- 1Department of Pharmacy, Personalized Drug Research and Therapy Key Laboratory of Sichuan Province, Sichuan Provincial People’s Hospital, School of Medicine, University of Electronic Science and Technology of China, Chengdu, China
- 2Department of Pharmacy, Chongqing Red Cross Hospital (Jiangbei District People’s Hospital), Chongqing, China
- 3Department of Pharmacy, Affiliated Hospital of Southwest Jiaotong University, The Third People’s Hospital of Chengdu, Chengdu, China
Background: The placental barrier is a critical interface that regulates drug transport between maternal and fetal circulation and is an important component in assessing fetal drug-exposure risk. Since pregnant women are often excluded from clinical trials, pharmacokinetic (PK) analysis data on placental drug transport remain limited. Currently, in vitro experiments and in silico simulation strategies are the primary and effective means for understanding drug transport across the placenta.
Method: Various in vitro experimental methods, including cell monolayer models, ex vivo placental perfusion, and organ-on-a-chip platforms, along with model-based computational simulations, were systematically reviewed. The advantages, limitations, and potential future applications of these methods were evaluated.
Result: A total of seven studies using cell models, 28 employing ex vivo perfusion, six utilizing placenta-on-a-chip technology, and 39 focusing on in silico simulations, were identified, involving 8, 34, 5, and 42 drugs, respectively. Antiviral agents, antibiotics, and opioids were the most frequently investigated drug types. Overall, in silico simulations informed by in vitro data as baseline parameters and constraints demonstrated higher predictive accuracy. Integrating multi-model data was shown to be a reliable strategy for improving the precision of placental PK studies.
Conclusion: This review highlights the current strategies in placental PK research and supports safer drug use during pregnancy. Multi-model data integration is essential for developing reliable and quantitative fetal drug-exposure assessment frameworks, thus addressing data gaps caused by the exclusion of pregnant women from clinical trials.
Highlights
This review systematically examined state-of-the-art in vitro modeling methods, including cell monolayer models, ex vivo placental perfusion experiments, organ-on-a-chip platforms, and model-guided computer simulations, and it critically appraised their respective strengths and limitations in quantifying transplacental drug transfer.
Physiologically based pharmacokinetic (PBPK) models simulate the absorption, distribution, metabolism, and excretion (ADME) of drugs and are a powerful tool for predicting fetal drug exposure during pregnancy.
The implementation of an integrated multi-model approach significantly enhances the reliability of maternal–fetal pharmacokinetic (PK) assessments, providing robust evidence to support guideline updates and clinical decision-making during pregnancy.
1 Introduction
Many pregnant women require one or more medications for health problems (Be et al., 2025; Lupattelli et al., 2014). Drug use during pregnancy is on the rise (Subramanian et al., 2023; Phillips et al., 2024), and recent studies have shown an increase in the use of at least one prescribed medication from 56.9% in 1998 to 63.3% in 2018 and an increase in the concurrent use of multiple medicines from 24.8% to 35.2% in the Danish population (Thunbo et al., 2024). Although drugs have therapeutic significance in clinical practice, their safety in mothers and fetuses remains unclear. Maternal drug use may affect the fetus, leading to developmental abnormalities (Foster et al., 2022; De Felice and Kane, 2021) or long-term health issues (Abolhassani et al., 2023; Owen et al., 2021). Therefore, fetal drug exposure and safety assessments have gradually become important topics in clinical pharmacology.
The effects of maternal drug use on the fetus are complex and sensitive and require a multifaceted assessment. The syncytiotrophoblast (STB) and its two polarized plasma membranes, the maternal-facing microvillous membrane (MVM) and fetal-facing basal membrane (BM), represent the primary barrier in the human placenta, controlling transplacental transfer of small solutes. Key factors, including the physicochemical properties of the drug (MW, logP, pKa, protein binding, transporters, and metabolism) and placental function at different time points, affect the ability of the drug to cross the placental barrier during pregnancy. Due to the complexity of the placental structure, the assessment of transplacental drug transfer is a long-standing and difficult issue. Understanding and quantifying drug transport through the placenta can inform the assessment of fetal drug exposure. However, pregnant women are often excluded from clinical and post-marketing studies. This has led to a lack of PK data regarding drug use during pregnancy, particularly data related to placental drug transfer (Pariente et al., 2016). Therefore, considering the mismatch between the available evidence from existing studies and real-world medication needs during pregnancy (Haas et al., 2018), PK research on placental drug transfer remains a challenging and long-term endeavor.
In recent years, with the advancement of computer and bioengineering technologies, methods for placental PK studies have developed considerably. In addition to the previously established in vitro methods (e.g., cell monolayer model, Figure 1a; ex vivo placental perfusion experiment; Figure 1b), several new attempts have been made (for example, organ-on-a-chip Figure 1c; PBPK model, Figure 1d; in silico models, Figure 1e). Multiple approaches have been attempted, including the combination of PBPK models with in vitro and ex vivo data. However, systematic evaluation and synergistic integration of these methods have not yet been fully explored. This review systematically discusses the evolution, application scenarios, and limitations of various placental drug transfer methods in the context of drug studies. We have also attempted to explore the future directions of these research methods. This study ultimately aims to contribute to the methodological advancement of placental pharmacokinetics and risk assessment for medication use during pregnancy.
Figure 1. Overview of methods for placental drug-transport studies. The main approaches of placental drug-transport studies include the BeWo/Caco-2 cell model (a), the ex vivo placental perfusion experiment (b), the organ-on-a-chip model (c), the PBPK model (d), and in silico models (e).
2 Literature review procedure
A systematic literature search was performed in PubMed (https://pubmed.ncbi.nlm.nih.gov/) from inception to February 2025 using the keywords “placental drug transfer,” “placental pharmacokinetics,” and “foetal drug exposure.” Two team members independently screened the titles and abstracts, and any controversial finding was resolved through discussion with a third researcher. Studies were included if they investigated drug transfer across the human placenta using cell monolayer models, ex vivo placental perfusion, organ-on-a-chip platforms, or in silico simulations and reported relevant methodological outcomes. Studies not related to the human placenta, not addressing drug transfer across the placenta, lacking original data, or with insufficient methodological details were excluded. Figure 1 summarizes the in vitro models applied in transplacental drug transfer research. Detailed information on the progress, advantages, and limitations of each method was reviewed and summarized to highlight the future research directions for in vitro human placental models.
3 Cell experimental models for placental drug-transfer
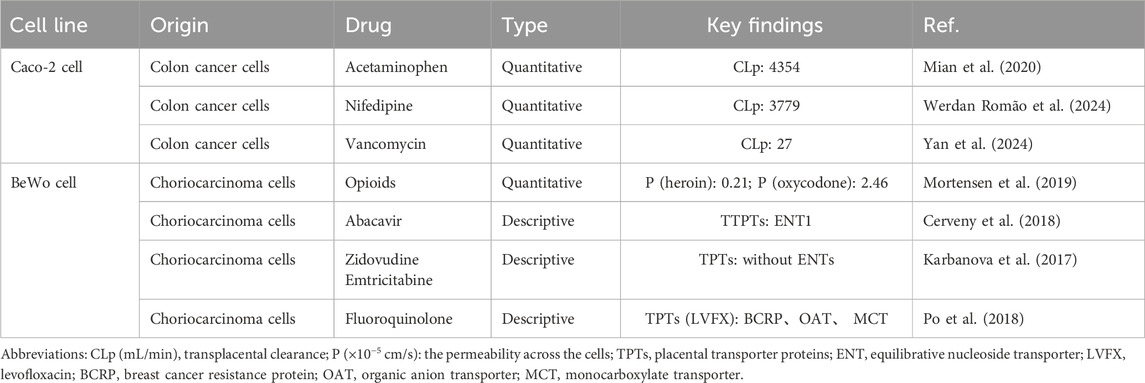
Current placental transport models include primary trophoblasts, trophoblast cell lines (BeWo b30, JEG-3, JAR, and HTR-8/SVneo), stem cell-derived systems, and intestinal Caco-2 (cancer coli-2) cells, with BeWo b30 and primary trophoblasts remaining the gold standards according to the FDA/ICH guidelines. We mainly focus on placental barrier studies, with studies on BeWo cells focusing on the mechanism of drug transport across the placenta and the role of transport proteins, while Caco-2 cells are mainly used to assess drug permeability across the placental barrier, and therefore, the two cellular studies will be described in detail below.
3.1 Trophoblast cell lines
The BeWo cell line serves as a valuable in vitro model for placental transport studies. Derived from human choriocarcinoma, these cells form functional syncytialized monolayers when properly differentiated upon cAMP induction (Drwal et al., 2018), exhibiting key characteristics of the placental barrier (Moore et al., 2021). Syncytialized BeWo cells were employed as an in vitro model of the placental barrier to investigate drug-transport kinetics, following verification of barrier integrity through transepithelial electrical resistance (TEER) measurements (≥80 Ω · cm2).
The transporters expressed in the STB, including the ABC family members (P-gp, BCRP, and MRP) and SLC family members (OCT and OAT), bidirectionally regulate placental substance exchange through efflux (e.g., P-gp/BCRP/MRP-mediated excretion into maternal circulation) and uptake (e.g., OCT/OAT-mediated substance intake) mechanisms (Bai et al., 2016). BeWo b30 cells, a subclone of the BeWo cell line, expresses important placental transporters (Han et al., 2018), particularly BCRP and P-gp (Ceckova et al., 2006), and it demonstrates compound permeability patterns that correlate well with ex vivo results (Correia Carreira et al., 2011; Li et al., 2013; Poulsen et al., 2009). Researchers have successfully utilized this model to initially elucidate the transplacental transport of clinically relevant compounds spanning antivirals, opioids, and fluoroquinolones (Table 1).
However, this model has notable limitations. First, a comparative transcriptomic study of human placental in vitro models revealed substantial numbers of differentially expressed genes across all in vitro placental models. To date, no placental cell line has been able to accurately mimic human placental tissue (Lapehn et al., 2024). Although steroidogenic enzymes such as 3β-hydroxysteroid dehydrogenase type 1 (3β-HSD1) and 11β-hydroxysteroid dehydrogenase type 2 (11β-HSD2) are highly expressed and functionally active in the in vivo placenta, their expression levels in trophoblast cell lines differ from those in the in vivo placenta (Karahoda et al., 2021). Similarly, cytochrome P450 enzymes, including CYP2C8, CYP2C9, and CYP2J2, are poorly expressed (Kammala et al., 2022). Second, the expression or localization of placental cellular transporters may also vary with temporal placental development; thus, the cell model cannot fully replicate the dynamic changes in transporter expression that occur during different gestational stages (Veiga-Lopez et al., 2025). Future research should focus on three key directions to enhance BeWo-based placental model characteristics (Be et al., 2025): developing microfluidic co-culture systems integrating BeWo cells with human umbilical vein endothelial cells (HUVECs) (Fuenzalida et al., 2024) under physiological shear stress; (Lupattelli et al., 2014) applying CRISPR editing to introduce disease-specific mutations; (Subramanian et al., 2023) establishing 3D organoid models through BeWo-fibroblast co-culture. These advanced models require standardized validation, including cAMP-induced syncytialization, TEER monitoring, and transporter functionality assays, to ensure regulatory acceptance and enhance the predictive accuracy in placental drug-transport studies.
3.2 Caco-2 cell
The Caco-2 cell model is widely used to study drug permeability (Artursson et al., 2001). Derived from human colon cancer cells, Caco-2 cells differentiate into polarized monolayers that resemble small intestinal epithelial cells and mimic the barrier function of the human intestine (Sambuy et al., 2005). Over the past 2 decades, the Caco-2 monolayer model has become a crucial tool for preclinical drug development, particularly for permeability screening (Panse and Gerk, 2022). The Caco-2 monolayer allows the study of all major absorption pathways, such as passive transcellular and paracellular diffusion and carrier-mediated active and facilitated transport (Volpe, 2020).
Caco-2 cells form a continuous monolayer barrier for intestinal barrier simulation, and similarly, researchers have adapted this well-characterized system as a functional surrogate for placental barrier studies, leveraging its reproducible tight junction formation and transport properties. Mian et al. utilized a confluent Caco-2 monolayer system in Transwell chambers, with pre-experimental TEER measurements confirming barrier integrity (Mian et al., 2020). This established model served as an effective placental barrier surrogate for characterizing acetaminophen transport and determining the apparent permeability coefficient (Papp) across the placental barrier. This experimental approach has been similarly applied to evaluate the transplacental permeability of various therapeutic agents, including nifedipine and vancomycin (Werdan Romão et al., 2024; Yan et al., 2024) (Table 1).
The utility of Caco-2 cells as an in vitro model for placental transport research is constrained by several inherent biological and methodological limitations. For instance, there are biological differences between Caco-2 cells and placental epithelial cells, which may lead to overestimation or underestimation of drug permeability. Caco-2 cells express key efflux transporters found in the STB, including P-glycoprotein (P-gp/ABCB1), BCRP (ABCG2), and certain MRPs (ABCC family) (Bai et al., 2016), but they show limited expression of uptake transporters such as OCTs and OATs (Wenzel et al., 2021). Therefore, Caco-2 cells are suitable for studying efflux-mediated transport but have limitations in modeling placental uptake processes. To bridge physiological gaps between models and human placenta, researchers employ genetic engineering strategies to upregulate key protein expression (Ohta et al., 2020) and apply biosimilar mucus to mimic native mucosal barriers (Panse and Gerk, 2022). Nevertheless, such models provide valuable preliminary data when access to more physiologically relevant systems is limited.
