- 1School of Medicine, Tehran University of Medical Sciences, Tehran, Iran
- 2Network of Immunity in Infection, Malignancy and Autoimmunity (NIIMA), Universal Scientific Education and Research Network (USERN), Tehran, Iran
- 3Research Center for Immunodeficiencies, Pediatrics Center of Excellence, Children’s Medical Center, Tehran University of Medical Sciences, Tehran, Iran
- 4Department of Immunology, School of Medicine, Tehran University of Medical Sciences, Tehran, Iran
Glioma is the most common malignant central nervous system tumor with significant mortality and morbidity. Despite considerable advances, the exact molecular pathways involved in tumor progression are not fully elucidated, and patients commonly face a poor prognosis. Long non-coding RNAs (lncRNAs) have recently drawn extra attention for their potential roles in different types of cancer as well as non-malignant diseases. More than 200 lncRNAs have been reported to be associated with glioma. We aimed to assess the roles of the most investigated lncRNAs in different stages of tumor progression and the mediating molecular pathways in addition to their clinical applications. lncRNAs are involved in different stages of tumor formation, invasion, and progression, including regulating the cell cycle, apoptosis, autophagy, epithelial-to-mesenchymal transition, tumor stemness, angiogenesis, the integrity of the blood-tumor-brain barrier, tumor metabolism, and immunological responses. The well-known oncogenic lncRNAs, which are upregulated in glioma, are H19, HOTAIR, PVT1, UCA1, XIST, CRNDE, FOXD2-AS1, ANRIL, HOXA11-AS, TP73-AS1, and DANCR. On the other hand, MEG3, GAS5, CCASC2, and TUSC7 are tumor suppressor lncRNAs, which are downregulated. While most studies reported oncogenic effects for MALAT1, TUG1, and NEAT1, there are some controversies regarding these lncRNAs. Expression levels of lncRNAs can be associated with tumor grade, survival, treatment response (chemotherapy drugs or radiotherapy), and overall prognosis. Moreover, circulatory levels of lncRNAs, such as MALAT1, H19, HOTAIR, NEAT1, TUG1, GAS5, LINK-A, and TUSC7, can provide non-invasive diagnostic and prognostic tools. Modulation of expression of lncRNAs using antisense oligonucleotides can lead to novel therapeutics. Notably, a profound understanding of the underlying molecular pathways involved in the function of lncRNAs is required to develop novel therapeutic targets. More investigations with large sample sizes and increased focus on in-vivo models are required to expand our understanding of the potential roles and application of lncRNAs in glioma.
Introduction
Glioma is the most common malignant central nervous system (CNS) tumor with significant mortality and morbidity (1). Glioblastoma is the most common and aggressive type of glioma with a median overall survival of less than two years (2). Notwithstanding substantial advances, the exact molecular pathways involved in tumorigenesis, tumor suppression, and treatment response are not fully elucidated in glioma, and patients commonly face a poor prognosis (3).
Non-coding ribonucleic acids (RNAs), comprising more than 97% of the human genome with various functions in physiological and pathological conditions, play a major role in glioma tumorigenesis (4). Non-coding RNAs are divided into the categories of short and long non-coding RNAs. The quintessential example of the former group are mi-RNAs, the role of which in glioma has been thoroughly investigated and reviewed (5, 6). In the past decade, long non-coding RNAs (lncRNAs) have drawn extra attention. More than 95% of the articles on lncRNAs and glioma retrieved from PubMed were published after 2017.
Lack of optimal treatment options in addition to specific and sensitive biomarkers (7) necessitates investigation of molecular pathways involved in glioma progression in the hope of finding novel therapeutic and diagnostic targets. LncRNAs may stand as prospective candidates for this purpose.
In this review, after providing a brief background on lncRNAs and their functions, we reviewed their role in various oncogenic processes. We also assessed their role in determining treatment response, survival, and prognosis. Lastly, the diagnostic and prognostic value of circulatory lncRNAs and potential therapeutic applications of modulation of lncRNAs expression in-vivo were investigated.
An Overview on LncRNAs
LncRNAs are non-protein-coding RNAs with more than 200 nucleotides that are transcribed mainly by RNA polymerase II. As a result, lncRNAs, like messenger (m)RNAs, are typically polyadenylated and capped (8). However, compared to mRNAs, they are more nuclear-localized, more scarce, less evolutionary conserved, and contain fewer exons (9).
LncRNAs can be categorized into six groups according to their location on the genome, namely (a) sense, (b) antisense, (c) bidirectional (d) intronic, (e), and (f) enhancer lncRNAs (Figure 1).
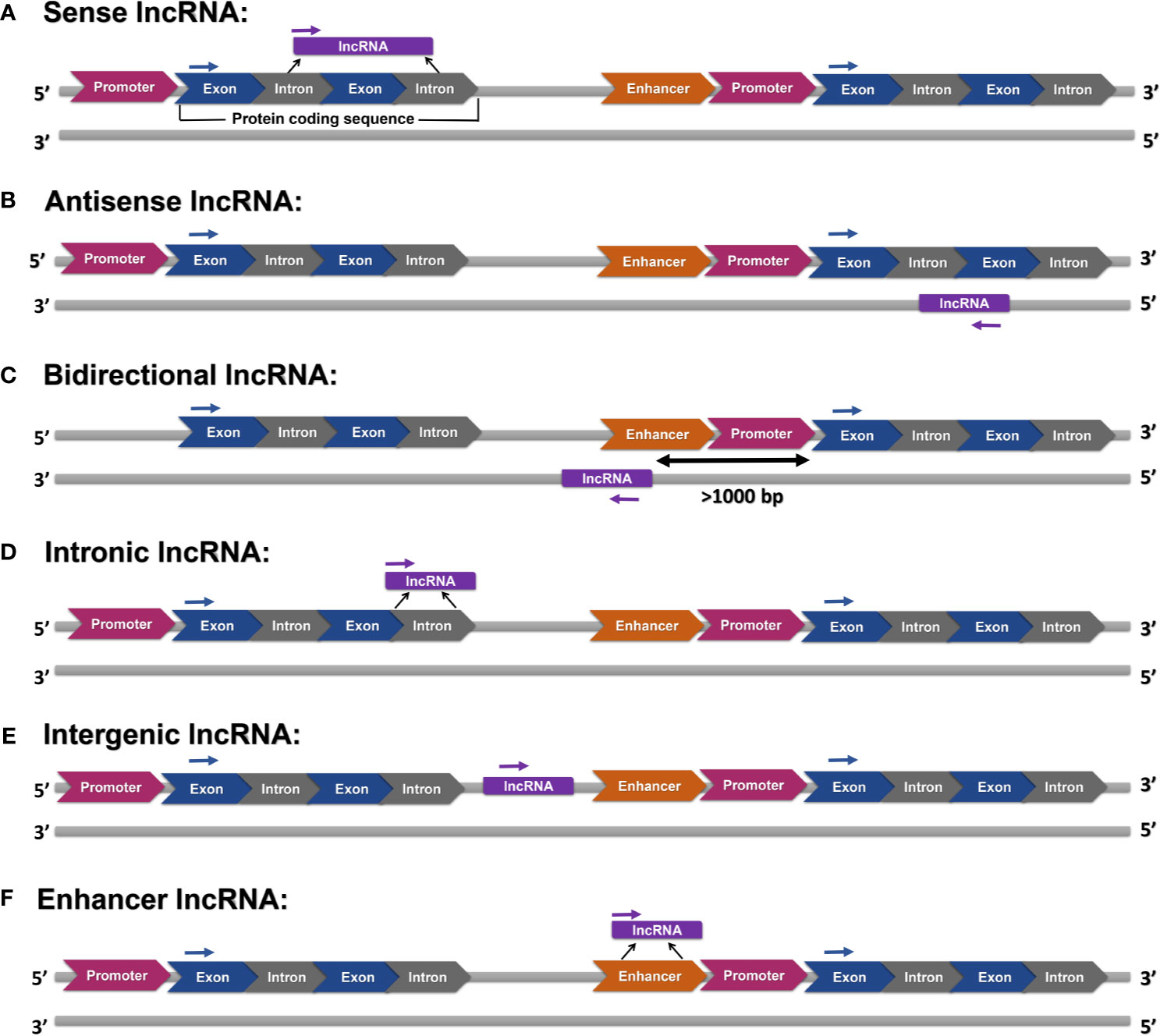
Figure 1 Various categories of lncRNAs: (A) sense lncRNAs are transcripts of one or more exons of protein-coding genes, (B) antisense lncRNAs are transcripts of the opposite strand of protein-coding or non-protein-coding genes, (C) bidirectional lncRNAs are transcribed in an opposite direction, and their transcription is initiated at more than 1000 base pairs (bp) far from the promoter region of a protein-coding gene, (D) intronic lncRNAs are transcribed from introns, (E) intergenic lncRNAs are transcribed from sequences without any overlap with annotated protein-coding genes, and (F) enhancer lncRNAs are produced from enhancer regions.
LncRNAs have various functions in the nucleus and cytoplasm. In the nucleus, they play a role in chromatin remodeling, modulating chromosomal interactions, transcription regulation, and regulation of gene expression at a post-transcriptional level by altering the function and integrity of nuclear bodies. In the cytoplasm, they are involved in mRNA turnover, translation, and post-translational modification regulation. To regulate mRNA stability, competing endogenous RNAs (ceRNA) can modulate mi-RNA availability via vying with mRNAs for mi-RNA and act as mi-RNA sponges. Moreover, lncRNAs can recruit mRNA degradation-associated proteins or act as decoys for RNA binding proteins involved in mRNA decay machinery. lncRNAs can affect translation through interacting with ribosomes or modifying mRNAs to activate their translation. lncRNAs are also involved in a variety of post-transcriptional modifications, most importantly phosphorylation and ubiquitination (9, 10).
LncRNAs in Glioma
MALAT1
Overview - Expression Pattern
Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), also known as nuclear enriched abundant transcript (NEAT)2, is an intergenic lncRNA located on chromosome 11q13. Originally, MALAT1 was introduced as a prognostic marker in non-small cell lung cancer. It is associated with several cancers such as breast, ovarian, prostate, pancreatic cancers, and leukemia (11).
Both higher (12, 13) and lower (14, 15) MALAT1 expression are found in glioma than non-neoplastic tissue. Similarly, among glioma cell lines, glioma stem cell lines showed either lower (14) or higher (15) MALAT1 expression than parental cells. Cancer stem cells showed upregulation of MALAT1 compared to differentiated cancer cells in glioblastoma (16). Notably, between different glioblastoma cell lines, MALAT1 expression was higher in U87 than U251 (15).
Role in Tumor Pathology
Cell cycle and proliferation: MALAT1 knockdown resulted in tumor growth inhibition (17) and induced cell cycle arrest at G1/S phase in glioblastoma (U251) cells putatively via regulating the miR-124/zinc finger E−box binding homeobox 2 (ZEB2) axis (18). Nano complexes of si-MALAT induced G2/M, in addition to G1, cell cycle arrest (16). Accordingly, MALAT1 expression enhanced tumor proliferation by upregulating Rap1b and zinc-fingers and homeoboxes 1 (ZHX1) by sponging miR-101 in glioma (19) and miR-199a in glioblastoma (20), respectively. Notably, ZHX1 plays a key role in glioblastoma progression (21).
In contrast to the above-mentioned mechanisms, Han et al. reported that knockdown of MALAT1 induced tumor proliferation in U87 and U252 cell lines, potentially by suppressing the extracellular signal-regulated kinases (ERK)/mitogen-activated protein kinase (MAPK) signaling pathway (22). In line with their finding, Cao et al. found that MALAT1 can have tumor-suppressing effects by reducing miR-155 expression and increasing expression of FBXW7 tumor suppressor, which interacts with several molecules involved in cellular growth, development, stemness, and cell cycle (14, 23, 24).
Apoptosis: MALAT1 knockdown increased apoptosis and expression of apoptotic regulators, including MYC and CCND1 (encoding cyclin D1) in glioma (13). Inhibition of apoptosis by MALAT1 can also be regulated via the MALAT1/miR-101/Rap1B axis (19) and the miR-124/ZEB2 axis (18). Moreover, inhibition of MALAT1 by si-MALAT1 resulted in a significant decrease in the levels of several molecules involved in apoptosis, such as Bcl-2, inhibitors of apoptosis proteins family, and heat shock protein (HSP) 70 (16). Additionally, MALAT-1 knockdown resulted in lower expression of Bax and higher expression of Bcl-2 via regulating the miR-199a/ZHX1 axis (20).
Autophagy: Although autophagy may induce cytotoxic effects, it has also been suggested to promote the progression and viability of glioma in stressful environments (25). MALAT1 is found to promote tumor progression by enhancing autophagy. Sponging miR-101 not only enhanced tumor proliferation by upregulating Rap1b (19), but also induced higher expression of autophagy-associated genes (Stathmin 1, RAB5A, and ATG4D) (26). MALAT1 acts as a sponge for miR-384 as well. Inhibition of miR-384 activity induced autophagy by putatively interfering with Golgi membrane protein 1 (GOLM1) and led to increased migration and invasion of glioma cells (27, 28).
Invasion and metastasis: Knockdown of MALAT1 suppressed migration and invasion of glioma cells via several mechanisms, such as inhibiting autophagy via regulating the miR-384/GOLM1 axis (27). MALAT1 played a critical role in tumor migration. Notably, Wnt inhibitory factor 1 (WIF1) regulated MALAT1 expression through the non-canonical Wnt signaling pathway (29). MALAT1 also promoted tumor invasiveness via regulating the miR-199a/ZHX1 axis (20).
Conversely, Han et al. found that MALAT1 knockdown increased invasion and proliferation of glioma cells in addition to inducing higher expression of matrix metalloproteinase (MMP)2 (22).
Stemness: MALAT1 overexpression promoted proliferation of glioma stem cells (30) by enhancing SRY-related HMG-box (SOX)-2 expression via inhibiting tumor suppressor miR-129, which led to increased tumor proliferation and viability (17). In addition to SOX-2, MALAT1 downregulation has shown inhibitory effects on the expression of Nestin (another stemness marker) and proliferation of glioma stem cell lines by activating the ERK/MAPK signaling pathway, which is a key pathway in tumor development (15, 31).
Blood tumor barrier (BTB): MALAT1 knockdown led to enhanced BTB permeability and reduced expression of tight junction proteins in glioma endothelial cells via upregulating miR-140. The effect of miR-140 on BTB is mediated by inhibiting expression of nuclear factor YA (NFYA), a regulator of BTB integrity, resulting in increased expression of tight junction proteins (12).
Immunology: In microglia, deactivating MALAT1 using si-MALAT1 modulated the miR-129-5p/high mobility group box 1 protein (HMGB1) axis resulting in a reduced inflammatory response (30).
Clinical Applications
Circulatory biomarker: Compared to healthy controls, glioma patients had lower serum MALAT1 levels (14). Serum levels of MALAT1 have also been used as diagnostic and prognostic biomarkers in some other cancers (32–34).
Prognostic value: Meta-analyses showed that increased MALAT1 expression could predict poor overall survival (35, 36) and higher “tumor, node, metastasis” (TNM) stage (37) in glioma patients. Moreover, tissue expression levels of MALAT1 positively correlated with tumor grade according to the world health organization (WHO) classification and tumor size (18, 38). Serum levels of MALAT1 were also positively associated with WHO grade, tumor size, functional impairment (14), and overall and recurrence-free survival (39). However, Shen et al. did not find a significant association between serum levels of MALAT1 and 2-year survival or disease-free survival (40).
Determining treatment response: MALAT1 plays a major role in tumor chemosensitivity with a higher expression in temozolomide (TMZ)-resistant glioblastoma cells. Its knockdown reduced TMZ resistance both in vivo and in vitro (16, 41). Additionally, elevated serum levels of MALAT1 predicted chemoresistance (39). MALAT1 can modulate treatment response via several mechanisms. It can inhibit the miR-101 pathway through direct binding, resulting in increased apoptosis and suppressed cell growth (41), downregulate miR-203 leading to upregulation of thymidylate synthase (39), and regulate ZEB1 expression (42). Furthermore, inhibition of MALAT1 resulted in decreased expression of multi-drug resistance (MDR)-associated protein 1 (MRP1), a drug efflux pump associated with TMZ resistance (16).
In-vivo therapeutic applications: Silencing MALAT1 suppressed proliferation and malignant behavior of glioma, leading to decreased tumor volume and increased survival (18) in vivo via regulating several pathways, including miR-199a/ZHX1 (20), miR-129/SOX2 (17), and miR-384/GOLM1 (27). In tumor xenograft models, nano complexes of si-MALAT1 targeting cancer stem cells TMZ sensitivity and survival in addition to proliferation inhibition (16, 41), while MALAT1 overexpression induced TMZ resistance (43).
H19
Overview - Expression Pattern
H19 is an imprinted intergenic lncRNA located on chromosome 11p15.5, which is generally expressed by the maternal allele. H19 has well-known oncogenic effects in several cancers, such as hepatocellular carcinoma, bladder, breast, gastric, and colorectal cancers (44).
The expression of H19 was higher in glioma tissue (low and high grade) (45, 46) and cell lines, including U251 and U87MG cells (47, 48).
Role in Tumor Pathology
Cell cycle and proliferation: Knockdown of H19 inhibited glioma cell growth, indicating that H19 interacts with the cell cycle and enhances glioma proliferation (45–47). H19 downregulation induced G0/G1 cell cycle arrest putatively via inhibiting the WNT/β-catenin signaling pathway (48). The oncogenic effects of H19 can be mediated via increased expression of miR-675, which regulates expression of cadherin 13 (45, 49) and vitamin D receptor (a transcriptional factor involved in several cell signaling pathways) (50). H19 also affects tumor proliferation through downregulating miR-152 (51) and upregulating tumor promoter inhibitor of apoptosis-stimulating protein of p53 (iASPP) via targeting miR-140 (52).
Apoptosis: Downregulation of H19 induced apoptosis and stopped the cell cycle (52) mainly by suppressing the Wnt/β-catenin signaling pathway (48), in addition to iASPP upregulation (52). Knockdown of H19 via siRNA resulted in increased TMZ-induced apoptosis rate in U87MG and U251 cell lines in glioblastoma (47).
Autophagy: H19 overexpression suppressed autophagy of glioma cells via regulating the mammalian target of rapamycin (mTOR)/Unc-51 like autophagy activating kinase 1 (ULK1) axis by inducing increased ULK1 phosphorylation and inhibiting mTOR phosphorylation (53).
Invasion and metastasis: in-vitro Matrigel invasion assay showed that overexpression of H19 enhanced the invasiveness of glioblastoma cells (46). Knockdown of H19 inhibited glioma metastasis in vivo and in vitro (52). H19 downregulation inhibited the Wnt/β-catenin signaling pathway (48). Additionally, H19 diminished the inhibitory effect of miR-181d on β-catenin by sponging this tumor suppressor miRNA (54). H19 also upregulated miR-675 (49) and downregulated tumor suppressor miR-152 (51).
