- 1Key Laboratory of Shaanxi Province for Craniofacial Precision Medicine Research, College of Stomatology, Xi’an Jiaotong University, Xi’an, Shaanxi, China
- 2Department of Oral and Maxillofacial Surgery, College of Stomatology, Xi’an Jiaotong University, Xi’an, Shaanxi, China
- 3State Key Laboratory of Oral Diseases and National Clinical Research Center for Oral Diseases, West China Hospital of Stomatology, Sichuan University, Chengdu, Sichuan, China
- 4Department of Oral and Maxillofacial Surgery, West China Hospital of Stomatology, Sichuan University, Chengdu, Sichuan, China
- 5Department of Pathology, College of Stomatology, Xi’an Jiaotong University, Xi’an, Shaanxi, China
Background: Mena, a cytoskeletal regulatory protein, is involved in actin-based regulation of cell motility and adhesion, and contributes to tumor invasion and metastasis. However, the role of Mena in oral squamous cell carcinoma remains unclear. This is the first research focusing on the prognostic value of Mena in OSCC. In this study, we aimed to investigate the correlation between Mena expression and clinicopathological significance, as well as prognostic value in OSCC.
Methods: Mena gene expression profiles of OSCC and normal tissues were collected from Oncomine, TCGA, and GEO databases. Biological function was analyzed through GO, KEGG and GSEA enrichment. Further, the expression level of Mena and tumor-related markers in 151 OSCC specimens was examined by IHC staining based on tissue microarray. Kaplan-Meier analysis was used to assess the prognostic performance of Mena in OSCC.
Result: Mena was generally upregulation in various malignancies, especially OSCC. The functional analyses indicated that Mena was involved in the assembly and regulation of actin, cell movement, and EMT. IHC staining revealed that high expression of Mena in OSCC was correlated with Lymphatic metastasis, TNM stage, E-cadherin, Vimentin, and MMP-2, but insignificantly Ki67. Kaplan-Meier analysis demonstrated that elevated expression of Mena was significantly associated with poor overall survival and disease-free survival of OSCC patients.
Conclusion: Mena could be a novel biomarker for predicting the prognosis of OSCC patients, which supports a theoretical basis for developing molecular target therapy.
1. Introduction
Oral squamous cell carcinoma (OSCC) is the most common histological type of cancer in the oral cavity, accounting for approximately 90% of oral cancer which is ranked the 15th leading cause of mortality and 16th most common malignancy worldwide (1, 2). Unfortunately, the estimated incidence of cases and the anticipated deaths attributable to OSCC in 2018 in America are 33,950 and 6,800 respectively (3). Despite substantial advances in diagnosis and treatment, OSCC still maintains a 5-year survival rate of 50% (4, 5). Local invasion and lymph node metastasis are the main factors of poor prognosis and death in patients with OSCC (6). Therefore, early prevention and detection is a vital component in the control of cancer. Highly invasive OSCC will be predicted by detecting its invasion and metastasis molecular biomarkers, which is helpful in preventing tumor progression and obtaining a better prognosis (7).
Mammalian-enabled (Mena) protein is a member of the Ena/VASP family, and encoded by the mammalian homolog gene ENAH (Enabled homolog) of Drosophila Ena on chromosome 1, and is associated with the formation of invasive membrane protruding (8). Mena contains 4 exons named +, ++, +++ and 11a (8–10). In the process of tumor progression, alternative splicing of Mena produced a variety of functional isoforms. Mena expressed in invasive tumor cells contained +++ exons but lacked 11a exons, called MenaINV. Mena expressed in non-invasive tumor cells lack +++ exons but express an 11a exons, called Mena11a (10). Isoforms that lack both 11a and +++ exons called Mena (11). Besides, another Splice Variant of Mena lacking internal exon 6 (MenaDv6) promotes invasive behavior in cancer cells (12).
As a cytoskeletal regulatory protein, Mena is also involved in the assembly and regulation of the cytoplasmic actin network and plays a role in actin-based regulation of cell motility and adhesion, which frequently contributes to tumor invasion and metastasis (13–18). Several studies have reported outcomes of differential expression of Mena protein in various cancers (19–21). In breast cancer, overexpression of Mena promotes the proliferation and invasion of tumor cells, but deletion of Mena inhibits the metastasis of breast cancer and upregulated Mena increases drug resistance to breast cancer (22, 23). Besides, elevated Mena is correlated with the tumor grade and stage of hepatocellular carcinoma (HCC), and high expression of Mena in HCC enhances the expression level of epithelial-mesenchymal transition (EMT) markers and improves the carcinogenicity of HCC (24, 25).
Although Mena protein is associated with a variety of cancers, the clinicopathological significance and prognostic value of Mena in OSCC remains unclear. Thus, the current study firstly sought to investigate the expression and clinical significance of Mena in OSCC, and its biological function. Besides, we also evaluated the prognostic value of Mena in patients with OSCC as per their clinicopathological factors.
2. Materials and methods
2.1. Bioinformatics analysis
2.1.1. Data collection
Mena expression levels in different types of cancer and normal tissues were compared by Oncomine (26, 27) (https://www.oncomine.org/resource/login.html) online analysis database. The gene expression profiles of OSCC samples and normal tissues were collected from The Cancer Genome Atlas (TCGA, https://portal.gdc.cancer.gov/), and Gene Expression Omnibus (GEO) database (GSE3524 (28), GSE74530 (29), GSE138206, http://www.ncbi.nlm.nih.gov/geo/).
2.1.2. ENAH differential expression analysis
The gene expression microarray data were processed by R software (version 4.0.4, https://www.r-project.org/), and the R package was used to convert probe symbols into gene symbols. The “Limma” package was used to identify differentially expressed genes between oral squamous cell carcinoma tissues and normal tissues. (Adjust P < 0.05 and absolute log2fold change > 1 were considered statistically significant). TIMER (https://cistrome.shinyapps.io/timer/) online database was used to analyze the differential expression of ENAH (encoding Mena protein) between tumor and adjacent normal tissues in all TCGA tumors studied by the Gene_DE module. GEPIA (http://gepia.cancer-pku.cn/index.html) database was used to compare the expression of ENAH in HNSC tumors and normal tissues. (P< 0.01 and absolute log2fold change > 1 were considered statistically significant).
2.1.3. Protein-protein interaction network and gene set enrichment analysis
In addition, STRING (http://string-db.org) online database was used to construct the Mena-related functional genes visualized as interactive networks, and gene set enrichment analysis (GSEA) was used to evaluate the gene biological function between high ENAH expression group and low ENAH expression group. Hallmark Gene sets were selected for enrichment analysis. (Genomes with the thresholds of FDR q <0.05 and NOM P<0.05 were considered statistically significant).
