- 1Saliva and Liquid Biopsy Translational Laboratory, The Centre for Biomedical Technologies, The School of Biomedical Sciences, Faculty of Health, Queensland University of Technology, Kelvin Grove, QLD, Australia
- 2Translational Research Institute, Brisbane, QLD, Australia
- 3School of Biomedical Engineering, University of Technology Sydney, Sydney, NSW, Australia
- 4The School of Biomedical Sciences, Faculty of Health, Queensland University of Technology, Brisbane, QLD, Australia
- 5Australian Prostate Cancer Research Centre, Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Princess Alexandra Hospital, Translational Research Institute, Brisbane, QLD, Australia
- 6Department of Urology, Princess Alexandra Hospital, Woolloongabba, QLD, Australia
- 7School of Medicine, University of Queensland, Royal Brisbane and Women’s Hospital, Central Integrated Regional Cancer Service, Queensland Health, Brisbane, QLD, Australia
- 8Department of Medical Oncology, Princess Alexandra Hospital, Woolloongabba, QLD, Australia
- 9School of Medicine, University of Queensland, Herston, QLD, Australia
- 10Saliva and Liquid Biopsy Translational Laboratory, Griffith Institute for Drug Discovery and Menzies Health Institute Queensland, Griffith University, Nathan, QLD, Australia
Despite efforts to improve earlier diagnosis of non-small cell lung cancer (NSCLC), most patients present with advanced stage disease, which is often associated with poor survival outcomes with only 15% surviving for 5 years from their diagnosis. Tumour tissue biopsy is the current mainstream for cancer diagnosis and prognosis in many parts of the world. However, due to tumour heterogeneity and accessibility issues, liquid biopsy is emerging as a game changer for both cancer diagnosis and prognosis. Liquid biopsy is the analysis of tumour-derived biomarkers in body fluids, which has remarkable advantages over the use of traditional tumour biopsy. Circulating tumour cells (CTCs) and circulating tumour DNA (ctDNA) are two main derivatives of liquid biopsy. CTC enumeration and molecular analysis enable monitoring of cancer progression, recurrence, and treatment response earlier than traditional biopsy through a minimally invasive liquid biopsy approach. CTC-derived ex-vivo cultures are essential to understanding CTC biology and their role in metastasis, provide a means for personalized drug testing, and guide treatment selection. Just like CTCs, ctDNA provides opportunity for screening, monitoring, treatment evaluation, and disease surveillance. We present an updated review highlighting the prognostic and therapeutic significance of CTCs and ctDNA in NSCLC.
Introduction
Lung cancer is currently the most common cause of cancer-related death worldwide (1) with a total of 1.80 million deaths in 2020 (2). Approximately 80% of lung cancer patients are diagnosed with non-small cell lung cancer (NSCLC), with only 15% surviving for 5 years (3, 4). In the past two decades, an improved understanding of oncogenic driver mutations, such as EFGR, ALK, and ROS1, has led to significant advancements in the treatment of NSCLC patients (5).
Tumour tissue biopsy is still the gold standard for clinical molecular analysis; however, collection of tumour biopsy is invasive and, in most cases, inaccessible due to the location of lung cancer and the ability to safely carry out a biopsy in an elderly patient population with multiple comorbidities. In addition, a comprehensive characterization of different regions of tumour obtained from the same patient has shown intratumour heterogeneity (spatial heterogeneity), as well as differences between serial biopsies over time (temporal heterogeneity) (6). Thus, inter- and intratumour heterogeneity poses a challenge to guide clinical decision as biopsies may be inaccurate in capturing the true genomic landscape of NSCLC. In contrast, the application of the analysis of tumour-derived material in body fluids (liquid biopsy) is currently gaining attention due to its non-invasiveness and the rapid, real-time application in NSCLC that has the potential to overcome tumour heterogeneity (7, 8).
The most widely studied liquid biopsy derivatives are circulating tumour cells (CTCs) and circulating tumour DNA (ctDNA). CTCs detach from either primary tumour or metastatic sites and are shed in the patient’s bloodstream, representing a relatively easily obtainable sample of cancer tissue. CTCs were first proposed by an Australian physician Thomas Ashworth in 1869. CTCs are very rare events in the bloodstream; therefore, various enrichment and isolation methods have been developed. The presence of nucleic acids in the circulation was first reported in 1948 by Mandel and Metais (9). Circulating cell-free DNA (cfDNA) is a common derivative found in body fluids such as saliva and blood and is present at a very low concentration (5–10 ng/ml) in healthy individuals (10). cfDNA levels were first demonstrated to be elevated in cancer patients in 1977 (11). While tissue biopsy sampling presents only a snapshot of the tumour at one time or location, the incorporation of CTCs and ctDNA has the potential to overcome tumour spatial and temporal heterogeneity and to provide real-time information relating to tumour burden. This review article highlights the recent advancements in the field of CTCs and ctDNA for the management of patients with NSCLC between 2018 and 2021. Circulating exosomes, microRNA, RNA, and tumour-educated platelets are other appealing tumour derivatives found in body fluids (12, 13) (Figure 1).
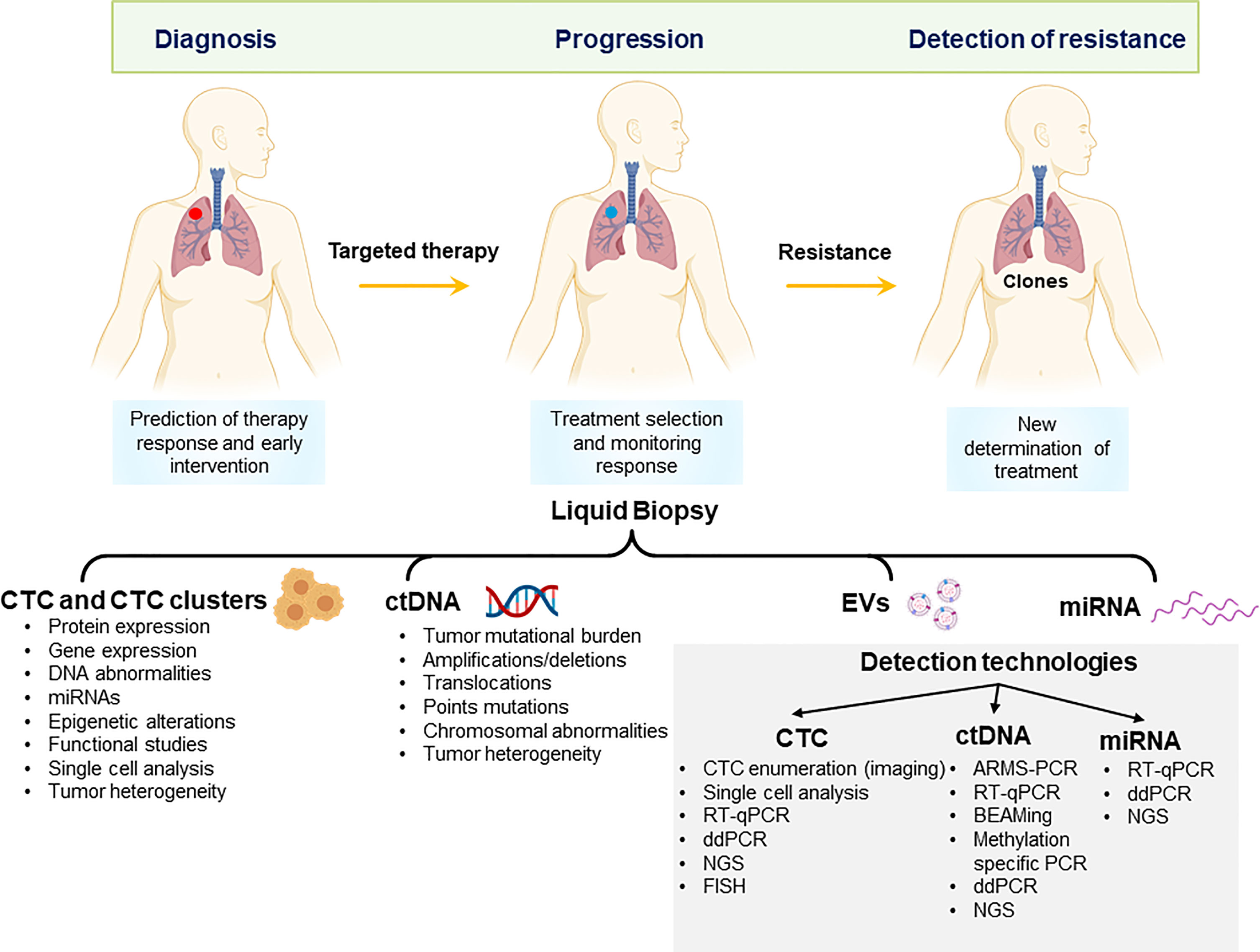
Figure 1 Potential applications of liquid biopsy in the management of non-small cell lung cancer. Liquid biopsy derivatives can be applied to early diagnosis, monitor of response to treatment and detection of resistance to treatment. CTCs, Circulating tumour cells and clusters; ctDNA, circulating tumour DNA; EV, extracellular vesicles; miRNA, microRNA; NGS, next-generation sequencing; ddPCR, digital droplet PCR; FISH, fluorescent in situ-hybridization.
Circulating Tumour Cells
Metastasis is a complex, multistage process which requires tumour cells to invade and move from the primary tumour into the circulation, intravasate, survive, extravasate into the bloodstream, and colonize at a distant site leading to a macroscopic metastatic lesion (14). CTCs represent an intermediate stage of metastasis. While rare (estimated to be as low as one to 10 cells per 10 ml of blood), they are uniquely accessible through simple non-invasive sampling of body fluids. To overcome the rarity of CTCs isolated from lung cancer patients’ blood samples, researchers have used pulmonary vein (PV) blood because tumour cells (besides the primary tumour cells) may circulate after passing through the PV (15–17). They found that CTCs were detected in 29 of 30 (96.7%) patients’ PV blood samples.
CTCs migrate as single cells (Figure 2A), clusters (a group of two or more CTCs, Figure 2B), and circulating tumour microemboli (CTM) (Figure 2C). CTM constitute cellular aggregates, which include platelets, stromal cells, and hematopoietic cells which ‘protect’ the tumour cells from undergoing apoptosis or being attacked by the immune system. As a result, CTM are likely to survive better in the bloodstream (19, 20). CTC clusters and CTM are subject to shear forces in the circulation with some of them having the ability to survive these forces (21). Therefore, they have shown to have a higher metastatic capacity compared to single CTCs (22, 23). In addition, CTCs that have undergone epithelial–mesenchymal transition (EMT) have shown to be associated with a poor survival in NSCLC patients (24–26).
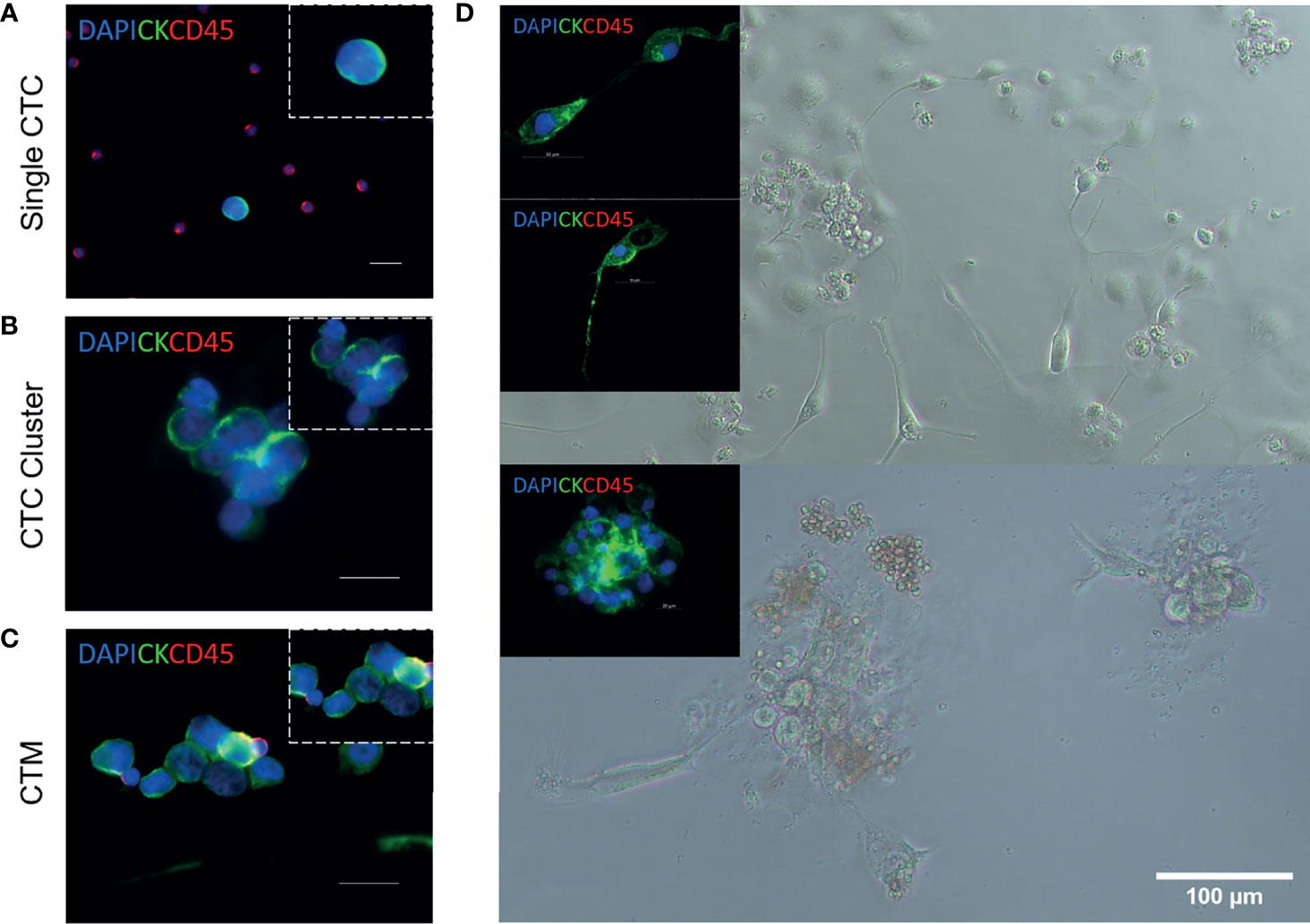
Figure 2 Circulating tumour cell isolation, expansion, and characterization from patients with NSCLC (18). Representative images of single circulating tumour cells (CTCs) (A), CTC clusters (B), and circulating tumour microemboli (CTM) (C). Immunofluorescent staining using anti-CK-FITC, anti-CD45-APC, and DAPI. Scale bar represents 20 μm. (D) Isolation, expansion, and characterization of CTCs from patients with advanced-stage NSCLC. Cultured CTCs derived from patient blood samples in a 96-well standard microplate. In-well staining of proliferating cells in culture at day 7. Immunofluorescent staining using anti-CK-FITC and anti-CD45-APC. Cells were shown to be CD45 negative.
