- 1Department of Orthopedic Surgery, The Second Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, China
- 2Department of Oncology Surgery, The Second Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, China
Osteosarcoma is the most common primary bone malignancy, typically occurring in childhood or adolescence. Unfortunately, the clinical outcomes of patients with osteosarcoma are usually poor because of the aggressive nature of this disease and few treatment advances in the past four decades. N6-methyladenosine (m6A) is one of the most extensive forms of RNA modification in eukaryotes found both in coding and non-coding RNAs. Accumulating evidence suggests that m6A-related factors are dysregulated in multiple osteosarcoma processes. In this review, we highlight m6A modification implicated in osteosarcoma, describing its pathophysiological role and molecular mechanism, as well as future research trends and potential clinical application in osteosarcoma.
Introduction
Osteosarcoma is a relatively rare bone malignancy that predominantly occurs in children and adolescents (1, 2). It is highly aggressive and difficult-to-treat (3, 4). Although the introduction of chemotherapy since the 1970s has increased the 5-year survival rate of patients with non-metastatic osteosarcoma to 70%, the 5-year survival rate of patients with metastatic osteosarcoma is only 20% (5, 6). More importantly, metastasis of osteosarcoma is not uncommon (7). Osteosarcoma is known to exhibit high heterogeneity and significant genome complexity (8–10); thus, tremendous efforts are needed to define the biology of osteosarcoma for developing new therapeutic alternatives.
The discovery of heritable alterations of chromatin structure and DNA modifications that do not change their DNA or RNA sequence itself pioneers a new field of epigenetics (11, 12). N6-methyladenosine (m6A) modification refers to the addition or deletion of the methyl group to/from the nitrogen on the 6th carbon of the adenine nucleotide, which is one of the most abundant epigenetic modifications found in RNA molecules (13). First discovered by Desrosiers in the 1970s, m6A modification has been found to be involved in almost all steps of RNA processing and metabolism, including pre-mRNA splicing, export, translation, stabilization, and degradation (14–19). With the burgeoning advances in molecular biology and sequencing, m6A modification has been reported to be implicated in virtually all cellular functions and multiple diseases (including osteosarcoma) (20–23). In this review, we intended to summarize the current advances in the pathophysiological roles and molecular mechanism of m6A modification in osteosarcoma and related diseases, and discuss the potential clinical application of m6A modification in osteosarcoma.
m6A Regulation
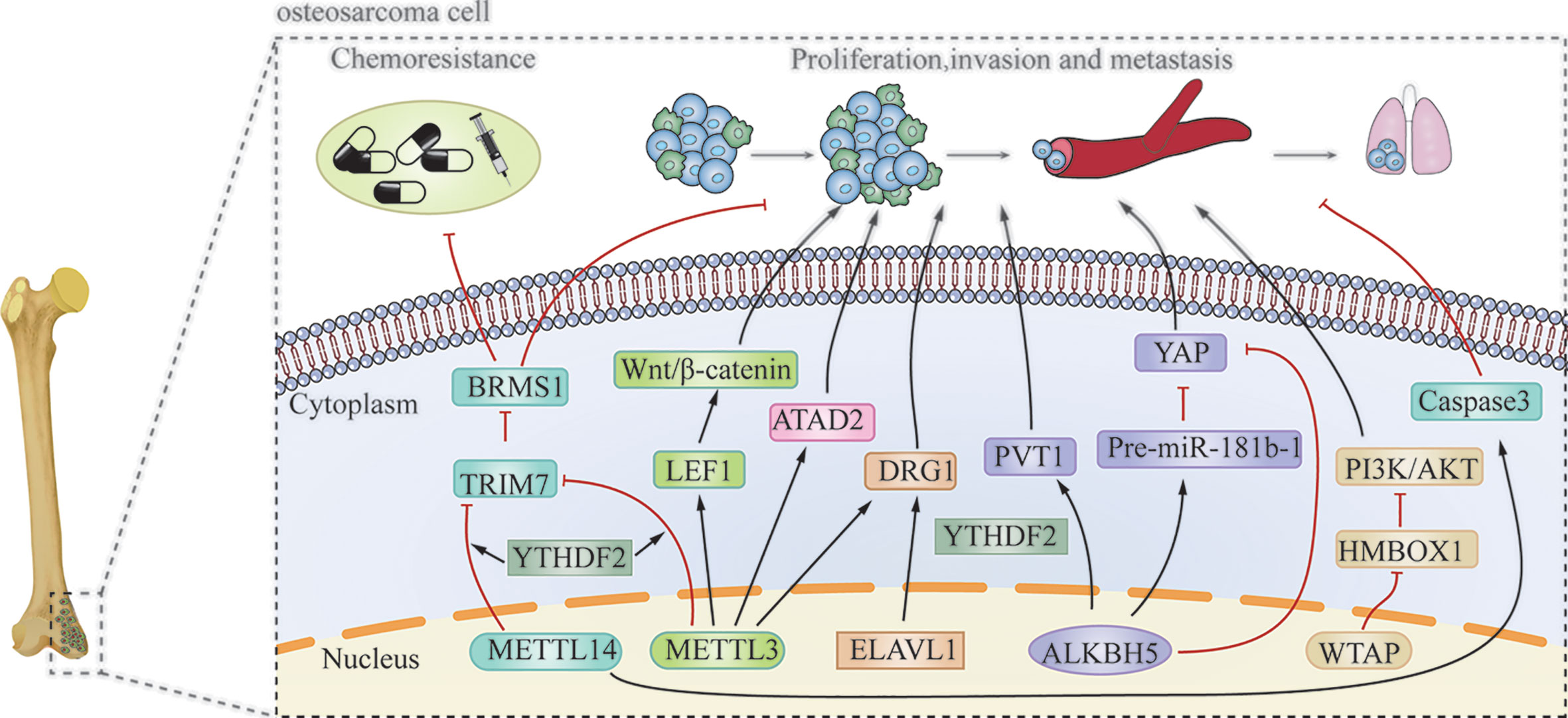
Similar to DNA and histone methylation, m6A modification is a dynamic and reversible process that is regulated by methylases and demethylases (Figure 1) (24, 25).
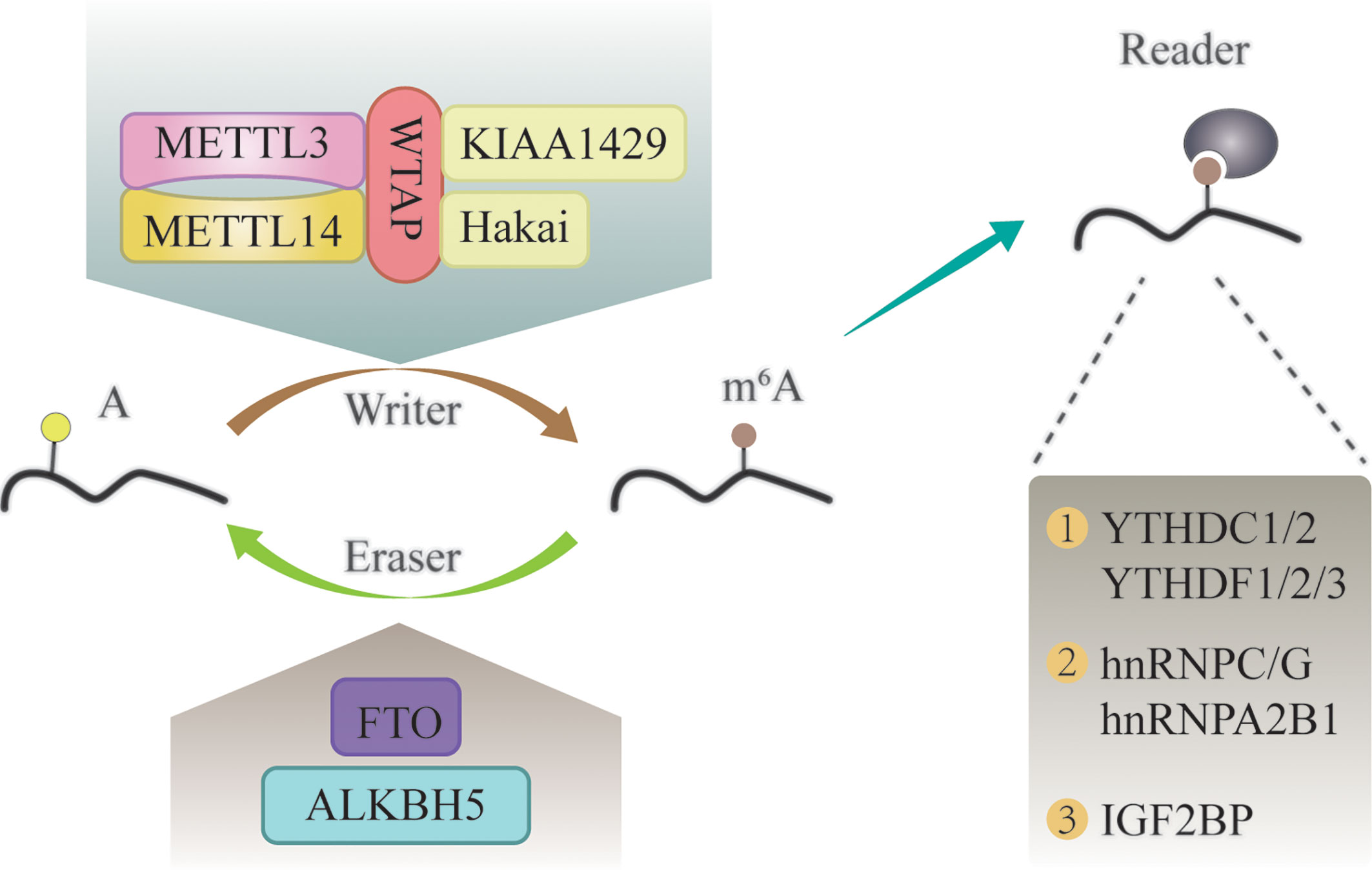
Figure 1 The molecular mechanism of m6A modification. It is a dynamic and reversible epigenetic modification that is regulated by “writers” and “erasers.” m6A markers in the RNA can be recognized by “readers”.
