Abstract
Background:
The genomic features of cancer cells may confer the metastatic ability of lung adenocarcinoma (LUAD) to metastasize to specific organs. We aimed to identify the differences in genomic alterations between patients with primary LUAD with and without metastases and to elucidate the metastatic biology that may help developing biomarker-directed therapies for advanced or metastatic disease.
Methods:
A retrospective cohort of 497 patients with LUAD including 388 primary tumors (PR), 53 bone metastases (MT-bone), 30 liver metastases (MT-liver), and 26 brain metastases (MT-brain) was tested for genomic alterations by a next-generation sequencing assay.
Results:
The EGFR, TP53, TERT, LRP1B, CDKN2A, ERBB2, ALK, and KMT2C genes had a high frequency of mutations, and the mutations were shared by PR and metastases groups. TP53 and EGFR were the most common mutated genes. In comparison with PR, KRAS, STK11, ATM, NPM1, and ROS1 were significantly mutated in MT-brain, and TP53, MYC, RSPO2, CDKN2a, and CDKN2B were significantly mutated in MT-liver. The frequencies of TP53, CDKN2A, MTAP, PRKCI, and APC mutations were higher in MT-bone than that in PR. The ERBB, phosphoinositide-3-kinase/protein kinase B (PI3K-AKT), cell cycle, Fibroblast growth factor (FGF), and homologous recombination deficiency signaling pathways were affected in both PR and metastases, and there is higher frequency of mutations in metastases. Moreover, the co-mutations in patients with PR and metastasis were respectively analyzed. In addition, the programmed death ligand 1 (PD-L1) level was obviously related to tumor stage and tumor metastases, and the tumor mutational burden was correlated to clinicopathological features including age, gender, pathological stages, and tumor metastases. FGFR1, KAT6A, MYC, RAD21, TP53, and DAXX were also dramatically correlated to the tumor mutational burden.
Conclusion:
Metastases are the most devastating stage of tumors and the main cause of cancer-related deaths. Our results provided a clinically relevant view of the tumor-intrinsic mutational landscape of patients with metastatic LUAD.
Introduction
Metastasis to distant organs remains the leading cause of cancer-related death (1). Metastasis is a dynamic process involving the dissemination of cancer cell from their primary site to a distant organ and the subsequent colonization of cancer cells at that distant site (1). Most cancers have specific metastatic patterns: a phenomenon known as “organotropism’’ or “organ-specific metastasis” (2). For lung cancer, the most common sites of metastasis are the contralateral lung, brain, bone, and liver (3, 4).
Lung cancer is the most common cancer in the world and the leading cause of cancer death (5), which has the highest morbidity and mortality rates in the world (6). For patients with metastatic lung cancer, the overall survival of 5-year was less than 5% until the last decade. Clinical observations suggested that metastatic sites may predict prognosis (3). The outcomes of the liver and bone metastases were inferior to brain metastasis (3). Lung adenocarcinoma (LUAD) is the most common subtype of lung cancer (7), preferentially metastasizes to the brain, liver, contralateral lung, bone, and adrenal system (3). Although targeted therapies have significantly improved the treatment of patients with LUAD for the past few years, the prognosis is still poor (8), and more investigation on the additional therapeutic targets for LUAD is needed.
Targeted therapy and immunotherapy often require genomic alteration or their derivatives to serve as biomarkers for the identification of appropriate patients (9). All malignant tumor is the result of genetic variation, and the development of tumor is a process of accumulation of genetic and epigenetic changes (10). Next-generation sequencing (NGS) of the primary tumor samples has been widely acted as a practical method for identifying genetic variation in patients with lung cancer (11). In general, metastatic cancers carry mutations similar to those of the primary cancer, but additional mutations occur after transformation (12). NGS has been widely used to identify genetic variation between groups of the primary tumor samples and metastases in LUAD, but the association of cancer-intrinsic mutational status with organ-specific metastases in LUAD remains unclear. Elucidating the metastatic biology of lung cancer may help developing biomarker-directed therapies and improving treatment strategies of advanced or metastatic disease.
To evaluate the genomic features of cancer cells that may confer the metastatic ability to specific organs, we investigated the association of cancer-intrinsic mutational status with organs of metastases. Understanding the genomic profiles of non-metastatic and metastatic tumors at different sites could help guide the treatment of LUAD and future drug development.
Material and Methods
Patients
A total of 497 Chinese patients with LUAD were selected from 959 patients for this study, whose tumor tissue and matched blood specimens were collected by authors from patients admitted to the Shandong Provincial Hospital Affiliated to Shandong First Medical University and Fourth Hospital of Hebei Medical University during December 2017 to March 2020. From the 497 eligible patients with LUAD, 388 patients failed to detect any tumor out of the primary lung adenocarcinoma (PR) and 109 patients had metastasis, including 30 patients with liver metastases (MT-liver), 26 patients with brain metastases (MT-brain), and 53 patients with bone metastases (MT-bone). This study was approved by the Institution Review Board according to the Declaration of Helsinki and obtained the informed consent from all enrolled patients.
Sample Preparation
The formalin-fixed paraffin-embedded (FFPE) tumor samples and matched blood samples were retrieved from the accredited clinical hospitals. The NGS results of these two (tissue and blood) sampling methods are not completely consistent, and when used in clinical practice, they can be conditionally collected simultaneously as a mutual supplement to make the genetic information of tumor cells obtained by analysis more complete. The diagnosis of the histologic subtyping was affirmed through independent pathologists from OrigiMed (Shanghai). The percentage of tumor cells in each sample was 20% or more, and at least 50 ng of tumor tissue DNA was extracted for subsequent genetic analysis.
Targeted NGS and Genetic Analysis
Genomic profiling was carried out using a targeted panel of 450+ cancer-related genes (Yuansu, Origimed Inc.) (13, 14). The FFPE samples and matched blood samples were obtained for genetic alteration testing. Tumor mutation burden (TMB) was defined as somatic mutation of genomic detection, including coding base substitutions and indel mutation per megabase (muts/Mb). We defined TMB ≥ 10 muts/Mb as TMB-High and <10 muts/Mb as TMB-Low (15).
PD-L1 Staining
The PD-L1 staining was performed as previously described (16). The FFPE samples were stained using anti–PD-L1 antibody (Abcam, Cambridge, UK), and the percentage of positive PD-L1 staining cells was counted. Positive membrane staining of 1% of tumor cells or tumor-infiltrating immune cells was defined as positive for PD-L1.
Statistical Analysis
The Fisher’s exact test was used to analyze the relationship between TMB or PD-L1 expression and clinical indexes. Wilcoxon test and T-test were performed to analyzed the genes correlated to TMB. The Fisher’s exact test and the Chi-square test were used to compare gene-level mutation frequency between PR and MT-bone, MT-liver, or MT-brain. A p-value ≤ 0.05 was recognized statistically significant.
Results
Patient Characteristics
A total of 497 patients with LUAD were investigated in this study. Among them, 388 (78%) samples were diagnosed as primary tumors, and 53 (10.7%), 30 (6.0%), and 26 (5.2%) samples were metastases in bone, liver, and brain, respectively. In patients with primary LUAD, there were 181 (46.6%) stage I, 95 (24.4%) stage IV, 67 (17.3%) stage III, and 40 (10.3%) stage II patients, respectively. The median age of patients with primary LUAD was 61, ranging from 32 to 81; and that of patients with bone metastasis was 59, ranging from 33 to 82; patients with liver metastasis was 63.5, ranging from 36 to 85; and patients with liver metastasis was 60, ranging from 26 to 72. Among the 388 patients with primary LUAD, 211 were female and 177 were male; among the 53 patients with bone metastasis, 23 were female and 30 were male; among the 30 patients with liver metastasis, 13 were female and 17 were male; and among the 26 patients with brain metastasis, 10 were female and 16 were male. In patients with primary LUAD, 116 patients had smoked and 254 never smoked. Among the patients with bone metastasis, 12 patients had smoked and 29 never smoked; among the patients with liver metastasis, 12 patients had smoked and 14 never smoked; and among the patients with brain metastasis, three had a history of smoking and 17 never smoked. The detailed clinical characteristics of these patients were listed in Table 1.
