- Biotechnology, LAL4Bsynbiotics L.L.C, Los Angeles, CA, United States
Colon cancer is one of the leading causes of cancer in the United States. Colon cancer develops from the many gene mutations found in the genomes of colon cancer cells. Long non-coding RNAs (lncRNAs) can cause the development and progression of many cancers, including colon cancer. LncRNAs have been and could be corrected through the gene-editing technology of the clustered repeats of the clustered regularly interspaced short palindromic repeats (CRISPR)-associated nuclease 9 (CRISPR/Cas9) system to reduce the proliferation of cancer cells in the colon. However, many current delivery systems for transporting CRISPR/Cas9-based therapeutics in vivo need more safety and efficiency. CRISPR/Cas9-based therapeutics require a safe and effective delivery system to more directly and specifically target cancer cells present in the colon. This review will present pertinent evidence for the increased efficiency and safety of using plant-derived exosome-like nanoparticles as nanocarriers for delivering CRISPR/Cas9-based therapeutics to target colon cancer cells directly.
Introduction
Colon cancer is a common gastrointestinal cancer in the world. In the United States, colon cancer frequently occurs more than breast, lung, and prostate cancers. Only lung cancer precedes colon cancer as the first leading cause of death. In the U.S., about 1.5 million individuals have colorectal cancer (CRC). Colonoscopies have decreased the rate of CRC. As a result of improved treatments, such as chemotherapy, immunotherapy, and colectomy, the 5-year survival rate of CRC patients is nearly 64% (1). The distribution of microorganisms and their metabolites are related to colon carcinogenesis; however, the CRC mechanisms of progression are still not as precise (2). Noncoding RNAs (ncRNAs) can cause the development of colon cancer. ncRNAs are part of a class of non-protein coding RNAs that do not become translated into proteins but can affect cellular processes. Non-coding RNAs internally residing in exosomes may be involved in the process of tumorigenesis and development, in which ncRNAs may also aid in significant intracellular communication within a tumor (3). Long non-coding RNAs (LncRNAs) have a DNA sequence length of 200 nucleotides and affect many biological processes, such as cell proliferation, differentiation, development, apoptosis, and metastasis (4).
LncRNAs bind to RNA, protein, and DNA, forming RNA-RNA, RNA-DNA, and RNA-protein structural complexes, which can lead to the control of gene expression through diverse mechanisms. These diverse mechanisms can consist of transcription, mRNA stability, and translation (5, 6). There is evidence emerging for lncRNAs being especially vital for colon cancer development and progression (7, 8). A study identified 200 lncRNAs expressed in colon tumors when analyzing data from RNA sequences in the TCGA dataset (9). LncRNAs also affect the prognoses of patients, the proliferation of cells, cell apoptosis, metastasis and invasion, the cycle of cellular division, epithelial-mesenchymal transition, drug resistance, and cancer stem cells. The expression of lncRNAs is associated with the pathogenicity of colon cancer (10). For example, the lncRNA zinc finger E-box binding homeobox 1 with antisense 1 expression is increasingly amplified in colon cancer tissues when compared to healthy colonocytes (11).
Although many improved cancer therapies exist, such as chemotherapy, targeted biological therapy, radiation therapy, and combination therapies, there is still a high propensity for relapse with increased resistance to chemo-radiation therapy. There are also many toxic side effects that occur from each therapy applied (12). Consequently, new and novel therapeutic strategies for cancer treatments are currently needed. The clustered regularly interspaced short palindromic repeats (CRISPR)-associated nuclease 9 (CRISPR/Cas9) system has provided an additional effective therapy for cancer. Presently, CRISPR/Cas9 is a type of molecular scissor with application in many studies, such as cancer research, the discovery of therapeutic drugs, treating cognitive illnesses, and being applied to plants. CRISPR is contained within the adaptive immune system of prokaryotes to provide immunity against viruses by cleaving foreign viral DNA (13, 14). Cancer is a genetic illness with multiple DNA and RNA mutations present in cellular genomes (15, 16).
These genetic mutations in the cancer cell genome can be corrected to overcome cancer (17). Much scientific evidence supports that CRISPR/Cas9 can correct cancer-causing genetic mutations (18). Essential data and evidence confirm the potential of the CRISPR/Cas9 system to target the protein-coding genome and emend lncRNAs present in humans (18–20). A few diseases have the potential to rapidly become treated with CRISPR-Cas9 technology via an ex vivo approach; however, for CRISPR-Cas9 therapeutics to achieve clinical success, the system must be directly applied to patients. A direct administration of CRISPR-Cas9 therapeutic requires a safe and precise delivery of its systems in vivo; however, these delivery aspects of CRISPR-Cas9 therapeutics are not wholly developed (21). Plant-derived exosome-like nanoparticles (PENs) may offer a promising delivery system for administering CRISPR-Cas9 therapeutics and treating colon cancer. PENs can potentially serve as effective nanocarriers of CRISPR-Cas9 therapeutics designed to target lncRNAs in human colon cancer cells through an oral administration. According to Kim et al. (22) Plant-derived exosome-like nanoparticles (PENs) are predicted and expected to develop into effective therapeutic techniques for treating diseases or delivering drugs. This study’s main focus and purpose was to describe PENs as effective delivery vehicles for drugs and biomolecules into the colon for treatment of colorectal cancer (CRC). This review will provide a brief background of the CRISPR-Cas9 structure and function, a survey of the current challenges of existing CRISPR-Cas delivery technology, and present supporting evidence for using PENs as efficient nanocarriers for CRISPR/Cas9-based therapeutics via an oral administration into the lower digestive tract.
Survey methodology
Structure and features of the CRISPR/Cas9 system
In 1987, Ishino and his colleagues first found CRISPR in Escherichia coli. They discovered that the clustered repeats had a series of spacer-type sequences, later called CRISPR. Since then, many researchers have found more varieties of CRISPR/Cas systems. There are three categories, including type I, II, and III, and many diverse subtypes divided based on their differing mechanisms (23, 24). The CRISPR/Cas9 system is a type II CRISPR system found in Streptococcus pyogenes. It is the most widely applied system in mammals since it is highly efficient and accurate. CRISPR/Cas9 is the first engineered CRISPR/Cas system for editing genomes since it contains single guide RNAs (sgRNA) that can be easily programmed with a recognition sequence of a short 20 nucleotides in length (13, 25, 26). The sgRNA is composed of CRISPR RNA (crRNA), which consists of a sequence complementary to the targeted site and a transactivating crRNA (tracrRNA), which is partially complementary to the crRNA (27–29). The Cas9 nuclease is also a component of the CRISPR/Cas9 system, in which the RNAs guide the Cas9 protein to the targeted sites while also activating the Cas9 nuclease activity. The sgRNA couples with the Cas9 protein, forming a more combined complex and recognizing the targeted site, which is a complementary DNA sequence flanked at the 3’ end and adjacent to the protospacer adjacent motif (PAM) (30) (Figure 1). The PAM primarily consists of NGG or NAG, in which N can be either A, T, G, or C, and the PAM assists with initiating the DNA double-stranded breaks (33).
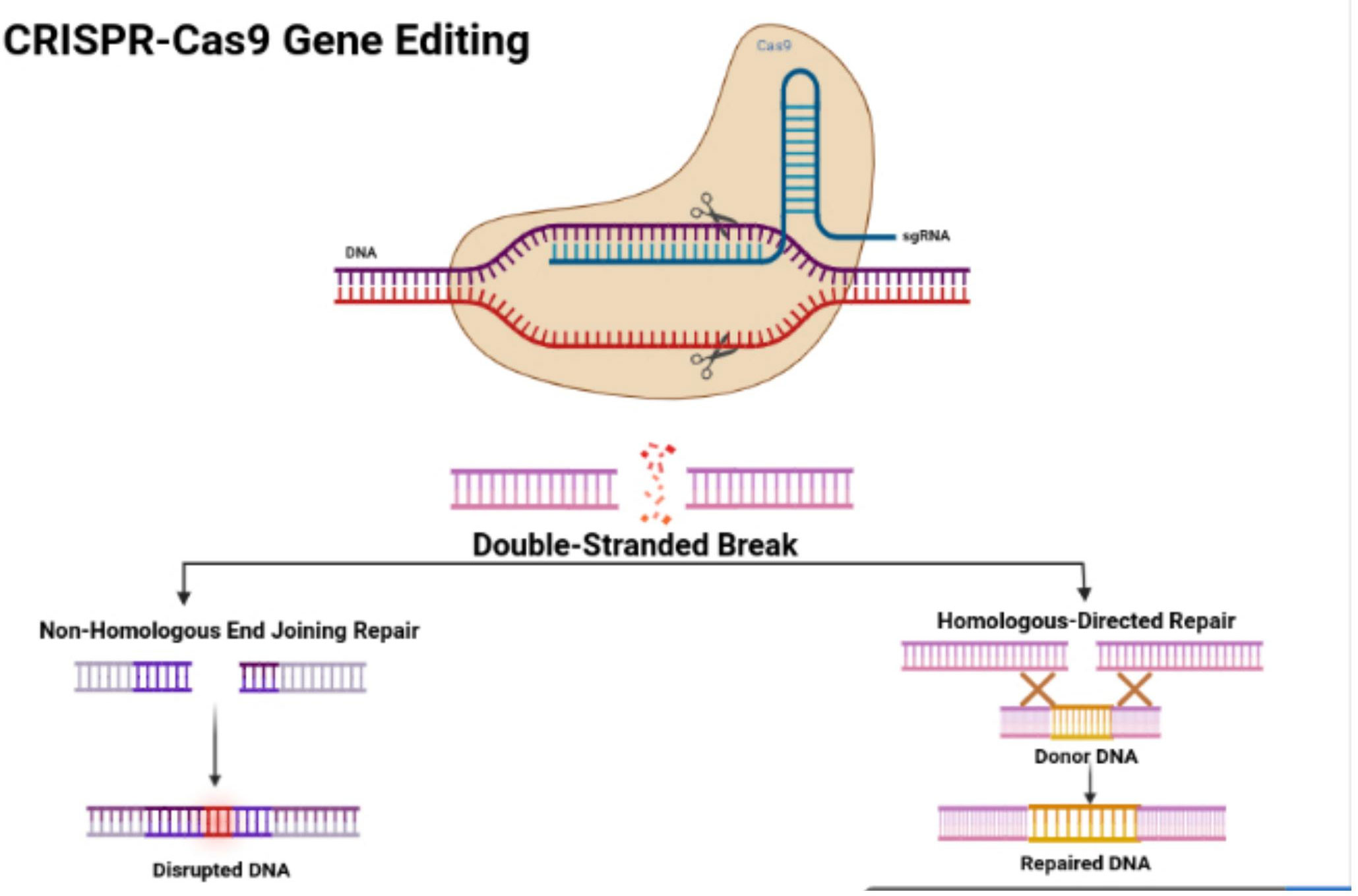
Figure 1 CRISPR/Cas9 structure and features. Figure displays the structure of the CRISPR-Cas9 protein with its complex containing the sgRNA bound to the target gene. The figure also shows the process of non-homologous end joining repair (NHEJ) and homologous directed (HDR). The figure was retrieved from Boti et al. (31, 32).
CRISPR and colon cancer
LncRNAs regulate cell proliferation, and LncRNA dysregulation controls the proliferation of colon cancer cells. The overexpression of ZEB1-AS1 modulates cell growth by increasing p21-activated kinases 2 (PAK2) expression by absorbing miR-455-3p into cancerous colon cells. LINC01082 becomes downregulated in cancerous colon tissues with the upregulation of LINC01082, causing the inhibition of cell proliferation in SW480 colon adenocarcinoma cells. Inhibiting gene expression reduced cell proliferation by suppressing LINC01296 gene expression in SW480 and SW620 colon cancer cells (31).
By silencing LINC01082, cell proliferation was inhibited by targeting the miR-21a of colon cancer cells (34). Increased taurine-upregulated gene 1 (TUG1) expression has been confirmed in the colon cancer tissues, with p63 downregulated, increasing TUG1 expression in HCT116 and LOVO cancerous colonocytes. Knocking down TUG1 expression inhibits the cell proliferation of HCT116 and LoVo cells (35).
By knocking down TUG1, the proliferation of colon cancer cells can be blocked while also reducing tumor growth in vivo (36). Since the dysregulation of cell growth causes the development of tumors by activating proto-oncogenes and inactivating tumor-suppressor genes (37), CRISPR/Cas9 is an effective genome engineering technology that offers new benefits for treating cancer with the potential to modify multiple genes. The CRISPR/Cas9 system can inhibit tumor growth by its knock-out of oncogenes (38) (Figure 2). Li et al. (39) applied CRISPR/Cas9 with the single-guide RNA (sgRNA) to build gene-modifying tools to edit mutations in the beta-catenin genes (40). The beta-catenin gene mutations are carcinogenic, in which correcting these genes can reduce their mutation rate, offering a new cancer gene therapy (41). Zhang et al. built HSV oncolytic viruses using the CRISPR/Cas9 system, and they genetically altered the HSV-1 genome to treat colon cancer (42). The secretory mucin termed MUC5AC causes the development of colon cancer due to drug resistance. Pothuraju et al. performed a gene knockout of MUC5AC for treating colorectal cancer cells via RNA interference and CRISPR/Cas9 modification in vitro and mouse models in vivo (43). Chakraborty D et al. discovered that targeting NPY/Y2R can treat colon cancer and help monitor angiogenesis. Using CRISPR/Cas9 gene modification, a knockout of the VEGF-A gene inhibited angiogenesis in mice treated with a Y2R antagonist (44). ELAVL1 is an RNA-binding protein of HuR that improves the expression of HuR. Using CRISPR/Cas9 technology to knockout HuR increased the activity of apoptosis. HuR can be a suitable therapeutic target for colon cancer (45). Takei et al. found that ERO1α had increased expression in colorectal cancer, causing a poor prognosis and increasing the development of colorectal cancer. ERO1α was knocked out, and the growth of the colon cancer was inhibited, with the cell proliferation and metastases being decreased with the reduced expression of the integrin-beta1 on the cells’ surface (46).