3.3 Primary trophoblasts
Primary cytotrophoblasts (CTBs) isolated from term human placentas that spontaneously fuse and differentiate into STB (Li and Schust, 2015)-like cells in vitro are used to investigate the functions of the STBs (Hawkins et al., 2025) and placentas with multiple modifications (Motomura et al., 2025). These cells maintain native transporter expression profiles (e.g., BCRP and P-gp) and metabolic activity, which are the most physiologically relevant in vitro cell model for studying human placental function, but unfortunately, CTBs are rarely used for placental barrier-related studies. In addition, trophoblast stem cells (hTSCs) were derived from human villous CTBs and human blastocysts (Okae et al., 2018). These long-term stem cell cultures may give rise to CTBs, EVTBs, and STB-like cells that show transcriptomes similar to those of the corresponding primary trophoblasts (Shannon et al., 2024), which have emerged as a powerful tool for modeling the placental cytotrophoblast in vitro (Karakis et al., 2025). Given that the primary focus of our investigation centers on placental barrier function, a comprehensive discussion of this aspect falls beyond the scope of this study.
4 Organ experimental models for placental transfer
Ex vivo placental perfusion experiments, first proposed by Professor Maurice Panigel in 1967, have been continuously refined to address challenges such as revascularization and placental hypoxia resistance (Schneider et al., 2021). As research has expanded in recent years, new methods of studying placental drug-transport have emerged. Organ-on-a-chip technology, which integrates microfluidics and cell culture, replicates the human organ microenvironment on a chip and accurately simulates the blood flow, oxygen exchange, and nutrient delivery.
4.1 Ex vivo placental perfusion experiments
Ex vivo placental perfusion experiments are crucial ex vivo models used to study the transport of exogenous substances and the effects of hormones (Page, 1991). This model simulates the exchange of substances between the mother and fetus by creating specific external conditions and perfusion systems that closely mimic physiological processes in the human body. This model allows researchers to study the mechanisms of exogenous substance transfer, fetal exposure, and associated diseases (Villalobos-Labra et al., 2023). In addition, this model can be used to study the effects of factors such as inflammation and oxidative stress on biochemical and physiological imbalances that are closely linked to conditions such as preeclampsia, fetal growth restriction, diabetes, and microbial infections (Koren and Ornoy, 2018).
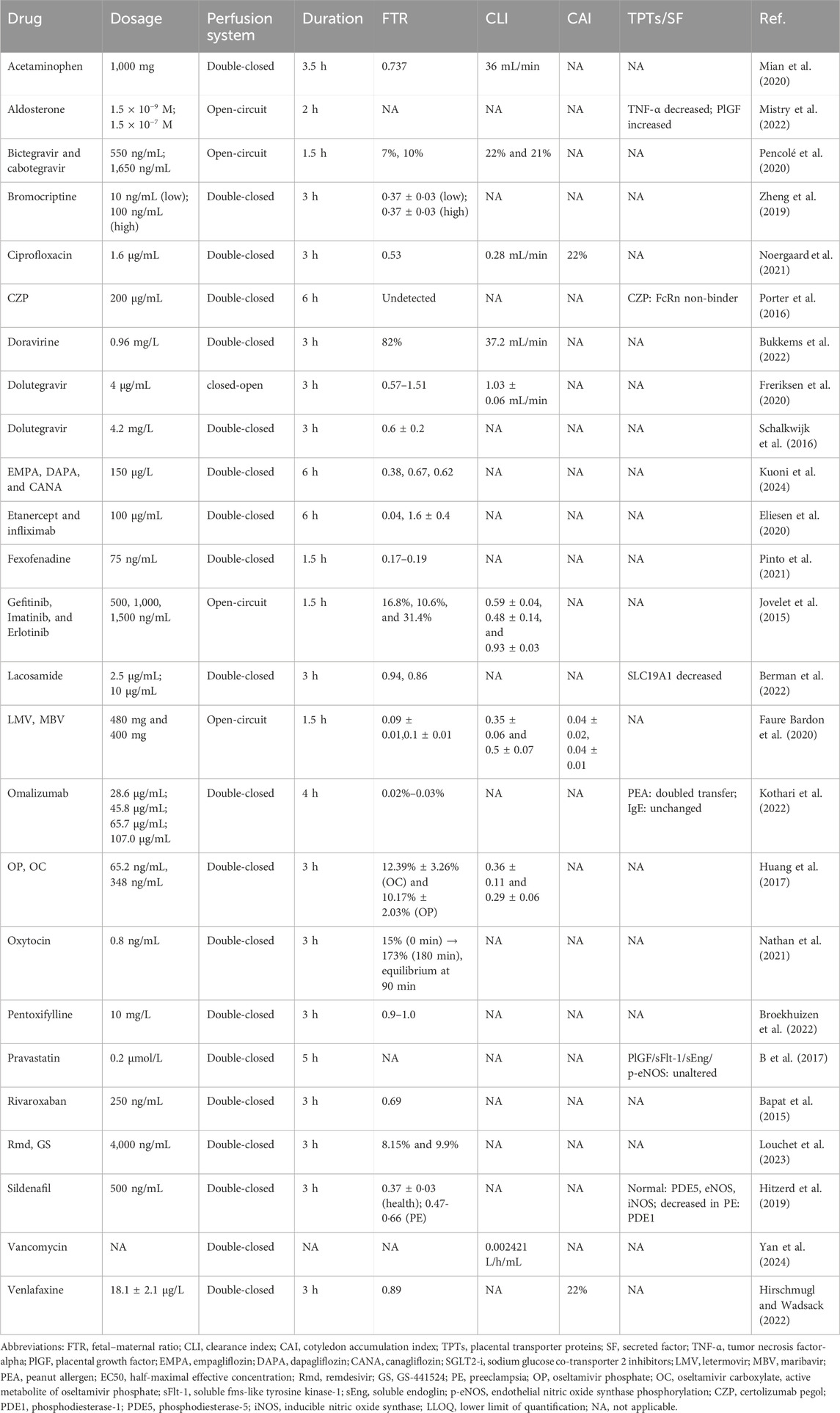
Ex vivo placental perfusion experiments comprised maternal and fetal reservoirs, the placenta, and a perfusion pump that simulated blood flow with controlled pressure and flow rate (Figure 2). A well-defined artery–vein pair on the fetal-side cotyledon with minimal branching was chosen for cannulation following the confirmation of intact placental integrity post-delivery (Jovelet et al., 2015). The system only remains patent for 2–6 h, so only short-term assessments can be made, and the circuit design (open, semi-closed, or closed loop) determines the physiological relevance, ranging from first-pass transfer simulation to steady-state PK assessment. The transfer process is mathematically described by Fick’s first law using the transfer equations outlined by Mian et al. (2020). Our review focused on 28 recent studies (2015–2025) employing ex vivo human placental perfusion to investigate transplacental kinetics of various therapeutics, including antivirals, psychotropics, and biologics (Table 2). This 10-year timeframe was selected to account for methodological advancements while ensuring data comparability. Notably, researchers exhibit a distinct preference for double-closed perfusion systems, which consistently operate for significantly longer durations than open-circuit configurations (avg. 3.6 h vs. 1.9 h). This operational divergence substantiates the superior suitability of double-closed systems for steady-state PK investigations. Small molecules dominate the studied drugs (30/34, 88%), exposing a critical evidence gap for biologics/antibodies despite their clinical relevance.
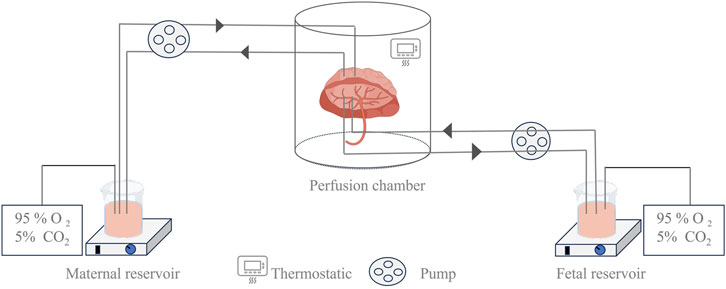
Figure 2. Illustration of the ex vivo dual-side cotyledon perfusion model. This device shows a schematic diagram of a dual closed-circuit extracorporeal placental perfusion test, which more closely simulates the process of maternal–fetal substance exchange and is widely used in the study of maternal–fetal transport mechanisms.
In addition, ex vivo placental perfusion models have emerged as pivotal tools for evaluating nanocarrier transplacental behavior, with recent studies demonstrating that targeted lipid nanoparticles (LNPs) remain largely confined to maternal circulation, exhibiting minimal fetal transfer, thereby suggesting their utility in preventing unintended drug passage across the placental barrier (van Kammen et al., 2024). Complementing these findings, a review of 16 perfusion studies further elucidates that nanoparticle (NP) transport efficiency is critically governed by physicochemical determinants, particularly sub-100-nm dimensions and surface modifications such as PEGylation, which collectively modulate placental uptake kinetics (Aengenheister et al., 2021). These insights establish that rational nanocarrier design represents a promising strategy for blocking unintended fetal drug exposure.
Despite being non-invasive, ex vivo placental perfusion experiments are constrained by the limited availability of human placental tissue (Parks, 2015) and the need to maintain strict physiological conditions (37 °C, fetal arterial pressure 30 mmHg–60 mmHg, 95% O2/5% CO2 (maternal), 95% N2/5% CO2 (fetal), oxygen consumption, glucose, lactate, and HCG indicators) (Schneider, 2015). Furthermore, the intricate vascular network of the placenta makes it challenging to accurately replicate blood flow and material exchange ex vivo, resulting in a low success rate and limiting its application (Mathiesen et al., 2010). Unfortunately, full-term placental extracts may not reflect the evolution of the placenta during pregnancy.
Future advancements should refine placental perfusion systems by implementing multi-cannula designs (e.g., 22 maternal cannulas) to replicate physiological oxygen gradients, which are confirmed by direct measurements to maintain the target median oxygen tension (∼50 mmHg) and reduce hypoxic heterogeneity in the intervillous space (Schneider, 2015). Concurrently, preserving endothelial integrity requires real-time monitoring of trophoblastic vacuolization, which is a histopathological marker strongly correlated with fetal circuit leakage, combined with mechanical stress reduction via low-pressure perfusion (<60 mmHg) and albumin-supplemented media (Maroun et al., 2014). Crucially, mitigating oxidative stress demands protocol modifications, such as pre-perfusion hypothermia (4 °C) and HO-1 inducers (e.g., hemin) to activate antioxidant pathways, pulsed reoxygenation to minimize ROS bursts, and antibiotic supplementation to prevent bacterial-induced inflammation (Schneider, 2009). Moreover, the standardization of placental perfusion techniques should be accelerated to ensure data consistency across research centers, facilitate cross-study validation, and promote broader adoption (Schneider et al., 2022). It is foreseeable that with continuous optimization, placental perfusion experiments will hold significant application in placental PK research.
4.2 Organ-on-a-chip
An organ-on-a-chip is an in vitro biomimetic microfluidic device that replicates the structure and function of human organs, typically at 10–6 to 10–4 times the size of the actual organ (Shuler, 2017). It provides a model that closely simulates the in vivo cellular environment, enabling cells and tissues to grow and complete their life cycles. This device uses microchip technology to create microfluidic cell culture chambers, where live cells are organized according to the organ physiology. By reproducing multicellular structures, tissue interfaces, and vascular perfusion, it mimics in vivo organ function and cellular environments (Figure 3) (Cavero et al., 2019). The basic organ-on-a-chip is fabricated using standard soft-lithography techniques, where a 10:1 mixture of polydimethylsiloxane (PDMS) substrate and curing agent is cast onto an SU-8 master mold to form a bilayer or multicavity microfluidic structure with precisely defined channels. A porous polyester track-etched membrane or polycarbonate membrane is then sandwiched between the microchannels as a biological barrier, which is subsequently assembled and selectively coated with extracellular matrix components to form a biomimetic placental barrier through plasma bonding.
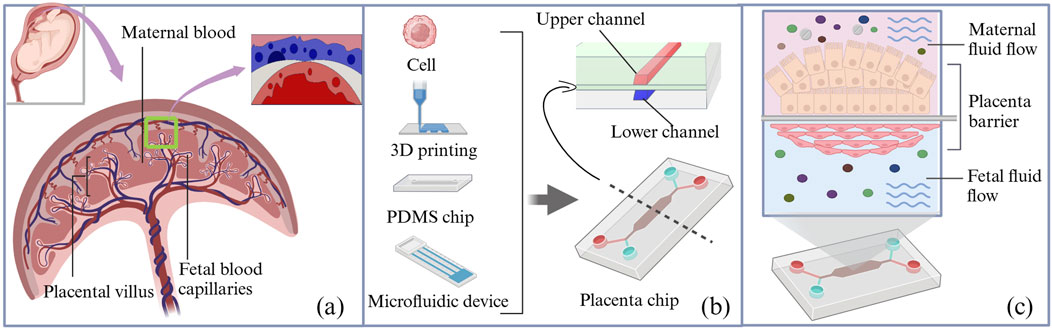
Figure 3. Schematic diagram of the structure of the placenta organ-on-a-chip device. (a) Schematic diagram of the placental barrier. (b) Schematic of composition and cross-sectional view of the organ-on-a-chip microfluidic device; a porous membrane separates the maternal and fetal channels in the microfluidic device. (c) The maternal and fetal sides were composed of BeWo cells and HUVECs, respectively. PDMS, polydimethylsiloxane.
