- 1 Chemical and Biochemical Engineering, The University of Iowa, Iowa City, IA, USA
- 2 Department of Psychiatry, The University of Iowa, Iowa City, IA, USA
- 3 Center for Family Research, University of Georgia, Athens, GA, USA
- 4 Neuroscience Program, The University of Iowa, Iowa City, IA, USA
Altered regulation of the serotonin transporter (SLC6A4) is hypothesized to be a key event in many forms of neuropsychiatric illness, yet our understanding of the molecular mechanisms through which changes in gene function could lead to illness remains incomplete. In prior studies, we and others have demonstrated that methylation of CpG residues in the promoter associated CpG island alters SLC6A4 gene expression, that the extent of that DNA methylation in child abuse is genotype dependent, and that adverse childhood experiences such as child sex abuse are related to methylation. However, we have not examined whether these effects are splice variant specific, whether the association of methylation to gene expression varies as a function of genotype, and whether methylation in other SLC6A4 gene regions are more likely candidates for GxE effects. In the current investigation we measured methylation in lymphoblast DNA from 158 female subjects in the Iowa Adoption Studies at 16 CpG residues spread across the SLC6A4 locus, and analyzed their relationship to gene expression for two SLC6A4 splice variants. Methylation of two CpG residues in the shore of the CpG island (cg22584138 and cg05951817), a location immediately upstream from exon 1A, predicted gene expression for the splice variant containing Exon 1A + 1B. Methylation at two residues in the CpG island itself (cg 25769822 and cg05016953) was associated with total SLC6A4 expression. Examination of these four CpG residues indicated that methylation of cg22584138 was influenced by both genotype and sex abuse, whereas methylation of cg05016953 was influenced only by sex abuse history. Factors influencing methylation at other CpG dinucleotide pairs were not identified. We conclude that methylation effects on transcription may vary as a function of underlying gene motif and splice variant, and that the shore of CpG islands, upstream of TSS, may be of particular interest in examining environmental effects on methylation.
Introduction
Altered serotonergic neurotransmission is postulated to play a critical role in the pathogenesis of a number of neuropsychiatric disorders with a key component of that modified transmission being altered serotonin transporter (SLC6A4) function (Ramamoorthy et al., 1993; Lesch et al., 1995). Over the past 10 years, a large body of evidence has accumulated which demonstrates that both genetic and environmental factors act through the serotonin transporter to alter the likelihood of behavioral illness (Caspi et al., 2003; Karg et al., 2011). At the current time, the exact mechanism(s) through which this differential vulnerability is enacted and the relative contribution of each of these mechanisms to illness remains unclear but it has been hypothesized that many of these changes serotonergic function involve altered epigenetic regulation, in particular altered DNA methylation.
These changes in epigenetic regulation of SLC6A4 are potentially genotype or position dependent. For example, methylation at another key regulator of neurotransmission, MAOA, is known to be genotype dependent (Philibert et al., 2008, 2010). But even so, not all changes in gene’s DNA methylation signature are equivalent with DNA methylation in the first exon of a gene tending to most highly associated with transcriptional silencing (Brenet et al., 2011).
These changes in SLC6A4 methylation may also result from developmental trauma. For example, methylation differences have been demonstrated in postmortem hippocampi obtained from suicide victims with a history of childhood abuse relative to those from either suicide victims with no childhood abuse or non-suicide controls (McGowan et al., 2008, 2009). Likewise, in animal models, early experience vis-à-vis the mother has been linked to lasting epigenetic change via CpG methylation (Liu et al., 1991; Kaffman and Meaney, 2007). Likewise in our prior work with women in the IAS sample, we found that sex abuse was associated with overall hypermethylation of the SLC6A4 promoter region (Beach et al., 2010).
Unfortunately, no matter what the source of altered DNA methylation, understanding the effects of those changes of DNA methylation on SLC6A4 or any other gene function in the brain is a difficult task. Even if individual specific changes in DNA methylation secondary to environmental stimuli are consistent between tissues (Monick et al., 2012), the effect of genotypic, trauma, or substance induced methylation differences may differ across tissues because most multi-exon genes have tissue specific splice variation (Johnson et al., 2003), and the expression of many, if not most genes, is under the regulation of tissue specific transcriptional factors (Heintzman et al., 2009). Hence, because the vast majority of gene expression studies of serotonergic function are performed using peripheral blood RNA, it very well may be that gene or trauma associated methylation patterns affect only some splice variants and not others, requiring attention to which splice variants are most relevant for CNS related gene expression.
This point may be particularly relevant for studies of the SLC6A4 locus. In prior examinations of this locus, we and others have demonstrated that SLC6A4 is under both genetic and epigenetic control and provided evidence that exposure to child abuse is associated with genotype contextual altered methylation of the promoter associated CpG island (Beach et al., 2010, 2011), a finding which is supported by similar findings in macaques (Kinnally et al., 2010). Unfortunately, while these findings of gene-environment (GxE) effects are interesting, they are incomplete because we only examined DNA methylation in the promoter region and did not examine these effects of abuse related methylation changes on gene transcription. Because Ozsarac et al. (2002) have demonstrated the existence of at least three SLC6A4 splice variants (Brenet et al., 2011) and the expression of each of those splice variants may be regulated differently, a more precise formulation of any role of DNA methylation in altered regulation of SLC6A4 function must include a more global assessment of the epigenetic status of the gene and the relationship of any abuse associated methylation changes to splice variant specific transcription.
