 10.1080/15548627.2019.1634945
172. Yu H, Chen Y, Jiang P. Circular RNA HIPK3 Exerts Oncogenic Properties Through Suppression of miR-124 in Lung Cancer. Biochem Biophys Res Commun (2018) 506:455–62. doi: 10.1016/j.bbrc.2018.10.087
173. Wang Y, Zhao W, Zhang S. STAT3-Induced Upregulation of Circccdc66 Facilitates the Progression of Non-Small Cell Lung Cancer by Targeting miR-33a-5p/KPNA4 Axis. Biomed Pharmacotherapy (2020) 126:110019. doi: 10.1016/j.biopha.2020.110019
174. Adams JM, Cory S. The Bcl-2 Protein Family: Arbiters of Cell Survival. Sci (New York NY) (1998) 281:1322–6. doi: 10.1126/science.281.5381.1322
175. Pattingre S, Levine B. Bcl-2 Inhibition of Autophagy: A New Route to Cancer? Cancer Res (2006) 66:2885–8. doi: 10.1158/0008-5472.CAN-05-4412
176. Su L, Suyila Q, Yang L, Li H, Xi Y, Su X. Bax is Involved in the Anticancer Activity of Velcade in Colorectal Cancer. Exp Ther Med (2017) 14:3179–83. doi: 10.3892/etm.2017.4857
177. Wang L, Ma H, Kong W, Liu B, Zhang X. Up-Regulated Circular RNA VANGL1 Contributes to Progression of Non-Small Cell Lung Cancer Through Inhibition of miR-195 and Activation of Bcl-2. Biosci Rep (2019) 39(6):BSR20182433. doi: 10.1042/BSR20182433
178. Bai Q, Li L, Chen F, Zhu J, Cao L, Yang Y, et al. Suppression of Circular RNA Hsa_circ_0109320 Attenuates Non-Small Cell Lung Cancer Progression via MiR-595/E2F7 Axis. Med Sci Monit (2020) 26:e921200. doi: 10.12659/MSM.921200
179. Geng Y, Bao Y, Deng L, Su D, Zheng H, Zhang W. Circular RNA Hsa_Circ_0014130 Inhibits Apoptosis in Non–Small Cell Lung Cancer by Sponging miR-136-5p and Upregulating Bcl2. Mol Cancer Res (2020) 18:748–56. doi: 10.1158/1541-7786.Mcr-19-0998
180. Qin S, Zhao Y, Lim G, Lin H, Zhang X, Zhang X. Circular RNA PVT1 Acts as a Competing Endogenous RNA for miR-497 in Promoting Non-Small Cell Lung Cancer Progression. BioMed Pharmacother (2019) 111:244–50. doi: 10.1016/j.biopha.2018.12.007
181. Chen J, Xu S, Chen S, Zong Z, Han X, Zhao Y, et al. CircPUM1 Promotes the Malignant Behavior of Lung Adenocarcinoma by Regulating miR-326. Biochem Biophys Res Commun (2019) 508:844–9. doi: 10.1016/j.bbrc.2018.11.176
182. McIlwain DR, Berger T, Mak TW. Caspase Functions in Cell Death and Disease. Cold Spring Harbor Perspect Biol (2013) 5(4):a008656. doi: 10.1101/cshperspect.a008656 a008656.
