- 1Institute of Hematology, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China
- 2Department of Clinical Laboratory, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China
- 3Collaborative Innovation Center of Hematology, Soochow University, Suzhou, China
Background: This study investigated the high-risk factors associated with the increased vulnerability for subsequent clinical CR-GNB infection in carbapenem-resistant Gram-negative bacteria (CR-GNB)-colonized hematological malignancy (HM) patients and built a statistical model to predict subsequent infection.
Method: All adult HM patients with positive rectoanal swabs culture for CR-GNB between January 2018 and June 2020 were prospectively followed to assess for any subsequent CR-GNB infections and to investigate the risk factors and clinical features of subsequent infection.
Results: A total of 392 HM patients were enrolled. Of them, 46.7% developed a subsequent clinical CR-GNB infection, with 42 (10.7%) cases of confirmed infection and 141 (36%) cases of clinically diagnosed infection. Klebsiella pneumoniae was the dominant species. The overall mortality rate of patients colonized and infected with CR-GNB was 8.6% and 43.7%. A multivariate analysis showed that remission induction chemotherapy and the duration of agranulocytosis, mucositis, and hypoalbuminemia were significant predictors of subsequent infection after CR-GNB colonization. According to our novel risk-predictive scoring model, the high-risk group were >3 times more likely to develop a subsequent infection in comparison with the low-risk group.
Conclusion: Our risk-predictive scoring model can early and accurately predict a subsequent CR-GNB infection in HM patients with CR-GNB colonization. The early administration of CR-GNB-targeted empirical therapy in the high-risk group is strongly recommended to decrease their mortality.
Introduction
Carbapenem-resistant Gram-negative bacteria (CR-GNB) are a major public health threat posed by the Centers for Disease Control and the World Health Organization (1, 2), Carbapenem-resistant Enterobacterales (CRE), Carbapenem-resistant Pseudomonas aeruginosa (CRPA), and Carbapenem-resistant Acinetobacter baumannii (CRA) are considered as carbapenem-resistant organisms (CROs) (3). In recent decades, their prevalence has increased year by year (4). The disease progresses rapidly, and the all-cause mortality of CR-GNB infected patients is high (ranging from 20% to 71%), which presents a tremendous challenge to clinicians. Limited treatment options are currently available, including ceftazidime/avibactam, aztreonam, fosfomycin, and polymyxin (5, 6). However, the delayed administration of these antibiotics still resulted in rapidly fatal outcomes.
Immunodeficiency due to primary diseases, neutropenia, high-dose chemotherapy, hematopoietic stem cell transplantation (HSCT), and the abuse of broad-spectrum antibiotics all lead to an increased risk of a CR-GNB infection in hematological malignancy (HM) patients, and the mortality rate of infection is significantly higher than that in other patients (7). More than half of HM patients are reported to die within 1 week after a CR-GNB infection, and their 30-day mortality rate is as high as 70.3% (8). Therefore, it has an extremely important clinical value to quantify the risk factors of a CR-GNB infection to guide the early diagnosis and appropriate target treatment in HM patients.
A pathogenic microorganism culture is the gold standard for a definite diagnosis of a CR-GNB infection, but a low positive rate of culture and long culture time directly delay the timing of medication and thus limit its therapeutic effect (9). Previous studies have shown that a CR-GNB infection is usually caused by colonized bacteria invading the body and colonization can early predict the existence of infection (10, 11). At present, the common methods for detecting CR-GNB colonization include a rectoanal swab culture, fecal culture, and pharyngeal wipe culture. Of them, the rectoanal swab culture is most widely adopted because the specimen is easily obtained without the contamination of miscellaneous bacteria and can accurately reflect the gastrointestinal bacterial status of the patient (12). Active surveillance of CR-GNB colonization has been demonstrated to help provide multimodal prevention and control intervention strategies and thus effectively control CR-GNB outbreaks (13). However, gut colonization patients are a heterogeneous population, with only a small proportion developing into a clinical infection. The administration of anti-CR-GNB target antibiotics only depending on CR-GNB colonization will result in the abuse of antibiotics and increased bacterial resistance. Therefore, identifying the factors that influence asymptomatic colonization to develop into a subsequent CR-GNB infection and establishing an accurate and convenient prediction model for early recognizing high-risk CR-GNB infected patients after colonization may improve the empiric antibiotic prescription and decrease the mortality rate and healthcare costs.
In this study, we compared the clinical characteristics between CR-GNB colonization patients and CR-GNB infection patients developed from CR-GNB colonization and tried to determine the high-risk factor of subsequent infection. Meanwhile, we sought to develop a novel predictive model to predict who among CR-GNB colonization patients is prone to have a subsequent clinical CR-GNB infection. This might be helpful for identifying the real patients who may benefit from the early application of an anti-CR-GNB-targeted regimen.
Patients and Methods
Study Design and Subjects
This study retrospectively reviewed the data of inpatients with HMs from January 2018 to June 2020 in Wuhan Union Hospital, Tongji Medical College, Huazhong University of Science and Technology. All hospitalized patients were screened for CR-GNB colonization on admission and weekly thereafter. Once positive CR-GNB screening was found, the patients were included in the study and followed up to assess subsequent infection until the patients were discharged from the hospital. Meanwhile, the clinical and microbiological data were collected. Duplicate infection from the same patient was identified as one infection, and the first culture-positive strain was recorded.
Data Collection
Demographic and clinical data, including gender, age, primary disease, the length of hospitalization, HSCT, pneumonia, the duration of agranulocytosis, hypoalbuminemia, mucositis, exposure to antimicrobial agents or special drugs [chemotherapy, immunosuppressant, glucocorticoid, proton pump inhibitors (PPIs)] before infection, invasive devices [central venous catheter (CVC), mechanical ventilation, sputum suction, bladder catheterization] before infection, and the survival status within 28 days after we acquired the first positive rectoanal swab culture for CR-GNB were collected.
Bacterial Identification and Drug Sensitivity Test
Once the patients demonstrated clinical infection symptoms, the biological samples including blood, rectoanal swabs, vein catheter samples, tracheal secretions, bronchoalveolar lavage fluid, intraperitoneal fluid, or pleural drainage fluid were collected for the microorganism culture according to the location of infection. Bacterial identification and drug sensitivity tests were performed using a Vitek® 2 automated system (France Biomerieux) and matrix-assisted laser-desorption ionization time-of-flight mass spectrometry (Bruker Daltonics Inc., Billerica, MA, USA). The disk diffusion method (K-B method) was used to determine minimal inhibitory concentrations (MICs). All antibiotics, except tigecycline and colistin, were interpreted according to the standard of the CLSI document (14). For tigecycline and colistin, the European Committee on Antimicrobial Susceptibility Testing (EUCAST) breakpoint was used (15). Enterobacteriaceae with an MIC ≥4 µg/ml were considered as resistance to carbapenem. Pseudomonas aeruginosa and Acinetobacter spp. with an MIC ≥8 µg/ml were considered as resistance to carbapenem.
Grouping
All CR-GNB-colonized patients were followed up to discharge and grouped based on clinical infection manifestation, microorganism evidence, and therapeutic reaction to antibiotics to determine any subsequent CR-GNB infections.
CR-GNB Colonization Group
The patients met one of the following criteria during the follow-up period:
① Non-infection: The patients had no fever and any clinical presentations of infection.