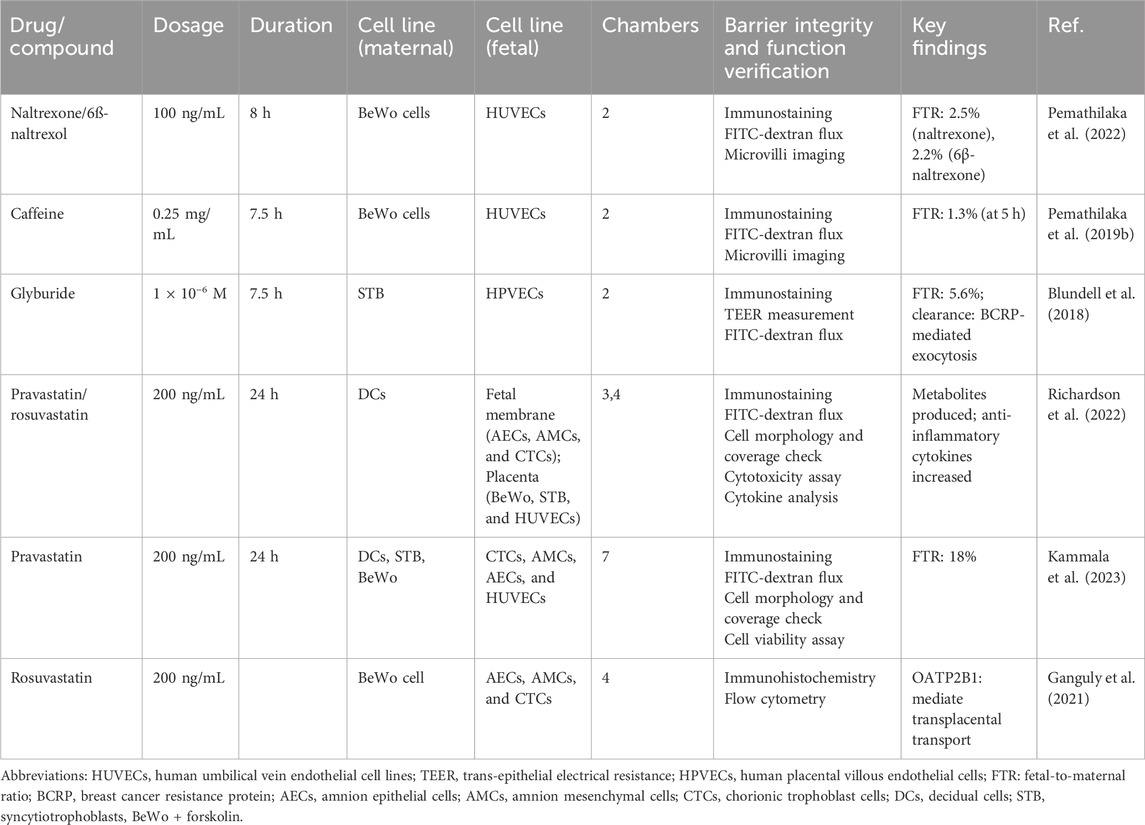
A comprehensive literature search revealed that, to date, placental drug transport has been evaluated and reported for only five pharmacological agents using the placenta-on-a-chip technology under controlled microphysiological conditions (Table 3), involving opioids, hypoglycemic agents, statins, and other drugs. Evidently, placenta-on-a-chip studies commonly use a core co-culture model of paired trophoblast cells (BeWo/STB) and endothelial cells (HUVECs/HPVECs) to replicate the maternal–fetal barrier. For statin (e.g., rosuvastatin)-specific studies, this basis can be extended to complex multicompartmental systems (3 layers–7 layers), which include fetal membrane cells (AECs, AMCs, and CTCs) and decidual cells (DCs) to mimic tissue interactions. Barrier integrity is generally verified by a tripartite protocol: immunostaining, FITC-dextran permeability assay, and morphological analysis, while some studies are supplemented with TEER measurements. Functionally, the simpler two-chamber design is sufficient for passive diffusion studies, whereas the ≥4-chamber configuration becomes crucial for drug metabolism studies, allowing important analyses of statin-derived metabolites and inflammatory cascades.
The development of three-dimensional (3D) placenta-on-a-chip models has been accelerated by the availability of microfluidic devices, commercial chips, and 3D bioprinting technologies. For instance, HUVECs and placental BeWo cells were co-cultured (1:3) for the first time to form spheroids (microtissues) on a 3D Petri Dish® mold, which mimicked in vivo responses more closely than 2D systems, in three main ways (Be et al., 2025): spontaneous self-organization into a vascularized histoarchitecture with HUVECs peripherally enveloping a BeWo core, mimicking placental villi (Lupattelli et al., 2014); enhanced intercellular junction formation (E-cadherin/ZO-1) and cytoskeletal reorganization yielding physiologically relevant TEER values (Subramanian et al., 2023); superior functional maturation evidenced by elevated β-hCG secretion and drug metabolic capacity that more accurately predicted in vivo pharmacological responses (Öztürk et al., 2024). Cao et al. developed a biomimetic placental barrier model that incorporated glucose transport and hCG secretion (Cao et al., 2023). Another study developed a second-trimester (14 W–28 W) placental model (Vidal et al., 2024) that offered valuable insights into placental drug transfer. These methodological advancements, when successfully implemented in transplacental drug studies, are anticipated to significantly enhance the quality of evidence regarding fetal drug exposure in future research endeavors.
The placenta-on-a-chip technology offers significant advantages over traditional trophoblast monolayer models by recreating the multilayer structure of the placental barrier and the hemodynamic environment (Zambuto et al., 2024). However, the model has several limitations. First, current microarrays struggle to replicate complex overall physiological environments, which hamper their ability to model systemic diseases. Second, current placenta-on-a-chip models are a poor representation of the human placental villous structure. Another limitation is that as the complexity of 3D co-culture models increases, it becomes more challenging to perform high-resolution screening and accurately localize the tissues (Bhatia and Ingber, 2014). Future placenta-on-a-chip development must, therefore, improve both physiological fidelity and microscale spatial resolution (Pemathilaka et al., 2019a; Esch et al., 2015), while incorporating gestational changes (Blundell et al., 2018). Critical advancements must target the following (Be et al., 2025): multi-lineage co-culture optimization (trophoblast/endothelial/stromal cell interactions) to maintain tissue-relevant cellular diversity (Lupattelli et al., 2014); sub-10 μm spatial resolution achievement through nanopatterned surface functionalization (Subramanian et al., 2023); hierarchical biomaterial development integrating decellularized ECM and precision-bioprinted villous structures; (Phillips et al., 2024) programmable microenvironment platforms with feedback-controlled gas/endocrine factor delivery, enabling phase-transition modeling between gestational periods. Following significant advancements in organ-on-a-chip technology, subsequent research efforts should prioritize the implementation of these systems for longitudinal exposure studies and polypharmacy interaction analyses that are specifically designed to characterize the teratogenic consequences of sustained drug administration during critical periods of fetal development.
5 In silico simulation for placental transfer
Animal studies remain difficult to interpret due to structural and functional inter-species placental differences. The ex vivo perfusion of the human placental cotyledon is the method of reference for studying the human placental transfer of drugs because it is thought to mimic the functional placental tissue. However, it is challenged by the constraints of limited placental tissue and low experimental success rates. Recently, in silico techniques have further been advanced as complementary tools to validate experimental placental transfer data, offering a promising alternative for high-throughput screening of potential fetotoxicity at the early stages of drug design. PBPK models are promising in silico simulation tools that provide valuable insights into drug PK, particularly those that are difficult to assess in vivo. Several other methods have also been used to investigate placental PK.
5.1 Physiologically based pharmacokinetics (PBPK)
The PBPK model uses mathematical simulations to describe the absorption, distribution, metabolism, and excretion (ADME) of drugs by integrating the anatomical, physiological, and drug-specific data with preclinical and clinical information to simulate PK (Dallmann et al., 2018). When applied to special populations, such as pregnant women and neonates, PBPK models incorporate organ development and individual variations. The key components of PBPK model development are shown in Figure 4a. The general workflow for developing a maternal–fetal PBPK model is shown (Figure 4c). First, a non-pregnant PBPK model was established and validated by comparison with the observed in vivo data. Once the non-pregnant PBPK model accurately captured PK, all drug-specific parameters were fixed. Pregnancy-specific changes [e.g., increase in blood volume, changes in metabolism, increased cardiac output, lower hypertension, increased renal blood flow, and variation with gestational week (Zhang et al., 2017; Zhang and Unadkat, 2017)] were then incorporated to extend the model to a pregnancy-specific version (Dallmann et al., 2017), which involved adding pregnancy-specific compartments to the 27-compartment model (Figure 4b). The maternal–fetal PBPK (M–F PBPK) model is linked via a placental permeability-limited model subdivided into maternal blood, placental tissue, and fetal blood compartments (Zhang and Unadkat, 2017). Transplacental clearance (Yan et al., 2024) and other key parameters were assessed using various methods (Figure 4d) and were included in the final formation of the M–F PBPK models. In addition, depending on the purpose of the study, researchers may selectively incorporate placental drug metabolism/efflux transporters into the model.
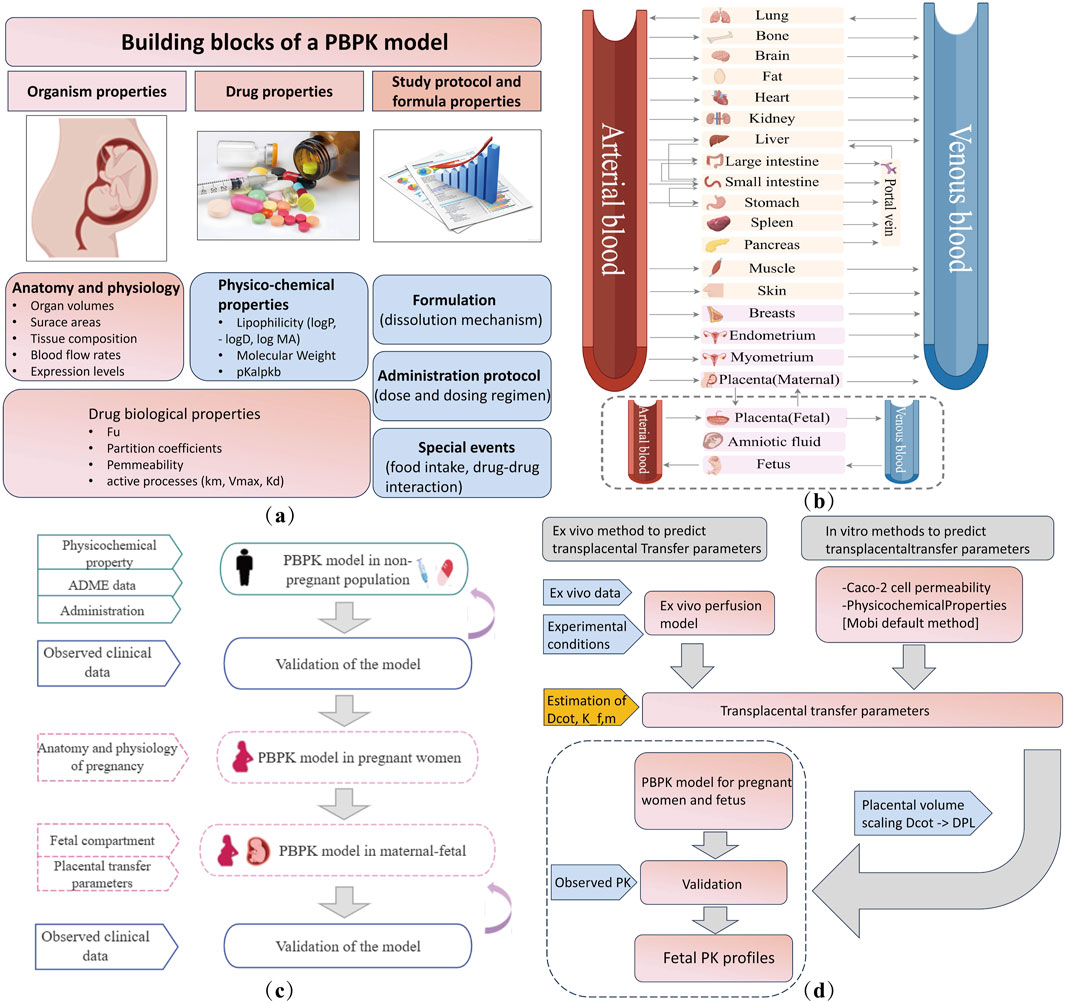
Figure 4. Maternal–fetal PBPK model details. (a) Building blocks for developing a PBPK model. (b) Structural diagram of the PBPK model of a pregnant woman in 27 compartments. The pink block section shows nine pregnancy-specific compartments. Fetal compartments can be refined according to the study objectives. (c) Workflow of maternal–fetal PBPK model development and evaluation. (d) Transplacental parameter estimation and integration. ADME, absorption, distribution, metabolism, and excretion. Dcot, transcotyledon passive diffusion clearance; Kf,m, partition coefficient between the fetal and maternal compartment; Dpl, transplacental passive diffusion clearance; PK, pharmacokinetics.
The M–F PBPK model was developed in 2015 (Dallmann et al., 2018) and has rapidly advanced since 2020. Some research groups have extensively worked on PBPK models to predict fetal drug exposure and showed very encouraging results. In Table 4, we summarize the developed M–F PBPK models encompassing 42 pharmacological agents from 39 studies, including antiviral agents, antibacterial, opioids, antipsychotics, hormones, analgesics, anti-hypertensives, and antineoplastic agents. The dominance of antivirals (39%) in all M–F PBPK studies is highlighted in Figure 5, which shows that the use of antiviral drugs in the treatment of HIV/AIDS and hepatitis B is a global concern. During our literature search, we found several interesting research areas, including PBPK models that focused on predicting maternal drug concentrations and fetal environmental toxin exposure. However, these topics were not the focus of this study and have not been discussed further.
