- Biophysics Department, Faculty of Science, Cairo University, Giza, Egypt
The human coronavirus (HCoV), SARS-CoV-2, caused more than 34 M confirmed infections from which more than 1 M deaths are reported until now (the WHO situation report-154). The current pandemic causes severe socio-economic burden. Due to the importance of understanding of the mode of recognition and viral entry, spike protein shed drug designers as the first look protein target with the first released solved structure on 26 February 2020 (PDB ID: 6VSB). It is proposed that the recognition site for GRP78 is found in SARS-CoV-2 and the immersed human coronaviruses but experimental validation is still required.
The human coronavirus (HCoV), SARS-CoV-2, caused more than 34 M confirmed infections from which ≥ 1 M deaths are reported until now (the WHO situation report-154). The current pandemic causes severe socio-economic burden (Nicola et al., 2020). Due to the importance of understanding of the mode of recognition and viral entry, spike protein sheds drug designers as the first look protein target. The first solved structure for the spike protein is released on February 26, 2020 (PDB ID: 6VSB) (Wrapp et al., 2020).
Heat Shock Protein A5 (HSPA5), or the Glucose Regulating Protein 78 (GRP78), was reported to be a possible route for SARS-CoV-2 attachment and entry (Elfiky, 2020; Ibrahim et al., 2020; Ha et al., 2020; Saghazadeh and Rezaei, 2020). The binding site in the SARS-CoV-2 spike is predicted to be the nine residues CNGVEGFNC (C480-C488 region) found in the S1 C-terminal domain, as shown in Figure 1 (see the enlarged panel) (Ibrahim et al., 2020). The prediction was based on the structural conservation between this region of the spike, and the cyclic Pep42 peptide (CTVALPGGYVRVC) that previously reported to target GRP78 (over cancer cells) selectively in vivo (Kim et al., 2006; Quinones et al., 2008; Braun et al., 2020). It was recently reported in Nature journal that SARS-CoV-2 spike has conserved motifs compared to previous strains of human coronaviruses (HKU1, 229E, NL63, OC43) (Braun et al., 2020) still experimental validation is required to prove the hypothesis.
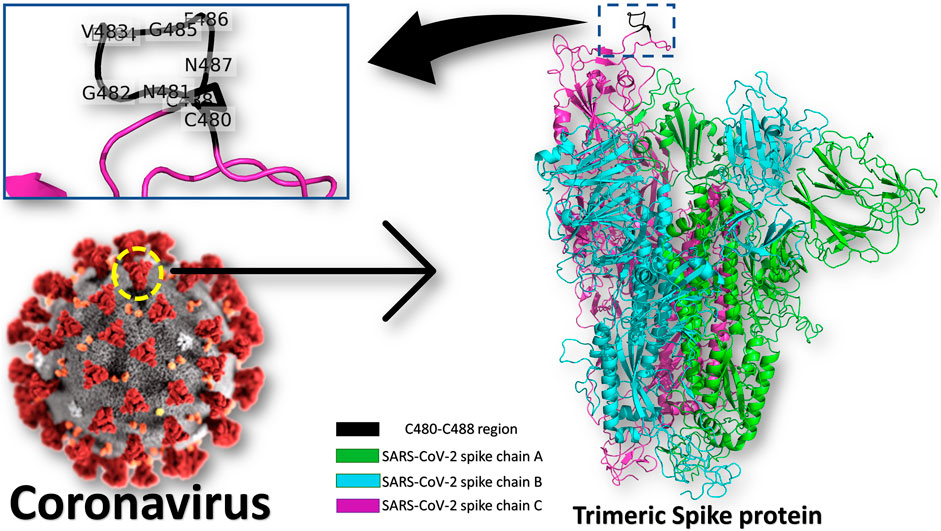
FIGURE 1. Coronavirus spike protein and GRP78 recognition site. The trimeric spike protein (green, cyan, and magenta cartoons) is shown in colored cartons, while the GRP78 recognition site (C480-C488 in SARS-CoV-2) is depicted in the black cartoon (see the enlarged panel). The GRP78 recognizing residues in SARS-CoV-2 are labeled in the enlarged panel.