Table 1
| Primary tumors (n = 388) | Bone metastases (n = 53) | Liver metastases (n = 30) | Brain metastases (n = 26) | |
|---|---|---|---|---|
| Age | ||||
| Median | 61 | 59 | 63.5 | 60 |
| Range | 32–81 | 33–82 | 36–85 | 26–72 |
| Gender | ||||
| Male | 177 (45.6%) | 30 (56.6%) | 17 (56.7%) | 16 (61.5%) |
| Female | 211 (54.4%) | 23 (43.4%) | 13 (43.3%) | 10 (38.5%) |
| Smoking status | ||||
| Smokers | 116 (29.9%) | 12 (22.65%) | 12 (40.0%) | 3 (11.5%) |
| Non-smokers | 254 (65.5%) | 29 (54.7%) | 14 (46.7%) | 17 (65.4%) |
| NA | 18 (4.6%) | 12 (22.65%) | 4 (13.3%) | 6 (23.1%) |
| Stage | ||||
| I | 182 (46.6%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| II | 42 (10.8%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| III | 67 (17.3%) | 2 (3.8%) | 0 (0.0%) | 0 (0.0%) |
| IV | 96 (24.7%) | 51 (96.2%) | 30 (100.0%) | 26 (100.0%) |
| NA | 1 (0.3%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| PD-L1 | ||||
| Negative | 117 (30.2%) | 27 (50.9%) | 10 (33.3%) | 9 (34.6%) |
| Positive | 26 (6.7%) | 6 (11.3%) | 5 (16.7%) | 6 (23.1%) |
| NA | 245 (63.1%) | 20 (37.7%) | 15 (50.0%) | 11 (42.3%) |
| TMB | ||||
| High | 67 (17.3%) | 8 (15.1%) | 6 (20.0%) | 13 (50.0%) |
| Low | 294 (75.8%) | 39 (73.6%) | 23 (76.7%) | 13 (50.0%) |
| NA | 27 (6.9%) | 6 (11.3%) | 1 (3.3%) | 0 (0.0%) |
Clinicopathological profile of patients.
The Mutational Landscape in Primary Tumors and Metastases
A total of 4843 mutations were found, including 209 fusions/rearrangements, 1,202 gene amplifications, 86 gene homozygous deletions, 1,627 substitutions, and 407 truncations, in the entire cohort: primary tumors (PR) and metastases (MT-liver, MT-bone, and MT-brain). The top 40 mutated genes in PR, MT-brain, MT-liver, and MT-bone were demonstrated in Figures 1A–D, respectively. The EGFR, TP53, TERT, LRP1B, CDKN2A, ERBB2, ALK, and KMT2C genes had a high frequency of mutations, and the mutations were shared by PR and metastases (MT-liver, MT-bone and MT-brain). TP53 and EGFR were the most frequently mutated genes, among which TP53 accounted for 83% of MT-liver and EGFR accounted for 70% of MT-bone. Notably, MAP3K13 mutation was found exclusively in MT-liver.
Figure 1
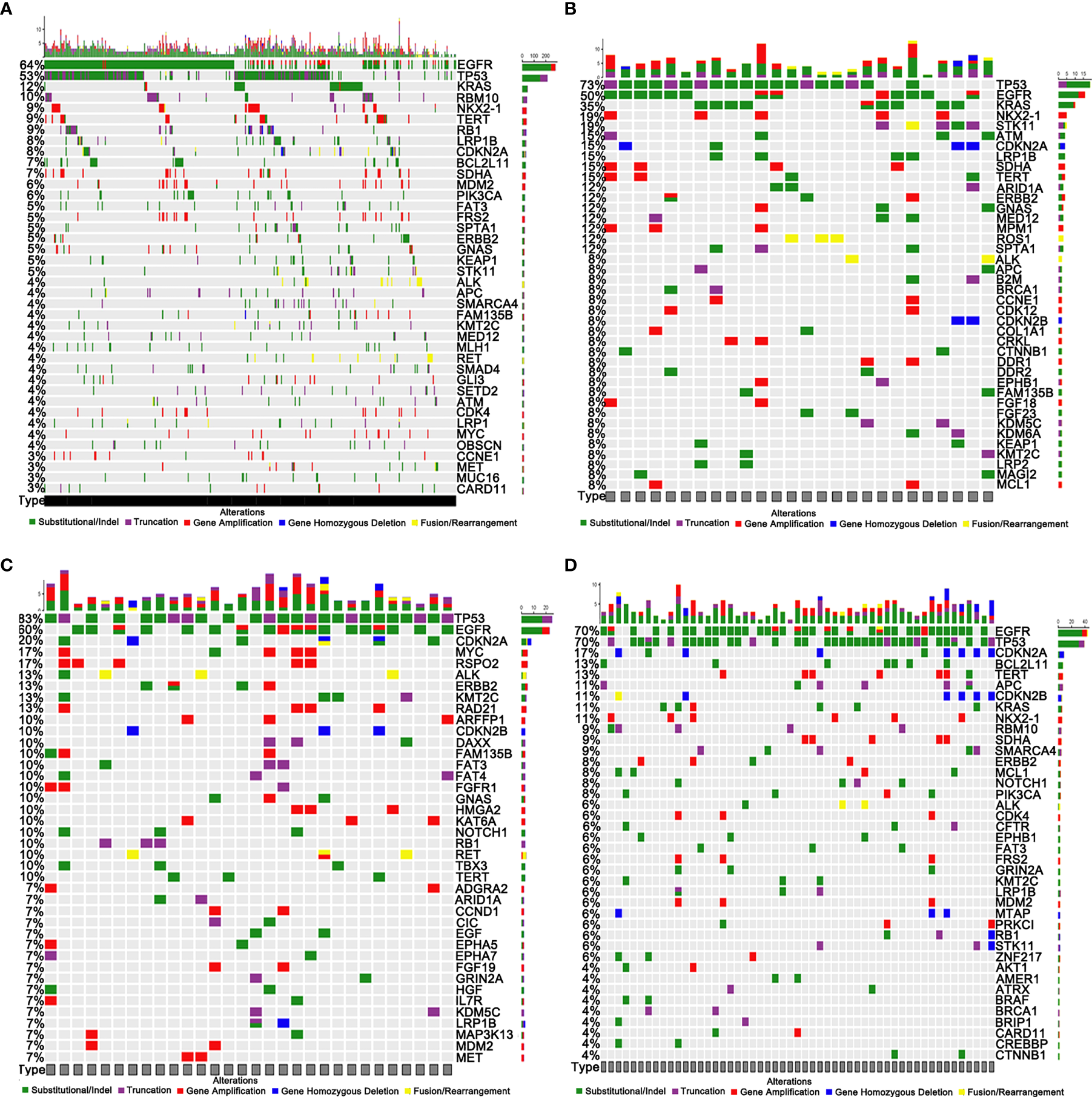
Genomic profiling of somatically altered genes and frequently mutated genes. (A) Primary tumors (PR). (B) Brain metastases (MT-brain). (C) Liver metastases (MT-liver). (D) Bone metastases (MT-bone).
Discrepancy of Mutation Frequency Between Primary Tumors and Metastases
The frequency of mutations was compared between PR and metastases by using the Fisher’s exact test. In comparison with PR, MT-brain, MT-liver, and MT-bone had more gene amplifications and gene homozygous deletions but fewer substitutions (Figures 2A–E). In addition, the frequently mutated genes in PR and metastases were displayed in Figures 3A–C. In comparison with PR, the mutations of ARID1A, ATM, B2M, COL1A1, CRKL, DDR1, KDM5C, KDM6A, KRAS, MTAP, NPM1, NRG3, ROS1, RUNX1T1, STK11, and VEGFA were significantly more frequent in MT-brain (Figure 3A). The mutation frequencies of ADGRA2, ARFRP1, CDKN2B, DAXX, EGF, EPHA7, FAT4, FGFR1, KAT6A, KMT2C, MSH2, MYC, NET1, NOTCH1, PRKDC, RAD21, RSPO2, TBX3, and TP53 were significantly different between PR and MT-liver (Figure 3B), whereas the mutation frequencies of APC, CDKN2A, CDKN2B, GRIN2A, MCL1, MTAP, PRKCI, and TP53 were significantly different between PR and MT-bone (Figure 3C).
