- 1Department of Chemistry, Bioscience and Environmental Engineering, Faculty of Science and Technology, University of Stavanger, Stavanger, Norway
- 2Environmental Engineering Department, Faculty of Civil Engineering, Istanbul Technical University, Istanbul, Türkiye
- 3Institute for Infection Prevention and Hospital Epidemiology, University Medical Center Freiburg, Freiburg, Germany
- 4The Science Academy, Istanbul, Türkiye
The aim of this study was to reveal the microbial and kinetic impacts of acute and chronic exposure to one of the frequently administered antibiotics, i.e., sulfamethoxazole, on an activated sludge biomass. Respirometric analysis and model evaluation of the oxygen utilization rate profiles were the backbone of this study. The results showed that continuous exposure to sulfamethoxazole resulted in the inhibition of substrate storage and an increase in the endogenous decay rates by twofold, which was supported by analysis of the resistance genes. A mild inhibition on the growth and hydrolysis kinetics was also observed. Moreover, sulfamethoxazole had a binding impact with available organic carbon, resulting in a slightly less oxygen consumption. DNA sequencing and antibiotic resistance gene analyses showed that continuous exposure to sulfamethoxazole caused a change in the community structure at the species level. Resistant bacteria including Arthrobacter sp. and members of the Chitinophagaceae and Intrasporangiaceae families were found to have dominated the bacterial community. The impact of intermittent exposure was also investigated, and the results indicated a drop in the severity of the impact after 20 days of intermittence.
1 Introduction
The contamination of water bodies with pharmaceuticals has been an issue for decades. Due to technical advances in analytical methods, many compounds can now be detected down to the nanogram per liter level. However, sulfamethoxazole (SMX) has a special place among the persistent organics present in water bodies because it has been used as human and veterinary medicine since 1969 and has been suggested to be persistent since 1985 (Straub, 2016). Due to its frequent use, bacterial resistance against SMX has developed; therefore, at present, it is administered in combination with trimethoprim (Drillia et al., 2005) to treat infections such as middle ear and urinary tract infections, bronchitis, and bacillary dysentery.
SMX belongs to the sulfonamide group of bacteriostatic antibiotics that prevent the formation of dihydrofolic acid (Sköld, 2001; Masters et al., 2003; Drillia et al., 2005), which leads to the reduction of tetrahydrofolic acid, a cofactor for nucleotide synthesis (Michelow and McCracken, 2009). It constitutes up to 21% of the antibiotic drugs used in humans (Göbel et al., 2005). Approximately 15% of the administered SMX remains unmetabolized and, hence, excreted unchanged from the body (Hirsch et al., 1999; Perez et al., 2005). In urban and hospital wastewaters, the concentration of SMX has been observed up to 66.4 μ/L; in Germany, SMX concentrations between 30 and 85 ng/L have been measured in surface waters (Hartig et al., 1999). SMX is among the most detected sulfonamide antibiotics in wastewater (Göbel et al., 2007; Choi et al., 2008; Le-Minh et al., 2010), and through cation exchange or bridging, it can bind to organic matter in the soil (Xu et al., 2011).
The sulfonamide resistance gene (sul) is coded on the dihydropteroate synthetase (DHPS) gene (Sköld, 2000) and is spread by mobile genetic elements (MGEs) (Huovinen, 2001; Antunes et al., 2007). Four different sulfonamide resistance genes, namely, sulA, sulI, sulII, and sulIII, have been defined in environmental bacteria. sulA is chromosomally coded on DHPS in Streptococcus pneumoniae, which was mutated by insertion, resulting in sulfonamide resistance (Maskell et al., 1997). sulI and sulII were detected in polluted sea and river samples (Lin and Biyela, 2005; Hu et al., 2008; Mohapatra et al., 2008), in cattle farm stool samples (Srinivasan et al., 2005), and in samples from wetland sediments (Agersø and Petersen, 2007; Akinbowale et al., 2007). sulI, being coded on class 1 integrons, can be transferred between bacteria in the water matrix (Tennstedt et al., 2003; Mukherjee and Chakraborty, 2006; Taviani et al., 2008).
In addition to resistance against SMX, Xu et al. (2011) found that SMX-resistant Bacillus cereus and Bacillus firmus also have the capacity to degrade SMX at high rates. Moreover, Thauera sp., bacteria known to have the ability to degrade aromatic compounds, were also assumed to metabolize SMX since they were abundantly detected under chronic exposure to SMX (Miran et al., 2018).
Yan et al. (2022) investigated the SMX biodegradation pathways and the fate of antibiotic-resistant genes (ARGs) in heterotrophic and autotrophic microorganisms by employing three different experimental setups: aerobic sludge, nitrifying sludge, and mixed sludge. They exposed the sludge to SMX in the range of 0.2–10 mg/L for 91 days and observed the removal of SMX in all experimental setups. Through sequencing and quantitative PCR (qPCR) analyses, they identified the microbial structure, quantified the sulI and sulII genes, and reported potential antibiotic-resistant bacteria (ARB) and SMX-utilizing bacteria after SMX exposure. The authors found that ammonia-oxidizing bacteria (AOB) could remove SMX quite efficiently, while the previously overlooked heterotrophic bacteria, although 15 times slower than the AOB, also displayed an important role in SMX removal in aerobic and mixed sludge reactors.
Cetecioglu et al. (2016) investigated the impact of SMX on the function of anaerobic processes. They loaded the system with up to 40 mg/L SMX, which was removed by biodegradation. It was found that 45 mg/L SMX was lethal for the anaerobic system. A shift in the microbial community under SMX pressure was also observed.
Müller et al. (2013) worked with a bench-scale activated sludge system, where they confirmed that the removal of SMX via adsorption was negligible. They also confirmed that SMX can be readily utilized as the energy, carbon, and/or nitrogen source for growth, presumably by the two groups of bacteria found in activated sludge communities: heterotrophic bacteria assimilating SMX as a carbon and/or nitrogen source and autotrophic nitrifying bacteria oxidizing the functional amino group on the aromatic ring of SMX. Furthermore, Müller et al. (2013) mentioned that the removal of SMX was enhanced under nitrogen-poor conditions, which is in line with the findings of Drillia et al. (2005). Based on their results, Drillia et al. (2005) concluded that SMX could be removed in systems with low readily biodegradable substrates, such as extended aeration systems.
Zhang et al. (2020) studied the removal of SMX in a long-term sulfur-based autotrophic denitrification reactor. Their results showed that SMX was removed by biodegradation and, at a concentration of 20 mg/L, did not have an impact on the nitrate removal performance of the reactor. However, SMX had a significant impact on the microbial community composition, which resulted in a shift in the autotrophic denitrifying microorganisms.
In current and previous studies (Orhon et al., 2009; Orhon and Sözen, 2012; Pala-Ozkok et al., 2014b), respirometry and the oxygen uptake rate (OUR) profiles have been used as essential tools to determine the kinetic impact of persistent pollutants on activated sludge systems with high precision. The chronic impact of other important antibiotics such as tetracycline and erythromycin was studied in the previous phase of the study on similar sludge systems at different sludge ages using respirometry as the main technique, which indicated that biodegradation, storage, and the hydrolysis kinetics were influenced by continuous antibiotic feeding. The impact of antibiotic exposure on the microbial community structure and the link between community structure and microbial kinetics have been successfully established (Pala-Ozkok and Orhon, 2013; Pala-Ozkok et al., 2019). Furthermore, Katipoglu-Yazan et al. (2016, 2018) investigated the impact of chronic exposure to SMX for 41 days on an enriched nitrifying microbial culture and showed that the nitratation and nitritation processes were significantly disturbed and the microbial community structure changed. In these previous works, respirometry was used to determine the substrate biodegradation and inhibition characteristics reflected in the OUR profiles without and with SMX. The kinetics were estimated and used for the interpretation of the short-/long-term influence of the antibiotic, and the kinetic impacts were identified on the enriched microbial cultures at the end of chronic exposure.
Existing studies on the influence of antibiotics on wastewater treatment systems have pointed out the importance of applied acclimation strategies, operational conditions, and microbial culture composition on the removal of substrates and the fate of pollutants. Studies that investigated the impact of exposure to various SMX concentrations underlined that the SMX utilization ability could be developed in activated sludge microbial populations (Drillia et al., 2005; Yan et al., 2022). Improvements in the treatment efficiencies with intermittent feeding of carbon sources were reported, particularly in anaerobic digestion for different types of substrates and wastewaters (Nadais et al., 2005; Bonk et al., 2018; Vikromvarasiri et al., 2023). However, the impact of the feeding regime, such as intermittent exposure to antibiotics, has yet to be studied on activated sludge systems.
The aim of this study was to explore the chronic impact of SMX feed, along with a peptone mixture, on a mixed microbial culture for a period of 30 days. The system was further operated without the addition of SMX followed by exposure to SMX 20 days later. In this respect, this study evaluated the acute and chronic impacts of SMX on the kinetics and the microbial community structure, as well as the SMX resistance genes (i.e., sulI, sulII, and sulIII), in a laboratory-scale activated sludge system. Moreover, for the first time, the kinetic effect of intermittent exposure to SMX on the same system was investigated. The OUR profiles obtained via respirometry were used for the simulation and calibration of the multicomponent models consisting of the related processes and parameters for the estimation of the process/inhibition kinetics, while high-throughput 454-pyrosequencing was used for the identification of the changes in the microbial community.
2 Materials and methods
2.1 Reactor operation and experimental setup
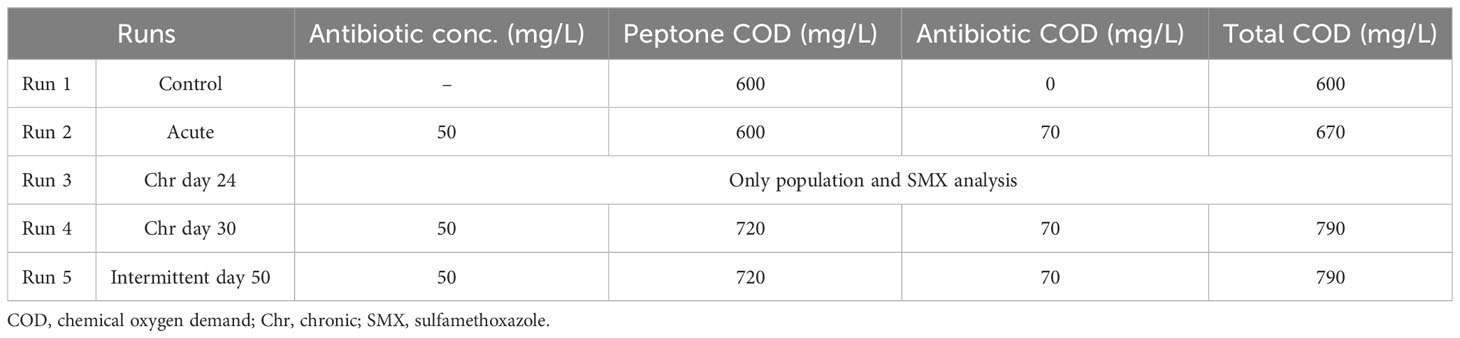
The control reactor was a 14-L laboratory-scale fill/draw reactor that was established using the seed sludge taken from the aeration tank of a full-scale domestic wastewater treatment plant in Istanbul. The hydraulic (θH) and sludge retention time (SRT, θX), i.e., sludge age, were 1 and 10 days, respectively. The dissolved oxygen (DO) concentration in the reactor was maintained above 3.0 mg/L at all times to ensure aerobic conditions. The reactor was acclimated to a peptone–meat extract mixture (peptone mixture), a recognized standard substrate for respirometric inhibition studies (Insel et al., 2006) that includes different fully biodegradable chemical oxygen demand (COD) fractions resembling real domestic wastewater. The peptone mixture per 1 L solution consisted of 16 g peptone [pancreatic digest of gelatin (Peptone G) 7182A], 3 g urea (Urea Agar Base 7226), 11 g beef extract (Beef Extract Powder 7228A; all from Acumedia, Lansing, MI, USA), 0.4 g CaCl2.2H2O, 0.7 g NaCl, 2.8 g K2HPO4, and 0.2 g MgSO4.7H2O. The reactor feed solution contained macro- and micronutrient solutions in addition to the peptone mixture, which consisted of K2HPO4, KH2PO4, FeSO4.7H2O, MgSO4.7H2O, MnSO4.H2O, ZnSO4.7H2O, and CaCl2.2H2O. For each addition of 1,000 mg COD peptone mixture, 20 mL from both solutions were added to the reactor. The control reactor was fed 600 mg COD/L of the peptone mixture every day, which reached a steady-state biomass concentration of 2,000 mg VSS/L.
