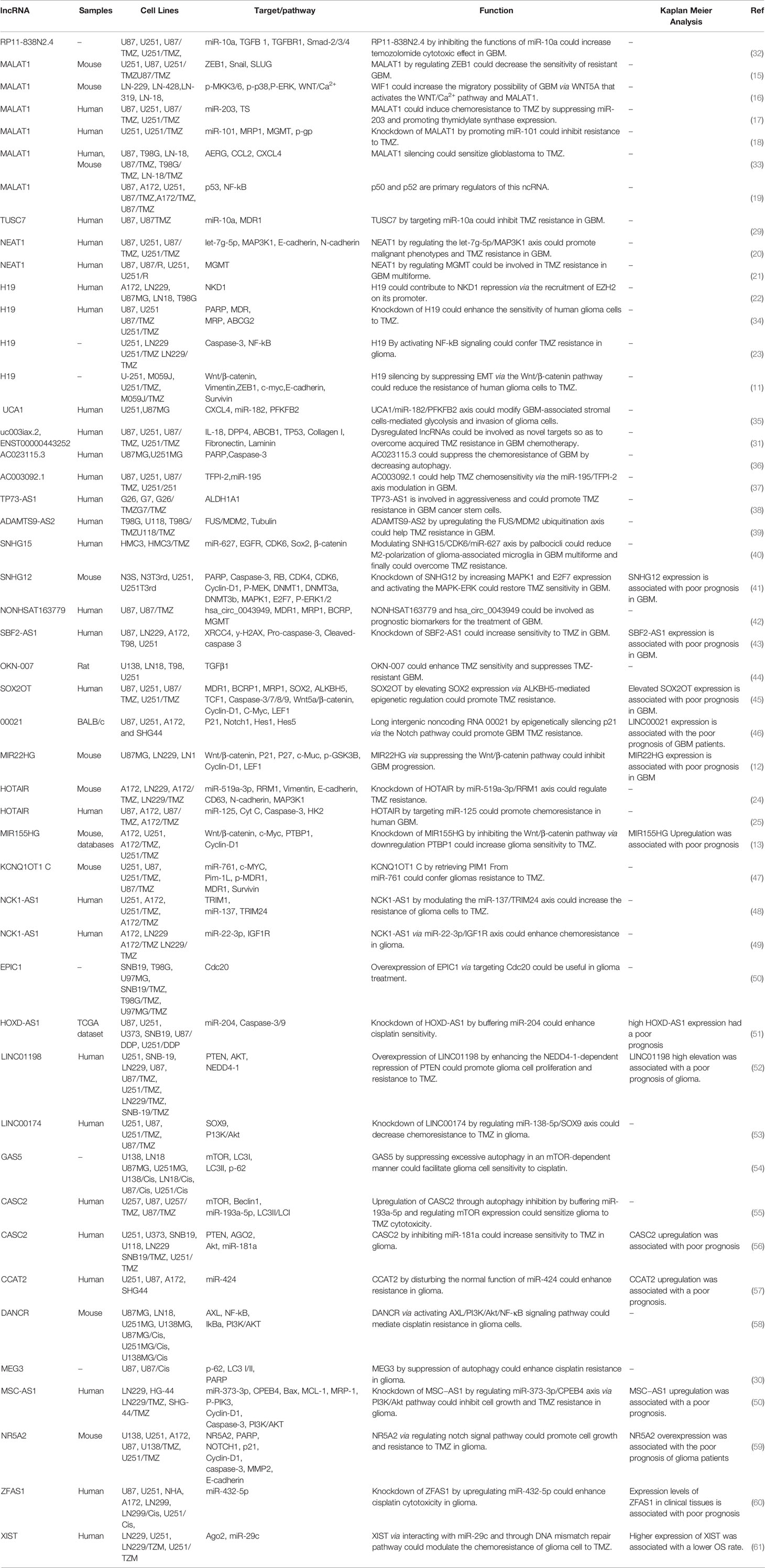
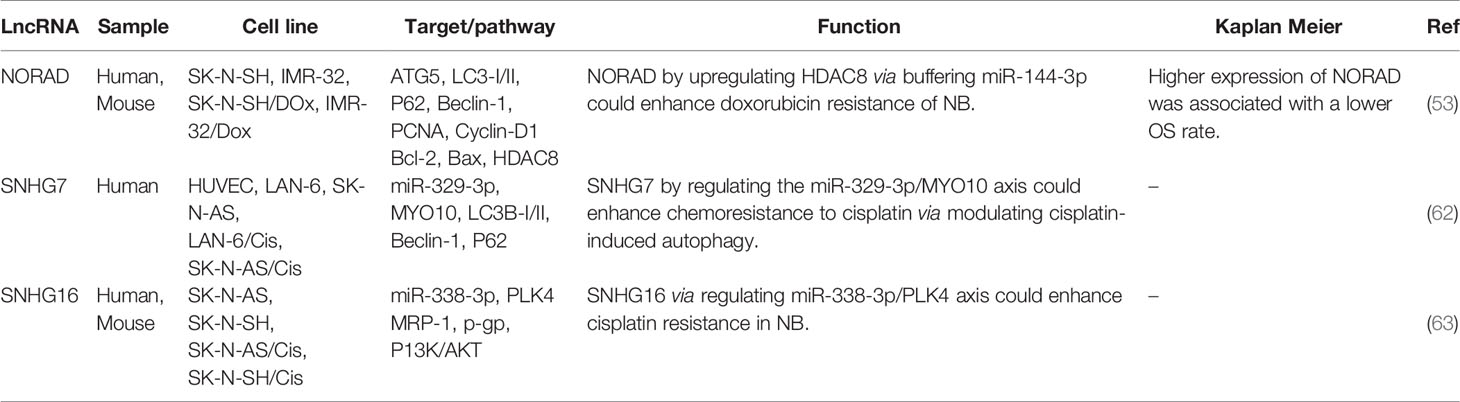
- 1Department of Medical Genetics, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- 2Department of Pharmacology, Tabriz University of Medical Sciences, Tabriz, Iran
- 3Department of Medical Genetics, Faculty of Medicine, Tabriz University of Medical Sciences, Tabriz, Iran
- 4Department of Anatomical Sciences, Faculty of Medicine, Birjand University of Medical Sciences, Birjand, Iran
- 5Urology and Nephrology Research Center, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- 6Skull Base Research Center, Loghman Hakim Hospital, Shahid Beheshti University of Medical Sciences, Tehran, Iran
Tumors of the nervous system can be originated from several locations. They mostly have high mortality and morbidity rate. The emergence of resistance to chemotherapeutic agents is a hurdle in the treatment of patients. Long non-coding RNAs (lncRNAs) have been shown to influence the response of glioblastoma/glioma and neuroblastoma to chemotherapeutic agents. MALAT1, NEAT1, and H19 are among lncRNAs that affect the response of glioma/glioblastoma to chemotherapy. As well as that, NORAD, SNHG7, and SNHG16 have been shown to be involved in conferring this phenotype in neuroblastoma. Prior identification of expression amounts of certain lncRNAs would help in the better design of therapeutic regimens. In the current manuscript, we summarize the impact of lncRNAs on chemoresistance in glioma/glioblastoma and neuroblastoma.