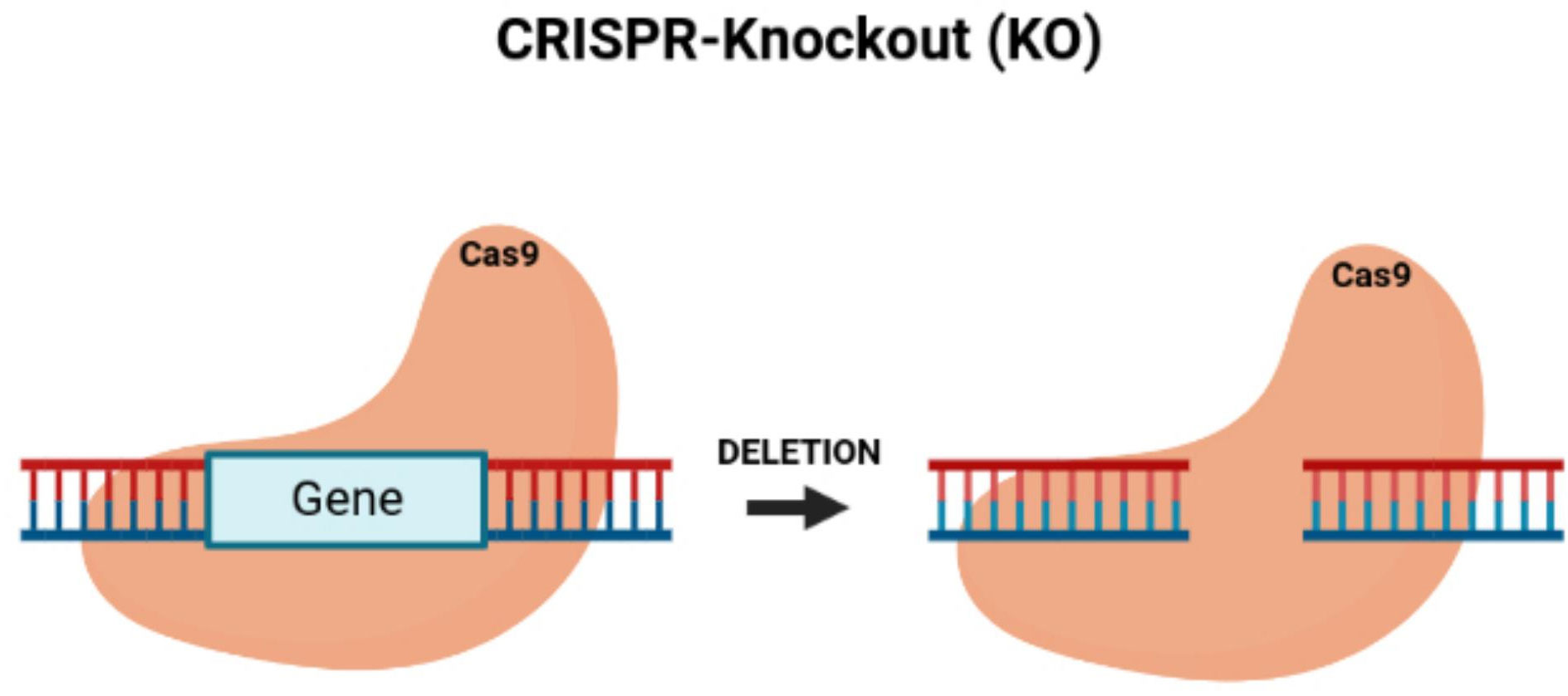
Figure 2 CRISPR/Cas9-knockout of targeted oncogenes. Figure shows the CRISPR/Cas9 deletion of a targeted gene, which represents an oncogene.
Pyruvate carboxylase (PC) increases the development and metastasis of colon cancer, causing a lesser time of survival with a poorer prognosis. A knockout of PC using CRISPR technology blocked and reduced tumor growth (47). Oh et al. confirmed that tubulin acetyltransferase αTAT1 controls Wnt1 expression to induce microtubule acetylation and enhances malignant tumor growth and invasion. CRISPR/Cas9 technology was used to knock out αTAT1, decreased tumor invasion, and inhibit colon cancer development (48). Membrane-associated loop CH protein 2 (MARCH 2) regulates autophagy and the transport of vesicles. Xia et al. discovered that MARCH 2 was increasingly expressed in colon cancer with a poorer prognosis and lower survival rate (49). MARCH 2 was knocked out, and the endoplasmic reticulum stress was stimulated with the inhibition of cancer cell growth and induction of apoptosis (50). Chen et al. confirmed that FAPP2 controls the Wnt/β-catenin signaling pathway and enhances the growth of cells of a tumor. FAPP2 is increasingly expressed in colon cancer cells and increases the expression of colon cancer cells. FAPP2 was knocked by CRISPR/Cas9 technology, the cell growth of tumors was blocked, and the tumorigenicity was decreased (51). CLCA1 was knocked out using the CRISPR system (49). After knocking out CLCA1, the proliferation, and migration of colon cancer tumor cells were increased, and the high level of expression of CLCA1 caused the inhibition of the Wnt signaling transduction and reduced the epithelial-mesenchymal-transition (EMT) process that blocked tumor growth (52).
Presently, researchers are investigating CRISPR-Cas9 gene-based therapeutics’ efficacy as a treatment for cancers of the lung, head, breast, liver, and of colon (53). ncRNAs, consisting of microRNAs, lncRNAs, and circularRNAs, all play a vital role in the development and progression of cancer. Approximately 99 percent of the human genome contains ncRNA regions with limited protein-coding activity (30). Therefore, CRISPR/Cas9 can target these non-coding regions and may become an efficient strategy for gene-based cancer therapy (30).
Current CRISPR delivery mechanisms
However, treating cancer will require the CRISPR/Cas9 components to interact directly with target cells by traversing multiple physical barriers (54, 55). Additionally, for the gene editing system to operate accurately, the Cas9 protein and the sgRNA must successfully enter the nucleus simultaneously at the same time (54). As a consequence, the type of delivery system is significant for applying gene editing therapies via the CRISPR/Cas9 technology. Microinjections, hydrodynamics, and electroporation delivered CRISPR/Cas9 components in vitro, yet these delivery modes are less effective in vivo. The use of viral vectors is also lacking in their capability to enter into the clinical use of therapeutics because viral vectors produce an immunogenic response, have increased off-target effect, are expensive, and have a smaller capacity for loading therapeutics (56). Viral vectors are most widely and commonly used for CRISPR/Cas9 delivery (49). However, adenoviral vectors cannot encapsulate the 10,000 base pairs of a CRISPR/Cas9 DNA plasmid vector. Using non-viral vectors, such as lipid nanoparticle carriers, can fully encapsulate the extensive and lengthy CRISPR/Cas9 DNA plasmids (49). For these reasons, applying non-viral vectors has created a new research field in which non-viral vectors are more advantageous than viral vectors. Non-viral vector systems include lipid nanoparticles and gold (Au) nanoparticles (49).
Non-viral vectors, through nanotechnology, can provide nanocarriers, such as polymers, lipids, and metal-organic structural complexes, to package cancer therapeutics, which have a low rate of causing an immune response, have sufficient delivery of cargo capabilities and have high biological compatibility (57, 58). Specifically, lipid-based delivery systems can transport through challenging physiological obstacles, are easy to synthesize, are frugal to compose, and can be mass-produced (57). When divided into different categories, lipid nanoparticles depend on their formation method and physicochemical properties, including solid lipid nanoparticles, nanostructured lipid carriers, niosomes, and liposomes. Solid lipid nanoparticles (SLNs) are formed from solid lipids (59), have a spherical shape, and have sizes of 50 and 1000 nm. Cancer therapeutics can be encapsulated in SLNs by encapsulation in the lipid matrix via a solid solution, a lipid shell model, and a drug-enriched shell (60). The solid solution model distributes the anticancer therapeutics into the lipid matrix. While forming the drug-enriched shell, the shaping of the shell occurs during the cooling of the SLNs in the phase separation of the drugs from the lipids. The drugs precipitate in the lipid shell model before the recrystallization of the lipid. As a result, SLNs can carry hydrophilic or hydrophobic drugs and more accurately deliver anticancer drugs.
Nanotechnology-based delivery systems for colon cancer
Through a ligand-receptor binding mechanism, ligand-altered nanoparticles can accumulate in a tumor. These types of nanoparticles are termed active targeting nanoparticles (61). Through passive targeting, these ligand-modified nanoparticles can gather in the tumor and then transit into tumor cells via active targeting, causing more specific and improved therapeutic effects. For the last three years, this active targeting strategy for nano-drug targeted delivery systems for colorectal cancer has implemented the receptor-ligand binding strategy, which included many increasingly expressed CRC receptors, such as a folate receptor, CD44, epithelial cell adhesion molecule (EpCAM), CD133, nucleolin, EGFR, αvβ3 integrin receptor, MUC1, P-selectin, SSTRs, glucocorticoid receptor, sigma-2 receptors, CXCR4+, checkpoint kinase 2, lipoprotein receptor-related protein-1, mannose receptor, carcinoembryonic antigen, hyaluronic acid receptor, and N-acetyl-D-glucosamine. A few nanoparticles can provide an accurately targeted therapy for CRC patients during stimulation conditions, such as external magnetic fields, reactive oxygen species (ROS), near-infrared (NR), and temperature. In recent years, much research has emphasized the usefulness of magnetic hyperthermia, in which magnetic nanoparticles are injected into a tumor during stimulation from external magnetic fields and then by giving patients a local radiofrequency with hyperthermia (62). During a research study, superparamagnetic chitosan-based nanocomposites were used to deliver SN-38 through a combination with hydrophilic polymeric prodrug poly (L-glutamic acid)-SN-38, which displayed an optimized aggregation in CRC and created a more facile internalization into cells with the aid of a topical magnetic field. Additionally, complexes of magnetic nanoparticles acquired a tumor inhibition ratio of 81% in a mice model with CRC xenografts (63). Dabaghi et al. confirmed that by combining magnetic hyperthermia with magnetic nanoparticles carrying the cancer drug, 5-FU which was more impactful for treating CRC and, more preferably, therapeutic in the mice model of CRC. The magnetic nano complex system can amplify tumor-targeted aggregation and enhance the inhibition of colorectal cancer progression. Because tumors produce excess amounts of ROS to transition the tumor from a state of inflammation into cancer, Zhang et al. formed a ROS-sensitive and hydrogen peroxide-eliminating matter through a cycle polysaccharide, then applied it to manufacture functional nanoparticles to carry the irinotecan as cargo. When a high concentration of ROS in a cancerous colon is stimulated, the irinotecan is released. An oral administration of the nanoparticles was given, and then the tumorigenesis with the development of colitis-induced CRC mice was inhibited.
NIR can be applied to function as an external stimulus to activate the drug release from nanoparticles at the target site. Yadav et al. (64) formulated a compacted shell-crosslinked micelle to carry and deliver indocyanine green (ICG) and doxorubicin (DOX), which could release the drugs when responding to NIR stimulation. When stimulated by NIR, the ICG produced ROS that fractured the micelle structure by degrading the disulfide bond in the micelles, and then an enlarged quantity of DOX would be released. Anugrah et al. (65) formed a hydrogel of alginate to encapsulate ICG and DOC, in which the diselenide bonds could be damaged by the ICG generation of the ROS and then release the DOX by a gel-sol transformation when stimulated by NIR light. Thus, NIR light-responsive drug delivery can be used as a targeted therapy for CRC. However, the immune system will identify and consume many nanomaterials after an intravenous and systemic administration that will cause negative side effects and decrease the therapeutic effects. Additionally, because CRC causes poor-quality vascularization, the number of nanoparticles injected intravenously can be reduced in amount before arriving at the colon with cancer, limiting the effectiveness of the nanoparticles (66). Consequently, most researchers are developing nanomaterials to be orally administered that can maintain a stable targeting of CRC. An oral administration can also augment compliance from patients.
A pH-dependent system can be used since the gastrointestinal tract is organized into the gastro, small intestines, and intestinal crissum, which differ in pH at each part of the GI tract. The stomach’s pH is approximately 1 to 3, the pH of the small intestines increases to 5.5 to 6.8, and then the pH of the colon is around 6 to 8 (67). Researchers were inspired by the enteric coating of tablets, and they initiated the development of a nano-drug delivery system consisting of enteric-coated materials to create pH-sensitive drugs to be released in the colon (68). When the pH values are low, the nanoparticles will remain complete and unbroken, but in high pH values, the dissolution of the coating materials would cause the nanoparticles to expand and attach to the colon, releasing the drug into the targeted area of interest. Eudragit and polysaccharides are used the most as coating materials.
Pectin can be extracted from polysaccharides to provide protection from the acidic gastrointestinal environment (69). Mohamed et al. (70) coated SLNs with pectin and dry skim milk, which released the curcumin cargo in the colon. As a result, the oral administration of curcumin was significantly increased and improved. An additional research study applied beta-lactoglobulin to carry irinotecan and prevent its destruction in the stomach, in which the drug was released in the small intestines. An MTT assay showed that these nanocarriers are more toxic to HT-29 cancer cell lines and AGS than a free drug. As more enteric-coated materials are found, the targeted nano-drug delivery systems established upon pH levels will be used more in targeted therapies for CRC. However, a pH-dependent system has limitations, such as the differing GI tracts between different individuals. Taymouri et al. (71) formulated a polymeric-coated capsule to carry and deliver simvastatin (SIM) into the colon, which was responsive to pH and time. To increase the solubility of the drug, a researcher used anti-solvent crystallization methods to form the nanosuspension of SIM and examined whether the nanosuspension could generate an improved anticancer effect for HT-29 versus the free drug. Subsequently, a capsule consisting of ethyl cellulose and Eudragit S100 was formulated, in which the ethyl cellulose produced a controlled release with a more time-dependent release, and the Eudragit had a solubility that was pH-dependent. This nanosuspension was infused with sodium dodecyl sulfate for freeze-drying to become loaded into the capsule. Their results confirmed that the SIM was not released into the stomach but into the colon. SIM loaded into nanoparticles increased cytotoxicity to HT-29 compared to free SIM. pH- or time-dependent nano-drug delivery systems have some limitations, such as the pH-dependent nanoparticles may not completely target the colon since the colon has a pH of 6.8, which is a similar pH present in the small intestines of ph 7.4, and the time is not fully certain for a gastrointestinal transit of the nanomaterials, which compels the time-dependent nanoparticles to sometimes neglect the targets (72). Designing nanoparticles to release drugs dependent on the degradation by microbes in the colon is an additional strategy for targeted CRC therapy. Because there are 400 classifications of microbial flora in the colon, including Escherichia coli and Clostridium (73), a few polysaccharides can only be broken down into smaller monosaccharides by the anaerobic microbiota of the colon and then implemented by the bacteria as their source of energy. However, these polysaccharides can become metabolized or consumed by gastric and intestinal enzymes (74). Therefore, polysaccharides are immensely resourceful in the enzyme-targeted therapies of CRC. Polysaccharides can regulate the location of the drug release and are biodegradable with much biocompatibility as natural polymers.
A study formulated a colonic enzyme-responsive dextran-based oligoester crosslinked with nanoparticles to deliver 5-FU. The nanoparticles released 75% of the 5-FU in vitro within 12 hours of its incubation with glucanase with no drug release under the pH-like conditions of the stomach and the small intestines (75). Tiryaki et al. (72) constructed nanoparticles to contain organic and inorganic materials, which included silica aerogels coated with dextran and dextran aldehyde. After coating with dextran and dextran aldehyde, the encapsulated drugs in silica aerogel particles became released in the colon as the dextran was degraded by the dextranase.
