- 1Hunan Cancer Hospital and the Affiliated Cancer Hospital of Xiangya School of Medicine, Central South University, Changsha, China
- 2Cancer Research Institute, School of Basic Medical Science, Central South University, Changsha, China
- 3Key Laboratory of Carcinogenesis and Cancer Invasion of Ministry of Education, China National Health Commission Key Laboratory of Carcinogenesis, Xiangya Hospital, Central South University, Changsha, China
Exosomes are natural nanoscale bilayer phospholipid vesicles that can be secreted by almost all types of cells and are detected in almost all types of body fluids. Exosomes are effective mediators of cell–cell signaling communication because of their ability to carry and transfer a variety of bioactive molecules, including non-coding RNAs. Non-coding RNAs have also been found to exert strong effects on a variety of biological processes, including tumorigenesis. Many researchers have established that exosomes encapsulate bioactive non-coding RNAs that alter the biological phenotype of specific target cells in an autocrine or a paracrine manner. However, the mechanism by which the producer cells package non-coding RNAs into exosomes is not well understood. This review focuses on the current research on exosomal non-coding RNAs, including the biogenesis of exosomes, the possible mechanism of sorting non-coding RNAs, their biological functions, and their potential for clinical application in the future.
Introduction
Since the discovery of extracellular vesicles (EVs), researchers have found EVs in almost all biological fluids. EVs have received unprecedented attention owing to their key roles, such as their potential as biomarkers for liquid biopsies and therapeutic applications (1, 2). Based on their formation, typical EVs are roughly divided into three categories: microvesicles (also called outer membrane vesicles, ectosomes, or shedding vesicles), exosomes, and apoptotic bodies. Microvesicles (100–1000-nm diameter) are released from the cell membrane through the blebbing and fission of the plasma membrane. Apoptotic bodies (50–5000-nm diameter) fall off during the process of cell apoptosis (3–6). Exosomes derived from the endosomal compartment are phospholipid bilayer vesicles with a diameter of 50–150 nm, generated by sequential invagination of the plasma membrane and endomembrane (7–9). They are specifically loaded with cargos, depending on the producer cell type and its homeostatic state. This review focuses on exosomes, which are the most studied type of EVs. Exosomes contain a variety of bioactive substances, including DNA, RNA, proteins, and lipids, which are secreted into the extracellular space and absorbed by target cells to change their phenotype. Functional exosomes can protect packaged cargos from being destroyed by various biological enzymes, extending the circulating half-life and enhancing exosomal RNA biological effects (10).
Accumulating evidence suggests that most pluricellular organisms sustain exosome-based communications via the intercellular exchange of non-coding RNAs (ncRNAs) between cells. Valadi et al. first reported exosomes containing mRNAs and microRNAs (miRNAs) (11). Subsequently, a large number of studies have confirmed the existence of many other ncRNA species in exosomes (12–15). More than 98% of the human genome is composed of ncRNAs that are involved in a broad range of physiological or pathophysiological processes in humans (16). NcRNAs play an indispensable role in genetic expression, epigenetic regulation, RNA splicing and translation processes, protein degradation, and transport, among others. Exosomal ncRNAs, especially those derived from tumors, are highly enriched and stable and act as messengers in cell–cell communications (17, 18). This review mainly focuses on three types of ncRNAs: miRNAs (average length of 22 nucleotides), long ncRNA (lncRNA) (>200 nucleotides in length), and circular RNA (circular molecule that has a covalently closed loop structure, lacking a poly A tail or 5′→3′ polarity) (19–21).
To advance the field of exosomal biology and understand the roles of exosomal ncRNAs, this review focuses on the cellular machinery and processes of exosome formation, sorting of ncRNAs into the exosome pathway, and biological roles of the three types of exosomal ncRNAs. The application of exosomal ncRNAs as biomarkers and the potential roles of exosomal ncRNAs as therapeutic agents are also discussed.
Biogenesis of Exosomes
Exosome biogenesis is closely related to the cellular endocytosis pathway. Endocytosis of the plasma membrane leads to the formation of early endosomes, which then bud inward and mature into late endosomes (LE). In turn, LEs develop into multivesicular bodies (MVBs) containing intraluminal vesicles (ILVs). Most MVBs fuse with lysosomes to degrade the contained cargo, whereas some bud and divide from the endosomal membrane to produce ILVs, which are released into the extracellular space when the endosomal membrane fuses with the cytoplasmic membrane, namely exosomes (22, 23). The biogenesis of exosomes has often been described to involve an endosomal sorting complex required for transport (ESCRT)-dependent or ESCRT-independent mechanism, although the pathways may not be entirely separated. In fact, the pathways may work synergistically, and different subpopulations of exosomes could depend on different machineries (Figure 1) (24–26).
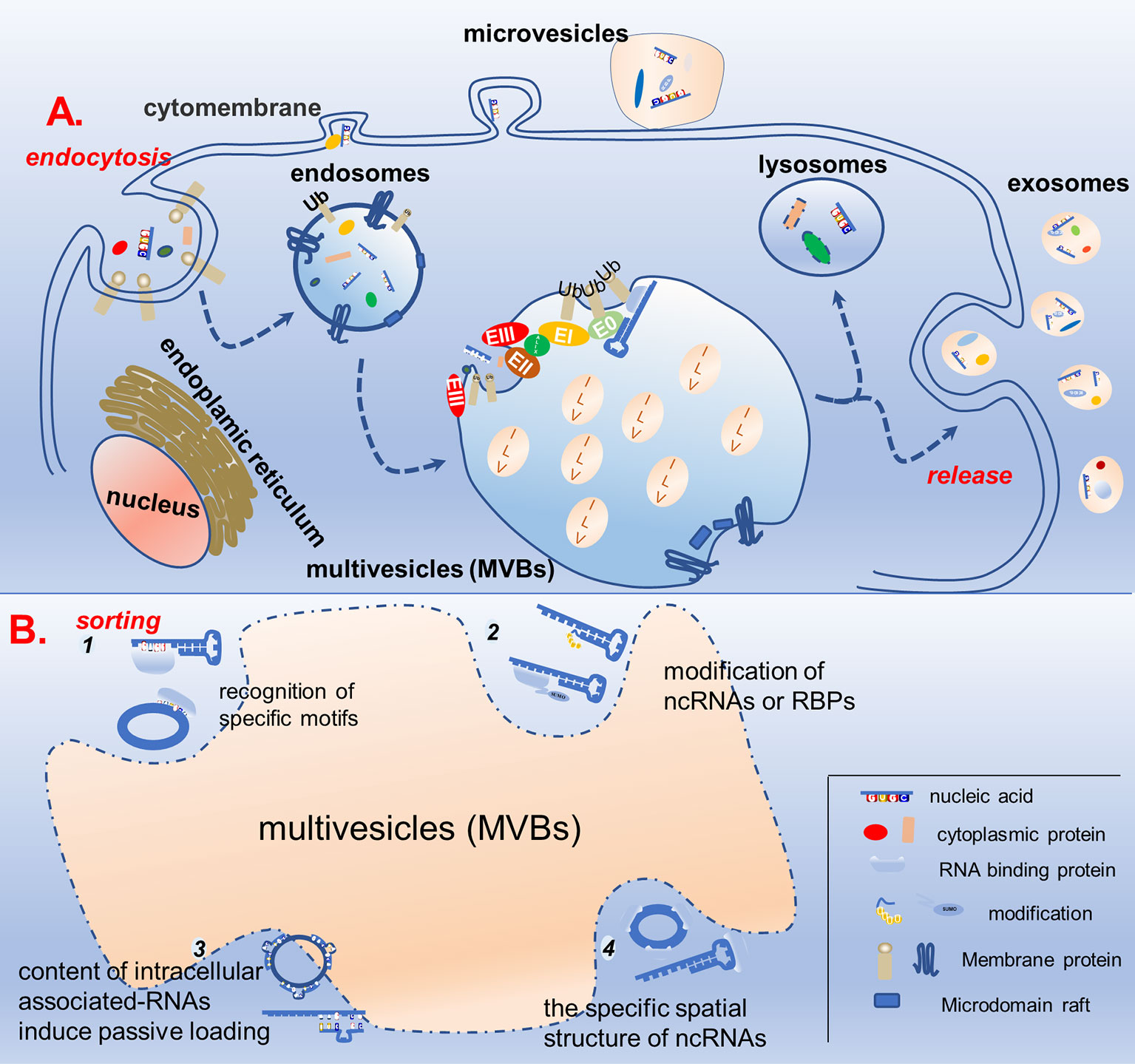
Figure 1 Schematic diagram of exosome biogenesis and potential mechanisms for sorting non-coding RNA. (A) Proteins and nucleic acids are secreted into extracellular vesicles through budding and fission of the cell membrane, while others enter the cell through the endocytosis pathway to form endosomes, which further mature into multivesicles bodies. In the multivesicles, membrane invagination, budding, and fission results in the formation of ILVs in ESCRT-dependent and ESCRT-independent manners. ILVs either fuse with lysosomes to degrade the cargos or dock with the cell membrane to be released, forming biological exosomes. (B) In the process of ILVs formation, specific non-coding RNAs are specifically sorted into ILVs. There are four possible mechanisms, as follows: (1) recognition of specific motifs on non-coding RNAs by RBP; (2) modification of non-coding RNAs or RBPs capable of binding to them to help them wrapped in ILVs; (3) the content of intracellular associated-RNAs modulate ncRNAs’ sorting into ILV; (4) the specific spatial structure of non-coding RNAs also affects its ability to enter ILVs.
ESCRT-Dependent Mechanism in the Biogenesis of Exosomes
The ESCRT pathway was initially defined in yeast genetic screens to identify the factors necessary to sort membrane proteins into intraluminal endosomal vesicles. ESCRT-dependent exosome biogenesis includes ubiquitin-dependent and ubiquitin-independent ESCRT sorting signals. The canonical mechanism by which ESCRT is activated is initiated by the recognition of ubiquitinated membrane proteins on cell membranes. Ubiquitinated endosomal proteins that are deposited into the lumens of MVBs are either sorted for lysosomal-mediated degradation or secreted as exosomes into the extracellular milieu. The ESCRT system includes four protein complexes: ESCRT-0, ESCRT-I, ESCRT-II, and ESCRT-III (26, 27). The ESCRT-0 complex is first activated by phosphatidylinositol 3-phosphate PI(3)P and ubiquitinated molecules on the multivesicular membrane (28). Subsequently, the ESCRT-I complex is recruited by ESCRT-0 with the help of the hepatocyte growth factor-regulated tyrosine kinase substrate prosaposin (HRS PSAP) subunits, which can be promoted by heparanase (endosomal enzyme) (29). The interaction between ESCRT-I and ESCRT-II is mediated through VPS28 and EAP45 (Vps36) subunits, which are essential for sorting cargo in the MVBs and deforming the membrane, resulting in bud formation (30–32). ESCRT-III aggregates at the neck of the bud and causes the sprout to shear, thereby releasing ILVs and driving vesicle scission from MVBs. The VPS4/VTAL complex is hydrolyzed by ATPase, which provides energy to depolymerize the polymerized ESCRT-III for recycling to complete further budding processes (33). The membrane protein syntenin combines with the cytoplasmic protein syndecan and ALIX accessory protein to form a ternary complex. This complex can interact with the TSG101 domain of ESCRT I and the CHMP4 domain of ESCRT III and participate in the ESCRT-dependent pathway for exosome secretion (34–36).
