- 1Department of Clinical Pharmacy, The First Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, China
- 2Laboratory Medicine Center, Department of Pathology, Zhejiang Provincial People’s Hospital, Affiliated People’s Hospital, Hangzhou Medical College, Hangzhou, China
- 3International Medical Department, The First Affiliated Hospital of University of Science and Technology of China, Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei, China
- 4Clinical Pharmacy Center, Department of Pharmacy, Zhejiang Provincial People’s Hospital, Affiliated People’s Hospital, Hangzhou Medical College, Hangzhou, China
- 5Key Laboratory of Endocrine Gland Diseases of Zhejiang Province, Zhejiang Provincial People’s Hospital, Affiliated People’s Hospital, Hangzhou Medical College, Hangzhou, China
Objective: Although the pathogenesis of hepatocellular carcinoma (HCC) is still unclear, hepatitis C virus (HCV) infection is considered a common cause of HCC. It has been reported that DDX60L can inhibit HCV replication, but its role in HCC is still poorly understood.
Methods: The expression levels of DDX60L in HCC tissues and in tissues adjacent to the tumor and their correlation with the clinicopathological features of patients were analyzed. We also used Kaplan–Meier curves of overall survival (OS) with Cox regression analysis and log-rank test to investigate the prognostic value of DDX60L in HCC. We further performed cell proliferation, Transwell, and wound healing assays to elucidate the role of DDX60L in HCC using the siRNA-DDX60L Hep3B or HCCLM3 cell line.
Results: Univariate analysis showed that sex, Edmondson grade, microvascular invasion, tumor stage (III–IV/I–II), AFP, and DDX60L expression were strongly associated with the prognosis of HCC patients. The results of multivariate analysis further suggested that DDX60L might be an independent prognostic factor for OS in patients with HCC (Pmoderate/low = 0.015, Phigh/low = 0.011). The low DDX60L expression in HCC patients with no-metastasis, age ≥55 years, tumor size <5 cm, Edmondson grade = I–II, microvascular invasion, no cirrhosis, HBV positivity, tumor stage = III–IV, AFP >20 μg/L, and multiple tumor was associated with poorer prognosis (P <0.05). Moreover, the expression of DDX60L was significantly lower in HCC samples (N = 285) than in the normal tissues adjacent to the tumor (N = 167, P <0.001). There were no HCV-related HCC patients in this study. Additionally, we found that DDX60L knockdown can promote the proliferation of Hep3B cells, migration and invasion ability of Hep3B and HCCLM3 cells.
Conclusion: We found that the downregulation of DDX60L expression correlated with poor prognosis in patients with HCC, which may be independent of the HCV-related pathway. Furthermore, DDX60L significantly inhibited the proliferation of Hep3B cells, migration and invasion of Hep3B and HCCLM3 cells. Therefore, DDX60L can serve as a prognostic biomarker and therapeutic target for HCC.
Introduction
Hepatocellular carcinoma (HCC) is the most common type of liver cancer and the fourth leading cause of cancer-related deaths worldwide, with 45.3% of HCC patients found in China (1). Additionally, HCC is an intractable tumor caused by multiple factors and shows the participation of multiple genes; the lack of specific and early diagnostic markers leads to a large proportion of patients showing advanced HCC during their diagnosis (2). Systemic therapy, namely, sorafenib, lenvatinib, regorafenib, ramucirumab, and cabozantinib have been recommended by the NCCN guidelines to treat patients with advanced HCC (3). Pembrolizumab is recommended for patients with advanced HCC who have received prior treatment with sorafenib (4). Despite these advances, the prognosis of HCC remains poor (5), and novel biomarkers and therapeutic targets need to be developed.
Viral infections, namely, hepatitis B virus (HBV) and/or hepatitis C virus (HCV), are specific risk factors for HCC development (6–8). In the United States, HCV-related HCC patients are common (1). Direct-acting antivirals (DAAs) may improve sustained virologic responses in patients with HCV (9, 10), which ultimately reduces the incidence of HCC (11–13). HCV induces an effective interferon (IFN) response early in the host infection, leading to interferon-stimulated gene (ISG) expression as the first line of defense against viral infection (14, 15). The DEAD-box helicase (DDX) family contains some ISG products that contribute to antiviral defense by sensing and fighting viral infections (16). In general, the DDX family has conserved structural domains and functions and is involved in the majority of RNA metabolic processes, from transcription to degradation (17). It has been shown that the DDX family plays an important role in tumor development. For example, high expression of DDX1 is present in neuroblastoma and retinoblastoma and promotes tumor progression, with effects on RNA transcription, export, and translation (18). The upregulated expression of DDX17 was closely associated with poor prognosis in HCC, and DDX17 knockout could inhibit the invasive capacity of HCC cells, indicating that DDX17 is a potential prognostic marker for HCC (19). Dead box polypeptide 60-like (DDX60L) is a member of the DDX family (20), and it was found that DDX60L is highly expressed in pancreatic ductal adenocarcinoma (PDAC) cells and that DDX60L knockdown significantly induced apoptosis and reduced the metastasis of PDAC cells (21). However, little is known about the correlation between DDX60L and the clinicopathological features of HCC. Therefore, in this study, we aimed to investigate the role of DDX60L and its clinical relevance in HCC.
Materials and Methods
Tissue Microarray and Cell Culture
HCC samples were obtained from the Zhejiang Provincial People’s Hospital (Hangzhou, China), and written informed consents were obtained from all patients. This study was approved by the Ethics Committee of the Zhejiang Provincial People’s Hospital. The tissue microarrays contained 285 HCC tissue samples and 167 tumor-adjacent normal tissue samples. The follow-up period was >5 years. Overall survival (OS) was calculated from the date of surgery to the end date of the follow-up or the date of death. The Hep3B cell line was purchased from the Hunan Fenghui Biological Technology Co., Ltd. HCCLM3 were obtained from the Zhongshan Hospital, Fudan University, Shanghai, China. Hep3B and HCCLM3 cells were cultured in DMEM with 10% FBS, 100 μg/ml penicillin, and 0.1 mg/ml streptomycin at 37°C and 5% CO2 in a humidified incubator.
Immunohistochemical Staining
Immunohistochemical staining was performed according to the standard methods. Briefly, 5 µm sections from the tissue microarrays were baked at 70°C for 2 h. Sections were then deparaffinized in xylene, rehydrated using an ethanol concentration gradient, and heated in a pressure cooker for 3 min in an antigen retrieval citra solution (pH 8.0, catalog number ZLI-9072, Zhongshan Golden Bridge Biotechnology) for antigen repair, and sealed with 3% hydrogen peroxide for 15 min to inhibit the activity of endogenous peroxidase. Subsequently, tissue microarray slices were incubated for 16 h at 4°C with the primary antibody against DDX60L (dilution, 1:400; catalog number bs-14236R; Beijing Biosynthesis Biotechnology Co., Ltd.), followed by incubation with EnVision+ System-HRP (catalog number K4001, Dako). Finally, the staining was visualized with the DAB Substrate kit (catalog number DA1010-2, Solarbio) and counterstained with hematoxylin (22).