Introduction
Tumors of the nervous system can be originated from several cellular compartments. The main classes of these tumors are glioma, meningioma, neuroblastoma, and spinal tumors (1). Although being quite rare, brain tumors are considered as high mortality cancers (2). Their protected position in the brain makes these neoplasms difficult to cure. Surgical removal of the tumor, radiotherapy, and chemotherapy are currently available therapeutic options for brain tumors. However, these options are associated with possible permanent morbidity for patients and incomplete cure of cancer (2). Inherent or attained chemoresistance is the chief reason for treatment failure in these patients (3). Alkylating agents constitute the backbone of chemotherapeutic regimens for brain tumors. These agents induce DNA damage and consequently activate apoptosis, yet their efficiency in killing cancer cells depends on the DNA repair system (3). As an example of an orally bioavailable alkylating agent, temozolomide (TMZ) has been used widely in the treatment of patients with brain tumors. This agent is spontaneously transformed to its active metabolite 5-(3-methyl triazen-1-yl) imidazole-4-carboxamide (MTIC) without requiring hepatic activation.). Furthermore, TMZ is an effective radiosensitizer and a vital constituent of chemoradiotherapy for patients with newly-diagnosed glioblastoma (4). Resistance to TMZ has been detected in about half of patients. Up-regulation of O6-methylguanine methyltransferase (MGMT) and defects in the DNA repair pathway are among possible mechanisms for resistance to this agent (5). Another recently acknowledged cause of chemoresistance in tumors of the nervous system and related cell lines is the aberrant expression of long non-coding RNAs (lncRNAs). In the current manuscript, we summarize the impact of lncRNAs on chemoresistance in glioma/glioblastoma and neuroblastoma.
LncRNAs Functions
Novel sequencing methods have enabled comprehensive genomic and transcriptomic analyses and shown transcription of a total of 85% of the human genome (6, 7). Based on the results of ENCODE projects, most human transcriptomes are non-coding RNAs (8). LncRNAs with sizes of more than 200 nucleotides constitute a major part of the transcriptome. These transcripts are considered essential regulators of gene transcription. Their functions as signals, decoys, scaffolds, guide transcripts, and enhancers have endowed them the aptitude to control gene expression via different routes. Through having “decoy” binding sites, they can sequester transcription factors, catalytic molecules, constituents of chromatin remodeling complexes, and microRNAs (miRNAs), thus decreasing their bioavailability (9). Dysregulation of lncRNAs has been noted in tumors of the nervous system (10). Figure 1 indicates the role of several lncRNAs in modulating the sensitivity of tumor cells to various chemotherapeutic agents via regulating the Wnt-β-catenin signaling pathway which is a highly conserved cascade and is activated in the development of glioma cells. Wnt/β-catenin signaling is an evolutionary conserved axis that controls important cellular functions, namely proliferation, differentiation, migratory potential, genetic stability, cell death and renewal of stem cells, thus it has important roles in the carcinogenesis (14).
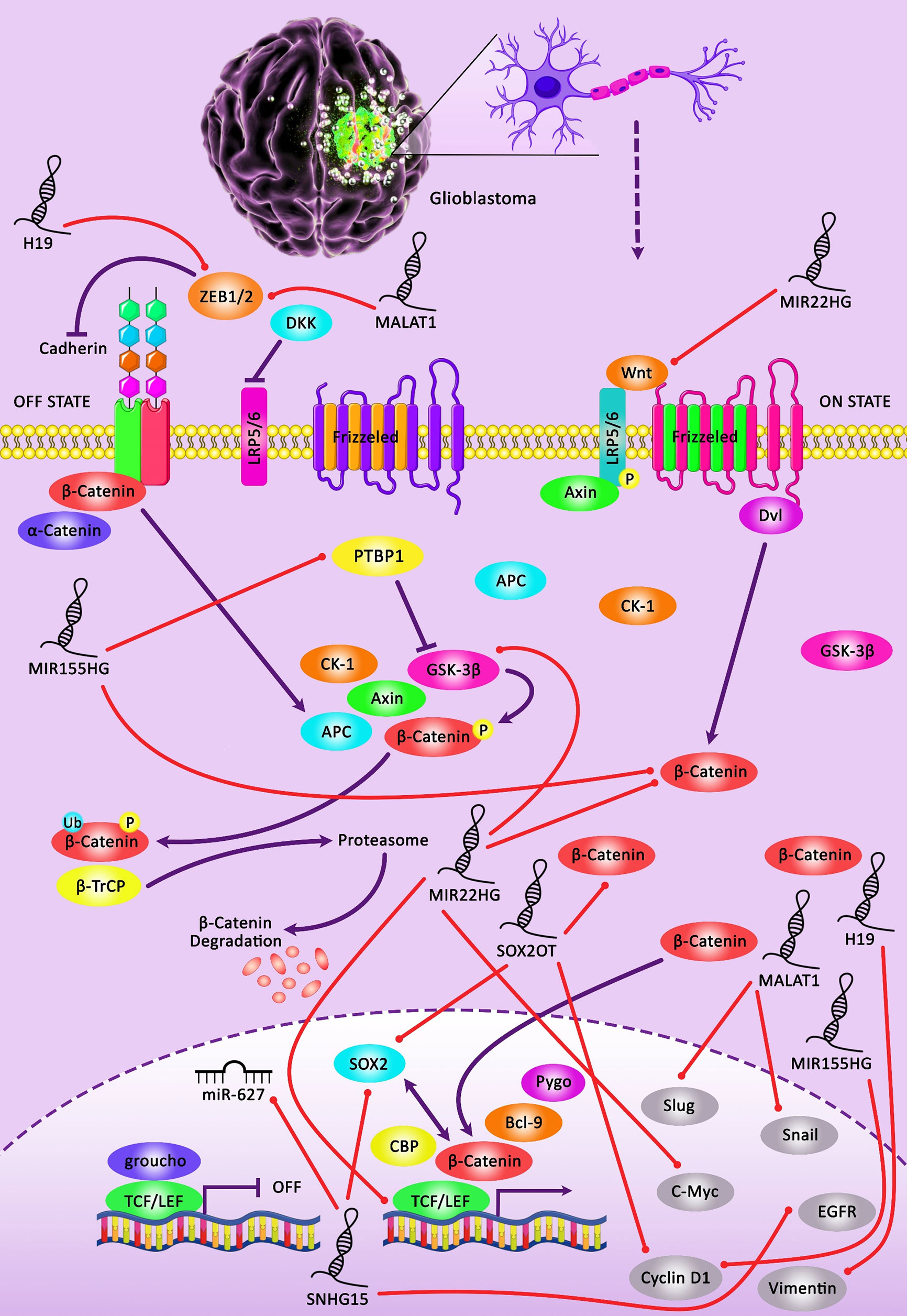
Figure 1 A schematic illustration of the crosstalk between lncRNAs and Wnt/β-catenin pathway involved in the modulation of the sensitivity of glioma cells to chemotherapeutic agents. Downregulation of lncRNA H19 could promote the sensitivity of glioma cells to temozolomide via inhibiting EMT through the suppression of the Wnt/β-Catenin signaling cascade. Silencing of H19 could downregulate the expression level of β-catenin and its downstream targets c-myc and Survivin in temozolomide-treated glioma cells (11). Besides, downregulating the expression of lncRNA MIR22HG could suppress the Wnt/β-catenin signaling pathway via loss of miR-22-3p and -5p. This could in turn lead to attenuating cell proliferation, invasion as well as tumor growth in glioma cells. MIR22HG silencing could result in downregulating the expression level of β-catenin, a key transcriptional regulator of Wnt, along with the inhibition of several Wnt downstream targets, containing c-Myc, cyclin D1, and LEF1, as well as a reduction in the expression of phospho-GSK3β (Ser9) in tumor cells (12). Besides, upregulation of lncRNA MIR155HG could promote temozolomide resistance in glioma cells through directly regulating canonical Wnt/β-catenin pathway activation via binding to PTBP1 in tumor cells (13).
