- 1Cell Death and Drug Resistance in Lymphoproliferative Disorders Team, Centre de Recherche des Cordeliers, INSERM UMRS 1138, Paris, France
- 2Sorbonne Université, Paris, France
- 3Service d’Hématologie Biologique, Hôpital Pitié-Salpêtrière, AP-HP, Paris, France
Although the 17p deletion [del(17p)] is rare in cases of treatment-naive chronic lymphocytic leukemia (CLL), its frequency is higher in refractory/relapsed CLL – particularly in patients undergoing chemo(immuno)therapy. TP53 disruption (deletion and/or mutation) is the strongest prognostic factor for refractoriness to chemotherapy; the use of Bruton tyrosine kinase inhibitors and BCL2 inhibitors is then indicated. Rare cases of CLL can also harbor translocation or gain of the MYC oncogene. “Double-hit CLL” (with del(17p) and MYC gain) is associated with a very poor prognosis. The prognostic impact of TP53 disruption with MYC aberrations in patients receiving targeted therapies must now be evaluated.
Introduction
Loss of the short arm of chromosome 17 [del(17p)] results from various chromosomal abnormalities, including deletions, translocations, isochromosomes, and ring chromosomes. All these chromosomal abnormalities lead to the loss of one copy of the TP53 gene (located at 17p13) in patients with chronic lymphocytic leukemia (CLL), and the remaining allele is mutated in more than 90% of cases. del(17p) is often associated with a complex karyotype (three or more chromosomal abnormalities) (1). Rare CLL cases can also harbor translocation or gain of the MYC gene, independently or in association with del(17p) (1–3).
MYC Translocation
The MYC oncogene (located at 8q24) is a transcription factor involved in many biological mechanisms, including as cell cycle control, apoptosis, cell growth, and cell differentiation. The translocation t(8;14)(q24;q32) and its variants t(8;22)(q24;q11) and t(2;8)(p11;q24) are typically associated with Burkitt lymphoma; MYC then comes under the control of an immunoglobulin heavy chain enhancer, a lambda light chain enhancer or a kappa light chain enhancer, respectively. MYC also has non-immunoglobulin gene partners. These translocations can be observed in other B cell neoplasms, such as diffuse large B-cell lymphoma (DLBCL), B-prolymphocytic leukemia (B-PLL) and CLL (4, 5). The World Health Organization’s classification of large B-cell lymphomas now includes a new entity called “double hit high-grade B cell lymphoma” (HGBL), in which MYC rearrangement is combined with a BCL2 and/or BCL6 rearrangement (6). This category of double- or triple-hit lymphomas only comprises translocations involving MYC and the two other genes; hence, lymphomas expressing MYC with BCL2 and/or BCL6 (according to immunochemical assessments) but that lack translocations are not encompassed by the definition (7).
MYC and Transformed Indolent B Cell Malignancies
MYC is often involved in transformed indolent mature B neoplasms, such as the transformations of follicular lymphoma (FL) to DLBCL and CLL to Richter syndrome (8). Transformation of FL occurs in 25-35% of cases. A very small proportion of cases of FL (<0.5%) harbor a t(MYC), and a progression to a HGBL double hit may occur in cases with both t(14;18) and t(MYC) (6). Extra copies of MYC can also be observed in FL but (unlike t(MYC)) do not appear to be associated with a risk of transformation (9). Although MYC translocation/activation is rare in FL, up to 75% of cases of transformed FL show a gain in MYC activity (8). With regard to DLBCL-type Richter syndrome, the MYC pathway is deregulated in about 70% of cases, and somatic structural MYC alterations are present in 30% of cases. MYC deregulation is often acquired upon transformation (10).
