- 1Department of Microbiology and Immunology, School of Medicine, University of Louisville, Louisville, KY, United States
- 2Department of Biology, School of Arts and Sciences, University of Louisville, Louisville, KY, United States
- 3Department of Dental Medicine, Karolinska Institutet, Huddinge, Sweden
- 4Department of Odontology, Umeå University, Umeå, Sweden
- 5Department of Oral Immunology and Infectious Diseases, School of Dentistry, University of Louisville, Louisville, KY, United States
Periodontitis is a dysbiotic disease caused by the interplay between the microbial ecosystem present in the disease with the dysregulated host immune response. The disease-associated microbial community is formed by the presence of established oral pathogens like Aggregatibacter actinomycetemcomitans as well as by newly dominant species like Filifactor alocis. These two oral pathogens prevail and grow within the periodontal pocket which highlights their ability to evade the host immune response. This review focuses on the virulence factors and potential pathogenicity of both oral pathogens in periodontitis, accentuating the recent description of F. alocis virulence factors, including the presence of an exotoxin, and comparing them with the defined factors associated with A. actinomycetemcomitans. In the disease setting, possible synergistic and/or mutualistic interactions among both oral pathogens might contribute to disease progression.
Introduction
Periodontitis is a multifactorial irreversible chronic inflammatory disease that affects the supporting structure of the teeth. Recent reports by the Centers for Disease Control and Prevention (CDC) show that 42.2% of adults 30 years and older develop some form of periodontitis [1]. Poor oral hygiene is the most common cause associated with periodontitis, but other factors such as age, gender, socioeconomic, and education status increase the risk to develop the disease [2]. Other risk factors such as smoking, diabetes, medications that cause dry mouth, stress, and genetics [1] can affect the oral cavity homeostasis and have a direct or indirect impact on the oral microbiome composition. This in turn promotes changes in the abundance and homeostatic relationships within the polymicrobial communities resulting in a dysbiotic ecosystem in disease. The disruption of tissue homeostasis is accompanied by microbial shifts or dysbiosis from indigenous symbionts (commensal bacteria) to predominantly pathogenic bacteria. The new state of polymicrobial dysbiosis promotes a dysregulated inflammatory state in the host that drives disease progression. Long studied microorganisms such as Porphyromonas gingivalis, Tannerella forsythia, Treponema denticola, and Aggregatibacter actinomycetemcomitans are now well-established “periodontal pathogens”, with evidenced involvement in disease initiation and progression. In the last decade, the advance in high throughput technology allowed us to obtain an in-depth characterization of the complexity of the oral microbiome both in health and in disease [3, 4]. As a result, several microorganisms were identified with high prevalence in disease sites compared to healthy sites, including Filifactor alocis, a newly appreciated microbial species.
A. actinomycetemcomitans is a non-motile gram-negative facultative anaerobe of the Pasteurellaceae family [5–8], and is known to contribute to gingival tissue inflammation, destruction, and bone resorption by expressing several virulence factors (Supplementary Table S1) such as cytolethal distending toxin (Cdt), leukotoxin A (LtxA) of the Repeats-In-Toxins (RTX) family of bacterial toxins, and collagenase [9–15]. On the other hand, F. alocis is a gram-positive anaerobic rod, and characterization of the organism's pathogenic credentials is still in its infancy. Some initial descriptions of the potential virulence factors of F. alocis include the presence of a moonlight surface protein that binds to and inhibits the complement component 3 (C3), a key step of the complement activation cascade; two enzymes that might provide oxidative stress resistance; and an exotoxin of the RTX family with an unknown biological function (Figure 1). Some recent reports describe that the presence of F. alocis increases A. actinomycetemcomitans total biomass when in co-infections with Veillonella sp [16, 17]. In this mini-review, we describe the virulence factors associated with A. actinomycetemcomitans and F. alocis, which are uniquely found in those species among all members of the oral microbial community.