Dos Santos et al. (76) made chitosan nanoparticles and loaded them with 5-FU, in which the nanoparticles were encapsulated with microparticles formed from decomposed starch and pectin. Retrograde starch can prevent degradation by enzymes of the upper digestive tract, and their results confirmed that fewer nanoparticles were released from the microparticles than from the nanoparticles in the gastrointestinal lumen. Therefore, polysaccharides are central to enzyme-responsive colon-targeted materials and preparations. Researchers have designed nanoparticles with many different targeting mechanisms and functions. Rajpoot and Jain (77) framed dual-targeted nanoparticles that consisted of folate-modified SLNs and enteric polymer-coated alginate microspheres that encapsulated the nanoparticles. Using the pH-sensitive enteric polymer allowed the enteric-coated microbeads to release the drug in the colon after an oral administration. Combining folate with the nanoparticles led to the nanoparticles targeting CRC. Another study used near-infrared, which could be tracked, persistent luminescence mesoporous silica nanoparticles coated with lactobacillus reuteri biofilm (LRM) to prevent the drug from digesting and transiting completely into the colon. The LRM lengthened the time of the release of the 5-FU and protected the 5-FU from digestion in the stomach to impel the nanoparticles for actively targeting the colon since the LRM could identify the biological components of CRC, including adhesin (78).
Drug delivery systems can be designed based on many different environmental signals. Ma et al. (79) designed a pH- and enzyme-dependent nanocarrier to carry and deliver chemotherapeutic drugs. Eudragit RS nanoparticles were used to encapsulate indomethacin, 5-FU, and curcumin, respectively, through nanoprecipitation and then inserted into biphasic microcapsules consisting of chitosan and hydroxypropyl methylcellulose by use of aerosolization. Coating the microcapsules with enteric Eudragit S100 could shield the microcapsules from degradation in the stomach. The microcapsules were released, and then their release would end when reaching the colon because chitosan was metabolized and consumed by bacterial enzymes, causing the drugs to accumulate in CRC. Since nanoparticles are highly effective in animal models with CRC, clinical applications with nanoparticles hold much promise and potential. However, there are still many limitations of nano-drug delivery systems. The main obstacle is producing nano-materials and formulations on a large industrial scale and examining their safety and efficacy in preclinical trials.
The compositions of nanomaterials for targeted CRC therapies are more complex, making the preparation process of these nano-materials more challenging to synthesize and replicate. Because the physical and chemical components of platforms should be regulated during the production process, manufacturing methods are increasingly made more difficult with a high cost of production. Through microfluidics, Valencia et al. (80) formed a self-assembly lipid-polymer and lipid-quantum dots (QDs), which formed stable and uniformly structured nanoparticles. However, this complex composite of nanoparticles causes much potential toxicity to patients. Therefore, it is required to choose a model that reflects the development of human CRC to examine the possibility of toxicity during preclinical studies (81). During clinical trials, it was discovered that nanomaterials tend to decrease the toxicity of drugs and not enhance the drugs’ efficacy (82). Many scholars have investigated and found that most nanoparticles compile at tumors utilizing the EPR effect, including after utilizing actively targeted nano preparations, but this EPR effect is more frequent in animals as CRC patients have different EPR effects, which impact the effectiveness of nano-preparations (83, 84). Thus, a more personalized treatment is essential that can use nanoparticles specific for patients with an intense EPR effect to improve efficacy (73). Preparations could be used to amass the tumor separate from the EPR effect, such as utilizing temperature-sensitive-based hydrogel for more localized and targeted treatment of CRC.
The CRISPR plasmid, mRNA, and gRNA of CRISPR-Cas9 technology that is negatively charged can be encapsulated in positively charged lipid nanoparticles during an electrostatic interaction (38). The lipid nanoparticles used as nanocarriers aid the CRISPR/Cas9 in being transported through the cell membrane, but they also protectively shield the CRISPR/Cas9 components from further degradation and immune response. Lipids that are more commercialized are solid and effective vehicles of delivery (38). Lipofectamine and RNAiMAX are formed to deliver many different CRISPR/Cas9 parts for gene therapy of cancer. Zuris et al. modified 80% of their gene targets in human cells using a lipid-based nanocarrier that completed the effective delivery of Cas9/sgRNA RNPs (38). Currently, pH-sensitive nanocarriers have been used for a controlled release of CRISPR/Cas9 when internal of a tumor, which has a low pH of 5 to 6.5 in the tumor cells’ organelles and an extracellular environment, cytosol, blood, and healthy tissues with pH values of 7.4 (85). A variety of pH-sensitive components, such as copolymers, inorganic nanoparticles of crystals, lipids, and liposomes, have been formed for delivering CRISPR/Cas9 into tumors via a response to a change in pH. When the pH is lowered, for instance, the structural components of the nanocarrier would become degraded, and then the CRISPR/Cas9 cargo is released into the tumor (38). Nanocarriers built with glutathione (GSH)-sensitive potential can be used for CRISPR/Cas9 delivery because GSH is more concentrated inside the cell at 2 to 10 µM (86). Recently, these GSH-triggered nanocarriers have been formulated to deliver gene therapies, which include biologically reducible lipid nanoparticles (85), copolymers (34), and phenylboronic acid-derived lipid nanoparticles (69).
Other nano-drug delivery systems for colorectal cancer
Porous nanoparticles are used more for delivering anticancer drugs because they have uniform pore sizes, have an organized physical shape, have alterable structures, and have larger surface areas (87). Porous nanoparticles as nanocarriers can produce an effective and controlled drug release that can be engineered by attaching stimuli-sensing pore-blockers or highly sensitive hybrid coats. Gold nanoparticles (AuNPs) have a large surface area. They can link with anionic drugs such as siRNA and plasmid DNA due to their having a cationic polymer surface via forming covalent bonds or an electrostatic interaction (88). AuNPs can increase cancer-targeting drug delivery by editing the cell-targeting components of the AuNPs, which can also improve passive permeation and retention effects (89). Applying a specific radiation wavelength can further spread the AuNPs for increased absorption and dispersion ability. Adjusting the AuNPs’ size, shape, and consistency can allow for effective biomedical diagnoses and examinations (90). Halal et al. used AuNPs to deliver cetuximab, and the AuNPs increased the endocytosis with significantly inhibiting downstream signaling pathways. Cell proliferation was reduced and the cell apoptosis amplified. However, many of these engineered nanoparticles may cause pulmonary inflammation by altering the membrane permeability, which may affect the particles ability to become dispersed beyond the lungs. A few of these chemically altered NPs may cause cardiovascular disease by impairing many vascular functions (91, 92). Gold nanoparticles (AuNPs) and carbon-based NPs can transit from the nose to the brain (91, 92). All chemically engineered NPs cause pulmonary inflammation (91, 92). Coated NPs can circulate systematically when inhaled. NPs can cause increased oxidative stress through inflammation and produce surface radicals (91, 92). NPs can cause the aggregation and increased accumulation of platelets, which lead to issues of blood clotting (91, 92). Additionally, because of the many and varied obstacles present in vivo, developing a nanocarrier for safely and efficiently delivering CRISPR/Cas9-based therapeutics continues to present many challenges.
Challenges of CRISPR delivery
The first challenge requires efficiently encapsulating the CRISPR/Cas9 complex. There are three main techniques for applying the CRISPR/Cas9 for gene editing, which include using a plasmid encoding the Cas9 protein and the sgRNA, utilizing Cas9 mRNA with a sgRNA mixture, and through editing, genes using the complete Cas9 protein with the sgRNA ribonucleoprotein (RNP) (55). It is challenging to simplify and package a large Cas9 protein of 160 kD, an RNP size of 10 nm, and a mainly negative charge of the sgRNA surface into a single carrier for delivery (22). Additionally, after administration, these delivery systems still need to circumvent many physiological barriers in vivo, with each barrier and obstacle affecting the therapeutic outcome of the tumor treatment (22). CRISPR/Cas9 delivered through nanocarriers by being loaded into nanoparticles must pass the blood barrier, interact with degrading the enzymes of the plasma (93), overcome potentially being cleared by the phagocytes and macrophages (94), bypass the opsonization limiting the dispersion of the nanoparticles, and prevent the filtration of nanocarriers in the glomerulus (95). When the CRISPR/Cas9 loaded nanocarriers enter blood circulation, the nanocarriers must permeate through the barrier of the tumor tissue and increase in concentration before interacting with the target cells. Expanded vascular endothelial cells will also present many physical barriers when the nanocarriers traverse the tumor tissue.
The blood vessels in tumors leak and cause the expulsion of delivery systems from blood vessels. Since the extracellular matrix has a negative charge, the positively charged nanocarriers become easily compacted in the interstitial region (96, 97). It is difficult for nanocarriers to enter the tumor because it has limited lymphatic drainage, high interstitial pressure, is highly acidic and hypoxic and has a highly dense extracellular stroma. A transcellular obstacle in the membrane acts as a barrier for the CRISPR/Cas9 delivery system from the tissue of the tumor into the cell. After the nanocarriers with the CRISPR/Cas9 disperse from the endosomes or the lysosomes, they must enter the nucleus to begin the process of gene editing (38). These intracellular blockades include the cell membrane, endosome, and nuclear membrane. For CRISPR to become a highly effective therapeutic, it is essential to bypass all these intracellular barriers (38). Consequently, designing many delivery vectors requires optimization and improvement for an efficient CRISPR/Cas9 delivery, which is significant for using CRISPR/Cas9 as a therapeutic (38).
Plant-derived exosome-like nanoparticles
Extracellular vesicles (EVs), such as exosomes, are nanoscale membrane-enclosed particles that help to orient the transport of proteins and genetic components (96–99). Exosomes can traverse the distance between cells, carry their cargo through the cell membrane, and deliver their contents that are biologically active (100). Since exosomes provide efficient and safe delivery of biomolecules, they have become increasingly attractive recently (101–103). Mammalian EVs have delivered siRNAs, miRNAs, drugs, proteins, and CRISPR/Cas9 molecules to formulate novel treatments and therapeutics (40). However, there are many limitations to the use of human exosomes as drug delivery carriers (102). One challenge includes fewer human exosomes that can be produced in vitro or collected from biological fluids. The production yield of exosomes affects the final cost of production and their practical applications in clinics (102).
Natural vesicles in plant cells can deliver agents and solve the present issues with existing nano-based delivery systems of therapeutics (102). PENs can be efficiently and frugally isolated from affordable edible plants in large quantities without toxicity. PENs innately carry different types of biomolecules, such as non-coding RNAs, and as a result, PENs can retain their stability when carrying their cargo from cell to cell (102). PENs are studied less than mammalian vesicles (102). Plant-derived exosome-like nanoparticles (PENS) are attracting increased attention to examine their application in disease therapy. Wang et al. were the first to develop PENs as nanovectors to deliver therapeutic drugs to brain tumors (104). They found evidence for the PENs’ ability to aggregate at specific tissues in vivo, in which the PENs could circulate long-term in the peripheral blood due to the high stability of the PENs. Researchers also discovered that plants could produce large-scale quantities of PENs. Although researchers are studying PENs at a higher rate, many previous studies describe the biogenesis and function of PENs. Early studies confirm plants’ production of exosome vesicles as a response to multiple biotic and abiotic stressors in the environment, which include pathogens and potential degradation from attack (105).
There is a need to improve the use of novel drug development strategies to treat diseases. Nanotechnology in drug development has become a promising approach; therefore, PEN-based therapies could become a new and novel strategy to treat cancers, inflammations, and immunological diseases. PENs are natural and innate nanoparticles released by edible plants such as grapes (106), grapefruit (107, 108) ginger, lemon (109), carrot (110), and many other plants. PENs provide plant chemical compounds that have many physiological and pathological activities. For example, ginger-derived exosomes (GDENs) decrease tumor cell proliferation, treat inflammatory bowel diseases and restore balance to the microbiota after the damage of tissues by monitoring gut bacteria (111–113). PENs are mostly non-toxic and do not cause inflammation. PENs are also safer than synthetic and artificial nanocarriers, including polymer-based nanoparticles, metal-based nanoparticles (gold or silver), and carbon-based nanoparticles (114–116).
Lipid extraction and PEN reassembly
Differential ultracentrifugation can isolate PENs from plants (117–119). In this process, freshly squeezed plant juice becomes centrifuged at a low speed of 8,000 to 10,000 x g (Figure 3). The supernatant is centrifuged at a higher speed of 150,000 x g for 1 hour or more to collect a pellet of nano-sized particles (Figure 3). Then, the pellet dissolved in phosphate buffer solution (PBS) enters homogenization, and the added sucrose produces a gradient of 8%, 15%, 30%, 45%, and then60% (Figure 3). The remaining solution enters centrifugation at a rate higher than 150,000 x g for 2 hours (Figure 3). Different types of nanoparticles can be isolated based on the g-force of sedimentation and the varied gradients of sucrose (Figure 3). The best source of PENs is the 30% to 45% sucrose layer (117, 120, 121). About 100 grams of edible plants can produce 350 mg to 450 mg of nanoparticles (117, 119). A limitation in producing PENs include that it is challenging to produce them uniformly. This is challenging because natural PENs have many sizes, from 50 nm to 500 nm (117, 119, 120, 122, 123). Many research studies report that PEN lipids should be isolated and assembled into a more uniform-sized nanoparticle before or after cargo loading (117, 124, 125).
