- Institute of Microbiology, University Hospital, University of Lausanne, Lausanne, Switzerland
Pharmacogenomics is moving from a candidate gene strategy to large scale approaches. This is in line with the new paradigm of linking a trait to (a) pathway(s) rather than to single genes. In addition, breakthroughs in genomics offer a non-a priori assessment of implicated genes, expanding the possibilities in pharmacogenomics research. In this review, we discuss the pros and cons of new concepts in study design and on high throughput approaches to be implemented in the near future.
Introduction
The concept that phenotypic traits are controlled by a single gene proved true for Mendelian diseases (Nebert et al., 2008). A similar paradigm applies to pharmacology: the single gene approach was useful for the understanding of quantitative traits characterized by a bimodal distribution, such as the metabolism of the antituberculosis drug isoniazid through N-acetyltransferase (Kinzig-Schippers et al., 2005), and the association of HLA*B5701 with the hypersensitivity reaction to the antiretroviral drug abacavir (Mallal et al., 2008). Where applicable, single gene association is a cost–effective and efficient approach in pharmacogenetics. On the other hand, since this particular approach relies on an a priori hypothesis of the implicated gene, it may be reductionist and may often result in non-replication of the findings across different studies; a well-known example is the genetic variation in the drug transporter ABCB1 and its effect in drug disposition (Leschziner et al., 2007).
Most diseases and phenotypic traits are thought to result from the interplay of several genes with environmental factors. Here, large scale candidate gene analyses aim at understanding the interactions among several genes and how these contribute to a particular trait. This strategy involves investigation of genes encoding proteins potentially involved in a given phenotype according to their described or supposed function. This is in line with the new paradigm of linking a trait to a pathway rather than to a single gene, but in return it does not allow an in-depth analysis of the investigated genes (Nebert et al., 2008; Lubomirov et al., 2010). In addition, like for single candidate gene analysis, there is an a priori hypothesis of the implicated genes.
Today, the new concept of study design that relies on a non-a priori assessment of implicated genes is applied in genome-wide association studies. In this review we discuss the pros and cons of this approach and other high throughput approaches likely to be used for pharmacogenomics research in the near future.
Genome-Wide Association Studies and Pharmacogenomics
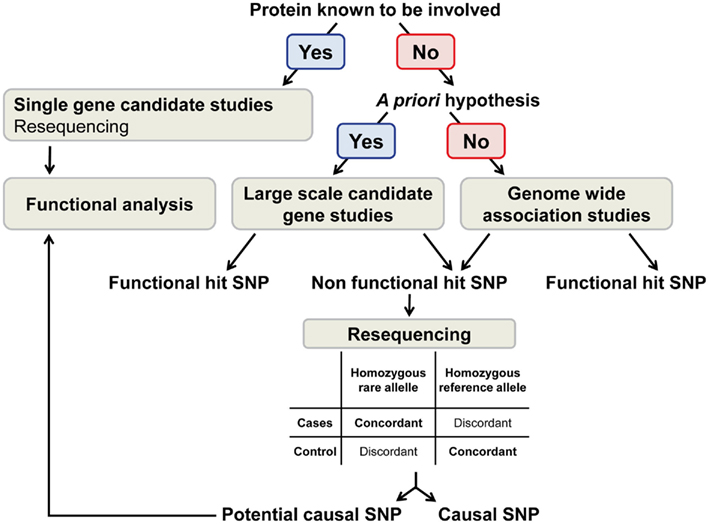
The genome-wide association study (GWAS) approach aims at discovering new biological pathways and further our understanding of complex phenotypic traits. This strategy consists in screening the genome to identify genetic variants associated with a given phenotype using high coverage genotyping arrays [from 100,000 to 2.3 million single nucleotide polymorphisms (SNPs) per array]. The essential genomic feature used in the design of GWAS arrays is that genetic variants are inherited together forming haplotypes (International HapMap Consortium, 2003). This characteristic allows assessment of SNPs that tag a haplotype (tag SNP) instead of genotyping all variants present in the haplotype. In return, tag SNPs do not necessarily allow the identification of the causal variant(s) responsible for the phenotypic trait (Figure 1).