del(17p) and MYC Aberrations in B-Prolymphocytic Leukemia
MYC translocations (t(MYC)) are frequent in B-PLL (4). In a recent study, we found that 21 of the 34 cases (62%) of B-PLL had a t(MYC). Furthermore, the translocated MYC gene was mutated in 3 of the 10 tested cases (30%). MYC gain was also observed in this disease, albeit at a lower frequency (5 out of 34, 15%) than t(MYC). Interestingly, t(MYC) and MYC gain were mutually exclusive; t(MYC) was present in the major clone, and MYC gain was mainly subclonal. It is noteworthy that MYC gain was associated with a highly complex karyotype, with five or more chromosomal abnormalities. We have shown that B-PLL patients with an MYC aberration (translocation or gain) and a del(17p) had the worse prognosis. In all evaluable del(17p) B-PLL cases, the remaining TP53 allele was mutated. However, the small sample size prevented a statistical analysis of TP53 mutational status and MYC aberration. Thus the combination of MYC and a TP53 aberration is associated with a very high-risk form of B-PLL (4).
del(17p) and MYC Aberrations in CLL
In contrast to B-PLL, translocations involving MYC are very infrequent (<0.5%) in CLL (2, 11). t(MYC) is often a secondary event in the course of the disease and is associated with a complex karyotype, an elevated prolymphocyte count, and an aggressive form of CLL (2). The MYC gene is also involved in 8q24 gain, which is detected in less than 0.5% or 3-4% of cases of CLL (using chromosome banding and microarrays analyses, respectively) (11–14). Gain of 8q can occur early in the course of CLL (15). It has been linked to a complex karyotype, a shorter overall survival time, and a shorter time to first treatment (11–14). Overall, MYC abnormalities – whether translocations or gains – are associated with a poor prognosis in CLL.
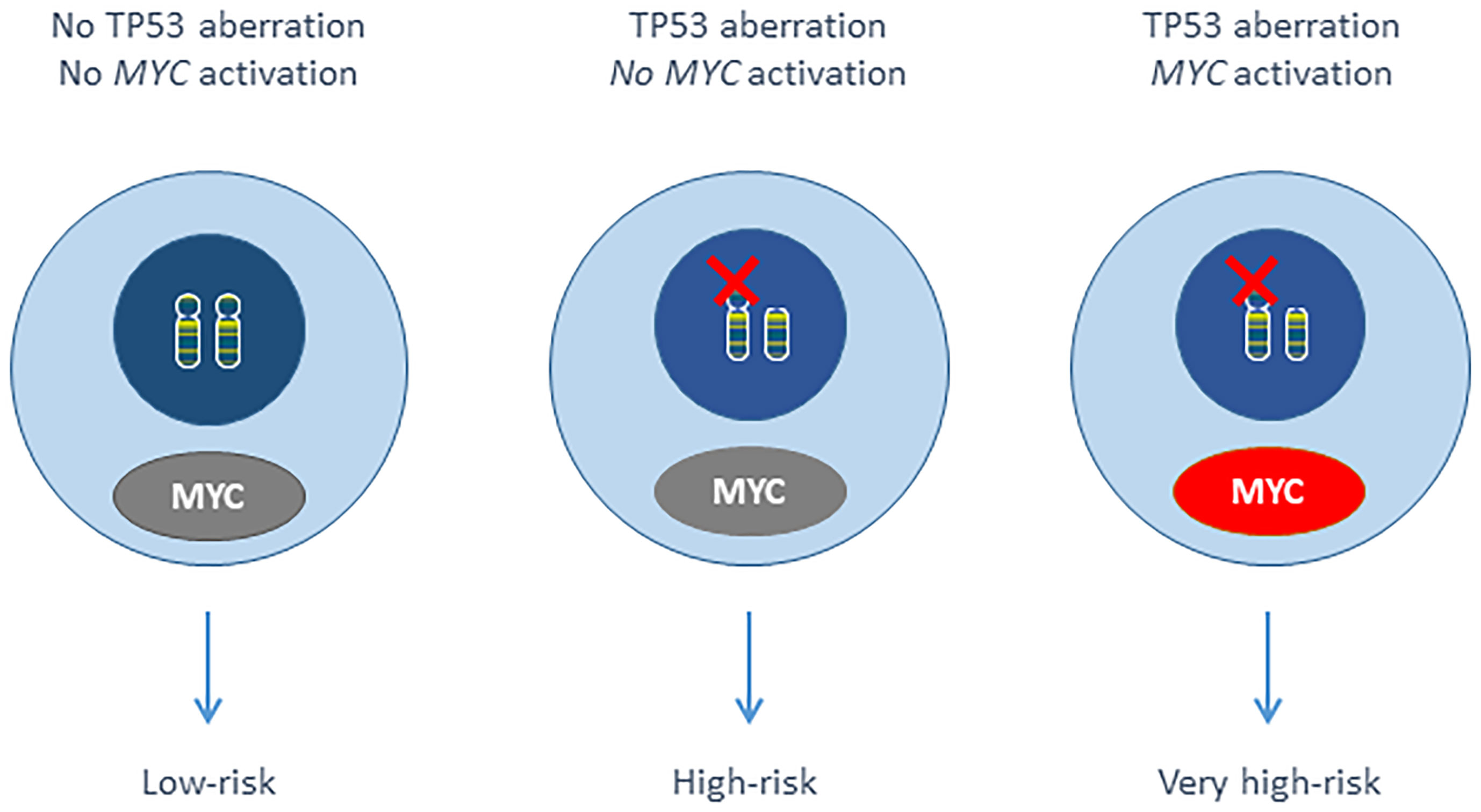
Harbel et al. showed that del(17p) occurred with a more than 3-fold increase in a cohort of 33 t(MYC) CLL compared to general CLL (3). The frequency of MYC gain is higher in CLL with del(17p) (ranging from 9% to 44%) (13, 14, 16–18), and we have demonstrated that the del(17p) + 8q24 gain combination (involving TP53 and MYC respectively) was associated with a very poor outcome within the del(17p) CLL. The remaining TP53 allele was mutated in 55 (92%) of the 60 evaluable del(17p) patients. The small number of cases prevented a statistical analysis of TP53 mutational status and MYC gain. It should be noted that there were not t(MYC) cases in our del(17p) CLL series (n=195). By analogy with double-hit HGBL, we identified double-hit CLL as an aggressive form of the disease (Figure 1) (1, 6).
