- 1Department of Medical Oncology, Affiliated Jinling Hospital, Medical School of Nanjing University, Nanjing, China
- 2Affiliated Jingling Hospital Research Institution of General Surgery, School of Medicine, Nanjing University, Nanjing, China
Hepatocellular carcinoma is the fifth-ranked cancer worldwide with a relatively low five-year survival rate. Long non-coding RNAs are a group of RNAs with remarkable aberrant expression which could act on multiple bioprocesses and ultimately impact upon tumor proliferation, invasion, migration, metastasis, apoptosis, and therapy resistance in cancer cells including hepatocellular carcinoma cells. In recent years, long non-coding RNAs have been reported to be indispensable targets in clinical target therapy to stop the growth of cancer and prolong the lifespan of patients with hepatocellular carcinoma. In this review, we enumerate the signaling pathways and life activities affected by long non-coding RNAs in hepatocellular carcinoma cells to illustrate the role of long non-coding RNAs in the development and therapy resistance of hepatocellular carcinoma.
Introduction
Liver cancers comprise diverse, histologically distinct primary hepatic neoplasms, which include hepatocellular carcinoma (HCC), intrahepatic bile duct carcinoma (holangio carcinoma), hepatoblastoma, bile duct cystadenocarcinoma, hemangiosarcoma, and epitheliod hemangioendothelioma (1). Among them, HCC is one of the most prevalent liver cancers worldwide (2, 3). The main risk factors for HCC vary from region to region. Chronic hepatitis B virus (HBV) infection and AFB1 exposure are major risk factors in most high-incidence regions like Asia and Africa. In contrast, HCV infection, excessive drinking, and diabetes/obesity/metabolic syndrome play a more important role in low-incidence areas, with the primary risk factor being HCV infection. In addition, some hereditary metabolic disorders such as hemochromatosis, α1 anti-trypsin deficiency, tyrosinemia, and several porphyrias also increase the risk (4–8). Till now, lots of treatments for HCC such as resection, ablation, transplantation, chemoembolization, and targeted drugs like sorafenib and lenvatinib are put into clinical practice (9–11). Despite all those treatments, the current five-year survival rate provided by the National Cancer Institute is still not satisfactory, so we still devote a lot of effort to create more effective methods (12). LncRNAs which are aberrant expressions are proved to be associated with tumorigenesis and poor prognosis of HCC. Given that, lncRNAs are promising potential biomarkers or targets of HCC.
LncRNAs belong to the ncRNA class, which consists of small interfering RNAs (siRNAs), microRNAs (miRNAs), PIWI-interacting RNAs (piRNAs), promoter-associated RNAs (PARNAs), small nucleolar RNAs (snoRNAs), and other recently described classes such as X-inactivation RNAs (xiRNAs), sno-derived RNAs (sdRNAs), microRNA-offset RNAs (moRNAs), tRNA-derived RNAs and MSY2-associated RNAs (MSY-RNAs) (13). LncRNAs are approximately 200 bp to 100 kb in length (14). Although most lncRNAs are not directly involved in gene coding and protein synthesis, they are indispensable in genomic imprinting, chromatin modification, post-transcriptional regulation, cleavage and modification (15–17). We screened lncRNAs in human HCC cells by using EVLncRNAs (a database) (Table 1) and selected some well-studied lncRNAs to describe in detail. Some well-known regulatory pathways of lncRNAs in HCC have also been demonstrated (Table 2).
FUNCTIONS of lncRNAs in HCC
LncRNAs and miRNAs
LncRNAs Acting as miRNA Sponges or miRNA Inhibitors
MALAT1 (LncRNA metastasis-associated lung adenocarcinoma transcript 1) binds to and inhibits miR-143-3p expression to decrease ZEB1 (zinc finger E-box binding home box 1) (21). Also, MALAT1 acts as molecular sponge of miR-146b-5p and miR-204 to facilitate HCC cells (22, 23). The UCA1 gene belongs to the HERV-H family. It contains the gag region, the protease–polymerase region, but no envelope region. In three regions, the UCA1 full-length cDNA consists of an unusual number of stop codons that transcribe non-coding RNA (45, 46). UCA1 facilitates FGFR1–ERK pathway by inhibiting expression of miR-216b (76). CCAT1 (The long non-coding RNA Colon Cancer Associated Transcript 1) contains two predicted let-7 targeting sites. It is reported that let-7 decreases tumor proliferation and induces apoptosis. Let-7 binds to CCAT1 but does not induce degradation of CCAT1. In other words, CCAT1 is physically related to let-7 and serves as a miRNA sponge for let-7. At the same time, CCAT1 regulates HMGA2 and c-Myc by competitively binding to let-7 (49). HOTAIR (Hox transcript antisense intergenic RNA), which is overexpressed in HCC tissues, enhances EMT by inhibiting miR-23b-3p, leading to malignant tumors of HCC and increased tumor metastasis (28–30). TUG1 is proved to act as a molecular sponge of miR-144. It interacts with miR-144 to promote proliferation and migration of HCC cells by activating the JAK2/STAT3 pathway. After knocking out TUG1 in tumor cells, the JAK2/STAT3 pathway is inactivated, and miR-144 is up-regulated to inhibit HCC tumor growth in vivo. To sum up, the interaction of TUG1 and miR-144 promotes proliferation, migration, and tumorigenesis by activating the JAK2/STAT3 pathway in HCC (54). PTENP1 regulates the PTEN/Akt pathway through interaction with miR-193a-3p (65). HULC promotes HCC via depleting miR-9-mediated RXRA signaling pathway (40). Overexpressed PTENP1 induces miR-17, miR-19b, and miR-20a, targeting PTEN, PHLPP (negative AKT regulatory factor) and autophagy genes such as ULK1, ATG7, and p62 (66).
LncRNAs Acting as Competing Endogenous RNAs of miRNAs
LncRNA-ATB (activated by TGF-β) which is up-regulated in HCC is activated by TGF-β and up-regulates ZEB1 and ZEB2 by competitive binding to the miR-200 family (77–79). ZEB1 gene encodes a zinc finger transcription factor that plays an important role in normal embryonic development which induce EMT (24). EMT converts cancerous epithelial cells into mesenchymal-like cells, confers migration and invasion properties, enables primary tumor cells to move, settles distant organs, and forms secondary tumors (18, 19, 80). As a result, the silence of ZEB1 hinders the metastasis and invasion of HCC through EMT inhibition. TUG1 promotes HCC development by competing with miR-132 to combine sonic hedgehog (Shh) as well as Kruppel-like factor 2 (KLF2) to combine polycomb repressive complex 2 (PRC2) (58, 59). Overexpression of MEG3 competitively inhibits miR-664, thereby releasing the inhibitory effect of miR-664 on ADH4 and promoting the expression of ADH4 (81).
MiRNAs Acting as lncRNA Inhibitors
HOTTIP (HOXA transcript at the distal tip) is mainly present in the nucleus, binds to AGO2 in the nucleus, and is regulated by some miRNAs. The level of HOTTIP is significantly decreased in cells when miR-192, miR-204, and miR-125b are overexpressed (82). MiR-192 and miR-204 inhibit the activity of HOTTIP through their target molecules such as DHFR, ZEB2, BCL2, and so on, thereby achieving the purpose of inhibiting the activity of HCC cells (55).
LncRNAs Acting on Proteins
HOTAIR up-regulates ATG3 and ATG7 to activate autophagy and promote HCC cell proliferation (31). In addition, OGFr (Opioid Growth Factor Receptor) which prominently impedes tumor growth is depleted in HCC because of HOTAIR (32). Besides, CCND1, RBM38, P14, P16, GLUT1, and mTOR signaling also participate in HOTAIR signaling to promote HCC progress (33–36). Meanwhile, HULC activates protective autophagy through Sirt1 (silent information regulator 1 protein)–USP22 (ubiquitin-specific peptidase 22) pathway and increases HCC proliferation through COX2–USP22 pathway. HULC reduces Sirt1 and COX2 degradation by elevating the expression of USP22 (83). TUC338 (Transcribed ncRNA encoding uc.338) post-transcriptionally regulates plasminogen activator inhibitor-1 RNA binding protein (PAI-RBP1), occupying a genomic region rich in unique or known motifs homologous to the tumor suppressor Pax6 and p53 (48). C-Myc directly binds to the promoter of CCAT1 and promotes HCC development (50). Activation of TCF/LEF by β-catenin is one of the most common molecular changes in HCC cells as well as a general regulator of stem cell self-renewal, tumorigenicity and tumor progression. DANCR (Differentiation antagonizing non-protein coding RNA) regulates the stability of above molecular changes to affect tumor proliferation (51, 84). Linc00152 (Long intergenic non-protein coding RNA 00152) prevents E-cadherin expression by interacting with EZH2 and promotes EMT in HCC cells (61). MEG3 (Maternally expressed gene 3) promotes p53 expression and inhibits MDM2 expression, and increasing p53 also inhibits the expression of MDM2. So that the ubiquitination of P53 by MDM2 is prevented, promoting apoptosis and inhibiting tumor proliferation (62, 63, 85, 86).
