- 1Department of Spleen and Stomach, Guangzhou University of Chinese Medicine Shenzhen Hospital (Futian), Shenzhen, China
- 2The Sixth Clinical Medical College, Guangzhou University of Chinese Medicine, Shenzhen, China
Purpose: Gastric cancer, characterised by a significant mortality rate, is one of the most prevalent malignant neoplasms globally. Exosomal non-coding RNAs play a key role in gastric carcinogenesis, metastasis and treatment resistance by regulating gene expression, remodelling the tumour microenvironment and mediating drug resistance. In the identification of early gastric cancer, these exosomal non-coding RNA molecules possess significant potential to be developed into biomarkers that do not entail invasive procedures.
Methods: Our study undertook a comprehensive and profound literature examination in core databases like PubMed, Web of Science, ScienceDirect, Embase, Scopus, and Medline, with the aim of precisely evaluating the potential effectiveness of exosomal miRNAs, lncRNAs, and circRNAs in the diagnosis of gastric cancer.
Results: A sum of 52 studies were incorporated, comprising 164 studies. These studies identified a total of 59 miRNAs, 17 lncRNAs, and 16 circRNAs. For miRNAs, sensitivity was estimated at 0.72 (95% CI, 0.69 - 0.76) and specificity at 0.80 (95% CI, 0.77 - 0.83). For lncRNAs, the sensitivity was 0.86 (95% CI, 0.84 - 0.87) and the specificity was 0.82 (95% CI, 0.80 - 0.83). In the case of circRNAs, the sensitivity and specificity were 0.71 (95% CI, 0.63 - 0.78) and 0.88 (95% CI, 0.81 - 0.93) respectively. The AUC for miRNAs was calculated as 0.83. As for lncRNAs, its AUC was established to be 0.89. Regarding circRNAs, the determined AUC was 0.86.
Conclusion: These results confirm the efficacy of exosomal ncRNAs as powerful biomarkers for the early diagnosis of gastric carcinomas, thereby laying a strong foundation for the advancement of novel diagnostic approaches.
1 Introduction
Gastric cancer (GC) ranks among the most prevalent malignant neoplasms globally. As per the latest data released by the International Agency for Research on Cancer (1, 2), the incidence of GC is the fifth highest among all malignant tumors, while its mortality rate is the fourth highest. The timely identification and management of gastric cancer are of paramount importance. However, due to the subtle nature of its early manifestations, a considerable percentage of patients get diagnosed during the intermediate or advanced stages. In cases where a patient is identified with GC at an advanced stage, the 5-year survival rate is notably low, typically below 10%. Conversely, when GC is detected in its early stages, the 5-year survival rate can be significantly higher, reaching up to 85% (3). The discovery of novel biomarkers holds substantial importance for the early-stage diagnosis and curative treatment of GC (4, 5). The diagnosis of GC routinely relies on gastroscopy, pathological evaluation, and imaging techniques (6), but these methods have certain limitations. For example, gastroscopy is an invasive procedure that involves obtaining mucosal tissue biopsies for pathological examination, which is not well accepted by patients (7), while imaging tests such as X-rays and CT scans have a low detection rate for early gastric cancer (8). Consequently, investigators have sought non-invasive or minimally invasive biomarkers to enhance the precision of GC detection.
In recent years, exosomes have attracted widespread attention as a new type of biomarker. Exosomes are double-layered lipid vesicles secreted by cells, serve as carriers for various molecular signals, such as nucleic acids and proteins, and play a crucial role in mediating intercellular communication (9). Studies (5, 10) have demonstrated that non-coding RNAs (ncRNAs) within exosomes, such as microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs), which exist in exosomes, play a crucial role in the onset and development of GC. These ncRNAs are consistently detected in bodily fluids such as blood and gastric juice, suggesting their prospective application as indicative markers for the diagnosis of GC. For instance, certain investigations have demonstrated that serum exosomal biomarkers, including miR-1246 and lncRNA-GCI, are capable of accurately differentiating cancer patients from healthy individuals (11–13). The abundance of these exosomal ncRNAs exhibits notable variations between GC sufferers and healthy people, indicating their possible value as markers for the identification of GC (5).
While the diagnostic potential of exosomal ncRNAs in gastric cancer has shown promise, current evidence remains fragmented and inconsistent across studies, highlighting the need for a comprehensive evaluation. This meta-analysis systematically assesses the diagnostic accuracy of exosomal ncRNAs—such as miRNAs, lncRNAs, and circRNAs—as biomarkers for gastric cancer, synthesizing existing data to quantify their pooled sensitivity, specificity, and clinical utility. By consolidating these findings, we aim to establish a robust evidence base that can guide the integration of these biomarkers into early diagnostic protocols, ultimately improving patient outcomes through more reliable and timely detection.
2 Research materials and methodologies
2.1 Statement of ethics considerations
The herein research work was executed and recorded in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses of Diagnostic Test Accuracy protocols (14), and the corresponding checklist is available in the Supplementary Material. The study has been registered in the PROSPERO database under the identification number CRD42024587170.
2.2 Search strategy
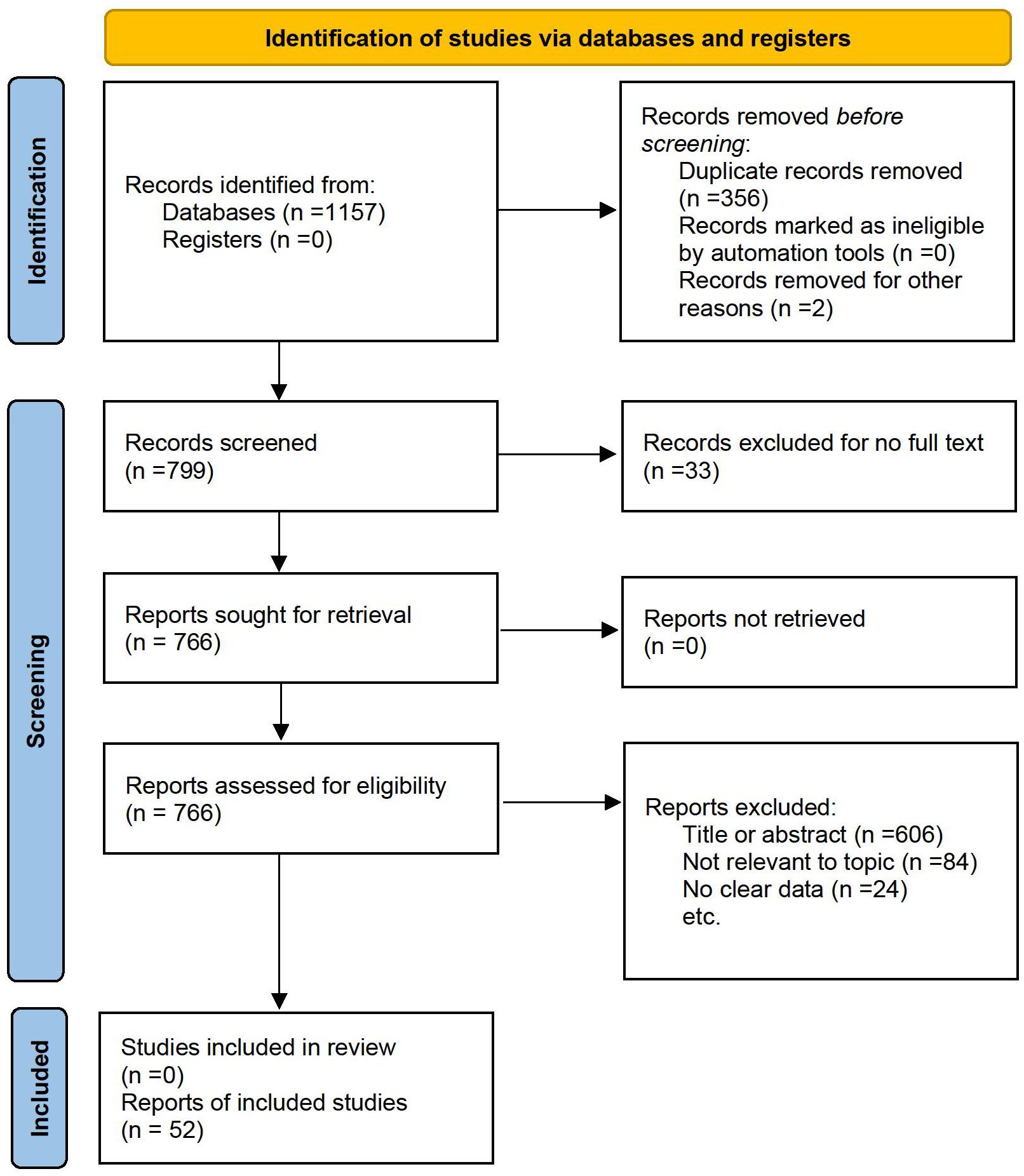
We began by identifying relevant search terms, including MeSH terms and keywords related to gastric tumors and exosomal non-coding RNAs. We conducted a systematic search across multiple databases including Embase, MEDLINE, PubMed, ScienceDirect, Scopus, and Web of Science, covering all available records through August 29, 2024 (see Supplementary Table S1). Following article collection, we excluded duplicates and studies lacking full-text access. We screened publications by title/abstract against predefined criteria, then progressed eligible studies to full-text review for final selection. This process identified the studies included in our final analysis.
