- 1Department of Anesthesiology, Second Affiliated Hospital of Zhejiang University School of Medicine, Hangzhou, China
- 2Institute of Pain Medicine and Special Environmental Medicine, Nantong University, Nantong, China
Nerve injury-induced gene expression change in the spinal cord is critical for neuropathic pain genesis. RNA N6-methyladenosine (m6A) modification represents an additional layer of gene regulation. We showed that spinal nerve ligation (SNL) upregulated the expression of matrix metallopeptidase 24 (MMP24) protein, but not Mmp24 mRNA, in the spinal cord neurons. Blocking the SNL-induced upregulation of spinal MMP24 attenuated local neuron sensitization, neuropathic pain development and maintenance. Conversely, mimicking MMP24 increase promoted the spinal ERK activation and produced evoked nociceptive hypersensitivity. Methylated RNA Immunoprecipitation Sequencing (MeRIP-seq) and RNA Immunoprecipitation (RIP) assay indicated the decreased m6A enrichment in the Mmp24 mRNA under neuropathic pain condition. Moreover, fat-mass and obesity-associated protein (FTO) was colocalized with MMP24 in spinal neurons and shown increased binding to the Mmp24 mRNA in the spinal cord after SNL. Overexpression or suppression of FTO correlates with promotion or inhibition of MMP24 expression in cultured spinal cord neurons. In conclusion, SNL promoted the m6A eraser FTO binding to the Mmp24 mRNA, which subsequently facilitated the translation of MMP24 in the spinal cord, and ultimately contributed to neuropathic pain genesis.
Introduction
Nerve injury-induced neuropathic pain is a refractory and debilitating disease (Mitka, 2003). Limited treatments are available for this disorder because most therapies work only symptomatically via neurotransmission blockade but ignore the underlying pain pathogenesis and its phasic progression. The development of more efficient treatment of this disease comes to a standstill due to our incomplete understanding of mechanisms underlying the neuropathic pain induction and maintenance (Ji et al., 2009). Abnormal gene expression changes in the dorsal root ganglion (DRG) and spinal cord are the crucial molecular basis for developing and maintaining neuropathic pain (Jiang et al., 2016; Pokhilko et al., 2020).
Epigenetic processes, such as DNA methylation, histone modifications, and non-coding RNAs, frequently regulate pain-related gene expression (Jiang et al., 2018; Wu et al., 2019; Lin et al., 2020). RNA methylation mediated RNA post-transcriptional modification has recently been emphasized as a significant epigenetic modification (He and He, 2021). N6-methyladenosine (m6A) is the most prevalent and dynamic modification in mRNA with a preference for the 3′-untranslated region (3′-UTR) and the transcription starting site (Dominissini et al., 2012; Meyer et al., 2012; Fu et al., 2014). m6A is installed by “writer” complex composed of methyltransferase-like 14 (METTL14), methyltransferase-like 3 (METTL3) and Wilms’ tumor 1-associating protein (WTAP), and removed by “eraser” demethylases fat-mass and obesity-associated protein (FTO) and AlkB homolog 5 (ALKBH5) (Dominissini et al., 2012; Zheng et al., 2013; Fu et al., 2014; Liu et al., 2015). This modification is recognized by m6A-binding proteins (Dominissini et al., 2012; Fu et al., 2014; Liu et al., 2015) to mediate various mRNA biogenesis, such as RNA stability, translation, splicing and export (Yang et al., 2018). In particular, recent evidence revealed that neuropathic pain could be partially attributed to the methyltransferases/demethylases-induced RNA m6A modification dysregulation of pain-associated genes in the DRG (Li et al., 2020; Albik and Tao, 2021). However, the role of spinal m6A modification in the neuropathic pain genesis remains largely unknown.
Increasing evidence suggests that proteases such as matrix metalloproteases (MMPs), caspases, and cathepsin S play a vital role in the genesis of neuropathic pain by regulating neuroinflammation in the central nervous system (Ji et al., 2009; Hannocks et al., 2019; Jiang et al., 2020a), through the cleavage of the extracellular matrix proteins, chemokines, and cytokines (Manicone and McGuire, 2008). The MMP family includes more than 20 members. Among them, MMP2 and MMP9 are involved in the regulation of neuropathic pain during the early and late phase, respectively, through the cleavage/activation of interleukin 1 beta (IL-1β) in the extracellular matrix and subsequent phosphorylation of extracellular signal-regulated kinase 1/2 [pERK1/2, a marker for spinal neuron hyper-sensitization (Zhuang et al., 2005)] (Kawasaki et al., 2008). MMP24 is abundantly expressed throughout the nervous system (Hayashita-Kinoh et al., 2001) and could degrade several extracellular matrix components, including cell-adhesion molecule N-cadherin to promote neurite outgrowth in cultured cells (Hayashita-Kinoh et al., 2001). Mouse strain lacking in MMP24 was found absent of thermal hyperalgesia with inflammation model (Folgueras et al., 2009). However, whether MMP24 in the spinal cord participates in the development or maintenance of neuropathic pain is still elusive.
Here, we report that spinal nerve ligation (SNL) leads to a significant increase in the level of MMP24 protein, but not Mmp24 mRNA, in the spinal cord. This increase contributes to the SNL-induced neuropathic pain induction and maintenance, possibly by the activation of ERK1/2. Mechanistically, SNL promoted FTO binding to the Mmp24 mRNA for the erasure of m6A enrichment and possibly accelerated the Mmp24 mRNA translation in the spinal cord.
Materials and Methods
Animals
Male C57BL/6J mice (6–8 weeks old for in vivo experiments) were purchased from SLAC Laboratory (Shanghai, CN). Rodents were hosted in a centralized location in the Second Affiliated Hospital of Zhejiang University (SAHZU), School of Medicine, with a standardized circadian cycle with 12 h of light and darkness. Mouse chow and water were provided, ad libitum. All experiments were approved by the Zhejiang Animal Care and Use Committee and the Ethics Committee of SAHZU, School of Medicine. Utmost care was taken to ensure the welfare of the rodents and kept their usage to a minimum. Behavioral experiments were undertaken with blinded investigators, with no knowledge of the viral content or other preparatory conditions.
Animal Model
L4 spinal nerve ligation (SNL) was carried out according to the methods previously described (Decosterd and Woolf, 2000; Wang and Wang, 2003; Rigaud et al., 2008). In brief, mice were anesthetized with Nembutal. The lower back was dissected until the transverse lumbar process was exposed. After the process was removed, the underneath L4 spinal nerve was ligated with a silk 6–0 thread. A slight distal location was chosen for transection around the ligation site. Subsequent layers of muscle and skin were closed. The sham groups undertook identical procedures, but without the transection or ligature of the corresponding nerve.
Behavioral Tests
Mechanical and thermal pain tests along with locomotor function assessments were performed as described in the previous studies (He et al., 2011; Zhao et al., 2013; Daskalaki et al., 2018). Each behavioral test was carried out at 1 h interval.
Paw withdrawal frequencies (PWF) were defined as a response to physical stimulation via von Frey filaments. In short, the animal was introduced to an individual Plexiglas chamber on an elevated mesh screen. Two calibrated von Frey filaments (0.07 and 0.4 g) (DanMic Global. Campbell, CA) were utilized to stimulate the hind paw for ∼1 s and the hind paw was stimulated repeatedly 10 times at 5 min interval. Paw withdrawal responses were aggregated, averaged over the number of trials, and calculated in percentage, which resulted in the PWF ((number of paw withdrawals/10 trials) × 100 = % response frequency).
A Model 336 Analgesia Meter (IITC Inc. Life Science Instruments. Woodland Hills, CA) was utilized to measure paw withdrawal latencies (PWL) to the noxious application of heat. In short, the rodent was introduced to an individual Plexiglas chamber on a glass plate. The analgesic meter's light emitter beamed through a keyhole onto the hind paw’s plantar surface, with the stimuli switched off upon paw withdrawal. The paw withdrawal latency was logged, defined as time passed between initiation of the stimuli to paw withdrawal. Five trials were undertaken with an interval of 5 min each. 20 s cut-off limit was defined to eliminate any tissue injury.
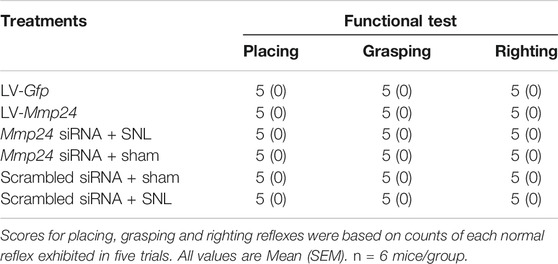
Locomotor function tests the righting, grasping, and placing reflexes after PWF and PWL. Righting reflex: rodent placed supine on a flat surface, the time it takes to upright itself to a normal position was recorded. Grasping reflex: rodent placed on a meshed wire, any contact or grasp of the wire was recorded. Placing reflex: rodent placed on the edge of a surface with the hind leg in a lower position than forelimbs, while leaving hindlimbs just off contact with the edge of a platform, and recorded whether hind paws reflexively placed on the platform. Each trial was repeated at a 5 min interval five times and the scores for each reflex were recorded on the basis of counts of each normal reflex.
Intraspinal and Intrathecal Injection
The intraspinal injection was performed as described previously (Jiang et al., 2016). In short, after anesthetized with Nembutal, mice underwent hemilaminectomy at the L1-L2 vertebral segments. The intraspinal injection was carried out ipsilaterally on the left side. By using a glass micropipette, each animal received two injections (5 × 105 TU per injection, 0.8 mm from the midline, 0.5 mm apart in rostrocaudal axis, 0.5 mm deep) of lentivirus following the L3-L4 dorsal root entry zone after exposure of spinal cord. The tip of glass micropipette should reach a depth of lamina II-IV of the spinal cord. Finally, the dorsal muscle and skin were sutured layer by layer.
Intrathecal injection of siRNA (20 μM, 10 μL) was performed daily for two to three consecutive days in sham or SNL mice. Mice were held firmly in place over the pelvic girdle and between L5 and L6 vertebrae inserted a 30-gauge needle attached to a 25 μL microsyringe. A slight flick of the tail after a sudden advancement of the needle confirmed the proper insertion of the needle into the subarachnoid space (Jiang et al., 2016). TurboFect in vivo transfection reagent (Thermo Fisher, R0541) was used to improve delivery and prevent siRNA degradation. Mmp24 siRNA1 (sense: 5′-GAG AUU CGU CUU CUU CAA ATT-3′, antisense: 5′-UUU GAA GAA GAC GAA UCU CTT-3′), Mmp24 siRNA2 (sense: 5′-GGA UAU UAC ACC UAC UUC UTT-3′, antisense: 5′-AGA AGU AGG UGU AAU AUC CTT-3′), Mmp24 siRNA3 (sense: 5′-CUA UCU UCC AAU UCA AGA ATT-3′, antisense: 5′-UUC UUG AAU UGG AAG AUA GTT-3′).