Interactions Within lncRNAs
It is reported that MALAT1/HULC is positively correlated with the expression of TRF2 in human hepatocellular carcinoma tissues. MALAT1 and TRF2 are highly expressed in HCC tissues and are positively correlated. The increased TRF2 binds to HULC and MALAT1 to form a complex, which is loaded into the telomere region of the chromosome. Therefore, the telomeres are greatly extended, leading to the rapid growth of HCC stem cells (25). Overexpression of HULC (highly up-regulated in liver cancer) prevents PTEN and miR15a, which leads to high expression of LC3I and LC3II (autophagy marker) and more autophagy in hepatoma cells. HULC inhibits PTEN by autophagy and P62-mediated ubiquitin–protein system and finally activates the AKT–PI3K–mTOR pathway to promote cell growth, colony-forming ability, and cell growth in vivo (41).
Roles of lncRNAs in HCC
LncRNAs as Tumor-Suppressive Genes in HCC
MEG3 expression is down-regulated in both HCC cell lines and tissues. Re-expression of MEG3 in HCC cells significantly reduces anchorage-dependent and independent cell growth and induces apoptosis (64, 87). Also, adenosine can resist HCC through up-regulating the expression of MEG3 (88).
Phosphatase and tensin homolog pseudogene 1 (PTENP1) is a pseudogene of the tumor suppressor gene phosphatase and tensin homolog deleted on chromosome ten (PTEN) (67, 68). It is shown that PTENP1 and PTEN are down-regulated in HCC cells. Over-expression of PTENP1 and PTEN in HCC cells can inhibit the oncogenic PI3K/AKT pathway, inhibit cell proliferation, migration, invasion, and induces autophagy, apoptosis, and inhibition of angiogenesis.
LncRNAs as Oncogenes in HCC
lncRNA-ATB promotes HCC colonization by inducing autocrine induction of IL-11 and activating STAT3 signaling (20). Plasma Linc00152 can be used as a potential non-invasive biomarker to predict the diagnosis of HCC (42). MALAT1 is up-regulated in HCC and plays an oncogenic role through activating the Wnt pathway and inducting oncogenic splicing factor SRSF1 to activate the mTOR pathway and resist apoptosis (26). HOTAIR with a length of 2,158 bp is remarkably associated with poor prognosis of HCC. It affects the histone H3 tri-methylated at lysine 27 (H3K27me3) by recruiting poly bulking inhibitor complex 2 (PRC2 complex) at the 5′ end, so that LSD1 (lysine-specific demethylase 1)/CoREST (RE1-silencing transcription factor co-repressor)/REST (RE1-silent transcription factor) complex promotes histone H3 Lysine 4 demethylation, eventually leading to gene silencing (37–39). UCA1 (urothelial cancer associated 1) is overexpressed in HCC, making it a potential biomarker to detect progression and prognosis in patients with HCC (89). Decreasing the expression level of UCA1 inhibits the proliferation, migration, and invasion of HCC cells and induces apoptosis. Moreover, bioinformatics analysis indicates that UCA1 may significantly disrupt the hippo signal pathway (47). TUC338 is a super-conservative lncRNA that contributes to the growth of transformed cells in HCC (HCC). TUC338 functions in a manner similar to transcription factors to regulate cell proliferation and transform cell growth in HCC. Overexpression of CCAT1 in hepatoma cells promotes proliferation, invasion, and metastasis of tumor cells (50). As a newly discovered cancer-associated lncRNA, HOTTIP is located at the 5′ end of the HOXA gene. Overexpression of HOTTIP could promote cell proliferation, migration, and invasion of HCC cells (54, 56, 90–92). Linc00152 is up-regulated in the human HCC cell line. Overexpressed Linc00152 in HCC cells increased cell proliferation and invasion. Knocking out Linc00152 inhibits the mTOR signaling pathway. The underlying mechanism is that expression of Linc00152 increases EpCAM levels, leading to activation of the mTOR signaling pathway and causing proliferation of HCC (49). DANCR positively regulates proliferation in cells by regulating miR-634, miR-496 miR-33a-5p, CDKN1A (cell cycle inhibitor p21), etc, indicating that it may be a carcinogenic lncRNA, and plays potential roles as an adenocarcinoma (ADC) biomarker and therapeutic target (52, 53). The combination of sorafenib-induced enhanced tumor growth inhibition and overexpression of RASAL1 in tumor xenografts suggests that the TUC338/RASAL1 axis may be a potential therapeutic target for current HCC treatment (93).
LncRNAs and Therapeutic Sensitivity in HCC
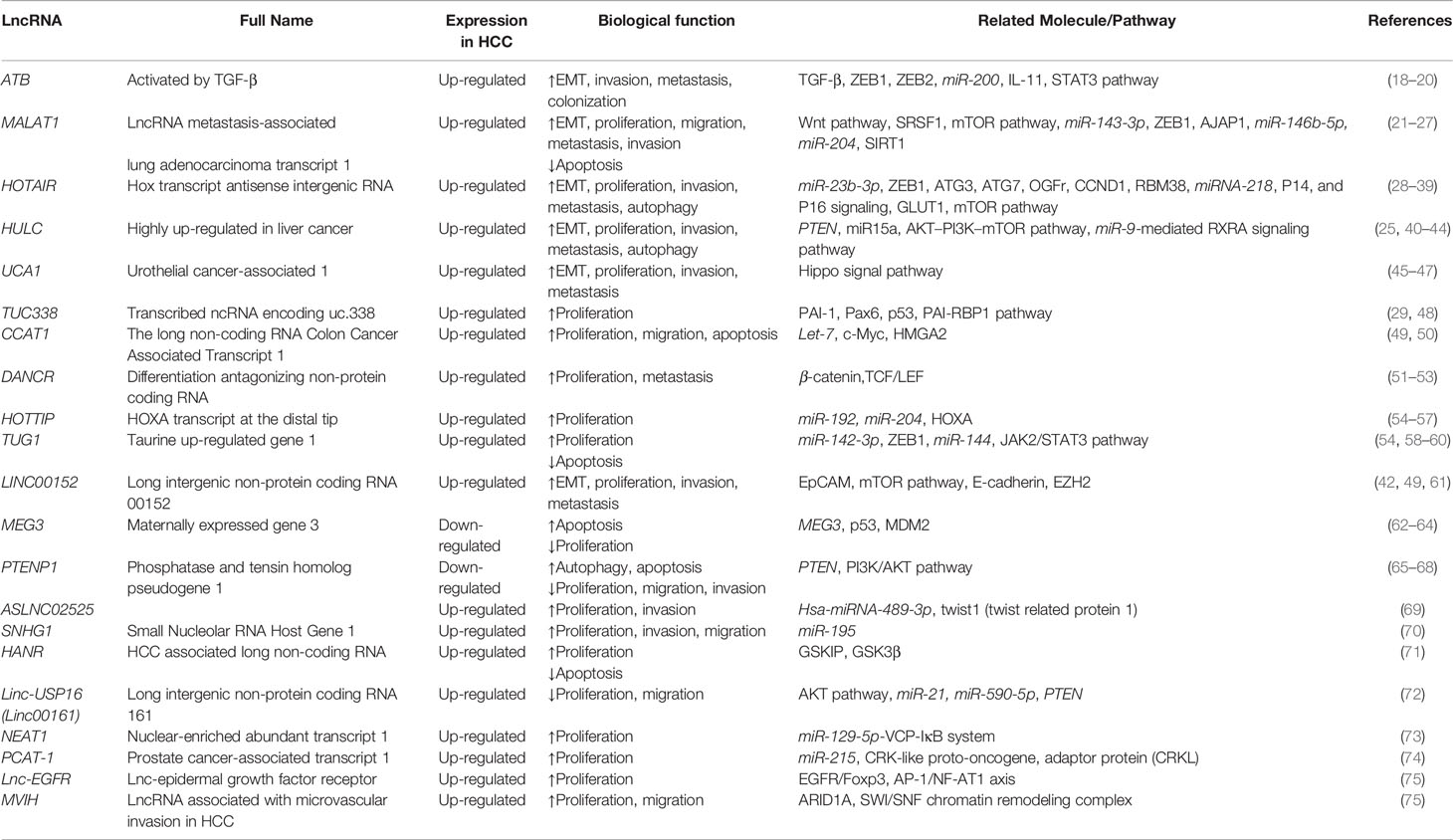
LncRNAs not only influence HCC proliferation, invasion, and migration through specific cell signaling pathways and molecules, but also induce HCC therapy resistance, which covers chemotherapy resistance and radiotherapy resistance. It has been found that lncRNA ROR reduces the sensitivity of HCC to radiotherapy (94). For chemotherapy, the most commonly used chemotherapy drugs for HCC include sorafenib, oxaliplatin, 5-fluorouracil, cisplatin, etc (95). Figure 1 is a schematic diagram of the relationship between lncRNAs and chemotherapeutic resistance.