In this communication, we attempt to address a portion of that knowledge deficit by examining mRNA splice variant expression and gene wide SLC6A4 methylation using clinical data and biomaterials from a subset of the subjects in whom we have previously shown significant environmental and GxE effects.
Materials and Methods
A detailed description of the methods and procedures used in the Iowa Adoption Study (IAS) has been previously published (Philibert, 2006). All procedures used in the IAS were approved by the University of Iowa Institutional Review Board.
The clinical data and biomaterial for the current studies were drawn from a set of 152 subjects examined previously in whom we have demonstrated GxE effects. In brief, the childhood abuse data is derived from Wave 4 of the IAS (1999–2004) during which were asked about a range of current symptoms and past childhood events. To assess familial childhood sex abuse, subjects were asked two questions. The first question was; “Before you were age 16, where there any sexual contact between you and any family member, like a parent or stepparent, grandparents, uncle, aunt, brother, sister, or cousin? By sexual contact I mean their touching your sexual parts, your touching their sexual parts, or sexual intercourse.” The second question was; “Was there sexual contact with a parent or grandparent?” The answers to these questions were combined to form the sex abuse variable. For the purposes of this study, and affirmative answer to either question was deemed a positive response. To assess for the presence of non-familial childhood sexual abuse, the subjects were asked an identically phrased question with the exception that “anyone other than a family member” was substituted for the text referring to family members in the two questions. Similarly, any affirmative response to either of these questions was deemed a positive response.
DNA and RNA for the current studies were prepared as previously described from EBV transformed lymphoblast cell lines (Philibert et al., 2007a). Genotyping of the 5HTTLPR variant was conducted as previously described.
The study attempted to measure the expression of three separate SLC6A4 spliced Exon 1 containing variants that include either (1) both Exon 1A and B (1A + 1B), (2) just Exon 1A or (3) just Exon 1C, immediately upstream of exon 2. We have already produced a probe specific to the first variant 1A + 1B; (Philibert et al., 2007b). Because was not possible to devise a Taqman Probe to assess the presence of the second variant which contains just exon 1A upstream of exon two (1A + 2), therefore, we assessed its presence using both conventional and Sybr green mediated PCR-based techniques. The reverse and forward primers were CCAGCCCGGGACCAGCCTCCCCGCGCAGCC and CCGAGCTCTCTATCGTCGGGATTGACACGTCGGGATTGA, respectively. The forward primer was situated on exon 2 and the reverse primer was situated on exon 1A. RTPCR was performed using the POWER SYBR Green Master Mix (ABI, Foster City, CA, USA) and two above primers and an annealing temperature of 70°C using a7900HT Fast Real-Time PCR System (ABI, Foster City, CA, USA). A third variant, which includes exon 1C, was assessed using the reverse primer for exon 2 listed above and a forward primer unique to exon 1C AGCCTCCACGGCGGTGAAATGAAG.
In order to determine the expression of total and (1A + 1B) SLC6A4 mRNA, total RNA was reverse transcribed to cDNA using an Applied Biosystems cDNA archiving kit (ABI, Foster City, CA, USA). Quantification of each 5HTTLPR variant was conducted with Taqman probe sets and amplification reagents from Applied Biosystems (Foster City, USA) using a Fluidigm BioMark Genetic Analysis System (South San Francisco, USA). The sequence of the 5HTT specific primer probe sets which bridge exons 1A and 1B (ABI, Hs00984354) and exons 8 and 9 (ABI, Hs00169010) have been described previously (Philibert et al., 2007b). The comparator housekeeping gene probe sets were for the following genes: CALR, RPL7A, RPS19, RPS 20, and UBC. After a brief pre-amplification procedure, RTPCR quantification of transcript levels was performed in triplicates in two separate runs (9216 reactions per run) on Biomark Fluidigm platform using default settings. CT counts were determined using proprietary Fluidigm software with the results being exported for analysis.
The SLC6A4 methylation values at 16 individual CpG residues for the subjects were extracted from a data set constructed for genome wide DNA methylation studies. In brief, methylation status at SLC6A4 and other loci were assessed using the Illumina HumanMethylation450 BeadChip under contract by the University of Minnesota Genome Center using the protocol specified by the manufacturer and the contractor using a random assignment paradigm. The resulting microarray data were inspected for complete bisulfite conversion of the DNA and the presence of chip or batch variation with no significant results after the average beta values (i.e., average methylation) for each CpG residue were determined using the GenomeStudio V2009.2; Methylation module Version 1.5.5., version 3.2 (Illumina, San Diego). The HumanMethylation450 BeadChip contains 485,577 probes that recognize at least 20,216 unique features. With respect to this sample, >99.76% of the 485,577 probes yielded statistically reliable data. The resulting beta values were then exported from the larger data set into Microsoft Excel (Microsoft, USA) for data management prior to data analysis.
Prior to analysis, all gene expression and methylation data were converted to z-scores with all data analysis being performed using the JMP version 9 (SAS Institute, Cary, SC, USA) and SPSS version 19 (IBM, New York).