In addition to the acute experiments, the control reactor also served as the source of the seed sludge for the chronic reactor, which was also fully aerated and operated at the same sludge age of 10 days. The concentration of the peptone mixture added to the reactor was adjusted to 720 mg COD/L for the chronic inhibition tests. In addition to the peptone mixture, the feed also included a daily dosing of 50 mg SMX/L (CAS no. 723-46-6) for 30 days. Furthermore, the chronic reactor was covered to avoid photodegradation of the antibiotic compound (Kümmerer, 2009).
The dose of 50 mg/L SMX utilized in this study was chosen as a representation of concentrated streams, such as pharmaceutical plants and hospitals (Chelliapan et al., 2006; Larsson et al., 2007). Moreover, previous studies by the group showed that, when applying the ISO 8192 method (ISO, 2007) at 50 mg/L concentration, the OUR value decreased from 90 to 57 mg O2/L.h after 30 min of exposure (Ozkok et al., 2011). Table 1 outlines the characteristics of the experimental runs.
2.2 Analytical procedures
Polyhydroxyalkanoate (PHA) samples were collected and analyzed according to Beun et al. (2000). Volatile suspended solids (VSS) and suspended solids (SS) were measured according to standard methods (APHA, 2012). Firstly, soluble COD samples were filtered through 0.45-μm Millipore membrane filters and the measurements then conducted according to the ISO 6060 procedure (ISO, 1989).
SMX measurements were conducted using an Agilent high-performance liquid chromatography (HPLC) equipped with a Nova-PaK C18 column (Waters, Milford, MA, USA) and a diode array detector at 280 nm. The mobile phase had a constant flow rate at 0.6 mL/min and consisted of 30:70 (v/v) methanol/water, which was acidified (pH 2.5) using 0.1% phosphoric acid (Beltran et al., 2008). The sample injection volume and the flow rate were 40 µL and 1 mL/min, respectively. The calibration curves for the SMX measurements are given in Figure 1.
Figure 1 Calibration curve of sulfamethoxazole. Reproduced from Pala-Ozkok et al. (2012).
2.3 Respirometric analysis and activated sludge modeling
The Ra-COMBO (Applitek Co., Nazareth, Belgium) continuous respirometer was used to produce the OUR profiles in all experimental runs. For this purpose, parallel reactors were set up to have identical conditions for all runs. To prevent any interference due to nitrification, a nitrification inhibitor (Formula 2533™; Hach, Loveland, CO, USA) was added into the OUR reactors during all analyses. The OUR tests were started and ended at endogenous decay level, at which no external carbon source was present. The respirometric reactor was continuously aerated, and the COD and polyhydroxybutyric acid (PHB) samples were collected in all experimental sets.
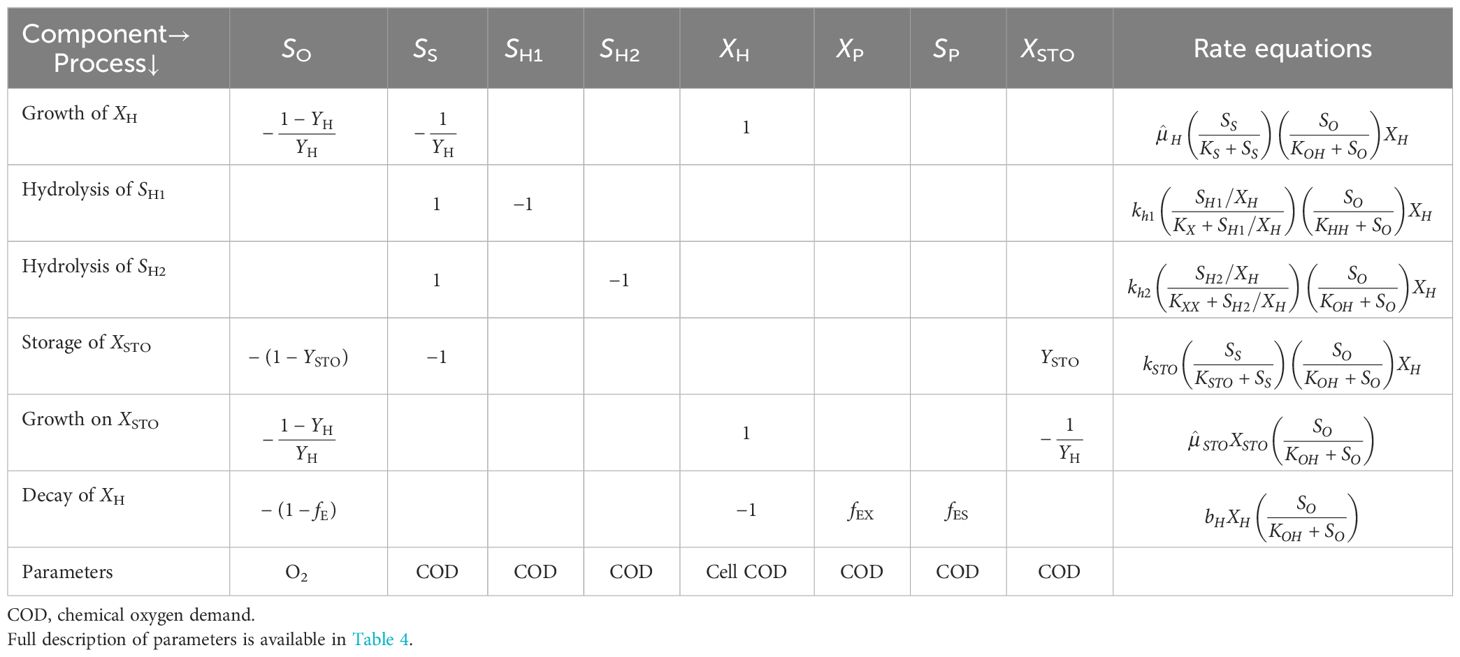
The model used in this study for the kinetic interpretation of the experimental data included the basic structure of the activated sludge model (ASM) for growth and storage (Gujer et al., 1999; Krishna and Van Loosdrecht, 1999). A matrix representation of the structure of the model is given in Table 2. Moreover, the AQUASIM simulation program was used for the modeling and simulation of the OUR data obtained from the different experimental runs (Reichert, 1994), where all model outputs were fitted on real-time data by manual calibration of the model components.
2.4 Activated sludge bacterial community and resistance gene analyses
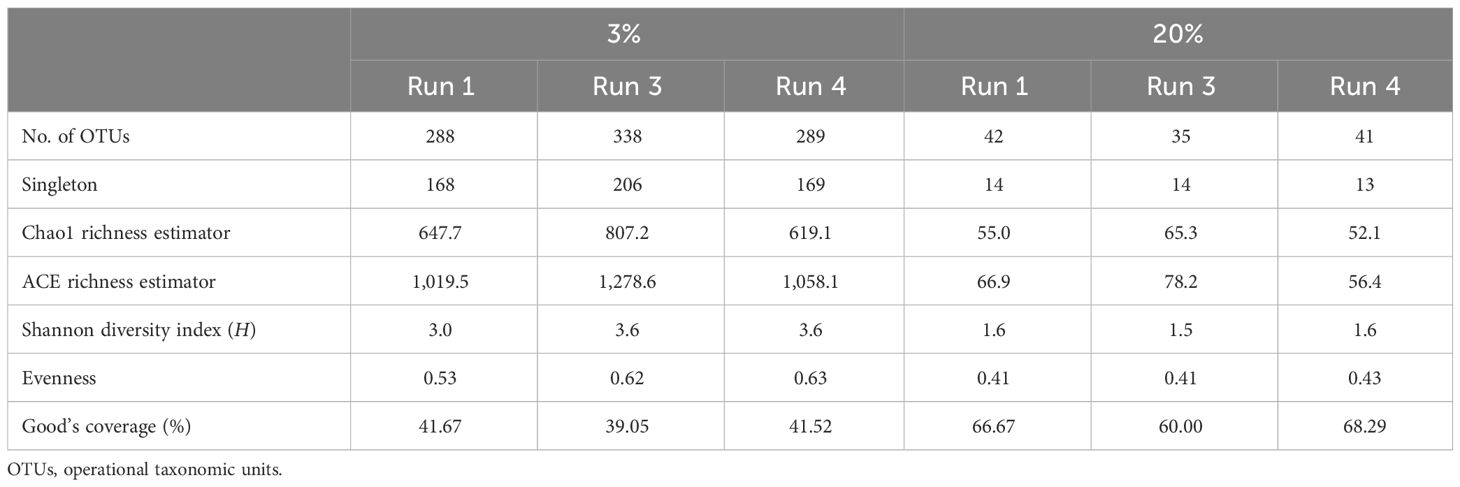
In order to determine the impact of chronic exposure to SMX on the microbial community structure, DNA samples were taken from the chronic reactor on day 0 (run 1), day 24 (run 3), and day 30 (run 4) and analyzed using 454-pyrosequencing. Details of the methodology for community analysis have been given in several previous publications of the group (Pala-Ozkok et al., 2012, 2014a, 2019). Genomic DNA was extracted from the activated sludge samples using the NucleoSpin Soil DNA extraction kit (Macherey-Nagel GmbH&Co, Düren, Germany). The 27F and 338R barcoded universal primers were used to amplify the V1–V2 hypervariable regions of the 16S rRNA gene. The amplified DNA from each sample was pooled in one sample and sent to the Institute for Clinical Molecular Biology at the University of Kiel for sequencing (Pala-Ozkok et al., 2014a). PANGEA PERL scripts were used to screen the obtained sequences for quality and length (Giongo et al., 2010). MOTHUR was used as the main tool for statistical and operational taxonomic unit (OTU)-based analyses. Moreover, significant changes at the species level were determined using the metastats command in MOTHUR, with a p-value threshold of 0.05 (Schloss et al., 2009). After the cleanup, the numbers of sequences obtained in each sample were 2,977, 3,118, and 2,752 for run 1, run 3, and run 4, respectively. The sequence reads were uploaded to the Sequence Read Archive (SRA) and made available under accession number SRA054103. The accession numbers for run 1, run 3, and run 4 are SRX155362/155363, SRX155377, and SRX155378, respectively. Moreover, the SMX resistance profile was analyzed using PCR of the sulI, sulII, and sulIII genes in the activated sludge samples according to the method described in Pei et al. (2006) and Pala-Ozkok (2012). The primer sequences for sulI, sulII, and sulIII and the annealing temperatures are given in Supplementary Table S1.