② Non-CR-GNB infection: The patients developed fever (temperature >38° C at three different times within a 12 h period or as a temperature >38.5° C in a single measurement) and/or clinical presentations of infection (16, 17). Meanwhile, the patients had a good response to antibiotics targeting non-CR-GNB (including third- and/or fourth-generation cephalosporin, carbapenem, aminoglycoside, quinolones, and vancomycin) and/or anti-fungal treatment no matter if they had positive cultures for non-CR-GNB microorganisms and fungals.
CR-GNB Infection Group
The patients subsequently developed fever and/or clinical presentations of infection and met one of the following criteria:
① Confirmed CR-GNB infections: Patients had the presence of CR-GNB microbiologically documented infection [the isolation of CR-GNB from blood cultures or from a well-defined site of infection (urine, respiratory secretions obtained using sterile procedures, or fluid collection) (17)].
② Probable CR-GNB infection: Patients had the absence of CR-GNB and non-CR-GNB microorganism evidence and fungal infection evidence. Conventional anti-infective treatments targeting all non-CR-GNBs and fungals were ineffective, and subsequent antibiotics targeting CR-GNB were effective.
Unidentified Group
The patients subsequently developed fever and/or clinical presentations of infection and met one of the following criteria. The patients in this group were finally excluded from our study.
① The patients had the absence of CR-GNB and non-CR-GNB microorganism evidence and fungal infection evidence, but the anti-infective treatments targeting all non-CR-GNB and CR-GNB were ineffective.
② The patients had confirmed non-CR-GNB or fungal infection, but non-CR-GNB targeted anti-infective treatments and anti-fungal treatments were ineffective (18).
Related Definition
Hypoproteinemia refers to the serum albumin <30 mg/L.
Immunosuppressant therapy was defined as the use of at least 1 of the following drugs within 30 days before a CR-GNB infection, including cyclosporine, tacrolimus, and antithymocyte globulin (ATG)/antilymphocyte globulin (ALG).
Glucocorticoid refers to the use of dexamethasone within 1 month before CR-GNB infection (dose ≥20 mg/d, duration ≥5 days).
Antimicrobial exposure was defined as the use of antibiotics for more than 72 h before CR-GNB infection.
Statistical Analysis
Categorical data were analyzed utilizing Pearson’s chi-square or Fisher’s exact test, and continuous data were analyzed utilizing the Mann–Whitney U test or Student’s t-test, as appropriate. Logistic regression (backward LR) methods (univariate, multivariate) were used to determine the infection risk factors for HM patients with CR-GNB colonization. Odds ratios (ORs) and their corresponding 95% confidence interval (CI) were calculated. The final model was constructed based on a forward stepwise method with the likelihood ratio test. To develop the risk score, variables that had statistical significance in the multivariate regression model were assigned a reference value (Wij) according to the regression coefficients (βi). The risk factors in this study were all categorical variables. Dummy variables were set for categories, coding “0” and “1,” and “0” (the base category) was taken as the referent risk factor profile (WiREF). How far each risk factor is from the base category in terms of regression units (D) was calculated. The formula was D=βi * (Wij - WiREF). The constant in the points system (B) for scoring one point was set as the minimum of regression coefficient (βi). Finally, the points associated with each category of each risk factor were calculated by the following: Points=D/B=βi * (Wij - WiREF)/B. The final calculation results were rounded to an integer to obtain the scores corresponding to each risk factor. The sum of the scores generated by the calculated risk factors is the predicted score for that patient. The discrimination of the model was assessed by the receiver-operator curve (ROC) characteristics and the area under the curve (AUC). An optimal breakpoint was assigned using the Youden index, and integer up. R 3.6.1 software was used for the analyses. Statistical significance was assigned to a P-value of less than 0.05.
Results
Microbiological Characteristics
The flow procedure of this study is shown in Figure 1. A total of 437 HM patients with positive CR-GNB colonization were firstly included in this study, and 45 patients were classified into the unidentified group and finally excluded from our study. The data from the remaining 392 patients were further analyzed, and the top three predominant pathogens of CR-GNB colonization were Klebsiella pneumoniae (32.1%), Escherichia coli (18.4%), and Acinetobacter baumannii complex (15.8%). The other strains were Enterobacter cloacae (12.8%), Klebsiella acidogenes (4.6%), Pseudomonas aeruginosa (3.6%), Klebsiella ozaenae (2.6%), Citrobacter freundi (2.3%), Enterobacter aerogenes (1.2%), Enterobacter polycluster (1.2%), Proteus (1.8%), and others (3.6%) (Figure 2A).
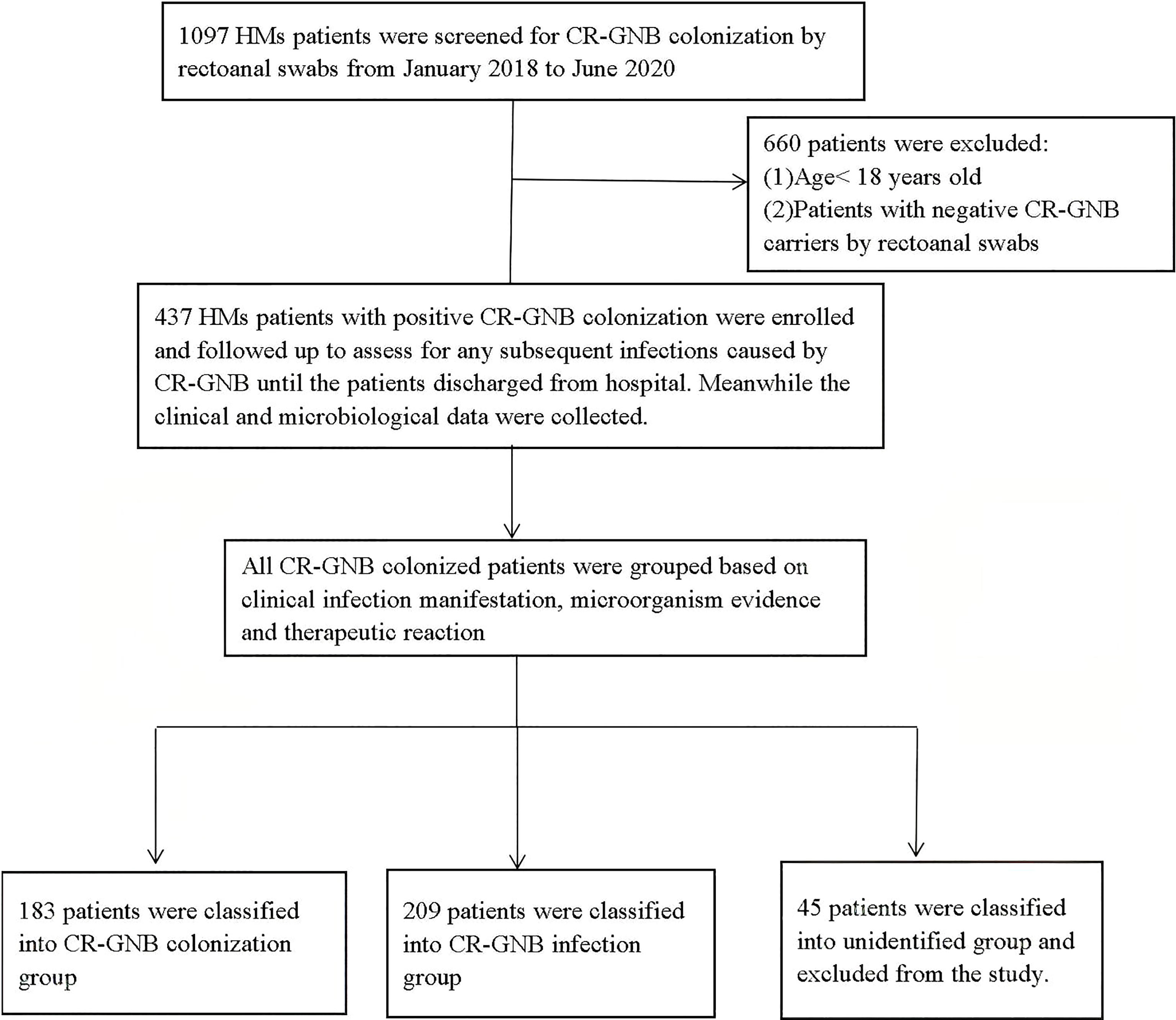
Figure 1 Screening algorithm of the patients with CR-GNB colonization. In all, 437 hospitalized patients had rectoanal swabs positive for CR-GNB, and a total of 392 eligible, unduplicated cases were recruited into this study.