M6A methylases, also called “writers”, can transfer a methyl group to the N-6 position of adenosine in the nucleic acid (26). It commonly works in the form of a multicomponent m6A methyltransferase complex (MTC) (27). Methyltransferase-like 3 (METTL3) has long been considered as the central catalytic subunit of MTC, while other components have recently been discovered (28). METTL14 is an allosteric adaptor of METTL3 that can stabilize METTL3 and recognize the substrate (29). In addition, Wilms tumor 1-associated protein (WTAP), Vir-like m6A methyltransferase associated (KIAA1429), and Cbl proto-oncogene-like 1 (Hakai) participate in the formation of MTC and play a vital role in m6A modification (30–32).
M6A demethylases, also named “erasers”, are proteins that remove the methyl groups from RNA; thus, conferring a reversible and dynamic nature to the regulation of m6A methylation (33). These enzymes mainly include fat mass and obesity-associated protein (FTO), and α-ketoglutarate-dependent dioxygenase alk B homolog 5 (ALKBH5) (34, 35). FTO was the first identified m6A demethylase (36). Since then, research on m6A modification has gained attention. FTO can demethylate m6A into N6-hydroxymethyladeosine (hm6A), which is further converted to N6-formyladenosine (f6A) before being hydrolyzed to the stable form adenine (37). The writers and erasers maintain the dynamic balance of m6A in the transcriptome, which is essential for normal physiological processes.
In addition to writers and erasers, “readers” are binding proteins that recognize the chemical signatures that are important for the regulation of m6A modification (38). The YTH domain-containing proteins are the first discovered readers, which interact with m6A through a “tryptophan cage” (39). The family of YTH domain-containing proteins consist of the following five proteins: YTHDC1, YTHDC2, YTHDF1, YTHDF2, and YTHDF3 (40). M6A can alter the structure of RNA substrates, and thereby, promote the binding of RNA-binding proteins (RBPs) to substrates (41). These RBPs are called “m6A structural switch” readers, and include heterogeneous nuclear ribonucleoprotein C (HNRNPC), HNRNPG, and HNRNPA2B1 (25, 41, 42). Insulin-like growth factor 2 mRNA-binding (IGF2BP) proteins (IGF2BP1, IGF2BP2, and IGF2BP3) are another category of m6A readers, which can stabilize RNA by interacting with YTHDF proteins or in an m6A structural switch manner (43, 44).
M6A can affect the whole process of gene expression, including transcription, post-transcription, translation, and post-translation (45). In the “life cycle” of m6A mRNA (Figure 2) (46, 47), the m6A writers and erasers first regulate the change in m6A during transcription in the nucleus. Then, m6A binds to specific nuclear readers and affect mRNA splicing, exporting, and other bioprocesses (48, 49). After being exported to the cytoplasm, the cytoplasm readers bind to m6A and influence mRNA decay, translation, and stabilization (46, 50). Through these processes, m6A modification regulates gene expression and exerts profound and diverse functions in cell differentiation, immune response, and disease development, especially tumorigenesis, progression, and cancer metastasis (51–54).
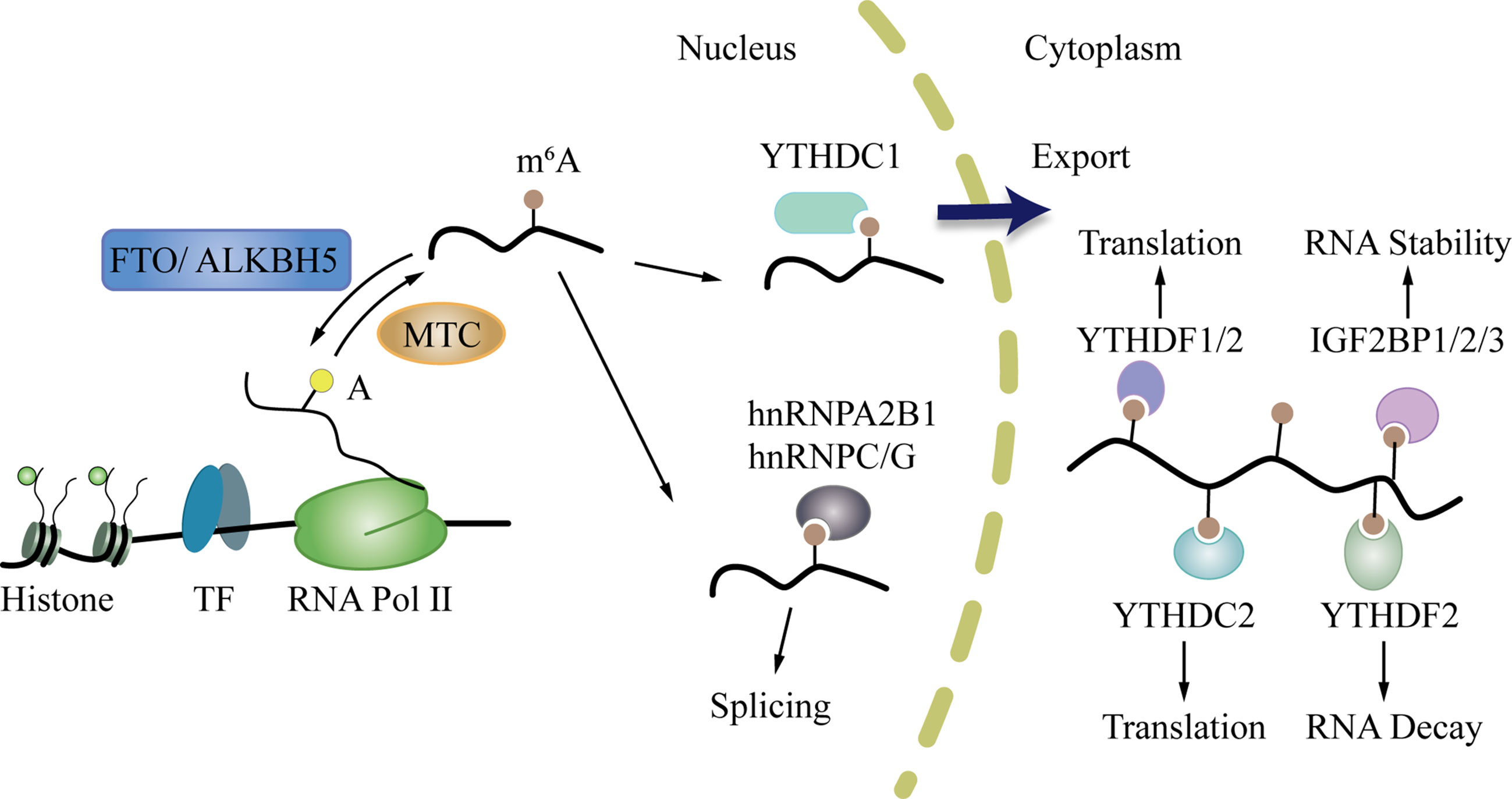
Figure 2 The life cycle of m6A mRNA. First, m6A writers and erasers regulate the change of m6A during transcription in the nucleus. After that, m6A can bind to specific nuclear readers, and influence mRNA splicing, exporting, and other bioprocesses. Then, m6A is exported to the cytoplasm where it binds to cytoplasm readers and influences mRNA decay, translation, and stabilization.