In order to identify functional loci for which methylation was associated with expression of different transcripts, regression analyses were conducted for each of the 16 methylation loci. We examined the effect of genotype and methylation jointly, controlling for the effect of level of alternate transcript. We first examined the association of methylation with transcription of Exon 1A + 1B controlling for transcription of Exon 8, and then examined transcription of Exon 8, controlling for transcription of Exon 1A + 1B. Following identification of functional regions with regard to each transcript, we then examined the role of sex abuse and genotype on methylation at functional loci. Regression of methylation level on sex abuse and genotype was conducted for each of the functional loci.
Results
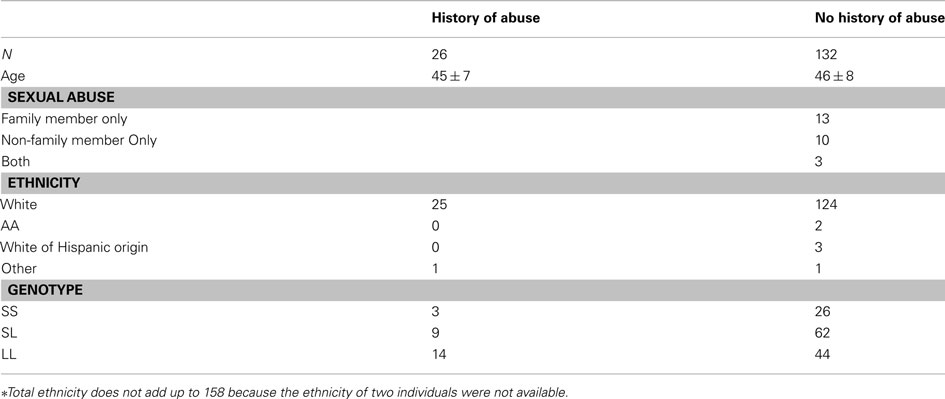
The clinical, demographic, and genetic characteristics of the 158 subjects included in this study are described in Table 1. Secondary the inability of the analytic algorithms to accommodate missing data, only 152 of these subjects were included in the GxE analyses but all subjects were included in the gene expression analyses. In summary, the subjects are all female and nearly all white. At the time of the blood draw for the preparation of the cell lines used in these epigenetic and gene expression and the subjects tended to be in their middle to late 40s. At the Wave 4 interview, 16 of the subjects reported sexual abuse by a family member with an additional 10 subjects denying sexual abuse by a family member but reporting sexual abuse by a non-family member.
The structure of the SLC6A4 gene and the placement of the 16 methylation probes are illustrated in Figure 1. SLC6A4 is a 25-kb, 14 exon gene located on chromosome 17. Exon one is subdivided into three segments, 1A, 1B, and 1C, that are differentially spliced to provide three known mRNA products that include exons 2-14 in addition to their unique portions of exon one (Exon 1A + B, 1A, and 1C).
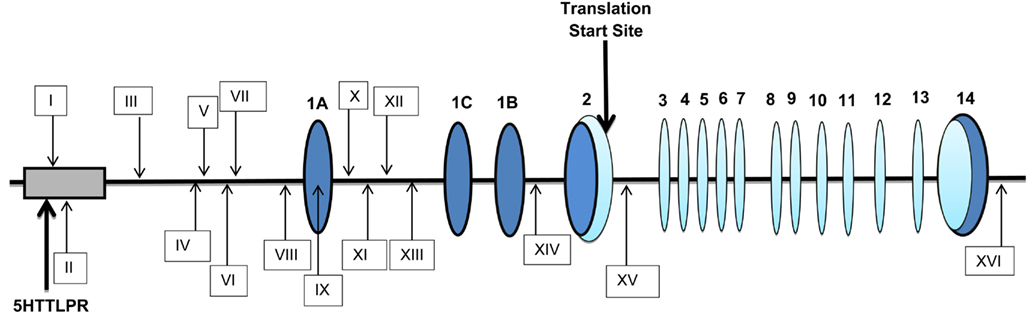
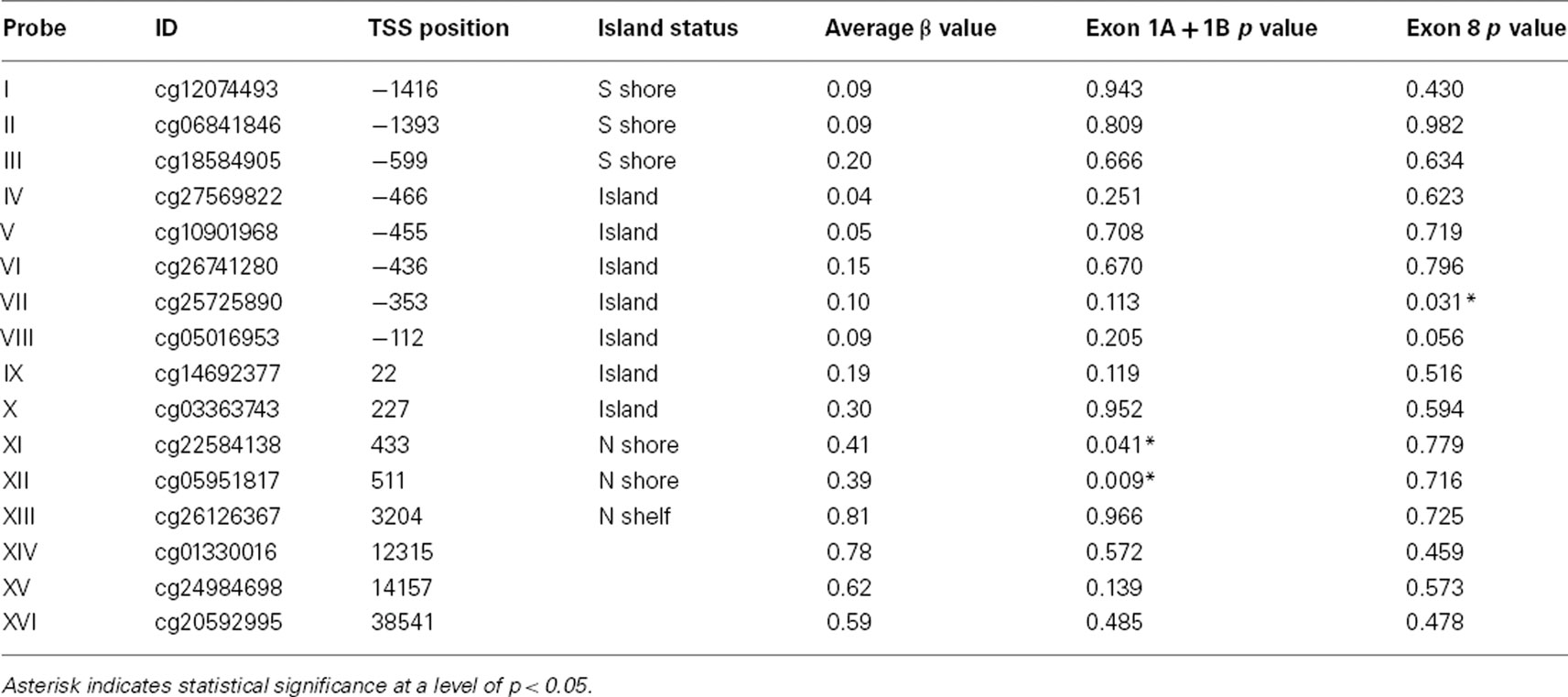
Figure 1. The position of the methylation probes relative to the structure of SLC6A4. Numbers in bold represents the 14 exons present in this gene. Dark blue ovals correspond to untranslated regions (UTR) of SLC6A4 while light blue ovals correspond to translated portions of the exons with the translation start site being contained within exon 2. The “first exon” actually contains three variably splice DNA segments labeled 1A, 1C, and 1B. The identity of the methylation probes is indicated by boxes with Roman numerals. Probes I (cg1207449), II (cg06841846), and III (cg18584905) are situated at the “south shore” of the gene upstream of the transcription start site (TSS). Probes IV (cg27569822), V (cg10901968), VI (cg26741280), VII (cg25725890), and VIII (cg05016953) are contained within the