Genome-wide association study approaches have been used for identification of genetic loci associated with disease susceptibility (Hindorff et al., 2009), and they are now applied to pharmacogenomics (Daly, 2010; Wang et al., 2011). Some studies confirmed previous results such as the association of variants in the genes CYP2C9, VKORC1, and CYP4F2 with the need of dose adjustment for the anticoagulant drug warfarin (Cooper et al., 2008; Takeuchi et al., 2009; Cha et al., 2010) and the decreased antiplatelet effect of clopidogrel with a genetic variant in CYP2C19 (Shuldiner et al., 2009). Other studies discovered new associations such as the enhanced risk of simvastatin-induced myopathy associated with the presence of the allele SLCO1B1*5 (Link et al., 2008), the drug associated liver injury of the antibiotic flucloxacillin with the presence of the allele HLA*B5701 (Daly et al., 2009) and finally, four GWAS have demonstrated that hepatitis C treatment-induced clearance was strongly dependent of genetic variants in IL28B, a gene encoding for the interferon-λ3. Two SNPs (rs12979860 and rs8099917) were significantly associated with response to pegylated interferon alpha and ribavirin in patients chronically infected with hepatitis C virus (HCV) genotype 1 (Ge et al., 2009; Suppiah et al., 2009; Tanaka et al., 2009; Rauch et al., 2010).
In summary, GWAS represent a breakthrough in the understanding of complex phenotypic traits. However, the SNPs identified in these studies as associated with the study phenotypes (hit SNPs) are not necessarily the causal variants. Therefore, comprehensive resequencing and functional analyses are needed to narrow down on the causal variants.
Resequencing as a Strategy to Improve Pharmacogenomic Studies
Resequencing aims at identifying the causal variant if the SNPs identified in the GWAS (hit SNPs) are unlikely to be functional (e.g., SNPs localized in intergenic regions). One approach consists in resequencing the chromosomic region of interest in individuals representing concordant and discordant genotype–phenotype associations. “Concordant” is defined in individuals homozygous for the rare allele of the hit SNP that present the associated phenotype, or in individuals that neither have the rare allele of the hit SNP nor the associated phenotype. “Discordant” is defined in individuals homozygous for the rare allele of the hit SNP without the study phenotype, or in individuals without the rare allele of the hit SNP but presenting the study phenotype. Haplotype inference in these individuals with extreme phenotype–genotype combinations allows the identification of genetic variants tagged by the hit SNP. The causal variants are expected to be as frequent as, or more frequent than the hit SNP in concordant individuals and/or less frequent in discordant individuals (Figure 1).
This approach has been successfully applied in an ADME pharmacogenetics study of the antiretroviral drug lopinavir coformulated with ritonavir (LPV/r). In this study, two of the hit SNPs associated with LPV/r clearance were located in SLCO1B1. One SNP was known to be a functional variant (rs4149056), the second was a tagSNP. Resequencing and haplotype inference identified a SNP leading to a non-synonymous amino acid change likely to be responsible for the detected effect (rs11045819; Lubomirov et al., 2010). The same approach aimed at determining the functional variant in IL28B that associates with HCV treatment and infection outcomes, and that is tagged by an intergenic SNP, rs8099917 (Suppiah et al., 2009; Tanaka et al., 2009; Rauch et al., 2010). Two distinct families of haplotypes and four candidate causal SNPs were identified on the basis of allelic frequency, genome location, and linkage disequilibrium pattern (Rauch et al., 2010). Haplotypes carrying rs8103142, one of the four candidate SNPs, increased the predictive value of HCV infection outcome, in comparison to the tag SNP rs8099917, initially identified in the GWAS (di Iulio et al., 2011). However, these haplotypes were highly linked to rs12979860, a tag SNP identified in other studies (Ge et al., 2009; Thomas et al., 2009) that had comparable predictive value, making the assignment of the true causal variant difficult.
In summary, resequencing approaches can identify the potential causal variant associated with a phenotype. However, functional analyses are still needed to assess the mechanism of action. In addition, since this strategy used non-automated, labor-intensive techniques, it can only be applied to small number of samples. This inconvenient may be solved by high throughput sequencing techniques.
How Can High Throughput Sequencing be Useful in Pharmacogenomic Studies?