Other groups of researchers recently reported that some ESCRT-related proteins can be recruited independently of ubiquitination to promote exosome secretion by cargos. The post-translational attachment of small ubiquitin-like modifier (SUMO) to proteins plays a vital role in mediating ESCRT-dependent sorting into EVs. SUMO is a small ubiquitin-like protein consisting of more than 100 amino acid residues with a molecular weight of approximately 12 kDa. It is named after ubiquitin, with which it shares a similar mode of action and spatial structure. SUMOylation is a covalent modification similar to ubiquitination in which the terminal diglycine configuration of SUMO forms an isopeptide bond with the E-amino group of the substrate protein lysine. Unlike ubiquitination, which often mediates the degradation of target proteins, SUMOylation modification is thought to enhance the stability of target proteins (37, 38). SUMO is recruited to ESCRT formation sites by interacting with phosphoinositols and requires the ESCRT subunits Tsg101 and VPS4, as well as the ESCRT-associated protein Alix. Furthermore, the release of the cytosolic protein α-synuclein within EVs in human cerebrospinal fluid is SUMO-dependent (39, 40). Liu et al. demonstrated the existence of a ubiquitin-independent pathway; for example, the COP9 signalosome (CSN)-associated protein CSN5 can regulate the sorting of exosomal proteins (such as HSP70 and HIV Gag protein) in both a deubiquitinating activity-dependent and -independent manner (41).
ESCRT-Independent Mechanism in the Biogenesis of Exosomes
Katarina et al. were the first to discover an ESCRT-independent exosome formation mechanism. They provided evidence that intracellular ceramide was deposited with the depletion of the neutral sphingomyelinase enzyme, which induces the coalescence of small raft-based microdomains into larger domains, promoting domain-induced budding (42). Subsequently, Aude et al. revealed the presence of lipid raft microdomains in exosomal membranes and indicated their possible involvement in vesicle formation and structure (43). Tetraspanins, a family of four transmembrane proteins, were shown to be responsible for exosome formation (44). Similarly, Van Niel found that tetraspanin CD63 directly participates in ESCRT-independent biogenesis of the PMEL (a component of melanocyte lysosome-related organelles) luminal domain, rather than traditional ESCRT-dependent cargoes, to ILVs (45). CD82 and CD9, which are also tetraspanin membrane proteins, induce beta-catenin export via exosomes, which are blocked by a sphingomyelinase inhibitor (46). Other tetraspanin membrane proteins, such as Tspan8, including CD106 and CD49d, as well as CD81 (RacGTPase), are also considered to be involved in the ESCRT-independent pathway (47, 48). Exosomal cholesterol secretion depends on the presence of flotillin protein (49). Many other molecules are also involved in the formation of exosomes, and further details need to be explored (50–52).
Another ESCRT-independent mechanism is RAB GTPase-dependent exosome generation. RAB GTPases are positioned on the surface of a specific membrane structure and regulate the vesicle transport of the corresponding membrane structure by recruiting effector factors. For example, RAB5 regulates the formation and mutual fusion of the endoplasmic reticulum. The transition from RAB5 to RAB7 on the endosome membrane regulates the transition from the early endosome to the late endosome. RAB7 regulates the fusion of late endosomes and lysosomes to degrade ILVs, whereas RAB27 regulates the docking and fusion of MVBs and membranes to release ILVs to form exosomes. The membrane proteins of endocytosis, especially the receptor tyrosine kinase family epidermal growth factor receptor (EGFR), are located in the endosome and MVBs and initiate lysosomal degradation through the fusion of MVBs and lysosomes. Kang et al. conducted experiments to knock down the ESCRT components HRS and TSG101 as well as the related protein Alix. As a result, they found that these were not involved in the formation of EGFR exosomes driven by RAB31Q65L. Active RAB31 drove EGFR into MVBs to form ILVs and exosomes, whereas EGFR phosphorylated RAB31 to drive homologous exosomes. Flotillin protein in the lipid raft microdomain was involved in the formation of ILVs driven by active RAB31, which was independent of the ESCRT mechanism. It was further proved that RAB31 recruited TBC1D2B to inactivate RAB7, inhibit the fusion of MVBs and lysosomes, and further promote the production of exosomes (53–55).
Possible Sorting Mechanism of Exosomal ncRNAs
The higher enrichment of certain ncRNAs in exosomes secreted by cells in specific states indicates that exosomal ncRNA encapsulation is an intense biological process that initiates exosomal ncRNA signaling. However, the exact cellular process responsible for selective specific exosomal ncRNA enrichment has not been well established in eukaryotic cells (Table 1).
RNA-Binding Proteins Mediate ncRNAs Sorting Depending on Their Characteristic Motif
It is worth noting that almost all RNAs in cells exists as ribonucleoprotein (RNP) complexes. As such, proteins capable of interacting with RNA (i.e., RBPs) can be critical factors for the promotion of ncRNA transmission in the parent cells and can serve as the intracellular inducers of ncRNA loading in exosomes in the recipient cell (76). It has also been reported that short nucleotide sequences on RNA can guide its transport to different subcellular compartments, including exosomes (77–80).
Proteomic analyses have detected the specific binding of heterogeneous nuclear RNP A2B1 (hnRNPA2B1) to exosomal miR-198 with the RTS motifs. hnRNPA2B1, which is present in exosomes, binds to exosomal miRNA directly and controls its loading into these microvesicles. Moreover, hnRNPA2B1 in exosomes is SUMOylated, and SUMOylation controls the binding of hnRNPA2B1 to miRNAs (77, 79, 81, 82). Another recent study found that epirubicin-treated endothelial cells specifically regulated the extracellular separation of miR-503 by disrupting hnRNPA2B1 (83). In addition to miRNAs, hnRNPA2B1 has been found to specifically regulate lncRNAs. There is a special motif at the 5′ end of lncARSR that can bind to the RBP hnRNPA2B1 and has been sorted into exosomes together with the target miR-198 of lncARSR (15). Lei et al. identified that the expression of lncRNA H19 was upregulated in gefitinib-resistant non-small cell lung cancer (NSCLC) cells and that there was a GGAG substrate in the 5′ terminal region, which was bound to hnRNPA2B1 protein to be specifically sorted into exosomes (73). In trastuzumab-resistant breast cancer cells, hnRNPA2B1 is overexpressed or silenced, and exosome AGAP2AS1 expression is upregulated or downregulated (74). Moreover, lncRNA LNMAT2 specifically binds to hnRNPA2B1 and is packaged into exosomes through its specific sequence of GGAG in the 1930–1960-nt region and the stem-loop structure in this region (75).
Serine and arginine rich splicing factor 1 has been identified as a mediator of exosomal miRNA enrichment in pancreatic cancer cells by binding to a specific miRNA sequence motif (64). Major vault protein can selectively enrich miR-193a to exosomes and reduce its intracellular content; however, this specific interaction region has not been studied. In turn, MiR-193a can affect its target GTPase Rab27B and exosome production (65, 66, 84, 85). Zhang et al. suggested that exosomes package circular RNAs (circRNAs) containing the purine-rich 5′-GMWGVWGRAG-3′ motif, with the characteristic “garbage dumping” and “intercellular signaling” functions (72). Zietzer et al. confirmed that the export of miRNAs into EVs depends on the binding efficiency of the respective miRNAs to hnRNPU. miR-30c-5p, the most significant miRNA regulated by hnRNPU, retains a significant enrichment of the sequence motif AAMRUGCU as a transport signal (56). Wozniak et al. identified a common short sequence of the “AAUGC” motif present in miRNAs that are selectively loaded into exosomes after RILP cleavage, which promotes the movement of MVBs toward the cell periphery and induces selective exosomal miRNA cargo loading. This motif binds the RBP FMR1 and directs miRNA loading into exosomes by interacting with components of the ESCRT pathway (57, 86). Syncrip/hnRNPQ, a highly conserved RBP, can identify hEXO (GGCU/A) sequences in target miRNAs and mediate exosome enrichment through the collaboration of the non-canonical N-terminal RNA recognition region NURR domain and the classical RRM domain (61, 62). Additionally, miR-133 was specifically sorted into H/R-induced EPC-derived exosomes via YBX-1 to increase fibroblast angiogenesis and MEndoT (87). Recent discoveries have indicated that circulating Ago2 complexes are responsible for the stability of plasma miRNAs through the KRAS-MEK-ERK signaling pathway, protecting miRNAs contained within EVs from RNase degradation (67, 88, 89). Considering RNA-binding ubiquitin E3 ligase (MEX3C) associates with Ago2 and the adaptor-related protein complex 2 (AP-2), which is involved in miRNA sorting, containing a C-terminal RING finger domain and the hnRNP K homology (KH) domain, miR-451a is specifically sorted into exosomes via a ceramide-dependent pathway (90, 91) In general, the concept that specific ncRNA motif binding to RBP is involved in exosomal ncRNA sorting has been confirmed in different types of cells.
Modification of ncRNAs or RBPs Guide Their Sorting Into Exosomes
Collectively, current research is beginning to uncover new mechanisms by which exosomes are involved in the post-transcriptional modification of ncRNA (92). Reports have shown that miRNAs are modified through a series of processing events after transcription, such as 5′-end phosphorylation, 3′-end adenylation or uridylation, and terminal nucleotide deletion. Khan et al. attempted to develop a method for competence-mass spectrometry, which can be used to perform a multiplex, direct analysis of miRNAs from biological samples, and revealed modifications of miRNAs in serum samples (93). Koppers et al. performed RNA sequencing and bioinformatics analysis and found a non-random distribution of miRNAs between B cells and exosomes. Subsequently, in the urine samples, the 3′-terminal adenylation miRNAs were relatively enriched in the cells, whereas the 3′-end uridine subtypes were found in excess in the exosomes, suggesting that post-transcriptional modifications, especially 3′-end adenylation and uridylation, play the opposite role and may at least partially guide the entry of ncRNAs into exosomes (69). Wani et al. revealed that post-transcriptional modifications, especially 3′-end adenylation and uridylation of miR-2909, exert opposing effects that may contribute to its sorting into exosomes secreted by cancer cells (70).
