- 1Center of Excellence in Genomic Medicine Research, Department of Medical Laboratory Technology, Faculty of Applied Medical Sciences, King Abdulaziz University, Jeddah, Saudi Arabia
- 2Center for Transdisciplinary Research, Department of Pharmacology, Saveetha Dental College and Hospitals, Saveetha Institute of Medical and Technical Sciences, Chennai, India
- 3Department of Obstetrics and Gynaecology, King Abdulaziz University Hospital, King Abdulaziz University, Jeddah, Saudi Arabia
Introduction: Alzheimer’s disease (AD) is a major cause of the development of cognitive decline and dementia. AD and associated dementias (ADRD) are the major contributors to the enormous burden of morbidity and mortality worldwide. To date, there are no robust therapies to alleviate or cure this debilitating disease. Most drug treatments focus on restoring the normal function of neurons and the cells that cause inflammation, such as microglia in the brain. However, the role of astrocytes, the brain’s housekeeping cells, in the development of AD and the initiation of dementia is still not well understood.
Objective: To decipher the role of astrocytes in the entorhinal cortex of AD patients using single nuclear RNA sequencing (snRNASeq) datasets from the Single Cell RNA-seq Database for Alzheimer’s Disease (scREAD). The datasets were originally derived from astrocytes, isolated from the entorhinal cortex of AD brain and healthy brain to decipher disease-specific signaling pathways as well as drugs and natural products that reverse AD-specific signatures in astrocytes.
Methods: We used snRNASeq datasets from the scREAD database originally derived from astrocytes isolated from the entorhinal cortex of AD and healthy brains from the Gene Expression Omnibus (GEO) (GSE138852 and GSE147528) and analyzed them using next-generation knowledge discovery (NGKD) platforms. scREAD is a user-friendly open-source interface available at https://bmbls.bmi.osumc.edu/scread/that enables more discovery-oriented strategies. snRNASeq data and metadata can also be visualized and downloaded via an interactive web application at adsn.ddnetbio.com. Differentially expressed genes (DEGs) for each snRNASeq dataset were analyzed using iPathwayGuide to compare and derive disease-specific pathways, gene ontologies, and in silico predictions of drugs and natural products that regulate AD -specific signatures in astrocytes. In addition, DEGs were analyzed using the L1000FWD and L1000CDS2 signature search programming interfaces (APIs) to identify additional drugs and natural products that mimic or reverse AD-specific gene signatures in astrocytes.
Results: We found that PI3K/AKT signaling, Wnt signaling, neuroactive ligand-receptor interaction pathways, neurodegeneration pathways, etc. were significantly impaired in astrocytes from the entorhinal cortex of AD patients. Biological processes such as glutamate receptor signaling pathway, regulation of synapse organization, cell-cell adhesion via plasma membrane adhesion molecules, and chylomicrons were negatively enriched in the astrocytes from the entorhinal cortex of AD patients. Gene sets involved in cellular components such as postsynaptic membrane, synaptic membrane, postsynapse, and synapse part were negatively enriched (p < 0.01). Moreover, molecular functions such as glutamate receptor activity, neurotransmitter receptor activity, and extracellular ligand-gated ion channels were negatively regulated in the astrocytes of the entorhinal cortex of AD patients (p < 0.01). Moreover, the application of NGKD platforms revealed that antirheumatic drugs, vitamin-E, emetine, narciclasine, cephaeline, trichostatin A, withaferin A, dasatinib, etc. can potentially reverse gene signatures associated with AD.
Conclusions: The present study highlights an innovative approach to use NGKD platforms to find unique disease-associated signaling pathways and specific synthetic drugs and natural products that can potentially reverse AD and ADRD-associated gene signatures.
1 Introduction
Alzheimer’s disease (AD) is a major cause of the development of cognitive decline and dementia in the elderly (Winblad et al., 2016; Matthews et al., 2019). AD-related dementias (ADRD) contribute to 50-70 percent of dementias worldwide (Winblad et al., 2016). AD and associated dementias (ADRD) are the largest contributors to the burden of morbidity and mortality and higher costs in health care systems worldwide (Hurd et al., 2013). Important risk factors for ADRD include ethnicity, age, and gender. Approximately 6.2 million Americans aged 65 years or older were affected by AD and this number is expected to double to 13.8 million by 2060 in the United States of America (United States) (Claxton et al., 2015; Matthews et al., 2019; Alzheimer’s Disease Facts and Figures, 2021). Therefore, ADRD has been declared a health priority worldwide (World Health Organization, 2012). In the United States of America (United States), AD is the sixth leading cause of death in the general population and the fifth leading cause of death in Americans aged 65 years and older. In contrast, reported deaths from other debilitating diseases such as stroke, heart disease, and HIV have declined, while deaths from AD have increased by more than 145% in the U.S. between 2000 and 2019 (Alzheimer’s Disease Facts and Figures, 2021).
AD is a neurodegenerative disease of the brain (Figure 1), and symptoms such as cognitive decline and language difficulties have slowly developed in AD patients in recent years. It is mostly diagnosed in the older population with an average age of 65 years or more and is referred to as late-onset AD (LOAD) (Gauthaman et al., 2014; Rasool et al., 2018; Rasool et al., 2021). The progressive damage to neurons from the aggregation of amyloid-beta (Ab) protein and tau protein, as well as neuroinflammation in certain parts of the brain, significantly impairs learning, speech, memory, and other cognitive abilities (Gauthaman et al., 2014; Rasool et al., 2018; Rasool et al., 2021). Importantly, the risk of ADRD is significantly increased in AD patients with diabetes mellitus (Gauthaman et al., 2014; Rasool et al., 2018; Rasool et al., 2021). Moreover, the cellular and molecular mechanisms of AD pathology and the role of specific cells in the brain in the development of ADRD are poorly understood (Rasool et al., 2021).
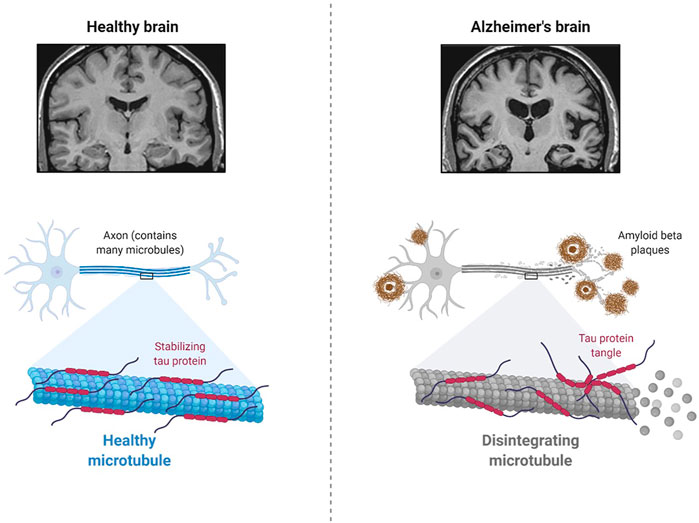
FIGURE 1. Pathology of Alzheimer’s Disease (Created using Biorender.com).
AD and ADRD pathology differ by brain region, cell type, age, and gender (Sala Frigerio et al., 2019; Rasool et al., 2021). Genome-wide association studies (GWAS) using genetic mapping concepts have revealed genes enriched in AD susceptibility loci, and transcriptomics of whole brain tissue using next-generation sequencing (NGS) platforms or microarray applications have shown an increase in microglial gene connectivity and impairment of neuronal connectivity in AD (Hitzemann et al., 2014). Although transcriptional network dynamics of mass analysis can provide more information about AD pathogenesis, it does not reveal all the dynamic changes at the cellular and molecular levels that contribute to AD pathology. A detailed understanding of the underlying role of individual cell types in AD patients is therefore essential for the development of new therapeutics to treat dementia.
Recent advances in NGS applications such as single-cell RNA sequencing (scRNA-Seq) have enabled researchers to study and understand the dynamic transcriptomic profile of individual cells in brain tissue or other biological samples. RNA-sequencing of posterior cingulate astrocytes (PC) in AD patients revealed differential expression of mitochondria-related genes, including TRMT61B, FASTKD2, and NDUFA4L2. In addition, immune response genes such as CLU, C3, and CD74 were identified to play a central role in the generation or clearance of amyloid-beta (Sekar et al., 2015). scRNASeq provides a higher resolution of cellular dynamics and a better understanding of individual cells in the tissue microenvironment (Grubman et al., 2019; Jiang et al., 2020; Wu and Zhang, 2020). Similarly, the single nucleus RNA sequencing (snRNA-Seq) technique is used to study frozen samples where dissociation of single cells becomes a problem and affects gene expression patterns. Although AD is one of the major reasons for the development of cognitive decline and dementia (Gauthaman et al., 2014; Rasool et al., 2018; Rasool et al., 2021), there are still no robust therapies to alleviate or cure this debilitating disease (Gao et al., 2016; Rasool et al., 2018) and most drug treatments focus on restoring normal function of cells that cause inflammation, such as microglia and neurons in the brain (Oksanen et al., 2017). However, the genetic basis of astrocytes in the development of AD, and the triggering of dementia is still not clearly understood (Oksanen et al., 2017; Kery et al., 2020). Therefore, a precise understanding of the underlying role of astrocytes in AD patients may provide clues for the development of effective therapies to treat dementia. Here, we used an innovative approach to leverage next-generation knowledge discovery (NGKD) platforms to decipher the AD -specific gene signatures in astrocytes isolated from the entorhinal cortex of AD patients and specific synthetic drugs and natural products to improve AD and associated disease pathologies such as dementia.
2 Materials and Methods
2.1 Ethical Statement
This study was exempt from Institutional Review Board (IRB) approval because it did not involve animal models or human subjects. It was performed using DEGs derived from the Single Cell RNA-seq Database for Alzheimer’s Disease (scREAD) based on publicly available and previously published single nucleus RNA sequencing datasets from the Gene Expression Omnibus (GEO).
2.2 Data Source
In the present study, we snRNASeq data from the scREAD, originally obtained from astrocytes isolated from the entorhinal cortex of AD brains and healthy brains from the Gene Expression Omnibus (GEO) (GSE138852 and GSE147528). scREAD is a user-friendly open-source interface available at https://bmbls.bmi.osumc.edu/scread/to enable more discovery-oriented strategies (Wu and Zhang, 2020; Jiang et al., 2020; Jiang et al., 2021) (Supplementary Figure S1). Datasets were filtered in scREAD by selecting the options for species (human), condition (all), region in the brain (entorhinal cortex), and gender (all), and are listed in Table 1 with the corresponding Braak levels (Braak and Braak, 1991). scREAD webtool was also used to visualize all cell types and sub-clusters of astrocytes in the entorhinal cortex region of the brain using Uniform Manifold Approximation and Projection (UMAP) (Becht et al., 2018). All snRNASeq data are freely available in the Gene Expression Omnibus (GEO) under accession numbers GSE138852 and GSE147528.
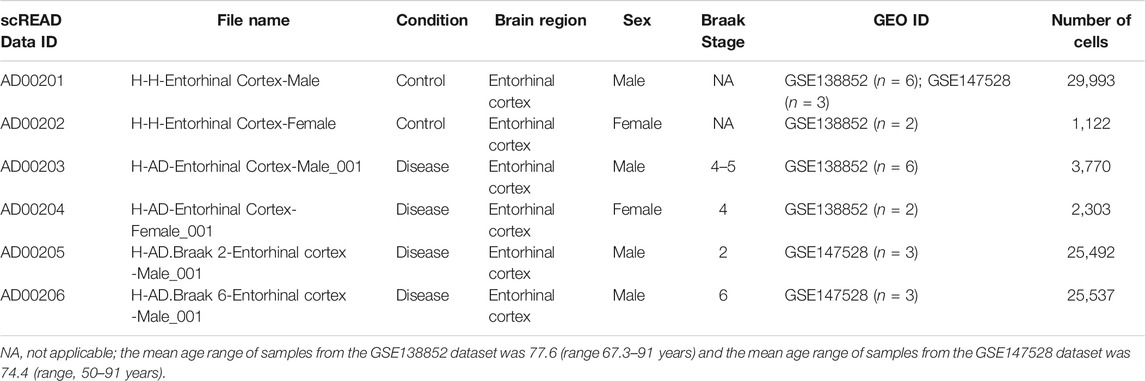
TABLE 1. Information on the snRNASeq datasets obtained from scREAD database for NGKD analysis (Human)*.