Epithelial-mesenchymal transition (EMT) process: EMT, a major role player in tumorigenesis by promoting metastasis, tumor stemness, and chemoresistance, is characterized by increased expression of epithelial markers, such as E-cadherin, and decreased mesenchymal markers, such as N-cadherin, vimentin, and ZEB1/2 (55).
H19 overexpression enhanced mesenchymal markers, namely N-cadherin and vimentin, expression. The potential underlying mechanism was sponging miR-130a-3p, which increased the expression of SOX4 (56), a critical transcription factor in the EMT process (57). Additionally, H19 silencing suppressed EMT (increased E-cadherin expression and decreased ZEB1 and vimentin expression) through inhibiting the Wnt/β-catenin pathway activity (58).
Stemness: H19 was highly expressed in glioblastoma stem cells (CD133+ cells) and promoted stemness (46). Accordingly, its knockdown led to decreased expression of stemness markers, including CD133, NANOG, Oct4, and SOX2 (47).
Angiogenesis: H19 plays a key role in angiogenesis in glioma via several mechanisms, including inhibiting miR-29a and miR-138. The former upregulated vasohibin-2 (VASH2) (an angiogenic factor) (59), and the latter induced higher expression of hypoxia-inducible factor (HIF)-1α and vascular endothelial growth factor (VEGF) (60). Furthermore, in glioblastoma, H19 reduced expression of Nkd1, which is a Wnt pathway inhibitor, via EZH2-mediated epigenetic regulations (61), and its overexpression increased angiogenesis in in-vitro investigations (46).
Clinical applications
Circulatory biomarker: While circulatory H19 levels were a reliable prognostic indicator, to the best of our knowledge, their diagnostic value has not been investigated in glioma (40, 62). Moreover, H19 plasma levels are proposed as a diagnostic biomarker for gastric cancer (63).
Prognostic value: H19 overexpression in glioma tissue was associated with poor overall and progression-free survival and more advanced tumor stage (45, 46, 48, 64). Moreover, serum levels of H19 showed a significant positive correlation with tumor grade (62). However, Shen et al. did not find a significant association between serum levels of H19 and 2-year or disease-free survival (40).
Determining treatment response: TMZ-resistant glioma cell lines had higher H19 expression (58, 65, 66). H19 induced chemoresistance by promoting EMT through the Wnt/B-catenin pathway (58). H19 silencing reduced chemoresistance and increased TMZ-induced apoptosis through inhibiting the NF-κB signaling pathway (47, 65) and downregulating chemoresistance-associated genes (MDR, MRP), and ATP-binding cassette subfamily G member 2 (ABCG2) (66).
In-vivo therapeutic applications: H19 promoted proliferation, migration, and angiogenesis in tumor xenograft investigations, while its knockdown inhibited tumor progression (46, 51, 52). Modulating the miR-342/Wnt5a/β-catenin axis is one of the proposed mechanisms for the oncogenic effect of H19 on tumor growth, metastasis, and angiogenesis in vivo (67). Notably, in a study assessing the therapeutic effects of phenformin in glioblastoma, phenformin was found to inhibit tumor stemness through downregulating H19 and high mobility group A (HMGA)2 (68).
MEG3
Overview - Expression Pattern
Maternally expressed gene 3 (MEG3), also known as gene-trap locus 2 (GTL2) in mice, is a maternally imprinted intergenic lncRNA (like H19) located on chromosome 14q32.3. MEG3 has shown anti-tumoral effects in several cancers, such as lung, breast, liver, gastric, colorectal, ovarian, and cervical, in addition to glioma (69).
MEG3 is downregulated in glioma tissue and cell lines (70–74). Its downregulation can be a result of hypermethylation (75).
Role in Tumor Pathology
Cell cycle and proliferation: MEG3 plays a substantial role in glioma proliferation and cell cycle regulation. Deletion of MEG3 increased tumor cell growth and enhanced cell proliferation in normal human astrocytes (76). MEG3 overexpression led to cell cycle arrest in the G2/M phase in U251 cells (77) and inhibited cell proliferation of glioma cells (71).
MEG3 upregulated key tumor suppressors mainly by interacting with the regulatory miRNAs (69). The p53 protein, encoded by the tumor suppressor protein p53 (TP53) gene, is involved in several cellular protective mechanisms, including inducing cell cycle arrest, DNA repair, and apoptosis (78). MEG3 is required for the activation of the p53 pathway (73). Decreased MEG3 expression due to DNA (cytosine-5)-methyltransferase 1 (DNMT1)- mediated hypermethylation inhibited the p53 pathway in glioma (75). Correspondingly, MEG3 overexpression increased TP53 mRNA levels and suppressed cell proliferation in U251 and U87 cell lines (73).
MEG3 is also associated with phosphatase and TENsin homolog (PTEN) expression, negatively regulating the phosphoinositide 3-kinase (PI3K). miR-19a is found to have repressive effects on PTEN expression. MEG3 acted as a ceRNA for miR-19a, recovering its inhibitory effects on PTEN expression. It resulted in decreased cell proliferation, cell cycle arrest at the G1/S phase, and increased apoptosis (79). Moreover, the regulatory role of the miR-377/PTEN axis was identified in U251 cells (77). MEG3 overexpression also upregulated metastasis suppressor 1 (MTSS1) by downregulating miR-96-5p (71).
Furthermore, EZH2-mediated H3K27me3 enrichment (tri-methylation of lysine 27 on histone H3 protein) of the MEG3 gene downregulated this lncRNA. MEG3 inhibited miR-21-3p expression resulting in reduced tumor proliferation and invasion (70).
MEG3 also modulated Wnt/β-catenin signaling, leading to enhanced tumor proliferation following MEG3 downregulation in glioma (76). MEG3 also increased the expression of SMARCB, which suppressed tumor proliferation and migration by sponging miR-6088 (80).
Apoptosis: MEG3 overexpression induced apoptosis in glioma cell lines, mainly regulated by the interaction of MEG3 and p53 activation (72, 73). Apoptosis was inhibited after silencing of MEG3 in U118 cells. At the same time, it was enhanced following MEG3 overexpression in U251 cells through induction of cell cycle arrest at G2/M phase and increasing mRNA levels of caspase 8/3 and TP53, both playing a crucial role in cell apoptosis (73, 77).
Autophagy: MEG3 overexpression promoted autophagy and induced higher expression of autophagy-associated proteins, including ATG3, ATG5, Beclin-1, LAMP1, and LC3 (72, 81).
Invasion and metastasis: Silencing of MEG3 increased migration and invasion in glioma (77). The interaction of MEG3 and tumor suppressors plays a key role in tumor invasion and metastasis. MEG3 upregulation inhibited metastasis via regulating the miR−96−5p/MTSS1 axis (71). Its downregulation promoted migration and invasion via modulating the miR-19a/PTEN axis through acting as a ceRNA for miR-19a (79). Downregulating miR-21-3p (70) and enhanced expression of SMARCB due to sponging miR-6088 (80) are among other proposed mechanisms by which MEG3 blocks tumor invasion and migration. Nevertheless, since MEG3 overexpression induced autophagy (72), it increased migration and invasion in U87 and U251 cells via this mechanism (81).
EMT: MEG3 overexpression led to reduced EMT with decreased expression of N-cadherin, vimentin, Snail-1, and -catenin (only reported in U251 cells) and increased expression of E-cadherin in U87 and U251 cells (77, 80). Accordingly, MEG3 silencing promoted EMT via regulating the miR-377/PTEN axis (77) in addition to inducing autophagy (81). However, in the U118 cell line, MEG3 overexpression did not significantly change the EMT markers (77). Conversely, Yang et al. reported that MEG3 overexpression induced a more mesenchymal cell-like morphology and increased expression of ZEB1/2. Notably, inhibition of autophagy suppressed MEG3-induced EMT (81).
Clinical Applications
Circulatory biomarker: To the best of our knowledge, the biomarker value of circulatory MEG3 has not been investigated in glioma. Although, circulatory MEG3 has shown biomarker value in other cancers, such as colorectal (82), gastric (83), breast (84), bladder (34), and pancreatic (85).
Prognostic value: Lower MEG3 expression was associated with higher WHO grade, older age at the time of diagnosis, low Karnofsky performance score (KPS), isocitrate dehydrogenase (IDH) wild-type, tumor recurrence, and poor overall survival (72, 74, 76, 86).
Determining treatment response: MEG3 also determined chemoresponse in glioma. TMZ-resistant glioblastoma had a lower MEG3 expression compared to TMZ responders (39). Moreover, enhanced MEG3 expression increased chemosensitivity to cisplatin while MEG3 silencing via si-RNA induced chemoresistance (87).
In-vivo therapeutic applications: Targeting epigenetic regulation of MEG3 expression can provide novel therapeutic choices for glioma. For example, the DNA methylation inhibitor 5-Aza-2’-deoxycytidine (5-AzadC) reduced the abnormal MEG3 promoter hypermethylation and prevented low MEG3 expression (75). Moreover, administration of synthetic miRNAs, such as miR-377 mimic, can help increase MEG3 expression and inhibit tumor migration and invasion (77). Notably, the anti-tumoral effect of tunicamycin was mediated through MEG3 upregulation (88).
HOTAIR
Overview - Expression Pattern
HOX transcript antisense intergenic RNA (HOTAIR), an oncogenic lncRNA located on chromosome 12q13.13, is the first identified trans-acting lncRNA with widely explored roles in breast, lung, cervical, colorectal, and bladder cancers, and glioma (89).
Glioma tissue (both low-grade and high-grade), as well as glioma cell lines (U867 and U251), had higher HOTAIR expression compared to non-neoplastic brain tissue (90–94). Investigating several datasets showed that DNA methylation, particularly methylation of CpG islands, regulated HOTAIR expression with demethylation resulting in increased transcription. Moreover, HOXA9, an oncogenic regulator in glioma (95), also induced HOTAIR expression via interacting with its promoter (91).
Role in Tumor Pathology
Cell cycle and proliferation: HOTAIR is required for the formation of glioblastoma (93) and influenced the cell cycle (96) by regulating molecules having a role in its different phases (97). Several mechanisms have been suggested for the involvement of HOTAIR in the cell cycle. HOTAIR can promote cell growth by suppressing EZH2 [the catalytic component of polycomb repressive complex 2 (PRC2)] activity, which leads to chromatin condensation by binding to the PRC2 complex (98). HOTAIR is found to have reciprocal interactions with miR-15-b and p53. miR-15-b positively regulated p53. Both of these molecules inhibit tumor proliferation and invasion, while HOTAIR activity can suppress their impact (99). Moreover, HOTAIR suppressed the β-catenin pathway, leading to cell cycle arrest and repression of invasion, putatively by downregulating Nemo-like kinase (NLK) in glioblastoma (93). HOTAIR silencing also decreased cyclin D1 expression by upregulating miR-219 (100). Additionally, HOTAIR promoted tumor proliferation by acting as a ceRNA for miR-218, resulting in upregulation of PDE7A (101). HOTAIR also activated the mTOR pathway via regulating miR-125a, resulting in increased tumor viability (102). HOTAIR also downregulated tumor suppressor programmed cell death 4 (PDCD4), leading to increased growth and proliferation of glioma stem cells (103).
Apoptosis: HOTAIR silencing induced apoptosis with several mechanisms. Upregulating PDE7A via decreasing miR-218 expression (101), enhancing miR-219 and Bax expression (100), regulating the miR-15-b/p53 axis (99), and activating the mTOR pathway (102) are among the possible underlying mechanisms.
Angiogenesis: HOTAIR induced angiogenesis via increasing expression of VEGFA in glioma, which was suppressed after HOTAIR silencing (104). Correspondingly, downregulation of HOTAIR inhibited the angiogenesis ability of human umbilical vein endothelial cells putatively by sponging miR-126-5p (105).
Invasion and metastasis: HOTAIR downregulation inhibited tumor invasiveness and migratory abilities. Several molecular mechanisms have been suggested for the positive effect of HOTAIR on tumor progression. Downregulating NLK, resulting in increased activation of the β-catenin pathway (93), suppressing miR-125a, leading to increased activity of the mTOR pathway (102), and inhibiting the tumor suppressor miR-15b/p53 axis (99) are among these mechanisms. Moreover, overexpression of HOTAIR was also associated with higher levels of MMP-7 and MMP-9 (106). Regulating the miR-218/PDE7A axis (101) and glutamine metabolism via downregulating miR-126-5p, which resulted in glutaminase upregulation (105), in addition to inhibiting tumor suppressor PDCD4 (103), are other proposed mechanisms.
BTB: HOTAIR knockdown decreased expression of tight junction proteins, including ZO-1, occludin, and claudin-5, and led to a discontinuous distribution pattern among them by negatively regulating miR-148b-3p and upregulating upstream stimulatory factor (USF)1 (107).
Metabolism: HOTAIR regulated glutamine metabolism, which is essential for glioma progression, by sponging miR-126-5p (105).
Clinical Applications
Circulatory biomarker: Serum HOTAIR levels were significantly higher in glioblastoma patients than controls and correlated with HOTAIR expression within the glioblastoma tissue and glioma grade (108). Circulatory HOTAIR has also been proposed as a potential biomarker for other cancers (109–111).
Prognostic value: In addition to the higher circulatory HOTAIR levels in higher glioma grades (108), several investigations, including those with large datasets, found that HOTAIR is far more expressed in high-grade than low-grade glioma tissue (91, 93, 96, 106). Moreover, IDH-wild type cases, which typically have a poor prognosis, had higher expression levels of HOTAIR (91). Higher HOTAIR expression was an independent predictor of reduced overall survival in glioblastoma (91). Additionally, two single-nucleotide polymorphisms (SNP) of HOTAIR (rs920778 CT and rs12826786 CT genotypes) were also associated with more prolonged overall survival in patients with WHO grade III anaplastic oligodendroglioma (112).
Determining treatment response: Expression of HOTAIR was higher in non-TMZ responder glioblastoma patients compared to responders (39). HOTAIR downregulation induced increased chemosensitivity to TMZ treatment, the underlying mechanism of which may be HOTAIR acting as ceRNA for miR-126-5p (105). Moreover, HOTAIR was found to induce higher expression of HK2 via downregulating miR-125. Increased hexokinase 2 (HK2) expression is associated with chemoresistance putatively through HK-2 mediated lactate production and mitochondria permeability transition pore opening (113). Notably, in addition to HOTAIR, the expression of HK-2 is also related to other lncRNAs, including MALAT1, UCA1, and PVT1.
In-vivo therapeutic applications: In vivo, knockdown of HOTAIR using shRNA inhibited tumor growth and invasiveness and enhanced chemosensitivity (93, 113). Notably, promoter demethylation using 5-Aza-2’-deoxycytidine, which is typically used in the treatment of leukemia, affected the expression of HOTAIR (91). The decreased HOTAIR expression was associated with inhibition of invasiveness, angiogenesis, and chemoresistance (105). Of note, HOTAIR downregulation can mediate the tumor-suppressive effects of some miRNAs, such as miR-326 (90).
PVT1
Overview - Expression Pattern
Plasmacytoma variant translocation 1 (PVT1) is an intergenic lncRNA located on chromosome 8q24, a well-known cancer-associated region. The role of PVT1 has been explored in several cancers such as leukemia, colon, hepatocellular, breast, lung, and ovarian cancers (114).
Several studies, including investigations of large datasets (115), found higher PVT1 expression in glioma tissue and cell lines than normal (116–120).
Role in Tumor Pathology
Cell cycle and proliferation: PVT1 downregulation inhibited tumor growth and expansion both in vitro and in vivo (117) and induced cell cycle arrest at the G1 phase (117–119, 121). The interactions of PVT1 with some miRNAs can mediate its positive effect on tumor proliferation. For instance, PVT1 downregulated miR-128-1-5p leading to increased polypyrimidine tract-binding protein 1 (PTBP1) expression (117). PVT1 silencing also modulated the miR-128-3p/Gremlin 1 (GREM1) axis resulting in inhibition of the bone morphogenetic protein (BMP) signaling pathway and tumor growth (121). Furthermore, PVT1 negatively regulated miR−200a, which has a critical role in glioma development (118). PVT1 also upregulated miR-190a-5p and miR-488-3p, resulting in inhibited expression of myocyte enhancer factor 2C (MEF2C), an oncogenic factor in glioma (119).
Apoptosis: Downregulation of PVT1 promoted apoptosis and DNA damage via increasing expression of Bax and cleaved caspase-3 protein and decreasing Bcl-2 expression. The stimulatory effect of PVT1 knockdown on apoptosis can be mediated via several pathways, including regulating the miR-128-1-5p/PTBP1 axis (117) and the expression of miR-128-3p (121), miR-190a-5p, and miR-488-3p (119).
Autophagy: PVT1 overexpression increased expression of autophagy-associated proteins, namely Atg7 and Beclin1, by inhibiting miR-187 in glioma vascular endothelial cells (122).
Invasion and metastasis: PVT1 induced tumor invasiveness via modulating several target molecules and signaling pathways (118). PVT1 silencing reduced tumor migration and invasiveness via sponging miR-128-3p, which inhibited GREM1 and inhibition of the BMP signaling pathway (121). Moreover, PVT1 silencing suppressed invasion, migration, and expression of MMP-2 and MMP-9 via upregulating miR-128-1-5p, which restrained expression of PTBP1 (117). In another proposed regulatory network, PVT1 knockdown reduced tumor invasiveness and migration via upregulating tumor suppressor miR-424 (120). PVT1 knockdown upregulated miR-190a-5p and miR-488-3p, resulting in inhibited expression of MEF2C. MEF2C upregulates promoter activity of JAGGED 1, which is involved in tumor malignant behavior (119). PVT1 upregulation accelerated migratory abilities of glioma via downregulating up-frameshift protein1 (UPF1), which is a key role player in the nonsense-mediated mRNA decay (NMD) (123).
Angiogenesis: Glioma vascular endothelial cells had a higher PVT1 expression. PVT1 overexpression promoted angiogenesis via degrading miR-186, resulting in upregulated Atg7 and Beclin1 expression (122). Moreover, PVT1 overexpression led to upregulation of connective tissue growth factor (CTGF) and angiopoietin 2 via targeting miR-26b (124).
Clinical Applications
Circulatory biomarker: To the best of our knowledge, the biomarker value of circulatory PVT1 has not been investigated in glioma. Nevertheless, circulatory PVT1 levels had diagnostic and prognostic value in some cancers (125, 126).
Prognostic value: Higher expression of PVT1 was an indicator of poor prognosis (116) and survival (127) in glioma. Patients with higher glioma grade, metastasis, or IDH wild type glioma had higher tissue expression of PVT1 (116, 119, 121, 123, 128). Higher PVT1 expression positively correlated with Ki-67 level and the number of TP53 mutations (127). However, PVT1 expression was not associated with gender, age, KPS score, or tumor size (116). Only a few studies have evaluated the prognostic role of PVT1 SNPs in glioma. Ding et al. reported that while rs13255292 and rs4410871 increased susceptibility to glioma in the Chinese Han population, they do not have a prognostic value (129).