m6A and Bone Development
The bone is a complex connective tissue, which is always under a dynamic balance between bone formation mediated by osteoblasts and bone resorption regulated by osteoclasts (55). If the bone homeostasis is disturbed, many bone metabolic diseases such as osteosarcoma, osteoporosis, and osteoarthritis may occur (56). M6A is reported to participate in the regulation of bone homeostasis (57). METTL3 is significantly increased during the process of osteogenic differentiation of the bone marrow stem cells (BMSCs). Specifically, silencing METTL3 can suppress the osteogenic differentiation by directly and indirectly regulating RUNX2 (METTL3/m6A-pre-miR-320/miR-320-RUNX2 Axis) in BMSCs (58). Remarkably, the expression of METTL3 was also increased during osteoclast differentiation. It can regulate the bone resorption by controlling Atp6v0d2 mRNA degradation and Traf6 mRNA nuclear export (59). METTL14 was found to be positively associated with bone formation in older women with fractures and ovariectomized mice. It promotes osteoblast activity by regulating miR-103-3p processing via microprocessor protein DGCR8 (60). Similarly, the FTO, represented as the RNA demethylase, is also closely related to the fate of BMSCs. Li et al. (61) reported that FTO could interact with miR-149-3p and promote osteogenic differentiation of BMSCs.
m6A and Osteosarcoma
Osteosarcoma always occurs where the bones are growing the fasted (62). The inseparable relationship between m6A regulators and bone development implies that m6A modification might contribute to the progression of osteosarcoma. Herein, we comprehensively review the current research on the association of m6A and osteosarcoma.
Dysregulation of m6A Writers in Osteosarcoma
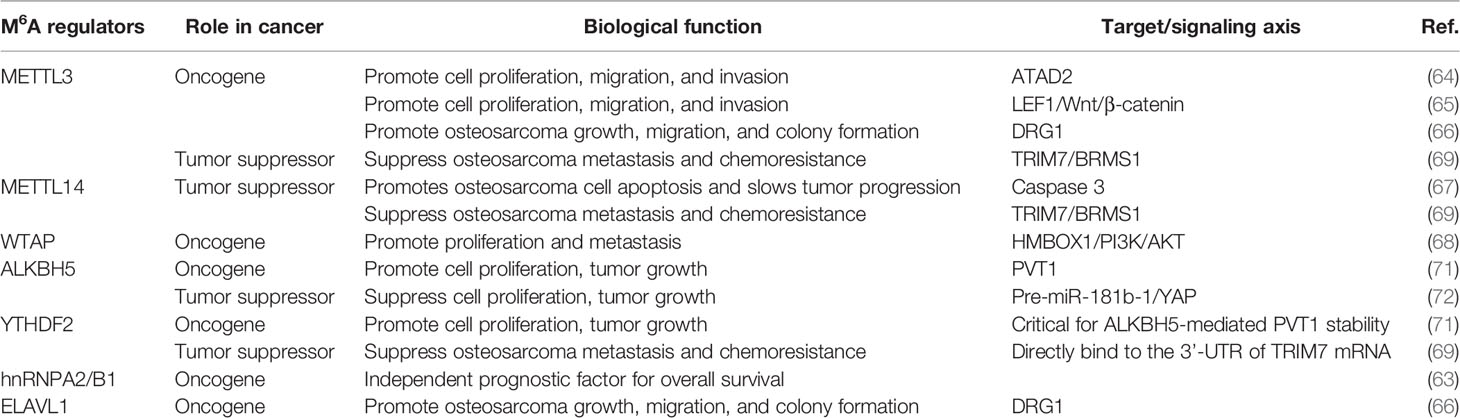
In osteosarcoma cells, m6A writers, including METTL3, METTL14, WTAP, and KIAA1429 are mostly present in the nucleus (63). Zhou et al. (64) revealed that METTL3 acts as an oncogene in osteosarcoma. Knockdown of METTL3 can suppress the proliferation, migration, and invasion of the human osteosarcoma cell lines SAOS-2 and MG63 by inhibiting the m6A methylation level and expression of the ATPase family AAA domain-containing protein 2 (ATAD2). Additionally, Miao et al. (65) found that METTL3 expression and m6A methylation levels were higher in human osteosarcoma tissues and osteosarcoma cells. METTL3 can promote osteosarcoma development by directly increasing the m6A methylation level and the expression of lymphoid enhancer-binding factor 1(LEF1), and by activating the Wnt/β-catenin signaling pathway. METTL3 has also been found to facilitate the methylation of GTP-binding protein (DRG) 1, and thereby, promoting osteosarcoma growth, migration, and colony formation (66). Conversely, METTL14 has an inhibitory effect in osteosarcoma. METTL14 overexpression promotes osteosarcoma cell apoptosis and slows tumor progression through caspase 3 activation (67). Recently, Chen et al. (68) demonstrated that another m6A writer, WTAP, can positively regulate osteosarcoma tumorigenesis and metastasis by reducing the stability of HMBOX1 in a m6A-dependent manner. Inhibiting PI3K/AKT pathways can partly reverse WTAP/HMBOX1- induced osteosarcoma progression.
METTL3/14 also plays an important role in modulating the chemoresistance of osteosarcoma. METTL3 and METTL14 decrease the RNA level of tripartite motif 7 (TRIM7), while the upregulation of TRIM7 can increase metastasis and the chemoresistance of osteosarcoma by regulating ubiquitination of breast cancer metastasis suppressor 1 (BRMS1) (69). The transcriptome-wide m6A sequencing result of chemoresistant osteosarcoma stem cells revealed that over-expression of METTL3 and low METTL14 expression are associated with doxorubicin chemoresistance and stemness of osteosarcoma cells (70). The regulatory mechanism of writer is shown in Table 1 and Figure 3.
Dysregulation of m6A Erasers in Osteosarcoma
The m6A erasers, FTO and ALKBH5, are distributed in both the nucleus and cytoplasm (63). The effect of ALKBH5 in osteosarcoma remains controversial. Chen et al. (71) revealed that ALKBH5 can promote osteosarcoma cell proliferation and tumor growth by decreasing the m6A modification of plasmacytoma variant translocation 1 (PVT1), subsequently impairing the binding of reader protein YTHDF2 in PVT1. However, Yuan et al. (72) showed that ALKBH5 can epigenetically silence pre-miR-181b-1/YAP signaling axis, and thus, suppress tumor progression in osteosarcoma. In addition, upregulation of ALKBH5 expression also contributes to chemoresistance and predicts worse metastasis-free survival in patients with osteosarcoma (70). The regulatory mechanism is shown in Table 1 and Figure 3.