Importantly, the snRNAseq datasets (GSE138852) are available via an interactive web application at adsn.ddnetbio.com (Grubman et al., 2019). The characteristics of all AD and healthy snRNASeq scREAD datasets used in this study are provided in Table 1. As of May 2021, the snRNASeq datasets used for this study had been already published and are publicly available (Barrett et al., 2013).
2.3 The snRNASeq Data Analysis Using iPathwayGuide
DEGs were obtained using scREAD analysis of snRNASeq data from astrocytes of AD groups (AD00203, AD00205, and AD00206) compared with the healthy control group (AD00201). DEGs of AD groups (AD00203, AD00205, and AD00206) were further filtered using a p-value cut-off of 0.05, and log2 fold change (Log2Fc) of ±0.3 in iPathwayGuide Software (Advaita Bioinformatics, United States) to obtain 739, 241, and 639 DEGs. Further analysis of these DEGs using iPathwayGuide software showed that 93 DEGs were commonly regulated in all disease groups (Figure 2). The Kyoto Encyclopedia of Genes and Genomes (KEGG) database was used to decipher differentially regulated pathways (Kanehisa and Goto, 2000; Kanehisa et al., 2002; Kanehisa et al., 2010; Kanehisa et al., 2012; Kanehisa et al., 2014), and the Gene Ontology Consortium database (Ashburner et al., 2000; Gene Ontology Consortium 2001) was used to identify the differentially regulated GO functions, and the Comparative Toxicogenomics Database was used to find the chemicals/drugs/toxicants (CDT and the KEGG database for diseases (Kanehisa and Goto, 2000; Kanehisa et al., 2002). The iPathwayGuide software used the Impact Analysis Method (IAM) (Draghici et al., 2003; Draghici et al., 2007; Draghici, 2011) to obtain significantly impacted DEGs and pathways compared with the corresponding control group; the p-value computed using Fisher’s method was used to determine the pathway score, and the p-value was adjusted based on the false discovery rate (FDR) (Benjamini and Hochberg, 1995; Benjamini and Yekutieli, 2001). and Bonferroni multiple testing corrections (Bonferroni, 1935). The p-values were computed based on the hypergeometric distribution in iPathwayGuide analysis and the FDR and Bonferroni methods for multiple testing corrections (Draghici et al., 2003; Draghici, 2011).
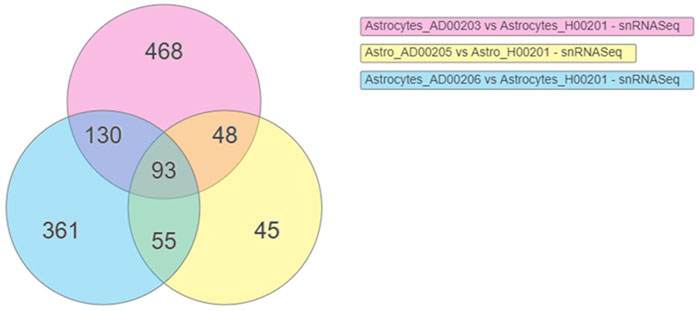
FIGURE 2. Venn diagram showing the DEGs obtained using the scREAD analysis of snRNASeq data from astrocytes of AD groups (AD00203, AD00205, and AD00206) compared to astrocytes of the healthy control group (AD00201) were 739, 241, and 639 respectively after filtering with a p-value cut off of 0.05 and log2 fold change (Log2Fc) of ±0.3. Further analysis of DEGs using iPathwayGuide software showed that 93 DEGs were commonly regulated in all the disease groups.
2.4 Determination of Upstream Drugs and Natural Products Using iPathwayGuide
The determination of upstream drugs or natural products was predicted based on the enrichment of DEGs and 2) a network of connections or interactions from the Advaita Knowledge Base (Draghici et al., 2003; Draghici, 2011). The iPathwayGuide analysis was based on two hypotheses (HP and HA). The overly abundant or present upstream chemical, drug, or toxicant (CDT) was predicted under the conditions analyzed under the first hypothesis called HP and the upstream CDT. is insufficient (or absent) was predicted under the conditions analyzed under the second hypothesis. HA. iPathwayGuide calculates a Z-score for each CDT z(u) by iterating over the genes in DT(u) and their incoming edges in (g) in testing both HP and HA. Subsequently, the p-value was computed corresponding to the z-score Pz (One-Tailed) under the probability density function for a normal distribution, N (0,1) (Draghici et al., 2003; Draghici, 2011).
2.4.1 Determination of Upstream Drugs and Natural Products Present or Overly Abundant Using iPathwayGuide
To determine the presence or abundance of CDTs based on the differentially expressed (DE) genes, CDT u, DE genes downstream of u, DTA (u) were compared to measured target genes predicted by chance to be both DE and consistent. An over-representation method was applied to calculate the statistical significance (p-value) based on the number of consistent DE genes in the iPathwayGuide analysis. The Ppres (p-value) was calculated based on the hypergeometric distribution (Draghici et al., 2003; Draghici, 2011). Then, the global probability value (PG) was computed by combining Pz and Ppres: and was used to rank the upstream regulators and test the HP research hypothesis. The p-values were combined into one test statistic using the standard Fisher’s method.
2.4.2 Determination of Upstream Drugs and Natural Products Absent or Insufficient Using iPathwayGuide
To determine the absence or insufficiency of CDTs based on the DE genes, Pabs was calculated using the iPathwayGuide analysis. The upstream CDTs that were absent or insufficient under the conditions investigated based on the number of consistent DE genes downstream of u, and DTI (u) was compared to the measured target genes predicted by chance to be both DE and consistent. The Pabs (p-value) was calculated based on the hypergeometric distribution (Draghici et al., 2003; Draghici, 2011). Then the PG was computed by combining Pz and Pabs and was used to rank the upstream regulators that were absent or insufficient and to test the research hypothesis HA. The analysis combines Pabs and Pz, using Fisher’s method as described previously, where Pz was measured only for significant negative z-scores (z ≤ −2) (Draghici et al., 2003; Draghici, 2011).
2.5 L1000FWD and L1000CDS2 Analyses
DEGs were subjected to L1000 Fire Works Display (L1000FWD) analysis using the L1000FWD signature search application programming interface (API) (Wang et al., 2018) to identify the top 50 drugs and natural products that have the potential to reverse AD-associated signaling. Similarly, the same set of DEGs was subjected to L1000 Characteristic Direction Signature Search Engine (L1000CDS2) analysis using the L1000CDS2 Signature Search API to identify the top 50 drugs and natural products with the potential to reverse AD-associated signaling (Duan et al., 2016).
3 Results
In the present study, snRNASeq datasets of astrocytes isolated from the entorhinal cortex region of AD patients and healthy brains were obtained from the scREAD database for NGKD platform analysis (Table 1). The scREAD web tool was used to visualize all cell types and sub-clusters of astrocytes in the entorhinal cortex region of the brain in AD and healthy snRNASeq datasets using UMAP (Supplementary Figure S1). A UMAP example for the healthy control and AD scREAD datasets is shown in Supplementary Figure S2.
DEGs in astrocytes from the entorhinal cortex compared to healthy controls were determined using paired comparisons with the healthy control (AD00201) and AD (AD00203, AD00205, and AD00206) datasets (Supplementary Tables S1–S3) The 93 DEGs common to all AD datasets can be found in Supplementary Table S4. The 15 pathways most affected by DEGs in the AD groups compared to healthy controls are listed in Tables 2–4. Based on the number of DEGs, the top signaling pathways differentially regulated in the astrocytes of AD patients in the context of neurodegeneration include Alzheimer’s disease, prion disease, Parkinson’s disease, Huntington’s disease, neurodegeneration signaling pathways (multiple diseases), amyotrophic lateral sclerosis, and the phosphatidylinositol 3-kinases/protein kinase B (PI3K/AKT) pathway. The differentially regulated KEGG pathways of Alzheimer’s disease in the groups of AD are shown in Figures 3–5, and the differentially regulated KEGG pathways of neurodegenerative degeneration (multiple diseases) are shown in Figures 6–8. Analysis of WNT pathway perturbation and PI3K/AKT pathways followed by iPathwayGuide coherent cascade activation revealed the dysregulation of these pathways in the astrocytes of AD patients from the entorhinal cortex (Supplementary Figure S3A and Supplementary Figure S4B). The differentially regulated genes in the WNT pathways and the PI3K/AKT pathways are also shown in Supplementary Figure S3B and Supplementary Figure S4B, respectively. In addition to the neurodegenerative diseases, we also observed the signaling pathways associated with Salmonella infection, human papillomavirus (HPV) infection, and human T-cell leukemia virus infection in the astrocytes of the severe AD groups (Table 2 and Table 4) compared with the healthy controls. Gene set enrichment analysis (GSEA) showed that gene sets involved in cellular components, such as postsynaptic membrane, synaptic membrane, postsynapse, synapse, and synapse, were negatively enriched (p < 0.01). Neuroactive ligand-receptor interaction based on KEGG pathways was significantly downregulated (p < 0.01), and cellular function of the transporter complex was also negatively enriched (p < 0.01). Similarly, genes associated with glutamate receptor activity, neurotransmitter receptor activity, glutamate receptor signaling, heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules, cell-cell adhesion via plasma membrane adhesion molecules, and behavior were also negatively enriched (p < 0.01) in astrocytes from AD patients (Table 5). Importantly, differential expression of GWAS genes in astrocytes from the entorhinal cortex in the brain of AD is listed in Table 6. The most downregulated GWAS genes in astrocytes from the entorhinal cortex associated with the pathogenesis of AD were NKAIN3, LRRC4C, CADM2, DLC1, APOE, TNIK, GADD45G, FRMD4A, CTNNA2, NPAS3, NCKAP5, and RORA.
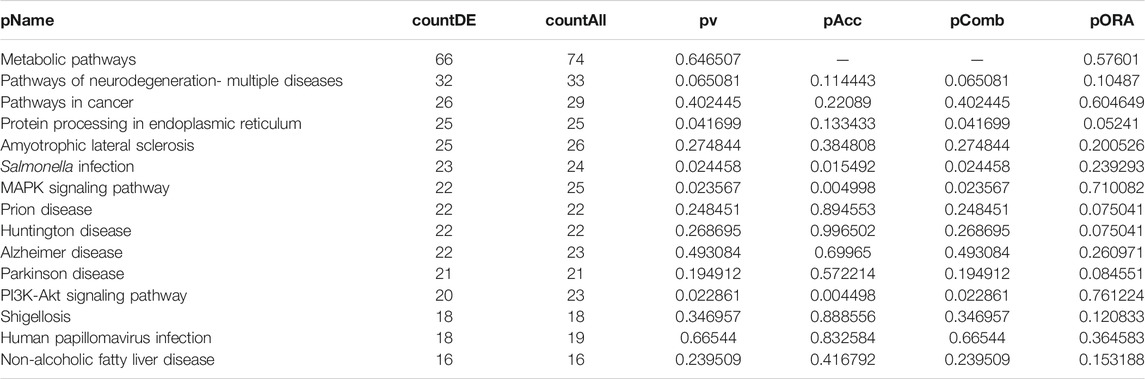
TABLE 2. Top 15 pathways ranked based on their associated differentially expressed genes derived from astrocytes based on the comparison AD00203 (disease) vs. AD00201 (control).