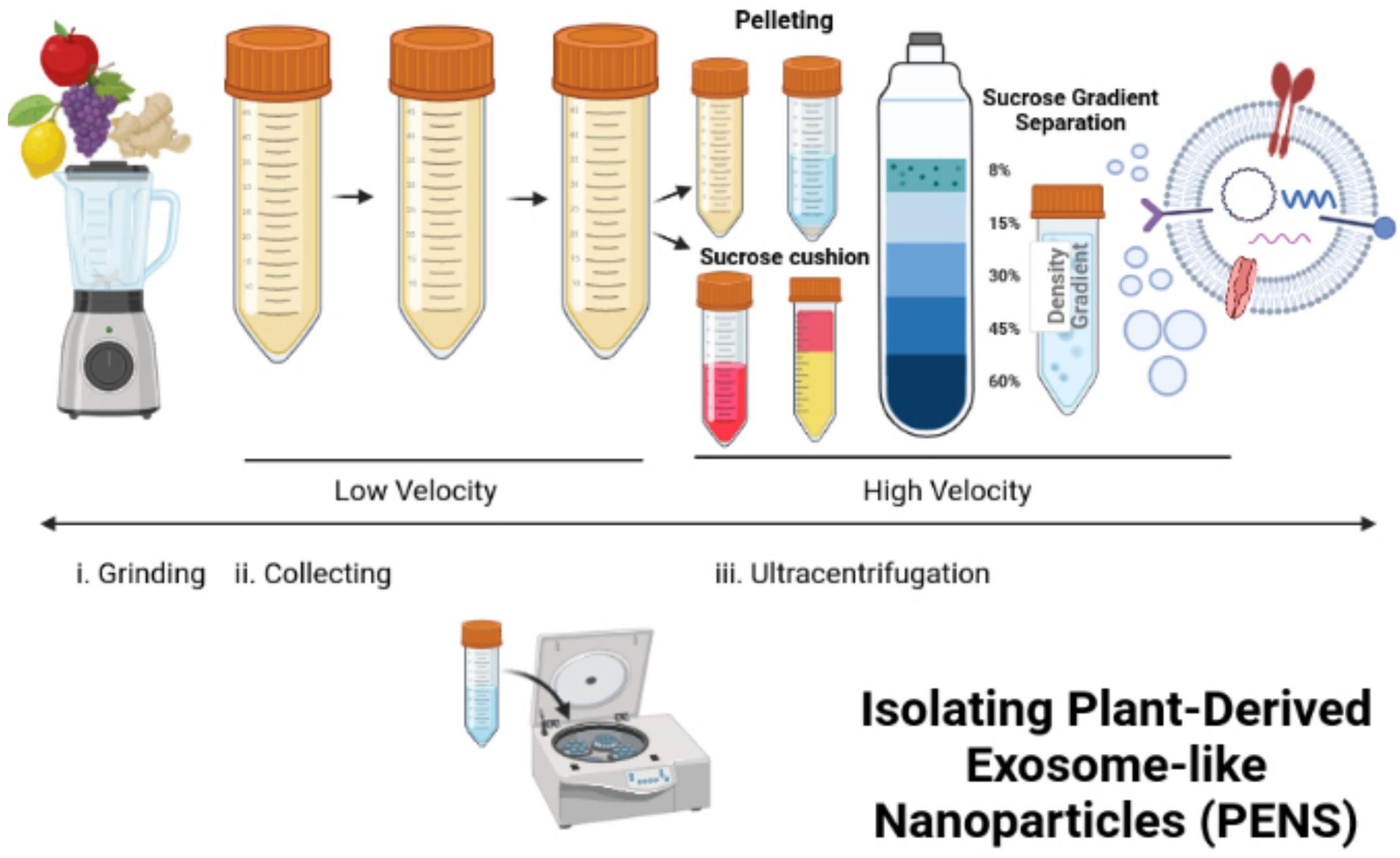
Figure 3 Isolation and reassembly of PENs. The figure presents the process of isolating plant-derived exosome-like nanoparticles (PENs) from fruits, such as apples, grapes, lemons, and ginger through low to high velocity ultracentrifugation and use of a sucrose gradient. Figure was retrieved from Kim et al. (22).
The two main types of lipids in PENs are phospholipids and glycerol lipids. These two lipids can gather in the organic phase during the two-phase liquid-liquid extraction (LLE). Zhang et al. (126) utilized the Bligh and Dyer method (127, 128), an LLE method, to isolate lipids from ginger nanoparticles and load siRNA-CD98 into the lipid/HEPES buffer solution. A 200-nm liposome extruder transfers the extracted lipids. An effective uniform-sized siRNA-CD98/ginger-derived lipid nanovesicle consists of an average diameter of 189.5 nm of size. The delivery of the nanoparticles to the colons of mice was efficient and successfully treated their ulcerative colitis. Wang et al. used the identical Bligh and Dyer procedure and extracted total lipids from grapefruit (117). After Wang et al. (2013) sonicated the lipids and passed them through a homogenizer, the lipids formed into flower-like nanoparticles of 200 nm in size.
Loading of cargo into PENs
Small molecular drugs, siRNAs, and DNA expression vectors can be loaded into PENs to help target many disease tissues (117, 118, 124, 126). Because PENs have a negative charge, positively charged compounds such as drugs can be quickly loaded into the PENs via sonication (117, 124). Neutral and negatively charged molecules loaded into PENs display no adverse effects on their biological functions (117, 118). Because PENs are highly hydrophobic, they can override the static electronic forces at the surface level, which guides them in the process of bioencapsulation.
PENS compared to other nanotechnologies combined with CRISPR therapy
The editing efficiency of lipid nanoparticles (LNPs) that carry CRISPR/Cas9 plasmids is low because their delivery efficiency into cells has been poor during in vivo experiments with animals (129). Thus, the LNP delivery of CRISPR/Cas9 plasmids has not fulfilled the clinical requirements due to their reduced editing and delivery efficiencies (129). Currently, it is challenging to deliver RNPs for targeting organs in the body because it is arduous to develop stable nanoparticles for systemic administration and delivery (129). Many researchers have established a universal procedure for engineering stable RNPs by adding cationic elements to ionizable LNP products. In these processes, the lipids guide theencapsulation of the RNPs by maintaining the activity and directing the DNA editing in the target tissues. LNPs have an 80% gene editing efficiency with a 70% editing efficiency for amino ionizable nanoparticles in vitro and in vivo (130). Polymeric nanoparticles (PNPs) such as Poly(lactic-co-glycolic acid) (PLGA) have an editing efficiency of 95%, and polyethylenimine (PEI)-β-cyclodextrin cationic polymers for nanoparticles edit at 19.1% in vitro (130). INPs edit at an efficiency of 60% in vivo and in vitro. NPs formed with DNA nanowires have an editing efficiency of 36% in vitro and in vivo (130). Gold Nanoparticles combined with lipids (lipid/AuNPs), forming complexes, were used to encapsulate the CRISPR-Cas9 system and then thermally stimulated to release the contents (131). The lipid/AuNPs encapsulated the CRISPR-Cas9 DNA plasmids at an efficiency of 97% with a release efficiency of 79.4% when triggered by irradiation from a pulse laser (131). The lipid/AuNP encapsulated CRISPR/Cas9 displayed high stability in the blood and exhibited more extended periods of circulation in the body (131).
Lipid nanoparticles can be formed with ionizable lipids that are biologically degradable, such as with PEG-DMG-lipid nanoparticles encapsulating Spy Cas9 mRNA and the sgRNA. These ionizable lipids can deliver the CRISPR/Cas9 units in vivo to edit genes, yielding a retained gene knockout for 52 weeks after a one-time administration (129). The LNP encapsulation of the CRISPR/Cas9 components reduced the protein expression of TTR in mice by 97% or more (129). Lipid nanoparticles should be further considered as effective delivery systems for gene therapy. Zwitterion amino lipids were formulated and delivered long RNAs of Cas9 mRNA and sgRNA in vitro and in vivo. The Zwitterion amino lipids maintained a 95% reduction in protein (129). The α-helical cationicpolyamine acid PPABLG was produced and displayed an increased potential to permeate through the cell membrane and yielded a high efficiency of endosomal escape. When combining CRISPR/sgRNA plasmids with copolymers, 60% of the Cas9 was expressed, and 35% of the polo-like kinase gene was knocked out (129).
DNA nano complexes are a new delivery system with an efficient loading capacity, high biocompatibility, biodegradability, a high level of cell uptake, an improved endosomal escape, and high efficiency of genome editing. Arginine-gold nanoparticles (ArgNPs) can deliver chemically altered Cas9 proteins with the targeting sgRNA, which resulted in a high delivery efficiency into the cytoplasm and the nucleus at approximately 90% (129). ArgNPs sustained genome editing with an efficiency of 23 to 30%. Most off-target effects were decreased (129). Poly(lactic-co-glycolic acid) (PLGA) nanoparticles were used as nanocarriers engineered to deliver a CRISPR/Cas9 plasmid into macrophages produced from bone marrow (132). These PLGA-nanocarriers had a size of 160 nm and encapsulated fluorophore 6,13-bis(triisopropylsilyl ethynyl) pentacene (TIPS pentacene). A PLGA nanocarrier capped with an amine group encapsulated about 1.6 wt% of DNA at an encapsulation efficiency of 80% (132). Most of the DNA was released within 24 hours, with 2 to 3 plasmid copies released from each nanoparticle (132).
In comparison, PENs have innate and specific-cell-targeting capacities. However, the mechanisms that underlie the targeting abilities of PENS are lacking and limited since PENS have not been as extensively studied as mammalian derived exosomes (MDE)s (22). The uptake of PENs by certain cells has been linked to genes encoding miRNA and siRNAs with small molecules localized in and on PENs that become extracellular ligands for the targeting components of the cells. PENs have high intrinsic and innate targeting potential, lowered off-targeting effects, and no identifiable toxicity compared to artificial nanoparticles (22). For these reasons, PENs do not require additional modifications to enhance their biocompatibility, in vivo adherence and stability, and pharmacokinetic properties. PENs have an assurance of high biocompatibility and stability when under many different physiological conditions, such as in the bloodstream and at different pH levels (22). PENs can function as vectors to be encapsulated with siRNAs and miRNAs, chemotherapeutic drugs, and hydrophobic compounds. PENs can circulate in the blood for long periods of time, which provides potential therapies for tumors and other chronic diseases (78).
There are many different methods of loading cargo into PENs. The passive loading procedure involving co-incubating exosomes and drugs allows PENs to become loaded with cargo molecules (22). Contrarily, passive loading procedures may not achieve an increased yield of encapsulation. Sonication, freeze-thaw cycling, and other manual processes can temporarily disrupt the cohesion of PEN membranes and improve cargo loading efficiency. To evade the potential of toxicity in vivo, which may be a cause of impurities, and to acquire nanocarriers of uniform sizes, nano vectors can be formed by using extracted lipids from PENs and then mixing the cargos with the extracted lipids while preparing the lipid thin film (22). This method can load more cargo, such as siRNA, antibodies, DNA expression vectors, and lipophilic drugs (22). For lipophilic drugs, such as doxorubicin, these encapsulation methods with PENs have provided high encapsulation rates of 95.9% ± 0.26%, yet the encapsulation efficiency of many other cargoes have not been found or explored (22). PENs in the colon
Ju et al. confirmed that grape-exosome-like nanoparticles induce the restoration of intestinal stem cells via the Wnt/β-catenin signaling pathway, which controls the genes of AXIN-2, Cyclin D1, c-MYC, and EGF (106). The grape-derived exosome-like nanoparticles (GELNs) showed a high cellular uptake by intestinal stem cells, in which a clathrin-mediated endocytosis inhibitor did not impact their uptake (106). It is presumed and accepted that there are specific ligands and routes of receptors between PENs and intestinal stem cells. Yet, it has been challenging to determine the functions of particular chemical molecules and ligands in PENs since the mechanisms of delivery and the internalization of PENs into cells still lack clarity. Although PENs are similar to mammalian-derived exosomes (MDEs), PENs differ from MDEs in many aspects. MDEs have lipid bilayers that consist of cholesterol, glycosphingolipids, ceramides, and phosphatidylserine, which give stability and a specific rigidity (133, 134). Contrastingly, the membranes of PENs contain phosphatidic acid (PA), phosphatidylcholines (PC), di galactosyl diacylglycerol (DGDG), and monogalactosyldiacylglycerol (MGDG) (135), in which these lipids provide intrinsic mammalian-cell-regulatory functions.
PA is the most well-known of the phospholipids for its potential to target and stimulate the mammalian target called rapamycin (mTOR), commonly identified in PENs. The mTOR pathway regulates cell growth, proliferation, and restoration of functions in many human health and disease processes. PC is a resource of choline in the body that may prevent damage to a cell wall in the large intestines by establishing cellular blockages in the cell membrane. Teng et al. purified GDENS and examined their genes, lipids, and proteins (135). Teng et al. discovered that phospholipid-enriched membranes of GDENS allowed their advantageous uptake by the microbiota that could help monitor the gut bacterial microbiome (135). Through the proteins and genes present in the GDENs compositions, the GDENS could assist with adjusting the intestinal microenvironment. To address the pharmacodynamics of PENs, because PENs are generated naturally by plants, give stability and rigidity, and have a well-suited morphology, they can encapsulate drugs within their lipid bilayer and target the tissues sought for treatment (135). PENs are membranous vesicles with site-specific targeting (136). These qualities of PENs, having a particular organization of proteins and lipids, allow them to alter genes for therapy, transfer drugs, prevent an immune response, and classify PEN as highly beneficial for future applications in medicine and treatment.
For the pharmacokinetics of PENs, they can safely transfer drugs and circulate in the blood for long periods after a systemic administration, offering a promising targeted delivery vehicle for disorders that cause tumors and other chronic diseases (136). To analyze the ability of PENs to cause toxicity, Zhang et al. used ginger-derived nanovectors (GDNVs) to treat tumors and evaluated whether the GDNVs could damage any tissues or organs (111). For the pharmacodynamics of PENs because the PENs were targeted entirely to the tumors, there was a reduction in the accumulation of GDNVs in the spleen and the liver, which lessened systemic toxicity from the drugs with a prolongation of the drugs circulating in the blood. The histological tests of the spleen, liver, lung, kidney, or heart showed no pronounced degradation to these organs compared to their control sample groups, which confirms that PENs can be used as a drug delivery nano-system, improving drug efficacy and lessening the toxicity of drugs (22). Zhang et al. (111) showed that GDNVs loaded with Doxorubicin (Dox) released their cargo in an acidic-like tumor microenvironment, and the GDNVs allowed an increased diffusion of the Dox more than the commonly used commercial liposomes. PENs can deliver chemical drugs, small molecules, and genes because they have high biocompatibility with immense biodistribution potential. As a consequence, PENs can deliver drugs with an accurate and specific targeting of tissues without causing systemic effects, causing an optimized therapeutic effect and lesser negative side effects (22).
Drug delivery systems (DDS) that carry siRNA and miRNA have low loading efficiencies with their less effective therapeutic effects, causing some adverse effects (137). To address these issues associated with GDENS, Zhang et al. extracted the lipids from the GDENS and then loaded them with the siRNA to target CD98 for treating ulcerative colitis (22). For the mechanism of action for PENs, Zhang et al. produced GDENs that had high biocompatibility without substantial toxicity, causing more apoptosis of macrophage and colon-26 cells in vitro than commercially used DC-Chol/DOPE liposomes (22). GDENs were transfected with siRNA to target CD98 using sonication and then applied to examine the mRNA expression of CD98 in an in vivo test. These siRNA-CD98-loaded GDENS were maintained in the gastrointestinal tract after oral administration and immensely lessened CD98 expression in the intestine compared with randomly siRNA-loaded GDENs. Ginger-derived lipid vectors (GDLVs) were discovered to efficiently carry divalent metal transporter 1 (DMT1)-siRNA blunts to intestinal epithelial cells to mediate the process of loading iron in the inherited hemochromatosis (136). To optimize the GDLV’s effectiveness in targeting the duodenum, the GDLVs were fused with folic acid (FA), allowing it to be interspersed into the duodenum and jejunum through the proton-coupled folate transporter. The siRNA-FA-GDLVs were given to mice, and the iron loading was diminished by a reduction of the Dmt1-mRNA expression, causing decreased levels of ferritin, TSAT, and non-heme Fe in many organs, which included the kidney, heart, pancreas, and the liver.