The sequence comparison between the known human coronavirus strains, including; alpha coronaviruses (NL63, and 229E), and beta coronaviruses (OC43, HKU1, MERS-CoV, SARS-CoV, and SARS-CoV-2), reveals exciting results in terms of C480-C488 region of the spike protein. Figure 2A shows part of the multiple sequence alignment, made by clustal omega webserver and viewed by ESpript 3, for the different human coronavirus strains at the GRP78 recognition site (C480-C488 in SARS-CoV-2). The recognition of the GRP78 by the peptide Pep42 is previously reported to be restricted for the cyclic form of the peptide (the first and last CYS residues form a disulfide bond) (Kim et al., 2006; Quinones et al., 2008). Surprisingly, the residues C480 and C488 is found in SARS-CoV-2 and the immersed human coronaviruses strains (NL63, 229E, OC43, and HKU1), but missing in SARS-CoV and MERS-CoV. Additionally, at least three identical (four similar) residues are found among the immersed human coronaviruses strains compared to SARS-CoV-2 (see Figure 2B). This implies the possibility of cross immune the SARS-CoV-2 and the immersed human coronaviruses strains (NL63, 229E, OC43, and HKU1).
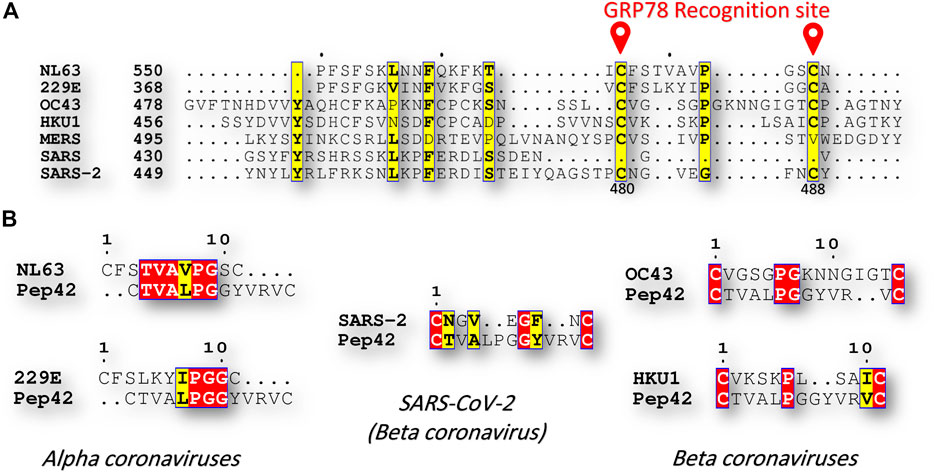
FIGURE 2. Sequence comparison (A) Part of the multiple sequence alignment for the spike protein of all of the currently reported human coronaviruses strains (NL63, 229E, OC43, HKU1, MERS-CoV, SARS-CoV, and SARS-CoV-2). The alignment is made using the Clustal Omega web server and is displayed by ESpript 3 software. The yellow highlighted residues are conserved among the seven HCoVs. GRP78 recognition site (C480-C488 in SARS-CoV-2) is marked in red (B) Pairwise sequence alignment between Pep42 from one side and NL63, 229E, OC43, HKU1, and SARS-CoV-2 from the other side. Red and yellow residues are identical and similar residues, respectively.
The human coronaviruses NL63, 229E, OC43, and HKU1, have been less impacted the human being (Zumla et al., 2016; van den Brand et al., 2015). People previously infected with these strains of human coronaviruses may develop immunity against SARS-CoV-2. Unfortunately, the immersed human coronavirus strains characterized by mild flu-like symptoms and information about viral distribution is rare (Huynh et al., 2012; Hilgenfeld and Peiris, 2013; Zumla et al., 2016).
Molecular dynamics study (using NAMD software) for the SARS-CoV-2 spike combined with molecular docking (Using AutoDock Vina software) revealed the existence of at least four interactions (H-bonds or hydrophobic contacts) between GRP78 and C480-C488 of SARS-CoV-2 spike. The hydrophobic contacts (two up to six) are found in all the docking experiments (seven replicas done at different dynamics states of SARS-CoV-2 spike after 100 ns MDS). This is in support of the previous reports about GRP78 recognition of hydrophobic patches in the unfolded proteins (Tsai and Lee, 2018; Li and Lee, 2006). Further experimental validation is required to prove this hypothesis.
Conclusively SARS-CoV-2 Spike-host cell recognition is crucial in fighting COVID-19 both as a therapeutic and prophylactic routes. The C480-C488 region is an essential viral spike epitope to be targeted by drugs, natural compounds, or antibodies to prevent or weaken the host cell recognition of SARS-CoV-2. This represents a hot topic that need further validation in laboratory.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
AE own the research idea and wrote the manuscript.
Conflict of Interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Ibrahim M. Ibrahim is appreciated for his help in MDS calculations.