Cooperation Between MYC and TP53 Defects
It has been shown that MYC and TP53 defects cooperate in MYC-induced murine lymphomas. In Eμ-MYC transgenic mice, MYC activation strongly selected for surviving cells, with inactivation of the ARF-Mdm2-p53 pathway (19). Thus lymphomagenesis in MYC mouse models requires additional genetic alterations - such as loss of p53 (20). In our del(17p) CLL series, MYC gain and del(17p) were in the same clone in 8 (62%) of the 13 evaluable cases, MYC was gained before the del(17p) in 3 cases (23%), and MYC was gained after the del(17p) in 2 cases (15%). It is noteworthy that 6 of the 13 (46%) cases carried the der(17)t(8;17) abnormality, with an unbalanced translocation between the short arm of chromosome 17 and the long arm of chromosome 8; this results in both MYC gain and del(17p) (1). Regarding t(MYC) in CLL, Put et al. described a case with t(MYC) before del(17p) and a case with t(MYC) and del(17p) in the same clone. In B-PLL, the majority of the 7 evaluable cases with t(MYC) and del(17p) in the literature had both abnormalities in the same clone (6/7); the last case had the del(17p) before t(MYC) (2, 4). There were two B-PLL cases with both MYC gain and del(17p): MYC gain and del(17p) were present in the same clone for one patient, and MYC was gained after del(17p) in the other patient (4). Overall, it is difficult to draw conclusions about the order of appearance of these two abnormalities, except in cases with the der(17)t(8;17). The two types of longitudinal event (MYC followed by TP53 aberrations, and TP53 followed by MYC aberrations) may exist.
BCR Signaling
Given that TP53 downregulates BCR signaling, and MYC represses downregulators of BCR signaling, both TP53 and MYC aberrations might results in elevated FOXP1 levels. One can reasonably hypothesize that a combination of an MYC-activating aberration (repressing miR-150 and miR-34a) and TP53 deletion/mutation (further repressing miR-34a) can lead to very prominent activation of FOXP1 and then the BCR. Both miR-150 and miR-34a target FOXP1, albeit at different positions (21–24).