An oral administration of PENs
Oral administration is the more selected and endorsed drug delivery route since it is the most facile and convenient for pharmaceuticals. An oral administration offers a low risk of infection, which differs from direct injective routes, enhances the permeability of the entire gastrointestinal tract, and can evade blood clearance (138). Oral administration is non-invasive and has been used for delivering MDEs. Lin et al. delivered bovine and porcine-milk-derived exosomes that contained miRNA through an oral administration, and they detected the bovine and porcine-milk-derived exosomes in the intestinal cells (139). There have not been many MDEs developed for oral administration (140). A majority of MDEs were not designed for oral delivery because of their low stability at many different pH levels and temperatures, their quick degradation in the digestive tract, and their limited ability to be produced at an industrial rate for oral dosing (141, 142). PENs have been developed for oral administration. PENs can be administered in many different methods because they have high tolerability, the potential to target specific tissues, and high biocompatibility. As a consequence, an oral administration of PENs can beget many rapid effects in pharmacotherapy (136). For example, grapefruit-derived nanovectors (GNVs) carried MTX to intestinal macrophages after an oral administration (22), in which these GNVs targeted the intestinal macrophages with greater efficiency than most common and commercial liposomes.
Researchers placed PENs in different pH levels of water, O.5 N of NaOH, and O.5 N HCl to examine the stability of PENS. The PENs were confirmed to have a reduced size when in acidic solutions (22), the PENs in alkaline solutions have no effect. PENs were then placed in solutions with gastric and intestinal enzymes to analyze the stability of PENs during digestion. The GNVs were increasingly repellent to digestion by gastric pepsin, the intestinal enzyme of pancreatin, and resistant to the bile solutions (22). The intestinal-like fluids had no effect on PENs derived from grapefruit, carrots, and grapes; however, the GDENs decreased the negative charges (110). In acidic conditions, PENs display a reduction in surface charge. For example, the surface charge on GDENs altered from negative to positive in the stomach acidic-like fluids and then returned to having the negative surface charge in the intestine-like solutions (143). For the proof of concept, after an oral administration of PENs delivering CRISPR/Cas9 components for targeting and treating CRC, they can be absorbed by target cells or tissues. For example, GDENs primarily accumulate in the liver after an oral administration, with none identified in the lungs, spleen, or other organs (143). This interesting propensity of PENs to be more concentrated in the liver may reduce the negative effects of anti-cancer drugs, which have a non-specific dispersion of therapeutics. GDENs were taken up by albumin plus hepatocytes; however, grapefruit-derived exosomes like nanovesicles were accumulated more in F4/80+liver Kupffer cells, showing that PENs that are derived from different plants have dissimilar and distinctive targeting capabilities. These findings also confirm the hypothesis that GDENs transit to the liver from the gut through vascular vessels (143).
PENs can deliver hydrophobic anticancer drugs and genes to target cells or tissues via oral administration. An anti-inflammatory drug termed MTX was loaded into grapefruit-derived exosome-like nanovesicles that targeted F4/80+ macrophages in the lamina propria and kept the MTX therapeutic effects. PENs transfected intestinal macrophages more efficiently than commercial liposomes after an oral administration. GDENs deliver RNA to target bacterial genes (135). The gut bacteria selectively interacted with and engaged the GDENs. After an oral administration of GDENs, genes were delivered into the intestines of mice, which were analyzed by qPCR tests of GDENs miRNA in the gut and feces after the treatment. GDENs also abated mouse colitis by impacting the distribution of the gut microbiota. Thus, PENs are favored as imperative candidates for oral-delivery materials. Many of the PENs discussed have been delivered through an oral administration, as most MDEs or liposomes have been commonly delivered by IV injection. An oral administration of PENs ensures the therapeutic effect is maintained with ameliorated targeting capability; however, PENs also guarantee a decreased risk of infection, whereby the therapeutic methods of using PENs via oral administration could be an excellent procedure to enhance patient participation and compliance in clinical trials.
In addition, plant-derived extracellular vesicles (PEV) can be extracted from plants, such as grapes, grapefruits, other fruits, vegetables, and spices (144). PEVs isolated from grapes and grapefruits are highly therapeutic (144). Ginger has a high efficiency in drug delivery with a substantial therapeutic ability (144). There are a few doubts about using natural plant vesicles as drug carriers because they can aggregate during purification and have a low cargo loading capacity. Since PEVs may aggregate during ultracentrifugation, PEVs are ineffective after intravenous administration. By adding multiple purification procedures when isolating the PEVs or reconstructing the plant nanovectors and then assembling the molecules that consist of their membranes, PEVs can become more efficacious (118). Nanovectors have been formed from grapefruit-derived lipid nanoparticles and utilized as a delivery system for chemotherapeutic drugs and siRNAs (145).
In a study by Garaeva et al. (102), the researchers used in vitro models to show the significant and efficient uptake of the fluorescently labeled proteins of HSP70-Af647 or BSA-AF647 packed into grape-fruit extracellular vesicles (GF-EVs) by human cells versus free proteins. Garaeva et al. (102) observed that the grapefruit vesicles were highly efficient when carrying the exogenous proteins into the human cells in vitro. For their in vivo analysis of the vesicles’ ability to distribute into the cells, they injected the 125I-BSA-loaded GF-EVs into mice intravenously. Garaeva et al. (102) found that loading the GF-EVs with the labeled protein did not alter the surface of the vesicles. The in vivo results showed that the animal tissues had a substantially effective uptake of the protein-loaded GF-EVs. The biodistribution patterns for the GF-EVs were similar to the human exosomes in mice (103). The results of the Garaeva et al. study succinctly demonstrated the high efficiency of native GF-EVs to deliver exogenous proteins into mammalian cells and tissues safely. The results from the Garaeva study and many preceding studies (118, 124, 126, 145, 146) establish a foundation and basis for further studies and the development of similar plant vesicle delivery systems for their application in novel therapeutics and medicine.
In slight contrast, this review and study suggest an oral administration of CRISPR/Cas9 plasmid DNA-loaded PEVs similar to the protein-loaded GF-EVs used in the Garaeva et al. study, rather than an intravenous administration, for the treatment of colon cancer cells (Figure 4). The CRISPR/Cas9 plasmid DNA-loaded PEVs could specifically target lncRNAs, such as LINC01296, to reduce the proliferation of SW480 and SW620 colon cancer cells. An oral administration of CRISPR/Cas9 plasmid DNA-loaded PEVs is recommended instead of an intravenous route because the CRISPR/Cas9 plasmid DNA-loaded PEVs can traverse through the digestive tract directly into the large intestines or the colon. PENs have a negative transmembrane potential on their surface from -12 mV to -17.1 mV (117). In the pH of the intestines, the PENs from grapes, grapefruit, and carrots have no change in the net charge on their surfaces. However, when these PENs enter the acidic pH of the stomach, the PENs’ negative charges are significantly reduced since the acidic conditions neutralize the negative charges (117).
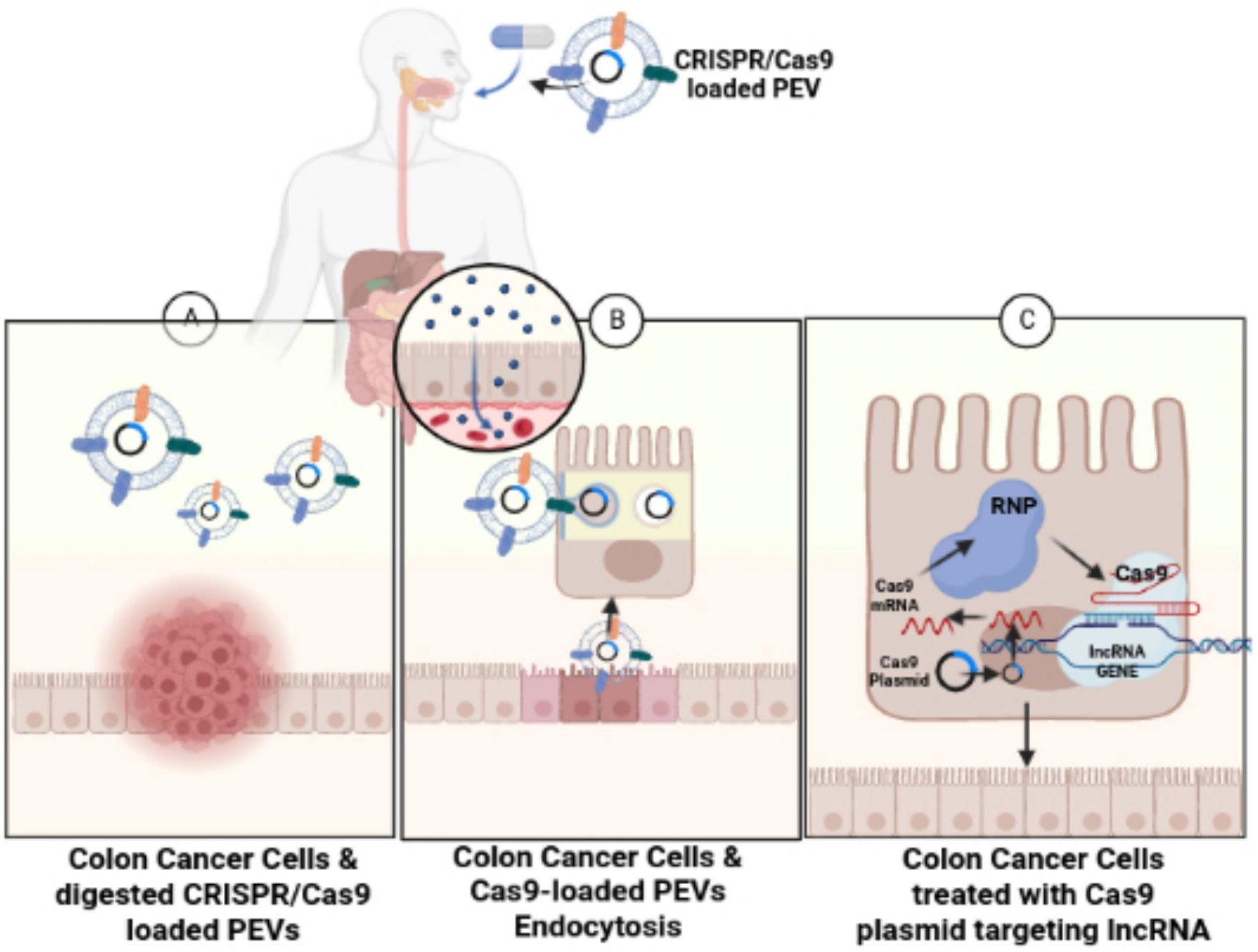
Figure 4 Treating colon cancer cells with CRISPR/Cas9-loaded PEVs. The figure conveys schematic representation for the oral administration of CRISPR/Cas9-loaded PEVs to the target lncRNAs present in colon cancer cells. (A) Figure shows the digestion of the CRISPR/Cas9-loaded PEVs into the colon. (B) The CRISPR/Cas9-loaded PEVs progress through endocytosis into the intestinal epithelial cells. (C) The Cas9-targeted treatment of lncRNA in colon cancer cells eliminates the cancerous colonocytes.
PENs are non-toxic and do not create a significant immune response, mainly because of their extraction from edible plants. Within 12 hours, 20% of B cells and 14% of T cells uptake the PENs, such as grapefruit-nanoparticles (GNPs), after treatment (117). The uptake of the GNPs also increased at a higher temperature. Wang et al. found grapefruit PENs are more stable than positively charged liposomes at 37°C in 10% bovine serum (117). The GNPs were stable at room temperature for more than a month, during which its loaded cargo was still bioactive during the month (117). Zhang et al. discovered that PENs from ginger-derived nanoparticles were increasingly more stable in stomach-like and intestine-like conditions and could withstand freeze with thaw cycles (117). Because PENs such as GNPs are more resistant to extreme temperatures and can endure stomach acids, PENs can deliver CRISPR-based gene therapies through and into the GI tract. An oral administration of CRISPR/Cas9 plasmid-DNA-loaded PEVs may ensure its direct delivery into the large intestines for treating colon cancer. Nevertheless, more research studies are required to confirm the safety and complete efficiency of this specific type of plant-based nanocarrier for application as a CRISPR/Cas9 oral delivery system.
Additionally, there are limitations for an oral administration of CRISPR/Cas9 DNA-loaded PEVs. In a neutral pH, the diameters of the plant-derived nanoparticles from ginger, grapefruit, grapes, and carrots are all lower than 300 nm (117). However, in an acidic pH, such as the pH of the acidic stomach, the size of the grape-derived nanoparticles dwindles, ginger-derived nanoparticles size increases, the grapefruit-derived nanoparticles divide into different sizes, and the two sizes of the carrot-derived nanoparticles become conjoined together in a single subset, which is larger (117). The sizes of these nanostructures vary since they alter their size in different solvents, with different buffer concentrations, and in extreme temperatures. Therefore, the plasmid-DNA encoding the CRISPR-Cas9 components should be loaded into the PEVs, since the plasmid-DNA of the CRISPR/Cas9 system will better fit into the PEVS that have sizes that can become altered by the acidic pH of the stomach. Another limitation is that siRNAs and DNAs have PEV loading efficiencies lower than positively charged therapeutics (109, 117). Sonication with freeze-thaw cycling may briefly deconstruct the PEN membranes and optimize the loading efficiency of the cargo. To prevent possible toxicity in vivo and to attain nanocarriers of uniform sizes, the formation of nanovectors can incorporate the lipids from PENs. Then the cargo is added when preparing the thin film of lipids (146). Utilizing this strategy of cargo loading can be effective for loading the lipophilic drug of doxorubicin, biomacromolecules such as siRNAs, antibodies, and DNA expression vectors (108, 145).
Discussion
PENs can offer tremendous therapeutic advantages compared to mammalian-derived exosomes (MDEs) or synthetic nanoparticles. The benefits of PENs include a less complicated method of mass production (113), less toxicity, decreased immunogenicity (147), an efficient uptake by cells (145), and high biocompatibility with increased stability (148). Many studies have confirmed the efficacy of PENs; however, many of the properties and bioactivities of PENs are not presently and wholly understood. Additional research studies are needed to improve our understanding of the functions and application of PENs.
Many previous studies report that PENs have similar properties to MDEs (149), in which PENs have been used for treating various diseases (150, 151). PENs can be acquired from large-scale manufacturing methods from many renewable sources (152), which can meet the demand for urgent production of high-quality exosomes. The natural components of PENs allow enhanced biocompatibility and increased safety with less cytotoxicity, and PENs also have reduced negative side effects. There are also many sources of PENs available, which many researchers can select from this diverse pool of nanovesicles depending on their applicability and potential to treat a disease. PENs have similar innate therapeutic materials and components to MDEs, which can be transferred and attached to the targeted cells. PENs can be used as nanocarriers because their lipid membranes are increasingly stable and can be easily altered to target specific ligands. Additionally, PENs can be rapidly examined by using eco-friendly procedures (153). The standardized process for producing PENs can become founded and organized by categorizing and characterizing PEN nanovesicle variations.