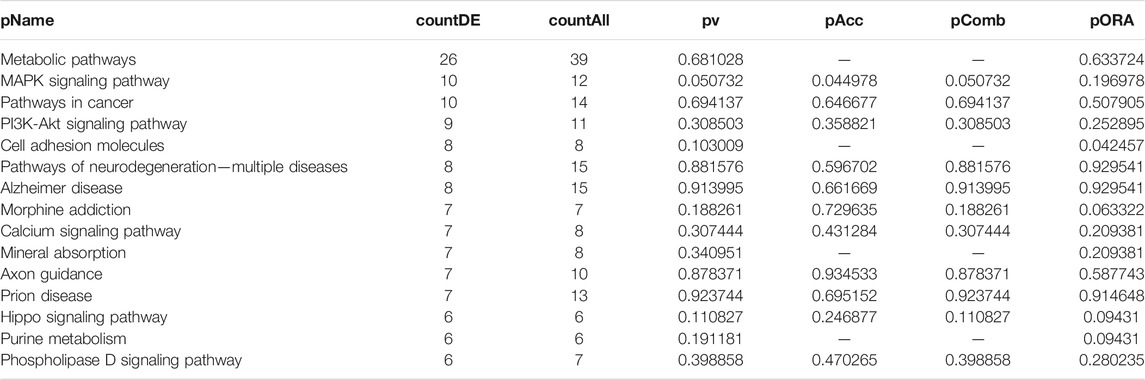
TABLE 3. Top 15 pathways ranked based on their associated differentially expressed genes derived from astrocytes based on the comparison AD00205 (disease) vs. AD00201 (control).
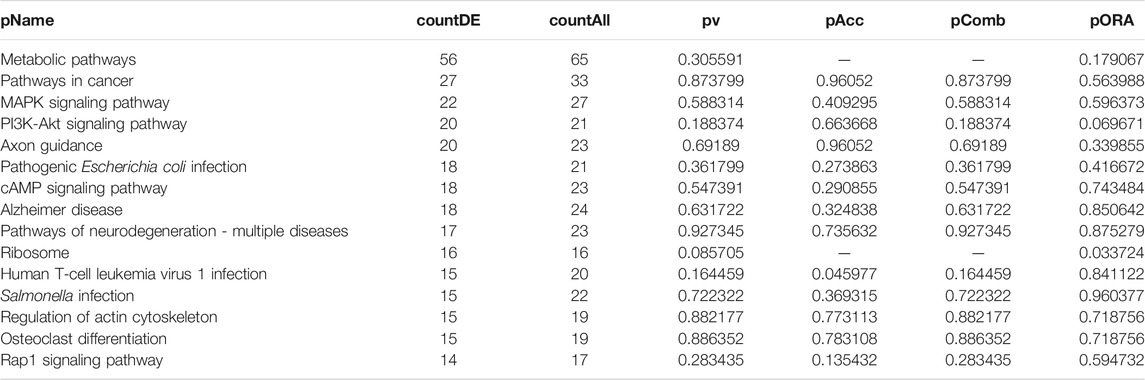
TABLE 4. Top 15 pathways ranked based on their associated differentially expressed genes derived from astrocytes based on the comparison AD00206 (disease) vs. AD00201 (control).
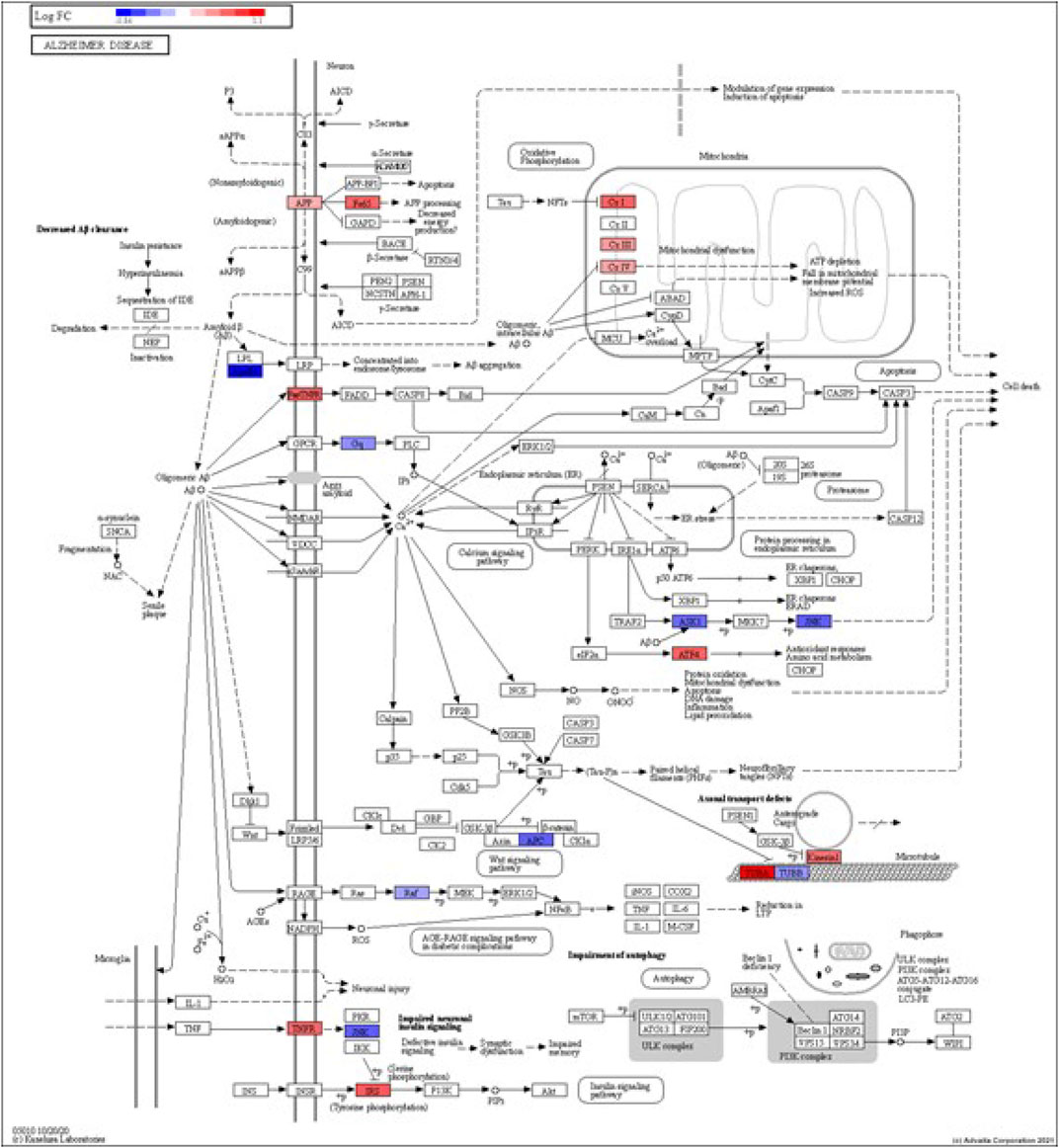
FIGURE 3. iPathwayGuide analysis shows the differentially regulated genes in the KEGG Alzheimer’s disease pathway in astrocytes from the AD group (AD00203) compared to astrocytes from the healthy control group (AD00201).
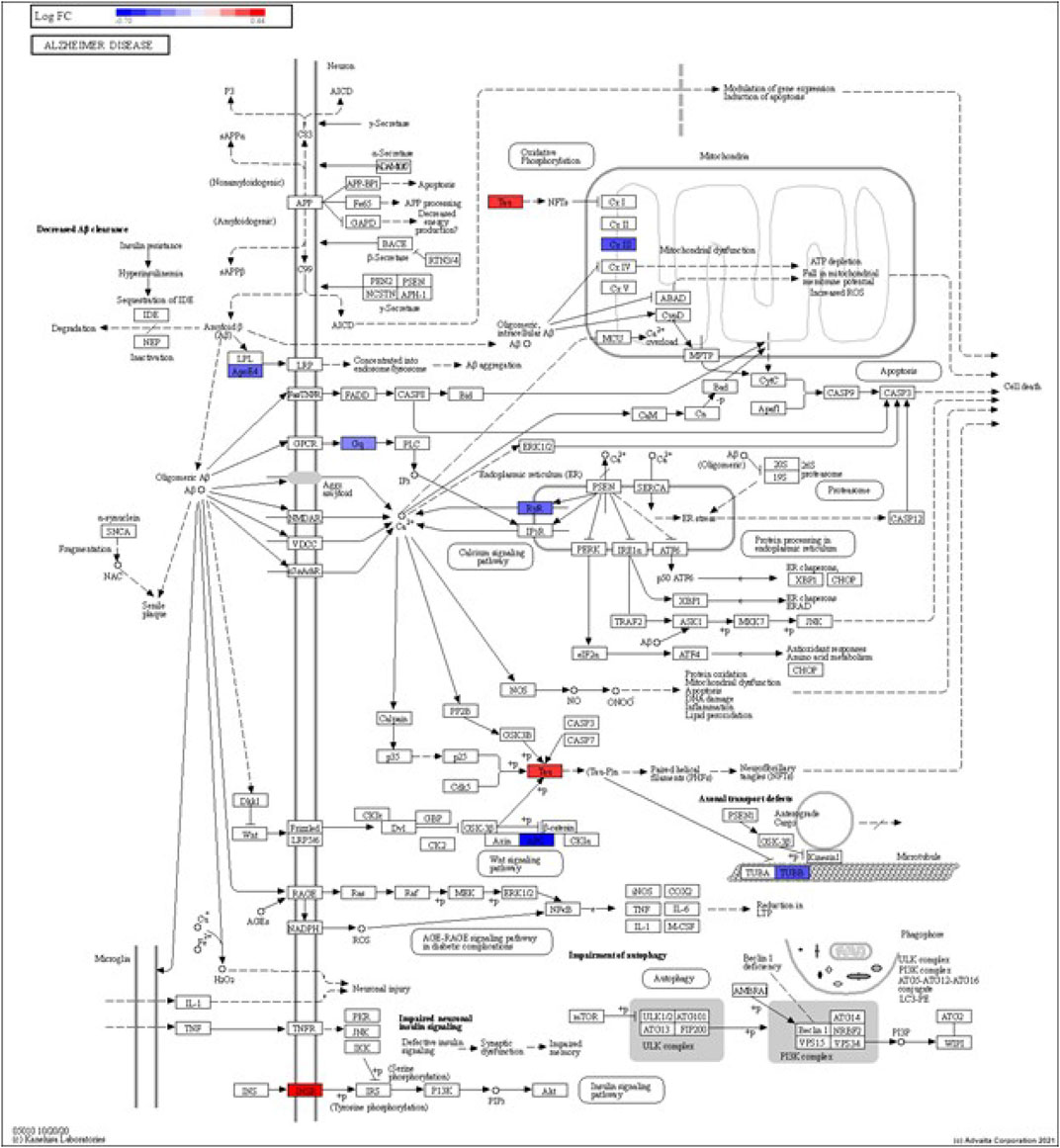
FIGURE 4. iPathwayGuide analysis shows the differentially regulated genes in the KEGG Alzheimer’s disease pathway in astrocytes from the AD group (AD00205) compared to astrocytes from the healthy control group (AD00201).
