- 1Department of Pathology, Ningbo Clinical Pathology Diagnosis Center, Ningbo, China
- 2Department of Pathology, Ruijin Hospital, Shanghai Jiaotong University School of Medicine, Shanghai, China
- 3Department of Pathology, Zhejiang Provincial People’s Hospital, People’s Hospital of Hangzhou Medical College, Hangzhou, China
Background: Microsatellite instability (MSI), or mismatch repair-deficiency (dMMR), is rare in prostate cancers (PCas). The histological and molecular features of PCas with MSI/dMMR are incompletely described. Thus, we sought to identify the characteristics of PCas with MSI/dMMR.
Methods and results: We analyzed 1,141 primary treatment-naive PCas by MMR-related protein immunohistochemistry (MLH1, PMS2, MSH2, and MSH6). We identified eight cases exhibiting MSI/dMMR (0.7%, 8/1141). Of these, six tumors had both MSH2 and MSH6 protein loss, one had both MLH1 and PMS2 protein loss, and one had only MSH6 loss. Histologically, MSI/dMMR-PCas frequently demonstrated high histological grade (Grade Group 4 or 5), ductal/intraductal histology (6/8 cases), pleomorphic giant-cell features (4/8 cases), and conspicuous tumor lymphocytic infiltration (8/8 cases). Polymerase chain reaction-based analysis of seven MSI/dMMR tumors revealed two MSI-H tumors with loss of both MSH2 and MSH6 proteins. Subsequently, the seven cases underwent next-generation sequencing (NGS) analysis with a highly validated targeted panel; four were MSI. All cases had a high tumor mutation burden (median: 45.3 mutations/Mb). Overall, the MSI/dMMR-PCas showed a high frequency of DNA damage-repair pathway gene changes, including five with pathogenic somatic or germline MMR gene mutations. Activating mutations in the MAPK pathway, PI3K pathway, and WNT/β-catenin pathway were common. TMPRSS2::ERG rearrangement was identified in one case (1/7, 14.3%).
Conclusions: Several pathological features are associated with MSI/dMMR in PCas. Identification of these features may help to select patients for genetic screening. As MSI/dMMR-PCas are enriched for actionable mutations, patients should be offered NGS to guide standard-of-care treatment.
Introduction
Mismatch repair (MMR) pathways play critical roles in maintaining genomic fidelity during DNA replication. MMR deficiency (dMMR) is triggered by germline, somatic, and epigenetic changes in MMR genes (most commonly MLH1, PMS2, MSH2, and MSH6), which inactivate these genes, causing loss of MMR-related protein expression, as noted by immunohistochemistry (IHC), and microsatellite instability (MSI) development (1–3). Most cancers with MSI occur sporadically, but about 16% result from inherited mutations (Lynch Syndrome) (4). Recent studies using large-scale genome analyses have found that MSI occurs in virtually all cancer types at some frequency (5, 6). MSI has been used as a surrogate marker for dMMR. Knowing the MSI status, or dMMR, can help to identify patients with susceptibility to immune checkpoint inhibitors (ICIs), which is associated with a greater and more durable treatment response (6, 7). Accordingly, in 2017, the US Food and Drug Administration approved the PD-1 inhibitor Pembrolizumab for the treatment of all advanced cancers with MSI-high (H) or dMMR, irrespective of the site of tumor origin (8). Therefore, MSI testing in all cancer types has become increasingly important. Recently, MSI-H/dMMR prostate cancer (PCa) patients have also been reported to benefit from treatment with ICIs inhibiting PD-1 (9, 10).
MSI/dMMR is a common and well-defined feature of colorectal and endometrial adenocarcinoma (11–13). Reports on MSI in PCa have come to different conclusions. Using different detection methods, several studies have shown that MSI/dMMR was observed in approximately 1.2%–12% of PCa patients (6, 9, 14, 15). The histological and molecular features of dMMR in PCa remain incompletely described. With advances in molecular sequencing, several investigators have correlated dMMR with an aggressive phenotype and late-stage PCas and have noted that patients with certain features may have MSI/dMMR, including unusual metastatic sites (such as the lungs), high-grade Gleason scores, or variant histologies, such as ductal/intraductal PCa (10, 14, 16–19). Currently, the clinicopathological significance of MSI/dMMR in PCa is not fully understood and the underlying mechanisms of MSI/dMMR in PCa deserve further investigation.
Here, we investigated the prevalence, and the clinicopathological and molecular characteristics of MSI/dMMR-PCa in the Chinese population, with particular attention to potential clinicopathological features that may alert clinicians to consider MSI or genetic investigations.
Material and methods
Patients and samples
We retrospectively reviewed cases of PCa evaluated by IHC to assess MMR-related proteins loss, from January 1, 2019, to January 31, 2021, at Ningbo Clinical Pathology Diagnosis Center, Shanghai Ruijin Hospital, and Zhejiang Provincial People’s Hospital. A total of 1141 primary treatment-naive PCas were identified. Overall, 665 PCa cases (including Grade group (GG) 1: 27, GG2: 155, GG3: 71, GG4: 247, GG5: 165) were identified at the Ningbo Clinical Pathology Diagnosis Center, 400 PCa cases were identified at the Department of Pathology of Shanghai Ruijin Hospital (including GG1: 47, GG2: 171, GG3: 100, GG4: 23, GG5: 59), and 76 PCa cases were identified at the Department of Pathology of Zhejiang Provincial People’s Hospital (including GG1: 14, GG2: 27, GG3: 19, GG4: 9, GG5: 7).
Eight PCa patients with loss of MMR-related proteins were identified (0.7%, 8/1,141). All cases involved primary tumors and were sampled on transurethral resection of the prostate (TURP) (n = 1), needle biopsy (n = 1), or radical prostatectomy (n = 6). Hematoxylin and eosin-stained sections of these eight cases were reviewed independently by three experienced urological pathologists (H.Z, X.Y, and M.Z) to assess the histological type, Gleason score, the presence of lymphovascular invasion (LVI) and perineural invasion (PNI), and the presence of ductal/intraductal histology, a pleomorphic giant-cell component, tumor infiltrating lymphocytes (TILs), and locoregional lymph node metastases. TILs were considered to be “significant” when 10 TILs were identified per high-power field.
For the eight MSI/dMMR-PCa patients, we collected the clinical history, surgical procedure, and clinicopathological data by review of the medical records and pathology reports. Follow-up data were collected by telephonic interview. The study was approved by our institutional review board.
Mismatch repair protein immunohistochemistry and interpretation
MMR-related protein IHC was performed using a BenchMark autostaining system (Roche, Basel, Switzerland), with appropriate controls. We used a mouse monoclonal antibody for MSH2 (clone MX061), a rabbit monoclonal antibody for MSH6 (clone EP49), a mouse monoclonal antibody for MLH-1 (clone ES05), and a rabbit monoclonal antibody for PMS2 (clone EP51). Immunostaining was assessed independently by three experienced urological pathologists. MMR-related protein loss was defined by MMR-related protein loss in any tumor cells in any tumor spot, with intact staining in admixed benign prostate gland and/or surrounding stromal cells, endothelial cells, or lymphocytes. MMR-related protein staining without internal control staining was considered ambiguous and was not scored. The tumors were defined as hypermutated if the tumor mutation burden (TMB) > 10 mutations/Mb. Tumors were classified as showing MSI/dMMR if they were found to harbor a deleterious germline or somatic alteration in an MMR-related gene or had MMR-related protein loss by IHC.
Microsatellite instability PCR
MSI-polymerase chain reaction (PCR) testing was performed by the genetics laboratory of the Department of Pathology of Ruijin Hospital, using the MSI analysis kit (AmoyDx, Xiamen, China) following the manufacturer instructions, for amplification of five mononucleotide repeat markers (CAT25, BAT-26, BAT-25, NR-24, MONO-27) and two pentanucleotide repeat loci (Penta-D and Penta-E), to confirm identity between the tumor and paired benign tissue. Status of MSI-high was given when ≥ two of these markers showed instability and status of MSI-low if only one marker showed instability, otherwise microsatellite stability (MSS) was assumed.