Dysregulation of m6A Readers in Osteosarcoma
The subcellular location of m6A readers in osteosarcoma cells is relatively complicated. YTHDF1 and YTHDF2 are mainly distributed in the cytoplasm. YTHDC1 is mainly located in the nucleus, while YTHDC2 is uniformly distributed in the nucleus and cytoplasm. Additionally, HNRNPC and HNRNPA2B1 are found only in the nucleus (63). YTHDF2 can directly bind to the 3’-UTR of TRIM7 mRNA and negatively regulate the expression of TRIM7 in osteosarcoma HOS and MG63 cells (69). ELAVL1 (also known as HuR) is a recently discovered m6A reader, which is located in the nucleus. It has been reported to regulate the stability of DRG1 mRNA. Silencing ELAVL1 inhibits osteosarcoma progression by decreasing the expression of DRG1 in an m6A-dependent pattern (66). These processes have been summarized in Table 1 and Figure 3.
m6A and Osteosarcoma-Related Diseases
m6A and Bone Disease
As an essential component of epigenetic regulation, m6A modification also has inextricable link with other bone diseases, such as intervertebral disc degeneration (IDD), osteoarthritis, and osteoporosis (73). In IDD, the microarray results showed that most of the dysregulated RNA was in a demethylated state. FTO and ZFP217 can demethylate LOC102555094 and activate downstream Wnt pathway, which may contribute to metabolic reprogramming of glucose metabolism in the IDD process (74). METTL14 can accelerate TNF-α-induced nucleus pulposus cell cycle arrest and senescence via processing miR-34a-5p (75). In osteoarthritis, METTL3 is suggested to be responsible for the development of the disease by regulating NF-κB signaling and extracellular matrix synthesis in chondrocytes (76). As for osteoporosis, Mo et al. (77) revealed that m6A-associated single nucleotide polymorphisms (SNPs) can affect bone mineral density (BMI). Their GWAS result showed that 138, 125, and 993 m6A-SNPs were associated with femoral neck, lumbar spine, and quantitative heel ultrasounds BMI respectively. Besides, METTL3 is found to be downregulated in human osteoporosis and the ovariectomized mouse model (58). The up-regulation of METTL3 in MSCs can prevent estrogen deficiency-induced osteoporosis (78).
m6A and Other Cancer
Since the mechanism of m6A in osteosarcoma is not well understood, we presumed that the mechanism of m6A in other cancer may provide some clue in this regard. Based on our review, we principally summarize the following three point:
First, the dysregulation of erasers and readers has been more investigated in other cancers. In pancreatic cancer, ALKBH5 acts as a tumor suppressor by decreasing WIF-1 RNA methylation and suppressing the Wnt pathway (79). It can also prevent the progression of pancreatic cancer by regulating the posttranscriptional activation of PER1 in an m6A-YTHDF2-dependent manner (80). In acute myeloid leukemia (AML), YTHDF2 decreases m6A RNA stability and is crucial for AML initiation and propagation (81). In breast cancer, hnRNPA2B1 can directly bind to the UAGGG locus of PFN2 mRNA to reduce its stability, and thereby, inhibit the metastasis of breast cancer (82). In light of the significant role of m6A erasers and readers in cancers, we look forward to more investigation about dysregulation of erasers and readers in osteosarcoma.
Second, m6A modification is associated with the molecular epidemiology and clinicopathology of cancer. Zeng et al. (83) performed a case-control study based on Chinese population, which showed that different SNPs of FTO were concerned with varying risk of breast cancer. For instance, presence of rs1477196, rs16953002, and TAC haplotype (rs9939609-rs1477196-rs1121980) in the FTO gene is associated with a high risk of breast cancer. Wu et al. (84) revealed that overexpression of METTL3, METTL14, FTO, and ALKBH5, and under-expression WTAP was closely related with luminal type breast cancer, while the expression level of FTO was significantly decreased in human epidermal growth factor receptor 2 (HER2) positive breast cancer. Xiao et al. (85) illustrated that detection of m6A combined with METTL14 and FTO expression in peripheral blood can diagnose breast cancer with a specificity of 97.4%.
Third, recent studies have found that the tumor microenvironment can induce the dysregulation of m6A regulators (86–88). For example, hypoxia can alter the level/activity of METTL14, ALKBH5, and YTHDF3, leading to decreased m6A modification in the target transcripts in breast cancer cells (86). Stress due to metabolic starvation can elevate the expression of FTO through the autophagy and NF-κB pathways (87). Inflammatory stimuli can induce YTHDF2 expression in hematopoietic stem cell (88). The bone microenvironment including mesenchymal stem cells (MSCs), hypoxia, acidic condition, chemokines and immune cells, is regarded as fertile soil for osteosarcoma (89, 90). Given this, the elucidation of m6A signatures and the bone microenvironment in osteosarcoma deserves further study.
Discussion
Clinical Implications of m6A for Osteosarcoma
Considering that the dysregulation of m6A modification has been linked to the initiation, metastasis, drug resistance, and other processes of osteosarcoma, m6A may bring new breakthroughs in the diagnosis and treatment of osteosarcoma.
On the one hand, m6A regulatory enzymes could be novel potential biomarkers for the early diagnosis and prognosis of osteosarcoma. High m6A methylation levels and dysregulated m6A enzymes always occur in patients with osteosarcoma. Based on two large-scale cohorts, HNRNPA2B1, HNRNPC, RBM15, YTHDF1, and YTHDC1 expression levels are upregulated in osteosarcoma tissues. Among them, HNRNPA2B1 was suggested to be an independent prognostic risk factor in patients with osteosarcoma and predict poor survival rates (63). On the other hand, m6A may also serve as a novel therapeutic target in osteosarcoma. M6A modification is pivotal in almost all pathophysiological processes of osteosarcoma, including tumorigenesis, invasion, and metastasis (64, 68, 69, 71). The small molecule inhibitor of m6A regulators has been regarded as a kind of potential anti-cancer agent (91, 92). Currently, several FTO and ALKBH5 inhibitors have been successfully identified, including N-oxalylglycine, entacapone, and meclofenamic acid (MA) (93–95). Furthermore, in vitro experiments have shown that these inhibitors can inhibit tumor growth (96). Encouragingly, entacapone and MA are already in the early phases of clinical trials for patients with late-stage cancer (97). Additionally, m6A also plays a key role in the resistance to chemotherapy of osteosarcoma. METTL3 and ALKBH5 expression levels are upregulated in doxorubicin-resistant osteosarcoma cells, while METTL14 expression is downregulated (70). YTHDF2 knockdown can significantly increase the expression of TRIM7 and cause resistance to doxorubicin and methotrexate treatment (69). Therefore, targeting dysregulated m6A enzymes represents an attractive strategy for cancer therapy. It can not only directly inhibit tumor growth but also sensitize cancer cells to anti-cancer agents.
Immunotherapy has emerged as a promising treatment modality that largely expands the therapeutic regimen for cancer (98). Bone is characterized as a highly specialized immune environment, and some immune-related factors are frequently dysregulated in osteosarcoma (99). Overall, the expression level of tumor-associated macrophages is reduced in metastatic osteosarcoma, while tumor-infiltrating lymphocytes appear to be associated with enhanced metastases in osteosarcoma (100). Programmed death ligand-1, one of the most effective immune checkpoint modulators, is expressed in approximately 25% of primary osteosarcoma tumors and is associated with poor prognosis (100, 101). However, immune checkpoint inhibitors are less effective in treating osteosarcoma (102). In the SARC028 trial, only 1 of the 22 patients with osteosarcoma, who received pembrolizumab (an anti-programmed death 1 (PD-1) antibody) showed a good response (103). In the PEMBROSARC study, 17 patients with advanced osteosarcoma received a combination of pembrolizumab and cyclophosphamide, and only 2 patients had significant clinical benefits (104). Interestingly, more studies have indicated that m6A regulators play an essential role in host immunity and may contribute to anticancer immunotherapy (105, 106). For instance, Han et al. (105) found that the m6A reader, YTHDF1, can promote the translation of mRNAs encoding lysosomal proteases. Downregulation of YTHDF1 results in the reduction of antigen cross-presentation and alleviation of the cytotoxic lymphocyte response against tumor antigens in dendritic cells, thereby enhancing the effectiveness of the PD-1 blockade therapy. Similarly, the m6A eraser, ALKBH5, can regulate the lactate content, tumor-infiltrating regulatory T cells, and myeloid-derived suppressor cell accumulation in the tumor microenvironment. In mouse melanoma, depleting ALKBH5 improved the efficacy of anti–PD-1 therapy and had survival benefits (106). In view of the promising effect of immunotherapy in cancers and the close association of m6A modification with immune response, there are reasons to believe that combining anticancer immunotherapy with m6A signatures may pave a way to improve the therapeutic effects of osteosarcoma.