LncRNAs and Chemoresistance in Glioma/Glioblastoma
Abnormal expression of several lncRNAs has been detected in resistant glioma/glioblastoma tumors or related cell lines. MALAT1, NEAT1, H19, and HOTAIR are among the mostly assessed lncRNAs in this field.
MALAT1
Li et al. have reported higher levels of MALAT1 in the U251/TMZ and U87/TMZ cells compared with the parental lines. Small interfering (si)RNA-mediated MALAT1 silencing has down-regulated expressions of MDR1, MRP5, and LRP1, increased sensitivity to TMZ, and decreased ZEB1 levels. In vivo experiments have also verified the impact of MALAT1 up-regulation in conferring TMZ resistance and upregulating ZEB1 levels. Taken together, MALAT1 can enhance the resistance of glioma cells to TMZ through regulating ZEB1 (15). Vassallo et al. have shown that MALAT1 silencing decreases migration of glioblastoma cells, without affecting proliferation. Meanwhile, down-regulation of WIF1 has been shown to enhance the migratory aptitude of glioblastoma via WNT5A that induces expression of MALAT1. They have suggested the contribution of canonical and non-canonical WNT routes in the pathogenesis of glioblastoma (16). Moreover, MALAT1 has been reported to induce chemoresistance to TMZ through suppressing miR-203 expression and promoting the expression of thymidylate synthase (17). Similarly, Cai et al. have reported up-regulation of MALAT1 in TMZ-resistant glioblastoma cells. MALAT1 silencing has reduced TMZ resistance of these cells as documented in cell lines and animal models. Functionally, MALAT1 confers this phenotype by inhibiting the miR-101 signaling pathway in glioblastoma cells (18). A clinical study in this field has shown the association between elevated serum levels of MALAT1 and poor response to TMZ and low survival rate of patients with glioblastoma (17). Notably, functional studies have verified that MALAT1 silencing reverses TMZ resistance in glioblastoma cell lines. MALAT1 exerts its function through modulating the expression of miR-203, thus reducing thymidylate synthase (TS) levels (17). Finally, Voce et al. have assessed the expression profile of glioblastoma cells to detect NF-κB-related transcripts whose expressions are changed following TMZ treatment. MALAT1 has been identified as one of the utmost elevated transcripts. Moreover, expression of MALAT1 has been simultaneously regulated by p50 and p53. TMZ has been shown to inhibit p50 recruitment to its cognate element following phosphorylation of Ser329. Administration of anti-MALAT1 siRNA via nanoparticles has enhanced response to TMZ in xenograft models of glioblastoma (19).
NEAT1
Expression of NEAT1 has been found to be elevated in serum samples of glioblastoma patients and glioma stem cells isolated from related cell lines. NEAT1 silencing has inhibited the malignant behaviors of these cells, as has been evident by the reduction of their proliferation, migration, and invasion. Functional studies have verified let-7g-5p as a target of NEAT1. Expression of MAP3K1, as a target of let-7g-5p, has been enhanced by NEAT1, Therefore, NEAT1 enhances malignant features of glioma stem cell and chemoresistant phenotype via let-7g-5p/MAP3K1 axis (20). Similarly, expression of NEAT1 has been lower in the TMZ-sensitive glioblastoma tissues and cell lines compared with TMZ-resistant ones. NEAT1 silencing has remarkably promoted TMZ-associated cell apoptosis in glioblastoma cells. Consistently, MGMT levels have been higher in TMZ-resistant cell lines. NEAT1 silencing has decreased mRNA and protein levels of MGMT (21).
H19
H19 is another oncogenic lncRNA in glioblastoma whose expression has been correlated with the expression of numerous genes participating in the growth and progression of this neoplasm. H19 silencing has reduced viability, migratory potential, and invasiveness of glioblastoma cells. Notably, H19 expression is inversely correlated with the expression of NKD1, an inhibitor of the Wnt pathway, thus H19 may modulate NKD1 expression via EZH2-associated H3K27 trimethylation. H19 binding with EZH2 has been verified in glioblastoma cells (22). H19 silencing has been shown to enhance TMZ cytotoxicity in glioma cells through inhibiting epithelial-mesenchymal transition (EMT) via the Wnt/β-catenin pathway (11) and inactivating NF-kB signaling (23).