Scoring of Immunohistochemical Staining
The pathologist scored the immunohistochemical staining of DDX60L based on the proportion and intensity of the positively stained cells. The staining intensity was assessed using a four-grade grading system: 3 (strong), 2 (moderate), 1 (weak), and 0 (negative). The percentage of DDX60L-positive cells was scored as follows: 0 (less than 5%), 1 (6–25%), 2 (26–50%), 3 (51–75%), and 4 (>75%) (22). The staining intensity scores and the percentage of positive cells were multiplied to obtain the final immunohistochemical staining score of DDX60L. Score ranges of 0–3, 4–8, and 9–12 were used to define low, moderate, and high expression levels of DDX60L, respectively.
Small Interfering RNA (siRNA) Transfection
Hep3B and HCCLM3 cells were transfected with a mixture of siRNA (100 nM) and Lipofectamine 2000 reagent (Guangzhou Ruibo Biological Technology Co., Ltd.) according to the manufacturer’s protocol. The siRNA sequences used were as follows: siRNA-DDX60L-1, sense: GGUAUUCCAGCAUAUUAAAdTdT, antisense: UUUAAUAUGCUGGAAUACCdTdT; and siRNA-DDX60L-2, sense: GAUUCAGAGUGCAUAUAAAdTdT, antisense: UUUAUAUGCACUCUGAAUCdTdT. Cells were collected 24 or 48 h after transfection for subsequent experiments. Moreover, the downregulation of DDX60L expression was confirmed via real-time PCR.
Cell Proliferation Assay
Briefly, 200 μl of Hep3B cells or siRNA-DDX60L Hep3B cells, and HCCLM3 cells or siRNA-DDX60L HCCLM3 cells, were seeded into 96-well cell culture-treated plates, and cultured for 12, 24, or 48 h. Then cell proliferation assay was performed using Cell Counting Kit-8 (CCK-8; Hangzhou Fude Biological Technology Co., Ltd.) according to the manufacturer’s protocol (23).
Transwell Assay and Wound Healing Assay
A total of 200 μl of Hep3B cells or siRNA-DDX60L Hep3B cells (2 × 104 cells/well), and also HCCLM3 cells or siRNA-DDX60L HCCLM3 cells, were seeded into the upper chamber of a Transwell (Corning, USA). DMEM supplemented with 10% FBS (600 μl) was added to the lower chamber and incubated at 37°C for 24 h. Cell migration was analyzed using methanol and 0.3% crystal violet staining.
Hep3B or siRNA-DDX60L Hep3B cells, and also HCCLM3 cells or siRNA-DDX60L HCCLM3 cells, were seeded at a density of 2 × 105 cells/cm2. Scratch wounds were made using a 10 µl pipette tip. Cells were washed twice with PBS and then cultured in serum-free DMEM at 37°C for 48 h. An inverted light microscope was used to observe the cells, and the ImageJ software (version 1.52) was used to analyze the effect of DDX60L on the migration ability of Hep3B or HCCLM3 cells.
Statistical Analysis
We used the IBM Statistical Package for Social Sciences (SPSS) software (version 20.0; SPSS Inc., Chicago, IL, USA) to perform the statistical analyses. Because the DDX60L expression was divided into low-, moderate-, and high-level expression groups, and we considered it as an ordinal categorical variable, the Mann–Whitney U test was applied separately to evaluate the difference in DDX60L expression in clinicopathological parameters. We used the Kaplan–Meier curve to compare the OS. Univariate and multivariate Cox regression models were used to analyze the independent risk factors associated with the OS. The mean and SD of at least three separate cell experiments were obtained. Unpaired t-tests were used to compare the differences between the two groups. Statistical significance was set at P <0.05.
Results
Clinical Characteristics and Prognosis of Patients With HCC
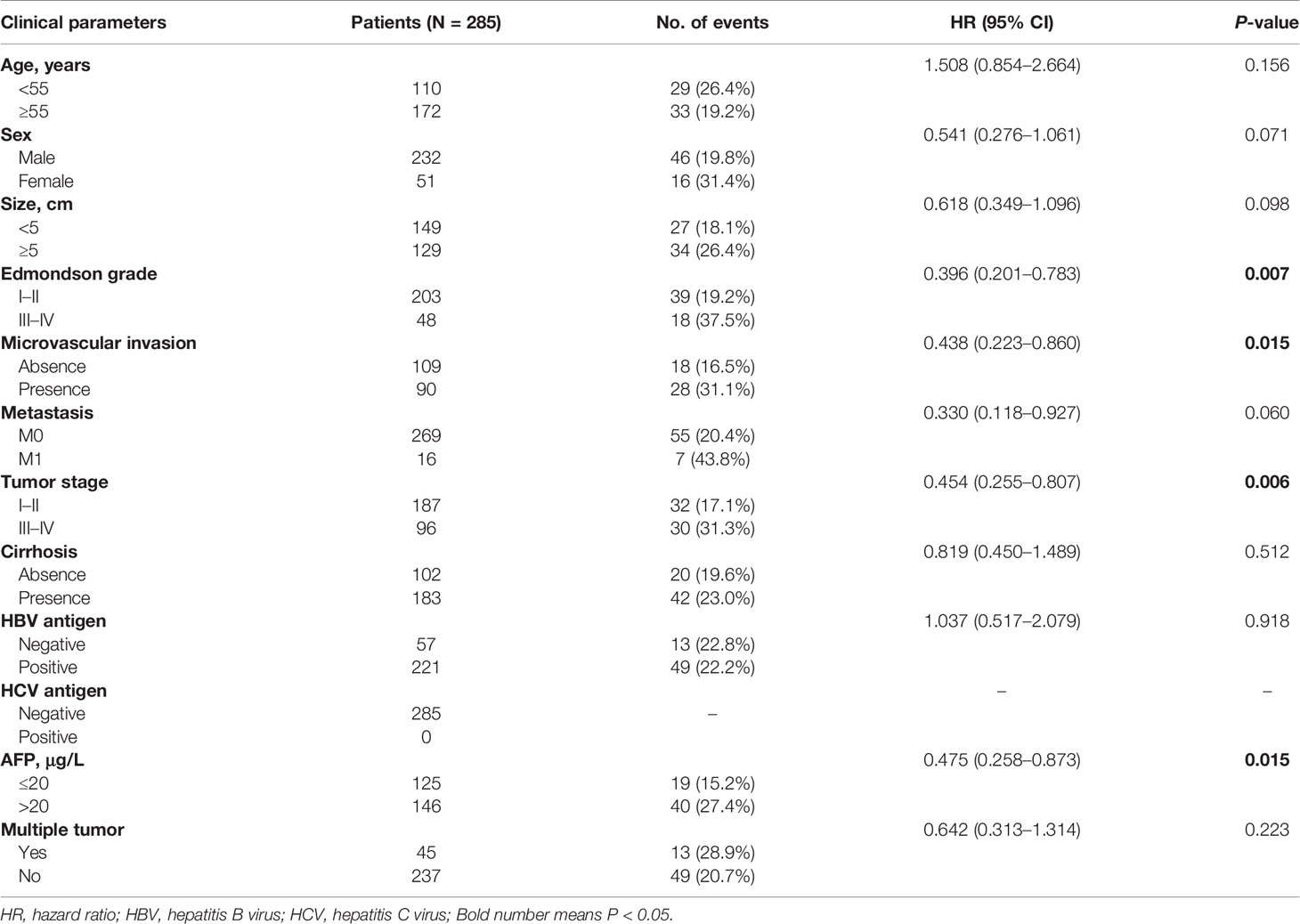
The clinical characteristics of HCC patients are shown in Table 1, with no significant differences except for the Edmondson grade (I–II/III–IV), microvascular invasion, tumor stage, and AFP (P = 0.007, 0.015, 0.006, and 0.015, respectively). We used a univariate Cox proportional model to analyze the correlation between the clinicopathological characteristics and OS in 285 patients with HCC. Furthermore, no history of HCV infection was found in any of the patients. As shown in Table 2, sex (female/male), Edmondson grade (III–IV/I–II), microvascular invasion (±), tumor stage (III–IV/I–II), AFP (>20/≤20), and DDX60L expression (moderate/low, and high/low) were significantly associated with OS (P <0.05), while age (≥55/<55), tumor size (≥5/<5), metastasis (M1/M0), HBV antigen (±), multiple tumor (±), and cirrhosis (±) were not significantly associated with OS (P >0.05). In addition, multivariate Cox regression models showed that DDX60L expression (moderate/low: HR = 0.248, 95% CI: 0.080–0.765, P = 0.015; high/low: HR = 0.153, 95% CI: 0.036–0.647, P = 0.011) was independent prognostic factor for HCC.