3 Results and discussions
3.1 Assessment of the oxygen uptake rate profiles and SMX utilization
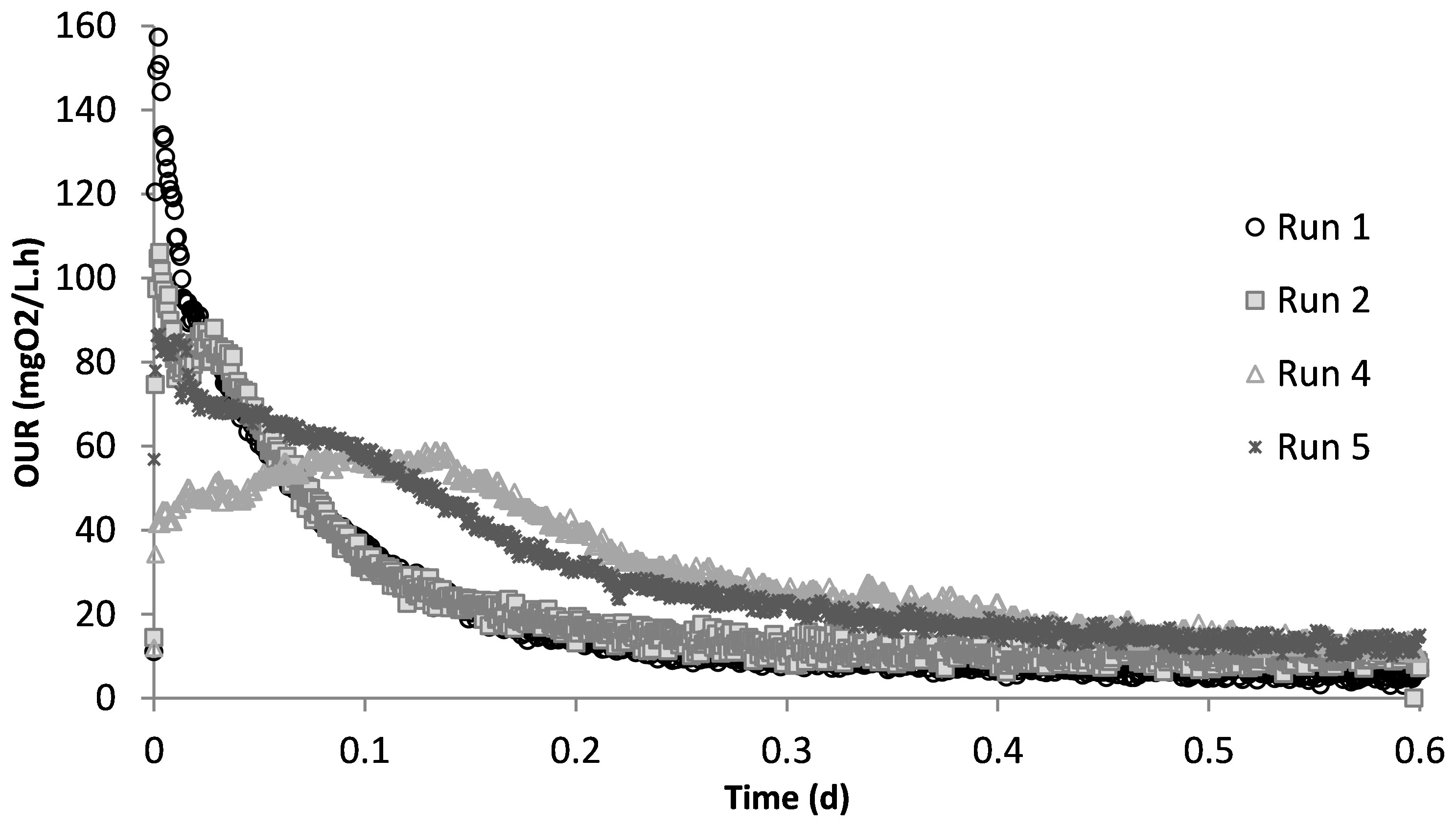
Run 1 served as the control test, where there was no impact of SMX on the biomass. Figure 2 displays the OUR curve for the peptone mixture biodegradation, which showed the peak of the curve at 160 mg/L.h and later dropping to the endogenous decay line. The curve presented the degradation of different COD fractions, of which the peptone mixture was included, with the corresponding degradation rates. The total oxygen consumption calculated from the area under the OUR curve and the corresponding COD removal efficiency were 211 mg/L and 94%, respectively (Pala-Ozkok et al., 2012; Pala-Ozkok and Orhon, 2013; Pala-Ozkok et al., 2019).
The acute impact of the addition of 50 mg/L SMX on the biomass from the control reactor was visualized using the obtained OUR profile (Figure 2). SMX addition caused the peak to drop from 160 mg/L.h in run 1 to 106 mg/L.h in run 2, and the oxygen consumed for microorganism growth in run 2 was calculated as 206 mg/L. In the chronic run (run 4), the total amount of oxygen consumed for growth was 278 mg/L. On day 50 (run 5), after 20 days of no SMX addition, the oxygen consumption was calculated as 275 mg/L.
Figure 3 shows the SMX concentrations measured in the samples taken during the acute and the chronic experimental runs. Additional measurements were performed on days 10 and 24 of chronic exposure. As can be seen, all of the SMX was measured in the liquid phase, which is in agreement with the literature indicating that SMX is not adsorbed onto the sludge (Yan et al., 2022). However, contrary to the findings by Yan et al. (2022), the results of the SMX measurements in this study showed that the biomass did not utilize SMX as a carbon or nitrogen source. Since all of the SMX was measured in the liquid phase, it was concluded that there was no contribution to oxygen consumption in the respirometric tests.
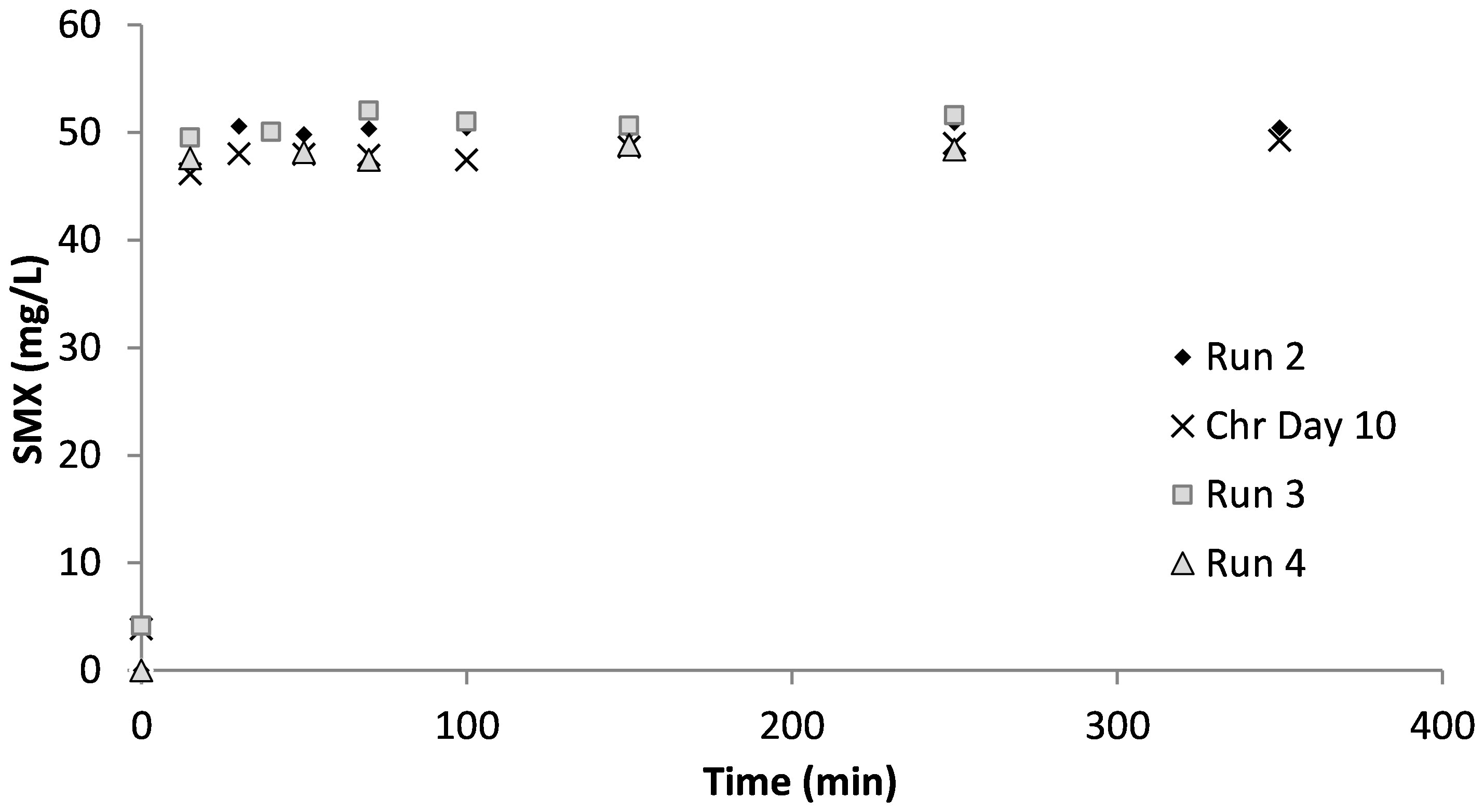
Figure 3 Effluent sulfamethoxazole (SMX) concentrations in the respirometric tests and in the chronic reactor.
3.2 Substrate binding impact of sulfamethoxazole
In previous works of the group, the substrate-binding feature of antibiotic substances has been explained and reported in detail for SMX, as well as for erythromycin and tetracycline, under different operating conditions, i.e., with and without nitrification, with different substrates, and at different sludge ages (Ozkok et al., 2011; Pala-Ozkok et al., 2014, 2014b; Kor-Bicakci et al., 2016; Katipoglu-Yazan et al., 2013, 2016, 2018, 2023). In all experimental runs, the OUR profiles were started from the endogenous decay line, in which there was no external substrate present in the medium. At the end of the experiment, the OUR profiles reached the endogenous decay line, indicating that all of the external biodegradable substrate was depleted. Therefore, the decrease in the amount of oxygen utilized during the experiments, i.e., the area under the OUR curve, indicated the utilization of less substrate by the biomass.
The data from the experimental results, including those obtained from previous studies by the group conducted on the same origin of activated sludge on different antibiotics (Katipoglu-Yazan et al., 2013; Pala-Ozkok and Orhon, 2013; Pala-Ozkok et al., 2014; Katipoglu-Yazan et al., 2016; Kor-Bicakci et al., 2016; Pala-Ozkok et al., 2019), suggest that the antibiotic compounds “blocked” a portion of the peptone mixture added to the reactor and prevented its utilization in the microbial growth process, resulting in less oxygen consumption and low effluent COD. Table 3 presents the mass balance created for the experimental sets. For the mass balance calculations, the yield for heterotrophic growth (YH) and that for the residual soluble microbial product, SP, (YSP) were determined from the control experiment (run 1) as 0.6 mg cell COD/mg COD and 0.06 mg COD/mg COD, respectively. These values were used for the rest of the experimental runs. In run 2, the COD initially available was 600 mg/L; however, the mass balance calculations showed that 85 mg COD/L (14%) of this initially available amount was not utilized for growth. In run 4 and run 5, the amounts of “blocked” COD, which was not used for growth, were calculated as 25 mg COD/L (3.6%) and 33 mg COD/L (4.6%), respectively.