portion of the CpG island proper upstream of the transcription start site (TSS) while Probe IX (cg14692377) is contained both within the CpG island proper and exon 1A. Probes X (cg03363743), XI (cg22584138), and XII (cg05951817) are situated at the north shore and Probe XIII (cg26126367) at the north shelf of the gene in intron 1A. Probe XIV (cg01330016) is in intron 1B while Probe XV (cg24984698) intron 2. Finally, Probe XVI (cg20592995) is immediately distal to the 3′UTR. The transcription start site is at exon 1A.
First, we examined the levels of total and splice variant specific SLC6A4 expression using RTPCR and primer probes sets designed to recognize the each of the three splice variants described by Ozsarac et al. (2002). Despite several attempts, we were unable to detect expression of the two variants containing exon 1C or just exon 1B. However, using a previously developed probe set which recognizes the exon 1A + B variant (the MGB probe bridges the junction between 1A and B) and a probe that recognizes all three variants (the MGB probe bridges exon 7 and 6). The relationship of the expression levels of each of these transcripts to 5HTTLPR genotype is given in Table 2. As the table indicates, although the directionality is consistent with prior findings, in this small group of samples 5HTTLPR genotype was only significantly associated with total SLC6A4 mRNA levels (Exon 8), but not with the levels of the Exon 1A + B containing transcript. In addition, consistent with prior assessments, the level of the Exon 1A + B containing transcript was only partially correlated with total mRNA levels (r = 0.70).
The relationship between methylation at each of the CpG residues and gene expression is delineated in Table 3. As the table indicates, the expression of the Exon 1A + B containing transcript was significantly associated with two consecutive CpG residues in the intronic region immediately adjacent to Exon 1C. In contrast, total mRNA expression was only significantly associated with a methylation at cg05016953, a probe found 375 bp upstream of the transcription start site (TSS). There were no significant rselationships between methylation status at any of the probes and 5HTTLPR genotype including at two probes (cg12074493 and cg06841846) which map to the 5HTTLPR repetitive region.
The methylation of the core portion of the CpG island has determined previously using MALDI-TOF mass spectroscopy. Therefore, in an attempt to understand the reliability of these two types of methylation measures, we compared the current values obtained using the Illumina arrays with previous values. Overall, 4 CpG residues (cg03363743, cg14692377, cg05016953, cg25725890) at this locus were surveyed by both approaches. At each CpG residue, the degree of methylation determined by each method was significantly correlated with the average adjusted r2 equaling ∼0.34.