2.2. The clinicopathological characteristics of Mena
2.2.1. Collection of clinical specimens and information
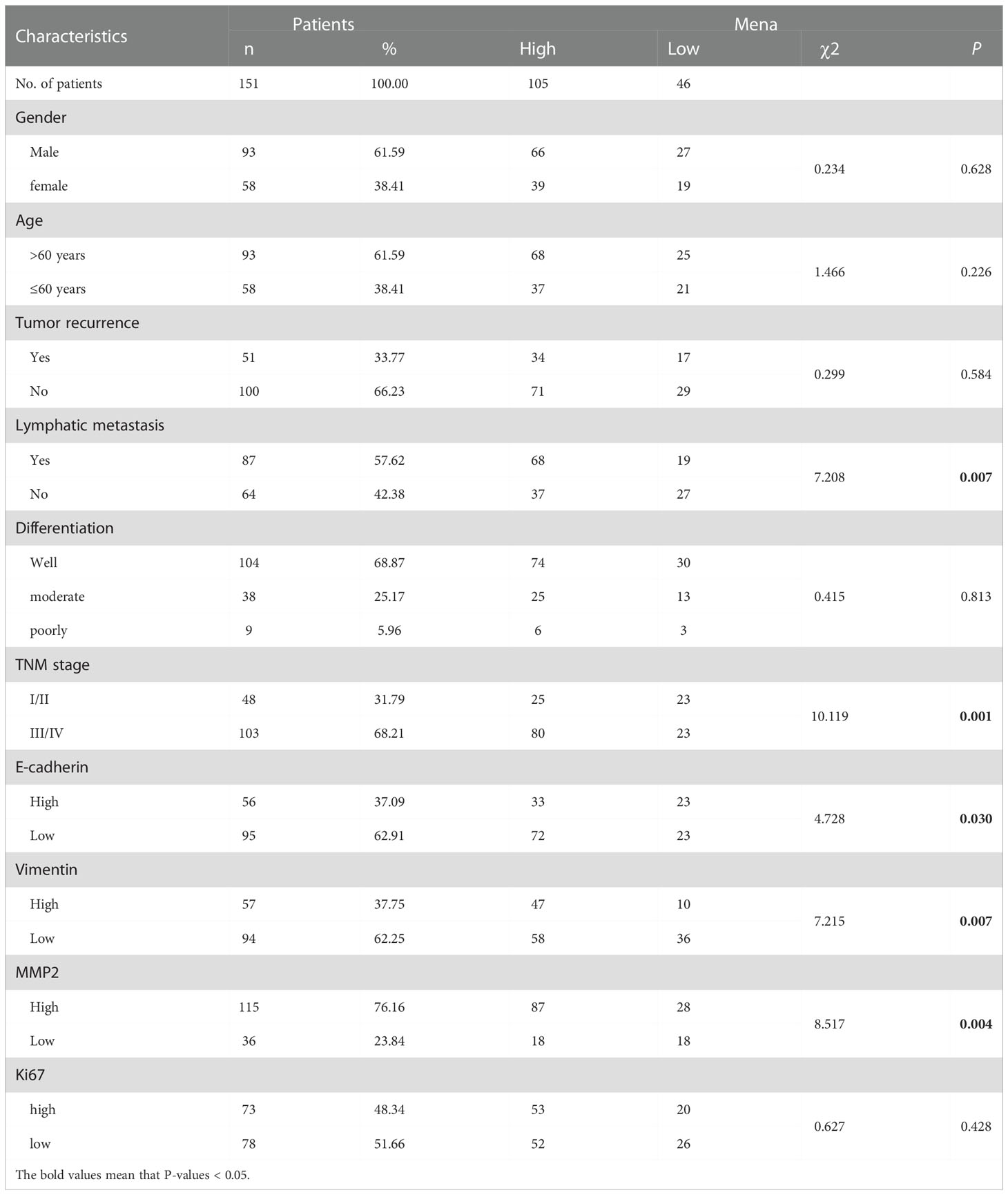
151 human OSCC specimens were obtained from the department of pathology, the Stomatological Hospital of Xi’an Jiaotong University from May 2013 to June 2020. All 151 patients with OSCC agreed and signed the informed consent. The research plan and purpose, for the collection of clinical information of OSCC patients, and the use of archived pathological specimens, have been approved by the Medical Ethics Committee of the Stomatological Hospital of Xi ‘an Jiaotong University (xjkqll [2022] NO.028). The screening criteria were as follows:(1) Patients were pathologically diagnosed as oral squamous cell carcinoma; (2) Patients had received surgical resection; (3) Patients have complete clinical data and follow-up data; (4) The patient does not have other malignant tumors. Clinical data of the patients were obtained from the Department of Pathology, Hospital of Stomatology, Xi’an Jiaotong University, including age, gender, tumor recurrence, tumor grade, clinical or pathological stage of tumor (cT/pT) and lymphatic status (cN/pN). Follow-up data were obtained by telephone or medical records review. The patient sample was comprised of 93 males and 58 females, with a mean age of 62 years (ranging from 27 to 85 years). Due to out-of-control situations, the postoperative follow-up was available only for 85 patients. In context, Kaplan-Meier curves were applied to analyze the overall survival and disease-free survival of those 85 patients, (43 patients with high expression of Mena and 42 ones with low expression of Mena).
2.2.2. Preparation of tissue microarray
All pathological paraffin blocks were stained with HE and checked diagnosis again. Select the effective area and mark it. According to the mark position, punch a 1.00mm diameter tissue core and placed it consecutively on the recipient blocks of approximately 2cm×2cm to make the tissue microarray (TMA) paraffin block.
2.2.3. Immunohistochemical analysis
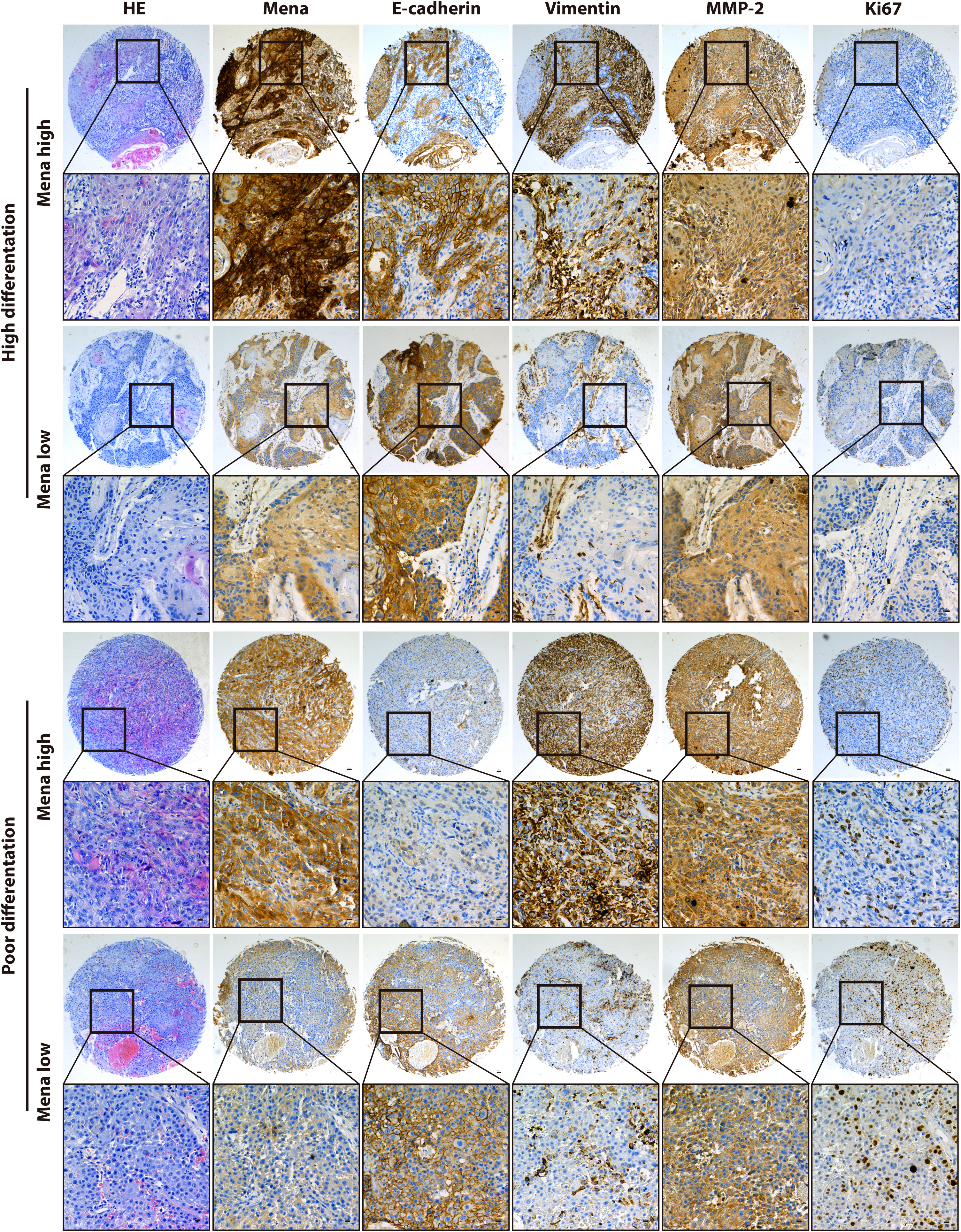
Immunohistochemistry (IHC) staining was performed using Leica Bond MAX automatic immunostaining machine (Leica Microsystems, Wetzlar, Germany). The tissue sections were deparaffinized and rehydrated with ethanol and heat-induced epitope retrieval at pH 9.0 for 20 minutes. Rabbit polyclonal anti-Mena (ab244417, Abcam, Cambridge, UK, 1:100), anti-E-cadherin (MAB-0738, Maixin, Fuzhou, CHN), anti-Vimentin (MAB-0735, Maixin, Fuzhou, CHN), anti-CD34 (Kit-0004, Maixin, Fuzhou, CHN), anti- MMP-2 (ab37150, Abcam, Cambridge, UK, 1:500), and Ki67 (MAB-0672, Maixin, Fuzhou, CHN) were used for IHC staining. Automatic staining was performed using a Bond Polymer Refine -HRP detection system (DS9800, Leica Microsystems, Wetzlar, Germany). It was developed with diaminobenzidine (DAB) and counterstained with hematoxylin. Images were analyzed by IHC profiler (30) of Image J for quantitative evaluation and automatic scoring. The software quantitatively determined the score as negative, low positive, positive, or high positive. Clinical and histopathological characteristics are shown in Table 1.
