- 1Department of Surgery, University of Nebraska Medical Center, Omaha, NE, United States
- 2Department of Surgery, VA Medical Center, Omaha, NE, United States
- 3Department of Genetics, Cell Biology and Anatomy, University of Nebraska Medical Center, Omaha, NE, United States
- 4Fred & Pamela Buffett Cancer Center, University of Nebraska Medical Center, Omaha, NE, United States
- 5Center for Advanced Surgical Technology, University of Nebraska Medical Center, Omaha, NE, United States
In this mini review the status, advantages, and disadvantages of large animal modeling of breast cancer (BC) will be discussed. While most older studies of large animal BC models utilized canine and feline subjects, more recently there has been interest in development of porcine BC models, with some early promising results for modeling human disease. Widely used rodent models of BC were briefly reviewed to give context to the work on the large animal BC models. Availability of large animal BC models could provide additional tools for BC research, including availability of human-sized subjects and BC models with greater biologic relevance.
Background: Breast Cancer Burden
The annual incidence of breast cancer (BC) in women (all ages and races) in the U.S. increased from 0.102% in 1980 to a peak of 0.142% in 1999, and then decreased slightly, plateauing at ~0.131% from 2011-2017 (1). As of 2017, a woman’s lifetime risk of developing BC in the U.S. is 12.9% (1). In 2021, the estimated number of new BC cases in the U.S. will be 281,550 (15.3% of all new cancer cases), with 43,600 estimated deaths (~20 per 100,000 in the general population, or 7% of all cancer deaths, or ~2% of all mortality in the U.S.) (1, 2).
All-stages 5-year survival for BC has improved from 75% in 1975 to 90% in 2016 (1), secondary to earlier diagnosis and more efficacious therapy (3). However, up to 50% of hormone dependent (estrogen/progesterone receptor positive or ER/PR+) BC patients acquire resistance under treatment, and 20% do not even respond to first-line hormone therapies (4). In addition, dormant ER/PR+ tumor cells can become reactivated and can cause disease relapse (5). Almost 50% of ER/PR+ BC tumors are also positive for Erb-B2 Receptor Tyrosine Kinase 2 (ERBB2), commonly referred to as HER2/NEU (human epidermal growth factor receptor 2) (6). While trastuzumab (a monoclonal antibody therapy targeting HER2) is standard treatment for HER2+ BC, almost all patients with metastatic HER2 + BC eventually develop treatment resistance (7, 8).
Survival with triple-negative BC (TNBC; minimal/nil expression of the estrogen receptor, progesterone receptor, and HER2), which accounts for 10-20% of all BC (9, 10), is 10, 20, and 30% lower at stages 2, 3, and 4, respectively, compared to non-TNBC (11). So, there remain a need for improved management of both receptor-positive BC and TNBC. The availability of validated, tractable large animal models that faithfully represent both (1) hormone and growth factor dependent human BC and (2) TNBC should allow development and testing of technologies and treatments that would not be possible in murine models, with resultant data that would be predictive of human tumor response.
Large Animal Models of BC
Justification for and Advantages of a Large Animal Model of BC
Breast cancer, and TNBC in particular, can be modeled with many of the murine and other small animal models. However, none of these models can overcome the limitation of inadequate subject size of small animals. Development of some diagnostic and interventional technologies require a human-sized model to understand how clinically relevant tumor size and tissue thickness affect the performance of the experimental technology, such as with three-dimensional scanning or with a tissue-ablation device. In addition, pharmacokinetic parameters (including absorption, distribution, metabolism, and excretion) can be vastly different between a 20 g mouse and a 70 kg patient (12). Testing the effect of an infused chemical entity (e.g., a novel anti-tumor agent) in a murine tumor model may result in an inaccurate conclusion secondary to these pharmacokinetic differences (Figure 1).
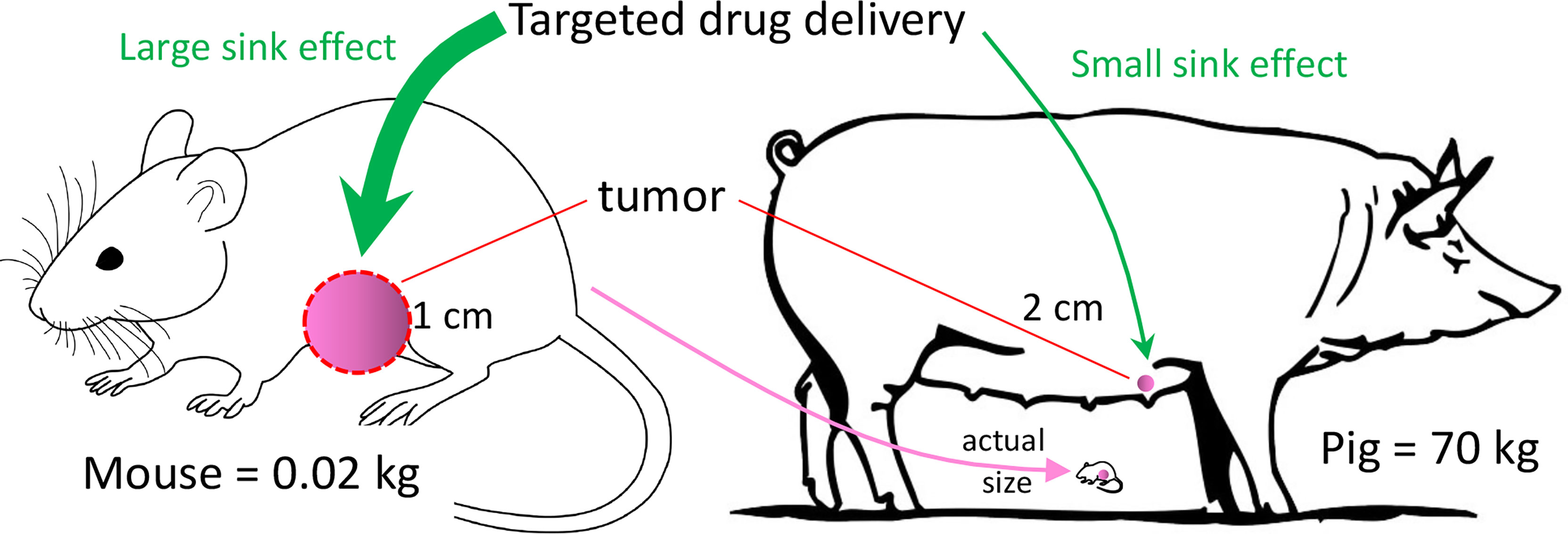
Figure 1 Tumor Sink. In this example, murine tumor:body mass ratio is 400x the porcine ratio, even though murine tumor mass is only ~12% that of the porcine tumor. If a targeted drug preferentially concentrates in the tumor (i.e., tumor sink), then a given drug dose in the mouse may have a relatively high concentration in the tumor with respect to the plasma. The effect of tumor sink on plasma concentration at the same dose in the pig would be negligible because of the large body size. Thus, the same dose in the pig would produce a higher plasma concentration, possibly producing greater systemic toxicity than in the mouse.
With regard to drug distribution, tumor burden in a 20 gram mouse with a 1-cm tumor is ~400-fold greater (mass:mass) than in a 70 kg subject with a 2-cm tumor, as diagramed in Figure 1. If the tumor acts as a sink for a candidate anti-tumor drug, then the tumor’s ability to decrease the drug’s plasma concentration would be much greater in the mouse. A consequence of this tumor sink effect (13) would be a gross underestimation of the drug’s toxicity from murine data. Alternatively, if the drug penetrates poorly into dense tumor stroma (14), then testing in a ~1 cm murine tumor may overestimate the drug’s anti-tumor efficacy, as opposed to testing in a large (≥4 cm) tumor. These drug distribution issues in a murine model could be minimized with a large animal model (e.g., a 70 kg pig) with clinically relevant tumor size. So, a primary justification for a large animal BC model would be its ability to replicate human tumor dimension and human PK parameters. Other advantages of large animal BC models (and large animal cancer models in general) have been listed in Table 1.