2.3 Criteria for inclusion and exclusion parameters
Two assessors separately appraised the qualified articles, and any discrepancies were settled via comprehensive deliberation with the participation of a third adjudicator. Inclusion criteria stipulated that the studies: (1) investigated the role of exosomal ncRNAs for the diagnosis of GC with the application of blood samples; (2) confirmed the diagnosis of GC patients through histopathological examination; and (3) provided adequate data to establish a 2 × 2 contingency table, incorporating true positives (TP), false positives (FP), true negatives (TN), and false negatives (FN). The criteria for exclusion encompassed: (1) research works that failed to focus on the diagnostic efficacy of exosomal ncRNAs regarding GC; (2) research works that were of poor quality, lacking sufficient data, or exhibiting duplication; (3) meta-studies, conference abstracts, review articles, case reports, or seminar papers; (4) articles from which the four-cell table could not be derived; and (5) studies involving co-diagnosis in conjunction with other neoplasm indicators.
2.4 Data retrieval and quality evaluation
Investigators independently retrieved the following data from all qualified studies using standardized forms: (1) fundamental study attributes, encompassing first author, publication year, study country, journal name, ethnic background, sample quantity, cancer classification, tumor stage level, tumor grade degree, average age, gender ratio, sample category, ncRNA examination, reference gene element, detection technique, as well as (2) diagnostic performance metrics, such as TP, FP, FN, TN, sensitivity (Sen), specificity (Spe), and area under the receiver operating characteristic (ROC) curve (AUC). Data not directly accessible were gathered using GetData and Origin software. Each included study was systematically assessed and independently scored by researchers based on the updated QUADAS-2 framework (15). QUADAS-2 comprises four domains: case selection, test evaluation, gold standard, and case progression. The assessment primarily focused on bias risk and clinical relevance. Divergences were ironed out through consultations with a third-party evaluator to attain unanimity.
In this meta-analysis, a “study” was defined as an independent dataset with non-overlapping samples to avoid unit-of-analysis bias. If a single article reported multiple datasets (e.g., a discovery cohort and a separate validation cohort), each dataset was treated as an independent study. This approach ensured that the analysis was comprehensive and unbiased, providing a robust evaluation of the diagnostic performance of the studied exosomal ncRNAs in the context of cancer diagnosis.
2.5 Statistical analysis
A threshold effect analysis, employing Spearman’s correlation of ranks coefficient, was executed to explore the association between the logarithm of sensitivity and that of 1-specificity within the included studies. A non-significant correlation (p > 0.05) suggests the absence of a threshold effect contributing to heterogeneity. Heterogeneity due to non-threshold factors was assessed with the application of Cochran’s Q examination and the I² evaluative index. Significant heterogeneity was indicated by p ≤ 0.1 and I² ≥ 50%, while p > 0.1 and I² < 50% suggested no significant heterogeneity. The diagnostic performance evaluation indicators covered sensitivity, specificity, positive diagnostic likelihood ratio (DLR+), negative diagnostic likelihood ratio (DLR-), AUC, and diagnostic odds ratio (DOR).
we evaluated the diagnostic performance of exosomal ncRNAs for GC using a comprehensive set of metrics, including TP, FP, FN, and TN. These values were used to calculate Sensitivity (TP/(TP + FN)), Specificity (TN/(FP + TN)), and the AUC to assess overall diagnostic accuracy. Additionally, DLR+ and DLR− were calculated to quantify the impact of positive and negative test results on disease probability, with DLR+ = Sensitivity/(1 − Specificity) and DLR− = (1 − Sensitivity)/Specificity. DOR was also determined using the formula DOR = (TP/FN)/(FP/TN) to provide a single measure of diagnostic efficacy. These metrics collectively highlight the robustness of exosomal ncRNAs as potential biomarkers for GC diagnosis, emphasizing their ability to accurately distinguish between patients with and without gastric cancer.
For the intention of probing into the sources of heterogeneity, both meta-regression and subgroup analyses were executed. Forest plots were generated for each evaluation metric, the assessment of publication bias was appraised by means of Deek’s funnel diagram and the evaluation of clinical effectiveness was evaluated through Fagan diagrams and likelihood scatter diagrams. Meta-Disc 1.4 and Stata 17.0 software were utilized to conduct the analysis.
3 Results
3.1 Literature screening results
The process of identifying suitable studies is illustrated in Figure 1. An exhaustive literature search was performed across six databases, resulting in a total of 1157 articles. Following the removal of duplicate entries and articles for which full texts were unavailable, 766 articles were subjected to further review. Ultimately, after evaluating the titles, abstracts, and full contents of these studies, 52 articles fulfilled the eligibility requirements and were incorporated into the meta-analysis. Thus, a sum of 52 eligible articles were brought into the meta-analysis.
3.2 Basic details of the incorporated literatures
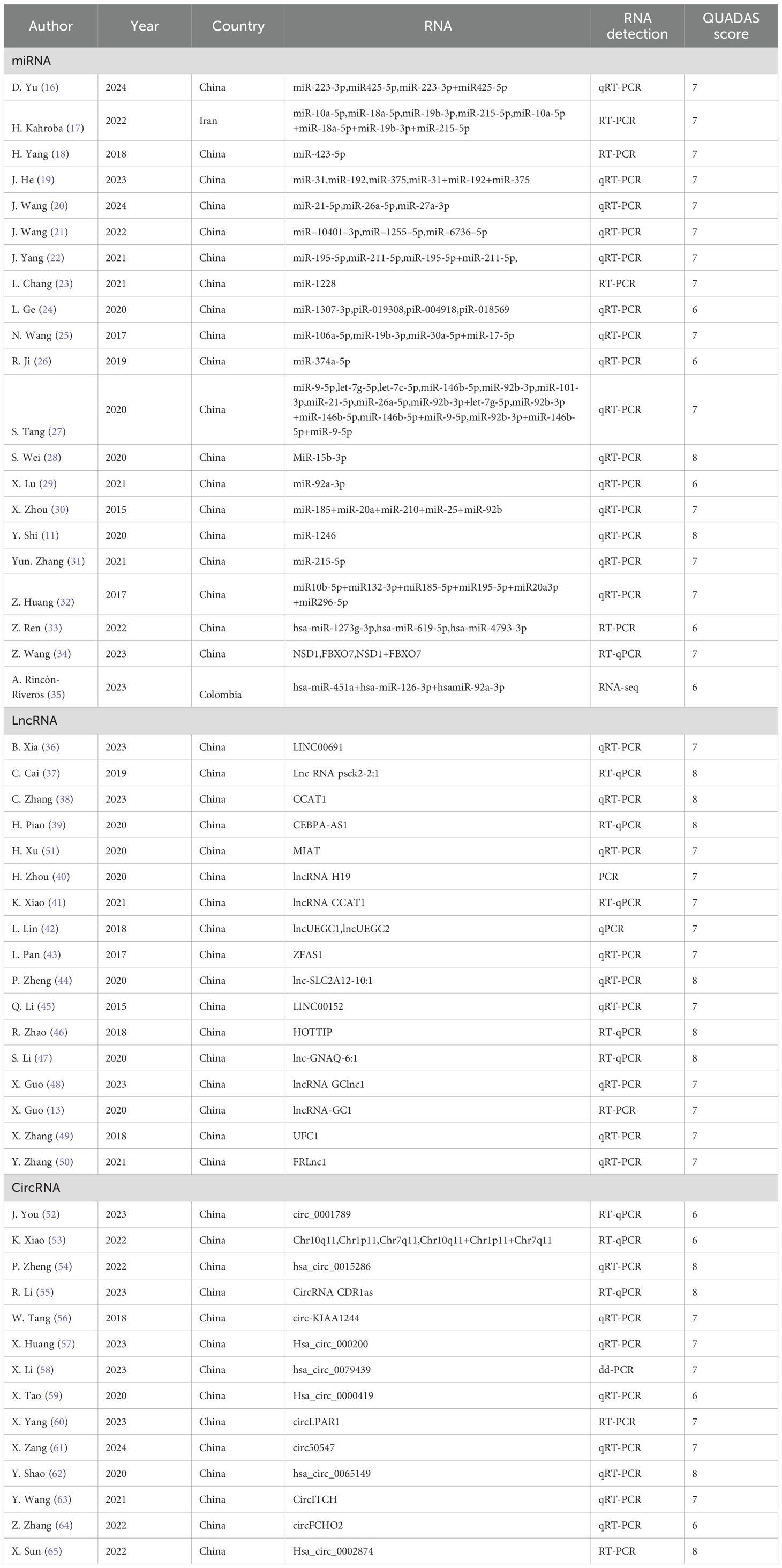
A total of 52 articles encompassing 164 studies were incorporated into the analysis. The included studies were conducted mainly in China, while the remaining two studies were conducted in Iran and Colombia, respectively. Specifically, 21 articles focusing on miRNAs (11, 16–35) comprised 84 studies, 17 articles on lncRNAs (13, 36–51) (37–53) included 60 studies, and 14 articles on circRNAs (52–65) involved 20 studies. The diagnostic performance metrics reported consisted of TP, FP, FN, TN, Sen, Spe, AUC, DLR+, DLR-, and DOR. Furthermore, with the aim of ensuring that the ncRNAs were sourced from exosomes, the exosomes were characterized using nanoparticle tracking assay (NTA), transmission electron microscopy (TEM), and detection of exosome-marker proteins. An overview of exosome-related information across the studies is presented in Supplementary Table S2. The vast majority of articles gave direct access to specific data, while the specificity and sensitivity of the remaining 17 articles were derived by analyzing the ROC curves using digital software such as GetData (11, 19–23, 25, 26, 32, 35, 38, 49, 51, 52, 58, 60, 64). The findings are detailed in Table 1; Supplementary Table S3.