Spinal Dorsal Horn Neuron Culture and Viral Transduction
Primary spinal neuronal cultures were prepared from 1 to 2 week old C57BL/6J mice using a procedure modified from our previously described method (Jiang et al., 2016). In short, a laminectomy was conducted and the spinal cord was carefully removed after decapitation. Superficial dorsal horn was isolated and then cut into several strips. The strips were incubated at 37°C for 45 min in Hanks’ balanced salt solution (HBSS, Invitrogen) containing papain (15 U/ml, Worthington Biochemical), then rinsed 3 times with HBSS, and placed in mixed Neurobasal Medium (Invitrogen) containing 5% fetal bovine serum (FBS, Invitrogen), heat-inactivated horse serum (5%, Invitrogen), B-27 (1%, Invitrogen), L-glutamax-1 (2 mM, Invitrogen), streptomycin (100 μg/ml, Invitrogen) and penicillin (100 U/ml, Invitrogen). A Fire-polished Pasteur pipette was used to dissociate fragments by gentle trituration mechanically. The resulting cell suspension was plated onto 6-well plates coated with poly-d-lysine and collagen. The cells were incubated at 37°C with 95% O2, 5% CO2. After the neurons were treated with cytosine arabinoside, 2–10 μL virus (titer ≥1 × 1013) was added to each well. The neurons were collected 3 days after virus transduction.
Reverse Transcription (RT)-PCR
Total RNA from the cultured samples or tissue was extracted and purified using miRNeasy kit with genome DNA Eliminator Columns (QIAGEN, Germany). The SuperScript™ First-Strand Synthesis System (Invitrogen/Thermo Fisher) was then used to reverse-transcribe RNA. Each sample was run in a 20 μL reaction with 20 ng cDNA, 10 µL SsoAdvanced Universal SYBR Green Supermix (Bio-Rad Laboratories, CA) and 250 nM forward and reverse primers. Tuba1a was used as an internal control for normalization. Real-time PCR was performed on the Applied Biosystems QuantStudio5 system (Applied Biosystems, CA). ΔCt method (2−ΔΔCt) was applied for mRNA levels calculation. All data were normalized to Tuba1a, which has been demonstrated to be stable even after peripheral nerve injury insult (Zhao et al., 2013; Wu et al., 2016). All the primers are listed in Table 1.
Plasmids Construction and Virus Production
The Mmp24 and Fto coding sequences were synthesized by Tsingke Biological Technology (Beijing, CN), and were further inserted into pro-viral plasmids pLV-CMV-MCS-Ubi-ZSGreen and pAAV-CMV-MCS-F2A-EGFP, respectively, using the Seamless Cloning and Assembly Kit (SunBio, CN). pLV-CMV-ZSGreen or pAAV-CMV-EGFP was used as the corresponding control (SunBio, CN). Lentiviruses and adeno-associated viruses packaging were completed by the Tsingke Biological Technology (Beijing, CN) and Sunbio Technology (Shanghai, CN). The Fto coding region's shRNA (GenBank accession number NM_011936.2) was designed to target the sequence 5′-GTC TCG TTG AAA TCC TTT GAT-3′. A scramble shRNA was used as control (5′-TTC TCC GAA CGT GTC ACG T-3′). Both Fto and scrambled shRNA oligonucleotides were inserted to pAAV-CAG-EGFP-U6-shRNA and packaged into adeno-associated virus by SunBio. All the constructs were sequenced to prove sequence integrity.
Immunohistochemistry
After anesthetized with Nembutal, mice were perfused with cold PBS and 4% paraformaldehyde through the ascending aorta. Following perfusion, the L4 spinal cord segments were removed, post-fixed in the same fixative at 4°C overnight and dehydrated. Spinal cord sections (25 μm) were cut in a cryostat and processed for immunofluorescence. The sections were blocked for 1 h at room temperature in PBS containing 5% goat or donkey serum and then incubated overnight at 4°C with the following primary antibodies: mouse anti-FTO (1:100, Abcam), rabbit anti-MMP24 (1:100, Affinity), mouse anti-NeuN (1:500, Abcam), rabbit anti-NeuN (1:500, Abcam), mouse anti-GFAP (1:500, EMD Millipore), rabbit anti-GFAP (1:500, Abcam), mouse anti-IBA1 (1:200, Sigma), rabbit anti-IBA1 (1:200, Wako). After wash, the sections were then incubated with corresponding second antibodies with either Alexa FluorTM 488- or Alexa FluorTM 594-labeled (1:500, Invitrogen) at room temperature for 1 h. 4′, 6-diamidino-2-phenylindole (DAPI) (Abcam) was finally utilized for slides mounting. Leica DMI4000 fluorescence microscope was used for immunofluorescence-labeled image examination (Leica, Germany).
Western Blotting
Cytosolic and nuclear proteins were extracted from the L4 spinal cord as previous study (Zhao et al., 2017). Ice-cold lysis buffer was utilized for the tissue homogenization, which contained 10 mM Tris, 5 mM EGTA, 2 mM MgCl2, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride, and 40 μM leupeptin. After centrifugation (4°C, 15 min, 1,500 g), the supernatants were gathered for cytosolic proteins and the pellets for nuclear proteins. The protein sample concentration was measured using Detergent Compatible Bradford Protein Assay Kit (Beyotime, CN). After heated at 99°C for 5 min, the samples were loaded onto an SDS-polyacrylamide gel (Genshare Biology, CN) and then electrophoretically transferred onto a polyvinylidene fluoride membrane (Millipore, Burlington, MA, United States). The membranes were blocked with 5% nonfat milk TBST for 1 h and then incubated overnight with the following antibodies: mouse anti-FTO antibody (1:1,000, Abcam), rabbit anti-METTL3 antibody (1:1,000, Abcam), rabbit anti-METTL14 antibody (1:1,000, Proteintech), rabbit anti-ALKBH5 antibody (1:1,000, Abcam), mouse anti-WTAP antibody (1:100, Santa Cruz), rabbit anti-MMP24 antibody (1:1,000, Affinity), mouse anti-histone H3 (1:3,000, Santa Cruz), mouse anti-GAPDH (1:1,000, Zhongshan Golden Bridge Bio-technology), rabbit anti-phospho-ERK1/2 (Thr202/Thy204, 1:2,000, CST), rabbit anti-ERK1/2 (1:2,000, CST). Horseradish peroxidase-conjugated anti-mouse or rabbit secondary antibody (1:5,000, Jackson ImmunoResearch) were utilized for protein detection and Clarity Western ECL Substrate (EMD Millipore) for visualization and ChemiDoc XRS + System (Bio-Rad) for exposure. NIH ImageJ software was utilized for quantification of the intensity of blots with densitometry. After each was normalized to the corresponding GAPDH or histone H3 (for nucleus proteins), the relative density values of the treated groups or different time points were determined by dividing the optical density values from these groups by the average value of the naïve/control groups.
RNA Immunoprecipitation Assay
Magna RIP Kit (EMD Millipore, Darmstadt, Germany) was used for the RNA Immunoprecipitation (RIP) assay. The spinal cord homogenates were suspended in the RIP lysis buffer containing RNase inhibitor and protease inhibitor cocktail. The RIP lysate was incubated on ice for 5 min and kept in −80°C. Use the RIP wash buffer to wash Magnetic Beads Protein A/G suspension for each IP. Magnetic Beads Protein A/G re-suspended in RIP wash buffer were incubated with Mouse anti-FTO antibody (4 μg; Santa Cruz), mouse anti-m6A antibody (4 μg; Abcam), or normal mouse IgG for 30 min at room temperature. The Beads Protein A/G-antibody complexes were re-suspended into the RIP immunoprecipitation buffer after being washed three times with RIP wash buffer. The RIP lysate supernatants were incubated with beads-antibody complex in the RIP buffer overnight at 4°C by rotating after being thawed and centrifuged at 14,000 rpm for 10 min at 4°C. RIP wash buffer was used to wash the samples six times. After incubating the beads in the proteinase K buffer at 55°C for 30 min by shaking, the RNA was eluted and purified by phenol/chloroform extraction from the beads. The RNA enrichment was analyzed by quantitative real-time PCR. The RIP lysate supernatant was used as input. All primers are listed in Table 1.
Methylated RNA Immunoprecipitation Sequencing (MeRIP-Seq) and MeRIP-Seq Libraries
Following the manufacturer’s procedure, Trizol reagent (Invitrogen, CA, United States) was used to extract total RNA. Bioanalyzer 2,100 and RNA 6,000 Nano LabChip Kit (Agilent, CA, United States) were utilized to analyze the total RNA quality and quantity with RIN number >7.0. To isolate Poly (A) mRNA, nearly more than 200 ug of total RNA were gathered and mixed with poly-T oligo attached magnetic beads (Invitrogen). After purification, divalent cations were applied to fragment the poly(A) mRNA fractions into 50–150 nt oligonucleotides under elevated temperature. The cleaved RNA fragments were then incubated in IP buffer (0.5% Igepal CA-630, 50 mM Tris-HCl and 750 mM NaCl) supplemented with BSA (0.5 μg/μL) for 2 h at 4°C with m6A-specific antibody (No.202003, Synaptic Systems, Germany). The mixture was then incubated together with protein-A beads and eluted with elution buffer (6.7 mM m6A and 1 × IP buffer). 75% ethanol was used to precipitate the eluted RNA. In conformity to a strand-specific library preparation by dUTP method, untreated input control fragments and eluted m6A-containing fragments (IP) are converted to the final cDNA library. The average insert size for the paired-end libraries was ∼100 bp. Following the vendor's recommended protocol, the paired-end 2 × 100 bp sequencing was then carried out on an Illumina Novaseq™ 6,000 platform at the LC-BIO Bio-tech Ltd. (Hangzhou, CN).
Firstly, to remove the reads that contained adaptor contamination, undetermined bases and low-quality bases, in-house Perl scripts and Cutadapt (Kechin et al., 2017) were applied. By using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/), the sequence quality was further validated. To map reads, bowtie (Langmead and Salzberg, 2012) were utilized to reference genome with default parameters. Mapped reads are then provided as input for MACS2 (Zhang et al., 2008), which helps to identify m6A peaks that can be adapted for visualization on the UCSC genome browser. De novo motif finding is carried out by using MEME (Bailey et al., 2009), followed by localization of the motif in respect of peak summit by in-house Perl scripts. By using ChIPseeker (Yu et al., 2015), called peaks are annotated by the intersection with gene architecture. MeRIP-seq data files were submitted to the GEO repository through the access code: GSE171004.
