- Department of Neurosurgery, The Second Affiliated Hospital of Harbin Medical University, Harbin, China
Accompanying the development of biomedicine, our knowledge of glioma, one of the most common primary intracranial carcinomas, is becoming more comprehensive. Unfortunately, patients with glioblastoma (GBM) still have a dismal prognosis and a high relapse rate, even with standard combination therapy, namely, surgical resection, postoperative radiotherapy and chemotherapy. The absence of validated biomarkers is responsible for the majority of these poor outcomes, and reliable therapeutic targets are indispensable for improving the prognosis of patients suffering from gliomas. Identification of both precise diagnostic and accurate prognostic markers and promising therapeutic targets has therefore attracted considerable attention from researchers. Encouragingly, accumulating evidence has demonstrated that long noncoding RNAs (lncRNAs) play important roles in the pathogenesis and oncogenesis of various categories of human tumors, including gliomas. Nevertheless, the underlying mechanisms by which lncRNAs regulate diverse biological behaviors of glioma cells, such as proliferation, invasion and migration, remain poorly understood. Consequently, this review builds on previous studies to further summarize the progress in the field of lncRNA regulation of gliomas over recent years and addresses the potential of lncRNAs as diagnostic and prognostic markers and therapeutic targets.
Introduction
Gliomas, originating from glial or precursor cells, which are categorized into astrocytomas, ependymomas and oligodendrogliomas, are the most common malignant primary tumors of the central nervous system (CNS) (1, 2). In addition, gliomas are graded by the World Health Organization (WHO) into four classifications based on their malignancy. Gliomas with WHO grades I-II are known as low-grade gliomas (LGGs), including angiocentric glioma and diffuse astrocytoma, while those with WHO grades III-IV are considered high-grade gliomas (HGGs), including mesenchymal astrocytoma and glioblastoma multiform gliomas (GBMs) (3, 4). In the 2016 WHO classification of CNS tumors, molecular parameters, including IDH, ATRX, TP53 and 1p/19, were considered in the classification of glioma subtypes, which is more detailed than its 2007 predecessor (5, 6). GBM has high mortality and recurrence rates and represents the most malignant CNS tumor (3). The present criteria for treating GBM continue to be neurosurgical resection of the neoplasm accompanied by chemotherapy with temozolomide (TMZ) and radiotherapy (7). Unfortunately, the median survival for GBM patients is only 15 months, even with this combination treatment (8). Therefore, exploring the specific mechanisms of the occurrence and progression of glioma has drawn widespread interest in recent years. Studies on precise biomarkers and reliable therapeutic targets are urgently needed.
Approximately 98% of transcripts do not encode proteins in the human genome, and this category of RNA is known as noncoding RNA (ncRNA). Long noncoding RNAs (lncRNAs), accounting for approximately 80-90% of ncRNAs, are transcripts consisting of more than 200 nucleotides that typically lack protein-coding capability and were once regarded as transcriptional noise (9). Open reading frames are generally absent in lncRNAs (10). Intriguingly, this “transcriptional noise” has been extensively researched and demonstrated to not only serve an important function in normal cellular physiological procedures but also play an invaluable role in regulating the malignant behavior of tumors (11). LncRNAs can be divided into sense lncRNAs, antisense lncRNAs, bidirectional lncRNAs, intronic lncRNAs and intergenic lncRNAs (LINCRNAs) based on genomic location (12). The order of nucleotide arrangement constitutes the primary structure of lncRNAs, and intricate secondary and tertiary structures guarantee the multiple functions of lncRNAs. However, the relationship between lncRNA secondary structure and functions remains unclear. Recent evidence has indicated that lncRNAs regulate gene expression at three levels: transcriptional, post-transcriptional and epigenetic modification (13).