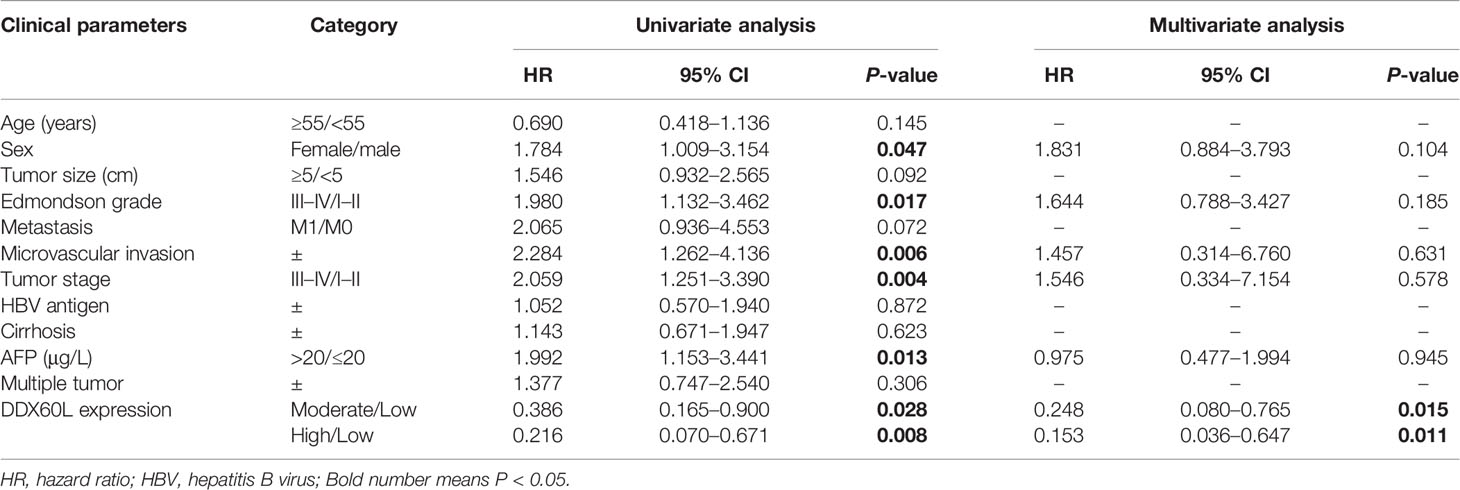
Table 2 Univariate and multivariate Cox regression analyses for the clinicopathological parameters in HCC patients.
DDX60L Expression Correlates With the Overall Survival and Clinicopathological Characteristics of Patients With HCC
Subsequently, the correlation between DDX60L expression and OS was assessed using Kaplan-Meier survival analysis and log-rank test. As shown in Figure 1A, HCC patients with low levels of DDX60L expression had a shorter OS (P = 0.017). In a stratified analysis (Figures 1B–V), the downregulation of DDX60L expression correlated with poor prognosis in HCC patients with non-metastasis (P = 0.013), age ≥55 (P = 0.009), tumor size <5 cm (P = 0.009), Edmondson grade = I–II (P = 0.042), microvascular invasion (P = 0.003), non-cirrhosis (P = 0.038), HBV positive (P = 0.017), tumor stages III–IV (P = 0.007), AFP >20 μg/L (P = 0.015), and multiple tumor (P = 0.009).
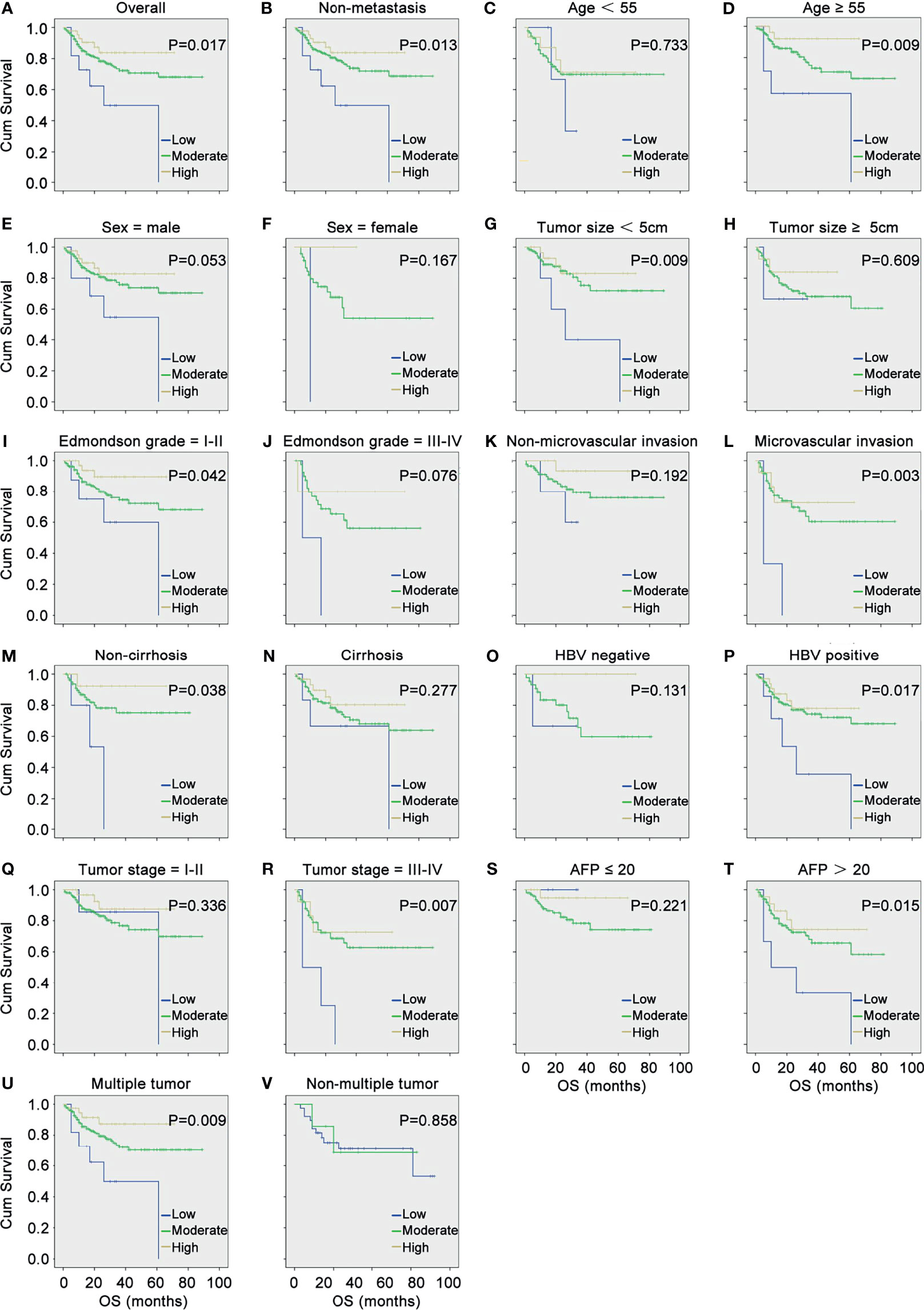
Figure 1 DDX60L expression correlated with the overall survival (OS) and clinicopathological features of HCC. (A) Kaplan–Meier survival analysis and log-rank test were used to compare the OS among different DDX60L expression groups; Kaplan–Meier survival analysis and log-rank test were used to analyze the correlation between DDX60L expression and OS stratified by non-metastasis (B); age (C, D); sex (E, F); tumor size (G, H); Edmondson grade (I, J); microvascular invasion (K, L); cirrhosis (M, N); HBV (O, P); tumor stage (Q, R); AFP (S, T); and multiple tumor (U, V).