Determining treatment response: In vitro, SHG-44 cells resistant to paclitaxel had higher PVT1 expression, and PVT1 knockdown enhanced chemoresponse (130).
In-vivo therapeutic applications: In vivo, silencing of PVT1 in nude mice with tumor xenograft resulted in decreased tumor volume and weight, which may be mediated via the interaction of PVT1 with miR-128-1-5p (117), miR-128-1-3p (121), and miR-424 (120). Additionally, silencing of PVT1 in addition to miR-190a-5p and miR-488-3p mimics prolonged survival and reduced tumor volume in mice with tumor xenograft (119).
UCA1
Overview - Expression Pattern
Urothelial carcinoma associated 1 (UCA1), located on chromosome 19p13.12, is an intergenic lncRNA involved in several cancers, such as lung, breast, gastric, and colorectal cancers, as well as glioma (131).
Upregulation of UCA1 is reported in glioma tissue and cell lines compared with the normal brain samples (132–136).
Role in Tumor Pathology
Cell cycle and proliferation: UCA1 interacted with the cell cycle. Its knockdown inhibited tumor proliferation, and its overexpression had the opposite effect both in vitro and in vivo (135, 136). UCA1 knockdown induced G0/G1 cell cycle arrest and downregulation of cyclin D1 (132). Cyclin D1 is also involved in the Wnt/β-catenin signaling, which is inhibited following UCA1 knockdown (135). Additionally, sponging tumor suppressor miR-122 (133), miR-135a (136), and enhancing iASSP expression via downregulating miR-182 (137) also contribute to the underlying mechanism of the positive effect of UCA1 expression on tumor proliferation.
Apoptosis: Silencing UCA1 facilitated apoptosis and reduced cell viability, and its overexpression had the opposite effect (135, 138). UCA1 enhanced CDK6 expression via sponging miR‐193a. Notably, CDK6 triggers PI3K/AKT, MAPK, and Notch signaling pathways (138).
Invasion and metastasis: UCA1 overexpression increased invasion and migration (139), while its silencing inhibited tumor progression via several mechanisms (134, 136, 138). UCA1 acted as an endogenous sponge for several tumor suppressor miRNAs, such as miR-122, miR-204-5p, and miR-135a. Therefore, UCA1 suppressed the inhibitory effect of miR-204-5p, miR-135a, and miR-2016 on ZEB1, HOXD9, and CLOCK, respectively, which resulted in their upregulation (133, 134, 136, 139). Moreover, some of the key signaling pathways for tumor progression, including the Wnt/β-catenin, PI3K/AKT, MAPK, and notch signaling pathways, were suppressed following UCA1 knockdown (135). Regulation of the miR-193/CDK6 axis mediated the positive effect of UCA1 on the three latter signaling pathways (138). Additionally, UCA1 enhanced the expression of tumor inducer iASSP expression via inhibiting miR-182 expression (137).
EMT: UCA1 knockdown inhibited the EMT process by increasing the expression of epithelial markers, i.e., E-cadherin, and decreasing the expression of mesenchymal markers, i.e., Slug, N‐cadherin, and vimentin (136, 139, 140). UCA1 upregulated Slug via acting as a ceRNA for miR-1 and miR-203 (140). Moreover, UCA1 upregulated EMT inducers, namely HOXD9, CLOCK, and ZEB1, via sponging miR-135a (136), miR-206 (134), and miR-204-5p, respectively (139). Furthermore, UCA1 is proposed to mediate the positive effect of TGF- β on EMT (140).
Stemness: Knockdown of UCA1 reduced expression of the stemness markers due to regulating the miR-1 and miR-203/Slug axis (140)
Metabolism: UCA1 may play a major role in glycolysis, a well-known characteristic of glioblastoma, via modulating the miR-182/6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 (PFKFB2) axis (141). Notably, the inhibition of glycolysis resulted in tumor-suppressive effects in glioma (142).
Clinical Applications
Circulatory biomarker: Diagnostic values of circulatory UCA1 has been reported in other cancers, such as bladder, gastric cancer, and colorectal cancer (143–145). However, we did not find any reports in patients with glioma.
Prognostic value: Higher expression of UCA1 was associated with higher glioma grade, poor prognosis, and survival (133, 135–137). However, it did not correlate with age, gender, tumor size, and KPS score (132).
Determining treatment response: UCA1 overexpression-induced chemoresistance (shown by an increased IC50) to cisplatin and TMZ in U87 and SHG139 cells. This effect was attenuated by inhibiting the Wnt/β-catenin signaling pathway. Notably, TMZ sensitivity increased after UCA1 knockdown (135).
In-vivo therapeutic applications: In addition to the in vitro tumor suppressor effects of si-UCA1 (132, 133), several studies showed that UCA1 knockdown suppressed tumor progression and reduced tumor volume and weight in tumor xenograft models while its overexpression promoted tumor growth (134–136, 139). miR135a/HOXD9 (136), miR-206/CLOCK (134), and miR-204-5p/ZEB1 (139) axes have been observed in vivo as well.
Other lncRNAs
More than two hundred lncRNAs have been identified to be associated with glioma (146). Providing a detailed review of all of them would be beyond the scope of this review. Therefore, in this section, we give an overview of the other most investigated lncRNAs in glioma.
Role in Tumor Pathology
The roles of the most investigated oncogene lncRNAs and the so far discovered intermediate molecular pathways, in addition to their clinical applications, are summarized in Table 1. Overall, almost all oncogene lncRNAs regulate the cell cycle, promote tumor proliferation, inhibit apoptosis, and induce tumor invasiveness and migration. While lncRNAs such as XIST (162–172), CRDNE (173–179), FOXD2-AS1 (186–194), ANRIL (195–197), HOXA11-AS (198–203), TP73-AS1 (204–206), and DANCR (207–210) are only known as tumor inducers, the ultimate function of some lncRNAs is controversial. For instance, for MALAT1, NEAT1, and TUG1, both oncogenic and tumor suppressor effects have been reported.

Table 1 The most investigated oncogene lncRNAs and the so far discovered intermediate molecular pathways, in addition to their clinical applications.
Multiple studies reported increased expression and an oncogenic effect for NEAT1 in glioma (147–160). NEAT1 increased the activity of several signaling pathways with key roles in the cell cycle, including WNT/β-Catenin and mTOR signaling, leading to increased proliferation, invasion, and metastasis and decreased apoptosis. The interaction of NEAT1 and EZH2, which mediates the trimethylation of H3K27 in their promoters, results in the activation of the WNT/β-Catenin pathway (156). Moreover, NEAT1 activated the mTOR signaling by acting as a ceRNA for miR-185-5p (158). NEAT1 also altered the activity of some cell cycle regulators, including CDK6 and CDK14, via regulating the expression of miR-107 and miR-139-5p, which led to promotion of tumor proliferation, invasion and stemness, and inhibition of apoptosis (148, 149, 154). NEAT1 also enhanced EMT via sponging miR-185-5p (158), and its knockdown resulted in increased BTB permeability by binding to miR-181d-5p. In contrast to these reports, Liu and colleagues found lower NEAT1 expression in glioma tissue compared to the adjacent tissue. They found that NEAT1 overexpression inhibited tumor proliferation and promoted apoptosis via regulating the miR-92b/DKK axis (161).
Moreover, several investigations found that TUG1 had a higher expression in glioma and promoted tumor proliferation, invasion, stemness, and angiogenesis (180–183). TUG1 knockdown resulted in increased apoptosis and induced cell cycle arrest at G0/G1 (180). However, in Li et al.’s study, TUG1 was downregulated in glioma (211). TUG1 acted as a tumor suppressor in few studies, with its downregulation inducing tumor proliferation and its overexpression resulting in increased apoptosis by triggering caspase-3 and caspase-9, inhibiting Bcl-2 (211), and upregulating PTEN (184). Additionally, TUG1 knockdown increased BTB permeability through binding to miR-144 (185).
XIST, another oncogenic lncRNA with increased expression in glioma, plays a major role in regulating the cell cycle leading to increased tumor proliferation and invasion and decreased apoptosis. Some of the underlying molecular mechanisms include regulating the expression of Bcl-2 via cross-talk with miR-204-5p (164), upregulating CREB1 via sponging miR-329 (167), and regulating the insulin receptor substrate 1 (IRS1)/PI3K/Akt pathway via acting as a ceRNA for miR-126. Additionally, XIST also promoted EMT and tumor stemness via regulating the miR-133a/SOX4 and miR-152-Krüppel-like factor 4 (KLF4) axes, respectively. XIST also induced tumor proliferation and angiogenesis via inversely regulating miR-429 (169) and miR-137. As a result of targeting miR-137, XIST knockdown also increased BTB permeability (168).
Unlike the oncogene lncRNAs, tumor suppressor lncRNAs are far less investigated. Table 2 summarizes the most investigated tumor suppressor lncRNAs and the so far discovered intermediate molecular pathways, in addition to their clinical applications. GAS5 (212–220), CASC2 (221–224), TUSC7 (225), and MATN-AS1 (226, 227) are among these lncRNAs.
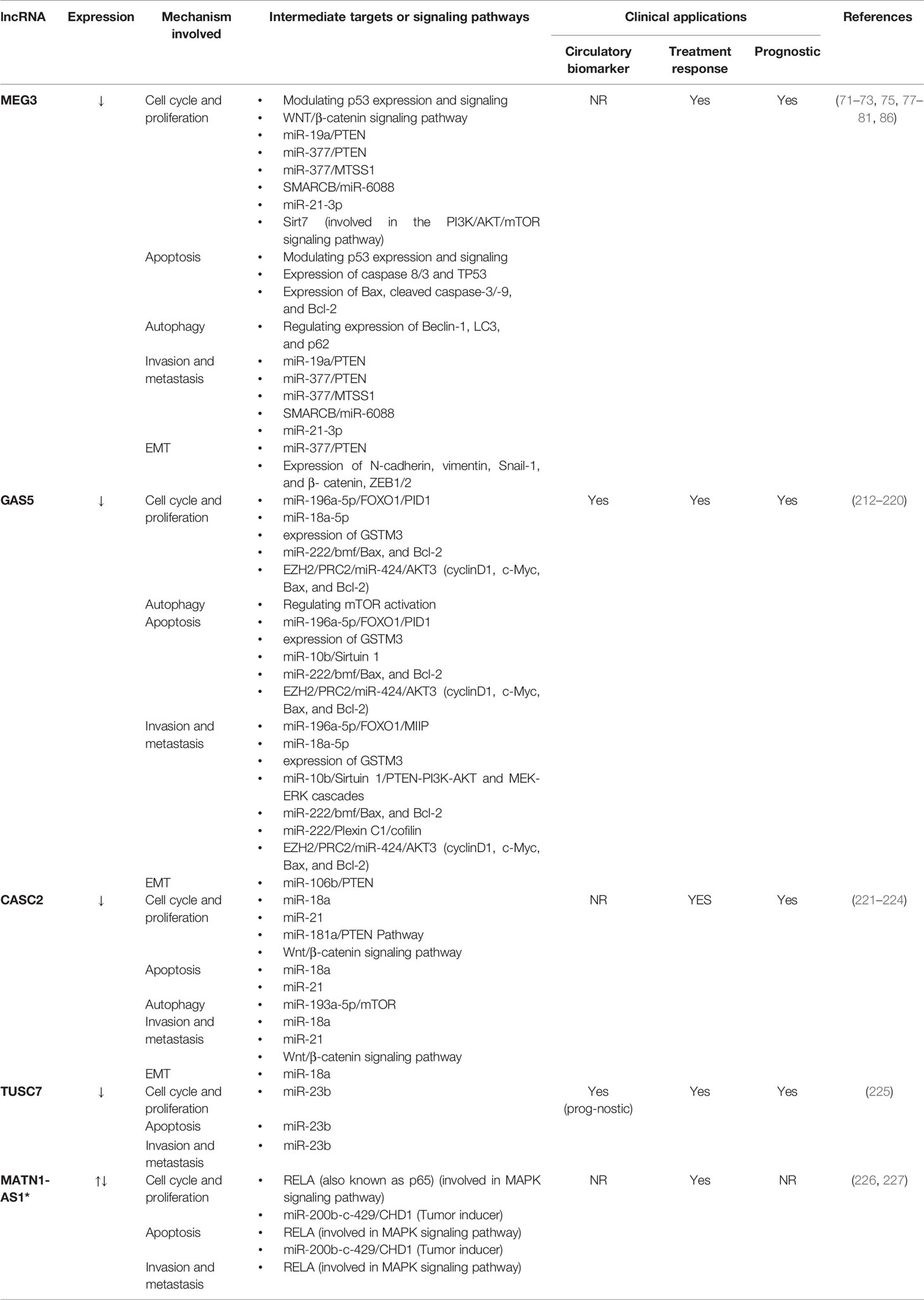
Table 2 The most investigated tumor suppressor lncRNAs and the so far discovered intermediate molecular pathways, in addition to their clinical applications.
Second to MEG3, the anti-tumoral effects of GAS5 are well investigated in glioma. GAS5 has a major role in cell cycle regulation with several mechanisms. For instance, GAS5 regulated the expression of tumor suppressors Bcl-2-modifying factor (Bmf) and Plexin C1 via targeting miR-222, which led to inhibition of tumor progression (215). GAS5 also inhibited tumor inducer miR-196-5p, which led to suppressed tumor growth by positive regulation of tumor suppressors forkhead box protein O1 (FOXO1) and phosphotyrosine interaction domain containing 1 (PID1) (213). Direct interaction of GAS5 and EZH2, in addition to the promotion of miR-424 expression, are among other putative mechanisms for the tumor-suppressing effect of GAS5. miR-424 inhibited AKT3 and regulated the expression of cyclin D1, c-Myc, Bax, and Bcl-2 (214). Furthermore, GAS5 was found to inhibit excessive autophagy in glioma (216).
Moreover, the function of MATN1-AS1 is controversial in glioma. Han et al. found lower expression of MATN1-AS1 in glioblastoma tissue compared to the adjacent tissue, acting as a tumor suppressor (226). Its overexpression inhibited proliferation and promoted apoptosis (226). On the other hand, Zhu and colleagues found higher MATN1-AS1 expression in glioma tissue and cell lines (227). They reported oncogene effects for this lncRNA, with its silencing inhibiting proliferation and promoting apoptosis./
As depicted in Figure 2, lncRNAs are involved in almost all stages of tumorigenesis (228). In addition to the lncRNAs described in Tables 1 and 2, we have included other oncogene lncRNAs, which are upregulated in glioma, such as CCAT1 (229), CCAT2 (230), SNHG16 (231, 232), MIAT (233, 234), DRAIC (235), and HCG11 (236) in this Figure.
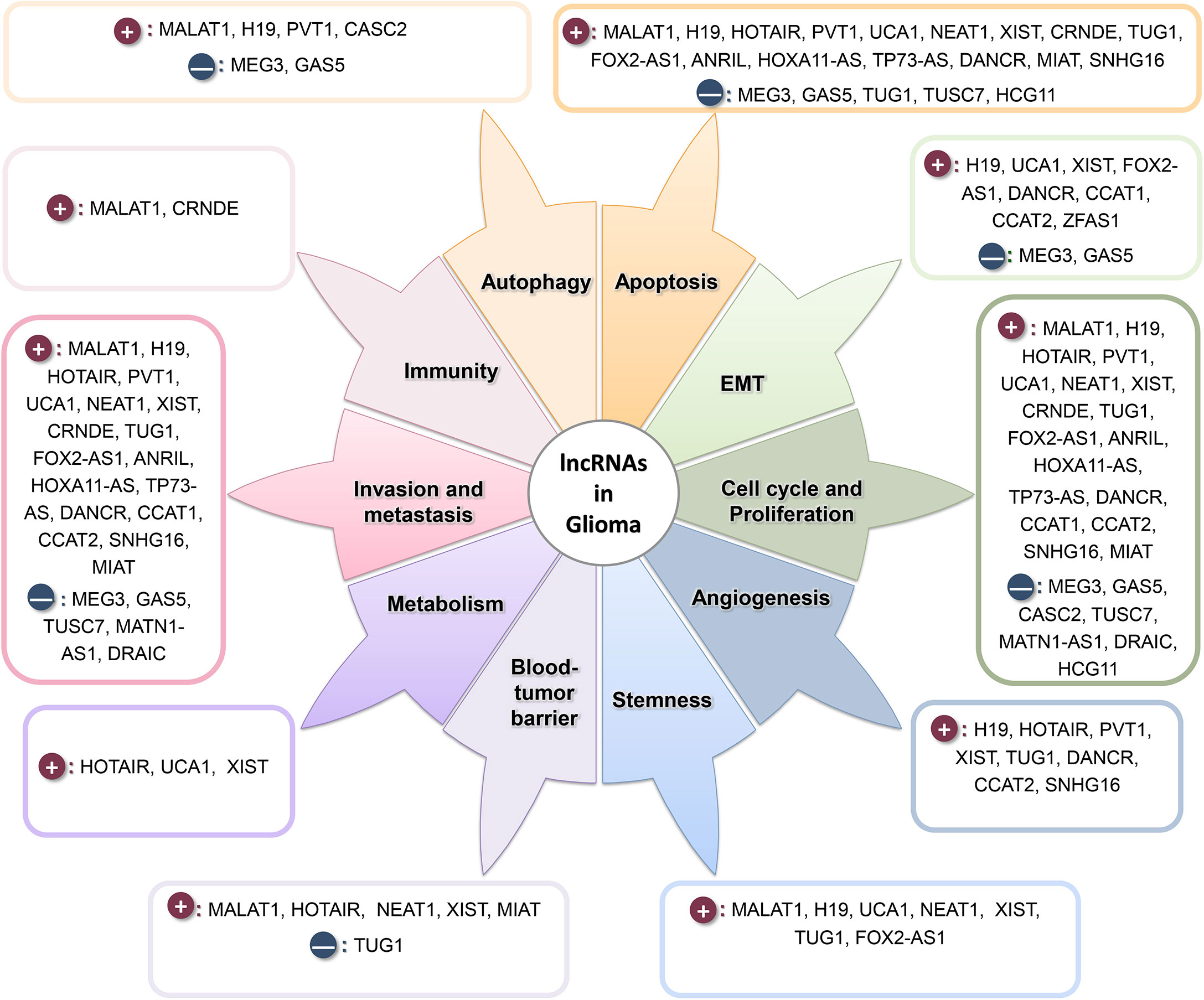
Figure 2 lncRNAs play a prominent role in various stages of tumor formation and progression. Dark-red circles show the oncogene lncRNAs, and navy circles show the tumor suppressor lncRNAs.
Clinical Applications
Circulatory biomarker: Higher circulatory levels of NEAT1 (151), LINK-A (237), and AWPPH (238), in addition to lower serum GASL1 levels (239), have shown considerable diagnostic value for glioma.
In addition to diagnosis, circulatory levels of lncRNAs can also aid in determining prognosis. Lower circulatory levels of GAS5 (40) and TUSC7 (240) and higher circulatory levels of AWPPH (238) were associated with poor prognosis. Moreover, levels of circulating lncRNAs may also illuminate treatment response. For instance, higher levels of lncSBF2-AS1 in serum exosomes were associated with poor TMZ-response (241). The most investigated lncRNAs with either diagnostic or prognostic value for their circulatory levels are described in Figure 3.