As mentioned above, lncRNAs are associated with both cellular physiology and disease origination and progression by regulating gene expression (11). Additionally, an increasing number of investigations have suggested that lncRNAs play pivotal roles in regulating the tumorigenesis, proliferation, aggression, metastasis, and drug resistance of gliomas. Consequently, as the molecular mechanism of lncRNA regulation of glioma is further investigated, the etiology of glioma will gradually be revealed. Furthermore, along with the advancement of sequencing technology, we will gradually recognize the entire spectrum of lncRNAs, implying that lncRNAs could be not only effective indicators for early diagnosis and determination of prognosis but also therapeutic targets for glioma.
The Functions and Mechanisms of lncRNAs in Gliomas
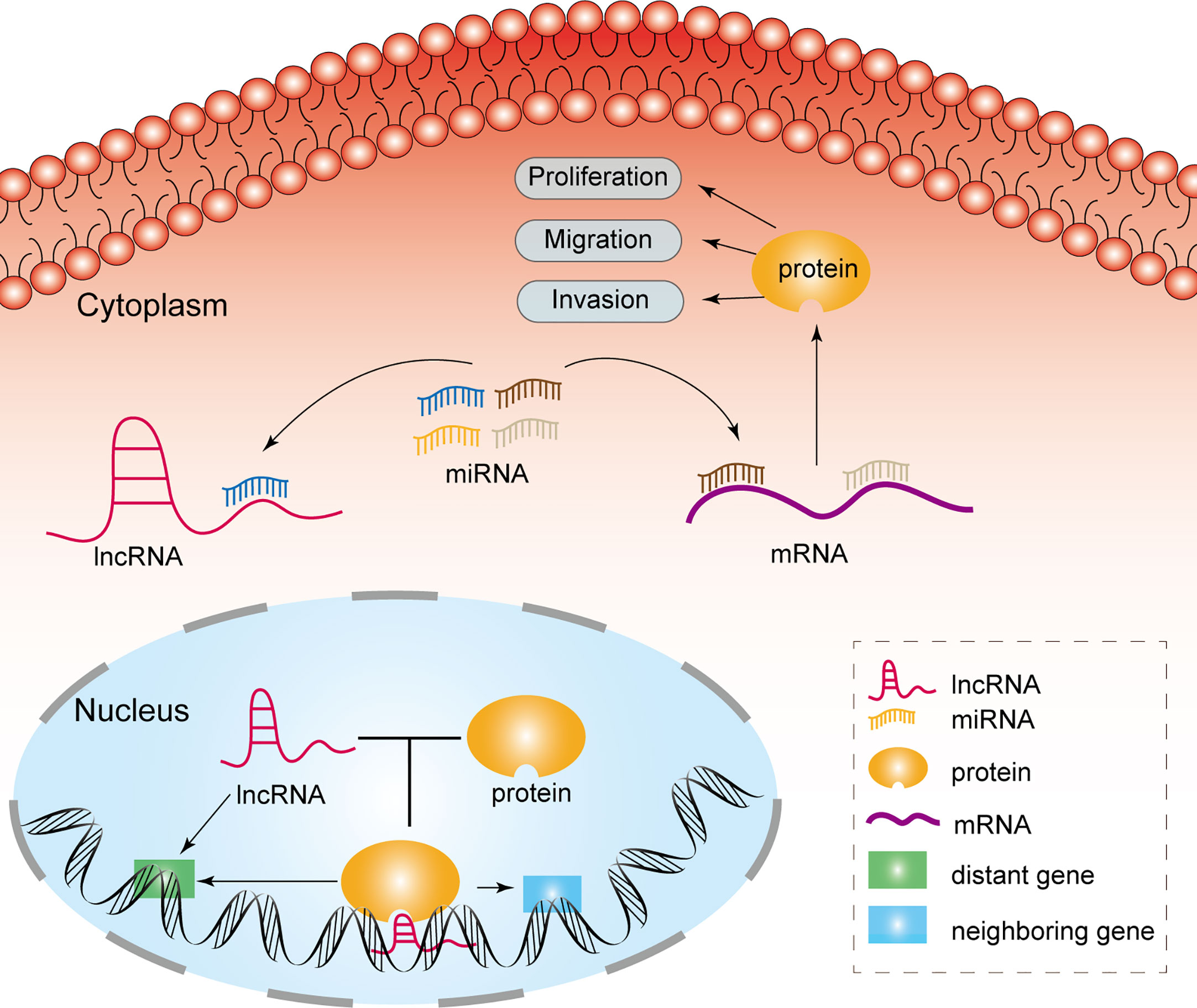
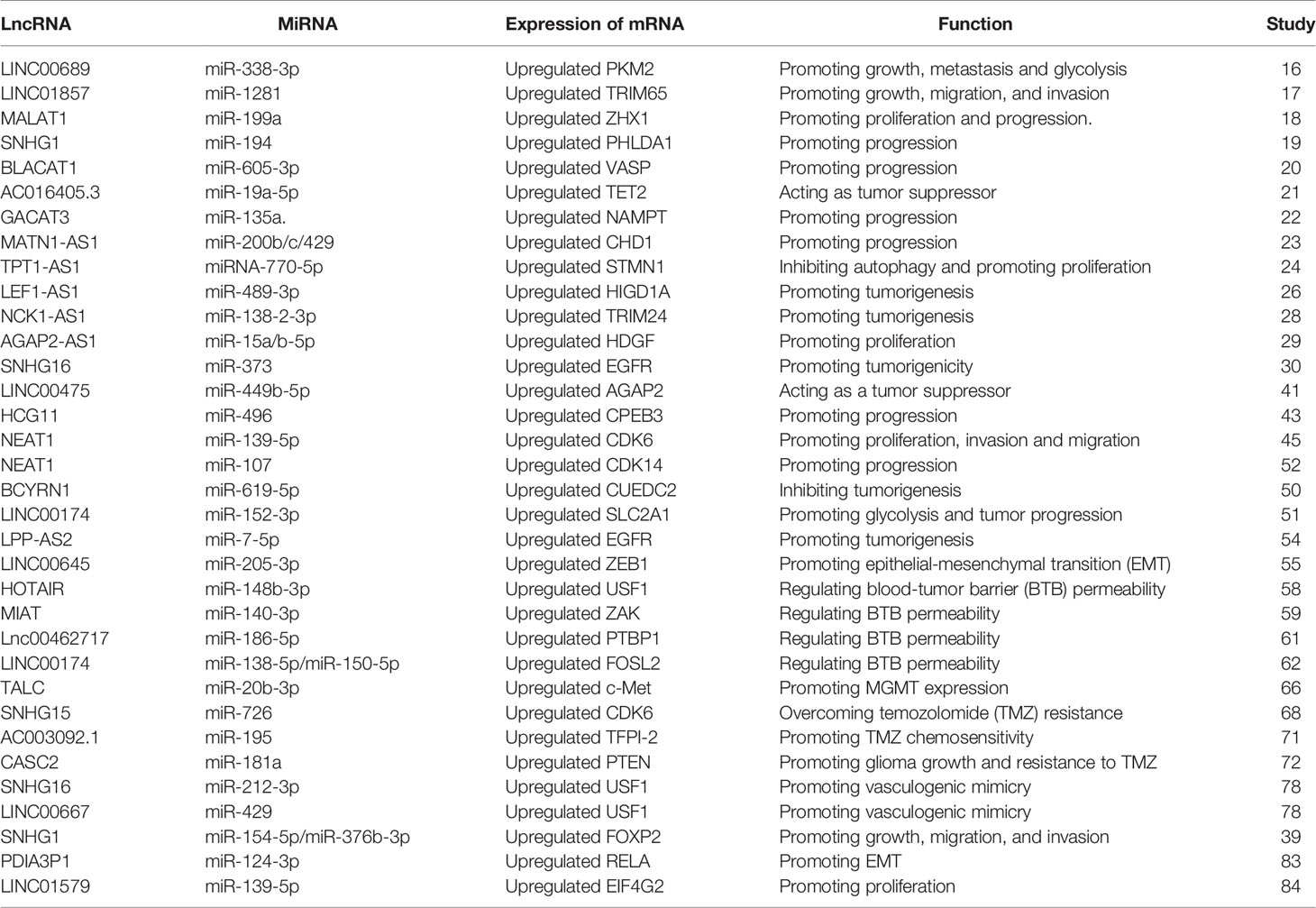
MiRNAs are a category of noncoding RNAs of approximately 20 nucleotides in length that can bind to target mRNAs via microRNA response elements (MREs) and thus perform negative regulatory functions, exerting critical post-transcriptional regulatory effects (14). MREs are short sequences of both lncRNAs and mRNAs that combine with miRNAs. Therefore, lncRNAs absorb miRNAs as sponges, enabling the expression of mRNAs that were previously repressed by miRNAs, and such lncRNAs are referred to as competitive endogenous RNAs (Figure 1) (15). Many studies have been conducted to demonstrate that lncRNAs, as ceRNAs, impact the progression of tumors at the post-transcriptional regulatory level (Table 1).
By analyzing GSE4290, Liu and colleagues showed that LINC00689 was highly expressed in glioma tissue compared to normal brain tissue. The expression of pyruvate kinase M2 (PKM2) was enhanced by LINC00689-mediated elimination of miR-338-3p, which facilitated malignant progression of glioma cells. As a consequence, the LINC00689/miR-338-3p/PKM2 axis functions as a carcinogenic driver in gliomas (16). Moreover, LINC01857 could promote tumorigenesis of glioma by sponging miR‐1281 to upregulate TRIM65 expression (17). MALAT1 has been proved to have a crucial role in the progression of multiple neoplasms such as lung, colorectal and gastric cancers, and shows a comparable regulatory role in glioma. MALAT1 promoted the level of ZHX1 by serving as a ceRNA of miR-199a, leading to augmented glioma development (18). LINC01579 accelerated cell proliferation and apoptosis of GBM by the competitive binding of miR-139-5p to affect EIF4G2 (19). Furthermore, the lncRNA SNHG1 is considered a sponge that absorbs miR-194 to promote glioma progression by regulating PHLDA1 expression (20). MiR-605-3p was eliminated by lncRNA BLACAT1 to accelerate VASP expression, contributing to glioma proliferation (21). The expression level of NAMPT was regulated by lncRNA-GACAT3 to promote glioma progression as a sponge for miR135a (22). LncRNA MATN1-AS1 competitively binding with miR-200b/c/429 also promoted the progression of glioma by modulating CHD1 expression (23). Chai et al. demonstrated that exosomal lncRNAe-ROR1-AS1 enhanced glioma progression by suppressing miR-4686 (24). LEF1-AS1 promoted glioma formation by competitively binding miR-489-3p to increase the expression of HIGD1A (25). Oncogenic lncRNA FOXD1-AS1 promoted the proliferation and metastasis of GBM cells by targeting miR339/342 (26). However, lncRNA could also act as a repressor to inhibit tumor progression. Zhen et al. indicated that NEAT1 sponged miR-107 to inhibit the expression of cyclin-dependent kinase 14 (CDK14) to repress the malignant progression of glioma (27). The anti-oncogene AC016405.3 restrained GBM cell proliferation and migration by sponging miR-19a through regulation of ten-eleven translocation-2 (TET2) (28). LncRNA TPT1-AS1 inhibited glioma cell autophagy by decreasing the expression of miR-110-5p, and upregulating STMN1 expression promoted the proliferation of glioma cells (29). In addition, mRNAs can indirectly regulate gene phenotypes by signaling pathways via the post-transcriptional regulation of ceRNAs. For instance, miR-183-2-3p was sponged by lncRNA NCK1-AS1. Low levels of miR-183-2-3p promoted TRIM24 expression and thereby activated the Wnt/β-catenin pathway to contribute to glioma progression (30). Correspondingly, the lncRNA AGAP2-AS1 exhibited analogous mechanisms in contributing to the development of glioma advancement via the miR-15a/b-5p/HDGF/WNT axis (31). The oncogene lncRNA SNHG16, in contrast, functioned in the proliferation, aggression and migration of glioma cells through the miR373/EGFR/PI3K/AKT axis (32). These researches illustrated that lncRNAs could not only promote but also inhibit tumor progression.