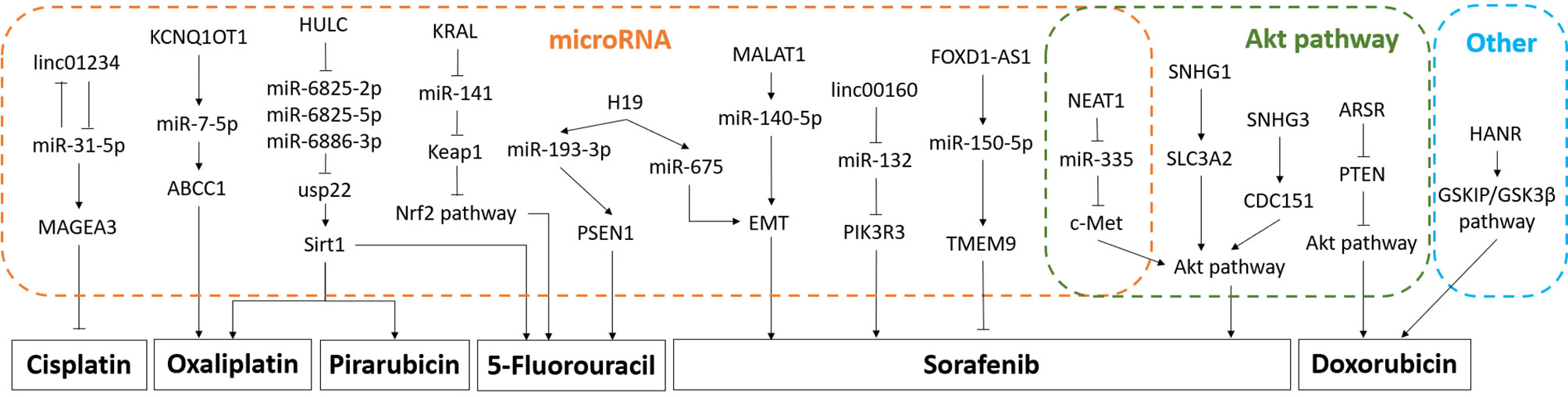
Figure 1 Diagram of the mechanism of lncRNA mediating chemotherapy resistance. Part of the mechanism is related to microRNAs, and part is related to Akt pathway. The promotion arrow in the panel refers to the promotion of drug resistance to the corresponding chemotherapeutic drug, and the inhibition arrow refers to the increase of the sensitivity to the chemotherapy drug.
Sorafenib
SNHG1, SNHG3, and SNHG16 in the small nucleolar RNA host gene (SNHG) family are related to sorafenib resistance in HCC cells (96–98). SNHG1 serves miR-21 as a mediator, as well as SNHG3 to produce positive feedback on the downstream Akt signaling pathway, and ultimately induces sorafenib resistance (99). Coincidentally, nuclear-enriched abundant transcript 1 (NEAT1) also acts on the downstream Akt signaling pathway through the miR-335/c-Met axis to induce sorafenib resistance (100). Other lncRNAs related to sorafenib resistance in HCC cells include linc00160, FOXD2-AS1, MALAT1, H19, ROR, etc (101). Linc00160 promotes phosphoinositide-3-kinase regulatory subunit 3 (PIK3R3) to induce sorafenib resistance by inhibiting miR-132 (102). FOXD2-AS1 acts as a competitive endogenous RNA of miR-150-5p to increase sensitivity to drugs (103). MALAT1 and H19 have similar mechanisms when inducing HCC resistance, which both promote EMT through intermediary molecules to induce resistance (104–106).
Oxaliplatin
The lncRNAs associated with oxaliplatin resistance contain KCNQ1OT1, HULC, and NR2F1-AS1 (107). Both KCNQ1OT1 and HULC act on resistance-related molecules through microRNA, which are miR-7-5p, miR-6825-2p, miR-6825-5p, and miR-6886-3p (83, 108).
Doxorubicin
The lncRNAs associated with oxaliplatin resistance contain HCC associated long non-coding RNA (HANR) and ARSR. HANR triggers GSKIP/GSK3β pathway (71). ARSR decreases the negative molecule PTEN of Akt pathway (109).
5-Fluorouracil
The above pathways of HULC also induce 5-fluorouracil and pirarubicin resistance (83). While H19 promotes presenilin-1 (PSEN1) through miR-193-3p, which distinguishes from the above mentioned, to achieve 5-fluorouracil resistance (110). lncRNA KRAL mediates 5-fluorouracil resistance in HCC by acting as ceRNA against miR-141 (111).
Cisplatin
Linc01234/miR-31-5p/melanoma-associated antigen A3 (MAGEA3) axis prompts cisplatin resistance in HCC cells when linc01234 is overexpressed (112).
Discussion
LncRNA-related clinical trials have not been conducted on hepatocellular carcinoma, but a clinical trial from Wuhan Union Hospital on lncRNA as a potential target for lung cancer diagnosis is underway. This clinical trial is mainly based on the identification of early lung-cancer-specific exosomal lncRNA biomarkers to improve the diagnosis rate of early lung cancer (113). Also, a clinical trial which is related to HOTAIR and thyroid cancer is being processed (114).
In this review, we discussed a variety of lncRNAs that are proved to involve in HCC (Figures 2–4). In the past few years, a series of studies have shown the essential role of lncRNAs on cell proliferation, invasion, migration, and therapy resistance through diverse signaling pathways and molecules. Based on current studies, lncRNAs are expected to be a marker for tumor diagnosis, prognosis, and expected therapeutic effects, while lncRNA-targeted drugs still have a long way to go. Especially, considering that lncRNAs are related to the sensitivity of HCC radiotherapy and chemotherapy, lncRNAs can be used as a molecular marker to predict the clinical treatment effect of therapy treatment, and it can also be used as a target in conjunction with chemotherapy or radiotherapy to increase treatment sensitivity. Our ideal expectation for lncRNA-targeted drugs is that lncRNA-targeted drugs will serve as independent targeted drugs to treat HCC, or they will be used as adjuvant drugs to increase the efficacy of existing chemotherapeutic drugs. First, given that more than a single lncRNA is confirmed to function in HCC, to make sure which type plays a leading role in development of HCC is fatal for further research. Not only in HCC, in tumors of various tissue sources, the tissue specificity of lncRNAs is also indispensable. If future research studies prove that it is unfeasible to aim at a single lncRNA to achieve better effect, perhaps using drug that has a targeted effect on several major lncRNAs with a certain commonality based on clinical research and individual differences in patients, or employ these drugs as adjuvants can be the solutions. Also, existing studies have shown that almost each lncRNA found is involved in a variety of cell signaling pathways, but these studies are far from sufficient. Moreover, the functions of lncRNAs remained undetected, which is also a burning question. Finally, due to the structural particularity of lncRNA molecules and the characteristics of being easily degraded, how to target lncRNA molecules is also an urgent problem to be solved. With further studies, the clinical transformation of lncRNA is bound to be more mature, bringing more effective treatments to patients suffering from HCC, and contributing to the fight against cancer in humans.
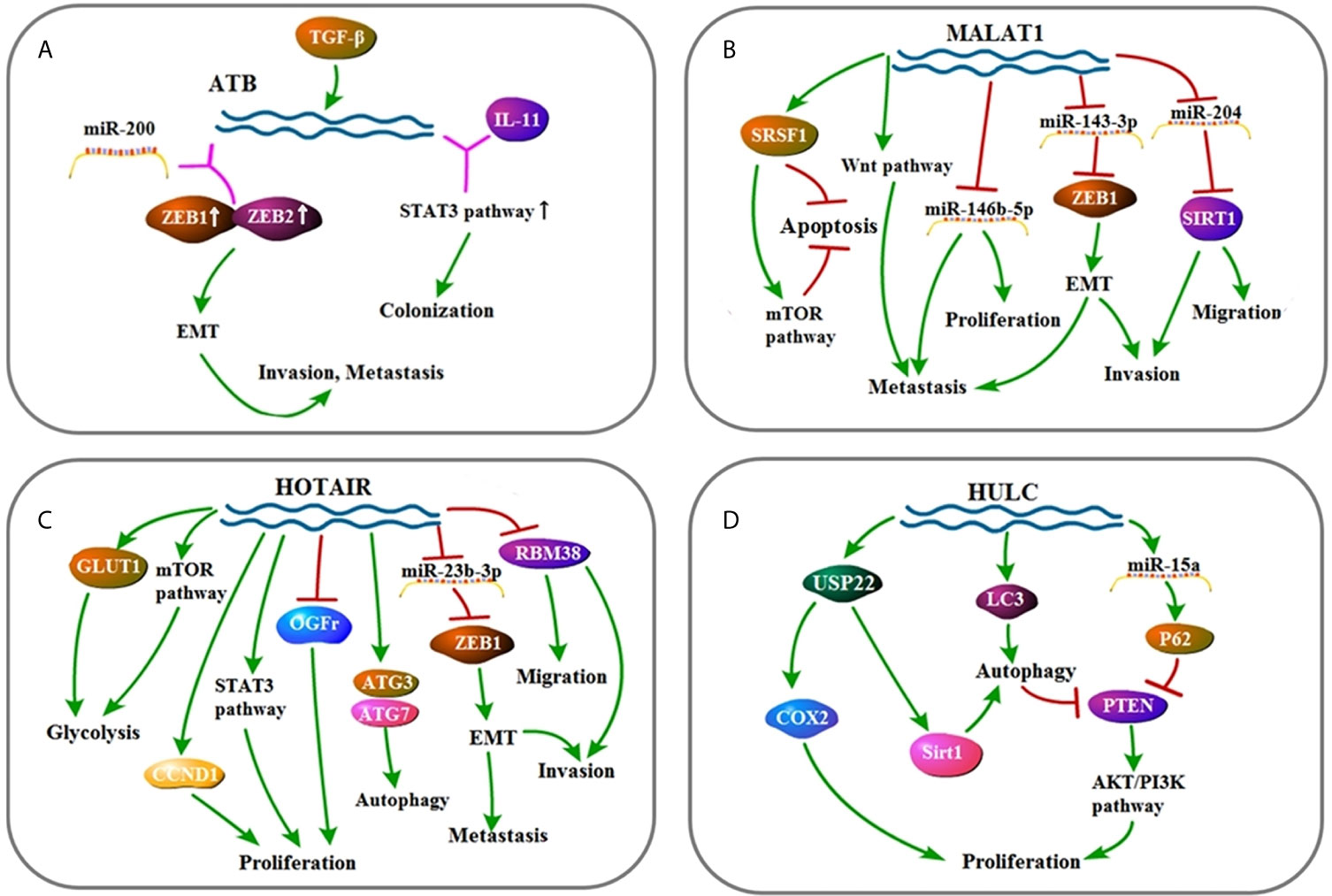
Figure 2 (A) Signaling pathway of ATB: ATB affects cell invasion, metastasis, and colonization by up-regulating EMT and STAT3 pathway. (B) Signaling pathway of MALAT1: MALAT1 affects cell invasion, metastasis, proliferation, migration, and apoptosis mainly through up-regulating EMT, Wnt pathway, STAT3 pathway, and mTOR pathway. (C) Signaling pathway of HOTAIR: HOTAIR affects cell invasion, migration, autophagy, proliferation, metastasis, and glycolysis by up-regulating EMT, mTOR pathway, and STAT3 pathway. (D) Signaling pathway of HULC: HULC affects cell proliferation and autophagy by up-regulating AKT/PI3K pathway.