Circulating Tumour Cell Technologies
Several technologies have been developed to capture CTCs, based on physical or biological properties of CTCs. The CellSearch® system remains the only FDA-approved technology for use in a clinical setting (27, 28) and has been validated as a prognostic and therapeutic tool for monitoring patients with metastatic colorectal (29), breast (28), and prostate cancer (30). CellSearch® is limited to capturing CTCs based on an epithelial phenotype, which is likely to underestimate other clinically relevant CTC phenotypes that have undergone EMT. To address this issue, numerous technologies have emerged as alternative platforms to capture CTCs in circulation. Technologies such as Parsortix, EPIC, RareCyte, and filtration have the potential to overcome the limitations of the CellSearch® system and can be utilized for downstream phenotyping or single-cell analysis (31). Microfluidics-based technologies enable the gentle capturing of CTCs, which then allow for further downstream analysis using cellular, microscopic, or molecular techniques (18, 32–37). Advances in integrated microfluidic platforms made possible the miniaturization of analytical techniques for automated, accurate, and high-throughput assays (38). Moreover, microfluidics presents an opportunity to integrate both isolation and detection in a single device, facilitating the development of true point-of-care diagnostic CTC devices (39). The microfluidic enrichment of CTCs can be utilized through positive [CTC-chip (32), GEDI chip (40)] or negative selection and biophysical properties [iChip (41), MCC (42), CROSS chip (43), spiral microfluidic biochip (44)]. Promising lab-on-a-chip approaches have been extensively reviewed in recent studies (38, 45). One of the drawbacks of microfluidic technology is the challenge of analysing large sample volumes to access key information about rare cells in the circulation.
A large number of isolation and enrichment methods have been used in the past decade with acceptable clinical sensitivity and specificity. However, there is discordance between various CTC isolation and enrichment platforms and cutoff thresholds for CTC enumeration in clinical applications. A study has compared the performance of two CTC detection systems based on the expression of the EpCAM antigen (CellSearch® assay) and on cell size (Isolation by Size of Epithelial Tumour Cells Technology ISET® assay). An underestimation of CTC numbers was determined when using the CellSearch® system in metastatic lung cancer patients (46, 47). A recent study by Papadaki and colleagues compared three CTC enrichment platforms (a. Ficoll density gradient centrifugation; b. ISET®, France; and c. Parsortix® technology, ANGLE plc, UK) and showed higher CTC counts when using the Parsortix system. Furthermore, comparison of various CTC enrichment approaches identified variations in CTC capture efficiency, disparities in the enrichment of distinct CTC subpopulations, and discordance in CTC counts (31, 48). Since CTCs are involved in tumour progression, they represent a window to identify mechanisms of metastases. Single CTC profiling may provide better understanding of the origin of tumour, the mechanisms of cancer metastasis, and the study of CTC heterogeneity (49). It is also known that single CTC sequencing and or analysis is important for the advancement of science as bulk analysis may not be representative of the actual tumour and may result in misinformation.
In a recent study by Zeinali et al., CTCs were detected in all 25 NSCLC patients (average of 417 CTC/ml) using a newly developed Labyrinth microfluidic device; they have also identified cells presenting mesenchymal phenotypes in 22 of 23 samples analysed (50). A study by Lim and colleagues was able to predict drug responsiveness in lung cancer patients based on the epithelial–mesenchymal transition (EMT) signature of baseline CTCs using a fluid-based separation FAST disc platform (51). This study highlights the importance of investigating the EMT phenotype on CTCs to investigate the dynamics of disease progression and predict the drug response in NSCLC. A recent study by Zhao et al. profiled the DNA methylome of CTCs from lung cancer patients and identified a unique CTC DNA methylation signature (52). Recent CTC studies in lung cancers are highlighted in Table 1.
Screening
The majority of lung cancer patients present with advanced disease; therefore, methods to facilitate earlier detection of lung cancer are of high importance. Primary methods of cancer prevention and early detection rely on the practice of screening and are currently available for several tumour types, particularly for colorectal, breast, and prostate cancer (72). Low-dose chest CT (LDCT) is currently used to screen individuals at a high risk of developing lung cancer. LDCT has shown to reduce mortality. However, LDCT as a screening tool has a high false-positive rate (73–75). Blood-based biomarkers might act as a standalone screening tool, refine the selection of patients at risk, or help to classify undetermined nodules detected on LDCT (76). Several published studies have validated CTC isolation using the ISET® technology with high sensitivity and specificity for many cancer types, including NSCLC (46, 47, 76, 77). Ilie et al. in 2014 used the ISET device to diagnose lung cancer nodules earlier in high-risk individuals. CTCs were detected in 3% (5 out of 168 patients) of the patients with chronic obstructive pulmonary disease (COPD) included in the study (76). Furthermore, the annual surveillance of the CTC-positive patients by CT-scan screening was able to detect lung nodules 1 to 4 years after CTC detection, potentially identifying lung cancers at an earlier stage. Interestingly, follow-up of the five patients by CT scan and ISET 12 months after surgery showed no tumour recurrence. In contrast, a prospective, multicentre cohort study by Charles-Hugo Marquette and colleagues enrolled 614 participants from 21 French university centres to test the performance of ISET to capture CTCs. Participants with chronic obstructive pulmonary disease (COPD) had three rounds of screening at 1-year intervals. Subsequently, participants underwent low-dose computed tomography (LDCT) and a matched blood test to detect CTCs using the ISET® platform. The authors concluded that the ISET® was not able to predict lung cancer or extrapulmonary cancer development (78). This is because the ISET® platform showed a low sensitivity to be used as a reliable platform to screen individuals at a higher risk of developing lung cancer. While age, smoking, and gender were comparable to the previous study by Ilie et al. in 2014, the baseline prevalence of lung cancer in the present study was about three times higher than in other studies. This study identified the presence of COPD as an independent risk factor (2–4 times higher in COPD patients than those without COPD) for lung cancer development in patients without clinically detectable lung cancer. The authors also suggest the use of alternative methods for CTC enrichment and detection which may provide better suitability for screening purposes (79). They concluded that a holistic approach of screening integrating clinical, biological, and radiological signatures may be the way forward for a lung cancer screening program.
Frick et al. in 2020 utilized a novel telomerase-based CTC assay and found that patients with elevated counts of CTCs prior to the start of stereotactic body radiotherapy (SBRT), as well as those patients whose CTCs remained persistently detectable after SBRT, were associated with increased regional and distant recurrence. Use of this CTC assay may translate clinically by helping to identify subsets of patients who may maximally benefit from systemic therapy after SBRT for early-stage NSCLC, and to help monitor for tumour recurrence or progression (61).
A study by Duan and colleagues used CTC as a prognostic biomarker to discriminate benign vs. malignant nodules as a means of early diagnosing lung cancer. They used a group of 44 patients and subcategorized them based on their pathological results and found CTC detection rates to be increased with the invasiveness of the nodules. While promising, this patient cohort was small and future prospective studies with larger cohorts are needed to confirm the benefits of this technique for detecting early-stage lung cancer nodules, especially in individuals at a higher risk of developing lung cancer (60). Further to this, studies evaluating screening potential of CTCs should also take into consideration the risk of the target population (80). It is difficult to verify that a single blood-based analyte will provide sufficient sensitivity and specificity for lung cancer screening, and therefore, it is likely that a combination approach may overcome the current challenges.
CTCs as Prognostic Biomarkers
A decrease in CTC counts after surgery and/or chemotherapy may indicate cancer remission while an increase in CTC counts may predict cancer progression. Changes in CTC counts provide important prognostic information (early detection of recurrence or relapse prior to clinical signs) for lung cancer patients and have been associated with poor outcomes with shorter disease-free survival (DFS) (81, 82), progression-free survival (PFS) (83), and overall survival (OS) (7, 84, 85). Recently, a meta-data analysis by Liang and colleagues using five studies involving 351 patients demonstrated the prognostic utility of postoperative liquid biopsy for early tumour recurrence and poor prognosis for DFS in NSCLC patients (86). A recent multicentre European study using the CellSearch® platform using 550 patients demonstrated that CTC counts of ≥2 and ≥ 5 per 7·5 ml are associated with reduced progression-free survival (PFS) and overall survival (OS), respectively (57). They concluded that CTCs can be used as independent prognostic indicators of PFS and OS.
Monitoring of Response to Treatment Using Circulating Tumour Cells
Monitoring CTC counts during therapy allows assessment of disease development in real time and in some cases may be demonstrated prior to obvious clinical signs of relapse. A decreased CTC count after surgery and/or chemotherapy may indicate cancer remission while an increased count may depict cancer progression. Despite the potential value, clinicians do not utilize CTC information to determine treatment decisions. One of the reasons for this is the lack of a standardized methodology for CTC enrichment and detection, the rarity of CTCs in circulation, and the lack of technology that can be easily integrated into a clinical setting (85, 87). Furthermore, optimal CTC cutoff values vary across isolation methods and clinical trials and therefore require standardization (88). However, there is currently no CTC assay in clinical use for monitoring response to therapy in lung cancer patients due to inconclusive clinical utility data.
In a prospective study with stage IV NSCLC patients (n = 81), 63% of patients initiating therapy had a change in CTC counts and was predictive of survival in patients receiving chemotherapy (89, 90). In a study by Huang et al., an EpCAM-independent method of subtraction enrichment and immunostaining-fluorescence in situ hybridization (SE-iFISH) (21) was used to enrich and characterize aneuploid CTCs in patients with surgically resectable NSCLC undergoing neoadjuvant chemotherapy (NAC). By monitoring CTCs in real time during the process of NAC, the correlations between CTC enumeration and karyotype with radiological and pathological responses were identified and investigated. The findings conclude that karyotyping aneuploid CTCs can serve as a surrogate marker for disease monitoring in NSCLC (63).
A study by Shishido and colleagues analysed a subset of patients to determine the significance of a high-definition single-cell assay (HD-SCA), to detect CTCs in stage IV NSCLC patients at the initiation of therapy. This non-enrichment-based workflow detects CTCs based on morphology which are identified as high-definition CTCs (HD-CTCs). CTC positivity based on the HD-SCA workflow is inclusive of the cellular morphology of all nucleated cells from the liquid biopsy. Positive HD-CTCs were identified in 51 (62.96%) of patients with a median of 2.20 (range 0–509.20) prior to the start of therapy (90). Patients with an increasing CTC count during the first 3 months post initiation of new treatment had better PFS and OS compared to the other groups. They also found a weak correlation between the absolute number of HD-CTCs at a single time point of therapy and patient outcomes (OS p value = 0.0754).
A case report by Horton et al. presented a 68-year-old male with stage III NSCLC whose primary tumour showed a response to chemoradiotherapy on CT imaging with no evidence of metastatic disease; however, an elevated CTC count was observed post treatment. The patient was later found to have liver metastasis at 3 months on routine imaging. Therefore, an elevated CTC count may have better sensitivity in detecting microscopic residual cancer following curative intent therapy (91).
Despite evidence that monitoring for CTCs during disease progression may provide predictive information (7, 85, 92), its uptake by the clinical community is low. To achieve clinical implementation, standardization of platforms and protocols for CTC isolation and characterization, optimal CTC cutoff values based on the cancer type and stage, and the need for multicentre, large prospective clinical trials are needed in the future (88).
Therapeutic Implications of CTCs
In the last decade, tyrosine kinase inhibitors (TKIs) have revolutionized the management of NSCLC patients who harbour oncogenic drivers (93). NSCLC, particularly adenocarcinoma, has been separated into molecular subgroups based on oncogenic driver alterations. Epidermal growth factor receptor (EGFR)-activating alterations (exon 19 deletion (E19del) and exon 20 insertion mutation) and anaplastic lymphoma kinase (ALK) fusion rearrangements are the two most studied driver genes for targeting patients for TKI therapies (94). Additionally, KRAS gene mutations and MET signalling are widely recognized alterations with important roles in both biological mechanisms and clinical sensitivity to lung cancer treatment (95). First-line EGFR mutation inhibitors include gefitinib and erlotinib. They have shown sensitivity in patients harbouring EGFR-activating mutations (L858R point mutation in exon 21 and exon 19 deletions). Approximately, 5% of NSCLC patients have these alterations (96). Furthermore, uncommon EGFR mutations collectively account for 10% of mutations (exons 18–21) with clinically variable responses to TKI therapies in NSCLC patients (97). Acquired resistance to first-line EGFR TKIs is common within 12 months of treatment, mainly due to the T790M mutation, which is detected in approximately 60% of patients (96, 98, 99). Rearrangements in the gene encoding ALK commonly involve the echinoderm microtubule-associated protein-like 4 (EML4) loci characterizing a unique molecular subgroup (4%) of NSCLC patients (100). ALK inhibitors such as crizotinib and alectinib are currently used for ALK-rearranged NSCLC (101). Despite advancements in TKI therapies, acquired resistance occurs in most cases. Limited tumour availability and heterogeneity within the primary site or between primary and metastatic sites are the major hurdles in identifying genetic alterations and selection of patients who are eligible for TKI therapies.
The ability of obtaining tumour cells through a simple blood draw of cancer patients allows for minimally invasive methods to monitor disease progression, treatment selection, and in the case of resistance to tailor treatment modification. Rihawi and colleagues have found the presence of ALK rearrangement coupled to MYC amplification in tumour and CTCs from the same patient, suggesting a role for MYC for primary resistance to crizotinib (102). Furthermore, a study by Pailler et al. highlights the genetic heterogeneity and clinical benefit of CTCs in identifying therapeutic resistance mutations in ALK-rearranged patients (103). Several mutations were detected in CTCs of crizotinib-resistant patients including EGFR, KRAS, BRAF, and TP53 (103). These findings suggest that CTCs provide clinically relevant molecular information and can be used in clinical practice as an alternative to traditional biopsy. Early detection of resistance and identification of acquired mutations in patients undergoing treatment are novel, given that sites of disease can be difficult to access/biopsy and therefore potential resistance mutations may be missed.