It would be interesting to evaluate the response to Bruton tyrosine kinase inhibitors (BTKi) in patients with double-hit CLL. As MYC acts as a key downstream BCR effector, its overexpression is known to rescue the absence of BCR activity in some B cells (8, 25). Indeed, upregulation of MYC has been observed in ibrutinib-resistant mantle cell lymphoma cell lines (26). Treating CLL with TP53 and MYC aberrations might be challenging. Intriguingly, it has been shown that in a context of chemotherapy in B-cell lymphoma with inactive p53, MYC gain can be used to over-activate cells and induce apoptosis (27).
“Double-Hit” CLL
The concept of a double hit involving the MYC gene in HGBL could be thus extended to other B cell malignancies in general and B-PLL and CLL in particular. When combined with del(17p) in B-PLL and CLL, MYC aberrations (translocations or gains) appeared to be associated with a very poor prognosis. However, only retrospective cohorts have been studied to date, and most patients were undergoing chemo(immuno)therapy. Moreover, TP53 mutational status must be further evaluated, in order to confirm that the combination of a TP53 mutation [and not only del(17p)] with a MYC aberration results in a poor prognosis. Given the low frequency of CLL cases with MYC aberrations, and the low proportion of cells with MYC aberrations (in case of a subclonal abnormality) and thus the requirement for systematic screening with a fluorescent in situ hybridization (FISH) probe, it will be challenging to evaluate the prognostic impact of these two abnormalities in prospective trials of targeted therapies (e.g. BTKi and BCl2 inhibitors). However, understand the mechanisms of resistance to new drugs is essential, and any aggressive abnormalities must be carefully analyzed. Although t(MYC) is easy to observe by karyotype, the MYC gain might be difficult to detect. In CLL, we recommend karyotyping and systematic FISH analysis with TP53 and MYC probes prior to the initiation of each line of treatment. It is noteworthy that MYC and TP53 aberrations can be present in a subclone and so might be overlooked by techniques like chromosomal microarrays, multiplex ligation-dependent probe amplification, massively parallel sequencing, and optical genome mapping. FISH is still the most sensitive technique for detecting chromosomal gains and losses. Of course, TP53 mutation analyses should (in addition to FISH) be performed in CLL (28).
In conclusion, the results of a retrospective study showed that del(17p) and 8q gain (involving TP53 and MYC, respectively) are associated with a very poor prognosis in CLL. This very high risk of double-hit CLL must now be confirmed (including the impact of TP53 mutation status and rare translocations involving MYC) for the targeted therapies (e.g. BTKi and BCL2 inhibitors) now used as first-line treatments.
Author Contributions
The author confirms being the sole contributor of this work and has approved it for publication.
Conflict of Interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The author thanks the reviewer for helpful comments.
References
1. Chapiro E, Lesty C, Gabillaud C, Durot E, Bouzy S, Armand M, et al. “Double-Hit” Chronic Lymphocytic Leukemia: An Aggressive Subgroup With 17p Deletion and 8q24 Gain. Am J Hematol (2018) 93(3):375–82. doi: 10.1002/ajh.24990