3.3 Evaluation of research quality
The results obtained from the QUADAS-2 quality assessment of the 52 incorporated articles are demonstrated in Supplementary Figure S1, with the QUADAS scores being shown in Table 1. The results of each study were assessed as “yes”, “no” or “unclear”. Specifically, “yes” was assigned 1 point, “no” was assigned 1 point and “unclear” was assigned 0 points. Studies with a score exceeding 4 were defined by us as high-quality studies, while those with a score below 4 were considered as low-quality studies. The overall quality of the included literature was satisfactory, and all of them were of high quality.
3.4 Heterogeneity analysis
Threshold effects represent fundamental sources of heterogeneity in diagnostic tests. Therefore, for diagnostic meta-analyses, the presence of threshold effects is initially evaluated. The Spearman correlation coefficients, calculated between the natural logarithms of sensitivity and specificity using Meta DiSc 14.0 software, were 0.426, 0.172, and 0.722 for exosomal miRNAs, lncRNAs, and circRNAs studies, respectively. Corresponding p-values were p < 0.001, p = 0.188 (p > 0.05), and p < 0.001, respectively. These findings indicate that threshold effects contribute to heterogeneity in exosomal miRNAs and circRNAs studies but not in lncRNAs studies. Heterogeneity across the included studies was further assessed using the Cochran’s Q test and the I² statistic in Stata software. For miRNAs, lncRNAs, and circRNAs, the results were: Cochran’s Q = 241.07 (p < 0.001), I² = 65.6% (95% CI, 49.8–74.9); Cochran’s Q = 425.58 (p < 0.001), I² = 86.1% (95% CI, 77.1–90.7); and Cochran’s Q = 62.85 (p < 0.001), I² = 69.8% (95% CI, 22.3–84.0), respectively. These results demonstrate significant heterogeneity among studies, attributable to factors other than threshold effects.
3.5 Diagnostic accuracy evaluation of exosomal ncRNAs
The investigators employed a random effects model to assess the diagnostic impact of exosomal miRNAs, lncRNAs, and circRNAs in GC. The overall sensitivity and specificity derived from the lncRNAs studies were 0.86 (95% CI, 0.84–0.87) and 0.82 (95% CI, 0.80–0.83), respectively. The aggregated diagnostic score and DOR were calculated as 3.27 (95% CI, 3.12 to 3.42) and 26.35 (95% CI, 22.76 to 30.51), respectively. A forest plot illustrating these findings is presented in Figure 2 from A to C. The included studies exhibited substantial heterogeneity in terms of sensitivity (p ≤ 0.001, I2 = 86.21%), specificity (p ≤ 0.001, I2 = 54.47%), DLR+ (p ≤ 0.001, I2 = 35.51%), DLR- (p ≤ 0.001, I2 = 87.42%), diagnostic score (p ≤ 0.001, I2 = 50.51%), and DOR (p ≤ 0.001, I2 = 100%). A summary receiver operating characteristic (SROC) curve was constructed, as depicted in Figure 2D, yielding an overall AUC of 0.89 (95% CI, 0.86–0.92). Furthermore, SROC curves for exosomal miRNAs and circRNAs were generated using a random effects model to evaluate their diagnostic potential in GC. The respective AUC were 0.83 (95% CI, 0.79–0.86) and 0.87 (95% CI, 0.84–0.90), as shown in Figure 3 from A to D and Figure 4 from A to D. By comparing high-frequency exosomal ncRNAs, lncRNA was found to be the most consistent biomarker, as shown in Supplementary Table S4, which suggests that among these ncRNAs, lncRNA has the most consistent diagnostic value in gastric cancer diagnosis.
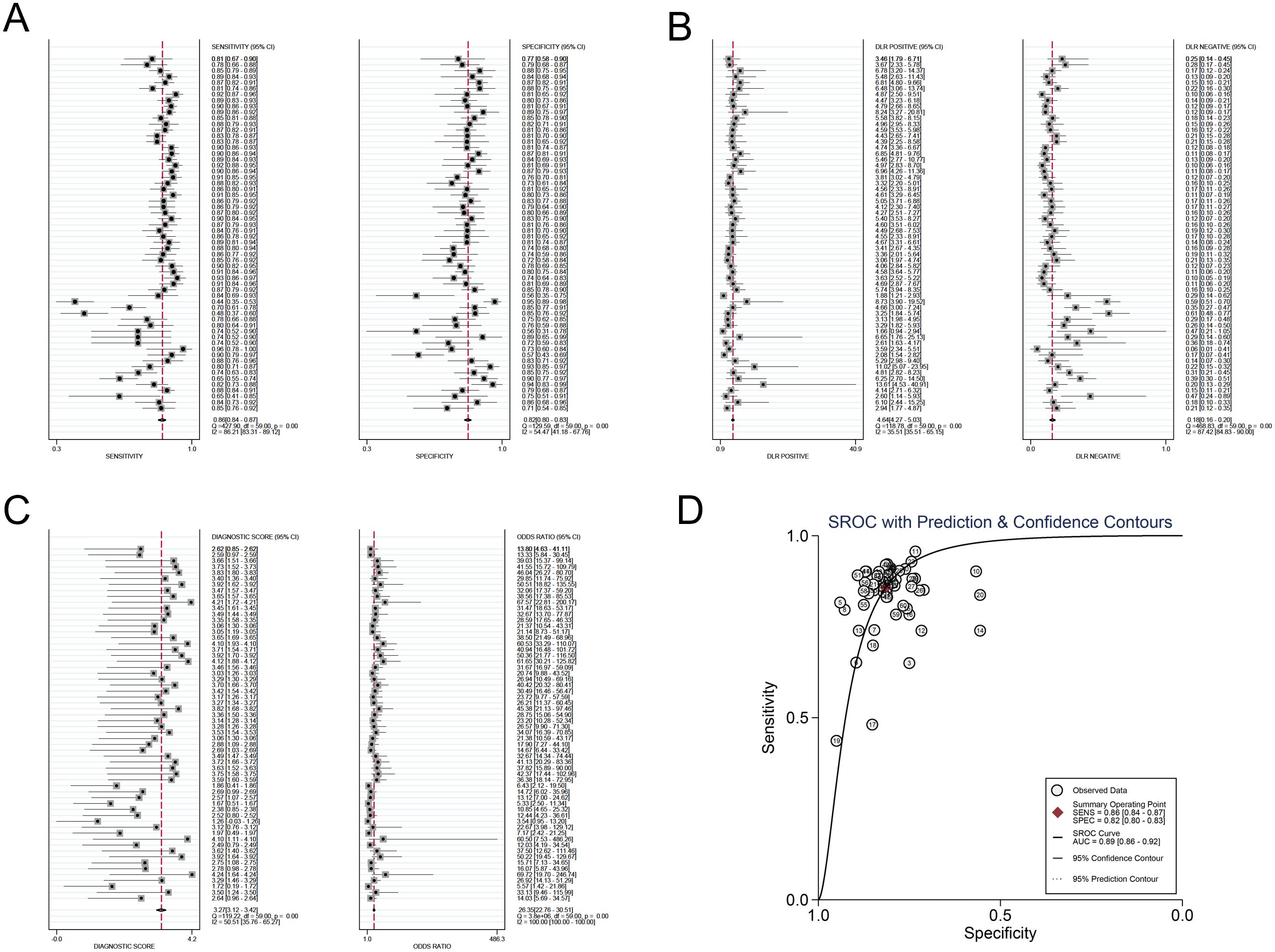
Figure 2. Diagnostic efficacy of exosomal lncRNA in gastric cancer patients. (A) Sensitivity and specificity. (B) Diagnostic likelihood ratios. (C) Diagnostic score and odds ratio. (D) SROC curve.