Current Status: Large Animal Models
Overview
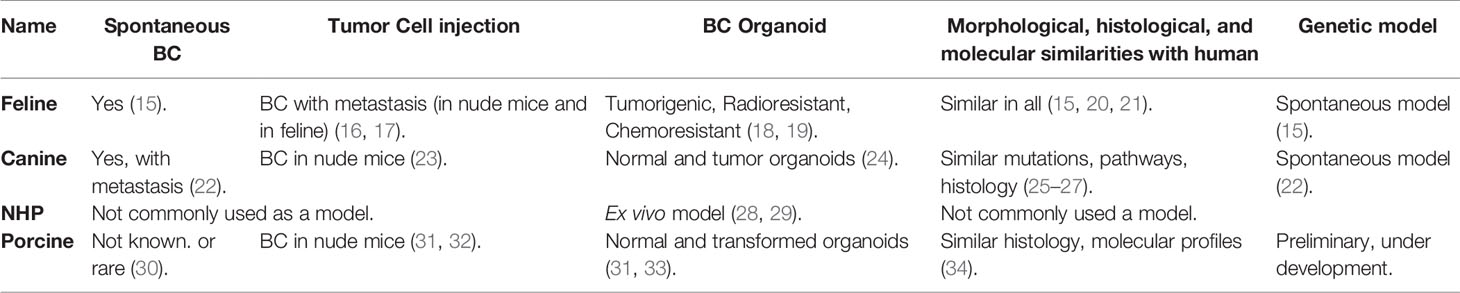
Most of the older work on large animal modeling of BC has focused on feline and canine subjects (Table 2). Of note, non-human primates (NHPs) have not been commonly utilized in BC or other cancer research. Recently, developmental work on porcine BC models has emerging, with promising data from both orthotopic implantation strategies and genetic editing. The current status of large animal BC models has been summarized in Table 2.
Feline BC Models
Feline mammary carcinomas (FMCs) are the third most common type of cancer in cats (35). In utero implantation of allogeneic mammary cancer cell lines into fetal cats (Felis catus) produced tumor at the injection site, followed by widespread metastasis after 6-10 weeks (16, 17). A nude mouse model of FMC demonstrated metastatic potential (bone, kidney, brain, lung, and liver; i.e., common sites of metastasis in human breast cancer) after injection to the primary site (36). Cancer stem cell-like populations in FMCs can form mammospheres (organoids) and are tumorigenic, radioresistant, and chemoresistant (18, 19). Mammospheres can be used a model to study feline breast cancer. Morphological, histological, and molecular to similarities between FMCs and human breast cancer have been described and discussed (15, 20, 21).
Canine BC Models
Similar to cats, canine mammary tumors (CMTs) occur spontaneously and are the second most common cancer in dogs (37). Development of CMTs is hormone dependent and showed dysregulated expression of BRCA1, BRCA2 and TP53 (25–27), analogous to human BC. Transcriptional analysis of CMTs demonstrated pathways that are active in human cancer, including those involved with cell cycle regulation, apoptotic signaling, immune functions, endoplasmic reticulum stress, angiogenesis, and cell migration (38). Approximately 25% of the genetic alterations in metastatic CMTs were associated with human mammary cancer (39). In addition, canine spontaneous mammary DCIS and invasive cancer shows similar histologic and molecular characteristics with DCIS and invasive cancer in humans (40). Spontaneous CMTs also can metastasize to lymph nodes and lung (22). HER2 overexpression in CMTs is controversial (41). Benign tumors and mesenchymal tumors are more prevalent in CMTs; the latter are rare with human breast cancer (42, 43).
Canine inflammatory BC cells were able to generate tumors in nude mice both ectopically and orthotopically (23). No morphological differences in the tumors were noted when human inflammatory BC cells were injected instead of canine cancer cells (23). There are several other canine BC cell lines that were characterized and used in development for BC in nude mice (44–47). A newly developed canine TNBC cell line, B-CMT, also showed BC in nude mice after 14 days of orthotopic inoculation (48). Canine normal mammary tissue and BC tissue were used to develop organoids, which showed multi lineage potential and existence of different population of self-renewing stem cells (24). These canine normal and BC organoids will serve as valuable models for future research with multiple species.
Non-Human Primate BC Models
Mammary gland tissue from common marmoset and rhesus macaques have been used in mammosphere culture, and can be used as an ex vivo model to study breast cancer (28, 29). However, due to the low incidence of spontaneous tumors, long incubation periods, and high costs, non-human primates have not been commonly used in breast cancer research.
Porcine BC Models
Swine have been used for decades as research subjects in diverse areas including transplantation, physiology, trauma, toxicology (49, 50), and recently cancer (30, 51–54). The porcine genome has been sequenced (55), and annotation is ongoing (56). Gene editing of pigs and creation of transgenic swine is now fairly common (52, 53, 57–60); some transgenics [such as the Oncopig (51)] are commercially available. With respect to breeding, the porcine gestation period (114 d) is relatively short (goat/sheep/cow = 150/152/283 d). In regard to pharmacokinetics, the pig has the most similarity to humans among studied mammals with respect to the cytochrome P450 enzymes (61, 62). Similar pharmacokinetic behavior between humans and pigs has been reported for a number of compounds (63), and pig is recognized as a model for enabling determinations of in vivo kinetics and drug metabolism in general (63, 64).
Early post-natal stem cell activity in the mammary development may be reactivated during initiation of human BC (34). RNA sequencing and immune histochemistry analysis showed that in neonatal pigs (4-39 days of age) mammary gland gene expression patterns were similar to that in human BC (34). Investigators have transformed porcine mammary epithelial cells in vitro with SV40 large T antigen insertion (31) or BRCA1 knockdown (65), with evidence of tumorigenicity in immunodeficient mice (31). In addition, a BRCA1 haploinsufficient Yucatan minipig was generated with somatic cell nuclear transfer (66), but postnatal survival of cloned piglets was ≤18 days for reasons that were unclear (BRCA1+/– mice are phenotypically normal) (67).
As a proof of principle, our group has transformed porcine mammary epithelial cells with KRASG12D and TP53R167H, and produced tumors in xenografted nude mice (32). Our follow-up work in this area currently is focused on using BC relevant genes in porcine mammary epithelial cells (68–70), to be utilized in an orthotopic porcine model of TNBC. In addition, our group is endeavoring to create a transgenic, inducible, restricted model of mammary ductal neoplasia. To be clear, however, no porcine model of BC currently exists. The KRAS/TP53 Oncopig (51) is not a BC model, but rather a “generic” porcine tumor model, potentially allowing transformation of all cell types. Of note, KRAS mutation is relatively uncommon (1-2%) in BC (68–70). Given the recent progress in porcine modeling of pancreatic cancer by multiple groups (32, 51, 54, 71–76), it is likely that the best candidate for a large animal BC model will be the pig.
Disadvantages of Using Large Animal Cancer Models
In order to illustrate the potential disadvantages of using a large animal model of breast cancer, a comparison of porcine with murine models is described below. This does reveal a number of disadvantages with the porcine models, including:
1. Costs. Transgenic pigs are more expensive. Transgenic mice for BC research generally are 200-300 USD per subject (Jackson Laboratory), while transgenic pigs (e.g., the Oncopig) can be in the range of 1-2K USD. Per diem cost for pigs are generally ~10x the murine per diem at most institutions. Drug costs are higher in pigs because they require 100-1,000x the amount of drug used by mice. Labor costs are higher with pigs because of physical handling required each time a procedure is done.
2. Husbandry. More than 100-fold mice than pigs can be housed in the same space, which can limit experimental planning at most institutions. A Sinclair mini-pig can still reach up to 50 kg at 1.5 year, so the research facility has to accommodate animals of this size. The murine gestation period (20 d) is <20% of porcine gestation period, so crossbreeding is much quicker with mice. A relatively simple maneuver of placing a 20 g mouse under general anesthesia becomes more complicated when dealing with a 30-50 kg pig.