Differential Expression of DDX60L in HCC Tissues and Normal Tissues Adjacent to Tumors
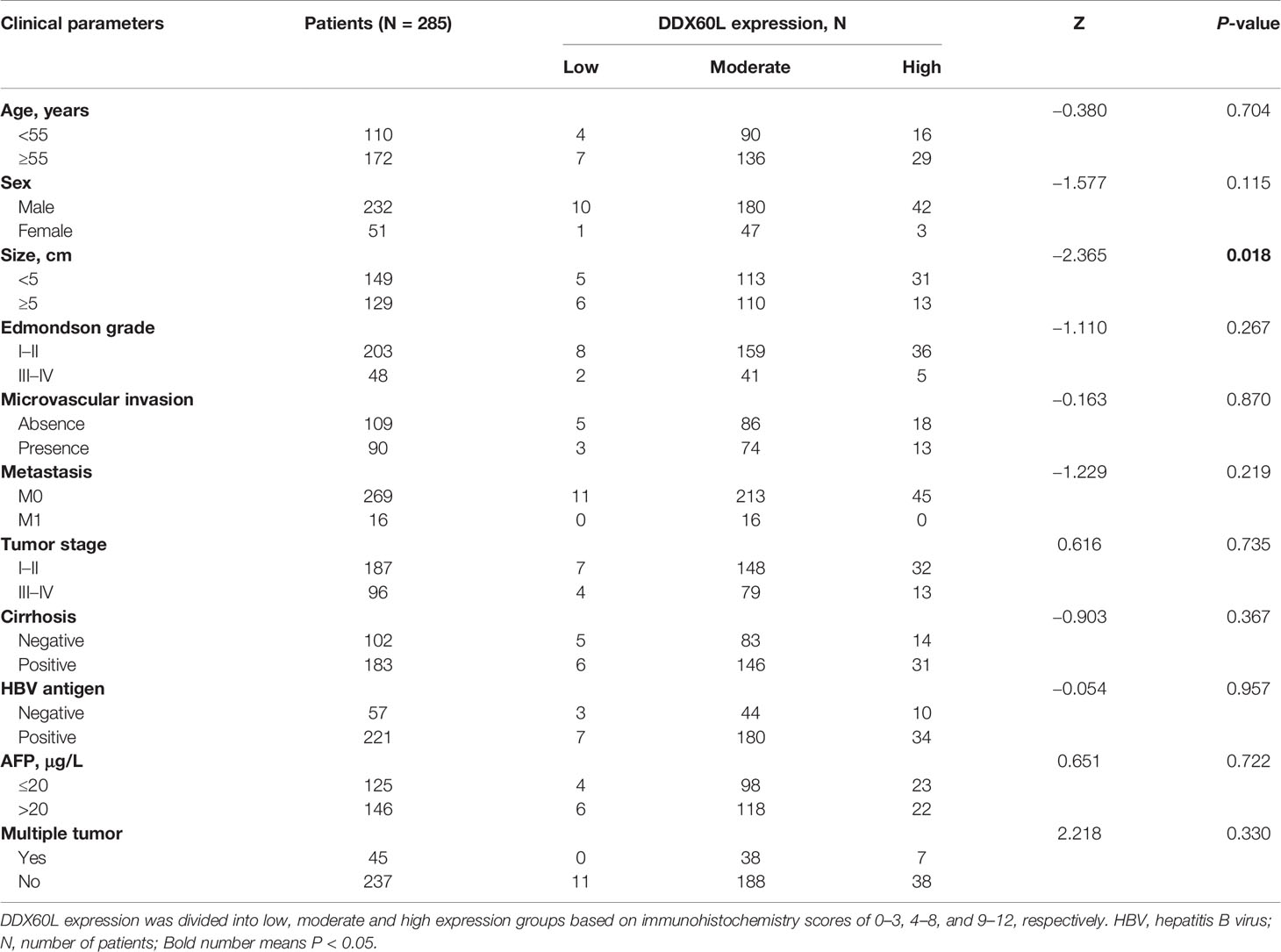
To assess the differences in DDX60L expression between HCC and tumor-adjacent normal tissues, we compared the final immunohistochemical scores of DDX60L in 285 HCC tissues and 167 tumor-adjacent normal tissues. Immunohistochemical results showed that DDX60L expression was significantly decreased in HCC tissues compared with that in adjacent normal tissues (Figure 2). As shown in Figure 3, the final immunohistochemical scores of DDX60L were significantly reduced in HCC (7.37 ± 2.79, N = 285) compared with normal tissue adjacent to the tumor (11.10 ± 1.96, N = 167, P <0.001). To explore the clinical significance of DDX60L expression, we further analyzed the correlation between the expression of DDX60L and different clinicopathological features of HCC patients. As shown in Table 3, DDX60L expression was significantly lower in patients with a tumor size ≥5 cm (N = 129) than in patients with a tumor size <5 cm (N = 149, P = 0.018). This suggests that tumor size is significantly associated with DDX60L expression in patients with HCC.