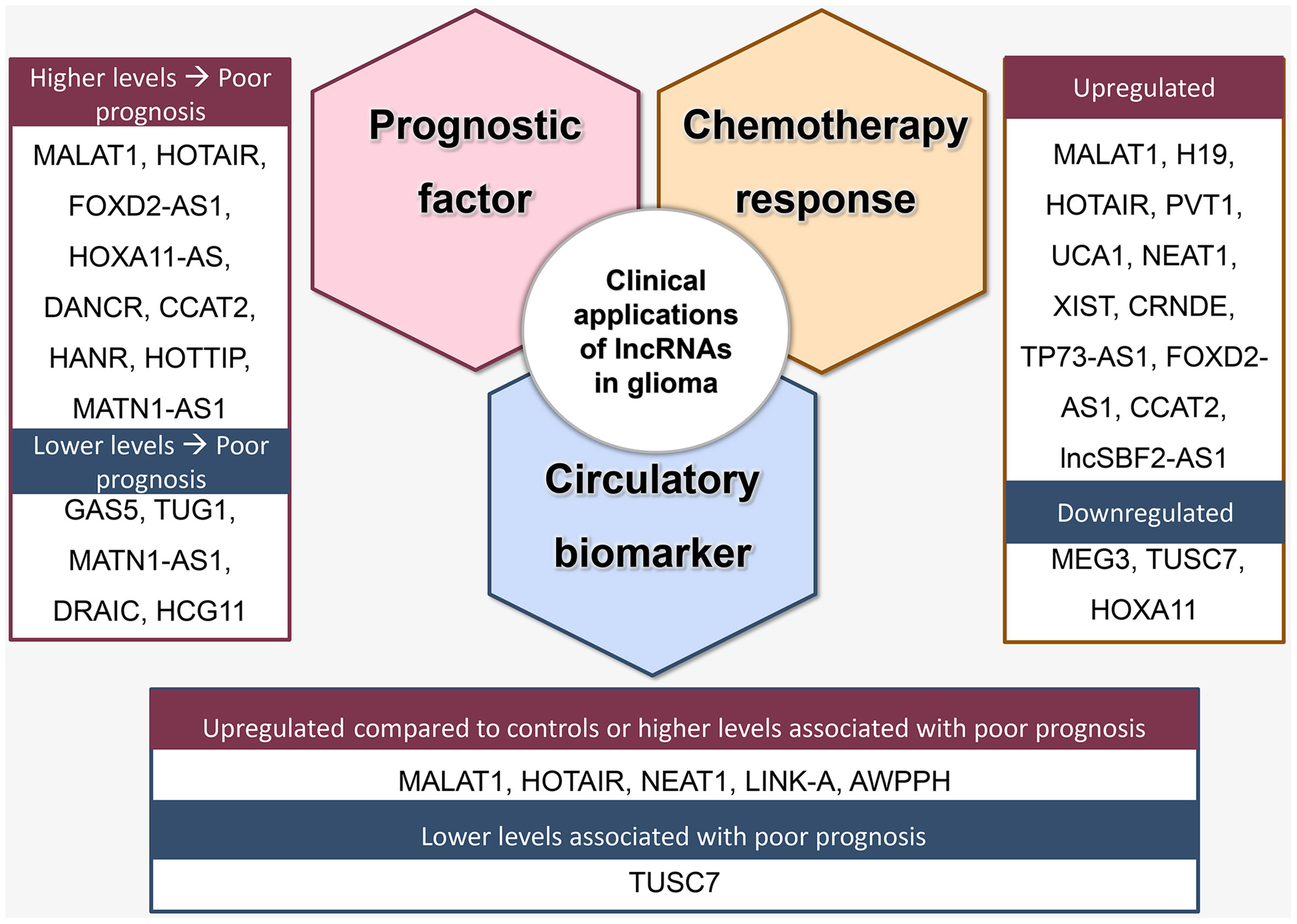
Figure 3 Clinical applications of lncRNA as prognostic factors, predictors of treatment response, and circulatory biomarkers.
Prognostic value: Higher expression of NEAT1 (147), XIST (164), CRNDE (173, 179), FOXD2-AS1 (186, 189, 192, 194), ANRIL (196), HOXA11-AS (198–200, 203), TP73-AS1 (204, 242), and DANCR (208, 210, 243), in addition to lower expression of GAS5 (220), CASC2 (222, 244), TUSC7 (240) correlated with a more advanced stage of disease or poor survival (Figure 3). Notably, controversial findings were reported for some lncRNAs. For MATN1-AS1, while Zhu et al. reported a positive association between its upregulation and tumor advancement and reduced overall survival (227), Han et al. reported that its downregulation was a poor outcome predictor (226). Moreover, while several studies reported an oncogenic effect for TUG1, Wang et al. found that TUG1 expression negatively correlated with tumor grade (245). In contrast to HOXA11-AS, for which several studies reported a positive correlation with poor prognosis, low expression of HOXA11, which is within the same family, was associated with poor outcome in glioblastoma (246). Additionally, higher levels of CCAT2 (247), HOTTIP, HANR, and lower levels of DRAIC and HCG11 were also associated with poor prognosis (248).
In addition to the expression level, SNPs of some lncRNAs may also provide prognostic information. For example, specific ANRIL SNPs were related to the susceptibility of glioma and patients’ overall survival (249, 250).
Determining treatment response: lncRNAs modulate treatment response by affecting sensitivity to chemotherapy drugs, mainly TMZ or cisplatin, or altering radiosensitivity. Higher NEAT1 (151, 160), XIST (172), FOXD2-AS1 (189, 192), TP73-AS1 (251), CCAT2 (252), and lncSBF2-AS1 (241) were associated with TMZ-resistance. These effects are mediated via several mechanisms. For instance, NEAT1 promoted glioma stem cell formation, which is critical for chemoresistance, via activating the Wnt/β-catenin pathway (160). In another example, FOXD2-AS1 reduced methylation and increased activity of O6-methylguanine-DNA methyltransferase (MGMT), which is a treatment response predictor in glioma (192). Furthermore, cisplatin resistance was associated with higher levels of DANCR (253), CRNDE (178), CCAT2 (252), and lower levels of GAS5 (216). Additionally, overexpression of XIST reduced radiosensitivity (167), while high expression of DRAIC was associated with a better prognosis of radiotherapy in low-grade glioma (254).
In-vivo therapeutic applications: For many oncogenic lncRNAs, including XIST (164, 169, 171), NEAT1 (150, 158), CRNDE (173, 175, 176), TUG1 (182, 183), FOXD2-AS1 (190), HOXA11-AS (199, 201), and TP73-AS1 (204, 208), and tumor suppressor lncRNAs, including GAS5 (214, 215), CASC2 (221), MATN1-AS1 (226), animal studies, which are more advanced stages of investigating roles of lncRNAs (255), validated their effect on glioma. Furthermore, lncRNAs can mediate the effects of anti-cancer drugs. For instance, the anti-tumoral effect of sevoflurane was mediated through regulating the ANRIL/let-7b-5p axis (195).
Discussion
Given the mounting and emerging evidence on the roles of lncRNAs in different cancers, including glioma, this review provided a comprehensive summary of the mechanisms of action and clinical relevance of the most investigated lncRNAs in glioma. A profound understanding of the underlying molecular pathways involved in the function of lncRNAs is required to develop novel therapeutic targets. As described earlier, several lncRNA/miRNA/mRNA axes have been proposed to mediate the oncogenic or tumor suppressor effects of lncRNAs. In addition to well-known roles and associations with prognosis and treatment response in various cancers, lncRNAs can provide clinical clues in several non-neoplastic diseases, such as neurodegenerative disease (256) and cardiovascular disease (257, 258). The disrupted pattern of lncRNAs in a wide spectrum of cancers raises the question of whether the roles and associations identified in a particular type of malignancy can be expanded to the other cancer types. A recent study found that while some lncRNAs are consistently associated with better or poorer prognosis across different cancer types, some other lncRNAs show different associations in various cancers (259).
LncRNAs hold promise for developing novel biomarkers and therapeutic targets. To the best of our knowledge, prostate cancer antigen 3 (PCA3) is the only lncRNA approved as a diagnostic biomarker for prostate cancer in clinical practice (260). All in all, given the high specificity of tissue/serum lncRNAs in glioma, they may be excellent candidates for novel biomarkers. LncRNAs secreted as exosomes in body fluids, especially serum, can provide novel non-invasive assessment tools (255). Two main approaches are commonly utilized to modulate the expression of lncRNAs, namely antisense oligonucleotides (ASOs) and duplex RNAs, such as siRNA. ASOs may be preferred for lncRNAs functioning in the nucleus, while siRNA may be selected for lncRNAs functioning in the cytoplasm (261). While oligonucleotide therapeutics provide an opportunity to target any gene of choice, there are several obstacles in their clinical use. As a consequence of their chemical structure, they are susceptible to rapid enzymatic and nonenzymatic degradation. Moreover, their relatively large size hinders their penetration through the blood-brain barrier and cellular uptake. Therefore, novel delivery systems, such as chemical engineering and nanoparticles, are needed to overcome these challenges (262, 263).
Despite considerable attention drawn to lncRNAs in cancer and the growing evidence, there are several gaps in the literature. The findings of some studies are not sufficiently reliable due to small sample sizes. Moreover, the majority of investigations are in-vitro assessments highlighting the need for further validation by nude animal models and clinical trials. However, the low conserveness of lncRNAs among different species may hinder using animal models since the function of a certain lncRNA can be different between humans and animal models. As a result, engineered models may be required (261).
Our study shed light on several directions for future studies. In addition to the need for increased in-vivo investigations and studies with larger sample sizes, it is not elucidated whether disruption in expression of lncRNAs is a culprit or consequence of the malignancy. Future studies need to address this question, particularly by investigating upstream regulators of the expression of lncRNAs. Moreover, the development of specific panels of lncRNAs for diagnostic or prognostic applications can lead to increased specificity and sensitivity. Several studies have already taken this approach and sought to find specific signatures of lncRNAs for glioma (264–266). Additionally, more studies are required on the therapeutic effects of combining the lncRNA-targeted therapies and conventional chemotherapy. For instance, a better outcome was achieved after administrating si-MALAT1 in addition to TMZ in vitro and in animal models (16). Additionally, detecting the disease-associated lncRNAs and their regulatory pathways can also lead to finding novel putative drugs (267). Lastly, the advancement of computational technologies and bioinformatics have provided novel opportunities for identifying new lncRNAs and their potential molecular mechanism (268). Applying artificial intelligence technology, including machine-learning and deep-learning models, can also aid in the identification of novel lncRNAs associated with a specific disease mainly via classification models (269).
To conclude, regulating diverse cellular signaling pathways and the expression of various proteins involved in different stages of tumor formation, proliferation, and invasion, lncRNAs are potential candidates for developing novel diagnostic, prognostic, and therapeutic approaches. The so far discovered associations between their expression in the tissue or circulatory exosomes with treatment response and prognosis boost hopes for their potential use in clinical practice. However, despite substantial advances, the role of lncRNAs in glioma remains fairly unknown. It is not well known whether the disruption in the expression of lncRNAs has a causal effect on glioma or is a consequence of the malignant process. Moreover, some controversial reports hinder drawing a concrete conclusion. More investigations with larger sample sizes and increased focus on in-vivo models are required to expand our understanding of the potential roles and application of lncRNAs in glioma.
Author Contributions
SM: Conceptualization, Investigation, Writing - Original Draft, and Visualization. NR: Conceptualization, Writing - Review & Editing, Supervision. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Glossary

References
1. Ostrom QT, Bauchet L, Davis FG, Deltour I, Fisher JL, Langer CE, et al. The Epidemiology of Glioma in Adults: A “State of the Science” Review. Neuro Oncol (2014) 16(7):896–913. doi: 10.1093/neuonc/nou087
2. Delgado-Lopez PD, Corrales-Garcia EM. Survival in Glioblastoma: A Review on the Impact of Treatment Modalities. Clin Transl Oncol (2016) 18(11):1062–71. doi: 10.1007/s12094-016-1497-x
3. Liang J, Lv X, Lu C, Ye X, Chen X, Fu J, et al. Prognostic Factors of Patients With Gliomas – An Analysis on 335 Patients With Glioblastoma and Other Forms of Gliomas. BMC Cancer (2020) 20(1):35. doi: 10.1186/s12885-019-6511-6
4. Latowska J, Grabowska A, Zarebska Z, Kuczynski K, Kuczynska B, Rolle K. Non-Coding RNAs in Brain Tumors, the Contribution of lncRNAs, circRNAs, and snoRNAs to Cancer Development-Their Diagnostic and Therapeutic Potential. Int J Mol Sci (2020) 21(19):7001. doi: 10.3390/ijms21197001
5. Zhou Q, Liu J, Quan J, Liu W, Tan H, Li W. MicroRNAs as Potential Biomarkers for the Diagnosis of Glioma: A Systematic Review and Meta-Analysis. Cancer Sci (2018) 109(9):2651–9. doi: 10.1111/cas.13714
6. Banelli B, Forlani A, Allemanni G, Morabito A, Pistillo MP, Romani M. MicroRNA in Glioblastoma: An Overview. Int J Genomics (2017) 2017:7639084. doi: 10.1155/2017/7639084
7. Linhares P, Carvalho B, Vaz R, Costa BM. Glioblastoma: Is There Any Blood Biomarker With True Clinical Relevance? Int J Mol Sci (2020) 21(16):5809. doi: 10.3390/ijms21165809
8. Cech TR, Steitz JA. The Noncoding RNA Revolution—Trashing Old Rules to Forge New Ones. Cell (2014) 157(1):77–94. doi: 10.1016/j.cell.2014.03.008
9. Yao RW, Wang Y, Chen LL. Cellular Functions of Long Noncoding RNAs. Nat Cell Biol (2019) 21(5):542–51. doi: 10.1038/s41556-019-0311-8
10. Zhang X, Wang W, Zhu W, Dong J, Cheng Y, Yin Z, et al. Mechanisms and Functions of Long Non-Coding RNAs at Multiple Regulatory Levels. Int J Mol Sci (2019) 20(22):5573. doi: 10.3390/ijms20225573
11. Sun Y, Ma L. New Insights Into Long Non-Coding RNA MALAT1 in Cancer and Metastasis. Cancers (Basel) (2019) 11(2):216. doi: 10.3390/cancers11020216
12. Ma J, Wang P, Yao Y, Liu Y, Li Z, Liu X, et al. Knockdown of Long non-Coding RNA MALAT1 Increases the Blood-Tumor Barrier Permeability by Up-Regulating miR-140. Biochim Biophys Acta (2016) 1859(2):324–38. doi: 10.1016/j.bbagrm.2015.11.008
13. Xiang J, Guo S, Jiang S, Xu Y, Li J, Li L, et al. Silencing of Long Non-Coding RNA MALAT1 Promotes Apoptosis of Glioma Cells. J Korean Med Sci (2016) 31(5):688–94. doi: 10.3346/jkms.2016.31.5.688
14. Cao S, Wang Y, Li J, Lv M, Niu H, Tian Y. Tumor-Suppressive Function of Long Noncoding RNA MALAT1 in Glioma Cells by Suppressing miR-155 Expression and Activating FBXW7 Function. Am J Cancer Res (2016) 6(11):2561–74.