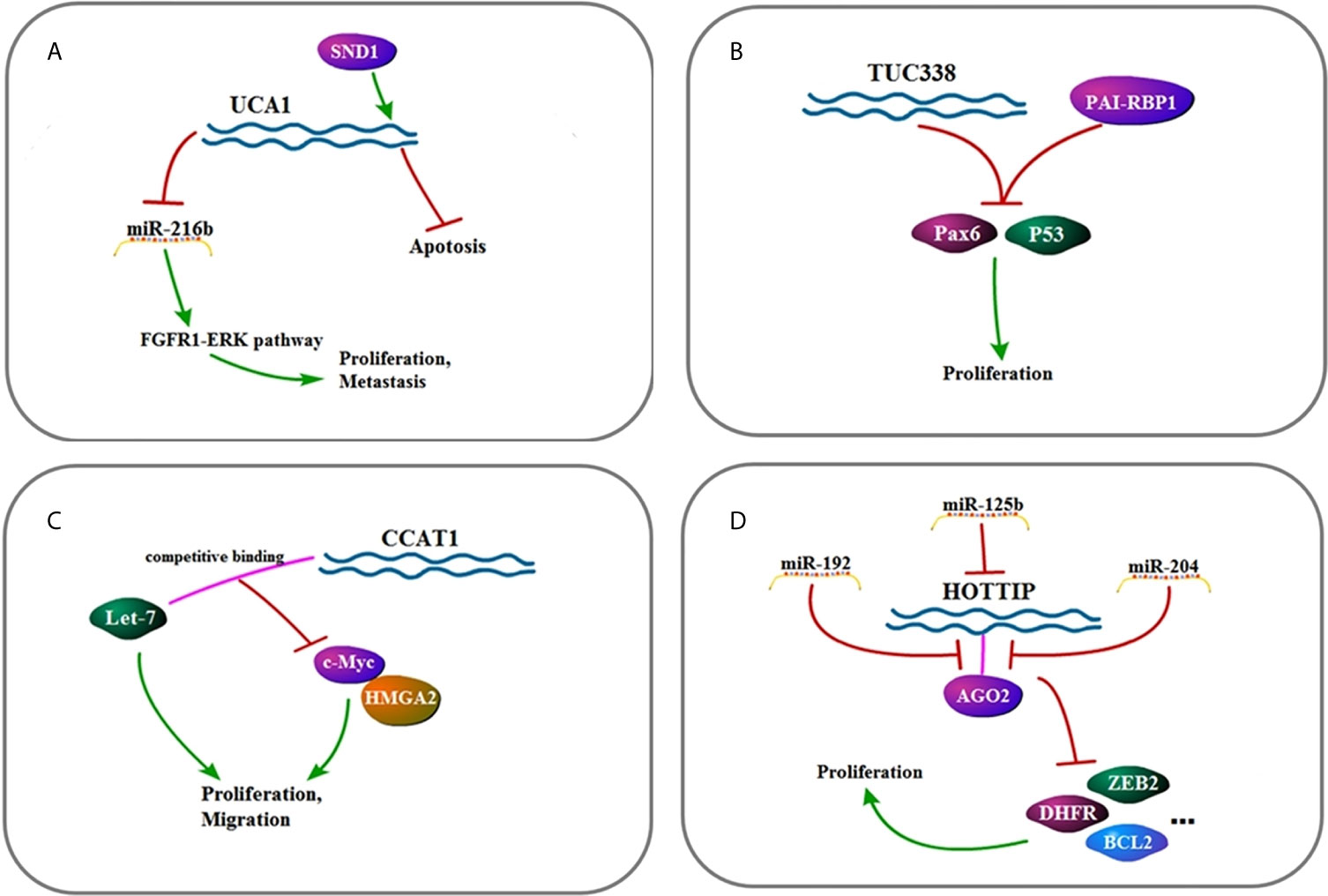
Figure 3 (A) Signaling pathway of UCA1: UCA1 affects cell metastasis, proliferation, and apoptosis by up-regulating FGFR1–ERK pathway. (B) Signaling pathway of TUC338: TUC338 affects cell proliferation by down-regulating Pax6 and P53. (C) Signaling pathway of CCAT1: CCAT1 affects cell proliferation and migration by regulating let-7, c-Myc, and HMGA2. (D) Signaling pathway of HOTTIP: HOTTIP affects cell proliferation by down-regulating AGO2.
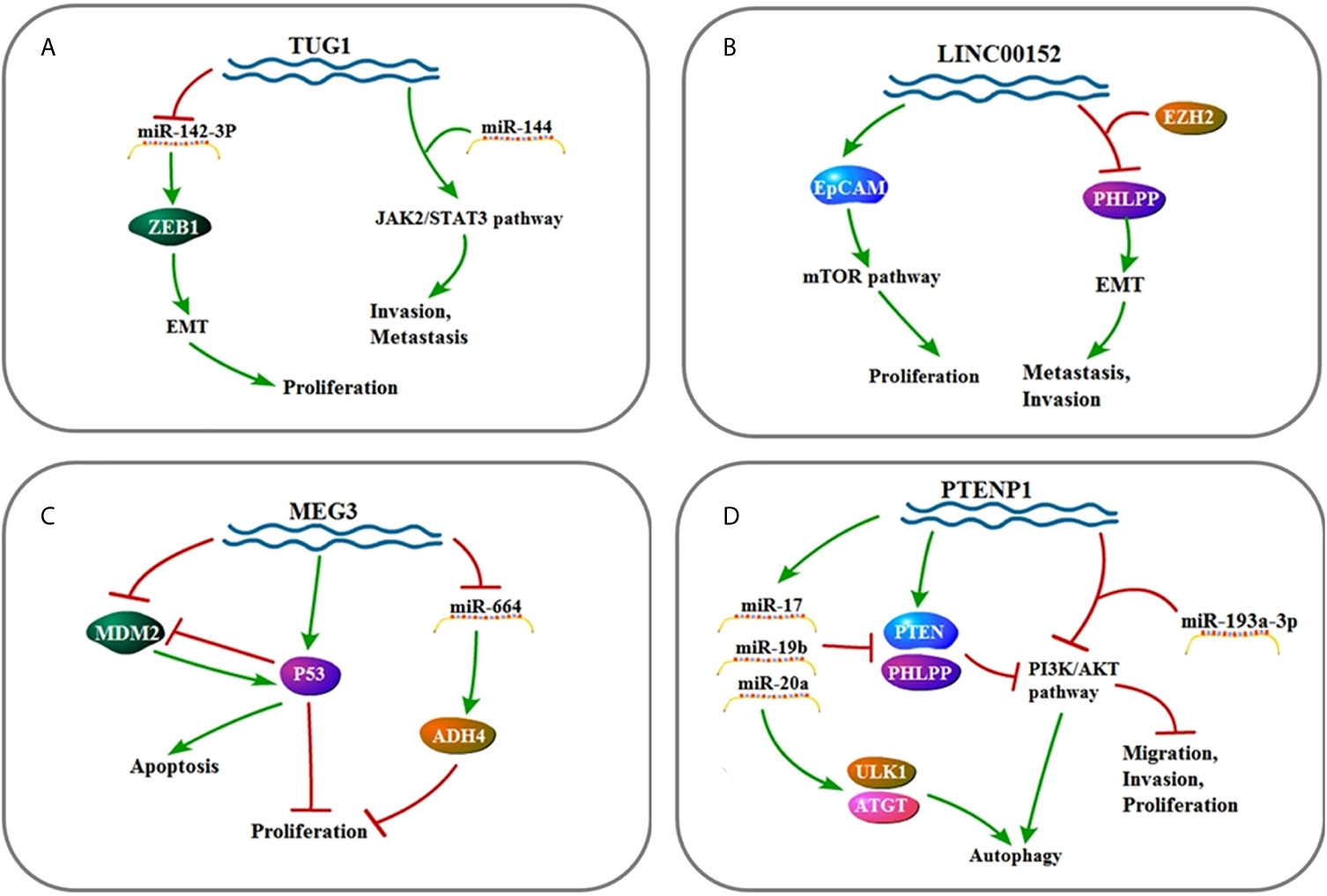
Figure 4 (A) Signaling pathway of TUG1: TUG1 affects cell invasion, metastasis, and proliferation by up-regulating EMT and JAK2/STAT3 pathway. (B) Signaling pathway of LINC00152: LINC00152 affects cell invasion, metastasis, and proliferation by up-regulating EMT and mTOR pathway. (C) Signaling pathway of MEG3: MEG3 affects cell proliferation and apoptosis by up-regulating P53 and ADH4. (D) Signaling pathway of PTENP1: PTENP1 affects cell invasion, proliferation, migration, and autophagy by up-regulating miR-17, miR-19b, and miR-20a as well as down-regulating the PI3K/AKT pathway.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
Author Contributions
XL and XX wrote the review article. RW reviewed the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This project was supported by grants from the National Natural Science Foundation of China (81472266 and 81772995) and the Excellent Youth Foundation of Jiangsu Province (BK20140032).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Farazi PA, DePinho RA. Hepatocellular Carcinoma Pathogenesis: From Genes to Environment. Nat Rev Cancer (2006) 6(9):674–87. doi: 10.1038/nrc1934
2. Smith RA, Andrews KS, Brooks D, Fedewa SA, Manassaram-Baptiste D, Saslow D, et al. Cancer Screening in the United States, 2018: A Review of Current American Cancer Society Guidelines and Current Issues in Cancer Screening. CA: A Cancer J Clin (2018) 68(4):297–316. doi: 10.3322/caac.21446
3. Au JS, Frenette CT. Management of Hepatocellular Carcinoma: Current Status and Future Directions. Gut Liver (2015) 9(4):437–48. doi: 10.5009/gnl15022
4. McGlynn KA, Pettrick J, London WT. Global Epidemiology of Hepatocellular Carcinoma: An Emphasis on Demographic and Regional Variability. Clinics Liver Dis (2015) 19(2):223–38. doi: 10.1016/j.cld.2015.01.001
5. Wu EM, Hernandez BY, Wong LL. Hepatocellular Carcinoma in Micronesians, a Growing Pacific Islander Population in the U.S. Open J Gastroenterol (2018) 08(06):223–33. doi: 10.4236/ojgas.2018.86025
6. Dutta R, Mahato RI. Recent Advances in Hepatocellular Carcinoma Therapy. Pharmacol Ther (2017) 173:106–17. doi: 10.1016/j.pharmthera.2017.02.010
7. Lee H, Yoon JH, Kim H, Yi N-J, Hong SK, Yoon KC, et al. False Positive Diagnosis of Hepatocellular Carcinoma in Liver Resection Patients. J Korean Med Sci (2017) 32(2):315. doi: 10.3346/jkms.2017.32.2.315
8. Coppola N, Alessio L, Onorato L, Sagnelli C, Macera M, Sagnelli E. Epidemiology and Management of Hepatitis C Virus Infections in Immigrant Populations. Infect Dis Poverty (2019)8(1):17. doi: 10.1186/s40249-019-0528-6
9. Hartke J, Johnson M, Ghabril M. The Diagnosis and Treatment of Hepatocellular Carcinoma. Semin Diagn Pathol (2017) 34(2):153–9. doi: 10.1053/j.semdp.2016.12.011
10. Forner A, Reig M, Bruix J. Hepatocellular Carcinoma. Lancet (2018) 391(10127):1301–14. doi: 10.1016/s0140-6736(18)30010-2
11. Benson AB, D’Angelica MI, Abbott DE, Abrams TA, Alberts SR, Anaya DA, et al. Guidelines Insights: Hepatobiliary Cancers, Version 2.2019. J Natl Compr Canc Netw (2019) 17(4):302–10. doi: 10.6004/jnccn.2019.0019
12. Budhu A, Chaisaingmongkol J, Dang H, Rabibhadana S, Pupacdi B, Kwon SM, et al. Common Molecular Subtypes Among Asian Hepatocellular Carcinoma and Cholangiocarcinoma. Cancer Cell (2017) 32(1):57–70. doi: 10.1016/j.ccell.2017.05.009
13. Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS. Non-Coding RNAs: Regulators of Disease. J Pathol (2010) 220(2):126–39. doi: 10.1002/path.2638
14. Xiaodong Song GC, Jing L, Lin S, Wang X, Zhang J, Wang M, et al. Analysing the Relationship Between lncRNA and Protein-Coding Gene and the Role of lncRNA as ceRNA in Pulmonary Fibrosis. Cell Mol Med (2014) 18:991–1103. doi: 10.1111/jcmm.12243
15. Gruhl F, Cheetham SW, Mattick JS, Dinger ME. Long Noncoding RNAs and the Genetics of Cancer. Br J Cancer (2013) 108:2419–25. doi: 10.1038/bjc.2013.233
16. Yuan W, Sun Y, Liu L, Zhou B, Wang S, Gu D. Circulating LncRNAs Serve as Diagnostic Markers for Hepatocellular Carcinoma. Cell Physiol Biochem (2017) 44:125–32. doi: 10.1159/000484589
17. Zaimy MA SN, Mohammadi A, Pourghadamyari H, Izadi P, Sarli A, Moghaddam LK, et al. New Methods in the Diagnosis of Cancer and Gene Therapy of Cancer Based on Nanoparticle. Cancer Gene Ther (2017) 24(6):233–43. doi: 10.1038/cgt.2017.16
18. Se Young Jang GK, Park SY, Lee YR, Kwon SH, Kim HS, Yoon JS, et al. Clinical Significance of lncRNA-ATB Expression in Human Hepatocellular Carcinoma. Oncotarget (2017) 8(45):78588–97. doi: 10.18632/oncotarget.21094
19. Iguchi T, Uchi R, Nambara S, Saito T, Komatsu H, Hirata H, et al. A Long Noncoding RNA, LncRNA-ATB, Is Involved in the Progression and Prognosis of Colorectal Cancer. Anticancer Res (2015) 35(3):1385–8.
20. Yuan J-h, Yang F, Wang F, Ma J-z, Guo Y-J, Tao Q-F, et al. A Long Noncoding RNA Activated by TGF-β Promotes the Invasion-Metastasis Cascade in Hepatocellular Carcinoma. Cancer Cell (2014) 25(5):666–81. doi: 10.1016/j.ccr.2014.03.010
21. Chen L, Yao H, Wang K, Liu X. Long Non-Coding RNA MALAT1 Regulates ZEB1 Expression by Sponging miR-143-3p and Promotes Hepatocellular Carcinoma Progression. J Cell Biochem (2017) 118(12):4836–43. doi: 10.1002/jcb.26158