By interacting with signaling pathways, lncRNAs can facilitate tumorigenesis. For example, silencing lncRNA MIR22HG inhibited GBM aggressiveness by suppressing the Wnt/β-catenin signaling pathway (33), while cancer susceptibility candidate 7 (CASC7) restrained the progression of glioma through the Wnt/β-catenin pathway (34). BCAR4 promoted glioma cell progression by stimulating the EGFR/PI3K/AKT pathway (35). LncRNA LPP-AS2 plays an important role in regulating the miR-7-5p/EGFR/PI3K/AKT/c-MYC feedback loop, which is correlated with glioma tumorigenesis (36). LncRNA BCYRN1 could suppress tumorigenesis of glioma as a molecular sponge of miR-619-5p to modulate the PTEN/AKT/p21 pathway and CUEDC2 expression (37). Furthermore, lncRNA MT1JP suppressed proliferation, invasion, and migration and promoted apoptosis of glioma cells through stimulation of the PTEN/Akt signaling pathway (38). LncRNA-THOR silencing accelerated human glioma cell apoptosis by activating the MAGEA6-AMPK signaling pathway (39). Accumulating studies have suggested that lncRNA is involved in the regulation of diverse biological behaviors in glioma through the regulation of signaling pathways including but not limited to Wnt/β, PI3K/AKT and NF-κB. Therefore, lncRNAs are promising biomarkers for glioma diagnosis, prognosis and treatment in theory.
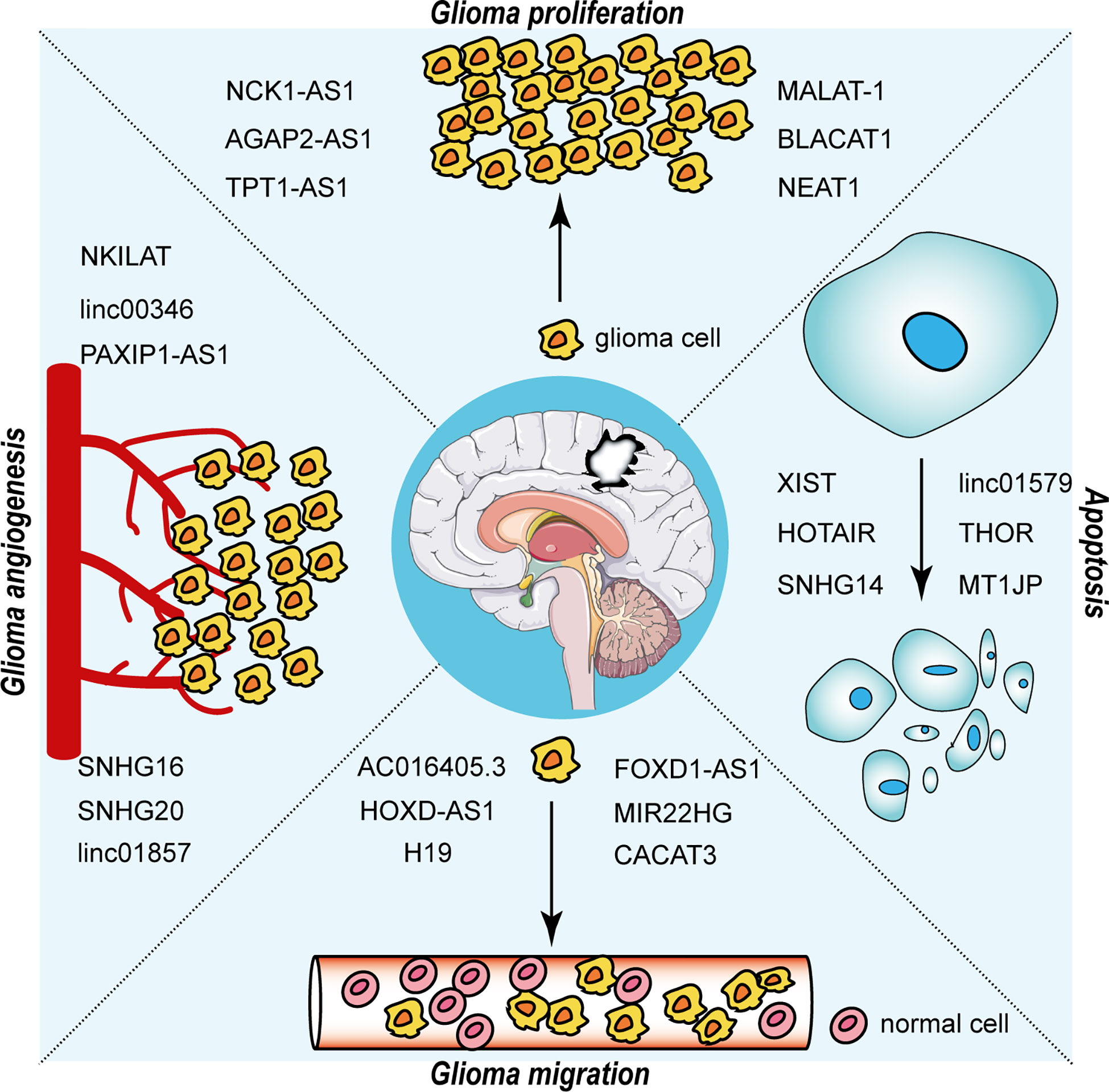
The interaction between RNA-binding proteins (RBPs) and lncRNAs plays a non-negligible role in the advancement of glioma. The expression level of EZH2 positively correlated with the malignancy of glioma and promoted the malignant behavior of glioma (40). Chen et al. first addressed the mechanism of the participation of lncRNA NEAT1 in tumorigenesis as a scaffold for EZH2. LncNEAT1 recruited EZH2 to interact with the promoter regions of downstream genes (Axin2, ICAT, GSK3B) to promote trimethylation modification of H3K27, thereby silencing these three genes. Further, the WNT/β-catenin pathway was activated, resulting in tumorigenesis (41). Moreover, RBP DGCR8 could bind with ZFAT-AS1, the interaction between DGCR8/ZFAT-AS1 and CDX2 contributed to the malignant progression of glioma (42). RBP, lncRNA and downstream gene could form negative or positive feedback loop to modulate biological behavior of glioma. SNHG1 regulated the miRNA154-5p/miR-376b-3p-FOXP2-KDM5B positive feedback loop to promote the malignant phenotype of glioma cells (43). LINC00475 silencing acted as a tumor suppressor in glioma under hypoxic conditions by impairing miRNA-449b-5p-dependent upregulation of AGAP2 expression (44). TRPM2‐AS inhibited the growth, migration, and invasion of gliomas through JNK, c‐Jun, and RGS4 (45). HCG11 inhibited glioma progression by modulating miR‐496 to upregulate cytoplasmic polyadenylation element binding protein 3 (CPEB3) expression (46). The lncRNA MNX1-AS1 reduced the level of miR-4443, leading to the promotion of proliferation, invasion and migration in glioma (47). NEAT1 and CDK6 could promote tumorigenesis of glioma cells; additionally, miR‐139‐5p restrained the biological functions of glioma cells (48). LncRNAs has diverse roles in glioma processes, such as proliferation, migration, apoptosis and angiogenesis, by distinct mechanisms, including ceRNA, interaction with RBPs and regulation of mRNA. Figure 2 portrays the lncRNAs associated with glioma proliferation, metastasis, apoptosis and angiogenesis.
LncRNAs as Diagnostic and Prognostic Biomarkers of Gliomas
Medical diagnosis in the twenty-first century is gradually moving from clinical pathology to molecular pathology. With the development of bioinformatics, an increasing number of studies have identified lncRNAs as biomarkers for glioma diagnosis and prognosis by exploring RNA microarrays, and advances in microarray and high-throughput RNA-seq technologies have provided numerous valuable lncRNAs for the diagnosis and prognosis of gliomas.