However, there are a few disadvantages of PENs. The main disadvantages include that PENs are heterogeneous in size and in physicality, which PENs may be considered and recognized as impurities by the body, which may cause unfavorable immune responses with other activities and mechanisms of regulation that have not been studied during treatment (154). The activities and roles of PENs are still not fully understood and lack much clarity; therefore, effects that have not yet been predicted can occur with the recognition of an unidentified biological material. During the application of the PENs, a few challenges to biosafety and toxicity can result from the unknown bioactive components of the plants. Because PENs are not developed from bodily fluids, tissues, or cells (155), PENs may have a reduction in targeting potential in specific tissues in the body. For future research studies to reduce and surmount these disadvantages of using PENs, their isolation processes should be enhanced to form more uniform nanovesicles. An evaluation of the morphologies, quantities, and chemical consistencies of PENs should be obtained to identify their functional roles and properties.
PENs have been tested in preclinical and clinical trials because they can transport cell-generated contents and are well-suited for industrial large-scale productive yields by standardized manufacturing processes (22). PENs are more underdeveloped than MDEs. PENs derived from grapes (NCT01668849), ginger, and aloe (NCT03493984) have been chosen and registered for clinical trials. Miller et al. affixed curcumin with plant exosomes and applied this treatment to participants with colon cancer (NCT01294072); however, this study has not initiated the recruitment phase (22). Although PENs are highly advantageous, their production processes during the stages of cultivation, isolation, and classification into clinical trials and manufacturing have not been completely executed (22). To subjugate these challenges in the clinical applications of PENs, the problems in good manufacturing practice (GMP) production of PENS should be considered, and GMP-quality grade PENs should be further examined and inspected.
This study’s main focus and purpose was to describe PENs as effective delivery vehicles of CRISPR/Cas9-based therapeutics for CRC. This review described the CRISPR/Cas9 system, targeting lnRNAs of colon cancer cells with CRISPR/Cas9 technology, discussed the challenges of the current delivery systems for CRISPR-based therapeutics, and provided established evidence for the use of PENs as nanocarriers to more efficiently deliver CRISPR-based therapeutics. This study examined the potential use of PENs and PEVs for delivering CRISPR-based therapeutics more directly into colon cancer cells via an oral administration versus the standard intravenous route. The intravenous route presents many barriers for nanocarriers to overcome for a final arrival into the digestive tract and the colon. The use of the oral route of administration also presents many obstacles and barriers, such as the stomach’s high acidity and interactions with the bile salts of the intestines. However, an oral administration may serve as an effective and more direct delivery of CRISPR-Cas9-loaded PEVs through the digestive tract into the colon for potentially treating rectal colon cancer. Because PEVs can resist degradation from stomach acids and high body temperatures, PEVs can successfully traverse through the digestive tract into the colon. Targeting the lncRNAs of colon cancer cells with CRISPR/Cas9-based therapeutics may become more efficient using PEVs as nanocarriers administered via an oral route through the digestive tract. The oral route may provide a plain and straightway delivery of CRISPR/Cas9 loaded-PEVs into the colon to eliminate colon cancer cells.
Research studies on the oral administration of CRISPR-Cas9-loaded PEVs are immensely needed. Therefore, this study is limited in describing the complete effects of CRISPR-cargo loaded PEVs processed through the digestive tract. Further research is needed to confirm the complete safety and efficiency of PEVs as a digestive delivery system for CRISPR/Cas9-based therapeutics to treat colon cancer cells. Future research studies could examine any safety concerns, and the successful delivery of CRISPR/Cas9 loaded PEVs into the colon via an oral route to more directly target colon cancer cells. This review may represent one of the many earlier studies of plant-derived exosome-like nanocarriers that may assist with furthering the investigation of PENS and PEVs as novel nanocarriers of CRISPR-based therapeutics.
Author contributions
The author confirms being the sole contributor of this work and has approved it for publication.
Acknowledgments
Many thanks are given to my mentor Dr. Tobin. Dr. Tobin provided lectures, workshops, seminars, and research opportunities, which helped to establish my understanding of the fundamentals of microbiology and genetic engineering.
Conflict of interest
Author TH was employed by the company LAL4Bsynbiotics L.L.C.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Miller KA, Siegel RL, Lin CC, Smith AB, Kramer JS, Rowland JH, et al. Cancer treatment and survivorship statistics, 2016. CA: A Cancer J Clin (2016) 66(4):271–89. doi: 10.3322/caac.21349
2. O’Keefe SJ. Diet, microorganisms and their metabolites, and colon cancer. Nat Rev Gastroenterol Hepatol (2016) 13(12):691–706. doi: 10.1038/nrgastro.2016.165
3. Chen Q, Li Y, Liu Y, Xu W, Zhu X. Exosomal non-coding RNAs-mediated crosstalk in the tumor microenvironment. Front Cell Dev Biol (2021) 9:646864. doi: 10.3389/fcell.2021.646864
4. Bhan A, Mandal SS. Long noncoding RNAs: emerging stars in gene regulation, epigenetics and human disease. ChemMedChem (2014) 9(9):1932–56. doi: 10.1002/cmdc.201300534
5. Gil N, Ulitsky I. Regulation of gene expression by cis-acting long non-coding RNAs. Nat Rev Genet (2020) 21(2):102–17. doi: 10.1038/s41576-019-0184-5
6. Yao R, Wang Y, Chen LQ. Cellular functions of long noncoding RNAs. Nat Cell Biol (2019) 21(5):542–51. doi: 10.1038/s41556-019-0311-8
7. Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res (2017) 77(15):3965–81. doi: 10.1158/0008-5472.CAN-16-2634
8. Deng H, Huang W, Zhang ZP. Nanotechnology-based CRISPR/Cas9 system delivery for genome editing: progress and prospect. Nano Res (2019) 12:2437–50. doi: 10.1007/s12274-019-2465-x
9. Forrest ME, Saiakhova A, Beard L, Buchner DM, Scacheri PC, LaFramboise T, et al. Colon cancer-upregulated long non-coding RNA lincDUSP regulates cell cycle genes and potentiates resistance to apoptosis. Sci Rep (2018) 8(1):1–12. doi: 10.1038/s41598-018-25530-5
10. Ni X, Ding Y, Yuan H, Shao J, Yan Y, Guo R, et al. Long non-coding RNA ZEB1-AS1 promotes colon adenocarcinoma malignant progression via miR-455-3p/PAK2 axis. Cell Proliferation (2020) 53(1):1–11. doi: 10.1111/cpr.12723
11. Zhao B, Qu X, Lv X, Wang Q, Bian D, Yang F, et al. Construction and characterization of a synergistic lncRNA–miRNA network reveals a crucial and prognostic role of lncRNAs in colon cancer. Front Genet (2020) 11:1–11. doi: 10.3389/fgene.2020.572983
12. Stupp R, Hegi ME, Mason WP, Van Den Bent MJ, Taphoorn MJB, Janzer RC, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol (2009) 10(5):459–66. doi: 10.1016/s1470-2045(09)70025-7
13. Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-Programmed genome editing in human cells. Elife (2013) 2:e00471. doi: 10.7554/eLife.00471
14. Barrangou R, Marraffini LA. CRISPR-cas systems: prokaryotes upgrade to adaptive immunity. Mol Cell (2014) 54(2):234–44. doi: 10.1016/j.molcel.2014.03.011
15. Hanahan D, Weinberg RA. The hallmarks of cancer. Cell (2000) 100(1):57–70. doi: 10.1016/s0092-8674(00)81683-9
16. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell (2011) 144(5):646–74. doi: 10.1016/j.cell.2011.02.013
17. White MK, Khalili K. CRISPR/Cas9 and cancer targets: future possibilities and present challenges. Oncotarget (2016) 7(11):12305–17. doi: 10.18632/oncotarget.7104
18. Fellmann C, Gowen BB, Lin P, Doudna JA, Corn JE. Cornerstones of CRISPR–cas in drug discovery and therapy. Nat Rev Drug Discov (2017) 16(2):89–100. doi: 10.1038/nrd.2016.238
19. Zhuo C, Hou W, Hu L, Lin C, Chen C, Lin X. Genomic editing of non-coding RNA genes with CRISPR/Cas9 ushers in a potential novel approach to study and treat schizophrenia. Front Mol Neurosci (2017) 10:1–l9. doi: 10.3389/fnmol.2017.00028
20. Canver MC, Bauer DE, Orkin SH. Functional interrogation of non-coding DNA through CRISPR genome editing. Methods (2017) 121–122:118–29. doi: 10.1016/j.ymeth.2017.03.008
21. Behr M, Zhou J, Xu B, Zhang H. In vivo delivery of CRISPR-Cas9 therapeutics: progress and challenges. Acta Pharm Sin B (2021) 11(8):2150–71. doi: 10.1016/j.apsb.2021.05.020
22. Kim J, Li S, Zhang S, Wang J. Plant-derived exosome-like nanoparticles and their therapeutic activities. Asian J Pharm Sci (2022) 17(1):53–69. doi: 10.1016/j.ajps.2021.05.006
23. Haft DH, Selengut JD, Mongodin EF, Nelson KE. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PloS Comput Biol (2005) 1(6):e60. doi: 10.1371/journal.pcbi.0010060
24. Makarova KS, Haft DH, Barrangou R, Brouns SJJ, Charpentier E, Horvath P, et al. Evolution and classification of the CRISPR–cas systems. Nat Rev Microbiol (2011) 9(6):467–77. doi: 10.1038/nrmicro2577
25. Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science (2013) 33:819–23. doi: 10.1126/science.1231143
26. Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9–crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci United States America (2012) 109(39):E2579–86. doi: 10.1073/pnas.1208507109
27. Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, et al. RNA-Guided human genome engineering via Cas9. Science (2013) 339:823–6. doi: 10.1126/science.1232033
28. Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature (2011) 471:602–7. doi: 10.1038/nature09886
29. Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science (2008) 321:960–4. doi: 10.1126/science.1159689
30. Yang J, Meng X, Pan J, Jiang N, Zhou C, Wu Z, et al. CRISPR/Cas9-mediated noncoding RNA editing in human cancers. RNA Biol 2 (2018) 15(1):35–43. doi: 10.1080/15476286.2017.1391443
31. Chen S, Shen X. Long noncoding RNAs: functions and mechanisms in colon cancer. Mol Cancer (2020) 19:1–3. doi: 10.1186/s12943-020-01287-2
32. Boti MA, Athanasopoulou K, Adamopoulos PG, Sideris DC, Scorilas A. Recent advances in genome-engineering strategies. Genes (2023) 14(1):129. doi: 10.3390/genes14010129
33. Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, et al. DNA Targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol (2013) 31:827–32. doi: 10.1038/nbt.2647
34. Chen G.J, Abdeen A.A, Wang YY, Shahi PK, Robertson S, Xie RS, et al. A biodegradable nanocapsule delivers a Cas9 ribonucleoprotein complex for in vivo genome editing. Nat Nanotechnol (2019) 14:974–+. doi: 10.1038/s41565-019-0539-2
35. Zhai HY, Sui MH, Yu X, Qu Z, Hu JC, Sun HQ, et al. Overexpression of long non-coding RNA TUG1 promotes colon cancer progression. Med Sci Monit (2016) 22:3281–7. doi: 10.12659/MSM.897072
36. Tian L, Zhao ZF, Xie L, Zhu JP. Taurine up-regulated 1 accelerates tumorigenesis colon cancer by regulating miR-26a-5p/MMP14/p38 MAPK/Hsp27 axis in vitro and in vivo. Life Sci (2019) 239:117035. doi: 10.1016/j.lfs.2019.117035
37. Skoulidis F, Heymach J.V. Co-Occurring genomic alterations in non-small-cell lung cancer biology and therapy. Nat Rev Cancer (2019) 19:495–509. doi: 10.1038/s41568-019-0179-8
38. Xu X, Liu C, Wang Y, Koivisto O, Zhou J, Shu Y, et al. Nanotechnology-based delivery of CRISPR/Cas9 for cancer treatment. Adv Drug Deliv Rev (2021) 176:113891. doi: 10.1016/j.addr.2021.113891
39. Liu J, Chang J, Jiang Y, Meng XD, Sun TM, Mao LQ, et al. Fast and efficient CRISPR/Cas9 genome editing in vivo enabled by bioreducible lipid and messenger RNA nanoparticles. Adv Mat (2019) 31:1902575. doi: 10.1002/adma.201902575
40. Meng W, He C, Hao Y, Wang L, Li L, Zhu G. Prospects and challenges of extracellular vesicle-based drug delivery system: considering cell source. Drug Deliv (2020) 27(1):585–98. doi: 10.1080/10717544.2020.1748758
41. Wu XH, Chen SP, Mao JY, Ji XX, Yao HT, Zhou SH. Expression and significance of hypoxia-inducible factor-1α and glucose transporter-1 in laryngeal carcinoma. Oncol Lett (2013) 5:261–6. doi: 10.3892/ol.2012.941
42. Lu ZJ, Yu Q, Zhou SH, Fan J, Shen LF, Bao YY, et al. Construction of a GLUT-1 and HIF-1α gene knockout cell model in HEp-2 cells using the CRISPR/Cas9 technique. Cancer Manag Res (2019) 11:2087–96. doi: 10.2147/CMAR.S183859
43. Gallipoli P, Giotopoulos G, Tzelepis K, Costa A, Vohra S, Medina-Perez P, et al. Glutaminolysis is a metabolic dependency in FLT3ITD acute myeloid leukemia unmasked by FLT3 tyrosine kinase inhibition. Blood (2018) 131:1639–53. doi: 10.1182/blood-2017-12-820035