15. Han Y, Zhou L, Wu T, Huang Y, Cheng Z, Li X, et al. Downregulation of lncRNA-MALAT1 Affects Proliferation and the Expression of Stemness Markers in Glioma Stem Cell Line SHG139S. Cell Mol Neurobiol (2016) 36(7):1097–107. doi: 10.1007/s10571-015-0303-6
16. Kim SS, Harford JB, Moghe M, Rait A, Pirollo KF, Chang EH. Targeted Nanocomplex Carrying siRNA Against MALAT1 Sensitizes Glioblastoma to Temozolomide. Nucleic Acids Res (2018) 46(3):1424–40. doi: 10.1093/nar/gkx1221
17. Xiong Z, Wang L, Wang Q, Yuan Y. LncRNA MALAT1/miR-129 Axis Promotes Glioma Tumorigenesis by Targeting SOX2. J Cell Mol Med (2018) 22:3929–40. doi: 10.1111/jcmm.13667
18. Cheng H, Zhao H, Xiao X, Huang Q, Zeng W, Tian B, et al. Long Non-Coding RNA MALAT1 Upregulates ZEB2 Expression to Promote Malignant Progression of Glioma by Attenuating miR-124. Mol Neurobiol (2020) 52:1006–16. doi: 10.1007/s12035-020-02165-0
19. Li Z, Xu C, Ding B, Gao M, Wei X, Ji N. Long Non-Coding RNA MALAT1 Promotes Proliferation and Suppresses Apoptosis of Glioma Cells Through Derepressing Rap1B by Sponging miR-101. J Neurooncol (2017) 134(1):19–28. doi: 10.1007/s11060-017-2498-5
20. Liao K, Lin Y, Gao W, Xiao Z, Medina R, Dmitriev P, et al. Blocking lncRNA MALAT1/miR-199a/ZHX1 Axis Inhibits Glioblastoma Proliferation and Progression. Mol Ther Nucleic Acids (2019) 18:388–99. doi: 10.1016/j.omtn.2019.09.005
21. Kwon RJ, Han ME, Kim YJ, Kim YH, Kim JY, Liu L, et al. Roles of Zinc-Fingers and Homeoboxes 1 During the Proliferation, Migration, and Invasion of Glioblastoma Cells. Tumour Biol (2017) 39(3):1010428317694575. doi: 10.1177/1010428317694575
22. Han Y, Wu Z, Wu T, Huang Y, Cheng Z, Li X, et al. Tumor-Suppressive Function of Long Noncoding RNA MALAT1 in Glioma Cells by Downregulation of MMP2 and Inactivation of ERK/MAPK Signaling. Cell Death Dis (2016) 7:e2123. doi: 10.1038/cddis.2015.407
23. Cao J, Ge MH, Ling ZQ. Fbxw7 Tumor Suppressor: A Vital Regulator Contributes to Human Tumorigenesis. Medicine (Baltimore) (2016) 95(7):e2496. doi: 10.1097/MD.0000000000002496
24. Yeh C-H, Bellon M, Nicot C. FBXW7: A Critical Tumor Suppressor of Human Cancers. Mol Cancer (2018) 17(1):115. doi: 10.1186/s12943-018-0857-2
25. Feng F, Zhang M, Yang C, Heng X, Wu X. The Dual Roles of Autophagy in Gliomagenesis and Clinical Therapy Strategies Based on Autophagic Regulation Mechanisms. BioMed Pharmacother (2019) 120:109441. doi: 10.1016/j.biopha.2019.109441
26. Fu Z, Luo W, Wang J, Peng T, Sun G, Shi J, et al. Malat1 Activates Autophagy and Promotes Cell Proliferation by Sponging miR-101 and Upregulating STMN1, RAB5A and ATG4D Expression in Glioma. Biochem Biophys Res Commun (2017) 492(3):480–6. doi: 10.1016/j.bbrc.2017.08.070
27. Ma R, Zhang BW, Zhang ZB, Deng QJ. LncRNA MALAT1 Knockdown Inhibits Cell Migration and Invasion by Suppressing Autophagy Through miR-384/GOLM1 Axis in Glioma. Eur Rev Med Pharmacol Sci (2020) 24(5):2601–15. doi: 10.26355/eurrev_202003_20529
28. Xu R, Ji J, Zhang X, Han M, Zhang C, Xu Y, et al. PDGFA/PDGFRalpha-Regulated GOLM1 Promotes Human Glioma Progression Through Activation of AKT. J Exp Clin Cancer Res (2017) 36(1):193. doi: 10.1186/s13046-017-0665-3
29. Vassallo I, Zinn P, Lai M, Rajakannu P, Hamou MF, Hegi ME. WIF1 Re-Expression in Glioblastoma Inhibits Migration Through Attenuation of non-Canonical WNT Signaling by Downregulating the lncRNA MALAT1. Oncogene (2016) 35(1):12–21. doi: 10.1038/onc.2015.61
30. Yang J, Sun G, Hu Y, Yang J, Shi Y, Liu H, et al. Extracellular Vesicle lncRNA Metastasis-Associated Lung Adenocarcinoma Transcript 1 Released From Glioma Stem Cells Modulates the Inflammatory Response of Microglia After Lipopolysaccharide Stimulation Through Regulating miR-129-5p/High Mobility Group Box-1 Protein Axis. Front Immunol (2019) 10:3161. doi: 10.3389/fimmu.2019.03161
31. Guo YJ, Pan WW, Liu SB, Shen ZF, Xu Y, Hu LL. ERK/MAPK Signalling Pathway and Tumorigenesis. Exp Ther Med (2020) 19(3):1997–2007. doi: 10.3892/etm.2020.8454
32. Huang SK, Luo Q, Peng H, Li J, Zhao M, Wang J, et al. A Panel of Serum Noncoding RNAs for the Diagnosis and Monitoring of Response to Therapy in Patients With Breast Cancer. Med Sci Monit (2018) 24:2476–88. doi: 10.12659/msm.909453
33. Zhang R, Xia Y, Wang Z, Zheng J, Chen Y, Li X, et al. Serum Long Non Coding RNA MALAT-1 Protected by Exosomes Is Up-Regulated and Promotes Cell Proliferation and Migration in Non-Small Cell Lung Cancer. Biochem Biophys Res Commun (2017) 490(2):406–14. doi: 10.1016/j.bbrc.2017.06.055
34. Duan W, Du L, Jiang X, Wang R, Yan S, Xie Y, et al. Identification of a Serum Circulating lncRNA Panel for the Diagnosis and Recurrence Prediction of Bladder Cancer. Oncotarget (2016) 7(48):78850–8. doi: 10.18632/oncotarget.12880
35. Zhou Q, Liu J, Quan J, Liu W, Tan H, Li W. lncRNAs as Potential Molecular Biomarkers for the Clinicopathology and Prognosis of Glioma: A Systematic Review and Meta-Analysis. Gene (2018) 668:77–86. doi: 10.1016/j.gene.2018.05.054
36. Li J, Cui Z, Li H, Lv X, Gao M, Yang Z, et al. Clinicopathological and Prognostic Significance of Long Noncoding RNA MALAT1 in Human Cancers: A Review and Meta-Analysis. Cancer Cell Int (2018) 18:109. doi: 10.1186/s12935-018-0606-z
37. Wang Y, Xue D, Li Y, Pan X, Zhang X, Kuang B, et al. The Long Noncoding RNA MALAT-1 Is A Novel Biomarker in Various Cancers: A Meta-Analysis Based on the GEO Database and Literature. J Cancer (2016) 7(8):991–1001. doi: 10.7150/jca.14663
38. Ma KX, Wang HJ, Li XR, Li T, Su G, Yang P, et al. Long Noncoding RNA MALAT1 Associates With the Malignant Status and Poor Prognosis in Glioma. Tumour Biol (2015) 36(5):3355–9. doi: 10.1007/s13277-014-2969-7
39. Chen W, Xu XK, Li JL, Kong KK, Li H, Chen C, et al. MALAT1 Is a Prognostic Factor in Glioblastoma Multiforme and Induces Chemoresistance to Temozolomide Through Suppressing miR-203 and Promoting Thymidylate Synthase Expression. Oncotarget (2017) 8(14):22783–99. doi: 10.18632/oncotarget.15199
40. Shen J, Hodges TR, Song R, Gong Y, Calin GA, Heimberger AB, et al. Serum HOTAIR and GAS5 Levels as Predictors of Survival in Patients With Glioblastoma. Mol Carcinog (2018) 57(1):137–41. doi: 10.1002/mc.22739
41. Cai T, Liu Y, Xiao J. Long Noncoding RNA MALAT1 Knockdown Reverses Chemoresistance to Temozolomide via Promoting microRNA-101 in Glioblastoma. Cancer Med (2018) 7(4):1404–15. doi: 10.1002/cam4.1384
42. Kruszka P, Addissie YA, McGinn DE, Porras AR, Biggs E, Share M, et al. 22q11.2 Deletion Syndrome in Diverse Populations. Am J Med Genet A (2017) 173(4):879–88. doi: 10.1002/ajmg.a.38199
43. Li H, Yuan X, Yan D, Li D, Guan F, Dong Y, et al. Long Non-Coding RNA MALAT1 Decreases the Sensitivity of Resistant Glioblastoma Cell Lines to Temozolomide. Cell Physiol Biochem (2017) 42(3):1192–201. doi: 10.1159/000478917
44. Ghafouri-Fard S, Esmaeili M, Taheri M. H19 lncRNA: Roles in Tumorigenesis. BioMed Pharmacother (2020) 123:109774. doi: 10.1016/j.biopha.2019.109774
45. Zhang T, Wang YR, Zeng F, Cao HY, Zhou HD, Wang YJ. LncRNA H19 Is Overexpressed in Glioma Tissue, Is Negatively Associated With Patient Survival, and Promotes Tumor Growth Through its Derivative miR-675. Eur Rev Med Pharmacol Sci (2016) 20(23):4891–7.
46. Jiang X, Yan Y, Hu M, Chen X, Wang Y, Dai Y, et al. Increased Level of H19 Long Noncoding RNA Promotes Invasion, Angiogenesis, and Stemness of Glioblastoma Cells. J Neurosurg (2016) 2016(1):129–36. doi: 10.3171/2014.12.JNS1426.test
47. Li W, Jiang P, Sun X, Xu S, Ma X, Zhan R. Suppressing H19 Modulates Tumorigenicity and Stemness in U251 and U87MG Glioma Cells. Cell Mol Neurobiol (2016) 36(8):1219–27. doi: 10.1007/s10571-015-0320-5
48. Guan N, Wang R, Guo WS, Lai YJ, Zhang YD, Cheng YY. Long Non-Coding RNA H19 Regulates the Development of Gliomas Through the Wnt/beta-Catenin Signaling Pathway. Eur Rev Med Pharmacol Sci (2019) 23(10):4243–53. doi: 10.26355/eurrev_201905_17929
49. Shi Y, Wang Y, Luan W, Wang P, Tao T, Zhang J, et al. Long Non-Coding RNA H19 Promotes Glioma Cell Invasion by Deriving miR-675. PloS One (2014) 9(1):e86295. doi: 10.1371/journal.pone.0086295
50. Pan JX, Chen TN, Ma K, Wang S, Yang CY, Cui GY. A Negative Feedback Loop of H19/miR-675/VDR Mediates Therapeutic Effect of Cucurmin in the Treatment of Glioma. J Cell Physiol (2020) 235(3):2171–82. doi: 10.1002/jcp.29127
51. Chen L, Wang Y, He J, Zhang C, Chen J, Shi D. Long Noncoding RNA H19 Promotes Proliferation and Invasion in Human Glioma Cells by Downregulating miR-152. Oncol Res (2018) 26(9):1419–28. doi: 10.3727/096504018X15178768577951
52. Zhao H, Peng R, Liu Q, Liu D, Du P, Yuan J, et al. The lncRNA H19 Interacts With miR-140 to Modulate Glioma Growth by Targeting iASPP. Arch Biochem Biophys (2016) 610:1–7. doi: 10.1016/j.abb.2016.09.014
53. Zhao W, Lin X, Han H, Zhang H, Li X, Jiang C, et al. Long Noncoding RNA H19 Contributes to the Proliferation and Autophagy of Glioma Cells Through mTOR/ULK1 Pathway. Neuroreport (2021) 32(5):352–8. doi: 10.1097/WNR.0000000000001602
54. Wu W, Hu Q, Nie E, Yu T, Wu Y, Zhi T, et al. Hypoxia Induces H19 Expression Through Direct and Indirect Hif-1alpha Activity, Promoting Oncogenic Effects in Glioblastoma. Sci Rep (2017) 7:45029. doi: 10.1038/srep45029
55. Cheng JT, Wang L, Wang H, Tang FR, Cai WQ, Sethi G, et al. Insights Into Biological Role of LncRNAs in Epithelial-Mesenchymal Transition. Cells (2019) 8(10):216. doi: 10.3390/cells8101178
56. Hu Q, Yin J, Zeng A, Jin X, Zhang Z, Yan W, et al. H19 Functions as a Competing Endogenous RNA to Regulate EMT by Sponging miR-130a-3p in Glioma. Cell Physiol Biochem (2018) 50(1):233–45. doi: 10.1159/000494002
57. Hanieh H, Ahmed EA, Vishnubalaji R, Alajez NM. SOX4: Epigenetic Regulation and Role in Tumorigenesis. Semin Cancer Biol (2020) 67(Pt 1):91–104. doi: 10.1016/j.semcancer.2019.06.022
58. Jia L, Tian Y, Chen Y, Zhang G. The Silencing of LncRNA-H19 Decreases Chemoresistance of Human Glioma Cells to Temozolomide by Suppressing Epithelial-Mesenchymal Transition via the Wnt/beta-Catenin Pathway. Onco Targets Ther (2018) 11:313–21. doi: 10.2147/OTT.S154339
59. Jia P, Cai H, Liu X, Chen J, Ma J, Wang P, et al. Long Non-Coding RNA H19 Regulates Glioma Angiogenesis and the Biological Behavior of Glioma-Associated Endothelial Cells by Inhibiting microRNA-29a. Cancer Lett (2016) 381(2):359–69. doi: 10.1016/j.canlet.2016.08.009
60. Liu ZZ, Tian YF, Wu H, Ouyang SY, Kuang WL. LncRNA H19 Promotes Glioma Angiogenesis Through miR-138/HIF-1alpha/VEGF Axis. Neoplasma (2020) 67(1):111–8. doi: 10.4149/neo_2019_190121N61
61. Fazi B, Garbo S, Toschi N, Mangiola A, Lombari M, Sicari D, et al. The lncRNA H19 Positively Affects the Tumorigenic Properties of Glioblastoma Cells and Contributes to NKD1 Repression Through the Recruitment of EZH2 on its Promoter. Oncotarget (2018) 9(21):15512–25. doi: 10.18632/oncotarget.24496
62. Shen L, Xu M, Wang Z, Yu Z. Prognostic Evaluation of Serum Long Non-Coding RNA H19 for Endoscopic Keyhole Surgery or Craniotomy in Glioma. Ann Clin Biochem (2020) 57(5):365–72. doi: 10.1177/0004563220941888
63. Zhou X, Yin C, Dang Y, Ye F, Zhang G. Identification of the Long Non-Coding RNA H19 in Plasma as a Novel Biomarker for Diagnosis of Gastric Cancer. Sci Rep (2015) 5:11516. doi: 10.1038/srep11516
64. Fawzy MS, Ellawindy A, Hussein MH, Khashana MS, Darwish MK, Abdel-Daim MM, et al. Long Noncoding RNA H19, and Not microRNA miR-326, Is Over-Expressed and Predicts Survival in Glioblastoma. Biochem Cell Biol (2018) 96(6):832–9. doi: 10.1139/bcb-2018-0122
65. Duan S, Li M, Wang Z, Wang L, Liu Y. H19 Induced by Oxidative Stress Confers Temozolomide Resistance in Human Glioma Cells via Activating NF-kappaB Signaling. Onco Targets Ther (2018) 11:6395–404. doi: 10.2147/OTT.S173244
66. Jiang P, Wang P, Sun X, Yuan Z, Zhan R, Ma X, et al. Knockdown of Long Noncoding RNA H19 Sensitizes Human Glioma Cells to Temozolomide Therapy. Onco Targets Ther (2016) 9:3501–9. doi: 10.2147/OTT.S96278
67. Zhou Q, Liu ZZ, Wu H, Kuang WL. LncRNA H19 Promotes Cell Proliferation, Migration, and Angiogenesis of Glioma by Regulating Wnt5a/beta-Catenin Pathway via Targeting miR-342. Cell Mol Neurobiol (2020). doi: 10.1007/s10571-020-00995-z
68. Jiang W, Finniss S, Cazacu S, Xiang C, Brodie Z, Mikkelsen T, et al. Repurposing Phenformin for the Targeting of Glioma Stem Cells and the Treatment of Glioblastoma. Oncotarget (2016) 7(35):56456–70. doi: 10.18632/oncotarget.10919
69. Al-Rugeebah A, Alanazi M, Parine NR. MEG3: An Oncogenic Long Non-Coding RNA in Different Cancers. Pathol Oncol Res (2019) 25(3):859–74. doi: 10.1007/s12253-019-00614-3
70. Qin WX, Shi Y, Zhu D, Li YP, Chen YH, Cui J, et al. EZH2-Mediated H3K27me3 Enrichment on the lncRNA MEG3 Promoter Regulates the Growth and Metastasis of Glioma Cells by Regulating miR-21-3p. Eur Rev Med Pharmacol Sci (2020) 24(6):3204–14. doi: 10.26355/eurrev_202003_20687
71. Zhang S, Guo W. Long Noncoding RNA MEG3 Suppresses the Growth of Glioma Cells by Regulating the miR965p/MTSS1 Signaling Pathway. Mol Med Rep (2019) 20(5):4215–25. doi: 10.3892/mmr.2019.10659
72. Zhao H, Wang X, Feng X, Li X, Pan L, Liu J, et al. Long Non-Coding RNA MEG3 Regulates Proliferation, Apoptosis, and Autophagy and Is Associated With Prognosis in Glioma. J Neurooncol (2018) 140(2):281–8. doi: 10.1007/s11060-018-2874-9
73. Wang P, Ren Z, Sun P. Overexpression of the Long Non-Coding RNA MEG3 Impairs In Vitro Glioma Cell Proliferation. J Cell Biochem (2012) 113(6):1868–74. doi: 10.1002/jcb.24055
74. Matjasic A, Tajnik M, Bostjancic E, Popovic M, Matos B, Glavac D. Identifying Novel Glioma-Associated Noncoding RNAs by Their Expression Profiles. Int J Genomics (2017) 2017:2312318. doi: 10.1155/2017/2312318
75. Li J, Bian EB, He XJ, Ma CC, Zong G, Wang HL, et al. Epigenetic Repression of Long Non-Coding RNA MEG3 Mediated by DNMT1 Represses the P53 Pathway in Gliomas. Int J Oncol (2016) 48(2):723–33. doi: 10.3892/ijo.2015.3285
76. Gong X, Huang M. Long Non-Coding RNA MEG3 Promotes the Proliferation of Glioma Cells Through Targeting Wnt/beta-Catenin Signal Pathway. Cancer Gene Ther (2017) 24(9):381–5. doi: 10.1038/cgt.2017.32
77. Wang D, Fu CW, Fan DQ. Participation of Tumor Suppressors Long Non-Coding RNA MEG3, microRNA-377 and PTEN in Glioma Cell Invasion and Migration. Pathol Res Pract (2019) 215(10):152558. doi: 10.1016/j.prp.2019.152558
78. Green DR, Kroemer G. Cytoplasmic Functions of the Tumour Suppressor P53. Nature (2009) 458(7242):1127–30. doi: 10.1038/nature07986
79. Qin N, Tong GF, Sun LW, Xu XL. Long Noncoding RNA MEG3 Suppresses Glioma Cell Proliferation, Migration, and Invasion by Acting as a Competing Endogenous RNA of miR-19a. Oncol Res (2017) 25(9):1471–8. doi: 10.3727/096504017X14886689179993
80. Gong X, Huang M-Y. Tumor-Suppressive Function of lncRNA-MEG3 in Glioma Cells by Regulating miR-6088/SMARCB1 Axis. BioMed Res Int (2020) 2020:4309161. doi: 10.1155/2020/4309161
81. Yang Z, Bian E, Xu Y, Ji X, Tang F, Ma C, et al. Meg3 Induces EMT and Invasion of Glioma Cells via Autophagy. Onco Targets Ther (2020) 13:989–1000. doi: 10.2147/OTT.S239648
82. Wang W, Xie Y, Chen F, Liu X, Zhong LL, Wang HQ, et al. LncRNA MEG3 Acts a Biomarker and Regulates Cell Functions by Targeting ADAR1 in Colorectal Cancer. World J Gastroenterol (2019) 25(29):3972–84. doi: 10.3748/wjg.v25.i29.3972
83. Ghaedi H, Mozaffari MAN, Salehi Z, Ghasemi H, Zadian SS, Alipoor S, et al. Co-Expression Profiling of Plasma miRNAs and Long Noncoding RNAs in Gastric Cancer Patients. Gene (2019) 687:135–42. doi: 10.1016/j.gene.2018.11.034
84. Ali MA, Shaker OG, Alazrak M, AbdelHafez MN, Khalefa AA, Hemeda NF, et al. Association Analyses of a Genetic Variant in Long Non-Coding RNA MEG3 With Breast Cancer Susceptibility and Serum MEG3 Expression Level in the Egyptian Population. Cancer Biomark (2020) 28(1):49–63. doi: 10.3233/CBM-191072
85. Permuth JB, Chen DT, Yoder SJ, Li J, Smith AT, Choi JW, et al. Linc-Ing Circulating Long Non-Coding RNAs to the Diagnosis and Malignant Prediction of Intraductal Papillary Mucinous Neoplasms of the Pancreas. Sci Rep (2017) 7(1):10484. doi: 10.1038/s41598-017-09754-5
86. Buccarelli M, Lulli V, Giuliani A, Signore M, Martini M, D’Alessandris QG, et al. Deregulated Expression of the Imprinted DLK1-DIO3 Region in Glioblastoma Stemlike Cells: Tumor Suppressor Role of lncRNA Meg3. Neuro Oncol (2020) 22(12):1771–84. doi: 10.1093/neuonc/noaa127
87. Ma B, Gao Z, Lou J, Zhang H, Yuan Z, Wu Q, et al. Long Noncoding RNA MEG3 Contributes to Cisplatininduced Apoptosis via Inhibition of Autophagy in Human Glioma Cells. Mol Med Rep (2017) 16(3):2946–52. doi: 10.3892/mmr.2017.6897
88. Li X, Xue L, Peng Q. Tunicamycin Inhibits Progression of Glioma Cells Through Downregulation of the MEG-3-Regulated Wnt/Beta-Catenin Signaling Pathway. Oncol Lett (2018) 15(6):8470–6. doi: 10.3892/ol.2018.8416
89. Angelopoulou E, Paudel YN, Piperi C. Critical Role of HOX Transcript Antisense Intergenic RNA (HOTAIR) in Gliomas. J Mol Med (Berl) (2020) 98(11):1525–46. doi: 10.1007/s00109-020-01984-x
90. Ke J, Yao YL, Zheng J, Wang P, Liu YH, Ma J, et al. Knockdown of Long non-Coding RNA HOTAIR Inhibits Malignant Biological Behaviors of Human Glioma Cells via Modulation of miR-326. Oncotarget (2015) 6(26):21934–49. doi: 10.18632/oncotarget.4290
91. Xavier-Magalhaes A, Goncalves CS, Fogli A, Lourenco T, Pojo M, Pereira B, et al. The Long Non-Coding RNA HOTAIR Is Transcriptionally Activated by HOXA9 and Is an Independent Prognostic Marker in Patients With Malignant Glioma. Oncotarget (2018) 9(21):15740–56. doi: 10.18632/oncotarget.24597
92. Balci T, Yilmaz Susluer S, Kayabasi C, Ozmen Yelken B, Biray Avci C, Gunduz C. Analysis of Dysregulated Long Non-Coding RNA Expressions in Glioblastoma Cells. Gene (2016) 590(1):120–2. doi: 10.1016/j.gene.2016.06.024
93. Zhou X, Ren Y, Zhang J, Zhang C, Zhang K, Han L, et al. HOTAIR Is a Therapeutic Target in Glioblastoma. Oncotarget (2015) 6(10):8353–65. doi: 10.18632/oncotarget.3229
94. Chen Y, Bian Y, Zhao S, Kong F, Li X. Suppression of PDCD4 Mediated by the Long Non-Coding RNA HOTAIR Inhibits the Proliferation and Invasion of Glioma Cells. Oncol Lett (2016) 12(6):5170–6. doi: 10.3892/ol.2016.5323
95. Pojo M, Goncalves CS, Xavier-Magalhaes A, Oliveira AI, Goncalves T, Correia S, et al. A Transcriptomic Signature Mediated by HOXA9 Promotes Human Glioblastoma Initiation, Aggressiveness and Resistance to Temozolomide. Oncotarget (2015) 6(10):7657–74. doi: 10.18632/oncotarget.3150
96. Zhang JX, Han L, Bao ZS, Wang YY, Chen LY, Yan W, et al. HOTAIR, a Cell Cycle-Associated Long Noncoding RNA and a Strong Predictor of Survival, Is Preferentially Expressed in Classical and Mesenchymal Glioma. Neuro Oncol (2013) 15(12):1595–603. doi: 10.1093/neuonc/not131
97. Huang K, Sun J, Yang C, Wang Y, Zhou B, Kang C, et al. HOTAIR Upregulates an 18-Gene Cell Cycle-Related mRNA Network in Glioma. Int J Oncol (2017) 4(2017):1271–8. doi: 10.3892/ijo.2017.3901
98. Zhang K, Sun X, Zhou X, Han L, Chen L, Shi Z, et al. Long Non-Coding RNA HOTAIR Promotes Glioblastoma Cell Cycle Progression in an EZH2 Dependent Manner. Oncotarget (2015) 6(1):537–46. doi: 10.18632/oncotarget.2681
99. Sun G, Wang Y, Zhang J, Lin N, You Y. MiR-15b/HOTAIR/p53 Form a Regulatory Loop That Affects the Growth of Glioma Cells. J Cell Biochem (2018) 119(6):4540–7. doi: 10.1002/jcb.26591
100. Li H, Guan C. HOTAIR Inhibits the Proliferation of Glioblastoma Cells by Targeting miR-219. Cancer Biomark (2020) 28(1):41–7. doi: 10.3233/CBM-190467
101. Wei Y, Zhou K, Wang C, Du X, Xiao Q, Chen C. Adsorption of miR-218 by lncRNA HOTAIR Regulates PDE7A and Affects Glioma Cell Proliferation, Invasion, and Apoptosis. Int J Clin Exp Pathol (2020) 13(12):2973–83.