Emerging evidence suggests that the post-translational modification of RBPs also has a considerable influence on the sorting of specific ncRNAs. Lee et al. showed that cav 1 14 (Y14)-tyrosine phosphorylation leads to interactions between caveolin 1 (cav 1) and hnRNPA2B1. Cav 1, as a lipid-raft scaffolding protein, has been proposed to induce local membrane composition and curvature, constitutes a complex with HNRNPA2B1-miRNAs, subsequently directs theirs their routing towards exosomes in lung epithelial cells. Oxidative stress induces the O-GlcNAcylation of hnRNPA2B1, resulting in a robustly altered hnRNPA2B1-bound miRNA repertoire. Notably, cav-1 pY14 also promoted hnRNPA2B1 O-GlcNAcylation. Functionally, macrophages serve as the principal recipients of epithelial EVs in the lungs. EV-containing cav-1/hnRNPA2B1 complex-bound miR-17/93 activates tissue macrophages (60). Furthermore, hnRNPA2B1 in exosomes is SUMOylated, which controls the binding of hnRNPA2B1 to miRNAs (58). The KRAS-MEK-ERK pathway-dependent phosphorylation of Ago2 has been demonstrated to exert specific control over the sorting of let-7a, miR-100, and miR-320a into exosomes (94).
Content of Intracellular Associated RNAs Modulates the Sorting of ncRNAs Into Exosomes
It has been reported that ncRNAs can be modulated by their upstream or downstream RNAs, thus being packaged into exosomes either via passive leakage or in an active secretion manner. The most classical is the RNA processing of miRNA, which affects miRNA enrichment in exosomes. It is known that miRNA maturation in the cytoplasm requires a type II endoribonuclease known as the Dicer enzyme. It cleaves the stem loop structure of pre-miRNA and produces ~22-nt miRNA double strands, which are unwound into mature single-stranded miRNAs and combined with RNA-induced silencing complex (RISC). The co-localization and accumulation or relocalization of miRISC components in MVBs is thought to facilitate the sorting of miRNAs into exosomes (95, 96).
It has been proven that ncRNAs can interact with their target RNAs to regulate the content of their own or target RNAs in cells, after which they are sorted into exosomes passively or actively. Squadrito et al. suggested that the occurrence of physiological (cell-activation-dependent) or artificial overexpression of miRNA target sequences (mRNA) in macrophages contributes to the enrichment of the corresponding miRNA exosomes and P-bodies. Perhaps, through such a mechanism, the miRNA loading in the producer cells can be reduced; however, through this mechanism, the activity of miRNA can be affected and cell homeostasis can be successfully achieved (97). In renal cell cancer cells, Qu et al. found a new lncRNA, lncARSR, which has a specific GGAG/CCCU at its 5′-end and can bind to the RBP hnRNPA2B1 and is selected as an exosome. Interestingly, the target of lncARSR, miR-198, rather than the target miR-18a, can be secreted into the exosomes along with the lncARSR-hnRNPA2B1 complex (15, 58). In HEK293T and MCF-7 cells, miR-7, the target of CDR1as, was artificially upregulated. Subsequently, Li et al. observed that CDR1as circRNA levels in exosomes were significantly downregulated. This result indicates that the sorting of circRNA to exosomes may be regulated by the level of related miRNAs in the production cells and may transfer biological activity to the recipient cells (98).
Secondary/Tertiary Structure of ncRNAs Regulates Their Sorting
These interactions could occur because the secondary/tertiary structures of ncRNAs, as opposed to those of other nucleic acid sequences, partially result in a greater protein binding capacity. Shurtleff et al. discovered that RBP Y-box protein I (YBX1) binds to and is required for the sorting of miR-223 in HEK293T cells. YBX1 interacts with miR-223 through its internal cold shock domain to form a hairpin-loop secondary structure, rather than a specific recognition motif, which promotes the separation of miR-223 into exosomes (63). Platelets are rich in circRNAs and seem to be more inclined to secrete smaller-sized circRNAs into exosomes than larger ones. It is reasonable to speculate that the mechanism by which circRNAs are sorted into exosomes is related to their circular tertiary structure, which needs to be further characterized (71).
Clinical Application of Exosomal ncRNAs
Biological Effects of Exosomal ncRNAs
Exosomes are known to be rich in biological information (proteins, nucleic acids, etc.). They are not only acting as tissue sampling to reflect the state of parent cells, but also representative tools to transmit cell-to-cell communication information. Exosomes play a vital role in mediating cell–cell communication and transporting cargo from donor cells to recipient cells, regardless of whether the recipient cells are located in distant or nearby tissues (7, 99). Bidirectional cell–cell communication involving exosome-borne cargos, such as ncRNAs, has emerged as a critical mechanism. As natural intercellular shuttles of ncRNAs, exosomes influence an array of developmental, physiological, and pathological processes in the recipient cell or tissue to which they can be selectively targeted (Figure 2) (100). For example, Exosomal ncRNAs could interact with many inflammatory factors and inflammatory cells to influence the progression of inflammatory diseases. The exosomes promoted macrophage M1 differentiation at least partially via transferring pro-inflammatory miRNAs, such as miR-155. Moreover, exosome-mediated miR-155 inhibitor delivery significantly prevented DSS-induced colitis (101). Hepatocyte exosomes induced by lipotoxic injury are rich in miR-192-5p. It can regulate the Rictor/Akt/FoxO1 signaling pathway, induce an increase in the expression of M1 macrophages and inflammatory factors, and affect the progression of steatohepatitis (102). Exosomes from patients with septic shock convey miRNAs and mRNAs related to inflammatory response for intercellular communication (103). In general, exosomal ncRNAs are essential as a major contributor that regulates delivery and reduces inflammatory response (59). The ncRNAs content of exosomes could be regulated by the physiological state of cells and may play a role in maintaining tissue homeostasis and synchronizing the functional state of cells. For example, miRNAs transferred from mother’s milk to the infant may play a crucial role in the development of the infant’s immune system (104). Exosomal miRNAs have been proven to act as regulators of neuron- astrocyte crosstalk, osteoblast differentiation, myoblast differentiation, and so on (105–108). Exosomal ncRNAs play a key role in premetastatic niche formation and metastasis (109, 110). Zhang et al. observed that the expression of circSATB2 in serum exosomes of patients with NSCLC was higher than that in non-cancerous donors, which was related to lymph node metastasis of lung cancer and could promote the proliferation of normal bronchial epithelial cells. This suggests that exosomal circSATB2 can transmit cell communication and promote the metastasis of tumor cells (111). Exosomes imposed by pathogenic microorganisms, such as viruses, bacteria, and parasites, may be exploited for the superior delivery of ncRNAs to evade host immune surveillance in vivo (112–114). Schistosoma japonicum egg-derived exosomal Sja-miR-71a attenuated pathological progression and liver fibrosis in S. japonicum infection (115). Adult Schistosoma secretes exosomal miRNAs that are internalized by Th cells to evade immune surveillance (116). Exosomes are soluble biological mediators obtained from mesenchymal stem cells (MSCs) cultured in vitro. MSC-derived exosomes produced under physiological or pathological conditions are central mediators of intercellular communication by transporting proteins, lipids, mRNAs, siRNA, rRNAs, and ncRNAs to neighboring or distant cells.
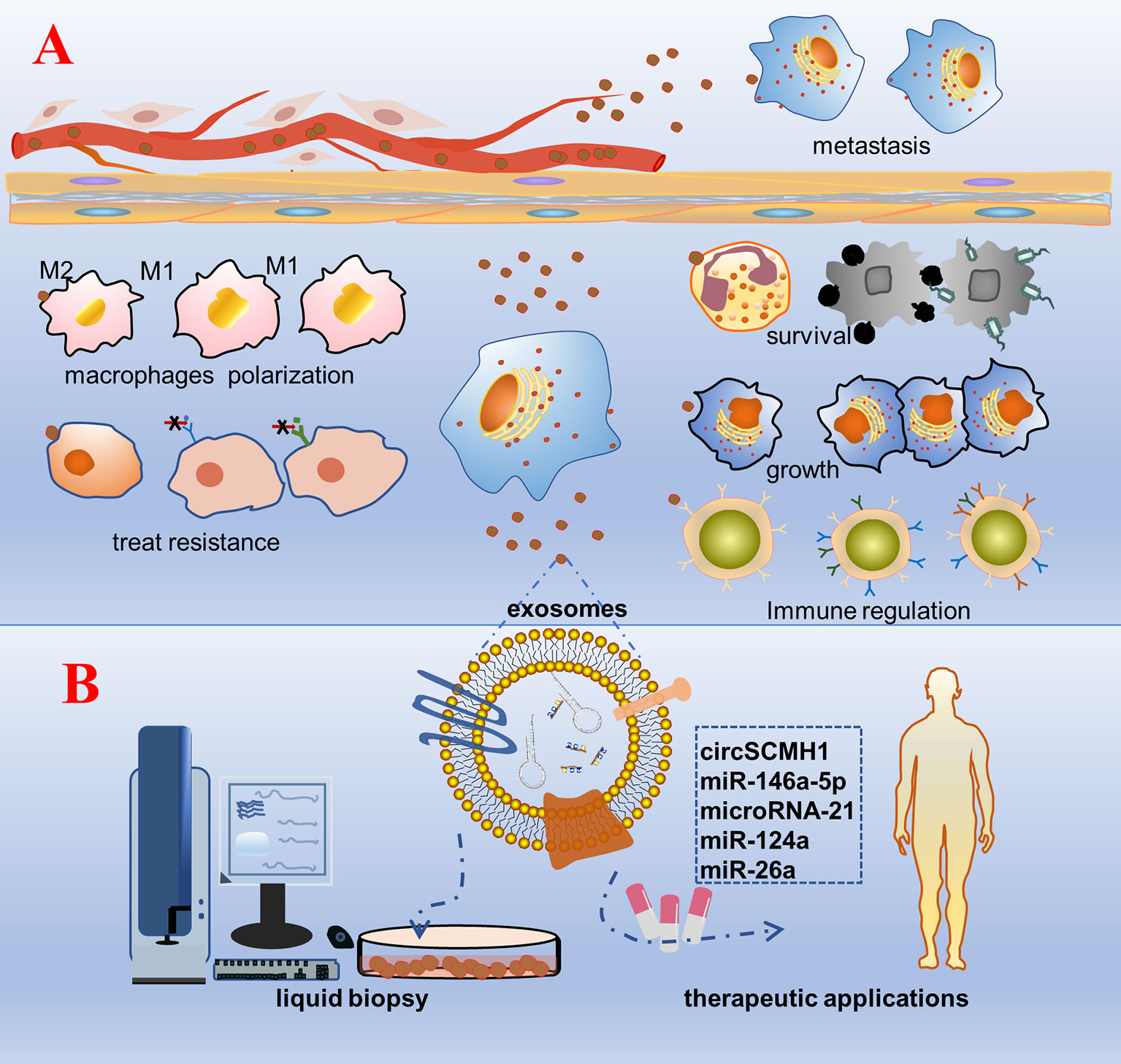
Figure 2 Schematic diagram of the biological function and clinical application prospects of exosomal non-coding RNAs. (A) Biological function: cells are stimulated by factors, such as tumorigenesis, secrete exosomes wrapped with bioactive non-coding RNAs, and are accepted by the recipient cell. As a result, a series of phenotypic changes occur: pathogenic microorganisms escape the body’s immune surveillance to survive; growth of specific recipient cells; regulation of the number and functions of immune cells, such as T cells and NK cells; polarization of macrophages and the inflammatory response; change in the tolerance of the recipient cells to treatment; transmission to distant tissue cells through body fluids inducing cancer metastasis. (B) Clinical application: purification, separation, and detection of exosomes in various body fluids, construction of a platform for rapid detection and analysis of diseases, and engineering of exosomal non-coding RNAs that are promising for treatment.