HOTAIR
Expression of HOTAIR has been elevated in TMZ-resistant glioblastoma cells and its silencing has suppressed proliferation, migration, invasion, and EMT in TMZ-resistant cells. Notably, exosomal transfer of this lncRNA HOTAIR has conferred TMZ resistance via modulating miR-519a-3p/RRM1 molecular route (24). HOTAIR silencing has also decreased HK2 expression, thus suppressing cell proliferation and enhancing sensitivity to TMZ both in vivo and in vitro. HOTAIR increases HK2 levels by influencing miR-125 levels, which suppresses cell proliferation and increases TMZ-associated cell death (25).
Other lncRNAs
Several other lncRNAs have also been shown to affect the response of glioblastoma/glioma cells to therapeutic agents. Some lncRNAs affect autophagy. Autophagy is a fundamental capability of cells to reinstate the energy equilibrium throughput the periods of fluctuating nutrient accessibility (26). During this evolutionarily conserved process, impaired or useless biomolecules, organelles, or other cytoplasmic elements are transferred to the lysosomal system be targeted for degradation (27). Dysregulation of autophagy is linked with tumorigenesis and resistance of cancer cells to therapeutics (28).
Expression of TUSC7 has been decreased in TMZ-resistant glioblastoma cells and tissues. Ectopic expression of TUSC7 has inhibited TMZ resistance and decreased expression of MDR1. TUSC7 exerts its function by suppressing miR-10a levels (29). Ma et al. have reported over-expression of MEG3 in glioma cells treated with cisplatin. Up-regulation of MEG3 has increased the sensitivity of glioblastoma cells to cisplatin. Functionally, MEG3 attenuates cisplatin-induced autophagy (30). In a high throughput study, Zeng et al. have compared the expression of mRNAs and lncRNAs between a TMZ-resistant glioblastoma cell line and parental cells. They have reported differential expression of more than 2000 lncRNAs between these cells. Notably, the ECM−receptor interaction pathway has been downregulated and ECM-related collagen I, fibronectin, laminin, and CD44 have been correlated with resistance phenotype in vitro (31). Table 1 shows the list of lncRNAs that modulate the response of glioblastoma/glioma to chemotherapy. Figure 2 demonstrates the role of various long noncoding RNAs including CASC2 and GAS5 in suppressing the autophagy pathway through regulating mTOR expression in glioma cells.
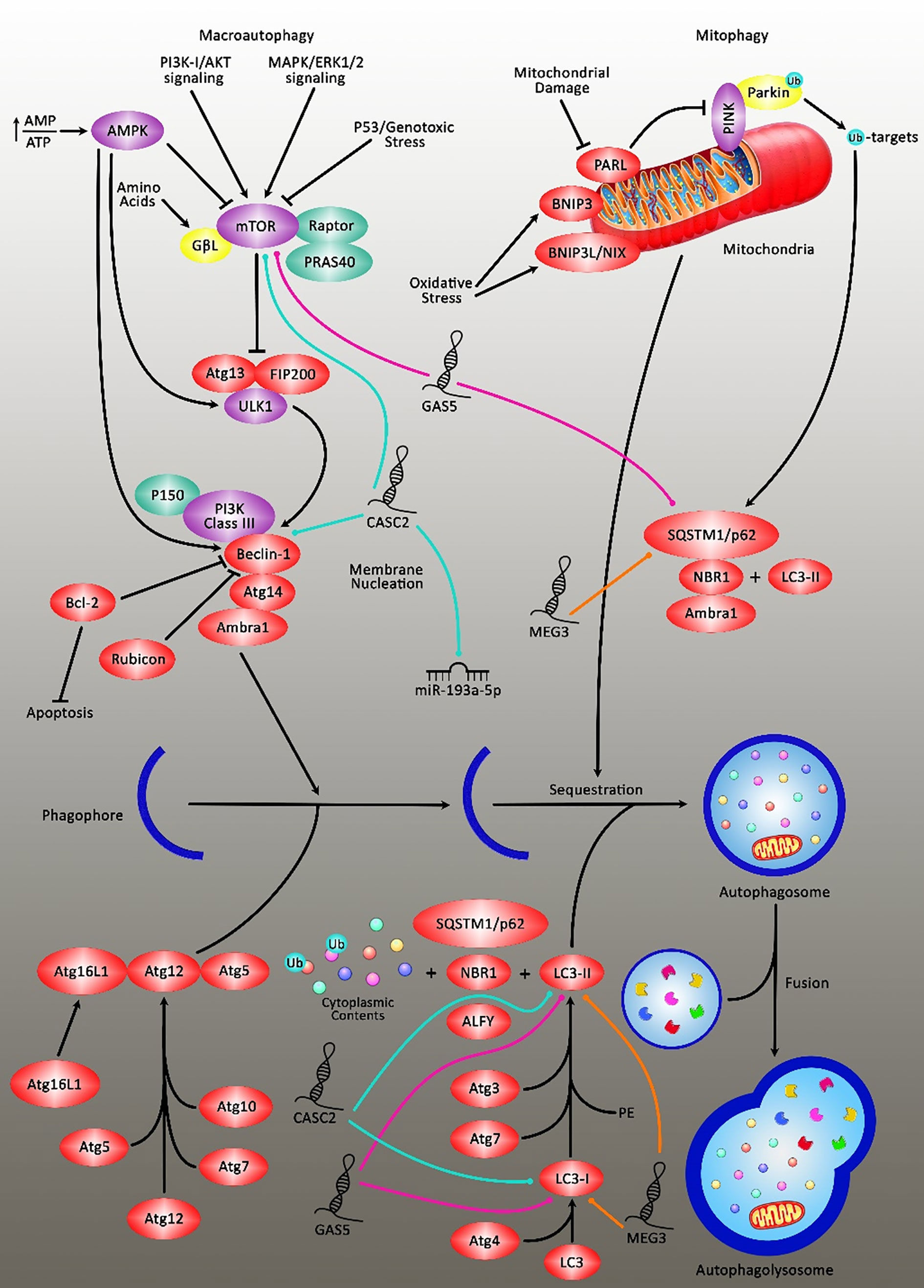
Figure 2 A schematic diagram of the inhibition of autophagy cascade via long noncoding RNAs in glioblastoma in an mTOR‐dependent manner. Overexpression of lncRNA CASC2 could downregulate the expression level of miR-193a-5p, which could, in turn, lead to reducing temozolomide-induced autophagy and promoting cell death through suppressing the expression level of mTOR, and thereby resulting in enhancing the sensitivity of glioma cells to temozolomide cytotoxicity to the large extent (55). Furthermore, the elevation of lncRNA GAS5 could enhance glioma cell sensitivity to cisplatin. Cisplatin could evoke excessive autophagy concomitant via promoting and suppressing the expression levels of LC3II and p62 respectively, which was negatively inhibited after GAS5 overexpression. Therefore, GAS5 could attenuate the resistance of glioma cells to cisplatin by restraining excessive autophagy through the activation of mTOR signaling (54). Also, upregulation of MEG3 could eliminate cisplatin-induced autophagy in glioma cells via directly targeting LC3II and p62 in tumor cells. The suppression of autophagy or knockdown of ATG5 could reverse the reduction in cell apoptosis caused by MEG3 knockdown in glioma cells treated with cisplatin (30).