44. Kanarek N, Keys HR, Cantor JR, Lewis CA, Chan SH, Kunchok T, et al. Histidine catabolism is a major determinant of methotrexate sensitivity. Nature (2018) 559:632–6. doi: 10.1038/s41586-018-0316-7
45. Papaccio F, Paino F, Regad T, Papaccio G, Desiderio V, Tirino V. Concise review: cancer cells, cancer stem cells, and mesenchymal stem cells: influence in cancer development. Stem Cells Transl Med (2017) 6:2115–25. doi: 10.1002/sctm.17-0138
46. Zomer A, Ellenbroek SI, Ritsma L, Beerling E, Vrisekoop N, Van Rheenen J. Intravital imaging of cancer stem cell plasticity in mammary tumors. Stem Cells (2013) 31:602–6. doi: 10.1002/stem.1296
47. Noh KH, Kim BW, Song KH, Cho H, Lee YH, Kim JH, et al. Nanog signaling in cancer promotes stem-like phenotype and immune evasion. J Clin Invest (2012) 122:4077–93. doi: 10.1172/JCI64057
48. Chang C, Lee SO, Yeh S, Chang TM. Androgen receptor (AR) differential roles in hormone-related tumors, including prostate, bladder, kidney, lung, breast, and liver. Oncogene (2014) 33:3225–34. doi: 10.1038/onc.2013.274
49. Meng H, Nan M, Li Y, Ding Y, Yin Y, Zhang M. Application of CRISPR-Cas9 gene editing technology in basic research, diagnosis, and treatment of colon cancer. Front Endocrinol (2023) 14. doi: 10.3389/fendo.2023.1148412
50. Ling K, Jiang L, Liang S, Kwong J, Yang L, Li Y, et al. Nanog interaction with the androgen receptor signaling axis induces ovarian cancer stem cell regulation: studies based on the CRISPR/Cas9 system. J Ovarian Res (2018) 11:36. doi: 10.1186/s13048-018-0403-2
51. Krausova M, Korinek V. Wnt signaling in adult intestinal stem cells and cancer. Cell Signal (2014) 26:570–9. doi: 10.1016/j.cellsig.2013.11.032
52. Zhan T, Ambrosi G, Wandmacher AM, Rauscher B, Betge J, Rindtorff N, et al. MEK inhibitors activate wnt signaling and induce stem cell plasticity in colorectal cancer. Nat Commun (2019) 10:2197. doi: 10.1038/s41467-019-09898-0
53. Wu XL, Ma WJ, Mei CJ, Chen X, Yao Y, Liu YY, et al. Description of CRISPR/Cas9 development and its prospect in hepatocellular carcinoma treatment. J Exp Clin Canc Res (2020) 39:1–11. doi: 10.1186/s13046-020-01603-0
54. Zhong Y, Du S, Dong Y. mRNA delivery in cancer immunotherapy. Acta Pharm Sinica B (2023) 5:1348–57. doi: 10.1016/j.apsb.2023.03.001
55. Xu XJ, Wan T, Xin HH, Li D, Pan HM, Wu J, et al. Delivery of CRISPR/Cas9 for therapeutic genome editing. J Gene Med (2019) 21:3107. doi: 10.1002/jgm.3107
56. Tong S, Moyo B, Lee CM, Leong K, Bao G. Engineered materials for in vivo delivery of genome-editing machinery. Nat Rev Mat (2019) 4:726–37. doi: 10.1038/s41578-019-0145-9
57. Khosa A, Reddi S, Saha RN. Nanostructured lipid carriers for site-specific drug delivery. Biomed Pharmacother (2018) 103:598–613. doi: 10.1016/j.biopha.2018.04.055
58. Parra-Nieto J, del Cid MAG, de Carcer IA, Baeza A. Inorganic porous nanoparticles for drug delivery in antitumoral therapy. Biotechnol J (2021) 16:2000150. doi: 10.1002/biot.202000150
59. Ghasemiyeh P, Mohammadi-Samani S. Solid lipid nanoparticles and nanostructured lipid carriers as novel drug delivery systems: applications, advantages and disadvantages. Res Pharm Sci (2018) 13:288–303. doi: 10.4103/1735-5362.235156
60. Battaglia L, Gallarate M. Lipid nanoparticles: state of the art, new preparation methods and challenges in drug delivery expert opin. Drug Del (2012) 9:497–508. doi: 10.1517/17425247.2012.673278
61. Hu Y, He Y, Ji J, Zheng S, Cheng Y. Tumor targeted curcumin delivery by folate-modified MPEG-PCL self-assembly micelles for colorectal cancer therapy. Int J Nanomed (2020) 15:1239–52. doi: 10.2147/IJN.S232777
62. Yusefi M, Lee-Kiun MS, Shameli K, Teow SY, Ali RR, Siew KK, et al. 5-fluorouracil loaded magnetic cellulose nanocomposites for potential colorectal cancer treatment. Carbohydr Polym (2021) 273:118523. doi: 10.1016/j.carbpol.2021.118523
63. Wu D, Li Y, Zhu LX, Zhang WY, Xu SM, Yang Y, et al. A biocompatible superparamagnetic chitosan-based nanoplatform enabling targeted SN-38 delivery for colorectal cancer therapy. Carbohydr Polym (2021) 274:118641. doi: 10.1016/j.carbpol.2021.118641
64. Yadav S, Ramesh K, Kumar P, Jo S-H, Yoo SI, Gal Y-S, et al. Near-InfraredLight-Responsive shell-CrosslinkedMicellesof poly(d,l-lactide)-b-poly((furfuryl methacrylate)-co-(N-acryloyl morpholine)) prepared by diels–alder reaction for the triggered release of doxorubicin. Materials (2021) 14:7913. doi: 10.3390/ma14247913
65. Anugrah DSB, Ramesh K, Kim M, Hyun K, Lim KT. Near-infrared light-responsive alginate hydrogels based on diselenide-containing cross-linkage for on-demand degradation and drug release. Carbohydr Polym (2019) 223:115070. doi: 10.1016/j.carbpol.2019.115070
66. Kumar B, Kulanthaivel S, Mondal A, Mishra S, Banerjee B, Bhaumik A, et al. Mesoporous silica nanoparticle-based enzyme responsive system for colon-specific drug delivery through guar gum capping. Colloids Surf B Biointerf (2017) 150:352–61. doi: 10.1016/j.colsurfb.2016.10.049
67. Koziolek M, Grimm M, Becker D, Iordanov V, Zou H, Shimizu J, et al. Investigation of pH and temperature profiles in the GI tract of fasted human subjects using the Intellicap((R)) system. J Pharm Sci (2015) 104:2855–63. doi: 10.1002/jps.24274
68. Wang M, Hao WS, Zhang L, Zhu YN, Chen K, Ma S, et al. Lipid-polymer nano core-shell type hybrid system for colon-specific drug delivery. J Drug Deliv Sci Technol (2021) 63:102540. doi: 10.1016/j.jddst.2021.102540
69. Tang Z, Jun T, Lv Y, Li Y, Zhang Z, Tao M, et al. Aptamer-conjugated and doxorubicin-loaded grapefruit-derived nanovectors for targeted therapy against HER2+ breast cancer. J Drug Target (2019) 28(2):186–94. doi: 10.1080/1061186x.2019.1624970
70. Mohamed JM, Alqahtani A, Ahmad F, Krishnaraju V, Kalpana K. Pectin co-functionalized dual layered solid lipid nanoparticle made by soluble curcumin for the targeted potential treatment of colorectal cancer. Carbohydr Polym (2021) 252:117180. doi: 10.1016/j.carbpol.2020.117180
71. Taymouri S, Ahmadi Z, Mirian M, Tavakoli N. Simvastatin nanosuspensions prepared using a combination of pH-sensitive and timed-release approaches for potential treatment of colorectal cancer. Pharm Dev Technol (2021) 26:335–48. doi: 10.1080/10837450.2021.1872086
72. Tiryaki E, Elalmis YB, Ikizler BK, Yucel S. Novel organic/inorganic hybrid nanoparticles as enzyme-triggered drug delivery systems: dextran and dextran aldehyde coated silica aerogels. J Drug Deliv Sci Technol (2020) 56:101517. doi: 10.1016/j.jddst.2020.101517
73. Wang K, Shen R, Meng T, Hu F, Yuan H. Nano-drug delivery systems based on different targeting mechanisms in the targeted therapy of colorectal cancer. Molecules (2022) 27(9):2981. doi: 10.3390/molecules27092981
74. Wang L, Rao Y, Liu X, Sun L, Gong J, Zhang H, et al. The administration route governs the therapeutic efficacy, biodistribution, and macrophage targeting of anti-inflammatory nanoparticles in the lung. J Nanobiotechnol (2021) 19(1):56. doi: 10.1186/s12951-021-00803-w
75. Abid M, Naveed M, Azeem I, Faisal A, Nazar MF, Yameen B. Colon specific enzyme responsive oligoester crosslinked dextran nanoparticles for controlled release of 5-fluorouracil. Int J Pharm (2020) 586:119605. doi: 10.1016/j.ijpharm.2020.119605
76. Dos Santos AM, Meneguin AB, Akhter DT, Fletcher N, Houston ZH, Bell C, et al. Understanding the role of colon-specific microparticles based on retrograded starch/pectin in the delivery of chitosan nanoparticles along the gastrointestinal tract. Eur J Pharm Biopharm (2021) 158:371–8. doi: 10.1016/j.ejpb.2020.12.004
77. Rajpoot K, Jain SK. Oral delivery of pH-responsive alginate microbeads incorporating folic acid-grafted solid lipid nanoparticles exhibits enhanced targeting effect against colorectal cancer: a dual-targeted approach. Int J Biol Macromol (2020) 151:830–44. doi: 10.1016/j.ijbiomac.2020.02.132
78. Wang ZH, Liu JM, Li CY, Wang D, Lv H, Lv SW, et al. Bacterial biofilm bioinspired persistent luminescence nanoparticles with gut-oriented drug delivery for colorectal cancer imaging and chemotherapy. ACS Appl Mater Interfaces (2019) 11:36409–19. doi: 10.1021/acsami.9b12853
79. Ma YM, Thurecht KJ, Coombes AGA. Development of enteric-coated, biphasic chitosan/HPMC microcapsules for colon-targeted delivery of anticancer drug-loaded nanoparticles. Int J Pharm (2021) 607:121026. doi: 10.1016/j.ijpharm.2021.121026
80. Valencia PM, Basto PA, Zhang LF, Rhee M, Langer R, Farokhzad OC, et al. Single-step assembly of homogenous lipid–polymeric and lipid–quantum dot nanoparticles enabled by microfluidic rapid mixing. ACS Nano (2010) 4:1671–9. doi: 10.1021/nn901433u
81. Choi SYC, Lin D, Gout PW, Collins CC, Xu Y, Wang YZ. Lessons from patient-derived xenografts for better in vitro modeling of human cancer. Adv Drug Deliv. Rev (2014) 79–80:222–37. doi: 10.1016/j.addr.2014.09.009
82. Min YZ, Caster JM, Eblan MJ, Wang AZ. Clinical translation of nanomedicine. Chem Rev (2015) 115:11147–90. doi: 10.1021/acs.chemrev.5b00116
83. Bertrand N, Wu J, Xu XY, Kamaly N, Farokhzad OC. Cancer nanotechnology: the impact of passive and active targeting in the era of modern cancer biology. Adv Drug Deliv. Rev (2014) 66:2–25. doi: 10.1016/j.addr.2013.11.009
84. Prabhakar U, Maeda H, Jain RK, Sevick-Muraca EM, Zamboni W, Farokhzad OC, et al. Challenges and key considerations of the enhanced permeability and retention effect for nanomedicine drug delivery in oncology. Cancer Res (2013) 73:2412–7. doi: 10.1158/0008-5472.CAN-12-4561
85. Liu G, Zhao X, Zhang Y, Xu J, Xu J, Li Y, et al. Engineering biomimetic platesomes for pH-responsive drug delivery and enhanced antitumor activity. Adv Mat (2019) 31:1900795 doi: 10.1002/adma.201900795
86. Mi P. Stimuli-responsive nanocarriers for drug delivery, tumor imaging, therapy and theranostics. Theranostics (2020) 10:4557–88. doi: 10.7150/thno.38069
87. Sayed E, Haj-Ahmad R, Ruparelia K, Arshad MS, Chang MW, Ahmad Z. Porous inorganic drug delivery systems-a review. AAPS Pharmscitech (2017) 18:1507–25. doi: 10.1208/s12249-017-0740-2
88. Zhang JJ, Mou L, Jiang XY. Surface chemistry of gold nanoparticles for health-related applications. Chem Sci (2020) 11:923–36. doi: 10.1039/C9SC06497D
89. Wei QL, Arami H, Santos HA, Zhang HB, Li YY, He J, et al. Zhou intraoperative assessment and photothermal ablation of the tumor margins using gold. Nanoparticles Adv Sci (2021) 8:1–15. doi: 10.1002/advs.202002788
90. Bouche M, Hsu JC, Dong YC, Kim J, Taing K, Cormode. DP. Recent advances in molecular imaging with gold nanoparticles. Bioconjug Chem (2020) 31:303–14. doi: 10.1021/acs.bioconjchem.9b00669
91. De Jong WH, Borm PJ. Drug delivery and nanoparticles:applications and hazards. Int J nanomed (2008) 3(2):133–49. doi: 10.2147/ijn.s596
92. Zielińska A, Carreiró F, Oliveira AM, Neves A, Pires B, Venkatesh DN, et al. Polymeric nanoparticles: production, characterization, toxicology and ecotoxicology. Molecules (2020) 25(16):3731. doi: 10.3390/molecules25163731
93. Kim JK, Howard MD, Dziubla TD, Rinehart JJ, Jay M, Lu. XL. Uniformity of drug payload and its effect on stability of solid lipid nanoparticles containing an ester prodrug. ACS Nano (2011) 5:209–16. doi: 10.1021/nn102357y
94. de Lazaro I, Mooney DJ. Obstacles and opportunities in a forward vision for cancer nanomedicine. Nat Mat (2021) 20:1469–79. doi: 10.1038/s41563-021-01047-7
95. Han J, Wang Q, Zhang Z, Gong T, Sun X. Cationic bovine serum albumin based self-assembled nanoparticles as siRNA delivery vector for treating lung metastatic cancer. Small (2014) 10:524–35. doi: 10.1002/smll.201301992
96. Wang HX, Zuo ZQ, Du JZ, Wang YC, Sun R, Cao ZT, et al. Surface charge critically affects tumor penetration and therapeutic efficacy of cancer nanomedicines. Nano Today (2016) 11:133–44. doi: 10.1016/j.nantod.2016.04.008
97. Van Niel G, D’Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol (2018) 19:213–28. doi: 10.1038/nrm.2017.125
98. Liao W, Du Y, Zhang L, Pan S, Yao Y, Zhuang T, et al. Exosomes: the next generation of endogenous nanomaterials for advanced drug delivery and therapy. Acta Biomat (2019) 86:1–14. doi: 10.1016/j.actbio.2018.12.045
99. El Andaloussi S, Mäger I, Breakefield XO, Wood MJA. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat Rev Drug Discov (2013) 12:347–57. doi: 10.1038/nrd3978
100. Hessvik NP, Llorente A. Current knowledge on exosoZXme biogenesis and release. Cell Mol Life Sci (2018) 75:193–208. doi: 10.1007/s00018-017-2595-9
101. Sil S, Dagur RS, Hu G, Liao K, Peeples E.S, Periyasamy P, et al. Strategies for the use of extracellular vesicles for the delivery of therapeutics. J Neuroimmune Pharmacol (2019) 15:422–42. doi: 10.1007/s11481-019-09873-y
102. Garaeva L, Kamyshinsky R, Kil Y, Varfolomeeva E, Verlov N, Komarova E, et al. Delivery of functional exogenous proteins by plant-derived vesicles to human cells in vitro. Sci Rep (2021) 11(1):6849.