Exosomal ncRNA as a Specific Biomarker for Liquid Biopsy
Liquid biopsy is a minimally invasive method for analyzing solid tissues, blood, and other body fluids. Exosomes can be conveniently detected in almost all human body fluids, making them an ideal indicator for liquid biopsy. Exosomes derived from different cell types and statuses have been shown to possess distinct RNA profiles, particularly ncRNAs. These new analytes represent an alternative tool to complement the diagnosis, monitoring, and prediction of response to treatment of tumor processes, as well as other human disease processes, such as those in viral and parasitic infections (117, 118). ncRNAs are important regulators of cellular signaling that can be detected and released into circulation with high stability via packing in exosomes (119–122). Therefore, the use of exosomal ncRNAs has promising prospects in liquid biopsy in case of diseases and may continue to be an exciting focus of research in the field.
Owing to the rapid development of various exosome detection technologies in recent years, exosomal ncRNAs have become a specific and effective biomarker for clinical liquid biopsy (123–125). Methods of exosome isolation to date include ultracentrifugation (mostly approved for exosome purity), ultrafiltration, size-exclusion chromatography, polymer precipitation, immunoaffinity chromatography, and microfluidics-based techniques (126–128). Wang et al. described the construction and testing of an electrochemical biosensor for the sensitive detection of exosomal miRNAs. The electrochemical biosensor exhibited good selectivity for miR-21 detection; showed benefits of simple operation, low cost, and portability; and provided a promising platform for the early diagnosis and screening of tumor biomarkers and the development of devices for point-of-care testing (129). Wu et al. established a platform for the simultaneous multiplex analysis of multiple exosome biomarkers (such as proteins and miRNAs) in clinical biological fluids, which not only allows for the observation of the tissue status of biomarkers in clinical samples but also shows that exosome subsets can more accurately distinguish the prognosis of patients (130). Zabegina et al. isolated exosomes with thyroid-specific surface molecules by immunobeads followed by miRNA analysis, demonstrating possibly improved diagnostic potency (130). Serum exosomal miRNA profiles of steroid-induced osteonecrosis of the femoral head (SONFH) and hsa-miR-135b-5p may be a unique diagnostic biomarker for SONFH (131). Zheng et al. found that the expression level of exosomal lnc-SLC2A12-10:1 was significantly correlated with tumor size, TNM stage, lymph node metastasis, and degree of differentiation, suggesting that exosomal lnc-SLC2A12-10:1 may be a potential noninvasive biomarker for the diagnosis and prognosis monitoring of gastric cancer (132). In summary, ncRNAs derived from exosomes are considered potential new biomarkers for various diseases, especially cancer, and can be easily detected in liquid biopsies.
Therapeutic Applications of Engineered Exosomal ncRNAs
A variety of ncRNA molecules are known to function in human diseases. However, safety issues with delivery systems have limited the exploration of the potential therapeutic roles of ncRNAs. Engineered EVs carrying therapeutic molecules are promising candidates for disease therapy. In recent years, exosomes have been discovered with low immunogenicity, positive safety in clinical trials, and the ability of selectively homing to inflammation and tumor sites. Therefore, engineered exosomal ncRNAs have great therapeutic potential (133–137).
Yang et al. found that in an ischemic stroke model, the engineered extracellular vesicular rabies virus glycoprotein-circSCMH1 selectively transmits circSCMH1 to the brain and mechanically binds to the transcription factor MeCP2, thus resulting in the inhibition of MeCP2 target gene transcription and promoting the functional recovery of stroke in animals (138). Exosomal miRNAs and proteins isolated from hiPSC-NSC cultures have many functions, such as neuroprotection and anti-inflammation. The intranasal administration of exosomes can be absorbed by a variety of nerve cells, which is beneficial for brain repair after injury or disease (139). A separate study showed that secreted exosomes coated with miR-146a-5p from MSCs relieved Group 2 innate lymphoid cells (ILC2s) in innate airway inflammation, showing significant advantages of low immunogenicity and high biosafety (140). Engineered exosomes loaded with anti-inflammatory agents, such as miRNA-21, could be used to target macrophages in the inflammatory region to regulate inflammatory responses for achieving the ability to regulate inflammatory responses when needed (141). Engineered exosomal ncRNAs have also shown great advantages in the treatment of tumors. In addition to the unique properties of MSCs, MSC-derived exosomal ncRNAs exert desirable therapeutic effects (142). Lang et al. reported that bone marrow-derived MSCs could encapsulate miRNAs, such as miR-124a, into exosomes, and these engineered exosomes could be used to treat mice harboring intracranial glioma stem cell. It was found that engineered exosomes could systematically transmit anti-glioma miRNAs to glioblastomas for longer survival (143). Liang et al. introduced a new approach for the targeted delivery of exosomes loaded with functional miR-26a to scavenger receptor class B type 1-expressing liver cancer cells, resulting in decreased rates of cell migration and proliferation (144).
Conclusion
Nanoscale exosomes encapsulating a variety of cargos, including ncRNAs, which protect the cargo from degradation by various enzymes in the extracellular space, are vital for intracellular and intercellular communication. Exosomes can be selectively secreted via ESCRT-dependent or ESCRT-independent pathways. Exosomes have multiple functions in physiological and pathological processes, and ncRNAs can also play an important regulatory role in these processes. The relationship between them has attracted great research interest in recent years. Subsequently, an increasing number of studies have revealed that exosomal ncRNAs are involved in pathogenic microbial infection and inflammatory disease, tumor invasion and metastasis, immunoregulation and immunotherapy, and resistance to treatment. However, how ncRNAs in exosomes are selectively packaged and then transported to target cells remains unclear, which hinders the prospects of clinical applications of exosomal ncRNAs as biomarkers. Elucidating the mechanism by which cells sort specific ncRNAs into the circulation will aid in the selection of more representative exosomal RNAs and pave the way for advances in the early diagnosis of diseases. Exosomal ncRNAs can transmit signals, such as those involved in growth promotion, invasion, metastasis, and angiogenesis. Moreover, animal experiments have confirmed their effect on the disease phenotype. Thus, we believe that understanding how these disease-causing or disease-suppressing exosomal ncRNAs are sorted, secreted, and spread to adjacent or distant cells in the initial stages of a disease will underlie the development of timely, accurate, and effective early intervention measures.
Although plenty of methods have been developed to isolate exosomes, it has been found that extracellular ncRNAs could be mixed with a variety of exosomal vectors such as lipoprotein and Argonaute protein, which obviously interferes with the accuracy of studies on exosomal ncRNAs. Therefore, researchers are making unremitting efforts to improve the technology of exosome purification and separation. In addition, exosomes have been recognized as a class of aggregated nanoparticles with different properties of extracellular vesicles. Currently, there is a lack of specific characterization of the heterogeneity of different exosome subtypes. Significant differences of the proteins, lipids, and nucleic acids contained in different exosome subtypes have been demonstrated. The heterogeneity of secreted nanoparticles is increasingly recognized. Numerous studies have exploited new technologies to characterize EV subtypes. Ayuko Hoshino and his team deconvolved the heterogeneity of extracellular nanoparticles and defined three distinct subsets: small exosomes (Exo-S, 50–70 nm), large exosomes (Exo-L, 90–120 nm) and exomeres. The exomeres obtained through sequential ultracentrifugation (SUC) were non-membranous particles (<50 nm). Many of the most abundant miRNAs are more associated with extracellular exomeres than parent cells or Exo-S components. Furthermore, some RBPs proved to be related to the mechanism of sorting ncRNA, such as YBX1 and MVP protein, were also strongly correlated with exomeres components. Therefore, there is an urgent need to identify signature proteins that can more effectively distinguish EV subtypes (145–149). Therefore, the application of the enriched ncRNAs in different exosome subtypes in liquid biopsy or clinical treatment also needs further study.
Technological advances have enabled researchers to opt for more effective methods for the separation of high-purity exosomes, allowing for the in-depth study of the mechanism underlying the non-random distribution of functional molecules to exosomes in cells. It is reasonable to speculate that hundreds of millions of patients, especially those with poor treatment outcomes, will greatly benefit from the in-depth study of the mechanism and role of exosomal ncRNAs.
Author Contributions
YQ and MW conceived, wrote, and edited the manuscript. YQ and PL designed figures and wrote and edited the manuscript. MW and ZZ wrote and edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work is supported by grants from the National Science Foundation for Young Scientists of China (Grant 81802871); Graduate Scientific Research and Innovation Project of Central South University (No. 1053320192778).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are very grateful to the many researchers working in this field.