Library preparation and targeted next-generation sequencing
Next-generation sequencing (NGS) was performed in a CLIA‐ and CAP‐accredited laboratory (Nanjing Geneseeq Technology Inc., Nanjing, China). Genomic DNA was extracted from formalin-fixed, paraffin-embedded (FFPE) samples using the QIAamp DNA FFPE Tissue Kit (Qiagen, Hilden, Germany), following the manufacturer’s instructions. Purified DNA was quantified by Qubit 3.0 using the dsDNA HS Assay Kit (Life Technologies, Carlsbad, CA, USA) and qualified by Nanodrop2000 (Thermo Fisher Scientific, Waltham, MA, USA). Sequencing libraries were prepared by KAPA Hyper Prep kit (KAPA Biosystems, Wilmington, MA, USA). A customized xGen lockdown-probe panel (Integrated DNA Technologies, Coralville, IA, USA) were used for selective enrichment for 437 predefined cancer-related genes (Geneseeq Technology, Inc., Toronto, ON, Canada; Prime panel). The capture reaction was performed with DynaBeads M-270 (Life Technologies) and xGen Lockdown Hybridization and Wash kit (Integrated DNA Technologies). The purified library was quantified by quantitative PCR using the KAPA Library Quantification kit (KAPA Biosystems), and its fragment size distribution was analyzed by Bioanalyzer 2100 (Agilent Technologies, Inc., Santa Clara, CA, USA). The target-enriched libraries were sequenced on a HiSeq4000 NGS platform (Illumina, San Diego, CA, USA) with 2 × 150-bp pair-end reads to primary coverage depths of 800×.
MSI was estimated based on 52 indel sites in the Geneseeq Prime panel. If > 40% of 52 sites showed unstable status (compared to the distribution in the 500 normal sample pools), the sample was defined as showing MSI. The TMB was defined as the number of somatic, coding, base substitution, and indel mutations per megabase of genome examined, and was calculated as previously described (20, 21).
Results
Prevalence and clinicopathological features of cases with MMR-related protein loss by immunohistochemistry
We screened for MMR-related protein loss in 1,141 primary PCas, including 88 GG1 tumors, 353 GG2 tumors, 190 GG3 tumors, 279 GG4 tumors, and 231 GG5 tumors. Altogether, 0.7% (8/1,141) of primary PCas had MMR-related protein loss (MSI-PCas), including 1.4% (4/279) GG4 and 1.5% (4/231) of GG5 tumors.
The clinicopathological characteristics of the eight MSI-PCa cases are detailed in Table 1. Patients’ age at diagnosis ranged from 63 to 91 years (median: 72 years). The MSI-PCa tumors were highly aggressive based on pathological features, including tumor grade and stage. All cases had Gleason scores of 8 or 9. In addition to usual acinar adenocarcinoma (AAC), ductal/intraductal histology was present in 6 of the 8 patients (ductal in 4, Figure 1A, and intraductal in 2. Figure 1B). Of the six cases with pathological stage information at radical prostatectomy available, three presented with pT2c and three with pT3b (2 with nodal involvement). PNI was observed in all MSI-PCa patients. Two cases had LVI (Figure 1C). Four cases demonstrate focal pleomorphic giant-cell features (Figure 1D). All cases had a higher density of TILs within the tumor (Figures 1E, F). By IHC, six cases had loss of both MSH2 and MSH6, one (case 4) had loss of both MLH1 and PMS2, and one (case 7) had loss of MSH6 protein only (Figure 2, case1-8). Interestingly, the AAC area in case 6 showed intact MMR-related protein immunostaining (Figure 2, case 6). Six patients had received standard androgen-deprivation therapy (ADT), and four had received first-line abiraterone or enzalutamide. One patient received anti-PD-1/PD-L1 therapy. One patient (case 4) progressed to prostatic small cell carcinoma after 10-month ADT with TURP. The PSCC showed MLH1 and PMS2 protein loss, in keeping with the needle biopsy specimen (Figure 2, case 4). One patient developed bone metastases 4 months after radical prostatectomy (case 2).
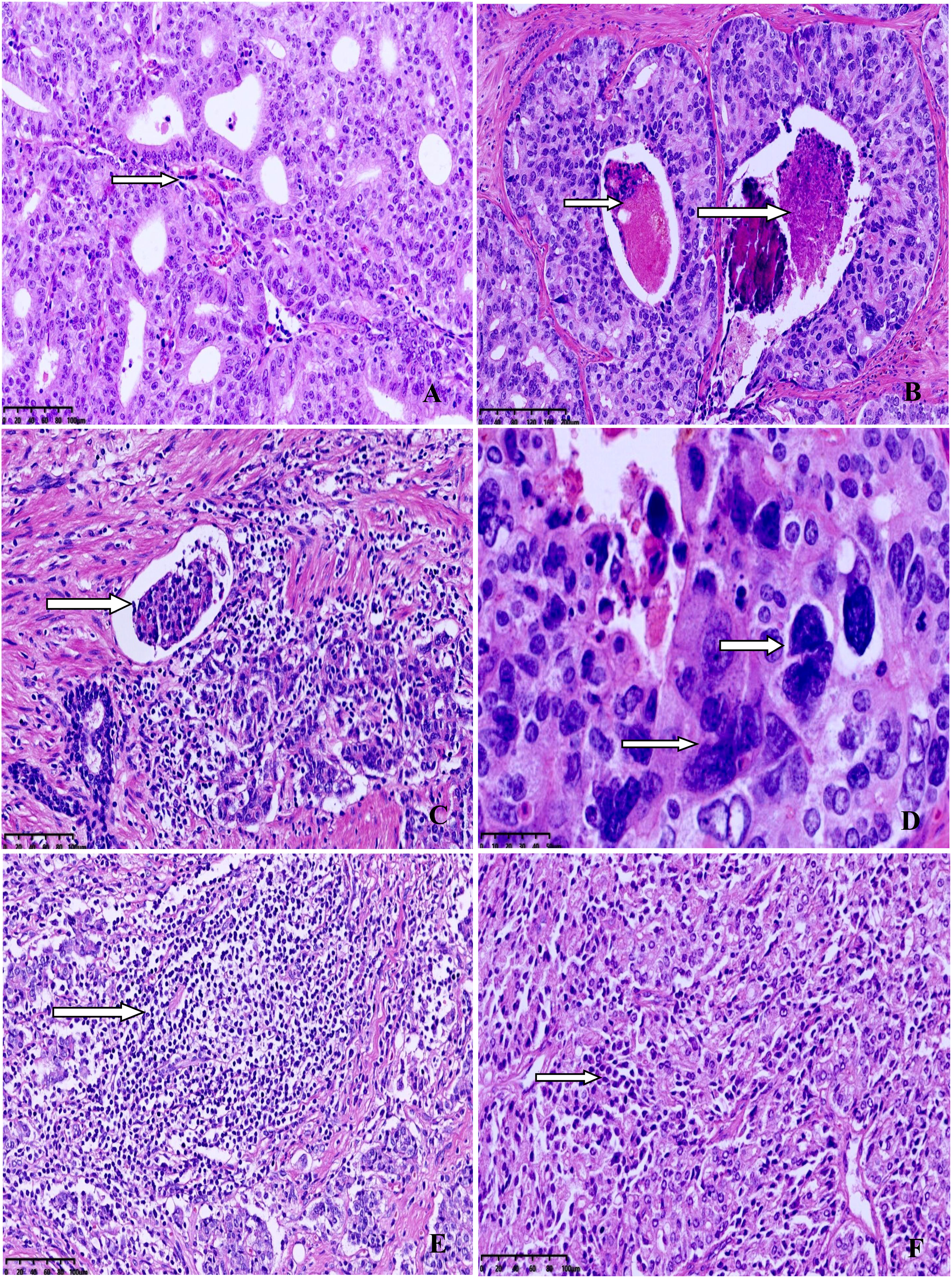
Figure 1 Representative morphology from selected cases. (A) Ductal adenocarcinoma with well-established fibrovascular cores. (B) Intraductal carcinoma with comedocecrosis. (C) Lymphovascular invasion. (D) Higher power view of pleomorphic giant-cells with bizarre atypia. (E) Dense lymphocytic infiltration in the tumor. (F) Diffuse lymphocytic infiltration between the tumor cells.