Statistical Analysis
The cells were suspended evenly and dispersed randomly in each well for in vitro trials. The animals were randomly distributed into various treatment groups for in vivo experiments. All of the results were specified as mean ± SEM. For the biochemical results, two-tailed, unpaired Student's t-test (two groups) and one-way ANOVA (> 2 groups) were utilized for the data analyses. For the behavioral results, two-way ANOVA was used for the data analyses. Pairwise comparisons between means were tested by the post hoc Tukey method (GraphPad Prism 8) when ANOVA showed significant differences. The detailed analyzed process utilized in each experiment was elaborated in the matching figure legends. The sample sizes were determined based on the previous reports, pilot studies in the field and power analyses (power of 0.90 at p < 0.05) (Ferreira et al., 2001; Zhao et al., 2013; Millard et al., 2016; Daskalaki et al., 2018; Sato et al., 2018). Significance was set at p < 0.05.
Results
MMP24 Protein Upregulation in the Spinal Cord after SNL
We first examined whether the MMP24 enrichment was altered in the spinal cord after SNL, a preclinical animal model that mimics nerve injury-induced neuropathic pain in the clinical setting (Rigaud et al., 2008). Unilateral SNL did not alter the level of Mmp24 mRNA in the spinal cord from day 3 to 14 post-SNL (Figure 1A). However, the expression of MMP24 protein was significantly increased in a time-dependent manner in the spinal cord after SNL (Figure 1B), but not after sham surgery (Figure 1C). Immunostaining further confirmed that the MMP24 protein was dramatically increased in the ipsilateral dorsal horn, but not in the contralateral dorsal horn, on day 7 post-SNL (Figures 1D–F). The distribution of MMP24 in the spinal dorsal horn was also checked by double immunostaining with different cell markers. As shown in Figures 1G–K, MMP24 was predominantly colocalized with the neuronal-specific nuclear protein (NeuN) (Figures 1G–I), and rarely with the glial fibrillary acidic protein (GFAP) (Figure 1J), or ionized calcium-binding adaptor molecule (IBA-1) (Figure 1K) in the spinal dorsal horn on day 14 after SNL. The above evidence suggests that the increased MMP24 protein in the spinal dorsal horn may involve neuropathic pain.

FIGURE 1. MMP24 protein is increased in the spinal cord after SNL (A). Expression of Mmp24 mRNA after SNL. n = 4 mice/group/time point. (B). Expression of MMP24 protein in the spinal cord after SNL surgery. n = 3–6 mice/time point. One-way ANOVA followed by post hoc Tukey test. Ftime (3, 8) = 29.00. *p < 0.05, **p < 0.01 vs. the corresponding control group (0 day). (C). Expression of MMP24 protein in the spinal cord after sham surgery. n = 3 mice/time point. (D–F). Representative images of MMP24 immunofluorescence in the L4 dorsal horn. MMP24 immunoreactivity was low in naïve mice, but increased in the ipsilateral dorsal horn compared with the contralateral dorsal horn 7 days after SNL. Scale bar: 50 μm; 25 μm (insets). (G–K). Double staining of MMP24 and markers of neuron, astrocytes, and microglia on day 14 after SNL. Scale bar: 50 μm.
Blocking Spinal MMP24 Increase Attenuates Neuropathic Pain Development and Maintenance
Does the increased MMP24 in the spinal cord participate in nerve injury-induced pain hypersensitivity? To this end, we first examined the effect of blocking spinal MMP24 increase on the development of SNL-induced pain hypersensitivity. As shown in Figure 2A, siRNA-2 displayed the best knockdown effect compared to the scrambled siRNA (Scram) in the cultured spinal cord cells. Thus, siRNA-2 (Mmp24 siRNA) and its control scrambled siRNA were intrathecally injected into the sham or SNL mice. Intrathecal injection of Mmp24 siRNA attenuated SNL-induced mechanical allodynia as demonstrated by a decrease in PWF to mechanical stimuli and ameliorated SNL-induced thermal hyperalgesia as indicated by the increase in PWL to heat stimulation from day 3 to 5 compared to the scrambled siRNA-treated SNL mice (Figures 2B–D). No changes were observed in basal mechanical and heat responses in sham mice following injection with either siRNA (Figures 2B–D). Moreover, we also observed the role of MMP24 in the maintenance phase of neuropathic pain through intrathecal injection of Mmp24 siRNA or scrambled siRNA into the sham or SNL mice 7 days after SNL. Consistently, blunted mechanical allodynia and heat hyperalgesia were observed on days 9 and 10 after SNL from the Mmp24 siRNA-treated mice compared to the scrambled siRNA-treated mice (Figures 2E–G). The basal pain behavior of the sham mice (Figures 2E–G) and the locomotor functions of the siRNA-treated mice were not affected (Table 2). As expected, a noticeable decrease in the amount of MMP24 protein was detected in the spinal cord from the Mmp24 siRNA-treated mice compared with the scrambled siRNA-treated mice on day 10 after SNL (Figure 2H). Consistently, intrathecal injection of Mmp24 siRNA dampened the SNL-induced spinal neuronal sensitization as indicated by abolishing the SNL-induced increase in pERK1/2 in the spinal cord (Figure 2I). Together, the results described above demonstrate that spinal MMP24 is necessary for SNL-induced central sensitization and pain hypersensitivities.
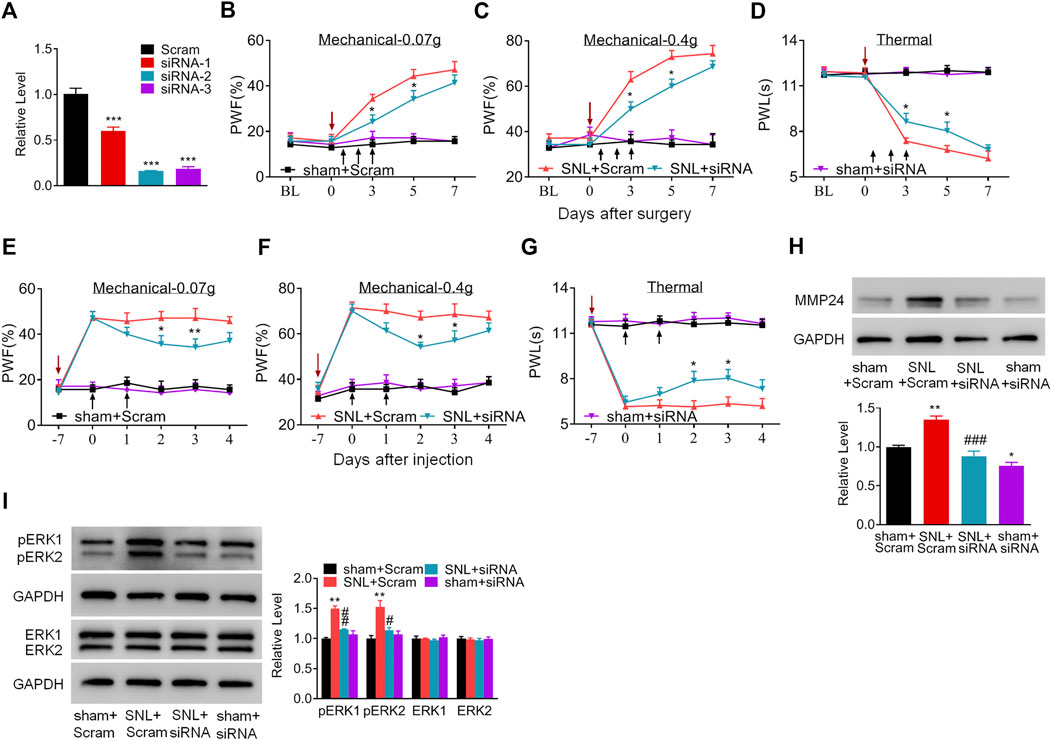
FIGURE 2. Blocking the increased spinal MMP24 attenuates SNL-induced pain hyperalgesia (A). Effect of three Mmp24 siRNAs on the expression of Mmp24 mRNA, among which siRNA-2 shows the best knockdown effect in the cultured spinal cord cells. n = 3 repeats, One-way ANOVA followed by post hoc Tukey test. Fgroup (3, 8) = 92.87, ***p < 0.001 vs. scrambled siRNA (Scram) group. (B–D). Effect of intrathecal injection of Mmp24 siRNA (siRNA) or control scrambled siRNA (Scram) into the sham or SNL mice on the paw withdrawal responses to mechanical (B, C) and heat stimuli (D) from day 3 to 7 after SNL as indicated. n = 7 mice/group. Two-way RM ANOVA followed by post hoc Tukey test. Fgroup (3, 90) = 48.41 for (B), Fgroup (3, 90) = 58.89 for (C), Fgroup (3, 90) = 120.0 for (D). *p < 0.05 vs. the SNL plus Scram group. The red arrow indicates surgery day and black arrow indicates siRNA intrathecal injection. (E–G). Effect of intrathecal injection of Mmp24 siRNA (siRNA) or control scrambled siRNA (Scram) into the sham or SNL mice on the paw withdrawal responses to mechanical (E, F) and heat stimuli (G) 7 days after SNL as indicated. n = 7 mice/group. Two-way RM ANOVA followed by post hoc Tukey test. Fgroup (3, 90) = 53.66 for (E), Fgroup (3, 90) = 61.53 for (F), Fgroup (3, 90) = 136.7 for (G). *p < 0.05, **p < 0.01 vs. the SNL plus Scram group. The red arrow indicates surgery day and black arrow indicates siRNA intrathecal injection. (H). MMP24 protein expression in the spinal cord of sham or SNL mice intrathecally injected with Mmp24 siRNA (siRNA) or scrambled siRNA (Scram) on day 10 after SNL (2 days after the last siRNA injection). n = 3 mice/group. One-way ANOVA followed by post hoc Tukey test. Fgroup (3, 8) = 29.00, *p < 0.05, **p < 0.01 vs. sham plus Scram group. ###p < 0.001 vs. SNL plus Scram group. (I). Effect of Mmp24 siRNA or scrambled siRNA on SNL-induced increase in the phosphorylation of ERK1/2 in the spinal cord on day 10 after SNL or sham surgery (2 days after the last siRNA injection). n = 3 mice/group. One-way ANOVA followed by post hoc Tukey test. Fgroup (3, 8) = 29.00 for pERK1, Fgroup (3, 8) = 11.72 for pERK2. **p < 0.01 vs. the sham plus Scram group; #p < 0.05, ##p < 0.01 vs. the SNL plus Scram group.