LncRNAs and Chemoresistance in Neuroblastoma
Wang et al. have demonstrated up-regulation of NORAD in neuroblastoma tissues and cell lines. Notably, NORAD expression has been inversely correlated with the survival of patients. NORAD has increased proliferation, metastatic ability, and resistance to doxorubicin while inhibiting apoptosis and autophagy in neuroblastoma cells through targeting miR-144-3p. HDAC8 has been identified as a direct target of miR-144-3p. NORAD up-regulation increases HDAC8 levels through suppression of miR-144-5p (53). SNHG7 is another lncRNA that modulates cisplatin-induced autophagy by regulating the miR-329-3p/MYO10 (62). Finally, SNHG16 regulates miR-338-3p/PLK4 axis to enhance cisplatin resistance in these cells (63). Table 2 shows lncRNAs that modulate the response of neuroblastoma to chemotherapy.
Discussion
LncRNAs have acknowledged roles in the pathogenesis of tumors of the nervous system through various mechanisms including suppression of apoptotic pathways, induction of cell cycle progression, and enhancement of cell proliferation (10). A more clinically important aspect of lncRNA participation in the pathogenesis of nervous system tumors is their influence on the response of these neoplastic cells to chemotherapeutic agents. TMZ, cisplatin, and doxorubicin are the most important chemotherapeutic agents that are influenced by lncRNAs. Cancer stem cells are possibly the most critical cell population within the tumors which are affected by lncRNAs in this context. The competing endogenous RNA (ceRNA) function of lncRNAs has endowed them the aptitude to sequester miRNA, thus enhancing the expression of miRNA targets. MALAT1/miR-101, MALAT1/miR-203, TUSC7/miR-10a, NEAT1/let-7g-5p, AC003092.1/miR-195, SNHG15/miR-627, HOTAIR/miR-519a-3p, HOTAIR/miR-125, KCNQ1OT1/miR-761, NCK1-AS1/miR-137, NCK1-AS1/miR-22-3p and HOXD-AS1/miR-204 are among lncRNA/miRNA pairs that regulate resistance to chemotherapeutic agents in glioma/glioblastoma. SNHG7/miR-329-3p and SNHG16/miR338-3p have similar roles in neuroblastoma.
Based on the prominent effects of lncRNAs in the modulation of response of tumors of the nervous system to chemotherapeutic agents, prior knowledge about the levels of these transcripts in the tumor tissues would help in the design of appropriate therapeutic regimens. However, the particular locations of these tumors preclude invasive sampling. Therefore, peripheral blood/serum is an alternative tissue for this purpose. Consistent with this speculation, elevated serum levels of MALAT1 have been associated with the poor response of patients with glioblastoma to TMZ (17). However, most studies in this field rely on cell line experiments or animal studies without assessing the impact of circulating levels of lncRNAs in the long-term survival of patients. Moreover, the impact of genomic variants within lncRNAs in the modulation of response of glioblastoma/neuroblastoma to chemotherapeutic agents has not been assessed either in cell lines or in clinical settings. Such data would facilitate understanding the underlying mechanism of resistance to these agents and subsequently would pave the way for the design of therapeutic options to combat this phenotype.
Taken together, the contribution of lncRNAs in chemoresistance of glioma and neuroblastoma tumors has been assessed in independent studies. Yet, the role of these transcripts in the modulation of resistance to these agents has not been evaluated in other types of nervous system tumors. The proposed lncRNAs in this study are putative candidates for expression assays in other types of nervous system tumors.