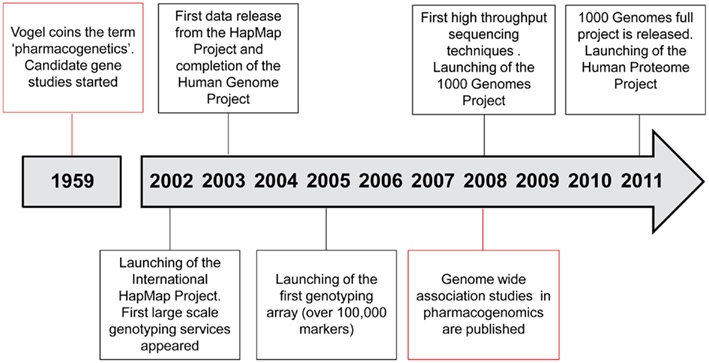
The expanding possibilities offered since the completion of the Human Genome Project1, the International HapMap Consortium (2003, 2005), and the 1000 Genomes Project Consortium (2010) together with the availability of new high throughput sequencing technologies, are the bases for future initiatives in pharmacogenomics.
High throughput sequencing allows whole genome/exome/transcriptome analyses. Variants can be identified independently of: (I) patterns of linkage dysequilibrium and tag SNPs; specifically, there is no need to customize the arrays according to the population studied (e.g., variants in African ancestry genomes are more difficult to tag because of higher recombination rates 2003, 2005); and (II) allelic frequency; it is thought that variants with high effect sizes will be rare as they may be more deleterious at a population level (Manolio et al., 2009).
High throughput sequencing can be performed either at the DNA or at the RNA level. RNA sequencing has the advantage to provide qualitative and quantitative information (genetic variation, gene expression, and alternative splicing). This is important since gene expression and alternative splicing are more directly correlated with the phenotype (Ozsolak and Milos, 2011).
In pharmacogenomics, high throughput sequencing may be very useful as changes in gene expression or splicing are important for drug disposition (Marez et al., 1995; Chou et al., 2001; Kuehl et al., 2001; Pitarque et al., 2001; Lang et al., 2004; Menard et al., 2011). However, the high cost and complexity of data analyses delay the implementation of this approach.
The Usefulness of the Next “Omics”: Proteomics in Pharmacogenomic Studies
The Human Proteome Project intends to create a comprehensive and public available map of the entire human proteome (Legrain et al., 2011) through mass spectrometry technology, a highly reproducible technique (Nilsson et al., 2010). Proteomics aims at studying the function and structure of proteins at large scale. It allows direct assessment of the abundance of main or rare isoforms. Proteomics is now on the early phase; evidence of expression at the protein level is still lacking for about 30% of the genes of the human genome2.
Specific to pharmacogenomics, proteomics may be essential in the identification of new players involved not only in drug pharmacokinetics but also pharmacodynamics. However, the cell proteome approach will be more challenging than the genome approach: the cell proteome is very dynamic (proteins are tissue specific and change over time), post-transcriptional modifications may affect protein function and splicing may result in isoforms with altered activity.
Conclusion
Breakthroughs in genomics influence pharmacogenomics studies (Figure 2). It is expected that pharmacogenomics will facilitate the deployment of personalized medicine and individualized drug therapy. The field now moves from the candidate gene strategies to larger scale approaches, including resequencing of candidate regions and high throughput exome/genome sequencing. The complexity of data handling and analysis will be a major difficulty to overcome, and functional studies will be required to decipher the underlying molecular mechanisms.
Glossary
• Pharmacogenetics: the study of how genetic variation in a single gene influences drug pharmacokinetics/pharmacodynamics.
• Pharmacogenomics: the study of how genetic variation throughout the genome influences drug pharmacokinetics/pharmacodynamics (same than pharmacogenetics but in large scale).
• Single nucleotide polymorphism: variation between members of the same species of a single nucleotide at a defined position in the genome, and occurring at a frequency higher than 1%.
• Genotype: the genetic background of a given individual.
• Phenotype: individual feature that can be measured or observed. It is influenced by the genotype and different environmental factors.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
References
Cha, P. C., Mushiroda, T., Takahashi, A., Kubo, M., Minami, S., Kamatani, N., and Nakamura, Y. (2010). Genome-wide association study identifies genetic determinants of warfarin responsiveness for Japanese. Hum. Mol. Genet. 19, 4735–4744.