2. Put N, Van Roosbroeck K, Konings P, Meeus P, Brusselmans C, Rack K, et al. Chronic Lymphocytic Leukemia and Prolymphocytic Leukemia With MYC Translocations: A Subgroup With an Aggressive Disease Course. Ann Hematol (2012) 91:863–73. doi: 10.1007/s00277-011-1393-y
3. Haberl S, Haferlach T, Stengel A, Jeromin S, Kern W, Haferlach C. MYC Rearranged B-Cell Neoplasms: Impact of Genetics on Classification. Cancer Genet (2016) 209(10):431–9. doi: 10.1016/j.cancergen.2016.08.007
4. Chapiro E, Pramil E, Diop M, Roos-Weil D, Dillard C, Gabillaud C, et al. Genetic Characterization of B-Cell Prolymphocytic Leukemia: A Prognostic Model Involving MYC and TP53. Blood (2019) 134(21):1821–31. doi: 10.1182/blood.2019001187
5. Klapproth K, Wirth T. Advances in the Understanding of MYC-Induced Lymphomagenesis. Br J Haematol (2010) 149(4):484–97. doi: 10.1111/j.1365-2141.2010.08159.x
6. Swerdlow SH, Campo E, Pileri SA, Harris NL, Stein H, Siebert R, et al. The 2016 Revision of the World Health Organization Classification of Lymphoid Neoplasms. Blood (2016) 127(20):2375–90. doi: 10.1182/blood-2016-01-643569
7. Collinge B, Ben-Neriah S, Chong L, Boyle M, Jiang A, Miyata-Takata T, et al. The Impact of MYC and BCL2 Structural Variants in Tumors of DLBCL Morphology and Mechanisms of False-Negative MYC IHC. Blood (2021) 137(16):2196–208. doi: 10.1182/blood.2020007193
8. Filip D, Mraz M. The Role of MYC in the Transformation and Aggressiveness of ‘Indolent’ B-Cell Malignancies. Leuk Lymphoma (2020) 61(3):510–24. doi: 10.1080/10428194.2019.1675877
9. Bussot L, Chevalier S, Cristante J, Grange B, Tesson B, Deteix-Santana C, et al. Adverse Outcome in Follicular Lymphoma Is Associated With MYC Rearrangements But Not MYC Extra Copies. Br J Haematol (2021) 194(2):382–92. doi: 10.1111/bjh.17550
10. Rossi D, Spina V, Gaidano G. Biology and Treatment of Richter Syndrome. Blood (2018) 131(25):2761–72. doi: 10.1182/blood-2018-01-791376
11. Haferlach C, Dicker F, Schnittger S, Kern W, Haferlach T. Comprehensive Genetic Characterization of CLL: A Study on 506 Cases Analysed With Chromosome Banding Analysis, Interphase FISH, IgV(H) Status and Immunophenotyping. Leukemia (2007) 21(12):2442–51. doi: 10.1038/sj.leu.2404935
12. Brown JR, Hanna M, Tesar B, Werner L, Pochet N, Asara JM, et al. Integrative Genomic Analysis Implicates Gain of PIK3CA at 3q26 and MYC at 8q24 in Chronic Lymphocytic Leukemia. Clin Cancer Res (2012) 18(14):3791–802. doi: 10.1158/1078-0432.CCR-11-2342
13. Rinaldi A, Mian M, Kwee I, Rossi D, Deambrogi C, Mensah AA, et al. Genome-Wide DNA Profiling Better Defines the Prognosis of Chronic Lymphocytic Leukaemia. Br J Haematol (2011) 154(5):590–9. doi: 10.1111/j.1365-2141.2011.08789.x
14. Houldsworth J, Guttapalli A, Thodima V, Yan XJ, Mendiratta G, Zielonka T, et al. Genomic Imbalance Defines Three Prognostic Groups for Risk Stratification of Patients With Chronic Lymphocytic Leukemia. Leuk Lymphoma (2014) 55(4):920–8. doi: 10.3109/10428194.2013.845882
15. Landau DA, Tausch E, Taylor-Weiner AN, Stewart C, Reiter JG, Bahlo J, et al. Mutations Driving CLL and Their Evolution in Progression and Relapse. Nature (2015) 526(7574):525–30. doi: 10.1038/nature15395
16. Delgado J, Salaverria I, Baumann T, Martinez-Trillos A, Lee E, Jimenez L, et al. Genomic Complexity and IGHV Mutational Status Are Key Predictors of Outcome of Chronic Lymphocytic Leukemia Patients With TP53 Disruption. Haematologica (2014) 99(11):e231–4. doi: 10.3324/haematol.2014.108365