The detection of serum HOTAIR levels can be employed for the clinical diagnosis of glioma, as reported by Tan et al. (49) These researchers also observed that the serum levels of HOTAIR were significantly higher in GBM patients than in controls, with a sensitivity of 86.1% and specificity of 87.5% (49). This report first showed that HOTAIR can function as a novel diagnostic and prognostic peripheral biomarker of GBM. Lin et al. screened six lncRNAs associated with low-grade glioma prognosis by TCGA and GTEx RNA-seq databases. These researchers constructed a prognostic risk signature with 6 lncRNAs in LGG, and this research illustrated that AL031722.1 and LINC00844 decreased when the risk score was increased, while the expression of AL354740.1, FGD5-AS1, and NEAT1 increased (50). The team of Li et al. indicated that the expression of LINC01060 was upregulated in glioma and significantly related to tumor grade and poor clinical prognosis (51). Furthermore, Liu et al. revealed that the level of RMST was related to histological grade, and 95.6% of HGGs had higher RMST expression (52). The lncRNA HOTAIRM1 was identified as a prognostic factor for glioma because it can maintain the tumorigenicity of GSCs by regulating HOX gene expression (53). And, LINC00115 was shown to act as a key role in GSC self-renewal and tumorigenicity by Tang et al. (54) LINC00174 accelerated glycolysis and tumor progression by competitively binding with miR-152-3p in glioma, indicating this molecule might act as a molecular target for glioma diagnosis (55). LncRNA H19, which mediates the effect of curcumin in treating glioma accompanied by miR-675 and VDR, could act as a novel diagnostic biomarker (56). Li et al. showed that LINC00645 could promote EMT, which was indispensable in the invasion and migration of glioma cells involving TGF-β by regulating the miR-205-3p-ZEB1 axis; thus, LINC00645 could be a prognostic indicator for glioma (57). A novel lncRNA-TOCN that targets the Smad2/PKCα signaling pathway to inhibit malignant progression of glioma was highlighted by Tang et al. and could serve as a prognostic indicator of GBM (58). The expression of multiple lncRNAs has been documented to correlate with the malignancy of gliomas and to be involved in their malignant progression, providing powerful theoretical evidence for their application as diagnostic and prognostic markers.
LncRNAs as Reliable Therapeutic Targets
Treatment Strategies Involving LncRNAs as Regulators Modulating the BTB
The BTB parallels the blood-brain barrier (BBB) and is comprised of vascular endothelial cells, basement membrane, and glioma cells. This structure can seriously impede the entry of drugs into the tumor microenvironment, resulting in poor drug efficacy and extremely unfavorable patient prognosis (59–61). Consequently, exploring lncRNAs that can regulate BTB permeability to promote chemotherapy and thus improve drug efficacy is one of the research directions for targeted glioma therapy. Li et al. showed that silencing HOTAIR could increase BTB permeability by eliminating miR-148b-3p, thereby further reducing the expression of glioma-microvascular endothelial cell tight junction (TJ)-related proteins by targeting USF1 (62). He et al. also indicated that MIAT regulated the expression of ZAK to promote the delivery efficiency of doxorubicin across the BTB (63). In addition, the IGF2BP2/FBXL19-AS1/ZNF765 axis could regulate the permeability of the BTB to improve the antitumor effect of doxorubicin (64). Lnc00462717 regulated BTB permeability by interacting with PTBP1 to restrain the miR-1865p/Occludin signaling pathway (65). Moreover, BTB permeability was shown to be augmented by silencing LINC00174 in glioma tissue (66). Overcoming the obstacle of the BTB to increase the local concentration of chemotherapeutic agents in glioma and then enhance therapeutic efficacy is a prospective strategy. As such, identifying appropriate targets has been a major concern. The formulation of individual drug delivery routes based on the corresponding targets is another strategy that can potentially enhance chemotherapeutic efficacy.
Treatment Strategies Involving LncRNAs Overcoming TMZ Resistance
Temozolomide (TMZ) is an oral alkylating agent that passes through the BBB, adding methyl groups to the purines of DNA to cause DNA damage and apoptosis for therapeutic effects (67, 68). Conversely, this process is reversed by the DNA damage repair enzyme O6-methyl-guanine-DNA methyltransferase (MGMT), which restores the damage caused by TMZ, resulting in the resistance of glioma cells to TMZ (69). A novel lncRNA, lnc-TALC, was found to be highly expressed in TMZ-resistant GBM cells by Wu et al. Lnc-TALC modulated the c-Met pathway by functioning as a ceRNA for miR-20b-3p, facilitating MGMT performance and in turn leading to TMZ resistance in GBM cells (70). Sun and colleagues revealed that the overexpression of miR-29c-3p could promote chemosensitivity to cisplatin, and CRNDE, which competitively binds with miR-29c-3p, plays a critical role in regulating the chemoresistance of medulloblastoma (71). In addition, tumorigenesis of glioma was attenuated with the downregulation of lncRNA-SNHG15 expression, and TMZ sensitivity was increased (72). Recently, a study reported that combining p50 and p53 with the proximal κB and p53 sites of the MALAT1 coding region, respectively, cooperatively downregulated MALAT1 expression, which in turn increased the chemosensitivity of GBM cells (73). The tumor microenvironment was remodeled with the secretion of oncogenic lncSBF2-AS1-enriched exosomes by GBM cells, resulting in tumor drug resistance (74). The lncRNA AC003092.1 inhibited miR-195, increasing the expression of tfpi-2, which promoted TMZ-induced apoptosis and thus made GBM cells more sensitive to TMZ (75). CASC2 has an essential function in the sensitivity of glioma to TMZ by upregulating PTEN expression through direct inhibition of miR-181a (76). High expression of SNHG12 in TMZ-resistant cells served as a molecular sponge for miR-129-5p to raise the levels of MAPK1 and E2F7 to enhance the sensitivity of GBM cells to TMZ. In contrast, knockdown of SNHG12 restored TMZ sensitivity (77). LncRNA SOX2OT activates the Wnt5a/β-catenin signaling pathway through upregulation of SOX2 expression, thereby inhibiting apoptosis, promoting cell proliferation, and resulting in resistance to TMZ (78). LncRNAs are not only linked to chemotherapy but also closely correlated with radioresistance. For instance, LINC-RA1 inhibited autophagy and enhanced radioresistance by inhibiting the H2Bub1/USP44 combination in glioma cells (79). In summary, lncRNA on the one hand can increase sensitivity of glioma to TMZ and on the other hand induce TMZ resistance of glioma. TMZ is currently the main chemotherapeutic agent for the treatment of glioma, however, glioma is prone to become resistance to it. Thus, it is feasible to target lncRNA to find drugs to overcome TMZ resistance in glioma.
Treatment Strategy Involving LncRNAs Mediating Angiogenesis
Angiogenesis is a requirement for the growth and metastasis of gliomas, which are solid tumors. Additionally, extensive evidence has demonstrated that the formation of novel blood vessels participates in the development and metabolic processes of tumors. Therefore, vasculogenic mimicry is considered a hallmark of malignant tumor development. Hence, antiangiogenic treatment is anticipated to be an additional efficacious strategy for glioma. Chen and colleagues demonstrated that overexpression of NKILAT was negatively correlated with survival time in glioma patients, and NKILAT augmented the Warburg effect and angiogenesis in glioma, suggesting that it may be a promising therapeutic strategy (80). In addition, Yang et al. elucidated the key role of the ANKHD1/LINC00346/ZNF655 feedback loop in regulating angiogenesis in glioma (81). Likewise, Wang et al. demonstrated that knockdown of USF1 suppressed angiogenesis in gliomas by stressing SNHG16/miR-212-3p and the LINC00667/miR-429 axis (82). Furthermore, overexpression of lncRNA PAXIP1-AS1 promoted glioma vasculogenic mimicry by recruiting the transcription factor EST to upregulate KIF4 expression (83). SNHG20 played a crucial role in the ZRANB2/SNHG20/FOXK1 axis to regulate vasculogenic mimicry of glioma (84). These studies provide compelling evidence that lncRNAs potentially act as therapeutic targets by regulating angiogenesis in gliomas.