Clinical Significance of PD-L1 Expression on CTCs
NSCLC patients treated with immune checkpoint inhibitors display durable responses in a subset of patients, but it is currently difficult to predict which patients will benefit from this expensive treatment using current tumour tissue biomarkers, such as the protein expression levels of programmed death ligand 1 (PD-L1). PD-L1 is currently the most commonly used method for predicting response to immune checkpoint inhibitors, but patients who express low or negative PD-L1 may still benefit from treatment. PD-L1 expression heterogeneity between primary and metastatic tumours as well as dynamic fluctuations at different time points creates uncertainty in relying on tumour tissue expression of PD-L1 for treatment selection, particularly for patients whose tumours harbour EGFR mutations (104, 105).
Overcoming tumour tissue heterogeneity and difficulties in obtaining longitudinal tumour samples has led researchers to focus on evaluating PD-L1 expression on CTCs for predictive and real-time monitoring of immune cell activation (106). The presence of PD-L1-positive CTCs has shown to correlate with the expression of EMT on CTCs, indicating a partial EMT phenotype (107). Recent studies have been hampered by a small number of patients, and as such data have been inconclusive. Therefore, multicentre prospective trials are required to support the potential clinical utility of PD-L1 expression levels on CTCs. A summary of the recent studies evaluating PD-L1 expression on CTCs in lung cancer is depicted in Table 2. However, integration of immunotherapies and immune checkpoint blockades targeting either the programmed cell death protein 1 (PD-1) or PD-L1 has been incorporated into routine clinical management of patients with NSCLC and has significantly improved patient outcomes (118–120). PD-L1 is a dynamic marker that has been shown to switch from negative to positive over the course of chemotherapy/radiotherapy. The increased expression of PD-L1 during chemotherapy or radiotherapy can be used as a predictor of benefit to immunotherapy, with clinical findings demonstrating efficacy in combining PD-1/PD-L1 inhibitors with chemotherapy and/or radiotherapy for improved therapeutic outcomes (121). Precision medicine is contributing significantly to improving life expectancy in a subset of patients with advanced NSCLC; however, intra-tumour heterogeneity and acquired resistance are known to significantly impact targeted-agent sensitivity (122). While PD-1/PD-L1 checkpoint inhibitors provide great benefit, randomized studies have known a lack of efficacy for single-agent checkpoint inhibitors (123). The addition of pembrolizumab to standard first-line chemotherapy provided significant survival benefit for EGFR/ALK wild-type patients (124). Evidence suggests that targeted therapy in combination with immunotherapy may benefit as a complementary approach for treatment of NSCLC patients.
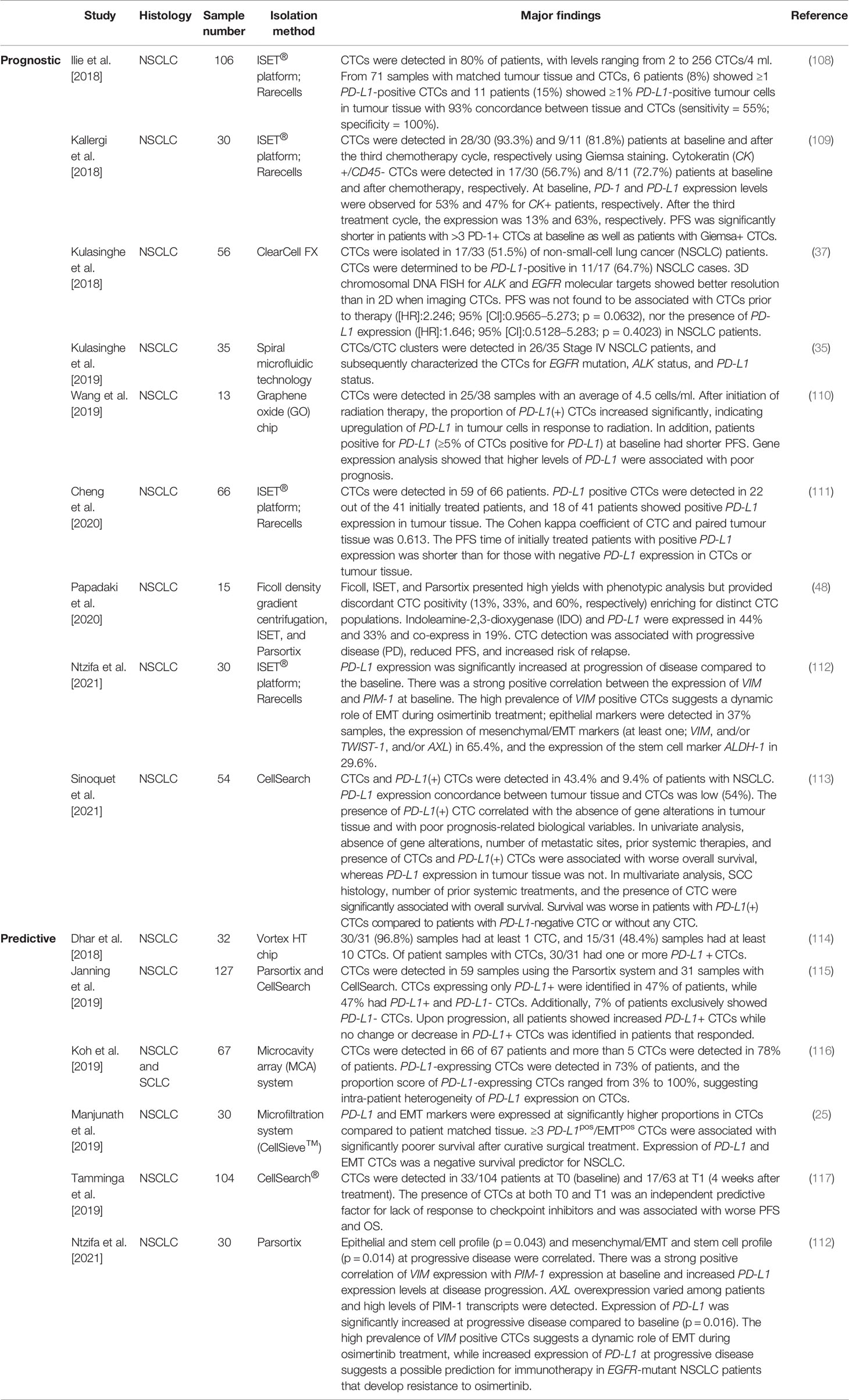
Table 2 Recent studies evaluating the expression of PD-L1 on lung cancer-derived circulating tumour cells.
Ex-Vivo Expansion of Circulating Tumour Cells
The expansion of CTCs ex-vivo provides a novel disease model to better understand metastasis and to identify drug susceptibility in a preclinical setting (Figure 2D). With the inevitable emergence of acquired drug resistance, preclinical models are becoming increasingly novel for individualized treatment. In recent years, several studies have developed methods to propagate CTCs outside of a patient’s body; however, optimal culture conditions are yet to be established. There has been limited success in immortalizing CTC cultures as CTC cell lines long-term, especially for NSCLC (7). Recent studies with successful short-term and long-term expansion are highlighted in Table 3.
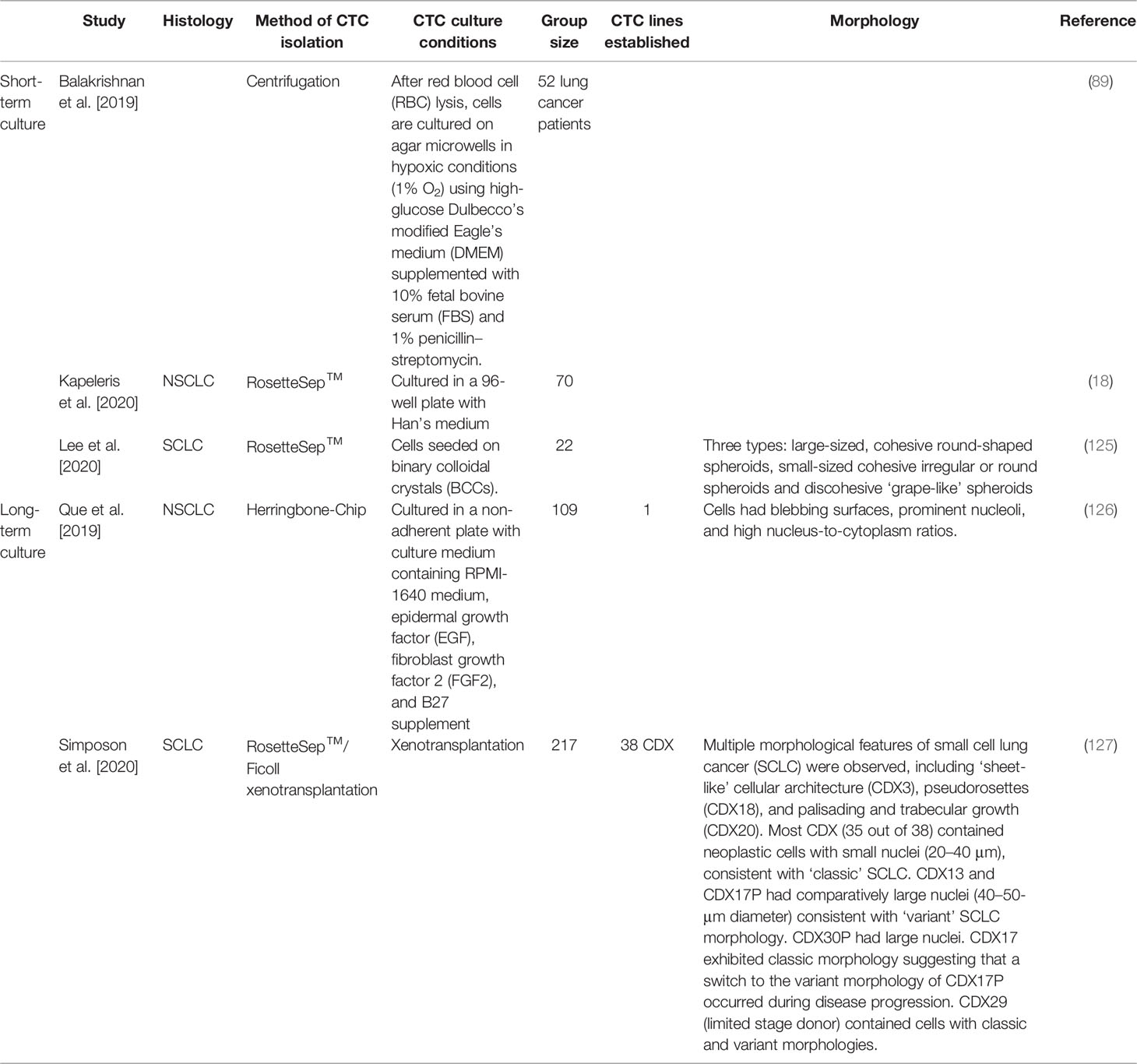
Table 3 Circulating tumour cell cultures derived from blood samples obtained from lung cancer patients.
Current studies have shown promise in ex-vivo cultures of CTCs; however, the success rate is still low, likely due to the small number of isolated CTCs from cancer patients’ blood sample. Higher success rates are observed when CTCs are derived from advanced-stage cancer patients reflecting higher tumour burden. While preclinical models have shown their potential, these models could be complementary to ex-vivo CTC cultures. Especially in patients where biopsies are difficult to obtain, CTC-derived explants (CDXs) provide an alternative source. In contrast, patient-derived explants (PDXs) could be utilized where more tumour tissue is available. CDXs can be derived from CTCs collected at different time points during patient follow-up, allowing the generation of paired models that recapitulate the patients’ tumour evolution (128).
With the rising interest in genomic profiling of CTCs to identify driver mutations and possible drug targets to alleviate drug resistance, expansion of CTCs in large quantities will enable identification of novel drug targets and methods for preclinical drug sensitivity testing. The successful short-term expansion of CTCs presents a novel opportunity to test therapies and to conduct functional analysis. However, in recent years, only a few short-term CTC cultures have been reported. Balakrishnan and colleagues cultured CTCs in laser-ablated microwells and reported that ex vivo, CTC cluster formation correlates with patients’ response to treatment (89). A recent study by the same authors was successful in expanding short-term CTC cultures from 9 NSCLC patients. They confirmed the presence of lung cancer-specific somatic mutations in CTC cultures using whole exome sequencing (WES) (18).
There has been limited success in long-term CTC cultures derived from NSCLC patients’ blood samples. Que et al. successfully established a CTC cell line from a patient with NSCLC where they tested chemotherapeutic drugs such as docetaxel and cisplatin. The authors identified an increased resistance to their CTCs compared to A549 and 95-D cell lines (126). CDX models bring promise to advancing cancer therapy (129). Expanding CTCs in vitro and in vivo show promise in better understanding tumour heterogeneity and to predict therapeutic responses; however, clinical trials are needed to validate clinical utility. Future studies should focus on the development of models that consider the tumour microenvironment for a more representative approach.
Circulating Cell-Free DNA
Like CTCs, the detection of cfDNA provides opportunity for screening, monitoring, treatment evaluation, and disease surveillance (130–134). cfDNA refers to small DNA fragments circulating in the blood that are released from apoptotic and/or necrotic cells (135). Increasing evidence has highlighted the clinical utility of detecting mutations in cfDNA, and the amount of cfDNA in circulation has been correlated with the tumour burden. In addition, cfDNA has been used to detect EGFR mutations (exon 19 deletion or exon 21 (L858R) mutations) in selecting NSCLC cancer patients who may benefit from treatment with EGFR-TKIs and in identifying drug resistance mutations in lung cancer patients (11, 136–143).