2.3. Statistical analysis
SPSS Statistics software 24.0 (IBM, Armonk, NY, United States) was used for all statistical analyses. Descriptive statistics were reported as mean or frequency and rate. The correlations of Mena with the clinicopathological factors and tumor-related markers were performed using the χ2 test. Kaplan-Meier method was used to plot the survival curve and compared by log-rank test. Cox regression analysis was used to determine the independent prognostic factor. The *P<0.05, **P<0.01 and ***P<0.001 represent statistical significance.
3. Results
3.1. The mRNA expression levels of mena in tumor and normal tissues
In order to explore Mena mRNA levels in different malignancies and corresponding normal tissues, we analyzed 393 studies using the Oncomine database. 43 studies showed higher Mena expression in the brain and central nervous system, breast, cervix, colorectum, esophagus, head and neck, liver, prostate, leukemia, lymphoma sarcoma, and other malignancies, compared to normal tissues (Figure 1A). Consistent with the above result, we compared RNA sequencing data from various malignancies in the TCGA database using the TIMER platform. Mena expression was found to be higher in most tumor tissues (including HNSC) than in normal tissues (Figure 1B).
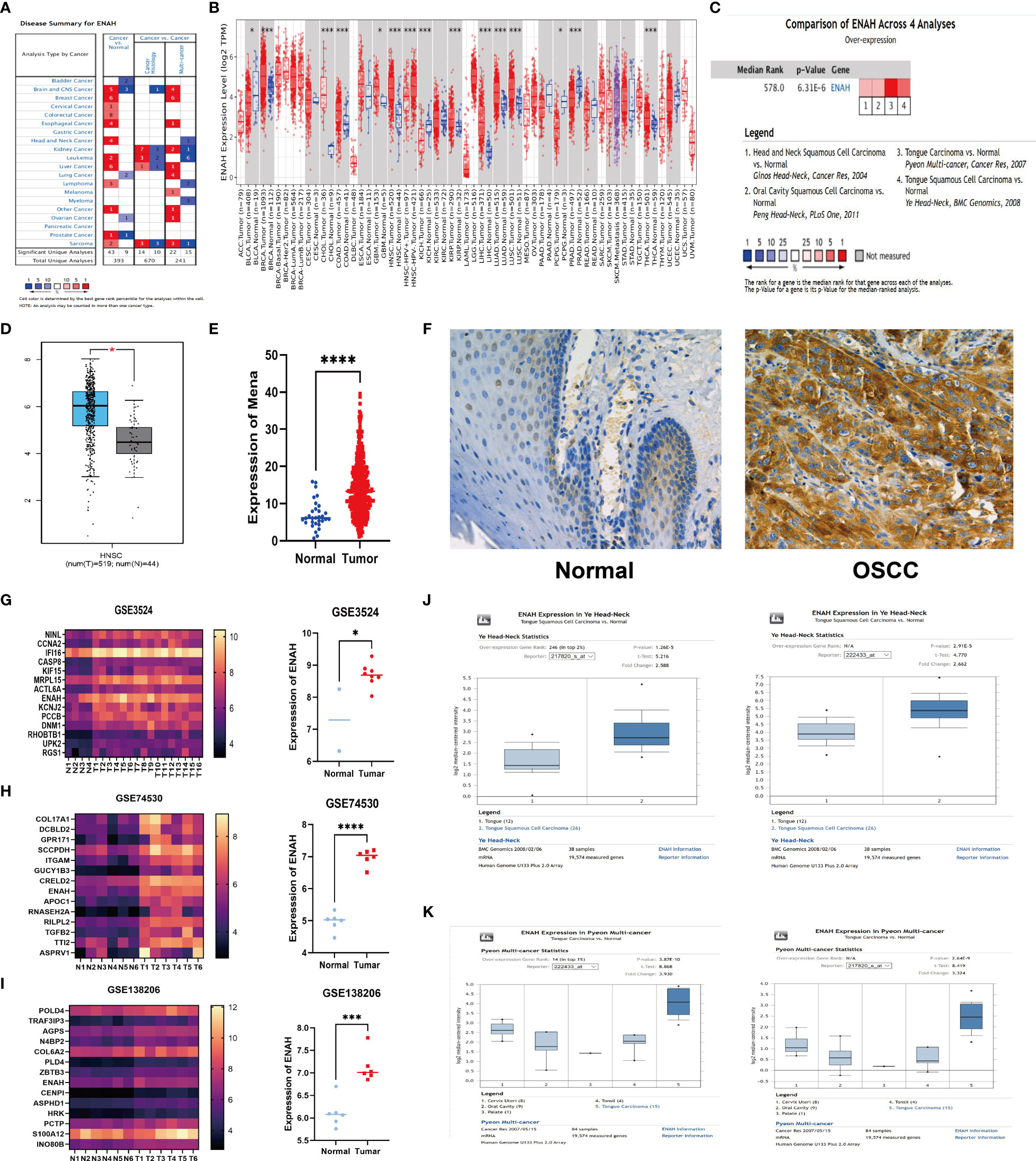
Figure 1 The abnormal expression of Mena mRNA between OSCC and normal tissues. (A) The expression of Mena mRNA in malignancies and normal tissues based on Oncomine database. (B) TIMER platform represents the expression of Mena mRNA in malignancies and normal tissues based on TCGA database. (C, D) Higher expression of ENAH (coded Mena) in HNSC tissues than normal tissues. (E) Mena expression between OSCC and normal specimens. (F) IHC staining shows lower or no expression of Mena in normal oral tissues. (G-K) The expression of Mena mRNA in OSCC and normal tissues based on GEO and Oncomine databases. *P < 0.05, ***P < 0.001, ****P < 0.0001 is mentioned in 2.3 statistical analysis.
Notably, Mena expression was significantly elevated in head and neck squamous carcinoma (HNSC) tissues compared with normal tissues in 4 studies of HNSC (Figure 1C). Similarly, the comparison of 519 tumors and 44 normal tissues of HNSC in TCGA and GTEx databases by Gene expression Profiling Interactive Analysis (GEPIA) showed that ENAH (Mena encoding gene) in tumor tissues was 1.555-fold higher than that in normal tissues, which were 64.960 in tumor tissues and 21.444 in normal tissues (P<2.33E-10) (Figure 1D).
To further explore the expression of Mena in OSCC, we analyzed RNA‐seq data of 399 TCGA patients with OSCC (tumor 369, normal 30) which was then used to identify differentially expressed genes (DEGs) and found that Mena was higher in tumor tissues (Average 7.232 in normal vs. 14.311 in tumor; P<0.0001) (Figure 1E). Meanwhile, the analysis of 3 GEO datasets related to OSCC (GSE3524 GSE74530 GSE138206) showed that the expression level of ENAH in tumor tissues was significantly higher than that in normal tissues (In GSE3524, average 7.288 in normal vs. 8.70 in OSCC, P=0.0162; In GSE74530, average 4.97 in normal vs. 6.98 in OSCC, P<0.0001; In GSE138206, average 6.11 in normal vs. 7.12 in OSCC, P=0.0003) (Figures 1G-I). Subsequently, we performed IHC staining on OSCC tissues and normal oral tissues. Weak or no staining of Mena was observed in epithelial cells of normal oral tissues and keratin pearls with severe keratosis (Figure 1F). The same results were verified in two OSCC Oncomine datasets (Ye and Pyeon) (Figures 1J, K).
3.2. The biological function of Mena in OSCC
DEGs of GSE3524, GSE74530, GSE138206, and 399 OSCC specimens of TCGA were analyzed by Limma package. The volcano plots showing significant DEGs were shown in Figures 2A–D. The Venn diagram showed a total of 171 common DEGs from 4 different datasets (Figure 2E). The data indicated that Mena was all in the up-regulated group of DEGs. The PPI of DEGs was constructed by STRING with a confidence score>0.6, and the PPI network was visualized using Cytoscape. 171 nodes and 1034 edges were covered in the PPI network, which is worth noting that Mena is a member of the common DEGs (Figure 2F).