MMP24 Overexpression Causes Neuropathic Pain-like Symptoms
We then asked whether the increased MMP24 in the spinal cord would be sufficient to induce neuropathic pain? To this end, we performed intraspinal lentivirus (LV) injection that expressed full-length Mmp24 (LV-Mmp24) into naïve mice. As shown in Figures 3A–C, LV-Mmp24, but not its control LV-Gfp, induced mechanical allodynia as indicated by an increase in PWF to mechanical stimulation and heat hyperalgesia as demonstrated by a decrease in PWL to heat stimulation from day 3 to 12 post-injection. The locomotor functions were not affected after viral injection (Table 2). Expectedly, the level of MMP24 protein in the L4 spinal cord was significantly increased 12 days after intraspinal injection with LV-Mmp24 compared to that of LV-Gfp (Figure 3D). These behavioral observations were further supported by the following evidence of spinal dorsal horn central sensitization. The level of pERK1/2 was markedly elevated in the L4 spinal dorsal horn on day 12 after intraspinal injection with LV-Mmp24 compared to that of LV-Gfp (Figure 3E).
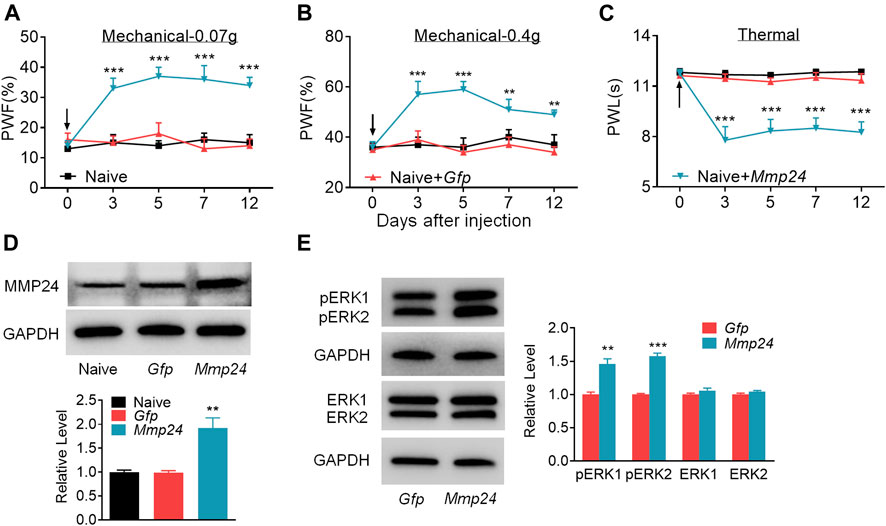
FIGURE 3. Spinal MMP24 overexpression triggers pain hyperalgesia (A–C). Effect of intraspinal injection of LV-Mmp24 (Mmp24) or LV-Gfp (Gfp) into the naïve mice on the paw withdrawal responses to mechanical (A, B) and heat stimuli (C) on day 3, 5, 7, and 12 after injection. n = 10 mice/group. Two-way RM ANOVA followed by post hoc Tukey test. Fgroup (2, 90) = 56.05 for (A), Fgroup (2, 90) = 29.72 for (B), Fgroup (2, 90) = 69.17 for (C). **p < 0.01, ***p < 0.001 vs. the Naive plus Gfp group. Black arrow indicates intraspinal virus injection. (D). MMP24 protein expression in the spinal cord of naïve mice 12 days after intraspinal injection with LV-Mmp24 (Mmp24) or LV-Gfp (Gfp). n = 3 mice/group. One-way ANOVA followed by post hoc Tukey test. Fgroup (2, 6) = 16.82, **p < 0.01 vs. Gfp group. (E). pERK1/2 and ERK1/2 expression in the spinal cord of naïve mice 12 days after intraspinal injection with LV-Mmp24 (Mmp24) or LV-Gfp (Gfp). n = 3 mice/group. **p < 0.01, ***p < 0.001 vs. the Gfp group by two-tailed unpaired Student’s t-test.
m6A Modification in the Spinal Mmp24 mRNA Was Decreased under Neuropathic Pain Condition
Although Mmp24 mRNA remained unchanged after SNL, the MMP24 protein was significantly increased in the spinal cord (Figures 1A,B), suggesting that the translation efficiency of spinal Mmp24 mRNA may be elevated under neuropathic pain condition. Of note, RNA m6A modification is known to play an important role in mRNA translation (He and He, 2021). To this end, we carried out methylated RNA immunoprecipitation sequencing (MeRIP-seq) assay to observe the changes of m6A sites across the transcriptome in the spinal cord on day 7 after peripheral nerve injury (Figure 4A). Consistent with previous studies in the DRG (Li et al., 2020), m6A sites change after nerve injury occurred predominantly at the 3′-UTR (55.09%) and to lesser extents at coding regions (29.61%), and 5′-UTR (15.3%) (Figures 4B,C). Approximately 55.6% (1,910/3,437) transcripts exhibited a loss of m6A sites, 33.9% (1,165/3,437) transcripts exhibited a gain of m6A sites, and 10.5% (362/3,437) transcripts exhibited both a loss and gain of m6A sites compared to the sham group (Figures 4B,C). Notably, Mmp24 mRNA exhibited a considerable loss of m6A sites at the 3′-UTR (Figure 4D). We then performed RIP-PCR to validate the MeRIP-seq results. The immunoprecipitation identified the m6A enrichment of the Mmp24 mRNA fragments with anti-m6A in the spinal cord (Figure 4E). However, the activity of immunoprecipitation from the spinal cord on day 7 post-SNL was significantly decreased compared to the sham group (Figure 4E). In summary, we found that m6A modification in the Mmp24 mRNA was decreased in the spinal cord under neuropathic pain conditions, suggesting the role of m6A modification in regulating MMP24 translation.
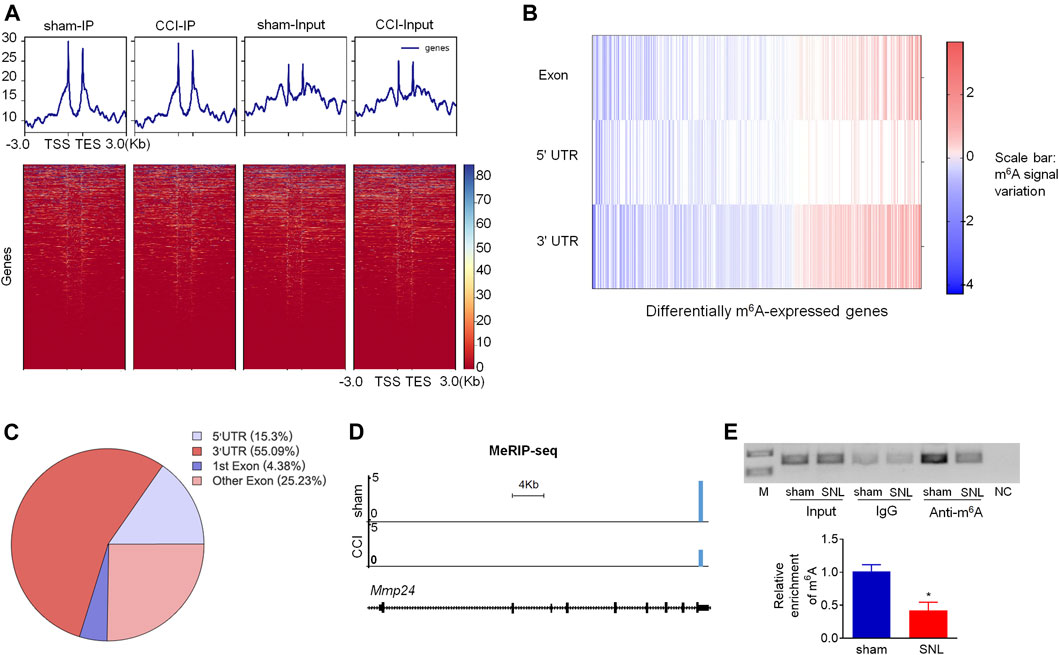
FIGURE 4. m6A modification in spinal Mmp24 mRNA is decreased after SNL (A). Heatmap of the MeRIP-seq signals from the spinal cord after chronic constriction injury (CCI) or sham surgery. Mean-normalized MeRIP-seq densities of equal bins along the gene. 3-kb region flanking the TSS or the TES are plotted. Red to blue color gradient of the heatmap represents the relative m6A level. (B–C). Dynamic changes (B) and distribution (C) of m6A sites across transcripts from the spinal cord on day 7 after CCI or sham surgery. (D). MeRIP-seq assay showed the example tracks of Mmp24 mRNA from the spinal cord on day 7 after CCI or sham surgery. m6A sites (blue) are indicated at the 3′-UTR. (E). Level of Mmp24 mRNA fragments immunoprecipitated by mouse anti-m6A in the spinal cord on day 7 after SNL or sham surgery. Input: total purified fragment. n = 4 biological repeats (4 mice/repeat). M: ladder marker. NC: negative control (H2O). *p < 0.05 vs. the sham group by two-tailed unpaired Student's t-test.
m6A Modification-Related Genes Expression in the Spinal Cord after SNL
To determine the specific regulators responsible for the m6A modification in the Mmp24 mRNA, we first examined the expression of m6A modification-related methyltransferases, including METTL3, METTL14, and WTAP, and demethylases, including FTO and ALKBH5 in the spinal cord under neuropathic pain condition. Unexpectedly, none of Mettl3, Mettl14, Wtap, Fto, and Alkbh5 mRNAs showed an obvious change in the spinal cord from day 3 to 14 after SNL (Figure 5A). To further check the nucleus and cytoplasm distribution of the protein above, we extracted the nucleus protein of the spinal cord and found that none of the METTL3, METTL14, WTAP, FTO, and ALKBH5 protein exhibited a significant change from day 3 to 14 after SNL (Figures 5B,C). FTO was recently reported to play a vital role in neuropathic pain genesis by diminishing the m6A enrichment in pain-related mRNAs in the DRG neurons (Li et al., 2020). Furthermore, FTO expression was found enriched in the spinal cord relative to that of DRG or cortex (Li et al., 2020), and higher than the other demethylase ALKBH5 in the spinal cord (Figure 5D), suggesting the possible role of spinal FTO in neuropathic pain genesis. Likewise, as shown in Figures 5E–I, FTO in the spinal dorsal horn was also expressed in the neurons but not in microglia or astrocytes. Therefore, we hypothesize that FTO may be accountable for the m6A modification of Mmp24 mRNA in the spinal neurons.