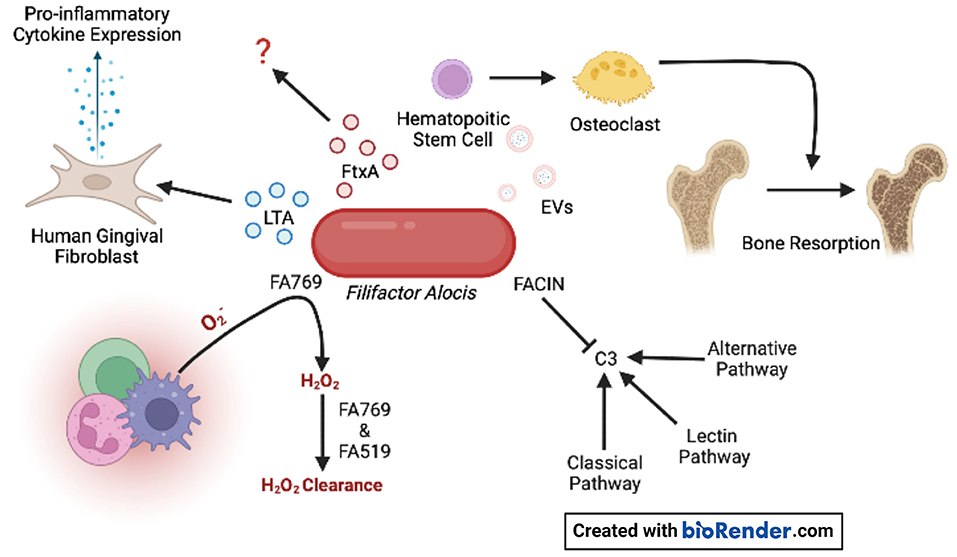
Figure 1. F. alocis virulence factors. Oxidative resistance is achieved by the conversion of superoxide generated by innate immune cells to H2O2 by FA769. The proteins FA769 and FA519 are associated with H2O2-induced stress resistance. F. alocis complement inhibitor (FACIN) binds complement component 3 (C3) which is essential to all three complement pathways. Extracellular vesicles (EVs) contain lipoproteins that stimulate osteoclastogenesis in committed osteoclast precursors via TLR2 which contributes to bone resorption. The novel RTX exotoxin, FtxA, is found in 60% of clinical isolates, but its biological effects are unknown. Lipoteichoic acid (LTA) induces the expression of pro-inflammatory cytokines by human gingival fibroblasts.
Periodontitis: Lessons learned from established and emerging periodontal pathogens
Subgingival plaque samples collected from periodontitis patients and periodontitis-free individuals differ from each other [18]. P. gingivalis, T. forsythia, and T. denticola, the so-called red complex, have shown the strongest association with periodontal disease [19]. Deepened periodontal pockets with an anaerobic environment, inflammatory conditions, and large access to substrates originating from tissue destruction all favor the growth of these pathogens and pathobionts. These gram-negative anaerobe pathobionts express virulence factors with the capacity to cause an imbalance in the host inflammatory response [20]. If these bacteria contribute to the degenerative process in periodontitis or if they are a result of the unique ecological niche in a periodontal pocket, it has not been fully investigated [21]. The study of the microbial composition shift from periodontally healthy toward disease onset can contribute to answer this challenging question. Longitudinal studies examined periodontally healthy adolescents that at baseline show that the presence of A. actinomycetemcomitans in the subgingival plaque is significantly associated with disease onset [22–25]. The prevalence of this bacterium varies on age, geographic origin, and periodontal status of the examined population [26]. A high intra-species genetic diversity exists, which resulted in the generation of highly virulent as well as harmless variants of this bacterium [27]. The most well-known virulent variant of A. actinomycetemcomitans is the JP2 genotype. This genotype expresses a high amount of LtxA and is often detected in young individuals with periodontitis [28, 29]. Interestingly, it has been shown that young individuals that carry A. actinomycetemcomitans in their subgingival plaque have an increased risk to develop attachment loss if F. alocis is detected in the same sample [17]. Based on these reports we propose a model in which A. actinomycetemcomitans initiates the degenerative process in the periodontium that creates an anaerobic environment attractive for translocation of F. alocis (Figure 2). In addition, it could be speculated that F. alocis manipulation of innate immune cells, like neutrophils, interferes with A. actinomycetemcomitans LtxA-induced inflammatory cell death [13, 30].