102. Jiang Y, Zhang Q, Bao J, Du C, Wang J, Tong Q, et al. Schisandrin B Inhibits the Proliferation and Invasion of Glioma Cells by Regulating the HOTAIR-micoRNA-125a-mTOR Pathway. Neuroreport (2017) 28(2):93–100. doi: 10.1097/wnr.0000000000000717
103. Fang K, Liu P, Dong S, Guo Y, Cui X, Zhu X, et al. Magnetofection Based on Superparamagnetic Iron Oxide Nanoparticle-Mediated Low lncRNA HOTAIR Expression Decreases the Proliferation and Invasion of Glioma Stem Cells. Int J Oncol (2016) 49(2):509–18. doi: 10.3892/ijo.2016.3571
104. Ma X, Li Z, Li T, Zhu L, Li Z, Tian N. Long Non-Coding RNA HOTAIR Enhances Angiogenesis by Induction of VEGFA Expression in Glioma Cells and Transmission to Endothelial Cells via Glioma Cell Derived-Extracellular Vesicles. Am J Transl Res (2017) 9(11):5012–21.
105. Liu L, Cui S, Wan T, Li X, Tian W, Zhang R, et al. Long Non-Coding RNA HOTAIR Acts as a Competing Endogenous RNA to Promote Glioma Progression by Sponging miR-126-5p. J Cell Physiol (2018) 233(9):6822–31. doi: 10.1002/jcp.26432
106. Zhao WH, Yuan HY, Ren XY, Huang K, Guo ZY. Association Between Expression of HOTAIR and Invasiveness of Gliomas, and its Predictive Value. Adv Clin Exp Med (2019) 28(9):1179–83. doi: 10.17219/acem/99527
107. Sa L, Li Y, Zhao L, Liu Y, Wang P, Liu L, et al. The Role of HOTAIR/miR-148b-3p/USF1 on Regulating the Permeability of BTB. Front Mol Neurosci (2017) 10:194. doi: 10.3389/fnmol.2017.00194
108. Tan SK, Pastori C, Penas C, Komotar RJ, Ivan ME, Wahlestedt C, et al. Serum Long Noncoding RNA HOTAIR as a Novel Diagnostic and Prognostic Biomarker in Glioblastoma Multiforme. Mol Cancer (2018) 17(1):74. doi: 10.1186/s12943-018-0822-0
109. Elsayed ET, Salem PE, Darwish AM, Fayed HM. Plasma Long Non-Coding RNA HOTAIR as a Potential Biomarker for Gastric Cancer. Int J Biol Markers (2018) 33(4):528–33. doi: 10.1177/1724600818760244
110. Zhang L, Song X, Wang X, Xie Y, Wang Z, Xu Y, et al. Circulating DNA of HOTAIR in Serum Is a Novel Biomarker for Breast Cancer. Breast Cancer Res Treat (2015) 152(1):199–208. doi: 10.1007/s10549-015-3431-2
111. Li N, Wang Y, Liu X, Luo P, Jing W, Zhu M, et al. Identification of Circulating Long Noncoding RNA HOTAIR as a Novel Biomarker for Diagnosis and Monitoring of Non-Small Cell Lung Cancer. Technol Cancer Res Treat (2017) 16(6):1060–6. doi: 10.1177/1533034617723754
112. Xavier-Magalhaes A, Oliveira AI, de Castro JV, Pojo M, Goncalves CS, Lourenco T, et al. Effects of the Functional HOTAIR Rs920778 and Rs12826786 Genetic Variants in Glioma Susceptibility and Patient Prognosis. J Neurooncol (2017) 132(1):27–34. doi: 10.1007/s11060-016-2345-0
113. Zhang J, Chen G, Gao Y, Liang H. HOTAIR/miR-125 Axis-Mediated Hexokinase 2 Expression Promotes Chemoresistance in Human Glioblastoma. J Cell Mol Med (2020) 24(10):5707–17. doi: 10.1111/jcmm.15233
114. Wang W, Zhou R, Wu Y, Liu Y, Su W, Xiong W, et al. PVT1 Promotes Cancer Progression via MicroRNAs. Front Oncol (2019) 9:609. doi: 10.3389/fonc.2019.00609
115. Zeng Y, Wang T, Liu Y, Su Z, Lu P, Chen X, et al. LncRNA PVT1 as an Effective Biomarker for Cancer Diagnosis and Detection Based on Transcriptome Data and Meta-Analysis. Oncotarget (2017) 8(43):75455–66. doi: 10.18632/oncotarget.20634
116. Fang J, Huang J. Clinical Significance of the Expression of Long Non-Coding RNA PVT1 in Glioma. Cancer Biomark (2019) 24(4):509–13. doi: 10.3233/CBM-182253
117. Dahai Z, Daliang C, Famu L, Xiang W, Lenian L, Jianmin C, et al. Lowly Expressed lncRNA PVT1 Suppresses Proliferation and Advances Apoptosis of Glioma Cells Through Up-Regulating microRNA-128-1-5p and Inhibiting PTBP1. Brain Res Bull (2020) 163:1–13. doi: 10.1016/j.brainresbull.2020.06.006
118. Zhang Y, Yang G, Luo Y. Long Non-Coding RNA PVT1 Promotes Glioma Cell Proliferation and Invasion by Targeting miR-200a. Exp Ther Med (2019) 17(2):1337–45. doi: 10.3892/etm.2018.7083
119. Xue W, Chen J, Liu X, Gong W, Zheng J, Guo X, et al. PVT1 Regulates the Malignant Behaviors of Human Glioma Cells by Targeting miR-190a-5p and miR-488-3p. Biochim Biophys Acta Mol Basis Dis (2018) 1864(5 Pt A):1783–94. doi: 10.1016/j.bbadis.2018.02.022
120. Han Y, Li X, He F, Yan J, Ma C, Zheng X, et al. Knockdown of lncRNA PVT1 Inhibits Glioma Progression by Regulating miR-424 Expression. Oncol Res (2019) 27(6):681–90. doi: 10.3727/096504018X15424939990246
121. Fu C, Li D, Zhang X, Liu N, Chi G, Jin X. LncRNA PVT1 Facilitates Tumorigenesis and Progression of Glioma via Regulation of MiR-128-3p/GREM1 Axis and BMP Signaling Pathway. Neurotherapeutics (2018) 15(4):1139–57. doi: 10.1007/s13311-018-0649-9
122. Ma Y, Wang P, Xue Y, Qu C, Zheng J, Liu X, et al. PVT1 Affects Growth of Glioma Microvascular Endothelial Cells by Negatively Regulating miR-186. Tumour Biol (2017) 39(3):1010428317694326. doi: 10.1177/1010428317694326
123. Lv ZH, Wang ZY, Li ZY. LncRNA PVT1 Aggravates the Progression of Glioma via Downregulating UPF1. Eur Rev Med Pharmacol Sci (2019) 23(20):8956–63. doi: 10.26355/eurrev_201910_19294
124. Zheng J, Hu L, Cheng J, Xu J, Zhong Z, Yang Y, et al. lncRNA PVT1 Promotes the Angiogenesis of Vascular Endothelial Cell by Targeting miR26b to Activate CTGF/ANGPT2. Int J Mol Med (2018) 42(1):489–96. doi: 10.3892/ijmm.2018.3595
125. Yang JP, Yang XJ, Xiao L, Wang Y. Long Noncoding RNA PVT1 as a Novel Serum Biomarker for Detection of Cervical Cancer. Eur Rev Med Pharmacol Sci (2016) 20(19):3980–6.
126. El-Fattah AAA, Sadik NAH, Shaker OG, Mohamed Kamal A, Shahin NN. Serum Long Non-Coding RNAs PVT1, HOTAIR, and NEAT1 as Potential Biomarkers in Egyptian Women With Breast Cancer. Biomolecules (2021) 11(2):301. doi: 10.3390/biom11020301
127. Zou H, Wu LX, Yang Y, Li S, Mei Y, Liu YB, et al. lncRNAs PVT1 and HAR1A Are Prognosis Biomarkers and Indicate Therapy Outcome for Diffuse Glioma Patients. Oncotarget (2017) 8(45):78767–80. doi: 10.18632/oncotarget.20226
128. Chen Y, Guo Y, Chen H, Ma F. Long Non-Coding RNA Expression Profiling Identifies a Four-Long Non-Coding RNA Prognostic Signature for Isocitrate Dehydrogenase Mutant Glioma. Front Neurol (2020) 11:573264. doi: 10.3389/fneur.2020.573264
129. Ding X, Zhao Y, Yuan H, Zhang Y, Gao Y. Role of PVT1 Polymorphisms in the Glioma Susceptibility and Prognosis. Eur J Cancer Prev (2021). doi: 10.1097/CEJ.0000000000000636
130. Song T, Yan L, Cai K, Zhao T, Xu M. Downregulation of Long Noncoding RNA PVT1 Attenuates Paclitaxel Resistance in Glioma Cells. Cancer Biomark (2018) 23(3):447–53. doi: 10.3233/CBM-181573
131. Yao F, Wang Q, Wu Q. The Prognostic Value and Mechanisms of lncRNA UCA1 in Human Cancer. Cancer Manag Res (2019) 11:7685–96. doi: 10.2147/CMAR.S200436
132. Zhao W, Sun C, Cui Z. A Long Noncoding RNA UCA1 Promotes Proliferation and Predicts Poor Prognosis in Glioma. Clin Transl Oncol (2017) 19(6):735–41. doi: 10.1007/s12094-016-1597-7
133. Sun Y, Jin JG, Mi WY, Zhang SR, Meng Q, Zhang ST. Long Noncoding RNA UCA1 Targets miR-122 to Promote Proliferation, Migration, and Invasion of Glioma Cells. Oncol Res (2018) 26(1):103–10. doi: 10.3727/096504017X14934860122864
134. Huang Z, Zhao X, Wu X, Xiang L, Yuan Y, Zhou S, et al. LncRNA UCA1 Facilitated Cell Growth and Invasion Through the miR-206/CLOCK Axis in Glioma. Cancer Cell Int (2019) 19:316. doi: 10.1186/s12935-019-1023-7
135. Zhang B, Fang S, Cheng Y, Zhou C, Deng F. The Long Non-Coding RNA, Urothelial Carcinoma Associated 1, Promotes Cell Growth, Invasion, Migration, and Chemo-Resistance in Glioma Through Wnt/beta-Catenin Signaling Pathway. Aging (Albany NY) (2019) 11(19):8239–53. doi: 10.18632/aging.102317
136. Li ZG, Xiang WC, Shui SF, Han XW, Guo D, Yan L. 11 Long Noncoding RNA UCA1 Functions as miR-135a Sponge to Promote the Epithelial to Mesenchymal Transition in Glioma. J Cell Biochem (2020) 121(3):2447–57. doi: 10.1002/jcb.29467
137. He Z, Wang Y, Huang G, Wang Q, Zhao D, Chen L. The lncRNA UCA1 Interacts With miR-182 to Modulate Glioma Proliferation and Migration by Targeting iASPP. Arch Biochem Biophys (2017) 623-624:1–8. doi: 10.1016/j.abb.2017.01.013
138. Xin H, Liu N, Xu X, Zhang J, Li Y, Ma Y, et al. Knockdown of lncRNA-UCA1 Inhibits Cell Viability and Migration of Human Glioma Cells by miR-193a-Mediated Downregulation of CDK6. J Cell Biochem (2019) 120(9):15157–69. doi: 10.1002/jcb.28777
139. Liang C, Yang Y, Guan J, Lv T, Qu S, Fu Q, et al. LncRNA UCA1 Sponges miR-204-5p to Promote Migration, Invasion and Epithelial-Mesenchymal Transition of Glioma Cells via Upregulation of ZEB1. Pathol Res Pract (2018) 214(9):1474–81. doi: 10.1016/j.prp.2018.07.036
140. Li Z, Liu H, Zhong Q, Wu J, Tang Z. LncRNA UCA1 Is Necessary for TGF-Beta-Induced Epithelial-Mesenchymal Transition and Stemness via Acting as a ceRNA for Slug in Glioma Cells. FEBS Open Bio (2018) 8(11):1855–65. doi: 10.1002/2211-5463.12533
141. He Z, You C, Zhao D. Long Non-Coding RNA UCA1/miR-182/PFKFB2 Axis Modulates Glioblastoma-Associated Stromal Cells-Mediated Glycolysis and Invasion of Glioma Cells. Biochem Biophys Res Commun (2018) 500(3):569–76. doi: 10.1016/j.bbrc.2018.04.091
142. Zhou W, Wahl DR. Metabolic Abnormalities in Glioblastoma and Metabolic Strategies to Overcome Treatment Resistance. Cancers (Basel) (2019) 11(9):1231. doi: 10.3390/cancers11091231
143. Pan J, Xie X, Li H, Li Z, Ren C, Ming L. Detection of Serum Long Non-Coding RNA UCA1 and Circular RNAs for the Diagnosis of Bladder Cancer and Prediction of Recurrence. Int J Clin Exp Pathol (2019) 12(8):2951–8.