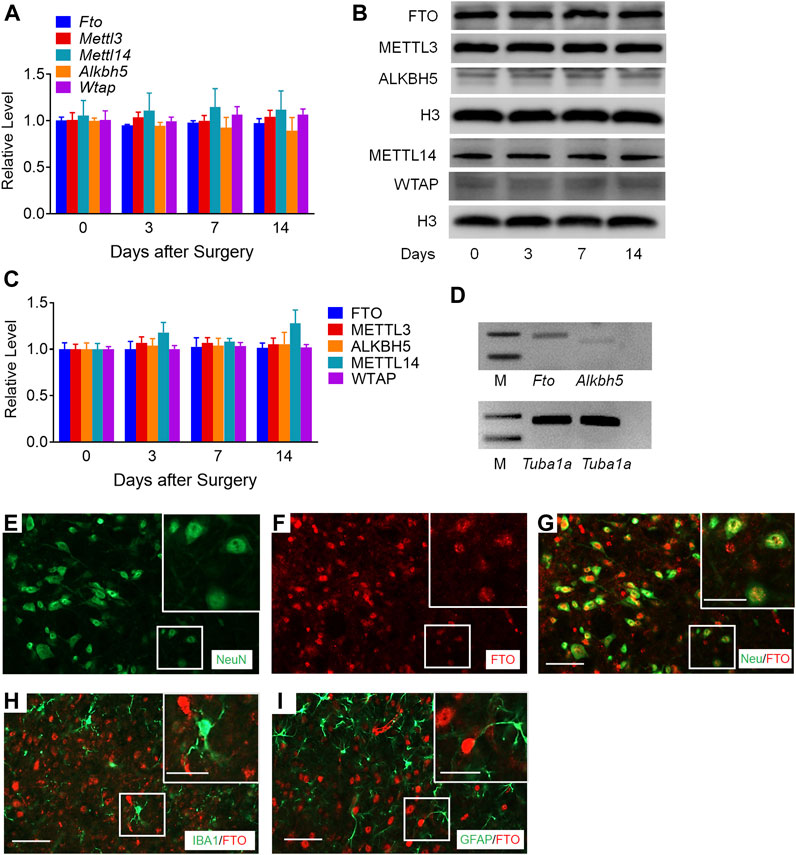
FIGURE 5. m6A-related genes expression in the spinal cord after SNL (A). Expression of Fto, Mettl3, Alkbh5, Mettl14, and Wtap mRNAs in the spinal cord from day 3 to 14 after SNL. n = 3–5 mice/group/time point. (B–C). Expression of nuclear FTO, METTL3, ALKBH5, METTL14 and WTAP proteins in the spinal cord from day 3 to 14 after SNL. n = 7–13 mice/group/time point. (D). The relative expression of Fto and Alkbh5 mRNA in the spinal cord. n = 3 repeats. M: ladder marker. (E–I). Representative images of double staining between FTO and markers of neuron, astrocytes and microglia in the spinal dorsal horn. Scale bar: 50 μm; 25 μm (insets).
Spinal FTO Is Responsible for the Decreased m6A Enrichment and Increased Translation of MMP24 after SNL in the Spinal Cord Neurons
To demonstrate the role of FTO in the regulation of MMP24, we first checked the distribution pattern of FTO and MMP24. Double immunostaining showed the colocalization of FTO and MMP24 in the spinal dorsal horn (Figures 6A–C). Moreover, the RIP assay further revealed that FTO could bind to the Mmp24 mRNA from the spinal cord (Figure 6D), and there was a significant elevation in the binding activity from the spinal cord 7 days after SNL (Figure 6D). It indicates that the SNL-induced decrease in the m6A enrichment in Mmp24 mRNA may be due to the increased FTO occupation in the Mmp24 mRNA after SNL.
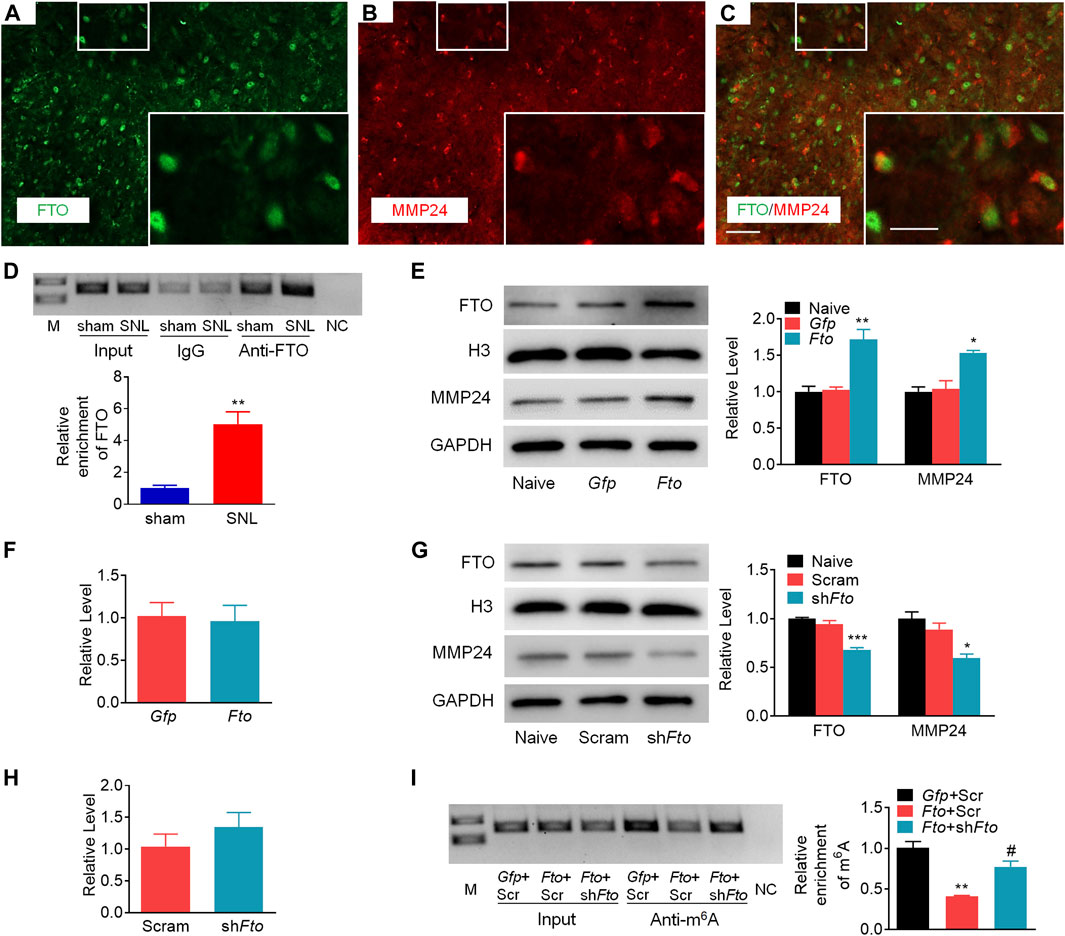
FIGURE 6. FTO promotes the expression of MMP24 in the spinal neurons (A–C). Representative images of the colocalization between FTO and MMP24 in the spinal dorsal horn. Scale bar: 50 μm; 25 μm (insets). (D). Level of Mmp24 mRNA fragments immunoprecipitated by mouse anti-FTO in the spinal cord on day 7 after SNL or sham surgery. Input: total purified fragment. n = 3 biological repeats (4 mice/repeat). M: ladder marker. NC: negative control (H2O). **p < 0.01 vs. the sham group by two-tailed unpaired Student’s t-test. (E). Expression of FTO and MMP24 in the cultured spinal neurons on day 3 after transduction with AAV5-Gfp (Gfp) or AAV5-Fto (Fto). n = 3 repeats. One-way ANOVA followed by post hoc Tukey test. Fgroup (2, 6) = 19.98 for FTO, Fgroup (2, 6) = 14.37 for MMP24. *p < 0.05, **p < 0.01 vs. the Gfp group. (F). Expression of Mmp24 mRNA in the cultured spinal neurons on day 3 after transduction with AAV5-Gfp (Gfp) or AAV5-Fto (Fto). n = 3 repeats (G). Expression of FTO and MMP24 in the cultured spinal neurons on day 3 after transduction with AAV5-Fto shRNA (shFto) or AAV5-scrambled shRNA (Scram). n = 3 repeats. One-way ANOVA followed by post hoc Tukey test. Fgroup (2, 6) = 46.71 for FTO, Fgroup (2, 6) = 12.07 for MMP24. *p < 0.05, ***p < 0.001 vs. the Scram group. (H). Expression of Mmp24 mRNA in the cultured spinal neurons on day 3 after transduction with AAV5-Fto shRNA (shFto) or AAV5-scrambled shRNA (Scram). n = 3 repeats (I). Enrichment of Mmp24 mRNA fragments immunoprecipitated by anti-m6A from the cultured spinal neurons transduced viruses as shown. Gfp: AAV5-Gfp; Scr: AAV5-scrambled shRNA; Fto: AAV5-Fto; shFto: AAV5-Fto shRNA. n = 3 biological repeats/group. One-way ANOVA followed by post hoc Tukey test. Fgroup (2, 6) = 24.41. **p < 0.01 vs. the Gfp plus Scr group. #p < 0.05 vs. the Fto plus Scr group.
We further examined the effect of FTO on the expression of MMP24 in the cultured spinal neurons. The expression of FTO was first verified 3 days after transduction with AAV5 that expressed full-length Fto (AAV5-Fto) or Gfp (AAV5-Gfp) (Figure 6E). A significant increase in the level of MMP24 protein was observed 3 days after transduction with AAV5-Fto compared to the AAV5-Gfp-treated group (Figure 6E). Interestingly, the level of Mmp24 mRNA was not altered on day 3 after transduction with AAV5-Fto compared to AAV5-Gfp (Figure 6F). We further checked the effect of FTO knockdown on the expression of MMP24 in the cultured spinal neurons through the transduction of AAV5 that expressed Fto shRNA (AAV5-Fto shRNA) or control shRNA (AAV5-scrambled shRNA). The knockdown effect of FTO was confirmed 3 days after transduction of AAV5-Fto shRNA (Figure 6G). As expected, the MMP24 protein was markedly downregulated 3 days after transduction with AAV5-Fto shRNA compared with that of AAV5-scrambled shRNA (Figure 6G). However, the level of Mmp24 mRNA was not significantly altered in AAV5-Fto shRNA-treated group compared to the AAV5-scrambled shRNA-treated group (Figure 6H), suggesting that FTO may promote the translation of Mmp24 mRNA in the spinal neurons. We further transduced AAV5-Fto into cultured spinal neurons and found that FTO overexpression produced a marked loss of m6A enrichment in the Mmp24 mRNA compared to the control group (Figure 6I). This decrease was reversed by blocking FTO overexpression in the cultured spinal neurons co-transduced with AAV5-Fto shRNA (Figure 6I). Given the role of m6A modification in mRNA translation (Jia et al., 2019; Zhang M. et al., 2020; Zhang Z. et al., 2020; Liu et al., 2020; Song et al., 2020), the evidence described above indicates that FTO likely erased the m6A enrichment in Mmp24 mRNA to promote the translation of Mmp24 mRNA in the spinal cord neurons under neuropathic pain condition.