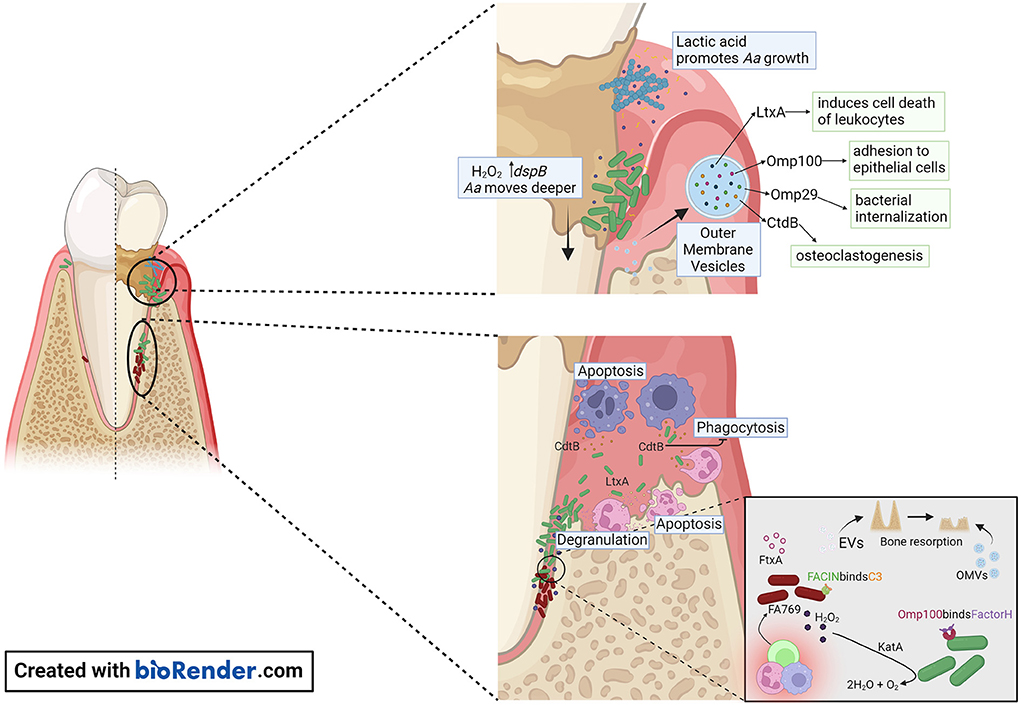
Figure 2. Proposed model of A. actinomycetemcomitans and F. alocis interactions with the host within the gingival pocket. In the early stages of periodontitis, A. actinomycetemcomitans makes use of lactic acid produced by Streptococcus sp. as a nutrient to increase its numbers. A. actinomycetemcomitans releases OMVs that are packed with outer membrane proteins (Omp) OmpA1, Omp 100, and the virulence factors CdtB, and LtxA. Omp100 mediates initial adhesion of the bacterium, and OmpA1 binds to its putative ligand on gingival epithelial cells and induces F-actin rearrangements resulting in A. actinomycetemcomitans cells being internalized. Production of H2O2 by Streptococcus sp. causes A. actinomycetemcomitans to migrate deeper in the gingival pocket, where the bacterial cells are exposed to the host immune response. The release of CtdB in this environment inhibits phagocytosis and LtxA release by A. actinomycetemcomitans will promote neutrophil degranulation or cell death when present at high concentrations. The release of OMVs by A. actinomycetemcomitans or EVs by F. alocis might contribute to the pathogenicity of these organisms by evasion of the host immune response and promoting bone resorption.