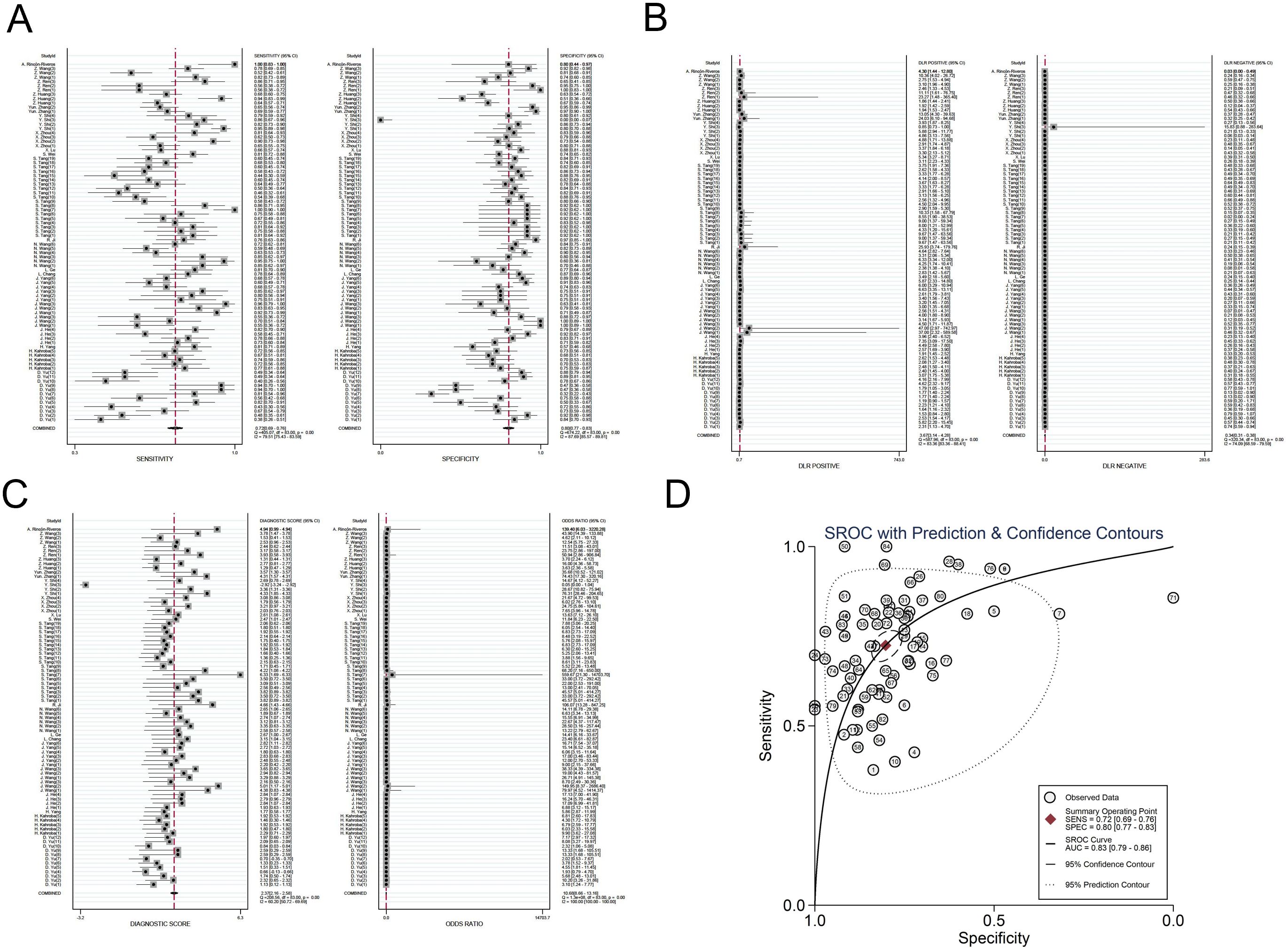
Figure 3. Diagnostic efficacy of exosomal miRNA in gastric cancer patients. (A) Sensitivity and specificity. (B) Diagnostic likelihood ratios. (C) Diagnostic score and odds ratio. (D) SROC curve.
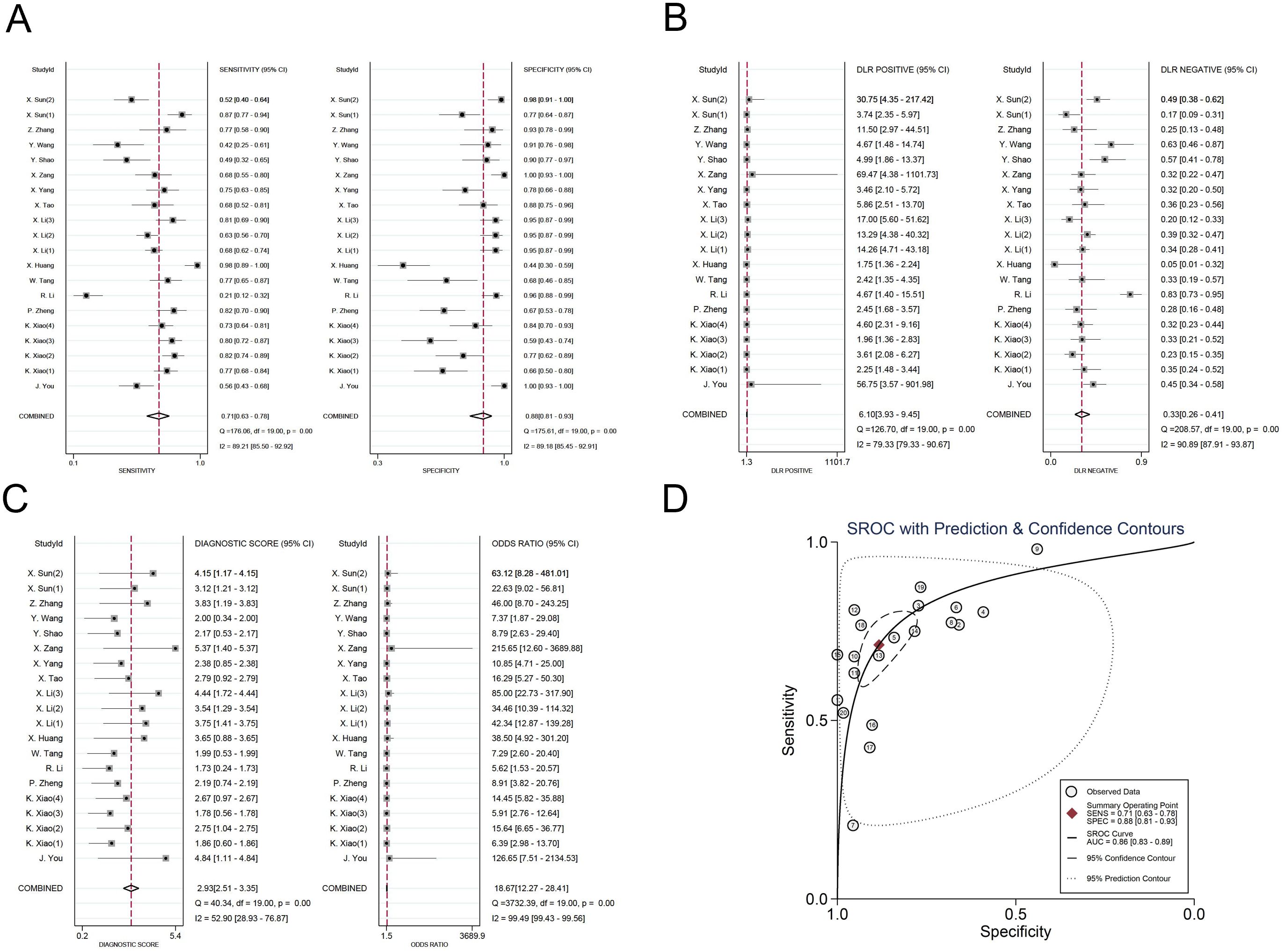
Figure 4. Diagnostic efficacy of exosomal circRNA in gastric cancer patients. (A) Sensitivity and specificity. (B) Diagnostic likelihood ratios. (C) Diagnostic score and odds ratio. (D) SROC curve.