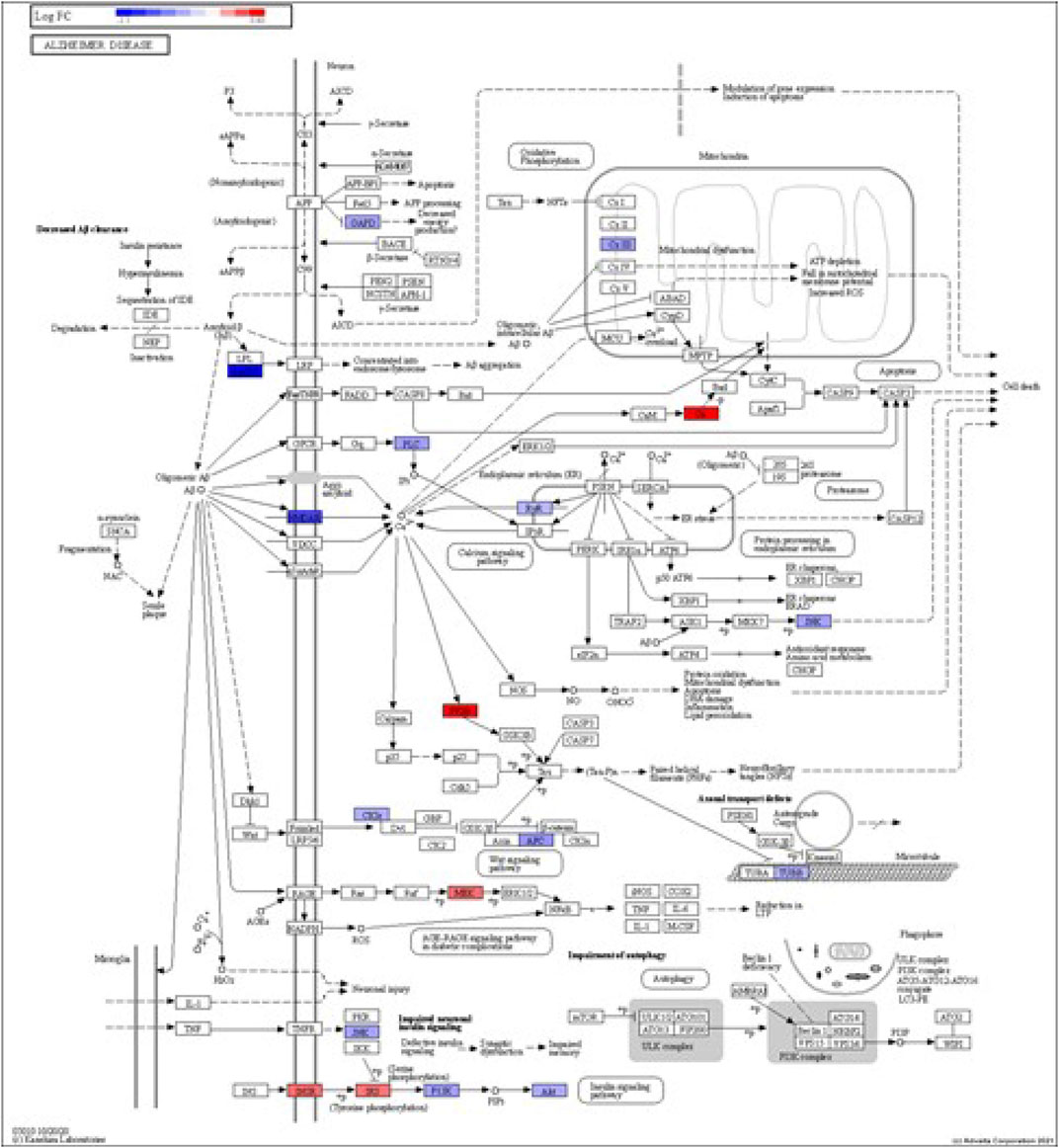
FIGURE 5. iPathwayGuide analysis shows the differentially regulated genes in the KEGG Alzheimer’s disease pathway in astrocytes from the AD group (AD00206) compared to astrocytes from the healthy control group (AD00201).
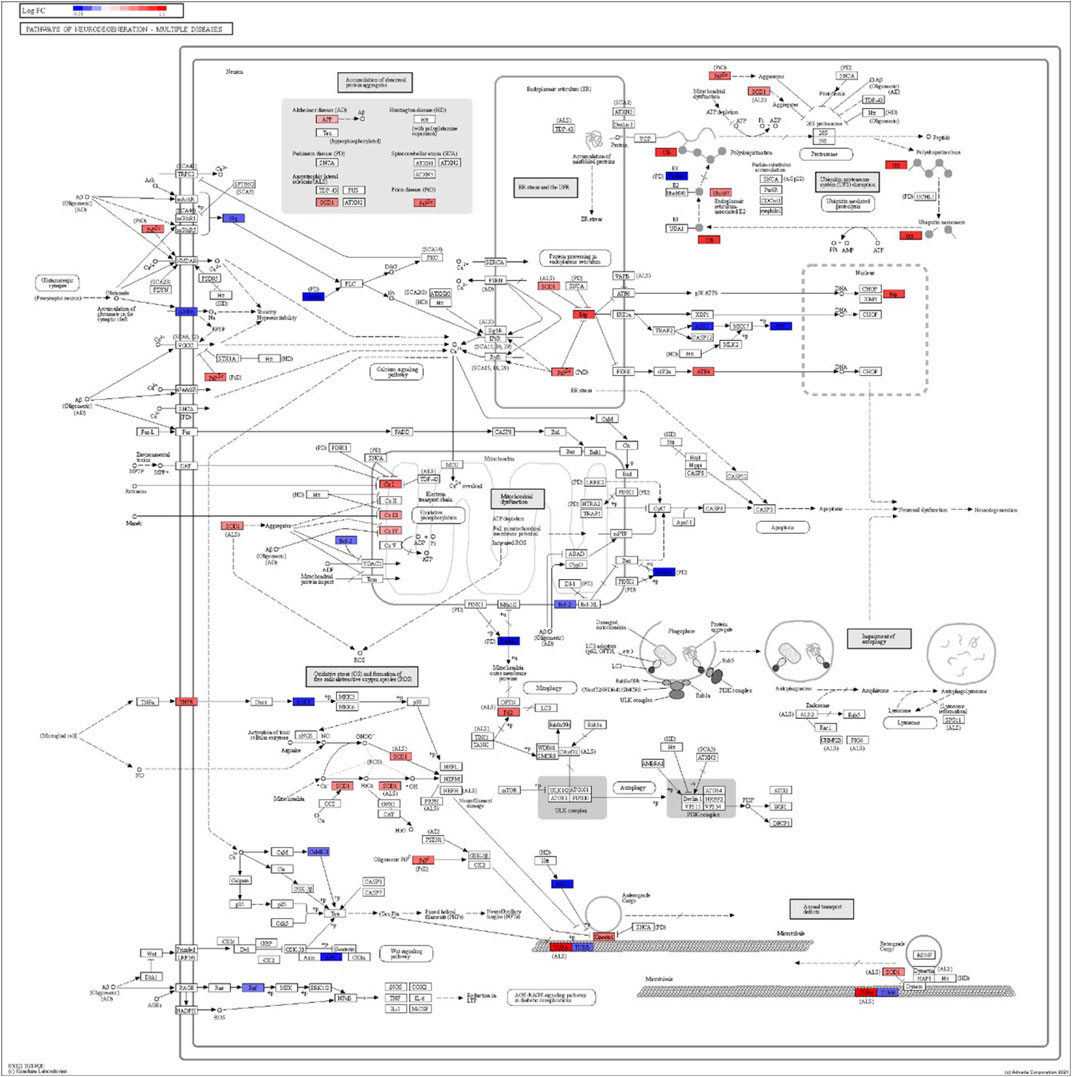
FIGURE 6. iPathwayGuide analysis shows the differentially regulated genes in the KEGG neurodegeneration pathway (multiple diseases) in the astrocytes of the AD group (AD00203) compared to the astrocytes of the healthy control group (AD00201).
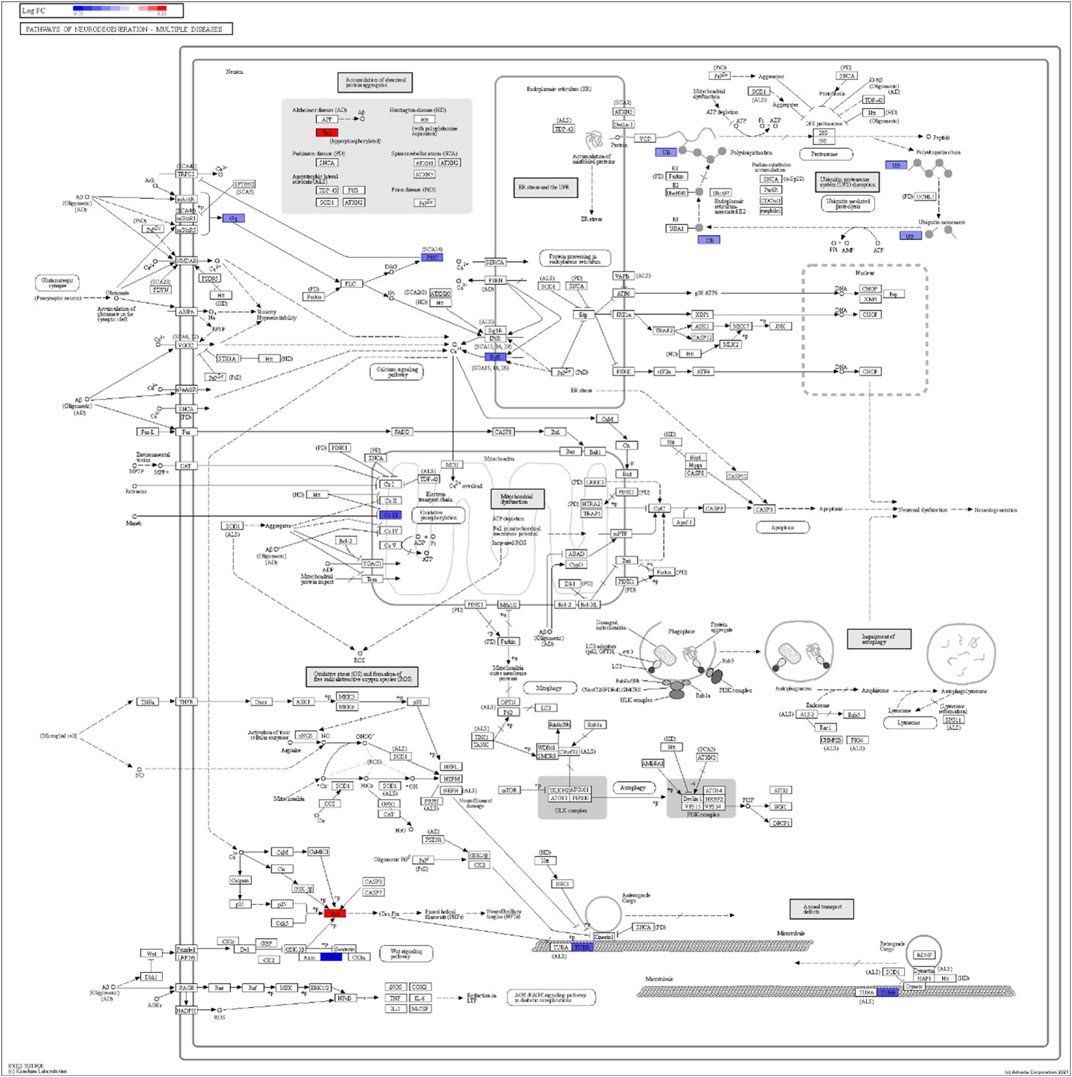
FIGURE 7. iPathwayGuide analysis shows the differentially regulated genes in the KEGG neurodegeneration pathway (multiple diseases) in the astrocytes of the AD group (AD00205) compared to the astrocytes of the healthy control group (AD00201).