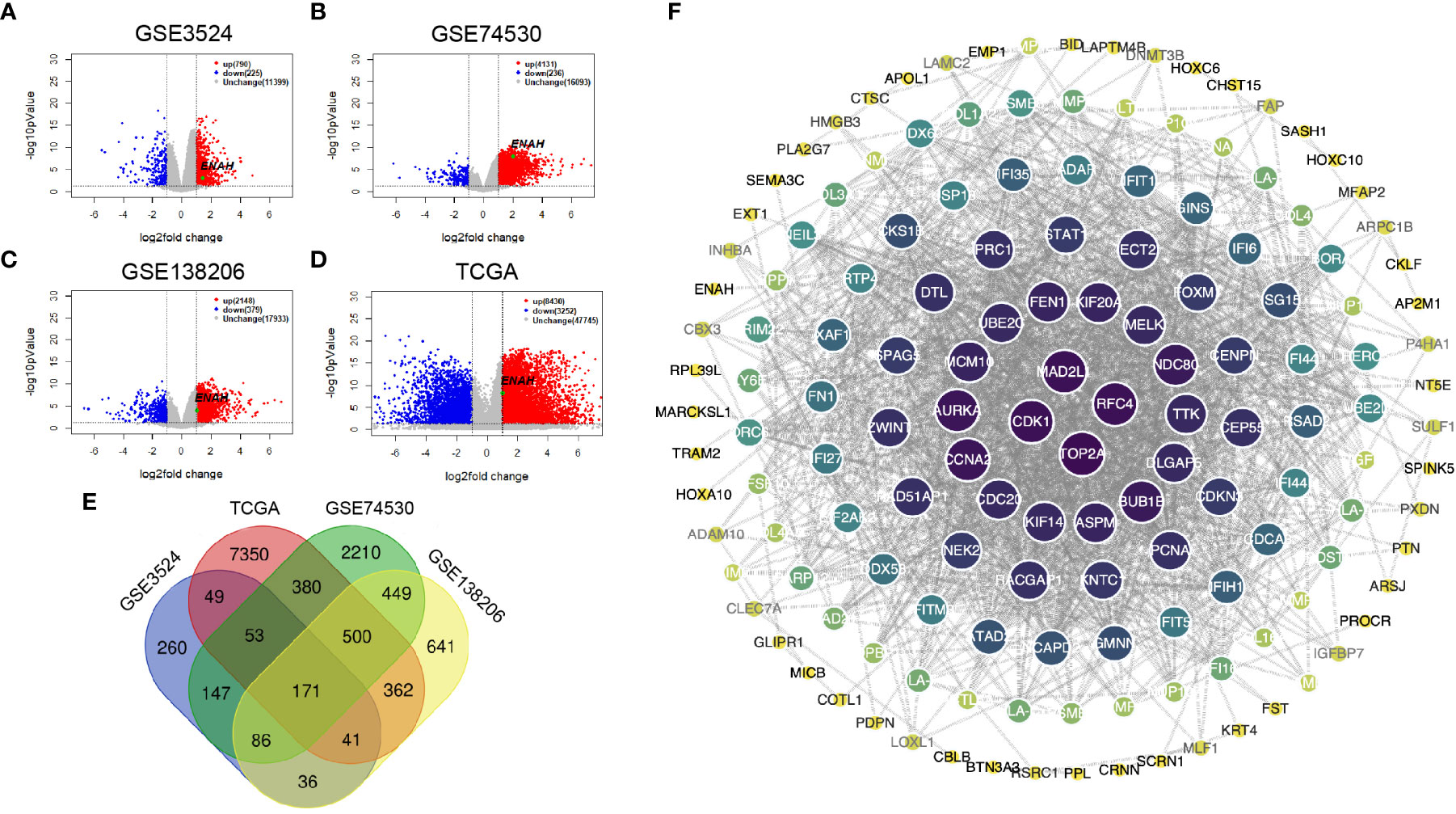
Figure 2 The construction and visualization PPI network of Mena based on DEGs from 3 GEO and 1 TCGA databases. (A-D) Volcano plots of DEGs from GSE3524, GSE74530, GSE138206, and OSCC-TCGA. (E) Venn diagram of 171 common DEGs in 3 GEO and 1 TCGA. (F) PPI network visualized by Cytoscape.
Since the differential expression of Mena was uncovered between OSCC and normal tissues, we next got an insight into the biological function of Mena in OSCC. By retrieving ENAH, we constructed a PPI network consisting of 11 nodes and 32 edges with a confidence score>0.7 utilizing the STRING database. The predicted functional genes associated with Mena mainly included ABL1, TES, VASP, EVL, APBB1IP, CTNND1, CDC42, BAIAP2, CTNNA1, and CTNNB1 (Figure 3A). Functional annotation including GO term and KEGG pathway analyses of these 11 Mena related genes were performed. GO annotation demonstrated that Mena-associated interacting proteins were mainly enriched in processes, such as “cytoskeleton organization”, “actin filament organization”, “cell junction organization”, “regulation of cytoskeleton organization”, “locomotion”, “movement of cell or subcellular component”, “positive regulation of cytoskeleton organization”, “regulation of actin filament organization”, “cell junction”, “cell leading edge”, “cytoskeleton, focal adhesion”, “lamellipodium”, “cell-cell junction”, “actin-based cell projection”, “filopodium”, “actin binding”, “profilin binding”, “proline-rich region binding” (Figure 3B). Meanwhile, KEGG pathway analysis indicated that the PPI network of Mena was significantly enriched in pathways like “pathways in cancer”, “endometrial cancer”, “focal adhesion”, “regulation of actin cytoskeleton”, “adherens junction”, “leukocyte transendothelial migration”, and “bacterial invasion of epithelial cells” (Figure 3C). The data of GO and KEGG annotation suggested that elevated expression of Mena may be involved in the cytoskeleton, regulation of actin assembly, and locomotion, which contributes to cell movement and migration, even tumor invasion and metastasis.
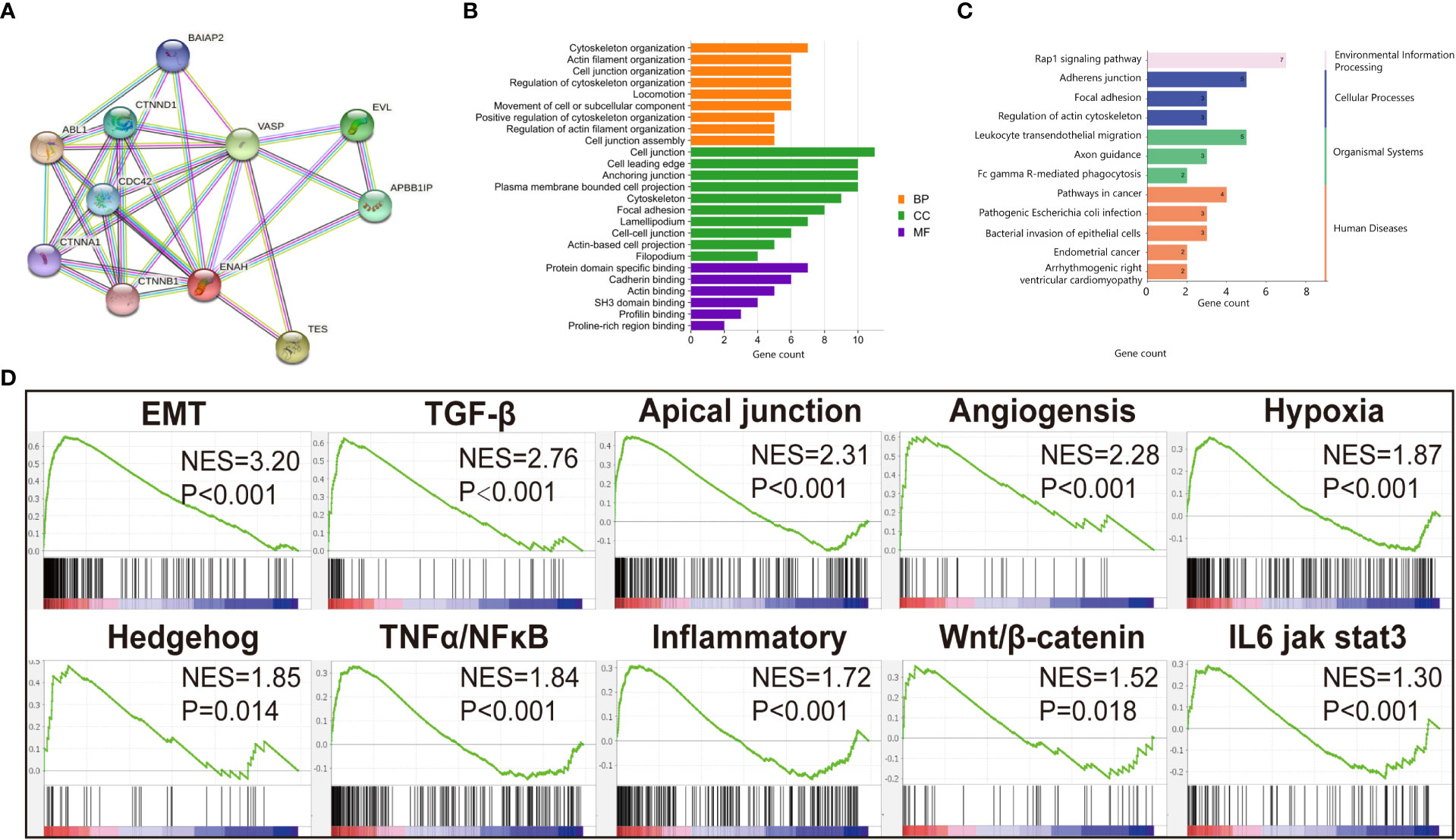
Figure 3 Functional annotation of Mena. (A) PPI network of Mena by STRING database. (B) Go enrichment analysis pf Mena related genes by STRING. (C) KEGG enrichment analysis of Mena related genes by STRING. (D) GSEA analysis to investigate the potential regulatory mechanisms with tumor hallmarks.