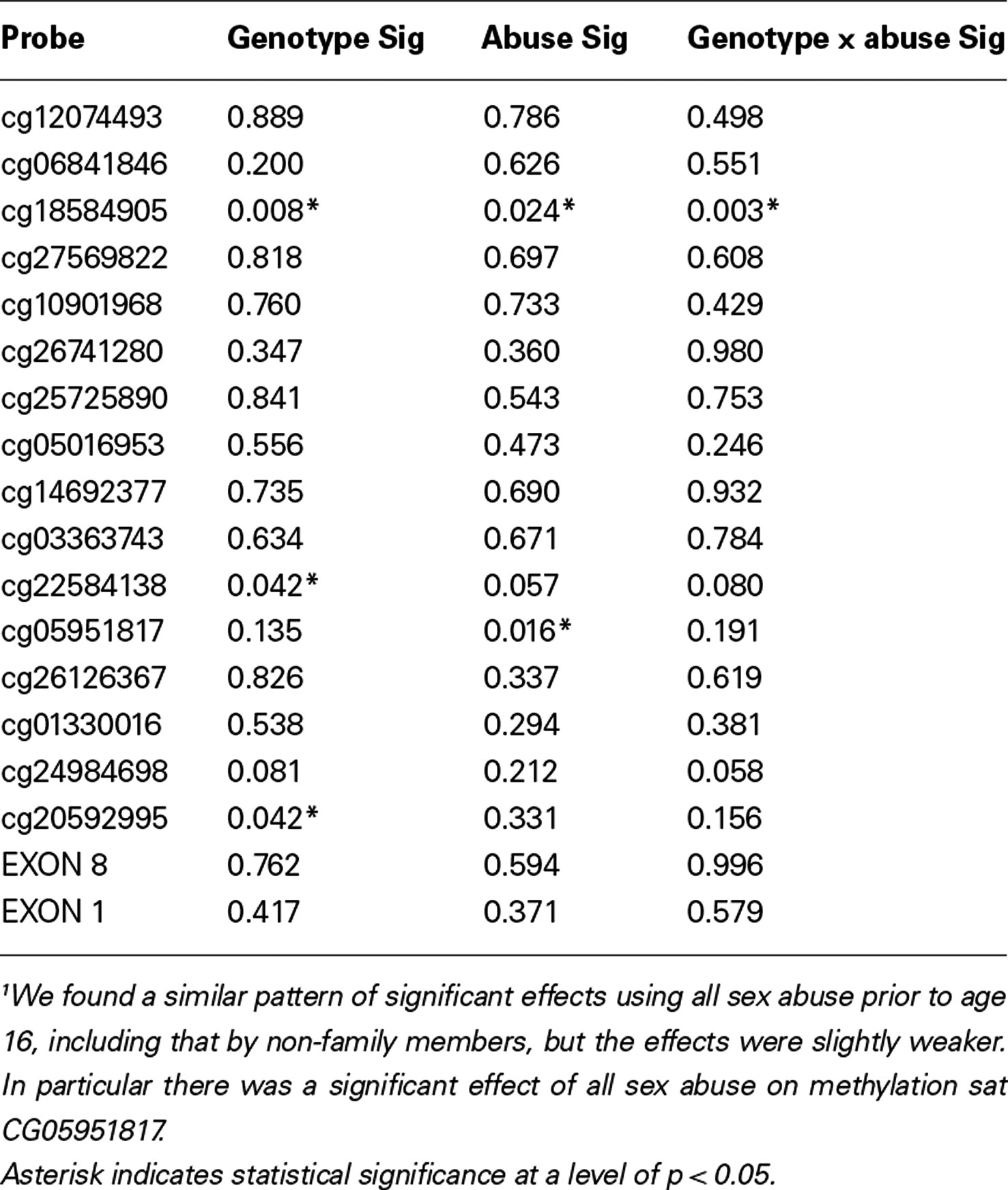
The relationship between methylation and gene expression to genotype, abuse, and the interaction effects of genotype and abuse is delineated in Table 4. Two distinct classes of sexual abuse were examined. However, since results were very similar we report only the analyses focused on sex abuse by a family member and will provide the parallel results including non-family member abuse upon request.
Discussion
In summary, we conducted iterative examinations of the relationships of gene methylation on SLC6A4 mRNA expression and of genotype and child abuse interaction effects with methylation. Limitations of this work include the small number of subjects, the lack of full correction for multiple comparisons, the use of non-CNS biomaterials and the self-report nature of the data.
The overarching goal of this project was to continue the biological conversation as the possible biological mechanisms through which childhood sex abuse leads to adult psychopathology. Whereas it is doubtless that a portion of this increased proclivity for depression and other forms of behavioral illness is engendered at the neuroanatomical level, one of the leading hypotheses is that childhood trauma also leaves an indelible mark on gene transcription. If so, the mechanisms underlying these changes should be demonstrable, and perhaps, remediable. In prior examinations, we demonstrated that changes in SLC6A4 genotypically contextual changes in promoter methylation were associated with self-reports of child abuse but did not analyze the relationship of those changes methylation to SLC6A4 transcription. The connection of changes in methylation to possible changes in gene transcription are critical to the hypotheses specifying molecular changes as a result of child abuse because of the tight connection between levels of mRNA and protein (Schwanhausser et al., 2011). But despite the relative ease of conducting examinations of peripheral SLC6A4 gene expression, in our published (Philibert, 2008) and unpublished examinations, we have not been able to demonstrate a significant relationship between SLC6A4 gene expression and depression and there is a relative paucity of publications in this area and we were led to conclude that on the basis of the extant literature, that there may not be a tight connection between the changes in gene methylation observed in these and other studies and peripheral SLC6A4 gene expression.