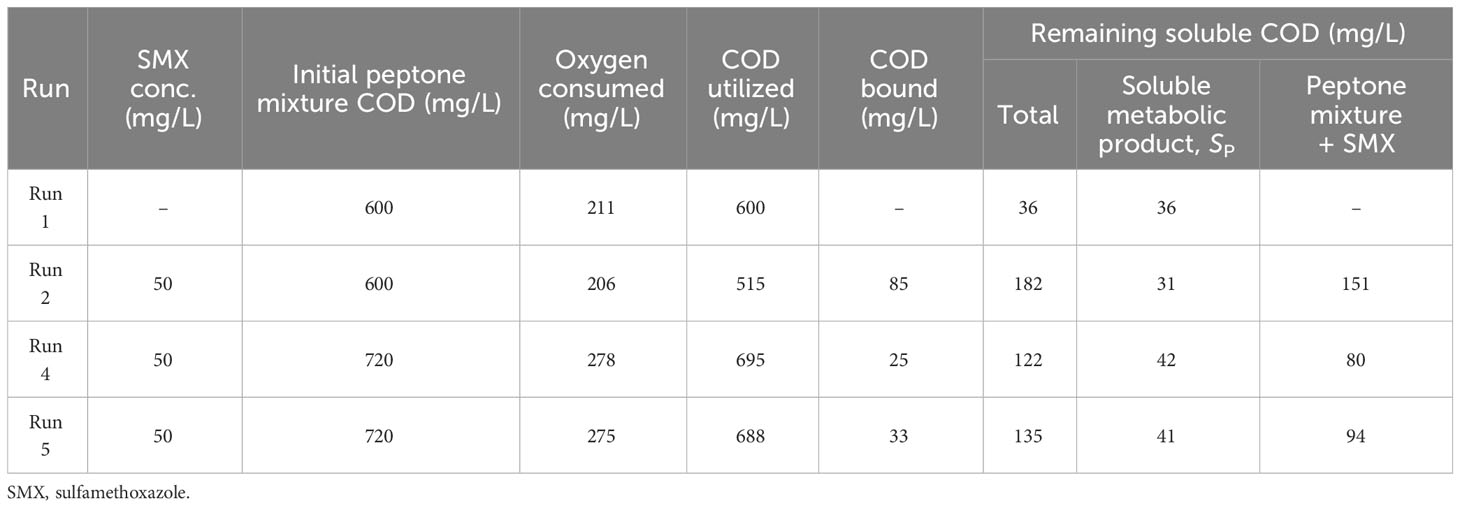
Table 3 Oxygen consumption and chemical oxygen demand (COD) utilization mass balances in the experimental runs.
Examination of the impact of other antibiotics in the studies conducted by the group (Pala-Ozkok and Orhon, 2013; Pala-Ozkok et al., 2014b; Katipoglu-Yazan et al., 2013; Pala-Ozkok et al., 2019) revealed different substrate binding impacts of acute and chronic exposures and differences in the type of antibiotics. For a sludge age of 10 days, where nitrification was inhibited, acute exposure to tetracycline resulted in 29% of the COD not being utilized for growth, while chronic exposure resulted in 30% of the COD not utilized. For erythromycin, the acute impact resulted in 52%, chronic exposure resulted in 18%, and intermittent exposure resulted in 25% of the COD not being used for growth. Compared with erythromycin and tetracycline, SMX showed definitely weaker substrate binding properties; nonetheless, it had an impact, and this impact was also seen in the kinetics of activated sludge.
Moreover, extensive work by the group showed that the type of carbon source, which the system was acclimated to, and the culture history (i.e., sludge age), together with the operational conditions (i.e., presence of nitrification), changed the outcome of the SMX impact (Katipoglu-Yazan et al., 2016, 2018). The acute impact of SMX was also shown to differ depending on the type of substrate (Pala-Ozkok et al., 2014), wherein, on an acetate-acclimated sludge at a sludge age of 2 days, SMX only exerted a slight inhibition on microbial growth, with full substrate utilization. In addition, the acute impact on a 10-day SRT system acclimated to acetate showed mild substrate binding, while the chronic impact showed SMX used as a secondary carbon source (Kor-Bicakci et al., 2016). Moreover, in another part of the study where nitrification was not prevented, it was seen that, after 50 mg/L acute SMX exposure, only half of the initial peptone COD was utilized, with the rest of the COD reported as “blocked” (Katipoglu-Yazan et al., 2023).
As a result of this evaluation, the substrate binding feature of antibiotics was taken into consideration. Moreover, the mass balances presented in Table 3 were created and used as the input data for activated sludge modeling studies.
3.3 Model evaluation of the impact of sulfamethoxazole
The peptone mixture consisted of three different COD fractions: the readily biodegradable (SS), readily hydrolysable (SH1), and hydrolysable (SH2) COD fractions. The model calibration of run 1 showed that the readily biodegradable and readily hydrolysable fractions constituted 9.5% and 56%, while the hydrolysable COD fraction constituted 34.5% of the total biodegradable COD added into the reactor. PHA analysis in run 1 showed that the control system had a PHA pool of 10 mg COD/L and a maximum PHA storage of 32 mg COD/L. However, in the acute and chronic experimental runs, the results of the PHA measurements showed that SMX inhibited the storage mechanism completely, which was also observed for tetracycline and erythromycin (Katipoglu-Yazan et al., 2013; Pala-Ozkok and Orhon, 2013; Pala-Ozkok et al., 2019). The OUR, COD, and PHA profiles and the model simulations of run 1 are given in Figure 4.
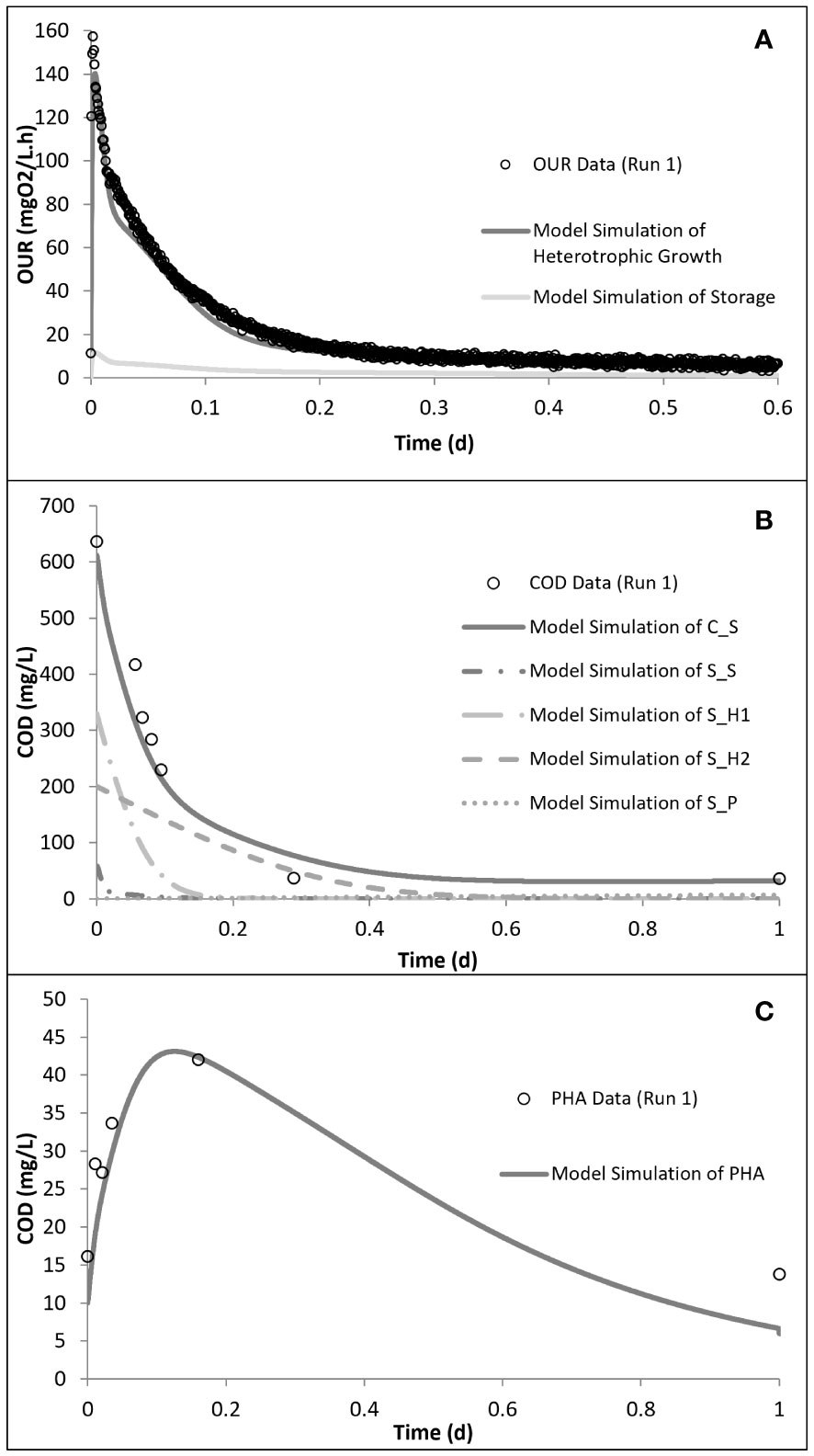
Figure 4 Model simulation of run 1: oxygen uptake rate (OUR) (A), chemical oxygen demand (COD) removal (B), and polyhydroxyalkanoate (PHA) storage (C) profiles. Reproduced from Pala-Ozkok et al. (2012, 2019).
Kinetic analysis of the impact of acute inhibition (run 2) on the storage mechanism showed that, although the PHA storage was inhibited, the biomass maintained the ability to grow on the stored PHA. In addition, it was shown that the addition of SMX increased the half-saturation constant (KS), making it less available for the growth of biomass (Table 4). It has also been determined that the biomass did not utilize all of the COD added into the reactor, but 85 mg COD/L less than the amount added for run 2. Furthermore, the addition of SMX doubled the endogenous decay level (bH) of the system (Figures 5, 6).
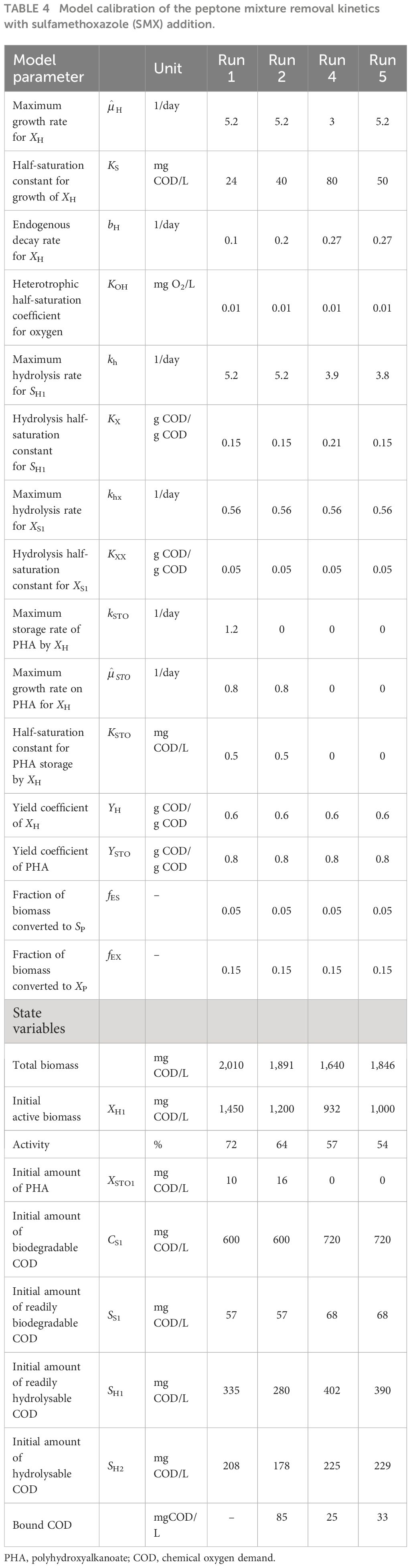
Table 4 Model calibration of the peptone mixture removal kinetics with sulfamethoxazole (SMX) addition.