In addition, lncRNAs that exert regulatory effects by binding to RBP also have the potential to become therapeutic targets. Lin28a elevated the expression and stability of SNHG14, while deletion of SNHG14 increased the expression of IRF6, which inhibited the transcription of PKM2 and GLUT1 and thus impaired glycolysis and proliferation of glioma cells and induced apoptosis. Therefore, considering the lin28a/SNHG14/IRF6 axis as a target provides novel insight for the treatment of glioma (85). Additionally, the TAF15/LINC00665/MTF1(YY2)/GTSE1 axis is crucial for regulating the malignant biological behaviors of glioma cells, which might help in the development of a novel therapeutic strategy for human glioma (86). The PABPC1-BDNF-AS-RAX2-DLG5 axis was shown to play the same role as the TAF15/LINC00665/MTF1(YY2)/GTSE1 axis in regulating the biological behavior of gliomas (87). LncRNA PDIA3P1 promoted glioma mesenchymal transition by competitively binding to miR-124-3p in a hypoxic environment to regulate RELA expression and activate the downstream NF-κB pathway. Consequently, the PDIA3P1-miR-124-3p-RELA axis is a possible target for glioma therapy (88, 89). SChLAP1 forms a complex with HNRNPL to maintain the stability of ACTN4 and thus activates the NF-κB pathway to promote the growth of GBM cells (90). The identification of this complex provided a new perspective for the treatment of glioma. The UPF1-LINC00313-miR-342-3p/miR-485-5p-Zic4-SHCBP1 positive feedback loop was capable of modulating the biological behaviors of glioma cells, demonstrating that this loop is probably a potential therapeutic target (91). The combination of lncRNA and RBP forms a complex that regulates downstream target gene to contribute to TMZ resistance of glioma. This regulatory model provides a novel insight into the therapeutic strategy for glioma. Thus, targeting lncRNA-regulated angiogenesis, BTB permeability and TMZ resistance of glioma as novel strategies for the treatment of glioma shows potential.
Conclusion
Over the previous decades, lncRNA studies have made major progress in the field of glioma research due to the rapid development of bioinformatics, and a series of lncRNAs have been found to act as indispensable factors in the occurrence and progression of glioma. LncRNAs act as ceRNAs in the cytoplasm to regulate glioma progression at the post-transcriptional level or to regulate gene expression through interactions with proteins in the nucleus. However, the mechanisms of most lncRNAs remain unclear. Therefore, the specific mechanisms of lncRNAs need to be further clarified. Glioma continues to be a major challenge to human health, and its advanced aggressiveness, chemoresistance and recurrence are the main factors contributing to the poor prognosis. Theoretically, there are numerous lncRNAs, such as HOTAIR, H19 and NEAT1, that can be applied as diagnostic and prognostic indicators of glioma. It is also possible that many lncRNAs can overcome TMZ resistance, modulate BTB permeability and control glioma angiogenesis, all of which are theoretically effective therapeutic strategies. Regrettably, no successful clinical use of lncRNAs has been achieved yet. In prospective research, lncRNA-centered gene regulatory networks should be constructed to elucidate the regulatory mechanisms of lncRNAs in tumor cells and then used as diagnostic, prognostic, and therapeutic indicators in clinical practice to improve the survival of glioma patients. One of the focuses of basic research is translation to the clinic to improve the survival of patients. Translating basic research into clinical strategies is a long road that will require generations of researchers to eventually understand the full picture of the function of lncRNAs in glioma at both the scientific and clinical levels.
Author Contributions
JQ and XM wrote the draft and revised it. JC and CJ designed the tables. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by The National Natural Science Foundation of China (No. 81874204, No. 81772666, No. 81972817, No. 82073298, No. 82003022), Excellent Young Talents Project of Central Government Supporting Local University Reform and Development Fund (0202-300011190006), Karolinska Institutet Research Foundation Grants 2020-2021 (No. FS-2020:0007), The Heilongjiang Postdoctoral Science Foundation (LBH-Z18103, LBH-Z19029), and The Research Project of the Health and Family Planning Commission of Heilongjiang Province (2019-102).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Chen Q, Han B, Meng X, Duan C, Yang C, Wu Z, et al. Immunogenomic Analysis Reveals LGALS1 Contributes to the Immune Heterogeneity and Immunosuppression in Glioma. Int J Cancer (2019) 145(2):517–30. doi: 10.1002/ijc.32102
2. Lapointe S, Perry A, Butowski NA. Primary Brain Tumours in Adults. Lancet (2018) 392(10145):432–46. doi: 10.1016/s0140-6736(18)30990-5
3. Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A Summary. Acta Neuropathol (2016) 131(6):803–20. doi: 10.1007/s00401-016-1545-1
4. Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, et al. The 2007 WHO Classification of Tumours of the Central Nervous System. Acta Neuropathol (2007) 114(2):97–109. doi: 10.1007/s00401-007-0243-4
5. Yang P, Cai J, Yan W, Zhang W, Wang Y, Chen B, et al. Classification Based on Mutations of TERT Promoter and IDH Characterizes Subtypes in Grade II/III Gliomas. Neuro Oncol (2016) 18(8):1099–108. doi: 10.1093/neuonc/now021
6. Han B, Cai J, Gao W, Meng X, Gao F, Wu P, et al. Loss of ATRX Suppresses ATM Dependent DNA Damage Repair by Modulating H3K9me3 to Enhance Temozolomide Sensitivity in Glioma. Cancer Lett (2018) 419:280–90. doi: 10.1016/j.canlet.2018.01.056
7. Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, et al. Radiotherapy Plus Concomitant and Adjuvant Temozolomide for Glioblastoma. N Engl J Med (2005) 352(10):987–96. doi: 10.1056/NEJMoa043330
8. Van Meir EG, Hadjipanayis CG, Norden AD, Shu HK, Wen PY, Olson JJ. Exciting New Advances in Neuro-Oncology: The Avenue to a Cure for Malignant Glioma. CA Cancer J Clin (2010) 60(3):166–93. doi: 10.3322/caac.20069
9. Ulitsky I, Bartel DP. Lincrnas: Genomics, Evolution, and Mechanisms. Cell (2013) 154(1):26–46. doi: 10.1016/j.cell.2013.06.020
10. Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. The GENCODE V7 Catalog of Human Long Noncoding RNAs: Analysis of Their Gene Structure, Evolution, and Expression. Genome Res (2012) 22(9):1775–89. doi: 10.1101/gr.132159.111