3.6 Meta-analysis and subgroup analysis
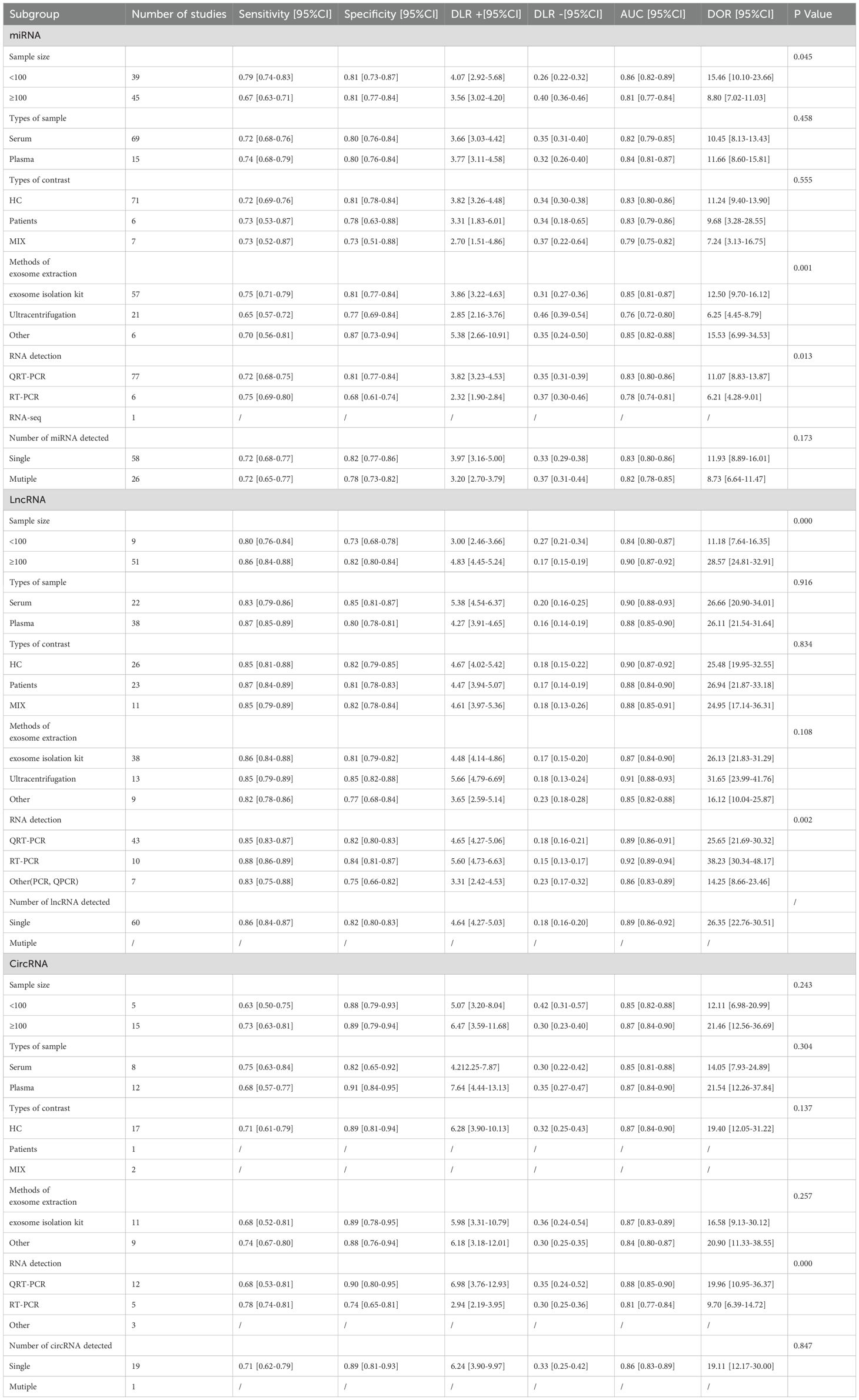
Given the observed heterogeneity, we conducted meta-regression and subgroup analyses to evaluate miRNAs, lncRNAs, and circRNAs. These analyses were performed to examine the influence of control source, exosome extraction method, and RNA detection method on miRNAs, lncRNAs, and circRNAs, considering factors such as sample size, sample type, control source, exosome extraction method, and RNA detection. The findings are summarized in Table 2. The meta-regression results indicated significant heterogeneity among studies investigating exosomal miRNAs, lncRNAs, and circRNAs. Specifically, the heterogeneity in exosomal miRNAs for diagnosing GC was influenced by sample size (p = 0.045), exosome extraction method (p = 0.001), and RNA detection method (p = 0.013). In contrast, the type of control (p = 0.555), sample type (p = 0.458), and the number of miRNAs detections (p = 0.173) did not significantly affect the overall outcomes. Subgroup analysis revealed that exosomal miRNAs derived from plasma exhibited higher sensitivity compared to those from serum. Additionally, exosomes isolated using exosome extraction kits demonstrated superior diagnostic performance relative to those obtained via ultracentrifugation. Furthermore, miRNAs detected by reverse transcription-polymerase chain reaction (RT-PCR) showed better diagnostic performance than those identified using quantitative real-time polymerase chain reaction (QRT-PCR).
For lncRNAs, subgroup analysis was conducted to evaluate the impact of various variables. The heterogeneity in exosomal lncRNAs for GC diagnosis was associated with sample size (p < 0.001) and lncRNA detection method (p = 0.002). However, sample type (p = 0.916), control source (p = 0.834), and exosome extraction method (p = 0.108) did not significantly influence the diagnostic outcomes.
Regarding circRNAs, the inter-study heterogeneity in gastric cancer diagnosis was not related to sample size (p = 0.243), sample type (p = 0.304), control type (p = 0.137), exosome extraction method (p = 0.257), or the number of circRNAs tests (p = 0.847). Instead, the heterogeneity was primarily attributed to the RNA detection method (p < 0.001). Subgroup analysis demonstrated that samples with a size exceeding 100 exhibited enhanced diagnostic sensitivity and specificity. Moreover, circRNAs detected by QRT-PCR showed improved diagnostic specificity, while those identified by RT-PCR exhibited superior diagnostic sensitivity.
3.7 Publication bias
Publication bias was assessed across the included studies through the application of the Deek funnel plot asymmetry test, as illustrated in Supplementary Figure S2. The analysis revealed no evidence of potential publication bias for circRNAs (p = 0.46). However, miRNAs (p = 0.04) and lncRNAs (p < 0.001) demonstrated statistically significant results, suggesting the presence of potential publication bias in these categories.
3.8 Clinical significance
To investigate the clinical relevance of miRNAs, lncRNAs, and circRNAs in the diagnosis of GC, we developed a Fagan diagram to illustrate the relationships among pre-test probability, likelihood ratios, and post-test probability. As depicted in Supplementary Figure S3, for exosomal miRNAs, with an assumed pre-test probability of 50%, the post-test probability reaches 79% based on the DLR+, suggesting that exosomal miRNAs exhibit strong clinical utility in diagnosing GC. Similarly, for exosomal lncRNAs, a pre-test probability of 50% yields a post-test probability of 82%, indicating that exosomal lncRNAs also demonstrate robust diagnostic potential for GC. For exosomal circRNAs, a pre-test probability of 50% results in a post-test probability of 85%, highlighting the significant diagnostic value of exosomal circRNAs in GC.
Additionally, a likelihood ratio scatter plot analysis was conducted. The scatter plot is segmented into four quadrants: the upper left quadrant (LUQ), upper right quadrant (RUQ), lower left quadrant (LLQ), and lower right quadrant (RLQ). In the LUQ, where DLR+ exceeds 10 and DLR- is below 0.1, the test is capable of both confirming and excluding gastric cancer. In the RUQ, with DLR+ greater than 10 and DLR- above 0.1, the test can only confirm gastric cancer. In the LLQ, where DLR+ is less than 10 and DLR- is below 0.1, the test is limited to excluding gastric cancer. In the RLQ, with DLR+ less than 10 and DLR- greater than 0.1, the test fails to either confirm or exclude gastric cancer.
As illustrated in Supplementary Figure S4, the diagnostic performance of exosomal miRNAs, lncRNAs, and circRNAs in both diagnosing and excluding GC is relatively similar. The likelihood ratio plot reveals that the summary point of the positive likelihood ratio (PLR) and negative likelihood ratio (NLR) falls within the lower right quadrant, indicating that exosomal miRNAs, lncRNAs, and circRNAs are not suitable as standalone diagnostic tools for either confirming or ruling out GC.
4 Disscussion
GC remains a significant global health challenge with a 5-year survival rate of only 20-30%, particularly for patients diagnosed at advanced stages (66). The poor prognosis and high mortality of GC are largely attributed to the lack of highly sensitive, specific and efficient early diagnostic strategies (7). Recent advances in liquid biopsy based on non-coding RNAs (ncRNAs), including miRNAs, lncRNAs and circRNAs, have shown great promise in addressing this issue (67). Exosomal ncRNAs, such as miR-1246, lncRNA GClnc1 and circ50547, have demonstrated high diagnostic accuracy and prognostic potential in GC, reflecting tumour heterogeneity and enabling non-invasive detection by blood tests (11, 13, 61). China has emerged as a leading contributor in exosomal ncRNA research for GC diagnosis, driven by its high epidemiological burden of GC and extensive clinical resources (68). In summarising the meta-analysis studies, we found that Chinese researchers have made full use of the rich clinical sample resources, combined with advanced technologies such as liquid biopsy and high-throughput sequencing, to publish a large number of high-quality research results. However, significant contributions have also come from other regions, including Japan, USA and Western countries, which have enriched our understanding of the molecular mechanisms and clinical applications of exosomal ncRNAs (69, 70). Exosomes are membrane-bound vesicles released following the fusion of multivesicular bodies with the cell membrane. These vesicles, approximately 30-150 nm in diameter, contain a variety of components, including proteins, lipids, nucleic acids, and metabolites (71, 72). Exosomes facilitate normal cellular functions, such as activating T cells during immune responses (72), and also contribute to tumor progression (73). Tumor-derived exosomes can transmit pro-tumorigenic signals, promoting cell proliferation, migration, invasion, and angiogenesis (74).
R. Ji et al. (26) demonstrated that exosomal miR-374a-5p achieved an AUC of 0.919 (95% CI: 0.866-0.972) in diagnosing GC, highlighting its high diagnostic accuracy, and the sensitivity was 0.761, meaning that this marker could correctly identify 76.1% of gastric cancer patients, which is slightly lower than the specificity, but still of high clinical value, especially in early diagnosis. The specificity was as high as 0.965, indicating that miR-374a-5p performed well in identifying non-gastric cancer patients with a very low misdiagnosis rate, which is important for reducing unnecessary treatment and psychological burden. In addition, miR-374a-5p had a low false positive rate (FP = 1), further confirming its reliability in diagnosis. Notably, miR-374a-5p exhibited a DOR of 102.398, outperforming other miRNAs such as miR-223-3p and miR-425-5p (16). Furthermore, miR-374a-5p, which is overexpressed in GC patients, was significantly associated with chemoresistance, suggesting its dual role as a diagnostic biomarker and a predictor of treatment response. These findings position miR-374a-5p as a promising non-invasive tool for GC diagnosis and therapeutic guidance.