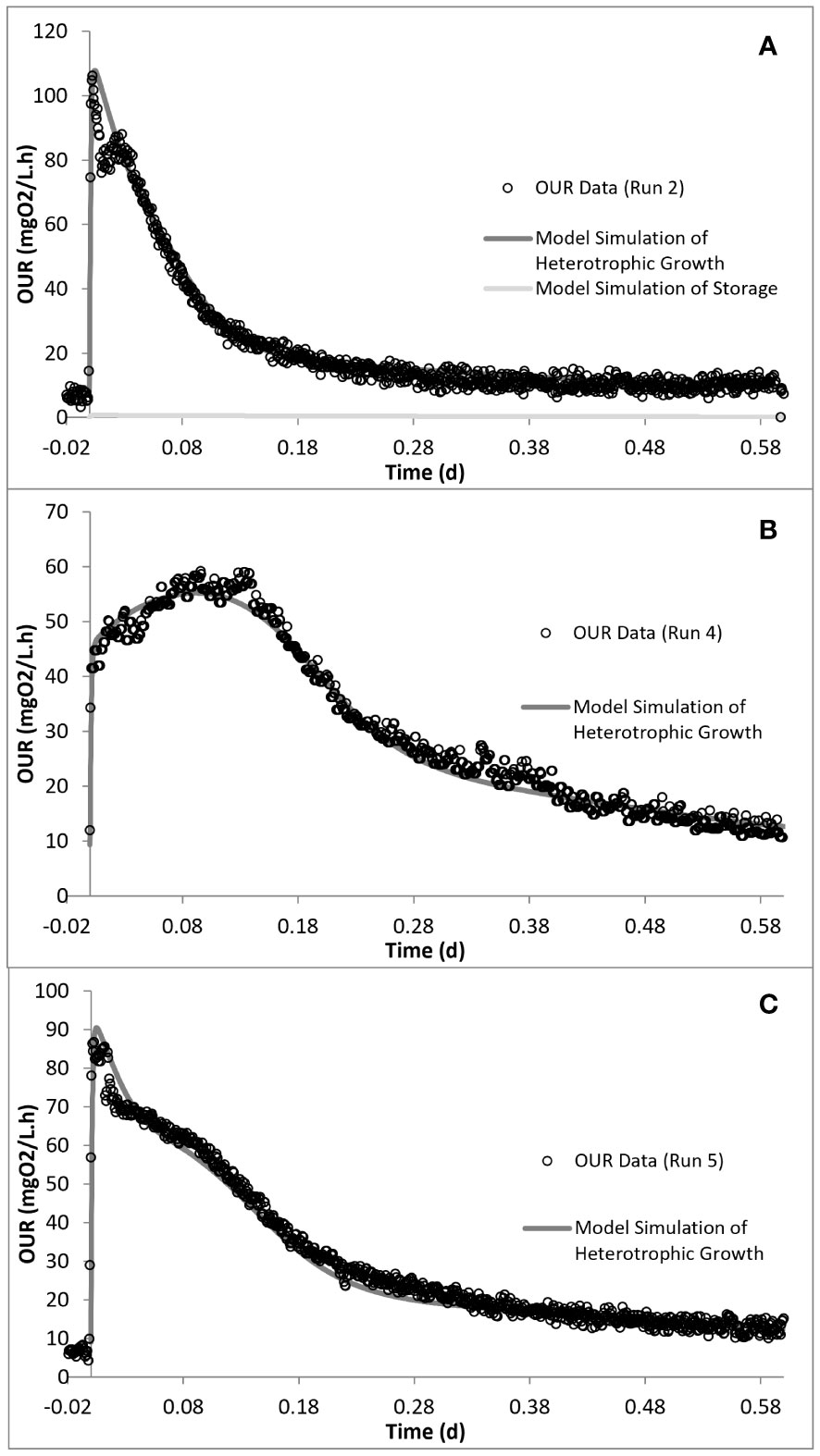
Figure 5 Model simulation of the oxygen uptake rate (OUR) profiles for run 2 (A), run 4 (B), and run 5 (C).
Simulation of the chronic exposure data (run 4) showed that exposure to 50 mg/L SMX for 30 days resulted in the increase of the half-saturation constant (KS) of the substrate from 24 mg COD/L in run 1 to 80 mg COD/L in run 4. Moreover, the maximum growth rate of the microorganisms decreased from 5.1 day−1 in run 1 to 3 day−1 in run 4, impacting both the growth and substrate degradation (Table 4). Moreover, chronic exposure to SMX for 30 days almost tripled the endogenous decay level, which increased from 0.1 day−1 in run 1 to 0.27 day−1 in run 4. The rate of hydrolysis (kh1) of SH1 decreased from 5.2 day−1 in run 1 to 3.9 day−1 in run 4, while the half-saturation constant for the hydrolysis (KX) of SH1 increased by 1.4-fold in run 4 compared with run 1. Finally, consistent with the stoichiometric calculations presented in Table 3, the model simulation showed that the system utilized 25 mg COD/L less than the amount provided (Figures 5, 6).
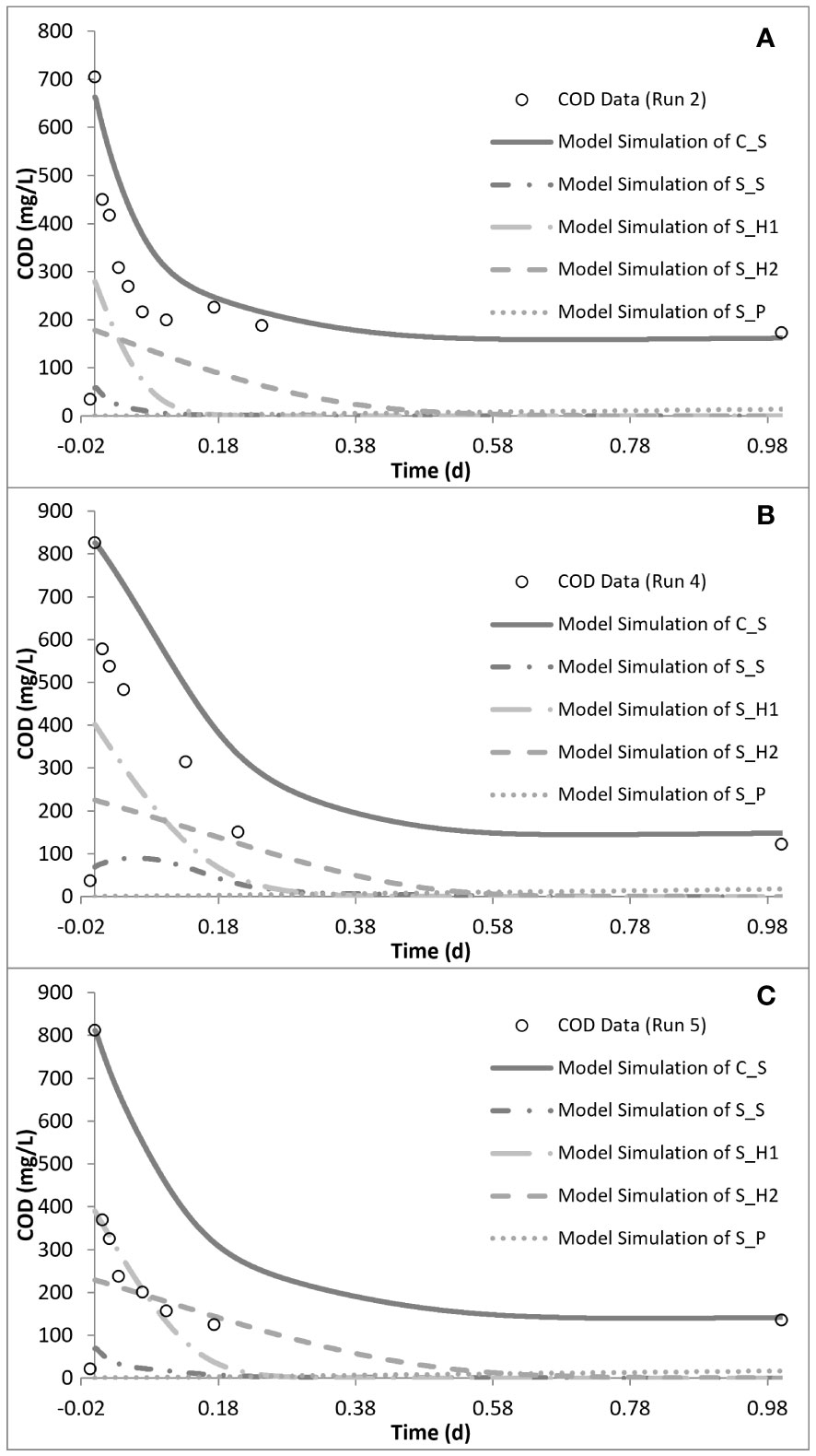
Figure 6 Model simulation of the chemical oxygen demand (COD) removal profiles for run 2 (A), run 4 (B), and run 5 (C).
In order to observe the effects of intermittent exposure to antibiotics, after 30 days of exposure to 50 mg/L SMX, the addition of SMX was stopped for 20 days and the reactor was fed only with the peptone mixture. On day 50, 50 mg/L SMX was again added to the system, with the responses obtained and simulated in run 5 (Figures 5, 6). The hydrolysis rate of SH1 remained the same when compared with that in run 4, while the half-saturation constant for SH1 hydrolysis recovered to its original level in run 1. The maximum heterotrophic growth rate, also recovered from 3 day−1 in run 4 to 5.2 day−1 in run 5, as it was in run 1. A recovery in the half-saturation constant (KS) for the growth of XH was also observed. Finally, the endogenous decay rate (bH) of the biomass remained at 0.27 day−1, as in day 30 (run 4), almost tripling the endogenous decay rate of the organisms. These results indicated some recovery in the 20 days of intermittence as the impact was not as severe as that on the 30th day (run 4). However, 33 mg COD/L of the amount provided was not utilized by the system. Finally, it was observed that, in both acute and chronic exposure runs, the hydrolysis of SH2 remained unaffected (Table 4).