References
Braun, J., Loyal, L., Frentsch, M., Wendisch, D., Georg, P., Kurth, F., et al. (2020). SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature 587 (7833), 270–274. doi:10.1038/s41586-020-2598-9
Elfiky, A. A. (2020). Natural products may interfere with SARS-CoV-2 attachment to the host cell. J. Biomol. Struct. Dyn. [Epub ahead of print]. doi:10.1080/07391102.2020.1761881
Ha, D. P., Van Krieken, R., Carlos, A., and Lee, A. S. (2020). The stress-inducible molecular chaperone GRP78 as potential therapeutic target for coronavirus infection. J. Infect. 81 (3), 452–482. doi:10.1016/j.jinf.2020.06.017
Hilgenfeld, R., and Peiris, M. (2013). From SARS to MERS: 10 years of research on highly pathogenic human coronaviruses. Antivir. Res. 100 (1), 286–295. doi:10.1016/j.antiviral.2013.08.015
Huynh, J., Li, S., Yount, B., Smith, A., Sturges, L., Olsen, J. C., et al. (2012). Evidence supporting a zoonotic origin of human coronavirus strain NL63. J. Virol. 86 (23), 12816–12825. doi:10.1128/JVI.00906-12
Ibrahim, I. M., Abdelmalek, D. H., Elshahat, M. E., and Elfiky, A. A. (2020). COVID-19 spike-host cell receptor GRP78 binding site prediction. J. Infect. 80 (5), 554–562. doi:10.1016/j.jinf.2020.02.026
Kim, Y., Lillo, A. M., Steiniger, S. C., Liu, Y., Ballatore, C., Anichini, A., et al. (2006). Targeting heat shock proteins on cancer cells: selection, characterization, and cell-penetrating properties of a peptidic GRP78 ligand. Biochemistry 45 (31), 9434–9444. doi:10.1021/bi060264j
Li, J., and Lee, A. S. (2006). Stress induction of GRP78/BiP and its role in cancer. Curr. Mol. Med. 6 (1), 45–54. doi:10.2174/156652406775574523
Nicola, M., Alsafi, Z., Sohrabi, C., Kerwan, A., Al-Jabir, A., Iosifidis, C., et al. (2020). The socio-economic implications of the coronavirus and COVID-19 pandemic: a review. Int. J. Surg. 78, 185–193. doi:10.1016/j.ijsu.2020.04.018
Quinones, Q. J., de Ridder, G. G., and Pizzo, S. V. (2008). GRP78: a chaperone with diverse roles beyond the endoplasmic reticulum. Histol. Histopathol. 23, 1409. doi:10.14670/HH-23.1409
Saghazadeh, A., and Rezaei, N. (2020). Towards treatment planning of COVID-19: rationale and hypothesis for the use of multiple immunosuppressive agents: anti-antibodies, immunoglobulins, and corticosteroids. Int. Immunopharm. 84, 106560. doi:10.1016/j.intimp.2020.106560
Tsai, Y-L., and Lee, A. S. (2018). “Cell surface GRP78: anchoring and translocation mechanisms and therapeutic potential in cancer,” in Cell surface GRP78, a new paradigm in signal transduction biology. Editor S. V. Pizzo (Chennai, India: Elsevier), 41–62.
van den Brand, J. M., Smits, S. L., and Haagmans, B. L. (2015). Pathogenesis of Middle East respiratory syndrome coronavirus. J. Pathol. 235 (2), 175–184. doi:10.1002/path.4458
Wrapp, D., Wang, N., Corbett, K. S., Goldsmith, J. A., Hsieh, C.-L., Abiona, O., et al. (2020). Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367 (6483), 1260–1263. doi:10.1126/science.abb2507
Keywords: GRP78, HSPA5, SARS-CoV-2, viral attachment, spike, host-cell recognition, COVID-19, cross-vaccination
Citation: Elfiky AA (2020) SARS-CoV-2 Spike-Heat Shock Protein A5 (GRP78) Recognition may be Related to the Immersed Human Coronaviruses. Front. Pharmacol. 11:577467. doi: 10.3389/fphar.2020.577467
Received: 29 June 2020; Accepted: 03 November 2020;
Published: 11 December 2020.
Edited by:
Roberto Paganelli, University of Studies G. d’Annunzio Chieti and Pescara, ItalyReviewed by:
Francesco Sessa, University of Foggia, ItalySalvatore Pizzo, Duke University, United States
Copyright © 2020 Elfiky. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Abdo A. Elfiky, YWJkb0BzY2kuY3UuZWR1LmVn
 Abdo A. Elfiky
Abdo A. Elfiky