Figure 2
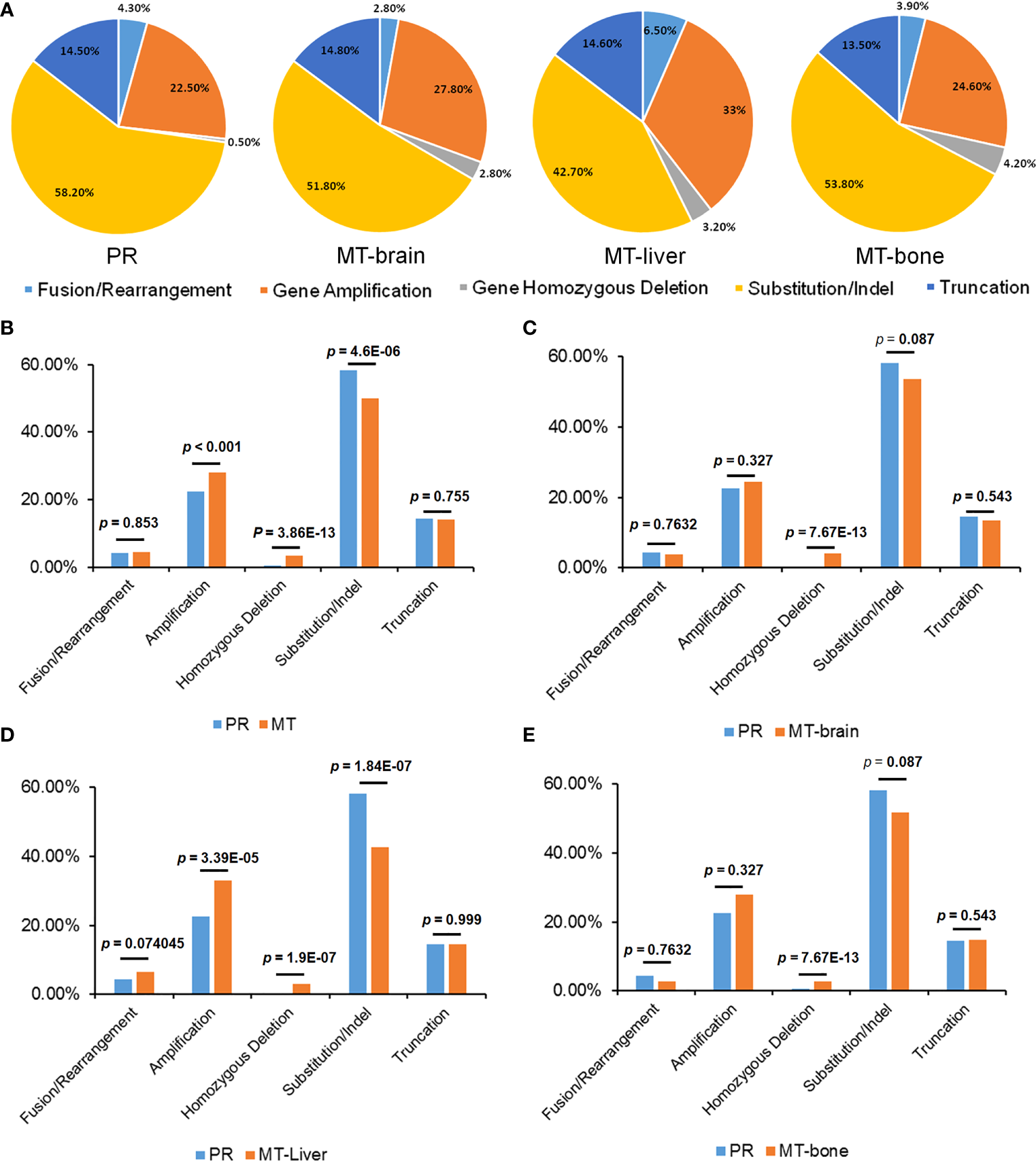
The distribution of mutation types of primary tumors and metastases. (A) The percentages of different mutation types in PR and metastases. (B) A comparison of the proportions of mutation types between patients with PR and metastasis, (C) PR and MT-brain, (D) PR and MT-liver, and (E) PR and MT-bone.
Figure 3
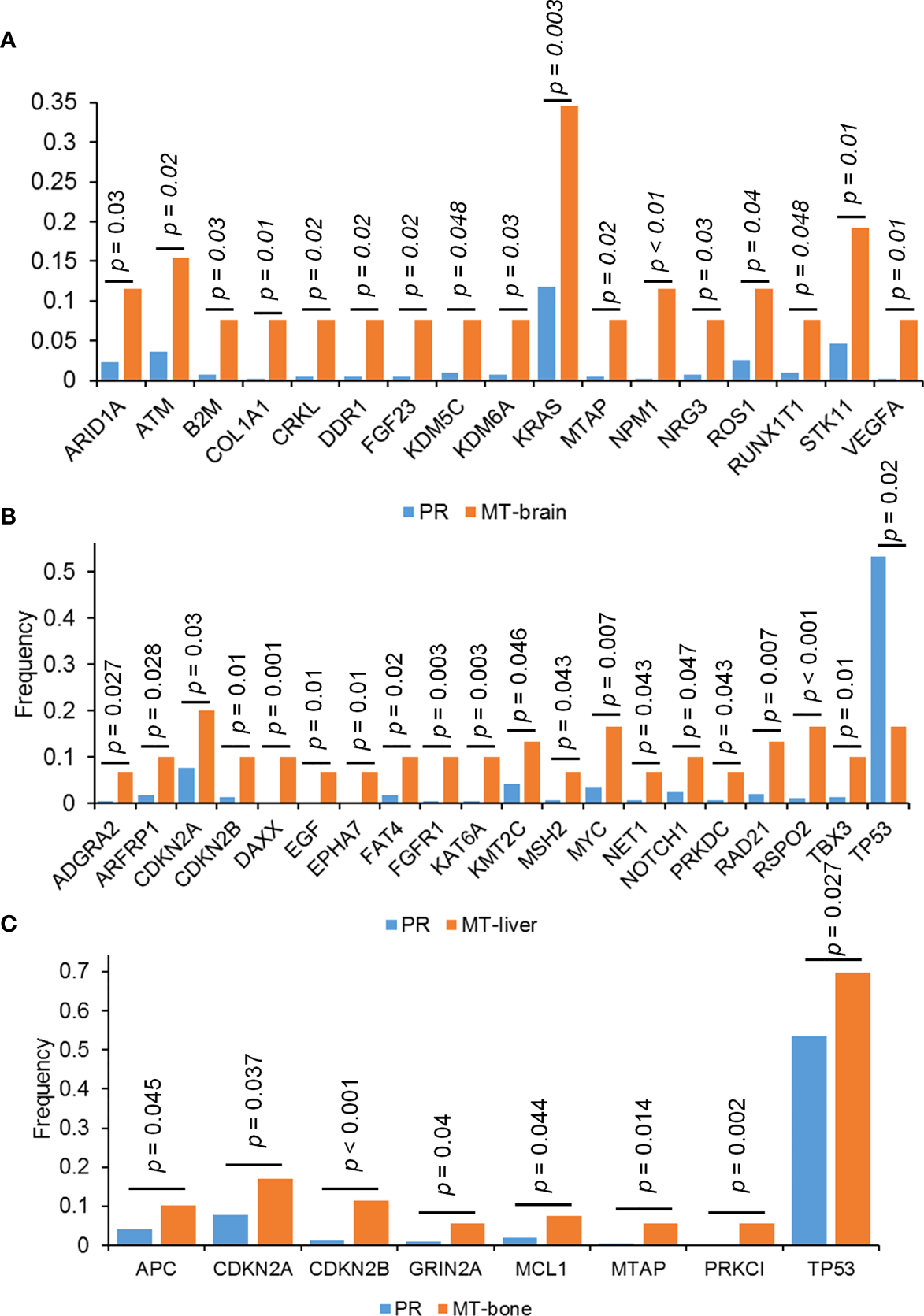
The differences of genomic mutations between PR and metastases. The differences of genomic mutations between (A) PR and MT-brain, (B) PR and MT-liver, and (C) PR and MT-bone.
To better discover the affected signaling pathways and guide the development of targeted therapy, we classified patients according to the presence and absence of mutation in each signaling pathway. The top five affected signaling pathways were ERBB, PI3K-AKT, cell cycle, FGF, and homologous recombination deficiency in PR and metastatic tumors, with a higher frequency of mutations in patients with metastasis (Figure 4).