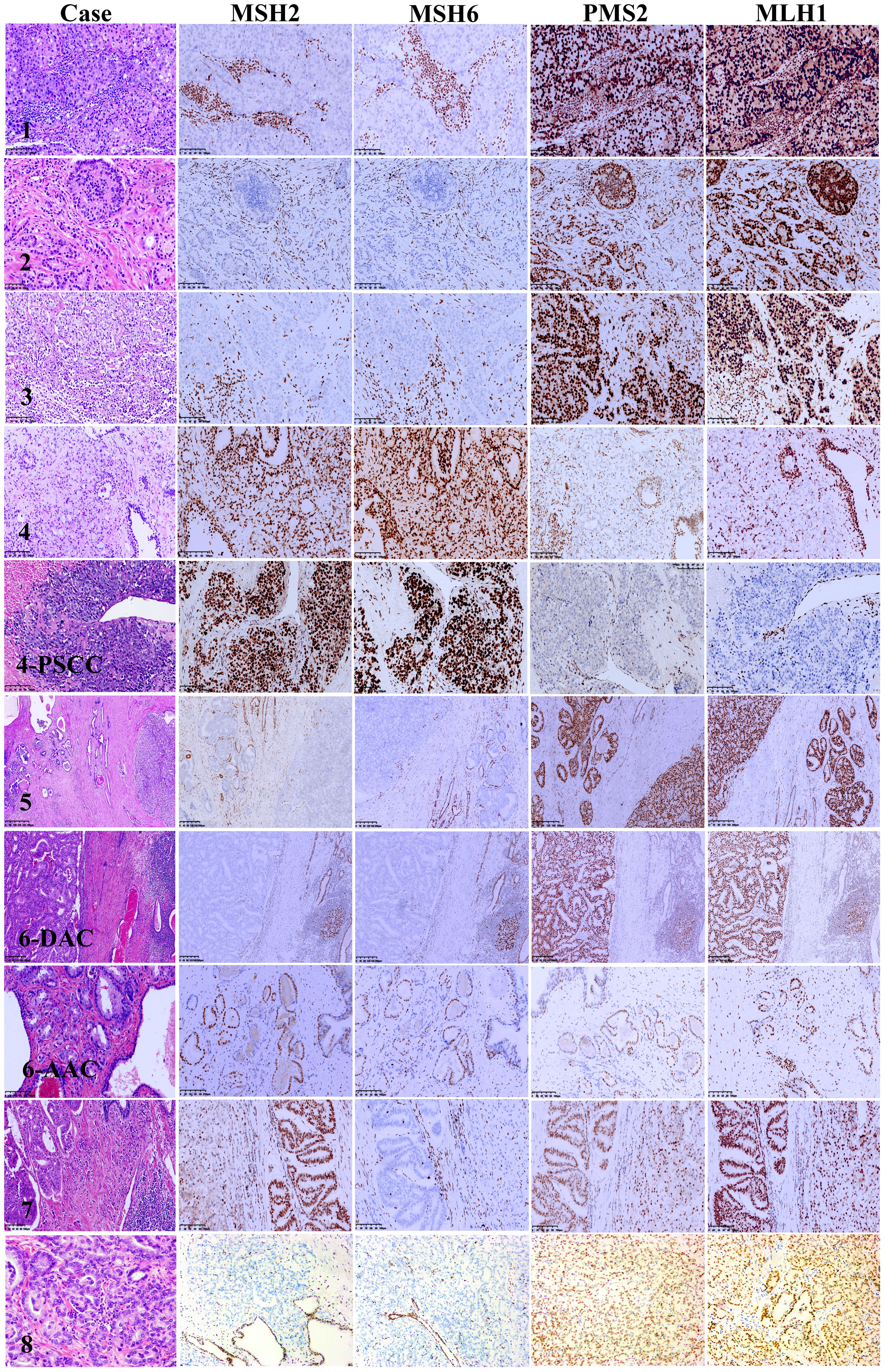
Figure 2 Mismatch repair protein immunohistochemistry in microsatellite instability prostate carcinoma. Case 1: MSH2 and MSH6 nuclear immunostaining are absent in tumor cells nuclei but present in infiltrating lymphocytes, PMS2 and MLH1 fully preserve expression. Case 2: MSH2 and MSH6 expression loss in both intraductal carcinoma and invasive carcinoma of the prostate, PMS2 and MLH1 overexpression in both intraductal carcinoma and invasive carcinoma of the prostate. Case 3: The high-grade invasive carcinoma of the prostate shows MSH2 and MSH6 loss with intact PMS2 and MLH1 nuclear expression. Case 4: Both the primary invasive carcinoma and small cell carcinomas after androgen deprivation therapy show intact MSH2 and MSH6 and lack of PMS2 and MLH1 nuclear staining. Case 5: Both intraductal carcinoma and invasive carcinoma of the prostate show MSH2 and MSH6 expression loss with PMS2 and MLH1 overexpression. Case 6: MSH2 and MSH6 expression loss in the ductal adenocarcinoma but not in the acinar adenocarcinoma. Both ductal adenocarcinoma and acinar adenocarcinoma show PMS2 and MLH1 overexpression. Case 7: Both the ductal adenocarcinoma and acinar adenocarcinoma show MSH6 loss with intact MSH2, PMS2 and MLH1 expression. Case 8: MSH2 and MSH6 expression loss, PMS2 and MLH1 overexpression. Lymphocytes, endothelial cells and stromal cells used as internal control.
Results of MSI-testing by PCR and Targeted NGS and correlation with IHC
Table 2 summarizes the MSI status and MMR gene alterations. For the eight samples with MMR-related protein loss by IHC, the MSI status of seven cases were also evaluated by PCR, which was not performed for case 1, due to the lack of control paired benign tissue. Of the seven tumors tested for MSI by PCR, two were MSI-H (Figures 3A, B) and both had loss of MSH2 and MSH6 expression by IHC. By targeted NGS, 4 of the seven cases showed MSI and also had loss of MMR-related protein expression by IHC (3 with MSH2 and MSH6 protein loss and 1 with MLH1 and PMS2 protein loss). Of the 4 MSI-postive cases by targeted NGS, 3 had pathogenic somatic mutations in MMR-associated genes, including one with mutation in MSH2 (case 3, Figure 4A), one with mutations in MSH2 and MSH6 (case 1, Figures 4B, C), and one with mutation in MLH3 (case 4). Three cases (case 2, 6, and 7) demonstrated MMR-related protein loss by IHC but showed MSS by both PCR and targeted NGS. Of these 3 cases, case 6 showed a pathogenic germline mutation of MSH6 (Figure 4D) and a pathogenic somatic mutation in MSH2, case 7 showed a pathogenic somatic mutation in MSH6, while case 2 demonstrated no mutations involving MMR genes.