In 2016, the FDA approved the ctDNA assay Cobas® EGFR Mutation Test v2 (Roche Diagnostics) for detection of EGFR mutations in NSCLC patients as a companion diagnostic to the EGFR inhibitor erlotinib. In addition, this year, the FDA also approved the Guardant Health’s Guardant360® companion diagnostic (CDx) liquid biopsy as the first pan-cancer, next-generation sequencing (NGS)-based comprehensive tumour mutation profiling test in patients with any solid cancer tumour. This is also used as a CDx to detect EGFR mutations in NSCLC patients who may benefit from treatment with AstraZeneca’s Tagrisso® (osimertinib) (144, 145). The recent TRACERx ongoing multicentre cohort study of 842 patients with NSCLC aims at tracking tumour evolution through longitudinal sampling and sequencing.
The role of ctDNA in molecular diagnosis and disease monitoring was investigated, detecting clonal and subclonal mutations which were present in matched tumour samples (146). The APPLE trial is a multicentre, 3-arm, phase II study evaluating a ctDNA test specific for T790M on a cohort of treatment-naïve EGFR-mutant NSCLC patients. The study aims at utilizing ctDNA to identify the optimal approach for sequencing of treatment with gefitinib and osimertinib in advanced NSCLC patients (147). ROS-1 rearranged in NSCLC, like other oncogene-driven cancers, will eventually develop resistance, highlighting the need for novel ROS-1 inhibitors (148). Molecular analysis of plasma samples from ROS-1-positive NSCLC patients identified seven distinct fusion partners representative of the primary tissue (149). Currently, the most common cfDNA detection platforms are next-generation sequencing and digital droplet PCR (ddPCR) (150–152). Genotyping cfDNA is a fast and accessible possibility to provide insights into tumour heterogeneity and detection of resistance, can allow non-invasive monitoring of disease, and has led to the development of technologies such as droplet ddPCR and NGS. ddPCR and NGS have shown high sensitivity and concordance to detect activating EGFR mutations and T790M mutations (153, 154). The NGS-based profiling of NSCLC parents recently has shown robustness in assessing KRAS (155) and ALK (156) mutational status in circulating DNA and may be valuable in future therapy decision making. The KWAY project assessed the economic sustainability of NGS technology of five Italian referral centres. Results highlighted that the adoption of NGS resulted in reductions of the overall cost of testing per patient (157). NGS enables the analysis of different biomarkers in different patients at the same time and can provide clinically relevant information both before and after targeted treatment, thus assisting treatment decision making in clinical practise (158, 159). Microfluidic-based strategies and lab-on-a-chip (LOC) devices for extraction of nucleic acids provide an opportunity for high-throughput screening with reduced sample volume and rapid quantification (38, 160, 161). An overview of current microfluidic technologies for cfDNA isolation and analysis has been recently reported (162). To demonstrate the clinical utility of ctDNA screening tests, prospective clinical trials comparing early detection strategies based on the ctDNA test with the standard of care are required.
GRAIL is currently conducting a study known as the Circulating Cell-Free Genome Atlas (CCGA) using an NGS approach to develop a reference library of mutations in the blood of patients with common cancers (163, 164). Recently, a novel technique ARMS-PCR has been reported to be highly sensitive and specific, providing a promising approach for the detection of EGFR T790 mutation in plasma of cfDNA (165, 166). The use of methylation-specific PCR that profiles epigenetic alterations has also gained attention in recent years. As ctDNA methylation occurs at early stages in lung carcinogenesis, it may assist in early diagnosis (167, 168).
ctDNA analyses provide an advantage with reduced costs and less complex extraction methods of tumour-derived nucleic acids compared to CTCs; however, in some cases, due to the uncertainty of the origin of cfDNA, they may provide inadequate and inaccurate tumour information. The emergence of phenotypic switching in drug resistance via non-genetic mechanisms signifies a major obstacle to treatment success (169). CTCs may present a larger research scope than ctDNA as they can maintain the intact genome of living cells and may provide significant information on tumour heterogeneity that ctDNA is unable to provide (130). Krug and colleagues have found higher sensitivity of detecting EGFR mutations in plasma when combining exosomal RNA (exoRNA) (98%) and ctDNA (90%) (170). A higher sensitivity for the detection of T790 mutation combining exoRNA and cfDNA (92%) compared to tumour biopsy (89%) confirms the potential clinical utility of liquid biopsy (171).
Additional Tumour-Derived Biomolecules
Circulating tumour cell-derived biomolecules such as exosomes and microRNA (miRNA) have shown promise as prognostic/predictive biomarkers. Exosomes are a subtype of extracellular vehicles (EVs) comprising nucleic acids, lipids, and metabolites and have been shown to play a role in facilitating tumorigenesis (171). Exosomes are released by all types of cells into extracellular space and once released can act as messengers, gaining traction as potential drug carriers and candidate biomarkers (172).
Circulating RNA molecules in particular miRNA have recently gained attention as potential biomarkers. miRNAs are short non-coding single-stranded RNA molecules (containing about 22 nucleotides) that regulate gene expression at the post-transcriptional level (173). Several studies have shown that circulating miRNAs may reflect tumour biology, with their expression relative to tumour development, progression, and metastases (174). There has been emphasis on their use as a screening tool to predict prognosis and therapy as well as predictors of survival in early-stage patients or patients with metastatic disease (175, 176). The use of exosomes and miRNA show promising results; however, major challenges with variability and lack of standardization have hampered their clinical application. In future, CTCs and ctDNA with the inclusion of additional sources of tumour biomarkers such as exosomes or miRNA may provide a complementary approach, thereby increasing the precision of information obtained through liquid biopsy (Figure 1).
Future Perspective
There is increasing evidence supporting the prognostic utility of CTCs in a number of solid tumour types. CTCs provide a mechanism to investigate tumour biology and to test and develop existing and novel biomarker-driven drugs, leading to precision medicine-based approaches for managing cancer patients (37, 177–180). Clinical implementation of targeted therapies has been hindered due to its accessibility and availability, quality, and quantity of tumour tissue from patients (181). In addition, tumour heterogeneity poses another challenge as a single biopsy may not represent the entire genomic landscape of a patient’s tumour (182). Integration of immune checkpoint inhibitors into clinical practice has presented a new era in immunotherapy; however, only a portion of patients respond to this form of therapy with some patients exhibiting immune-related adverse reactions (183). Recent clinical trials have shown that targeted therapies have improved PFS and OS compared to chemotherapy or immunotherapy (184–187). Identification of predictive biomarkers is an unmet clinical need to stratify patients who may benefit from immunotherapy or combination approaches.
Isolation and enrichment of CTCs remains as one of the main challenges that need to be addressed before CTC workflow can be implemented in a clinical setting (188). Inconsistencies between CTC data across similar patient cohorts and multiple CTC detection platforms, as well as the difficulty in reproducing CTC studies, hinder the translation of CTC into clinics (189). Successful ex-vivo short-term cultures of CTCs and long-term establishment of CTC cell lines provide a powerful tool to model tumour heterogeneity in vitro at clinically relevant time points and enable preclinical personalized drug testing. Furthermore, future application of CTCs may assist patients through the entire course of disease from diagnosis through to treatment selection, monitoring, and follow-up assessment. A recent meta-analysis evaluated the diagnostic accuracy for CTCs for the clinical determination of lung cancer assessment in 21 studies with 3,997 subjects. The pooled sensitivity and specificity were 0.72 (95% CI: 0.65–0.79) and 0.96 (95% CI: 0.91–0.98), respectively, and the pooled positive and negative likelihood ratios were 16.86 (95% CI: 7.65–37.12) and 0.29 (95% CI: 0.23–0.37), respectively. Zhao and colleagues concluded that CTCs had good diagnostic value for detecting lung cancer (190).
The involvement of ctDNA mutation data to predict acquired treatment resistance and to facilitate therapeutic decision making is becoming increasingly popular (191, 192). This is due to its ease of use, analysis being less expensive, addressing of intra-tumour heterogeneity, and high sensitivity for detecting tumour burden potential. As an example, the androgen receptor splice variant 7 (AR-V7) CTC liquid biopsy test by Epic Sciences is the first clinically validated test for castration-resistant prostate cancer (mCRPC) (193). Currently, no similar tests exist for NSCLC.
A recent meta-data study analysed the diagnostic utility of both CTCs and ctDNA for number of gene mutations in lung cancer, highlighting improved diagnostic performance compared to ctDNA analysis alone (150). While CTCs hold promise as prognostic and predictive biomarkers for the management of patients with lung cancer, the lack of standardised CTC isolation and enrichment platforms, the discrepancy of available data, and the lack of agreed cutoff levels make CTCs more difficult to be integrated into clinical practise (194). Future focus on harmonization of studies and data sets may improve the diagnostic and prognostic utility of CTCs (195). Public–private partnerships such as CANCER-ID (https://www.cancer-id.eu/) and the US-based Blood Profiling Atlas in Cancer (BloodPAC) consortium (https://www.bloodpac.org/) are focused on standardizing methods and technologies for circulating blood-based biomarkers. The integration of a complementary approach using additional liquid biopsy derivatives such as ctDNA and exosomes may provide a more comprehensive representation of the tumour genomic landscape, thereby overcoming current challenges with tumour heterogeneity and identifying new targetable mutations (130). Large sample, multicentre cohort, and prospective clinical trials will advance the current understanding of tumour evolution and its effect on cancer biology and patient outcomes.
Author Contributions
Idea: JK, KO’B, CP. Preparation of figures and tables: JK, ME. All authors were involved in the preparation, review, and editing of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
CP is funded by Cancer Australia (APP 1145657), a NHMRC Ideas Grant (APP 2002576), and the Royal Brisbane Women’s Hospital Foundation and National Institute of Health.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer Statistics, 2022. CA: A Cancer J Clin (2022) 72(1):7–33. doi: 10.3322/caac.21708
2. Available at: https://www.who.int/news-room/fact-sheets/detail/cancer.
3. Sun S, Schiller JH, Gazdar AF. Lung Cancer in Never Smokers–a Different Disease. Nat Rev Cancer (2007) 7(10):778–90. doi: 10.1038/nrc2190
4. Yuan M, Huang L-L, Chen J-H, Wu J, Xu Q. The Emerging Treatment Landscape of Targeted Therapy in Non-Small-Cell Lung Cancer. Signal Transduct Target Ther (2019) 4(1):61. doi: 10.1038/s41392-019-0099-9
5. Herbst RS, Morgensztern D, Boshoff C. The Biology and Management of Non-Small Cell Lung Cancer. Nature (2018) 553:446. doi: 10.1038/nature25183
6. Gerlinger M, Rowan AJ, Horswell S, Math M, Larkin J, Endesfelder D, et al. Intratumor Heterogeneity and Branched Evolution Revealed by Multiregion Sequencing. N Engl J Med (2012) 366(10):883–92. doi: 10.1056/NEJMoa1113205
7. Kapeleris J, Kulasinghe A, Warkiani ME, Vela I, Kenny L, O'Byrne K, et al. The Prognostic Role of Circulating Tumor Cells (CTCs) in Lung Cancer. Front Oncol (2018) 8:311. doi: 10.3389/fonc.2018.00311
8. Guibert N, Pradines A, Favre G, Mazieres J. Current and Future Applications of Liquid Biopsy in Nonsmall Cell Lung Cancer From Early to Advanced Stages. Eur Respir Rev (2020) 29(155). doi: 10.1183/16000617.0052-2019
9. Mandel PPM. Les Acides Nucléiques Du Plasma Sanguin Chez L’homme [Nuclear Acids In Human Blood Plasma]. Acad Sci Paris (1948) 142:241–3.
10. El Messaoudi S, Rolet F, Mouliere F, Thierry AR. Circulating Cell Free DNA: Preanalytical Considerations. Clin Chim Acta (2013) 424:222–30. doi: 10.1016/j.cca.2013.05.022
11. Leon SA, Shapiro B, Sklaroff DM, Yaros MJ. Free DNA in the Serum of Cancer Patients and the Effect of Therapy. Cancer Res (1977) 37(3):646–50.