22. Hou Z, Xu X, Zhou L, Fu X, Tao S, Zhou J, et al. The Long Non-Coding RNA MALAT1 Promotes the Migration and Invasion of Hepatocellular Carcinoma by Sponging miR-204 and Releasing SIRT1. Tumour Biol (2017) 39(7):1010428317718135. doi: 10.1177/1010428317718135
23. Chao Li RM, Liu S, Wan Y, Zhang S, Deng Y, Bi J, et al. : Down-Regulation of miR-146b-5p by Long Noncoding RNA MALAT1 in Hepatocellular Carcinoma Promotes Cancer Growth and Metastasis. Oncotarget (2017) 8(17):28683–95. doi: 10.18632/oncotarget.15640
24. Sun X LY, Yu J, Pei H, Luo P, Zhang J. miR-128 Modulates Chemosensitivity and Invasion of Prostate Cancer Cells Through Targeting ZEB1. Clinics Liver Dis (2015) 45(5):474–82. doi: 10.1093/jjco/hyv027
25. Wu M, Lin Z, Li X, Xin X, An J, Zheng Q, et al. HULC Cooperates With MALAT1 to Aggravate Liver Cancer Stem Cells Growth Through Telomere Repeat-Binding Factor 2. Sci Rep (2016) 6(1):36045. doi: 10.1038/srep36045
26. Malakar P, Shilo A, Mogilevsky A, Stein I, Pikarsky E, Nevo Y, et al. Long Noncoding RNA MALAT1 Promotes Hepatocellular Carcinoma Development by SRSF1 Upregulation and mTOR Activation. Cancer Res (2017) 77(5):1155–67. doi: 10.1158/0008-5472.CAN-16-1508
27. Han J, Xie C, Pei T, Wang J, Lan Y, Huang K, et al. Deregulated Ajap1/β-Catenin/ZEB1 Signaling Promotes Hepatocellular Carcinoma Carcinogenesis and Metastasis. Cell Death Dis (2017) 8(4):e2736–6. doi: 10.1038/cddis.2017.126
28. TaoYang X, Chen A, Tan K, Du X. LncRNA HOTAIR Contributes to the Malignancy of Hepatocellular Carcinoma by Enhancing Epithelial-Mesenchymal Transition Via Sponging miR-23b-3p From ZEB1. Gene (2018) 670(5):114–22. doi: 10.1016/j.gene.2018.05.061
29. Li J, Wang J, Zhong Y, Guo R, Chu D, Qiu H, et al. HOTAIR: A Key Regulator in Gynecologic Cancers. Cancer Cell Int (2017) 17(1):65. doi: 10.1186/s12935-017-0434-6
30. Zhong DN, Luo YH, Mo WJ, Zhang X, Tan Z, Zhao N, et al. High Expression of Long Non−Coding HOTAIR Correlated With Hepatocarcinogenesis and Metastasis. Mol Med Rep (2018)17(1):1148–56. doi: 10.3892/mmr.2017.7999
31. Yang L, Zhang X, Li H, Liu J. The Long Noncoding RNA HOTAIR Activates Autophagy by Upregulating ATG3 and ATG7 in Hepatocellular Carcinoma. Mol Biosyst (2016) 12(8):2605–12. doi: 10.1039/c6mb00114a
32. Ying Wu QX, Li S, Yang X, Ge F. Integrated Proteomic and Transcriptomic Analysis Reveals Long Noncoding RNA HOX Transcript Antisense Intergenic RNA (Hotair) Promotes Hepatocellular Carcinoma Cell Proliferation by Regulating Opioid Growth Factor Receptor (Ogfr). Mol Cell Proteomics (2018)17(1):146–59. doi: 10.1074/mcp.RA117.000277
33. Zhou JJ, Cheng D, He XY, Meng Z, Li WZ, Chen RF. Knockdown of Hotair Suppresses Proliferation and Cell Cycle Progression in Hepatocellular Carcinoma Cell by Downregulating CCND1 Expression. Mol Med Rep (2017) 16(4):4980–6. doi: 10.3892/mmr.2017.7162
34. Ding C, Cheng S, Yang Z, Lv Z, Xiao H, Du C, et al. Long Non-Coding RNA HOTAIR Promotes Cell Migration and Invasion Via Down-Regulation of RNA Binding Motif Protein 38 in Hepatocellular Carcinoma Cells. Int J Mol Sci (2014) 15(3):4060–76. doi: 10.3390/ijms15034060
35. Fu WM, Zhu X, Wang WM, Lu YF, Hu BG, Wang H, et al. Hotair Mediates Hepatocarcinogenesis Through Suppressing miRNA-218 Expression and Activating P14 and P16 Signaling. J Hepatol (2015) 63(4):886–95. doi: 10.1016/j.jhep.2015.05.016
36. Wei S, Fan Q, Yang L, Zhang X, Ma Y, Zong Z, et al. Promotion of Glycolysis by HOTAIR Through GLUT1 Upregulation Via mTOR Signaling. Oncol Rep (2017) 38(3):1902–8. doi: 10.3892/or.2017.5840
37. Liu Z, Liu X-H, Sun M, Liu J, Wang Z-X, De W. The Long non-Coding RNA HOTAIR Indicates a Poor Prognosis and Promotes Metastasis in Non-Small Cell Lung Cancer. BMC Cancer (2013) 13:464. doi: 10.1186/1471-2407-13-464
38. Wang X, Liu W, Wang P, Li S. RNA Interference of Long Noncoding RNA HOTAIR Suppresses Autophagy and Promotes Apoptosis and Sensitivity to Cisplatin in Oral Squamous Cell Carcinoma. J Oral Pathol Med (2018) 47(10):930–7. doi: 10.1111/jop.12769
39. Ishibashi M, Kogo R, Shibata K, Sawada G, Takahashi Y, Kurashige J, et al. Clinical Significance of the Expression of Long Non-Coding RNA HOTAIR in Primary Hepatocellular Carcinoma. Oncol Rep (2013) 29(3):946–50. doi: 10.3892/or.2012.2219
40. Cui M, Xiao Z, Wang Y, Zheng M, Song T, Cai X, et al. Long Noncoding RNA HULC Modulates Abnormal Lipid Metabolism in Hepatoma Cells Through an Mir-9-Mediated RXRA Signaling Pathway. Cancer Res (2015) 75(5):846–57. doi: 10.1158/0008-5472.can-14-1192
41. Xin X, Wu M, Meng Q, Wang C, Lu Y, Yang Y, et al. Long Noncoding RNA HULC Accelerates Liver Cancer by Inhibiting PTEN Via Autophagy Cooperation to Mir15a. Mol Cancer (2018) 17(1):94. doi: 10.1186/s12943-018-0843-8
42. Li J, Wang X, Tang J, Jiang R, Zhang W, Ji J, et al. HULC and Linc00152 Act as Novel Biomarkers in Predicting Diagnosis of Hepatocellular Carcinoma. Cell Physiol Biochem (2015) 37(2):687–96. doi: 10.1159/000430387
43. Wang BG, Lv Z, Ding HX, Fang XX, Wen J, Xu Q, et al. The Association of lncRNA-HULC Polymorphisms With Hepatocellular Cancer Risk and Prognosis. Gene (2018) 670:148–54. doi: 10.1016/j.gene.2018.05.096
44. Yang-Hua Fan M-JW, Jiang Y, Ye M, Lu S-G, Wu L, Zhu X-G. Long Non-Coding RNA HULC as a Potential Prognostic Biomarker in Human Cancers: A Meta-Analysis. Oncotarget (2017) 8(13):21410–7. doi: 10.18632/oncotarget.15247
45. Wang XS. Rapid Identification of UCA1 as a Very Sensitive and Specific Unique Marker for Human Bladder Carcinoma. Clin Cancer Res (2006) 12(16):4851–8. doi: 10.1158/1078-0432.ccr-06-0134