3. Tools & Reagents. The availability of antibodies, reagents, and other species-specific research tools is much greater in mice compared to pigs, though the availability for pigs has improved in the past decade.
4. Subject Age. In general, investigators only have access to young (<1 year) pigs; however, modeling epithelial tumors with young pigs may not be optimal.
5. Research Community. The number of investigators who utilize swine in cancer research is still relatively small, so other investigators may be reluctant to consider porcine experimentation.
6. Social Issues. Public reaction to swine use in research could be more negative compared to the reaction against rodent use. This possibility may demand more investigator effort devoted to public education.
7. Ethical Issues. On the surface, ethical issues with large animal research tend to be more complex than with small animal research. Among large animals, NHPs and dogs appear to be more protected by ethical standards compared to pigs in the research environment, but criteria have been difficult to quantify. In general, having strong institutional oversight of any animal research, with careful, intentional, and transparent regard for animal welfare, has been the most valuable consideration in the ethical performance of research with animal subjects.
Other Animal Models of BC
Overview
Rodent models of BC are available in a wide variety of genotypes and phenotypes, and have been an essential tool in preclinical BC research for decades. In order to provide historical context and relevance to the development of large animal BC models, the status of rodent BC modeling is briefly reviewed below.
Murine BC Models
Commonly utilized murine BC models include chemically induced, cell-line derived xenograft (CDX), patient derived xenograft (PDX), humanized CDX and PDX and genetically engineered murine (GEM) models (77–80).
Polycyclic aromatic hydrocarbons such as 7,12-Dimethylbenz(a)-anthracene (DMBA) and methylcholanthrene (MC) compounds have been used in mice to induce BC (81–85). DMBA has been studied in knockout, hemizygous and SENCAR (SENsitivity to CARcinogenesis) mice, producing breast tumors after 3-34 weeks (81–84). MC has produced tumors after 7 months (85). Cell-line derived xenograft (CDX) murine models are based off the transplantation of human cell lines into immunocompromised animals (86, 87). However, a major limitation associated with these models is the lack of a functional host immune system, which means that CDX tumors do not undergo any appreciative immunoediting (88, 89). They also have reduced intra-tumoral heterogeneity which does not optimally represent a human breast tumor (86). In addition, CDX tumors are frequently derived from highly aggressive malignant tumors or pleural effusions and thus are less useful in studying the early stages of disease (86).
PDX BC models, which involve the transplantation of fragmented primary human tumor into immune deficient mice, have been shown to conserve ER, PR, and HER2 expression, particularly when grafted directly into mammary ducts (90, 91), and have shown similar metastatic progression compared to human tumors (92, 93). However, the prolonged length of time it takes to generate tumors does not always match clinical or research needs (86, 87, 94). Another concern with PDX BC modeling is that by the third in vivo passage murine stroma replaces human stroma, which may result in changes in paracrine regulation as well as in physical properties (95, 96). The humanized mouse model (in which an immunocompromised mouse is engrafted with components of the human immune system) has been utilized in preclinical studies on immunotherapies and BC, particularly TNBC and HER2+ cancers (97). Humanized BC models have shown clinically relevant reduction of tumor growth in response to therapy (98, 99). However, major concerns of these models include (1) a lack GM-CSF, which is important for the differentiation and maturation of the myeloid lineage; and (2) xenograft-versus-host disease, in which mature human T cells attack their murine host secondary to HLA mismatching between the hNSG and PDX components (99, 100).
There are several types of GEM models, including conventional, knockout, and conditional. Conventional GEM models are typically driven by mammary-specific promoters that direct expression of specific oncogenes (transgenes) which may not be specific to mammary epithelial cells (101–104). Expression of C-MYC, V-HRAS, WNT, PyMT, and HER2/NEU/ERBB2 through these promotes resulted in mammary tumorigenesis and metastatic lesions (105–111). Homozygous knockout of TP53 GEM mice developed lymphomas and died at around 4-6 months of age (112, 113). To overcome problem with non-specificity to certain cell lineage and prevent early embryonic death, conditional GEM models have been developed (86). WAP-Cre and MMTV-Cre mice have been able to generate hereditary breast cancer models specifically by modeling the heterozygous mutations observed in the BRCA1/BRCA2 genes (114–116). GEM models that contained a conditional mutant BRCA1 allele and a disruption in TP53 have accelerated mammary tumor development (116). TP53 in combination with other genes has been extensively studied using conditional GEM models (117).
Rat BC Models
Rats have been considered a suitable animal model to study breast cancer due to their similarity with human mammary cancer in terms of histology, immunocytochemical markers and biological behavior of tumors (118). The histologic characteristics of normal mammary luminal epithelium and myoepithelium is similar between rats and human (119). Long term studies have shown that some rats can develop breast tumors spontaneously (118). Use of a chemical carcinogen in rats can result in a shorter latency period to tumor development (119–123). Recently, 17b-estradiol (E2) was used to induce breast tumors in August Copenhagen Irish-rats by modulating estrogen mediated mechanisms in breast cancer development (121).
In xenograft-Matrigel implantation experiments, younger rats have experienced greater tumor growth compared to older rats (124, 125). A bone metastasis immunodeficient rat model has been developed in which human breast cancer cells (MDA-MB-231) were intra-arterially injected into a hindlimb artery (126). Genetically engineered rat models of breast cancer have been developed in which HER2 and TGF-α were overexpressed through the mouse mammary tumor virus (MMTV) promoter (127). This model stochastically produced a variety of benign, hyperplastic, and malignant lesions, including ductal carcinoma in situ and carcinoma within a year.
In a rat model with three copies of human HRAS proto-oncogene, induction of carcinogenesis with nitrosomethylurea resulted in large mammary tumors within 8 weeks (128). Mammary carcinogenesis in rats was induced through injection of high-titer, Neu-containing, replication-defective retrovirus which produced hormonally responsive in situ carcinomas within 15-days post infusion, and regressed spontaneously after 20-days post infusion (129). Injection of human adenovirus type 9 (Ad9) also is known to induce estrogen-dependent mammary tumors in rats within 7-12 months (130). Overall, however, use of rats in BC research has lagged far behind the broad use of mice.
Hamster BC Models
Similar to rats, nitrosomethylurea can induce mammary carcinoma in Syrian hamsters (Mesocricetus auratus), producing high-grade poorly differentiated mammary adenocarcinomas (131). Subcutaneous allogenic implantation of cell lines established from these primary tumors generated secondary tumors. Hamster models can be useful in studies on oncolytic adenoviruses, a self-replicating cancer cell-killing virus. Oncolytic adenoviruses can replicate in immunocompetent hamsters, making the hamster model of cancer a suitable non-immunocompromised model to study therapeutic potential of these viruses (132).
Tree Shrew BC Models
The tree shrew (Tupaia belangeri chinensis) can develop spontaneous mammary tumors, which are similar to human papillary tumors in terms of morphology and pathology (133, 134). Chemical induction with DMBA plus medroxyprogesterone acetate (MPA) also produced breast cancer in tree shrews (135). Injection of a lentiviral vector with PyMT (polyomavirus middle T antigen, an oncogene that activates c-Src), into the mammary duct of tree shrews resulted in tumor development in all subjects by 7 weeks post-injection (136).
Conclusions and Future Directions
The current landscape of animal modeling for breast cancer is dominated by murine models, which have developed into powerful and multi-faceted tools for the BC researcher. It would be difficult to improve on the utility that murine BC models have provided. However, there remain certain areas of research, such as device development and drug testing, which could benefit from the availability of a large animal model of BC. These BC models are still in their infancy, essentially at the point murine models were in the 1980’s. While there have been a number of principle-proving studies involving BC and large animals, a validated and tractable large animal model of BC is not yet available, necessitating that additional work needs to be done in this area if the advantages of large animal BC modeling are to be realized. While large animal BC models likely will never be able to match the proven utility and ease-of-use of murine models, the availability of validated large animal BC models could provide additional tools to the BC researcher that would address specific BC questions or BC-relevant technology development, such as those requiring a human sized subject for generation of relevant data.