Chou, F. C., Tzeng, S. J., and Huang, J. D. (2001). Genetic polymorphism of cytochrome P450 3A5 in Chinese. Drug Metab. Dispos. 29, 1205–1209.
Cooper, G. M., Johnson, J. A., Langaee, T. Y., Feng, H., Stanaway, I. B., Schwarz, U. I., Ritchie, M. D., Stein, C. M., Roden, D. M., Smith, J. D., Veenstra, D. L., Rettie, A. E., and Rieder, M. J. (2008). A genome-wide scan for common genetic variants with a large influence on warfarin maintenance dose. Blood 112, 1022–1027.
Daly, A. K. (2010). Genome-wide association studies in pharmacogenomics. Nat. Rev. Genet. 11, 241–246.
Daly, A. K., Donaldson, P. T., Bhatnagar, P., Shen, Y., Pe’er, I., Floratos, A., Daly, M. J., Goldstein, D. B., John, S., Nelson, M. R., Graham, J., Park, B. K., Dillon, J. F., Bernal, W., Cordell, H. J., Pirmohamed, M., Aithal, G. P., and Day, C. P. (2009). HLA-B*5701 genotype is a major determinant of drug-induced liver injury due to flucloxacillin. Nat. Genet. 41, 816–819.
di Iulio, J., Ciuffi, A., Fitzmaurice, K., Kelleher, D., Rotger, M., Fellay, J., Martinez, R., Pulit, S., Furrer, H., Gunthard, H. F., Battegay, M., Bernasconi, E., Schmid, P., Hirschel, B., Barnes, E., Klenerman, P., Telenti, A., and Rauch, A. (2011). Estimating the net contribution of interleukin-28B variation to spontaneous hepatitis C virus clearance. Hepatology 53, 1446–1454.
Ge, D., Fellay, J., Thompson, A. J., Simon, J. S., Shianna, K. V., Urban, T. J., Heinzen, E. L., Qiu, P., Bertelsen, A. H., Muir, A. J., Sulkowski, M., Mchutchison, J. G., and Goldstein, D. B. (2009). Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature 461, 399–401.
1000 Genomes Project Consortium. (2010). A map of human genome variation from population-scale sequencing. Nature 467, 1061–1073.
Hindorff, L. A., Sethupathy, P., Junkins, H. A., Ramos, E. M., Mehta, J. P., Collins, F. S., and Manolio, T. A. (2009). Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. Sci. U.S.A. 106, 9362–9367.
International HapMap Consortium. (2005). A haplotype map of the human genome. Nature 437, 1299–1320.
Kinzig-Schippers, M., Tomalik-Scharte, D., Jetter, A., Scheidel, B., Jakob, V., Rodamer, M., Cascorbi, I., Doroshyenko, O., Sorgel, F., and Fuhr, U. (2005). Should we use N-acetyltransferase type 2 genotyping to personalize isoniazid doses? Antimicrob. Agents Chemother. 49, 1733–1738.
Kuehl, P., Zhang, J., Lin, Y., Lamba, J., Assem, M., Schuetz, J., Watkins, P. B., Daly, A., Wrighton, S. A., Hall, S. D., Maurel, P., Relling, M., Brimer, C., Yasuda, K., Venkataramanan, R., Strom, S., Thummel, K., Boguski, M. S., and Schuetz, E. (2001). Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nat. Genet. 27, 383–391.
Lang, T., Klein, K., Richter, T., Zibat, A., Kerb, R., Eichelbaum, M., Schwab, M., and Zanger, U. M. (2004). Multiple novel nonsynonymous CYP2B6 gene polymorphisms in Caucasians: demonstration of phenotypic null alleles. J. Pharmacol. Exp. Ther. 311, 34–43.
Legrain, P., Aebersold, R., Archakov, A., Bairoch, A., Bala, K., Beretta, L., Bergeron, J., Borchers, C. H., Corthals, G. L., Costello, C. E., Deutsch, E. W., Domon, B., Hancock, W., He, F., Hochstrasser, D., Marko-Varga, G., Salekdeh, G. H., Sechi, S., Snyder, M., Srivastava, S., Uhlen, M., Wu, C. H., Yamamoto, T., Paik, Y. K., and Omenn, G. S. (2011). The human proteome project: current state and future direction. Mol. Cell Proteomics 10, M111 009993.