144. Shan L, Liu C, Ma C. High Expression of Serum UCA1 may be a Potential Biomarker for Clinical Diagnosis of Gastric Cancer. Clin Lab (2019) 65(9). doi: 10.7754/Clin.Lab.2019.190317
145. Galamb O, Bartak BK, Kalmar A, Nagy ZB, Szigeti KA, Tulassay Z, et al. Diagnostic and Prognostic Potential of Tissue and Circulating Long Non-Coding RNAs in Colorectal Tumors. World J Gastroenterol (2019) 25(34):5026–48. doi: 10.3748/wjg.v25.i34.5026
146. Ning S, Zhang J, Wang P, Zhi H, Wang J, Liu Y, et al. Lnc2Cancer: A Manually Curated Database of Experimentally Supported lncRNAs Associated With Various Human Cancers. Nucleic Acids Res (2016) 44(D1):D980–D5. doi: 10.1093/nar/gkv1094
147. He C, Jiang B, Ma J, Li Q. Aberrant NEAT1 Expression Is Associated With Clinical Outcome in High Grade Glioma Patients. APMIS (2016) 124(3):169–74. doi: 10.1111/apm.12480
148. Yang X, Xiao Z, Du X, Huang L, Du G. Silencing of the Long Non-Coding RNA NEAT1 Suppresses Glioma Stem-Like Properties Through Modulation of the miR-107/CDK6 Pathway. Oncol Rep (2017) 37(1):555–62. doi: 10.3892/or.2016.5266
149. Zhen Y, Nan Y, Guo S, Zhang L, Li G, Yue S, et al. Knockdown of NEAT1 Repressed the Malignant Progression of Glioma Through Sponging miR-107 and Inhibiting CDK14. J Cell Physiol (2019) 234(7):10671–9. doi: 10.1002/jcp.27727
150. Li Y, Wang X, Zhao Z, Shang J, Li G, Zhang R. LncRNA NEAT1 Promotes Glioma Cancer Progression via Regulation of miR-98-5p/BZW1. Biosci Rep (2021). doi: 10.1042/BSR20200767
151. Bi CL, Liu JF, Zhang MY, Lan S, Yang ZY, Fang JS. LncRNA NEAT1 Promotes Malignant Phenotypes and TMZ Resistance in Glioblastoma Stem Cells by Regulating Let-7g-5p/MAP3K1 Axis. Biosci Rep (2020) 40(10). doi: 10.1042/BSR20201111
152. Li B, Lu X, Ma C, Sun S, Shu X, Wang Z, et al. Long Non-Coding RNA NEAT1 Promotes Human Glioma Tumor Progression via miR-152-3p/CCT6A Pathway. Neurosci Lett (2020) 732:135086. doi: 10.1016/j.neulet.2020.135086
153. Gong W, Zheng J, Liu X, Ma J, Liu Y, Xue Y. Knockdown of NEAT1 Restrained the Malignant Progression of Glioma Stem Cells by Activating microRNA Let-7e. Oncotarget (2016) 7(38):62208–23. doi: 10.18632/oncotarget.11403
154. Wu DM, Wang S, Wen X, Han XR, Wang YJ, Fan SH, et al. Long Noncoding RNA Nuclear Enriched Abundant Transcript 1 Impacts Cell Proliferation, Invasion, and Migration of Glioma Through Regulating miR-139-5p/CDK6. J Cell Physiol (2019) 234(5):5972–87. doi: 10.1002/jcp.27093
155. Zhou K, Zhang C, Yao H, Zhang X, Zhou Y, Che Y, et al. Knockdown of Long Non-Coding RNA NEAT1 Inhibits Glioma Cell Migration and Invasion via Modulation of SOX2 Targeted by miR-132. Mol Cancer (2018) 17(1):105. doi: 10.1186/s12943-018-0849-2
156. Chen Q, Cai J, Wang Q, Wang Y, Liu M, Yang J, et al. Long Noncoding RNA NEAT1, Regulated by the EGFR Pathway, Contributes to Glioblastoma Progression Through the WNT/beta-Catenin Pathway by Scaffolding EZH2. Clin Cancer Res (2018) 24(3):684–95. doi: 10.1158/1078-0432.CCR-17-0605
157. Zhen L, Yun-Hui L, Hong-Yu D, Jun M, Yi-Long Y. Long Noncoding RNA NEAT1 Promotes Glioma Pathogenesis by Regulating miR-449b-5p/C-Met Axis. Tumour Biol (2016) 37(1):673–83. doi: 10.1007/s13277-015-3843-y
158. Yu H, Xu A, Wu B, Wang M, Chen Z. Long Noncoding RNA NEAT1 Promotes Progression of Glioma as a ceRNA by Sponging miR-185-5p to Stimulate DNMT1/mTOR Signaling. J Cell Physiol (2021) 236(1):121–30. doi: 10.1002/jcp.29644
159. Guo J, Cai H, Zheng J, Liu X, Liu Y, Ma J, et al. Long Non-Coding RNA NEAT1 Regulates Permeability of the Blood-Tumor Barrier via miR-181d-5p-Mediated Expression Changes in ZO-1, Occludin, and Claudin-5. Biochim Biophys Acta Mol Basis Dis (2017) 1863(9):2240–54. doi: 10.1016/j.bbadis.2017.02.005
160. Gao XY, Zang J, Zheng MH, Zhang YF, Yue KY, Cao XL, et al. Temozolomide Treatment Induces HMGB1 to Promote the Formation of Glioma Stem Cells via the TLR2/NEAT1/Wnt Pathway in Glioblastoma. Front Cell Dev Biol (2021) 9:620883. doi: 10.3389/fcell.2021.620883
161. Liu D, Zou Z, Li G, Pan P, Liang G. Long Noncoding RNA NEAT1 Suppresses Proliferation and Promotes Apoptosis of Glioma Cells Via Downregulating MiR-92b. Cancer Control (2020) 27(1):1073274819897977. doi: 10.1177/1073274819897977
162. Luo C, Quan Z, Zhong B, Zhang M, Zhou B, Wang S, et al. lncRNA XIST Promotes Glioma Proliferation and Metastasis Through miR-133a/SOX4. Exp Ther Med (2020) 19(3):1641–8. doi: 10.3892/etm.2020.8426
163. Wang Z, Yuan J, Li L, Yang Y, Xu X, Wang Y. Long Non-Coding RNA XIST Exerts Oncogenic Functions in Human Glioma by Targeting miR-137. Am J Transl Res (2017) 9(4):1845–55.
164. Shen J, Xiong J, Shao X, Cheng H, Fang X, Sun Y, et al. Knockdown of the Long Noncoding RNA XIST Suppresses Glioma Progression by Upregulating miR-204-5p. J Cancer (2020) 11(15):4550–9. doi: 10.7150/jca.45676
165. Yao Y, Ma J, Xue Y, Wang P, Li Z, Liu J, et al. Knockdown of Long Non-Coding RNA XIST Exerts Tumor-Suppressive Functions in Human Glioblastoma Stem Cells by Up-Regulating miR-152. Cancer Lett (2015) 359(1):75–86. doi: 10.1016/j.canlet.2014.12.051
166. Li HL, Han PH, Pan DQ, Chen G, Lu XH, Li J. LncRNA XIST Regulates Cell Proliferation, Migration and Invasion of Glioblastoma via Regulating miR-448 and ROCK1. J Biol Regul Homeost Agents (2020) 34(6):2049–58. doi: 10.23812/20-558-L
167. Wang YP, Li HQ, Chen JX, Kong FG, Mo ZH, Wang JZ, et al. Overexpression of XIST Facilitates Cell Proliferation, Invasion and Suppresses Cell Apoptosis by Reducing Radio-Sensitivity of Glioma Cells via miR-329-3p/CREB1 Axis. Eur Rev Med Pharmacol Sci (2020) 24(6):3190–203. doi: 10.26355/eurrev_202003_20686
168. Yu H, Xue Y, Wang P, Liu X, Ma J, Zheng J, et al. Knockdown of Long Non-Coding RNA XIST Increases Blood-Tumor Barrier Permeability and Inhibits Glioma Angiogenesis by Targeting miR-137. Oncogenesis (2017) 6(3):e303. doi: 10.1038/oncsis.2017.7
169. Cheng Z, Li Z, Ma K, Li X, Tian N, Duan J, et al. Long Non-Coding RNA XIST Promotes Glioma Tumorigenicity and Angiogenesis by Acting as a Molecular Sponge of miR-429. J Cancer (2017) 8(19):4106–16. doi: 10.7150/jca.21024
170. Gong M, Wang X, Mu L, Wang Y, Pan J, Yuan X, et al. Steroid Receptor Coactivator-1 Enhances the Stemness of Glioblastoma by Activating Long Noncoding RNA XIST/miR-152/KLF4 Pathway. Cancer Sci (2021) 112(2):604–18. doi: 10.1111/cas.14685
171. Cheng Z, Luo C, Guo Z. LncRNA-XIST/microRNA-126 Sponge Mediates Cell Proliferation and Glucose Metabolism Through the IRS1/PI3K/Akt Pathway in Glioma. J Cell Biochem (2020) 121(3):2170–83. doi: 10.1002/jcb.29440
172. Du P, Zhao H, Peng R, Liu Q, Yuan J, Peng G, et al. LncRNA-XIST Interacts With miR-29c to Modulate the Chemoresistance of Glioma Cell to TMZ Through DNA Mismatch Repair Pathway. Biosci Rep (2017) 37(5). doi: 10.1042/BSR20170696
173. Kiang KM, Zhang XQ, Zhang GP, Li N, Cheng SY, Poon MW, et al. CRNDE Expression Positively Correlates With EGFR Activation and Modulates Glioma Cell Growth. Target Oncol (2017) 12(3):353–63. doi: 10.1007/s11523-017-0488-3
174. Li DX, Fei XR, Dong YF, Cheng CD, Yang Y, Deng XF, et al. The Long Non-Coding RNA CRNDE Acts as a ceRNA and Promotes Glioma Malignancy by Preventing miR-136-5p-Mediated Downregulation of Bcl-2 and Wnt2. Oncotarget (2017) 8(50):88163–78. doi: 10.18632/oncotarget.21513
175. Zheng J, Li XD, Wang P, Liu XB, Xue YX, Hu Y, et al. CRNDE Affects the Malignant Biological Characteristics of Human Glioma Stem Cells by Negatively Regulating miR-186. Oncotarget (2015) 6(28):25339–55. doi: 10.18632/oncotarget.4509
176. Zheng J, Liu X, Wang P, Xue Y, Ma J, Qu C, et al. CRNDE Promotes Malignant Progression of Glioma by Attenuating miR-384/PIWIL4/STAT3 Axis. Mol Ther (2016) 24(7):1199–215. doi: 10.1038/mt.2016.71
177. Li H, Li Q, Guo T, He W, Dong C, Wang Y. LncRNA CRNDE Triggers Inflammation Through the TLR3-NF-kappaB-Cytokine Signaling Pathway. Tumour Biol (2017) 39(6):1010428317703821. doi: 10.1177/1010428317703821
178. Sun XH, Fan WJ, An ZJ, Sun Y. Inhibition of Long Noncoding RNA CRNDE Increases Chemosensitivity of Medulloblastoma Cells by Targeting miR-29c-3p. Oncol Res (2020) 28(1):95–102. doi: 10.3727/096504019X15742472027401
179. Jing SY, Lu YY, Yang JK, Deng WY, Zhou Q, Jiao BH. Expression of Long Non-Coding RNA CRNDE in Glioma and its Correlation With Tumor Progression and Patient Survival. Eur Rev Med Pharmacol Sci (2016) 20(19):3992–6.
180. Zhao Z, Wang B, Hao J, Man W, Chang Y, Ma S, et al. Downregulation of the Long Non-Coding RNA Taurine-Upregulated Gene 1 Inhibits Glioma Cell Proliferation and Invasion and Promotes Apoptosis. Oncol Lett (2018) 15(3):4026–32. doi: 10.3892/ol.2018.7784
181. Yu G, Li S, Liu P, Shi Y, Liu Y, Yang Z, et al. LncRNA TUG1 Functions as a ceRNA for miR-6321 to Promote Endothelial Progenitor Cell Migration and Differentiation. Exp Cell Res (2020) 388(1):111839. doi: 10.1016/j.yexcr.2020.111839
182. Katsushima K, Natsume A, Ohka F, Shinjo K, Hatanaka A, Ichimura N, et al. Targeting the Notch-Regulated Non-Coding RNA TUG1 for Glioma Treatment. Nat Commun (2016) 7:13616. doi: 10.1038/ncomms13616
183. Cai H, Liu X, Zheng J, Xue Y, Ma J, Li Z, et al. Long Non-Coding RNA Taurine Upregulated 1 Enhances Tumor-Induced Angiogenesis Through Inhibiting microRNA-299 in Human Glioblastoma. Oncogene (2017) 36(3):318–31. doi: 10.1038/onc.2016.212
184. Li J, An G, Zhang M, Ma Q. Long Non-Coding RNA TUG1 Acts as a miR-26a Sponge in Human Glioma Cells. Biochem Biophys Res Commun (2016) 477(4):743–8. doi: 10.1016/j.bbrc.2016.06.129
185. Cai H, Xue Y, Wang P, Wang Z, Li Z, Hu Y, et al. The Long Noncoding RNA TUG1 Regulates Blood-Tumor Barrier Permeability by Targeting miR-144. Oncotarget (2015) 6(23):19759–79. doi: 10.18632/oncotarget.4331
186. Zhao QS, Ying JB, Jing JJ, Wang SS. LncRNA FOXD2-AS1 Stimulates Glioma Progression Through Inhibiting P53. Eur Rev Med Pharmacol Sci (2020) 24(8):4382–8. doi: 10.26355/eurrev_202004_21019
187. Wang J, Li B, Wang C, Luo Y, Zhao M, Chen P. Long Noncoding RNA FOXD2-AS1 Promotes Glioma Cell Cycle Progression and Proliferation Through the FOXD2-AS1/miR-31/CDK1 Pathway. J Cell Biochem (2019) 120(12):19784–95. doi: 10.1002/jcb.29284
188. Ni W, Xia Y, Bi Y, Wen F, Hu D, Luo L. FoxD2-AS1 Promotes Glioma Progression by Regulating miR-185-5p/HMGA2 Axis and PI3K/AKT Signaling Pathway. Aging (Albany NY) (2019) 11(5):1427–39. doi: 10.18632/aging.101843
189. Gu N, Wang X, Di Z, Xiong J, Ma Y, Yan Y, et al. Silencing lncRNA FOXD2-AS1 Inhibits Proliferation, Migration, Invasion and Drug Resistance of Drug-Resistant Glioma Cells and Promotes Their Apoptosis via microRNA-98-5p/CPEB4 Axis. Aging (Albany NY) (2019) 11(22):10266–83. doi: 10.18632/aging.102455
190. Shen F, Chang H, Gao G, Zhang B, Li X, Jin B. Long Noncoding RNA FOXD2-AS1 Promotes Glioma Malignancy and Tumorigenesis via Targeting miR-185-5p/CCND2 Axis. J Cell Biochem (2019) 120(6):9324–36. doi: 10.1002/jcb.28208
191. Zhao J, Zeng XB, Zhang HY, Xiang JW, Liu YS. Long Non-Coding RNA FOXD2-AS1 Promotes Cell Proliferation, Metastasis and EMT in Glioma by Sponging miR-506-5p. Open Med (Wars) (2020) 15(1):921–31. doi: 10.1515/med-2020-0175
192. Shangguan W, Lv X, Tian N. FoxD2-AS1 Is a Prognostic Factor in Glioma and Promotes Temozolomide Resistance in a O(6)-Methylguanine-DNA Methyltransferase-Dependent Manner. Korean J Physiol Pharmacol (2019) 23(6):475–82. doi: 10.4196/kjpp.2019.23.6.475
193. Dong H, Cao W, Xue J. Long Noncoding FOXD2-AS1 Is Activated by CREB1 and Promotes Cell Proliferation and Metastasis in Glioma by Sponging miR-185 Through Targeting AKT1. Biochem Biophys Res Commun (2019) 508(4):1074–81. doi: 10.1016/j.bbrc.2018.12.050
194. Zhang Y, Liang C, Zhang Y, Wang Z, Li R, Wei Z, et al. The Role of FOXD2-AS1 in Cancer: A Comprehensive Study Based on Data Mining and Published Articles. Biosci Rep (2020) 40(11). doi: 10.1042/BSR20190372
195. Gao Y, Ma H, Hou D. Sevoflurane Represses Proliferation and Migration of Glioma Cells by Regulating the ANRIL/let-7b-5p Axis. Cancer Biother Radiopharm (2020). doi: 10.1089/cbr.2020.3596
196. Dai W, Tian C, Jin S. Effect of lncRNA ANRIL Silencing on Anoikis and Cell Cycle in Human Glioma via microRNA-203a. Onco Targets Ther (2018) 11:5103–9. doi: 10.2147/OTT.S169809
197. Dong X, Jin Z, Chen Y, Xu H, Ma C, Hong X, et al. Knockdown of Long Non-Coding RNA ANRIL Inhibits Proliferation, Migration, and Invasion But Promotes Apoptosis of Human Glioma Cells by Upregulation of miR-34a. J Cell Biochem (2018) 119(3):2708–18. doi: 10.1002/jcb.26437
198. Xu CH, Xiao LM, Liu Y, Chen LK, Zheng SY, Zeng EM, et al. The lncRNA HOXA11-AS Promotes Glioma Cell Growth and Metastasis by Targeting miR-130a-5p/HMGB2. Eur Rev Med Pharmacol Sci (2019) 23(1):241–52. doi: 10.26355/eurrev_201901_16770
199. Jiang J, Wang X, Gao G, Liu X, Chang H, Xiong R, et al. Silencing of lncRNA HOXA11-AS Inhibits Cell Migration, Invasion, Proliferation, and Promotes Apoptosis in Human Glioma Cells via Upregulating microRNA-125a: In Vitro and In Vivo Studies. Am J Transl Res (2019) 11(10):6382–92.