183. Dong L, Zheng J, Gao Y, Zhou X, Song W, Huang J. The Circular RNA NT5E Promotes Non-Small Cell Lung Cancer Cell Growth via Sponging microRNA-134. Aging (2020) 12:3936–49. doi: 10.18632/aging.102861
184. Chen L, Nan A, Zhang N, Jia Y, Li X, Ling Y, et al. Circular RNA 100146 Functions as an Oncogene Through Direct Binding to miR-361-3p and miR-615-5p in Non-Small Cell Lung Cancer. Mol Cancer (2019) 18(1):13. doi: 10.1186/s12943-019-0943-0
185. Yang J, Jia Y, Wang B, Yang S, Du K, Luo Y, et al. Circular RNA TUBA1C Accelerates the Progression of Non-Small-Cell Lung Cancer by Sponging miR-143-3p. Cell Signalling (2020) 74:109693. doi: 10.1016/j.cellsig.2020.109693
186. Biomarkers Definitions Working Group. Biomarkers and Surrogate Endpoints: Preferred Definitions and Conceptual Framework. Clin Pharmacol Ther (2001) 69:89–95. doi: 10.1067/mcp.2001.113989
187. Tarro G, Perna A, Esposito C. Early Diagnosis of Lung Cancer by Detection of Tumor Liberated Protein. J Cell Physiol (2005) 203:1–5. doi: 10.1002/jcp.20195
188. Li Y, Tian X, Gao L, Jiang X, Fu R, Zhang T, et al. Clinical Significance of Circulating Tumor Cells and Tumor Markers in the Diagnosis of Lung Cancer. Cancer Med (2019) 8:3782–92. doi: 10.1002/cam4.2286
189. Liu X-X, Yang Y-E, Liu X, Zhang M-Y, Li R, Yin Y-H, et al. A Two-Circular RNA Signature as a Noninvasive Diagnostic Biomarker for Lung Adenocarcinoma. J Trans Med (2019) 17:50. doi: 10.1186/s12967-019-1800-z
190. Zhu X, Wang X, Wei S, Chen Y, Chen Y, Fan X, et al. Hsa_Circ_0013958: A Circular RNA and Potential Novel Biomarker for Lung Adenocarcinoma. FEBS J (2017) 284:2170–82. doi: 10.1111/febs.14132
191. Zhang N, Nan A, Chen L, Li X, Jia Y, Qiu M, et al. Circular RNA Circsatb2 Promotes Progression of Non-Small Cell Lung Cancer Cells. Mol Cancer (2020) 19:101. doi: 10.1186/s12943-020-01221-6
192. Hang D, Zhou J, Qin N, Zhou W, Ma H, Jin G, et al. A Novel Plasma Circular RNA circFARSA Is a Potential Biomarker for Non-Small Cell Lung Cancer. Cancer Med (2018) 7:2783–91. doi: 10.1002/cam4.1514
193. Zhou X, Liu H-Y, Wang W-Y, Zhao H, Wang T. Hsa_circ_0102533 Serves as a Blood-Based Biomarker for Non-Small-Cell Lung Cancer Diagnosis and Regulates Apoptosis In Vitro. Int J Clin Exp Pathol (2018) 11:4395–404.
194. Wang Y, Zhang H, Wang J, Li B, Wang X. Circular RNA Expression Profile of Lung Squamous Cell Carcinoma: Identification of Potential Biomarkers and Therapeutic Targets. Biosci Rep (2020) 40(4):BSR20194512. doi: 10.1042/BSR20194512
195. Wang J, Zhao X, Wang Y, Ren F, Sun D, Yan Y, et al. circRNA-002178 Act as a ceRNA to Promote PDL1/PD1 Expression in Lung Adenocarcinoma. Cell Death Dis (2020) 11:32. doi: 10.1038/s41419-020-2230-9
196. Zhang Y, Zhao H, Zhang L. Identification of the Tumor−Suppressive Function of Circular RNA FOXO3 in Non−Small Cell Lung Cancer Through Sponging Mir−155. Mol Med Rep (2018) 17:7692–700. doi: 10.3892/mmr.2018.8830