11. Zhang XQ, Leung GK. Long Non-Coding RNAs in Glioma: Functional Roles and Clinical Perspectives. Neurochem Int (2014) 77:78–85. doi: 10.1016/j.neuint.2014.05.008
12. Li J, Zhu Y, Wang H, Ji X. Targeting Long Noncoding RNA in Glioma: A Pathway Perspective. Mol Ther Nucleic Acids (2018) 13:431–41. doi: 10.1016/j.omtn.2018.09.023
13. Bonasio R, Shiekhattar R. Regulation of Transcription by Long Noncoding Rnas. Annu Rev Genet (2014) 48:433–55. doi: 10.1146/annurev-genet-120213-092323
14. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA Hypothesis: The Rosetta Stone of a Hidden RNA Language? Cell (2011) 146(3):353–8. doi: 10.1016/j.cell.2011.07.014
15. Tay Y, Rinn J, Pandolfi PP. The Multilayered Complexity of ceRNA Crosstalk and Competition. Nature (2014) 505(7483):344–52. doi: 10.1038/nature12986
16. Liu X, Zhu Q, Guo Y, Xiao Z, Hu L, Xu Q. Lncrna LINC00689 Promotes the Growth, Metastasis and Glycolysis of Glioma Cells by Targeting miR-338-3p/PKM2 Axis. BioMed Pharmacother (2019) 117:109069. doi: 10.1016/j.biopha.2019.109069
17. Hu G, Liu N, Wang H, Wang Y, Guo Z. Lncrna LINC01857 Promotes Growth, Migration, and Invasion of Glioma by Modulating miR-1281/TRIM65 Axis. J Cell Physiol (2019) 234(12):22009–16. doi: 10.1002/jcp.28763
18. Liao K, Lin Y, Gao W, Xiao Z, Medina R, Dmitriev P, et al. Blocking Lncrna MALAT1/miR-199a/ZHX1 Axis Inhibits Glioblastoma Proliferation and Progression. Mol Ther Nucleic Acids (2019) 18:388–99. doi: 10.1016/j.omtn.2019.09.005
19. Chai Y, Xie M. LINC01579 Promotes Cell Proliferation by Acting as a ceRNA of miR-139-5p to Upregulate EIF4G2 Expression in Glioblastoma. J Cell Physiol (2019) 234(12):23658–66. doi: 10.1002/jcp.28933
20. Liu L, Shi Y, Shi J, Wang H, Sheng Y, Jiang Q, et al. The Long non-Coding RNA SNHG1 Promotes Glioma Progression by Competitively Binding to miR-194 to Regulate PHLDA1 Expression. Cell Death Dis (2019) 10(6):463. doi: 10.1038/s41419-019-1698-7
21. Liu N, Hu G, Wang H, Wang Y, Guo Z. Lncrna BLACAT1 Regulates VASP Expression Via Binding to miR-605-3p and Promotes Giloma Development. J Cell Physiol (2019) 234(12):22144–52. doi: 10.1002/jcp.28778
22. Wang J, Zhang M, Lu W. Long Noncoding RNA GACAT3 Promotes Glioma Progression by Sponging Mir-135a. J Cell Physiol (2019) 234(7):10877–87. doi: 10.1002/jcp.27946
23. Zhu J, Gu W, Yu C. Matn1-AS1 Promotes Glioma Progression by Functioning as ceRNA of miR-200b/c/429 to Regulate CHD1 Expression. Cell Prolif (2020) 53(1):e12700. doi: 10.1111/cpr.12700
24. Chai Y, Wu HT, Liang CD, You CY, Xie MX, Xiao SW. Exosomal Lncrna ROR1-AS1 Derived From Tumor Cells Promotes Glioma Progression Via Regulating Mir-4686. Int J Nanomed (2020) 15:8863–72. doi: 10.2147/IJN.S271795
25. Cheng Z, Wang G, Zhu W, Luo C, Guo Z. Lef1-AS1 Accelerates Tumorigenesis in Glioma by Sponging miR-489-3p to Enhance HIGD1A. Cell Death Dis (2020) 11(8):690. doi: 10.1038/s41419-020-02823-0
26. Gao YF, Liu JY, Mao XY, He ZW, Zhu T, Wang ZB, et al. Lncrna FOXD1-AS1 Acts as a Potential Oncogenic Biomarker in Glioma. CNS Neurosci Ther (2020) 26(1):66–75. doi: 10.1111/cns.13152
27. Zhen Y, Nan Y, Guo S, Zhang L, Li G, Yue S, et al. Knockdown of NEAT1 Repressed the Malignant Progression of Glioma Through Sponging miR-107 and Inhibiting CDK14. J Cell Physiol (2019) 234(7):10671–9. doi: 10.1002/jcp.27727
28. Ren S, Xu Y. Ac016405.3, a Novel Long Noncoding RNA, Acts as a Tumor Suppressor Through Modulation of TET2 by microRNA-19a-5p Sponging in Glioblastoma. Cancer Sci (2019) 110(5):1621–32. doi: 10.1111/cas.14002
29. Jia L, Song Y, Mu L, Li Q, Tang J, Yang Z, et al. Long Noncoding RNA Tpt1-AS1 Downregulates the microRNA-770-5p Expression to Inhibit Glioma Cell Autophagy and Promote Proliferation Through STMN1 Upregulation. J Cell Physiol (2020) 235(4):3679–89. doi: 10.1002/jcp.29262
30. Huang L, Li X, Ye H, Liu Y, Liang X, Yang C, et al. Long non-Coding RNA Nck1-AS1 Promotes the Tumorigenesis of Glioma Through Sponging microRNA-138-2-3p and Activating the TRIM24/Wnt/beta-Catenin Axis. J Exp Clin Cancer Res (2020) 39(1):63. doi: 10.1186/s13046-020-01567-1
31. Zheng Y, Lu S, Xu Y, Zheng J. Long non-Coding RNA Agap2-AS1 Promotes the Proliferation of Glioma Cells by Sponging miR-15a/b-5p to Upregulate the Expression of HDGF and Activating Wnt/beta-catenin Signaling Pathway. Int J Biol Macromol (2019) 128:521–30. doi: 10.1016/j.ijbiomac.2019.01.121
32. Zhou XY, Liu H, Ding ZB, Xi HP, Wang GW. LncRNA SNHG16 Promotes Glioma Tumorigenicity Through miR-373/EGFR Axis by Activating PI3K/AKT Pathway. Genomics (2020) 112(1):1021–9. doi: 10.1016/j.ygeno.2019.06.017
33. Han M, Wang S, Fritah S, Wang X, Zhou W, Yang N, et al. Interfering With Long Non-Coding RNA MIR22HG Processing Inhibits Glioblastoma Progression Through Suppression of Wnt/beta-catenin Signalling. Brain (2020) 143(2):512–30. doi: 10.1093/brain/awz406
34. Gong X, Liao X, Huang M. Lncrna CASC7 Inhibits the Progression of Glioma Via Regulating Wnt/beta-Catenin Signaling Pathway. Pathol Res Pract (2019) 215(3):564–70. doi: 10.1016/j.prp.2019.01.018
35. Wei L, Yi Z, Guo K, Long X. Long Noncoding RNA BCAR4 Promotes Glioma Cell Proliferation Via EGFR/PI3K/AKT Signaling Pathway. J Cell Physiol (2019) 234(12):23608–17. doi: 10.1002/jcp.28929
36. Zhang X, Niu W, Mu M, Hu S, Niu C. Long non-Coding RNA Lpp-AS2 Promotes Glioma Tumorigenesis Via miR-7-5p/EGFR/PI3K/AKT/c-MYC Feedback Loop. J Exp Clin Cancer Res (2020) 39(1):196. doi: 10.1186/s13046-020-01695-8
37. Mu M, Niu W, Zhang X, Hu S, Niu C. Lncrna BCYRN1 Inhibits Glioma Tumorigenesis by Competitively Binding With miR-619-5p to Regulate CUEDC2 Expression and the PTEN/AKT/p21 Pathway. Oncogene (2020) 39(45):6879–92. doi: 10.1038/s41388-020-01466-x
38. Zhang Y, Sui R, Chen Y, Liang H, Shi J, Piao H. Long Noncoding RNA MT1JP Inhibits Proliferation, Invasion, and Migration While Promoting Apoptosis of Glioma Cells Through the Activation of PTEN/Akt Signaling Pathway. J Cell Physiol (2019) 234(11):19553–64. doi: 10.1002/jcp.28553
39. Xue J, Zhong S, Sun BM, Sun QF, Hu LY, Pan SJ. Lnc-THOR Silencing Inhibits Human Glioma Cell Survival by Activating MAGEA6-AMPK Signaling. Cell Death Dis (2019) 10(11):866. doi: 10.1038/s41419-019-2093-0
40. Han B, Meng X, Wu P, Li Z, Li S, Zhang Y, et al. ATRX/EZH2 Complex Epigenetically Regulates FADD/PARP1 Axis, Contributing to TMZ Resistance in Glioma. Theranostics (2020) 10(7):3351–65. doi: 10.7150/thno.41219