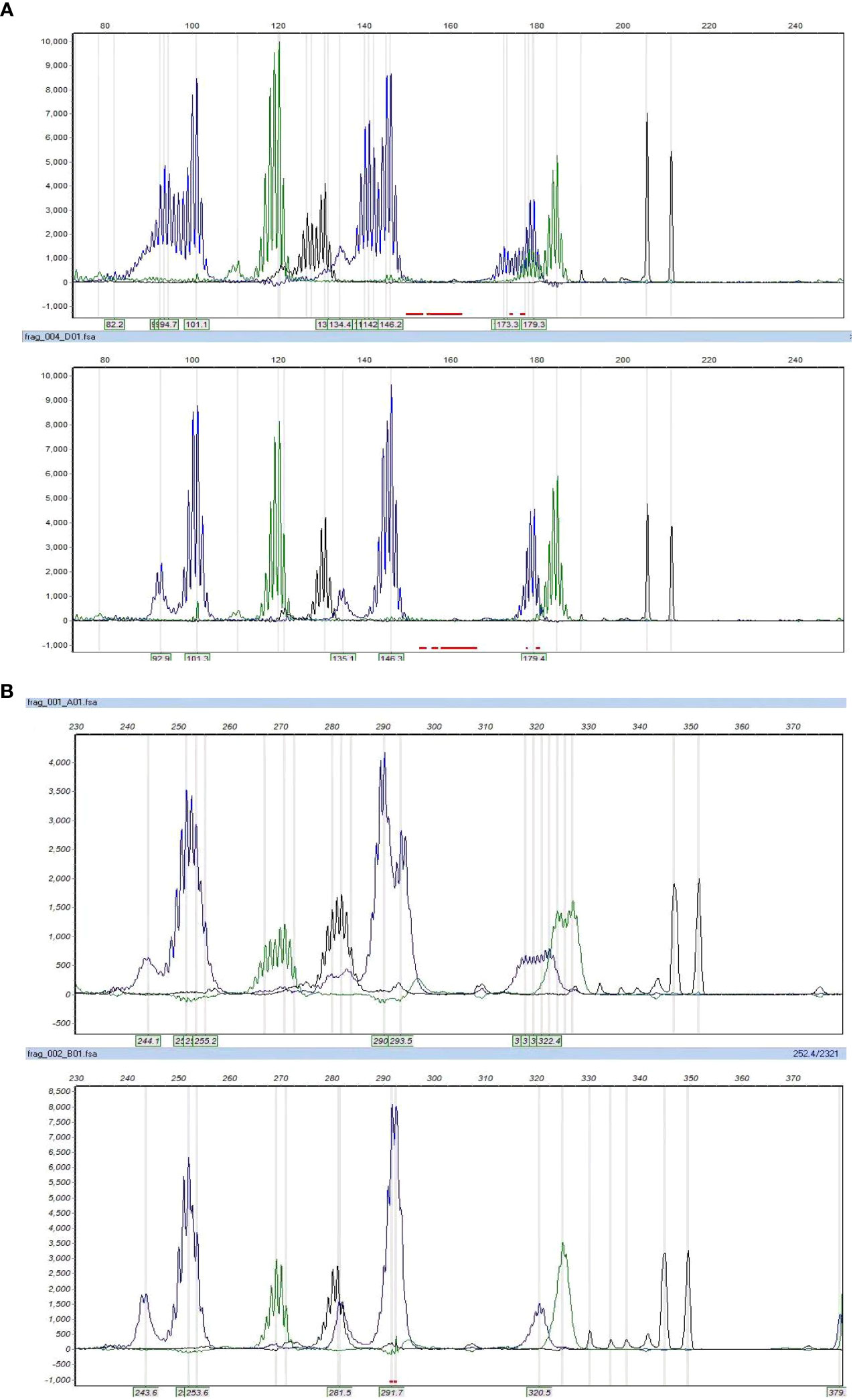
Figure 3 The microsatellite instability status by PCR. Cases 3 and 8 showed MSI-H, with allelic shifts in 4 of 7 markers in case 3 (A) and 7 of 7 markers in case 8 (B).
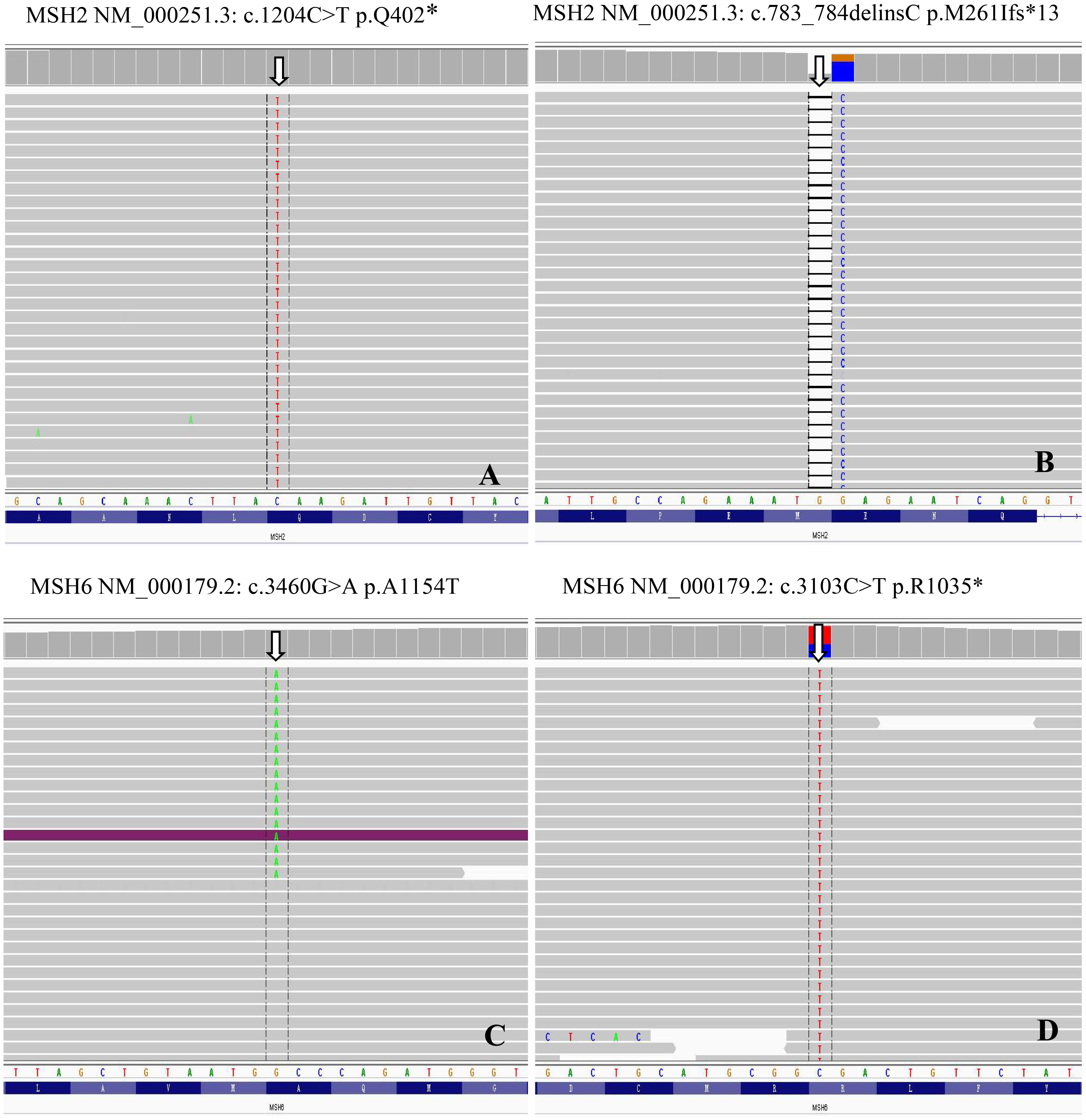
Figure 4 MMR genes mutations in MSI prostate cancer. Genomics Viewer screenshot of the representative MMR genes mutations, including MSH2 (A), c.1204C>T p.Q402*; (B), c.783_784delinsC p.M261Ifs*13) and MSH6 (C), c.3460G>A p.A1154T; (D), c.3103C>T p.R1035*).
Comprehensive analysis of somatic and germline alterations
Tables 2, 3 summarize the genomic characteristics. All cases with targeted NGS showed high TMB. The median TMB was 45.3 mutations/Mb (range, 25.7–71.1 mutations/Mb). Interestingly, the AAC component in case 6 had no tumor mutation burden. Somatic and germline alterations of relevant genes in MSI/dMMR-PCas are shown in Figure 5, and grouped in pathways that are potentially clinically actionable. Overall, MSI/dMMR-PCa cases showed a high frequency of alterations in genes in DNA damage repair (DDR) pathways and of germline mutations in DDR genes, including the homologous recombination repair (HRR), MMR, and other DDR pathways. Six patients (6/7) had a known or likely pathogenic somatic mutation in DDR pathway genes, including in MSH2 (3/6), MSH6 (3/6), ATR (2/6), ATM (2/6), TP53 (2/6), PTEN (2/6), MLH3 (1/6), RAD50 (1/6), PALB2 (1/6), ERCC2 (1/6), RAD54L (1/6), and ATRX (1/6). Among seven patients carrying the pathogenic or likely pathogenic DDR gene mutations, five (71.4%) were involved in the MMR pathway, two (28.6%) were in the HRR pathway, and four (57.1%) were in other DDR pathways. Five patients (5/7) harbored a germline alteration in a gene that is involved in HRR pathway, including BRCA2, RECQL4, PALB2, RAD54L, BRCA1, POLH, and RAD51D. In total, 71.4% of patients (5/7) had pathogenic somatic or germline alterations in MMR genes, including MSH2 (3/5), MSH6 (3/5), POLD1 (3/5), and MLH3 (2/5). Two patients without evidence of MSI by PCR and targeted NGS had pathogenic or likely pathogenic germline and somatic MMR gene alterations One patient with MSI (case 5) had no pathogenic or likely pathogenic MMR genes and other DDR pathway gene alterations, however, the patient had likely pathogenic mutations in ARID1A, JAK1, and APC. The ductal adenocarcinoma (DAC) of case 6 also had a likely pathogenic mutation in ARID1A and pathogenic mutations in BRAF and JAK3.