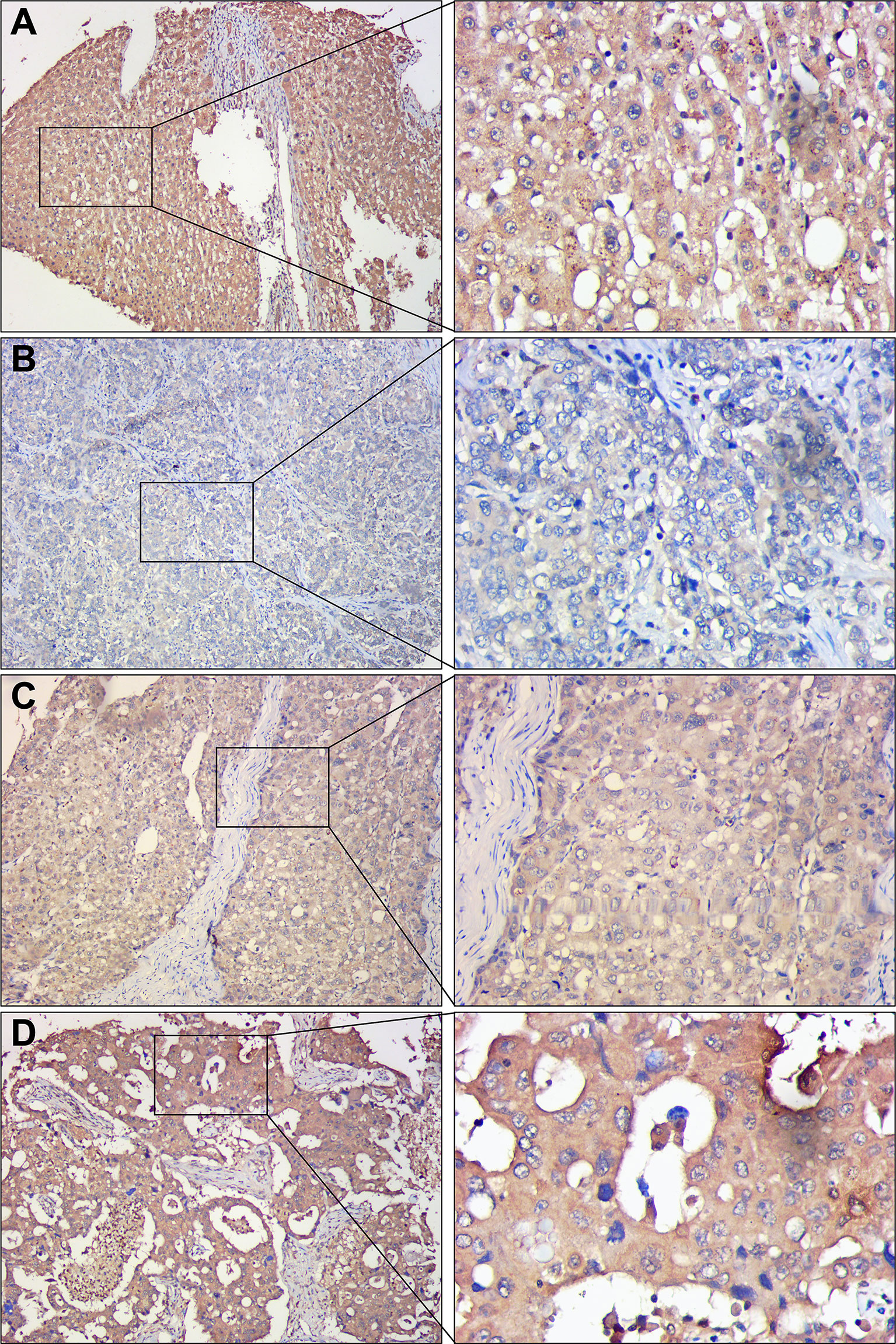
Figure 2 Representative immunohistochemical staining of DDX60L in HCC tissues and tumor-adjacent normal tissues. Magnification in the left column, ×100; magnification in the right column, ×400. (A) tumor-adjacent normal tissues, showing high DDX60L expression in cell membrane and cytoplasm; (B) Hepatocellular carcinoma tissues, showing a low DDX60L expression in the cell membrane and cytoplasm; (C) Hepatocellular carcinoma tissues, showing moderate DDX60L expression in cell membrane and cytoplasm; (D) Hepatocellular carcinoma tissues, showing high DDX60L expression in the cell membrane and cytoplasm.
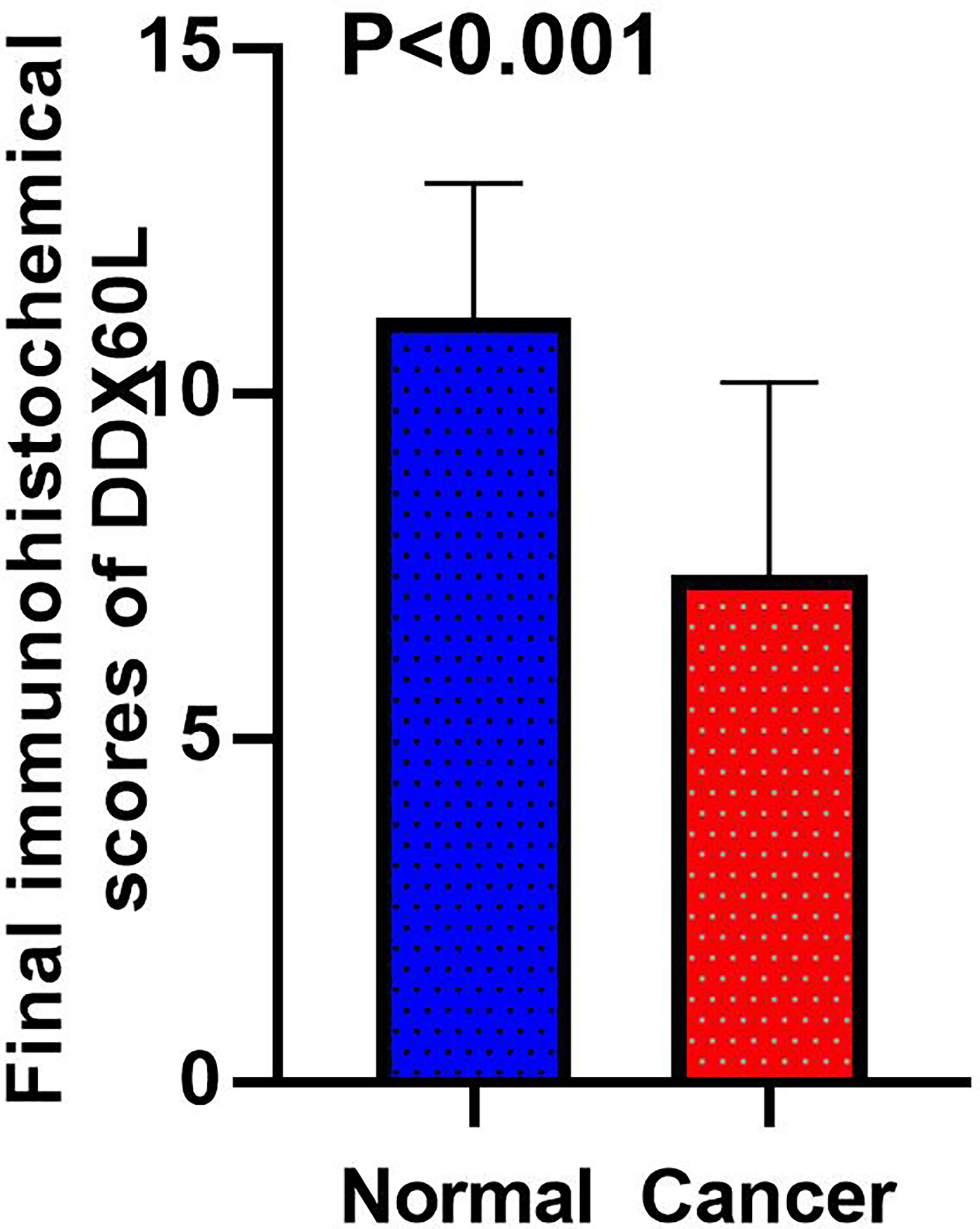
Figure 3 Final immunohistochemical scores of DDX60L in HCC tissues (N = 285) and tumor-adjacent normal tissues (N = 167).