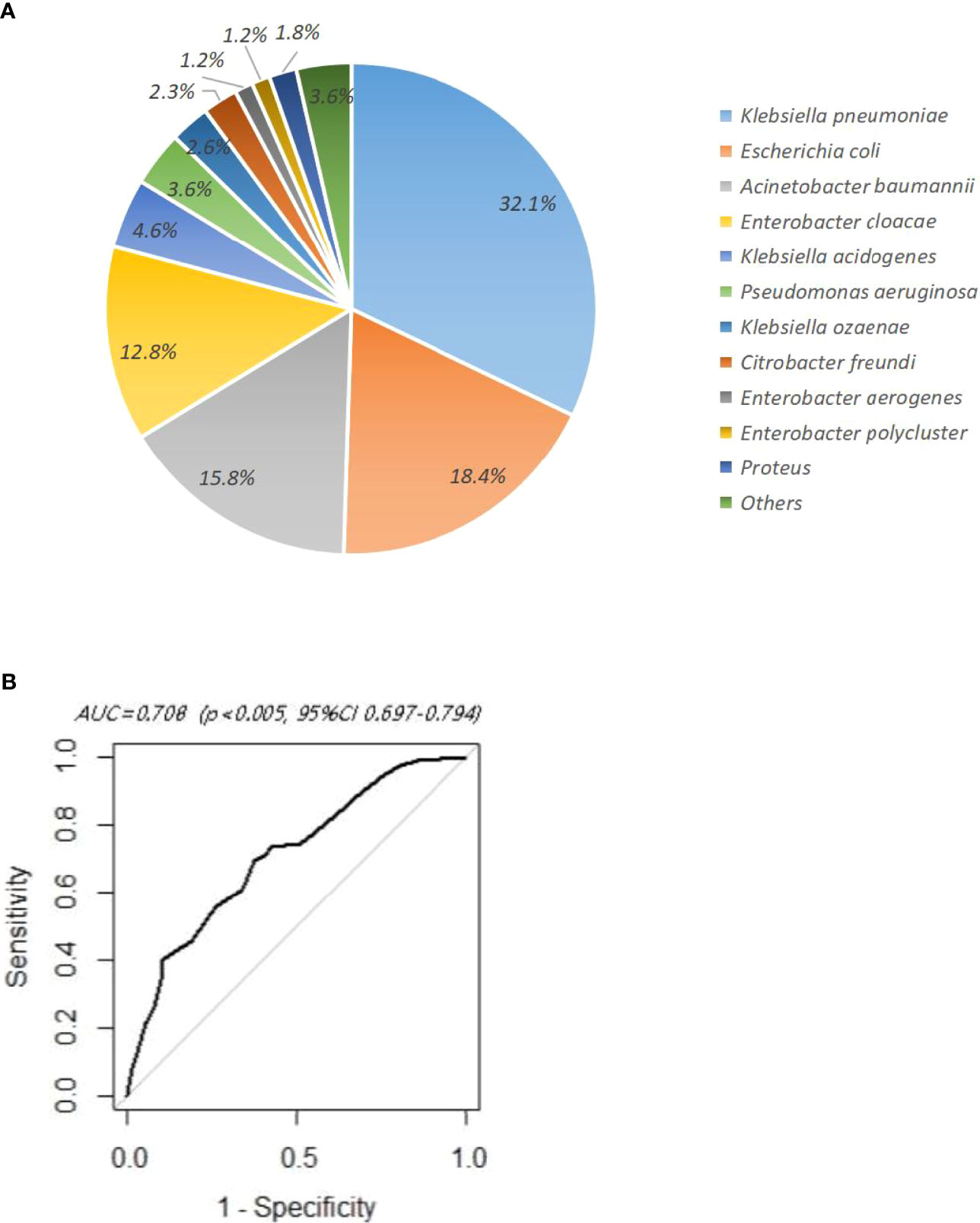
Figure 2 (A) Distribution of carbapenem-resistant Gram-negative bacteria in rectoanal swabs. (B) ROC curve of multivariate logistic regression analysis.
Clinical Characteristic Analysis
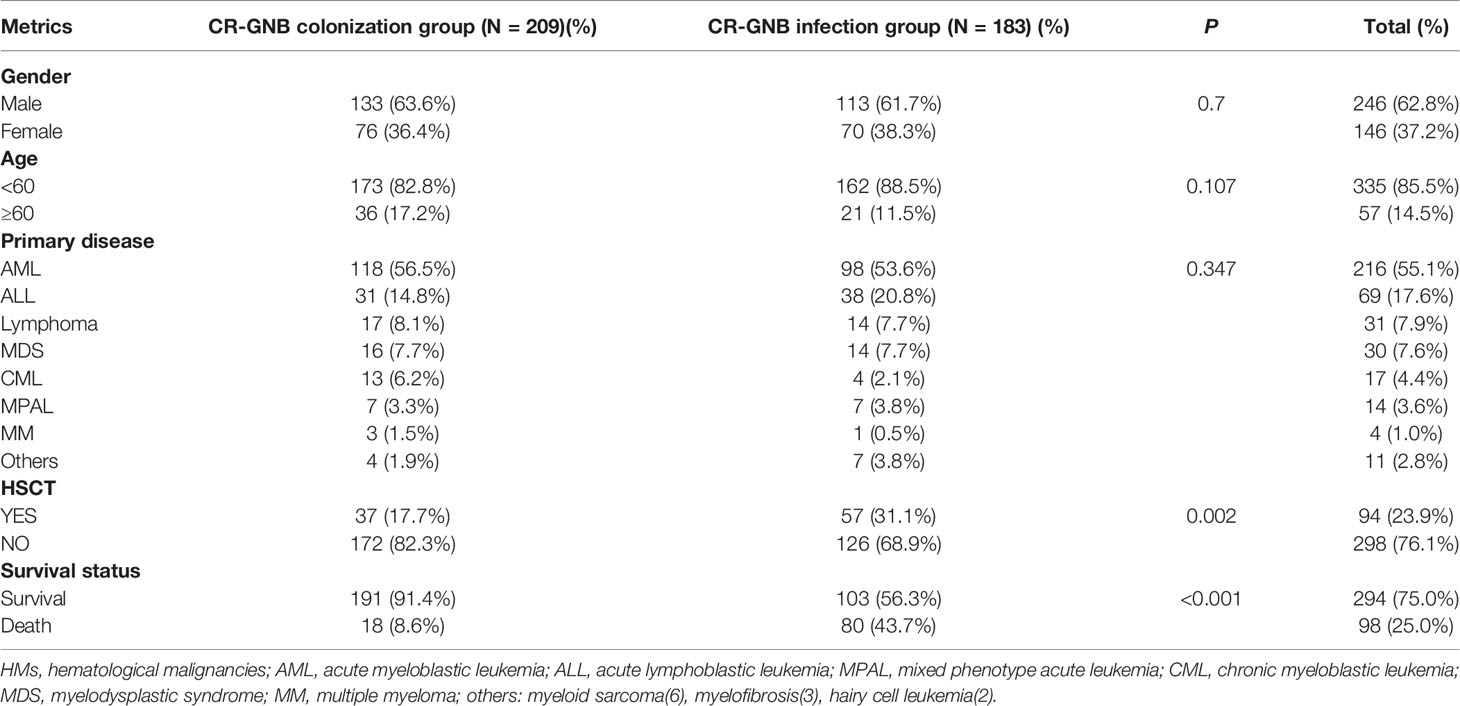
As shown in Table 1, among the 392 CR-GNB colonized patients, 183 (46.7%) subsequently developed a CR-GNB infection including 42 (10.7%) confirmed infections and 142 (36.2%) clinically diagnosed infections. The CR-GNB colonization group and infection group did not differ significantly in age and gender. In terms of disease distribution, acute myelocytic leukemia (AML) was the predominant primary disease (n=216, 55.1%), followed by acute lymphocytic leukemia (ALL) (n=69, 17.6%). CR-GNB infected patients had a longer length of hospitalization (27 d vs. 23 d, p=0.028) and significantly higher mortality rate (43.7% vs. 8.6%, P < 0.001).