Figure 4
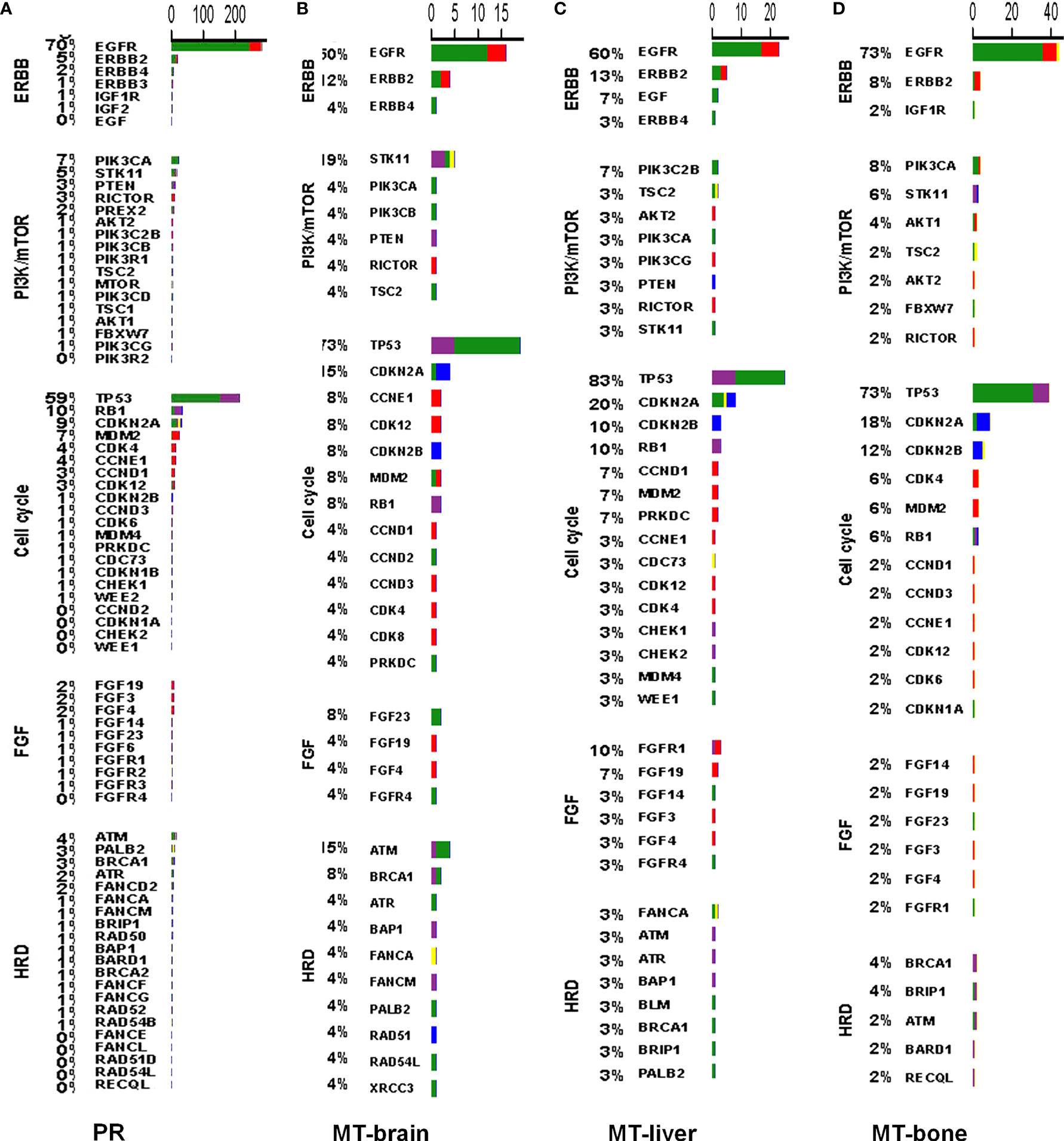
The mutation profiles according to signaling pathways in PR and metastases. The mutation profiles of the top five affected signaling pathways in (A) PR, (B) MT-brain, (C) MT-liver, and (D) MT-bone.
Co-Mutation Analysis in Primary Tumors and Metastases
Co-mutations could provide information for drug combination therapy and medication instruction. In PR, EGFR mutations and SMARCA4, ALK, STK11, KEAP1, SPTA1, LRP1B, and KRAS mutations were mutually exclusive, but EGFR mutations were found to be co-occurred with BCL2L11, RB1, and TP53 mutations. TP53 mutations were significantly co-occurred with LRP1B and RB1 mutations. KRAS, KEAP1, and FAM135B mutations were co-occurred with STK11 mutations, and NKX2-1 mutations were co-occurred with ERBB2, FRS2, and RB1 mutations, respectively. SDHA mutations were significantly co-occurred with TERT and RB1 mutations. MDM2 mutations were significantly co-occurred with FRS2 mutations. SMARCA4 mutations were co-occurred with FAT3 and SPTA1 mutations (Figure 5A). In MT-brain, LRP1B mutations were significantly co-occurred with SPTA1 mutations, ERBB2 mutations were co-occurred with CDK12 mutations, and SPTA1 mutations were significantly co-occurred with CCNE1 mutations (Figure 5B). In MT-liver, CDKN2B mutations were significantly co-occurred with CDKN2A mutations, and RAD21 mutations were obviously co-occurred with MYC and RSPO2 mutations. HGF mutations were co-occurred with FAM135B mutations, and CIC mutations were co-occurred with GNAS mutations (Figure 5C). In MT-bone, MDM2 and GRIN2A mutations were both significantly co-occurred with CDK4, FRS2, and GRIN2A mutations, and CDKN2B mutations were co-occurred with CDKN2A mutations. SDHA mutations were co-occurred with TERT mutations, and LRP1B mutations were co-occurred with KMT2C mutations (Figure 5D).
Figure 5
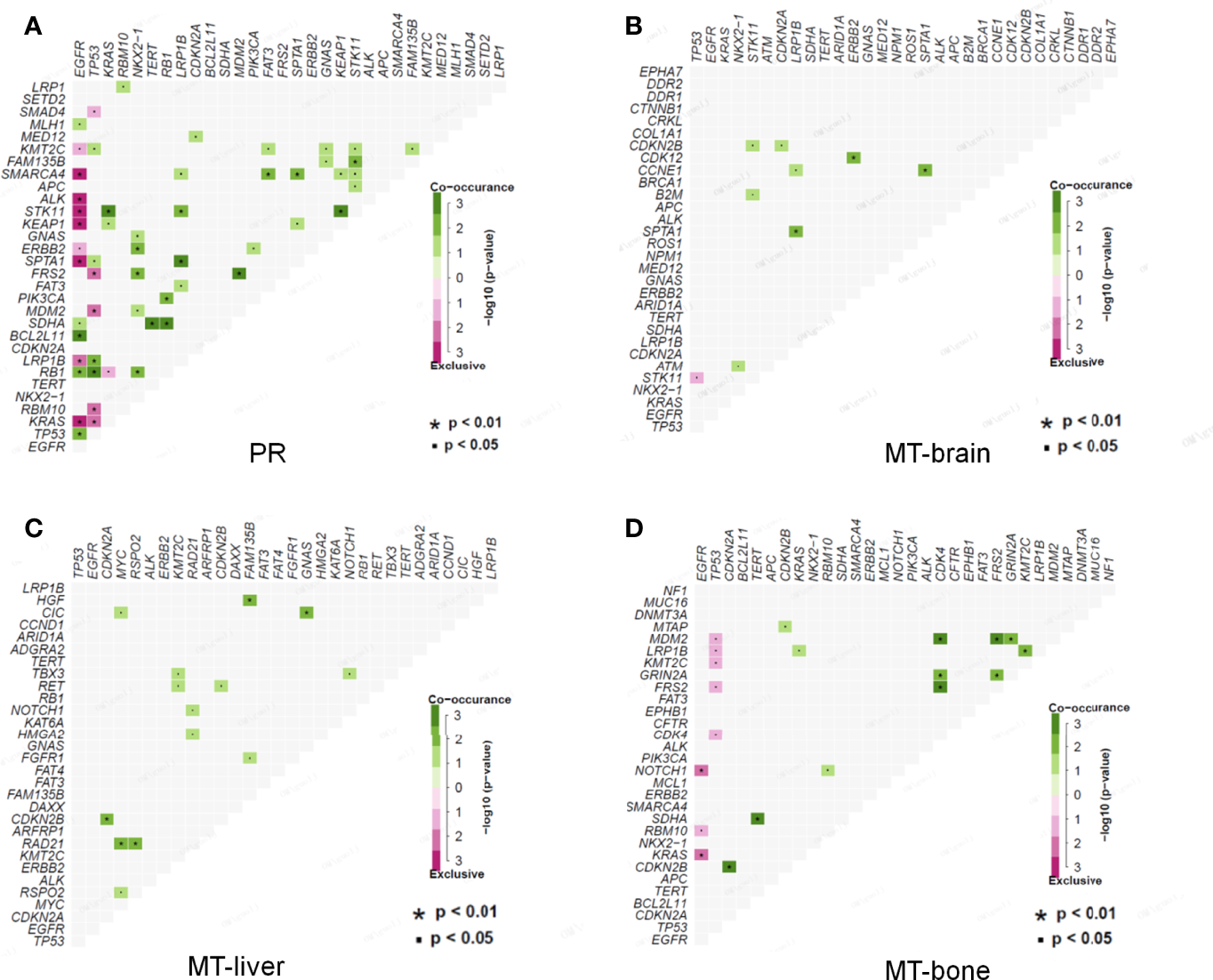
Co-occurrence of genomic alterations in PR and metastases. (A) The co-occurrence relationship between genomic mutations in PR is displayed in heatmap. (B–D) The co-occurrence relationships between genomic mutations in MT-brain, MT-liver, and MT-bone are shown in corresponding heatmaps, respectively.