Author Contributions
SG-F and MT wrote the draft and revised it. HS, GS, AAg, AAb, and MMHT collected the data, designed the tables, and figures. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
2. Reilly KM. Brain Tumor Susceptibility: The Role of Genetic Factors and Uses of Mouse Models to Unravel Risk. Brain Pathol (2009) 19(1):121–31. doi: 10.1111/j.1750-3639.2008.00236.x
3. Sarkaria JN, Kitange GJ, James CD, Plummer R, Calvert H, Weller M, et al. Mechanisms of Chemoresistance to Alkylating Agents in Malignant Glioma. Clin Cancer Res (2008) 14(10):2900–8. doi: 10.1158/1078-0432.CCR-07-1719
4. Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, et al. Radiotherapy Plus Concomitant and Adjuvant Temozolomide for Glioblastoma. New Engl J Med (2005) 352(10):987–96. doi: 10.1056/NEJMoa043330
5. Lee SY. Temozolomide Resistance in Glioblastoma Multiforme. Genes Dis (2016) 3(3):198–210. doi: 10.1016/j.gendis.2016.04.007
6. Hangauer MJ, Vaughn IW, McManus MT. Pervasive Transcription of the Human Genome Produces Thousands of Previously Unidentified Long Intergenic Noncoding RNAs. PloS Genet (2013) 9(6):e1003569. doi: 10.1371/journal.pgen.1003569
7. Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, et al. Landscape of Transcription in Human Cells. Nature (2012) 489(7414):101–8. doi: 10.1038/nature11233
8. Bernstein B, Birney E, Dunham I, Green E, Gunter C, Snyder M. Consortium EP. An Integrated Encyclopedia of DNA Elements in the Human Genome. Nature (2012) 489(7414):57–74. doi: 10.1038/nature11247
9. Fang Y, Fullwood MJ. Roles, Functions, and Mechanisms of Long non-Coding RNAs in Cancer. Genomics Proteomics Bioinf (2016) 14(1):42–54. doi: 10.1016/j.gpb.2015.09.006
10. Rezaei O, Tamizkar KH, Sharifi G, Taheri M, Ghafouri-Fard S. Emerging Role of Long Non-Coding RNAs in the Pathobiology of Glioblastoma. Front Oncol (2021) 10:3381. doi: 10.3389/fonc.2020.625884
11. Jia L, Tian Y, Chen Y, Zhang G. The Silencing of LncRNA-H19 Decreases Chemoresistance of Human Glioma Cells to Temozolomide by Suppressing Epithelial-Mesenchymal Transition Via the Wnt/β-Catenin Pathway. OncoTargets Ther (2018) 11:313. doi: 10.2147/OTT.S154339
12. Han M, Wang S, Fritah S, Wang X, Zhou W, Yang N, et al. Interfering With Long Non-Coding RNA MIR22HG Processing Inhibits Glioblastoma Progression Through Suppression of Wnt/β-Catenin Signalling. Brain (2020) 143(2):512–30. doi: 10.1093/brain/awz406
13. Lin Y, Pan X, Chen Z, Lin S, Chen S. Identification of an Immune-Related Nine-lncRNA Signature Predictive of Overall Survival in Colon Cancer. Front Genet (2020) 11:318. doi: 10.3389/fgene.2020.00318
14. Pai SG, Carneiro BA, Mota JM, Costa R, Leite CA, Barroso-Sousa R, et al. Wnt/Beta-Catenin Pathway: Modulating Anticancer Immune Response. J Hematol Oncol (2017) 10(1):1–12. doi: 10.1186/s13045-017-0471-6
15. Li H, Yuan X, Yan D, Li D, Guan F, Dong Y, et al. Long Non-Coding RNA MALAT1 Decreases the Sensitivity of Resistant Glioblastoma Cell Lines to Temozolomide. Cell Physiol Biochem (2017) 42(3):1192–201. doi: 10.1159/000478917
16. Vassallo I, Zinn P, Lai M, Rajakannu P, Hamou M, Hegi MJO. WIF1 Re-Expression in Glioblastoma Inhibits Migration Through Attenuation of Non-Canonical WNT Signaling by Downregulating the lncRNA Malat1. (2016) 35(1):12–21. doi: 10.1038/onc.2015.61
17. Chen W, Xu X-K, Li J-L, Kong K-K, Li H, Chen C, et al. MALAT1 is a Prognostic Factor in Glioblastoma Multiforme and Induces Chemoresistance to Temozolomide Through Suppressing miR-203 and Promoting Thymidylate Synthase Expression. Oncotarget (2017) 8(14):22783. doi: 10.18632/oncotarget.15199
18. Cai T, Liu Y, Xiao J. Long Noncoding RNA MALAT 1 Knockdown Reverses Chemoresistance to Temozolomide Via Promoting Micro RNA-101 in Glioblastoma. Cancer Med (2018) 7(4):1404–15. doi: 10.1002/cam4.1384
19. Voce DJ, Bernal GM, Wu L, Crawley CD, Zhang W, Mansour NM, et al. Temozolomide Treatment Induces Lncrna MALAT1 in an NF-κb and p53 Codependent Manner in Glioblastoma. Cancer Res (2019) 79(10):2536–48. doi: 10.1158/0008-5472.CAN-18-2170
20. Mohebi M, Ghafouri-Fard S, Modarressi MH, Dashti S, Zekri A, Kholghi-Oskooei V, et al. Expression Analysis of Vimentin and the Related lncRNA Network in Breast Cancer. Exp Mol Pathol (2020) 115:104439. doi: 10.1016/j.yexmp.2020.104439
21. Xu K-l, Xu Q-s, Wu Z-x, Xu S-j, Shen J. Lncrna NEAT1 is Involved in Temozolomide Resistance by Regulating MGMT in Glioblastoma Multiforme. Clin Surg Res Commun (2018) 2(1):24–30. doi: 10.31491/CSRC.2018.3.011
22. Fazi B, Garbo S, Toschi N, Mangiola A, Lombari M, Sicari D, et al. The Lncrna H19 Positively Affects the Tumorigenic Properties of Glioblastoma Cells and Contributes to NKD1 Repression Through the Recruitment of EZH2 on Its Promoter. Oncotarget (2018) 9(21):15512. doi: 10.18632/oncotarget.24496
23. Duan S, Li M, Wang Z, Wang L, Liu Y. H19 Induced by Oxidative Stress Confers Temozolomide Resistance in Human Glioma Cells Via Activating NF-κb Signaling. OncoTargets Ther (2018) 11:6395. doi: 10.2147/OTT.S173244