Author Contributions
All authors participated in the drafting and editing of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by a grant from the National Cancer Institute (R01CA222907) to MC.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Howlader N, Noone A, Krapcho M, Miller D, Brest A, Yu M, et al. Surveillance, Epidemiology, and End Results (SEER) Cancer Statistics Review, 1975-2017. Bethesda, MD: National Cancer Institute (2020).
2. Kochanek KD, Murphy SL, Xu J, Arias E. Deaths: Final Data For 2017. Natl Vital Stat Rep (2019) 68(9):1–76.
3. NCCN, National Comprehensive Cancer Network. NCCN Clinical Practice Guidelines in Oncology: Breast Cancer. Version 3.2020 (2020). Available at: www.nccn.org.
4. Murphy CG, Dickler MN. Endocrine Resistance in Hormone-Responsive Breast Cancer: Mechanisms and Therapeutic Strategies. Endocr Relat Cancer (2016) 23(8):R337–52. doi: 10.1530/ERC-16-0121
5. Zhang XH, Giuliano M, Trivedi MV, Schiff R, Osborne CK. Metastasis Dormancy in Estrogen Receptor-Positive Breast Cancer. Clin Cancer Res (2013) 19(23):6389–97. doi: 10.1158/1078-0432.CCR-13-0838
6. Vernieri C, Milano M, Brambilla M, Mennitto A, Maggi C, Cona MS, et al. Resistance Mechanisms to Anti-HER2 Therapies in HER2-Positive Breast Cancer: Current Knowledge, New Research Directions and Therapeutic Perspectives. Crit Rev Oncol Hematol (2019) 139:53–66. doi: 10.1016/j.critrevonc.2019.05.001
7. Pohlmann PR, Mayer IA, Mernaugh R. Resistance to Trastuzumab in Breast Cancer. Clin Cancer Res (2009) 15(24):7479–91. doi: 10.1158/1078-0432.CCR-09-0636
8. Vu T, Claret FX. Trastuzumab: Updated Mechanisms of Action and Resistance in Breast Cancer. Front Oncol (2012) 2:62. doi: 10.3389/fonc.2012.00062
9. Denkert C, Liedtke C, Tutt A, von Minckwitz G. Molecular Alterations in Triple-Negative Breast Cancer-the Road to New Treatment Strategies. Lancet (2017) 389(10087):2430–42. doi: 10.1016/S0140-6736(16)32454-0
10. Lebert JM, Lester R, Powell E, Seal M, McCarthy J. Advances in the Systemic Treatment of Triple-Negative Breast Cancer. Curr Oncol (2018) 25(Suppl 1):S142–S50. doi: 10.3747/co.25.3954
11. Li X, Yang J, Peng L, Sahin AA, Huo L, Ward KC, et al. Triple-Negative Breast Cancer has Worse Overall Survival and Cause-Specific Survival Than Non-Triple-Negative Breast Cancer. Breast Cancer Res Treat (2017) 161(2):279–87. doi: 10.1007/s10549-016-4059-6
12. Lin JH. Species Similarities and Differences in Pharmacokinetics. Drug Metab Dispos (1995) 23(10):1008–21.
13. Chu KS, Hasan W, Rawal S, Walsh MD, Enlow EM, Luft JC, et al. Plasma, Tumor and Tissue Pharmacokinetics of Docetaxel Delivered via Nanoparticles of Different Sizes and Shapes in Mice Bearing SKOV-3 Human Ovarian Carcinoma Xenograft. Nanomedicine (2013) 9(5):686–93. doi: 10.1016/j.nano.2012.11.008
14. Tanaka HY, Kano MR. Stromal Barriers to Nanomedicine Penetration in the Pancreatic Tumor Microenvironment. Cancer Sci (2018) 109(7):2085–92. doi: 10.1111/cas.13630
15. De Maria R, Olivero M, Iussich S, Nakaichi M, Murata T, Biolatti B, et al. Spontaneous Feline Mammary Carcinoma is a Model of HER2 Overexpressing Poor Prognosis Human Breast Cancer. Cancer Res (2005) 65(3):907–12.
16. Smith BF, Migone FK, Cox NR, Baker HJ. An In Utero Allotransplantation Model of Metastatic Breast Cancer in the Cat. In Vivo (2003) 17(1):35–9.
17. Minke JM, Weijer K, Misdorp W. Allotransplantation of K248 Feline Mammary Carcinoma Cell Line in Cats. A Model for Monoclonal Antibody Guided Detection and Therapy of Human Breast Cancer. Lab Invest (1991) 65(4):421–32.
18. Barbieri F, Wurth R, Ratto A, Campanella C, Vito G, Thellung S, et al. Isolation of Stem-Like Cells From Spontaneous Feline Mammary Carcinomas: Phenotypic Characterization and Tumorigenic Potential. Exp Cell Res (2012) 318(7):847–60. doi: 10.1016/j.yexcr.2012.02.008
19. Pang LY, Blacking TM, Else RW, Sherman A, Sang HM, Whitelaw BA, et al. Feline Mammary Carcinoma Stem Cells Are Tumorigenic, Radioresistant, Chemoresistant and Defective in Activation of the ATM/p53 DNA Damage Pathway. Vet J (2013) 196(3):414–23. doi: 10.1016/j.tvjl.2012.10.021
20. Wiese DA, Thaiwong T, Yuzbasiyan-Gurkan V, Kiupel M. Feline Mammary Basal-Like Adenocarcinomas: A Potential Model for Human Triple-Negative Breast Cancer (TNBC) With Basal-Like Subtype. BMC Cancer (2013) 13:403. doi: 10.1186/1471-2407-13-403
21. Burrai GP, Mohammed SI, Miller MA, Marras V, Pirino S, Addis MF, et al. Spontaneous Feline Mammary Intraepithelial Lesions as a Model for Human Estrogen Receptor- and Progesterone Receptor-Negative Breast Lesions. BMC Cancer (2010) 10:156. doi: 10.1186/1471-2407-10-156
22. Misdorp W, Hart AA. Canine Mammary Cancer. II. Therapy and Causes of Death. J Small Anim Pract (1979) 20(7):395–404. doi: 10.1111/j.1748-5827.1979.tb06744.x
23. Caceres S, Alonso-Diez A, Crespo B, Pena L, Illera MJ, Silvan G, et al. Tumor Growth Progression in Ectopic and Orthotopic Xenografts From Inflammatory Breast Cancer Cell Lines. Vet Sci (2021) 8(9):194. doi: 10.3390/vetsci8090194
24. Cocola C, Molgora S, Piscitelli E, Veronesi MC, Greco M, Bragato C, et al. FGF2 and EGF Are Required for Self-Renewal and Organoid Formation of Canine Normal and Tumor Breast Stem Cells. J Cell Biochem (2017) 118(3):570–84. doi: 10.1002/jcb.25737