X. Guo et al. (13) investigated exosomal lncRNA-GC1, which exhibits elevated expression in individuals with early-stage GC. The diagnostic performance of exosomal lncRNA-GC1, assessed in both the combined test set and the validation set, demonstrated a sensitivity of 0.8824, a specificity of 0.8229, and an AUC of 0.8905 (95% CI: 0.8371–0.9438) for detecting early-stage GC. Similarly, the aggregated sensitivity, specificity, and AUC values for lncRNAs across the 68 studies we reviewed were consistent, with respective values of 0.86, 0.82, and 0.89 (95% CI: 0.86–0.92). K. Xiao et al. (41) identified that the exosomal long non-coding RNA CCAT1 exhibited elevated expression levels in gastric cancer patients, demonstrating an AUC value of 0.89, a sensitivity of 0.80, and a specificity of 0.93. These findings underscore the potential of exosomal lncRNAs, such as lncRNA-GC1 and CCAT1, as reliable biomarkers for the early detection of GC, offering high diagnostic accuracy and consistency across studies. Comparable diagnostic performance was also noted for exosomal circRNAs (58). X. Yang et al. (60) demonstrated that the expression level of exosomal circLPAR1 in the serum of gastric cancer patients was significantly decreased, exhibiting an AUC of 0.836 (95% CI: 0.765-0.906), a sensitivity of 0.748, and a specificity of 0.780. These findings underscore the potential diagnostic significance of exosomal ncRNAs in GC.
This meta-analysis reveals both the diagnostic promise and current limitations of exosomal ncRNAs for gastric cancer detection through a comprehensive evaluation of 52 studies. The findings demonstrate robust performance across ncRNA classes, with lncRNAs showing the highest diagnostic accuracy (pooled AUC 0.89; 95% CI 0.86-0.92), followed by circRNAs (0.86; 0.83-0.89) and miRNAs (0.83; 0.79-0.86). Among individual biomarkers, lncRNA GClnc1 (48) emerged as particularly noteworthy, maintaining consistent accuracy (AUC 0.84-0.94) across 28 independent studies, while miR-374a-5p (26) (AUC 0.92) and miR-1246 (11) (sensitivity 0.79-0.95) showed excellent but less validated performance in specific cohorts.
The analysis uncovered several critical insights regarding biomarker performance. Multi-marker panels surprisingly failed to outperform single biomarkers, likely due to overfitting in small discovery cohorts. Certain circRNAs, exemplified by hsa_circ_0079439 (58), demonstrated exceptional specificity exceeding 95% despite more modest sensitivity (63-81%), suggesting their potential as complementary diagnostic tools. The superior consistency observed for lncRNAs may reflect their stable packaging in exosomes and tissue-specific expression patterns.
However, substantial heterogeneity challenges the interpretation of these findings, stemming from multiple sources. The evaluation of 59 distinct miRNAs, 17 lncRNAs, and 16 circRNAs across studies, with minimal target overlap, introduces significant variability in reported diagnostic performance. Meta-regression identified technical factors contributing to this heterogeneity, particularly RNA detection methods and exosome extraction protocols, though the fundamental issue of non-overlapping ncRNA targets remains unresolved. This variability limits the generalizability of pooled results, as diagnostic accuracy often appears dependent on specific RNA targets rather than reflecting consistent biological signals.
The most promising biomarkers, including GClnc1, miR-374a-5p, lncRNA-GC1, and CCAT1, demonstrated consistently high diagnostic accuracy (AUCs 0.88-0.92) across multiple reports (13, 26, 38, 41, 48). Nevertheless, the field faces critical challenges requiring immediate attention. Standardized methodologies for exosome isolation and ncRNA detection must be developed to reduce technical variability, while consensus frameworks are needed for biomarker prioritization and validation. Future research should prioritize multicenter prospective studies to validate these candidates, addressing current limitations in study design and methodological consistency to advance the most robust biomarkers toward clinical implementation.
Our synthesis of the literature revealed an interesting pattern regarding single versus multiple ncRNA markers, though we emphasize these observations derive from cross-study comparisons rather than direct experimental comparisons. Several individual studies reported superior diagnostic performance for single ncRNA markers compared to multi-marker combinations. For example, miR-374a-5p (26) (AUC = 0.919, sensitivity = 0.761, specificity = 0.965) and lncRNA-GC1 (13) (AUC = 0.8905, sensitivity = 0.8824, specificity = 0. 8229), tended to have higher AUC and DOR than multiple marker combinations. For instance, the combination of miR-223-3p and miR-425-5p had a lower AUC (0.707) and DOR (17.191) compared to a single marker (16). However, these comparisons must be interpreted cautiously as they originate from distinct studies employing different methodologies, patient cohorts, and experimental conditions. The apparent advantage of single markers may reflect study-specific factors rather than inherent biological superiority. This observation underscores the need for future studies specifically designed to directly compare single and combinatorial ncRNA approaches within standardized experimental frameworks, which would provide more definitive evidence for optimizing diagnostic strategies.
5 Limitations
The research presents several limitations. Initially, the inclusion of numerous retrospective investigations may introduce selection bias. Secondly, the exclusion of non-English publications could lead to information bias, as valuable data might be overlooked. Thirdly, variations in study design and execution may introduce confounding variables, potentially influencing the outcomes. The substantial heterogeneity among the studies poses a challenge to the reliability and reproducibility of the meta-analysis findings.
A comprehensive meta-analysis encompassing numerous studies has revealed that exosomal ncRNAs are pivotal in the diagnostic process of gastric cancer. Specifically, miRNAs, lncRNAs, and circRNAs exhibit substantial clinical relevance, as evidenced by their favorable positive diagnostic likelihood ratios. To translate these findings into clinical practice, it is imperative to elucidate the underlying mechanisms through which these ncRNAs contribute to the diagnosis and physiological aspects of GC. This understanding should be substantiated through rigorous experimental validation and in vivo model studies.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
QX: Conceptualization, Data curation, Investigation, Methodology, Visualization, Writing – original draft. QY: Data curation, Writing – original draft. ZD: Data curation, Writing – original draft. XC: Data curation, Writing – original draft. TL: Data curation, Writing – original draft. CP: Data curation, Writing – original draft. GH: Project administration, Writing – review & editing. XL: Project administration, Writing – review & editing. JW: Funding acquisition, Project administration, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the Futian District Health and Public Welfare Research Project of Shenzhen City (FTWS2022033, FTWS2023099, FTWS054); Construction of Clinical Key Specialties in Futian District, Shenzhen.
Acknowledgments
The authors thank all patients and clinical investigators who participated in the original studies analyzed here.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2025.1570020/full#supplementary-material
References
1. Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I, et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2024) 74:229–63. doi: 10.3322/caac.21834
2. Thrift AP, Wenker TN, and El-Serag HB. Global burden of gastric cancer: epidemiological trends, risk factors, screening and prevention. Nat Rev Clin Oncol. (2023) 20:338–49. doi: 10.1038/s41571-023-00747-0