3.4 Microbial community analysis
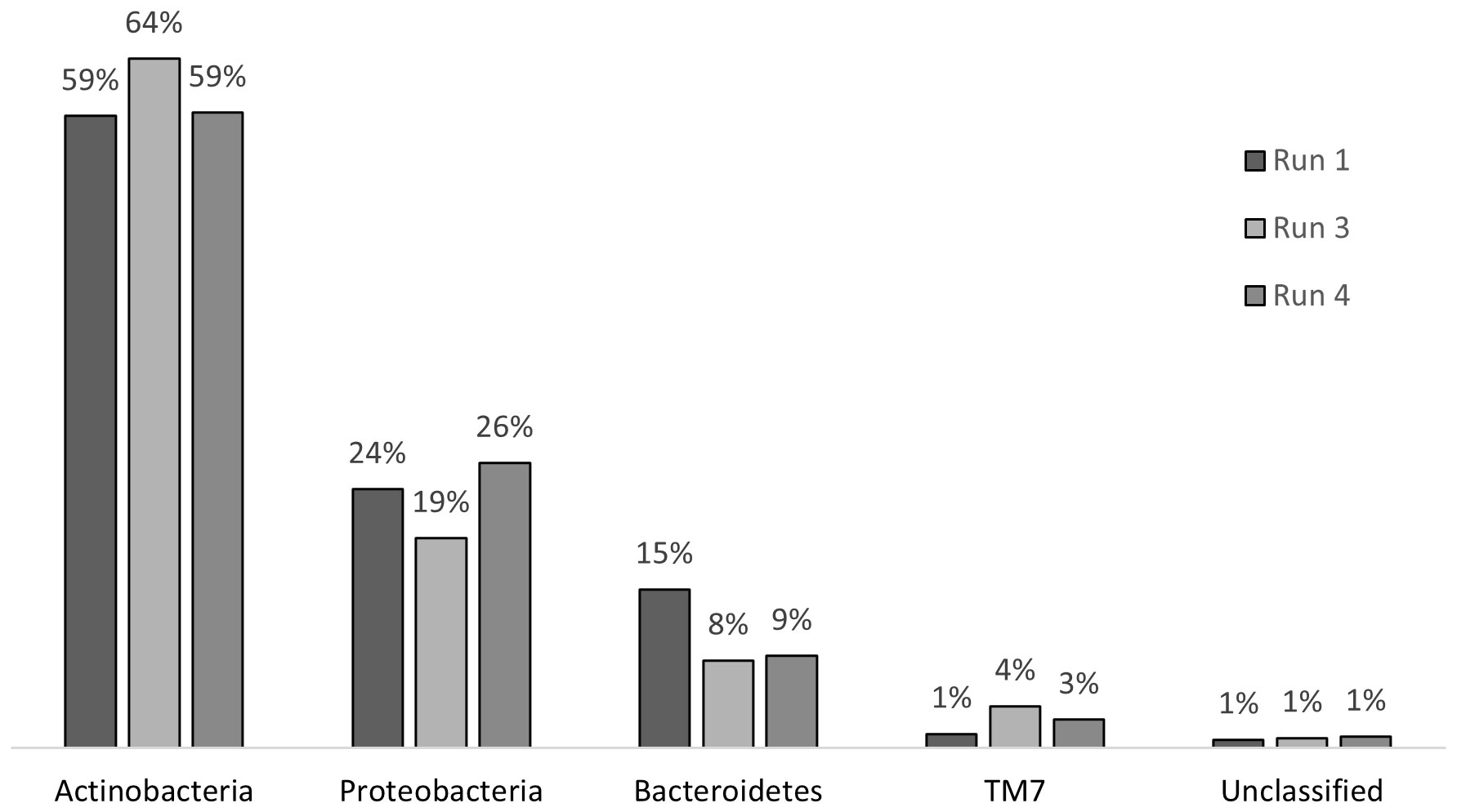
Chronic exposure to SMX caused stress on the microbial community; however, this stress did not result in a shift in the community structure at the phylum level (Figure 7). In run 1, where there was no impact of SMX, the phylum structure of the community consisted of 59% Actinobacteria, 15% Bacteroidetes, 24% Proteobacteria, and 1% TM7. After 24 days of exposure in run 3, the abundance rates changed to 64%, 8%, 19%, and 4% for Actinobacteria, Bacteroidetes, Proteobacteria, and TM7, respectively. In run 4, there was a change back to the original structure with regard to the abundance of Actinobacteria and Proteobacteria. However, the abundance of the phyla Bacteroidetes and TM7 remained the same as that in run 3. The change in the distribution of phyla is presented in Figure 7. The changes in Actinobacteria and Proteobacteria between run 1 and run 3 were also determined to be significant when compared with the RDP library, which also showed that, throughout the treatment, the decrease in the abundance of Bacteroidetes was significant. However, as a result of the pressure of SMX, Proteobacteria, Actinobacteria, and Bacteroidetes continued to dominate the community as the most abundant phyla in the reactor, which is in accordance with the findings of Zhang et al. (2016).
The change in richness due to SMX exposure was visualized using rarefaction curves. At the species level, the richness showed an increasing trend, with run 3 having the highest richness (Figure 8). According to the data in the Venn diagrams, the total richness of all groups combined was calculated to consist of 636 species-level OTUs (Figure 8), while the total shared richness was calculated as consisting of 82 species-level OTUs. At the species level, run 1, run 3, and run 4 contained 288, 338, and 289 species-level OTUs, respectively. Run 1 and run 3 exclusively shared 34 species-level OTUs, while run 1 and run 3 exclusively shared 20 and 63 species-level OTUs, respectively. Moreover, the ACE and Chao1 richness estimators indicated that the richness increased by the 24th day of exposure (run 3); however, this slightly decreased back to the level of run 1 by the 30th day (run 4) (Table 5). Assessment of the Shannon diversity index (H) revealed that, at the species level, the diversity increased with the addition of SMX into the system (Table 5). An increase in the Shannon diversity index was also observed in the study of Zhang et al. (2016), where activated sludge was exposed to 10 mg/L SMX.
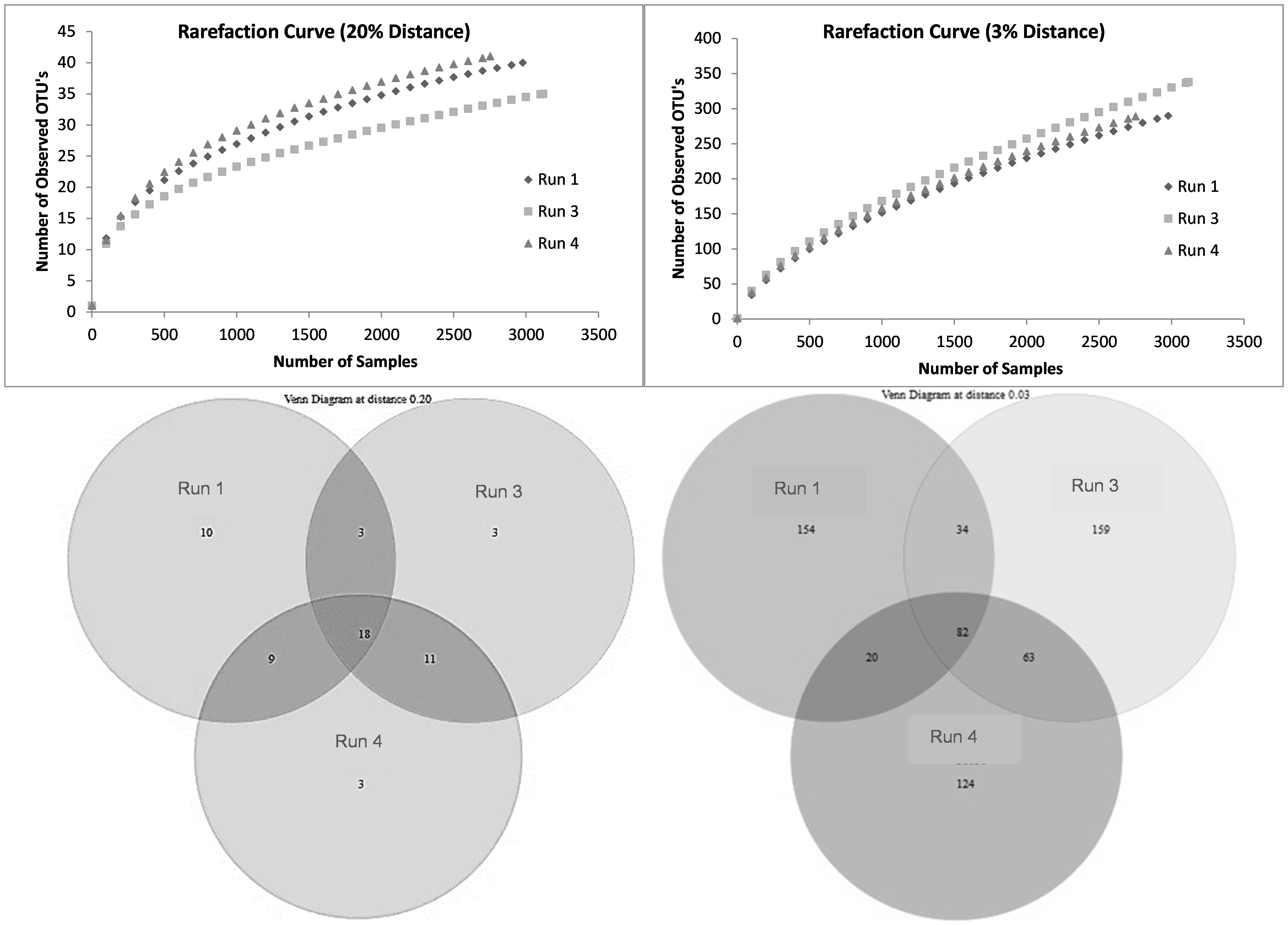
Figure 8 Rarefaction curves and Venn diagrams at the phylum (20%) and species (3%) levels for run 1, run 3, and run 4.
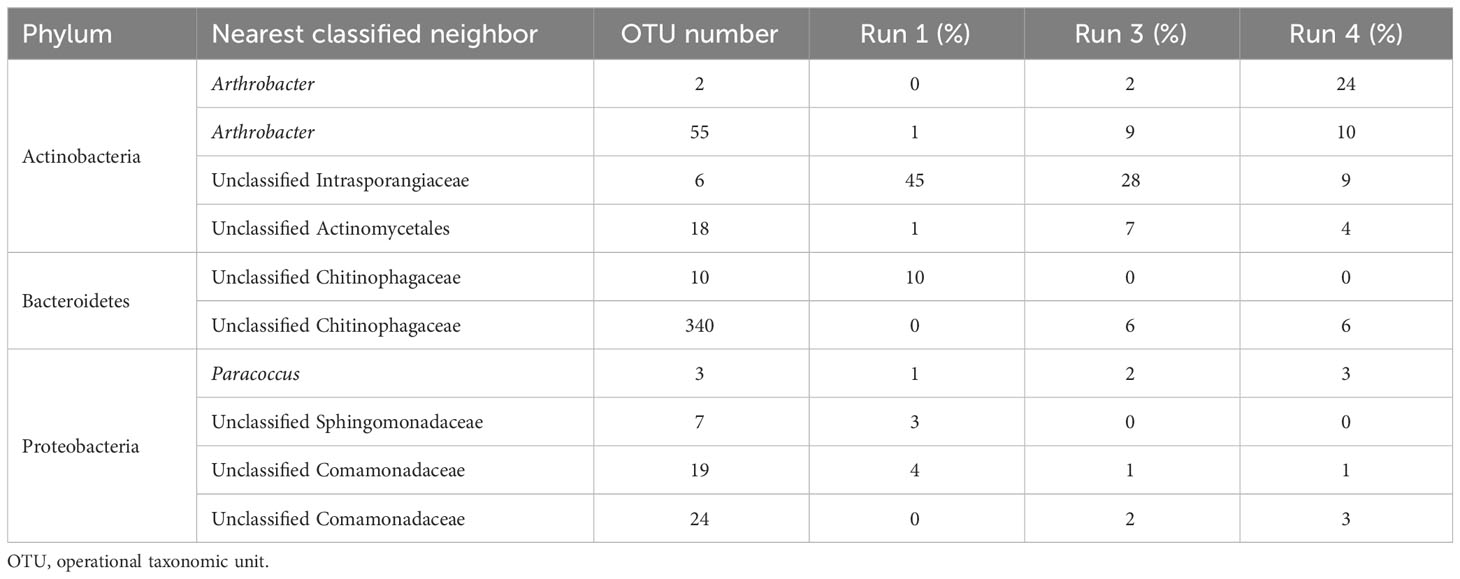
Table 6 outlines the significant changes in the community structure at the species level. It can be seen that an unclassified member of class Actinobacteria (OTU#6) and an unclassified member of class Chitinophagaceae (OTU#10) were the most abundant species in run 1, with 45% and 10% abundance, respectively. However, in run 3 (after 24 days of SMX exposure), OTU#10 vanished and did not reappear (p< 0.05, q > 0.05). On the other hand, OTU#6 gradually decreased from 45% down to 9% in run 4 (30 days of exposure). Moreover, bacteria that were in very low abundance in run 1 increased significantly and dominated the community. Species of the genus Arthrobacter OTU#2 and OUT#55 became gradually abundant, reaching 24% and 10% in run 4 (30 days of exposure), with total abundance of 34% for this genus. In addition, an unclassified member of class Chitinophagaceae (OTU#340) showed increased abundance in 30 days and reached 6% abundance in run 4.