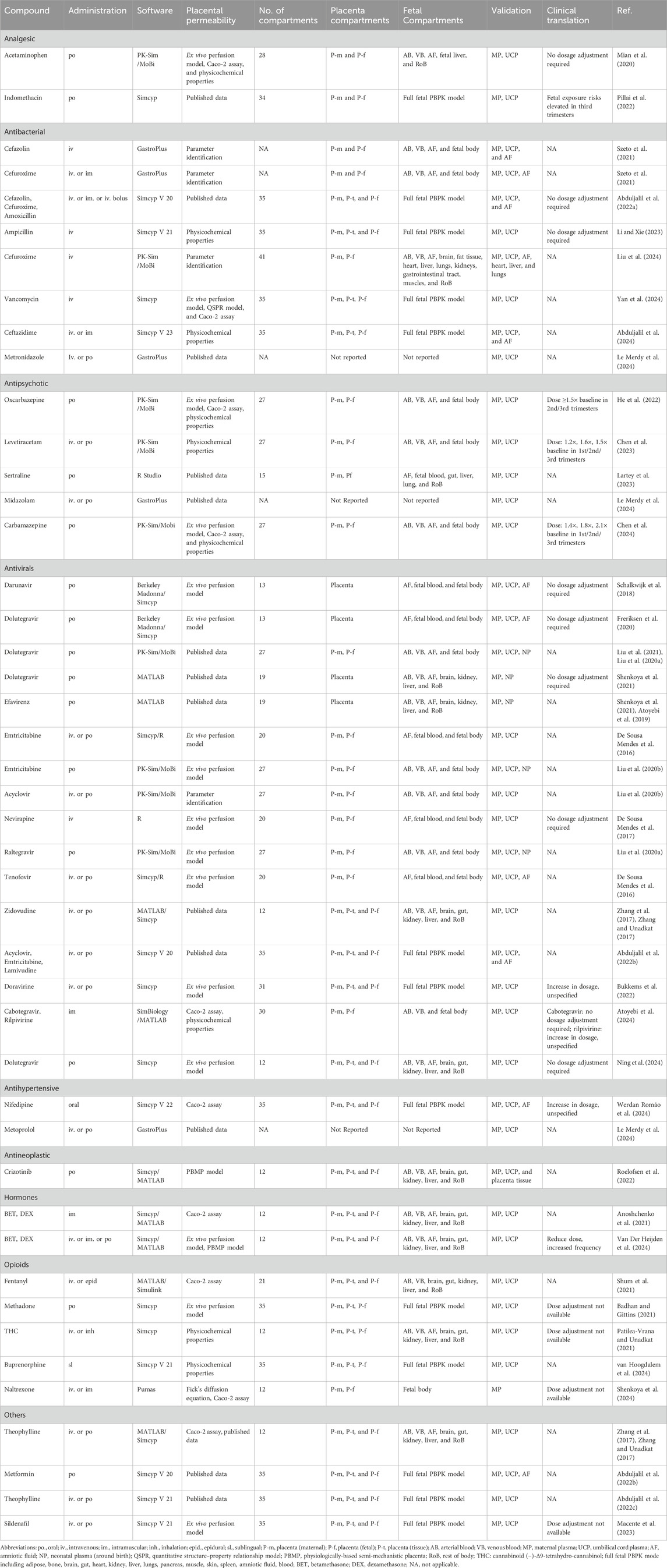
Table 4. Overview of reported fetal drug exposure studies for M–F PBPK (categorized by the pharmacological effect).
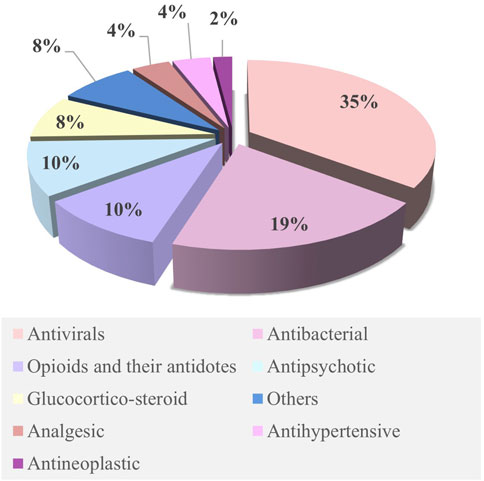
Figure 5. Ratios of various drugs studied in the M–F PBPK model. Publications focusing on M–F PBPK were identified by systematically searching the scientific literature for peer-reviewed articles using PubMed. Relevant key terms were used to identify publications (e.g., “Physiologically based pharmacokinetic model,” “Pregnancy,” and “Foetal”). No date restriction was set. Collected publications were scrutinized for possible additional references not covered by this search.
Although PBPK is a physiological PK simulation, it still relies on mathematical and mechanistic modeling. The goal of PBPK model development is to accurately reflect the in vivo physiological conditions. Studies have incorporated the fetal liver as an independent metabolic organ (Mian et al., 2020), considering the scaling of fetal drug-unbound fractions (Roelofsen et al., 2022) and adjusted parameters, such as fetal liver and kidney volumes and cytochrome P450 enzyme levels, to model fetal drug metabolism and clearance (Szeto et al., 2021). These efforts were aimed at improving the precision of fetal drug concentration predictions. Moreover, given the dynamic development of the placenta (Aplin et al., 2020) and the challenges in obtaining early pregnancy samples, future research should prioritize drug safety studies during early and mid-pregnancy to better guide maternal medication use. Further exploration of these approaches is crucial for future research as it will provide valuable evidence for clinical decision-making.
5.2 Computational approaches
Diverting from mechanistic approaches, novel mathematical techniques such as artificial intelligence and machine learning (ML) methods ranging from regression analysis to deep learning and neural networks have recently been utilized for the investigation of placental PK. One study used ML to stratify high-risk pregnant women based on biomarkers and electronic health data, providing insights into maternal vaccination and placental antibody transfer for personalized immunization (Wessel and Dolatshahi, 2024). Another study explored the placental transport of toxic substances using a uterine perfusion model and molecular docking (Zhang et al., 2024). Additionally, a quantitative structure–property relationship (QSPR) model optimized using Monte Carlo methods was developed for high-throughput screening of placental drug permeability (Vukomanović et al., 2024). Similarly, researchers have used eigenvalue analysis, a chiral-sensitive QSPR method combined with ML, to predict placental drug transfer, focusing on chirality and stereochemistry (Gomatam and Coutinho, 2024). These efforts highlight the growing role of computational approaches in advancing our understanding of placental PK and enhancing drug safety during pregnancy. Although the application of these techniques for the prediction of placental drug transfer is in its infancy, they are slowly gaining traction as powerful PK tools. However, it is apparent that novel mathematical techniques such as these have great potential to fill the knowledge gap in underrepresented populations and clinical scenarios, and future research initiatives should encompass these techniques.
6 Discussion
Placental pharmacokinetics refers to the study of how drugs are absorbed, distributed, metabolized, and excreted as they pass from the maternal to the fetal circulation via the placenta. As a selective barrier and active transport interface, the placenta plays a pivotal role in determining fetal drug exposure. Understanding placental pharmacokinetics is essential for optimizing medication use during pregnancy, ensuring maternal efficacy while minimizing fetal risk. To address the complexity of maternal–fetal drug transfer, a range of experimental and computational models have been developed to investigate the placental transport mechanisms and predict fetal exposure. Recent advances in ex vivo placental perfusion, organ-on-a-chip platforms, and in silico simulations offer promising avenues to improve the accuracy of fetal drug-exposure predictions.
6.1 Placental pharmacokinetic models: characteristics and applications
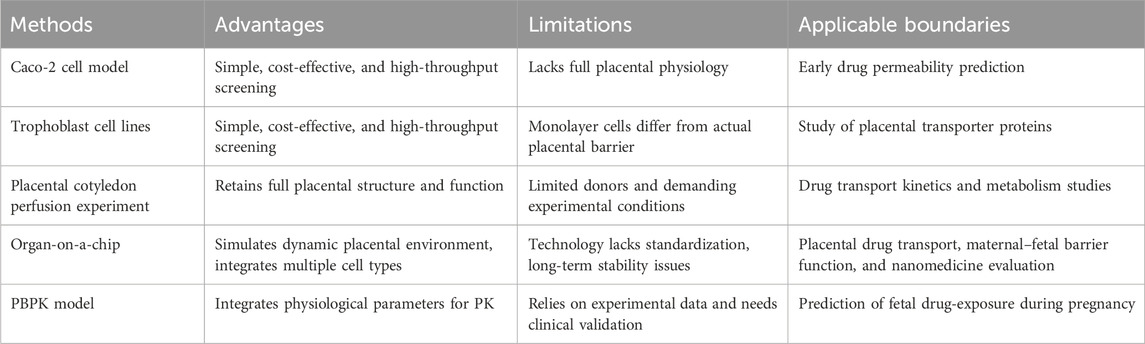
Current methods for placental pharmacokinetics include cell models, organ models, and in silico-guided models, each with its own distinct characteristics (Table 5). Monolayer cell models are convenient and rapid, but they oversimplify placental dynamics (Birch et al., 2018). Ex vivo and organ-on-chip systems offer greater physiological relevance, yet they face practical constraints (Pemathilaka et al., 2019a). In silico models, such as PBPK, allow quantitative predictions but rely on experimental data, highlighting the need for integrated approaches. A review of current studies shows that monolayer cell models are used to evaluate placental drug permeability and transporter-mediated transfer (Mian et al., 2020; Po et al., 2018), ex vivo and organ-on-a-chip systems quantify fetal drug exposure and pharmacokinetics (Freriksen et al., 2020; Zhang and Unadkat, 2017), and PBPK models support clinical dosing during pregnancy while predicting fetal drug exposure (Schneider et al., 2022; Abduljalil et al., 2022b).
Based on the comparative evaluation of each method, model selection in practical research should be guided by the research objective, the required physiological relevance, and the available resources. Simplified in vitro systems, such as cell monolayers or co-cultures, are appropriate for high-throughput screening or preliminary mechanistic studies because of their reproducibility and efficiency. Studies that demand physiologically relevant transport dynamics benefit from ex vivo placental perfusion or organ-on-a-chip models, which preserve tissue architecture and flow conditions absent in conventional in vitro systems. Computational approaches, including PBPK modeling, can supplement experimental data by providing quantitative predictions across different scenarios and species. Careful alignment of model choice with study goals and experimental constraints enhances both the reliability and translational value.
6.2 Multi-model integration: strengthening predictive power and translational relevance
The integration of multiple models holds significant potential in enhancing predictive accuracy and individual risk assessment. For instance, multiple studies demonstrate that incorporating ex vivo placental perfusion parameters into PBPK modeling yields predictions within 2-fold of clinical observations, significantly improving fetal drug-exposure estimates compared to non-integrated models (Mian et al., 2020; Roelofsen et al., 2022; Schalkwijk et al., 2018; Van Der Heijden et al., 2024). Another study showed that incorporating BeWo cell transporter kinetics and placental transporter abundance (particularly efflux transporters) into PBPK models can improve the physiological relevance of fetal drug-exposure predictions, as evidenced by the maternal-to-fetal plasma ratio shifting from approximately 1.0 to 0.1 (Al-Majdoub et al., 2025). In addition, integrating omics data (transcriptomics and proteomics) with dynamic placenta-on-a-chip platforms could enhance our understanding of drug–nutrient interactions, transporter regulation, and placental metabolism. The synergy between these methods facilitates a more accurate representation of the complex physiological processes involved in drug transfer, metabolism, and response, ultimately paving the way for tailored, safer treatments, especially in sensitive populations such as pregnant women. Beyond studies focused solely on placental drug-transfer, modeling approaches developed in this domain may offer significant insights into fetal drug-exposure arising from maternal vaccination, prolonged pharmacotherapy, or environmental toxin exposure during pregnancy.
Interestingly, the integration of data-driven approaches with mechanistic PBPK models also represents a highly promising strategy in PK research, particularly for pregnant populations. ML and QSPR methods can estimate parameters that are difficult to obtain experimentally; these parameters can be incorporated into PBPK models to enhance physiological realism and predictive accuracy. For example, a hybrid deep learning-PBPK model has successfully predicted human PK profiles from chemical structures, generating reliable concentration–time curves (Li et al., 2024). AI-assisted PBPK models have also been applied in the delivery of nanomedicines, producing predictions that closely match experimental PK data (Chou et al., 2023). These studies collectively demonstrate that utilizing ML or QSPR to inform mechanistic models can strengthen model development, optimize predictive performance, and enhance translational relevance. The application of such integrative strategies to placental pharmacokinetics holds considerable promise for advancing the field.
These technologies can help clarify how drugs cross the placenta and guide safer treatment for pregnant women with conditions such as epilepsy, hypertension, or HIV, where treatment is often unavoidable. The valuable results obtained from these methods could not only support drug label modifications but also inform the development of evidence-based treatment guidelines. Placental perfusion studies integrated with PBPK modeling reveal that doravirine exhibits a 55% reduction in maternal AUC during late pregnancy (40 weeks) (Bukkems et al., 2022), elevating treatment failure risk while predicting substantial fetal exposure. These findings have prompted multiple guideline bodies to re-evaluate its gestational use, shifting from “insufficient data, not recommended” toward “use with monitoring.” Similarly, quantified placental transfer kinetics of dexamethasone/betamethasone have enabled the precise prediction of maternal–fetal exposure dynamics (Van Der Heijden et al., 2024), advancing guidelines from fixed-dose to gestational age-adjusted regimens for optimal risk–benefit balance. In addition, a study combining Caco-2 cell models and placental perfusion experiments with PBPK modeling predicted that arterial umbilical concentrations of acetaminophen remain well below the recommended postnatal ductus arteriosus closure threshold (24.47 mg/L) (Mian et al., 2020). Regulatory agencies such as the Food and Drug Administration (FDA) and European Medicines Agency (EMA) increasingly accept PBPK modeling and may require placental transport assessment for drugs used in pregnancy in the future.