3. Yang L, Ying X, Liu S, Lyu G, Xu Z, Zhang X, et al. Gastric cancer: Epidemiology, risk factors and prevention strategies. Chin J Cancer Res. (2020) 32:695–704. doi: 10.21147/j.issn.1000-9604.2020.06.03
4. Yuan L, Xu ZY, Ruan SM, Mo S, Qin JJ, and Cheng XD. Long non-coding RNAs towards precision medicine in gastric cancer: early diagnosis, treatment, and drug resistance. Mol Cancer. (2020) 19:96. doi: 10.1186/s12943-020-01219-0
5. Tang XH, Guo T, Gao XY, Wu XL, Xing XF, Ji JF, et al. Exosome-derived noncoding RNAs in gastric cancer: functions and clinical applications. Mol Cancer. (2021) 20:99. doi: 10.1186/s12943-021-01396-6
6. Ajani JA, D’Amico TA, Bentrem DJ, Chao J, Cooke D, Corvera C, et al. Gastric cancer, version 2.2022, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. (2022) 20:167–92. doi: 10.6004/jnccn.2022.0008
7. Ma S, Zhou M, Xu Y, Gu X, Zou M, Abudushalamu G, et al. Clinical application and detection techniques of liquid biopsy in gastric cancer. Mol Cancer. (2023) 22:7. doi: 10.1186/s12943-023-01715-z
8. Shi H, Li T, Liu Z, Zhao J, and Qi F. Early detection of gastric cancer via high-resolution terahertz imaging system. Front Bioeng Biotechnol. (2022) 10:1052069. doi: 10.3389/fbioe.2022.1052069
9. Fu M, Gu J, Jiang P, Qian H, Xu W, and Zhang X. Exosomes in gastric cancer: roles, mechanisms, and applications. Mol Cancer. (2019) 18:41. doi: 10.1186/s12943-019-1001-7
10. Zhang X, Wang S, Wang H, Cao J, Huang X, Chen Z, et al. Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol Cancer. (2019) 18:20. doi: 10.1186/s12943-018-0935-5
11. Shi Y, Wang Z, Zhu X, Chen L, Ma Y, Wang J, et al. Exosomal miR-1246 in serum as a potential biomarker for early diagnosis of gastric cancer. Int J Clin Oncol. (2020) 25:89–99. doi: 10.1007/s10147-019-01532-9
12. Zhang Y, Wu Y, Luo S, Yang C, Zhong G, Huang G, et al. DNA nanowire guided-catalyzed hairpin assembly nanoprobe for in situ profiling of circulating extracellular vesicle-associated microRNAs. ACS Sens. (2022) 7:1075–85. doi: 10.1021/acssensors.1c02717
13. Guo X, Lv X, Ru Y, Zhou F, Wang N, Xi H, et al. Circulating exosomal gastric cancer-associated long noncoding RNA1 as a biomarker for early detection and monitoring progression of gastric cancer: A multiphase study. JAMA Surg. (2020) 155:572–79. doi: 10.1001/jamasurg.2020.1133
14. McInnes MDF, Moher D, Thombs BD, McGrath TA, Bossuyt PM, Clifford T, et al. Preferred reporting items for a systematic review and meta-analysis of diagnostic test accuracy studies: the PRISMA-DTA statement. Jama. (2018) 319:388–96. doi: 10.1001/jama.2017.19163
15. Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. (2011) 155:529–36. doi: 10.7326/0003-4819-155-8-201110180-00009
16. Yu D, Zhang J, Wang M, Ji R, Qian H, Xu W, et al. Exosomal miRNAs from neutrophils act as accurate biomarkers for gastric cancer diagnosis. Clin Chim Acta. (2024), 554. doi: 10.1016/j.cca.2024.117773
17. Kahroba H, Samadi N, Mostafazadeh M, Hejazi MS, Sadeghi MR, Hashemzadeh S, et al. Evaluating the presence of deregulated tumoral onco-microRNAs in serum-derived exosomes of gastric cancer patients as noninvasive diagnostic biomarkers. BioImpacts. (2022) 12:127–38. doi: 10.34172/bi.2021.22178
18. Yang H, Fu H, Wang B, Zhang X, Mao J, Li X, et al. Exosomal miR-423-5p targets SUFU to promote cancer growth and metastasis and serves as a novel marker for gastric cancer. Mol Carcinogene. (2018) 57:1223–36. doi: 10.1002/mc.22838
19. He J, Wu J, Dong S, Xu J, Wang J, Zhou X, et al. Exosome-Encapsulated miR-31, miR-192, and miR-375 Serve as Clinical Biomarkers of Gastric Cancer. J Oncol. (2023) 2023. doi: 10.1155/2023/7335456
20. Wang JH, Bai ZZ, Niu XD, Zhu CL, Liang T, Hu YL, et al. Serum extracellular vesicle-derived miR-21-5p and miR-26a-5p as non-invasive diagnostic potential biomarkers for gastric cancer: A preliminary study. Int J Biol Markers. (2024) 39(3):217–25. doi: 10.1177/03936155241261390
21. Wang JF, Jiang YM, Zhan WH, Ye SP, Li TY, and Zhang JN. Screening of serum exosomal miRNAs as diagnostic biomarkers for gastric cancer using small RNA sequencing. J Oncol. (2022), 2022. doi: 10.1155/2022/5346563
22. Yang J, Li X, Wei S, Peng L, Sang H, Jin D, et al. Evaluation of the diagnostic potential of a plasma exosomal miRNAs panel for gastric cancer. Front Oncol. (2021), 11. doi: 10.3389/fonc.2021.683465
23. Chang L, Gao H, Wang L, Wang N, Zhang S, Zhou X, et al. Exosomes derived from miR-1228 overexpressing bone marrow-mesenchymal stem cells promote growth of gastric cancer cells. Aging. (2021) 13:11808–21. doi: 10.18632/aging.202878
24. Ge L, Zhang N, Li D, Wu Y, Wang H, and Wang J. Circulating exosomal small RNAs are promising non-invasive diagnostic biomarkers for gastric cancer. J Cell Mol Med. (2020) 24:14502–13. doi: 10.1111/jcmm.16077
25. Wang N, Wang L, Yang Y, Gong L, Xiao B, and Liu X. A serum exosomal microRNA panel as a potential biomarker test for gastric cancer. Biochem Biophys Res Commun. (2017) 493:1322–28. doi: 10.1016/j.bbrc.2017.10.003
26. Ji R, Zhang X, Gu H, Ma J, Wen X, Zhou J, et al. miR-374a-5p: A new target for diagnosis and drug resistance therapy in gastric cancer. Mol Ther Nucleic Acids. (2019) 18:331. doi: 10.1016/j.omtn.2019.07.025
27. Tang S, Cheng J, Yao Y, Lou C, Wang L, Huang X, et al. Combination of four serum exosomal miRNAs as novel diagnostic biomarkers for early-stage gastric cancer. Front Genet. (2020), 11. doi: 10.3389/fgene.2020.00237
28. Wei S, Peng L, Yang J, Sang H, Jin D, Li X, et al. Exosomal transfer of miR-15b-3p enhances tumorigenesis and Malignant transformation through the DYNLT1/Caspase-3/Caspase-9 signaling pathway in gastric cancer. J Exp Clin Cancer Res. (2020) 39. doi: 10.1186/s13046-019-1511-6
29. Lu X, Lu J, Wang S, Zhang Y, Ding Y, Shen X, et al. Circulating serum exosomal miR-92a-3p as a novel biomarker for early diagnosis of gastric cancer. Future Oncol. (2021) 17:907–19. doi: 10.2217/fon-2020-0792
30. Zhou X, Zhu W, Li H, Wen W, Cheng W, Wang F, et al. Diagnostic value of a plasma microRNA signature in gastric cancer: a microRNA expression analysis. Sci Rep. (2015) 5:11251. doi: 10.1038/srep11251
31. Zhang Y, Huang F, Xu N, Wang J, Li D, and Yin L. Overexpression of serum extracellular vesicle microRNA-215-5p is associated with early tumor recurrence and poor prognosis of gastric cancer. Clinics (Sao Paulo Brazil). (2021) 76:e2081. doi: 10.6061/clinics/2021/e2081
32. Huang Z, Zhu D, Wu L, He M, Zhou X, Zhang L, et al. Six serum-based miRNAs as potential diagnostic biomarkers for gastric cancer. Cancer Epidemiol Biomarkers Prev. (2017) 26:188–96. doi: 10.1158/1055-9965.EPI-16-0607
33. Ren ZJ, Zhao Y, Wang G, Miao LL, Zhang ZC, Ma L, et al. Identification of differentially expressed miRNAs derived from serum exosomes associated with gastric cancer by microarray analysis. Clin Chim Acta. (2022) 531:25–35. doi: 10.1016/j.cca.2022.03.010
34. Wang Z, Ding J, Xiao Y, Xiao K, Su P, Dong Z, et al. Serum extracellular vesicles with NSD1 and FBXO7 mRNA as novel biomarkers for gastric cancer. Clin Biochem. (2023) 120:110653. doi: 10.1016/j.clinbiochem.2023.110653
35. Rincón-Riveros A, Villegas VE, Motta NSQ, López-Kleine L, and Rodríguezand JA. Exosomal microRNA signature from plasma-derived extracellular vesicles in gastric cancer. bioRxiv. (2023) 2023:2023.04.28.538562. doi: 10.1101/2023.04.28.538562
36. Xia B, Gu X, Xu T, Yan M, Huang L, Jiang C, et al. Exosomes-mediated transfer of LINC00691 regulates the formation of CAFs and promotes the progression of gastric cancer. BMC Cancer. (2023) 23. doi: 10.1186/s12885-023-11373-5
37. Cai C, Zhang H, Zhu Y, Zheng P, Xu Y, Sun J, et al. Serum exosomal long noncoding RNA Pcsk2-2:1 as a potential novel diagnostic biomarker for gastric cancer. OncoTargets Ther. (2019) 12:10035–41. doi: 10.2147/OTT.S229033
38. Zhang C, Wang H, Liu Q, Dai S, Tian G, Wei X, et al. LncRNA CCAT1 facilitates the progression of gastric cancer via PTBP1-mediated glycolysis enhancement. J Exp Clin Cancer Res. (2023) 42:246. doi: 10.1186/s13046-023-02827-6
39. Piao HY, Guo S, Wang Y, and Zhang J. Exosomal long non-coding RNA CEBPA-AS1 inhibits tumor apoptosis and functions as a non-invasive biomarker for diagnosis of gastric cancer. OncoTargets Ther. (2020) 13:1365–74. doi: 10.2147/OTT.S238706