197. Geng QQ, Wu QF, Zhang Y, Zhang GJ, Fu JK, Chen NZ. Clinical Significance of Circ-MTHFD2 in Diagnosis, Pathological Staging and Prognosis of NSCLC. Eur Rev Med Pharmacol Sci (2020) 24:9473–9. doi: 10.26355/eurrev_202009_23032
198. Li S, Sun X, Miao S, Lu T, Wang Y, Liu J, et al. Hsa_Circ_0000729, a Potential Prognostic Biomarker in Lung Adenocarcinoma. Thoracic Cancer (2018) 9:924–30. doi: 10.1111/1759-7714.12761
199. Zhang S, Zeng X, Ding T, Guo L, Li Y, Ou S, et al. Microarray Profile of Circular RNAs Identifies Hsa_Circ_0014130 as a New Circular RNA Biomarker in Non-Small Cell Lung Cancer. Sci Rep (2018) 8:2878. doi: 10.1038/s41598-018-21300-5
200. Wang C, Tan S, Liu W-R, Lei Q, Qiao W, Wu Y, et al. RNA-Seq Profiling of Circular RNA in Human Lung Adenocarcinoma and Squamous Cell Carcinoma. Mol Cancer (2019) 18:134. doi: 10.1186/s12943-019-1061-8
201. Fu Y, Huang L, Tang H, Huang R. Hsa_circRNA_012515 Is Highly Expressed in NSCLC Patients and Affects Its Prognosis. Cancer Manage Res (2020) 12:1877–86. doi: 10.2147/CMAR.S245525
202. Zong L, Sun Q, Zhang H, Chen Z, Deng Y, Li D, et al. Increased Expression of circRNA_102231 in Lung Cancer and its Clinical Significance. Biomed Pharmacother = Biomedecine Pharmacotherapie (2018) 102:639–44. doi: 10.1016/j.biopha.2018.03.084
203. Ying X, Zhu J, Zhang Y. Circular RNA Circ-TSPAN4 Promotes Lung Adenocarcinoma Metastasis by Upregulating ZEB1 via Sponging miR-665. Mol Genet Genomic Med (2019) 7:e991. doi: 10.1002/mgg3.991
204. Zhang X, Yang D, Wei Y. Overexpressed CDR1as Functions as an Oncogene to Promote the Tumor Progression via miR-7 in Non-Small-Cell Lung Cancer. OncoTargets Ther Volume (2018) 11:3979–87. doi: 10.2147/ott.S158316
205. Wang Y, Li Y, He H, Wang F. Circular RNA Circ-PRMT5 Facilitates Non-Small Cell Lung Cancer Proliferation Through Upregulating EZH2 via Sponging miR-377/382/498. Gene (2019) 720:144099. doi: 10.1016/j.gene.2019.144099
206. Yan B, Zhang W, Mao XW, Jiang LY. Circular RNA ciRS-7 Correlates With Advance Disease and Poor Prognosis, and its Down-Regulation Inhibits Cells Proliferation While Induces Cells Apoptosis in Non-Small Cell Lung Cancer. Eur Rev Med Pharmacol Sci (2018) 22:8712–21. doi: 10.26355/eurrev_201812_16636
207. Wang J, Li H. CircRNA Circ_0067934 Silencing Inhibits the Proliferation, Migration and Invasion of NSCLC Cells and Correlates With Unfavorable Prognosis in NSCLC. Eur Rev Med Pharmacol Sci (2018) 22:3053–60. doi: 10.26355/eurrev_201805_15063
208. Zou Q, Wang T, Li B, Li G, Zhang L, Wang B, et al. Overexpression of Circ-0067934 Is Associated With Increased Cellular Proliferation and the Prognosis of Non-Small Cell Lung Cancer. Oncol Lett (2018) 16:5551–6. doi: 10.3892/ol.2018.9357