To further investigate the potential mechanisms of this hypothesis, 369 OSCC samples retrieved from TCGA database were divided into high Mena expression group and low expression group for gene set enrichment analysis (GSEA). We found that Hallmarks of tumor such as “epithelial-mesenchymal transition (EMT)”, “TGF-β pathway”, “Apical junction”, “Angiogenesis”, “Hypoxia”, “Hedgehog pathway”, “TNFα/NF-κB pathway”, “Inflammatory response”, “Wnt/β-catenin pathway”, and “IL6 jak stat3” were dynamically enriched in samples with high expression of Mena (Figure 3D). The result of GSEA analysis showed that Mena may promote tumor progression by participating in various cancer invasion-related signaling pathways, especially EMT and its related pathways.
3.3. The correlation between Mena and clinicopathological characteristics in OSCC
Regarding Mena expression, there were 105 cases (69.5%) with high expression Mena, while 46 cases (30.5%) with low expression Mena (Table 1). The highest expression of Mena was predominantly present in the cytoplasm, followed by higher expression in the cell membrane than in the nucleus (Figure 4). These data showed that Mena was significantly increased in patients with OSCC.
In term of association with clinicopathological factors, Mena expression was significantly correlated with Lymphatic metastasis (χ2 = 7.208, P=0.007) and TNM stage (χ2 = 10.119, P=0.001) (Table 1). However, no significant correlation was found between Mena expression and other clinicopathological parameters, including Gender (χ2 = 0.234, P=0.628), Age (χ2 = 1.466, P=0.226), Tumor recurrence (χ2 = 0.299, P=0.584), and Differentiation (χ2 = 0.415, P=0.813) (Table 1).
The biological function and Mena expression in OSCC suggest that Mena expression may be closely associated with tumor invasion and metastasis. To prove this presumption, we analyzed the correlation between Mena and tumor-related markers. The results indicated that Mena expression was statistically associated with the expression of E-cadherin (χ2 = 4.728, P=0.03), Vimentin (χ2 = 7.215, P=0.007), and MMP-2 (χ2 = 8.517, P=0.004) but not Ki67 (χ2 = 0.627, P=0.428) (Table 1). Interestingly, Vimentin staining was stronger in most OSCC specimens with high-expression Mena, while E-cadherin staining was weaker. However, the staining results of Vimentin and E-cadherin were opposite in OSCC patients with low expression of Mena. Taken all, the evidences reveal that Mena may promote tumor progression through the EMT process.
3.4. Elevated expression of mena predicts the unfavorable prognosis of OSCC patients
The prognostic value of Mena in OSCC patients was estimated by Kaplan-Meier survival analysis. OSCC patients with high Mena expression significantly shortened overall survival (OS) (P=0.037) and disease-free survival (DFS) (P=0.0031) (Figure 5A). Cox regression analysis revealed that elevated Mena expression was adverse prognostic factors for OS (HR 2.517, 95%CI 1.025-6.181; P=0.044) and DFS (HR 2.353, 95%CI 1.312-4.221; P=0.004) (Figure 5A). Increased Vimentin expression leaded to shorter OS (P=0.0264) and DFS (P=0.0036) in OSCC patients, and gave rise to worse OS (HR 2.500, 95%CI 1.083-5.773; P=0.032) and DFS (HR 2.290, 95%CI 1.290-4.066; P=0.005) (Figure 5B). The ROC curve of Mena presented diagnostic accuracy (area under the curve, 0.6461; P=0.001), compared with Vimentin (a typical EMT marker during metastasis progression) (Figure 5C). These results indicated that Mena played a negative role in the prognosis of OSCC patients, and it could be an optimal diagnostic biomarker for identifying OSCC invasion and metastasis.
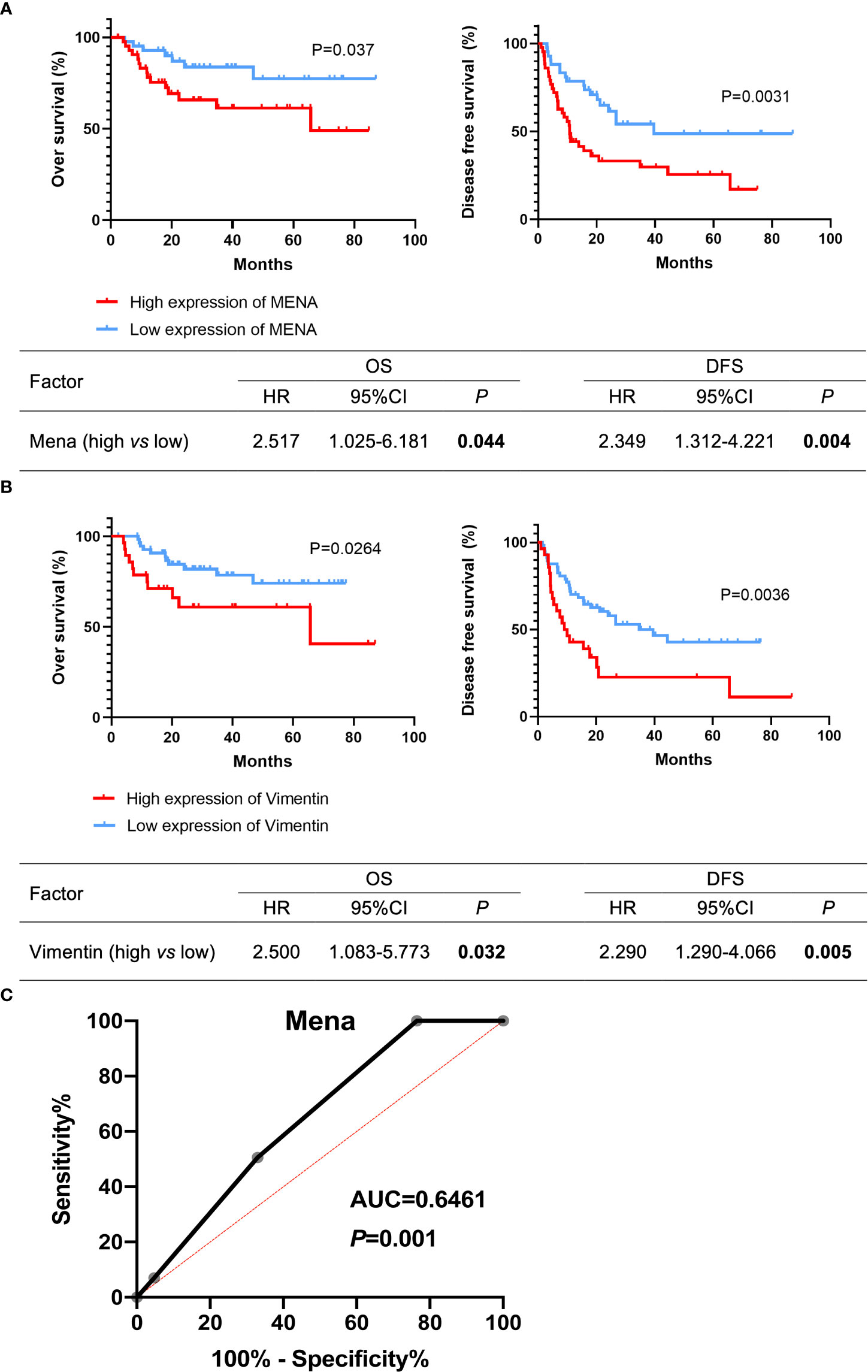
Figure 5 Potential prognostic value of Mena expression in OSCC patients. (A, B) Kaplan-Meier survival curves. (C) Receiver operating curves. The bold values mean that P-values < 0.05.