40. Zhou H, Shen WF, Zou HX, Lv QS, and Shao PY. Circulating exosomal long non-coding RNA H19 as a potential novel diagnostic and prognostic biomarker for gastric cancer. J Int Med Res. (2020) 48:11. doi: 10.1177/0300060520934297
41. Xiao K, Dong Z, Wang D, Liu M, Ding J, Chen W, et al. Clinical value of lncRNA CCAT1 in serum extracellular vesicles as a potential biomarker for gastric cancer. Oncol Lett. (2021) 21. doi: 10.3892/ol.2021.12708
42. Lin L-Y, Yang L, Zeng Q, Wang L, Chen M-L, Zhao Z-H, et al. Tumor-originated exosomal lncUEGC1 as a circulating biomarker for early-stage gastric cancer. Mol Cancer. (2018) 17:84. doi: 10.1186/s12943-018-0834-9
43. Pan L, Liang W, Fu M, Huang ZH, Li X, Zhang W, et al. Exosomes-mediated transfer of long noncoding RNA ZFAS1 promotes gastric cancer progression. J Cancer Res Clin Oncol. (2017) 143:991–1004. doi: 10.1007/s00432-017-2361-2
44. Zheng P, Zhang H, Gao H, Sun J, Li J, Zhang X, et al. Plasma exosomal long noncoding RNA lnc-SLC2A12-10:1 as a novel diagnostic biomarker for gastric cancer. OncoTargets Ther. (2020) 13:4009–18. doi: 10.2147/OTT.S253600
45. Li Q, Shao Y, Zhang X, Zheng T, Miao M, Qin L, et al. Plasma long noncoding RNA protected by exosomes as a potential stable biomarker for gastric cancer. Tumor Biol. (2015) 36:2007–12. doi: 10.1007/s13277-014-2807-y
46. Zhao R, Zhang Y, Zhang X, Yang Y, Zheng X, Li X, et al. Exosomal long noncoding RNA HOTTIP as potential novel diagnostic and prognostic biomarker test for gastric cancer. Mol Cancer. (2018) 17. doi: 10.1186/s12943-018-0817-x
47. Li S, Zhang M, Zhang H, Hu K, Cai C, Wang J, et al. Exosomal long noncoding RNA lnc-GNAQ-6:1 may serve as a diagnostic marker for gastric cancer. Clin Chim Acta. (2020) 501:252–57. doi: 10.1016/j.cca.2019.10.047
48. Guo X, Peng Y, Song Q, Wei J, Wang X, Ru Y, et al. A liquid biopsy signature for the early detection of gastric cancer in patients. Gastroenterology. (2023) 165:402–13.e13. doi: 10.1053/j.gastro.2023.02.044
49. Zhang X, Liang W, Liu J, Zang X, Gu J, Pan L, et al. Long non-coding RNA UFC1 promotes gastric cancer progression by regulating miR-498/Lin28b. J Exp Clin Cancer Res. (2018) 37. doi: 10.1186/s13046-018-0803-6
50. Zhang Y, Chen L, Ye X, Wu Z, Zhang Z, Sun B, et al. Expression and mechanism of exosome-mediated A FOXM1 related long noncoding RNA in gastric cancer. J Nanobiotechnol. (2021) 19. doi: 10.1186/s12951-021-00873-w
51. Xu H, Zhou J, Tang J, Min X, Yi T, Zhao J, et al. Identification of serum exosomal lncRNA MIAT as a novel diagnostic and prognostic biomarker for gastric cancer. J Clin Lab Anal. (2020) 34. doi: 10.1002/jcla.23323
52. You J, Chen Y, Chen D, Li Y, Wang T, Zhu J, et al. Circular RNA 0001789 sponges miR-140-3p and regulates PAK2 to promote the progression of gastric cancer. J Trans Med. (2023) 21. doi: 10.1186/s12967-022-03853-2
53. Xiao K, Li SR, Ding J, Wang Z, Wang D, Cao XT, et al. Expression and clinical value of circRNAs in serum extracellular vesicles for gastric cancer. Front Oncol. (2022) 12:962831. doi: 10.3389/fonc.2022.962831
54. Zheng P, Gao H, Xie X, and Lu P. Plasma exosomal hsa_circ_0015286 as a potential diagnostic and prognostic biomarker for gastric cancer. Pathol Oncol Res. (2022) 28:1610446. doi: 10.3389/pore.2022.1610446
55. Li R, Tian X, Jiang J, Qian H, Shen H, and Xu W. CircRNA CDR1as: a novel diagnostic and prognostic biomarker for gastric cancer. Biomarkers. (2023) 28:448–57. doi: 10.1080/1354750X.2023.2206984
56. Tang W, Fu K, Sun H, Rong D, Wang H, and Cao H. CircRNA microarray profiling identifies a novel circulating biomarker for detection of gastric cancer. Mol Cancer. (2018) 17. doi: 10.1186/s12943-018-0888-8
57. Huang XJ, Wang Y, Wang HT, Liang ZF, Ji C, Li XX, et al. Exosomal hsa_circ_000200 as a potential biomarker and metastasis enhancer of gastric cancer via miR-4659a/b-3p/HBEGF axis. Cancer Cell Int. (2023) 23. doi: 10.1186/s12935-023-02976-w
58. Li X, Lin Y-L, Shao J-K, Wu X-J, Li X, Yao H, et al. Plasma exosomal hsa_circ_0079439 as a novel biomarker for early detection of gastric cancer. World J Gastroenterol. (2023) 29:3482–96. doi: 10.3748/wjg.v29.i22.3482
59. Tao X, Shao Y, Lu R, Ye Q, Xiao B, Ye G, et al. Clinical significance of hsa_circ_0000419 in gastric cancer screening and prognosis estimation. Pathol Res Pract. (2020) 216:152763. doi: 10.1016/j.prp.2019.152763
60. Yang XB, Xia J, Peng CS, and Cai WP. Expression of plasma exosomal circLPAR1 in patients with gastric cancer and its clinical application value. Am J Cancer Res. (2023) 13:4269–76.
61. Zang X, Wang R, Wang Z, Qiu S, Zhang F, Zhou L, et al. Exosomal circ50547 as a potential marker and promotor of gastric cancer progression via miR-217/HNF1B axis. Trans Oncol. (2024), 45. doi: 10.1016/j.tranon.2024.101969
62. Shao Y, Tao X, Lu R, Zhang H, Ge J, Xiao B, et al. Hsa_circ_0065149 is an indicator for early gastric cancer screening and prognosis prediction. Pathol Oncol Res. (2020) 26:1475–82. doi: 10.1007/s12253-019-00716-y
63. Wang Y, Wang H, Zheng R, Wu P, Sun Z, Chen J, et al. Circular RNA ITCH suppresses metastasis of gastric cancer via regulating miR-199a-5p/Klotho axis. Cell Cycle. (2021) 20:522–36. doi: 10.1080/15384101.2021.1878327
64. Zhang Z, Sun C, Zheng Y, and Gong Y. circFCHO2 promotes gastric cancer progression by activating the JAK1/STAT3 pathway via sponging miR-194-5p. Cell Cycle. (2022) 21:2145–64. doi: 10.1080/15384101.2022.2087280
65. Sun X, Kong S, Jiang C, Jing R, Ju S, and Cong H. Diagnostic value of circular RNA hsa_circ_0002874 expression in peripheral blood of patients with gastric cancer. Lab Med. (2022) 53:65–70. doi: 10.1093/labmed/lmab062
66. Ilic M and Ilic I. Epidemiology of stomach cancer. World J Gastroenterol. (2022) 28:1187–203. doi: 10.3748/wjg.v28.i12.1187
67. Zhang Z, Wu H, Chong W, Shang L, Jing C, and Li L. Liquid biopsy in gastric cancer: predictive and prognostic biomarkers. Cell Death Dis. (2022) 13:903. doi: 10.1038/s41419-022-05350-2
68. Chen Y, Chen T, and Fang J-Y. Burden of gastrointestinal cancers in China from 1990 to 2019 and projection through 2029. Cancer Lett. (2023) 560:216127. doi: 10.1016/j.canlet.2023.216127
69. Kogure A, Yoshioka Y, and Ochiya T. Extracellular vesicles in cancer metastasis: potential as therapeutic targets and materials. Int J Mol Sci. (2020) 21. doi: 10.3390/ijms21124463
70. Munagala R, Aqil F, Jeyabalan J, Kandimalla R, Wallen M, Tyagi N, et al. Exosome-mediated delivery of RNA and DNA for gene therapy. Cancer Lett. (2021) 505:58–72. doi: 10.1016/j.canlet.2021.02.011
71. Liu C, Liu D, Wang S, Gan L, Yang X, and Ma C. Identification of the SNARE complex that mediates the fusion of multivesicular bodies with the plasma membrane in exosome secretion. J Extracell Vesicles. (2023) 12:e12356. doi: 10.1002/jev2.12356
72. Kalluri R and LeBleu VS. The biology, function, and biomedical applications of exosomes. Science. (2020) 367. doi: 10.1126/science.aau6977
73. Dai J, Su Y, Zhong S, Cong L, Liu B, Yang J, et al. Exosomes: key players in cancer and potential therapeutic strategy. Signal Transduct Target Ther. (2020) 5:145. doi: 10.1038/s41392-020-00261-0
Keywords: exosomes, noncoding RNA, gastric cancer, biomarker, meta-analysis
Citation: Xu Q, Yan Q, Dai Z, Chen X, Liu T, Peng C, Huang G, Liu X and Wang J (2025) Diagnostic value of exosome non-coding RNAs as non-invasive biomarkers in gastric cancer: a meta-analysis. Front. Oncol. 15:1570020. doi: 10.3389/fonc.2025.1570020
Received: 02 February 2025; Accepted: 28 May 2025;
Published: 12 June 2025.
Edited by:
Yuriy Gusev, Georgetown University, United StatesReviewed by:
Satyajit Ghosh, Indian Institute of Technology Jodhpur, IndiaFarimah Fayyaz, Johns Hopkins University, United States
Copyright © 2025 Xu, Yan, Dai, Chen, Liu, Peng, Huang, Liu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jingbin Wang, d2FuZ2ppbmdiaW5AZ3p1Y20uZWR1LmNu
 Qiumiao Xu1,2
Qiumiao Xu1,2 Qinghong Yan
Qinghong Yan Jingbin Wang
Jingbin Wang