6.3 Future perspectives: gestational age and pathophysiology considerations
We have previously emphasized the need to investigate dynamic changes in the placenta during pregnancy, which are mainly driven by alterations in blood volume, blood flow velocity, tissue distribution, and the expression of metabolic enzymes and transporters. Nevertheless, few studies have specifically addressed this aspect, thus requiring greater attention in future research. Reviewing existing methodologies and models, several potential strategies may address this challenge, including (Be et al., 2025) conducting ex vivo placental perfusion using tissue samples from different gestational stages (early, mid, and late pregnancy) under ethically permissible conditions (Goeden and Bonnin, 2013); (Lupattelli et al., 2014) inducing pluripotent stem cells (iPSCs) to differentiate into trophoblasts, simulating developmental states across pregnancy stages (early trophoblasts vs. mature STBs) (Horii et al., 2016); (Subramanian et al., 2023) placenta-on-a-chip platforms, simulating the endocrine environment of different gestational stages by regulating key hormone levels (progesterone, estrogen, and hCG) in culture media (Mathiesen et al., 2025; Shojaei et al., 2021; Phillips et al., 2024). PBPK modeling can incorporate time-dependent parameters such as placental blood flow and transporter expression profiles to construct precise drug-use models for different gestational periods (Zhang et al., 2017) (Zhang and Unadkat, 2017). Furthermore, integrating in vitro data from different gestational stages with PBPK simulations offers a promising strategy to account for pregnancy-related variability.
In addition to gestational age-related dynamic changes, various pathological conditions can also markedly alter the placental structure and function, thereby influencing drug transfer. Preeclampsia [reduced placental perfusion and abnormal spiral artery remodeling (Roberts and Escudero, 2012; Yagel et al., 2022; Harmon et al., 2016)], gestational diabetes mellitus [hyperglycemia-induced upregulation of inflammatory mediators and altered transporter expression (Calvo et al., 2024)], and intrauterine infection [enhanced inflammatory response and increased tissue permeability (Meg et al., 2022)] are among the most common pregnancy complications associated with changes in villous stroma and vascular architecture. These pathological alterations can further modify the expression and activity of transporters and metabolic enzymes, ultimately affecting maternal–fetal drug transfer. To reproduce these pathological phenotypes in vitro or through modeling, several feasible approaches have been explored. For instance, ex vivo placental perfusion using tissues from pathological pregnancies [e.g., preeclampsia or GDM (Villota et al., 2021)] allows direct measurement of permeability differences under disease conditions. In cell-based and organ-on-a-chip models, preeclampsia, infection, or hyperglycemia can be mimicked by inducing hypoxia, adding pro-inflammatory cytokines (such as TNF-α or IL-6), or applying high-glucose culture conditions (Elzinga et al., 2023). Moreover, chip platforms can be adapted to modify shear stress, perfusion rates, or introduce pathological flow patterns to reflect abnormal spiral artery remodeling and reduced perfusion (Mosavati et al., 2020). Together, incorporating both pathophysiological states and gestational dynamics into placental models would greatly enhance their translational relevance for predicting drug disposition during pregnancy.
7 Conclusion
In this review, we describe the cell monolayer model, ex vivo placental perfusion experiments, placental-on-a-chip, PBPK model, and other in vitro research methods and discuss their advantages and disadvantages. The approach of a multi-model data fusion strategy (e.g., integrating a placenta perfusion experiment and PBPK modeling) is promising for the investigation of drug pharmacokinetics during pregnancy early in drug development, but further standardization and extension is needed. As technological fidelity improves and validation expands, placental drug modeling may become an essential, not optional, component of maternal–fetal pharmacokinetics.
Author contributions
ZL: Conceptualization, Methodology, Visualization, Writing – original draft, Writing – review and editing. YW: Conceptualization, Supervision, Validation, Writing – review and editing. SZ: Formal Analysis, Investigation, Visualization, Writing – original draft. FW: Data curation, Formal Analysis, Investigation, Writing – original draft. JZ: Conceptualization, Data curation, Supervision, Writing – original draft. SL: Formal Analysis, Resources, Visualization, Writing – original draft. YoY: Conceptualization, Funding acquisition, Resources, Supervision, Writing – review and editing. YuY: Writing – review and editing, Conceptualization, Project administration, Supervision.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the Scientific Research and Development Center for Higher Education, Ministry of Education; under Grant 2023HT070 and the Association for Promoting Sichuan Development by Sci-Tech and Education under Grant KJXC24-0201.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that Generative AI was used in the creation of this manuscript. Generative AI and AI-assisted technologies should only be used in the writing process to improve the readability and language of the manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abduljalil, K., Ning, J., Pansari, A., Pan, X., and Jamei, M. (2022a). Prediction of maternal and fetoplacental concentrations of cefazolin, cefuroxime, and amoxicillin during pregnancy using Bottom-Up physiologically based pharmacokinetic models. Drug Metab. Dispos. 50 (4), 386–400. doi:10.1124/dmd.121.000711
Abduljalil, K., Pansari, A., Ning, J., and Jamei, M. (2022b). Prediction of maternal and fetal Acyclovir, emtricitabine, lamivudine, and metformin concentrations during pregnancy using a physiologically based pharmacokinetic modeling approach. Clin. Pharmacokinet. 61 (5), 725–748. doi:10.1007/s40262-021-01103-0
Abduljalil, K., Gardner, I., and Jamei, M. (2022c). Application of a physiologically based pharmacokinetic approach to predict theophylline pharmacokinetics using virtual Non-Pregnant, pregnant, fetal, Breast-Feeding, and neonatal populations. Front. Pediatr. 10, 840710. doi:10.3389/fped.2022.840710
Abduljalil, K., Gardner, I., and Jamei, M. (2024). An application of a physiologically based pharmacokinetic approach to predict ceftazidime pharmacokinetics in a pregnant population. Pharmaceutics 16 (4), 474. doi:10.3390/pharmaceutics16040474
Abolhassani, N., Winterfeld, U., Kaplan, Y. C., Jaques, C., Minder Wyssmann, B., Del, G. C., et al. (2023). Major malformations risk following early pregnancy exposure to metformin: a systematic review and meta-analysis. BMJ Open Diabetes Res. Care 11 (1), e002919. doi:10.1136/bmjdrc-2022-002919
Aengenheister, L., Favaro, R. R., Morales-Prieto, D. M., Furer, L. A., Gruber, M., Wadsack, C., et al. (2021). Research on nanoparticles in human perfused placenta: state of the art and perspectives. Placenta 104, 199–207. doi:10.1016/j.placenta.2020.12.014
Al-Majdoub, Z. M., Freriksen, J. J. M., Colbers, A., van den Heuvel, J., Koenderink, J., Abduljalil, K., et al. (2025). Absolute membrane protein abundance of P-glycoprotein, breast cancer resistance protein, and multidrug resistance proteins in term human placenta tissue and commonly used cell systems: application in physiologically based pharmacokinetic modeling of placental drug disposition. Drug Metab. Dispos. 53 (1), 100007. doi:10.1124/dmd.124.001824
Anoshchenko, O., Milad, M. A., and Unadkat, J. D. (2021). Estimating fetal exposure to the P-gp substrates, corticosteroids, by PBPK modeling to inform prevention of neonatal respiratory distress syndrome. CPT Pharmacometrics Syst. Pharmacol. 10 (9), 1057–1070. doi:10.1002/psp4.12674
Aplin, J. D., Myers, J. E., Timms, K., and Westwood, M. (2020). Tracking placental development in health and disease. Nat. Rev. Endocrinol. 16 (9), 479–494. doi:10.1038/s41574-020-0372-6
Artursson, P., Palm, K., and Luthman, K. (2001). Caco-2 monolayers in experimental and theoretical predictions of drug transport. Adv. Drug Deliv. Rev. 46 (1-3), 27–43. doi:10.1016/s0169-409x(00)00128-9
Atoyebi, S. A., Rajoli, R. K. R., Adejuyigbe, E., Owen, A., Bolaji, O., Siccardi, M., et al. (2019). Using mechanistic physiologically-based pharmacokinetic models to assess prenatal drug exposure: thalidomide versus efavirenz as case studies. Eur. J. Pharm. Sci. 140, 105068. doi:10.1016/j.ejps.2019.105068
Atoyebi, S., Bunglawala, F., Cottura, N., Grañana-Castillo, S., Montanha, M. C., Olagunju, A., et al. (2024). Physiologically-based pharmacokinetic modelling of long-acting injectable cabotegravir and rilpivirine in pregnancy. Br. J. Clin. Pharmacol. 91, 989–1002. doi:10.1111/bcp.16006
Balan, A., Szaingurten-Solodkin, I., Swissa, S. S., Feinshtein, V., Huleihel, M., Holcberg, G., et al. (2017). The effects of pravastatin on the normal human placenta: lessons from ex-vivo models. PLoS One 12 (2), e0172174. doi:10.1371/journal.pone.0172174
Badhan, R. K. S., and Gittins, R. (2021). Precision dosing of methadone during pregnancy: a pharmacokinetics virtual clinical trials study. J. Subst. Abuse Treat. 130, 108521. doi:10.1016/j.jsat.2021.108521
Bai, M. R., Sun, D. L., Jiang, H. D., and Zheng, C. H. (2016). Expression and regulation of drug transporters in placenta. Yao Xue Xue Bao 51 (6), 879–885. doi:10.16438/j.0513-4870.2015-0861
Bapat, P., Pinto, L. S., Lubetsky, A., Berger, H., and Koren, G. (2015). Rivaroxaban transfer across the dually perfused isolated human placental cotyledon. Am. J. Obstet. Gynecol. 213 (5), 710.e1–e6. doi:10.1016/j.ajog.2015.06.065
Belsti, Y., Mousa, A., Jackson, H., Moran, L. J., Palmer, K. R., Dhungana, R. R., et al. (2025). The use of multiple medications during pregnancy among an ethnically diverse population in South-Eastern Melbourne: a retrospective analysis to explore potential risks and complications. Drug Saf. 48 (1), 87–97. doi:10.1007/s40264-024-01482-w
Berman, E., Kohn, E., Berkovitch, M., Kovo, M., and Eyal, S. (2022). Lacosamide effects on placental carriers of essential compounds in comparison with valproate: studies in perfused human placentas. Epilepsia 63 (11), 2949–2957. doi:10.1111/epi.17395
Bhatia, S. N., and Ingber, D. E. (2014). Microfluidic organs-on-chips. Nat. Biotechnol. 32 (8), 760–772. doi:10.1038/nbt.2989
Birch, D., Diedrichsen, R. G., Christophersen, P. C., Mu, H., and Nielsen, H. M. (2018). Evaluation of drug permeation under fed state conditions using mucus-covered Caco-2 cell epithelium. Eur. J. Pharm. Sci. 118, 144–153. doi:10.1016/j.ejps.2018.02.032
Blundell, C., Yi, Y. S., Ma, L., Tess, E. R., Farrell, M. J., Georgescu, A., et al. (2018). Placental Drug Transport-on-a-Chip: a microengineered in vitro model of transporter-mediated Drug efflux in the human placental barrier. Adv. Healthc. Mater 7 (2), 1700786. doi:10.1002/adhm.201700786
Broekhuizen, M., de Vries, R., Smits, M. A. W., Dik, W. A., Schoenmakers, S., Koch, B. C. P., et al. (2022). Pentoxifylline as a therapeutic option for pre-eclampsia: a study on its placental effects. Br. J. Pharmacol. 179 (22), 5074–5088. doi:10.1111/bph.15931
Bukkems, V. E., van Hove, H., Roelofsen, D., Freriksen, J. J. M., van Ewijk-Beneken Kolmer, E. W. J., Burger, D. M., et al. (2022). Prediction of maternal and fetal doravirine exposure by integrating physiologically based pharmacokinetic modeling and human placenta perfusion experiments. Clin. Pharmacokinet. 61 (8), 1129–1141. doi:10.1007/s40262-022-01127-0
Calvo, M. J., Parra, H., Santeliz, R., Bautista, J., Luzardo, E., Villasmil, N., et al. (2024). The placental role in gestational diabetes mellitus: a molecular perspective. touchREV Endocrinol. 20 (1), 10–18. doi:10.17925/EE.2024.20.1.5