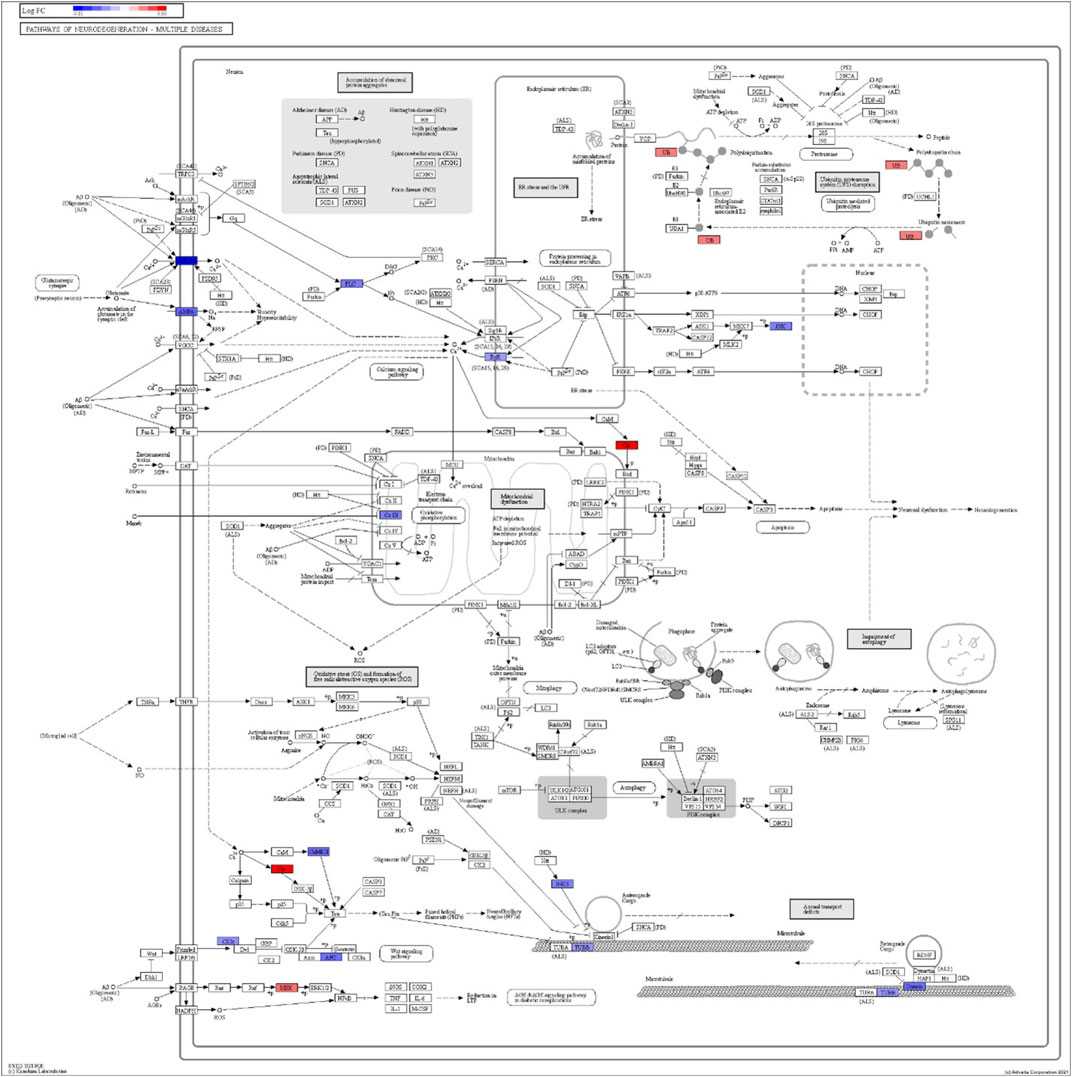
FIGURE 8. iPathwayGuide analysis shows the differentially regulated genes in the KEGG neurodegeneration pathway (multiple diseases) in the astrocytes of the AD group (AD00206) compared to the astrocytes of the healthy control group (AD00201).

TABLE 5. Top 25 Impacted Pathways obtained using Gene Set Enrichment Analysis (GSEA) based on Normalized Enrichment Score (NES) and False Discovery Rate (FDR) using the web tool available at http://adsn.ddnetbio.com/
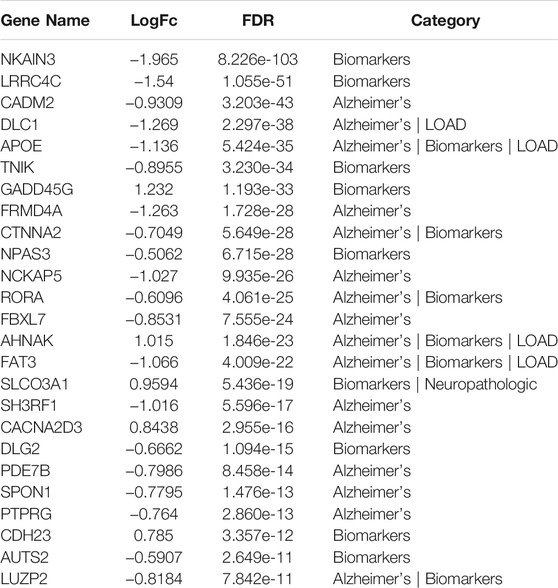
TABLE 6. Top 25 differentially expressed GWAS genes in astrocytes from entorhinal cortex in AD brain based on analysis using the web tool available at http://adsn.ddnetbio.com/.
Comparative analysis of the AD datasets from the scREAD based on the DEGs with iPathwayGuide showed that the antirheumatic drugs, vitamin E, salinomycin, and clorgyline have insufficient (p < 0.05) signaling effect in the astrocytes of AD patients (Table 7). In addition, Tables 8–10 list the drugs or natural products that could potentially reverse the gene signatures of astrocytes in the AD groups (AD00203, AD00205, and AD00206) based on the L1000FWD web tool analysis. L1000FWD analysis revealed that natural products such as emetine, cephaeline, homoharringtonine, narciclasine, withaferin A and several synthetic drugs such as dasatinib can significantly reverse gene signatures associated with AD pathology.
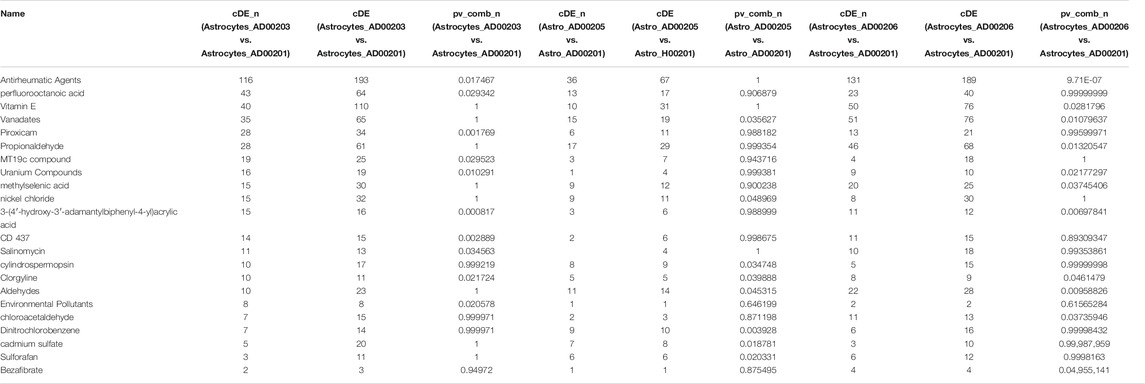
TABLE 7. The upstream Chemicals, Drugs, or Toxicants (CDTs) were predicted as absent (or insufficient) in the astrocytes of AD based on the number of DEGs significantly impacted in each category.
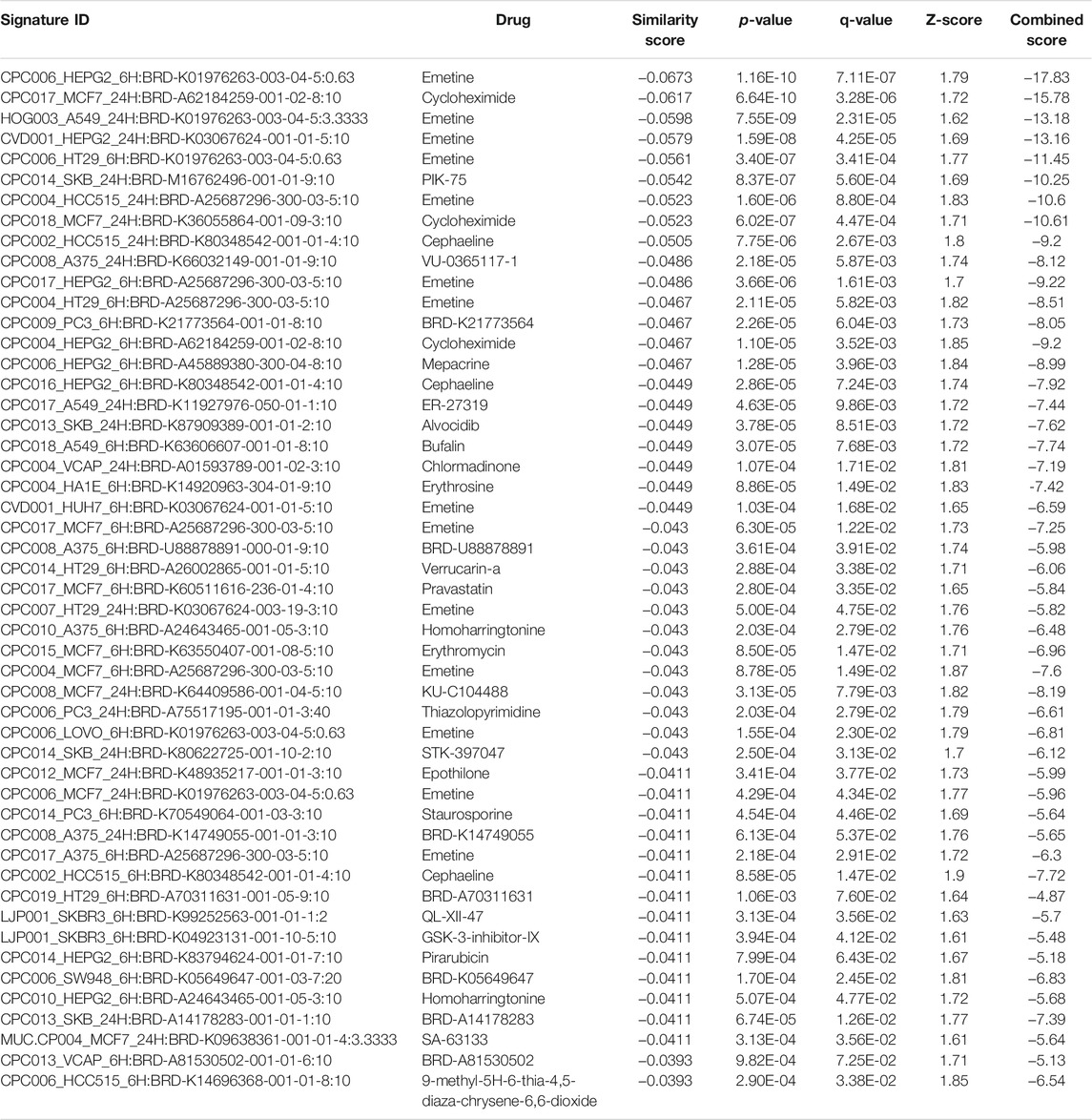
TABLE 8. The top 50 drugs or natural products that reverse DEGs of astrocytes from entorhinal cortex in AD (AD00203 (disease) vs AD00201 (control) based on L1000FWD analysis.