12. Freitas C, Sousa C, Machado F, Serino M, Santos V, Cruz-Martins N, et al. The Role of Liquid Biopsy in Early Diagnosis of Lung Cancer. Front Oncol (2021) 11:634316. doi: 10.3389/fonc.2021.634316
13. Zhou B, Xu K, Zheng X, Chen T, Wang J, Song Y, et al. Application of Exosomes as Liquid Biopsy in Clinical Diagnosis. Signal Transduct Target Ther (2020) 5(1):144. doi: 10.1038/s41392-020-00258-9
14. Mittal V. Epithelial Mesenchymal Transition in Tumor Metastasis. Annu Rev Pathol (2018) 13:395–412. doi: 10.1146/annurev-pathol-020117-043854
15. Okumura Y, Tanaka F, Yoneda K, Hashimoto M, Takuwa T, Kondo N, et al. Circulating Tumor Cells in Pulmonary Venous Blood of Primary Lung Cancer Patients. Ann Thorac Surg (2009) 87(6):1669–75. doi: 10.1016/j.athoracsur.2009.03.073
16. Murlidhar V, Reddy RM, Fouladdel S, Zhao L, Ishikawa MK, Grabauskiene S, et al. Poor Prognosis Indicated by Venous Circulating Tumor Cell Clusters in Early-Stage Lung Cancers. Cancer Res (2017) 77(18):5194–206. doi: 10.1158/0008-5472.CAN-16-2072
17. Crosbie PAJ, Shah R, Krysiak P, Zhou C, Morris K, Tugwood J, et al. Circulating Tumor Cells Detected in the Tumor-Draining Pulmonary Vein Are Associated With Disease Recurrence After Surgical Resection of NSCLC. J Thorac Oncol (2016) 11(10):1793–7. doi: 10.1016/j.jtho.2016.06.017
18. Kapeleris J, Kulasinghe A, Warkiani ME, Oleary C, Vela I, Leo P, et al. Ex Vivo Culture of Circulating Tumour Cells Derived From Non-Small Cell Lung Cancer. Transl Lung Cancer Res (2020) 9(5):1795–809. doi: 10.21037/tlcr-20-521
19. Dive C, Brady G. SnapShot: Circulating Tumor Cells. Cell (2017) 168(4):742–742.e1. doi: 10.1016/j.cell.2017.01.026
20. Szczerba BM, Castro-Giner F, Vetter M, Krol I, Gkountela S, Landin J, et al. Neutrophils Escort Circulating Tumour Cells to Enable Cell Cycle Progression. Nature (2019) 566(7745):553–7. doi: 10.1038/s41586-019-0915-y
21. Wang W-C, Zhang X-F, Peng J, Li X-F, Wang A-L, Bie Y-Q, et al. Survival Mechanisms and Influence Factors of Circulating Tumor Cells. BioMed Res Int (2018) 2018:6304701. doi: 10.1155/2018/6304701
22. Aceto N, Bardia A, Miyamoto DT, Donaldson MC, Wittner BS, Spencer JA, et al. Circulating Tumor Cell Clusters Are Oligoclonal Precursors of Breast Cancer Metastasis. Cell (2014) 158(5):1110–22. doi: 10.1016/j.cell.2014.07.013
23. Aceto N, Toner M, Maheswaran S, Haber DA. En Route to Metastasis: Circulating Tumor Cell Clusters and Epithelial-To-Mesenchymal Transition. Trends Cancer (2015) 1(1):44–52. doi: 10.1016/j.trecan.2015.07.006
24. Lecharpentier A, Vielh P, Perez-Moreno P, Planchard D, Soria JC, Farace F. Detection of Circulating Tumour Cells With a Hybrid (Epithelial/Mesenchymal) Phenotype in Patients With Metastatic Non-Small Cell Lung Cancer. Br J Cancer (2011) 105(9):1338–41. doi: 10.1038/bjc.2011.405
25. Manjunath Y, Upparahalli SV, Avella DM, Deroche CB, Kimchi ET, Staveley-O'Carroll KF, et al. PD-L1 Expression With Epithelial Mesenchymal Transition of Circulating Tumor Cells Is Associated With Poor Survival in Curatively Resected Non-Small Cell Lung Cancer. Cancers (2019) 11(6):806. doi: 10.3390/cancers11060806
26. Milano A, Mazzetta F, Valente S, Ranieri D, Leone L, Botticelli A, et al. Molecular Detection of EMT Markers in Circulating Tumor Cells From Metastatic Non-Small Cell Lung Cancer Patients: Potential Role in Clinical Practice. Anal Cell Pathol (2018) 2018:3506874. doi: 10.1155/2018/3506874
27. Andree KC, van Dalum G, Terstappen LWMM. Challenges in Circulating Tumor Cell Detection by the Cell Search System. Mol Oncol (2016) 10(3):395–407. doi: 10.1016/j.molonc.2015.12.002
28. Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, et al. Circulating Tumor Cells, Disease Progression, and Survival in Metastatic Breast Cancer. N Engl J Med (2004) 351(8):781–91. doi: 10.1056/NEJMoa040766
29. Cohen SJ, Punt CJA, Iannotti N, Saidman BH, Sabbath KD, Gabrail NY, et al. Relationship of Circulating Tumor Cells to Tumor Response, Progression-Free Survival, and Overall Survival in Patients With Metastatic Colorectal Cancer. J Clin Oncol (2008) 26(19):3213–21. doi: 10.1200/JCO.2007.15.8923
30. de Bono JS, Scher HI, Montgomery RB, Parker C, Miller MC, Tissing H, et al. Circulating Tumor Cells Predict Survival Benefit From Treatment in Metastatic Castration-Resistant Prostate Cancer. Clin Cancer Res (2008) 14(19):6302–9. doi: 10.1158/1078-0432.CCR-08-0872
31. Ferreira MM, Ramani VC, Jeffrey SS. Circulating Tumor Cell Technologies. Mol Oncol (2016) 10(3):374–94. doi: 10.1016/j.molonc.2016.01.007
32. Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, et al. Isolation of Rare Circulating Tumour Cells in Cancer Patients by Microchip Technology. Nature (2007) 450(7173):1235–9. doi: 10.1038/nature06385
33. Dong Y, Skelley AM, Merdek KD, Sprott KM, Jiang C, Pierceall WE, et al. Microfluidics and Circulating Tumor Cells. J Mol Diagn (2013) 15(2):149–57. doi: 10.1016/j.jmoldx.2012.09.004
34. Warkiani ME, Khoo BL, Wu L, Tay AKP, Bhagat AAS, Han J, et al. Ultra-Fast, Label-Free Isolation of Circulating Tumor Cells From Blood Using Spiral Microfluidics. Nat Protoc (2016) 11(1):134–48. doi: 10.1038/nprot.2016.003
35. Kulasinghe A, Kapeleris J, Cooper C, Warkiani ME, O'Byrne K, Punyadeera C. Phenotypic Characterization of Circulating Lung Cancer Cells for Clinically Actionable Targets. Cancers (2019) 11(3):380. doi: 10.3390/cancers11030380
36. Khoo BL, Grenci G, Lim YB, Lee SC, Han J, Lim CT. Expansion of Patient-Derived Circulating Tumor Cells From Liquid Biopsies Using a CTC Microfluidic Culture Device. Nat Protoc (2018) 13(1):34–58. doi: 10.1038/nprot.2017.125
37. Kulasinghe A, Kapeleris J, Kimberley R, Mattarollo SR, Thompson EW, Thiery JP, et al. The Prognostic Significance of Circulating Tumor Cells in Head and Neck and Non-Small-Cell Lung Cancer. Cancer Med (2018) 7(12):5910–9. doi: 10.1002/cam4.1832
38. Carvalho Â., Ferreira G, Seixas D, Guimarães-Teixeira C, Henrique R, Monteiro FJ, et al. Emerging Lab-On-a-Chip Approaches for Liquid Biopsy in Lung Cancer: Status in CTCs and ctDNA Research and Clinical Validation. Cancers (2021) 13(9):2101. doi: 10.3390/cancers13092101
39. Pandey CM, Augustine S, Kumar S, Kumar S, Nara S, Srivastava S, et al. Microfluidics Based Point-Of-Care Diagnostics. Biotechnol J (2018) 13(1). doi: 10.1002/biot.201700047
40. Gleghorn JP, Pratt ED, Denning D, Liu H, Bander NH, Tagawa ST, et al. Capture of Circulating Tumor Cells From Whole Blood of Prostate Cancer Patients Using Geometrically Enhanced Differential Immunocapture (GEDI) and a Prostate-Specific Antibody. Lab Chip (2010) 10(1):27–9. doi: 10.1039/B917959C
41. Ozkumur E, Shah AM, Ciciliano JC, Emmink BL, Miyamoto DT, Brachtel E, et al. Inertial Focusing for Tumor Antigen-Dependent and -Independent Sorting of Rare Circulating Tumor Cells. Sci Transl Med (2013) 5(179):179ra47. doi: 10.1126/scitranslmed.3005616
42. Casavant BP, Mosher R, Warrick JW, Maccoux LJ, Berry SM, Becker JT, et al. A Negative Selection Methodology Using a Microfluidic Platform for the Isolation and Enumeration of Circulating Tumor Cells. Methods (2013) 64(2):137–43. doi: 10.1016/j.ymeth.2013.05.027
43. Ribeiro-Samy S, Oliveira MI, Pereira-Veiga T, Muinelo-Romay L, Carvalho S, Gaspar J, et al. Fast and Efficient Microfluidic Cell Filter for Isolation of Circulating Tumor Cells From Unprocessed Whole Blood of Colorectal Cancer Patients. Sci Rep (2019) 9(1):8032. doi: 10.1038/s41598-019-44401-1
44. Warkiani ME, Khoo BL, Tan DS, Bhagat AA, Lim WT, Yap YS, et al. An Ultra-High-Throughput Spiral Microfluidic Biochip for the Enrichment of Circulating Tumor Cells. Analyst (2014) 139(13):3245–55. doi: 10.1039/C4AN00355A
45. Zhu S, Jiang F, Han Y, Xiang N, Ni Z. Microfluidics for Label-Free Sorting of Rare Circulating Tumor Cells. Analyst (2020) 145(22):7103–24. doi: 10.1039/D0AN01148G
46. Farace F, Massard C, Vimond N, Drusch F, Jacques N, Billiot F, et al. A Direct Comparison of CellSearch and ISET for Circulating Tumour-Cell Detection in Patients With Metastatic Carcinomas. Br J Cancer (2011) 105(6):847–53. doi: 10.1038/bjc.2011.294
47. Hofman V, Ilie MI, Long E, Selva E, Bonnetaud C, Molina T, et al. Detection of Circulating Tumor Cells as a Prognostic Factor in Patients Undergoing Radical Surgery for Non-Small-Cell Lung Carcinoma: Comparison of the Efficacy of the CellSearch Assay™ and the Isolation by Size of Epithelial Tumor Cell Method. Int J Cancer (2011) 129(7):1651–60. doi: 10.1002/ijc.25819
48. Papadaki MA, Sotiriou AI, Vasilopoulou C, Filika M, Aggouraki D, Tsoulfas PG, et al. Optimization of the Enrichment of Circulating Tumor Cells for Downstream Phenotypic Analysis in Patients With Non-Small Cell Lung Cancer Treated With Anti-PD-1 Immunotherapy. Cancers (2020) 12(6):1556. doi: 10.3390/cancers12061556
49. Song Y, Tian T, Shi Y, Liu W, Zou Y, Khajvand T, et al. Enrichment and Single-Cell Analysis of Circulating Tumor Cells. Chem Sci (2017) 8(3):1736–51. doi: 10.1039/C6SC04671A
50. Zeinali M, Lee M, Nadhan A, Mathur A, Hedman C, Lin E, et al. High-Throughput Label-Free Isolation of Heterogeneous Circulating Tumor Cells and CTC Clusters From Non-Small-Cell Lung Cancer Patients. Cancers (2020) 12(1):127. doi: 10.3390/cancers12010127
51. Lim M, Park J, Lowe AC, Jeong H-O, Lee S, Park HC, et al. A Lab-on-a-Disc Platform Enables Serial Monitoring of Individual CTCs Associated With Tumor Progression During EGFR-Targeted Therapy for Patients With NSCLC. Theranostics (2020) 10(12):5181–94. doi: 10.7150/thno.44693
52. Zhao L, Wu X, Zheng J, Dong D. DNA Methylome Profiling of Circulating Tumor Cells in Lung Cancer at Single Base-Pair Resolution. Oncogene (2021) 40(10):1884–95. doi: 10.1038/s41388-021-01657-0
53. Chen L, Peng M, Li N, Song Q, Yao Y, Xu B, et al. Combined Use of EpCAM and Frα Enables the High-Efficiency Capture of Circulating Tumor Cells in Non-Small Cell Lung Cancer. Sci Rep (2018) 8(1):1188. doi: 10.1038/s41598-018-19391-1
54. Turetta M, Bulfoni M, Brisotto G, Fasola G, Zanello A, Biscontin E, et al. Assessment of the Mutational Status of NSCLC Using Hypermetabolic Circulating Tumor Cells. Cancers (2018) 10(8):270. doi: 10.3390/cancers10080270
55. Chinniah C, Aguarin L, Cheng P, Decesaris C, Cutillo A, Berman AT, et al. Early Detection of Recurrence in Patients With Locally Advanced Non-Small-Cell Lung Cancer via Circulating Tumor Cell Analysis. Clin Lung Cancer (2019) 20(5):384–90. doi: 10.1016/j.cllc.2019.04.011
56. Li Y, Tian X, Gao L, Jiang X, Fu R, Zhang T, et al. Clinical Significance of Circulating Tumor Cells and Tumor Markers in the Diagnosis of Lung Cancer. Cancer Med (2019) 8(8):3782–92. doi: 10.1002/cam4.2286
57. Lindsay CR, Blackhall FH, Carmel A, Fernandez-Gutierrez F, Gazzaniga P, Groen HJM, et al. EPAC-Lung: Pooled Analysis of Circulating Tumour Cells in Advanced Non-Small Cell Lung Cancer. Eur J Cancer (2019) 117:60–8. doi: 10.1016/j.ejca.2019.04.019