References
1. Hoshino A, Kim HS, Bojmar L, Gyan KE, Cioffi M, Hernandez J, et al. Extracellular Vesicle and Particle Biomarkers Define Multiple Human Cancers. Cell (2020) 182(4):1044–61.e18. doi: 10.1016/j.cell.2020.07.009
2. de Jong OG, Kooijmans SAA, Murphy DE, Jiang L, Evers MJW, Sluijter JPG, et al. Drug Delivery With Extracellular Vesicles: From Imagination to Innovation. Acc Chem Res (2019) 52(7):1761–70. doi: 10.1021/acs.accounts.9b00109
3. Mateescu B, Kowal EJ, van Balkom BW, Bartel S, Bhattacharyya SN, Buzas EI, et al. Obstacles and Opportunities in the Functional Analysis of Extracellular Vesicle RNA - an ISEV Position Paper. J Extracell Vesicles (2017) 6(1):1286095. doi: 10.1080/20013078.2017.1286095
4. Anand S, Samuel M, Kumar S, Mathivanan S. Ticket to a Bubble Ride: Cargo Sorting Into Exosomes and Extracellular Vesicles. Biochim Biophys Acta Proteins Proteom (2019) 1867(12):140203. doi: 10.1016/j.bbapap.2019.02.005
5. Janas T, Janas MM, Sapon K, Janas T. Mechanisms of RNA Loading Into Exosomes. FEBS Lett (2015) 589(13):1391–8. doi: 10.1016/j.febslet.2015.04.036
6. Villarroya-Beltri C, Baixauli F, Gutierrez-Vazquez C, Sanchez-Madrid F, Mittelbrunn M. Sorting it Out: Regulation of Exosome Loading. Semin Cancer Biol (2014) 28:3–13. doi: 10.1016/j.semcancer.2014.04.009
7. O’Brien K, Breyne K, Ughetto S, Laurent LC, Breakefield XO. RNA Delivery by Extracellular Vesicles in Mammalian Cells and its Applications. Nat Rev Mol Cell Biol (2020) 21(10):585–606. doi: 10.1038/s41580-020-0251-y
8. Kalluri R, LeBleu VS. The Biology, Function, and Biomedical Applications of Exosomes. Science (2020) 367(6478):eaau6977. doi: 10.1126/science.aau6977
9. Battaglia R, Musumeci P, Ragusa M, Barbagallo D, Scalia M, Zimbone M, et al. Ovarian Aging Increases Small Extracellular Vesicle CD81(+) Release in Human Follicular Fluid and Influences miRNA Profiles. Aging (Albany NY) (2020) 12(12):12324–41. doi: 10.18632/aging.103441
10. Mathieu M, Martin-Jaular L, Lavieu G, Thery C. Specificities of Secretion and Uptake of Exosomes and Other Extracellular Vesicles for Cell-to-Cell Communication. Nat Cell Biol (2019) 21(1):9–17. doi: 10.1038/s41556-018-0250-9
11. Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-Mediated Transfer of mRNAs and microRNAs is a Novel Mechanism of Genetic Exchange Between Cells. Nat Cell Biol (2007) 9(6):654–9. doi: 10.1038/ncb1596
12. Yin Y, Cai X, Chen X, Liang H, Zhang Y, Li J, et al. Tumor-Secreted miR-214 Induces Regulatory T Cells: A Major Link Between Immune Evasion and Tumor Growth. Cell Res (2014) 24(10):1164–80. doi: 10.1038/cr.2014.121
13. Yin J, Zeng A, Zhang Z, Shi Z, Yan W, You Y. Exosomal Transfer of miR-1238 Contributes to Temozolomide-Resistance in Glioblastoma. EBioMedicine (2019) 42:238–51. doi: 10.1016/j.ebiom.2019.03.016
14. Shao N, Xue L, Wang R, Luo K, Zhi F, Lan Q. Mir-454-3p Is an Exosomal Biomarker and Functions as a Tumor Suppressor in Glioma. Mol Cancer Ther (2019) 18(2):459–69. doi: 10.1158/1535-7163.mct-18-0725
15. Qu L, Ding J, Chen C, Wu ZJ, Liu B, Gao Y, et al. Exosome-Transmitted Lncarsr Promotes Sunitinib Resistance in Renal Cancer by Acting as a Competing Endogenous Rna. Cancer Cell (2016) 29(5):653–68. doi: 10.1016/j.ccell.2016.03.004
16. Slack FJ, Chinnaiyan AM. The Role of Non-coding Rnas in Oncology. Cell (2019) 179(5):1033–55. doi: 10.1016/j.cell.2019.10.017
17. Hong BS, Cho JH, Kim H, Choi EJ, Rho S, Kim J, et al. Colorectal Cancer Cell-Derived Microvesicles are Enriched in Cell Cycle-Related mRNAs That Promote Proliferation of Endothelial Cells. BMC Genomics (2009) 10:556. doi: 10.1186/1471-2164-10-556
18. Spinelli C, Adnani L, Choi D, Rak J. Extracellular Vesicles as Conduits of Non-Coding Rna Emission and Intercellular Transfer in Brain Tumors. Noncoding RNA (2018) 5(1):1. doi: 10.3390/ncrna5010001
19. Geng X, Lin X, Zhang Y, Li Q, Guo Y, Fang C, et al. Exosomal Circular RNA Sorting Mechanisms and Their Function in Promoting or Inhibiting Cancer. Oncol Lett (2020) 19(5):3369–80. doi: 10.3892/ol.2020.11449
20. Zhang WL, Liu Y, Jiang J, Tang YJ, Tang YL, Liang XH. Extracellular Vesicle Long non-Coding RNA-mediated Crosstalk in the Tumor Microenvironment: Tiny Molecules, Huge Roles. Cancer Sci (2020) 111(8):2726–35. doi: 10.1111/cas.14494
21. Vignard V, Labbé M, Marec N, André-Grégoire G, Jouand N, Fonteneau JF, et al. MicroRNAs in Tumor Exosomes Drive Immune Escape in Melanoma. Cancer Immunol Res (2020) 8(2):255–67. doi: 10.1158/2326-6066.cir-19-0522
22. Hanson PI, Cashikar A. Multivesicular Body Morphogenesis. Annu Rev Cell Dev Biol (2012) 28:337–62. doi: 10.1146/annurev-cellbio-092910-154152
23. Bebelman MP, Bun P, Huveneers S, van Niel G, Pegtel DM, Verweij FJ. Real-Time Imaging of Multivesicular Body-Plasma Membrane Fusion to Quantify Exosome Release From Single Cells. Nat Protoc (2020) 15(1):102–21. doi: 10.1038/s41596-019-0245-4
24. Palmulli R, van Niel G. To be or Not to be... Secreted as Exosomes, a Balance Finely Tuned by the Mechanisms of Biogenesis. Essays Biochem (2018) 62(2):177–91. doi: 10.1042/EBC20170076
25. Hessvik NP, Llorente A. Current Knowledge on Exosome Biogenesis and Release. Cell Mol Life Sci (2018) 75(2):193–208. doi: 10.1007/s00018-017-2595-9
26. Vietri M, Radulovic M, Stenmark H. The Many Functions of Escrts. Nat Rev Mol Cell Biol (2020) 21(1):25–42. doi: 10.1038/s41580-019-0177-4
27. Han Q, Lv L, Wei J, Lei X, Lin H, Li G, et al. Vps4A Mediates the Localization and Exosome Release of β-Catenin to Inhibit Epithelial-Mesenchymal Transition in Hepatocellular Carcinoma. Cancer Lett (2019) 457:47–59. doi: 10.1016/j.canlet.2019.04.035
28. Tamai K, Tanaka N, Nakano T, Kakazu E, Kondo Y, Inoue J, et al. Exosome Secretion of Dendritic Cells is Regulated by Hrs, an ESCRT-0 Protein. Biochem Biophys Res Commun (2010) 399(3):384–90. doi: 10.1016/j.bbrc.2010.07.083
29. David G, Zimmermann P. Heparanase Involvement in Exosome Formation. Adv Exp Med Biol (2020) 1221:285–307. doi: 10.1007/978-3-030-34521-1_10
30. Schoneberg J, Lee IH, Iwasa JH, Hurley JH. Reverse-Topology Membrane Scission by the ESCRT Proteins. Nat Rev Mol Cell Biol (2017) 18(1):5–17. doi: 10.1038/nrm.2016.121
31. Kostelansky MS, Sun J, Lee S, Kim J, Ghirlando R, Hierro A, et al. Structural and Functional Organization of the ESCRT-I Trafficking Complex. Cell (2006) 125(1):113–26. doi: 10.1016/j.cell.2006.01.049
32. Hierro A, Sun J, Rusnak AS, Kim J, Prag G, Emr SD, et al. Structure of the ESCRT-II Endosomal Trafficking Complex. Nature (2004) 431(7005):221–5. doi: 10.1038/nature02914
33. Larios J, Mercier V, Roux A, Gruenberg J. ALIX- and ESCRT-III-dependent Sorting of Tetraspanins to Exosomes. J Cell Biol (2020) 219(3):e201904113. doi: 10.1083/jcb.201904113
34. Sun R, Liu Y, Lu M, Ding Q, Wang P, Zhang H, et al. ALIX Increases Protein Content and Protective Function of iPSC-derived Exosomes. J Mol Med (Berl) (2019) 97(6):829–44. doi: 10.1007/s00109-019-01767-z
35. Ghossoub R, Lembo F, Rubio A, Gaillard CB, Bouchet J, Vitale N, et al. Syntenin-ALIX Exosome Biogenesis and Budding Into Multivesicular Bodies are Controlled by ARF6 and PLD2. Nat Commun (2014) 5:3477. doi: 10.1038/ncomms4477
36. Baietti MF, Zhang Z, Mortier E, Melchior A, Degeest G, Geeraerts A, et al. Syndecan-syntenin-ALIX Regulates the Biogenesis of Exosomes. Nat Cell Biol (2012) 14(7):677–85. doi: 10.1038/ncb2502
37. Isik S, Sano K, Tsutsui K, Seki M, Enomoto T, Saitoh H, et al. The SUMO Pathway is Required for Selective Degradation of DNA Topoisomerase IIbeta Induced by a Catalytic Inhibitor ICRF-193(1). FEBS Lett (2003) 546(2-3):374–8. doi: 10.1016/s0014-5793(03)00637-9
38. Matic I, van Hagen M, Schimmel J, Macek B, Ogg SC, Tatham MH, et al. In Vivo Identification of Human Small Ubiquitin-Like Modifier Polymerization Sites by High Accuracy Mass Spectrometry and an In Vitro to In Vivo Strategy. Mol Cell Proteomics (2008) 7(1):132–44. doi: 10.1074/mcp.M700173-MCP200
39. Kunadt M, Eckermann K, Stuendl A, Gong J, Russo B, Strauss K, et al. Extracellular Vesicle Sorting of α-Synuclein is Regulated by Sumoylation. Acta Neuropathol (2015) 129(5):695–713. doi: 10.1007/s00401-015-1408-1
40. Chang HM, Yeh ETH. Sumo: From Bench to Bedside. Physiol Rev (2020) 100(4):1599–619. doi: 10.1152/physrev.00025.2019