Risk Factors for Subsequent Infection After CR-GNB Colonization
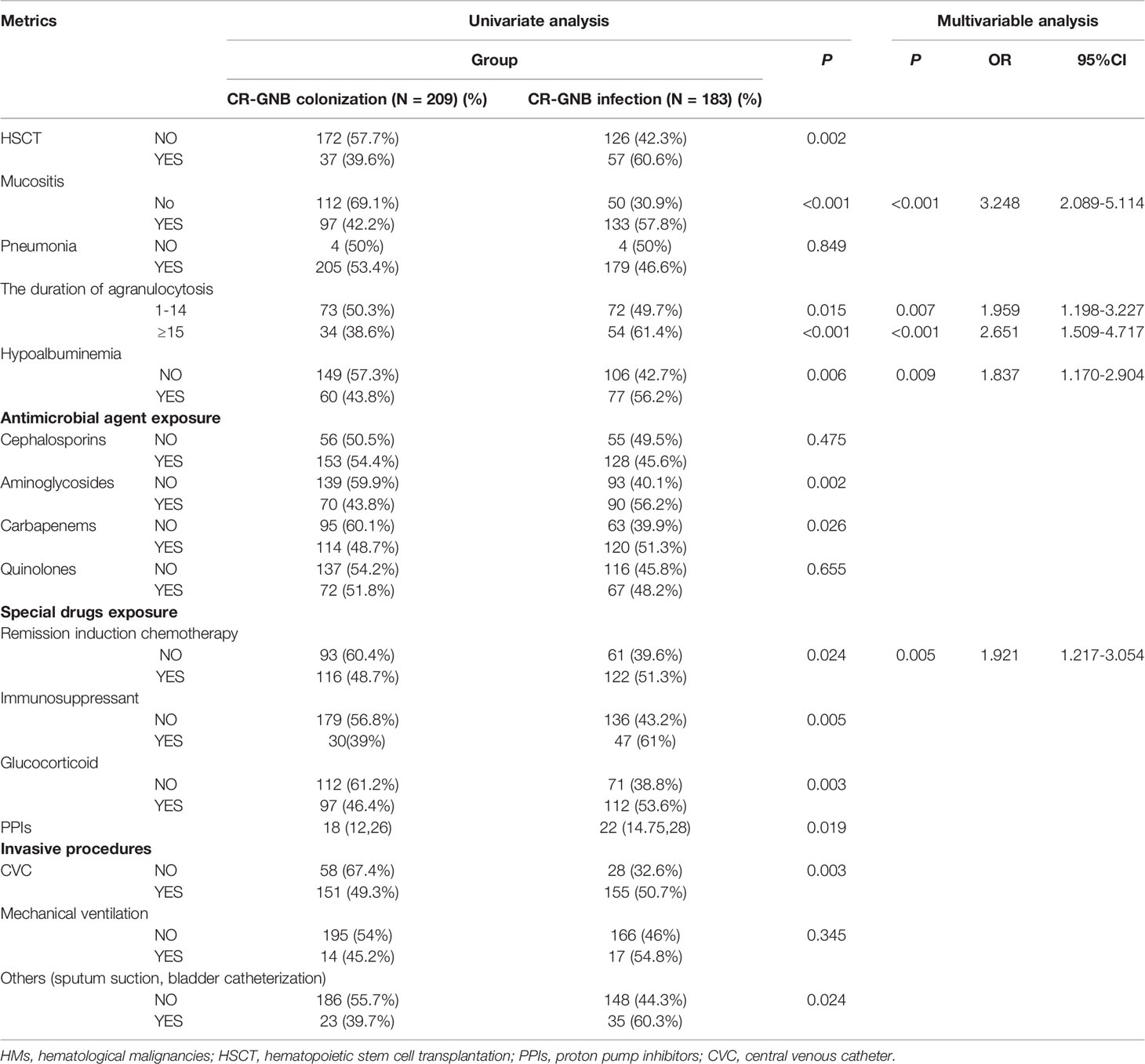
A univariate analysis (Table 2) showed that the risk factors for subsequent CR-GNB infection after CR-GNB colonization included HCST, mucositis, the duration of agranulocytosis, hypoalbuminemia, pre-exposure to specific agents (carbapenem antibiotic, aminoglycoside antibiotic, remission induction chemotherapy, immunosuppressant, glucocorticoids, PPIs), and invasive procedures (CVC, sputum suction, bladder catheterization) (p<0.05).
In order to avoid the interaction among the above risk factors and correct the bias, the factors with significant differences in univariate analysis were further included for multivariate logistic regression analysis. The results showed that remission induction chemotherapy (OR 1.921, 95%CI 1.217–3.054, p=0.005), the duration of agranulocytosis 1–14 days (OR 1.959, 95%CI 1.198–3.227, p=0.007), the duration of agranulocytosis ≥15 days (OR 2.651, 95%CI 1.509–4.717, p<0.001), mucositis (OR 3.248, 95%CI 2.089-5.114, p<0.001), and hypoalbuminemia (OR 1.837, 95% CI 1.170-2.904, p=0.009) were independent risk factors. The ROC curve showed that AUC=0.708>0.7 (p<0.005, 95%CI 0.697–0.794), indicating that multivariate analysis displayed an acceptable goodness of fit (Figure 2B).