Correlation Between Expression of PD-L1, TMB, and Clinicopathological Characteristics
TMB is a promising prognostic biomarker for immunotherapy across multiple cancer types. The median TMB for the cohort was 4.3 muts/Mb, with a range from 0.5 to 55.7. TMB-High was seen in 18.9% of patients, and TMB-Low was seen in 74.2% of patients. Higher percentages of patients had the late tumor stage and tumor metastasis condition, which were also displayed in the TMB-High group, suggesting that these clinical factors might be relate to higher TMB to some degree (Figure 6A). We determined the relationship between TMB level and the clinicopathological characteristics of LUAD. The results observed that the level of TMB was obviously correlated to the age, gender, pathological stages, and tumor metastasis (Figures 6B–F). TMB was also significantly associated with APC, KRAS, NOTCH1, SMARCA4, STK11, and ATRX mutations (Table 2).
Figure 6
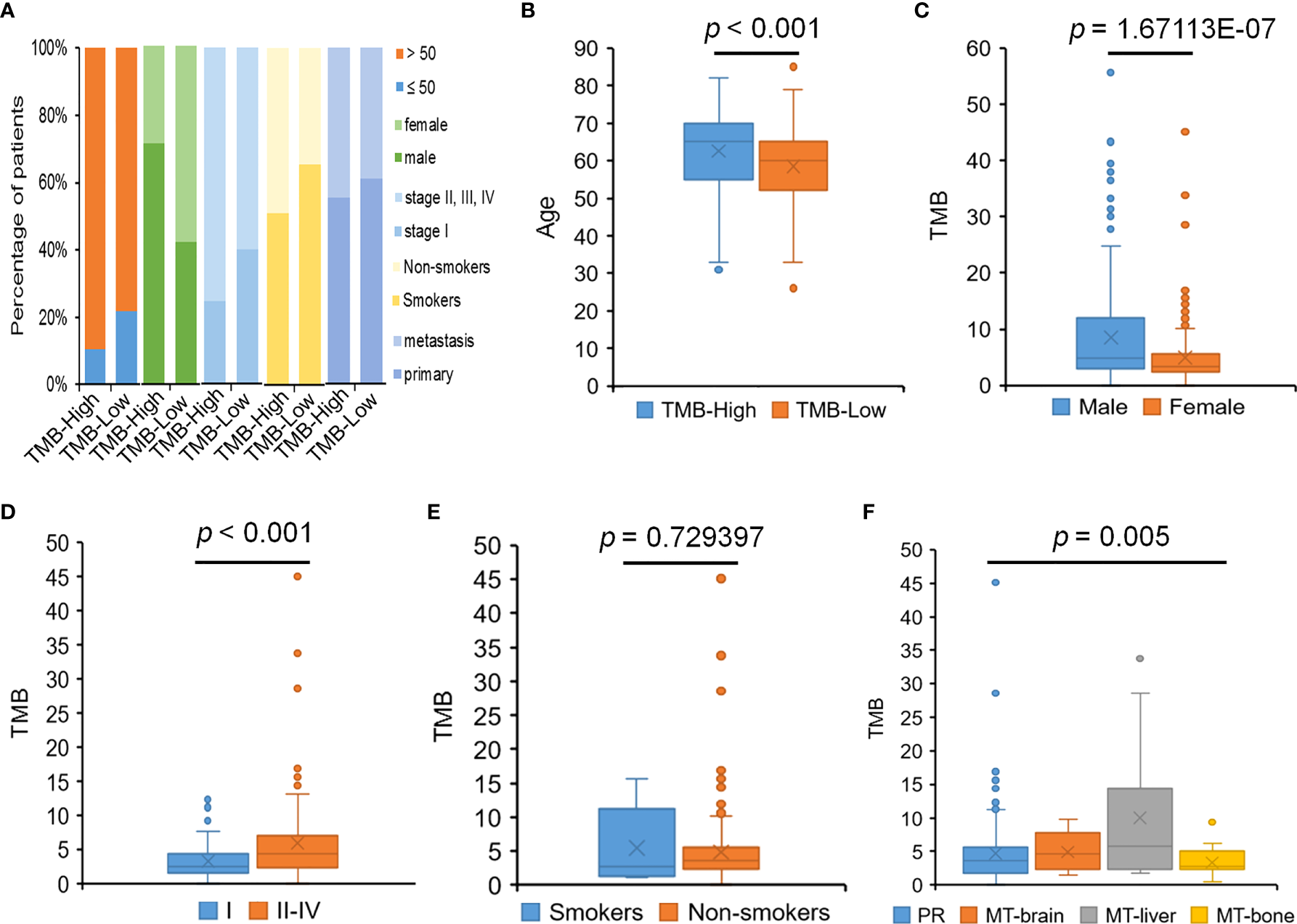
The relationship between TMB and clinical features. (A) The percentage of patients with different clinical features in the TMB-High and TMB-Low groups. (B) The relationship between TMB and age. (C) The relationship between TMB and gender. (D) The relationship between TMB and pathological stages. (E) The relationship between TMB and smoking history. (F) The relationship between TMB and tumor metastasis.
Table 2
| Gene | T-testp-value | Wilcox test p-value | Gene | T-testp-value | Wilcox test p-value |
|---|---|---|---|---|---|
| APC | 0.041871 | 0.004278 | KMT2C | 0.003101 | 3.32E−05 |
| ATRX | 0.029324 | 0.002635 | KRAS | 0.01462 | 8.58E−04 |
| BRCA1 | 0.014862 | 3.69E−04 | LRP1B | 2.19E−04 | 1.96E−08 |
| CIC | 0.04393 | 0.001201 | LRP2 | 0.046485 | 0.010335 |
| DICER1 | 0.04464 | 0.004705 | MAGI2 | 0.020854 | 4.43E−04 |
| EGFR | 9.56E−06 | 1.05E−04 | MUC16 | 0.002019 | 4.93E−06 |
| ERBB4 | 0.021124 | 0.004603 | NOTCH1 | 0.029449 | 0.002045 |
| ERRFI1 | 0.04559 | 0.00235 | NOTCH2 | 0.045895 | 0.002165 |
| FAM135B | 0.005188 | 1.26E−04 | NTRK2 | 0.015291 | 0.004905 |
| FANCA | 0.049024 | 2.57E−04 | PLCG2 | 0.041594 | 0.002491 |
| FAT3 | 0.014584 | 2.93E−04 | POLE | 0.00878 | 8.55E−06 |
| FAT4 | 0.009024 | 3.19E−04 | RB1 | 0.03535 | 8.56E−04 |
| GATA1 | 0.016516 | 9.58E−04 | SMARCA4 | 0.001403 | 2.43E−06 |
| GATA3 | 0.036069 | 0.037538 | SOX9 | 0.048958 | 0.011621 |
| GLI1 | 0.029308 | 0.008542 | SPEN | 0.020829 | 6.66E−04 |
| GNAS | 0.024932 | 0.014035 | SPTA1 | 4.57E−04 | 1.03E−06 |
| IL7R | 0.027336 | 8.68E−04 | STK11 | 0.003398 | 2.95E−06 |
| KDR | 0.036724 | 0.009621 | TP53 | 1.40E−06 | 1.94E−07 |
| KEAP1 | 0.02231 | 0.00137 |
The mutated genes associated with TMB.