3.5 Evaluation of changes in the microbial community
Comparison of the results of the pyrosequencing analysis with those in the available literature indicated that the dominant bacteria in this system were resistant to SMX, which was also cross-checked with the results of the resistance gene analysis of the system. The results showed that the system harbored two sulfonamide resistance genes, namely, sulI and sulII, but not sulIII. sulI and sulII were detected in all samples in run 1, run 3, and run 4, whereas sulIII was not found in any of the samples (Supplementary Table S2). It was also shown that these sulfonamide resistance genes, particularly sulI, are located on mobile genetic elements, and the class 1 integron has been known to harbor sulI (Liebert et al., 1999; Carattoli, 2001; Byrne-Bailey et al., 2009; Baran et al., 2011). Zhang et al. (2016) showed that, under tetracycline and SMX pressure, the total number of the antibiotic resistance genes they investigated (i.e., tetA, tetC, tetG, tetK, tetM, and sulI) increased. Furthermore, the authors revealed that the change in the antibiotic resistance genes was significantly correlated with the change in the class 1 integron. Moreover, Hoa et al. (2008) showed that Arthrobacter sp., found abundantly in the SMX reactor, was positive for all three sul genes. A positive correlation with multidrug resistance genes was also found for Chitinophagaceae (Shen et al., 2019). In addition, members of the family Intrasporangiaceae were found to assimilate SMX in soil, as discovered by Ouyang et al. (2019). Since SMX was not degraded in this study, it is safe to assume that the members of Intrasporangiaceae found in this study are resistant to SMX. Yan et al. (2022) determined the functional bacteria potentially carrying the sul resistance genes in their systems under chronic exposure to SMX. The authors found, among others, Comamonas and Taibaiella species as two of the potential ARB, which is consistent with the results of this study as these species belong to the Comamonadaceae and Chitinophagaceae families, respectively, the members of which were also identified in this study (Table 6), investigated the sul resistance genes.
4 Conclusions
This study investigated the acute and chronic impacts of the addition of SMX on an activated sludge system. The impact of intermittent exposure was also explored for the first time. These impacts were evaluated from different points of view: microbial kinetics, antibiotic biodegradation, microbial community structure, and antibiotic resistance. The results indicated that SMX was not used as a carbon or nitrogen source by the biomass, but had a mild impact on the substrate degradation kinetics of the activated sludge biomass, which included an increase in the half-saturation constant for heterotrophic growth (KS) and the endogenous decay level (bH). This impact was more pronounced in the chronic exposure studies. However, when compared with previous studies conducted using erythromycin and tetracycline, the impact is at a lower level, with SMX also having an impact on substrate binding. All the kinetic and stoichiometric effects of SMX on the biomass decreased after 20 days of intermittence. The microbial community structure was not severely affected by SMX; however, potentially resistant species were identified, where a mild community shift at the species level occurred.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
IP-O: Investigation, Methodology, Writing – original draft, Writing – review & editing. TK-Y: Investigation, Writing – review & editing. TO-H: Methodology, Writing – review & editing. DJ: Supervision, Writing – review & editing. EU-C: Supervision, Writing – review & editing. DO: Funding acquisition, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported by Turkish Academy of Sciences as part of Fellowship Program for Integrated Doctoral Studies. Authors thank the University of Stavanger for the open access funding, the Scientific Research Fund of Istanbul Technical University (ProjectNr: 33680 and ProjectNr: 33742) and The Scientific and Technological Research Council of Turkey.
Acknowledgments
Authors thank University of Kiel Institute of Clinical Molecular Biology for their technical assistance.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/frabi.2024.1335654/full#supplementary-material
References
Agersø Y., Petersen A. (2007). The tetracycline resistance determinant Tet 39 and the sulphonamide resistance gene sulII are common among resistant Acinetobacter spp. isolated from integrated fish farms in Thailand. J. Antimicrob. Chemother. 59, 23–27. doi: 10.1093/jac/dkl419
Akinbowale O. L., Peng H., Barton M. D. (2007). Class 1 integron mediates antibiotic resistance in Aeromonas spp. from rainbow trout farms in Australia. Int. J. Antimicrob. Agents. 29, S113. doi: 10.1016/S0924-8579(07)70357-6
Antunes P., MaChado J., Peixe L. (2007). Dissemination of sul3-containing elements linked to class 1 integrons with an unusual 3’ conserved sequence region among Salmonella isolates. Antimicrob. Agents. Ch. 51, 1545–1548. doi: 10.1128/AAC.01275-06
APHA. (2012). Standard methods for the examination of water and wastewater, 22nd edition. Edited by E. W. Rice, R. B. Baird, A. D. Eaton and L. S. Clesceri. (Washington, DC: American Public Health Association (APHA), American Water Works Association (AWWA) and Water Environment Federation (WEF)).
Baran W., Adamek E., Ziemianska J., Sobczak A. (2011). Effects of the presence of sulfonamides in the environment and their influence on human health. J. Hazard. Mater. 196, 1–15. doi: 10.1016/j.jhazmat.2011.08.082
Beltran F. J., Aguinaco A., Garcia-Araya J. F., Oropesa A. (2008). Ozone and photocatalytic processes to remove the antibiotic sulfamethoxazole from water. Water Res. 42, 3799–3808. doi: 10.1016/j.watres.2008.07.019
Beun J. J., Paletta F., van Loosdrecht M. C. M., Heijnen J. J. (2000). Stoichiometry and kinetics of poly-beta-hydroxybutyrate metabolism in aerobic, slow growing, activated sludge cultures. Biotechnol. Bioeng. 67, 379–389. doi: 10.1002/(SICI)1097-0290(20000220)67:4<379::AID-BIT1>3.0.CO;2-2
Bonk F., Popp D., Weinrich Ş., Sträuber H., Kleinsteuber S., Harms H., et al. (2018). Intermittent fasting for microbes: how discontinuous feeding increases functional stability in anaerobic digestion. Biotechnol. Biofuels Bioprod. 11, 274. doi: 10.1186/s13068-018-1279-5
Byrne-Bailey K. G., Gaze W. H., Kay P., Boxall A. B. A., Hawkey P. M., Wellington E. M. H. (2009). Prevalence of sulfonamide resistance genes in bacterial isolates from manured agricultural soils and pig slurry in the United Kingdom. Antimicrob. Agents. Chemother. 53, 696–702. doi: 10.1128/AAC.00652-07
Carattoli A. (2001). Importance of integrons in the diffusion of resistance, Vet. Res. 32, 243–259. doi: 10.1051/vetres:2001122
Cetecioglu Z., Ince B., Orhon D., Ince O. (2016). Anaerobic sulfamethoxazole degradation is driven by homoacetogenesis coupled with hydrogenotrophic methanogenesis. Water Res. 90, 79e89. doi: 10.1016/j.watres.2015.12.013
Chelliapan S., Wilby T., Sallis P. J. (2006). Performance of an up-flow anaerobic stage reactor (UASR) in the treatment of pharmaceutical wastewater containing macrolide antibiotics. Water Res. 40, 507–516. doi: 10.1016/j.watres.2005.11.020
Choi K. J., Kim S. G., Kim S. H. (2008). Removal of tetracycline and sulfonamide classes of antibiotic compound by powdered activated carbon. Environmental Technology 29(3), 333–342. doi: 10.1080/09593330802102223
Drillia P., Dokianakis S. N., Fountoulakis M. S., Kornaros M., Stamatelatou K., Lyberatos G. (2005). On the occasional biodegradation of pharmaceu- ticals in the activated sludge process: the example of the antibiotic sulfamethoxazole. J. Hazard. Mater. 122, 259–265. doi: 10.1016/j.jhazmat.2005.03.009
Giongo A., Crabb D. B., Davis-Richardson A. G., Chauliac D., Mobberley J. M., Gano K. A., et al. (2010). PANGEA: pipeline for analysis of next generation amplicons. ISME J. 4, 852–861. doi: 10.1038/ismej.2010.16
Göbel A., McArdell C. S., Joss A., Siegrist H., Giger W. (2007). Fate of sulfonamides, macrolides, and trimethoprim in different wastewater treatment technologies. Sci. Total Environ. 372, 361–371. doi: 10.1016/j.scitotenv.2006.07.039
Göbel A., Thomsen A., McArdell C. S., Joss A., Giger W. (2005). Occurrence and sorption behavior of sulfonamides, macrolides, and trimethoprim in activated sludge treatment. Environ. Sci. Technol. 39, 3981–3989. doi: 10.1021/es048550a
Gujer W., Henze M., Mino T., van Loosdrecht M. (1999). Activated sludge model No. 3. Water Sci. Technol. 39, 183–193. doi: 10.2166/wst.1999.0039
Hartig C., Storm T., Jekel M. (1999). Detection and identification of sulphonamide drugs in municipal wastewater by liquid chromatography coupled with electrospray ionization tandem mass spectrometry. J. Chromatogr. A 854, 163–173. doi: 10.1016/S0021-9673(99)00378-7
Hirsch R., Ternes T., Haberer K., Kratz K.-L. (1999). Occurrence of antibiotics in the aquatic environment. Sci. Total Environ. 225, 109–118. doi: 10.1016/S0048-9697(98)00337-4
Hoa P. T. P., Nonaka L., Viet P. H., Suzuki S. (2008). Detection of the sul1, sul2 and sul3 genes in sulfonamide resistant bacteria from wastewater and shrimp ponds of North Vietnam. Sci. Total Environ. 405, 377–384. doi: 10.1016/j.scitotenv.2008.06.023
Hu J. Y., Shi J. C., Chang H., Li D., Yang M., Kamagata Y. C. (2008). Phenotyping and genotyping of antihiotic-resistant Escherichia coli isolated from a natural river basin. Environ. Sci. Technol. 42, 3415–3420. doi: 10.1021/es7026746
Huovinen P. (2001). Resistance to trimethoprim-sulfamethoxazole. Clin. Infect. Dis. 32, 1608–1614. doi: 10.1086/320532
Insel G., Karahan O., Ozdemir S., Pala I., Katipoglu T., Cokgor E. U., et al. (2006). Unified basis for the respirometric evaluation of inhibition for activated sludge. J. Environ. Sci. Health A 41, 1763–1780. doi: 10.1080/10934520600778895
ISO. (1989). Water quality – Determination of the chemical oxygen demand. ISO 6060. (Switzerland: International Organization for Standardization).
ISO. (2007). Water quality – test for inhibition of oxygen consumption by activated sludge for carbonaceous and ammonium oxidation. ISO 8192. (Switzerland: International Organization for Standardization).