41. Liu Y, Shah SV, Xiang X, Wang J, Deng ZB, Liu C, et al. COP9-Associated CSN5 Regulates Exosomal Protein Deubiquitination and Sorting. Am J Pathol (2009) 174(4):1415–25. doi: 10.2353/ajpath.2009.080861
42. Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, Wieland F, et al. Ceramide Triggers Budding of Exosome Vesicles Into Multivesicular Endosomes. Science (2008) 319(5867):1244–7. doi: 10.1126/science.1153124
43. de Gassart A, Geminard C, Fevrier B, Raposo G, Vidal M. Lipid Raft-Associated Protein Sorting in Exosomes. Blood (2003) 102(13):4336–44. doi: 10.1182/blood-2003-03-0871
44. Rana S, Zoller M. Exosome Target Cell Selection and the Importance of Exosomal Tetraspanins: A Hypothesis. Biochem Soc Trans (2011) 39(2):559–62. doi: 10.1042/BST0390559
45. van Niel G, Charrin S, Simoes S, Romao M, Rochin L, Saftig P, et al. The Tetraspanin CD63 Regulates ESCRT-independent and -Dependent Endosomal Sorting During Melanogenesis. Dev Cell (2011) 21(4):708–21. doi: 10.1016/j.devcel.2011.08.019
46. Chairoungdua A, Smith DL, Pochard P, Hull M, Caplan MJ. Exosome Release of Beta-Catenin: A Novel Mechanism That Antagonizes Wnt Signaling. J Cell Biol (2010) 190(6):1079–91. doi: 10.1083/jcb.201002049
47. Nazarenko I, Rana S, Baumann A, McAlear J, Hellwig A, Trendelenburg M, et al. Cell Surface Tetraspanin Tspan8 Contributes to Molecular Pathways of Exosome-Induced Endothelial Cell Activation. Cancer Res (2010) 70(4):1668–78. doi: 10.1158/0008-5472.CAN-09-2470
48. Perez-Hernandez D, Gutierrez-Vazquez C, Jorge I, Lopez-Martin S, Ursa A, Sanchez-Madrid F, et al. The Intracellular Interactome of Tetraspanin-Enriched Microdomains Reveals Their Function as Sorting Machineries Toward Exosomes. J Biol Chem (2013) 288(17):11649–61. doi: 10.1074/jbc.M112.445304
49. Strauss K, Goebel C, Runz H, Mobius W, Weiss S, Feussner I, et al. Exosome Secretion Ameliorates Lysosomal Storage of Cholesterol in Niemann-Pick Type C Disease. J Biol Chem (2010) 285(34):26279–88. doi: 10.1074/jbc.M110.134775
50. Sahu R, Kaushik S, Clement CC, Cannizzo ES, Scharf B, Follenzi A, et al. Microautophagy of Cytosolic Proteins by Late Endosomes. Dev Cell (2011) 20(1):131–9. doi: 10.1016/j.devcel.2010.12.003
51. Zhu H, Guariglia S, Yu RY, Li W, Brancho D, Peinado H, et al. Mutation of SIMPLE in Charcot-Marie-Tooth 1C Alters Production of Exosomes. Mol Biol Cell (2013) 24(11):1619–37. doi: 10.1091/mbc.E12-07-0544
52. Laulagnier K, Grand D, Dujardin A, Hamdi S, Vincent-Schneider H, Lankar D, et al. PLD2 is Enriched on Exosomes and its Activity is Correlated to the Release of Exosomes. FEBS Lett (2004) 572(1-3):11–4. doi: 10.1016/j.febslet.2004.06.082
53. Worst TS, Meyer Y, Gottschalt M, Weis CA, von Hardenberg J, Frank C, et al. Rab27a, RAB27B and VPS36 are Downregulated in Advanced Prostate Cancer and Show Functional Relevance in Prostate Cancer Cells. Int J Oncol (2017) 50(3):920–32. doi: 10.3892/ijo.2017.3872
54. Wei D, Zhan W, Gao Y, Huang L, Gong R, Wang W, et al. RAB31 Marks and Controls an ESCRT-independent Exosome Pathway. Cell Res (2020) 31(2):157–77. doi: 10.1038/s41422-020-00409-1
55. Kenific CM, Zhang H, Lyden D. An Exosome Pathway Without an ESCRT. Cell Res (2020) 31(2):105–6. doi: 10.1038/s41422-020-00418-0
56. Zietzer A, Hosen MR, Wang H, Goody PR, Sylvester M, Latz E, et al. The RNA-binding Protein hnRNPU Regulates the Sorting of microRNA-30c-5p Into Large Extracellular Vesicles. J Extracell Vesicles (2020) 9(1):1786967. doi: 10.1080/20013078.2020.1786967
57. Wozniak AL, Adams A, King KE, Dunn W, Christenson LK, Hung WT, et al. The RNA Binding Protein FMR1 Controls Selective Exosomal miRNA Cargo Loading During Inflammation. J Cell Biol (2020) 219(10):e201912074. doi: 10.1083/jcb.201912074
58. Villarroya-Beltri C, Gutierrez-Vazquez C, Sanchez-Cabo F, Perez-Hernandez D, Vazquez J, Martin-Cofreces N, et al. Sumoylated hnRNPA2B1 Controls the Sorting of miRNAs Into Exosomes Through Binding to Specific Motifs. Nat Commun (2013) 4:2980. doi: 10.1038/ncomms3980
59. Zhou X, Brown BA, Siegel AP, El Masry MS, Zeng X, Song W, et al. Exosome-Mediated Crosstalk Between Keratinocytes and Macrophages in Cutaneous Wound Healing. ACS Nano (2020) 14(10):12732–48. doi: 10.1021/acsnano.0c03064
60. Lee H, Li C, Zhang Y, Zhang D, Otterbein LE, Jin Y. Caveolin-1 Selectively Regulates microRNA Sorting Into Microvesicles After Noxious Stimuli. J Exp Med (2019) 216(9):2202–20. doi: 10.1084/jem.20182313
61. Santangelo L, Giurato G, Cicchini C, Montaldo C, Mancone C, Tarallo R, et al. The RNA-Binding Protein SYNCRIP is a Component of the Hepatocyte Exosomal Machinery Controlling MicroRNA Sorting. Cell Rep (2016) 17(3):799–808. doi: 10.1016/j.celrep.2016.09.031
62. Hobor F, Dallmann A, Ball NJ, Cicchini C, Battistelli C, Ogrodowicz RW, et al. A Cryptic RNA-binding Domain Mediates Syncrip Recognition and Exosomal Partitioning of miRNA Targets. Nat Commun (2018) 9(1):831. doi: 10.1038/s41467-018-03182-3
63. Shurtleff MJ, Temoche-Diaz MM, Karfilis KV, Ri S, Schekman R. Y-Box Protein 1 is Required to Sort microRNAs Into Exosomes in Cells and in a Cell-Free Reaction. Elife (2016) 5:e19276. doi: 10.7554/eLife.19276
64. Xu YF, Xu X, Gin A, Nshimiyimana JD, Mooers BHM, Caputi M, et al. SRSF1 Regulates Exosome microRNA Enrichment in Human Cancer Cells. Cell Commun Signal (2020) 18(1):130. doi: 10.1186/s12964-020-00615-9
65. Teng Y, Ren Y, Hu X, Mu J, Samykutty A, Zhuang X, et al. MVP-Mediated Exosomal Sorting of miR-193a Promotes Colon Cancer Progression. Nat Commun (2017) 8:14448. doi: 10.1038/ncomms14448
66. Statello L, Maugeri M, Garre E, Nawaz M, Wahlgren J, Papadimitriou A, et al. Identification of RNA-binding Proteins in Exosomes Capable of Interacting With Different Types of RNA: RBP-Facilitated Transport of RNAs Into Exosomes. PloS One (2018) 13(4):e0195969. doi: 10.1371/journal.pone.0195969
67. McKenzie AJ, Hoshino D, Hong NH, Cha DJ, Franklin JL, Coffey RJ, et al. Kras-Mek Signaling Controls Ago2 Sorting Into Exosomes. Cell Rep (2016) 15(5):978–87. doi: 10.1016/j.celrep.2016.03.085
68. Bakirtzi K, Man Law IK, Fang K, Iliopoulos D, Pothoulakis C. MiR-21 in Substance P-Induced Exosomes Promotes Cell Proliferation and Migration in Human Colonic Epithelial Cells. Am J Physiol Gastrointest Liver Physiol (2019) 317(6):G802–10. doi: 10.1152/ajpgi.00043.2019
69. Koppers-Lalic D, Hackenberg M, Bijnsdorp IV, van Eijndhoven MAJ, Sadek P, Sie D, et al. Nontemplated Nucleotide Additions Distinguish the Small RNA Composition in Cells From Exosomes. Cell Rep (2014) 8(6):1649–58. doi: 10.1016/j.celrep.2014.08.027
70. Wani S, Kaul D. Cancer Cells Govern miR-2909 Exosomal Recruitment Through its 3’-End Post-Transcriptional Modification. Cell Biochem Funct (2018) 36(2):106–11. doi: 10.1002/cbf.3323
71. Preußer C, Hung LH, Schneider T, Schreiner S, Hardt M, Moebus A, et al. Selective Release of circRNAs in Platelet-Derived Extracellular Vesicles. J Extracell Vesicles (2018) 7(1):1424473. doi: 10.1080/20013078.2018.1424473
72. Zhang J, Zhang X, Li C, Yue L, Ding N, Riordan T, et al. Circular RNA Profiling Provides Insights Into Their Subcellular Distribution and Molecular Characteristics in HepG2 Cells. RNA Biol (2019) 16(2):220–32. doi: 10.1080/15476286.2019.1565284
73. Lei Y, Guo W, Chen B, Chen L, Gong J, Li W. Tumorreleased Lncrna H19 Promotes Gefitinib Resistance Via Packaging Into Exosomes in Nonsmall Cell Lung Cancer. Oncol Rep (2018) 40(6):3438–46. doi: 10.3892/or.2018.6762
74. Zheng Z, Chen M, Xing P, Yan X, Xie B. Increased Expression of Exosomal Agap2-AS1 (Agap2 Antisense Rna 1) In Breast Cancer Cells Inhibits Trastuzumab-Induced Cell Cytotoxicity. Med Sci Monit (2019) 25:2211–20. doi: 10.12659/MSM.915419
75. Chen C, Luo Y, He W, Zhao Y, Kong Y, Liu H, et al. Exosomal Long Noncoding RNA LNMAT2 Promotes Lymphatic Metastasis in Bladder Cancer. J Clin Invest (2020) 130(1):404–21. doi: 10.1172/JCI130892
76. Zang J, Lu D, Xu A. The Interaction of circRNAs and RNA Binding Proteins: An Important Part of circRNA Maintenance and Function. J Neurosci Res (2020) 98(1):87–97. doi: 10.1002/jnr.24356
77. Hoek KS, Kidd GJ, Carson JH, Smith R. Hnrnp A2 Selectively Binds the Cytoplasmic Transport Sequence of Myelin Basic Protein Mrna. Biochemistry (1998) 37(19):7021–9. doi: 10.1021/bi9800247