41. Chen Q, Cai J, Wang Q, Wang Y, Liu M, Yang J, et al. Long Noncoding Rna NEAT1, Regulated by the EGFR Pathway, Contributes to Glioblastoma Progression Through the WNT/beta-Catenin Pathway by Scaffolding Ezh2. Clin Cancer Res (2018) 24(3):684–95. doi: 10.1158/1078-0432.CCR-17-0605
42. Zhang F, Ruan X, Ma J, Liu X, Zheng J, Liu Y, et al. Dgcr8/Zfat-As1 Promotes Cdx2 Transcription in a PRC2 Complex-Dependent Manner to Facilitate the Malignant Biological Behavior of Glioma Cells. Mol Ther (2020) 28(2):613–30. doi: 10.1016/j.ymthe.2019.11.015
43. Li H, Xue Y, Ma J, Shao L, Wang D, Zheng J, et al. SNHG1 Promotes Malignant Biological Behaviors of Glioma Cells Via Microrna-154-5p/Mir-376b-3p- FOXP2- KDM5B Participating Positive Feedback Loop. J Exp Clin Cancer Res (2019) 38(1):59. doi: 10.1186/s13046-019-1063-9
44. Yu L, Gui S, Liu Y, Qiu X, Qiu B, Zhang X, et al. Long Intergenic non-Protein Coding RNA 00475 Silencing Acts as a Tumor Suppressor in Glioma Under Hypoxic Condition by Impairing Microrna-449b-5p-Dependent AGAP2 Up-Regulation. Ther Adv Med Oncol (2020) 12:1758835920940936. doi: 10.1177/1758835920940936
45. Bao MH, Lv QL, Szeto V, Wong R, Zhu SZ, Zhang YY, et al. Trpm2-AS Inhibits the Growth, Migration, and Invasion of Gliomas Through JNK, c-Jun, and RGS4. J Cell Physiol (2020) 235(5):4594–604. doi: 10.1002/jcp.29336
46. Chen Y, Bao C, Zhang X, Lin X, Huang H, Wang Z. Long non-Coding RNA HCG11 Modulates Glioma Progression Through Cooperating With miR-496/CPEB3 Axis. Cell Prolif (2019) 52(5):e12615. doi: 10.1111/cpr.12615
47. Gao Y, Xu Y, Wang J, Yang X, Wen L, Feng J. Lncrna MNX1-AS1 Promotes Glioblastoma Progression Through Inhibition of Mir-4443. Oncol Res (2019) 27(3):341–7. doi: 10.3727/096504018X15228909735079
48. Wu DM, Wang S, Wen X, Han XR, Wang YJ, Fan SH, et al. Long Noncoding RNA Nuclear Enriched Abundant Transcript 1 Impacts Cell Proliferation, Invasion, and Migration of Glioma Through Regulating miR-139-5p/ Cdk6. J Cell Physiol (2019) 234(5):5972–87. doi: 10.1002/jcp.27093
49. Tan SK, Pastori C, Penas C, Komotar RJ, Ivan ME, Wahlestedt C, et al. Serum Long Noncoding RNA HOTAIR as a Novel Diagnostic and Prognostic Biomarker in Glioblastoma Multiforme. Mol Cancer (2018) 17(1):74. doi: 10.1186/s12943-018-0822-0
50. Lin JZ, Lin N, Zhao WJ. Identification and Validation of a six-lncRNA Prognostic Signature With its ceRNA Networks and Candidate Drugs in Lower-Grade Gliomas. Genomics (2020) 112(5):2990–3002. doi: 10.1016/j.ygeno.2020.05.016
51. Li J, Liao T, Liu H, Yuan H, Ouyang T, Wang J, et al. Hypoxic Glioma Stem Cell-Derived Exosomes Containing LINC01060 Promote Progression of Glioma by Regulating the MZF1/C-Myc/HIF-1alpha. Cancer Res (2021) 81(1):114–28. doi: 10.1158/0008-5472
52. Liu C, Peng Z, Li P, Fu H, Feng J, Zhang Y, et al. Lncrna RMST Suppressed GBM Cell Mitophagy Through Enhancing FUS Sumoylation. Mol Ther Nucleic Acids (2020) 19:1198–208. doi: 10.1016/j.omtn.2020.01.008
53. Xia H, Liu Y, Wang Z, Zhang W, Qi M, Qi B, et al. Long Noncoding Rna HOTAIRM1 Maintains Tumorigenicity of Glioblastoma Stem-Like Cells Through Regulation of HOX Gene Expression. Neurotherapeutics (2020) 17(2):754–64. doi: 10.1007/s13311-019-00799-0
54. Tang J, Yu B, Li Y, Zhang W, Alvarez AA, Hu B, et al. TGF-Beta-Activated Lncrna LINC00115 Is a Critical Regulator of Glioma Stem-Like Cell Tumorigenicity. EMBO Rep (2019) 20(12):e48170. doi: 10.15252/embr.201948170
55. Shi J, Zhang Y, Qin B, Wang Y, Zhu X. Long Non-Coding RNA LINC00174 Promotes Glycolysis and Tumor Progression by Regulating miR-152-3p/SLC2A1 Axis in Glioma. J Exp Clin Cancer Res (2019) 38(1):395. doi: 10.1186/s13046-019-1390-x
56. Pan JX, Chen TN, Ma K, Wang S, Yang CY, Cui GY. A Negative Feedback Loop of H19/miR-675/VDR Mediates Therapeutic Effect of Cucurmin in the Treatment of Glioma. J Cell Physiol (2020) 235(3):2171–82. doi: 10.1002/jcp.29127
57. Li C, Zheng H, Hou W, Bao H, Xiong J, Che W, et al. Long Non-Coding RNA LINC00645 Promotes TGF-Beta-Induced Epithelial-Mesenchymal Transition by Regulating miR-205-3p-ZEB1 Axis in Glioma. Cell Death Dis (2019) 10(10):717. doi: 10.1038/s41419-019-1948-8
58. Tang C, Wang Y, Zhang L, Wang J, Wang W, Han X, et al. Identification of Novel LncRNA Targeting Smad2/PKCalpha Signal Pathway to Negatively Regulate Malignant Progression of Glioblastoma. J Cell Physiol (2020) 235(4):3835–48. doi: 10.1002/jcp.29278
59. Vogelbaum MA. Targeted Therapies for Brain Tumors: Will They Ever Deliver? Clin Cancer Res (2018) 24(16):3790–1. doi: 10.1158/1078-0432.CCR-18-0855
60. Wang X, Sun N, Meng X, Chen M, Jiang C, Cai J. Review of Clinical Nerve Repair Strategies for Neurorestoration of Central Nervous System Tumor Damage. J Neurorestoratol (2020) 8(3):172–81. doi: 10.26599/jnr.2020.9040018
61. Huang H, Chen L, Mao G, Sharma HS. Clinical Neurorestorative Cell Therapies: Developmental Process, Current State and Future Prospective. J Neurorestoratol (2020) 8(2):61–82. doi: 10.26599/jnr.2020.9040009
62. Sa L, Li Y, Zhao L, Liu Y, Wang P, Liu L, et al. The Role of HOTAIR/miR-148b-3p/USF1 on Regulating the Permeability of BTB. Front Mol Neurosci (2017) 10:194. doi: 10.3389/fnmol.2017.00194
63. He J, Xue Y, Wang Q, Zhou X, Liu L, Zhang T, et al. Long non-Coding RNA MIAT Regulates Blood Tumor Barrier Permeability by Functioning as a Competing Endogenous RNA. Cell Death Dis (2020) 11(10):936. doi: 10.1038/s41419-020-03134-0
64. Liu X, Wu P, Su R, Xue Y, Yang C, Wang D, et al. IGF2BP2 Stabilized FBXL19-AS1 Regulates the Blood-Tumour Barrier Permeability by Negatively Regulating ZNF765 by STAU1-mediated mRNA Decay. RNA Biol (2020) 17(12):1777–88. doi: 10.1080/15476286.2020.1795583
65. Zhang C, Zhang X, Wang J, Di F, Xue Y, Lin X, et al. Lnc00462717 Regulates the Permeability of the Blood-Brain Tumor Barrier Through Interaction With PTBP1 to Inhibit the miR-186-5p/Occludin Signaling Pathway. FASEB J (2020) 34(8):9941–58. doi: 10.1096/fj.202000045R
66. Guo J, Shen S, Liu X, Ruan X, Zheng J, Liu Y, et al. Role of LINC00174/miR-138-5p (Mir-150-5p)/FOSL2 Feedback Loop on Regulating the Blood-Tumor Barrier Permeability. Mol Ther Nucleic Acids (2019) 18:1072–90. doi: 10.1016/j.omtn.2019.10.031
67. Karachi A, Dastmalchi F, Mitchell DA, Rahman M. Temozolomide for Immunomodulation in the Treatment of Glioblastoma. Neuro Oncol (2018) 20(12):1566–72. doi: 10.1093/neuonc/noy072