46. Han Y, Yang Y-N, Yuan H-h, Zhang T-T, Sui H, Wei X-L, et al. UCA1, a Long Non-Coding RNA Up-Regulated in Colorectal Cancer Influences Cell Proliferation, Apoptosis and Cell Cycle Distribution. Pathology (2014) 46(5):396–401. doi: 10.1097/pat.0000000000000125
47. Qin L-T, Tang R-X, Lin P, Li Q, Yang H, Luo D-Z, et al. Biological Function of UCA1 in Hepatocellular Carcinoma and its Clinical Significance: Investigation With In Vitro and Meta-Analysis. Pathol - Res Pract (2018) 214(9):1260–72. doi: 10.1016/j.prp.2018.03.025
48. Wen H-J, Walsh MP, Yan IK, Takahashi K, Fields A, Patel T. Functional Modulation of Gene Expression by Ultraconserved Long Non-coding Rna TUC338 During Growth of Human Hepatocellular Carcinoma. iScience (2018) 2:210–20. doi: 10.1016/j.isci.2018.03.004
49. Jie Ji JT, Deng L, Xie Y, Jiang R, Li G, Sun B. LINC00152 Promotes Proliferation in Hepatocellular Carcinoma by Targeting EpCAM Via the mTOR Signaling Pathway. Oncotarget (2015) 6(40):42813–24. doi: 10.18632/oncotarget.5970
50. Huaqiang Zhu XZ, Chang H, Li H, Liu F, Ma C, Lu J. CCAT1 Promotes Hepatocellular Carcinoma Cell Proliferation and Invasion. Int J Clin Exp Pathol (2015) 8(5):5427–34.
51. Yuan S-X, Wang J, Yang F, Tao Q-F, Zhang J, Wang L-L, et al. Long Noncoding RNADANCRincreases Stemness Features of Hepatocellular Carcinoma by Derepression Ofctnnb1. Hepatology (2016) 63(2):499–511. doi: 10.1002/hep.27893
52. Thin KZ, Liu X, Feng X, Raveendran S, Tu JC. LncRNA-DANCR: A Valuable Cancer Related Long non-Coding RNA for Human Cancers. Pathol - Res Pract (2018) 214(6):801–5. doi: 10.1016/j.prp.2018.04.003
53. Lu Y, Hu Z, Mangala LS, Stine ZE, Hu X, Jiang D, et al. Myc Targeted Long Noncoding RNA DANCR Promotes Cancer in Part by Reducing p21 Levels. Cancer Res (2018) 78(1):64–74. doi: 10.1158/0008-5472.CAN-17-0815
54. Lv J, Kong Y, Gao Z, Liu Y, Zhu P, Yu Z. LncRNA TUG1 Interacting With miR-144 Contributes to Proliferation, Migration and Tumorigenesis Through Activating the JAK2/STAT3 Pathway in Hepatocellular Carcinoma. Int J Biochem Cell Biol (2018) 101:19–28. doi: 10.1016/j.biocel.2018.05.010
55. Terracciano L, Ge Y, Yan X, Jin Y, Yang X, Yu X, et al. fMiRNA-192 and Mirna-204 Directly Suppress Lncrna HOTTIP and Interrupt Gls1-Mediated Glutaminolysis in Hepatocellular Carcinoma. PloS Genet (2015) 11(12):e1005726. doi: 10.1371/journal.pgen.1005726
56. Liu Y-X, Sun Y. LncRNA HOTTIP Improves Diabetic Retinopathy by Regulating the P38-MAPK Pathway. Eur Rev Med Pharmacol Sci (2018) 22:2941–8. doi: 10.26355/eurrev_201805_1504
57. Wang X, Peng J, Wang J, Li M, Wu D, Wu S, et al. Hepatitis C Virus Core Impacts Expression of miR122 and miR204 Involved in Carcinogenic Progression Via Regulation of TGFBRAP1 and HOTTIP Expression. OncoTargets Therapy Vol (2018) 11:1173–82. doi: 10.2147/ott.s149254
58. Jingjing Li QZ, Fan X, Mo W, Dai W, Feng J, Wu L, et al. The Long Noncoding RNA TUG1 Acts as a Competing Endogenous RNA to Regulate the Hedgehog Pathway by Targeting miR-132 in Hepatocellular Carcinoma. Oncotarget (2017) 8(39):65932–45. doi: 10.18632/oncotarget.19582
59. Huang M-D, Chen W-M, Qi F-Z, Sun M, Xu T-P, Ma P, et al. Long Non-Coding RNA TUG1 Is Up-Regulated in Hepatocellular Carcinoma and Promotes Cell Growth and Apoptosis by Epigenetically Silencing of KLF2. Mol Cancer (2015) 14(1):165. doi: 10.1186/s12943-015-0431-0
60. He C, Liu Z, Jin L, Zhang F, Peng X, Xiao Y, et al. Lncrna TUG1-Mediated Mir-142-3p Downregulation Contributes to Metastasis and the Epithelial-to-Mesenchymal Transition of Hepatocellular Carcinoma by Targeting Zeb1. Cell Physiol Biochem (2018) 48(5):1928–41. doi: 10.1159/000492517
61. Deng X, Zhao XF, Liang XQ, Chen R, Pan YF, Liang J. Linc00152 Promotes Cancer Progression in Hepatitis B Virus-Associated Hepatocellular Carcinoma. Biomed Pharmacother (2017) 90:100–8. doi: 10.1016/j.biopha.2017.03.031
62. Le Chang GW, Jia T, Zhang L, Li Y, Han Y, Zhang K, et al. Armored Long Non-Coding RNA MEG3 Targeting EGFR Based on Recombinant MS2 Bacteriophage Virus-Like Particles Against Hepatocellular Carcinoma. Oncotarget (2016) 7(17):23988–4004. doi: 10.18632/oncotarget.8115
63. Zhou Y, Zhong Y, Wang Y, Zhang X, Batista DL, Gejman R, et al. Activation of p53 by MEG3 Non-Coding RNA. J Biol Chem (2007) 282(34):24731–42. doi: 10.1074/jbc.M702029200
64. Braconi C, Kogure T, Valeri N, Huang N, Nuovo G, Costinean S, et al. microRNA-29 Can Regulate Expression of the Long Non-Coding RNA Gene MEG3 in Hepatocellular Cancer. Oncogene (2011) 30(47):4750–6. doi: 10.1038/onc.2011.193
65. Yu-Yuan Qian KL, Liu Q-Y, Liu Z-S. Long Non-Coding RNA PTENP1 Interacts With miR-193a-3p to Suppress Cell Migration and Invasion Through the PTEN Pathway in Hepatocellular Carcinoma. Oncotarget (2017) 8(64):107859–69. doi: 10.18632/oncotarget.22305
66. Chen C-L, Tseng Y-W, Wu J-C, Chen G-Y, Lin K-C, Hwang S-M, et al. Suppression of Hepatocellular Carcinoma by Baculovirus-Mediated Expression of Long Non-Coding RNA PTENP1 and MicroRNA Regulation. Biomaterials (2015) 44:71–81. doi: 10.1016/j.biomaterials.2014.12.023
67. Chen S, Wang Y, Zhang J-H, Xia Q-J, Sun Q, Li Z-K, et al. Long Non-Coding RNA PTENP1 Inhibits Proliferation and Migration of Breast Cancer Cells Via AKT and MAPK Signaling Pathways. Oncol Lett (2017) 14(4):4659–62. doi: 10.3892/ol.2017.6823
68. Rupeng Zhang YG, Ma Z, Ma G, Xue Q, Li F. Liren Liu: Long Non-Coding RNA PTENP1 Functions as a ceRNA to Modulate PTEN Level by Decoying miR-106b and miR-93 in Gastric Cancer. Oncotarget (2017) 8(16):26079–89. doi: 10.18632/oncotarget.15317
69. Chen Z, Xu D, Zhang T. Inhibition of Proliferation and Invasion of Hepatocellular Carcinoma Cells by lncRNA-ASLNC02525 Silencing and the Mechanism. Int J Oncol (2017) 51(3):851–8. doi: 10.3892/ijo.2017.4069