Meanwhile, the relationships between the PD-L1 level and the clinicopathological features of LUAD were also analyzed. A total of 206 patients with LUAD enrolled in our study had PD-L1 staining results, and the proportion of PD-L1–positive cases accounted for 79.1%. The 206 patients included 15 patients with MT-brain (PD-L1 negative, 6; PD-L1 positive, 9), 33 patients with MT-bone (PD-L1 negative, 6; PD-L1 positive, 27), and 15 patients with MT-liver (PD-L1 negative, 5; PD-L1 positive, 10). We observed that PD-L1 expression was dramatically related to tumor metastases and pathological stages, whereas there was no significant relationship between the PD-L1 level and the other clinicopathological indexes including age, gender, and smoking history (Figure 7).
Figure 7
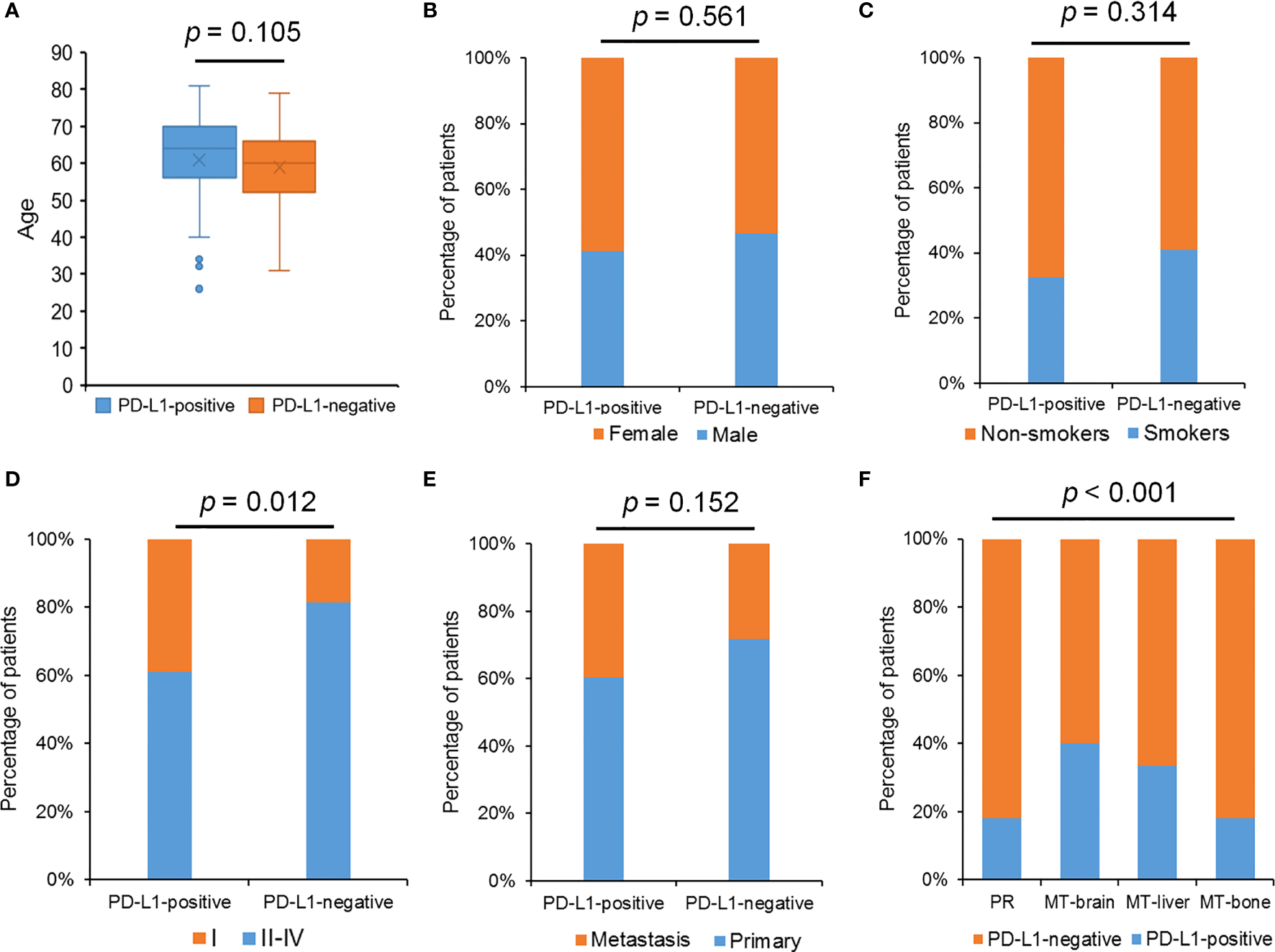
The relationship between PD-L1 expression and clinical features. (A) The relationship between PD-L1 expression and age. (B) The percentage of patients with different gender in the PD-L1–positive group and the PD-L1–negative group. (C) The percentage of patients with smoking history in the PD-L1–positive group and the PD-L1–negative group. (D) The percentage of patients with different pathological stages in the PD-L1–positive group and the PD-L1–negative group. (E) The percentage of patients with different tumor status in the PD-L1–positive group and the PD-L1–negative group. (F) The percentage of patients with different tumor metastasis status in the PD-L1–positive group and the PD-L1–negative group. The Chi-square test was used to detect p-values, and p-value ≤ 0.05 was recognized statistically significant.
Discussion
Most lung cancers have already metastasized at the time of initial diagnosis, and the survival rate of 5-year is poor (17). LUAD accounts for nearly 61% of the pathological subtypes of lung cancer, so it is of great research value to improve the survival rate of LUAD (18). Although more attention has been paid on LUAD gene sequencing, the genetic profile and the underlying mechanisms of metastatic cancer progression are still poorly understood. We hope that the discovery of the differences in genomic landscape between patients with primary LUAD with and without metastases in our study could provide guidance for the treatment and future drug development of LUAD.
Our study found that the frequently mutated genes including EGFR, TP53, KRAS, TERT, LRP1B, CDKN2A, ERBB2, ALK, and KMT2C were shared by PR and metastases. We found that EGFR mutations were frequently mutated in PR (64%), MT-brain (50%), MT-liver (50%), and MT-bone (70%). The results was consistent with a previous study reporting that EGFR was the most frequently shared driver gene, which accounted for more than 40%–50% of the whole LUAD population (19). KRAS mutations have been observed to be related to poor prognosis in resected lung cancer, lack of survival benefit from adjuvant chemotherapy, and resistance to erlotinib or gefitinib (20–22). The results in the study of Pao et al. have indicated that, by determining the mutant status of EGFR and KRAS, treatment decisions for the use of these kinase inhibitors can be improved (22). Gefitinib/erlotinib targeting EGFR mutations and crizotinib targeting ALK translocations have shown clinical benefit and approved for clinical use (23). telomerase reverse transcriptase (TERT) inhibition has been acted as a promising therapeutic strategy for LUAD (24, 25). These mutated genes of LUAD may serve as potential targets, providing more possibilities and strategies for the treatment of LUAD.