Cao, R., Wang, Y., Liu, J., Rong, L., and Qin, J. (2023). Self-assembled human placental model from trophoblast stem cells in a dynamic organ-on-a-chip system. Cell. Prolif. 56 (5), e13469. doi:10.1111/cpr.13469
Cavero, I., Guillon, J. M., and Holzgrefe, H. H. (2019). Human organotypic bioconstructs from organ-on-chip devices for human-predictive biological insights on drug candidates. Expert Opin. Drug Saf. 18 (8), 651–677. doi:10.1080/14740338.2019.1634689
Ceckova, M., Libra, A., Pavek, P., Nachtigal, P., Brabec, M., Fuchs, R., et al. (2006). Expression and functional activity of breast cancer resistance protein (BCRP, ABCG2) transporter in the human choriocarcinoma cell line BeWo. Clin. Exp. Pharmacol. Physiol. 33 (1-2), 58–65. doi:10.1111/j.1440-1681.2006.04324.x
Cerveny, L., Ptackova, Z., Ceckova, M., Karahoda, R., Karbanova, S., Jiraskova, L., et al. (2018). Equilibrative nucleoside transporter 1 (ENT1, SLC29A1) facilitates transfer of the antiretroviral drug abacavir across the placenta. Drug Metab. Dispos. 46 (11), 1817–1826. doi:10.1124/dmd.118.083329
Chen, J., You, X., Wu, W., Guo, G., Lin, R., Ke, M., et al. (2023). Application of PBPK modeling in predicting maternal and fetal pharmacokinetics of levetiracetam during pregnancy. Eur. J. Pharm. Sci. 181, 106349. doi:10.1016/j.ejps.2022.106349
Chen, Y., Ke, M., Fang, W., Jiang, Y., Lin, R., Wu, W., et al. (2024). Physiologically based pharmacokinetic modeling to predict maternal pharmacokinetics and fetal carbamazepine exposure during pregnancy. Eur. J. Pharm. Sci. 194, 106707. doi:10.1016/j.ejps.2024.106707
Chou, W. C., Chen, Q., Yuan, L., Cheng, Y. H., He, C., Monteiro-Riviere, N. A., et al. (2023). An artificial intelligence-assisted physiologically-based pharmacokinetic model to predict nanoparticle delivery to tumors in mice. J. Control Release 361, 53–63. doi:10.1016/j.jconrel.2023.07.040
Correia Carreira, S., Cartwright, L., Mathiesen, L., Knudsen, L. E., and Saunders, M. (2011). Studying placental transfer of highly purified non-dioxin-like PCBs in two models of the placental barrier. Placenta 32 (3), 283–291. doi:10.1016/j.placenta.2010.12.024
Dallmann, A., Ince, I., Meyer, M., Willmann, S., Eissing, T., and Hempel, G. (2017). Gestation-Specific changes in the anatomy and physiology of healthy pregnant women: an extended repository of model parameters for physiologically based pharmacokinetic modeling in pregnancy. Clin. Pharmacokinet. 56 (11), 1303–1330. doi:10.1007/s40262-017-0539-z
Dallmann, A., Pfister, M., van den Anker, J., and Eissing, T. (2018). Physiologically based pharmacokinetic modeling in pregnancy: a systematic review of published models. Clin. Pharmacol. Ther. 104 (6), 1110–1124. doi:10.1002/cpt.1084
De Felice, K. M., and Kane, S. (2021). Safety of anti-TNF agents in pregnancy. J. Allergy Clin. Immunol. 148 (3), 661–667. doi:10.1016/j.jaci.2021.07.005
De Sousa Mendes, M., Hirt, D., Vinot, C., Valade, E., Lui, G., Pressiat, C., et al. (2016). Prediction of human fetal pharmacokinetics using ex vivo human placenta perfusion studies and physiologically based models. Br. J. Clin. Pharmacol. 81 (4), 646–657. doi:10.1111/bcp.12815
De Sousa Mendes, M., Lui, G., Zheng, Y., Pressiat, C., Hirt, D., Valade, E., et al. (2017). A physiologically-based pharmacokinetic model to predict human fetal exposure for a drug metabolized by several CYP450 pathways. Clin. Pharmacokinet. 56 (5), 537–550. doi:10.1007/s40262-016-0457-5
Drwal, E., Rak, A., and Gregoraszczuk, E. (2018). Co-culture of JEG-3, BeWo and syncBeWo cell lines with adrenal H295R cell line: an alternative model for examining endocrine and metabolic properties of the fetoplacental unit. Cytotechnology 70 (1), 285–297. doi:10.1007/s10616-017-0142-z
Eliesen, G. A. M., van Drongelen, J., van Hove, H., Kooijman, N. I., van den Broek, P., de Vries, A., et al. (2020). Assessment of placental disposition of Infliximab and etanercept in women with autoimmune diseases and in the Ex Vivo perfused placenta. Clin. Pharmacol. Ther. 108 (1), 99–106. doi:10.1002/cpt.1827
Elzinga, F. A., Khalili, B., Touw, D. J., Prins, J. R., Olinga, P., Leuvenink, H. G. D., et al. (2023). Placenta-on-a-Chip as an in vitro approach to evaluate the physiological and structural characteristics of the human placental barrier upon drug exposure: a systematic review. J. Clin. Med. 12 (13), 4315. doi:10.3390/jcm12134315
Esch, E. W., Bahinski, A., and Huh, D. (2015). Organs-on-chips at the frontiers of drug discovery. Nat. Rev. Drug Discov. 14 (4), 248–260. doi:10.1038/nrd4539
Faure Bardon, V., Peytavin, G., Lê, M. P., Guilleminot, T., Elefant, E., Stirnemann, J., et al. (2020). Placental transfer of Letermovir and Maribavir in the ex vivo human cotyledon perfusion model. New perspectives for in utero treatment of congenital cytomegalovirus infection. PLoS One 15 (4), e0232140. doi:10.1371/journal.pone.0232140
Foster, E. G., Gendelman, H. E., and Bade, A. N. (2022). HIV-1 integrase strand transfer inhibitors and neurodevelopment. Pharm. (Basel). 15 (12), 1533. doi:10.3390/ph15121533
Freriksen, J. J. M., Schalkwijk, S., Colbers, A. P., Abduljalil, K., Russel, F. G. M., Burger, D. M., et al. (2020). Assessment of maternal and fetal Dolutegravir exposure by integrating Ex Vivo placental perfusion data and physiologically-based pharmacokinetic modeling. Clin. Pharmacol. Ther. 107 (6), 1352–1361. doi:10.1002/cpt.1748
Fuenzalida, B., Basler, V., Koechli, N., Yi, N., Staud, F., and Albrecht, C. (2024). Modelling the maternal-fetal interface: an in vitro approach to investigate nutrient and drug transport across the human placenta. J. Cell. Mol. Med. 28 (20), e70151. doi:10.1111/jcmm.70151
Ganguly, E., Kammala, A. K., Benson, M., Richardson, L. S., Han, A., and Menon, R. (2021). Organic anion transporting polypeptide 2B1 in human fetal membranes: a novel gatekeeper for drug transport during pregnancy? Front. Pharmacol. 12, 771818. doi:10.3389/fphar.2021.771818
Goeden, N., and Bonnin, A. (2013). Ex vivo perfusion of mid-to-late-gestation mouse placenta for maternal-fetal interaction studies during pregnancy. Nat. Protoc. 8 (1), 66–74. doi:10.1038/nprot.2012.144
Gomatam, A., and Coutinho, E. (2024). A chirality-sensitive approach to predict chemical transfer across the human placental barrier. Toxicol. Lett. 394, 66–75. doi:10.1016/j.toxlet.2024.02.012
Haas, D. M., Marsh, D. J., Dang, D. T., Parker, C. B., Wing, D. A., Simhan, H. N., et al. (2018). Prescription and other medication use in pregnancy. Obstet. Gynecol. 131 (5), 789–798. doi:10.1097/AOG.0000000000002579
Han, L. W., Gao, C., and Mao, Q. (2018). An update on expression and function of P-gp/ABCB1 and BCRP/ABCG2 in the placenta and fetus. Expert Opin. Drug Metab. Toxicol. 14 (8), 817–829. doi:10.1080/17425255.2018.1499726
Harmon, A. C., Cornelius, D. C., Amaral, L. M., Faulkner, J. L., Cunningham, M. W., Wallace, K., et al. (2016). The role of inflammation in the pathology of preeclampsia. Clin. Sci. (Lond). 130 (6), 409–419. doi:10.1042/CS20150702
Hawkins, A., Pantazi, P., Yang, L., Coyne, C. B., Bokun, V., Lemme-Dumit, J. M., et al. (2025). Long-term culture and passaging of term trophoblast for the investigation of syncytiotrophoblast function. Placenta 166, 25–32. doi:10.1016/j.placenta.2024.08.014
He, L., Ke, M., Wu, W., Chen, J., Guo, G., Lin, R., et al. (2022). Application of physiologically based pharmacokinetic modeling to predict maternal pharmacokinetics and fetal exposure to oxcarbazepine. Pharmaceutics 14 (11), 2367. doi:10.3390/pharmaceutics14112367
Hirschmugl, B., and Wadsack, C. (2022). Transplacental transfer of venlafaxine evaluated by ex vivo perfusion. Placenta 117, 150–153. doi:10.1016/j.placenta.2021.12.007
Hitzerd, E., Broekhuizen, M., Mirabito Colafella, K. M., Glisic, M., de Vries, R., Koch, B. C. P., et al. (2019). Placental effects and transfer of sildenafil in healthy and preeclamptic conditions. EBioMedicine 45, 447–455. doi:10.1016/j.ebiom.2019.06.007
Horii, M., Li, Y., Wakeland, A. K., Pizzo, D. P., Nelson, K. K., Sabatini, K., et al. (2016). Human pluripotent stem cells as a model of trophoblast differentiation in both normal development and disease. Proc. Natl. Acad. Sci. U. S. A. 113 (27), E3882–E3891. doi:10.1073/pnas.1604747113
Huang, H., Wang, J., Li, Q., Duan, J., Yao, Q., Zheng, Q., et al. (2017). Transplacental transfer of oseltamivir phosphate and its metabolite oseltamivir carboxylate using the ex vivo human placenta perfusion model in Chinese Hans population. J. Matern. Fetal Neonatal Med. 30 (11), 1288–1292. doi:10.1080/14767058.2016.1211634
Jovelet, C., Seck, A., Mir, O., Simasotchi, C., Broutin, S., Goffinet, F., et al. (2015). Variation in transplacental transfer of tyrosine kinase inhibitors in the human perfused cotyledon model. Ann. Oncol. 26 (7), 1500–1504. doi:10.1093/annonc/mdv172
Kammala, A. K., Lintao, R. C. V., Vora, N., Mosebarger, A., Khanipov, K., Golovko, G., et al. (2022). Expression of CYP450 enzymes in human fetal membranes and its implications in xenobiotic metabolism during pregnancy. Life Sci. 307, 120867. doi:10.1016/j.lfs.2022.120867
Kammala, A. K., Richardson, L. S., Radnaa, E., Han, A., and Menon, R. (2023). Microfluidic technology and simulation models in studying pharmacokinetics during pregnancy. Front. Pharmacol. 14, 1241815. doi:10.3389/fphar.2023.1241815
Karahoda, R., Kallol, S., Groessl, M., Ontsouka, E., Anderle, P., Fluck, C., et al. (2021). Revisiting steroidogenic pathways in the human placenta and primary human trophoblast cells. Int. J. Mol. Sci. 22 (4), 1704. doi:10.3390/ijms22041704
Karakis, V., Britt, J. W., Jabeen, M., San Miguel, A., and Rao, B. M. (2025). Derivation of human trophoblast stem cells from placentas at birth. J. Biol. Chem. 301 (6), 108505. doi:10.1016/j.jbc.2025.108505
Karbanova, S., Cerveny, L., Ceckova, M., Ptackova, Z., Jiraskova, L., Greenwood, S., et al. (2017). Role of nucleoside transporters in transplacental pharmacokinetics of nucleoside reverse transcriptase inhibitors zidovudine and emtricitabine. Placenta 60, 86–92. doi:10.1016/j.placenta.2017.10.011
Koren, G., and Ornoy, A. (2018). The role of the placenta in drug transport and fetal drug exposure. Expert Rev. Clin. Pharmacol. 11 (4), 373–385. doi:10.1080/17512433.2018.1425615
Kothari, A., Hirschmugl, B., Lee, J. S., Pfaller, B., Schmidthaler, K., Szépfalusi, Z., et al. (2022). The impact of maternal-fetal omalizumab transfer on peanut-specific responses in an ex vivo placental perfusion model. Allergy 77 (12), 3684–3686. doi:10.1111/all.15468
Kuoni, S., Steiner, R., Saleh, L., Lehmann, R., Ochsenbein-Kölble, N., and Simões-Wüst, A. P. (2024). Safety assessment of the SGLT2 inhibitors empagliflozin, dapagliflozin and canagliflozin during pregnancy: an ex vivo human placenta perfusion and in vitro study. Biomed. Pharmacother. 171, 116177. doi:10.1016/j.biopha.2024.116177
Lapehn, S., Nair, S., Firsick, E. J., MacDonald, J., Thoreson, C., Litch, J. A., et al. (2024). A transcriptomic comparison of in vitro models of the human placenta. Placenta 159, 52–61. doi:10.1016/j.placenta.2024.11.007
Lartey, D., Jateng, D., Li, M., Nguyen, C., Crentsil, V., Beitz, J., et al. (2023). Quantification of sertraline maternal/fetal ratio and amniotic fluid concentration using a pregnancy physiologically based pharmacokinetic model. Br. J. Clin. Pharmacol. 91, 1003–1015. doi:10.1111/bcp.15826
Le Merdy, M., Szeto, K. X., Perrier, J., Bolger, M. B., and Lukacova, V. (2024). PBPK modeling approach to predict the behavior of drugs cleared by metabolism in pregnant subjects and fetuses. Pharmaceutics 16 (1), 96. doi:10.3390/pharmaceutics16010096
Li, L., and Schust, D. J. (2015). Isolation, purification and in vitro differentiation of cytotrophoblast cells from human term placenta. Reprod. Biol. Endocrinol. 13, 71. doi:10.1186/s12958-015-0070-8