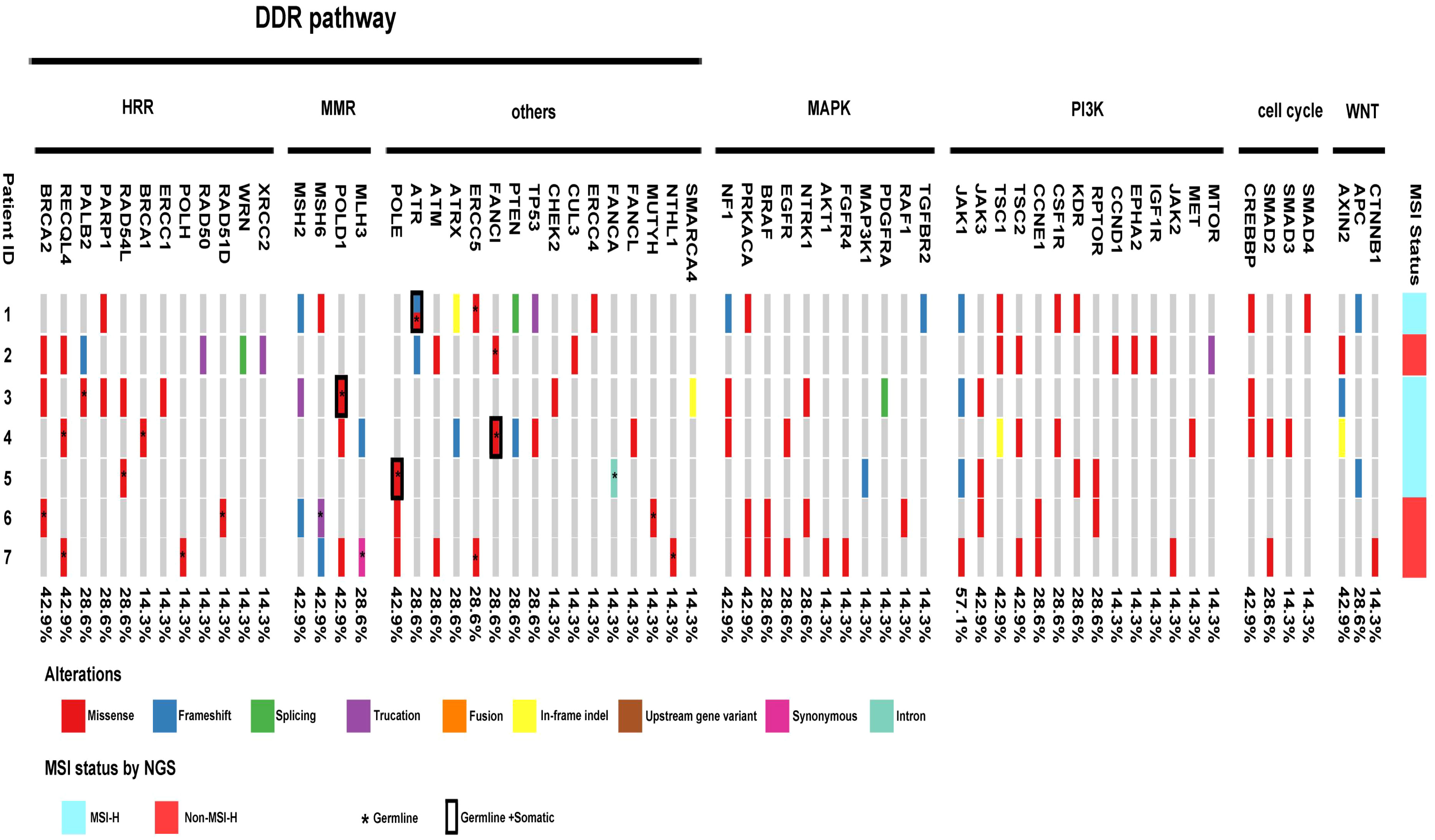
Figure 5 Landscape of genomic alterations across MSI prostate cancer, including alterations in genes involved in DDR pathway, MAPK pathway, PI3K pathway, Cell cycle and Wnt-βcatenin pathway. Among seven patients carrying the pathogenic or likely pathogenic DDR gene mutations, five (71.4%) were involved in the MMR pathway, two (28.6%) were in the HRR pathway, and four (57.1%) were in other DDR pathways. 85.7% of patients (6/7) harbored somatic alterations in MAPK pathway genes. All patients carried somatic alterations in the PI3K pathway. Four of the seven patients harbored somatic alterations in cell-cycle pathway genes. Six of the seven patients carried somatic alterations in the WNT/β-catenin pathway.
In addition, 85.7% of patients (6/7) harbored somatic alterations in mitogen-activated protein kinase (MAPK) pathway genes, including mutations in NF1 (3/6), PRKACA (3/6), BRAF(2/6), EGFR (2/6), NTRK1 (2/6), AKT1 (1/6), FGFR4 (1/6), MAP3K1 (1/6), PDGFRA (1/6), RAF1 (1/6), and TGFBR2 (1/6). All patients carried somatic alterations in the PI3K pathway, including hot spot mutations in JAK1, JAK3, TSC1, and TSC2. Four of the seven patients harbored somatic alterations in cell-cycle pathway genes, including CREBBP, SMAD2, SMAD3, and SMAD4. Six of the seven patients carried somatic alterations in the WNT/β-catenin pathway, including AXIN2, APC, and CTNNB1. Notably, one patient (1/7, case 4) had TMPRSS2::ERG fusion.
Discussion
In the current study, we reported a 0.7% prevalence rate of MSI/dMMR tumors among primary PCa patients in the Chinese population. The identified MSI/dMMR-PCas showed a high frequency of DDR pathway gene mutations, including five cases with pathogenic somatic or germline MMR gene mutations. Activating mutations in the MAPK pathway, PI3K pathway, and WNT/β-catenin pathway were common, while a TMPRSS2::ERG rearrangement was identified in one case (case 4).
MSI/dMMR has been detected in 3% of unselected PCa cases (9, 22) and in 5% of metastatic PCa cases (10, 14). In a study of 60 advanced PCas, 12% (7/60) had MMR gene mutations and MSI (16). The MSI-H/dMMR frequency was found to be 3.1% in a large series of study that included 1,033 PCa patients and used NGS (9). Guedes et al. (15) reported MSH2 protein loss in primary PCas, whereas MLH1, PMS2, and MSH6 were not evaluated. In their study, 1.2% (14/1,176) of PCas had MSH2 loss, including 1% (12/1133) of primary adenocarcinomas and 5% (2/43) of PSCCs (15). In a recent study by Fraune et al. (22), 3.5% (7/200) of advanced PCas were MSI/dMMR based on IHC. Recently, Kagawa et al. (23) investigated the prevalence of dMMR-PCas in the Japanese population. They found that 1.2% (4/337) showed loss of MMR-related protein expression by IHC. The overall prevalence of MSI/dMMR-PCas in the Chinese population is thus extremely low, probably due to differences in the cohorts selected (our study has included all Gleason score groups) or racial differences.