DDX60L Inhibits the Proliferation, Invasion, and Migration of HCC Cells In Vitro
We found that si-DDX60L-1 and si-DDX60L-2 had a good inhibitory effect in Hep3B and HCCLM3 cells (Figure 4A). The results of the cell proliferation assay showed that the downregulation of DDX60L expression promoted the proliferation of Hep3B cells, but had little effect on proliferation of HCCLM3 cells (Figure 4B). Subsequently, we found that DDX60L knockdown could promote the invasion ability of Hep3B and HCCLM3 cells (Figures 4C, E). Similar to results of Transwell assay, we also found that DDX60L knockdown significantly increased the migration of Hep3B and HCCLM3 cells (Figures 4D, F). Overall, these findings suggest that DDX60L significantly inhibited the proliferation of Hep3B cells, migration and invasion of Hep3B and HCCLM3 cells in vitro.
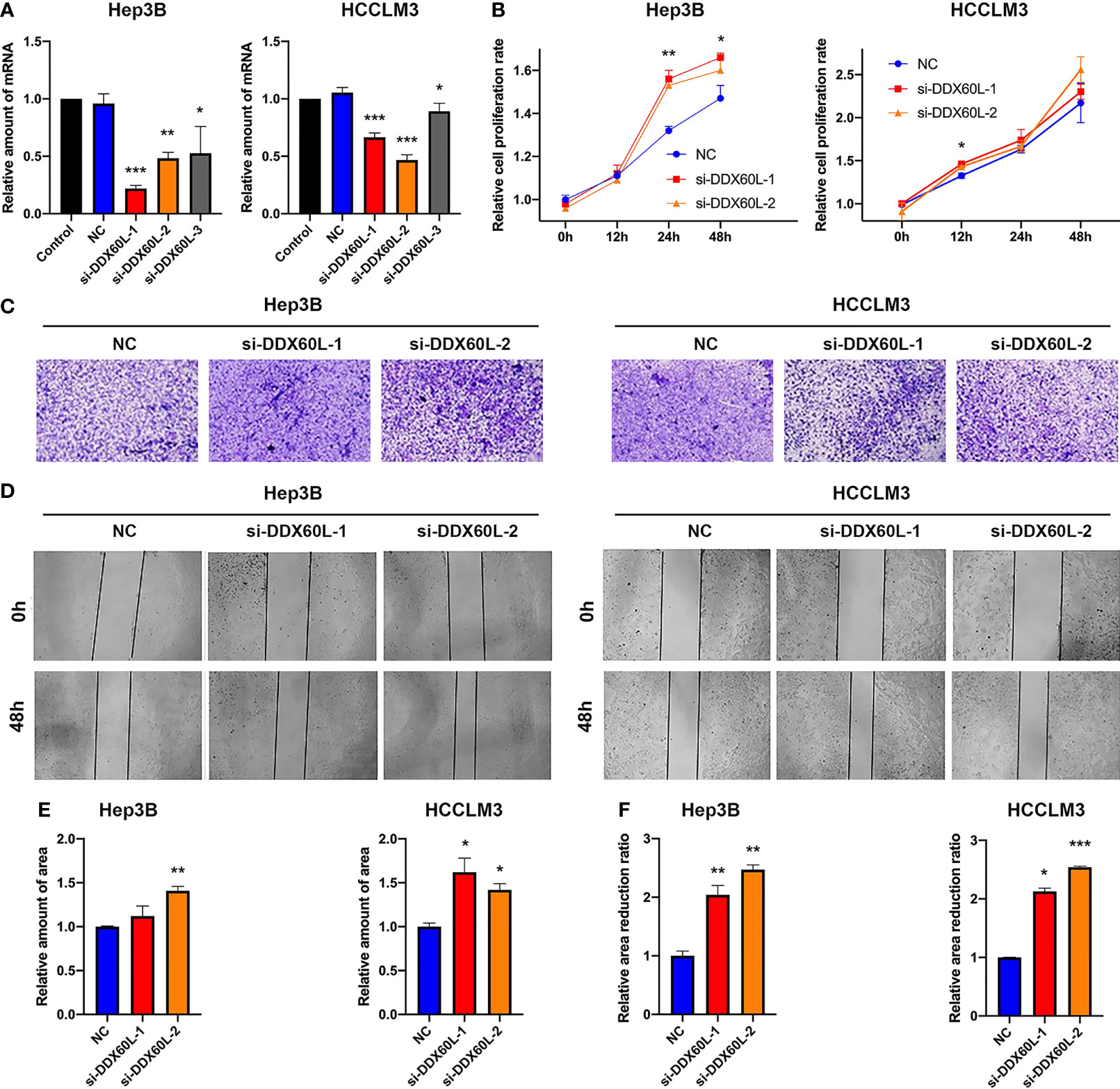
Figure 4 DDX60L inhibited the proliferation of Hep3B, the invasion and migration of Hep3B and HCCLM3 cells. (A) si-DDX60L-1 and si-DDX60L-2 had good inhibitory effects; (B) The results of CCK-8 assays showed that the DDX60L knockdown could promote the proliferation of Hep3B cells, but hardly affected the proliferation of HCCLM3 cells; (C, E) The results of Transwell assays showed that DDX60L knockdown could promote the invasion of Hep3B and HCCLM3 cells. Magnification, ×10; (D, F) Wound healing assays demonstrated that DDX60L knockdown could increase the migration of Hep3B and HCCLM3 cells. Magnification, ×10. *P < 0.05 and **P < 0.01, compared with NC. NC, negative control. ***P < 0.001
Discussion
Prior studies have noted the importance of the DDX family in HCC (24). For example, DDX3 is deregulated in HCV-associated HCC and plays an important role in cell growth (24); DDX5 suppressed HCC by inducing autophagy (25), and there was a reduced expression of DDX5 in HBV-related HCC patients, who had a poor prognosis (26). Additionally, DDX5 inhibits HBV biosynthesis and Wnt signaling in HBV-related HCC (27). Oncogene DDX17 is a potential prognostic marker for HCC (19), while DDX20 inhibited HCC by regulating the CDC42-integrin pathway (28). As a member of the DDX family, the functions of DDX60L in HCC are still unknown. There is evidence that DDX60L may inhibit viral RNA replication (29). It is well known that HCV infection is related to the prognosis of HCC, because HCV infection induced cirrhosis, and then contribute to the increased incidence of HCC (30, 31).