Discussion
This study demonstrates that SNL results in an accelerated translation of Mmp24 mRNA in the spinal cord. Increased spinal MMP24 contributes to the SNL-induced central sensitization and nociceptive hyperalgesia. Mechanistically, SNL leads to an elevation of FTO occupation in the Mmp24 mRNA. Increased occupation of FTO contributes to reducing m6A enrichment in Mmp24 mRNA in the spinal neurons after SNL, which subsequently accelerates the translation of MMP24 and eventually contributes to neuropathic pain by activating ERK (Figure 7).
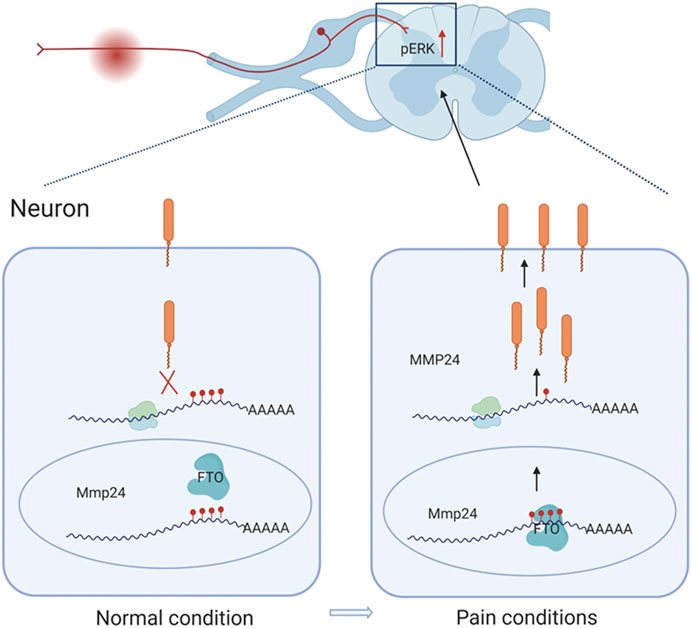
FIGURE 7. Schematic representation of the mechanism of MMP24 participation in neuropathic pain. Black arrows depict promotion. Red arrows represent upregulation. Never injury can facilitate the binding of FTO to Mmp24 mRNA in spinal neurocytes nuclei and subsequently lead to the reduced level of m6A modification followed by the enhanced translation of its mRNA. Increased MMP24 can facilitate ERK phosphorylation in the spinal cord and induce neuropathic pain.
MMP24 was explicitly detected in neurons of both the central and peripheral nervous systems (Hayashita-Kinoh et al., 2001; Sekine-Aizawa et al., 2001) and identified as a brain-specific MMP (Llano et al., 1999; Wang and Pei, 2001). The present study showed that MMP24 was mainly expressed in the spinal neurons as it was co-expressed with NeuN-labeled individual neurons. Meanwhile, our results also supported the existence of MMP24 in the spinal astrocytes and microglia after SNL even though at a low abundance. Besides, the morphologic observation of the MMP24-labeled cells proved the presence of MMP24 in the matrix of the dorsal horn, supporting the role of MMP24, as an extracellular matrix metalloproteinase, in regulating cleavage of the cell-cell adhesion molecule N-cadherin to influence the neuronal circuit formation and plasticity (Komori et al., 2004; Folgueras et al., 2009).
Previous studies showed that mice with genetically ablated MMP24 exhibited a lowered severity of mechanical allodynia after partial sciatic nerve injury or spinal cord transection (Komori et al., 2004) through the lowered degradation of chondroitin sulfate proteoglycans, which are abundant in neuronal tissues and inhibit neurite outgrowth (Dou and Levine, 1994). Deletion of Mmp24 also relieved thermal pain after the inflammation model through increased interaction between mast cells and nociceptive neurites (Folgueras et al., 2009). Our study further demonstrated the involvement of spinal MMP24 in the different phases of neuropathic pain. Specifically, blocking the SNL-induced increase in MMP24 in the spinal cord attenuates both the mechanical allodynia and heat hyperalgesia in both development and maintenance phases, accompanied by lessened activation ERK1/2. Moreover, intraspinal specific overexpression of MMP24 by LV persistently induced both the mechanical allodynia and heat hyperalgesia with the activation of ERK1/2 absence of SNL. MMP24 was reported to show the ability to proteolytically activate MMP2 (Llano et al., 1999), which could cleave pro-IL-1β to activate ERK in the spinal cord (Kawasaki et al., 2008). Furthermore, MMP24 was required for the inflammatory response to TNF-α and IL-1β in the peripheral nervous system (Folgueras et al., 2009). Thus, MMP24 is likely to activate ERK through inflammatory mediators like IL-1β and TNF-α in the spinal cord. ERK1/2 phosphorylation has been widely considered in recent years as a spinal neuron sensitization marker (Zhuang et al., 2005; Gu et al., 2018). Its activation could subsequently induce the expression and release of various downstream pain-related genes like Cxcl10, Ccl2, and Ccl7, or affect neuronal excitability of the central neurons, and eventually trigger the pain behaviors (Jiang et al., 2020a; Jiang et al., 2020b).
Mounting evidence indicates that epigenetic alterations play a critical role in neuropathic pain induction and maintenance, including histone acetylation/methylation, DNA methylation and non-coding RNAs (Penas and Navarro, 2018). Spinal coactivator-associated arginine methyltransferase 1 (CARM1) regulated histone methylation of the potassium channel promoter to participate in neuropathic pain development (Hsieh et al., 2021). Also, spinal ten-eleven translocation methylcytosine dioxygenase 1 (Tet1) decreased the DNA methylation state in metabotropic glutamate receptor subtype 5 (mGluR5) promoter to mediate spinal plasticity and pain hypersensitivity (Hsieh et al., 2017). Epigenetic modification at the transcriptional level has achieved much progress in neuropathic pain genesis these years, while the translational level epigenetic regulation is still incomplete. Interestingly, the current study demonstrates that although the level of Mmp24 mRNA was not altered in SNL, the expression of MMP24 protein was significantly increased in the spinal cord in a time-dependent manner, suggesting the accelerated translation of spinal Mmp24 mRNA after SNL. A previous study showed that m6A enrichment in many transcripts was changed in the DRG upon peripheral nerve injury. This change contributed to increased translation during axon regeneration, revealing the critical role of m6A modification in response to nerve injury (Weng et al., 2018). Our MeRIP-seq and RIP-PCR assay showed a considerable loss of m6A sites in Mmp24 mRNA in the spinal cord under neuropathic pain. It suggests that m6A modification in Mmp24 mRNA may be accountable for the accelerated translation of Mmp24 mRNA. Indeed, we found that after SNL, the increased level of MMP24 protein (not Mmp24 mRNA) was accompanied by a higher occupancy of FTO in the Mmp24 mRNA and the consequently lessened enrichment of m6A. Moreover, FTO overexpression erased the m6A sites in the Mmp24 mRNA and increased the level of MMP24 protein. Conversely, FTO knockdown markedly increased the m6A enrichment in the Mmp24 mRNA and reduced the expression of MMP24 protein, but not Mmp24 mRNA, in the spinal neurons. Together, the accelerated Mmp24 mRNA translation after SNL in the spinal cord was likely resulted from the lowered FTO-mediated m6A modification in Mmp24 mRNA.
Previous studies displayed that FTO was localized in the nucleus in neurons in the central nervous system, including the hippocampus and midbrain (Hess et al., 2013; Li et al., 2017). Functionally, FTO bound to the m6A sites and reduced the m6A enrichment to mediate the biogenesis of its target transcripts in the central neurons (Hess et al., 2013; Leonetti et al., 2020). Consistently, the present study revealed that FTO was co-expressed with NeuN in nuclei of the spinal neurons and colocalized with MMP24. However, the expression of spinal FTO was not altered after SNL. How the FTO preferentially binds to Mmp24 mRNA under the neuropathic pain condition is unclear. This could be due to that the binding of FTO and Mmp24 mRNA in the spinal cord is regulated by nerve injury-sensitive signaling pathways (Bresson et al., 2020), such as the upregulation of mediators in promoting the interaction of FTO and Mmp24 mRNA or downregulation of competitors in competing for the binding between FTO and Mmp24 mRNA (Song et al., 2019; Ontiveros et al., 2020). Although demethylase FTO, as an “eraser”, removed the m6A in Mmp24 mRNA after SNL, the specific “reader” decoding the m6A modification in the Mmp24 mRNA remains unknown. Previous studies found that m6A promoted translation efficiency via the reader proteins (Roundtree et al., 2017), like YTHDF1 (Jia et al., 2019). However, recent studies from C.H. et al. found that the effect of m6A on translation was highly heterogeneous and depended on the binding of specific RNA binding proteins (Zhang Z. et al., 2020). For example, the newly identified m6A effector YBX3, opposite to YTHDF1 in function, mediated m6A effect by repressing the translation of YBX3-bound m6A transcripts (Snyder et al., 2015; Zhang Z. et al., 2020). Here, the m6A loss-mediated promotion in Mmp24 mRNA translation possibly depends on the effectors like YBX3, but not YTHDF1. Further investigation is warrented for its identification.
In summary, our study reveals an FTO-triggered epigenetic mechanism of MMP24 upregulation in the spinal cord after SNL. Blocking the SNL-induced increase of MMP24 in the spinal cord mitigated pain hypersensitivity both in the development and maintenance phase without altering the basal/acute responses or locomotor functions. MMP24 may be an endogenous initiator of neuropathic pain and could be a potential target for this disorder's prevention and treatment.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The animal study was reviewed and approved by the Zhejiang Animal Care and Use Committee and the Ethics Committee of SAHZU, School of Medicine.