Li, S., and Xie, F. (2023). Foetal and neonatal exposure prediction and dosing evaluation for ampicillin using a physiologically-based pharmacokinetic modelling approach. Br. J. Clin. Pharmacol. 89 (4), 1402–1412. doi:10.1111/bcp.15589
Li, H., van Ravenzwaay, B., Rietjens, I. M., and Louisse, J. (2013). Assessment of an in vitro transport model using BeWo b30 cells to predict placental transfer of compounds. Arch. Toxicol. 87 (9), 1661–1669. doi:10.1007/s00204-013-1074-9
Li, Y., Wang, Z., Li, Y., Du, J., Gao, X., Li, Y., et al. (2024). A combination of machine learning and PBPK modeling approach for pharmacokinetics prediction of small molecules in humans. Pharm. Res. 41 (7), 1369–1379. doi:10.1007/s11095-024-03725-y
Liu, X. I., Momper, J. D., Rakhmanina, N. Y., Green, D. J., Burckart, G. J., Cressey, T. R., et al. (2020a). Prediction of maternal and fetal pharmacokinetics of Dolutegravir and Raltegravir using physiologically based pharmacokinetic modeling. Clin. Pharmacokinet. 59 (11), 1433–1450. doi:10.1007/s40262-020-00897-9
Liu, X. I., Momper, J. D., Rakhmanina, N., van den Anker, J. N., Green, D. J., Burckart, G. J., et al. (2020b). Physiologically based pharmacokinetic models to predict maternal pharmacokinetics and fetal exposure to emtricitabine and acyclovir. J. Clin. Pharmacol. 60 (2), 240–255. doi:10.1002/jcph.1515
Liu, X. I., Momper, J. D., Rakhmanina, N. Y., Green, D. J., Burckart, G. J., Cressey, T. R., et al. (2021). Physiologically based pharmacokinetic modeling framework to predict neonatal pharmacokinetics of transplacentally acquired emtricitabine, Dolutegravir, and Raltegravir. Clin. Pharmacokinet. 60 (6), 795–809. doi:10.1007/s40262-020-00977-w
Liu, X. I., Green, D. J., van den Anker, J., Ahmadzia, H. K., Burckart, G. J., and Dallmann, A. (2024). Development of a generic fetal physiologically based pharmacokinetic model and prediction of human maternal and fetal organ concentrations of cefuroxime. Clin. Pharmacokinet. 63 (1), 69–78. doi:10.1007/s40262-023-01323-6
Louchet, M., Ribot, M., Bouazza, N., Foissac, F., Froelicher, L., Buth, V., et al. (2023). Transplacental transfer of Remdesivir and GS-441524: an ex vivo perfusion study. Health Sci. Rep. 6 (7), e1144. doi:10.1002/hsr2.1144
Lupattelli, A., Spigset, O., Twigg, M. J., Zagorodnikova, K., Mårdby, A. C., Moretti, M. E., et al. (2014). Medication use in pregnancy: a cross-sectional, multinational web-based study. BMJ Open 4 (2), e004365. doi:10.1136/bmjopen-2013-004365
Macente, J., Nauwelaerts, N., Russo, F. M., Deprest, J., Allegaert, K., Lammens, B., et al. (2023). PBPK-based dose finding for sildenafil in pregnant women for antenatal treatment of congenital diaphragmatic hernia. Front. Pharmacol. 14, 1068153. doi:10.3389/fphar.2023.1068153
Maroun, L. L., Mathiesen, L., Hedegaard, M., Knudsen, L. E., and Larsen, L. G. (2014). Pathologic evaluation of normal and perfused term placental tissue. Pediatr. Dev. Pathol. 17 (5), 330–338. doi:10.2350/12-08-1243-OA.1
Mathiesen, L., Mose, T., Mørck, T. J., Nielsen, J. K., Nielsen, L. K., Maroun, L. L., et al. (2010). Quality assessment of a placental perfusion protocol. Reprod. Toxicol. 30 (1), 138–146. doi:10.1016/j.reprotox.2010.01.006
Mathiesen, L., Sandal, D., Hammer, I. E. M., Styrishave, B., and Knudsen, L. E. (2025). Models of endocrine-disrupting effects: human placental steroidogenesis. Basic Clin. Pharmacol. Toxicol. 137 (2), e70073. doi:10.1111/bcpt.70073
Megli, C. J., and Coyne, C. B. (2022). Infections at the maternal-fetal interface: an overview of pathogenesis and defence. Nat. Rev. Microbiol. 20 (2), 67–82. doi:10.1038/s41579-021-00610-y
Mian, P., Allegaert, K., Conings, S., Annaert, P., Tibboel, D., Pfister, M., et al. (2020). Integration of placental transfer in a fetal–maternal physiologically based pharmacokinetic model to characterize acetaminophen exposure and metabolic clearance in the fetus. Clin. Pharmacokinet. 59 (7), 911–925. doi:10.1007/s40262-020-00861-7
Mistry, H. D., Klossner, R., Kallol, S., Lüthi, M. P., Moser, R., Schneider, H., et al. (2022). Effects of aldosterone on the human placenta: insights from placental perfusion studies. Placenta 123, 32–40. doi:10.1016/j.placenta.2022.03.129
Moore, K. H., Murphy, H. A., Chapman, H., and George, E. M. (2021). Syncytialization alters the extracellular matrix and barrier function of placental trophoblasts. Am. J. Physiol. Cell. Physiol. 321 (4), C694–c703. doi:10.1152/ajpcell.00177.2021
Mortensen, N. P., Caffaro, M. M., Snyder, R. W., Yueh, Y. L., and Fennell, T. R. (2019). Placental trophoblast transfer of opioids following exposures to individual or mixtures of opioids in vitro. Exp. Biol. Med. (Maywood) 244 (10), 846–849. doi:10.1177/1535370219851109
Mosavati, B., Oleinikov, A. V., and Du, E. (2020). Development of an Organ-on-a-Chip-Device for study of placental pathologies. Int. J. Mol. Sci. 21 (22), 8755. doi:10.3390/ijms21228755
Motomura, K., Morita, H., Yamamoto, H., Wada, S., Sago, H., Takahashi, H., et al. (2025). Isolation of pure primary term human cytotrophoblasts and their differentiation into syncytiotrophoblast-like cells as an ex vivo model of the human placenta. Placenta 166, 8–16. doi:10.1016/j.placenta.2024.07.307
Nathan, N. O., Hedegaard, M., Karlsson, G., Knudsen, L. E., and Mathiesen, L. (2021). Intrapartum transfer of oxytocin across the human placenta: an ex vivo perfusion experiment. Placenta 112, 105–110. doi:10.1016/j.placenta.2021.07.289
Ning, J., Pansari, A., Rowland Yeo, K., Heikkinen, A. T., Waitt, C., and Almond, L. M. (2024). Using PBPK modeling to supplement clinical data and support the safe and effective use of dolutegravir in pregnant and lactating women. CPT Pharmacometrics Syst. Pharmacol. 13 (11), 1924–1938. doi:10.1002/psp4.13251
Noergaard, M., Jensen, P. B., Resendal Gotfredsen, D., Bergholt, T., Trærup Andersen, J., and Mathiesen, L. (2021). Therapeutic concentration of ciprofloxacin and transfer across the human term placenta. Am. J. Obstet. Gynecol. 225 (6), 670.e1–670.e9. doi:10.1016/j.ajog.2021.05.032
Ohta, Y., Kazuki, K., Abe, S., Oshimura, M., Kobayashi, K., and Kazuki, Y. (2020). Development of Caco-2 cells expressing four CYPs via a Mammalian artificial chromosome. BMC Biotechnol. 20 (1), 44. doi:10.1186/s12896-020-00637-8
Okae, H., Toh, H., Sato, T., Hiura, H., Takahashi, S., Shirane, K., et al. (2018). Derivation of human trophoblast stem cells. Cell. Stem Cell. 22 (1), 50–63. doi:10.1016/j.stem.2017.11.004
Owen, M. D., Baker, B. C., Scott, E. M., and Forbes, K. (2021). Interaction between metformin, folate and vitamin B(12) and the potential impact on fetal growth and long-term metabolic health in diabetic pregnancies. Int. J. Mol. Sci. 22 (11), 5759. doi:10.3390/ijms22115759
Öztürk, S., Demir, M., Koçkaya, E. A., Karaaslan, C., and Süloğlu, A. K. (2024). Establishment of a 3D multicellular placental microtissues for investigating the effect of antidepressant vortioxetine. Reprod. Toxicol. 123, 108519. doi:10.1016/j.reprotox.2023.108519
Page, K. R. (1991). Perfusion of isolated human placenta. Proc. Nutr. Soc. 50 (2), 345–347. doi:10.1079/pns19910044
Panse, N., and Gerk, P. M. (2022). The Caco-2 model: modifications and enhancements to improve efficiency and predictive performance. Int. J. Pharm. 624, 122004. doi:10.1016/j.ijpharm.2022.122004
Pariente, G., Leibson, T., Carls, A., Adams-Webber, T., Ito, S., and Koren, G. (2016). Pregnancy-Associated changes in pharmacokinetics: a systematic review. PLoS Med. 13 (11), e1002160. doi:10.1371/journal.pmed.1002160
Parks, W. T. (2015). Placental hypoxia: the lesions of maternal malperfusion. Semin. Perinatol. 39 (1), 9–19. doi:10.1053/j.semperi.2014.10.003
Patilea-Vrana, G. I., and Unadkat, J. D. (2021). Development and verification of a linked Δ (9)-THC/11-OH-THC physiologically based pharmacokinetic model in healthy, nonpregnant population and extrapolation to pregnant women. Drug Metab. Dispos. 49 (7), 509–520. doi:10.1124/dmd.120.000322
Pemathilaka, R. L., Reynolds, D. E., and Hashemi, N. N. (2019a). Drug transport across the human placenta: review of placenta-on-a-chip and previous approaches. Interface Focus 9 (5), 20190031. doi:10.1098/rsfs.2019.0031
Pemathilaka, R. L., Caplin, J. D., Aykar, S. S., Montazami, R., and Hashemi, N. N. (2019b). Placenta-on-a-Chip: in vitro Study of caffeine transport across placental barrier using liquid chromatography Mass spectrometry. Glob. Chall. 3 (3), 1800112. doi:10.1002/gch2.201800112
Pemathilaka, R. L., Alimoradi, N., Reynolds, D. E., and Hashemi, N. N. (2022). Transport of maternally administered pharmaceutical agents across the placental barrier in vitro. ACS Appl. Bio Mater 5 (5), 2273–2284. doi:10.1021/acsabm.2c00121
Pencolé, L., Lê, M. P., Bouchet-Crivat, F., Duro, D., Peytavin, G., and Mandelbrot, L. (2020). Placental transfer of the integrase strand inhibitors cabotegravir and bictegravir in the ex-vivo human cotyledon perfusion model. Aids 34 (14), 2145–2149. doi:10.1097/QAD.0000000000002637
Phillips, K., Nirantharakumar, K., Wakerley, B. R., and Crowe, F. L. (2024). Trends in the prevalence and pharmacological management of migraine during pregnancy in the UK, 2000-2018. J. Neurol. Neurosurg. Psychiatry 95 (10), 938–946. doi:10.1136/jnnp-2024-333530
Pillai, V. C., Shah, M., Rytting, E., Nanovskaya, T. N., Wang, X., Clark, S. M., et al. (2022). Prediction of maternal and fetal pharmacokinetics of indomethacin in pregnancy. Br. J. Clin. Pharmacol. 88 (1), 271–281. doi:10.1111/bcp.14960
Pinto, L., Bapat, P., de Lima Moreira, F., Lubetsky, A., de Carvalho Cavalli, R., Berger, H., et al. (2021). Chiral transplacental pharmacokinetics of fexofenadine: impact of P-Glycoprotein inhibitor fluoxetine using the human placental perfusion model. Pharm. Res. 38 (4), 647–655. doi:10.1007/s11095-021-03035-7
Polachek, H., Debotton, N., Feinshtein, V., Rubin, M., Ben-Zvi, Z., Holcberg, G., et al. (2018). The role of various transporters in the placental uptake of ofloxacin in an in vitro model of human villous trophoblasts. Drug Des. Devel Ther. 12, 4129–4138. doi:10.2147/DDDT.S181493
Porter, C., Armstrong-Fisher, S., Kopotsha, T., Smith, B., Baker, T., Kevorkian, L., et al. (2016). Certolizumab pegol does not bind the neonatal Fc receptor (FcRn): consequences for FcRn-mediated in vitro transcytosis and ex vivo human placental transfer. J. Reprod. Immunol. 116, 7–12. doi:10.1016/j.jri.2016.04.284
Poulsen, M. S., Rytting, E., Mose, T., and Knudsen, L. E. (2009). Modeling placental transport: correlation of in vitro BeWo cell permeability and ex vivo human placental perfusion. Toxicol Vitro 23 (7), 1380–1386. doi:10.1016/j.tiv.2009.07.028
Richardson, L. S., Kammala, A. K., Costantine, M. M., Fortunato, S. J., Radnaa, E., Kim, S., et al. (2022). Testing of drugs using human feto-maternal interface organ-on-chips provide insights into pharmacokinetics and efficacy. Lab. Chip 22 (23), 4574–4592. doi:10.1039/d2lc00691j
Roberts, J. M., and Escudero, C. (2012). The placenta in preeclampsia. Pregnancy Hypertens. 2 (2), 72–83. doi:10.1016/j.preghy.2012.01.001
Roelofsen, D., van Hove, H., Bukkems, V., Russel, F., Eliesen, G., and Greupink, R. (2022). Predicting fetal exposure of crizotinib during pregnancy: combining human ex vivo placenta perfusion data with physiologically-based pharmacokinetic modeling. Toxicol Vitro 85, 105471. doi:10.1016/j.tiv.2022.105471
Sambuy, Y., De Angelis, I., Ranaldi, G., Scarino, M. L., Stammati, A., and Zucco, F. (2005). The Caco-2 cell line as a model of the intestinal barrier: influence of cell and culture-related factors on Caco-2 cell functional characteristics. Cell. Biol. Toxicol. 21 (1), 1–26. doi:10.1007/s10565-005-0085-6