In the current study, we again demonstrate significant GxE effects but do not find a robust connection between these methylation changes and SLC6A4 gene expression. A possible explanation this observation is that the changes in peripheral blood methylation do effect gene transcription, but only affect transcription in the brain. In prior decades, this possibility would be deemed unlikely but over the past several years it has become increasingly clear that although individual variance in methylation may be conserved across tissues (Mill et al., 2011; Monick et al., 2012), secondary to the effects of tissue specific transcription factors and multiply spliced genes, these harmonized changes in methylation may not have synonymous effects on gene transcription (Fan and Zhang, 2009). Since SLC6A4 has three known splice variants (Ozsarac et al., 2002) and we have repeatedly demonstrated that the expression of the Exon 1A + B variant is only partially correlated with total mRNA as measured by the probe set that spans the juncture of exon 7 and 8 (Philibert et al., 2007b; Philibert, 2008), if the changes observed in DNA methylation in association with child abuse are real, they may be exerting their effects through changes in other, more CNS specific SLC6A4 splice variants.
Unfortunately, to the best of our knowledge, the transcriptional repertoire of CNS serotonergic neurons has not been described thus making it difficult to make more exact comparisons of lymphoblast and CNS specific SLC6A4 transcription. However in rats, the 1A + 2, which was not detected at all in our efforts, was expressed more abundantly in the CNS than 1A + 1B + 2 variant (Ozsarac et al., 2002). Therefore, studies to examine the relationship of methylation to expression of CNS specific splice variants, preferably using native human cells would be enlightening.
In this regard, it is important to also discuss the strengths and weaknesses of the lymphoblast cell lines used in the current studies. Without a doubt, lymphoblasts are valuable models for investigations of the relationship of genetic changes to gene expression and have been used in thousands of previous studies. However, it is also clear that these cell lines have weaknesses when examining methylation. An early study by Grafodatskaya et al. (2009) using four lymphoblast cell lines demonstrated that the signatures in lymphoblasts are not the same as native lymphocytes and that extended serial passage of these lines further exacerbated these differences. However, a recent study by Sun et al. (2010) using the Illumina 27K platform of 34 matched sets of lymphoblasts showed that the vast majority of cell lines had correlation coefficients of greater than 0.9 with the vast majority of all differences in methylation between the cell types being identifiable by factor analysis. Given these findings and the findings by Vawter and colleagues which demonstrated the high correlation of gene expression between these two types of cell preparations (Rollins et al., 2010), the extant literature indicates that the current findings are reliable but needing further confirmation.
Of significant relevance to the hypothesis that child abuse exerts a part of its effects thorough altered SLC6A4 methylation is that the CpG residue immediately downstream of the 5HTTLPR is the most significantly associated residue in our analyses. Given the wealth of prior analyses that demonstrate the effects of this motif at the transcriptional and interactive levels, this seems only logical. However, the failure of the two immediately upstream (more 5′) residues is difficult to understand at first glance. However, it is important to note that these two residues (cg12074493 and cg06841846) map to the repetitive element themselves and the molecular mechanism through which increased VNTR number confers increased transcription is unknown. Therefore, it is quite possible that these probes do not accurately interrogate the methylation status at CpG residues these degenerate sequences or that they do not interrogate the most relevant portion of the VNTR associated haplotype.
Overall, 26 of the 158 (16%) female subjects who participated in the study reported childhood sexual abuse which is somewhat lower than the 22% rate reported in the general population (Gorey and Leslie, 1997). An additional consideration in this regard is the reliability of this type of adult recollections of childhood abuse. A study by Widom and Morris examine the accuracy of these recollections in a sample of subjects with documented histories of sex abuse. Surprisingly, she found that only two thirds of those with documented histories of sexual abuse as children reported this abuse when interviewed as an adult some 20 years later (Widom and Morris, 1997). Conversely, Loftus and colleagues have demonstrated the tendency of individuals to falsely report memories of childhood abuse (Loftus, 1996). Hence, it is reasonable to be suspicious of both Type I and Type II errors in these analyses.
Somewhat surprisingly, the expansion of the affected status in the sexual abuse analyses to include those 10 individuals who were abused by non-family members but not by family members did not increase the significance of the results. Reassuringly, however, in this second expanded analysis the results continued to identify differential methylation at cg18584905 continued to be significant in all aspects of analysis. Although there are a number of ways to interpret these results, one possible conclusion from these results is that consistent with previous findings (Bulik et al., 2001), the impact of sexual abuse by non-family members in this population is not as severe as the impact of sexual abuse by members of one’s own family.
This is one of few studies to directly compare the results obtained from the Sequenom mass spectroscopy and Illumina single base extension technologies using aliquots of the same samples of DNA. The results, which show modest concordance between the results at the four residues, suggest that the current results are reliable but suggest that there is room for improvement in at least one of the approaches.