Author Contributions
MY, BJ, and LM conceived the project and designed the experiments. MY, FZ, and LY coordinated and supervised all experiments. LM, YH, NS, JR, DG, and XP conducted most of the molecular, biochemical, and morphological experiments. LM, YH, FZ, JL, and LY performed most of the animal surgery and behavioral experiments. LM, YH, FZ, SX, JL, and LY analyzed the data and did the statistical analysis. LM, MY, BJ, and DG wrote and revised the manuscript. All authors have read and agreed to submit this manuscript for publication.
Funding
This study was supported by the National Natural Science Foundation of China (NSFC 81471128, 81771197, 81771189, 82071227), Zhejiang Provincial Natural Science Foundation of China (LZ18H090001, LY18H090006), grants for Scientific Research from the Chinese Ministry of Health-Zhejiang Health Department, China (2018272998).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank XZ, CL, and SC (the Second Affiliated Hospital of Zhejiang University) for technical support. Figure 7 of this paper was created with the aid of BioRender.
References
Albik, S., and Tao, Y.-X. (2021). Emerging Role of RNA m6A Modification in Chronic Pain. Pain, Publish Ahead of Print. doi:10.1097/j.pain.0000000000002219
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: Tools for Motif Discovery and Searching. Nucleic Acids Res. 37, W202–W208. doi:10.1093/nar/gkp335
Bresson, S., Shchepachev, V., Spanos, C., Turowski, T. W., Rappsilber, J., and Tollervey, D. (2020). Stress-Induced Translation Inhibition through Rapid Displacement of Scanning Initiation Factors. Mol. Cell 80 (3), 470–484. doi:10.1016/j.molcel.2020.09.021
Daskalaki, M. G., Tsatsanis, C., and Kampranis, S. C. (2018). Histone Methylation and Acetylation in Macrophages as a Mechanism for Regulation of Inflammatory Responses. J. Cell Physiol. 233 (9), 6495–6507. doi:10.1002/jcp.26497
Decosterd, I., and Woolf, C. J. (2000). Spared Nerve Injury: an Animal Model of Persistent Peripheral Neuropathic Pain. Pain 87 (2), 149–158. doi:10.1016/S0304-3959(00)00276-1
Dominissini, D., Moshitch-Moshkovitz, S., Schwartz, S., Salmon-Divon, M., Ungar, L., Osenberg, S., et al. (2012). Topology of the Human and Mouse m6A RNA Methylomes Revealed by m6A-Seq. Nature 485 (7397), 201–206. doi:10.1038/nature11112
Dou, C., and Levine, J. (1994). Inhibition of Neurite Growth by the NG2 Chondroitin Sulfate Proteoglycan. J. Neurosci. 14 (12), 7616–7628. doi:10.1523/jneurosci.14-12-07616.1994
Ferreira, R., Naguibneva, I., Pritchard, L. L., Ait-Si-Ali, S., and Harel-Bellan, A. (2001). The Rb/chromatin Connection and Epigenetic Control: Opinion. Oncogene 20 (24), 3128–3133. doi:10.1038/sj.onc.1204337
Folgueras, A. R., Valdes-Sanchez, T., Llano, E., Menendez, L., Baamonde, A., Denlinger, B. L., et al. (2009). Metalloproteinase MT5-MMP Is an Essential Modulator of Neuro-Immune Interactions in Thermal Pain Stimulation. Proc. Natl. Acad. Sci. 106 (38), 16451–16456. doi:10.1073/pnas.0908507106
Fu, Y., Dominissini, D., Rechavi, G., and He, C. (2014). Gene Expression Regulation Mediated through Reversible m6A RNA Methylation. Nat. Rev. Genet. 15 (5), 293–306. doi:10.1038/nrg3724
Gu, P., Pan, Z., Wang, X.-M., Sun, L., Tai, L. W., and Cheung, C. W. (2018). Histone Deacetylase 5 (HDAC5) Regulates Neuropathic Pain through SRY-Related HMG-Box 10 (SOX10)-dependent Mechanism in Mice. Pain 159 (3), 526–539. doi:10.1097/j.pain.0000000000001125
Hannocks, M.-J., Zhang, X., Gerwien, H., Chashchina, A., Burmeister, M., Korpos, E., et al. (2019). The Gelatinases, MMP-2 and MMP-9, as Fine Tuners of Neuroinflammatory Processes. Matrix Biol. 75-76, 102–113. doi:10.1016/j.matbio.2017.11.007
Hayashita-Kinoh, H., Kinoh, H., Okada, A., Komori, K., Itoh, Y., Chiba, T., et al. (2001). Membrane-type 5 Matrix Metalloproteinase Is Expressed in Differentiated Neurons and Regulates Axonal Growth. Cell Growth Differ. 12 (11), 573–580.
He, P. C., and He, C. (2021). m(6) A RNA Methylation: from Mechanisms to Therapeutic Potential. EMBO J. 40 (3), e105977. doi:10.15252/embj.2020105977
He, X.-B., Yi, S.-H., Rhee, Y.-H., Kim, H., Han, Y.-M., Lee, S.-H., et al. (2011). Prolonged Membrane Depolarization Enhances Midbrain Dopamine Neuron Differentiation via Epigenetic Histone Modifications. Stem Cells 29 (11), 1861–1873. doi:10.1002/stem.739
Hess, M. E., Hess, S., Meyer, K. D., Verhagen, L. A. W., Koch, L., Brönneke, H. S., et al. (2013). The Fat Mass and Obesity Associated Gene (Fto) Regulates Activity of the Dopaminergic Midbrain Circuitry. Nat. Neurosci. 16 (8), 1042–1048. doi:10.1038/nn.3449
Hsieh, M.-C., Ho, Y.-C., Lai, C.-Y., Chou, D., Wang, H.-H., Chen, G.-D., et al. (2017). Melatonin Impedes Tet1-dependent mGluR5 Promoter Demethylation to Relieve Pain. J. Pineal Res. 63 (4), e12436. doi:10.1111/jpi.12436
Hsieh, M.-C., Ho, Y.-C., Lai, C.-Y., Wang, H.-H., Yang, P.-S., Cheng, J.-K., et al. (2021). Blocking the Spinal Fbxo3/CARM1/K+ Channel Epigenetic Silencing Pathway as a Strategy for Neuropathic Pain Relief. Neurotherapeutics. doi:10.1007/s13311-020-00977-5
Ji, R.-R., Xu, Z.-Z., Wang, X., and Lo, E. H. (2009). Matrix Metalloprotease Regulation of Neuropathic Pain. Trends Pharmacol. Sci. 30 (7), 336–340. doi:10.1016/j.tips.2009.04.002
Jia, R., Chai, P., Wang, S., Sun, B., Xu, Y., Yang, Y., et al. (2019). m6A Modification Suppresses Ocular Melanoma through Modulating HINT2 mRNA Translation. Mol. Cancer 18 (1), 161. doi:10.1186/s12943-019-1088-x
Jiang, B.-C., Cao, D.-L., Zhang, X., Zhang, Z.-J., He, L.-N., Li, C.-H., et al. (2016). CXCL13 Drives Spinal Astrocyte Activation and Neuropathic Pain via CXCR5. J. Clin. Invest. 126 (2), 745–761. doi:10.1172/JCI81950
Jiang, B.-C., Liu, T., and Gao, Y.-J. (2020a). Chemokines in Chronic Pain: Cellular and Molecular Mechanisms and Therapeutic Potential. Pharmacol. Ther. 212, 107581. doi:10.1016/j.pharmthera.2020.107581
Jiang, B.-C., Zhang, J., Wu, B., Jiang, M., Cao, H., Wu, H., et al. (2020b). G Protein-Coupled Receptor GPR151 Is Involved in Trigeminal Neuropathic Pain through the Induction of Gβγ/extracellular Signal-Regulated Kinase-Mediated Neuroinflammation in the Trigeminal Ganglion. Pain Publish Ahead of Print. doi:10.1097/j.pain.0000000000002156
Jiang, B.-C., Zhang, W.-W., Yang, T., Guo, C.-Y., Cao, D.-L., Zhang, Z.-J., et al. (2018). Demethylation of G-Protein-Coupled Receptor 151 Promoter Facilitates the Binding of Krüppel-like Factor 5 and Enhances Neuropathic Pain after Nerve Injury in Mice. J. Neurosci. 38 (49), 10535–10551. doi:10.1523/JNEUROSCI.0702-18.2018
Kawasaki, Y., Xu, Z.-Z., Wang, X., Park, J. Y., Zhuang, Z.-Y., Tan, P.-H., et al. (2008). Distinct Roles of Matrix Metalloproteases in the Early- and Late-phase Development of Neuropathic Pain. Nat. Med. 14 (3), 331–336. doi:10.1038/nm1723
Kechin, A., Boyarskikh, U., Kel, A., and Filipenko, M. (2017). cutPrimers: A New Tool for Accurate Cutting of Primers from Reads of Targeted Next Generation Sequencing. J. Comput. Biol. 24 (11), 1138–1143. doi:10.1089/cmb.2017.0096
Komori, K., Nonaka, T., Okada, A., Kinoh, H., Hayashita-Kinoh, H., Yoshida, N., et al. (2004). Absence of Mechanical Allodynia and Abeta-Fiber Sprouting after Sciatic Nerve Injury in Mice Lacking Membrane-type 5 Matrix Metalloproteinase. FEBS Lett. 557 (1-3), 125–128. doi:10.1016/s0014-5793(03)01458-3
Langmead, B., and Salzberg, S. L. (2012). Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 9 (4), 357–359. doi:10.1038/nmeth.1923
Leonetti, A. M., Chu, M. Y., Ramnaraign, F. O., Holm, S., and Walters, B. J. (2020). An Emerging Role of m6A in Memory: A Case for Translational Priming. Int. J. Mol. Sci. 21 (20), 7447. doi:10.3390/ijms21207447
Li, L., Zang, L., Zhang, F., Chen, J., Shen, H., Shu, L., et al. (2017). Fat Mass and Obesity-Associated (FTO) Protein Regulates Adult Neurogenesis. Hum. Mol. Genet. 26 (13), 2398–2411. doi:10.1093/hmg/ddx128
Li, Y., Guo, X., Sun, L., Xiao, J., Su, S., Du, S., et al. (2020). N 6 ‐Methyladenosine Demethylase FTO Contributes to Neuropathic Pain by Stabilizing G9a Expression in Primary Sensory Neurons. Adv. Sci. 7 (13), 1902402. doi:10.1002/advs.201902402
Lin, T.-B., Lai, C.-Y., Hsieh, M.-C., Ho, Y.-C., Wang, H.-H., Yang, P.-S., et al. (2020). Inhibiting MLL1-WDR5 Interaction Ameliorates Neuropathic Allodynia by Attenuating Histone H3 Lysine 4 Trimethylation-dependent Spinal mGluR5 Transcription. Pain 161 (9), 1995–2009. doi:10.1097/j.pain.0000000000001898
Liu, L., Wu, Y., Li, Q., Liang, J., He, Q., Zhao, L., et al. (2020). METTL3 Promotes Tumorigenesis and Metastasis through BMI1 m6A Methylation in Oral Squamous Cell Carcinoma. Mol. Ther. 28 (10), 2177–2190. doi:10.1016/j.ymthe.2020.06.024
Liu, N., Dai, Q., Zheng, G., He, C., Parisien, M., and Pan, T. (2015). N6-methyladenosine-dependent RNA Structural Switches Regulate RNA-Protein Interactions. Nature 518 (7540), 560–564. doi:10.1038/nature14234
Llano, E., Pendás, A. M., Freije, J. P., Nakano, A., Knäuper, V., Murphy, G., et al. (1999). Identification and Characterization of Human MT5-MMP, a New Membrane-Bound Activator of Progelatinase a Overexpressed in Brain Tumors. Cancer Res. 59 (11), 2570–2576.