The Establishment of a Risk Prediction Score Model for Subsequent CR-GNB Infection After CR-GNB Colonization
In order to further quantify the proportion of independent risk factors, a scoring table was established. As shown in Table 3, the assignment of points based on the ORs and the regression coefficients (βi) for these four independent variables generated an individual risk score ranging from 0 to 6 (AUC=0.697, p<0.05, 95%CI: 0.647–0.747). The maximum value of the Youden index and integer up was taken as the optimal cutoff value for the scoring model. The score of independent risk factors are as follows: remission induction chemotherapy (score 1), the duration of agranulocytosis 1–14 days (score 1), the duration of agranulocytosis ≥15 days (score 2), mucositis (score 2), and hypoalbuminemia (score 1).

Table 3 Predictive scoring table for risk of subsequent infection after CR-GNB colonization in HM patients.
According to the coordinates of the ROC curve, the optimal cutoff value was determined to be 4. Colonized patients with a total score <4 were defined as the low-risk infection group and a total score ≥4 were defined as the high-risk infection group. All colonized patients were validated in this model. Among them, 209 cases were classified into the low-risk group, and 68 cases (32.54%) had a subsequent CR-GNB infection. The rest of 183 cases were classified into the high-risk group, including 113 cases (61.75%) with a subsequent CR-GNB infection. There was a significant difference in the subsequent CR-GNB infection rates between the two groups (p<0.001). The OR was 3.347 (95% CI 2.218–5.094), suggesting that the risk of subsequent infection in the high-risk group was more than 3 times that in the low-risk group (Table 4). The sensitivity, specificity, positive predictive value, and negative predictive value were 62.4%, 67.1%, 0.6234, and 0.6682, respectively.
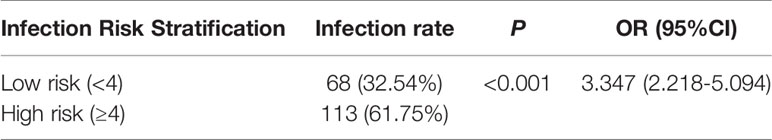
Table 4 Comparison of incidence of subsequent infection after CR-GNB colonization in different risk stratification groups.
Discussion
In this retrospective study, we found that 46.7% of HM patients developed a CR-GNB infection after CR-GNB colonization, including confirmed infections (10.7%) and clinical infections (36.2%). Mucositis and the duration of agranulocytosis ≥15 days were the strongest predictors. According to our predictive scoring model including remission induction chemotherapy (score 1), the duration of agranulocytosis 1–14 days (score 1), the duration of agranulocytosis ≥15 days (score 2), mucositis (score 2), and hypoalbuminemia (score 1), the total score ≥4 suggests that HM patients with CR-GNB colonization will develop a CR-GNB infection.
The overall prevalence of CR-GNB colonization varies between 18.1% and 30.4% in different geographical regions and diseases (19–24). In our study, the CR-GNB colonization rate in HM patients was 35.7%, which was higher than that in the above study. HM patients are prone to multiple drug-resistant bacterial infections, especially during febrile neutropaenic episodes (25, 26). It has been reported that the main CR-GNB strains include K. pneumoniae, K. acidogenes, Citrobacter freundi in Europe, and K. pneumoniae, Enterobacter cloacae, and Escherichia coli in Asia (21, 27–34). In our study, the dominant strains were K. pneumoniae (32.1%), E. coli (18.4%), and Acinetobacter baumanii (15.8%), which was consistent with previous reports.
Multiple risk factors were reported to be associated with increased vulnerability for a CR-GNB infection including immunocompromise, central venous catheter, chemotherapy or radiation therapy, neutropenia, carbapenems exposure, and prior colonization (21, 22, 34, 35). Similarly, in our univariate analysis, HCST, mucositis, the duration of agranulocytosis, hypoalbuminemia, pre-exposure to specific agents (carbapenem antibiotic, aminoglycoside antibiotic, remission induction chemotherapy, immunosuppressant, glucocorticoids, PPIs), and invasive procedures (CVC, sputum suction, bladder catheterization) may be risk factors for a subsequent CR-GNB infection after CR-GNB colonization. However, in our multivariate logistic regression analysis, only remission induction chemotherapy, the duration of agranulocytosis, mucositis, and hypoalbuminemia were independent risk factors.
During remission induction chemotherapy, extremely severe immunodeficiency is most common due to both high tumor burden and potent high-dose chemotherapeutic drugs. Furthermore, the incidence of febrile granulocytopenia is highest during the first cycle of anticancer chemotherapy (36). If the patients cannot receive the remission of primary disease after induction chemotherapy, this population has a longer neutropenia duration and especially a higher risk of CR-GNB infection (37). This is in accordance with our result that remission induction chemotherapy is an independent risk factor for CR-GNB infection. In addition, transplantation patients are another population with a higher colonization prevalence due to previous chemotherapy history compared with newly diagnosed HM patients (38, 39). However, stem cell transplantation and isolation in a laminar air-flow room have been also thought to be significant factors protecting against the occurrence of Gram-negative bacterial infections (40). These can explain that transplantation was no longer a significant risk factor in the multivariate analysis in our study.
Compared with short-term agranulocytosis in patients with other malignant tumors, a long duration of agranulocytosis is more common in HM patients due to an underlying disease and high-intensity chemotherapy and leads to their significantly reduced ability to resist pathogenic microorganisms (41). Studies have shown that more than 80% of HM patients will develop infections related to neutropenia after more than 1 course of chemotherapy, compared with 10%–50% of patients with solid tumors (42). In HM patients, leukemia patients undergoing intensive induction chemotherapy have especially prolonged episodes of neutropenia. The presence of febrile neutropenia was independently associated with increased mortality in infections caused by carbapenem-resistant Enterobacteriaceae in HM patients in a Latin American study (43). In this cohort, we also found that the duration of agranulocytosis (≥15 days) independently increases the risk of a subsequent infection due to CR-GNB colonization and thus negatively affects their clinical course.
Mucositis is a serious and debilitating side effect of cytotoxic chemotherapy and persistent reduction of neutrophils (44, 45). Herein, we found that mucositis is another independent risk factor for a CR-GNB infection. Oral and gut microbiome alterations are prevalent in HM patients due to the administration of chemotherapeutic drugs and broad-spectrum antibiotics, which favor the colonization or excessive growth of CR- GNB. Oral and gastrointestinal mucositis result in the damage of the mucous membrane barrier and promote colonized CR-GNB to enter into the blood circulation (46, 47). In HM patients with mucositis, CR-GNB-colonized patients may require increased vigilance for sepsis detection, owing to an increased risk for gut translocation and endogenous infection development.
Hypoalbuminemia is a common complication in hematological malignancies due to inadequate nutrition intake and cachexia, which are correlated with increased vascular permeability and interstitial volume. Furthermore, serum albumin has the effects of anti- oxidation and anti-apoptosis. Its reduction will cause low host immunity, a delayed repair of microcirculatory mucosal injury, and increased infection (48). A retrospective chart review confirmed that hypoalbuminemia is a clinical predictor of early infection in HM patients (49). In this study, a multivariate logistic showed that hypoalbuminemia was an independent risk factor for subsequent CR-GNB infection after CR-GNB colonization and accounted for 1 point in our predictive scoring model.