58. Pailler E, Faugeroux V, Oulhen M, Mezquita L, Laporte M, Honoré A, et al. Acquired Resistance Mutations to ALK Inhibitors Identified by Single Circulating Tumor Cell Sequencing in ALK-Rearranged Non–Small-Cell Lung Cancer. Clin Cancer Res (2019) 25(22):6671–82. doi: 10.1158/1078-0432.CCR-19-1176
59. Scharpenseel H, Hanssen A, Loges S, Mohme M, Bernreuther C, Peine S, et al. EGFR and HER3 Expression in Circulating Tumor Cells and Tumor Tissue From Non-Small Cell Lung Cancer Patients. Sci Rep (2019) 9(1):7406–6. doi: 10.1038/s41598-019-43678-6
60. Duan G-C, Zhang X-P, Wang H-E, Wang Z-K, Zhang H, Yu L, et al. Circulating Tumor Cells as a Screening and Diagnostic Marker for Early-Stage Non-Small Cell Lung Cancer. OncoTargets Ther (2020) 13:1931–9. doi: 10.2147/OTT.S241956
61. Frick MA, Feigenberg SJ, Jean-Baptiste SR, Aguarin LA, Mendes A, Chinniah C, et al. Circulating Tumor Cells Are Associated With Recurrent Disease in Patients With Early-Stage Non–Small Cell Lung Cancer Treated With Stereotactic Body Radiotherapy. Clin Cancer Res (2020) 26(10):2372–80. doi: 10.1158/1078-0432.CCR-19-2158
62. Ichimura H, Nawa T, Yamamoto Y, Shimizu K, Kobayashi K, Kitazawa S, et al. Detection of Circulating Tumor Cells in Patients With Lung Cancer Using Metallic Micro−Cavity Array Filter: A Pilot Study. Mol Clin Oncol (2020) 12(3):278–83. doi: 10.3892/mco.2020.1973
63. Katz RL, Zaidi TM, Pujara D, Shanbhag ND, Truong D, Patil S, et al. Identification of Circulating Tumor Cells Using 4-Color Fluorescence in Situ Hybridization: Validation of a Noninvasive Aid for Ruling Out Lung Cancer in Patients With Low-Dose Computed Tomography–Detected Lung Nodules. Cancer Cytopathol (2020) 128(8):553–62. doi: 10.1002/cncy.22278
64. Kulasinghe A, Lim Y, Kapeleris J, Warkiani M, O’Byrne K, Punyadeera C. The Use of Three-Dimensional DNA Fluorescent In Situ Hybridization (3d DNA FISH) for the Detection of Anaplastic Lymphoma Kinase (ALK) in Non-Small Cell Lung Cancer (NSCLC) Circulating Tumor Cells. Cells (2020) 9(6):1465. doi: 10.3390/cells9061465
65. Zhou J, Kulasinghe A, Bogseth A, O’Byrne K, Punyadeera C, Papautsky I. Isolation of Circulating Tumor Cells in Non-Small-Cell-Lung-Cancer Patients Using a Multi-Flow Microfluidic Channel. Microsys Nanoeng (2019) 5(1):8. doi: 10.1038/s41378-019-0045-6
66. Huang M, Ma Y, Lv C, Li S, Lu F, Zhang S, et al. Aneuploid Circulating Tumor Cells as a Predictor of Response to Neoadjuvant Chemotherapy in Non-Small Cell Lung Cancer. Int J Gen Med (2021) 14:6609–20. doi: 10.2147/IJGM.S330361
67. Kong SL, Liu X, Tan SJ, Tai JA, Phua LY, Poh HM, et al. Complementary Sequential Circulating Tumor Cell (CTC) and Cell-Free Tumor DNA (ctDNA) Profiling Reveals Metastatic Heterogeneity and Genomic Changes in Lung Cancer and Breast Cancer. Front Oncol (2021) 11:698551. doi: 10.3389/fonc.2021.698551
68. Wan L, Liu Q, Liang D, Guo Y, Liu G, Ren J, et al. Circulating Tumor Cell and Metabolites as Novel Biomarkers for Early-Stage Lung Cancer Diagnosis. Front Oncol (2021) 11:630672. doi: 10.3389/fonc.2021.630672
69. Wang H, Wu J, Zhang Q, Hao J, Wang Y, Li Z, et al. A Modified Method to Isolate Circulating Tumor Cells and Identify by a Panel of Gene Mutations in Lung Cancer. Technol Cancer Res Treat (2021) 20:1533033821995275. doi: 10.1177/1533033821995275
70. Yang L, Yan X, Chen J, Zhan Q, Hua Y, Xu S, et al. Hexokinase 2 Discerns a Novel Circulating Tumor Cell Population Associated With Poor Prognosis in Lung Cancer Patients. Proc Natl Acad Sci USA (2021) 118(11):e2012228118. doi: 10.1073/pnas.2012228118
71. Zhang W, Duan X, Zhang Z, Yang Z, Zhao C, Liang C, et al. Combination of CT and Telomerase+ Circulating Tumor Cells Improves Diagnosis of Small Pulmonary Nodules. JCI Insight (2021) 6(11):e148182. doi: 10.1172/jci.insight.148182
72. Loomans-Kropp HA, Umar A. Cancer Prevention and Screening: The Next Step in the Era of Precision Medicine. NPJ Precis Oncol (2019) 3:3–3. doi: 10.1038/s41698-018-0075-9
73. Pinsky PF, Bellinger CR, Miller DP Jr. False-Positive Screens and Lung Cancer Risk in the National Lung Screening Trial: Implications for Shared Decision-Making. J Med Screen (2018) 25(2):110–2. doi: 10.1177/0969141317727771
74. Aberle DR, Adams AM, Berg CD, Black WC, Clapp JD, Fagerstrom RM, et al. Reduced Lung-Cancer Mortality With Low-Dose Computed Tomographic Screening. N Engl J Med (2011) 365(5):395–409. doi: 10.1056/NEJMoa1102873
75. Patz EF Jr, Pinsky P, Gatsonis C, Sicks JD, Kramer BS, Tammemägi MC, et al. Overdiagnosis in Low-Dose Computed Tomography Screening for Lung Cancer. JAMA Intern Med (2014) 174(2):269–74. doi: 10.1001/jamainternmed.2013.12738
76. Ilie M, Hofman V, Long-Mira E, Selva E, Vignaud JM, Padovani B, et al. "Sentinel" Circulating Tumor Cells Allow Early Diagnosis of Lung Cancer in Patients With Chronic Obstructive Pulmonary Disease. PloS One (2014) 9(10):e111597. doi: 10.1371/journal.pone.0111597
77. Fiorelli A, Accardo M, Carelli E, Angioletti D, Santini M, Di Domenico M. Circulating Tumor Cells in Diagnosing Lung Cancer: Clinical and Morphologic Analysis. Ann Thorac Surg (2015) 99(6):1899–905. doi: 10.1016/j.athoracsur.2014.11.049
78. Marquette C-H, Boutros J, Benzaquen J, Ferreira M, Pastre J, Pison C, et al. Circulating Tumour Cells as a Potential Biomarker for Lung Cancer Screening: A Prospective Cohort Study. Lancet Respir Med (2020) 8(7):709–16. doi: 10.1016/S2213-2600(20)30081-3
79. Gonzalez J, Marín M, Sánchez-Salcedo P, Zulueta JJ. Lung Cancer Screening in Patients With Chronic Obstructive Pulmonary Disease. Ann Transl Med (2016) 4(8):160. doi: 10.21037/atm.2016.03.57
80. Millner LM, Linder MW, Valdes R Jr. Circulating Tumor Cells: A Review of Present Methods and the Need to Identify Heterogeneous Phenotypes. Ann Clin Lab Sci (2013) 43(3):295–304.
81. Dong J, Zhu D, Tang X, Qiu X, Lu D, Li B, et al. Detection of Circulating Tumor Cell Molecular Subtype in Pulmonary Vein Predicting Prognosis of Stage I-III Non-Small Cell Lung Cancer Patients. Front Oncol (2019) 9:1139. doi: 10.3389/fonc.2019.01139
82. Hofman V, Bonnetaud C, Ilie MI, Vielh P, Vignaud JM, Flejou JF, et al. Preoperative Circulating Tumor Cell Detection Using the Isolation by Size of Epithelial Tumor Cell Method for Patients With Lung Cancer Is a New Prognostic Biomarker. Clin Cancer Res an Off J Am Assoc Cancer Res (2011) 17(4):827–35. doi: 10.1158/1078-0432.CCR-10-0445
83. Pailler E, Oulhen M, Borget I, Remon J, Ross K, Auger N, et al. Circulating Tumor Cells With Aberrant ALK Copy Number Predict Progression-Free Survival During Crizotinib Treatment in ALK-Rearranged Non-Small Cell Lung Cancer Patients. Cancer Res (2017) 77(9):2222–30. doi: 10.1158/0008-5472.CAN-16-3072
84. Muinelo-Romay L, Vieito M, Abalo A, Nocelo MA, Barón F, Anido U, et al. Evaluation of Circulating Tumor Cells and Related Events as Prognostic Factors and Surrogate Biomarkers in Advanced NSCLC Patients Receiving First-Line Systemic Treatment. Cancers (2014) 6(1):153–65. doi: 10.3390/cancers6010153
85. Krebs MG, Sloane R, Priest L, Lancashire L, Hou JM, Greystoke A, et al. Evaluation and Prognostic Significance of Circulating Tumor Cells in Patients With Non-Small-Cell Lung Cancer. J Clin Oncol (2011) 29(12):1556–63. doi: 10.1200/JCO.2010.28.7045
86. Liang H, Huang J, Wang B, Liu Z, He J, Liang W. The Role of Liquid Biopsy in Predicting Post-Operative Recurrence of Non-Small Cell Lung Cancer. J Thorac Dis (2018) 10(Suppl 7):S838–45. doi: 10.21037/jtd.2018.04.08
87. Punnoose EA, Atwal S, Liu W, Raja R, Fine BM, Hughes BG, et al. Evaluation of Circulating Tumor Cells and Circulating Tumor DNA in Non-Small Cell Lung Cancer: Association With Clinical Endpoints in a Phase II Clinical Trial of Pertuzumab and Erlotinib. Clin Cancer Res an Off J Am Assoc Cancer Res (2012) 18(8):2391–401. doi: 10.1158/1078-0432.CCR-11-3148
88. Mamdouhi T, Twomey JD, McSweeney KM, Zhang B. Fugitives on the Run: Circulating Tumor Cells (CTCs) in Metastatic Diseases. Cancer Metastasis Rev (2019) 38(1):297–305. doi: 10.1007/s10555-019-09795-4
89. Balakrishnan A, Koppaka D, Anand A, Deb B, Grenci G, Viasnoff V, et al. Circulating Tumor Cell Cluster Phenotype Allows Monitoring Response to Treatment and Predicts Survival. Sci Rep (2019) 9(1):7933. doi: 10.1038/s41598-019-44404-y
90. Shishido S, Carlsson A, Nieva J, Bethel K, Hicks J, Bazhenova L, et al. Circulating Tumor Cells as a Response Monitor in Stage IV Non-Small Cell Lung Cancer. J Transl Med (2019) 17(1):294. doi: 10.1186/s12967-019-2035-8
91. Horton CE, Kamal M, Leslie M, Zhang R, Tanaka T, Razaq M. Circulating Tumor Cells Accurately Predicting Progressive Disease After Treatment in a Patient With Non-Small Cell Lung Cancer Showing Response on Scans. Anticancer Res (2018) 38(2):1073–6. doi: 10.21873/anticanres.12325
92. Gallo M, De Luca A, Frezzetti D, Passaro V, Maiello MR, Normanno N. The Potential of Monitoring Treatment Response in Non-Small Cell Lung Cancer Using Circulating Tumour Cells. Expert Rev Mol Diagn (2019) 19(8):683–94. doi: 10.1080/14737159.2019.1640606
93. Rossi E, Zamarchi R. Single-Cell Analysis of Circulating Tumor Cells: How Far Have We Come in the -Omics Era? Front Genet (2019) 10:958–8. doi: 10.3389/fgene.2019.00958
94. Lindeman NI, Cagle PT, Beasley MB, Chitale DA, Dacic S, Giaccone G, et al. Molecular Testing Guideline for Selection of Lung Cancer Patients for EGFR and ALK Tyrosine Kinase Inhibitors: Guideline From the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology. J Thorac Oncol (2013) 8(7):823–59. doi: 10.1097/JTO.0b013e318290868f
95. Luo SY, Lam DCL. Oncogenic Driver Mutations in Lung Cancer. Trans Respir Med (2013) 1(1):6. doi: 10.1186/2213-0802-1-6
96. Rosell R, Carcereny E, Gervais R, Vergnenegre A, Massuti B, Felip E, et al. Erlotinib Versus Standard Chemotherapy as First-Line Treatment for European Patients With Advanced EGFR Mutation-Positive Non-Small-Cell Lung Cancer (EURTAC): A Multicentre, Open-Label, Randomised Phase 3 Trial. Lancet Oncol (2012) 13(3):239–46. doi: 10.1016/S1470-2045(11)70393-X
97. Gristina V, Malapelle U, Galvano A, Pisapia P, Pepe F, Rolfo C, et al. The Significance of Epidermal Growth Factor Receptor Uncommon Mutations in Non-Small Cell Lung Cancer: A Systematic Review and Critical Appraisal. Cancer Treat Rev (2020) 85:101994. doi: 10.1016/j.ctrv.2020.101994
98. Cortot AB, Janne PA. Molecular Mechanisms of Resistance in Epidermal Growth Factor Receptor-Mutant Lung Adenocarcinomas. Eur Respir Rev (2014) 23(133):356–66. doi: 10.1183/09059180.00004614
99. Sacher AG, Janne PA, Oxnard GR. Management of Acquired Resistance to Epidermal Growth Factor Receptor Kinase Inhibitors in Patients With Advanced Non-Small Cell Lung Cancer. Cancer (2014) 120(15):2289–98. doi: 10.1002/cncr.28723
100. Du X, Shao Y, Qin HF, Tai YH, Gao HJ. ALK-Rearrangement in Non-Small-Cell Lung Cancer (NSCLC). Thorac Cancer (2018) 9(4):423–30. doi: 10.1111/1759-7714.12613