In the present study, we found that the expression of DDX60L was strongly associated with poor OS, suggesting that DDX60L could be an independent risk factor for HCC. Additionally, the results of functional studies showed that DDX60L knockdown significantly promoted the proliferation of Hep3B cells, migration and invasion of Hep3B and HCCLM3 cells. Nevertheless, very little is currently known about whether DDX60L inhibits HCC progression by only depending on its role in the HCV infection pathway. Thus, we tried to explore the influence of HCV on the role of DDX60L, and found DDX60L suppressed the development of HCC independent of the HCV-related pathway, because there were no HCV-related HCC patients in this study. Furthermore, recent studies suggest the DDX family was one of the typical splicing regulators that played a central role in RNA metabolism and usually functions as part of the spliceosome (32). A comprehensive analysis showed that tumor samples had 30% more AS events than normal samples (33), and abnormal AS was strongly associated with invasion, metastasis, poor prognosis of tumors (34). DDX60L is one of the important members of the DDX family, so we speculate that it may play a role in HCC by regulating AS, but the conclusion deserves further confirmation in the future.
To the best of our knowledge, this is the first study about the role of DDX60L in HCC, which will be very helpful to understand other molecular mechanisms of HCC. However, there are some limitations. For instance, the relationship between disease-free survival (DFS) and DDX60L, the influence of sorafenib treatment and alcohol intake on the role of DDX60L, the specific regulatory mechanism of DDX60L in HCC, have not been elucidated. Therefore, further studies need to be carried out to address these limitations and clarify the specific mechanism of DDX60L in HCC.
Conclusion
In summary, we observed the downregulation of DDX60L is closely associated with poor prognosis of HCC patients, and DDX60L may be involved in HCC independent of the HCV-related pathway. Then, we found DDX60L inhibited the proliferation, migration, and invasion of HCC cells. Taken together, our findings might highlight the significance of DDX60L in the tumorigenesis, development and clinical outcome of HCC, and suggest that DDX60L can serve as a prognostic biomarker and therapeutic target for HCC.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.
Ethics Statement
The studies involving human participants were reviewed and approved by the Ethics Committee of Zhejiang Provincial People’s Hospital. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
ZY and ZX designed this study. XZ and LL collected the data of the patients and performed the experiment of immunohistochemical staining. YZ and ZY contributed to the data analysis. ZY, ZX and PH participated in the writing and revision of the manuscript. All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Funding
This study was supported by the Natural Science Foundation of Zhejiang Province (Grant Number LYY21H310008) and the Fundamental Research Funds for the Central Universities (Grant Number WK9110000183).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Marrero JA, Kulik LM, Sirlin CB, Zhu AX, Finn RS, Abecassis MM, et al. Diagnosis, Staging, and Management of Hepatocellular Carcinoma: 2018 Practice Guidance by the American Association for the Study of Liver Diseases. Hepatology (2018) 68(2):723–50. doi: 10.1002/hep.29913
2. Zhao P, Malik S, Xing S. Epigenetic Mechanisms Involved in HCV-Induced Hepatocellular Carcinoma (HCC). Front Oncol (2021) 11:677926. doi: 10.3389/fonc.2021.677926
3. Benson AB, D'Angelica MI, Abbott DE, Anaya DA, Anders R, Are C, et al. Hepatobiliary Cancers, Version 2.2021, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw (2021) 19(5):541–65. doi: 10.6004/jnccn.2021.0022
4. Finn RS, Ryoo BY, Merle P, Kudo M, Bouattour M, Lim HY, et al. Pembrolizumab As Second-Line Therapy in Patients With Advanced Hepatocellular Carcinoma in KEYNOTE-240: A Randomized, Double-Blind, Phase III Trial. J Clin Oncol (2020) 38(3):193–202. doi: 10.1200/jco.19.01307
5. Man S, Luo C, Yan M, Zhao G, Ma L, Gao W. Treatment for Liver Cancer: From Sorafenib to Natural Products. Eur J Med Chem (2021) 224:113690. doi: 10.1016/j.ejmech.2021.113690
6. de Martel C, Maucort-Boulch D, Plummer M, Franceschi S. World-Wide Relative Contribution of Hepatitis B and C Viruses in Hepatocellular Carcinoma. Hepatology (2015) 62(4):1190–200. doi: 10.1002/hep.27969
7. Ryerson AB, Eheman CR, Altekruse SF, Ward JW, Jemal A, Sherman RL, et al. Annual Report to the Nation on the Status of Cance-2012, Featuring the Increasing Incidence of Liver Cancer. Cancer (2016) 122(9):1312–37. doi: 10.1002/cncr.29936
8. Ganne-Carrie N, Nahon P. Hepatocellular Carcinoma in the Setting of Alcohol-Related Liver Disease. J Hepatol (2019) 70(2):284–93. doi: 10.1016/j.jhep.2018.10.008
9. Suwanthawornkul T, Anothaisintawee T, Sobhonslidsuk A, Thakkinstian A, Teerawattananon Y. Efficacy of Second Generation Direct-Acting Antiviral Agents for Treatment Naive Hepatitis C Genotype 1: A Systematic Review and Network Meta-Analysis. PLoS One (2015) 10(12):e0145953. doi: 10.1371/journal.pone.0145953
10. Falade-Nwulia O, Suarez-Cuervo C, Nelson DR, Fried MW, Segal JB, Sulkowski MS. Oral Direct-Acting Agent Therapy for Hepatitis C Virus Infection: A Systematic Review. Ann Intern Med (2017) 166(9):637–48. doi: 10.7326/M16-2575
11. Ogata F, Kobayashi M, Akuta N, Osawa M, Fujiyama S, Kawamura Y, et al. Outcome of All-Oral Direct-Acting Antiviral Regimens on the Rate of Development of Hepatocellular Carcinoma in Patients With Hepatitis C Virus Genotype 1-Related Chronic Liver Disease. Oncology (2017) 93(2):92–8. doi: 10.1159/000470910