Katipoglu-Yazan T., Merlin C., Pons M. N., Ubay-Cokgor E., Orhon D. (2016). Chronic impact of sulfamethoxazole on the metabolic activity and composition of enriched nitrifying microbial culture. Water Res. 100, 546–555. doi: 10.1016/j.watres.2016.05.043
Katipoglu-Yazan T., Pala-Ozkok I., Ubay-Cokgor E., Orhon D. (2013). Acute impact of erythromycin and tetracycline on the kinetics of nitrification and organic carbon removal in mixed microbial culture. Bioresour. Technol. 144, 410–419. doi: 10.1016/j.biortech.2013.06.121
Katipoglu-Yazan T., Ubay-Cokgor E., Orhon D. (2018). Chronic impact of sulfamethoxazole: How does process kinetics relate to metabolic activity and composition of enriched nitrifying microbial culture? J. Chem. Technol. Biot. 93, 1722–1732. doi: 10.1002/jctb.5545
Katipoglu-Yazan T., Ubay-Cokgor E., Orhon D. (2023). Significance of enriched culture on the assessment of the acute inhibitory impact of sulfamethoxazole on nitrifying biomass. J. Chem. Technol. Biot. 98, 706–717. doi: 10.1002/jctb.7274
Kor-Bicakci G., Pala-Ozkok I., Ural A., Jonas D., Orhon D., Ubay-Cokgor E. (2016). Is the chronic impact of sulfamethoxazole different for slow growing culture? effect culture history Bioresour. Technol. 206, 65–76. doi: 10.1016/j.biortech.2016.01.069
Krishna C., Van Loosdrecht M. C. M. (1999). Substrate flux into storage and growth in relation to activated sludge modeling. Water Res. 33, 3149–3161. doi: 10.1016/S0043-1354(99)00031-7
Kümmerer K. (2009). The presence of pharmaceuticals in the environment due to human use–present knowledge and future challenges. J. Environ. Manage. 90, 2354–2366. doi: 10.1016/j.jenvman.2009.01.023
Larsson D. G. J., de Pedro C., Paxeus N. (2007). Effluent from drug manufactures contains extremely high levels of pharmaceuticals. J. Hazard. Mater. 148, 751–755. doi: 10.1016/j.jhazmat.2007.07.008
Le-Minh N., Khan S., Drewes J., Stuetz R. (2010). Fate of antibiotics during municipal water recycling treatment processes. Water Res. 44, 4295–4323. doi: 10.1016/j.watres.2010.06.020
Liebert C. A., Hall R. M., Summers A. O. (1999). Transposon Tn21, flagship of the floating genome. Microbiol. Mol. Biol. Rev. 63, 507–522. doi: 10.1128/MMBR.63.3.507-522.1999
Lin J., Biyela P. T. (2005). Convergent acquisition of antibiotic resistance determinants amongst the Enterobacteriaceae sp. isolates of the Mhlathuze River, KwaZulu-Natal (RSA). Water SA. 31, 257–260. doi: 10.4314/wsa.v31i2.5193
Maskell J. P., Sefton A. M., Hall L. M. C. (1997). Mechanism of sulfonamide resistance in clinical isolates of streptococcus pneumonia. J. Antimicrob. Chemother. 41, 2121–2126. doi: 10.1128/AAC.41.10.2121
Masters P. A., O'Bryan T. A., Zurlo J., Miller D. Q., Joshi N. (2003). Trimethoprim-sulfamethoxazole revisited. Arch. Intern. Med. 163, 402–410. doi: 10.1001/archinte.163.4.402
Michelow I. C., McCracken G. H. (2009). “Antibacterial therapeutic agents,” in Feigin and Cherry's Textbook of Pediatric Infectious Diseases, Sixth Edition (United States of America: Elsevier), 3178–3227. doi: 10.1016/B978-1-4160-4044-6.50253-3
Miran W., Jang J., Nawaz M., Shahzad A., Lee D. S. (2018). Biodegradation of the sulfonamide antibiotic sulfamethoxazole by sulfamethoxazole acclimatized cultures in microbial fuel cells. Sci. Total Environ. 627, 1058–1065. doi: 10.1016/j.scitotenv.2018.01.326
Mohapatra H., Mohapatra S. S., Mantri C. K., Colwell R. R., Singh D. V. (2008). Vibrio cholerae non-O1, non-O139 strains isolated before 1992 from Varanasi, India are multiple drug resistant, contain intSXT, dfr18 and aadA5 genes. Environ. Microbiol. 10, 866–873. doi: 10.1111/j.1462-2920.2007.01502.x
Mukherjee S., Chakraborty R. (2006). Incidence of class 1 integrons in multiple antibiotic-resistant Gram-negative copiotrophic bacteria rom the River Torsa in India. Res. Microbiol. 157, 220–226. doi: 10.1016/j.resmic.2005.08.003
Müller E., Schüssler W., Horn H., Lemmer H. (2013). Aerobic biodegradation of the sulfonamide antibiotic sulfamethoxazole by activated sludge applied as co-substrate and sole carbon and nitrogen source. Chemosphere 92, 969–978. doi: 10.1016/j.chemosphere.2013.02.070
Nadais H., Capela I., Arroja L., Duarte A. (2005). Optimum cycle time for intermittent UASB reactors treating dairy wastewater. Water Res. 39, 1511–1518. doi: 10.1016/j.watres.2005.01.020
Orhon D., Sözen S. (2012). Fate and effect of xenobiotics on biodegradation processes: basis for respirometric appraisal. Environ. Technol. 33, 1517–1522. doi: 10.1080/09593330.2012.655323
Orhon D., Ubay Cokgor E., Insel G., Karahan O., Katipoglu T. (2009). Validity of monod kinetics at different sludge ages – peptone biodegradation under aerobic conditions. Bioresour. Technol. 100, 5678–5686. doi: 10.1016/j.biortech.2009.06.046
Ouyang W.-Y., Su J.-Q., Richnow H. H., Adrian L. (2019). Identification of dominant sulfamethoxazole-degraders in pig farm-impacted soil by DNA and protein stable isotope probing. Environ. Int. 126, 118–126. doi: 10.1016/j.envint.2019.02.001
Ozkok I. P., Yazan T. K., Cokgor E. U., Insel G., Talinli I., Orhon D. (2011). Respirometric assessment of substrate binding by antibiotics in peptone biodegradation. J. Environ. Sci. Health A 46, 1588–1597. doi: 10.1080/10934529.2011.609442
Pala-Ozkok I. (2012). Inhibitory Impact of Selected Antibiotics on Biodegradation Characteristics and Microbial Population Under Aerobic Conditions. Istanbul Technical University, Turkey. PhD Thesis.
Pala-Ozkok I., Kor-Bicakci G., Ural A., Katipoglu-Yazan T., Yağcı N., Ubay-Çokgor E., et al. (2014). Modeling acute impact of sulfamethoxazole on the utilization of simple and complex substrates by fast growing microbial culture. J. Chem. Technol. Biotechnol. 89, 603–615. doi: 10.1002/jctb.4165
Pala-Ozkok I., Orhon D. (2013). Chronic effect of erythromycin on substrate biodegradation kinetics of activated sludge. Biochem. Eng. J. 81, 29–39. doi: 10.1016/j.bej.2013.10.002
Pala-Ozkok I., Rehman A., Ubay-Cokgor E., Jonas D., Orhon D. (2014a). Pyrosequencing reveals the inhibitory impact of chronic exposure to erythromycin on activated sludge bacterial community structure. Biochem. Eng. J. 90, 195–205. doi: 10.1016/j.bej.2014.06.003
Pala-Ozkok I., Rehman A., Yagci N., Ubay-Cokgor E., Jonas D., Orhon D. (2012). Characteristics of mixed microbial culture at different sludge ages: effect on variable kinetics for substrate utilization. Bioresour. Technol. 126, 274–282. doi: 10.1016/j.biortech.2012.08.115
Pala-Ozkok I., Ubay-Cokgor E., Cakar Z. P., Orhon D. (2014b). Acute impact of erythromycin on substrate utilization by activated sludge: effect of sludge age, J. Chem. Technol. Biotechnol. 89, 1091–1102. doi: 10.1002/jctb.4208
Pala-Ozkok I., Ubay-Cokgor E., Jonas D., Orhon D. (2019). Kinetic and microbial response of activated sludge community to acute and chronic exposure to tetracycline. J. Hazard. Mater. 367, 418–426. doi: 10.1016/j.jhazmat.2018.12.094
Pei R., Kim S.-C., Carlson K. H., Pruden A. (2006). Effect of River Landscape on the sediment concentrations of antibiotics and corresponding antibiotic resistance genes (ARG). Water Res. 40, 2427–2435. doi: 10.1016/j.watres.2006.04.017
Perez S., Eichhorn P., Aga D. S. (2005). Evaluating the biodegradability of sulfamethazine, sulfamethoxazole, sulfathiazole, and trimethoprim at different stages of sewage treatment. Environ. Toxicol. Chem. 24, 1361–1367. doi: 10.1897/04-211R.1
Reichert P. (1994). Concepts underlying a computer program for the identification and simulation of aquatic systems, vol. 7. Dübendorf, Switzerland: Schriftenreihe der Eawag.
Schloss P. D., Westcott S. L., Ryabin T., Hall J. R., Hartmann M., Hollister E. B., et al. (2009). Introducing mothur: Open-source, platform-independent, community-supported software for describing and com- paring microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Shen Y., Stedtfeld R. D., Guo X., Bhalsod G. D., Jeon S., Tiedje J. M., et al. (2019). Pharmaceutical exposure changed antibiotic resistance genes and bacterial communities in soil-surface- and overhead-irrigated greenhouse lettuce. Environ. Int. 131, 105031. doi: 10.1016/j.envint.2019.105031
Sköld O. (2000). Sulfonamide resistance: mechanisms and trends. Drug Resist. Updat. 3, 155–160. doi: 10.1054/drup.2000.0146
Sköld O. (2001). Resistance to trimethoprim and sulfonamides. Vet. Res. 32, 261–273. doi: 10.1051/vetres:2001123
Srinivasan V., Nam H. M., Nguyen L. T., Tamilselvam B., Murinda S. E., Oliver S. P. (2005). Prevalence of antimicrobial resistance genes in Listeria monocytogenes isolated from dairy farms. Foodborne Patho. Dis. 2, 201–211. doi: 10.1089/fpd.2005.2.201
Straub J. O. (2016). Aquatic environmental risk assessment for human use of the old antibiotic sulfamethoxazole in Europe. Crit. Review. Environ. Toxicol. Chem. 35, 767–779. doi: 10.1002/etc.2945
Taviani E., Ceccarelli D., Lazaro N., Bani S., Cappuccinelli P., Colwell R. R., et al. (2008). Environmental Vibrio spp., isolated in Mozambique, contain a polymorphic group of integrative conjugative elements and class 1 integrons. FEMS Microbiol. Ecol. 64, 45–54. doi: 10.1111/j.1574-6941.2008.00455.x
Tennstedt T., Szczepanowski R., Braun S., Pühler A., Schlüter A. (2003). Occurrence of integron-associated resistance gene cassettes located on antibiotic resistance plasmids isolated from a wastewater treatment plant. FEMS Microbiol. Ecol. 45, 239–252. doi: 10.1016/S0168-6496(03)00164-8
Vikromvarasiri N., Koyama M., Kurniawan W., Pisutpaisal N., Nakasaki K. (2023). Enhancing methane recovery by intermittent substrate feeding and microbial community response in anaerobic digestion of glycerol. Renew. Energ. 204, 106–113. doi: 10.1016/j.renene.2023.01.002
Xu B., Mao D., Luo Y., Xu L. (2011). Sulfamethoxazole biodegradation and biotransformation in the water–sediment system of a natural river. Bioresour. Technol. 102, 7069–7076. doi: 10.1016/j.biortech.2011.04.086
Yan R., Wang Y., Li J., Wang X., Wanga Y. (2022). Determination of the lower limits of antibiotic biodegradation and the fate of antibiotic resistant genes in activated sludge: Both nitrifying bacteria and heterotrophic bacteria matter. J. Hazard. Mater. 425, 127764. doi: 10.1016/j.jhazmat.2021.127764
Zhang Y., Geng J., Ma H., Ren H., Xu K., Ding L. (2016). Characterization of microbial community and antibiotic resistance genes in activated sludge under tetracycline and sulfamethoxazole selection pressure. Sci. Total Environ. 571, 479–486. doi: 10.1016/j.scitotenv.2016.07.014
Keywords: acute inhibition, activated sludge, antibiotic resistance genes, modeling, chronic exposure, 454-pyro-sequencing, sulfamethoxazole
Citation: Pala-Ozkok I, Katipoglu-Yazan T, Olmez-Hanci T, Jonas D, Ubay-Cokgor E and Orhon D (2024) Impact of acute and chronic exposure to sulfamethoxazole on the kinetics and microbial structure of an activated sludge community. Front. Antibiot. 3:1335654. doi: 10.3389/frabi.2024.1335654
Received: 09 November 2023; Accepted: 05 March 2024;
Published: 02 April 2024.
Edited by:
Stephen Henry Gillespie, University of St Andrews, United KingdomReviewed by:
Mikaeel Young, Baylor University, United StatesMojtaba Akbari, Isfahan University of Medical Sciences, Iran
Copyright © 2024 Pala-Ozkok, Katipoglu-Yazan, Olmez-Hanci, Jonas, Ubay-Cokgor and Orhon. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ilke Pala-Ozkok, aWxrZS5wYWxhb3prb2tAdWlzLm5v
 Ilke Pala-Ozkok
Ilke Pala-Ozkok Tugce Katipoglu-Yazan
Tugce Katipoglu-Yazan Tugba Olmez-Hanci
Tugba Olmez-Hanci Daniel Jonas
Daniel Jonas Emine Ubay-Cokgor2
Emine Ubay-Cokgor2 Derin Orhon
Derin Orhon