101. Camidge DR, Bang YJ, Kwak EL, Iafrate AJ, Varella-Garcia M, Fox SB, et al. Activity and Safety of Crizotinib in Patients With ALK-Positive Non-Small-Cell Lung Cancer: Updated Results From a Phase 1 Study. Lancet Oncol (2012) 13(10):1011–9. doi: 10.1016/S1470-2045(12)70344-3
102. Rihawi K, Alfieri R, Fiorentino M, Fontana F, Capizzi E, Cavazzoni A, et al. MYC Amplification as a Potential Mechanism of Primary Resistance to Crizotinib in ALK-Rearranged Non-Small Cell Lung Cancer: A Brief Report. Transl Oncol (2019) 12(1):116–21. doi: 10.1016/j.tranon.2018.09.013
103. Pailler E, Mezquita L, Faugeroux V, Oulhen M, Queffelec P, Remon J, et al. Analysis of Single Circulating Tumor Cells (CTCs) to Identify Resistance Mutations to ALK-Inhibitors in Both ALK-Gene and Bypass Oncogenic Pathways. J Clin Oncol (2018) 36(15_suppl):12038–8. doi: 10.1200/JCO.2018.36.15_suppl.12038
104. Lisberg A, Cummings A, Goldman JW, Bornazyan K, Reese N, Wang T, et al. A Phase II Study of Pembrolizumab in EGFR-Mutant, PD-L1+, Tyrosine Kinase Inhibitor Naïve Patients With Advanced NSCLC. J Thorac Oncol (2018) 13(8):1138–45. doi: 10.1016/j.jtho.2018.03.035
105. Yi M, Niu M, Xu L, Luo S, Wu K. Regulation of PD-L1 Expression in the Tumor Microenvironment. J Hematol Oncol (2021) 14(1):10. doi: 10.1186/s13045-020-01027-5
106. Kloten V, Lampignano R, Krahn T, Schlange T. Circulating Tumor Cell PD-L1 Expression as Biomarker for Therapeutic Efficacy of Immune Checkpoint Inhibition in NSCLC. Cells (2019) 8(8):809. doi: 10.3390/cells8080809
107. Raimondi C, Carpino G, Nicolazzo C, Gradilone A, Gianni W, Gelibter A, et al. PD-L1 and Epithelial-Mesenchymal Transition in Circulating Tumor Cells From Non-Small Cell Lung Cancer Patients: A Molecular Shield to Evade Immune System? Oncoimmunology (2017) 6(12):e1315488. doi: 10.1080/2162402X.2017.1315488
108. Ilié M, Szafer-Glusman E, Hofman V, Chamorey E, Lalvée S, Selva E, et al. Detection of PD-L1 in Circulating Tumor Cells and White Blood Cells From Patients With Advanced Non-Small-Cell Lung Cancer. Ann Oncol (2018) 29(1):193–9. doi: 10.1093/annonc/mdx636
109. Kallergi G, Vetsika EK, Aggouraki D, Lagoudaki E, Koutsopoulos A, Koinis F, et al. Evaluation of PD-L1/PD-1 on Circulating Tumor Cells in Patients With Advanced Non-Small Cell Lung Cancer. Ther Adv Med Oncol (2018) 10:1758834017750121. doi: 10.1177/1758834017750121
110. Wang Y, Kim TH, Fouladdel S, Zhang Z, Soni P, Qin A, et al. PD-L1 Expression in Circulating Tumor Cells Increases During Radio(chemo)therapy and Indicates Poor Prognosis in Non-Small Cell Lung Cancer. Sci Rep (2019) 9(1):566. doi: 10.1038/s41598-018-36096-7
111. Cheng Y, Wang T, Lv X, Li R, Yuan L, Shen J, et al. Detection of PD-L1 Expression and Its Clinical Significance in Circulating Tumor Cells From Patients With Non-Small-Cell Lung Cancer. Cancer Manag Res (2020) 12:2069–78. doi: 10.2147/CMAR.S245425
112. Ntzifa A, Strati A, Kallergi G, Kotsakis A, Georgoulias V, Lianidou E. Gene Expression in Circulating Tumor Cells Reveals a Dynamic Role of EMT and PD-L1 During Osimertinib Treatment in NSCLC Patients. Sci Rep (2021) 11(1):2313. doi: 10.1038/s41598-021-82068-9
113. Sinoquet L, Jacot W, Gauthier L, Pouderoux S, Viala M, Cayrefourcq L, et al. Programmed Cell Death Ligand 1-Expressing Circulating Tumor Cells: A New Prognostic Biomarker in Non-Small Cell Lung Cancer. Clin Chem (2021) 67(11):1503–12. doi: 10.1093/clinchem/hvab131
114. Dhar M, Wong J, Che J, Matsumoto M, Grogan T, Elashoff D, et al. Evaluation of PD-L1 Expression on Vortex-Isolated Circulating Tumor Cells in Metastatic Lung Cancer. Sci Rep (2018) 8(1):2592. doi: 10.1038/s41598-018-19245-w
115. Janning M, Kobus F, Babayan A, Wikman H, Velthaus J-L, Bergmann S, et al. Determination of PD-L1 Expression in Circulating Tumor Cells of NSCLC Patients and Correlation With Response to PD-1/PD-L1 Inhibitors. Cancers (2019) 11(6):835. doi: 10.3390/cancers11060835
116. Koh Y, Yagi S, Akamatsu H, Kanai K, Hayata A, Tokudome N, et al. Heterogeneous Expression of Programmed Death Receptor-Ligand 1 on Circulating Tumor Cells in Patients With Lung Cancer. Clin Lung Cancer (2019) 20(4):270–7.e1. doi: 10.1016/j.cllc.2019.03.004
117. Tamminga M, de Wit S, Hiltermann TJN, Timens W, Schuuring E, Terstappen LWMM, et al. Circulating Tumor Cells in Advanced Non-Small Cell Lung Cancer Patients Are Associated With Worse Tumor Response to Checkpoint Inhibitors. J ImmunoTher Cancer (2019) 7(1):173. doi: 10.1186/s40425-019-0649-2
118. Borghaei H, Paz-Ares L, Horn L, Spigel DR, Steins M, Ready NE, et al. Nivolumab Versus Docetaxel in Advanced Nonsquamous Non-Small-Cell Lung Cancer. N Engl J Med (2015) 373(17):1627–39. doi: 10.1056/NEJMoa1507643
119. Brahmer J, Reckamp KL, Baas P, Crinò L, Eberhardt WE, Poddubskaya E, et al. Nivolumab Versus Docetaxel in Advanced Squamous-Cell Non-Small-Cell Lung Cancer. N Engl J Med (2015) 373(2):123–35. doi: 10.1056/NEJMoa1504627
120. Herbst RS, Baas P, Kim DW, Felip E, Pérez-Gracia JL, Han JY, et al. Pembrolizumab Versus Docetaxel for Previously Treated, PD-L1-Positive, Advanced Non-Small-Cell Lung Cancer (KEYNOTE-010): A Randomised Controlled Trial. Lancet (2016) 387(10027):1540–50. doi: 10.1016/S0140-6736(15)01281-7
121. Lazzari C, Karachaliou N, Bulotta A, Viganó M, Mirabile A, Brioschi E, et al. Combination of Immunotherapy With Chemotherapy and Radiotherapy in Lung Cancer: Is This the Beginning of the End for Cancer? Ther Adv Med Oncol (2018) 10:1758835918762094. doi: 10.1177/1758835918762094
122. Listì A, Barraco N, Bono M, Insalaco L, Castellana L, Cutaia S, et al. Immuno-Targeted Combinations in Oncogene-Addicted Non-Small Cell Lung Cancer. Trans Cancer Res (2018) 8(Suppl 1):S55–63. doi: 10.21037/tcr.2018.10.04
123. Lee CK, Man J, Lord S, Links M, Gebski V, Mok T, et al. Checkpoint Inhibitors in Metastatic EGFR-Mutated Non-Small Cell Lung Cancer-A Meta-Analysis. J Thorac Oncol (2017) 12(2):403–7. doi: 10.1016/j.jtho.2016.10.007
124. Gandhi L, Rodríguez-Abreu D, Gadgeel S, Esteban E, Felip E, De Angelis F, et al. Pembrolizumab Plus Chemotherapy in Metastatic Non-Small-Cell Lung Cancer. N Engl J Med (2018) 378(22):2078–92. doi: 10.1056/NEJMoa1801005
125. Lee HL, Chiou JF, Wang PY, Lu LS, Shen CN, Hsu HL, et al. Ex Vivo Expansion and Drug Sensitivity Profiling of Circulating Tumor Cells From Patients With Small Cell Lung Cancer. Cancers (2020) 12(11):3394. doi: 10.3390/cancers12113394
126. Que Z, Luo B, Zhou Z, Dong C, Jiang Y, Wang L, et al. Establishment and Characterization of a Patient-Derived Circulating Lung Tumor Cell Line In Vitro and In Vivo. Cancer Cell Int (2019) 19(1):21. doi: 10.1186/s12935-019-0735-z
127. Simpson KL, Stoney R, Frese KK, Simms N, Rowe W, Pearce SP, et al. A Biobank of Small Cell Lung Cancer CDX Models Elucidates Inter- and Intratumoral Phenotypic Heterogeneity. Nat Cancer (2020) 1(4):437–51. doi: 10.1038/s43018-020-0046-2
128. Lallo A, Gulati S, Schenk MW, Khandelwal G, Berglund UW, Pateras IS, et al. Ex Vivo Culture of Cells Derived From Circulating Tumour Cell Xenograft to Support Small Cell Lung Cancer Research and Experimental Therapeutics. Br J Pharmacol (2018) 176(3):436–50. doi: 10.1111/bph.14542
129. Smit DJ, Pantel K, Jücker M. Circulating Tumor Cells as a Promising Target for Individualized Drug Susceptibility Tests in Cancer Therapy. Biochem Pharmacol (2021) 188:114589. doi: 10.1016/j.bcp.2021.114589
130. Heitzer E, Haque IS, Roberts CES, Speicher MR. Current and Future Perspectives of Liquid Biopsies in Genomics-Driven Oncology. Nat Rev Genet (2019) 20(2):71–88. doi: 10.1038/s41576-018-0071-5
131. Su P-L, Yang S-C, Chen Y-L, Wu Y-L, Lin C-Y, Chang W-Y, et al. Real-World Outcomes of NSCLC Patients Receiving Tissue or Circulating Tumor DNA-Guided Osimertinib Treatment. Cancer Med (2019) 8(13):5939–47. doi: 10.1002/cam4.2485
132. Buder A, Hochmair MJ, Schwab S, Bundalo T, Schenk P, Errhalt P, et al. Cell-Free Plasma DNA-Guided Treatment With Osimertinib in Patients With Advanced EGFR-Mutated NSCLC. J Thorac Oncol (2018) 13(6):821–30. doi: 10.1016/j.jtho.2018.02.014
133. Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, et al. Detection of Circulating Tumor DNA in Early- and Late-Stage Human Malignancies. Sci Trans Med (2014) 6(224):224ra24–224ra24. doi: 10.1126/scitranslmed.3007094
134. Galarza Fortuna GM, Dvir K, Circulating tumor DNA. Where Are We Now? A Mini Review of the Literature. World J Clin Oncol (2020) 11(9):723–31. doi: 10.5306/wjco.v11.i9.723
135. Jahr S, Hentze H, Englisch S, Hardt D, Fackelmayer FO, Hesch RD, et al. DNA Fragments in the Blood Plasma of Cancer Patients: Quantitations and Evidence for Their Origin From Apoptotic and Necrotic Cells. Cancer Res (2001) 61(4):1659–65.
136. Chaudhuri AA, Chabon JJ, Lovejoy AF, Newman AM, Stehr H, Azad TD, et al. Early Detection of Molecular Residual Disease in Localized Lung Cancer by Circulating Tumor DNA Profiling. Cancer Discov (2017) 7(12):1394–403. doi: 10.1158/2159-8290.CD-17-0716
137. Gautschi O, Bigosch C, Huegli B, Jermann M, Marx A, Chassé E, et al. Circulating Deoxyribonucleic Acid as Prognostic Marker in Non-Small-Cell Lung Cancer Patients Undergoing Chemotherapy. J Clin Oncol (2004) 22(20):4157–64. doi: 10.1200/JCO.2004.11.123
138. Fernandez-Cuesta L, Perdomo S, Avogbe PH, Leblay N, Delhomme TM, Gaborieau V, et al. Identification of Circulating Tumor DNA for the Early Detection of Small-Cell Lung Cancer. EBioMedicine (2016) 10:117–23. doi: 10.1016/j.ebiom.2016.06.032
139. Abbosh C, Birkbak NJ, Wilson GA, Jamal-Hanjani M, Constantin T, Salari R, et al. Phylogenetic ctDNA Analysis Depicts Early-Stage Lung Cancer Evolution. Nature (2017) 545(7655):446–51. doi: 10.1038/nature22364
140. Jamal-Hanjani M, Wilson GA, Horswell S, Mitter R, Sakarya O, Constantin T, et al. Detection of Ubiquitous and Heterogeneous Mutations in Cell-Free DNA From Patients With Early-Stage Non-Small-Cell Lung Cancer. Ann Oncol (2016) 27(5):862–7. doi: 10.1093/annonc/mdw037
141. Jia J, Huang B, Zhuang Z, Chen S. Circulating Tumor DNA as Prognostic Markers for Late Stage NSCLC With Bone Metastasis. Int J Biol Markers (2018) 33(2):222–30. doi: 10.1177/1724600817753576
142. Harrison PT, Vyse S, Huang PH. Rare Epidermal Growth Factor Receptor (EGFR) Mutations in Non-Small Cell Lung Cancer. Semin Cancer Biol (2020) 61:167–79. doi: 10.1016/j.semcancer.2019.09.015
143. Goss G, Tsai CM, Shepherd FA, Bazhenova L, Lee JS, Chang GC, et al. Osimertinib for Pretreated EGFR Thr790Met-Positive Advanced Non-Small-Cell Lung Cancer (AURA2): A Multicentre, Open-Label, Single-Arm, Phase 2 Study. Lancet Oncol (2016) 17(12):1643–52. doi: 10.1016/S1470-2045(16)30508-3
144. Guardant Health, I. Guardant Health Guardant360® CDx First FDA-Approved Liquid Biopsy for Comprehensive Tumor Mutation Profiling Across All Solid Cancers. Redwood City: Guardant Health, Inc. (2020).
145. Administration, U. S. F. a. D. FDA Approves First Liquid Biopsy Next-Generation Sequencing Companion Diagnostic Test. (2020).