209. Wan J, Hao L, Zheng X, Li Z. Circular RNA Circ_0020123 Promotes Non-Small Cell Lung Cancer Progression by Acting as a ceRNA for miR-488–3p to Regulate ADAM9 Expression. Biochem Biophys Res Commun (2019) 515:303–9. doi: 10.1016/j.bbrc.2019.05.158
210. Gu X, Wang G, Shen H, Fei X. Hsa_circ_0033155: A Potential Novel Biomarker for Non-Small Cell Lung Cancer. Exp Ther Med (2018) 16:3220–6. doi: 10.3892/etm.2018.6565
211. Yao J-T, Zhao S-H, Liu Q-P, Lv M-Q, Zhou D-X, Liao Z-J, et al. Over-Expression of CircRNA_100876 in Non-Small Cell Lung Cancer and its Prognostic Value. Pathol Res Pract (2017) 213:453–6. doi: 10.1016/j.prp.2017.02.011
212. Lim Z-F, Ma PC. Emerging Insights of Tumor Heterogeneity and Drug Resistance Mechanisms in Lung Cancer Targeted Therapy. J Hematol Oncol (2019) 12:134. doi: 10.1186/s13045-019-0818-2
213. Terlizzi M, Colarusso C, Pinto A, Sorrentino R. Drug Resistance in Non-Small Cell Lung Cancer (NSCLC): Impact of Genetic and Non-Genetic Alterations on Therapeutic Regimen and Responsiveness. Pharmacol Ther (2019) 202:140–8. doi: 10.1016/j.pharmthera.2019.06.005
214. Rotow J, Bivona TG. Understanding and Targeting Resistance Mechanisms in NSCLC. Nat Rev Cancer (2017) 17:637–58. doi: 10.1038/nrc.2017.84
215. Wu S-G, Shih J-Y. Management of Acquired Resistance to EGFR TKI-Targeted Therapy in Advanced Non-Small Cell Lung Cancer. Mol Cancer (2018) 17:38. doi: 10.1186/s12943-018-0777-1
216. Altorki NK, Markowitz GJ, Gao D, Port JL, Saxena A, Stiles B, et al. The Lung Microenvironment: An Important Regulator of Tumour Growth and Metastasis. Nat Rev Cancer (2019) 19(1):9–31. doi: 10.1038/s41568-018-0081-9
217. Amable L. Cisplatin Resistance and Opportunities for Precision Medicine. Pharmacol Res (2016) 106:27–36. doi: 10.1016/j.phrs.2016.01.001
218. Sun C-Y, Nie J, Huang J-P, Zheng G-J, Feng B. Targeting STAT3 Inhibition to Reverse Cisplatin Resistance. Biomed Pharmacother = Biomedecine Pharmacotherapie (2019) 117:109135. doi: 10.1016/j.biopha.2019.109135
219. Dong Y, Xu T, Zhong S, Wang B, Zhang H, Wang X, et al. Circ_0076305 Regulates Cisplatin Resistance of Non-Small Cell Lung Cancer via Positively Modulating STAT3 by Sponging miR-296-5p. Life Sci (2019) 239:116984. doi: 10.1016/j.lfs.2019.116984
220. Xu Y, Jiang T, Wu C, Zhang Y. CircAKT3 Inhibits Glycolysis Balance in Lung Cancer Cells by Regulating miR-516b-5p/STAT3 to Inhibit Cisplatin Sensitivity. Biotechnol Lett (2020) 42:1123–35. doi: 10.1007/s10529-020-02846-9
221. Hong W, Xue M, Jiang J, Zhang Y, Gao X. Circular RNA Circ-CPA4/let-7 miRNA/PD-L1 Axis Regulates Cell Growth, Stemness, Drug Resistance and Immune Evasion in Non-Small Cell Lung Cancer (NSCLC). J Exp Clin Cancer Res (2020) 39:149. doi: 10.1186/s13046-020-01648-1
222. Harrison EB, Porrello A, Bowman BM, Belanger AR, Yacovone G, Azam SH, et al. A Circle RNA Regulatory Axis Promotes Lung Squamous Metastasis via CDR1-Mediated Regulation of Golgi Trafficking. Cancer Res (2020) 80(22):4972–85. doi: 10.1158/0008-5472.CAN-20-1162