25. Uva P, Aurisicchio L, Watters J, Loboda A, Kulkarni A, Castle J, et al. Comparative Expression Pathway Analysis of Human and Canine Mammary Tumors. BMC Genomics (2009) 10:135. doi: 10.1186/1471-2164-10-135
26. Lee CH, Kim WH, Lim JH, Kang MS, Kim DY, Kweon OK. Mutation and Overexpression of P53 as a Prognostic Factor in Canine Mammary Tumors. J Vet Sci (2004) 5(1):63–9. doi: 10.4142/jvs.2004.5.1.63
27. Chang CC, Tsai MH, Liao JW, Chan JP, Wong ML, Chang SC. Evaluation of Hormone Receptor Expression for Use in Predicting Survival of Female Dogs With Malignant Mammary Gland Tumors. J Am Vet Med Assoc (2009) 235(4):391–6. doi: 10.2460/javma.235.4.391
28. Wu A, Dong Q, Gao H, Shi Y, Chen Y, Zhang F, et al. Characterization of Mammary Epithelial Stem/Progenitor Cells and Their Changes With Aging in Common Marmosets. Sci Rep (2016) 6:32190. doi: 10.1038/srep32190
29. Mariya S, Dewi FN, Suparto IH, Wilkerson GK, Cline MJ, Iskandriati D, et al. Mammosphere Culture of Mammary Cells From Cynomolgus Macaques (Macaca Fascicularis). Comp Med (2019) 69(2):144–50. doi: 10.30802/AALAS-CM-18-000030
30. Flisikowska T, Kind A, Schnieke A. The New Pig on the Block: Modelling Cancer in Pigs. Transgenic Res (2013) 22(4):673–80. doi: 10.1007/s11248-013-9720-9
31. Rowson-Hodel AR, Manjarin R, Trott JF, Cardiff RD, Borowsky AD, Hovey RC. Neoplastic Transformation of Porcine Mammary Epithelial Cells In Vitro and Tumor Formation In Vivo. BMC Cancer (2015) 15:562. doi: 10.1186/s12885-015-1572-7
32. Remmers N, Carlson MA. Ectopic Expression of KRASG12D and P53r167h in Porcine Mammary Epithelial Cells Results in Transformation and Tumorigenesis. bioRxiv (2021). doi: 10.1101/2021.06.20.449198
33. Bartlett AP, Van de Walle GR. Establishment and Characterization of Mammary Organoids From Non-Traditional Model Organisms. bioRxiv (2021). doi: 10.1101/2021.01.15.426833
34. Smith S, Stone A, Oswalt H, Vaughan L, Ferdous F, Scott T, et al. Evaluation of Early Post-Natal Pig Mammary Gland Development and Human Breast Cancer Gene Expression. Dev Biol (2022) 481:95–103. doi: 10.1016/j.ydbio.2021.10.004
35. Thomas R. Cytogenomics of Feline Cancers: Advances and Opportunities. Vet Sci (2015) 2(3):246–58. doi: 10.3390/vetsci2030246
36. Hassan BB, Elshafae SM, Supsavhad W, Simmons JK, Dirksen WP, Sokkar SM, et al. Feline Mammary Cancer. Vet Pathol (2017) 54(1):32–43. doi: 10.1177/0300985816650243
37. Pinho SS, Carvalho S, Cabral J, Reis CA, Gärtner F. Canine Tumors: A Spontaneous Animal Model of Human Carcinogenesis. Transl Res (2012) 159(3):165–72. doi: 10.1016/j.trsl.2011.11.005
38. Graim K, Gorenshteyn D, Robinson DG, Carriero NJ, Cahill JA, Chakrabarti R, et al. Modeling Molecular Development of Breast Cancer in Canine Mammary Tumors. Genome Res (2020) 31(2):337–47. doi: 10.1101/gr.256388.119
39. Klopfleisch R, Lenze D, Hummel M, Gruber AD. Metastatic Canine Mammary Carcinomas can be Identified by a Gene Expression Profile That Partly Overlaps With Human Breast Cancer Profiles. BMC Cancer (2010) 10:618. doi: 10.1186/1471-2407-10-618
40. Mohammed SI, Utturkar S, Lee M, Yang HH, Cui Z, Atallah Lanman N, et al. Ductal Carcinoma In Situ Progression in Dog Model of Breast Cancer. Cancers (2020) 12(2):418. doi: 10.3390/cancers12020418
41. Burrai GP, Tanca A, De Miglio MR, Abbondio M, Pisanu S, Polinas M, et al. Investigation of HER2 Expression in Canine Mammary Tumors by Antibody-Based, Transcriptomic and Mass Spectrometry Analysis: Is the Dog a Suitable Animal Model for Human Breast Cancer? Tumour Biol (2015) 36(11):9083–91. doi: 10.1007/s13277-015-3661-2
42. Salas Y, Márquez A, Diaz D, Romero L. Epidemiological Study of Mammary Tumors in Female Dogs Diagnosed During the Period 2002-2012: A Growing Animal Health Problem. PloS One (2015) 10(5):e0127381. doi: 10.1371/journal.pone.0127381
43. Goldschmidt M, Peña L, Rasotto R, Zappulli V. Classification and Grading of Canine Mammary Tumors. Vet Pathol (2011) 48(1):117–31. doi: 10.1177/0300985810393258
44. Else RW, Norval M, Neill WA. The Characteristics of a Canine Mammary Carcinoma Cell Line, REM 134. Br J Cancer (1982) 46(4):675–81. doi: 10.1038/bjc.1982.254
45. Hellmen E. Characterization of Four In Vitro Established Canine Mammary Carcinoma and One Atypical Benign Mixed Tumor Cell Lines. In Vitro Cell Dev Biol (1992) 28A(5):309–19. doi: 10.1007/BF02877054
46. van der Burg B, van Selm-Miltenburg AJ, van Maurik P, Rutteman GR, Misdorp W, de Laat SW, et al. Isolation of Autonomously Growing Dog Mammary Tumor Cell Lines Cultured in Medium Supplemented With Serum Treated to Inactivate Growth Factors. J Natl Cancer Inst (1989) 81(20):1545–51. doi: 10.1093/jnci/81.20.1545
47. Wolfe LG, Smith BB, Toivio-Kinnucan MA, Sartin EA, Kwapien RP, Henderson RA, et al. Biologic Properties of Cell Lines Derived From Canine Mammary Carcinomas. J Natl Cancer Inst (1986) 77(3):783–92. doi: 10.1093/jnci/77.3.783
48. Li R, Wu H, Sun Y, Zhu J, Tang J, Kuang Y, et al. A Novel Canine Mammary Cancer Cell Line: Preliminary Identification and Utilization for Drug Screening Studies. Front Vet Sci (2021) 8:665906. doi: 10.3389/fvets.2021.665906
49. Lunney JK. Advances in Swine Biomedical Model Genomics. Int J Biol Sci (2007) 3(3):179–84. doi: 10.7150/ijbs.3.179
50. Swindle MM, Smith AC. Swine in the Laboratory: Surgery, Anesthesia, Imaging, and Experimental Techniques. 3rd ed. Boca Raton, FL: CRC Press (2016).