The prediction score model of infection for HM patients with CR-GNB colonization was seldomly reported. Recently, a risk prediction model for CRE bloodstream infection (BSI) in intestinal carriers in the hematology department and intensive care unit (ICU) was established in a retrospective study (50). Gastrointestinal injury, tigecycline exposure, and carbapenem resistance score were chosen as valuable markers for the risk prediction model of CRE BSI in intestinal carriers. However, the BSI rates are very low for HM patients (4.7%~23.1% in Europe and America and 4.6%-8.9% in China) (21, 51, 52), these prediction score models may miss some clinically infected patients with negative blood culture. Therefore, in this study, the patients were grouped by combining clinical infection manifestation, the microorganism evidence, and therapeutic reaction to antibiotics. Both CR-GNB microbiologically documented infections and clinical CR-GNB infections were included in CR-GNB infection group; only 10.2% of patients had positive blood cultures. We selected four independent variables including remission induction chemotherapy (score 1), the duration of neutropenia (score 1 for 1–14 days and score 2 for ≥15 days), mucositis (score 2), and hypoalbuminemia (score 1) to establish our predicting model and divided all the colonized patients into a low-risk group (<4 points) and a high-risk group (≥4 points). Our model has a certain sensitivity and specificity and is in high accordance with subsequent CR-GNB infection rates, and the high-risk group was >3 times more likely to develop a subsequent infection in comparison with the low-risk group (OR3.347, 95%CI 2.218–5.094, p<0.001). Therefore, the early administration of CR-GNB-targeted empirical therapy in a high-risk group is strongly recommended to decrease their mortality. Meanwhile, for those patients during low-risk periods, anti-non-CR-GNB treatment and close monitoring might be enough.
This study has several important limitations including its retrospective, observational design. Due to species preservation and equipment, we did not identify the enzyme type of CR-GNB strain and there is no information on the specific carbapenemases (KPC, OXA, etc.) identified in our studied population. An external validation cohort is needed to assess its discriminatory ability and goodness of fit. Additionally, the sample size of the transplantation group was still relatively small, limiting our ability to conduct a statistically significant comparison between the transplantation group and the nontransplantation group. Finally, it is needed to determine whether the established scoring model is reproducible through relevant prospective studies.
In conclusion, a CRGNB infection has become a major threat to public health, especially in HM patients. Not all CR-GNB colonized patients subsequently develop a CR-GNB clinical infection. Therefore, our model is of great clinical value to guide the clinicians for the selection of early prophylactic anti-CR-GNB treatment to reduce mortality during high-risk periods. Meanwhile for those patients during low-risk periods, anti-non-CR-GNB treatment and close monitoring might be enough, which can decrease the mortality rate, bacterial resistance rate, and healthcare costs.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The studies involving human participants were reviewed and approved by Independent Ethics Committee of Union Hospital, Tongji Medical College, Huazhong University of Science and Technology. Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
Author Contributions
MH conceived and designed the study. QW, CQ, HY, FL, YW, WL, LX, and LM collected and analyzed data. QW and CQ wrote the paper. MH, LX, and LM reviewed and edited the manuscript. All authors read and approved the manuscript.
Funding
This study was supported by the National Natural Science Foundation of PR China (no. 81570193 and no. 81770219, for QW).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Tacconelli E, Carrara E, Savoldi A, Harbarth S, Mendelson M, Monnet DL, et al. Discovery, Research, and Development of New Antibiotics: The WHO Priority List of Antibiotic-Resistant Bacteria and Tuberculosis. Lancet Infect Dis (2018) 18:318–27. doi: 10.1016/S1473-3099(17)30753-3
2. Centers for Disease Control and Prevention, USA. Antibiotic Resistance Threats in the United States. Atlanta, GA: Centers for Disease Control and Prevention (2013). Availabe at: http://www.cdc.gov/drugresistance/pdf/ar-threats-2013-508.
3. Kois AK, Nicolau DP, Kuti JL. Unresolved Issues in the Identification and Treatment of Carbapenem-Resistant Gram-Negative Organisms. Curr Opin Infect Dis (2020) 33:482–94. doi: 10.1097/QCO.0000000000000682
4. Yigit H, Queenan AM, Anderson GJ, Domenech-Sanchez A, Biddle JW, Steward CD, et al. Novel Carbapenem-Hydrolyzing Beta-Lactamase, KPC-1, From a Carbapenem-Resistant Strain of Klebsiella Pneumoniae. Antimicrob Agents Chemother (2001) 45:1151–61. doi: 10.1128/AAC.45.4.1151-1161.2001
5. Karakonstantis S, Kritsotakis EI, Gikas A. Pandrug-Resistant Gram-Negative Bacteria: A Systematic Review of Current Epidemiology, Prognosis and Treatment Options. J Antimicrob Chemother (2020) 75:271–82. doi: 10.1093/jac/dkz401
6. Karakonstantis S, Kritsotakis EI, Gikas A. Treatment Options for K. Pneumoniae, P. Aeruginosa and A. Baumannii Co-Resistant to Carbapenems, Aminoglycosides, Polymyxins and Tigecycline: An Approach Based on the Mechanisms of Resistance to Carbapenems. Infection (2020) 48:835–51. doi: 10.1007/s15010-020-01520-6
7. Torres-Gonzalez P, Cervera-Hernandez ME, Niembro-Ortega MD, Leal-Vega F, Cruz-Hervert LP, García-García L, et al. Factors Associated to Prevalence and Incidence of Carbapenem-Resistant Enterobacteriaceae Fecal Carriage: A Cohort Study in a Mexican Tertiary Care Hospital. PloS One (2015) 10:e0139883. doi: 10.1371/journal.pone.0139883
8. Lee JY, Kang CI, Ko JH, Lee WJ, Seok HR, Park GE, et al. Clinical Features and Risk Factors for Development of Breakthrough Gram-Negative Bacteremia During Carbapenem Therapy. Antimicrob Agents Chemother (2016) 60:6673–8. doi: 10.1128/AAC.00984-16
9. Asai N, Sakanashi D, Suematsu H, Kato H, Hagihara M, Nishiyama N, et al. The Epidemiology and Risk Factor of Carbapenem-Resistant Enterobacteriaceae Colonization and Infections: Case Control Study in a Single Institute in Japan. J Infect Chemother (2018) 24:505–9. doi: 10.1016/j.jiac.2018.02.005
10. Averbuch D, Tridello G, Hoek J, Mikulska M, Akan H, Yanez San Segundo L, et al. Antimicrobial Resistance in Gram-Negative Rods Causing Bacteremia in Hematopoietic Stem Cell Transplant Recipients: Intercontinental Prospective Study of the Infectious Diseases Working Party of the European Bone Marrow Transplantation Group. Clin Infect Dis (2017) 65:1819–28. doi: 10.1093/cid/cix646
11. Lübbert C, Becker-Rux D, Rodloff AC, Laudi S, Busch T, Bartels M, et al. Colonization of Liver Transplant Recipients With KPC-Producing Klebsiella Pneumoniae is Associated With High Infection Rates and Excess Mortality: A Case-Control Analysis. Infection (2014) 42:309–16. doi: 10.1007/s15010-013-0547-3
12. Wiener-Well Y, Rudensky B, Yinnon AM, Kopuit P, Schlesinger Y, Broide E, et al. Carriage Rate of Carbapenem-Resistant Klebsiella Pneumoniae in Hospitalised Patients During a National Outbreak. J Hosp Infect (2010) 74:344–9. doi: 10.1016/j.jhin.2009.07.022
13. Dai Y, Meng T, Wang X, Tang B, Wang F, Du Y, et al. Validation and Extrapolation of a Multimodal Infection Prevention and Control Intervention on Carbapenem-Resistant Klebsiella Pneumoniae in an Epidemic Region: A Historical Control Quasi-Experimental Study. Front Med (Lausanne) (2021) 8:692813. doi: 10.3389/fmed.2021.692813
14. C.L.S.I. (CLSI). Performance Standards for Antimicrobial Susceptibility Testing: Twenty-Four Informational Supplement. Wayne: Clinical and Laboratory Standards Institute (2020).