223. Zhao Y, Zheng R, Chen J, Ning D. CircRNA CDR1as/miR-641/HOXA9 Pathway Regulated Stemness Contributes to Cisplatin Resistance in Non-Small Cell Lung Cancer (NSCLC). Cancer Cell Int (2020) 20:289. doi: 10.1186/s12935-020-01390-w
224. Huang M-S, Yuan F-Q, Gao Y, Liu J-Y, Chen Y-X, Wang C-J, et al. Circular RNA Screening From EIF3a in Lung Cancer. Cancer Med (2019) 8:4159–68. doi: 10.1002/cam4.2338
225. Pang J, Ye L, Zhao D, Zhao D, Chen Q. Circular RNA PRMT5 Confers Cisplatin-Resistance via miR-4458/REV3L Axis in Non-Small-Cell Lung Cancer. Cell Biol Int (2020) 44(12):2416–26. doi: 10.1002/cbin.11449
226. Li H, Liu F, Qin W. Circ_0072083 Interference Enhances Growth-Inhibiting Effects of Cisplatin in Non-Small-Cell Lung Cancer Cells via miR-545-3p/CBLL1 Axis. Cancer Cell Int (2020) 20:78. doi: 10.1186/s12935-020-1162-x
227. Lu H, Xie X, Wang K, Chen Q, Cai S, Liu D, et al. Circular RNA Hsa_Circ_0096157 Contributes to Cisplatin Resistance by Proliferation, Cell Cycle Progression, and Suppressing Apoptosis of Non-Small-Cell Lung Carcinoma Cells. Mol Cell Biochem (2020) 475(1–2):63–77. doi: 10.1007/s11010-020-03860-1
228. Kong R. Circular RNA Hsa_Circ_0085131 is Involved in Cisplatin-Resistance of Non-Small-Cell Lung Cancer Cells by Regulating Autophagy. Cell Biol Int (2020) 44:1945–56. doi: 10.1002/cbin.11401
229. Sun H, Chen Y, Fang Y-Y, Cui T-Y, Qiao X, Jiang C-Y, et al. Circ_0000376 Enhances the Proliferation, Metastasis, and Chemoresistance of NSCLC Cells via Repressing miR-384. Cancer Biomarkers: Section A Dis Markers (2020) 29(4):463–73. doi: 10.3233/CBM-190912
230. Xiao G, Huang W, Zhan Y, Li J, Tong W. CircRNA_103762 Promotes Multidrug Resistance in NSCLC by Targeting DNA Damage Inducible Transcript 3 (CHOP). J Clin Lab Anal (2020) 34:e23252. doi: 10.1002/jcla.23252
231. Huang M-S, Liu J-Y, Xia X-B, Liu Y-Z, Li X, Yin J-Y, et al. Hsa_circ_0001946 Inhibits Lung Cancer Progression and Mediates Cisplatin Sensitivity in Non-Small Cell Lung Cancer via the Nucleotide Excision Repair Signaling Pathway. Front Oncol (2019) 9:508. doi: 10.3389/fonc.2019.00508
232. Huang W, Yang Y, Wu J, Niu Y, Yao Y, Zhang J, et al. Circular RNA Cesrp1 Sensitises Small Cell Lung Cancer Cells to Chemotherapy by Sponging miR-93-5p to Inhibit TGF-β Signalling. Cell Death Differ (2020) 27:1709–27. doi: 10.1038/s41418-019-0455-x
233. Adjei AA. Pemetrexed (Alimta): A Novel Multitargeted Antifolate Agent. Expert Rev Anticancer Ther (2003) 3:145–56. doi: 10.1586/14737140.3.2.145
234. Adjei AA. Pharmacology and Mechanism of Action of Pemetrexed. Clin Lung Cancer (2004) 5(Suppl 2):S51–5. doi: 10.3816/CLC.2004.s.003
235. Mao Y, Xu R. Circular RNA CDR1-AS Contributes to Pemetrexed and Cisplatin Chemoresistance Through EGFR/PI3K Signaling Pathway in Lung Adenocarcinoma. Biomed Pharmacother = Biomedecine Pharmacotherapie (2020) 123:109771. doi: 10.1016/j.biopha.2019.109771