4. Discussion
Oral squamous cell carcinoma remains a fatal and teratogenic disease. Tumor invasion and metastasis are the main factors of poor prognosis and death in patients with OSCC. Therefore, early prediction and treatment of invasive OSCC are of great significance to improve the prognosis of patients with OSCC.
Mena is a cytoskeletal regulatory protein, and it is first identified through its genetic interaction with Drosophila Abelson tyrosine kinase (D-Abl) (31, 32). Mena is located in the tips of filopodia, the lamellipodial leading edge, cell-cell contacts, and focal adhesions and actin stress fibers, and Mena protein is related to actin network remodeling and cell motility, and cytoskeleton regulation (8, 33–36). Cytoskeleton and actin dynamics of tumor cells play a critical role in this process of invasion and metastasis (37, 38). In the invasive tumor cells, Mena and other proteins (such as Arp2/3 complex (39, 40), Cofilin (41), and mDia2 (42) promote cytoskeleton and motor to drive the formation of membrane protrusions for invasion and motility (43, 44). Significantly increased Mena level has been revealed in a series of human cancers, such as breast cancer (45, 46), gastric cancer (47), hepatocellular carcinoma cells (28, 29), and esophageal carcinoma (48), highlighting the effect on promoting tumor cell proliferation and invasion. Nevertheless, there is a lack of studies on the relationship between Mena expression and oral squamous cell carcinoma.
In this study, bioinformatics analysis showed that in the four datasets (GSE3524, GSE74530, GSE138206, and 399 TCGA in OSCC patients), the expression of ENAH (the gene encoding MENA protein) in tumor tissues was significantly higher than that in normal tissues. The present results of immunohistochemical staining showed high expression of Mena in 105 (69.5%) OSCC patients. Further, elevated Mena expression was correlated with Lymphatic metastasis and TNM stage. Similar results have been reported in gastric cancer (47), cervical caner (20), and thyroid cancer (49). These results suggest that Mena increases the risk of tumor invasion and metastasis. The Mena PPI network was mainly enriched in the dynamic actin formation region, such as cell leading edge, cytoskeleton, focal adhesion, lamellipodium, actin-based cell projection, and filopodium. Their functions are related to the assembly and regulation of actin cytoskeleton and Locomotion, while Mena plays an important role in cell movement and migration through enhancing actin aggregation (13). As previously reported, hyperactivity of cell movement and migration leads to tumor invasion and metastasis and is involved in the progression of multiple cancers (50).
In order to further investigate the role of Mena in tumor invasion and progression, we performed biological function analysis by GSEA. The data showed that EMT, TGF-β signaling pathway, Apical junction, Angiogenesis, Hypoxia, etc. were enriched in high expression Mena phenotype. In the previous literature, TGF-β signaling pathway triggers the EMT process as an inducer (51), and the apical junction complex, including the tight junction and the adherens junction, plays a key role in tumor metastasis induced by EMT (52). E-cadherin, a vital apical junction complex, is a transmembrane protein responsible for anchoring neighboring cells to one another and forming adherens junctions. Decreased E-cadherin is necessary for the EMT process to occur, which promotes tumor metastasis (53). Additionally, the Mena intensity was associated with vascular invasion, and intensity of angiogenesis in colorectal carcinoma (20). Fibronectin (FN) levels are high around blood vessels and at invasive edges in tumors (54). Mena, as an actin regulatory protein, is an intrinsic mechanism driving tumor cells, which promotes haptotaxis along FN gradients (55). Thus, high expression levels of MenaINV and FN are correlated with increased recurrence and poor prognosis in breast cancer. Hypoxia is a feature of solid tumors, and contributes to EMT, invasion and metastasis. Hypoxia regulated human Mena alternative splicing and promoted EMT through activating TGF-β-RBFOX2-ESRP1 axis in breast cancer (56). In addition, HIF-2α promoted glioma cells migration under hypoxia by activating an Oct-4/Sox-2-MenaINV axis (57). Furthermore, our IHC results proved that high expression level of Mena was significantly correlated with EMT markers E-cadherin and Vimentin, and tumor invasive marker MMP-2, but uncorrelated with Ki67. In addition to E-cadherin, Vimentin, another key EMT marker widely expressed in mesenchymal and mesenchymal-derived cells, plays a role in maintaining cellular integrity and contributes to cell migration and invasion during EMT (58). Additionally, MMP-2 not only plays a key effect on ECM degradation in cancer progression, which contributes to cancer cell migration out of primary tumor to form metastases but also promotes EMT through activating TGF-β pathway (59, 60). As previously reported, EMT was initially described in the literature on normal cell differentiation in early development (61). In recent years, more and more studies found that EMT is the primary mechanism by which epithelial cancer cells acquire malignant phenotypes that promote metastasis (62). In pancreatic cancer, upregulation of Mena activates EMT induced by TGF-β1/Smad2 pathway (63). Overexpression of Mena in HCC has been reported to up-regulate the expression of EMT markers (24). All the above findings suggested that Mena may activate the invasion and metastasis of OSCC and accelerate the cancer progression by inducing the EMT process.
Survival analyses of OS and DFS uncovered that Mena is associated with poor prognosis in patients with OSCC. ROC curve analysis proved the diagnostic value of Mena for OSCC invasion and metastasis. The findings suggest that Mena may be helpful in decisions for therapeutic strategies. Further research will be conducted to investigate the molecular mechanism of Mena in the development and progression of OSCC.
In conclusion, this is the first study to reveal that increased expression of Mena in OSCC patients is associated with tumor invasion and metastasis resulting from EMT, as well as poor prognosis of OSCC. Therefore, Mena may serve as a novel biomarker to predict the prognosis of OSCC, which provides a theoretical basis for developing a molecular target therapy.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, (Raw data included https://www.jianguoyun.com/c/sd/160b878/24df4ead91ec7249) further inquiries can be directed to the corresponding author/s.
Ethics statement
This research has been reviewed and approved by the Medical Ethics Committee of the Stomatological Hospital of Xi ‘an Jiaotong University (xjkqll [2022] NO.028). The patients/participants provided their written informed consent to participate in this study.
Author contributions
SN, HC, ZG, XL, and KS designed the study, analyzed data, and wrote the manuscript. JT and SN provided funding acquisition. XG, XQL, LX, and YZ performed the experiments and analyzed the data. JT, SN, and HQ supervised the research, analyzed data, and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
Key Research and Development Projects of Shaanxi Province (2020SF-182), Central University Basic Scientific Research Business Expenses Special Funds (xzy012020048).