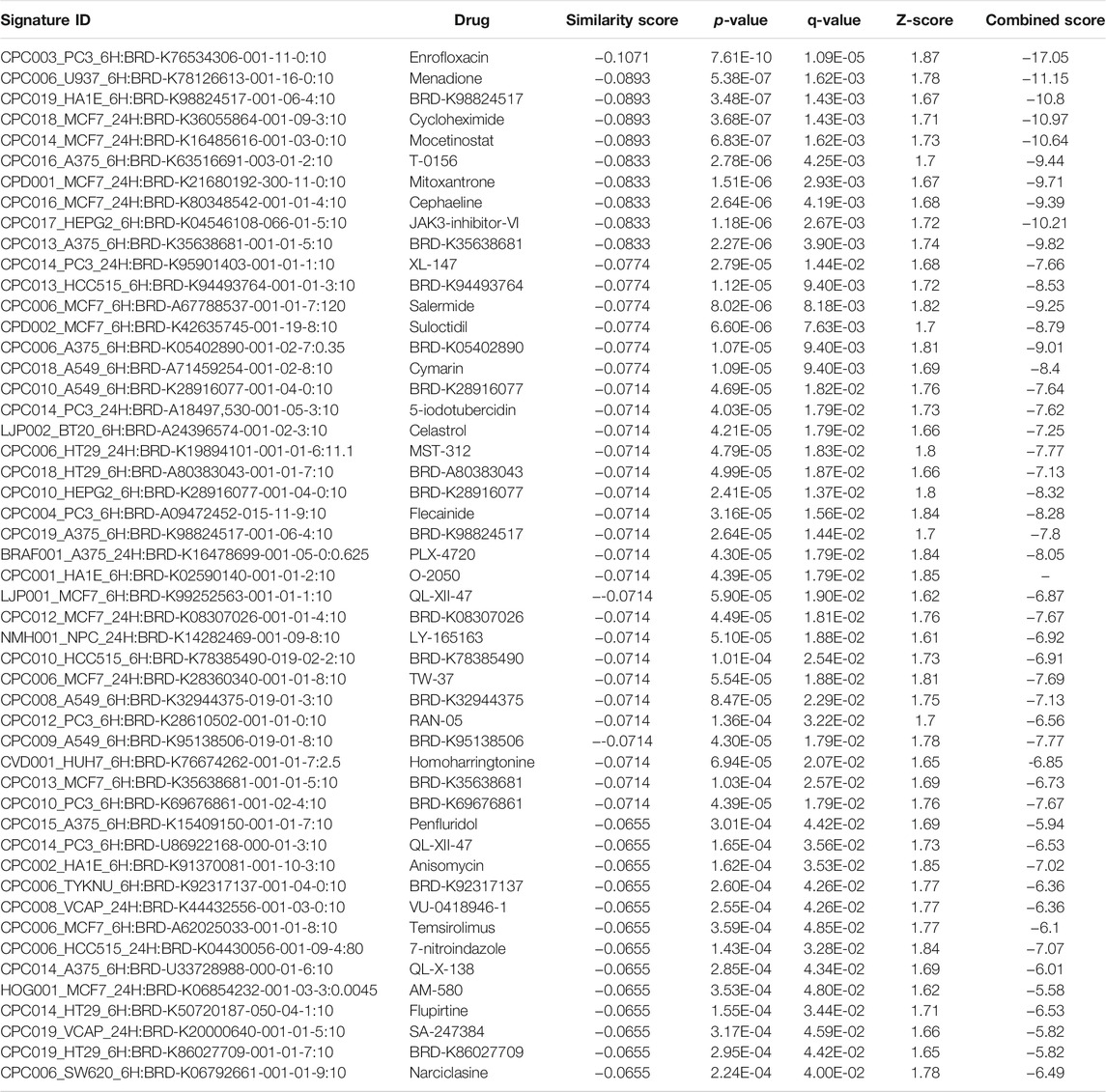
TABLE 9. The top 50 drugs or natural products that reverse DEGs of astrocytes from entorhinal cortex in AD (AD00205 (disease) vs AD00201 (control) based on L1000FWD analysis.
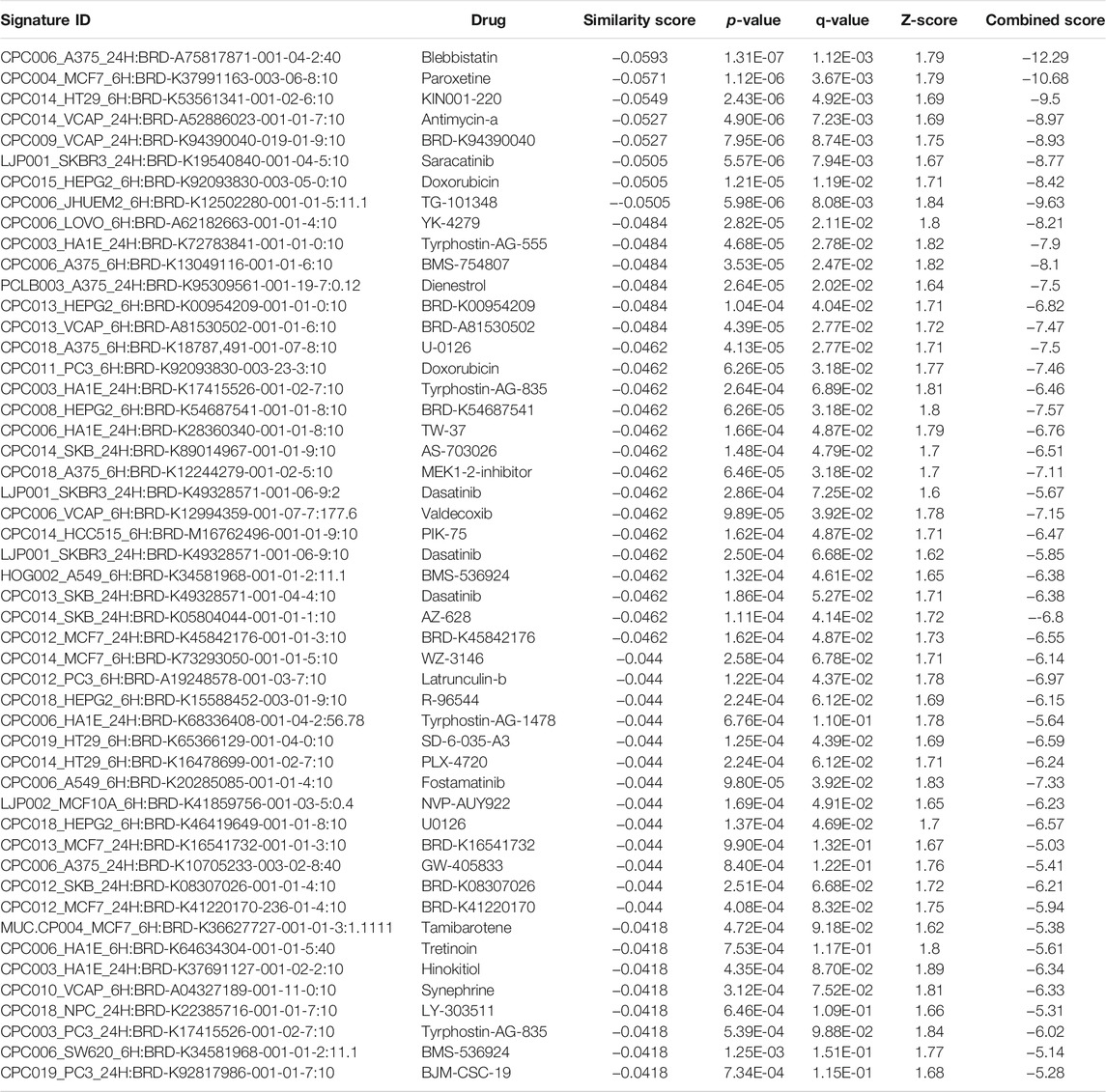
TABLE 10. The top 50 drugs or natural products that reverse DEGs of astrocytes from entorhinal cortex in AD (AD00206 (disease) vs AD00201 (control) based on L1000FWD analysis.
The drugs or natural products that could potentially reverse the gene signatures of astrocytes in AD groups (AD00203, AD00205, and AD00206) based on the L1000CDS2 web tool are provided in Supplementary Tables S1–S3, respectively. The L1000CDS2 analysis uncovered the natural products emetine, narciclasine, trichostatin A, homoharringtonine, ouabain, bufalin, and withaferin A, as well as synthetic drugs, such as dasatinib, that have the potential to reverse AD-associated gene signatures in astrocytes from AD patients.
4 Discussion
AD is a neurodegenerative disease of the brain and a major cause of the development of cognitive decline and dementia in the elderly (Winblad et al., 2016; Matthews et al., 2019). ADRD contributes to the majority of dementia cases worldwide (Winblad et al., 2016). Recent advances in genome sequencing technologies such as scRNA-Seq and snRNASeq are critical for deciphering the roles of heterogeneous cell populations in the brain at the single-cell level, and subsequent dissecting of these datasets using high throughput knowledge discovery platforms may provide clues as to why a particular group of cells is susceptible to AD and ADRD (Jiang et al., 2020; Wu and Zhang, 2020; Wang et al., 2021). Here, snRNASeq datasets of astrocytes isolated from the entorhinal cortex region of AD patients and healthy brains were obtained and analyzed using scREAD web-tool. scREAD includes 73 datasets from 16 studies, 10 brain regions, and 713,640 cells, and provides cell type and sub-cluster predictions, decipherment of DEGs, and discovery of cell type-specific regulons (Jiang et al., 2020; Wu and Zhang, 2020; Wang et al., 2021).
We observed that Wnt signaling and PI3K/AKT signaling pathways were dysregulated or impaired in astrocytes from the entorhinal cortex of AD patients. Wnt signaling is very important at the synapse and necessary for synaptic plasticity and maintenance in the brain (Palomer et al., 2019). The PI3K/AKT pathway regulates apoptosis, cell proliferation, and metabolism and is essential for protection against amyloid protein (Aβ)-induced neurotoxicity (Long et al., 2021). Neuroactive ligand-receptor interaction, axon guidance, Alzheimer’s disease, GABAergic synapse, glutamatergic synapse, etc. were negatively enriched or dysregulated in astrocytes from AD patients. GABAergic transmission is essential for all central nervous system functions (Luscher et al., 2011) and the GABAergic synapse pathway is impaired in the astrocytes of AD. This was also confirmed by GSEA analysis, which showed that the sets of genes involved in cellular components such as postsynaptic membrane, synaptic membrane, postsynapse, transporter complex, and interaction between neuroactive ligands and receptors were negatively enriched in the astrocytes of AD patients. Similarly, genes associated with glutamate receptor activity, neurotransmitter receptor activity, glutamate receptor signaling, heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules, cell-cell adhesion via plasma membrane adhesion molecules, and behavior were also negatively enriched in the astrocytes of AD patients. Importantly, the downregulated GWAS genes in astrocytes derived from the entorhinal cortex, such as NKAIN3, LRRC4C, CADM2, DLC1, APOE, TNIK, GADD45G, FRMD4A, CTNNA2, NPAS3, NCKAP5, RORA, etc., associated with AD pathogenesis, can be used either as biomarkers for neuropathology, AD or LOAD (Riaz et al., 2021). Interestingly, we found signaling pathways associated with Salmonella infection, HPV infection, and human T-cell leukemia virus infection in the astrocytes of severe AD groups. Previous studies have shown that infections with Salmonella (Himmelhoch et al., 1947), HPV (Lin et al., 2020), and human T-cell leukemia virus (Lycke et al., 1993) are associated with dementia and cognitive decline in humans.