Schalkwijk, S., Greupink, R., Colbers, A. P., Wouterse, A. C., Verweij, V. G., van Drongelen, J., et al. (2016). Placental transfer of the HIV integrase inhibitor dolutegravir in an ex Vivo human cotyledon perfusion model. J. Antimicrob. Chemother. 71 (2), 480–483. doi:10.1093/jac/dkv358
Schalkwijk, S., Buaben, A. O., Freriksen, J. J. M., Colbers, A. P., Burger, D. M., Greupink, R., et al. (2018). Prediction of fetal Darunavir exposure by integrating human Ex-Vivo placental transfer and physiologically based pharmacokinetic modeling. Clin. Pharmacokinet. 57 (6), 705–716. doi:10.1007/s40262-017-0583-8
Schneider, H. (2009). Tolerance of human placental tissue to severe hypoxia and its relevance for dual ex vivo perfusion. Placenta 30 (Suppl. A), S71–S76. doi:10.1016/j.placenta.2008.11.004
Schneider, H. (2015). IFPA senior award lecture: energy metabolism of human placental tissue studied by ex vivo perfusion of an isolated cotyledon. Placenta 36 (Suppl. 1), S29–S34. doi:10.1016/j.placenta.2014.11.022
Schneider, H., Brownbill, P., and Albrecht, C. (2021). Ex vivo dual perfusion of an isolated cotyledon of human placenta: history and future challenges. Placenta 107, 8–12. doi:10.1016/j.placenta.2021.02.017
Schneider, H., Albrecht, C., Ahmed, M. S., Broekhuizen, M., Aengenheister, L., Buerki-Thurnherr, T., et al. (2022). Ex vivo dual perfusion of an isolated human placenta cotyledon: towards protocol standardization and improved inter-centre comparability. Placenta 126, 83–89. doi:10.1016/j.placenta.2022.05.003
Shannon, M. J., McNeill, G. L., Koksal, B., Baltayeva, J., Wächter, J., Castellana, B., et al. (2024). Single-cell assessment of primary and stem cell-derived human trophoblast organoids as placenta-modeling platforms. Dev. Cell. 59 (6), 776–92.e11. doi:10.1016/j.devcel.2024.01.023
Shenkoya, B., Atoyebi, S., Eniayewu, I., Akinloye, A., and Olagunju, A. (2021). Mechanistic modeling of maternal lymphoid and fetal plasma antiretroviral exposure during the third trimester. Front. Pediatr. 9, 734122. doi:10.3389/fped.2021.734122
Shenkoya, B., Gopalakrishnan, M., and Eke, A. C. (2024). Physiologically based pharmacokinetic modeling of long-acting extended-release naltrexone in pregnant women with opioid use disorder. CPT Pharmacometrics Syst. Pharmacol. 13 (11), 1939–1952. doi:10.1002/psp4.13252
Shojaei, S., Ali, M. S., Suresh, M., Upreti, T., Mogourian, V., Helewa, M., et al. (2021). Dynamic placenta-on-a-chip model for fetal risk assessment of nanoparticles intended to treat pregnancy-associated diseases. Biochim. Biophys. Acta Mol. Basis Dis. 1867 (7), 166131. doi:10.1016/j.bbadis.2021.166131
Shuler, M. L. (2017). Organ-body- and disease-on-a-chip systems. Lab. Chip 17 (14), 2345–2346. doi:10.1039/c7lc90068f
Shum, S., Shen, D. D., and Isoherranen, N. (2021). Predicting maternal-fetal disposition of fentanyl following intravenous and epidural administration using physiologically based pharmacokinetic modeling. Drug Metab. Dispos. 49 (11), 1003–1015. doi:10.1124/dmd.121.000612
Subramanian, A., Azcoaga-Lorenzo, A., Anand, A., Phillips, K., Lee, S. I., Cockburn, N., et al. (2023). Polypharmacy during pregnancy and associated risk factors: a retrospective analysis of 577 medication exposures among 1.5 million pregnancies in the UK, 2000-2019. BMC Med. 21 (1), 21. doi:10.1186/s12916-022-02722-5
Szeto, K. X., Le Merdy, M., Dupont, B., Bolger, M. B., and Lukacova, V. (2021). PBPK modeling approach to predict the behavior of drugs cleared by kidney in pregnant subjects and fetus. Aaps J. 23 (4), 89. doi:10.1208/s12248-021-00603-y
Thunbo, M., Vendelbo, J. H., Witte, D. R., Larsen, A., and Pedersen, L. H. (2024). Use of medication in pregnancy on the rise: study on 1.4 million Danish pregnancies from 1998 to 2018. Acta Obstet. Gynecol. Scand. 103 (6), 1210–1223. doi:10.1111/aogs.14805
Van Der Heijden, J. E. M., Van Hove, H., Van Elst, N. M., Van Den Broek, P., Van Drongelen, J., Scheepers, H. C. J., et al. (2024). Optimization of the betamethasone and dexamethasone dosing regimen during pregnancy: a combined placenta perfusion and pregnancy physiologically based pharmacokinetic modeling approach. Am. J. Obstet. Gynecol. 232, 228.e1–228.e9. doi:10.1016/j.ajog.2024.05.012
van Hoogdalem, M. W., Tanaka, R., Abduljalil, K., Johnson, T. N., Wexelblatt, S. L., Akinbi, H. T., et al. (2024). Forecasting fetal buprenorphine exposure through maternal-fetal physiologically based pharmacokinetic modeling. Pharmaceutics 16 (3), 375. doi:10.3390/pharmaceutics16030375
van Kammen, C., van Hove, H., Kapsokalyvas, D., Greupink, R., Schiffelers, R., Lely, T., et al. (2024). Targeted lipid nanoparticles to prevent trans-placental passage in the ex vivo human placental cotyledon perfusion model. Drug Deliv. Transl. Res. 15, 1985–1993. doi:10.1007/s13346-024-01715-6
Veiga-Lopez, A., Elkin, E. R., Harris, S. M., Blake, B. E., Paquette, A. G., Aleksunes, L. M., et al. (2025). Current approaches and advances in placental toxicology. Trends Endocrinol. Metab. doi:10.1016/j.tem.2025.05.001
Vidal, M. S., Richardson, L. S., Kumar Kammala, A., Kim, S., Lam, P. Y., Cherukuri, R., et al. (2024). Endocrine-disrupting compounds and their impact on human placental function: evidence from placenta organ-on-chip studies. Lab. Chip 24 (6), 1727–1749. doi:10.1039/d3lc00998j
Villalobos-Labra, R., Liu, R., Spaans, F., Sáez, T., Semeria Maitret, T., Quon, A., et al. (2023). Placenta-Derived extracellular vesicles from preeclamptic pregnancies impair vascular endothelial function via lectin-like oxidized LDL Receptor-1. Hypertension 80 (10), 2226–2238. doi:10.1161/HYPERTENSIONAHA.123.21205
Villota, S. D., Toledo-Rodriguez, M., and Leach, L. (2021). Compromised barrier integrity of human feto-placental vessels from gestational diabetic pregnancies is related to downregulation of occludin expression. Diabetologia 64 (1), 195–210. doi:10.1007/s00125-020-05290-6
Volpe, D. A. (2020). Advances in cell-based permeability assays to screen drugs for intestinal absorption. Expert Opin. Drug Discov. 15 (5), 539–549. doi:10.1080/17460441.2020.1735347
Vukomanović, P., Stefanović, M., Stevanović, J. M., Petrić, A., Trenkić, M., Andrejević, L., et al. (2024). Monte Carlo optimization method based QSAR modeling of placental barrier permeability. Pharm. Res. 41 (3), 493–500. doi:10.1007/s11095-024-03675-5
Wenzel, C., Drozdzik, M., and Oswald, S. (2021). Organic cation transporter 1 an intestinal uptake transporter: fact or fiction? Front. Pharmacol. 12, 648388. doi:10.3389/fphar.2021.648388
Werdan Romão, M. A., Pinto, L., Cavalli, R. C., Duarte, G., de Moraes, N. V., Abduljalil, K., et al. (2024). Mechanistic framework to predict maternal-placental-fetal pharmacokinetics of nifedipine employing physiologically based pharmacokinetic modeling approach. J. Clin. Pharmacol. 64 (5), 568–577. doi:10.1002/jcph.2404
Wessel, R. E., and Dolatshahi, S. (2024). Regulators of placental antibody transfer through a modeling lens. Nat. Immunol. 25 (11), 2024–2036. doi:10.1038/s41590-024-01971-1
Yagel, S., Cohen, S. M., and Goldman-Wohl, D. (2022). An integrated model of preeclampsia: a multifaceted syndrome of the maternal cardiovascular-placental-fetal array. Am. J. Obstet. Gynecol. 226 (2s), S963–S972. doi:10.1016/j.ajog.2020.10.023
Yan, Y., Wang, Q., Wu, W., Yi, H., and Xie, F. (2024). Evaluation of various approaches to estimate transplacental clearance of Vancomycin for predicting fetal concentrations using a maternal-fetal physiologically based pharmacokinetic model. Pharm. Res. 41 (5), 899–910. doi:10.1007/s11095-024-03705-2
Zambuto, S. G., Scott, A. K., and Oyen, M. L. (2024). Beyond 2D: novel biomaterial approaches for modeling the placenta. Placenta 157, 55–66. doi:10.1016/j.placenta.2024.03.006
Zhang, Z., and Unadkat, J. D. (2017). Development of a Novel Maternal-Fetal Physiologically Based Pharmacokinetic Model II: verification of the model for passive placental permeability drugs. Drug Metab. Dispos. 45 (8), 939–946. doi:10.1124/dmd.116.073957
Zhang, Z., Imperial, M. Z., Patilea-Vrana, G. I., Wedagedera, J., Gaohua, L., and Unadkat, J. D. (2017). Development of a novel maternal-fetal physiologically based pharmacokinetic model I: insights into factors that determine fetal drug exposure through simulations and sensitivity analyses. Drug Metab. Dispos. 45 (8), 920–938. doi:10.1124/dmd.117.075192
Zhang, S., Cheng, Z., Li, X., Shi, Y., Zhu, H., Zhang, T., et al. (2024). Trans-Placental transfer mechanisms of aromatic amine antioxidants (AAs) and p-Phenylenediamine quinones (PPD-Qs): evidence from human gestation exposure and the Rat Uterine perfusion model. Environ. Sci. Technol. 58 (48), 21166–21176. doi:10.1021/acs.est.4c09416
Zheng, Q., Zhou, Q., Li, J., Tian, Y., Huang, H., Yao, Q., et al. (2019). Placental transfer of bromocriptine in an ex vivo human placental perfusion model. J. Matern. Fetal Neonatal Med. 32 (7), 1155–1159. doi:10.1080/14767058.2017.1402000
Glossary
AB Arterial blood
ABC ATP-binding cassette
ADME Absorption, distribution, metabolism, and excretion
AF Amniotic fluid
AECs Amniotic epithelial cells
AMCs Amniotic mesenchymal cells
BCRP Breast cancer resistance protein (ABCG2)
BeWo Human choriocarcinoma trophoblast cell line BeWo
BET Betamethasone
CANA Canagliflozin
CAI Cellular accumulation index
CLI Clearance index
CLp Transplacental clearance
CTBs Cytotrophoblasts
CTCs Cytotrophoblast cells
CZP Certolizumab pegol
DAPA Dapagliflozin
DCs Decidual cells
DEX Dexamethasone
Dcot Cotyledon clearance
EMA European Medicines Agency
EMPA Empagliflozin
ENT Equilibrative nucleoside transporter
EVTBs Extravillous trophoblasts
FDA Food and Drug Administration
FTR Fetal transfer rate
GS GS-441524
HUVECs Human umbilical vein endothelial cells
HPVECs Human placental vascular endothelial cells
hTSCs Human trophoblast stem cells
iNOS Inducible nitric oxide synthase
IV Intravenous
LMV Lamivudine
LLOQ Lower limit of quantification
LVFX Levofloxacin
MBV Maribavir
MCT Monocarboxylate transporter
MP Maternal plasma
MRPs Multidrug resistance-associated proteins (ABCC family)
MVM Microvillous membrane
NA Not applicable
NP Nanoparticle
OATs Organic anion transporters
OATP2B1 Organic anion transporting polypeptide 2B1
OCTs Organic cation transporters
OC Oseltamivir carboxylate
OP Oseltamivir phosphate
PBMP Physiologically based mechanistic placenta
PBPK Physiologically based pharmacokinetic
PEA Peanut allergen
PDE1 Phosphodiesterase 1
PDE5 Phosphodiesterase 5
P-gp P-glycoprotein (ABCB1)
PK Pharmacokinetics
PlGF Placental growth factor
po Oral administration
P-m Placenta (maternal)
P-f placenta (fetal)
P-t placenta (tissue)
QSPR Quantitative structure–property relationship
Rmd Remdesivir
RoB Rest of body
SF Surfactant
sEng Soluble endoglin
sFlt-1 Soluble fms-like tyrosine kinase-1
SGLT2-i Sodium-glucose cotransporter-2 inhibitor
sl Sublingual
STB/STBs Syncytiotrophoblasts
TEER Transepithelial electrical resistance
THC Cannabinoid (-)-Δ9-tetrahydro-cannabinol
TNF-α Tumor necrosis factor alpha
TPTs Transporter proteins
UCP Umbilical cord plasma
VB Venous blood
Keywords: pharmacokinetics, placental drug transfer, in vitro model, ex vivo placental perfusion, in silico simulation
Citation: Li Z, Wu Y, Zeng S, Wang F, Zhang J, Li S, Yang Y and Yang Y (2025) Strategy advancements in placental pharmacokinetics: from in vitro experiments to in silico prediction. Front. Pharmacol. 16:1694886. doi: 10.3389/fphar.2025.1694886
Received: 29 August 2025; Accepted: 07 October 2025;
Published: 27 October 2025.
Edited by:
Jiangxin Wang, Shenzhen University, ChinaReviewed by:
Zixi Chen, Shenzhen University, ChinaZheng Yuan, China Academy of Chinese Medical Sciences, China
Copyright © 2025 Li, Wu, Zeng, Wang, Zhang, Li, Yang and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yong Yang, eXl4cG93ZXJAMTYzLmNvbQ==; Yujie Yang, c2N1eWFuZ3l1amllQDE2My5jb20=
 Zhimin Li
Zhimin Li Yue Wu1
Yue Wu1 Siyu Zeng
Siyu Zeng Fei Wang
Fei Wang Jiao Zhang
Jiao Zhang Shiran Li
Shiran Li Yong Yang
Yong Yang Yujie Yang
Yujie Yang