78. Wang G, Chen HW, Oktay Y, Zhang J, Allen EL, Smith GM, et al. : PNPASE Regulates RNA Import Into Mitochondria. Cell (2010) 142(3):456–67. doi: 10.1016/j.cell.2010.06.035
79. Hwang HW, Wentzel EA, Mendell JT. A Hexanucleotide Element Directs microRNA Nuclear Import. Science (2007) 315(5808):97–100. doi: 10.1126/science.1136235
80. Groot M, Lee H. Sorting Mechanisms for MicroRNAs Into Extracellular Vesicles and Their Associated Diseases. Cells (2020) 9(4):1044. doi: 10.3390/cells9041044
81. Michlewski G, Caceres JF. Antagonistic Role of Hnrnp A1 and KSRP in the Regulation of let-7a Biogenesis. Nat Struct Mol Biol (2010) 17(8):1011–8. doi: 10.1038/nsmb.1874
82. Li T, Evdokimov E, Shen RF, Chao CC, Tekle E, Wang T, et al. Sumoylation of Heterogeneous Nuclear Ribonucleoproteins, Zinc Finger Proteins, and Nuclear Pore Complex Proteins: A Proteomic Analysis. Proc Natl Acad Sci USA (2004) 101(23):8551–6. doi: 10.1073/pnas.0402889101
83. Pérez-Boza J, Boeckx A, Lion M, Dequiedt F, Struman I. hnRNPA2B1 Inhibits the Exosomal Export of miR-503 in Endothelial Cells. Cell Mol Life Sci (2020) 77(21):4413–28. doi: 10.1007/s00018-019-03425-6
84. Pu Y, Zhao F, Cai W, Meng X, Li Y, Cai S. MiR-193a-3p and miR-193a-5p Suppress the Metastasis of Human Osteosarcoma Cells by Down-Regulating Rab27B and SRR, Respectively. Clin Exp Metastasis (2016) 33(4):359–72. doi: 10.1007/s10585-016-9783-0
85. Ostrowski M, Carmo NB, Krumeich S, Fanget I, Raposo G, Savina A, et al. Rab27a and Rab27b Control Different Steps of the Exosome Secretion Pathway. Nat Cell Biol (2010) 12(1):19–30. doi: 10.1038/ncb2000
86. Willson J. RILP Gets Cleaved and Exosomes Leave. Nat Rev Mol Cell Biol (2020) 21(11):658–9. doi: 10.1038/s41580-020-00299-6
87. Lin F, Zeng Z, Song Y, Li L, Wu Z, Zhang X, et al. YBX-1 Mediated Sorting of miR-133 Into Hypoxia/Reoxygenation-Induced EPC-derived Exosomes to Increase Fibroblast Angiogenesis and Mendot. Stem Cell Res Ther (2019) 10(1):263. doi: 10.1186/s13287-019-1377-8
88. Li L, Zhu D, Huang L, Zhang J, Bian Z, Chen X, et al. Argonaute 2 Complexes Selectively Protect the Circulating microRNAs in Cell-Secreted Microvesicles. PloS One (2012) 7(10):e46957. doi: 10.1371/journal.pone.0046957
89. Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, et al. Argonaute2 Complexes Carry a Population of Circulating microRNAs Independent of Vesicles in Human Plasma. Proc Natl Acad Sci USA (2011) 108(12):5003–8. doi: 10.1073/pnas.1019055108
90. Buchet-Poyau K, Courchet J, Le Hir H, Seraphin B, Scoazec JY, Duret L, et al. Identification and Characterization of Human Mex-3 Proteins, a Novel Family of Evolutionarily Conserved RNA-binding Proteins Differentially Localized to Processing Bodies. Nucleic Acids Res (2007) 35(4):1289–300. doi: 10.1093/nar/gkm016
91. Lu P, Li H, Li N, Singh RN, Bishop CE, Chen X, et al. MEX3C Interacts With Adaptor-Related Protein Complex 2 and Involves in miR-451a Exosomal Sorting. PloS One (2017) 12(10):e0185992. doi: 10.1371/journal.pone.0185992
92. Cerutti H, Ibrahim F. Turnover of Mature miRNAs and siRNAs in Plants and Algae. Adv Exp Med Biol (2011) 700:124–39. doi: 10.1007/978-1-4419-7823-3_11
93. Khan N, Mironov G, Berezovski MV. Direct Detection of Endogenous MicroRNAs and Their Post-Transcriptional Modifications in Cancer Serum by Capillary Electrophoresis-Mass Spectrometry. Anal Bioanal Chem (2016) 408(11):2891–9. doi: 10.1007/s00216-015-9277-y
94. Cha DJ, Franklin JL, Dou Y, Liu Q, Higginbotham JN, Demory Beckler M, et al. KRAS-Dependent Sorting of miRNA to Exosomes. Elife (2015) 4:e07197. doi: 10.7554/eLife.07197
95. Fatima F, Nawaz M. Long Distance Metabolic Regulation Through Adipose-Derived Circulating Exosomal Mirnas: A Trail for RNA-Based Therapies? Front Physiol (2017) 8:545. doi: 10.3389/fphys.2017.00545
96. Fatima F, Nawaz M. Vesiculated Long non-Coding RNAs: Offshore Packages Deciphering Trans-Regulation Between Cells, Cancer Progression and Resistance to Therapies. Noncoding RNA (2017) 3(1):10. doi: 10.3390/ncrna3010010
97. Squadrito ML, Baer C, Burdet F, Maderna C, Gilfillan GD, Lyle R, et al. Endogenous RNAs Modulate microRNA Sorting to Exosomes and Transfer to Acceptor Cells. Cell Rep (2014) 8(5):1432–46. doi: 10.1016/j.celrep.2014.07.035
98. Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, et al. Circular RNA is Enriched and Stable in Exosomes: A Promising Biomarker for Cancer Diagnosis. Cell Res (2015) 25(8):981–4. doi: 10.1038/cr.2015.82
99. Kim KM, Abdelmohsen K, Mustapic M, Kapogiannis D, Gorospe M. RNA in Extracellular Vesicles. Wiley Interdiscip Rev RNA (2017) 8(4):e1413. doi: 10.1002/wrna.1413
100. Fanale D, Taverna S, Russo A, Bazan V. Circular RNA in Exosomes. Adv Exp Med Biol (2018) 1087:109–17. doi: 10.1007/978-981-13-1426-1_9
101. Wei M, Gao X, Liu L, Li Z, Wan Z, Dong Y, et al. Visceral Adipose Tissue Derived Exosomes Exacerbate Colitis Severity Via Pro-inflammatory MiRNAs in High Fat Diet Fed Mice. ACS Nano (2020) 14(4):5099–110. doi: 10.1021/acsnano.0c01860
102. Liu XL, Pan Q, Cao HX, Xin FZ, Zhao ZH, Yang RX, et al. Lipotoxic Hepatocyte-Derived Exosomal MicroRNA 192-5p Activates Macrophages Through Rictor/Akt/Forkhead Box Transcription Factor O1 Signaling in Nonalcoholic Fatty Liver Disease. Hepatology (2020) 72(2):454–69. doi: 10.1002/hep.31050
103. Real JM, Ferreira LRP, Esteves GH, Koyama FC, Dias MVS, Bezerra-Neto JE, et al. Exosomes From Patients With Septic Shock Convey miRNAs Related to Inflammation and Cell Cycle Regulation: New Signaling Pathways in Sepsis? Crit Care (2018) 22(1):68. doi: 10.1186/s13054-018-2003-3
104. Zhou Q, Li M, Wang X, Li Q, Wang T, Zhu Q, et al. Immune-Related microRNAs are Abundant in Breast Milk Exosomes. Int J Biol Sci (2012) 8(1):118–23. doi: 10.7150/ijbs.8.118
105. Yanez-Mo M, Siljander PR, Andreu Z, Zavec AB, Borras FE, Buzas EI, et al. Biological Properties of Extracellular Vesicles and Their Physiological Functions. J Extracell Vesicles (2015) 4:27066. doi: 10.3402/jev.v4.27066
106. Xu JF, Yang GH, Pan XH, Zhang SJ, Zhao C, Qiu BS, et al. Altered microRNA Expression Profile in Exosomes During Osteogenic Differentiation of Human Bone Marrow-Derived Mesenchymal Stem Cells. PloS One (2014) 9(12):e114627. doi: 10.1371/journal.pone.0114627
107. Morel L, Regan M, Higashimori H, Ng SK, Esau C, Vidensky S, et al. Neuronal Exosomal miRNA-dependent Translational Regulation of Astroglial Glutamate Transporter GLT1. J Biol Chem (2013) 288(10):7105–16. doi: 10.1074/jbc.M112.410944
108. Forterre A, Jalabert A, Chikh K, Pesenti S, Euthine V, Granjon A, et al. Myotube-Derived Exosomal miRNAs Downregulate Sirtuin1 in Myoblasts During Muscle Cell Differentiation. Cell Cycle (2014) 13(1):78–89. doi: 10.4161/cc.26808
109. Sun Z, Yang S, Zhou Q, Wang G, Song J, Li Z, et al. Emerging Role of Exosome-Derived Long non-Coding RNAs in Tumor Microenvironment. Mol Cancer (2018) 17(1):82. doi: 10.1186/s12943-018-0831-z
110. Hannafon BN, Ding WQ. Functional Role of miRNAs in the Progression of Breast Ductal Carcinoma in Situ. Am J Pathol (2019) 189(5):966–74. doi: 10.1016/j.ajpath.2018.06.025
111. Zhang N, Nan A, Chen L, Li X, Jia Y, Qiu M, et al. Circular RNA circSATB2 Promotes Progression of non-Small Cell Lung Cancer Cells. Mol Cancer (2020) 19(1):101. doi: 10.1186/s12943-020-01221-6
112. Fukayama M, Kunita A, Kaneda A. Gastritis-Infection-Cancer Sequence of Epstein-Barr Virus-Associated Gastric Cancer. Adv Exp Med Biol (2018) 1045:437–57. doi: 10.1007/978-981-10-7230-7_20
113. Koppers-Lalic D, Hogenboom MM, Middeldorp JM, Pegtel DM. Virus-Modified Exosomes for Targeted RNA Delivery; a New Approach in Nanomedicine. Adv Drug Delivery Rev (2013) 65(3):348–56. doi: 10.1016/j.addr.2012.07.006
114. Feng Z, Hirai-Yuki A, McKnight KL, Lemon SM. Naked Viruses That Aren’t Always Naked: Quasi-Enveloped Agents of Acute Hepatitis. Annu Rev Virol (2014) 1(1):539–60. doi: 10.1146/annurev-virology-031413-085359
115. Wang L, Liao Y, Yang R, Yu Z, Zhang L, Zhu Z, et al. Sja-miR-71a in Schistosome Egg-Derived Extracellular Vesicles Suppresses Liver Fibrosis Caused by Schistosomiasis Via Targeting Semaphorin 4D. J Extracell Vesicles (2020) 9(1):1785738. doi: 10.1080/20013078.2020.1785738