We have previously shown that natural products such as albiziasaponin-A, iso-orientin, and salvadorin can ameliorate the pathologies associated with AD in vivo (Rasool et al., 2018) and that the natural products could be useful for the treatment of age-related degenerative diseases (Kalamegam et al., 2020). In addition, we have recently shown that NGKD platforms can be successfully used to find drugs and natural products that may reverse disease-specific gene signatures (Pushparaj et al., 2021). Therefore, NGKD platforms can be used to find drugs and natural products that can potentially reverse AD-associated gene signatures in astrocytes. Here, we used iPathwayGuide, L1000FWD, and L1000CDS2 tools to identify promising drug-responsive molecules for ADRD. Comparative analysis of AD datasets using iPathwayGuide showed that antirheumatic drugs have insufficient signaling in astrocytes from AD patients. Disease-modifying antirheumatic drugs (DMARDs) are used to treat patients with rheumatoid arthritis (Bahlas et al., 2019) and recent studies have found that patients with rheumatoid arthritis taking antirheumatic drugs have a lower risk of developing dementia (Judge et al., 2017; Huang et al., 2019). Our finding is consistent with these studies that antirheumatic drugs can reverse AD-associated gene signatures in astrocytes. Similarly, vitamin E gene signatures were absent in astrocytes from AD. The role of vitamin E in the treatment of AD remains a controversial topic to date (Browne et al., 2019) and our results provide some evidence for the importance of vitamin E in the treatment of AD and ADRD. A recent study found that emetine may have the potential to clear amyloid-beta plaques in AD (Ahmad et al., 2019). The isoquinoline alkaloids emetine and its desmethyl analog cepaheline have been predicted to be protective against cognitive decline and AD (Fernández-Martínez et al., 2020). Withaferin A is a steroidal lactone and a withanolide found in the medicinal plant Withania somnifera, and a number of studies have shown that it plays a neuroprotective role in AD (Das et al., 2021). Narciclasine is an active constituent of the Lycoris radiata (L'Her.) herb. It is used in traditional Chinese medicine for the treatment of AD (Shen et al., 2019). A recent study found that senolytic therapy with a combination of dasatinib and quercetin reduced Aβ-associated oligodendrocyte progenitor cell senescence and cognitive decline in an AD model (Zhang et al., 2019). The histone deacetylase inhibitor trichostatin A (Hsing et al., 2015) increased albumin expression and Aβ clearance in APP/PS1 mice and improved cognitive deficits (Su et al., 2021). Trichostatin A increased the antioxidant capacity and cell viability of SH -SY5Y cells by enhancing Keap1-mediated inhibition of the Nrf2 pathway, thereby reducing amyloid-β peptide-mediated cell damage (Li et al., 2020). Importantly, we recently predicted the potential of withaferin A, narciclasine, and trichostatin A to reverse gene signatures in neuro- COVID (Pushparaj et al., 2021). However, the effects of natural products such as emetine, cephaeline, narciclasine, withaferin A, trichostatin A and drugs such as DMARDs and dasatinib which may be able to reverse AD gene signatures in astrocytes should be validated with appropriate experimental models from AD before being used for further clinical testing.
5 Conclusion
The present study provides a valuable method for analyzing snRNASeq datasets deposited in open-source repositories with NGKD platforms to decipher AD -specific pathways, genes, and drugs from synthetic and natural sources for the amelioration of AD-related disease pathologies such as ADRD. However, further studies are required to confirm these drugs and natural products that reverse the gene signatures of AD using appropriate experimental models to deduce the precise mechanisms of action, followed by appropriate clinical trials to evaluate the safety and efficacy of the likely therapeutic interventions for AD and ADRD in a typical clinical milieu. Our innovative approach of applying NGKD platforms to uncover AD-specific pathways and potential drugs and natural products that reverse the AD-specific signatures could be useful in the future for developing personalized medicine for AD patient care.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
PP, KG, and MR were involved in conceptualization, intellectual contribution, statistical evaluation, and manuscript writing. PP was involved in the NGKD work and data analysis. PP, GK, KHWS and MR were involved in the coordination of the work, review, and editing of the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We acknowledge this project funded by the King Abdulaziz City for Science and Technology (KACST), Riyadh, Kingdom of Saudi Arabia, under grant number 13-MED2437-03. We acknowledge with thanks the KACST and the Science and Technology Unit, King Abdulaziz University for technical and financial support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2021.720170/full#supplementary-material
Supplementary Figure S1 | scREAD. A Single Cell RNA Sequencing Database for Alzheimer’s Disease (Adapted from Jiang et al., 2020 and shared under CC BY-NC-ND 4.0 license)
Supplementary Figure S2 | An example UMAP of healthy control and AD scREAD datasets. scREAD web tool was used to visualize all the cell types and sub-clusters of astrocytes in the entorhinal cortex region of the brain in AD and Healthy snRNASeq datasets using UMAP.
Supplementary Figure S3 | (A) The iPathwayGuide analysis showing the differentially regulated genes in the Wnt signaling KEGG pathway in the astrocytes of the AD group (AD00203) compared to astrocytes of the healthy control group (AD00201). (B) Bar graph depicting the differentially regulated genes in the Wnt signaling pathway in the astrocytes of AD compared to healthy control.
Supplementary Figure S4 | (A) The iPathwayGuide perturbation analysis based on the differentially regulated genes in the PI3K/AKT KEGG pathway in the astrocytes of the AD group (AD00203) compared to astrocytes of the healthy control group (AD00201). (B) Bar graph depicting the differentially regulated genes in the PI3K/AKT signaling pathway in the astrocytes of AD compared to healthy control.
References
Ahmad, S. S., Khan, H., Danish Rizvi, S. M., Ansari, S. A., Ullah, R., Rastrelli, L., et al. (2019). Computational Study of Natural Compounds for the Clearance of Amyloid-Βeta: A Potential Therapeutic Management Strategy for Alzheimer's Disease. Molecules 24 (18), 3233. doi:10.3390/molecules24183233
Alzheimer's Disease Facts and Figures (2021). 2021 Alzheimer's Disease Facts and Figures. Alzheimers Demen. 17 (3), 327–406. doi:10.1002/alz.12328
Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., et al. (2000). Gene Ontology: Tool for the Unification of Biology. The Gene Ontology Consortium. Nat. Genet. 25 (1), 25–29. doi:10.1038/75556
Bahlas, S., Damiati, L., Dandachi, N., Sait, H., Alsefri, M., and Pushparaj, P. N. (2019). Rapid Immunoprofiling of Cytokines, Chemokines and Growth Factors in Patients with Active Rheumatoid Arthritis Using Luminex Multiple Analyte Profiling Technology for Precision Medicine. Clin. Exp. Rheumatol. 37 (1), 112–119.
Barrett, T., Wilhite, S. E., Ledoux, P., Evangelista, C., Kim, I. F., Tomashevsky, M., et al. (2013). NCBI GEO: Archive for Functional Genomics Data Sets-Uupdate. Nucleic Acids Res. 41 (Database issue), D991–D995. doi:10.1093/nar/gks1193
Becht, E., McInnes, L., Healy, J., Dutertre, C.-A., Kwok, I. W. H., Ng, L. G., et al. (2018). Dimensionality Reduction for Visualizing Single-Cell Data Using UMAP. Nat. Biotechnol. 37, 38–44. doi:10.1038/nbt.4314
Benjamini, Y., and Hochberg, Y. (1995). Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Methodol. 57 (1), 289–300. doi:10.1111/j.2517-6161.1995.tb02031.x
Benjamini, Y., and Yekutieli, D. (2001). The Control of the False Discovery Rate in Multiple Testing under Dependency. Ann. Stat. 29 (4), 1165–1188. doi:10.1214/aos/1013699998
Bonferroni, C. E. (1935). “Il calcolo delle assicurazioni su gruppi di teste,” in Studi in Onore del Professore Salvatore Ortu Carboni (Rome: Tipografia del Senato), 13–60.
Braak, H., and Braak, E. (1991). Neuropathological Stageing of Alzheimer-Related Changes. Acta Neuropathol. 82 (4), 239–259. doi:10.1007/BF00308809
Browne, D., McGuinness, B., Woodside, J. V., and McKay, G. J. (2019). Vitamin E and Alzheimer's Disease: what Do We Know So Far? Clin. Interv. Aging 14, 1303–1317. doi:10.2147/CIA.S186760
Claxton, A., Baker, L. D., Hanson, A., Trittschuh, E. H., Cholerton, B., Morgan, A., et al. (2015). Long Acting Intranasal Insulin Detemir Improves Cognition for Adults with Mild Cognitive Impairment or Early-Stage Alzheimer's Disease Dementia. J. Alzheimers Dis. 45 (4), 1269–1270. doi:10.3233/JAD-159002
Das, R., Rauf, A., Akhter, S., Islam, M. N., Emran, T. B., Mitra, S., et al. (2021). Role of Withaferin A and its Derivatives in the Management of Alzheimer's Disease: Recent Trends and Future Perspectives. Molecules 26 (12), 3696. doi:10.3390/molecules26123696
Draghici, S., Khatri, P., Martins, R. P., Ostermeier, G. C., and Krawetz, S. A. (2003). Global Functional Profiling of Gene Expression. Genomics 81 (2), 98–104. doi:10.1016/s0888-7543(02)00021-6
Draghici, S., Khatri, P., Tarca, A. L., Amin, K., Done, A., Voichita, C., et al. (2007). A Systems Biology Approach for Pathway Level Analysis. Genome Res. 17 (10), 1537–1545. doi:10.1101/gr.6202607
Draghici, S. (2011). Statistics and Data Analysis for Microarrays Using R and Bioconductor. 2nd edition. London: Chapman and Hall/CRC.
Duan, Q., Reid, S. P., Clark, N. R., Wang, Z., Fernandez, N. F., Rouillard, A. D., et al. (2016). L1000CDS2: LINCS L1000 Characteristic Direction Signatures Search Engine. NPJ Syst. Biol. Appl. 2, 16015. doi:10.1038/npjsba.2016.15
Fernández-Martínez, J. L., Álvarez-Machancoses, Ó., deAndrés-Galiana, E. J., Bea, G., and Kloczkowski, A. (2020). Robust Sampling of Defective Pathways in Alzheimer's Disease. Implications in Drug Repositioning. Int. J. Mol. Sci. 21 (10), 3594. doi:10.3390/ijms21103594
Gao, L. B., Yu, X. F., Chen, Q., and Zhou, D. (2016). Alzheimer's Disease Therapeutics: Current and Future Therapies. Minerva Med. 107 (2), 108–113.