103. Yi YW, Lee JH, Kim SY, Pack CG, Ha DH, Park SR, et al. Strategies for the use of extracellular vesicles for the delivery of therapeutics. J Neuroimmune Pharmacol (2019) 15:422–42. doi: 10.1007/s11481-019-09873-y
104. Wang Q, Zhuang K, Mu J, Deng ZB, Jiang YH, Zhang H, et al. Delivery of therapeutic agents by nanoparticles made of grapefruit-derived lipids. Nat Commun (2013) 4:1811–67. doi: 10.1038/ncomms3358
105. An Q, Huckelhoven R, Kogel KH, van Bel AJ. Multivesicular bodies participate in a cell wall-associated defence response in barley leaves attacked by the pathogenic powdery mildew fungus. Cell Microbiol (2006) 8(6):1009–19. doi: 10.1111/j.1462-5822.2006.00683.x
106. Ju S, Mu J, Dokland T, Zhuang X, Wang Q, Jiang H, et al. Grape exosome-like nanoparticles induce intestinal stem cells and protect mice from DSS-induced colitis. Mol Ther (2013) 21(7):1345–57. doi: 10.1038/mt.2013.64
107. Wang B, Zhuang X, Deng ZB, Jiang H, Mu J, Wang Q, et al. Targeted drug delivery to intestinal macrophages by bioactive nanovesicles released from grapefruit. Mol Ther (2014) 22(3):522–34. doi: 10.1038/mt.2013.190
108. Wang Q. Y, Mu J, Egilmez NK, Zhuang X, Deng Z, Zhang L, et al. Grapefruit-derived nanovectors use an activated leukocyte trafficking pathway to deliver therapeutic agents to inflammatory tumor sites. Cancer Res (2015) 75(12):2520–9. doi: 10.1158/0008-5472.CAN-14-3095
109. Raimondo S, Naselli F, Fontana S, Monteleone F, Lo Dico A, Saieva L, et al. Citrus limon-derived nanovesicles inhibit cancer cell proliferation and suppress CML xenograft growth by inducing TRAIL-mediated cell death. Oncotarget (2015) 6(23):19514–27. doi: 10.18632/oncotarget.4004
110. Mu J, Zhuang X, Wang Q, Jiang H, Deng ZB, Wang B, et al. Interspecies communication between plant and mouse gut host cells through edible plant derived exosome-like nanoparticles. Mol Nutr Food Res (2014) 58(7):1561–73. doi: 10.1002/mnfr.201300729
111. Zhang M, Xiao B, Wang H, Han MK, Zhang Z, Viennois E, et al Edible ginger-derived nano-lipids loaded with doxorubicin as a novel drug-delivery approach for colon cancer therapy. Mol Ther (2016) 24(10):1783–96. doi: 10.1038/mt.2016.159
112. Zhang M, Wang X, Han MK, Collins JF, Merlin D. Oral administration of ginger-derived nanolipids loaded with siRNA as a novel approach for efficient siRNA drug delivery to treat ulcerative colitis. Nanomedicine (2017) 12(16):1927–43. doi: 10.2217/nnm-2017-0196
113. Li Z, Wang H, Yin H, Bennett C, Zhang HG, Guo P. Arrowtail RNA for ligand display on ginger exosome-like nanovesicles to systemic deliver siRNA for cancer suppression. Sci Rep (2018) 8(1):14644. doi: 10.1038/s41598-018-32953-7
114. Wolfram J, Zhu M, Yang Y, Shen J, Gentile E, Paolino D, et al. Safety of nanoparticles in medicine. Curr Drug Targets (2015) 16(14):1671–81. doi: 10.2174/1389450115666140804124808
115. Nelemans LC, Gurevich L. Drug delivery with polymeric nanocarriers-cellular uptake mechanisms. Materials (2020) 13(2):366. doi: 10.3390/ma13020366
116. Zhou M, Huang H, Wang D, Lu H, Chen J, Chai Z, et al. Light-triggered PEGylation/dePEGylation of the nanocarriers for enhanced tumor penetration. Nano Lett (2019) 19(6):3671–5. doi: 10.1021/acs.nanolett.9b00737
117. Yang C, Zhang M. Merlin D.(2018).Advances in plant-derived edible nanoparticle-based lipid nano-drug delivery systems as therapeutic nanomedicines. J Mat Chem B 6(9):1312–21. doi: 10.1039/C7TB03207B
118. Wang Q, Zhuang X, Mu J, Deng ZB, Jiang H, Zhang L, et al. Delivery of therapeutic agents by nanoparticles made of grapefruit-derived lipids. Nat Commu (2013) 4:1867ñ1877. doi: 10.1038/ncomms2886
119. Mu J, Zhuang X, Wang Q, Jiang H, Deng ZB, Wang B, et al. Interspecies communication between plant and mouse gut host cells through edible plant derived exosome-like nanoparticles. Mol Nutr Food Res (2014) 58:1561ñ1573. doi: 10.1002/mnfr.201300729
120. Zhang M, Viennois E, Prasad M, Zhang Y, Wang L, Zhang Z, et al. Edible ginger-derived nanoparticles: A novel therapeutic approach for the prevention and treatment of inflammatory bowel disease and colitis-associated cancer. Biomaterials (2016) 101:321ñ340. doi: 10.1016/j.biomaterials.2016.06.018
121. Deng Z, Rong Y, Teng Y, Mu J, Zhuang X, Tseng M, et al. Broccoli-derived nanoparticle inhibits mouse colitis by activating dendritic cell amp-activated protein kinase. Mol Ther (2017) 25:1641ñ1654. doi: 10.1016/j.ymthe.2017.01.025
122. Zhuang X, Deng ZB, Mu J, Zhang L, Yan J, Miller D, et al. Ginger-derived nanoparticles protect against alcohol-induced liver damage. Extracell Vesicles (2015) 4:28713ñ28730. doi: 10.3402/jev.v4.28713
123. Zhang M, Xiao B, Wang H, Han MK, Zhang Z, Viennois E, et al. Edible ginger-derived nano-lipids loaded with doxorubicin as a novel drug-delivery approach for colon cancer therapy. Mol Ther (2016) 24:1783ñ1796. doi: 10.1038/mt.2016.159
124. Wang Q, Ren Y, Mu J, Egilmez NK, Zhuang X, Deng Z, et al. Grapefruit-derived nanovectors use an activated leukocyte trafficking pathway to deliver therapeutic agents to inflammatory tumor sites. Cancer Res (2015) 75:2520ñ2529. doi: 10.1158/0008-5472.CAN-14-3095
125. Loureiro JA, Andrade S, Duarte A, Neves AR, Queiroz JF, Nunes C, et al. Resveratrol and grape extract-loaded solid lipid nanoparticles for the treatment of alzheimer’s disease. Molecules (2017) 22:277ñ292. doi: 10.3390/molecules22020277
126. Wang B, Zhuang X, Deng ZB, Jiang H, Mu J, Wang Q, et al. Targeted drug delivery to intestinal macrophages by bioactive nanovesicles released from grapefruit. Mol Ther (2014) 22:522ñ534. doi: 10.1038/mt.2013.190
127. Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Phys (1959) 37:911ñ917. doi: 10.1139/o59-099
128. Ulmer CZ, Jones CM, Yost RA, Garrett TJ, Bowden JA. Optimization of folch, Bligh-dyer, and matyash sample-to-extraction solvent ratios for human plasma-based lipidomics studies. Anal chimica Acta (2018) 1037:351–7. doi: 10.1016/j.aca.2018.08.004
129. Duan L, Ouyang K, Xu X, Xu L, Wen C, Zhou X, et al. Nanoparticle delivery of CRISPR/Cas9 for genome editing. Front Genet (2021) 12:673286. doi: 10.3389/fgene.2021.673286
130. Nie D, Guo T, Yue M, Li W, Zong X, Zhu Y, et al. Research progress on nanoparticles-based CRISPR/Cas9 system for targeted therapy of tumors. Biomolecules (2022) 12(9):1239. doi: 10.3390/biom12091239
131. Wang P, Zhang L, Zheng W, Cong L, Guo Z, Xie Y, et al. Thermo-triggered release of CRISPR-Cas9 system by lipid-encapsulated gold nanoparticles for tumor therapy. Angew. Chem Int Ed (2017) 57:1491–6. doi: 10.1002/anie.201708689
132. Jo A, Ringel-Scaia VM, McDaniel DK, Thomas CA, Zhang R, Riffle JS, et al. Fabrication and characterization of PLGA nanoparticles encapsulating large CRISPR–Cas9 plasmid. J Nanobiotechnol (2020) 18:16. doi: 10.1186/s12951-019-0564-1
133. Stremersch S, De Smedt SC, Raemdonck K. Therapeutic and diagnostic applications of extracellular vesicles. J Control Release (2016) 244:167–83. doi: 10.1016/j.jconrel.2016.07.054
134. Mashouri L, Yousefi H, Aref AR, Ahadi AM, Molaei F, Alahari SK. Exosomes: composition, biogenesis, and mechanisms in cancer metastasis and drug resistance. Mol Cancer (2019) 18:75. doi: 10.1186/s12943-019-0991-5
135. Teng Y, Ren Y, Sayed M, Hu X, Lei C, Kumar A, et al. Plant-derived exosomal microRNAs shape the gut microbiota. Cell Host Microbe (2018) 24(5):637–52. doi: 10.1016/j.chom.2018.10.001
136. Wang X, Zhang M, Flores SRL, Woloshun RR, Yang C, Yin L, et al. Oral gavage of ginger nanoparticle-derived lipid vectors carrying dmt1 siRNA blunts iron loading in murine hereditary hemochromatosis. Mol Ther (2019) 27(3):493–506. doi: 10.1016/j.ymthe.2019.01.003
137. Mainini MR. Eccles Lipid and polymer-based nanoparticle siRNA delivery systems for cancer therapy. Molecules (2020) 25(11):2692. doi: 10.3390/molecules25112692
138. Durand-Reville TF, Comita-Prevoir J, Zhang J, Wu X, May-Dracka TL, Romero JAC, et al. Discovery of an orally available diazabicyclooctane inhibitor (ETX0282) of class a, c, and d serine beta-lactamases. J Med Chem (2020) 63(21):12511–25. doi: 10.1021/acs.jmedchem.0c00579
139. Lin D, Chen T, Xie M, Li M, Zeng B, Sun R, et al. Oral administration of bovine and porcine milk exosomes alter miRNAs profiles in piglet serum. Sci Rep (2020) 10(1):6983. doi: 10.1038/s41598-020-63485-8
140. Arntz OJ, Pieters BC, Oliveira MC, Broeren MG, Bennink MB, de Vries M, et al. Oral administration of bovine milk-derived extracellular vesicles attenuates arthritis in two mouse models. Mol Nutr Food Res (2015) 59(9):1701–12. doi: 10.1002/mnfr.201500222
141. Cheng Y, Zeng Q, Han Q, Xia W. Effect of pH, temperature, and freezing-thawing on quantity changes and cellular uptake of exosomes. Protein Cell (2019) 10(4):295–9. doi: 10.1007/s13238-018-0529-4
142. Title AC, Denzler R, Stoffel M. Uptake and function studies of maternal milk-derived microRNAs. J Biol Chem (2015) 290(39):23680–91. doi: 10.1074/jbc.M115.676734
143. Zhuang X, Deng ZB, Mu J, Zhang L, Yan J, Miller D, et al. Ginger-derived nanoparticles protect against alcohol-induced liver damage. J Extracell Vesicles (2015) 4(1):28713. doi: 10.3402/jev.v4.28713
144. Karamanidou T, Tsouknidas A. Plant-derived extracellular vesicles as therapeutic nanocarriers. Int J Mol Sci (2021) 23(1):191. doi: 10.3390/ijms23010191
145. Wang Q, Zhuang X, Mu J, Deng Z, Jiang H, Xiang X, et al. Delivery of therapeutic agents by nanoparticles made of grapefruit-derived lipids. Nat Commun (2013) 4:11. doi: 10.1038/ncomms3358
146. Li Z, Wang H, Yin H, Bennett C, Zhang HG, Guo P. Arrowtail. (2018).RNA for ligand display on ginger exosome-like nanovesicles to systemic deliver siRNA for cancer suppression. Sci Rep 8(1):14644. doi: 10.1038/s41598-018-32953-7
147. Deng Z, Rong Y, Teng Y, Mu J, Zhuang X, Tseng M, et al. Broccoli-derived nanoparticle inhibits mouse colitis by activating dendritic cell AMP-activated protein kinase. Mol Ther (2017) 25(7):1641–54. doi: 10.1016/j.ymthe.2017.01.025
148. Zhang M, Viennois E, Prasad M, Zhang Y, Wang L, Zhang Z, et al. Edible ginger-derived nanoparticles. a novel therapeutic approach for the prevention and treatment of inflammatory bowel disease and colitis-associated. Cancer Biomat (2016) 101:321–40. doi: 10.1016/j.biomaterials.2016.06.018
149. Rome S. Biological properties of plant-derived extracellular vesicles. Food Funct (2019) 10(2):529–38. doi: 10.1039/C8FO02295J
150. Cai Q, Qiao L, Wang M, He B, Lin F-M, Palmquist J, et al. Plants send small RNAs in extracellular vesicles to the fungal pathogen to silence virulence genes. Science (2018) 360(6393):1126–9. doi: 10.1126/science.aar4142
151. Regente M, Pinedo M, San Clemente H, Balliau T, Jamet E, de La L. Canal plant extracellular vesicles are incorporated by a fungal pathogen and inhibit its growth. J Exp Bot (2017) 68(20):5485–95. doi: 10.1093/jxb/erx355
152. Wiklander OPB, Brennan MA, Lotvall J, Breakefield XO, El Andaloussi S. Advances in therapeutic applications of extracellular vesicles. Sci Transl Med (2019) 11(492):eaav8521. doi: 10.1126/scitranslmed.aav8521
153. Pocsfalvi G, Turiak L, Ambrosone A, Del Gaudio P, Puska G, Fiume I, et al. Protein biocargo of citrus fruit-derived vesicles reveals heterogeneous transport and extracellular vesicle populations. J Plant Physiol (2018) 229:111–21. doi: 10.1016/j.jplph.2018.07.006
154. Li L. S, Hu XY. Chen Non-viral delivery systems for CRISPR/Cas9-based genome editing: challenges and opportunities. Biomaterials (2018) 171:207–18. doi: 10.1016/j.biomaterials.2018.04.031
155. Tang Q, Liu J, Jiang Y, Zhang MN, Mao LQ, Wang M. Cell-selective messenger RNA delivery and CRISPR/Cas9 genome editing by modulating the interface of phenylboronic acid-derived lipid nanoparticles and cellular surface sialic acid, ACS appl. Mater Inter (2019) 11:46585–90. doi: 10.1021/acsami.9b17749
Keywords: CRISPR, exosomes, nanocarrier, colon cancer, gene-based therapeutics
Citation: Hillman T (2023) The use of plant-derived exosome-like nanoparticles as a delivery system of CRISPR/Cas9-based therapeutics for editing long non-coding RNAs in cancer colon cells. Front. Oncol. 13:1194350. doi: 10.3389/fonc.2023.1194350
Received: 28 March 2023; Accepted: 16 May 2023;
Published: 14 June 2023.
Edited by:
Matiullah Khan, AIMST University, MalaysiaReviewed by:
Shiyan Dong, University of Texas MD Anderson Cancer Center, United StatesMehdi Soleymani-Goloujeh, Royan institute for Stem Cell Biology and Technology (RI-SCBT), Iran
Copyright © 2023 Hillman. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tatiana Hillman, dGhpbGxtYW5AdGhlbDRiaW5jLm9yZw==
 Tatiana Hillman
Tatiana Hillman