Considering the spatial heterogeneity of the tumor, the genetic mutation status of a few tumor cells in the primary site may not represent the mutation status of distant metastasis (26). ARID1A, ATM, B2M, COL1A1, CRKL, DDR1, KDM5C, KDM6A, KRAS, MTAP, NPM1, NRG3, ROS1, RUNX1T1, STK11, and VEGFA were significantly mutated in the 26 patients with brain metastases in our cohort, compared with PR. Shih et al. have shown that the most frequently mutated genes (MYC, TERT, MDM2, CDK4, CCND1, and NKX2–2) in both the brain metastasis–LUAD and The Cancer Genome Atlas (TCGA)-LUAD cohorts (27). A previous study revealed that more ARID1A mutations were observed in MT-brain than that in PR (28). The deficiency of MTAP to predict better treatment response in patients with advanced LUAD who receive early pemetrexed platin chemotherapy and bevacizumab (29). KRAS mutation was significantly more common in MT-brain than described for extracranial tumor manifestations (30). Whether the newly identified genes highly mutated in both MT-brain and PR can be used as predictive biomarkers and provide more guidance for the treatment of MT-brain remains to be validated. The mutation frequencies of ADGRA2, ARFRP1, CDKN2B, DAXX, EGF, EPHA7, FAT4, FGFR1, KAT6A, KMT2C, MSH2, MYC, NET1, NOTCH1, PRKDC, RAD21, RSPO2, TBX3, and TP53 were significantly different between PR and MT-liver. Emerging evidence supported that TP53 mutations augmented the metastatic potential of tumors (31). The underlying mechanism might be correlated to chromosomal instability or drug resistance but still remained to be further elucidated (32). In a study reported by Liao et al., in 2018, APC was only mutated in PR but not in MT-brain, whereas our study showed that APC was mutated not only in PR but also in MT-brain and MT-bone (33). Our results indicated that the mutation frequencies of APC, CDKN2A, CDKN2B, GRIN2A, MCL1, MTAP, PRKCI, and TP53 were significantly different between PR and MT-bone. CDKN2A/B were more abundant across all three metastatic cohorts. The mutations of CDKN2A/B were frequently involved in genomic deletions. CDKN2A/B are frequently mutated and tested in various tumors. Several studies indicated CDKN2A/B deleted or mutated patients can benefit from CDK4/6 inhibitors (34–37). In a phase II clinical trial (NCT01291017), advanced non–small cell lung cancer patients with wild-type RB and inactive CDKN2A can benefit from palbociclib treatment. Eight of the 16 patients have achieved stable disease for more than 4 months. Emergence of new genes with significantly higher mutation frequency in each organ metastasis compared with the primary tumors suggests that the different immunotherapeutic responses of each organ metastasis to highly mutated tumors and their sensitivity to PARP inhibitors may be a potential treatment for a specific metastatic organ.
In the past, it is generally accepted that lung cancer drive gene mutations were mutually exclusive (38). With the development of gene detection technology, cases of co-existing driver gene mutations were explored. In PR, KRAS, KEAP1, and FAM135B mutations were co-occurred with STK11 mutations. This finding was similar to previously result that co-mutations of both STK11 and KEAP1 were a strong determinant of unfavorable prognosis with currently available therapies (39). NKX2-1 mutation was co-occurred with ERBB2, FRS2, and RB1 mutations, which was different from a previous study reported a co-mutation of NKX2-1 and NEKB1A in PR (9). Patients with metastasis had different co-mutation profiles from PR. In MT-brain, LRP1B mutation was significantly co-occurred with SPTA1 mutation, and ERBB2 mutation was co-occurred with CDK12 mutation. RAD21 mutation was obviously co-occurred with MYC and RSPO2 mutations in MT-liver. In MT-bone, MDM2 mutation was significantly co-occurred with CDK4 mutation, and LRP1B mutation was co-occurred with KMT2C mutation. Previous studies have shown that co-mutations of TP53 and KRAS can function as potential biomarkers for immune checkpoint blockade in lung cancer (40) and that co-mutations of KRAS and TP53 could identify long-term responders to first-line palliative treatment with pembrolizumab from patients with LUAD with high PD-L1 level (41). In our study, we drafted the co-mutation profiles of LUAD with different site of metastasis, which might provide potential biomarkers for immune checkpoint blockade in LUAD with different organ metastases.
In conclusion, we analyzed the mutational profiles that represent the tumor-intrinsic factors of LUAD metastases. Metastatic cancer is a highly devastating disease. The development of novel systemic treatments of metastatic cancer depends on the insight into the therapeutic implications of metastatic heterogeneity. Our findings on genomic characterization of PR and metastasis provide a viable strategy for discovering potential pathways to prevent and treat metastatic LUAD.
Funding
This study was supported in part by the Natural Science Foundation of Shandong (Grant Nos. ZR2020MH229 and ZR2020QH200), the Radiation Oncology Translational Medicine Foundation for Scientific Research of Betthune (Grant No. flzh202123), and the special foundation for CSCO Cancer Research (Grant Nos. Y-QL2019-0149 and Y-2019AZMS-0522).
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Statements
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The studies involving human participants were reviewed and approved by Fourth Hospital of Hebei Medical University. The patients/participants provided their written informed consent to participate in this study.
Author contributions
AF and DJ contributed to conception and design of the study. LG, SZ, and XL performed the statistical analysis. AF, YL, GL, YW, QW, ZY, KT, HL, and DJ wrote the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
Acknowledgments
We would like to thank OrigiMed for great support.
Conflict of interest
Author AF was employed by Shandong Qidu Pharmaceutical Co. Ltd and LG, SZ and XL are employees of OrigiMed.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships.
References
1
Chaffer CL Weinberg RA . A Perspective on Cancer Cell Metastasis. Sci (New York NY) (2011) 331(6024):1559–64. doi: 10.1126/science.1203543
2
Gao Y Bado I Wang H Zhang W Rosen JM Zhang XH . Metastasis Organotropism: Redefining the Congenial Soil. Dev Cell (2019) 49(3):375–91. doi: 10.1016/j.devcel.2019.04.012
3
Riihimaki M Hemminki A Fallah M Thomsen H Sundquist K Sundquist J et al . Metastatic Sites and Survival in Lung Cancer. Lung Cancer (2014) 86(1):78–84. doi: 10.1016/j.lungcan.2014.07.020
4
Hess KR Varadhachary GR Taylor SH Wei W Raber MN Lenzi R et al . Metastatic Patterns in Adenocarcinoma. Cancer (2006) 106(7):1624–33. doi: 10.1002/cncr.21778
5
Bray F Ferlay J Soerjomataram I Siegel RL Torre LA Jemal A . Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA: Cancer J Clin (2018) 68(6):394–424. doi: 10.3322/caac.21492
6
Li Y Dong R Lu M Cheng C Feng Z Zhao R et al . Let-7b-3p Inhibits Tumor Growth and Metastasis by Targeting the BRF2-Mediated MAPK/ERK Pathway in Human Lung Adenocarcinoma. Transl Lung Cancer Res (2021) 10(4):1841–56. doi: 10.21037/tlcr-21-299
7
Zhang J Li C Zhang L Heng Y Xu T Zhang Y et al . Andrographolide Induces Noxa-Dependent Apoptosis by Transactivating ATF4 in Human Lung Adenocarcinoma Cells. Front Pharmacol (2021) 12:680589. doi: 10.3389/fphar.2021.680589
8
Arbour KC Riely GJ . Systemic Therapy for Locally Advanced and Metastatic Non-Small Cell Lung Cancer: A Review. JAMA (2019) 322(8):764–74. doi: 10.1001/jama.2019.11058
9
Chen X Bu Q Yan X Li Y Yu Q Zheng H et al . Genomic Mutations of Primary and Metastatic Lung Adenocarcinoma in Chinese Patients. J Oncol (2020) (2020):6615575. doi: 10.1155/2020/6615575