24. Yuan Z, Yang Z, Li W, Wu A, Su Z, Jiang B. Exosome-Mediated Transfer of Long Noncoding RNA HOTAIR Regulates Temozolomide Resistance by miR-519a-3p/RRM1 Axis in Glioblastoma. Cancer Biotherapy Radiopharmaceuticals (2020). doi: 10.1089/cbr.2019.3499
25. Zhang J, Chen G, Gao Y, Liang H. Hotair/miR-125 Axis-Mediated Hexokinase 2 Expression Promotes Chemoresistance in Human Glioblastoma. J Cell Mol Med (2020) 24(10):5707–17. doi: 10.1111/jcmm.15233
26. Green DR, Levine B. To be or Not to be? How Selective Autophagy and Cell Death Govern Cell Fate. Cell (2014) 157(1):65–75. doi: 10.1016/j.cell.2014.02.049
27. Janku F, McConkey DJ, Hong DS, Kurzrock R. Autophagy as a Target for Anticancer Therapy. Nat Rev Clin Oncol (2011) 8(9):528. doi: 10.1038/nrclinonc.2011.71
28. Li X, Zhou Y, Li Y, Yang L, Ma Y, Peng X, et al. Autophagy: A Novel Mechanism of Chemoresistance in Cancers. Biomed Pharmacotherapy (2019) 119:109415. doi: 10.1016/j.biopha.2019.109415
29. Shang C, Tang W, Pan C, Hu X, Hong Y. Long Non-Coding RNA TUSC7 Inhibits Temozolomide Resistance by Targeting miR-10a in Glioblastoma. Cancer Chemotherapy Pharmacol (2018) 81(4):671–8. doi: 10.1007/s00280-018-3522-y
30. Ma B, Gao Z, Lou J, Zhang H, Yuan Z, Wu Q, et al. Long non−Coding RNA MEG3 Contributes to Cisplatin−Induced Apoptosis Via Inhibition of Autophagy in Human Glioma Cells. Mol Med Rep (2017) 16(3):2946–52. doi: 10.3892/mmr.2017.6897
31. Zeng H, Xu N, Liu Y, Liu B, Yang Z, Fu Z, et al. Genomic Profiling of Long Non-Coding RNA and mRNA Expression Associated With Acquired Temozolomide Resistance in Glioblastoma Cells. Int J Oncol (2017) 51(2):445–55. doi: 10.3892/ijo.2017.4033
32. Liu Y, Xu N, Liu B, Huang Y, Zeng H, Yang Z, et al. Long Noncoding RNA Rp11-838N2.4 Enhances the Cytotoxic Effects of Temozolomide by Inhibiting the Functions of miR-10a in Glioblastoma Cell Lines. Oncotarget (2016) 7(28):43835–51. doi: 10.18632/oncotarget.9699
33. Kim S-S, Harford JB, Moghe M, Rait A, Pirollo KF, Chang E. Targeted Nanocomplex Carrying siRNA Against MALAT1 Sensitizes Glioblastoma to Temozolomide. Nucleic Acids Res (2018) 46(3):1424–40. doi: 10.1093/nar/gkx1221
34. Jiang P, Wang P, Sun X, Yuan Z, Zhan R, Ma X, et al. Knockdown of Long Noncoding RNA H19 Sensitizes Human Glioma Cells to Temozolomide Therapy. OncoTargets Ther (2016) 9:3501. doi: 10.2147/OTT.S96278
35. He Z, You C, Zhao DJB. Communications BrLong Non-Coding RNA Uca1/miR-182/PFKFB2 Axis Modulates Glioblastoma-Associated Stromal Cells-Mediated Glycolysis and Invasion of Glioma Cells. Biochem Biophys Res Commun (2018) 500(3):569–76. doi: 10.1016/j.bbrc.2018.04.091
36. Ma B, Yuan Z, Zhang L, Lv P, Yang T, Gao J, et al. Long non-Coding RNA AC023115. 3 Suppresses Chemoresistance of Glioblastoma by Reducing Autophagy. Biochim Biophys Acta Mol Cell Res (2017) 1864(8):1393–404. doi: 10.1016/j.bbamcr.2017.05.008
37. Xu N, Liu B, Lian C, Doycheva DM, Fu Z, Liu Y, et al. Long Noncoding RNA AC003092. 1 Promotes Temozolomide Chemosensitivity Through miR-195/TFPI-2 Signaling Modulation in Glioblastoma. Cell Death Dis (2018) 9(12):1–16. doi: 10.1038/s41419-018-1183-8
38. Mazor G, Levin L, Picard D, Ahmadov U, Carén H, Borkhardt A, et al. The Lncrna TP73-AS1 Is Linked to Aggressiveness in Glioblastoma and Promotes Temozolomide Resistance in Glioblastoma Cancer Stem Cells. Cell Death Dis (2019) 10(3):1–14. doi: 10.1038/s41419-019-1477-5
39. Yan Y, Xu Z, Chen X, Wang X, Zeng S, Zhao Z, et al. Novel Function of Lncrna ADAMTS9-AS2 in Promoting Temozolomide Resistance in Glioblastoma Via Upregulating the FUS/MDM2 Ubiquitination Axis. Front Cell Dev Biol (2019) 7:217. doi: 10.3389/fcell.2019.00217
40. Li Z, Zhang J, Zheng H, Li C, Xiong J, Wang W, et al. Modulating Lncrna SNHG15/CDK6/miR-627 Circuit by Palbociclib, Overcomes Temozolomide Resistance and Reduces M2-Polarization of Glioma Associated Microglia in Glioblastoma Multiforme. J Exp Clin Cancer Res (2019) 38(1):1–13. doi: 10.1186/s13046-019-1371-0
41. Lu C, Wei Y, Wang X, Zhang Z, Yin J, Li W, et al. DNA-Methylation-Mediated Activating of Lncrna SNHG12 Promotes Temozolomide Resistance in Glioblastoma. Mol Cancer (2020) 19(1):28. doi: 10.1186/s12943-020-1137-5
42. Zhao C, Gao Y, Guo R, Li H, Yang B. Microarray Expression Profiles and Bioinformatics Analysis of mRNAs, lncRNAs, and circRNAs in the Secondary Temozolomide-Resistant Glioblastoma. Invest New Drugs (2019) 10:1–9. doi: 10.1007/s10637-019-00884-3
43. Zhang Z, Yin J, Lu C, Wei Y, Zeng A, You Y. Exosomal Transfer of Long non-Coding RNA Sbf2-AS1 Enhances Chemoresistance to Temozolomide in Glioblastoma. J Exp Clin Cancer Res (2019) 38(1):1–16. doi: 10.1186/s13046-019-1139-6
44. Towner RA, Smith N, Saunders D, Brown CA, Cai X, Ziegler J, et al. Okn-007 Increases Temozolomide (TMZ) Sensitivity and Suppresses TMZ-resistant Glioblastoma (GBM) Tumor Growth. Trans Oncol (2019) 12(2):320–35. doi: 10.1016/j.tranon.2018.10.002
45. Liu B, Zhou J, Wang C, Chi Y, Wei Q, Fu Z, et al. Lncrna SOX2OT Promotes Temozolomide Resistance by Elevating SOX2 Expression Via Mediated Epigenetic Regulation in Glioblastoma. Cell Death Dis (2020) 11(5):1–18. doi: 10.1038/s41419-020-2540-y