51. Schook LB, Collares TV, Hu W, Liang Y, Rodrigues FM, Rund LA, et al. A Genetic Porcine Model of Cancer. PloS One (2015) 10(7):e0128864. doi: 10.1371/journal.pone.0128864
52. Schook LB, Rund L, Begnini KR, Remiao MH, Seixas FK, Collares T. Emerging Technologies to Create Inducible and Genetically Defined Porcine Cancer Models. Front Genet (2016) 7:28. doi: 10.3389/fgene.2016.00028
53. Kalla D, Kind A, Schnieke A. Genetically Engineered Pigs to Study Cancer. Int J Mol Sci (2020) 21(2):1–21. doi: 10.3390/ijms21020488
54. Patel NS, Bailey KL, Lazenby AJ, Carlson MA. Induction of Pancreatic Neoplasia in the KRAS/TP53 Oncopig: Preliminary Report. bioRxiv (2020). doi: 10.1101/2020.05.29.123547
55. Groenen MA, Archibald AL, Uenishi H, Tuggle CK, Takeuchi Y, Rothschild MF, et al. Analyses of Pig Genomes Provide Insight Into Porcine Demography and Evolution. Nature (2012) 491(7424):393–8. doi: 10.1038/nature11622
56. Giuffra E, Tuggle CK, Consortium F. Functional Annotation of Animal Genomes (FAANG): Current Achievements and Roadmap. Annu Rev Anim Biosci (2019) 7:65–88. doi: 10.1146/annurev-animal-020518-114913
57. Niu D, Wei HJ, Lin L, George H, Wang T, Lee IH, et al. Inactivation of Porcine Endogenous Retrovirus in Pigs Using CRISPR-Cas9. Science (2017) 357(6357):1303–7. doi: 10.1126/science.aan4187
58. Prather RS, Lorson M, Ross JW, Whyte JJ, Walters E. Genetically Engineered Pig Models for Human Diseases. Annu Rev Anim Biosci (2013) 1:203–19. doi: 10.1146/annurev-animal-031412-103715
59. Rogers CS, Stoltz DA, Meyerholz DK, Ostedgaard LS, Rokhlina T, Taft PJ, et al. Disruption of the CFTR Gene Produces a Model of Cystic Fibrosis in Newborn Pigs. Science (2008) 321(5897):1837–41. doi: 10.1126/science.1163600
60. Whitworth KM, Lee K, Benne JA, Beaton BP, Spate LD, Murphy SL, et al. Use of the CRISPR/Cas9 System to Produce Genetically Engineered Pigs From In Vitro-Derived Oocytes and Embryos. Biol Reprod (2014) 91(3):78. doi: 10.1095/biolreprod.114.121723
61. Soucek P, Zuber R, Anzenbacherova E, Anzenbacher P, Guengerich FP. Minipig Cytochrome P450 3A, 2A and 2C Enzymes Have Similar Properties to Human Analogs. BMC Pharmacol (2001) 1:11. doi: 10.1186/1471-2210-1-11
62. Dalgaard L. Comparison of Minipig, Dog, Monkey and Human Drug Metabolism and Disposition. J Pharmacol Toxicol Methods (2015) 74:80–92. doi: 10.1016/j.vascn.2014.12.005
63. Petri N, Bergman E, Forsell P, Hedeland M, Bondesson U, Knutson L, et al. First-Pass Effects of Verapamil on the Intestinal Absorption and Liver Disposition of Fexofenadine in the Porcine Model. Drug Metab Dispos (2006) 34(7):1182–9. doi: 10.1124/dmd.105.008409
64. Tannergren C, Evilevitch L, Pierzynowski S, Piedra JV, Westrom B, Erlwanger K, et al. The Effect of Pancreatic and Biliary Depletion on the In Vivo Pharmacokinetics of Digoxin in Pigs. Eur J Pharm Sci (2006) 29(3-4):198–204. doi: 10.1016/j.ejps.2006.06.009
65. Donninger H, Hobbing K, Schmidt ML, Walters E, Rund L, Schook L, et al. A Porcine Model System of BRCA1 Driven Breast Cancer. Front Genet (2015) 6:269. doi: 10.3389/fgene.2015.00269
66. Luo Y, Li J, Liu Y, Lin L, Du Y, Li S, et al. High Efficiency of BRCA1 Knockout Using rAAV-Mediated Gene Targeting: Developing a Pig Model for Breast Cancer. Transgenic Res (2011) 20(5):975–88. doi: 10.1007/s11248-010-9472-8
67. Gowen LC, Johnson BL, Latour AM, Sulik KK, Koller BH. Brca1 Deficiency Results in Early Embryonic Lethality Characterized by Neuroepithelial Abnormalities. Nat Genet (1996) 12(2):191–4. doi: 10.1038/ng0296-191
68. Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, et al. Mutational Landscape and Significance Across 12 Major Cancer Types. Nature (2013) 502(7471):333–9. doi: 10.1038/nature12634
69. Nik-Zainal S, Davies H, Staaf J, Ramakrishna M, Glodzik D, Zou X, et al. Landscape of Somatic Mutations in 560 Breast Cancer Whole-Genome Sequences. Nature (2016) 534(7605):47–54. doi: 10.1038/nature17676
70. Stephens PJ, Tarpey PS, Davies H, Van Loo P, Greenman C, Wedge DC, et al. The Landscape of Cancer Genes and Mutational Processes in Breast Cancer. Nature (2012) 486(7403):400–4. doi: 10.1038/nature11017
71. Bailey KL, Carlson MA. Porcine Models of Pancreatic Cancer. Front Oncol (2019) 9:144. doi: 10.3389/fonc.2019.00144
72. Bailey KL, Cartwright SB, Patel NS, Remmers N, Lazenby AJ, Hollingsworth MA, et al. Porcine Pancreatic Ductal Epithelial Cells Transformed With KRASG12D and SV40T Are Tumorigenic. Sci Rep (2021) 11:13436. doi: 10.1038/s41598-021-92852-2
73. Adam SJ, Rund LA, Kuzmuk KN, Zachary JF, Schook LB, Counter CM. Genetic Induction of Tumorigenesis in Swine. Oncogene (2007) 26(7):1038–45. doi: 10.1038/sj.onc.1209892
74. Boas FE, Nurili F, Bendet A, Cheleuitte-Nieves C, Basturk O, Askan G, et al. Induction and Characterization of Pancreatic Cancer in a Transgenic Pig Model. PloS One (2020) 15(9):e0239391. doi: 10.1371/journal.pone.0239391
75. Principe DR, Overgaard NH, Park AJ, Diaz AM, Torres C, McKinney R, et al. KRAS(G12D) and TP53(R167H) Cooperate to Induce Pancreatic Ductal Adenocarcinoma in Sus Scrofa Pigs. Sci Rep (2018) 8(1):12548. doi: 10.1038/s41598-018-30916-6
76. Schachtschneider KM, Schwind RM, Newson J, Kinachtchouk N, Rizko M, Mendoza-Elias N, et al. The Oncopig Cancer Model: An Innovative Large Animal Translational Oncology Platform. Front Oncol (2017) 7:190. doi: 10.3389/fonc.2017.00190
77. Day CP, Merlino G, Van Dyke T. Preclinical Mouse Cancer Models: A Maze of Opportunities and Challenges. Cell (2015) 163(1):39–53. doi: 10.1016/j.cell.2015.08.068
78. Gengenbacher N, Singhal M, Augustin HG. Preclinical Mouse Solid Tumour Models: Status Quo, Challenges and Perspectives. Nat Rev Cancer (2017) 17(12):751–65. doi: 10.1038/nrc.2017.92
79. Park MK, Lee CH, Lee H. Mouse Models of Breast Cancer in Preclinical Research. Lab Anim Res (2018) 34(4):160–5. doi: 10.5625/lar.2018.34.4.160
80. Zitvogel L, Pitt JM, Daillere R, Smyth MJ, Kroemer G. Mouse Models in Oncoimmunology. Nat Rev Cancer (2016) 16(12):759–73. doi: 10.1038/nrc.2016.91
81. Day JK, Besch-Williford C, McMann TR, Hufford MG, Lubahn DB, MacDonald RS, et al. Dietary Genistein Increased DMBA-Induced Mammary Adenocarcinoma in Wild-Type, But NotMice. Nutr Cancer (2001) 39(2):226–32. doi: 10.1207/S15327914nc392_11
82. Fischer SM, Conti CJ, Locniskar M, Belury MA, Maldve RE, Lee ML, et al. The Effect of Dietary Fat on the Rapid Development of Mammary Tumors Induced by 7,12-Dimethylbenz(a)Anthracene in SENCAR Mice. Cancer Res (1992) 52(3):662–6.