12. Carrat F, Fontaine H, Dorival C, Simony M, Diallo A, Hezode C, et al. Clinical Outcomes in Patients With Chronic Hepatitis C After Direct-Acting Antiviral Treatment: A Prospective Cohort Study. Lancet (2019) 393(10179):1453–64. doi: 10.1016/S0140-6736(18)32111-1
13. Ma YJ, Du LY, Yan LB, Liao J, Cheng X, Xie WW, et al. Long-Term Follow-Up of HCV Patients With Sustained Virological Response After Treatment With Pegylated Interferon Plus Ribavirin. Hepatobiliary Pancreat Dis Int (2021) 20(2):137–41. doi: 10.1016/j.hbpd.2020.02.004
14. Bigger CB, Brasky KM, Lanford RE. DNA Microarray Analysis of Chimpanzee Liver During Acute Resolving Hepatitis C Virus Infection. J Virol (2001) 75(15):7059–66. doi: 10.1128/JVI.75.15.7059-7066.2001
15. Su AI, Pezacki JP, Wodicka L, Brideau AD, Supekova L, Thimme R, et al. Genomic Analysis of the Host Response to Hepatitis C Virus Infection. Proc Natl Acad Sci USA (2002) 99(24):15669–74. doi: 10.1073/pnas.202608199
16. Fullam A, Schroder M. DExD/H-Box RNA Helicases as Mediators of Anti-Viral Innate Immunity and Essential Host Factors for Viral Replication. Biochim Biophys Acta (2013) 1829(8):854–65. doi: 10.1016/j.bbagrm.2013.03.012
17. Fuller-Pace FV. DExD/H Box RNA Helicases: Multifunctional Proteins With Important Roles in Transcriptional Regulation. Nucleic Acids Res (2006) 34(15):4206–15. doi: 10.1093/nar/gkl460
18. Godbout R, Li L, Liu RZ, Roy K. Role of DEAD Box 1 in Retinoblastoma and Neuroblastoma. Future Oncol (2007) 3(5):575–87. doi: 10.2217/14796694.3.5.575
19. Zhou HZ, Li F, Cheng ST, Xu Y, Deng HJ, Gu DY, et al. DDX17-Regulated Alternative Splicing That Produced a Novel Oncogenic Isoform of PXN-AS1 to Promote HCC Metastasis. Hepatology (2021). doi: 10.1002/hep.32195
20. Miyashita M, Oshiumi H, Matsumoto M, Seya T. DDX60, a DEXD/H Box Helicase, Is a Novel Antiviral Factor Promoting RIG-I-Like Receptor-Mediated Signaling. Mol Cell Biol (2011) 31(18):3802–19. doi: 10.1128/MCB.01368-10
21. Wu H, Tian W, Tai X, Li X, Li Z, Shui J, et al. Identification and Functional Analysis of Novel Oncogene DDX60L in Pancreatic Ductal Adenocarcinoma. BMC Genomics (2021) 22(1):833. doi: 10.1186/s12864-021-08137-5
22. Ye Z, Wang S, Chen W, Zhang X, Chen J, Jiang J, et al. Fat Mass and Obesity-Associated Protein Promotes the Tumorigenesis and Development of Liver Cancer. Oncol Lett (2020) 20(2):1409–17. doi: 10.3892/ol.2020.11673
23. Rong Z-X, Li Z, He J-J, Liu L-Y, Ren X-X, Gao J, et al. Downregulation of Fat Mass and Obesity Associated (FTO) Promotes the Progression of Intrahepatic Cholangiocarcinoma. Front Oncol (2019) 9:369. doi: 10.3389/fonc.2019.00369
24. Chang PC, Chi CW, Chau GY, Li FY, Tsai YH, Wu JC, et al. DDX3, a DEAD Box RNA Helicase, Is Deregulated in Hepatitis Virus-Associated Hepatocellular Carcinoma and Is Involved in Cell Growth Control. Oncogene (2006) 25(14):1991–2003. doi: 10.1038/sj.onc.1209239
25. Zhang H, Zhang Y, Zhu X, Chen C, Zhang C, Xia Y, et al. DEAD Box Protein 5 Inhibits Liver Tumorigenesis by Stimulating Autophagy via Interaction With P62/SQSTM1. Hepatology (2019) 69(3):1046–63. doi: 10.1002/hep.30300
26. Zhang H, Xing Z, Mani SK, Bancel B, Durantel D, Zoulim F, et al. RNA Helicase DEAD Box Protein 5 Regulates Polycomb Repressive Complex 2/Hox Transcript Antisense Intergenic RNA Function in Hepatitis B Virus Infection and Hepatocarcinogenesis. Hepatology (2016) 64(4):1033–48. doi: 10.1002/hep.28698
27. Mani SKK, Yan B, Cui Z, Sun J, Utturkar S, Foca A, et al. Restoration of RNA Helicase DDX5 Suppresses Hepatitis B Virus (HBV) Biosynthesis and Wnt Signaling in HBV-Related Hepatocellular Carcinoma. Theranostics (2020) 10(24):10957–72. doi: 10.7150/thno.49629
28. Huang Y, Wang C, Li K, Ye Y, Shen A, Guo L, et al. Death-Associated Protein Kinase 1 Suppresses Hepatocellular Carcinoma Cell Migration and Invasion by Upregulation of DEAD-Box Helicase 20. Cancer Sci (2020) 111(8):2803–13. doi: 10.1111/cas.14499
29. Grunvogel O, Esser-Nobis K, Reustle A, Schult P, Muller B, Metz P, et al. DDX60L Is an Interferon-Stimulated Gene Product Restricting Hepatitis C Virus Replication in Cell Culture. J Virol (2015) 89(20):10548–68. doi: 10.1128/JVI.01297-15
30. Goodgame B, Shaheen NJ, Galanko J, El-Serag HB. The Risk of End Stage Liver Disease and Hepatocellular Carcinoma Among Persons Infected With Hepatitis C Virus: Publication Bias? Am J Gastroenterol (2003) 98(11):2535–42. doi: 10.1111/j.1572-0241.2003.07678.x
31. Ahmad J, Eng FJ, Branch AD. HCV and HCC: Clinical Update and a Review of HCC-Associated Viral Mutations in the Core Gene. Semin Liver Dis (2011) 31(4):347–55. doi: 10.1055/s-0031-1297924
32. Linder P, Jankowsky E. From Unwinding to Clamping - the DEAD Box RNA Helicase Family. Nat Rev Mol Cell Biol (2011) 12(8):505–16. doi: 10.1038/nrm3154
33. Kahles A, Lehmann KV, Toussaint NC, Huser M, Stark SG, Sachsenberg T, et al. Comprehensive Analysis of Alternative Splicing Across Tumors From 8,705 Patients. Cancer Cell (2018) 34(2):211–24.e216. doi: 10.1016/j.ccell.2018.07.001
Keywords: hepatocellular carcinoma, DDX60L, prognosis, HCV infection, HCC
Citation: Ye Z, Zhang X, Zhang Y, Liu L, Xuan Z and Huang P (2022) Associations of DDX60L With the Clinical Features and Prognosis of Hepatocellular Carcinoma. Front. Oncol. 12:761021. doi: 10.3389/fonc.2022.761021
Received: 19 August 2021; Accepted: 18 January 2022;
Published: 11 February 2022.
Edited by:
Yu-Yun Shao, National Taiwan University Hospital, TaiwanReviewed by:
Kai Wang, Southwest Medical University, ChinaYang Deng, Shandong First Medical University, China
Copyright © 2022 Ye, Zhang, Zhang, Liu, Xuan and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zixue Xuan, eHVhbnppeHVlMDIyMkAxNjMuY29t; Ping Huang, aHVhbmdwaW5nMTg0MUB6amNjLm9yZy5jbg==
 Ziqi Ye
Ziqi Ye Xin Zhang2
Xin Zhang2 Zixue Xuan
Zixue Xuan