46. Zhang S, Guo S, Liang C, Lian M. Long Intergenic Noncoding RNA 00021 Promotes Glioblastoma Temozolomide Resistance by Epigenetically Silencing p21 Through Notch Pathway. IUBMB Life (2020) 72(8):1747–56. doi: 10.1002/iub.2301
47. Wang W, Han S, Gao W, Feng Y, Li K, Wu D. Long Noncoding Rna KCNQ1OT1 Confers Gliomas Resistance to Temozolomide and Enhances Cell Growth by Retrieving Pim1 From Mir-761. Cell Mol Neurobiol (2020) 1–14. doi: 10.1007/s10571-020-00958-4
48. Chen M, Cheng Y, Yuan Z, Wang F, Yang L, Zhao H. Nck1-As1 Increases Drug Resistance of Glioma Cells to Temozolomide by Modulating Mir-137/TRIM24. Cancer Biother Radiopharm (2020) 35(2):101–8. doi: 10.1089/cbr.2019.3054
49. Wang B, Wang K, Jin T, Xu Q, He Y, Cui B, et al. Nck1-AS1 Enhances Glioma Cell Proliferation, Radioresistance and Chemoresistance Via miR-22-3p/IGF1R ceRNA Pathway. Biomed Pharmacotherapy (2020) 129:110395. doi: 10.1016/j.biopha.2020.110395
50. Wang J, Yang S, Ji Q, Li Q, Zhou F, Li Y, et al. Long Non-Coding RNA EPIC1 Promotes Cell Proliferation and Motility and Drug Resistance in Glioma. Mol Therapy-Oncolytics (2020) 17:130–7. doi: 10.1016/j.omto.2020.03.011
51. Zhou H, Ma Y, Zhong D, Yang L. Knockdown of Lncrna HOXD-AS1 Suppresses Proliferation, Migration and Invasion and Enhances Cisplatin Sensitivity of Glioma Cells by Sponging Mir-204. Biomed Pharmacotherapy (2019) 112:108633. doi: 10.1016/j.biopha.2019.108633
52. Chen W-L, Chen H-J, Hou G-Q, Zhang X-H, Ge J-W. LINC01198 Promotes Proliferation and Temozolomide Resistance in a NEDD4-1-Dependent Manner, Repressing PTEN Expression in Glioma. Aging (Albany NY) (2019) 11(16):6053. doi: 10.18632/aging.102162
53. Li B, Zhao H, Song J, Wang F, Chen M. LINC00174 Down-Regulation Decreases Chemoresistance to Temozolomide in Human Glioma Cells by Regulating miR-138-5p/SOX9 Axis. Hum Cell (2020) 33(1):159–74. doi: 10.1007/s13577-019-00281-1
54. Huo JF, Chen XB. Long Noncoding RNA Growth Arrest-Specific 5 Facilitates Glioma Cell Sensitivity to Cisplatin by Suppressing Excessive Autophagy in an mTOR-Dependent Manner. J Cell Biochem (2019) 120(4):6127–36. doi: 10.1002/jcb.27900
55. Jiang C, Shen F, Du J, Fang X, Li X, Su J, et al. Upregulation of CASC2 Sensitized Glioma to Temozolomide Cytotoxicity Through Autophagy Inhibition by Sponging miR-193a-5p and Regulating mTOR Expression. Biomed Pharmacotherapy (2018) 97:844–50. doi: 10.1016/j.biopha.2017.10.146
56. Liao Y, Shen L, Zhao H, Liu Q, Fu J, Guo Y, et al. Lncrna CASC2 Interacts With miR-181a to Modulate Glioma Growth and Resistance to TMZ Through PTEN Pathway. J Cell Biochem (2017) 118(7):1889–99. doi: 10.1002/jcb.25910
57. Ding J, Zhang L, Chen S, Cao H, Xu C, Wang X. Lncrna CCAT2 Enhanced Resistance of Glioma Cells Against Chemodrugs by Disturbing the Normal Function of Mir-424. OncoTargets Ther (2020) 13:1431. doi: 10.2147/OTT.S227831
58. Ma Y, Zhou G, Li M, Hu D, Zhang L, Liu P, et al. Long Noncoding RNA DANCR Mediates Cisplatin Resistance in Glioma Cells Via Activating AXL/PI3K/Akt/NF-κb Signaling Pathway. Neurochem Int (2018) 118:233–41. doi: 10.1016/j.neuint.2018.03.011
59. Yang Q, Deng L, Li J, Miao P, Liu W, Huang Q. Nr5a2 Promotes Cell Growth and Resistance to Temozolomide Through Regulating Notch Signal Pathway in Glioma. OncoTargets Ther (2020) 13:10231. doi: 10.2147/OTT.S243833
60. Yang G, Han B, Feng T. ZFAS1 Knockdown Inhibits Viability and Enhances Cisplatin Cytotoxicity by Up-Regulating miR-432-5p in Glioma Cells. Basic Clin Pharmacol Toxicol (2019) 125(6):518–26. doi: 10.1111/bcpt.13286
61. Du P, Zhao H, Peng R, Liu Q, Yuan J, Peng G, et al. LncRNA-XIST Interacts With miR-29c to Modulate the Chemoresistance of Glioma Cell to TMZ Through DNA Mismatch Repair Pathway. Bioscience Rep (2017) 37(5):BSR20170696. doi: 10.1042/BSR20170696
62. Wang SY, Wang X, Zhang CY. LncRNA SNHG7 Enhances Chemoresistance in Neuroblastoma Through Cisplatin-Induced Autophagy by Regulating miR-329-3p/MYO10 Axis. Eur Rev Med Pharmacol Sci (2020) 24(7):3805–17. doi: 10.26355/eurrev_202004_20847
Keywords: long non-coding RNA, lncRNA, chemoresistance, brain tumor, expression
Citation: Ghafouri-Fard S, Agabalazadeh A, Abak A, Shoorei H, Hassanzadeh Taheri MM, Taheri M and Sharifi G (2021) Role of Long Non-Coding RNAs in Conferring Resistance in Tumors of the Nervous System. Front. Oncol. 11:670917. doi: 10.3389/fonc.2021.670917
Received: 22 February 2021; Accepted: 12 May 2021;
Published: 09 June 2021.
Edited by:
Palmiro Poltronieri, Italian National Research Council, ItalyReviewed by:
Ilaria Genovese, University of Ferrara, ItalyJuliano Andreoli Miyake, Federal University of Santa Catarina, Brazil
Copyright © 2021 Ghafouri-Fard, Agabalazadeh, Abak, Shoorei, Hassanzadeh Taheri, Taheri and Sharifi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mohammad Taheri, bW9oYW1tYWRfODIzQHlhaG9vLmNvbQ==; Guive Sharifi, Z2libm93QHlhaG9vLmNvbQ==
 Soudeh Ghafouri-Fard
Soudeh Ghafouri-Fard Amin Agabalazadeh
Amin Agabalazadeh Atefe Abak
Atefe Abak Hamed Shoorei
Hamed Shoorei Mohammad Mehdi Hassanzadeh Taheri4
Mohammad Mehdi Hassanzadeh Taheri4 Mohammad Taheri
Mohammad Taheri Guive Sharifi
Guive Sharifi