Conclusions and Future Perspective
M6A modification has emerged as an indispensable factor that accounts for tumor initiation and progression in osteosarcoma. The function of m6A modification is just like that of a “double-edged sword”, by which it can either accelerate or inhibit the progression of osteosarcoma via different modes. Undoubtedly, the advent of m6A regulation has provided new insight into the molecular mechanism of osteosarcoma and will potentially help develop new more effective therapies.
However, a full understanding of the mechanism underlying m6A modification is still in its infancy, several knowledge gaps remain. First, existing studies on m6A in osteosarcoma mainly focused on writers; the dysregulation of m6A erasers and readers in osteosarcoma require further study. Second, although m6A is considered a potential biomarker for the diagnosis and prognosis of osteosarcoma, only few studies have yet elucidated the relationship between m6A-related factors and the molecular epidemiology as well as clinicopathology of osteosarcoma. Third, the clinical guidance of sequencing data of small osteosarcoma samples is limited due to the high genomic heterogeneity of this disease. Therefore, a large sample of sequencing database for m6A-related factors and osteosarcoma is warranted. Fourth, researchers have noted the potential of m6A as a therapeutic target for osteosarcoma, but few studies have focused on the application of potent and specific drugs that target m6A enzymes in osteosarcoma. In addition, compared with other cancer, osteosarcoma has its own biological and clinical features, such as the close connection with bone microenvironment and poor immunotherapy effect. Hence, combining m6A factors with tumor microenvironment and anticancer immunotherapy in osteosarcoma must be explored.
Author Contributions
YZ, YYW, and LY conceived and designed the research. YZ, YYW, and ST searched the literature and analyzed the data. YZ and YYW wrote the manuscript and created the figures. YXW, PL, MS, and BH reviewed and made significant revisions to the manuscript. YZ, YYW, and LY contributed equally to this work as the first authors. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Nature Science Foundation of Zhejiang Province (LQ19H060002, LQ19H160041 and LY18H060004) and the Medical and Health Science and Technology Project of Zhejiang Province (2020KY143).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Mirabello L, Troisi RJ, Savage SA. Osteosarcoma Incidence and Survival Rates From 1973 to 2004: Data From the Surveillance, Epidemiology, and End Results Program. Cancer (2009) 115:1531–43. doi: 10.1002/cncr.24121
2. Ward E, Desantis C, Robbins A, Kohler B, Jemal A. Childhood and Adolescent Cancer Statistics. CA Cancer J Clin (2014) 64:83–103. doi: 10.3322/caac.21219
3. Whelan JS, Davis LE. Osteosarcoma, Chondrosarcoma, and Chordoma. J Clin Oncol (2018) 36:188–93. doi: 10.1200/JCO.2017.75.1743
4. Kansara M, Teng MW, Smyth MJ, Thomas DM. Translational Biology of Osteosarcoma. Nat Rev Cancer (2014) 14:722–35. doi: 10.1038/nrc3838
5. Link MP, Goorin AM, Miser AW, Green AA, Pratt CB, Belasco JB, et al. The Effect of Adjuvant Chemotherapy on Relapse-Free Survival in Patients With Osteosarcoma of the Extremity. N Engl J Med (1986) 314:1600–6. doi: 10.1056/NEJM198606193142502
6. Meyers PA, Healey JH, Chou AJ, Wexler LH, Merola PR, Morris CD, et al. Addition of Pamidronate to Chemotherapy for the Treatment of Osteosarcoma. Cancer (2011) 117:1736–44. doi: 10.1002/cncr.25744
7. Miller BJ, Cram P, Lynch CF, Buckwalter JA. Risk Factors for Metastatic Disease At Presentation With Osteosarcoma: An Analysis of the SEER Database. J Bone Joint Surg Am (2013) 95:e89. doi: 10.2106/JBJS.L.01189
8. Mirabello L, Zhu B, Koster R, Karlins E, Dean M, Yeager M, et al. Frequency of Pathogenic Germline Variants in Cancer-Susceptibility Genes in Patients With Osteosarcoma. JAMA Oncol (2020) 6:724–34. doi: 10.1001/jamaoncol.2020.0197
9. Savage SA, Mirabello L. Bone Cancer: Is the Osteosarcoma Genome Targetable? Nat Rev Endocrinol (2017) 13:506–8. doi: 10.1038/nrendo.2017.101
10. Smida J, Xu H, Zhang Y, Baumhoer D, Ribi S, Kovac M, et al. Genome-Wide Analysis of Somatic Copy Number Alterations and Chromosomal Breakages in Osteosarcoma. Int J Cancer (2017) 141:816–28. doi: 10.1002/ijc.30778
11. Perez MF, Lehner B. Intergenerational and Transgenerational Epigenetic Inheritance in Animals. Nat Cell Biol (2019) 21:143–51. doi: 10.1038/s41556-018-0242-9
12. Frye M, Jaffrey SR, Pan T, Rechavi G, Suzuki T. RNA Modifications: What Have We Learned and Where are We Headed? Nat Rev Genet (2016) 17:365–72. doi: 10.1038/nrg.2016.47
13. Roundtree IA, Evans ME, Pan T, He C. Dynamic RNA Modifications in Gene Expression Regulation. Cell (2017) 169:1187–200. doi: 10.1016/j.cell.2017.05.045
14. Desrosiers R, Friderici K, Rottman F. Identification of Methylated Nucleosides in Messenger RNA From Novikoff Hepatoma Cells. Proc Natl Acad Sci USA (1974) 71:3971–5. doi: 10.1073/pnas.71.10.3971
15. Goh YT, Koh CWQ, Sim DY, Roca X, Goh WSS. METTL4 Catalyzes m6Am Methylation in U2 snRNA to Regulate pre-mRNA Splicing. Nucleic Acids Res (2020) 48:9250–61. doi: 10.1093/nar/gkaa684
16. Chen RX, Chen X, Xia LP, Zhang JX, Pan ZZ, Ma XD, et al. N(6)-Methyladenosine Modification of circNSUN2 Facilitates Cytoplasmic Export and Stabilizes HMGA2 to Promote Colorectal Liver Metastasis. Nat Commun (2019) 10:4695. doi: 10.1038/s41467-019-12651-2
17. Chang G, Shi L, Ye Y, Shi H, Zeng L, Tiwary S, et al. Ythdf3 Induces the Translation of M(6)a-Enriched Gene Transcripts to Promote Breast Cancer Brain Metastasis. Cancer Cell (2020) 38:857–71.e857. doi: 10.1016/j.ccell.2020.10.004
18. Dixit D, Prager BC, Gimple RC, Poh HX, Wang Y, Wu Q, et al. The RNA m6A Reader YTHDF2 Maintains Oncogene Expression and is a Targetable Dependency in Glioblastoma Stem Cells. Cancer Discovery (2020) 11:480–99. doi: 10.1158/2159-8290.CD-20-0331
19. Chen X, Xu M, Xu X, Zeng K, Liu X, Pan B, et al. METTL14-Mediated N6-methyladenosine Modification of SOX4 mRNA Inhibits Tumor Metastasis in Colorectal Cancer. Mol Cancer (2020) 19:106. doi: 10.1186/s12943-020-01220-7
20. Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, et al. Topology of the Human and Mouse M6a RNA Methylomes Revealed by M6a-Seq. Nature (2012) 485:201–6. doi: 10.1038/nature11112
21. Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive Analysis of mRNA Methylation Reveals Enrichment in 3’ UTRs and Near Stop Codons. Cell (2012) 149:1635–46. doi: 10.1016/j.cell.2012.05.003
22. Huang H, Weng H, Chen J. M(6)a Modification in Coding and Non-coding Rnas: Roles and Therapeutic Implications in Cancer. Cancer Cell (2020) 37:270–88. doi: 10.1016/j.ccell.2020.02.004
23. Chen X, Hua W, Huang X, Chen Y, Zhang J, Li G. Regulatory Role of RNA N(6)-Methyladenosine Modification in Bone Biology and Osteoporosis. Front Endocrinol (Lausanne) (2019) 10:911. doi: 10.3389/fendo.2019.00911
24. Wu R, Jiang D, Wang Y, Wang X. N (6)-Methyladenosine (M(6)a) Methylation in mRNA With A Dynamic and Reversible Epigenetic Modification. Mol Biotechnol (2016) 58:450–9. doi: 10.1007/s12033-016-9947-9
25. Zaccara S, Ries RJ, Jaffrey SR. Reading, Writing and Erasing mRNA Methylation. Nat Rev Mol Cell Biol (2019) 20:608–24. doi: 10.1038/s41580-019-0168-5
26. Wang X, Huang J, Zou T, Yin P. Human M(6)a Writers: Two Subunits, 2 Roles. RNA Biol (2017) 14:300–4. doi: 10.1080/15476286.2017.1282025
27. Sledz P, Jinek M. Structural Insights Into the Molecular Mechanism of the M(6)a Writer Complex. Elife (2016) 5:e18434. doi: 10.7554/eLife.18434
28. Wang P, Doxtader KA, Nam Y. Structural Basis for Cooperative Function of Mettl3 and Mettl14 Methyltransferases. Mol Cell (2016) 63:306–17. doi: 10.1016/j.molcel.2016.05.041
29. Wang X, Feng J, Xue Y, Guan Z, Zhang D, Liu Z, et al. Structural Basis of N(6)-adenosine Methylation by the METTL3-METTL14 Complex. Nature (2016) 534:575–8. doi: 10.1038/nature18298
30. Bertero A, Brown S, Madrigal P, Osnato A, Ortmann D, Yiangou L, et al. The SMAD2/3 Interactome Reveals That TGFbeta Controls M(6)a mRNA Methylation in Pluripotency. Nature (2018) 555:256–9. doi: 10.1038/nature25784
31. Guo J, Tang HW, Li J, Perrimon N, Yan D. Xio is a Component of the Drosophila Sex Determination Pathway and RNA N(6)-Methyladenosine Methyltransferase Complex. Proc Natl Acad Sci USA (2018) 115:3674–9. doi: 10.1073/pnas.1720945115
32. Ruzicka K, Zhang M, Campilho A, Bodi Z, Kashif M, Saleh M, et al. Identification of Factors Required for M(6) A mRNA Methylation in Arabidopsis Reveals a Role for the Conserved E3 Ubiquitin Ligase HAKAI. New Phytol (2017) 215:157–72. doi: 10.1111/nph.14586
33. Yang Y, Hsu PJ, Chen YS, Yang YG. Dynamic Transcriptomic M(6)a Decoration: Writers, Erasers, Readers and Functions in RNA Metabolism. Cell Res (2018) 28:616–24. doi: 10.1038/s41422-018-0040-8
34. Tang C, Klukovich R, Peng H, Wang Z, Yu T, Zhang Y, et al. ALKBH5-Dependent m6A Demethylation Controls Splicing and Stability of Long 3’-UTR mRNAs in Male Germ Cells. Proc Natl Acad Sci USA (2018) 115:E325–33. doi: 10.1073/pnas.1717794115
35. Xiang Y, Laurent B, Hsu CH, Nachtergaele S, Lu Z, Sheng W, et al. RNA M(6)a Methylation Regulates the Ultraviolet-Induced DNA Damage Response. Nature (2017) 543:573–6. doi: 10.1038/nature21671
36. Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, et al. N6-Methyladenosine in Nuclear RNA is a Major Substrate of the Obesity-Associated FTO. Nat Chem Biol (2011) 7:885–7. doi: 10.1038/nchembio.687
37. Fu Y, Jia G, Pang X, Wang RN, Wang X, Li CJ, et al. FTO-Mediated Formation of N6-hydroxymethyladenosine and N6-formyladenosine in Mammalian RNA. Nat Commun (2013) 4:1798. doi: 10.1038/ncomms2822
38. Zhao Y, Shi Y, Shen H, Xie W. M(6)a-Binding Proteins: The Emerging Crucial Performers in Epigenetics. J Hematol Oncol (2020) 13:35. doi: 10.1186/s13045-020-00872-8
39. Theler D, Dominguez C, Blatter M, Boudet J, Allain FH. Solution Structure of the YTH Domain in Complex With N6-methyladenosine RNA: A Reader of Methylated RNA. Nucleic Acids Res (2014) 42:13911–9. doi: 10.1093/nar/gku1116
40. Liao S, Sun H, Xu C. Yth Domain: A Family of N(6)-methyladenosine (M(6)a) Readers. Genomics Proteomics Bioinf (2018) 16:99–107. doi: 10.1016/j.gpb.2018.04.002
41. Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T. N(6)-Methyladenosine-Dependent RNA Structural Switches Regulate RNA-protein Interactions. Nature (2015) 518:560–4. doi: 10.1038/nature14234
42. Wu B, Su S, Patil DP, Liu H, Gan J, Jaffrey SR, et al. Molecular Basis for the Specific and Multivariant Recognitions of RNA Substrates by Human hnRNP A2/B1. Nat Commun (2018) 9:420. doi: 10.1038/s41467-017-02770-z
43. Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, et al. Recognition of RNA N(6)-Methyladenosine by IGF2BP Proteins Enhances mRNA Stability and Translation. Nat Cell Biol (2018) 20:285–95. doi: 10.1038/s41556-018-0045-z
44. Sun L, Fazal FM, Li P, Broughton JP, Lee B, Tang L, et al. RNA Structure Maps Across Mammalian Cellular Compartments. Nat Struct Mol Biol (2019) 26:322–30. doi: 10.1038/s41594-019-0200-7
45. Min KW, Zealy RW, Davila S, Fomin M, Cummings JC, Makowsky D, et al. Profiling of M6a RNA Modifications Identified an Age-Associated Regulation of AGO2 mRNA Stability. Aging Cell (2018) 17:e12753. doi: 10.1111/acel.12753
46. Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ, et al. YTHDF3 Facilitates Translation and Decay of N(6)-methyladenosine-modified Rna. Cell Res (2017) 27:315–28. doi: 10.1038/cr.2017.15
47. Meyer KD, Jaffrey SR. Rethinking M(6)a Readers, Writers, and Erasers. Annu Rev Cell Dev Biol (2017) 33:319–42. doi: 10.1146/annurev-cellbio-100616-060758
48. Kan L, Grozhik AV, Vedanayagam J, Patil DP, Pang N, Lim KS, et al. The M(6)a Pathway Facilitates Sex Determination in Drosophila. Nat Commun (2017) 8:15737. doi: 10.1038/ncomms15737
49. Roundtree IA, Luo GZ, Zhang Z, Wang X, Zhou T, Cui Y, et al. YTHDC1 Mediates Nuclear Export of N(6)-methyladenosine Methylated Mrnas. Elife (2017) 6:e31311. doi: 10.7554/eLife.31311
50. Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, et al. N6-Methyladenosine-Dependent Regulation of Messenger RNA Stability. Nature (2014) 505:117–20. doi: 10.1038/nature12730
51. Weng H, Huang H, Wu H, Qin X, Zhao BS, Dong L, et al. Mettl14 Inhibits Hematopoietic Stem/Progenitor Differentiation and Promotes Leukemogenesis Via mRNA M(6)a Modification. Cell Stem Cell (2018) 22:191–205.e199. doi: 10.1016/j.stem.2017.11.016