17. Blanco G, Puiggros A, Baliakas P, Athanasiadou A, Garcia-Malo M, Collado R, et al. Karyotypic Complexity Rather Than Chromosome 8 Abnormalities Aggravates the Outcome of Chronic Lymphocytic Leukemia Patients With TP53 Aberrations. Oncotarget (2016) 7(49):80916–24. doi: 10.18632/oncotarget.13106
18. Forconi F, Rinaldi A, Kwee I, Sozzi E, Raspadori D, Rancoita PM, et al. Genome-Wide DNA Analysis Identifies Recurrent Imbalances Predicting Outcome in Chronic Lymphocytic Leukaemia With 17p Deletion. Br J Haematol (2008) 143(4):532–6. doi: 10.1111/j.1365-2141.2008.07373.x
19. Eischen CM, Weber JD, Roussel MF, Sherr CJ, Cleveland JL. Disruption of the ARF-Mdm2-P53 Tumor Suppressor Pathway in Myc-Induced Lymphomagenesis. Genes Dev (1999) 13(20):2658–69. doi: 10.1101/gad.13.20.2658
20. Schuster C, Berger A, Hoelzl MA, Putz EM, Frenzel A, Simma O, et al. The Cooperating Mutation or "Second Hit" Determines the Immunologic Visibility Toward MYC-Induced Murine Lymphomas. Blood (2011) 118(17):4635–45. doi: 10.1182/blood-2010-10-313098
21. Cerna K, Oppelt J, Chochola V, Musilova K, Seda V, Pavlasova G, et al. MicroRNA miR-34a Downregulates FOXP1 During DNA Damage Response to Limit BCR Signalling in Chronic Lymphocytic Leukaemia B Cells. Leukemia (2019) 33(2):403–14. doi: 10.1038/s41375-018-0230-x
22. Mraz M, Chen L, Rassenti LZ, Ghia EM, Li H, Jepsen K, et al. miR-150 Influences B-Cell Receptor Signaling in Chronic Lymphocytic Leukemia by Regulating Expression of GAB1 and FOXP1. Blood (2014) 124(1):84–95. doi: 10.1182/blood-2013-09-527234
23. Craig VJ, Cogliatti SB, Imig J, Renner C, Neuenschwander S, Rehrauer H, et al. Myc-Mediated Repression of microRNA-34a Promotes High-Grade Transformation of B-Cell Lymphoma by Dysregulation of Foxp1. Blood (2011) 117(23):6227–36. doi: 10.1182/blood-2010-10-312231
24. Musilova K, Devan J, Cerna K, Seda V, Pavlasova G, Sharma S, et al. miR-150 Downregulation Contributes to the High-Grade Transformation of Follicular Lymphoma by Upregulating FOXP1 Levels. Blood (2018) 132(22):2389–400. doi: 10.1182/blood-2018-06-855502
25. Varano G, Raffel S, Sormani M, Zanardi F, Lonardi S, Zasada C, et al. The B-Cell Receptor Controls Fitness of MYC-Driven Lymphoma Cells via GSK3beta Inhibition. Nature (2017) 546(7657):302–6. doi: 10.1038/nature22353
26. Lee J, Zhang LL, Wu W, Guo H, Li Y, Sukhanova M, et al. Activation of MYC, a Bona Fide Client of HSP90, Contributes to Intrinsic Ibrutinib Resistance in Mantle Cell Lymphoma. Blood Adv (2018) 2(16):2039–51. doi: 10.1182/bloodadvances.2018016048
27. Harrington CT, Sotillo E, Robert A, Hayer KE, Bogusz AM, Psathas J, et al. Transient Stabilization, Rather Than Inhibition, of MYC Amplifies Extrinsic Apoptosis and Therapeutic Responses in Refractory B-Cell Lymphoma. Leukemia (2019) 33(10):2429–41. doi: 10.1038/s41375-019-0454-4
Keywords: TP53, MYC, 17p deletion, chronic lymphocytic leukemia, MYC gain, MYC translocation
Citation: Nguyen-Khac F (2022) “Double-Hit” Chronic Lymphocytic Leukemia, Involving the TP53 and MYC Genes. Front. Oncol. 11:826245. doi: 10.3389/fonc.2021.826245
Received: 30 November 2021; Accepted: 21 December 2021;
Published: 13 January 2022.
Edited by:
John Riches, Queen Mary University of London, United KingdomReviewed by:
Marek Mraz, Central European Institute of Technology (CEITEC), CzechiaCopyright © 2022 Nguyen-Khac. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Florence Nguyen-Khac, ZmxvcmVuY2Uubmd1eWVuLWtoYWNAYXBocC5mcg==
 Florence Nguyen-Khac
Florence Nguyen-Khac