15. Åkerlund A, Jonasson E, Matuschek E, Serrander L, Sundqvist M, Kahlmeter G. EUCAST Rapid Antimicrobial Susceptibility Testing (RAST) in Blood Cultures: Validation in 55 European Laboratories. J Antimicrob Chemother (2020) 75:3230–8. doi: 10.1093/jac/dkaa333
16. Castagnola E, Mikulska M, Viscoli CJM, Douglas BS. Prophylaxis and Empirical Therapy of Infection in Cancer Patients. In: Mandell, Douglas, and Bennett's Principles and Practice of Infectious Diseases (2015). 3395–413.e2. doi: 10.1016/B978-1-4557-4801-3.00310-6
17. Nesher L, Rolston KV. The Current Spectrum of Infection in Cancer Patients With Chemotherapy Related Neutropenia. Infection (2014) 42:5–13. doi: 10.1007/s15010-013-0525-9
18. De Pauw B, Walsh TJ, Donnelly JP, Stevens DA, Edwards JE, Calandra T, et al. Revised Definitions of Invasive Fungal Disease From the European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group and the National Institute of Allergy and Infectious Diseases Mycoses Study Group (EORTC/MSG) Consensus Group. Clin Infect Dis (2008) 46:1813–21. doi: 10.1086/588660
19. Lee CM, Lai CC, Chiang HT, Lu MC, Wang LF, Tsai TL, et al. Presence of Multidrug-Resistant Organisms in the Residents and Environments of Long-Term Care Facilities in Taiwan. J Microbiol Immunol Infect (2017) 50:133–44. doi: 10.1016/j.jmii.2016.12.001
20. Le MN, Kayama S, Yoshikawa M, Hara T, Kashiyama S, Hisatsune J, et al. Oral Colonisation by Antimicrobial-Resistant Gram-Negative Bacteria Among Long-Term Care Facility Residents: Prevalence, Risk Factors, and Molecular Epidemiology. Antimicrob Resist Infect Control (2020) 9:45. doi: 10.1186/s13756-020-0705-1
21. Andria N, Henig O, Kotler O, Domchenko A, Oren I, Zuckerman T, et al. Mortality Burden Related to Infection With Carbapenem-Resistant Gram-Negative Bacteria Among Haematological Cancer Patients: A Retrospective Cohort Study. J Antimicrob Chemother (2015) 70:3146–53. doi: 10.1093/jac/dkv218
22. Mohan B, Prasad A, Kaur H, Hallur V, Gautam N, Taneja N. Fecal Carriage of Carbapenem-Resistant Enterobacteriaceae and Risk Factor Analysis in Hospitalised Patients: A Single Centre Study From India. Indian J Med Microbiol (2017) 35:555–62. doi: 10.4103/ijmm.IJMM_17_144
23. Schechner V, Kotlovsky T, Kazma M, Mishali H, Schwartz D, Navon-Venezia S, et al. Asymptomatic Rectal Carriage of blaKPC Producing Carbapenem-Resistant Enterobacteriaceae: Who Is Prone to Become Clinically Infected? Clin Microbiol Infect (2013) 19:451–6. doi: 10.1111/j.1469-0691.2012.03888.x
24. Averbuch D, Cordonnier C, Livermore DM, Mikulska M, Orasch C, Viscoli C, et al. Targeted Therapy Against Multi-Resistant Bacteria in Leukemic and Hematopoietic Stem Cell Transplant Recipients: Guidelines of the 4th European Conference on Infections in Leukemia (ECIL-4, 2011). Haematologica (2013) 98:1836–47. doi: 10.3324/haematol.2013.091330
25. Freifeld AG, Bow EJ, Sepkowitz KA, Boeckh MJ, Ito JI, Mullen CA, et al. Clinical Practice Guideline for the Use of Antimicrobial Agents in Neutropenic Patients With Cancer: 2010 Update by the Infectious Diseases Society of America. Clin Infect Dis (2011) 52:e56–93. doi: 10.1093/cid/cir073
26. Xing C, Ge B, Yu K, Gao S, Liang B, Ye H. Bloodstream Infections Caused by Klebsiella Pneumoniae Carbapenemase 2-Producing K. pneumoniae at a Hematology Department in Wenzhou, China. Intern Med (Tokyo Japan) (2016) 55:2087–91. doi: 10.2169/internalmedicine.55.6369
27. Asai S, Ohshima T, Iwashita H, Ishii Y, Aoki K, Minakawa Y, et al. Carbapenem-Resistant Klebsiella Pneumoniae in a Febrile Neutropenia Patient With Acute Myelogenous Leukemia After Hematopoietic Stem Cell Transplantation. Infect Dis Clin Pract (Baltimore Md) (2018) 26:e38–9. doi: 10.1097/IPC.0000000000000633
28. Zhang R, Dong N, Huang Y, Zhou H, Xie M, Chan EW, et al. Evolution of Tigecycline- and Colistin-Resistant CRKP (Carbapenem-Resistant Klebsiella Pneumoniae) In Vivo and its Persistence in the GI Tract. Emerg Microbes Infect (2018) 7:127. doi: 10.1038/s41426-018-0129-7
29. Huang L, Wang X, Feng Y, Xie Y, Xie L, Zong Z. First Identification of an IMI-1 Carbapenemase-Producing Colistin-Resistant Enterobacter Cloacae in China. Ann Clin Microbiol Antimicrob (2015) 14:51. doi: 10.1186/s12941-015-0112-2
30. Glasner C, Albiger B, Buist G, Tambić Andrasević A, Canton R, Carmeli Y, et al. Carbapenemase-Producing Enterobacteriaceae in Europe: A Survey Among National Experts From 39 Countries, February 2013. Euro Surveill (2013) 18(28):20525. doi: 10.2807/1560-7917.ES2013.18.28.20525
31. Martin RM, Cao J, Brisse S, Passet V, Wu W, Zhao L, et al. Molecular Epidemiology of Colonizing and Infecting Isolates of Klebsiella Pneumoniae. mSphere (2016) 1(5):e00261–16. doi: 10.1128/mSphere.00261-16
32. Mochon AB, Garner OB, Hindler JA, Krogstad P, Ward KW, Lewinski MA, et al. New Delhi Metallo-β-Lactamase (NDM-1)-Producing Klebsiella Pneumoniae: Case Report and Laboratory Detection Strategies. J Clin Microbiol (2011) 49:1667–70. doi: 10.1128/JCM.00183-11
33. Elias E, Targownik LE. The Clinician's Guide to Proton Pump Inhibitor Related Adverse Events. Drugs (2019) 79:715–31. doi: 10.1007/s40265-019-01110-3
34. Ho SW, Tsai MC, Teng YH, Yeh YT, Wang YH, Yang SF, et al. Population-Based Cohort Study on the Risk of Pneumonia in Patients With non-Traumatic Intracranial Haemorrhage Who Use Proton Pump Inhibitors. BMJ Open (2014) 4:e006710. doi: 10.1136/bmjopen-2014-006710
35. Ho SW, Teng YH, Yang SF, Yeh HW, Wang YH, Chou MC, et al. Association of Proton Pump Inhibitors Usage With Risk of Pneumonia in Dementia Patients. J Am Geriatr Soc (2017) 65:1441–7. doi: 10.1111/jgs.14813