Acknowledgments
The authors would like to thank the Oncomine, TCGA and GEO databases for the availability of the data.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Dalanon J, Matsuka Y. Decreased global interest in oral cancer during the covid-19 pandemic. Asian Pac J Cancer Prev (2021) 22(7):2117–24. doi: 10.31557/apjcp.2021.22.7.2117
2. Thompson L. World health organization classification of tumours: Pathology and genetics of head and neck tumours. Ear Nose Throat J (2006) 85(2):74. doi: 10.1177/014556130608500201
3. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin (2018) 68(1):7–30. doi: 10.3322/caac.21442
4. Na S, Zhang J, Zhou X, Tang A, Huang D, Xu Q, et al. Plumbagin-mediating Glut1 suppresses the growth of human tongue squamous cell carcinoma. Oral Dis (2018) 24(6):920–9. doi: 10.1111/odi.12799
5. Kim JW, Park Y, Roh JL, Cho KJ, Choi SH, Nam SY, et al. Prognostic value of glucosylceramide synthase and p-glycoprotein expression in oral cavity cancer. Int J Clin Oncol (2016) 21(5):883–9. doi: 10.1007/s10147-016-0973-1
6. Zanoni DK, Montero PH, Migliacci JC, Shah JP, Wong RJ, Ganly I, et al. Survival outcomes after treatment of cancer of the oral cavity (1985-2015). Oral Oncol (2019) 90:115–21. doi: 10.1016/j.oraloncology.2019.02.001
7. Sasahira T, Kirita T. Hallmarks of cancer-related newly prognostic factors of oral squamous cell carcinoma. Int J Mol Sci (2018) 19(8):2413. doi: 10.3390/ijms19082413
8. Gertler FB, Niebuhr K, Reinhard M, Wehland J, Soriano P. Mena, a relative of vasp and drosophila enabled, is implicated in the control of microfilament dynamics. Cell (1996) 87(2):227–39. doi: 10.1016/S0092-8674(00)81341-0
9. Di Modugno F, DeMonte L, Balsamo M, Bronzi G, Nicotra MR, Alessio M, et al. Molecular cloning of hMena (ENAH) and its splice variant hMena+11a: epidermal growth factor increases their expression and stimulates hMena+11a phosphorylation in breast cancer cell lines. Cancer Res (2007) 67(6):2657–65. doi: 10.1158/0008-5472.CAN-06-1997
10. Goswami S, Philippar U, Sun D, Patsialou A, Avraham J, Wang W, et al. Identification of invasion specific splice variants of the cytoskeletal protein mena present in mammary tumor cells during invasion. vivo Clin Exp Metastasis (2009) 26(2):153–9. doi: 10.1007/s10585-008-9225-8
11. Philippar U, Roussos ET, Oser M, Yamaguchi H, Kim HD, Giampieri S, et al. A mena invasion isoform potentiates EGF-induced carcinoma cell invasion and metastasis. Dev Cell (2008) 15(6):813–28. doi: 10.1016/j.devcel.2008.09.003
12. Di Modugno F, Iapicca P, Boudreau A, Mottolese M, Terrenato I, Perracchio L, et al. Splicing program of human MENA produces a previously undescribed isoform associated with invasive, mesenchymal-like breast tumors. Proc Natl Acad Sci U.S.A. (2012) 109(47):19280–5. doi: 10.1073/pnas.1214394109
13. Skruber K, Warp PV, Shklyarov R, Thomas JD, Swanson MS, Henty-Ridilla JL, et al. Arp2/3 and Mena/Vasp require profilin 1 for actin network assembly at the leading edge. Curr Biol (2020) 30(14):2651–64 e5. doi: 10.1016/j.cub.2020.04.085
14. Bear JE, Svitkina TM, Krause M, Schafer DA, Loureiro JJ, Strasser GA, et al. Antagonism between Ena/Vasp proteins and actin filament capping regulates fibroblast motility. Cell (2002) 109(4):509–21. doi: 10.1016/s0092-8674(02)00731-6
15. Vasioukhin V, Bauer C, Yin M, Fuchs E. Directed actin polymerization is the driving force for epithelial cell–cell adhesion. Cell (2000) 100(2):209–19. doi: 10.1016/s0092-8674(00)81559-7
16. Krause M, Dent EW, Bear JE, Loureiro JJ, Gertler FB. Ena/Vasp proteins: Regulators of the actin cytoskeleton and cell migration. Annu Rev Cell Dev Biol (2003) 19:541–64. doi: 10.1146/annurev.cellbio.19.050103.103356
17. Barzik M, Kotova TI, Higgs HN, Hazelwood L, Hanein D, Gertler FB, et al. Ena/Vasp proteins enhance actin polymerization in the presence of barbed end capping proteins. J Biol Chem (2005) 280(31):28653–62. doi: 10.1074/jbc.M503957200
18. Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell (2003) 112(4):453–65. doi: 10.1016/S0092-8674(03)00120-X
19. Toyoda A, Kawana H, Azuhata K, Yu J, Omata A, Kishi H, et al. Aberrant expression of human ortholog of mammalian enabled (Hmena) in human colorectal carcinomas: Implications for its role in tumor progression. Int J Oncol (2009) 34(1):53–60.
20. Gurzu S, Jung I, Prantner I, Chira L, Ember I. The immunohistochemical aspects of protein mena in cervical lesions. Rom J Morphol Embryol (2009) 50(2):213–6.
21. Pino MS, Balsamo M, Di Modugno F, Mottolese M, Alessio M, Melucci E, et al. Human Mena+11a isoform serves as a marker of epithelial phenotype and sensitivity to epidermal growth factor receptor inhibition in human pancreatic cancer cell lines. Clin Cancer Res (2008) 14(15):4943–50. doi: 10.1158/1078-0432.CCR-08-0436
22. Roussos ET, Wang Y, Wyckoff JB, Sellers RS, Wang W, Li J, et al. Mena deficiency delays tumor progression and decreases metastasis in polyoma middle-T transgenic mouse mammary tumors. Breast Cancer Res (2010) 12(6):R101. doi: 10.1186/bcr2784
23. Oudin MJ, Barbier L, Schafer C, Kosciuk T, Miller MA, Han S, et al. Mena confers resistance to paclitaxel in triple-negative breast cancer. Mol Cancer Ther (2017) 16(1):143–55. doi: 10.1158/1535-7163.MCT-16-0413
24. Hu K, Huang P, Luo H, Yao Z, Wang Q, Xiong Z, et al. Mammalian-enabled (Mena) protein enhances oncogenic potential and cancer stem cell-like phenotype in hepatocellular carcinoma cells. FEBS Open Bio (2017) 7(8):1144–53. doi: 10.1002/2211-5463.12254
25. Hu K, Wang J, Yao Z, Liu B, Lin Y, Liu L, et al. Expression of cytoskeleton regulatory protein mena in human hepatocellular carcinoma and its prognostic significance. Med Oncol (2014) 31(5):939. doi: 10.1007/s12032-014-0939-y
26. Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, et al. Oncomine: A cancer microarray database and integrated data-mining platform. Neoplasia (2004) 6(1):1–6. doi: 10.1016/S1476-5586(04)80047-2
27. Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, et al. Oncomine 3.0: Genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia (2007) 9(2):166–80. doi: 10.1593/neo.07112
28. Toruner GA, Ulger C, Alkan M, Galante AT, Rinaggio J, Wilk R, et al. Association between gene expression profile and tumor invasion in oral squamous cell carcinoma. Cancer Genet Cytogenet (2004) 154(1):27–35. doi: 10.1016/j.cancergencyto.2004.01.026
29. Oghumu S, Knobloch TJ, Terrazas C, Varikuti S, Ahn-Jarvis J, Bollinger CE, et al. Deletion of macrophage migration inhibitory factor inhibits murine oral carcinogenesis: Potential role for chronic pro-inflammatory immune mediators. Int J Cancer (2016) 139(6):1379–90. doi: 10.1002/ijc.30177
30. Varghese F, Bukhari AB, Malhotra R, De A. Ihc profiler: An open source plugin for the quantitative evaluation and automated scoring of immunohistochemistry images of human tissue samples. PloS One (2014) 9(5):e96801. doi: 10.1371/journal.pone.0096801
31. Gertler FB, Doctor JS, Hoffmann FM. Genetic suppression of mutations in the drosophila abl proto-oncogene homolog. Science (1990) 248(4957):857–60. doi: 10.1126/science.2188361
32. Gertler FB, Comer AR, Juang JL, Ahern SM, Clark MJ, Liebl EC, et al. Enabled, a dosage-sensitive suppressor of mutations in the drosophila abl tyrosine kinase, encodes an abl substrate with Sh3 domain-binding properties. Genes Dev (1995) 9(5):521–33. doi: 10.1101/gad.9.5.521
33. Lambrechts A, Kwiatkowski AV, Lanier LM, Bear JE, Vandekerckhove J, Ampe C, et al. Camp-dependent protein kinase phosphorylation of evl, a Mena/Vasp relative, regulates its interaction with actin and Sh3 domains. J Biol Chem (2000) 275(46):36143–51. doi: 10.1074/jbc.M006274200