52. Gao Y, Vasic R, Song Y, Teng R, Liu C, Gbyli R, et al. M(6)a Modification Prevents Formation of Endogenous Double-Stranded RNAs and Deleterious Innate Immune Responses During Hematopoietic Development. Immunity (2020) 52:1007–21. doi: 10.1016/j.immuni.2020.05.003
53. Liu J, Harada BT, He C. Regulation of Gene Expression by N(6)-methyladenosine in Cancer. Trends Cell Biol (2019) 29:487–99. doi: 10.1016/j.tcb.2019.02.008
54. Cui X, Wang Z, Li J, Zhu J, Ren Z, Zhang D, et al. Cross Talk Between RNA N6-Methyladenosine Methyltransferase-Like 3 and miR-186 Regulates Hepatoblastoma Progression Through Wnt/β-Catenin Signalling Pathway. Cell Prolif (2020) 53:e12768. doi: 10.1111/cpr.12768
55. Kim JM, Lin C, Stavre Z, Greenblatt MB, Shim JH. Osteoblast-Osteoclast Communication and Bone Homeostasis. Cells (2020) 9:2073. doi: 10.3390/cells9092073
56. Song W, Xie J, Li J, Bao C, Xiao Y. The Emerging Roles of Long Noncoding RNAs in Bone Homeostasis and Their Potential Application in Bone-Related Diseases. DNA Cell Biol (2020) 39:926–37. doi: 10.1089/dna.2020.5391
57. Zhang Y, Gu X, Li D, Cai L, Xu Q. Mettl3 Regulates Osteoblast Differentiation and Inflammatory Response Via Smad Signaling and MAPK Signaling. Int J Mol Sci (2019) 21:199. doi: 10.3390/ijms21010199
58. Yan G, Yuan Y, He M, Gong R, Lei H, Zhou H, et al. M(6)a Methylation of Precursor-miR-320/RUNX2 Controls Osteogenic Potential of Bone Marrow-Derived Mesenchymal Stem Cells. Mol Ther Nucleic Acids (2020) 19:421–36. doi: 10.1016/j.omtn.2019.12.001
59. Li D, Cai L, Meng R, Feng Z, Xu Q. Mettl3 Modulates Osteoclast Differentiation and Function by Controlling Rna Stability and Nuclear Export. Int J Mol Sci (2020) 21:1660. doi: 10.3390/ijms21051660
60. Sun Z, Wang H, Wang Y, Yuan G, Yu X, Jiang H, et al. MiR-103-3p Targets the M(6) A Methyltransferase METTL14 to Inhibit Osteoblastic Bone Formation. Aging Cell (2021) 20:e13298. doi: 10.1111/acel.13298
61. Li Y, Yang F, Gao M, Gong R, Jin M, Liu T, et al. Mir-149-3p Regulates the Switch Between Adipogenic and Osteogenic Differentiation of BMSCs by Targeting Fto. Mol Ther Nucleic Acids (2019) 17:590–600. doi: 10.1016/j.omtn.2019.06.023
62. Gianferante DM, Mirabello L, Savage SA. Germline and Somatic Genetics of Osteosarcoma - Connecting Aetiology, Biology and Therapy. Nat Rev Endocrinol (2017) 13:480–91. doi: 10.1038/nrendo.2017.16
63. Li J, Rao B, Yang J, Liu L, Huang M, Liu X, et al. Dysregulated M6a-Related Regulators are Associated With Tumor Metastasis and Poor Prognosis in Osteosarcoma. Front Oncol (2020) 10:769. doi: 10.3389/fonc.2020.00769
64. Zhou L, Yang C, Zhang N, Zhang X, Zhao T, Yu J. Silencing METTL3 Inhibits the Proliferation and Invasion of Osteosarcoma by Regulating ATAD2. BioMed Pharmacother (2020) 125:109964. doi: 10.1016/j.biopha.2020.109964
65. Miao W, Chen J, Jia L, Ma J, Song D. The m6A Methyltransferase METTL3 Promotes Osteosarcoma Progression by Regulating the m6A Level of LEF1. Biochem Biophys Res Commun (2019) 516:719–25. doi: 10.1016/j.bbrc.2019.06.128
66. Ling Z, Chen L, Zhao J. m6A-dependent Up-Regulation of DRG1 by METTL3 and ELAVL1 Promotes Growth, Migration, and Colony Formation in Osteosarcoma. Biosci Rep (2020) 40:BSR20200282. doi: 10.1042/bsr20200282
67. Liu Z, Liu N, Huang Z, Wang W. Mettl14 Overexpression Promotes Osteosarcoma Cell Apoptosis and Slows Tumor Progression Via Caspase 3 Activation. Cancer Manag Res (2020) 12:12759–67. doi: 10.2147/CMAR.S284273
68. Chen S, Li Y, Zhi S, Ding Z, Wang W, Peng Y, et al. WTAP Promotes Osteosarcoma Tumorigenesis by Repressing HMBOX1 Expression in an M(6)a-Dependent Manner. Cell Death Dis (2020) 11:659. doi: 10.1038/s41419-020-02847-6
69. Zhou C, Zhang Z, Zhu X, Qian G, Zhou Y, Sun Y, et al. N6-Methyladenosine Modification of the TRIM7 Positively Regulates Tumorigenesis and Chemoresistance in Osteosarcoma Through Ubiquitination of BRMS1. EBioMedicine (2020) 59:102955. doi: 10.1016/j.ebiom.2020.102955
70. Wang Y, Zeng L, Liang C, Zan R, Ji W, Zhang Z, et al. Integrated Analysis of Transcriptome-Wide M(6)a Methylome of Osteosarcoma Stem Cells Enriched by Chemotherapy. Epigenomics (2019) 11:1693–715. doi: 10.2217/epi-2019-0262
71. Chen S, Zhou L, Wang Y. ALKBH5-Mediated M(6)a Demethylation of Lncrna PVT1 Plays an Oncogenic Role in Osteosarcoma. Cancer Cell Int (2020) 20:34. doi: 10.1186/s12935-020-1105-6
72. Yuan Y, Yan G, He M, Lei H, Li L, Wang Y, et al. ALKBH5 Suppresses Tumor Progression Via an M(6)a-Dependent Epigenetic Silencing of pre-miR-181b-1/YAP Signaling Axis in Osteosarcoma. Cell Death Dis (2021) 12:60. doi: 10.1038/s41419-020-03315-x
73. Zhang W, He L, Liu Z, Ren X, Qi L, Wan L, et al. Multifaceted Functions and Novel Insight Into the Regulatory Role of RNA N(6)-Methyladenosine Modification in Musculoskeletal Disorders. Front Cell Dev Biol (2020) 8:870. doi: 10.3389/fcell.2020.00870
74. Wang X, Chen N, Du Z, Ling Z, Zhang P, Yang J, et al. Bioinformatics Analysis Integrating Metabolomics of M(6)a RNA Microarray in Intervertebral Disc Degeneration. Epigenomics (2020) 12:1419–41. doi: 10.2217/epi-2020-0101
75. Zhu H, Sun B, Zhu L, Zou G, Shen Q. N6-Methyladenosine Induced Mir-34a-5p Promotes Tnf-α-Induced Nucleus Pulposus Cell Senescence by Targeting Sirt1. Front Cell Dev Biol (2021) 9:642437. doi: 10.3389/fcell.2021.642437
76. Liu Q, Li M, Jiang L, Jiang R, Fu B. METTL3 Promotes Experimental Osteoarthritis Development by Regulating Inflammatory Response and Apoptosis in Chondrocyte. Biochem Biophys Res Commun (2019) 516:22–7. doi: 10.1016/j.bbrc.2019.05.168
77. Mo XB, Zhang YH, Lei SF. Genome-Wide Identification of M(6)a-Associated SNPs as Potential Functional Variants for Bone Mineral Density. Osteoporos Int (2018) 29:2029–39. doi: 10.1007/s00198-018-4573-y
78. Wu Y, Xie L, Wang M, Xiong Q, Guo Y, Liang Y, et al. Mettl3-mediated M(6)a RNA Methylation Regulates the Fate of Bone Marrow Mesenchymal Stem Cells and Osteoporosis. Nat Commun (2018) 9:4772. doi: 10.1038/s41467-018-06898-4
79. Tang B, Yang Y, Kang M, Wang Y, Wang Y, Bi Y, et al. M(6)a Demethylase ALKBH5 Inhibits Pancreatic Cancer Tumorigenesis by Decreasing WIF-1 RNA Methylation and Mediating Wnt Signaling. Mol Cancer (2020) 19:3. doi: 10.1186/s12943-019-1128-6
80. Guo X, Li K, Jiang W, Hu Y, Xiao W, Huang Y, et al. RNA Demethylase ALKBH5 Prevents Pancreatic Cancer Progression by Posttranscriptional Activation of PER1 in an M6a-YTHDF2-dependent Manner. Mol Cancer (2020) 19:91. doi: 10.1186/s12943-020-01158-w