36. Wan W, Wang J, Dong F, Zhao W, Tian L, Hu K, et al. Clinical Characteristics and Prognosis of 84 Elderly Patients With Acute Myeloid Leukemia. Zhongguo Shi Yan Xue Ye Xue Za Zhi (2019) 27:692–701. doi: 10.19746/j.cnki.issn.1009-2137.2019.03.009
37. Mills JP, Talati NJ, Alby K, Han JH. The Epidemiology of Carbapenem-Resistant Klebsiella Pneumoniae Colonization and Infection Among Long-Term Acute Care Hospital Residents. Infect Control Hosp Epidemiol (2016) 37:55–60. doi: 10.1017/ice.2015.254
38. Bilinski J, Robak K, Peric Z, Marchel H, Karakulska-Prystupiuk E, Halaburda K, et al. Impact of Gut Colonization by Antibiotic-Resistant Bacteria on the Outcomes of Allogeneic Hematopoietic Stem Cell Transplantation: A Retrospective, Single-Center Study. Biol Blood Marrow Transplant (2016) 22:1087–93. doi: 10.1016/j.bbmt.2016.02.009
39. Cordonnier C, Herbrecht R, Buzyn A, Leverger G, Leclercq R, Nitenberg G, et al. Risk Factors for Gram-Negative Bacterial Infections in Febrile Neutropenia. Haematologica (2005) 90:1102–9. doi: 10.3324/%25x
40. Freifeld AG, Bow EJ, Sepkowitz KA, Boeckh MJ, Ito JI, Mullen CA, et al. Clinical Practice Guideline for the Use of Antimicrobial Agents in Neutropenic Patients With Cancer: 2010 Update by the Infectious Diseases Society of America. Clin Infect Dis (2011) 52:427–31. doi: 10.1093/cid/ciq147
41. Klastersky J. Management of Fever in Neutropenic Patients With Different Risks of Complications. Clin Infect Dis (2004) 39 Suppl 1:S32–7. doi: 10.1086/383050
42. Gualtero S, Valderrama S, Valencia M, Rueda D, Muñoz-Velandia O, Ariza B, et al. Factors Associated With Mortality in Infections Caused by Carbapenem-Resistant Enterobacteriaceae. J Infect Dev Ctries (2020) 14:654–9. doi: 10.3855/jidc.12267
43. Niscola P. Mucositis in Malignant Hematology. Expert Rev Hematol (2010) 3:57–65. doi: 10.1586/ehm.09.71
44. Yarom N, Hovan A, Bossi P, Ariyawardana A, Jensen SB, Gobbo M, et al. Systematic Review of Natural and Miscellaneous Agents, for the Management of Oral Mucositis in Cancer Patients and Clinical Practice Guidelines - Part 2: Honey, Herbal Compounds, Saliva Stimulants, Probiotics, and Miscellaneous Agents. Support Care Cancer (2020) 28:2457–72. doi: 10.1007/s00520-019-05256-4
45. Guastalegname M, Russo A, Falcone M, Giuliano S, Venditti M. Candidemia Subsequent to Severe Infection Due to Clostridium Difficile: Is There a Link? Clin Infect Dis (2013) 57:772–4. doi: 10.1093/cid/cit362
46. Papadimitriou-Olivgeris M, Spiliopoulou A, Fligou F, Manolopoulou P, Spiliopoulou I, Vrettos T, et al. Association of KPC-Producing Klebsiella Pneumoniae Colonization or Infection With Candida Isolation and Selection of non-Albicans Species. Diagn Microbiol Infect Dis (2014) 80:227–32. doi: 10.1016/j.diagmicrobio.2014.07.012
47. Ballmer PE. Causes and Mechanisms of Hypoalbuminaemia. Clin Nutr (Edinburgh Scotland) (2001) 20:271–3. doi: 10.1054/clnu.2001.0439
48. Zhang S, Kobayashi K, Faridnia M, Skummer P, Zhang D, Karmel MI. Clinical Predictors of Port Infections in Adult Patients With Hematologic Malignancies. J Vasc Interv Radiol (2018) 29:1148–55. doi: 10.1016/j.jvir.2018.04.014
49. Wang Y, Lin Q, Chen Z, Hou H, Shen N, Wang Z, et al. Construction of a Risk Prediction Model for Subsequent Bloodstream Infection in Intestinal Carriers of Carbapenem-Resistant Enterobacteriaceae: A Retrospective Study in Hematology Department and Intensive Care Unit. Infect Drug Resist (2021) 14:815–24. doi: 10.2147/IDR.S286401
50. Satlin MJ, Cohen N, Ma KC, Gedrimaite Z, Soave R, Askin G, et al. Bacteremia Due to Carbapenem-Resistant Enterobacteriaceae in Neutropenic Patients With Hematologic Malignancies. J Infect (2016) 73:336–45. doi: 10.1016/j.jinf.2016.07.002
51. Zhang L, Lu HW, Liu HL, Zhu XY, Tang BL, Zheng CC, et al. Pathogens and Clinical Characteristics of Bacterial Infection in Hematology Department Between 2010 and 2014. Zhonghua Xue Ye Xue Za Zhi (2016) 37:383–7. doi: 10.3760/cma.j.issn.0253-2727.2016.05.006
52. Xu CH, Zhu GQ, Lin QS, Wang LL, Wang XX, Gong JY, et al. A Single-Center Study on the Distribution and Antibiotic Resistance of Pathogens Causing Bloodstream Infection in Adult Patients With Hematological Disease During the Period 2014-2018. Zhonghua Xue Ye Xue Za Zhi (2020) 41:643–8. doi: 10.3760/cma.j.issn.0253-2727.2020.08.005
Keywords: carbapenem-resistant Gram-negative bacteria, hematologic malignancies, colonization, infection, rectoanal swabs, predictive model
Citation: Wu Q, Qian C, Yin H, Liu F, Wu Y, Li W, Xia L, Ma L and Hong M (2022) A Novel Risk Predictive Scoring Model for Predicting Subsequent Infection After Carbapenem-Resistant Gram-Negative Bacteria Colonization in Hematological Malignancy Patients. Front. Oncol. 12:897479. doi: 10.3389/fonc.2022.897479
Received: 16 March 2022; Accepted: 12 April 2022;
Published: 11 May 2022.
Edited by:
J. Luis Espinoza, Kanazawa University, JapanReviewed by:
Hisham N. Altayb, King Abdulaziz University, Saudi ArabiaKasturi Banerjee, Stony Brook University, United States
Copyright © 2022 Wu, Qian, Yin, Liu, Wu, Li, Xia, Ma and Hong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Linghui Xia, bGluZ2h1aXhpYUBodXN0LmVkdS5jbg==; Ling Ma, bWFsaW5neGhAMTYzLmNvbQ==; Mei Hong, bWVpaG9uZ2NuY25AYWxpeXVuLmNvbQ==
†These authors have contributed equally to this work and share first authorship
 Qiuling Wu
Qiuling Wu Chenjing Qian
Chenjing Qian Hua Yin
Hua Yin Fang Liu1
Fang Liu1 Weiming Li
Weiming Li Linghui Xia
Linghui Xia Mei Hong
Mei Hong