83. Huang MT, Lou YR, Xie JG, Ma W, Lu YP, Yen P, et al. Effect of Dietary Curcumin and Dibenzoylmethane on Formation of 7,12-Dimethylbenz[a]Anthracene-Induced Mammary Tumors and Lymphomas/Leukemias in Sencar Mice. Carcinogenesis (1998) 19(9):1697–700. doi: 10.1093/carcin/19.9.1697
84. Jerry DJ, Butel JS, Donehower LA, Paulson EJ, Cochran C, Wiseman RW, et al. Infrequent P53 Mutations in 7,12-Dimethylbenz[a]Anthracene-Induced Mammary Tumors in BALB/c and P53 Hemizygous Mice. Mol Carcinog (1994) 9(3):175–83. doi: 10.1002/mc.2940090309
85. Kouri RE, Ratrie H, Whitmire CE. Evidence of a Genetic Relationship Between Susceptibility to 3-Methyl-Cholanthrene-Induced Subcutaneous Tumors and Inducibility of Aryl Hydrocarbon Hydroxylase. J Natl Cancer Inst (1973) 51(1):197–200. doi: 10.1093/jnci/51.1.197
86. Holen I, Speirs V, Morrissey B, Blyth K. In Vivo Models in Breast Cancer Research: Progress, Challenges and Future Directions. Dis Model Mech (2017) 10(4):359–71. doi: 10.1242/dmm.028274
87. Kawaguchi T, Foster BA, Young J, Takabe K. Current Update of Patient-Derived Xenograft Model for Translational Breast Cancer Research. J Mammary Gland Biol Neoplasia (2017) 22(2):131–9. doi: 10.1007/s10911-017-9378-7
88. Vesely MD, Kershaw MH, Schreiber RD, Smyth MJ. Natural Innate and Adaptive Immunity to Cancer. Annu Rev Immunol (2011) 29:235–71. doi: 10.1146/annurev-immunol-031210-101324
89. Wagner M, Koyasu S. Cancer Immunoediting by Innate Lymphoid Cells. Trends Immunol (2019) 40(5):415–30. doi: 10.1016/j.it.2019.03.004
90. Dobrolecki LE, Airhart SD, Alferez DG, Aparicio S, Behbod F, Bentires-Alj M, et al. Patient-Derived Xenograft (PDX) Models in Basic and Translational Breast Cancer Research. Cancer Metastasis Rev (2016) 35(4):547–73. doi: 10.1007/s10555-016-9653-x
91. Richard E, Grellety T, Velasco V, MacGrogan G, Bonnefoi H, Iggo R. The Mammary Ducts Create a Favourable Microenvironment for Xenografting of Luminal and Molecular Apocrine Breast Tumours. J Pathol (2016) 240(3):256–61. doi: 10.1002/path.4772
92. DeRose YS, Wang G, Lin YC, Bernard PS, Buys SS, Ebbert MT, et al. Tumor Grafts Derived From Women With Breast Cancer Authentically Reflect Tumor Pathology, Growth, Metastasis and Disease Outcomes. Nat Med (2011) 17(11):1514–20. doi: 10.1038/nm.2454
93. Roarty K, Echeverria GV. Laboratory Models for Investigating Breast Cancer Therapy Resistance and Metastasis. Front Oncol (2021) 11:645698. doi: 10.3389/fonc.2021.645698
94. Paez-Ribes M, Man S, Xu P, Kerbel RS. Development of Patient Derived Xenograft Models of Overt Spontaneous Breast Cancer Metastasis: A Cautionary Note. PloS One (2016) 11(6):e0158034. doi: 10.1371/journal.pone.0158034
95. De Wever O, Mareel M. Role of Tissue Stroma in Cancer Cell Invasion. J Pathol (2003) 200(4):429–47. doi: 10.1002/path.1398
96. Junttila MR, de Sauvage FJ. Influence of Tumour Micro-Environment Heterogeneity on Therapeutic Response. Nature (2013) 501(7467):346–54. doi: 10.1038/nature12626
97. Jin KT, Du WL, Lan HR, Liu YY, Mao CS, Du JL, et al. Development of Humanized Mouse With Patient-Derived Xenografts for Cancer Immunotherapy Studies: A Comprehensive Review. Cancer Sci (2021) 112(7):2592–606. doi: 10.1111/cas.14934
98. Compte M, Harwood SL, Erce-Llamazares A, Tapia-Galisteo A, Romero E, Ferrer I, et al. An Fc-Free EGFR-Specific 4-1BB-Agonistic Trimerbody Displays Broad Antitumor Activity in Humanized Murine Cancer Models Without Toxicity. Clin Cancer Res (2021) 27(11):3167–77. doi: 10.1158/1078-0432.CCR-20-4625
99. Rosato RR, Davila-Gonzalez D, Choi DS, Qian W, Chen W, Kozielski AJ, et al. Evaluation of Anti-PD-1-Based Therapy Against Triple-Negative Breast Cancer Patient-Derived Xenograft Tumors Engrafted in Humanized Mouse Models. Breast Cancer Res (2018) 20(1):108. doi: 10.1186/s13058-018-1037-4
100. Morton JJ, Alzofon N, Jimeno A. The Humanized Mouse: Emerging Translational Potential. Mol Carcinog (2020) 59(7):830–8. doi: 10.1002/mc.23195
101. Asch BB. Tumor Viruses and Endogenous Retrotransposons in Mammary Tumorigenesis. J Mammary Gland Biol Neoplasia (1996) 1(1):49–60. doi: 10.1007/BF02096302
102. Green JE, Shibata MA, Yoshidome K, Liu ML, Jorcyk C, Anver MR, et al. The C3(1)/SV40 T-Antigen Transgenic Mouse Model of Mammary Cancer: Ductal Epithelial Cell Targeting With Multistage Progression to Carcinoma. Oncogene (2000) 19(8):1020–7. doi: 10.1038/sj.onc.1203280
103. Webster J, Wallace RM, Clark AJ, Whitelaw CB. Tissue-Specific, Temporally Regulated Expression Mediated by the Proximal Ovine Beta-Lactoglobulin Promoter in Transgenic Mice. Cell Mol Biol Res (1995) 41(1):11–5.
104. Wen J, Kawamata Y, Tojo H, Tanaka S, Tachi C. Expression of Whey Acidic Protein (WAP) Genes in Tissues Other Than the Mammary Gland in Normal and Transgenic Mice Expressing mWAP/hGH Fusion Gene. Mol Reprod Dev (1995) 41(4):399–406. doi: 10.1002/mrd.1080410402
105. Stewart TA, Pattengale PK, Leder P. Spontaneous Mammary Adenocarcinomas in Transgenic Mice That Carry and Express MTV/myc Fusion Genes. Cell (1984) 38(3):627–37. doi: 10.1016/0092-8674(84)90257-5
106. Guy CT, Cardiff RD, Muller WJ. Activated Neu Induces Rapid Tumor Progression. J Biol Chem (1996) 271(13):7673–8. doi: 10.1074/jbc.271.13.7673
107. Muller WJ, Sinn E, Pattengale PK, Wallace R, Leder P. Single-Step Induction of Mammary Adenocarcinoma in Transgenic Mice Bearing the Activated C-Neu Oncogene. Cell (1988) 54(1):105–15. doi: 10.1016/0092-8674(88)90184-5
108. Sinn E, Muller W, Pattengale P, Tepler I, Wallace R, Leder P. Coexpression of MMTV/v-Ha-Ras and MMTV/c-Myc Genes in Transgenic Mice: Synergistic Action of Oncogenes In Vivo. Cell (1987) 49(4):465–75. doi: 10.1016/0092-8674(87)90449-1
109. Tsukamoto AS, Grosschedl R, Guzman RC, Parslow T, Varmus HE. Expression of the Int-1 Gene in Transgenic Mice is Associated With Mammary Gland Hyperplasia and Adenocarcinomas in Male and Female Mice. Cell (1988) 55(4):619–25. doi: 10.1016/0092-8674(88)90220-6
110. Guy CT, Cardiff RD, Muller WJ. Induction of Mammary Tumors by Expression of Polyomavirus Middle T Oncogene: A Transgenic Mouse Model for Metastatic Disease. Mol Cell Biol (1992) 12(3):954–61. doi: 10.1128/mcb.12.3.954-961.1992
111. Christenson JL, Butterfield KT, Spoelstra NS, Norris JD, Josan JS, Pollock JA, et al. MMTV-PyMT and Derived Met-1 Mouse Mammary Tumor Cells as Models for Studying the Role of the Androgen Receptor in Triple-Negative Breast Cancer Progression. Horm Cancer (2017) 8(2):69–77. doi: 10.1007/s12672-017-0285-6
112. Jerry DJ, Kittrell FS, Kuperwasser C, Laucirica R, Dickinson ES, Bonilla PJ, et al. A Mammary-Specific Model Demonstrates the Role of the P53 Tumor Suppressor Gene in Tumor Development. Oncogene (2000) 19(8):1052–8. doi: 10.1038/sj.onc.1203270
113. Kuperwasser C, Hurlbut GD, Kittrell FS, Dickinson ES, Laucirica R, Medina D, et al. Development of Spontaneous Mammary Tumors in BALB/c P53 Heterozygous Mice. A Model for Li-Fraumeni Syndrome. Am J Pathol (2000) 157(6):2151–9. doi: 10.1016/S0002-9440(10)64853-5
114. Wagner KU, McAllister K, Ward T, Davis B, Wiseman R, Hennighausen L. Spatial and Temporal Expression of the Cre Gene Under the Control of the MMTV-LTR in Different Lines of Transgenic Mice. Transgenic Res (2001) 10(6):545–53. doi: 10.1023/A:1013063514007
115. Wagner KU, Wall RJ, St-Onge L, Gruss P, Wynshaw-Boris A, Garrett L, et al. Cre-Mediated Gene Deletion in the Mammary Gland. Nucleic Acids Res (1997) 25(21):4323–30. doi: 10.1093/nar/25.21.4323
116. Trusler O, Goodwin J, Laslett AL. BRCA1 and BRCA2 Associated Breast Cancer and the Roles of Current Modelling Systems in Drug Discovery. Biochim Biophys Acta Rev Cancer (2021) 1875(1):188459. doi: 10.1016/j.bbcan.2020.188459
117. Cressman VL, Backlund DC, Hicks EM, Gowen LC, Godfrey V, Koller BH. Mammary Tumor Formation in P53- and BRCA1-Deficient Mice. Cell Growth Differ (1999) 10(1):1–10.