116. Meningher T, Barsheshet Y, Ofir-Birin Y, Gold D, Brant B, Dekel E, et al. Schistosomal Extracellular Vesicle-Enclosed miRNAs Modulate Host T Helper Cell Differentiation. EMBO Rep (2020) 21(1):e47882. doi: 10.15252/embr.201947882
117. Hu W, Liu C, Bi ZY, Zhou Q, Zhang H, Li LL, et al. Comprehensive Landscape of Extracellular Vesicle-Derived RNAs in Cancer Initiation, Progression, Metastasis and Cancer Immunology. Mol Cancer (2020) 19(1):102. doi: 10.1186/s12943-020-01199-1
118. Cheng J, Meng J, Zhu L, Peng Y. Exosomal Noncoding RNAs in Glioma: Biological Functions and Potential Clinical Applications. Mol Cancer (2020) 19(1):66. doi: 10.1186/s12943-020-01189-3
119. Zeuschner P, Linxweiler J, Junker K. Non-Coding RNAs as Biomarkers in Liquid Biopsies With a Special Emphasis on Extracellular Vesicles in Urological Malignancies. Expert Rev Mol Diagn (2020) 20(2):151–67. doi: 10.1080/14737159.2019.1665998
120. Wang J, Ni J, Beretov J, Thompson J, Graham P, Li Y. Exosomal microRNAs as Liquid Biopsy Biomarkers in Prostate Cancer. Crit Rev Oncol Hematol (2020) 145:102860. doi: 10.1016/j.critrevonc.2019.102860
121. Visci G, Tolomeo D, Agostini A, Traversa D, Macchia G, Storlazzi CT. CircRNAs and Fusion-circRNAs in Cancer: New Players in an Old Game. Cell Signal (2020) 75:109747. doi: 10.1016/j.cellsig.2020.109747
122. Wang Y, Liu J, Ma J, Sun T, Zhou Q, Wang W, et al. Exosomal circRNAs: Biogenesis, Effect and Application in Human Diseases. Mol Cancer (2019) 18(1):116. doi: 10.1186/s12943-019-1041-z
123. Chen Z, Yang Y, Yamaguchi H, Hung MC, Kameoka J. Isolation of Cancer-Derived Extracellular Vesicle Subpopulations by a Size-Selective Microfluidic Platform. Biomicrofluidics (2020) 14(3):034113. doi: 10.1063/5.0008438
124. Kang YT, Hadlock T, Jolly S, Nagrath S. Extracellular Vesicles on Demand (EVOD) Chip for Screening and Quantification of Cancer-Associated Extracellular Vesicles. Biosens Bioelectron (2020) 168:112535. doi: 10.1016/j.bios.2020.112535
125. Zhou Y, Ma Z, Tayebi M, Ai Y. Submicron Particle Focusing and Exosome Sorting by Wavy Microchannel Structures Within Viscoelastic Fluids. Anal Chem (2019) 91(7):4577–84. doi: 10.1021/acs.analchem.8b05749
126. Yang XX, Sun C, Wang L, Guo XL. New Insight Into Isolation, Identification Techniques and Medical Applications of Exosomes. J Control Release (2019) 308:119–29. doi: 10.1016/j.jconrel.2019.07.021
127. Li P, Kaslan M, Lee SH, Yao J, Gao Z. Progress in Exosome Isolation Techniques. Theranostics (2017) 7(3):789–804. doi: 10.7150/thno.18133
128. Gheinani AH, Vogeli M, Baumgartner U, Vassella E, Draeger A, Burkhard FC, et al. Improved Isolation Strategies to Increase the Yield and Purity of Human Urinary Exosomes for Biomarker Discovery. Sci Rep (2018) 8(1):3945. doi: 10.1038/s41598-018-22142-x
129. Wang LL, Chen WQ, Wang YR, Zeng LP, Chen TT, Chen GY, et al. Numerous Long Single-Stranded DNAs Produced by Dual Amplification Reactions for Electrochemical Detection of Exosomal Micrornas. Biosens Bioelectron (2020) 169:112555. doi: 10.1016/j.bios.2020.112555
130. Wu X, Zhao H, Natalia A, Lim CZJ, Ho NRY, Ong CJ, et al. Exosome-Templated Nanoplasmonics for Multiparametric Molecular Profiling. Sci Adv (2020) 6(19):eaba2556. doi: 10.1126/sciadv.aba2556
131. Narita M, Nishida H, Asahina R, Nakata K, Yano H, Dickinson PJ, et al. Expression of microRNAs in Plasma and in Extracellular Vesicles Derived From Plasma for Dogs With Glioma and Dogs With Other Brain Diseases. Am J Vet Res (2020) 81(4):355–60. doi: 10.2460/ajvr.81.4.355
132. Zheng P, Zhang H, Gao H, Sun J, Li J, Zhang X, et al. Plasma Exosomal Long Noncoding Rna lnc-SLC2A12-10:1 as a Novel Diagnostic Biomarker for Gastric Cancer. Onco Targets Ther (2020) 13:4009–18. doi: 10.2147/ott.s253600
133. Lee H, Park H, Noh GJ, Lee ES. pH-responsive Hyaluronate-Anchored Extracellular Vesicles to Promote Tumor-Targeted Drug Delivery. Carbohydr Polym (2018) 202:323–33. doi: 10.1016/j.carbpol.2018.08.141
134. Sancho-Albero M, Navascués N, Mendoza G, Sebastián V, Arruebo M, Martín-Duque P, et al. Exosome Origin Determines Cell Targeting and the Transfer of Therapeutic Nanoparticles Towards Target Cells. J Nanobiotechnol (2019) 17(1):16. doi: 10.1186/s12951-018-0437-z
135. Perets N, Betzer O, Shapira R, Brenstein S, Angel A, Sadan T, et al. Golden Exosomes Selectively Target Brain Pathologies in Neurodegenerative and Neurodevelopmental Disorders. Nano Lett (2019) 19(6):3422–31. doi: 10.1021/acs.nanolett.8b04148
136. Morales-Kastresana A, Musich TA, Welsh JA, Telford W, Demberg T, Wood JCS, et al. High-Fidelity Detection and Sorting of Nanoscale Vesicles in Viral Disease and Cancer. J Extracell Vesicles (2019) 8(1):1597603. doi: 10.1080/20013078.2019.1597603
137. Kibria G, Ramos EK, Wan Y, Gius DR, Liu H. Exosomes as a Drug Delivery System in Cancer Therapy: Potential and Challenges. Mol Pharm (2018) 15(9):3625–33. doi: 10.1021/acs.molpharmaceut.8b00277
138. Yang L, Han B, Zhang Z, Wang S, Bai Y, Zhang Y, et al. Extracellular Vesicle-Mediated Delivery of Circular Rna SCMH1 Promotes Functional Recovery in Rodent and Nonhuman Primate Ischemic Stroke Models. Circulation (2020) 142(6):556–74. doi: 10.1161/circulationaha.120.045765
139. Upadhya R, Madhu LN, Attaluri S, Gitaí DLG, Pinson MR, Kodali M, et al. Extracellular Vesicles From Human iPSC-derived Neural Stem Cells: miRNA and Protein Signatures, and Anti-Inflammatory and Neurogenic Properties. J Extracell Vesicles (2020) 9(1):1809064. doi: 10.1080/20013078.2020.1809064
140. Fang SB, Zhang HY, Wang C, He BX, Liu XQ, Meng XC, et al. Small Extracellular Vesicles Derived From Human Mesenchymal Stromal Cells Prevent Group 2 Innate Lymphoid Cell-Dominant Allergic Airway Inflammation Through Delivery of Mir-146a-5p. J Extracell Vesicles (2020) 9(1):1723260. doi: 10.1080/20013078.2020.1723260
141. Dou G, Tian R, Liu X, Yuan P, Ye Q, Liu J, et al. Chimeric Apoptotic Bodies Functionalized With Natural Membrane and Modular Delivery System for Inflammation Modulation. Sci Adv (2020) 6(30):eaba2987. doi: 10.1126/sciadv.aba2987
142. Xie M, Tao L, Zhang Z, Wang W. Mesenchymal Stem Cells Mediated Drug Delivery in Tumor-Targeted Therapy. Curr Drug Delivery (2020). doi: 10.2174/1567201817999200819140912
143. Lang FM, Hossain A, Gumin J, Momin EN, Shimizu Y, Ledbetter D, et al. Mesenchymal Stem Cells as Natural Biofactories for Exosomes Carrying miR-124a in the Treatment of Gliomas. Neuro Oncol (2018) 20(3):380–90. doi: 10.1093/neuonc/nox152
144. Liang G, Kan S, Zhu Y, Feng S, Feng W, Gao S. Engineered Exosome-Mediated Delivery of Functionally Active miR-26a and its Enhanced Suppression Effect in HepG2 Cells. Int J Nanomed (2018) 13:585–99. doi: 10.2147/ijn.s154458
145. Zhang H, Freitas D, Kim HS, Fabijanic K, Li Z, Chen H, et al. Identification of Distinct Nanoparticles and Subsets of Extracellular Vesicles by Asymmetric Flow Field-Flow Fractionation. Nat Cell Biol (2018) 20(3):332–43. doi: 10.1038/s41556-018-0040-4
146. Jeppesen DK, Fenix AM, Franklin JL, Higginbotham JN, Zhang Q, Zimmerman LJ, et al. Reassessment of Exosome Composition. Cell (2019) 177(2):428–445.e18. doi: 10.1016/j.cell.2019.02.029
147. Zhang Q, Higginbotham JN, Jeppesen DK, Yang YP, Li W, McKinley ET, et al. Transfer of Functional Cargo in Exomeres. Cell Rep (2019) 27(3):940–954.e6. doi: 10.1016/j.celrep.2019.01.009
148. Anand S, Samuel M, Mathivanan S. Exomeres: A New Member of Extracellular Vesicles Family. Subcell Biochem (2021) 97:89–97. doi: 10.1007/978-3-030-67171-6_5
Keywords: exosome, extracellular vesicles, non-coding RNAs, sorting mechanism, cell-cell communication, ncRNAs
Citation: Qiu Y, Li P, Zhang Z and Wu M (2021) Insights Into Exosomal Non-Coding RNAs Sorting Mechanism and Clinical Application. Front. Oncol. 11:664904. doi: 10.3389/fonc.2021.664904
Received: 06 February 2021; Accepted: 07 April 2021;
Published: 27 April 2021.
Edited by:
Marco Tafani, Sapienza University of Rome, ItalyReviewed by:
Muhammad Nawaz, University of Gothenburg, SwedenWeifeng He, Army Medical University, China
Reza Rahbarghazi, Tabriz University of Medical Sciences, Iran
Copyright © 2021 Qiu, Li, Zhang and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Minghua Wu, d3VtaW5naHVhNTU0QGFsaXl1bi5jb20=; Zuping Zhang, emhhbmd6cDc0QDEyNi5jb20=
 Yi Qiu
Yi Qiu Peiyao Li
Peiyao Li Zuping Zhang
Zuping Zhang Minghua Wu
Minghua Wu