34. Laurent V, Loisel TP, Harbeck B, Wehman A, Gröbe L, Jockusch BM, et al. Role of proteins of the Ena/Vasp family in actin-based motility of listeria monocytogenes. J Cell Biol (1999) 144(6):1245–58. doi: 10.1083/jcb.144.6.1245
35. Ahern-Djamali SM, Bachmann C, Hua P, Reddy SK, Kastenmeier AS, Walter U, et al. Identification of profilin and src homology 3 domains as binding partners for drosophila enabled. Proc Natl Acad Sci U.S.A. (1999) 96(9):4977–82. doi: 10.1073/pnas.96.9.4977
36. Bachmann C, Fischer L, Walter U, Reinhard M. The Evh2 domain of the vasodilator-stimulated phosphoprotein mediates tetramerization, f-actin binding, and actin bundle formation. J Biol Chem (1999) 274(33):23549–57. doi: 10.1074/jbc.274.33.23549
37. Izdebska M, Zielińska W, Hałas-Wiśniewska M, Grzanka A. Involvement of actin and actin-binding proteins in carcinogenesis. Cells (2020) 9(10):2245. doi: 10.3390/cells9102245
38. Datta A, Deng S, Gopal V, Yap KC, Halim CE, Lye ML, et al. Cytoskeletal dynamics in epithelial-mesenchymal transition: Insights into therapeutic targets for cancer metastasis. Cancers (Basel) (2021) 13(8):1882. doi: 10.3390/cancers13081882
39. Nürnberg A, Kitzing T, Grosse R. Nucleating actin for invasion. Nat Rev Cancer (2011) 11(3):177–87. doi: 10.1038/nrc3003
40. Wang W, Wyckoff JB, Goswami S, Wang Y, Sidani M, Segall JE, et al. Coordinated regulation of pathways for enhanced cell motility and chemotaxis is conserved in rat and mouse mammary tumors. Cancer Res (2007) 67(8):3505–11. doi: 10.1158/0008-5472.CAN-06-3714
41. Sidani M, Wessels D, Mouneimne G, Ghosh M, Goswami S, Sarmiento C, et al. Cofilin determines the migration behavior and turning frequency of metastatic cancer cells. J Cell Biol (2007) 179(4):777–91. doi: 10.1083/jcb.200707009
42. Yang C, Czech L, Gerboth S, Kojima S, Scita G, Svitkina T. Novel roles of formin mDia2 in lamellipodia and filopodia formation in motile cells. PloS Biol (2007) 5(11):e317. doi: 10.1371/journal.pbio.0050317
43. Wang W, Mouneimne G, Sidani M, Wyckoff J, Chen X, Makris A, et al. The activity status of cofilin is directly related to invasion, intravasation, and metastasis of mammary tumors. J Cell Biol (2006) 173(3):395–404. doi: 10.1083/jcb.200510115
44. Wang W, Goswami S, Lapidus K, Wells AL, Wyckoff JB, Sahai E, et al. Identification and testing of a gene expression signature of invasive carcinoma cells within primary mammary tumors. Cancer Res (2004) 64(23):8585–94. doi: 10.1158/0008-5472.CAN-04-1136
45. Di Modugno F, Mottolese M, Di Benedetto A, Conidi A, Novelli F, Perracchio L, et al. The cytoskeleton regulatory protein hmena (Enah) is overexpressed in human benign breast lesions with high risk of transformation and human epidermal growth factor receptor-2-Positive/Hormonal receptor-negative tumors. Clin Cancer Res (2006) 12(5):1470–8. doi: 10.1158/1078-0432.CCR-05-2027
46. Berger AJ, Renner CM, Hale I, Yang X, Ponik SM, Weisman PS, et al. Scaffold stiffness influences breast cancer cell invasion via egfr-linked mena upregulation and matrix remodeling. Matrix Biol (2020) 85-86:80–93. doi: 10.1016/j.matbio.2019.07.006
47. Xu L, Tan H, Liu R, Huang Q, Zhang N, Li X, et al. Expression of the cytoskeleton regulatory protein mena in human gastric carcinoma and its prognostic significance. Oncol Lett (2017) 14(5):6024–30. doi: 10.3892/ol.2017.6974
48. Wang C, Tang D, Wang H, Hu G, Hu S, Li L, et al. Circular rna Hsa_Circ_0030018 acts as a sponge of mir-599 to aggravate esophageal carcinoma progression by regulating enah expression. J Cell Biochem (2019). doi: 10.1002/jcb.29507
49. Liu R, Liu R, Li J, Liu Y, Jiang H, Wang S. Expression status of cytoskeleton regulatory protein mena as a prognostic marker for human thyroid carcinoma. Int J Clin Exp Pathol (2018) 11(3):1546–53.
50. Lambrechts A, Van Troys M, Ampe C. The actin cytoskeleton in normal and pathological cell motility. Int J Biochem Cell Biol (2004) 36(10):1890–909. doi: 10.1016/j.biocel.2004.01.024
51. Horiguchi K, Shirakihara T, Nakano A, Imamura T, Miyazono K, Saitoh M. Role of ras signaling in the induction of snail by transforming growth factor-beta. J Biol Chem (2009) 284(1):245–53. doi: 10.1074/jbc.M804777200
52. Takahashi H, Oshi M, Yan L, Endo I, Takabe K. Gastric cancer with enhanced apical junction pathway has increased metastatic potential and worse clinical outcomes. Am J Cancer Res (2022) 12(5):2146–59.
53. Onder TT, Gupta PB, Mani SA, Yang J, Lander ES, Weinberg RA. Loss of e-cadherin promotes metastasis Via multiple downstream transcriptional pathways. Cancer Res (2008) 68(10):3645–54. doi: 10.1158/0008-5472.Can-07-2938
54. Astrof S, Crowley D, George EL, Fukuda T, Sekiguchi K, Hanahan D, et al. Direct test of potential roles of EIIIA and EIIIB alternatively spliced segments of fibronectin in physiological and tumor angiogenesis. Mol Cell Biol (2004) 24(19):8662–70. doi: 10.1128/MCB.24.19.8662-8670.2004
55. Oudin MJ, Jonas O, Kosciuk T, Broye LC, Guido BC, Wyckoff J, et al. Tumor cell-driven extracellular matrix remodeling drives haptotaxis during metastatic progression. Cancer Discovery (2016) 6(5):516–31. doi: 10.1158/2159-8290.CD-15-1183
56. Ahuja N, Ashok C, Natua S, Pant D, Cherian A, Pandkar MR, et al. Hypoxia-induced TGF-β-RBFOX2-ESRP1 axis regulates human MENA alternative splicing and promotes EMT in breast cancer. NAR Cancer (2020) 2(3):zcaa021. doi: 10.1093/narcan/zcaa021
57. Bhagat M, Palanichamy JK, Ramalingam P, Mudassir M, Irshad K, Chosdol K, et al. HIF-2α mediates a marked increase in migration and stemness characteristics in a subset of glioma cells under hypoxia by activating an Oct-4/Sox-2-Mena (INV) axis. Int J Biochem Cell Biol (2016) 74:60–71. doi: 10.1016/j.biocel.2016.02.017
58. Fuchs E, Weber K. Intermediate filaments: Structure, dynamics, function, and disease. Annu Rev Biochem (1994) 63:345–82. doi: 10.1146/annurev.bi.63.070194.002021
59. Jacob A, Prekeris R. The regulation of mmp targeting to invadopodia during cancer metastasis. Front Cell Dev Biol (2015) 3:4. doi: 10.3389/fcell.2015.00004
60. Gialeli C, Theocharis AD, Karamanos NK. Roles of matrix metalloproteinases in cancer progression and their pharmacological targeting. FEBS J (2011) 278(1):16–27. doi: 10.1111/j.1742-4658.2010.07919.x
61. Micalizzi DS, Farabaugh SM, Ford HL. Epithelial-mesenchymal transition in cancer: Parallels between normal development and tumor progression. J Mammary Gland Biol Neoplasia (2010) 15(2):117–34. doi: 10.1007/s10911-010-9178-9
62. Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer (2002) 2(6):442–54. doi: 10.1038/nrc822
Keywords: mena, oral squamous cell carcinoma, clinicopathological significance, prognosis, epithelial-mesenchymal transition
Citation: Na S, Cui H, Guo Z, Liang X, Sakran KA, Guo X, Li X, Xie L, Zhu Y, Qi H and Tu J (2022) Overexpression of Mena is associated with tumor progression and poor prognosis in oral squamous cell carcinoma via EMT. Front. Oncol. 12:1052375. doi: 10.3389/fonc.2022.1052375
Received: 23 September 2022; Accepted: 05 December 2022;
Published: 23 December 2022.
Edited by:
Ling Gao, The Affiliated Hospital of Qingdao University, ChinaCopyright © 2022 Na, Cui, Guo, Liang, Sakran, Guo, Li, Xie, Zhu, Qi and Tu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hong Qi, cWlob25nQG1haWwueGp0dS5lZHUuY24=; Junbo Tu, dHVqdW5ib0BtYWlsLnhqdHUuZWR1LmNu
†These authors have contributed equally to this work
 Sijia Na
Sijia Na Hao Cui
Hao Cui Zhichen Guo
Zhichen Guo Xiang Liang1,2
Xiang Liang1,2 Karim Ahmed Sakran
Karim Ahmed Sakran Linyang Xie
Linyang Xie