Manicone, A., and McGuire, J. (2008). Matrix Metalloproteinases as Modulators of Inflammation. Semin. Cell Dev. Biol. 19 (1), 34–41. doi:10.1016/j.semcdb.2007.07.003
Meyer, K. D., Saletore, Y., Zumbo, P., Elemento, O., Mason, C. E., and Jaffrey, S. R. (2012). Comprehensive Analysis of mRNA Methylation Reveals Enrichment in 3′ UTRs and Near Stop Codons. Cell 149 (7), 1635–1646. doi:10.1016/j.cell.2012.05.003
Millard, C. J., Varma, N., Saleh, A., Morris, K., Watson, P. J., Bottrill, A. R., et al. (2016). The Structure of the Core NuRD Repression Complex Provides Insights into its Interaction with Chromatin. Elife 5, e13941. doi:10.7554/eLife.13941
Mitka, M. (2003). Virtual Textbook" on Pain Developed: Effort Seeks to Remedy Gap in Medical Education. JAMA 290 (18), 2395. doi:10.1001/jama.290.18.239510.1001/jama.290.1.27
Ontiveros, R. J., Shen, H., Stoute, J., Yanas, A., Cui, Y., Zhang, Y., et al. (2020). Coordination of mRNA and tRNA Methylations by TRMT10A. Proc. Natl. Acad. Sci. USA 117 (14), 7782–7791. doi:10.1073/pnas.1913448117
Penas, C., and Navarro, X. (2018). Epigenetic Modifications Associated to Neuroinflammation and Neuropathic Pain after Neural Trauma. Front. Cell Neurosci. 12, 158. doi:10.3389/fncel.2018.00158
Pokhilko, A., Nash, A., and Cader, M. Z. (2020). Common Transcriptional Signatures of Neuropathic Pain. Pain 161 (7), 1542–1554. doi:10.1097/j.pain.0000000000001847
Rigaud, M., Gemes, G., Barabas, M.-E., Chernoff, D. I., Abram, S. E., Stucky, C. L., et al. (2008). Species and Strain Differences in Rodent Sciatic Nerve Anatomy: Implications for Studies of Neuropathic Pain. Pain 136 (1-2), 188–201. doi:10.1016/j.pain.2008.01.016
Roundtree, I. A., Evans, M. E., Pan, T., and He, C. (2017). Dynamic RNA Modifications in Gene Expression Regulation. Cell 169 (7), 1187–1200. doi:10.1016/j.cell.2017.05.045
Sato, T., Chang, H.-C., Bayeva, M., Shapiro, J. S., Ramos-Alonso, L., Kouzu, H., et al. (2018). mRNA-binding Protein Tristetraprolin Is Essential for Cardiac Response to Iron Deficiency by Regulating Mitochondrial Function. Proc. Natl. Acad. Sci. USA 115 (27), E6291–E6300. doi:10.1073/pnas.1804701115
Sekine-Aizawa, Y., Hama, E., Watanabe, K., Tsubuki, S., Kanai-Azuma, M., Kanai, Y., et al. (2001). Matrix Metalloproteinase (MMP) System in Brain: Identification and Characterization of Brain-specific MMP Highly Expressed in Cerebellum. Eur. J. Neurosci. 13 (5), 935–948. doi:10.1046/j.0953-816x.2001.01462.x
Snyder, E., Soundararajan, R., Sharma, M., Dearth, A., Smith, B., and Braun, R. E. (2015). Compound Heterozygosity for Y Box Proteins Causes Sterility Due to Loss of Translational Repression. PLoS Genet. 11 (12), e1005690, doi:10.1371/journal.pgen.1005690
Song, P., Feng, L., Li, J., Dai, D., Zhu, L., Wang, C., et al. (2020). β-Catenin Represses miR455-3p to Stimulate m6A Modification of HSF1 mRNA and Promote its Translation in Colorectal Cancer. Mol. Cancer 19 (1), 129. doi:10.1186/s12943-020-01244-z
Song, T., Yang, Y., Wei, H., Xie, X., Lu, J., Zeng, Q., et al. (2019). Zfp217 Mediates m6A mRNA Methylation to Orchestrate Transcriptional and Post-transcriptional Regulation to Promote Adipogenic Differentiation. Nucleic Acids Res. 47 (12), 6130–6144. doi:10.1093/nar/gkz312
Wang, L. X., and Wang, Z. J. (2003). Animal and Cellular Models of Chronic Pain. Adv. Drug Deliv. Rev. 55 (8), 949–965. doi:10.1016/s0169-409x(03)00098-x
Wang, X., and Pei, D. (2001). Shedding of Membrane Type Matrix Metalloproteinase 5 by a Furin-type Convertase. J. Biol. Chem. 276 (38), 35953–35960. doi:10.1074/jbc.M103680200
Weng, Y.-L., Wang, X., An, R., Cassin, J., Vissers, C., Liu, Y., et al. (2018). Epitranscriptomic m6A Regulation of Axon Regeneration in the Adult Mammalian Nervous System. Neuron 97 (2), 313–325. doi:10.1016/j.neuron.2017.12.036
Wu, S., Bono, J., and Tao, Y.-X. (2019). Long Noncoding RNA (lncRNA): a Target in Neuropathic Pain. Expert Opin. Ther. Targets 23 (1), 15–20. doi:10.1080/14728222.2019.1550075
Wu, S., Marie Lutz, B., Miao, X., Liang, L., Mo, K., Chang, Y.-J., et al. (2016). Dorsal Root Ganglion Transcriptome Analysis Following Peripheral Nerve Injury in Mice. Mol. Pain 12, 174480691662904. doi:10.1177/1744806916629048
Yang, Y., Hsu, P. J., Chen, Y.-S., and Yang, Y.-G. (2018). Dynamic Transcriptomic m6A Decoration: Writers, Erasers, Readers and Functions in RNA Metabolism. Cell Res 28 (6), 616–624. doi:10.1038/s41422-018-0040-8
Yu, G., Wang, L.-G., and He, Q.-Y. (2015). ChIPseeker: an R/Bioconductor Package for ChIP Peak Annotation, Comparison and Visualization. Bioinformatics 31 (14), 2382–2383. doi:10.1093/bioinformatics/btv145
Zhang, M., Zhai, Y., Zhang, S., Dai, X., and Li, Z. (2020). Roles of N6-Methyladenosine (m6A) in Stem Cell Fate Decisions and Early Embryonic Development in Mammals. Front. Cel Dev. Biol. 8, 782. doi:10.3389/fcell.2020.00782
Zhang, Y., Liu, T., Meyer, C. A., Eeckhoute, J., Johnson, D. S., Bernstein, B. E., et al. (2008). Model-based Analysis of ChIP-Seq (MACS). Genome Biol. 9 (9), R137. doi:10.1186/gb-2008-9-9-r137
Zhang, Z., Luo, K., Zou, Z., Qiu, M., Tian, J., Sieh, L., et al. (2020). Genetic Analyses Support the Contribution of mRNA N6-Methyladenosine (m6A) Modification to Human Disease Heritability. Nat. Genet. 52 (9), 939–949. doi:10.1038/s41588-020-0644-z
Zhao, J.-Y., Liang, L., Gu, X., Li, Z., Wu, S., Sun, L., et al. (2017). DNA Methyltransferase DNMT3a Contributes to Neuropathic Pain by Repressing Kcna2 in Primary Afferent Neurons. Nat. Commun. 8, 14712. doi:10.1038/ncomms14712
Zhao, X., Tang, Z., Zhang, H., Atianjoh, F. E., Zhao, J.-Y., Liang, L., et al. (2013). A Long Noncoding RNA Contributes to Neuropathic Pain by Silencing Kcna2 in Primary Afferent Neurons. Nat. Neurosci. 16 (8), 1024–1031. doi:10.1038/nn.3438
Zheng, G., Dahl, J. A., Niu, Y., Fedorcsak, P., Huang, C.-M., Li, C. J., et al. (2013). ALKBH5 Is a Mammalian RNA Demethylase that Impacts RNA Metabolism and Mouse Fertility. Mol. Cell 49 (1), 18–29. doi:10.1016/j.molcel.2012.10.015
Keywords: neuropathic pain, matrix metallopeptidase 24, fat-mass and obesity-associated protein, N6-methyladenosine, spinal cord
Citation: Ma L, Huang Y, Zhang F, Gao DS, Sun N, Ren J, Xia S, Li J, Peng X, Yu L, Jiang B-C and Yan M (2021) MMP24 Contributes to Neuropathic Pain in an FTO-Dependent Manner in the Spinal Cord Neurons. Front. Pharmacol. 12:673831. doi: 10.3389/fphar.2021.673831
Received: 28 February 2021; Accepted: 06 April 2021;
Published: 29 April 2021.
Edited by:
Serena Boccella, University of Campania Luigi Vanvitelli, ItalyReviewed by:
Yuan-Xiang Tao, Rutgers, The State University of New Jersey, United StatesTzer-Bin Lin, Taipei Medical University, Taiwan
Copyright © 2021 Ma, Huang, Zhang, Gao, Sun, Ren, Xia, Li, Peng, Yu, Jiang and Yan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bao-Chun Jiang, amlhbmdiYW9jaHVuQG50dS5lZHUuY24=; Min Yan, enJ5YW5taW5Aemp1LmVkdS5jbg==
†These authors have contributed equally to this work and share first authorship
 Longfei Ma
Longfei Ma Yangyuxin Huang
Yangyuxin Huang Fengjiang Zhang1†
Fengjiang Zhang1† Lina Yu
Lina Yu Bao-Chun Jiang
Bao-Chun Jiang Min Yan
Min Yan