10
Shen H Laird PW . Interplay Between the Cancer Genome and Epigenome. Cell (2013) 153(1):38–55. doi: 10.1016/j.cell.2013.03.008
11
Zhang Y Wang DC Shi L Zhu B Min Z Jin J . Genome Analyses Identify the Genetic Modification of Lung Cancer Subtypes. Semin Cancer Biol (2017) 42:20–30. doi: 10.1016/j.semcancer.2016.11.005
12
Talmadge JE Fidler IJ . AACR Centennial Series: The Biology of Cancer Metastasis: Historical Perspective. Cancer Res (2010) 70(14):5649–69. doi: 10.1158/0008-5472.CAN-10-1040
13
Lin J Shi J Guo H Yang X Jiang Y Long J et al . Alterations in DNA Damage Repair Genes in Primary Liver Cancer. Clin Cancer Res (2019) 25(15):4701–11. doi: 10.1158/1078-0432.CCR-19-0127
14
Cao J Chen L Li H Chen H Yao J Mu S et al . An Accurate and Comprehensive Clinical Sequencing Assay for Cancer Targeted and Immunotherapies. Oncologist (2019) 24(12):e1294–e1302. doi: 10.1634/theoncologist.2019-0236
15
Hellmann MD Ciuleanu TE Pluzanski A Lee JS Otterson GA Audigier-Valette C et al . Nivolumab Plus Ipilimumab in Lung Cancer With a High Tumor Mutational Burden. N Engl J Med (2018) 378(22):2093–104. doi: 10.1056/NEJMoa1801946
16
Li L Wang Y Shi W Zhu M Liu Z Luo N et al . Serial Ultra-Deep Sequencing of Circulating Tumor DNA Reveals the Clonal Evolution in non-Small Cell Lung Cancer Patients Treated With Anti-PD1 Immunotherapy. Cancer Med (2019) 8(18):7669–78. doi: 10.1002/cam4.2632
17
Chen HJ Shen JM Lin YN Du W Zhou JP Li QY . Long-Term Survival of Advanced Lung Adenocarcinoma by Maintenance Chemotherapy Followed by EGFR-TKI: A Case Report. Med (Baltimore) (2021) 100(6):e24688. doi: 10.1097/MD.0000000000024688
18
Yu Y Tian X . Analysis of Genes Associated With Prognosis of Lung Adenocarcinoma Based on GEO and TCGA Databases. Med (Baltimore) (2020) 99(19):e20183. doi: 10.1097/MD.0000000000020183
19
Li Y Tan Y Hu S Xie J Yan Z Zhang X et al . Targeted Sequencing Analysis of Predominant Histological Subtypes in Resected Stage I Invasive Lung Adenocarcinoma. J Cancer (2021) 12(11):3222–9. doi: 10.7150/jca.51405
20
Graziano SL Gamble GP Newman NB Abbott LZ Rooney M Mookherjee S et al . Prognostic Significance of K-Ras Codon 12 Mutations in Patients With Resected Stage I and II Non-Small-Cell Lung Cancer. J Clin Oncol (1999) 17(2):668–75. doi: 10.1200/JCO.1999.17.2.668
21
Winton T Livingston R Johnson D Rigas J Johnston M Butts C et al . Vinorelbine Plus Cisplatin vs. Observation in Resected non-Small-Cell Lung Cancer. N Engl J Med (2005) 352(25):2589–97. doi: 10.1056/NEJMoa043623
22
Pao W Wang TY Riely GJ Miller VA Pan Q Ladanyi M et al . KRAS Mutations and Primary Resistance of Lung Adenocarcinomas to Gefitinib or Erlotinib. PloS Med (2005) 2(1):e17. doi: 10.1371/journal.pmed.0020017
23
Zer A Leighl N . Promising Targets and Current Clinical Trials in Metastatic Non-Squamous NSCLC. Front Oncol (2014) 4:329. doi: 10.3389/fonc.2014.00329
24
Baginski M Serbakowska K . In Silico Design of Telomerase Inhibitors. Drug Discovery Today (2020) 25(7):1213–22. doi: 10.1016/j.drudis.2020.04.024
25
Hu Z Wu C Shi Y Guo H Zhao X Yin Z et al . A Genome-Wide Association Study Identifies Two New Lung Cancer Susceptibility Loci at 13q12.12 and 22q12.2 in Han Chinese. Nat Genet (2011) 43(8):792–6. doi: 10.1038/ng.875
26
Dagogo-Jack I Shaw AT . Tumour Heterogeneity and Resistance to Cancer Therapies. Nat Rev Clin Oncol (2018) 15(2):81–94. doi: 10.1038/nrclinonc.2017.166
27
Shih DJH Nayyar N Bihun I Dagogo-Jack I Gill CM Aquilanti E et al . Genomic Characterization of Human Brain Metastases Identifies Drivers of Metastatic Lung Adenocarcinoma. Nat Genet (2020) 52(4):371–7. doi: 10.1038/s41588-020-0592-7
28
Dono A Takayasu T Yan Y Bundrant BE Arevalo O Lopez-Garcia CA et al . Differences in Genomic Alterations Between Brain Metastases and Primary Tumors. Neurosurgery (2021) 88(3):592–602. doi: 10.1093/neuros/nyaa471
29
Jing W Zhu H Liu W Zhai X Tian H Yu J . MTAP-Deficiency Could Predict Better Treatment Response in Advanced Lung Adenocarcinoma Patients Initially Treated With Pemetrexed-Platinum Chemotherapy and Bevacizumab. Sci Rep (2020) 10(1):843. doi: 10.1038/s41598-020-57812-2
30
Hagemann IS Devarakonda S Lockwood CM Spencer DH Guebert K Bredemeyer AJ et al . Clinical Next-Generation Sequencing in Patients With Non-Small Cell Lung Cancer. Cancer (2015) 121(4):631–9. doi: 10.1002/cncr.29089
31
Powell E Piwnica-Worms D Piwnica-Worms H . Contribution of P53 to Metastasis. Cancer Discovery (2014) 4(4):405–14. doi: 10.1158/2159-8290.CD-13-0136
32
Birkbak NJ McGranahan N . Cancer Genome Evolutionary Trajectories in Metastasis. Cancer Cell (2020) 37(1):8–19. doi: 10.1016/j.ccell.2019.12.004
33
Liao L Ji X Ge M Zhan Q Huang R Liang X et al . Characterization of Genetic Alterations in Brain Metastases From Non-Small Cell Lung Cancer. FEBS Open Bio (2018) 8(9):1544–52. doi: 10.1002/2211-5463.12501
34
Gao J Adams RP Swain SM . Does CDKN2A Loss Predict Palbociclib Benefit? Curr Oncol (Toronto Ont) (2015) 22(6):e498–501. doi: 10.3747/co.22.2700
35
Infante JR Cassier PA Gerecitano JF Witteveen PO Chugh R Ribrag V et al . A Phase I Study of the Cyclin-Dependent Kinase 4/6 Inhibitor Ribociclib (LEE011) in Patients With Advanced Solid Tumors and Lymphomas. Clin Cancer Res (2016) 22(23):5696–705. doi: 10.1158/1078-0432.CCR-16-1248
36
Patnaik A Rosen LS Tolaney SM Tolcher AW Goldman JW Gandhi L et al . Efficacy and Safety of Abemaciclib, an Inhibitor of CDK4 and CDK6, for Patients With Breast Cancer, Non-Small Cell Lung Cancer, and Other Solid Tumors. Cancer Discovery (2016) 6(7):740–53. doi: 10.1158/2159-8290.CD-16-0095
37
Sherr CJ Beach D Shapiro GI . Targeting CDK4 and CDK6: From Discovery to Therapy. Cancer Discovery (2016) 6(4):353–67. doi: 10.1158/2159-8290.CD-15-0894
38
Sholl LM Aisner DL Varella-Garcia M Berry LD Dias-Santagata D Wistuba II et al . Multi-Institutional Oncogenic Driver Mutation Analysis in Lung Adenocarcinoma: The Lung Cancer Mutation Consortium Experience. J Thorac Oncol (2015) 10(5):768–77. doi: 10.1097/JTO.0000000000000516
39
Shen R Martin A Ni A Hellmann M Arbour KC Jordan E et al . Harnessing Clinical Sequencing Data for Survival Stratification of Patients With Metastatic Lung Adenocarcinomas. JCO Precis Oncol (2019) 3:PO.18.00307. doi: 10.1200/PO.18.00307
40
Fang C Zhang C Zhao WQ Hu WW Wu J Ji M . Co-Mutations of TP53 and KRAS Serve as Potential Biomarkers for Immune Checkpoint Blockade in Squamous-Cell Non-Small Cell Lung Cancer: A Case Report. BMC Med Genomics (2019) 12(1):136. doi: 10.1186/s12920-019-0592-6
41
Frost N Kollmeier J Vollbrecht C Grah C Matthes B Pultermann D et al . KRAS(G12C)/TP53 Co-Mutations Identify Long-Term Responders to First Line Palliative Treatment With Pembrolizumab Monotherapy in PD-L1 High (>/=50%) Lung Adenocarcinoma. Transl Lung Cancer Res (2021) 10(2):737–52. doi: 10.21037/tlcr-20-958
Summary
Keywords
lung adenocarcinoma, bone metastases, liver metastases, brain metastases, next-generation sequencing
Citation
Feng A, Li Y, Li G, Wang Y, Wen Q, Yang Z, Tian K, Lv H, Guo L, Zhang S, Liu X and Jiang D (2022) Genomic Features of Organ-Specific Metastases in Lung Adenocarcinoma. Front. Oncol. 12:908759. doi: 10.3389/fonc.2022.908759
Received
31 March 2022
Accepted
09 June 2022
Published
14 July 2022
Volume
12 - 2022
Edited by
Xiao Zhu, Guangdong Medical University, China
Reviewed by
Jing Guo, Stanford University, United States; Qinglan Li, University of Pennsylvania, United States
Updates
Copyright
© 2022 Feng, Li, Li, Wang, Wen, Yang, Tian, Lv, Guo, Zhang, Liu and Jiang.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Da Jiang, jiangda139@163.com
This article was submitted to Cancer Genetics, a section of the journal Frontiers in Oncology
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.