118. Russo J. Significance of Rat Mammary Tumors for Human Risk Assessment. Toxicol Pathol (2015) 43(2):145–70. doi: 10.1177/0192623314532036
119. Singh M, McGinley JN, Thompson HJ. A Comparison of the Histopathology of Premalignant and Malignant Mammary Gland Lesions Induced in Sexually Immature Rats With Those Occurring in the Human. Lab Investigation J Tech Methods Pathol (2000) 80(2):221–31. doi: 10.1038/labinvest.3780025
120. C R. Animal Models of Breast Cancer: Their Diversity and Role in Biomedical Research. Breast Cancer Res Treat (1996) 39(1):1–6. doi: 10.1007/BF01806073
121. Dennison KL, Samanas NB, Harenda QE, Hickman MP, Seiler NL, Ding L, et al. Development and Characterization of a Novel Rat Model of Estrogen-Induced Mammary Cancer. Endocrine-Related Cancer (2015) 22(2):239–48. doi: 10.1530/ERC-14-0539
122. Gao D, Liu J, Yuan J, Wu J, Kuang X, Kong D, et al. Intraductal Administration of N-Methyl-N-Nitrosourea as a Novel Rodent Mammary Tumor Model. Ann Trans Med (2021) 9(7):576. doi: 10.21037/atm-21-1540
123. Banerjee S, Bueso-Ramos C, Aggarwal BB. Suppression of 7,12-Dimethylbenz(a)Anthracene-Induced Mammary Carcinogenesis in Rats by Resveratrol: Role of Nuclear Factor-Kappab, Cyclooxygenase 2, and Matrix Metalloprotease 9. Cancer Res (2002) 62(17):4945–54.
124. Mullen P, Ritchie A, Langdon SP, Miller WR. Effect of Matrigel on the Tumorigenicity of Human Breast and Ovarian Carcinoma Cell Lines. Int J Cancer (1996) 67(6):816–20. doi: 10.1002/(SICI)1097-0215(19960917)67:6<816::AID-IJC10>3.0.CO;2-#
125. Aliaga A, Rousseau JA, Ouellette R, Cadorette J, van Lier JE, Lecomte R, et al. Breast Cancer Models to Study the Expression of Estrogen Receptors With Small Animal PET Imaging. Nucl Med Biol (2004) 31(6):761–70. doi: 10.1016/j.nucmedbio.2004.02.011
126. Bäuerle T, Adwan H, Kiessling F, Hilbig H, Armbruster FP, Berger MR. Characterization of a Rat Model With Site-Specific Bone Metastasis Induced by MDA-MB-231 Breast Cancer Cells and Its Application to the Effects of an Antibody Against Bone Sialoprotein. Int J Cancer (2005) 115(2):177–86. doi: 10.1002/ijc.20840
127. Davies BR, Platt-Higgins AM, Schmidt G, Rudland PS. Development of Hyperplasias, Preneoplasias, and Mammary Tumors in MMTV-c-erbB-2 and MMTV-TGFalpha Transgenic Rats. Am J Pathol (1999) 155(1):303–14. doi: 10.1016/S0002-9440(10)65124-3
128. Asamoto M, Ochiya T, Toriyama-Baba H, Ota T, Sekiya T, Terada M, et al. Transgenic Rats Carrying Human C-Ha-Ras Proto-Oncogenes Are Highly Susceptible to N-Methyl-N-Nitrosourea Mammary Carcinogenesis. Carcinogenesis (2000) 21(2):243–9. doi: 10.1093/carcin/21.2.243
129. Woditschka S, Haag JD, Sullivan R, Gould MN. A Short-Term Rat Mammary Carcinogenesis Model for the Prevention of Hormonally Responsive and Nonresponsive In Situ Carcinomas. Cancer Prev Res (Phila) (2009) 2(2):153–60. doi: 10.1158/1940-6207.CAPR-08-0114
130. Javier R, Shenk T. Mammary Tumors Induced by Human Adenovirus Type 9: A Role for the Viral Early Region 4 Gene. Breast Cancer Res Treat (1996) 39(1):57–67. doi: 10.1007/BF01806078
131. Coburn MA, Brueggemann S, Bhatia S, Cheng B, Li BD, Li XL, et al. Establishment of a Mammary Carcinoma Cell Line From Syrian Hamsters Treated With N-Methyl-N-Nitrosourea. Cancer Lett (2011) 312(1):82–90. doi: 10.1016/j.canlet.2011.08.003
132. Li X, Wang P, Li H, Du X, Liu M, Huang Q, et al. The Efficacy of Oncolytic Adenovirus Is Mediated by T-Cell Responses Against Virus and Tumor in Syrian Hamster Model. Clin Cancer Res (2017) 23(1):239–49. doi: 10.1158/1078-0432.CCR-16-0477
133. Xia HJ, Wang CY, Zhang HL, He BL, Jiao JL, Chen CS. Characterization of Spontaneous Breast Tumor in Tree Shrews (Tupaia Belangeri Chinenesis). Dong Wu Xue Yan Jiu Zoological Res (2012) 33(1):55–9. doi: 10.3724/SP.J.1141.2012.01055
134. Elliot OS, Elliot MW, Lisco H. Breast Cancer in a Tree Shrew (Tupaia Glis). Nature (1966) 211(5053):1105. doi: 10.1038/2111105a0
135. Xia HJ, He BL, Wang CY, Zhang HL, Ge GZ, Zhang YX, et al. PTEN/PIK3CA Genes Are Frequently Mutated in Spontaneous and Medroxyprogesterone Acetate-Accelerated 7,12-Dimethylbenz(a)Anthracene-Induced Mammary Tumours of Tree Shrews. Eur J Cancer (2014) 50(18):3230–42. doi: 10.1016/j.ejca.2014.10.012
Keywords: breast cancer, large animal models, animal models, porcine model, BRCA1, TP53
Citation: Mondal P, Bailey KL, Cartwright SB, Band V and Carlson MA (2022) Large Animal Models of Breast Cancer. Front. Oncol. 12:788038. doi: 10.3389/fonc.2022.788038
Received: 01 October 2021; Accepted: 18 January 2022;
Published: 04 February 2022.
Edited by:
Kyle Schachtschneider, University of Illinois at Chicago, United StatesReviewed by:
Thomas Liehr, Friedrich Schiller University Jena, GermanyAhmet Acar, Middle East Technical University, Turkey
Copyright © 2022 Mondal, Bailey, Cartwright, Band and Carlson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mark A. Carlson, bWFjYXJsc29AdW5tYy5lZHU=
 Pinaki Mondal
Pinaki Mondal Katie L. Bailey
Katie L. Bailey Sara B. Cartwright
Sara B. Cartwright Vimla Band
Vimla Band Mark A. Carlson
Mark A. Carlson