The role of MMR-related proteins has been extensively studied in colorectal and endometrial cancer. Few reports have analyzed dMMR-PCa by IHC (15, 22, 24–28). We here analyzed the immunohistochemical expression of MMR-related proteins in a large series of PCas. To our knowledge, no previous study has examined the phenotype of sporadic primary PCas with dMMR in a large number of Chinese specimens. Among the MSI/dMMR patients in our study (8/1,141), six of eight patients demonstrated combined loss of MSH2 and MSH6, which was consistent with the studies by Kagawa et al. (23) (4/4) and Fraune et al. (22) (6/7).The combined loss pattern of MSH2 and MSH6 may be more frequent in PCas, while it is rather infrequent in colorectal and endometrial cancer (29). All of the dMMR-PCa cases in our series were high-grade PCas, including 1.4% (4/279) of tumors with GG4 and 1.5% (4/231) of tumors with GG5. The high incidence of dMMR in higher grade PCas in the present study also agrees with previous findings. As reported by Guedes et al. (15), 8% (7/91) of tumors with primary Gleason pattern 5 (5 + 4 = 9 or 5 + 5 = 10) PCas had MSH2 loss, while <1% of tumors from lower Gleason score groups had MSH2 loss. Kagawa et al. (23) also reported patients with dMMR PCas were at a significantly higher stage and had a greater Gleason score (≥8) than those with proficient MMR PCas. Recently, Wyvekens et al. (30) evaluated the histopathological features of 19 dMMR PCas and found that all treatment-naive dMMR cases (11/11, 100%) were GG 4 or 5. These findings suggest that patients with high-grade primary PCas (Gleason score ≥ 8) are more likely to harbor dMMR/MSI, which argues the need for routine clinical screening for MMR gene loss using IHC.
The morphological and molecular correlates of dMMR have been recognized in several tumor types, such as colorectal adenocarcinoma, endometrial adenocarcinoma, and upper tract urothelial carcinoma (11, 31, 32), whereas the histopathological features of dMMR primary PCas remain unclear. Recently, several morphological features have been reported as associated with dMMR-PCa, such as ductal/intraductal carcinoma, pleomorphic giant-cell features, LVI, cribriform and/or solid growth patterns, and TILs (15, 18, 19, 30, 33). Ductal/intraductal carcinoma (60%) and pleomorphic giant-cell features (50%) were frequently present in dMMR PCas. Prominent TILs were identified in all our dMMR-PCa cases. Guedes et al. (15) found that tumors with MSH2-loss had a higher density of infiltrating CD8+-lymphocytes than did grade-matched controls without MSH2-loss. Recently, Wyvekens et al. (30) also reported that TILs was a histopathological feature of dMMR-PCas (7/11, 64%).As we know, prominent intra- and peritumoral lymphocytic infiltrates are common in dMMR colorectal carcinomas (31). TILs might be a useful histopathological feature for identifying selected PCas for MMR-related protein IHC and genetic testing. Recently, TMB has also been proposed as a biomarker of response to immunotherapy (34). All of dMMR PCa patients in our study showed high TMB (median: 45.3 mutations/Mb, range: 25.7–71.1 mutations/Mb). Further studies examining the role of varied histological features, TIL density, or TMB in predicting sensitivity to immunotherapy should be considered.
We confirmed that dMMR-PCa cases are enriched for actionable mutations and found a significantly higher frequency of changes in DDR pathway-related genes, including mutations in MMR-related genes. Targeted NGS revealed pathogenic or pathogenic somatic mutations in MMR genes, including MSH2 (3/5) and MSH6 (2/5) in five of 7 dMMR-PCa cases. One dMMR PCa patient was identified as having a pathogenic germline MSH6 mutation. Pritchard et al. (16) demonstrated that MSI-advanced PCas are frequently driven by complex structural MSH2 or MSH6 rearrangements, rather than by MLH1 epigenetic silencing. None of the MMR mutations in our study were inherited in the germline. In a larger study of 1,133 primary PCas, a small percentage (1.2%) demonstrated MSH2 loss by IHC, with confirmation by NGS, while only three patients had germline mutations (15). Recently, Abida et al. (9) revealed genetic alterations in MSH2 and in MSH6 in 46% each of MSI-H/dMMR PCas, whereas the mutations rates for PMS2 and MLH1 were significantly lower (< 20% each), and seven of the 32 MSI-H/dMMR patients (21.9%) had a pathogenic or likely pathogenic germline mutation in MMR genes, including five in MSH2, one in MSH6, and one in PMS2. Furthermore, Wyvekens et al. (30) evaluated the molecular features of 19 dMMR-PCas and demonstrated that dMMR was secondary to functional loss of MSH2/MSH6 and MLH1/PMS2 in 15 (79%) and four cases (21%), respectively, while germline mutations were present in four cases (4 of 19, 21%). Our study further suggests that somatic MMR mutations are more common than germline mutations in PCas, and that inactivation of MSH2 and MSH6 appears to be the main cause of MSI in PCas, in contrast to colorectal and endometrial cancer, where MSI is most often due to epigenetic silencing of MLH1 (11, 12).
Most interestingly, in addition to MMR gene alterations, other pathogenic or likely pathogenic somatic mutations in DDR pathway genes in our dMMR-PCa cases included mutations in TP53, PTEN, ATR, RAD50, PALB2, ATM, XRCC2, RAD54L, and ATRX. Furthermore, some genes were recurrently mutated, including those involved in the MAPK and PI3K signaling pathways. These findings were consistent with a recent report examining genomic characterization of DAC: three of seven patients with MMR alterations also had concurrent secondary mutations in HRR pathway-related genes (35). Approximately 50% of primary PCas harbor ETS rearrangements, most commonly TMPRSS2::ERG. Interestingly, only one case in our MSI-PCas group showed TMPRSS2::ERG rearrangement (35). In a recent study of 19 dMMR-PCa cases, TMPRSS2::ERG fusions were detected in only two cases (2/19, 11%) (30). These results suggested that TMPRSS2::ERG fusions are infrequent in MSI/dMMR-PCas. TMPRSS2::ERG fusion is dependent on androgen receptor (AR) signaling and is thought to be restricted to the prostate (36). These data may suggest that AR-directed therapy is not absolutely required for MSI/dMMR-PCa patients.
Recent studies have shown a meaningful relationship of ductal histology with DDR genes alterations as well as MSI/dMMR status (18, 19, 35, 37). A relatively large sample size-based study by Schweizer et al. (35) showed that approximately 50% (25/51) of DAC patients demonstrated DDR pathway alterations. In our case 6, the DAC component accounted for about 60% of the tumor, and the DAC and AAC components were clearly separated. We separately sequenced the AAC and DAC component and the results showed that only the DAC component had somatic pathogenic mutations in MMR genes, with a higher frequency of gene alterations. This case suggested that concurrent DAC and AAC may share different gene alterations, as well as different changes in DDR genes between these components.
The MSI/dMMR status may be discordant when different assessment methods are used. In our study, eight PCas demonstrated loss of MMR-related protein expression by IHC. However, only two of the seven PCas showed MSI-H by multiplex PCR-based testing. MSI-PCR typically targets five informative microsatellite markers in the standard molecular diagnosis of MSI, although MSI-PCR has been validated for colorectal tumors and may have poor performance in other cancer types (38). In a previous study, MSI-PCR achieved only 81.8% sensitivity for PCas and 75.0% sensitivity for endometrial tumors (39). Similar observations have been made by Guedes et al. (15) and Fraune et al. (22), who found that only 61% (8/13) and 66.7% (4/6) of advanced dMMR-PCas exhibited MSI by MSI-PCR. Moreover, TILs may lead to false-negative MSI-PCR results. Two of the five discrepant PCas in our cohort evidenced MSI by NGS-based MSI testing, indicating false negative MSI-PCR results. In addition, two prostate tumors carried somatic pathogenic or likely pathogenic mutations in MMR genes by NGS, similarly indicating MSI-PCR was inaccurate. Interestingly, one PCa showed MMR-related protein loss, but no MSI by MSI-PCR or NGS. The optimal method for determining MSI/dMMR status in PCa patients remains unknown, however, our study suggests that NGS may represent a robust and efficient strategy to identify the subset of PCa patients.