146. Negrao MV, Quek K, Zhang J, Sepesi B. TRACERx: Tracking Tumor Evolution to Impact the Course of Lung Cancer. J Thorac Cardiovasc Surg (2018) 155(3):1199–202. doi: 10.1016/j.jtcvs.2017.10.134
147. Remon J, Menis J, Hasan B, Peric A, De Maio E, Novello S, et al. The APPLE Trial: Feasibility and Activity of AZD9291 (Osimertinib) Treatment on Positive PLasma T790M in EGFR-Mutant NSCLC Patients. EORTC 1613. Clin Lung Cancer (2017) 18(5):583–8. doi: 10.1016/j.cllc.2017.02.005
148. Lin JJ, Shaw AT. Recent Advances in Targeting ROS1 in Lung Cancer. J Thorac Oncol (2017) 12(11):1611–25. doi: 10.1016/j.jtho.2017.08.002
149. Dagogo-Jack I, Rooney M, Nagy RJ, Lin JJ, Chin E, Ferris LA, et al. Molecular Analysis of Plasma From Patients With ROS1-Positive NSCLC. J Thorac Oncol (2019) 14(5):816–24. doi: 10.1016/j.jtho.2019.01.009
150. Zhu G, Ye X, Dong Z, Lu YC, Sun Y, Liu Y, et al. Highly Sensitive Droplet Digital PCR Method for Detection of EGFR-Activating Mutations in Plasma Cell-Free DNA From Patients With Advanced Non-Small Cell Lung Cancer. J Mol Diagn (2015) 17(3):265–72. doi: 10.1016/j.jmoldx.2015.01.004
151. Xiang Z, Wan R, Zou B, Qi X, Huang Q, Kumar S, et al. Highly Sensitive and Specific Real-Time PCR by Employing Serial Invasive Reaction as a Sequence Identifier for Quantifying EGFR Mutation Abundance in cfDNA. Anal Bioanal Chem (2018) 410(26):6751–9. doi: 10.1007/s00216-018-1316-z
152. Bennett CW, Berchem G, Kim YJ, El-Khoury V, Cell-free DNA. And Next-Generation Sequencing in the Service of Personalized Medicine for Lung Cancer. Oncotarget (2016) 7(43):71013–35. doi: 10.18632/oncotarget.11717
153. Ding PN, Becker T, Bray V, Chua W, Ma Y, Xu B, et al. Plasma Next Generation Sequencing and Droplet Digital PCR-Based Detection of Epidermal Growth Factor Receptor (EGFR) Mutations in Patients With Advanced Lung Cancer Treated With Subsequent-Line Osimertinib. Thorac Cancer (2019) 10(10):1879–84. doi: 10.1111/1759-7714.13154
154. Guo Q-M, Wang L, Yu W-J, Qiao L-H, Zhao M-N, Hu X-M, et al. Detection of Plasma EGFR Mutations in NSCLC Patients With a Validated ddPCR Lung cfDNA Assay. J Cancer (2019) 10(18):4341–9. doi: 10.7150/jca.31326
155. Nacchio M, Sgariglia R, Gristina V, Pisapia P, Pepe F, De Luca C, et al. KRAS Mutations Testing in Non-Small Cell Lung Cancer: The Role of Liquid Biopsy in the Basal Setting. J Thorac Dis (2020) 12(7):3836–43. doi: 10.21037/jtd.2020.01.19
156. Sánchez-Herrero E, Serna-Blasco R, Ivanchuk V, García-Campelo R, Dómine Gómez M, Sánchez JM, et al. NGS-Based Liquid Biopsy Profiling Identifies Mechanisms of Resistance to ALK Inhibitors: A Step Toward Personalized NSCLC Treatment. Mol Oncol (2021) 15(9):2363–76. doi: 10.1002/1878-0261.13033
157. Pisapia P, Pepe F, Baggi A, Barberis M, Galvano A, Gristina V, et al. Next Generation Diagnostic Algorithm in Non-Small Cell Lung Cancer Predictive Molecular Pathology: The KWAY Italian Multicenter Cost Evaluation Study. Crit Rev Oncol Hematol (2022) 169:103525. doi: 10.1016/j.critrevonc.2021.103525
158. Chen M, Zhao H. Next-Generation Sequencing in Liquid Biopsy: Cancer Screening and Early Detection. Hum Genomics (2019) 13(1):34. doi: 10.1186/s40246-019-0220-8
159. Papadopoulou E, Tsoulos N, Tsantikidi K, Metaxa-Mariatou V, Stamou PE, Kladi-Skandali A, et al. Clinical Feasibility of NGS Liquid Biopsy Analysis in NSCLC Patients. PloS One (2019) 14(12):e0226853. doi: 10.1371/journal.pone.0226853
160. Xu Y, Zhang Z, Su Z, Zhou X, Han X, Liu Q. Continuous Microfluidic Purification of DNA Using Magnetophoresis. Micromachines (Basel) (2020) 11(2):187. doi: 10.3390/mi11020187
161. Xu M, Zhao H, Chen J, Liu W, Li E, Wang Q, et al. An Integrated Microfluidic Chip and Its Clinical Application for Circulating Tumor Cell Isolation and Single-Cell Analysis. Cytometry A (2020) 97(1):46–53. doi: 10.1002/cyto.a.23902
162. Xu Z, Qiao Y, Tu J. Microfluidic Technologies for cfDNA Isolation and Analysis. Micromachines (2019) 10(10):672. doi: 10.3390/mi10100672
163. Aravanis AM, Lee M, Klausner RD. Next-Generation Sequencing of Circulating Tumor DNA for Early Cancer Detection. Cell (2017) 168(4):571–4. doi: 10.1016/j.cell.2017.01.030
164. Morrison C. Search for Liquid Biopsy Grail Points the Way to Drug Discovery and Development Gems. Nat Rev Drug Discov (2017) 16(6):373–4. doi: 10.1038/nrd.2017.96
165. Wang W, Song Z, Zhang Y. A Comparison of ddPCR and ARMS for Detecting EGFR T790M Status in ctDNA From Advanced NSCLC Patients With Acquired EGFR-TKI Resistance. Cancer Med (2017) 6(1):154–62. doi: 10.1002/cam4.978
166. Li Y, Xu Y, Wu X, He C, Liu Q, Wang F. Comprehensive Analysis of EGFR T790M Detection by ddPCR and ARMS-PCR and the Effect of Mutant Abundance on the Efficacy of Osimertinib in NSCLC Patients. J Thorac Dis (2019) 11(7):3004–14. doi: 10.21037/jtd.2019.07.42
167. Li L, Fu K, Zhou W, Snyder M. Applying Circulating Tumor DNA Methylation in the Diagnosis of Lung Cancer. Precis Clin Med (2019) 2(1):45–56. doi: 10.1093/pcmedi/pbz003
168. Kang G, Chen K, Yang F, Chuai S, Zhao H, Zhang K, et al. Monitoring of Circulating Tumor DNA and Its Aberrant Methylation in the Surveillance of Surgical Lung Cancer Patients: Protocol for a Prospective Observational Study. BMC Cancer (2019) 19(1):579–9. doi: 10.1186/s12885-019-5751-9
169. Gunnarsson EB, De S, Leder K, Foo J. Understanding the Role of Phenotypic Switching in Cancer Drug Resistance. J Theor Biol (2020) 490:110162. doi: 10.1016/j.jtbi.2020.110162
170. Krug AK, Enderle D, Karlovich C, Priewasser T, Bentink S, Spiel A, et al. Improved EGFR Mutation Detection Using Combined Exosomal RNA and Circulating Tumor DNA in NSCLC Patient Plasma. Ann Oncol (2018) 29(3):700–6. doi: 10.1093/annonc/mdx765
171. Castellanos-Rizaldos E, Grimm DG, Tadigotla V, Hurley J, Healy J, Neal PL, et al. Exosome-Based Detection of EGFR T790M in Plasma From Non-Small Cell Lung Cancer Patients. Clin Cancer Res an Off J Am Assoc Cancer Res (2018) 24(12):2944–50. doi: 10.1158/1078-0432.CCR-17-3369
172. Masaoutis C, Mihailidou C, Tsourouflis G, Theocharis S. Exosomes in Lung Cancer Diagnosis and Treatment. From the Translating Research Into Future Clinical Practice. Biochimie (2018) 151:27–36. doi: 10.1016/j.biochi.2018.05.014
173. Ambros V. The Functions of Animal microRNAs. Nature (2004) 431(7006):350–5. doi: 10.1038/nature02871
174. Wu K-L, Tsai Y-M, Lien C-T, Kuo P-L, Hung, Jen Y. The Roles of MicroRNA in Lung Cancer. Int J Mol Sci (2019) 20(7):1611. doi: 10.3390/ijms20071611
175. Zhang L, Lin J, Ye Y, Oba T, Gentile E, Lian J, et al. Serum MicroRNA-150 Predicts Prognosis for Early-Stage Non-Small Cell Lung Cancer and Promotes Tumor Cell Proliferation by Targeting Tumor Suppressor Gene Srcin1. Clin Pharmacol Ther (2018) 103(6):1061–73. doi: 10.1002/cpt.870
176. Zhang Y, Roth JA, Yu H, Ye Y, Xie K, Zhao H, et al. A 5-microRNA Signature Identified From Serum microRNA Profiling Predicts Survival in Patients With Advanced Stage Non-Small Cell Lung Cancer. Carcinogenesis (2019) 40(5):643–50. doi: 10.1093/carcin/bgy132
177. Cohen SJ, Punt CJ, Iannotti N, Saidman BH, Sabbath KD, Gabrail NY, et al. Prognostic Significance of Circulating Tumor Cells in Patients With Metastatic Colorectal Cancer. Ann Oncol (2009) 20(7):1223–9. doi: 10.1093/annonc/mdn786
178. Naito T, Tanaka F, Ono A, Yoneda K, Takahashi T, Murakami H, et al. Prognostic Impact of Circulating Tumor Cells in Patients With Small Cell Lung Cancer. J Thorac Oncol (2012) 7(3):512–9. doi: 10.1097/JTO.0b013e31823f125d
179. Peeters DJE, van Dam PJ, Van den Eynden GGM, Rutten A, Wuyts H, Pouillon L, et al. Detection and Prognostic Significance of Circulating Tumour Cells in Patients With Metastatic Breast Cancer According to Immunohistochemical Subtypes. Br J Cancer (2014) 110(2):375–83. doi: 10.1038/bjc.2013.743
180. Gadgeel SM, Garassino MC, Esteban E, Speranza G, Felip E, Hochmair MJ, et al. KEYNOTE-189: Updated OS and Progression After the Next Line of Therapy (PFS2) With Pembrolizumab (Pembro) Plus Chemo With Pemetrexed and Platinum vs Placebo Plus Chemo for Metastatic Nonsquamous NSCLC. J Clin Oncol (2019) 37(15_suppl):9013–3. doi: 10.1200/JCO.2019.37.15_suppl.9013
181. Malone ER, Oliva M, Sabatini PJB, Stockley TL, Siu LL. Molecular Profiling for Precision Cancer Therapies. Genome Med (2020) 12(1):8. doi: 10.1186/s13073-019-0703-1
182. Bedard PL, Hansen AR, Ratain MJ, Siu LL. Tumour Heterogeneity in the Clinic. Nature (2013) 501(7467):355–64. doi: 10.1038/nature12627
183. Baxi S, Yang A, Gennarelli RL, Khan N, Wang Z, Boyce L, et al. Immune-Related Adverse Events for Anti-PD-1 and Anti-PD-L1 Drugs: Systematic Review and Meta-Analysis. Bmj (2018) 360:k793. doi: 10.1136/bmj.k793
184. Shaw AT, Riely GJ, Bang YJ, Kim DW, Camidge DR, Solomon BJ, et al. Crizotinib in ROS1-Rearranged Advanced Non-Small-Cell Lung Cancer (NSCLC): Updated Results, Including Overall Survival, From PROFILE 1001. Ann Oncol Off J Eur Soc Med Oncol (2019) 30(7):1121–6. doi: 10.1093/annonc/mdz131
185. Ramalingam SS, Gray J, Ohe Y, Cho B, Vansteenkiste J, Zhou C, et al. Osimertinib vs Comparator EGFR-TKI as First-Line Treatment for EGFRm Advanced NSCLC (FLAURA): Final Overall Survival Analysis. Ann Oncol (2019) 30(s5):914–+. doi: 10.1093/annonc/mdz394.076
186. Garon EB, Hellmann MD, Rizvi NA, Carcereny E, Leighl NB, Ahn MJ, et al. Five-Year Overall Survival for Patients With Advanced Non-Small-Cell Lung Cancer Treated With Pembrolizumab: Results From the Phase I KEYNOTE-001 Study. J Clin Oncol (2019) 37(28):2518–27. doi: 10.1200/JCO.19.00934
187. Camidge DR, Dziadziuszko R, Peters S, Mok T, Noe J, Nowicka M, et al. Updated Efficacy and Safety Data and Impact of the EML4-ALK Fusion Variant on the Efficacy of Alectinib in Untreated ALK-Positive Advanced Non–Small Cell Lung Cancer in the Global Phase III ALEX Study. J Thorac Oncol (2019) 14(7):1233–43. doi: 10.1016/j.jtho.2019.03.007
188. Eslami-S Z, Cortés-Hernández LE, Alix-Panabières C. Circulating Tumor Cells: Moving Forward Into Clinical Applications. Precis Cancer Med (2019) 3:4. doi: 10.21037/pcm.2019.11.07
189. Geeurickx E, Hendrix A. Targets, Pitfalls and Reference Materials for Liquid Biopsy Tests in Cancer Diagnostics. Mol Aspects Med (2020) 72:100828. doi: 10.1016/j.mam.2019.10.005
190. Zhao Q, Yuan Z, Wang H, Zhang H, Duan G, Zhang X. Role of Circulating Tumor Cells in Diagnosis of Lung Cancer: A Systematic Review and Meta-Analysis. J Int Med Res (2021) 49(3):300060521994926. doi: 10.1177/0300060521994926
191. Gallo M, Luca AD, Maiello MR, D’Alessio A, Esposito C, Chicchinelli N, et al. Clinical Utility of Circulating Tumor Cells in Patients With Non-Small-Cell Lung Cancer. Transl Lung Cancer Res (2017) 6(4):486–98. doi: 10.21037/tlcr.2017.05.07
192. Keller L, Belloum Y, Wikman H, Pantel K. Clinical Relevance of Blood-Based ctDNA Analysis: Mutation Detection and Beyond. Br J Cancer (2021) 124(2):345–58. doi: 10.1038/s41416-020-01047-5
193. Zhang T, Karsh LI, Nissenblatt MJ, Canfield SE. Androgen Receptor Splice Variant, AR-V7, as a Biomarker of Resistance to Androgen Axis-Targeted Therapies in Advanced Prostate Cancer. Clin Genitourin Cancer (2020) 18(1):1–10. doi: 10.1016/j.clgc.2019.09.015
194. Lyu M, Zhou J, Ning K, Ying B. The Diagnostic Value of Circulating Tumor Cells and ctDNA for Gene Mutations in Lung Cancer. OncoTargets Ther (2019) 12:2539–52. doi: 10.2147/OTT.S195342
Keywords: lung cancer, NSCLC, circulating tumour DNA (ctDNA), circulating tumour cell (CTC), liquid biopsy
Citation: Kapeleris J, Ebrahimi Warkiani M, Kulasinghe A, Vela I, Kenny L, Ladwa R, O’Byrne K and Punyadeera C (2022) Clinical Applications of Circulating Tumour Cells and Circulating Tumour DNA in Non-Small Cell Lung Cancer—An Update. Front. Oncol. 12:859152. doi: 10.3389/fonc.2022.859152
Received: 21 January 2022; Accepted: 14 February 2022;
Published: 15 March 2022.
Edited by:
Pasquale Pisapia, University of Naples Federico II, ItalyCopyright © 2022 Kapeleris, Ebrahimi Warkiani, Kulasinghe, Vela, Kenny, Ladwa, O’Byrne and Punyadeera. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chamindie Punyadeera, Yy5wdW55YWRlZXJhQGdyaWZmaXRoLmVkdS5hdQ==; Kenneth O’Byrne, ay5vYnlybmVAcXV0LmVkdS5hdQ==
 Joanna Kapeleris
Joanna Kapeleris Majid Ebrahimi Warkiani
Majid Ebrahimi Warkiani Arutha Kulasinghe
Arutha Kulasinghe Ian Vela4,5,6
Ian Vela4,5,6 Liz Kenny
Liz Kenny