Leschziner, G. D., Andrew, T., Pirmohamed, M., and Johnson, M. R. (2007). ABCB1 genotype and PGP expression, function and therapeutic drug response: a critical review and recommendations for future research. Pharmacogenomics J. 7, 154–179.
Link, E., Parish, S., Armitage, J., Bowman, L., Heath, S., Matsuda, F., Gut, I., Lathrop, M., and Collins, R. (2008). SLCO1B1 variants and statin-induced myopathy – a genomewide study. N. Engl. J. Med. 359, 789–799.
Lubomirov, R., di Iulio, J., Fayet, A., Colombo, S., Martinez, R., Marzolini, C., Furrer, H., Vernazza, P., Calmy, A., Cavassini, M., Ledergerber, B., Rentsch, K., Descombes, P., Buclin, T., Decosterd, L. A., Csajka, C., and Telenti, A. (2010). ADME pharmacogenetics: investigation of the pharmacokinetics of the antiretroviral agent lopinavir coformulated with ritonavir. Pharmacogenet. Genomics 20, 217–230.
Mallal, S., Phillips, E., Carosi, G., Molina, J. M., Workman, C., Tomazic, J., Jagel-Guedes, E., Rugina, S., Kozyrev, O., Cid, J. F., Hay, P., Nolan, D., Hughes, S., Hughes, A., Ryan, S., Fitch, N., Thorborn, D., and Benbow, A. (2008). HLA-B*5701 screening for hypersensitivity to abacavir. N. Engl. J. Med. 358, 568–579.
Manolio, T. A., Collins, F. S., Cox, N. J., Goldstein, D. B., Hindorff, L. A., Hunter, D. J., Mccarthy, M. I., Ramos, E. M., Cardon, L. R., Chakravarti, A., Cho, J. H., Guttmacher, A. E., Kong, A., Kruglyak, L., Mardis, E., Rotimi, C. N., Slatkin, M., Valle, D., Whittemore, A. S., Boehnke, M., Clark, A. G., Eichler, E. E., Gibson, G., Haines, J. L., Mackay, T. F., Mccarroll, S. A., and Visscher, P. M. (2009). Finding the missing heritability of complex diseases. Nature 461, 747–753.
Marez, D., Sabbagh, N., Legrand, M., Lo-Guidice, J. M., Boone, P., and Broly, F. (1995). A novel CYP2D6 allele with an abolished splice recognition site associated with the poor metabolizer phenotype. Pharmacogenetics 5, 305–311.
Menard, V., Eap, O., Roberge, J., Harvey, M., Levesque, E., and Guillemette, C. (2011). Transcriptional diversity at the UGT2B7 locus is dictated by extensive pre-mRNA splicing mechanisms that give rise to multiple mRNA splice variants. Pharmacogenet. Genomics 21, 631–641.
Nebert, D. W., Zhang, G., and Vesell, E. S. (2008). From human genetics and genomics to pharmacogenetics and pharmacogenomics: past lessons, future directions. Drug Metab. Rev. 40, 187–224.
Nilsson, T., Mann, M., Aebersold, R., Yates, J. R. III., Bairoch, A., and Bergeron, J. J. (2010). Mass spectrometry in high-throughput proteomics: ready for the big time. Nat. Methods 7, 681–685.
Ozsolak, F., and Milos, P. M. (2011). RNA sequencing: advances, challenges and opportunities. Nat. Rev. Genet. 12, 87–98.
Pitarque, M., Von Richter, O., Oke, B., Berkkan, H., Oscarson, M., and Ingelman-Sundberg, M. (2001). Identification of a single nucleotide polymorphism in the TATA box of the CYP2A6 gene: impairment of its promoter activity. Biochem. Biophys. Res. Commun. 284, 455–460.