Data regarding the response of dMMR PCa to standard treatment are conflicting. Some studies reported response and survival outcomes to standard therapies, which were similar to those reported in unselected patients, while other studies showed poor response to hormonal treatments in dMMR PCa patients (40). Considering the low number of patients included in these studies and the different methods used to assess MMR/MSI status, no firm conclusion can be drawn regarding the response to standard treatments of dMMR PCa. Immunotherapy-based approaches have enriched the therapeutical opportunities of many cancer types, improving patient survival; and in advanced PCa patients, there were several retrospective small series data on the treatment response of dMMR/MSI-H PCas to immunotherapy. Abida et al. (9) reported a PSA50 response rate of 54.5% in 11 patients with dMMR/MSI-H PCa with a durable response. A PSA50 response of 50% was also found by Antonarakis et al. (10) in 4 patients with MMR-mutated advanced PCa treated with anti-PD1 immunotherapy. Graham et al. (41) documented a similar PSA50 response rate of 53%. In addition, Nava Rodrigues et al. (42) have demonstrated a higher frequency of immunopositivity to antibody against to PD-L1 carboxy-terminal domain in dMMR PCas (50%) than in pMMR PCas (9.8%). Overall, these studies suggest that immunotherapy has promising prospects for treating advanced, dMMR/MSI-H PCas.
Our study had several limitations. First, it was a retrospective study exclusively from ethnic Chinese sample, and the number of patients with MSI was limited. Furthermore, it was not designed to assess the diagnostic accuracy of MSI in MMR-related proteins by IHC, which is prone to technical artifacts and may cause false-negative results when point mutations, missense mutations, or some protein-truncating mutations affect MMR genes. Finally, information on responses to anti-PD-1/PD-L1 immunotherapy was unavailable.
In conclusion, our study demonstrated that the MSI/dMMR phenotype is uncommon in PCas and that several clinicopathological features may be associated with the presence of MSI/dMMR or DDR mutations, in particular TILs. We propose that genetic testing should be considered for all PCas with TILs. NGS assays perform better than MSI-PCR for detecting MSI in PCas. Although MSI/dMMR-PCas are uncommon, they are clinically important, as this study strongly suggests that MSI/dMMR-PCas are enriched for actionable mutations. Cases with PCas with some clinicopathological features (such as intraductal/ductal histology and TILs) should be offered NGS to guide treatment.
Data availability statement
The datasets presented in this study can be found in online repositories. The name of the repository and accession number can be found below: NCBI Sequence Read Archive; PRJNA1010485.
Author contributions
HZ: Writing – original draft. XY: Writing – original draft. JX: Writing – original draft. XC: Writing – review & editing. JC: Conceptualization, Writing – review & editing. MS: Conceptualization, Writing – original draft. WD: Conceptualization, Writing – review & editing. SW: Conceptualization, Writing – review & editing. ZZ: Conceptualization, Writing – review & editing. CW: Writing – review & editing. MZ: Conceptualization, Validation, Writing – review & editing, Writing – original draft.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by Ningbo Medical Science and Technology Project (2020Y30), the Project of NINGBO Leading Medical&Health Discipline (2022-F30), Zhejiang Provincial Natural Science Foundation (LY21H160052) and Zhejiang Provincial Medicine and Health Research Foundation (2023KY040). The funder did not have any role in the design and conduct of the study, the analysis and interpretation of the data, and preparation of the manuscript.
Acknowledgments
The authors thank Nanjing Geneseeq Technology Inc., Nanjing, China, for the targeted NGS service.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Li K, Luo H, Huang L, Luo H, Zhu X. Microsatellite instability: A review of what the oncologist should know. Cancer Cell Int (2020) 20:16. doi: 10.1186/s12935-019-1091-8
2. Malapelle U, Parente P, Pepe F, De Luca C, Pisapia P, Sgariglia R, et al. Evaluation of microsatellite instability and mismatch repair status in different solid tumors: a multicenter analysis in a real world setting. Cells (2021) 10(8):1878. doi: 10.3390/cells10081878
3. Beltran H. DNA mismatch repair in prostate cancer. J Clin Oncol (2013) 31(14):1782–4. doi: 10.1200/jco.2012.48.4667
4. Latham A, Srinivasan P, Kemel Y, Shia J, Bandlamudi C, Mandelker D, et al. Microsatellite instability is associated with the presence of Lynch syndrome pan-Cancer. J Clin Oncol (2019) 37(4):286–95. doi: 10.1200/JCO.18.00283
5. Hause RJ, Pritchard CC, Shendure J, Salipante SJ. Classification and characterization of microsatellite instability across 18 cancer types. Nat Med (2016) 22(11):1342–50. doi: 10.1038/nm.4191
6. Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science (2017) 357(6349):409–13. doi: 10.1126/science.aan6733
7. Zhao P, Li L, Jiang X, Li Q. Mismatch repair deficiency/microsatellite instability-high as a predictor for anti-PD-1/PD-L1 immunotherapy efficacy. J Hematol Oncol (2019) 12(1):54. doi: 10.1186/s13045-019-0738-1
8. US Food and Drug Administration. FDA grants accelerated approval to pembrolizumab for first tissue/site agnostic indication. US Food & Drug Administration (2017). Available at: https://www.fda.gov/drugs/resources-information-approveddrugs/fda-grants-accelerated-approval-pembrolizumab-first-tissuesite-agnosticindication.
9. Abida W, Cheng ML, Armenia J, Middha S, Autio KA, Vargas HA, et al. Analysis of the prevalence of microsatellite instability in prostate cancer and response to immune checkpoint blockade. JAMA Oncol (2019) 5(4):471–8. doi: 10.1001/jamaoncol.2018.5801
10. Antonarakis ES, Shaukat F, Isaacsson Velho P, Kaur H, Shenderov E, Pardoll DM, et al. Clinical features and therapeutic outcomes in men with advanced prostate cancer and DNA mismatch repair gene mutations. Eur Urol (2019) 75(3):378–82. doi: 10.1016/j.eururo.2018.10.009
11. Cancer Genome Atlas Network, Kandoth C, Schultz N, Cherniack AD, Akbani R, Liu Y, et al. Integrated genomic characterization of endometrial carcinoma. Nature (2013) 497(7447):67–73. doi: 10.1038/nature12113
12. Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature (2012) 487(7407):330–7. doi: 10.1038/nature11252
13. Lynch HT, Snyder CL, Shaw TG, Heinen CD, Hitchins MP. Milestones of lynch syndrome: 1895–2015. Nat Rev Cancer (2015) 15(3):181–94. doi: 10.1038/nrc3878
14. Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, et al. Integrative clinical genomics of advanced prostate cancer. Cell (2015) 161(5):1215–28. doi: 10.1016/j.cell.2015.05.001
15. Guedes LB, Antonarakis ES, Schweizer MT, Mirkheshti N, Almutairi F, Park JC, et al. MSH2 loss in primary prostate cancer. Clin Cancer Res (2017) 23(22):6863–74. doi: 10.1158/1078-0432.CCR-17-0955
16. Pritchard CC, Morrissey C, Kumar A, Zhang X, Smith C, Coleman I, et al. Complex MSH2 and MSH6 mutations in hypermutated microsatellite unstable advanced prostate cancer. Nat Commun (2014) 5:4988. doi: 10.1038/ncomms5988
17. Shenderov E, Isaacsson Velho P, Awan AH, Wang H, Mirkheshti N, Lotan TL, et al. Genomic and clinical characterization of pulmonary-only metastatic prostate cancer: a unique molecular subtype. Prostate (2019) 79(13):1572–9. doi: 10.1002/pros.23881
18. Schweizer MT, Cheng HH, Tretiakova MS, Vakar-Lopez F, Klemfuss N, Konnick EQ, et al. Mismatch repair deficiency may be common in ductal adenocarcinoma of the prostate. Oncotarget (2016) 7(50):82504–10. doi: 10.18632/oncotarget.12697
19. Isaacsson Velho P, Silberstein JL, Markowski MC, Luo J, Lotan TL, Isaacs WB, et al. Intraductal/ductal histology and lymphovascular invasion are associated with germline DNA-repair gene mutations in prostate cancer. Prostate (2018) 78:401–7. doi: 10.1002/pros.23484
20. Zhang X, Liu F, Bao H, Wang A, Han M, Wu X, et al. Distinct genomic profile in h. pylori-associated gastric cancer. Cancer Med (2021) 10(7):2461–9. doi: 10.1002/cam4.3765
21. Fang W, Ma Y, Yin JC, Hong S, Zhou H, Wang A, et al. Comprehensive genomic profiling identifies novel genetic predictors of response to anti-PD-(L)1 therapies in non-small cell lung cancer. Clin Cancer Res (2019) 25(16):5015–26. doi: 10.1158/1078-0432.CCR-19-0585
22. Fraune C, Simon R, Höflmayer D, Möller K, Dum D, Büscheck F, et al. High homogeneity of mismatch repair deficiency in advanced prostate cancer. Virchows Arch (2020) 476(5):745–52. doi: 10.1007/s00428-019-02701-x
23. Kagawa M, Kawakami S, Yamamoto A, Suzuki O, Eguchi H, Okazaki Y, et al. Prevalence and clinicopathological/molecular characteristics of mismatch repair protein-deficient tumours among surgically treated patients with prostate cancer in a Japanese hospital-based population. Jpn J Clin Oncol (2021) 51(4):639–45. doi: 10.1093/jjco/hyaa207
24. Chen Y, Wang J, Fraig MM, Henderson K, Bissada NK, Watson DK, et al. Alterations in PMS2, MSH2 and MLH1 expression in human prostate cancer. Int J Oncol (2003) 22(5):1033–43. doi: 10.3892/ijo.22.5.1033
25. Chen Y, Wang J, Fraig MM, Metcalf J, Turner WR, Bissada NK, et al. Defects of DNA mismatch repair in human prostate cancer. Cancer Res (2001) 61(10):4112–21.