In our original analyses, we only examined the effects of child abuse by family members. In this study, we expand our analyses to include sexual abuse by non-family members. Despite the increased number of affected individuals, the strength of the findings at cg18584905 did not increase. Although many explanations for this are possible, consistent with the findings of Sen and colleagues who found that the severity of stressors had significant effects on the strength of the GxE effects (Karg et al., 2011), it may well be that sexual abuse by family members has much more severe effects on young women than abuse by non-family members.
In summary, in this communication, using a different technique, we examine the effects of childhood sexual abuse on methylation at 16 residues spanning the SLC6A4 gene and, among functional loci, demonstrate a significant genotype dependent effect at cg22584138 and a significant effect of sex abuse at the adjacent loci cg05951817, residues that are downstream of exon 1A.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by grants to Dr. Philibert (DA 015789 and MH0809898). On behalf of Drs. Philibert and Madan, the University of Iowa has filed an intellectual property claim on the CpG island. This work is dedicated to the continuing memory of Dr. Remi Cadoret, Founder of the Iowa Adoption Studies who strongly believed in the role of the environment in shaping behaviors.
References
Beach, S. R., Brody, G. H., Todorov, A. A., Gunter, T. D., and Philibert, R. A. (2010). Methylation at SLC6A4 is linked to family history of child abuse: an examination of the Iowa adoptee sample. Am. J. Med. Genet. B Neuropsychiatr. Genet. 153, 710–713.
Beach, S. R., Brody, G. H., Todorov, A. A., Gunter, T. D., and Philibert, R. A. (2011). Methylation at 5HTT mediates the impact of child sex abuse on women’s antisocial behavior: an examination of the Iowa adoptee sample. Psychosom. Med. 73, 83–87.
Brenet, F., Moh, M., Funk, P., Feierstein, E., Viale, A. J., Socci, N. D., and Scandura, J. M. (2011). DNA methylation of the first exon is tightly linked to transcriptional silencing. PLoS ONE 6, e14524. doi:10.1371/journal.pone.0014524
Bulik, C. M., Prescott, C. A., and Kendler, K. S. (2001). Features of childhood sexual abuse, and the development of psychiatric, and substance use disorders. Br. J. Psychiatry Suppl. 179, 444–449.
Caspi, A., Sugden, K., Moffitt, T. E., Taylor, A., Craig, I. W., Harrington, H. L., McClay, J., Mill, J., Martin, J., Braithwaite, A., and Poulton, R. (2003). Influence of life stress on depression: moderation by a polymorphism in the 5-HTT gene. Science 301, 386–389.
Fan, S., and Zhang, X. (2009). CpG island methylation pattern in different human tissues, and its correlation with gene expression. Biochem. Biophys. Res. Commun. 383, 421–425.
Gorey, K., and Leslie, D. (1997). The prevalence of child sexual abuse: Integrative review adjustment for potential response, and measurement biases. Child Abuse Negl. 21, 391–398.
Grafodatskaya, D., Choufani, S., Ferreira, J. C., Butcher, D. T., Lou, Y., Zhao, C., Scherer, S. W., and Weksberg, R. (2009). EBV transformation, and cell culturing destabilizes DNA methylation in human lymphoblastoid cell lines. Genomics 95, 73–83.
Heintzman, N. D., Hon, G. C., Hawkins, R. D., Kheradpour, P., Stark, A., Harp, L. F., Ye, Z., Lee, L. K., Stuart, R. K., Ching, C. W., Ching, K. A., Antosiewicz-Bourget, J. E., Liu, H., Zhang, X., Green, R. D., Lobanenkovm, V. V., Stewart, R., Thomson, J. A., Crawford, G. E., Kellis, M., and Ren, B. (2009). Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459, 108–112.
Johnson, J. M., Castle, J., Garrett-Engele, P., Kan, Z., Loerch, P. M., Armour, C. D., Santos, R., Schadt, E. E., Stoughton, R., and Shoemaker, D. D. (2003). Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science 302, 2141–2144.
Kaffman, A., and Meaney, M. J. (2007). Neurodevelopmental sequelae of postnatal maternal care in rodents: clinical, and research implications of molecular insights. J. Child Psychol. Psychiatry 48, 224–244.
Karg, K., Burmeister, M., Shedden, K., and Sen, S. (2011). The serotonin transporter promoter variant (5-HTTLPR), stress, and depression meta-analysis revisited: evidence of genetic moderation. Arch. Gen. Psychiatry 68, 444–454.
Kinnally, E. L., Capitanio, J. P., Leibel, R., Deng, L., Le Duc, C., Haghighi, F., and Mann, J. J. (2010). Epigenetic regulation of serotonin transporter expression, and behavior in infant rhesus macaques. Genes Brain Behav. 9, 575–582.
Lesch, K. P., Gross, J., Franzek, E., Wolozin, B. L., Riederer, P., and Murphy, D. L. (1995). Primary structure of the serotonin transporter in unipolar depression, and bipolar disorder. Biol. Psychiatry 37, 215–223.
Liu, D., Diorio, J., Tannenbaum, B., Caldji, C., Francis, D., Freedman, A., Sharma, S., Pearson, D., Plotsky, P. M., and Meaney, M. J. (1991). Maternal care, hippocampal glucocorticoid receptors, and hypothalamic-pituitary-adrenal responses to stress. Science 277, 1659–1662.
Loftus, E. F. (1996). Memory distortion and false memory creation. Bull. Am. Acad. Psychiatry Law 24, 281–295.