200. Cui Y, Yi L, Zhao JZ, Jiang YG. Long Noncoding RNA HOXA11-AS Functions as miRNA Sponge to Promote the Glioma Tumorigenesis Through Targeting miR-140-5p. DNA Cell Biol (2017) 36(10):822–8. doi: 10.1089/dna.2017.3805
201. Xu C, He T, Li Z, Liu H, Ding B. Regulation of HOXA11-AS/miR-214-3p/EZH2 Axis on the Growth, Migration and Invasion of Glioma Cells. BioMed Pharmacother (2017) 95:1504–13. doi: 10.1016/j.biopha.2017.08.097
202. Yang JX, Liu B, Yang BY, Meng Q. Long Non-Coding RNA Homeobox (HOX) A11-AS Promotes Malignant Progression of Glioma by Targeting miR-124-3p. Neoplasma (2018) 65(4):505–14. doi: 10.4149/neo_2018_170705N462
203. Wang Q, Zhang J, Liu Y, Zhang W, Zhou J, Duan R, et al. A Novel Cell Cycle-Associated lncRNA, HOXA11-AS, Is Transcribed From the 5-Prime End of the HOXA Transcript and is a Biomarker of Progression in Glioma. Cancer Lett (2016) 373(2):251–9. doi: 10.1016/j.canlet.2016.01.039
204. Wang JB, Chen XL, Han ZB, Wang HW, Wang ZH, Li NN, et al. Long Non-Coding RNA TP73-AS1 Contributes to Glioma Tumorigenesis by Sponging the miR-103a/GALNT7 Pathway. Brain Res (2020) 1741:146886. doi: 10.1016/j.brainres.2020.146886
205. Xiao S, Wang R, Wu X, Liu W, Ma S. The Long Noncoding RNA TP73-AS1 Interacted With miR-124 to Modulate Glioma Growth by Targeting Inhibitor of Apoptosis-Stimulating Protein of P53. DNA Cell Biol (2018) 37(2):117–25. doi: 10.1089/dna.2017.3941
206. Zhang R, Jin H, Lou F. The Long Non-Coding RNA TP73-AS1 Interacted With miR-142 to Modulate Brain Glioma Growth Through HMGB1/RAGE Pathway. J Cell Biochem (2018) 119(4):3007–16. doi: 10.1002/jcb.26021
207. Feng L, Lin T, Che H, Wang X. Long Noncoding RNA DANCR Knockdown Inhibits Proliferation, Migration and Invasion of Glioma by Regulating miR-135a-5p/BMI1. Cancer Cell Int (2020) 20:53. doi: 10.1186/s12935-020-1123-4
208. Yang JX, Sun Y, Gao L, Meng Q, Yang BY. Long Non-Coding RNA DANCR Facilitates Glioma Malignancy by Sponging miR-33a-5p. Neoplasma (2018) 65(5):790–8. doi: 10.4149/neo_2018_170724N498
209. Wang W, Li Y, Ma Q, Yan H, Su W. Differentiation Antagonizing Non-Protein Coding RNA Modulates the Proliferation, Migration, and Angiogenesis of Glioma Cells by Targeting the miR-216a/LGR5 Axis and the PI3K/AKT Signaling Pathway. Onco Targets Ther (2019) 12:2439–49. doi: 10.2147/OTT.S196851
210. Li J, Zhou L. Overexpression of lncRNA DANCR Positively Affects Progression of Glioma via Activating Wnt/beta-Catenin Signaling. BioMed Pharmacother (2018) 102:602–7. doi: 10.1016/j.biopha.2018.03.116
211. Li J, Zhang M, An G, Ma Q. LncRNA TUG1 Acts as a Tumor Suppressor in Human Glioma by Promoting Cell Apoptosis. Exp Biol Med (Maywood) (2016) 241(6):644–9. doi: 10.1177/1535370215622708
212. Li G, Cai Y, Wang C, Huang M, Chen J. LncRNA GAS5 Regulates the Proliferation, Migration, Invasion and Apoptosis of Brain Glioma Cells Through Targeting GSTM3 Expression. The Effect of LncRNA GAS5 on Glioma Cells. J Neurooncol (2019) 143(3):525–36. doi: 10.1007/s11060-019-03185-0
213. Zhao X, Liu Y, Zheng J, Liu X, Chen J, Liu L, et al. GAS5 Suppresses Malignancy of Human Glioma Stem Cells via a miR-196a-5p/FOXO1 Feedback Loop. Biochim Biophys Acta Mol Cell Res (2017) 1864(10):1605–17. doi: 10.1016/j.bbamcr.2017.06.020
214. Jin C, Zhao J, Zhang ZP, Wu M, Li J, Xiao GL, et al. Long Non-Coding RNA GAS5, by Up-Regulating PRC2 and Targeting the Promoter Methylation of miR-424, Suppresses Multiple Malignant Phenotypes of Glioma. J Neurooncol (2020) 148(3):529–43. doi: 10.1007/s11060-020-03544-2
215. Zhao X, Wang P, Liu J, Zheng J, Liu Y, Chen J, et al. Gas5 Exerts Tumor-Suppressive Functions in Human Glioma Cells by Targeting miR-222. Mol Ther (2015) 23(12):1899–911. doi: 10.1038/mt.2015.170
216. Huo JF, Chen XB. Long Noncoding RNA Growth Arrest-Specific 5 Facilitates Glioma Cell Sensitivity to Cisplatin by Suppressing Excessive Autophagy in an mTOR-Dependent Manner. J Cell Biochem (2019) 120(4):6127–36. doi: 10.1002/jcb.27900
217. Liu Q, Yu W, Zhu S, Cheng K, Xu H, Lv Y, et al. Long Noncoding RNA GAS5 Regulates the Proliferation, Migration, and Invasion of Glioma Cells by Negatively Regulating miR-18a-5p. J Cell Physiol (2018) 234(1):757–68. doi: 10.1002/jcp.26889
218. Ding Y, Wang J, Zhang H, Li H. Long Noncoding RNA-GAS5 Attenuates Progression of Glioma by Eliminating microRNA-10b and Sirtuin 1 in U251 and A172 Cells. Biofactors (2020) 46(3):487–96. doi: 10.1002/biof.1604
219. Zhu XP, Pan SA, Chu Z, Zhou YX, Huang YK, Han DQ. LncRNA GAS5 Regulates Epithelial-Mesenchymal Transition and Viability of Glioma Cells by Targeting microRNA-106b and Regulating PTEN Expression. Neurosci Res (2020). doi: 10.1016/j.neures.2020.08.009
220. Wang Y, Xin S, Zhang K, Shi R, Bao X. Low GAS5 Levels as a Predictor of Poor Survival in Patients With Lower-Grade Gliomas. J Oncol (2019) 2019:1785042. doi: 10.1155/2019/1785042
221. Wang J, Qin C, Zhong C, Wen Y, Ke S, Liao BO. Long Non-Coding RNA CASC2 Targeting miR-18a Suppresses Glioblastoma Cell Growth, Metastasis and EMT In Vitro and In Vivo. J Biosci (2020) 45. doi: 10.1007/s12038-020-00077-8
222. Liao Y, Shen L, Zhao H, Liu Q, Fu J, Guo Y, et al. LncRNA CASC2 Interacts With miR-181a to Modulate Glioma Growth and Resistance to TMZ Through PTEN Pathway. J Cell Biochem (2017) 118(7):1889–99. doi: 10.1002/jcb.25910
223. Wang P, Liu YH, Yao YL, Li Z, Li ZQ, Ma J, et al. Long Non-Coding RNA CASC2 Suppresses Malignancy in Human Gliomas by miR-21. Cell Signal (2015) 27(2):275–82. doi: 10.1016/j.cellsig.2014.11.011
224. Jiang C, Shen F, Du J, Fang X, Li X, Su J, et al. Upregulation of CASC2 Sensitized Glioma to Temozolomide Cytotoxicity Through Autophagy Inhibition by Sponging miR-193a-5p and Regulating mTOR Expression. BioMed Pharmacother (2018) 97:844–50. doi: 10.1016/j.biopha.2017.10.146
225. Shang C, Guo Y, Hong Y, Xue YX. Long Non-Coding RNA TUSC7, a Target of miR-23b, Plays Tumor-Suppressing Roles in Human Gliomas. Front Cell Neurosci (2016) 10:235. doi: 10.3389/fncel.2016.00235
226. Han N, Yang L, Zhang X, Zhou Y, Chen R, Yu Y, et al. LncRNA MATN1-AS1 Prevents Glioblastoma Cell From Proliferation and Invasion via RELA Regulation and MAPK Signaling Pathway. Ann Transl Med (2019) 7(23):784. doi: 10.21037/atm.2019.11.36
227. Zhu J, Gu W, Yu C. MATN1-AS1 Promotes Glioma Progression by Functioning as ceRNA of miR-200b/C/429 to Regulate CHD1 Expression. Cell Prolif (2020) 53(1):e12700. doi: 10.1111/cpr.12700
228. Xin S, Huang K, Zhu XG. Non-Coding RNAs: Regulators of Glioma Cell Epithelial-Mesenchymal Transformation. Pathol Res Pract (2019) 215(9):152539. doi: 10.1016/j.prp.2019.152539
229. Cui B, Li B, Liu Q, Cui Y. lncRNA CCAT1 Promotes Glioma Tumorigenesis by Sponging miR-181b. J Cell Biochem (2017) 118(12):4548–57. doi: 10.1002/jcb.26116
230. Sun SL, Shu YG, Tao MY. LncRNA CCAT2 Promotes Angiogenesis in Glioma Through Activation of VEGFA Signalling by Sponging miR-424. Mol Cell Biochem (2020) 468(1-2):69–82. doi: 10.1007/s11010-020-03712-y
231. Wang D, Zheng J, Liu X, Xue Y, Liu L, Ma J, et al. Knockdown of USF1 Inhibits the Vasculogenic Mimicry of Glioma Cells via Stimulating SNHG16/miR-212-3p and Linc00667/miR-429 Axis. Mol Ther Nucleic Acids (2019) 14:465–82. doi: 10.1016/j.omtn.2018.12.017
232. Zhou XY, Liu H, Ding ZB, Xi HP, Wang GW. lncRNA SNHG16 Exerts Oncogenic Functions in Promoting Proliferation of Glioma Through Suppressing P21. Pathol Oncol Res (2020) 26(2):1021–8. doi: 10.1007/s12253-019-00648-7
233. He J, Xue Y, Wang Q, Zhou X, Liu L, Zhang T, et al. Long Non-Coding RNA MIAT Regulates Blood Tumor Barrier Permeability by Functioning as a Competing Endogenous RNA. Cell Death Dis (2020) 11(10):936. doi: 10.1038/s41419-020-03134-0
234. Bountali A, Tonge DP, Mourtada-Maarabouni M. RNA Sequencing Reveals a Key Role for the Long Non-Coding RNA MIAT in Regulating Neuroblastoma and Glioblastoma Cell Fate. Int J Biol Macromol (2019) 130:878–91. doi: 10.1016/j.ijbiomac.2019.03.005
235. Li C, Feng SY, Chen L. SET7/9 Promotes H3K4me3 at lncRNA DRAIC Promoter to Modulate Growth and Metastasis of Glioma. Eur Rev Med Pharmacol Sci (2020) 24(23):12241–50. doi: 10.26355/eurrev_202012_24016
236. Chen Y, Bao C, Zhang X, Lin X, Huang H, Wang Z. Long Non-Coding RNA HCG11 Modulates Glioma Progression Through Cooperating With miR-496/CPEB3 Axis. Cell Prolif (2019) 52(5):e12615. doi: 10.1111/cpr.12615
237. Hua X, Li G, Liu Z, Niu Z. LINK-A lncRNA Participates in the Pathogenesis of Glioma by Interacting With Survivin. Exp Ther Med (2019) 18(3):1581–6. doi: 10.3892/etm.2019.7716
238. Zhang T, Wang F, Liao Y, Yuan L, Zhang B. LncRNA AWPPH Promotes the Invasion and Migration of Glioma Cells Through the Upregulation of HIF1alpha. Oncol Lett (2019) 18(6):6781–6. doi: 10.3892/ol.2019.11018
239. Hu Y, Jiao B, Chen L, Wang M, Han X. Long Non-Coding RNA GASL1 may Inhibit the Proliferation of Glioma Cells by Inactivating the TGF-Beta Signaling Pathway. Oncol Lett (2019) 17(6):5754–60. doi: 10.3892/ol.2019.10273
240. Ma XL, Zhu WD, Tian LX, Sun WD, Shang F, Lin QT, et al. Long Non-Coding RNA TUSC7 Expression is Independently Predictive of Outcome in Glioma. Eur Rev Med Pharmacol Sci (2017) 21(16):3605–10.
241. Zhang Z, Yin J, Lu C, Wei Y, Zeng A, You Y. Exosomal Transfer of Long Non-Coding RNA SBF2-AS1 Enhances Chemoresistance to Temozolomide in Glioblastoma. J Exp Clin Cancer Res (2019) 38(1):166. doi: 10.1186/s13046-019-1139-6
242. Zhang B, Li Q, Wu B, Zhang S, Li L, Jin K, et al. Long Non-Coding RNA TP73-AS1 Is a Potential Immune Related Prognostic Biomarker for Glioma. Aging (Albany NY) (2021) 13(4):5638–49. doi: 10.18632/aging.202490
243. Xu D, Yu J, Gao G, Lu G, Zhang Y, Ma P. LncRNA DANCR Functions as a Competing Endogenous RNA to Regulate RAB1A Expression by Sponging miR-634 in Glioma. Biosci Rep (2018) 38(1). doi: 10.1042/BSR20171664
244. Skiriute D, Stakaitis R, Steponaitis G, Tamasauskas A, Vaitkiene P. The Role of CASC2 and miR-21 Interplay in Glioma Malignancy and Patient Outcome. Int J Mol Sci (2020) 21(21):7962. doi: 10.3390/ijms21217962
245. Wang W, Zhao Z, Yang F, Wang H, Wu F, Liang T, et al. An Immune-Related lncRNA Signature for Patients With Anaplastic Gliomas. J Neurooncol (2018) 136(2):263–71. doi: 10.1007/s11060-017-2667-6
246. Se YB, Kim SH, Kim JY, Kim JE, Dho YS, Kim JW, et al. Underexpression of HOXA11 Is Associated With Treatment Resistance and Poor Prognosis in Glioblastoma. Cancer Res Treat (2017) 49(2):387–98. doi: 10.4143/crt.2016.106
247. Zeng J, Du T, Song Y, Gao Y, Li F, Wu R, et al. Knockdown of Long Noncoding RNA CCAT2 Inhibits Cellular Proliferation, Invasion, and Epithelial-Mesenchymal Transition in Glioma Cells. Oncol Res (2017) 25(6):913–21. doi: 10.3727/096504016X14792098307036
248. Zhang Y, Xiao Y, Li GC, Gong FY, Zhang XN, Hou K. Long Non-Coding RNAs as Epigenetic Mediator and Predictor of Glioma Progression, Invasiveness, and Prognosis. Semin Cancer Biol (2020). doi: 10.1016/j.semcancer.2020.08.016
249. Deng Y, Zhou L, Li N, Wang M, Yao L, Dong S, et al. Impact of Four lncRNA Polymorphisms (Rs2151280, Rs7763881, Rs1136410, and Rs3787016) on Glioma Risk and Prognosis: A Case-Control Study. Mol Carcinog (2019) 58(12):2218–29. doi: 10.1002/mc.23110
250. Cunnington MS, Santibanez Koref M, Mayosi BM, Burn J, Keavney B. Chromosome 9p21 SNPs Associated With Multiple Disease Phenotypes Correlate With ANRIL Expression. PloS Genet (2010) 6(4):e1000899. doi: 10.1371/journal.pgen.1000899
251. Mazor G, Levin L, Picard D, Ahmadov U, Caren H, Borkhardt A, et al. The lncRNA TP73-AS1 Is Linked to Aggressiveness in Glioblastoma and Promotes Temozolomide Resistance in Glioblastoma Cancer Stem Cells. Cell Death Dis (2019) 10(3):246. doi: 10.1038/s41419-019-1477-5
252. Ding J, Zhang L, Chen S, Cao H, Xu C, Wang X. lncRNA CCAT2 Enhanced Resistance of Glioma Cells Against Chemodrugs by Disturbing the Normal Function of miR-424. Onco Targets Ther (2020) 13:1431–45. doi: 10.2147/OTT.S227831
253. Ma Y, Zhou G, Li M, Hu D, Zhang L, Liu P, et al. Long Noncoding RNA DANCR Mediates Cisplatin Resistance in Glioma Cells via Activating AXL/PI3K/Akt/NF-kappaB Signaling Pathway. Neurochem Int (2018) 118:233–41. doi: 10.1016/j.neuint.2018.03.011
254. Li Z, Cai S, Li H, Gu J, Tian Y, Cao J, et al. Developing a lncRNA Signature to Predict the Radiotherapy Response of Lower-Grade Gliomas Using Co-Expression and ceRNA Network Analysis. Front Oncol (2021) 11:622880. doi: 10.3389/fonc.2021.622880
255. Gao N, Li Y, Li J, Gao Z, Yang Z, Li Y, et al. Long Non-Coding RNAs: The Regulatory Mechanisms, Research Strategies, and Future Directions in Cancers. Front Oncol (2020) 10:598817. doi: 10.3389/fonc.2020.598817
256. Wu Y-Y, Kuo H-C. Functional Roles and Networks of Non-Coding RNAs in the Pathogenesis of Neurodegenerative Diseases. J Biomed Sci (2020) 27(1):49. doi: 10.1186/s12929-020-00636-z
257. Sallam T, Sandhu J, Tontonoz P. Long Noncoding RNA Discovery in Cardiovascular Disease: Decoding Form to Function. Circ Res (2018) 122(1):155–66. doi: 10.1161/CIRCRESAHA.117.311802
258. Yeh C-F, Chang Y-CE, Lu C-Y, Hsuan C-F, Chang W-T, Yang K-C. Expedition to the Missing Link: Long Noncoding RNAs in Cardiovascular Diseases. J Biomed Sci (2020) 27(1):48. doi: 10.1186/s12929-020-00647-w
259. Huang Y, Ling A, Pareek S, Huang RS. Oncogene or Tumor Suppressor? Long Noncoding RNAs Role in Patient’s Prognosis Varies Depending on Disease Type. Transl Res (2021) 230:98–110. doi: 10.1016/j.trsl.2020.10.011
260. Lemos AEG, Matos ADR, Ferreira LB, Gimba ERP. The Long Non-Coding RNA PCA3: An Update of its Functions and Clinical Applications as a Biomarker in Prostate Cancer. Oncotarget (2019) 10(61):6589–603. doi: 10.18632/oncotarget.27284
261. Matsui M, Corey DR. Non-Coding RNAs as Drug Targets. Nat Rev Drug Discov (2017) 16(3):167–79. doi: 10.1038/nrd.2016.117
262. Aigner A, Kogel D. Nanoparticle/siRNA-Based Therapy Strategies in Glioma: Which Nanoparticles, Which siRNAs? Nanomedicine (Lond) (2018) 13(1):89–103. doi: 10.2217/nnm-2017-0230
263. Roberts TC, Langer R, Wood MJA. Advances in Oligonucleotide Drug Delivery. Nat Rev Drug Discov (2020) 19(10):673–94. doi: 10.1038/s41573-020-0075-7
264. Zhou M, Zhang Z, Zhao H, Bao S, Cheng L, Sun J. An Immune-Related Six-lncRNA Signature to Improve Prognosis Prediction of Glioblastoma Multiforme. Mol Neurobiol (2018) 55(5):3684–97. doi: 10.1007/s12035-017-0572-9
265. Gong Z, Hong F, Wang H, Zhang X, Chen J. An eight-mRNA Signature Outperforms the lncRNA-Based Signature in Predicting Prognosis of Patients With Glioblastoma. BMC Med Genet (2020) 21(1):56. doi: 10.1186/s12881-020-0992-7
266. Cheng M, Sun L, Huang K, Yue X, Chen J, Zhang Z, et al. A Signature of Nine lncRNA Methylated Genes Predicts Survival in Patients With Glioma. Front Oncol (2021) 11:646409. doi: 10.3389/fonc.2021.646409
267. Lin JZ, Lin N, Zhao WJ. Identification and Validation of a six-lncRNA Prognostic Signature With its ceRNA Networks and Candidate Drugs in Lower-Grade Gliomas. Genomics (2020) 112(5):2990–3002. doi: 10.1016/j.ygeno.2020.05.016
268. Qian Y, Shi L, Luo Z. Long Non-Coding RNAs in Cancer: Implications for Diagnosis, Prognosis, and Therapy. Front Med (2020) 7:612393. doi: 10.3389/fmed.2020.612393
Keywords: biomarker, glioma, glioblasoma, long non coding RNA, micro RNA, prognosis, survival, treatment
Citation: Momtazmanesh S and Rezaei N (2021) Long Non-Coding RNAs in Diagnosis, Treatment, Prognosis, and Progression of Glioma: A State-of-the-Art Review. Front. Oncol. 11:712786. doi: 10.3389/fonc.2021.712786
Received: 21 May 2021; Accepted: 25 June 2021;
Published: 12 July 2021.
Edited by:
Xiao Zhu, Guangdong Medical University, ChinaReviewed by:
Qi Shengcai, Shanghai Stomatology Prevention Hospital, ChinaYing Wang, Eastern Hepatobiliary Surgery Hospital, China
Copyright © 2021 Momtazmanesh and Rezaei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nima Rezaei, cmV6YWVpX25pbWFAdHVtcy5hYy5pcg==
 Sara Momtazmanesh
Sara Momtazmanesh Nima Rezaei
Nima Rezaei