26. Albero-González R, Hernández-Llodrà S, Juanpere N, Lorenzo M, Lloret A, Segalés L, et al. Immunohistochemical expression of mismatch repair proteins(MSH2, MSH6, MLH1, and PMS2) in prostate cancer: correlation with grade groups (WHO 2016) and ERG and PTEN status. Virchows Arch (2019) 475(2):223–31. doi: 10.1007/s00428-019-02591-z
27. Burger M, Denzinger S, Hammerschmied CG, Tannapfel A, Obermann EC, Wieland WF, et al. Elevated microsatellite alterations at selected tetranucleotides (EMAST) and mismatch repair gene expression in prostate cancer. J Mol Med (Berl) (2006) 84(10):833–41. doi: 10.1007/s00109-006-0074-0
28. Wilczak W, Rashed S, Hube-Magg C, Kluth M, Simon R, Büscheck F, et al. Up-regulation of mismatch repair genes MSH6, PMS2 and MLH1 parallels development of genetic instability and is linked to tumor aggressiveness and early PSA recurrence in prostate cancer. Carcinogenesis (2017) 38(1):19–27. doi: 10.1093/carcin/bgw116
29. Mills AM, Liou S, Ford JM, Berek JS, Pai RK, Longacre TA. Lynch syndrome screening should be considered for all patients with newly diagnosed endometrial cancer. Am J Surg Pathol (2014) 38(11):1501–9. doi: 10.1097/PAS.0000000000000321
30. Wyvekens N, Tsai HK, Sholl LM, Tucci J, Giannico GA, Gordetsky JB, et al. Histopathological and genetic features of mismatch repairdeficient high-grade prostate cancer. Histopathology (2022) 80(7):1050–60. doi: 10.1111/his.14645
31. Wright CL, Stewart ID. Histopathology and mismatch repair status of 458 consecutive colorectal carcinomas. Am J Surg Pathol (2003) 27(11):1393–406. doi: 10.1097/00000478-200311000-00001
32. Schneider B, Glass Ä, Jagdmann S, Hühns M, Claus J, Zettl H, et al. Loss of mismatch repair protein expression and microsatellite instability in upper tract urothelial carcinoma and clinicopathologic implications. Clin Genitourin Cancer (2020) 18(5):e563–72. doi: 10.1016/j.clgc.2020.03.006
33. Lotan TL, Kaur HB, Alharbi AM, Pritchard CC, Epstein JI. DNA damage repair alterations are frequent in prostatic adenocarcinomas with focal pleomorphic giant-cell feature. Histopathology (2019) 74(6):836–43. doi: 10.1111/his.13806
34. Goodman AM, Kato S, Bazhenova L, Patel SP, Frampton GM, Miller V, et al. Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Mol Cancer Ther (2017) 16(11):2598–608. doi: 10.1158/1535-7163.MCT-17-0386
35. Schweizer MT, Antonarakis ES, Bismar TA, Guedes LB, Cheng HH, Tretiakova MS, et al. Genomic characterization of prostatic ductal adenocarcinoma identifies a high prevalence of DNA repair gene mutations. JCO Precis Oncol (2019), 00327. doi: 10.1200/PO.18.00327
36. Mani RS, Tomlins SA, Callahan K, Ghosh A, Nyati MK, Varambally S, et al. Induced chromosomal proximity and gene fusions in prostate cancer. Science (2009) 326(5957):1230. doi: 10.1126/science.1178124
37. Seipel AH, Whitington T, Delahunt B, Samaratunga H, Mayrhofer M, Wiklund P, et al. Genetic profile of ductal adenocarcinoma of the prostate. Hum Pathol (2017) 69:1–7. doi: 10.1016/j.humpath.2017.04.015
38. de la Chapelle A, Hampel H. Clinical relevance of microsatellite instability in colorectal cancer. J Clin Oncol (2010) 28(20):3380–7. doi: 10.1200/JCO.2009.27.0652
39. Waalkes A, Smith N, Penewit K, Hempelmann J, Konnick EQ, Hause RJ, et al. Accurate pan-cancer molecular diagnosis of microsatellite instability by single molecule molecular inversion probe capture and high throughput sequencing. Clin Chem (2018) 64(6):950–8. doi: 10.1373/clinchem.2017.285981
40. Rebuzzi SE, Rescigno P, Catalano F, Mollica V, Vogl UM, Marandino L, et al. Immune checkpoint inhibitors in advanced prostate cancer: current data and future perspectives. Cancers (Basel) (2022) 14(5):1245. doi: 10.3390/cancers14051245
41. Graham LS, Montgomery B, Cheng HH, Yu EY, Nelson PS, Pritchard C, et al. Mismatch repair deficiency in metastatic prostate cancer: Response to PD-1 blockade and standard therapies. PloS One (2020) 15(5):e0233260. doi: 10.1371/journal.pone.0233260
Keywords: DNA repair, DNA mismatch repair, high throughput nucleotide sequencing, microsatellite instability, prostatic neoplasms
Citation: Zhang H, Yang X, Xie J, Cheng X, Chen J, Shen M, Ding W, Wang S, Zhang Z, Wang C and Zhao M (2023) Clinicopathological and molecular analysis of microsatellite instability in prostate cancer: a multi-institutional study in China. Front. Oncol. 13:1277233. doi: 10.3389/fonc.2023.1277233
Received: 14 August 2023; Accepted: 26 September 2023;
Published: 13 October 2023.
Edited by:
Goutam Chakraborty, Icahn School of Medicine at Mount Sinai, United StatesReviewed by:
Michele Moschetta, Novartis Institutes for BioMedical Research, SwitzerlandFabio Catalano, Department of Oncology and Hematology, San Martino Hospital (IRCCS), Italy
Copyright © 2023 Zhang, Yang, Xie, Cheng, Chen, Shen, Ding, Wang, Zhang, Wang and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ming Zhao, emhhb21pbmdwYXRob2xAMTYzLmNvbQ==; Chaofu Wang, d2FuZ2NoYW9mdUAxMjYuY29t
†These authors have contributed equally to this work
 Huizhi Zhang
Huizhi Zhang Xiaoqun Yang
Xiaoqun Yang Jialing Xie2†
Jialing Xie2† Xiao Cheng
Xiao Cheng Miaomiao Shen
Miaomiao Shen Chaofu Wang
Chaofu Wang Ming Zhao
Ming Zhao