70. Zhang H, Zhou D, Ying M, Chen M, Chen P, Chen Z, et al. Expression of Long Non-Coding RNA (LncRNA) Small Nucleolar RNA Host Gene 1 (Snhg1) Exacerbates Hepatocellular Carcinoma Through Suppressing Mir-195. Med Sci Monit (2016) 22:4820–9. doi: 10.12659/msm.898574
71. Xiao J, Lv Y, Jin F, Liu Y, Ma Y, Xiong Y, et al. Lncrna HANR Promotes Tumorigenesis and Increase of Chemoresistance in Hepatocellular Carcinoma. Cell Physiol Biochem (2017) 43(5):1926–38. doi: 10.1159/000484116
72. Sui J, Yang X, Qi W, Guo K, Gao Z, Wang L, et al. Long Non-Coding Rna Linc-USP16 Functions As a Tumour Suppressor in Hepatocellular Carcinoma by Regulating Pten Expression. Cell Physiol Biochem (2017) 44(3):1188–98. doi: 10.1159/000485449
73. Fang L, Sun J, Pan Z, Song Y, Zhong L, Zhang Y, et al. Long non-Coding RNA NEAT1 Promotes Hepatocellular Carcinoma Cell Proliferation Through the Regulation of Mir-129-5p-VCP-Ikappab. Am J Physiol Gastrointest Liver Physiol (2017) 313(2):G150–6. doi: 10.1152/ajpgi.00426.2016
74. Ren Y, Shang J, Li J, Liu W, Zhang Z, Yuan J, et al. The Long Noncoding RNA PCAT-1 Links the microRNA miR-215 to Oncogene CRKL-Mediated Signaling in Hepatocellular Carcinoma. J Biol Chem (2017) 292(43):17939–49. doi: 10.1074/jbc.M116.773978
75. Runqiu Jiang JT, Chen Y, Deng L, Ji J, Xie Y, Wang K, et al. : The Long Noncoding RNA lnc-EGFR Stimulates T-Regulatory Cells Differentiation Thus Promoting Hepatocellular Carcinoma Immune Evasion. Nat Commun (2017) 25(8):15129. doi: 10.1038/ncomms15129
76. Feng Wang H-QY, He B-S, Pan Y-Q, Deng Q-W, Sun H-L, Chen J, et al. Upregulated lncRNA-UCA1 Contributes to Progression of Hepatocellular Carcinoma Through Inhibition of miR-216b and Activation of FGFR1/ERK Signaling Pathway. Oncotarget (2015) 6(10):7899–917. doi: 10.18632/oncotarget.3219
77. Xiangling Feng ZW, Fillmore R, Xi Y. MiR-200, a New Star miRNA in Human Cancer. Cancer Lett (2014) 344(2):166–73. doi: 10.1016/j.canlet.2013.11.004
78. Bracken CP, Khew-Goodall Y, Goodall GJ. Network-Based Approaches to Understand the Roles of miR-200 and Other microRNAs in Cancer. Cancer Res (2015) 75(13):2594–9. doi: 10.1158/0008-5472.CAN-15-0287
79. Kang Y, Li W. A New lnc in Metastasis: Long Noncoding RNA Mediates the ProMetastatic Functions of TGF-β. Cancer Cell (2014) 25:557–9. doi: 10.1016/j.ccr.2014.04.014
80. Singh M, Yelle N, Venugopal C, Singh SK. Emt: Mechanisms and Therapeutic Implications. Pharmacol Ther (2018) 182:80–94. doi: 10.1016/j.pharmthera.2017.08.009
81. He JH, Han ZP, Liu JM, Zhou JB, Zou MX, Lv YB, et al. Overexpression of Long non-Coding RNA Meg3 Inhibits Proliferation of Hepatocellular Carcinoma Huh7 Cells Via Negative Modulation of Mirna-664. J Cell Biochem (2017) 118(11):3713–21. doi: 10.1002/jcb.26018
82. Tsang FH AS, Wei L, Fan DN, Lee JM, Wong CC, Ng IO, et al. Long non-Coding RNA HOTTIP is Frequently Up-Regulated in Hepatocellular Carcinoma and Is Targeted by Tumor Suppressive Mir-125b. Liver Int (2015) 35(5):1597–606. doi: 10.1111/liv.12746
83. Xiong H, Ni Z, He J, Jiang S, Li X, He J, et al. LncRNA HULC Triggers Autophagy Via Stabilizing Sirt1 and Attenuates the Chemosensitivity of HCC Cells. Oncogene (2017) 36(25):3528–40. doi: 10.1038/onc.2016.521
84. Maimaiti Y, Jie T, Jing Z, Changwen W, Pan Y, Chen C, et al. Aurora Kinase A Induces Papillary Thyroid Cancer Lymph Node Metastasis by Promoting Cofilin-1 Activity. Biochem Biophys Res Commun (2016) 473(1):212–8. doi: 10.1016/j.bbrc.2016.03.081
85. Cui S ZK, Li C, Chen J, Pan Y, Feng B, Lu L, et al. Methylation-Associated Silencing of microRNA-129-3p Promotes Epithelial-Mesenchymal Transition, Invasion and Metastasis of Hepatocelluar Cancer by Targeting Aurora-a. Oncotarget (2016) 7(47):78009–28. doi: 10.18632/oncotarget.12870
86. Zhu J, Liu S, Ye F, Shen Y, Tie Y, Zhu J, et al. Long Noncoding Rna MEG3 Interacts With P53 Protein and Regulates Partial P53 Target Genes in Hepatoma Cells. PloS One (2015) 10(10):e0139790. doi: 10.1371/journal.pone.0139790
87. Zhuo H, Tang J, Lin Z, Jiang R, Zhang X, Ji J, et al. The Aberrant Expression of MEG3 Regulated by UHRF1 Predicts the Prognosis of Hepatocellular Carcinoma. Mol Carcinog (2016) 55(2):209–19. doi: 10.1002/mc.22270
88. Liu L-X, Deng WEI, Zhou X-T, Chen R-P, Xiang M-Q, Guo Y-T, et al. The Mechanism of Adenosine-Mediated Activation of Lncrna MEG3 and its Antitumor Effects in Human Hepatoma Cells. Int J Oncol (2016) 48(1):421–9. doi: 10.3892/ijo.2015.3248
89. Zheng ZK, Pang C, Yang Y, Duan Q, Zhang J, Liu WC. Serum Long Noncoding RNA Urothelial Carcinoma-Associated 1: A Novel Biomarker for Diagnosis and Prognosis of Hepatocellular Carcinoma. J Int Med Res (2018) 46(1):348–56. doi: 10.1177/0300060517726441
90. Quagliata L, Quintavalle C, Lanzafame M, Matter MS, Novello C, di Tommaso L, et al. High Expression of HOXA13 Correlates With Poorly Differentiated Hepatocellular Carcinomas and Modulates Sorafenib Response in In Vitro Models. Lab Invest (2018) 98(1):95–105. doi: 10.1038/labinvest.2017.107
91. Zhang Y, Huang JC, Cai KT, Yu XB, Chen YR, Pan WY, et al. Long Noncoding RNA HOTTIP Promotes Hepatocellular Carcinoma Tumorigenesis and Development: A Comprehensive Investigation Based on Bioinformatics, qRTPCR and Metaanalysis of 393 Cases. Int J Oncol (2017) 51(6):1705–21. doi: 10.3892/ijo.2017.4164
92. Quagliata L, Matter MS, Piscuoglio S, Arabi L, Ruiz C, Procino A, et al. Long Noncoding RNA HOTTIP/HOXA13 Expression Is Associated With Disease Progression and Predicts Outcome in Hepatocellular Carcinoma Patients. Hepatology (2014) 59(3):911–23. doi: 10.1002/hep.26740