Gauthaman, K., Pushparaj, P. N., Rajeshkumar, M., Narasimhan, K., Al-Qahtani, M., Cheung, N. S., et al. (2014). Common Cellular and Molecular Mechanisms Underlying Alzheimer's Disease and Type 2 Diabetes: a Knowledge-Driven Approach. CNS Neurol. Disord. Drug Targets 13 (2), 247–258. doi:10.2174/18715273113126660138
Gene Ontology Consortium (2001). Creating the Gene Ontology Resource: Design and Implementation. Genome Res. 11 (8), 1425–1433. doi:10.1101/gr.180801
Grubman, A., Chew, G., Ouyang, J. F., Sun, G., Choo, X. Y., McLean, C., et al. (2019). A Single-Cell Atlas of Entorhinal Cortex from Individuals with Alzheimer's Disease Reveals Cell-type-specific Gene Expression Regulation. Nat. Neurosci. 22 (12), 2087–2097. doi:10.1038/s41593-019-0539-4
Himmelhoch, E., Latham, O., and Mcdonald, C. G. (1947). Alzheimer's Disease Complicated by a Terminal salmonella Infection. Med. J. Aust. 1 (23), 701–703. doi:10.5694/j.1326-5377.1947.tb94344.x
Hitzemann, R., Darakjian, P., Walter, N., Iancu, O. D., Searles, R., and McWeeney, S. (2014). Introduction to Sequencing the Brain Transcriptome. Int. Rev. Neurobiol. 116, 1–19. doi:10.1016/B978-0-12-801105-8.00001-1
Hsing, C. H., Hung, S. K., Chen, Y. C., Wei, T. S., Sun, D. P., Wang, J. J., et al. (2015). Histone Deacetylase Inhibitor Trichostatin A Ameliorated Endotoxin-Induced Neuroinflammation and Cognitive Dysfunction. Mediators Inflamm. 2015, 163140. doi:10.1155/2015/163140
Huang, L. C., Chang, Y. H., and Yang, Y. H. (2019). Can Disease-Modifying Anti-Rheumatic Drugs Reduce the Risk of Developing Dementia in Patients with Rheumatoid Arthritis? Neurotherapeutics. 16 (3), 703–709. doi:10.1007/s13311-019-00715-6
Hurd, M. D., Martorell, P., Delavande, A., Mullen, K. J., and Langa, K. M. (2013). Monetary Costs of Dementia in the United States. New Engl. J. Med. 368 (14), 1326–1334. doi:10.1056/NEJMsa1204629
Jiang, J., Wang, C., Qi, R., Fu, H., and Ma, Q. (2020). scREAD: A Single-Cell RNA-Seq Database for Alzheimer's Disease. iScience 23 (11), 101769. doi:10.1016/j.isci.2020.101769
Judge, A., Garriga, C., Arden, N. K., Lovestone, S., Prieto-Alhambra, D., Cooper, C., et al. (2017). Protective Effect of Antirheumatic Drugs on Dementia in Rheumatoid Arthritis Patients. Alzheimers Demen. 3 (4), 612–621. doi:10.1016/j.trci.2017.10.002
Kalamegam, G., Alfakeeh, S. M., Bahmaid, A. O., AlHuwait, E. A., Gari, M. A., Abbas, M. M., et al. (2020). In Vitro Evaluation of the Anti-inflammatory Effects of Thymoquinone in Osteoarthritis and In Silico Analysis of Inter-Related Pathways in Age-Related Degenerative Diseases. Front. Cel Develop. Biol. 8, 646. doi:10.3389/fcell.2020.00646
Kanehisa, M., and Goto, S. (2000). KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 28 (1), 27–30. doi:10.1093/nar/28.1.27
Kanehisa, M., Goto, S., Kawashima, S., and Nakaya, A. (2002). The KEGG Databases at GenomeNet. Nucleic Acids Res. 30 (1), 42–46. doi:10.1093/nar/30.1.42
Kanehisa, M., Goto, S., Furumichi, M., Tanabe, M., and Hirakawa, M. (2010). KEGG for Representation and Analysis of Molecular Networks Involving Diseases and Drugs. Nucleic Acids Res. 38 (Database issue), D355–D360. doi:10.1093/nar/gkp896
Kanehisa, M., Goto, S., Sato, Y., Furumichi, M., and Tanabe, M. (2012). KEGG for Integration and Interpretation of Large-Scale Molecular Data Sets. Nucleic Acids Res. 40 (Database issue), D109–D114. doi:10.1093/nar/gkr988
Kanehisa, M., Goto, S., Sato, Y., Kawashima, M., Furumichi, M., and Tanabe, M. (2014). Data, Information, Knowledge and Principle: Back to Metabolism in KEGG. Nucleic Acids Res. 42 (Database issue), D199–D205. doi:10.1093/nar/gkt1076
Kery, R., Chen, A., and Kirschen, G. W. (2020). Genetic Targeting of Astrocytes to Combat Neurodegenerative Disease. Neural Regen. Res. 15 (2), 199–211. doi:10.4103/1673-5374.265541
Li, L. H., Peng, W. N., Deng, Y., Li, J. J., and Tian, X. R. (2020). Action of Trichostatin A on Alzheimer's Disease-like Pathological Changes in SH-SY5Y Neuroblastoma Cells. Neural Regen. Res. 15 (2), 293–301. doi:10.4103/1673-5374.265564
Lin, C. H., Chien, W. C., Chung, C. H., Chiang, C. P., Wang, W. M., Chang, H. A., et al. (2020). Increased Risk of Dementia in Patients with Genital Warts: A Nationwide Cohort Study in Taiwan. J. Dermatol. 47 (5), 503–511. doi:10.1111/1346-8138.15277
Long, H. Z., Cheng, Y., Zhou, Z. W., Luo, H. Y., Wen, D. D., and Gao, L. C. (2021). PI3K/AKT Signal Pathway: A Target of Natural Products in the Prevention and Treatment of Alzheimer's Disease and Parkinson's Disease. Front. Pharmacol. 12, 648636. doi:10.3389/fphar.2021.648636
Luscher, B., Fuchs, T., and Kilpatrick, C. L. (2011). GABAA Receptor Trafficking-Mediated Plasticity of Inhibitory Synapses. Neuron 70 (3), 385–409. doi:10.1016/j.neuron.2011.03.024
Lycke, J., Svennerholm, B., Svenningsson, A., Horal, P., Nordqvist-Brandt, E., and Andersen, O. (1993). Possible Association of HTLV-I Infection and Dementia. Acta Neurol. Scand. 88 (3), 199–203. doi:10.1111/j.1600-0404.1993.tb04216.x
Matthews, K. A., Xu, W., Gaglioti, A. H., Holt, J. B., Croft, J. B., Mack, D., et al. (2019). Racial and Ethnic Estimates of Alzheimer's Disease and Related Dementias in the United States (2015-2060) in Adults Aged ≥65 Years. Alzheimers Demen. 15 (1), 17–24. doi:10.1016/j.jalz.2018.06.3063
Oksanen, M., Petersen, A. J., Naumenko, N., Puttonen, K., Lehtonen, Š., Gubert Olivé, M., et al. (2017). PSEN1 Mutant iPSC-Derived Model Reveals Severe Astrocyte Pathology in Alzheimer's Disease. Stem Cel Rep. 9 (6), 1885–1897. doi:10.1016/j.stemcr.2017.10.016
Palomer, E., Buechler, J., and Salinas, P. C. (2019). Wnt Signaling Deregulation in the Aging and Alzheimer's Brain. Front. Cell Neurosci. 13, 227. doi:10.3389/fncel.2019.00227
Pushparaj, P. N., Abdulkareem, A. A., and Naseer, M. I. (2021). Identification of Novel Gene Signatures Using Next-Generation Sequencing Data from COVID-19 Infection Models: Focus on Neuro-COVID and Potential Therapeutics. Front. Pharmacol. 12, 688227. doi:10.3389/fphar.2021.688227
Rasool, M., Malik, A., Waquar, S., Tul-Ain, Q., Jafar, T. H., Rasool, R., et al. (2018). In-Silico Characterization and In-Vivo Validation of Albiziasaponin-A, Iso-Orientin, and Salvadorin Using a Rat Model of Alzheimer's Disease. Front. Pharmacol. 9, 730. doi:10.3389/fphar.2018.00730
Rasool, M., Malik, A., Waquar, S., Zaheer, A., Asif, M., Iqbal, Z., et al. (2021). Cellular and Molecular Mechanisms of Dementia: Decoding the Causal Link of Diabetes Mellitus in Alzheimer's Disease. CNS Neurol. Disord. Drug Targets 20, 602. doi:10.2174/1871527320666210212114116
Riaz, M., Huq, A., Ryan, J., Orchard, S. G., Tiller, J., Lockery, J., et al. (2021). Effect of APOE and a Polygenic Risk Score on Incident Dementia and Cognitive Decline in a Healthy Older Population. Aging Cell 20, e13384. doi:10.1111/acel.13384
Sala Frigerio, C., Wolfs, L., Fattorelli, N., Thrupp, N., Voytyuk, I., Schmidt, I., et al. (2019). The Major Risk Factors for Alzheimer's Disease: Age, Sex, and Genes Modulate the Microglia Response to Aβ Plaques. Cel Rep. 27 (4), 1293–1306. doi:10.1016/j.celrep.2019.03.099
Sekar, S., McDonald, J., Cuyugan, L., Aldrich, J., Kurdoglu, A., Adkins, J., et al. (2015). Alzheimer's Disease Is Associated with Altered Expression of Genes Involved in Immune Response and Mitochondrial Processes in Astrocytes. Neurobiol. Aging 36 (2), 583–591. doi:10.1016/j.neurobiolaging.2014.09.027
Shen, C. Y., Xu, X. L., Yang, L. J., and Jiang, J. G. (2019). Identification of Narciclasine from Lycoris Radiata (L'Her.) Herb. And its Inhibitory Effect on LPS-Induced Inflammatory Responses in Macrophages. Food Chem. Toxicol. 125, 605–613. doi:10.1016/j.fct.2019.02.003
Su, Q., Li, T., He, P. F., Lu, X. C., Yu, Q., Gao, Q. C., et al. (2021). Trichostatin A Ameliorates Alzheimer's Disease-Related Pathology and Cognitive Deficits by Increasing Albumin Expression and Aβ Clearance in APP/PS1 Mice. Alzheimers Res. Ther. 13 (1), 7. doi:10.1186/s13195-020-00746-8
Wang, Z., Lachmann, A., Keenan, A. B., and Ma'ayan, A. (2018). L1000FWD: Fireworks Visualization of Drug-Induced Transcriptomic Signatures. Bioinformatics 34 (12), 2150–2152. doi:10.1093/bioinformatics/bty060
Wang, C., Xiang, Y., Fu, H., and Ma, Q. (2021). Use of scREAD to Explore and Analyze Single-Cell and Single-Nucleus RNA-Seq Data for Alzheimer's Disease. STAR Protoc. 2 (2), 100513. doi:10.1016/j.xpro.2021.100513
Winblad, B., Amouyel, P., Andrieu, S., Ballard, C., Brayne, C., Brodaty, H., et al. (2016). Defeating Alzheimer's Disease and Other Dementias: a Priority for European Science and societyThe Lancet. Neurology 15 (5), 455–532. doi:10.1016/S1474-4422(16)00062-4
World Health Organization (2012). Dementia: a public health priority. World Health Organization. Available at: https://apps.who.int/iris/handle/10665/75263 (Accessed June 1, 2021).
Wu, Y., and Zhang, K. (2020). Tools for the Analysis of High-Dimensional Single-Cell RNA Sequencing Data. Nat. Rev. Nephrol. 16 (7), 408–421. doi:10.1038/s41581-020-0262-0
Zhang, P., Kishimoto, Y., Grammatikakis, I., Gottimukkala, K., Cutler, R. G., Zhang, S., et al. (2019). Senolytic Therapy Alleviates Aβ-Associated Oligodendrocyte Progenitor Cell Senescence and Cognitive Deficits in an Alzheimer's Disease Model. Nat. Neurosci. 22 (5), 719–728. doi:10.1038/s41593-019-0372-9
Keywords: astrocytes, alzheimer’s disease and dementia, scREAD, single-nucleus RNA sequencing, in silico tools, anti-rheumatic agents, dasatinib, natural products
Citation: Pushparaj PN, Kalamegam G, Wali Sait KH and Rasool M (2022) Decoding the Role of Astrocytes in the Entorhinal Cortex in Alzheimer’s Disease Using High-Dimensional Single-Nucleus RNA Sequencing Data and Next-Generation Knowledge Discovery Methodologies: Focus on Drugs and Natural Product Remedies for Dementia. Front. Pharmacol. 12:720170. doi: 10.3389/fphar.2021.720170
Received: 03 June 2021; Accepted: 10 December 2021;
Published: 28 February 2022.
Edited by:
Tahir Ali, University of Calgary, CanadaReviewed by:
Luc Ver Donck, Janssen Research and Development, BelgiumSatoshi Saito, National Cerebral and Cardiovascular Center, Japan
Copyright © 2022 Pushparaj, Kalamegam, Wali Sait and Rasool. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Peter Natesan Pushparaj, cGV0ZXIubi5wdXNocGFyYWpAZ21haWwuY29t; Mahmood Rasool, bWFobW9vZHJhc29vbEB5YWhvby5jb20=
 Peter Natesan Pushparaj
Peter Natesan Pushparaj Gauthaman Kalamegam
Gauthaman Kalamegam Khalid Hussain Wali Sait3
Khalid Hussain Wali Sait3 Mahmood Rasool
Mahmood Rasool