68. Zhang J, Stevens MF, Bradshaw TD. Temozolomide: Mechanisms of Action, Repair and Resistance. Curr Mol Pharmacol (2012) 5(1):102–14. doi: 10.2174/1874467211205010102
69. Banerjee A, Vest KE, Pavlath GK, Corbett AH. Nuclear Poly(a) Binding Protein 1 (PABPN1) and Matrin3 Interact in Muscle Cells and Regulate RNA Processing. Nucleic Acids Res (2017) 45(18):10706–25. doi: 10.1093/nar/gkx786
70. Wu P, Cai J, Chen Q, Han B, Meng X, Li Y, et al. Lnc-TALC Promotes O(6)-Methylguanine-DNA Methyltransferase Expression Via Regulating the c-Met Pathway by Competitively Binding With Mir-20b-3p. Nat Commun (2019) 10(1):2045. doi: 10.1038/s41467-019-10025-2
71. Sun XH, Fan WJ, An ZJ, Sun Y. Inhibition of Long Noncoding RNA Crnde Increases Chemosensitivity of Medulloblastoma Cells by Targeting Mir-29c-3p. Oncol Res (2020) 28(1):95–102. doi: 10.3727/096504019X15742472027401
72. Li Z, Zhang J, Zheng H, Li C, Xiong J, Wang W, et al. Modulating Lncrna SNHG15/CDK6/miR-627 Circuit by Palbociclib, Overcomes Temozolomide Resistance and Reduces M2-Polarization of Glioma Associated Microglia in Glioblastoma Multiforme. J Exp Clin Cancer Res (2019) 38(1):380. doi: 10.1186/s13046-019-1371-0
73. Voce DJ, Bernal GM, Wu L, Crawley CD, Zhang W, Mansour NM, et al. Temozolomide Treatment Induces Lncrna MALAT1 in an NF-kappaB and P53 Codependent Manner in Glioblastoma. Cancer Res (2019) 79(10):2536–48. doi: 10.1158/0008-5472.CAN-18-2170
74. Zhang Z, Yin J, Lu C, Wei Y, Zeng A, You Y. Exosomal Transfer of Long Non-Coding RNA Sbf2-AS1 Enhances Chemoresistance to Temozolomide in Glioblastoma. J Exp Clin Cancer Res (2019) 38(1):166. doi: 10.1186/s13046-019-1139-6
75. Xu N, Liu B, Lian C, Doycheva DM, Fu Z, Liu Y, et al. Long Noncoding RNA AC003092.1 Promotes Temozolomide Chemosensitivity Through miR-195/TFPI-2 Signaling Modulation in Glioblastoma. Cell Death Dis (2018) 9(12):1139. doi: 10.1038/s41419-018-1183-8
76. Liao Y, Shen L, Zhao H, Liu Q, Fu J, Guo Y, et al. Lncrna CASC2 Interacts With miR-181a to Modulate Glioma Growth and Resistance to TMZ Through PTEN Pathway. J Cell Biochem (2017) 118(7):1889–99. doi: 10.1002/jcb.25910
77. Lu C, Wei Y, Wang X, Zhang Z, Yin J, Li W, et al. DNA-Methylation-Mediated Activating of Lncrna SNHG12 Promotes Temozolomide Resistance in Glioblastoma. Mol Cancer (2020) 19(1):28. doi: 10.1186/s12943-020-1137-5
78. Liu B, Zhou J, Wang C, Chi Y, Wei Q, Fu Z, et al. Lncrna SOX2OT Promotes Temozolomide Resistance by Elevating SOX2 Expression Via ALKBH5-mediated Epigenetic Regulation in Glioblastoma. Cell Death Dis (2020) 11(5):384. doi: 10.1038/s41419-020-2540-y
79. Zheng J, Wang B, Zheng R, Zhang J, Huang C, Zheng R, et al. Linc-RA1 Inhibits Autophagy and Promotes Radioresistance by Preventing H2Bub1/USP44 Combination in Glioma Cells. Cell Death Dis (2020) 11(9):758. doi: 10.1038/s41419-020-02977-x
80. Chen Z, Li S, Shen L, Wei X, Zhu H, Wang X, et al. NF-Kappa B Interacting Long Noncoding RNA Enhances the Warburg Effect and Angiogenesis and Is Associated With Decreased Survival of Patients With Gliomas. Cell Death Dis (2020) 11(5):323. doi: 10.1038/s41419-020-2520-2
81. Yang C, Zheng J, Liu X, Xue Y, He Q, Dong Y, et al. Role of ANKHD1/LINC00346/ZNF655 Feedback Loop in Regulating the Glioma Angiogenesis Via Staufen1-Mediated mRNA Decay. Mol Ther Nucleic Acids (2020) 20:866–78. doi: 10.1016/j.omtn.2020.05.004
82. Wang D, Zheng J, Liu X, Xue Y, Liu L, Ma J, et al. Knockdown of USF1 Inhibits the Vasculogenic Mimicry of Glioma Cells Via Stimulating Snhg16/miR-212-3p and LINC00667/miR-429 Axis. Mol Ther Nucleic Acids (2019) 14:465–82. doi: 10.1016/j.omtn.2018.12.017
83. Xu H, Zhao G, Zhang Y, Jiang H, Wang W, Zhao D, et al. Long Non-Coding RNA Paxip1-AS1 Facilitates Cell Invasion and Angiogenesis of Glioma by Recruiting Transcription Factor ETS1 to Upregulate KIF14 Expression. J Exp Clin Cancer Res (2019) 38(1):486. doi: 10.1186/s13046-019-1474-7
84. Li X, Xue Y, Liu X, Zheng J, Shen S, Yang C, et al. Zranb2/Snhg20/Foxk1 Axis Regulates Vasculogenic Mimicry Formation in Glioma. J Exp Clin Cancer Res (2019) 38(1):68. doi: 10.1186/s13046-019-1073-7
85. Lu J, Liu X, Zheng J, Song J, Liu Y, Ruan X, et al. Lin28A Promotes IRF6-Regulated Aerobic Glycolysis in Glioma Cells by Stabilizing SNHG14. Cell Death Dis (2020) 11(6):447. doi: 10.1038/s41419-020-2650-6
86. Ruan X, Zheng J, Liu X, Liu Y, Liu L, Ma J, et al. Lncrna LINC00665 Stabilized by TAF15 Impeded the Malignant Biological Behaviors of Glioma Cells Via STAU1-Mediated mRNA Degradation. Mol Ther Nucleic Acids (2020) 20:823–40. doi: 10.1016/j.omtn.2020.05.003
87. Su R, Ma J, Zheng J, Liu X, Liu Y, Ruan X, et al. PABPC1-Induced Stabilization of BDNF-AS Inhibits Malignant Progression of Glioblastoma Cells Through STAU1-Mediated Decay. Cell Death Dis (2020) 11(2):81. doi: 10.1038/s41419-020-2267-9
88. Wang S, Qi Y, Gao X, Qiu W, Liu Q, Guo X, et al. Hypoxia-Induced Lncrna PDIA3P1 Promotes Mesenchymal Transition Via Sponging of miR-124-3p in Glioma. Cell Death Dis (2020) 11(3):168. doi: 10.1038/s41419-020-2345-z
89. Zhang M-B, Song C-C, Li G-Z, Chen L-F, Ma R, Yu X-H, et al. Transplantation of Umbilical Cord Blood Mononuclear Cells Attenuates the Expression of IL-1β Via the TLR4/NF-κb Pathway in Hypoxic-Ischemic Neonatal Rats. J Neurorestoratol (2020) 8(2):122–30. doi: 10.26599/jnr.2020.9040015
90. Ji J, Xu R, Ding K, Bao G, Zhang X, Huang B, et al. Long Noncoding Rna SChLAP1 Forms a Growth-Promoting Complex With HNRNPL in Human Glioblastoma Through Stabilization of ACTN4 and Activation of NF-kappaB Signaling. Clin Cancer Res (2019) 25(22):6868–81. doi: 10.1158/1078-0432.CCR-19-0747
Keywords: lncRNAs, glioma, biomarker, therapeutic target, prognosis, chemoresistance
Citation: Qin J, Jiang C, Cai J and Meng X (2021) Roles of Long Noncoding RNAs in Conferring Glioma Progression and Treatment. Front. Oncol. 11:688027. doi: 10.3389/fonc.2021.688027
Received: 30 March 2021; Accepted: 26 May 2021;
Published: 11 June 2021.
Edited by:
Yaohua Liu, Shanghai First People’s Hospital, ChinaReviewed by:
Karanbir Brar, University of Toronto, CanadaYe Zhang, China Medical University, China
Wenhua Liang, First Affiliated Hospital of Guangzhou Medical University, China
Copyright © 2021 Qin, Jiang, Cai and Meng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinquan Cai, Y2FpamlucXVhbjY2Njc3N0AxMjYuY29t; Xiangqi Meng, bmVwdHVuZV9teHFAMTI2LmNvbQ==
 Jie Qin
Jie Qin Chuanlu Jiang
Chuanlu Jiang Jinquan Cai
Jinquan Cai Xiangqi Meng
Xiangqi Meng