McGowan, P. O., Sasaki, A., D’Alessio, A. C., Dymov, S., Labonté, B., Szyf, M., Turecki, G., and Meaney, M. J. (2009). Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat. Neurosci. 12, 342–348.
McGowan, P. O., Sasaki, A., Huang, T. C., Unterberger, A., Suderman, M., Ernst, C., Meaney, M. J., Turecki, G., and Szyf, M. (2008). Promoter-wide hypermethylation of ribosomal RNA gene promoter in the suicide brain (electronic article). PLoS ONE 3, e2085. doi: :10.1371/journal.pone.0002085
Mill, J., Davies, M., Volta, M., Dobson, R., Meaburn, E., and Schalkwyk, L. (2011). Methylome brain, and blood: brain areas-specific differentially methylated regions, individual differences, and allele specific DNA methylation. Presented in American Society for Human Genetics, Montréal, QC.
Monick, M. M., Beach, S. R., Plume, J., Sears, R., Gerrard, M., Brody, G. H., and Philibert, R. A. (2012). Coordinated changes in AHRR methylation in lymphoblasts, and pulmonary macrophages from smokers. Am. J. Med. Genet. B Neuropsychiatr. Genet. 159, 141–151.
Ozsarac, N., Santha, E., and Hoffman, B. J. (2002). Alternative non-coding exons support serotonin transporter mRNA expression in the brain, and gut. J. Neurochem. 82, 336–344.
Philibert, R. (2006). Merging genetic, and environmental effects in the Iowa adoption studies: focus on depression. Ann. Clin. Psychiatry 18, 219–222.
Philibert, R. A. (2008). The relationship of 5HTT (SLC6A4) methylation and genotype on mRNA expression and liability to major depression and alcohol dependence in subjects from the Iowa adoption studies. Am. J. Med. Genet. B Neuropsychiatr. Genet. 147, 543–549.
Philibert, R. A., Beach, S. R., Gunter, T. D., Brody, G. H., Madan, A., and Gerrard, M. (2010). The effect of smoking on MAOA promoter methylation in DNA prepared from lymphoblasts, and whole blood. Am. J. Med. Genet. 153, 619–628.
Philibert, R. A., Gunter, T. D., Beach, S. R., Brody, G. H., and Madan, A. (2008). MAOA methylation is associated with nicotine, and alcohol dependence in women. Am. J. Med. Genet. 147, 565–570.
Philibert, R. A., Ryu, G. Y., Yoon, J. G., Sandhu, H., Hollenbeck, N., Gunter, T., Barkhurst, A., Adams, W., and Madan, A. (2007a). Transcriptional profiling of subjects from the Iowa adoption studies. Am. J. Med. Genet. B Neuropsychiatr. Genet. 144, 683–690.
Philibert, R., Madan, A., Andersen, A., Cadoret, R., Packer, H., and Sandhu, H. (2007b). Serotonin transporter mRNA levels are associated with the methylation of an upstream CpG island. Am. J. Med. Genet. 144B, 101–105.
Ramamoorthy, S., Baumant, A., Mooret, K. R., Han, H., Yang-feng, T., Chang, A. S., Ganapathy, V., and Blakelyt, R. D. (1993). Antidepressant-, and cocaine-sensitive human serotonin transporter: molecular cloning, expression, and chromosomal localization. Proc. Natl. Acad. Sci. U.S.A. 90, 2542–2546.
Rollins, B., Martin, M. V., Morgan, L., and Vawter, M. P. (2010). Analysis of whole genome biomarker expression in blood, and brain. Am. J. Med. Genet. B Neuropsychiatr. Genet. 153, 919–936.
Schwanhausser, B., Busse, D., Li, N., Dittmar, G., Schuchhardt, J., Wolf, J., Chen, W., and Selbach, M. (2011). Global quantification of mammalian gene expression control. Nature 473, 337–342.
Sun, Y., Turner, S. T., Smith, J. A., Hammond, P. I., Lazarus, A., Van De Rostyne, J. L., Cunningham, J. M., and Kardia, S. L. (2010). Comparison of the DNA methylation profiles of human peripheral blood cells, and transformed B-lymphocytes. Hum. Genet. 127, 651–658.
Keywords: serotonin transporter, methylation, mRNA expression, child abuse, epigenetics
Citation: Vijayendran M, Beach SRH, Plume JM, Brody GH and Philibert RA (2012) Effects of genotype and child abuse on DNA methylation and gene expression at the serotonin transporter. Front. Psychiatry 3:55. doi: 10.3389/fpsyt.2012.00055
Received: 03 January 2012; Accepted: 18 May 2012;
Published online: 13 June 2012.
Edited by:
Alan Apter, Schneider Children’s Medical Center of Israel, IsraelReviewed by:
Bart P. F. Rutten, Maastricht University, NetherlandsMunis Dundar, Erciyes University, Turkey
Doron Gothelf, Sheba Medical Center, Israel
Copyright: © 2012 Vijayendran, Beach, Plume, Brody and Philibert. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Robert A. Philibert, Department of Psychiatry, The University of Iowa, Room 2-126 MEB Psychiatry Research/MEB, Iowa City, IA 52242-1000, USA. e-mail:cm9iZXJ0LXBoaWxpYmVydEB1aW93YS5lZHU=