Rauch, A., Kutalik, Z., Descombes, P., Cai, T., di Iulio, J., Mueller, T., Bochud, M., Battegay, M., Bernasconi, E., Borovicka, J., Colombo, S., Cerny, A., Dufour, J. F., Furrer, H., Gunthard, H. F., Heim, M., Hirschel, B., Malinverni, R., Moradpour, D., Mullhaupt, B., Witteck, A., Beckmann, J. S., Berg, T., Bergmann, S., Negro, F., Telenti, A., and Bochud, P. Y. (2010). Genetic variation in IL28B is associated with chronic hepatitis C and treatment failure: a genome-wide association study. Gastroenterology 138, 1338–1345.
Shuldiner, A. R., O’connell, J. R., Bliden, K. P., Gandhi, A., Ryan, K., Horenstein, R. B., Damcott, C. M., Pakyz, R., Tantry, U. S., Gibson, Q., Pollin, T. I., Post, W., Parsa, A., Mitchell, B. D., Faraday, N., Herzog, W., and Gurbel, P. A. (2009). Association of cytochrome P450 2C19 genotype with the antiplatelet effect and clinical efficacy of clopidogrel therapy. JAMA 302, 849–857.
Suppiah, V., Moldovan, M., Ahlenstiel, G., Berg, T., Weltman, M., Abate, M. L., Bassendine, M., Spengler, U., Dore, G. J., Powell, E., Riordan, S., Sheridan, D., Smedile, A., Fragomeli, V., Muller, T., Bahlo, M., Stewart, G. J., Booth, D. R., and George, J. (2009). IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat. Genet. 41, 1100–1104.
Takeuchi, F., Mcginnis, R., Bourgeois, S., Barnes, C., Eriksson, N., Soranzo, N., Whittaker, P., Ranganath, V., Kumanduri, V., Mclaren, W., Holm, L., Lindh, J., Rane, A., Wadelius, M., and Deloukas, P. (2009). A genome-wide association study confirms VKORC1, CYP2C9, and CYP4F2 as principal genetic determinants of warfarin dose. PLoS Genet. 5, e1000433. doi: 10.1371/journal.pgen.1000433
Tanaka, Y., Nishida, N., Sugiyama, M., Kurosaki, M., Matsuura, K., Sakamoto, N., Nakagawa, M., Korenaga, M., Hino, K., Hige, S., Ito, Y., Mita, E., Tanaka, E., Mochida, S., Murawaki, Y., Honda, M., Sakai, A., Hiasa, Y., Nishiguchi, S., Koike, A., Sakaida, I., Imamura, M., Ito, K., Yano, K., Masaki, N., Sugauchi, F., Izumi, N., Tokunaga, K., and Mizokami, M. (2009). Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat. Genet. 41, 1105–1109.
Thomas, D. L., Thio, C. L., Martin, M. P., Qi, Y., Ge, D., O’huigin, C., Kidd, J., Kidd, K., Khakoo, S. I., Alexander, G., Goedert, J. J., Kirk, G. D., Donfield, S. M., Rosen, H. R., Tobler, L. H., Busch, M. P., Mchutchison, J. G., Goldstein, D. B., and Carrington, M. (2009). Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature 461, 798–801.
Keywords: pharmacogenomics, GWAS, next generation sequencing
Citation: di Iulio J and Rotger M (2011) Pharmacogenomics: what is next? Front. Pharmacol. 2:86. doi: 10.3389/fphar.2011.00086
Received: 16 November 2011;
Paper pending published: 05 December 2011;
Accepted: 14 December 2011;
Published online: 13 January 2012.
Edited by:
George P. Patrinos, University of Patras School of Health Sciences, GreeceReviewed by:
Naohiko Anzai, Dokkyo Medical University School of Medicine, JapanHortensia Alonso, Hospital del Sureste, Spain
Sotiria Boukouvala, Democritus University of Thrace, Greece
Alfonso Carvajal, Universidad de Valladolid, Spain
Copyright: © 2012 di Iulio and Rotger. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Julia di Iulio and Margalida Rotger, Institute of Microbiology, Bugnon 48, CHUV, 1011 Lausanne, Switzerland. e-mail:anVsaWEuZGktaXVsaW9AY2h1di5jaA==;bWFyZ2FsaWRhLnJvdGdlckBjaHV2LmNo
 Julia di Iulio*
Julia di Iulio*