236. Zheng F, Xu R. CircPVT1 Contributes to Chemotherapy Resistance of Lung Adenocarcinoma Through miR-145-5p/ABCC1 Axis. Biomed Pharmacother = Biomedecine Pharmacotherapie (2020) 124:109828. doi: 10.1016/j.biopha.2020.109828
237. Mini E, Nobili S, Caciagli B, Landini I, Mazzei T. Cellular Pharmacology of Gemcitabine. Ann Oncol: Off J Eur Soc Med Oncol (2006) 17(Suppl 5):v7–12. doi: 10.1093/annonc/mdj941
238. Cappuzzo F, De Marinis F, Nelli F, Calandri C, Maestri A, Benedetti G, et al. Phase II Study of Gemcitabine-Cisplatin-Paclitaxel Triplet as Induction Chemotherapy in Inoperable, Locally-Advanced Non-Small Cell Lung Cancer. Lung Cancer (Amsterdam Neth) (2003) 42:355–61. doi: 10.1016/S0169-5002(03)00365-9
239. Lu H, Xie X, Chen Q, Cai S, Liu S, Bao C, et al. Clinical Significance of Circpvt1 in Patients With Non-Small Cell Lung Cancer Who Received Cisplatin Combined With Gemcitabine Chemotherapy. Tumori (2020) 107(3):204–8. doi: 10.1177/0300891620941940 300891620941940
240. Tong S. Circular RNA SMARCA5 may Serve as a Tumor Suppressor in Non-Small Cell Lung Cancer. J Clin Lab Anal (2020) 34:e23195. doi: 10.1002/jcla.23195
241. Marupudi NI, Han JE, Li KW, Renard VM, Tyler BM, Brem H. Paclitaxel: A Review of Adverse Toxicities and Novel Delivery Strategies. Expert Opin Drug Saf (2007) 6:609–21. doi: 10.1517/14740338.6.5.609
242. Guo C, Wang H, Jiang H, Qiao L, Wang X. Circ_0011292 Enhances Paclitaxel Resistance in Non-Small Cell Lung Cancer by Regulating miR-379-5p/TRIM65 Axis. Cancer Biother Radiopharm (2020). doi: 10.1089/cbr.2019.3546
243. Li X, Yang B, Ren H, Xiao T, Zhang L, Li L, et al. Hsa_circ_0002483 Inhibited the Progression and Enhanced the Taxol Sensitivity of Non-Small Cell Lung Cancer by Targeting miR-182-5p. Cell Death Dis (2019) 10:953. doi: 10.1038/s41419-019-2180-2
244. Hsu WH, Yang JCH, Mok TS, Loong HH. Overview of Current Systemic Management of EGFR-Mutant NSCLC. Ann Oncol: Off J Eur Soc Med Oncol (2018) 29:i3–9. doi: 10.1093/annonc/mdx702
245. Zhou Y, Zheng X, Xu B, Chen L, Wang Q, Deng H, et al. Circular RNA Hsa_Circ_0004015 Regulates the Proliferation, Invasion, and TKI Drug Resistance of Non-Small Cell Lung Cancer by miR-1183/PDPK1 Signaling Pathway. Biochem Biophys Res Commun (2019) 508:527–35. doi: 10.1016/j.bbrc.2018.11.157
246. Li D, Mullinax JE, Aiken T, Xin H, Wiegand G, Anderson A, et al. Loss of PDPK1 Abrogates Resistance to Gemcitabine in Label-Retaining Pancreatic Cancer Cells. BMC Cancer (2018) 18:772. doi: 10.1186/s12885-018-4690-1
247. Wen C, Xu G, He S, Huang Y, Shi J, Wu L, et al. Screening Circular RNAs Related to Acquired Gefitinib Resistance in Non-Small Cell Lung Cancer Cell Lines. J Cancer (2020) 11:3816–26. doi: 10.7150/jca.39783