93. Jin W, Chen L, Cai X, Zhang Y, Zhang J, Ma D, et al. Long Non-Coding RNA TUC338 is Functionally Involved in Sorafenib-Sensitized Hepatocarcinoma Cells by Targeting RASAL1. Oncol Rep (2017) 37(1):273–80. doi: 10.3892/or.2016.5248
94. Chen Y, Shen Z, Zhi Y, Zhou H, Zhang K, Wang T, et al. Long non-Coding RNA ROR Promotes Radioresistance in Hepatocelluar Carcinoma Cells by Acting as a ceRNA for microRNA-145 to Regulate RAD18 Expression. Arch Biochem Biophys (2018) 645:117–25. doi: 10.1016/j.abb.2018.03.018
95. Llovet JM, Montal R, Sia D, Finn RS. Molecular Therapies and Precision Medicine for Hepatocellular Carcinoma. Nat Rev Clin Oncol (2018) 15(10):599–616. doi: 10.1038/s41571-018-0073-4
96. Guo Z, Zhang J, Fan L, Liu J, Yu H, Li X, et al. Long Noncoding RNA (LncRNA) Small Nucleolar RNA Host Gene 16 (Snhg16) Predicts Poor Prognosis and Sorafenib Resistance in Hepatocellular Carcinoma. Med Sci Monit (2019) 25:2079–86. doi: 10.12659/MSM.915541
97. Ye J, Zhang R, Du X, Chai W, Zhou Q. Long Noncoding RNA SNHG16 Induces Sorafenib Resistance in Hepatocellular Carcinoma Cells Through Sponging Mir-140-5p. Oncol Targets Ther (2019) 12:415–22. doi: 10.2147/OTT.S175176
98. Zhang PF, Wang F, Wu J, Wu Y, Huang W, Liu D, et al. Lncrna SNHG3 Induces EMT and Sorafenib Resistance by Modulating the miR-128/CD151 Pathway in Hepatocellular Carcinoma. J Cell Physiol (2019) 234(3):2788–94. doi: 10.1002/jcp.27095
99. Li W, Dong X, He C, Tan G, Li Z, Zhai B, et al. Lncrna SNHG1 Contributes to Sorafenib Resistance by Activating the Akt Pathway and Is Positively Regulated by miR-21 in Hepatocellular Carcinoma Cells. J Exp Clin Cancer Res (2019) 38(1):183. doi: 10.1186/s13046-019-1177-0
100. Chen S, Xia X. Long Noncoding RNA NEAT1 Suppresses Sorafenib Sensitivity of Hepatocellular Carcinoma Cells Via Regulating Mir-335-C-Met. J Cell Physiol (2019). doi: 10.1002/jcp.27567
101. Zhi Y, Abudoureyimu M, Zhou H, Wang T, Feng B, Wang R, et al. Foxm1-Mediated LINC-ROR Regulates the Proliferation and Sensitivity to Sorafenib in Hepatocellular Carcinoma. Mol Ther Nucleic Acids (2019) 16:576–88. doi: 10.1016/j.omtn.2019.04.008
102. Zhang W, Liu Y, Fu Y, Han W, Xu H, Wen L, et al. Long Non-Coding RNA LINC00160 Functions as a Decoy of microRNA-132 to Mediate Autophagy and Drug Resistance in Hepatocellular Carcinoma Via Inhibition of PIK3R3. Cancer Lett (2020) 478:22–33. doi: 10.1016/j.canlet.2020.02.014
103. Sui C, Dong Z, Yang C, Zhang M, Dai B, Geng L, et al. LncRNA FOXD2-AS1 as a Competitive Endogenous RNA Against miR-150-5p Reverses Resistance to Sorafenib in Hepatocellular Carcinoma. J Cell Mol Med (2019) 23(9):6024–33. doi: 10.1111/jcmm.14465
104. Fan L, Huang X, Chen J, Zhang K, Gu YH, Sun J, et al. Long Noncoding RNA MALAT1 Contributes to Sorafenib Resistance by Targeting Mir-140-5p/Aurora-a Signaling in Hepatocellular Carcinoma. Mol Cancer Ther (2020) 19(5):1197–209. doi: 10.1158/1535-7163.MCT-19-0203
105. Xu Y, Liu Y, Li Z, Li H, Li X, Yan L, et al. Long Noncoding RNA H19 Is Involved in Sorafenib Resistance in Hepatocellular Carcinoma by Upregulating Mir675. Oncol Rep (2020) 44(1):165–73. doi: 10.3892/or.2020.7608
106. Schultheiss CS, Laggai S, Czepukojc B, Hussein UK, List M, Barghash A, et al. The Long Non-Coding RNA H19 Suppresses Carcinogenesis and Chemoresistance in Hepatocellular Carcinoma. Cell Stress (2017) 1(1):37–54. doi: 10.15698/cst2017.10.105
107. Huang H, Chen J, Ding CM, Jin X, Jia ZM, Peng J. Lncrna NR2F1-AS1 Regulates Hepatocellular Carcinoma Oxaliplatin Resistance by Targeting ABCC1 Via Mir-363. J Cell Mol Med (2018) 22(6):3238–45. doi: 10.1111/jcmm.13605
108. Hu H, Yang L, Li L, Zeng C. Long non-Coding RNA KCNQ1OT1 Modulates Oxaliplatin Resistance in Hepatocellular Carcinoma Through Mir-7-5p/ ABCC1 Axis. Biochem Biophys Res Commun (2018) 503(4):2400–6. doi: 10.1016/j.bbrc.2018.06.168
109. Li Y, Ye Y, Feng B, Qi Y. Long Noncoding RNA Lncarsr Promotes Doxorubicin Resistance in Hepatocellular Carcinoma Via Modulating Pten-PI3K/Akt Pathway. J Cell Biochem (2017) 118(12):4498–507. doi: 10.1002/jcb.26107
110. Ma H, Yuan L, Li W, Xu K, Yang L. The LncRNA H19/miR-193a-3p Axis Modifies the Radio-Resistance and Chemotherapeutic Tolerance of Hepatocellular Carcinoma Cells by Targeting PSEN1. J Cell Biochem (2018) 119(10):8325–35. doi: 10.1002/jcb.26883
111. Wu L, Pan C, Wei X, Shi Y, Zheng J, Lin X, et al. LncRNA KRAL Reverses 5-Fluorouracil Resistance in Hepatocellular Carcinoma Cells by Acting as a ceRNA Against Mir-141. Cell Commun Signal (2018) 16(1):47. doi: 10.1186/s12964-018-0260-z
112. Chen Y, Zhao H, Li H, Feng X, Tang H, Qiu C, et al. LINC01234/MicroRNA-31-5p/MAGEA3 Axis Mediates the Proliferation and Chemoresistance of Hepatocellular Carcinoma Cells. Mol Ther Nucleic Acids (2020) 19:168–78. doi: 10.1016/j.omtn.2019.10.035
113. Available at: https://clinicaltrials.gov/ct2/show/NCT03469544?cond=lncRNA&rank=8.
Keywords: long non-coding RNA, hepatocellular carcinoma, signaling pathway, therapy resistance, clinical transformation
Citation: Lin X, Xiang X, Feng B, Zhou H, Wang T, Chu X and Wang R (2021) Targeting Long Non-Coding RNAs in Hepatocellular Carcinoma: Progress and Prospects. Front. Oncol. 11:670838. doi: 10.3389/fonc.2021.670838
Received: 22 February 2021; Accepted: 07 June 2021;
Published: 25 June 2021.
Edited by:
Aamir Ahmad, University of Alabama at Birmingham, United StatesReviewed by:
Silvia Zappavigna, University of Campania Luigi Vanvitelli, ItalyXiaoli Ma, Jinan Central Hospital, China
Copyright © 2021 Lin, Xiang, Feng, Zhou, Wang, Chu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rui Wang, d2FuZ3J1aTIxOEAxNjMuY29t
†These authors have contributed equally to this work
 Xinrong Lin
Xinrong Lin Xiaosong Xiang2†
Xiaosong Xiang2† Rui Wang
Rui Wang