81. Paris J, Morgan M, Campos J, Spencer GJ, Shmakova A, Ivanova I, et al. Targeting the RNA M(6)a Reader Ythdf2 Selectively Compromises Cancer Stem Cells in Acute Myeloid Leukemia. Cell Stem Cell (2019) 25:137–48. doi: 10.1016/j.stem.2019.03.021
82. Liu Y, Li H, Liu F, Gao LB, Han R, Chen C, et al. Heterogeneous Nuclear Ribonucleoprotein A2/B1 is a Negative Regulator of Human Breast Cancer Metastasis by Maintaining the Balance of Multiple Genes and Pathways. EBioMedicine (2020) 51:102583. doi: 10.1016/j.ebiom.2019.11.044
83. Zeng X, Ban Z, Cao J, Zhang W, Chu T, Lei D, et al. Association of FTO Mutations With Risk and Survival of Breast Cancer in a Chinese Population. Dis Markers (2015) 2015:101032. doi: 10.1155/2015/101032
84. Wu L, Wu D, Ning J, Liu W, Zhang D. Changes of N6-methyladenosine Modulators Promote Breast Cancer Progression. BMC Cancer (2019) 19:326. doi: 10.1186/s12885-019-5538-z
85. Xiao H, Fan X, Zhang R, Wu G. Upregulated N6-Methyladenosine RNA in Peripheral Blood: Potential Diagnostic Biomarker for Breast Cancer. Cancer Res Treat (2021) 53:399–408. doi: 10.4143/crt.2020.870
86. Panneerdoss S, Eedunuri VK, Yadav P, Timilsina S, Rajamanickam S, Viswanadhapalli S, et al. Cross-Talk Among Writers, Readers, and Erasers of M(6)a Regulates Cancer Growth and Progression. Sci Adv (2018) 4:eaar8263. doi: 10.1126/sciadv.aar8263
87. Yang S, Wei J, Cui YH, Park G, Shah P, Deng Y, et al. M(6)a mRNA Demethylase FTO Regulates Melanoma Tumorigenicity and Response to anti-PD-1 Blockade. Nat Commun (2019) 10:2782. doi: 10.1038/s41467-019-10669-0
88. Mapperley C, Van De Lagemaat LN, Lawson H, Tavosanis A, Paris J, Campos J, et al. The mRNA m6A Reader YTHDF2 Suppresses Proinflammatory Pathways and Sustains Hematopoietic Stem Cell Function. J Exp Med (2021) 218:e20200829. doi: 10.1084/jem.20200829
89. Yang C, Tian Y, Zhao F, Chen Z, Su P, Li Y, et al. Bone Microenvironment and Osteosarcoma Metastasis. Int J Mol Sci (2020) 21:6985. doi: 10.3390/ijms21196985
90. Zhou Y, Yang D, Yang Q, Lv X, Huang W, Zhou Z, et al. Single-Cell RNA Landscape of Intratumoral Heterogeneity and Immunosuppressive Microenvironment in Advanced Osteosarcoma. Nat Commun (2020) 11:6322. doi: 10.1038/s41467-020-20059-6
91. Su R, Dong L, Li Y, Gao M, Han L, Wunderlich M, et al. Targeting FTO Suppresses Cancer Stem Cell Maintenance and Immune Evasion. Cancer Cell (2020) 38:79–96. doi: 10.1016/j.ccell.2020.04.017
92. Huang Y, Su R, Sheng Y, Dong L, Dong Z, Xu H, et al. Small-Molecule Targeting of Oncogenic Fto Demethylase in Acute Myeloid Leukemia. Cancer Cell (2019) 35:677–91. doi: 10.1016/j.ccell.2019.03.006
93. Peng S, Xiao W, Ju D, Sun B, Hou N, Liu Q, et al. Identification of Entacapone as a Chemical Inhibitor of FTO Mediating Metabolic Regulation Through FOXO1. Sci Transl Med (2019) 11:eaau7116. doi: 10.1126/scitranslmed.aau7116
94. Huang Y, Yan J, Li Q, Li J, Gong S, Zhou H, et al. Meclofenamic Acid Selectively Inhibits FTO Demethylation of m6A Over ALKBH5. Nucleic Acids Res (2015) 43:373–84. doi: 10.1093/nar/gku1276
95. Islam MS, Leissing TM, Chowdhury R, Hopkinson RJ, Schofield CJ. 2-Oxoglutarate-Dependent Oxygenases. Annu Rev Biochem (2018) 87:585–620. doi: 10.1146/annurev-biochem-061516-044724
96. Niu Y, Wan A, Lin Z, Lu X, Wan G. N (6)-Methyladenosine Modification: A Novel Pharmacological Target for Anti-Cancer Drug Development. Acta Pharm Sin B (2018) 8:833–43. doi: 10.1016/j.apsb.2018.06.001
97. Li B, Jiang J, Assaraf YG, Xiao H, Chen ZS, Huang C. Surmounting Cancer Drug Resistance: New Insights From the Perspective of N(6)-methyladenosine RNA Modification. Drug Resist Update (2020) 53:100720. doi: 10.1016/j.drup.2020.100720
98. Galluzzi L, Chan TA, Kroemer G, Wolchok JD, López-Soto A. The Hallmarks of Successful Anticancer Immunotherapy. Sci Transl Med (2018) 10:eaat7807. doi: 10.1126/scitranslmed.aat7807
100. Sundara YT, Kostine M, Cleven AH, Bovée JV, Schilham MW, Cleton-Jansen AM. Increased PD-L1 and T-cell Infiltration in the Presence of HLA Class I Expression in Metastatic High-Grade Osteosarcoma: A Rationale for T-cell-based Immunotherapy. Cancer Immunol Immunother (2017) 66:119–28. doi: 10.1007/s00262-016-1925-3
101. Koirala P, Roth ME, Gill J, Piperdi S, Chinai JM, Geller DS, et al. Immune Infiltration and PD-L1 Expression in the Tumor Microenvironment are Prognostic in Osteosarcoma. Sci Rep (2016) 6:30093. doi: 10.1038/srep30093
102. Heymann MF, Schiavone K, Heymann D. Bone Sarcomas in the Immunotherapy Era. Br J Pharmacol (2020) 178:1955–72. doi: 10.1111/bph.14999
103. Keung EZ, Burgess M, Salazar R, Parra ER, Rodrigues-Canales J, Bolejack V, et al. Correlative Analyses of the SARC028 Trial Reveal an Association Between Sarcoma-Associated Immune Infiltrate and Response to Pembrolizumab. Clin Cancer Res (2020) 26:1258–66. doi: 10.1158/1078-0432.Ccr-19-1824
104. Le Cesne A, Marec-Berard P, Blay JY, Gaspar N, Bertucci F, Penel N, et al. Programmed Cell Death 1 (PD-1) Targeting in Patients With Advanced Osteosarcomas: Results From the PEMBROSARC Study. Eur J Cancer (2019) 119:151–7. doi: 10.1016/j.ejca.2019.07.018
105. Han D, Liu J, Chen C, Dong L, Liu Y, Chang R, et al. Anti-Tumour Immunity Controlled Through mRNA M(6)a Methylation and YTHDF1 in Dendritic Cells. Nature (2019) 566:270–4. doi: 10.1038/s41586-019-0916-x
Keywords: N6-methyladenosine (m6A), molecular mechanisms, osteosarcoma, biomarker, therapeutic target
Citation: Zhang Y, Wang Y, Ying L, Tao S, Shi M, Lin P, Wang Y and Han B (2021) Regulatory Role of N6-methyladenosine (m6A) Modification in Osteosarcoma. Front. Oncol. 11:683768. doi: 10.3389/fonc.2021.683768
Received: 22 March 2021; Accepted: 30 April 2021;
Published: 19 May 2021.
Edited by:
Yubing Zhou, First Affiliated Hospital of Zhengzhou University, ChinaReviewed by:
Xin Huang, Guangdong Second Provincial General Hospital, ChinaBashdar Mahmud Hussen, Hawler Medical University, Iraq
Wenjie Zheng, Affiliated Hospital of Nantong University, China
Copyright © 2021 Zhang, Wang, Ying, Tao, Shi, Lin, Wang and Han. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanyan Wang, cnVpeGl3eXlAemp1LmVkdS5jbg==
†These authors have contributed equally to this work
 Yujie Zhang1†
Yujie Zhang1† Yanyan Wang
Yanyan Wang Mingmin Shi
Mingmin Shi Peng Lin
Peng Lin