248. Liu Y-T, Han X-H, Xing P-Y, Hu X-S, Hao X-Z, Wang Y, et al. Circular RNA Profiling Identified as a Biomarker for Predicting the Efficacy of Gefitinib Therapy for Non-Small Cell Lung Cancer. J Thoracic Dis (2019) 11:1779–87. doi: 10.21037/jtd.2019.05.22
249. Joseph NA, Chiou S-H, Lung Z, Yang C-L, Lin T-Y, Chang H-W, et al. The Role of HGF-MET Pathway and CCDC66 cirRNA Expression in EGFR Resistance and Epithelial-to-Mesenchymal Transition of Lung Adenocarcinoma Cells. J Hematol Oncol (2018) 11:74. doi: 10.1186/s13045-018-0557-9
250. Ma J, Qi G, Li L. A Novel Serum Exosomes-Based Biomarker Hsa_Circ_0002130 Facilitates Osimertinib-Resistance in Non-Small Cell Lung Cancer by Sponging miR-498. OncoTargets Ther (2020) 13:5293–307. doi: 10.2147/OTT.S243214
251. Chen T, Luo J, Gu Y, Huang J, Luo Q, Yang Y. Comprehensive Analysis of Circular RNA Profiling in AZD9291-Resistant Non-Small Cell Lung Cancer Cell Lines. Thoracic Cancer (2019) 10:930–41. doi: 10.1111/1759-7714.13032
252. Duma N, Santana-Davila R, Molina JR. Non-Small Cell Lung Cancer: Epidemiology, Screening, Diagnosis, and Treatment. Mayo Clinic Proc (2019) 94:1623–40. doi: 10.1016/j.mayocp.2019.01.013
253. Hsu P-C, Jablons DM, Yang C-T, You L. Epidermal Growth Factor Receptor (EGFR) Pathway, Yes-Associated Protein (YAP) and the Regulation of Programmed Death-Ligand 1 (PD-L1) in Non-Small Cell Lung Cancer (NSCLC). Int J Mol Sci (2019) 20(15):3821. doi: 10.3390/ijms20153821
254. Luo Y-H, Yang Y-P, Chien C-S, Yarmishyn AA, Ishola AA, Chien Y, et al. Plasma Level of Circular RNA Hsa_Circ_0000190 Correlates With Tumor Progression and Poor Treatment Response in Advanced Lung Cancers. Cancers (2020) 12(7):1740. doi: 10.3390/cancers12071740
255. Uzel EK, Figen M, Uzel Ö. Radiotherapy in Lung Cancer: Current and Future Role. Sisli Etfal Hastanesi Tip Bulteni (2019) 53:353–60. doi: 10.14744/SEMB.2019.25991
256. Ni J, Bucci J, Malouf D, Knox M, Graham P, Li Y. Exosomes in Cancer Radioresistance. Front Oncol (2019) 9:869. doi: 10.3389/fonc.2019.00869
Keywords: circular RNA, lung cancer, biomarker, mechanisms, drug-resistance, radioresistance
Citation: Chen H-H, Zhang T-N, Wu Q-J, Huang X-M and Zhao Y-H (2021) Circular RNAs in Lung Cancer: Recent Advances and Future Perspectives. Front. Oncol. 11:664290. doi: 10.3389/fonc.2021.664290
Received: 04 February 2021; Accepted: 21 June 2021;
Published: 06 July 2021.
Edited by:
Antonio Calles, Gregorio Marañón Hospital, SpainReviewed by:
Azhar Ali, National University of Singapore, SingaporeJia Huang, Shanghai Jiaotong University, China
Copyright © 2021 Chen, Zhang, Wu, Huang and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yu-Hong Zhao, emhhb3l1aG9uZ0Bzai1ob3NwaXRhbC5vcmc=
†These authors have contributed equally to this work and share first authorship
 Huan-Huan Chen1,2,3†
Huan-Huan Chen1,2,3† Tie-Ning Zhang
Tie-Ning Zhang Qi-Jun Wu
Qi-Jun Wu Xin-Mei Huang
Xin-Mei Huang Yu-Hong Zhao
Yu-Hong Zhao