More recently the anaerobic gram-positive bacterium F. alocis has attracted interest in the etiology and pathogenesis of periodontitis [31]. This bacterium is often detected in periodontal pockets of different individuals [32, 33]. The role of F. alocis in the pathogenicity of periodontitis is still not known, however, recent studies indicate a capacity to dysregulate the innate immune response [34]. To begin to characterize F. alocis' potential virulence factors, we screened the whole genome of the F. alocis reference strain ATCC 35896 (also known as CCUG 47790), for novel identification and deeper characterization of virulence elements. We discovered that the reference strain encodes a hitherto unrecognized RTX toxin member, which we designated as “FtxA” for consistency with the nomenclature of other RTX toxin-gene encoding operons [35]. We have used ATCC 35896, and our clinical collection of nine additional F. alocis strains, isolated from different oral infections, to further characterize the FtxA protein, and whether the gene encoding it may be conserved in the phylogenetic lineage(s) of F. alocis, and hence might represent a candidate diagnostic marker for more virulent strains. According to PCR, ftxA was encoded by five of the ten tested strains [35]. To corroborate the PCR results, and for subsequent multi-locus sequence typing (MLST), all nine strains that had been isolated at the clinical laboratory were then subject to whole-genome sequencing. Extraction of the genome sequence data essentially confirmed the PCR findings, with highly conserved FtxA protein sequences, encoded in apparent ftxABD operons. However, one of the ftxA-negative strains according to PCR, 854G-16U, was found to encode an FtxA homolog. Relative to the ATCC 35896 FtxA protein, it was only ~46% identical at the amino acid sequence level, consistent with sequence variability among the FtxA proteins in F. alocis [35]. Taken together, ftxA was carried by six of the ten tested strains and is therefore not a universal property of this bacterium. The expression of this gene and its role in F. alocis virulence is still not known, yet proteomic characterization of available strains has identified intra-species differences, as well as clustering of FtxA with another six intracellular proteins [36].
A. actinomycetemcomitans and F. alocis virulence factors: Contributions to microbial pathogenicity and host immune evasion
Exotoxins
The different virulence factors expressed by A. actinomycetemcomitans are well studied [37] (Supplementary Table S1). Like other gram-negative bacteria, A. actinomycetemcomitans releases endotoxins and exotoxins that activate inflammatory response through interaction with the Toll-like receptors 4 (TLR4) [38]. Unique for this bacterium among the inhabitants in the oral microbiota is the expression of two exotoxins. One is LtxA, that is closely associated to disease onset and progression, as shown by a strong correlation to disease onset in carriers of highly leukotoxic variants of A. actinomycetemcomitans [24, 25]. LtxA is secreted from the bacterium by a Type I secretion system [39]. This exotoxin induces cell death of human defense cells in an active pro-inflammatory process named pyroptosis [13, 40]. It is well-established that the dysbiotic immune response in pyroptosis is linked to the pathogenesis of periodontitis [41]. These cellular mechanisms also function as a tool for accessing host-derived nutrients for the invading bacteria [42] (Figure 2). The second exotoxin is the cytolethal distending toxin (Cdt), particularly the active unit CdtB, that enters the nucleus of target cells and induces double-strand breaks in chromosomal DNA of proliferating cells [15] (Figure 2). This toxin induces cellular mechanisms involved in the pathogenesis of periodontitis, but its role in disease initiation and progression is unclear [43, 44]. F. alocis virulence factors are beginning to be characterized (Figure 1). Most recently, FtxA, has been identified as a putative ~250 kDa exotoxin of F. alocis [35], and similar to LtxA, belongs to the large family of RTX proteins, found in both gram-negative and gram-positive bacteria [45]. The activity of FtxA is not yet known; interestingly it appeared to be encoded and expressed only by six of the 10 assessed F. alocis strains [35, 36], suggesting potentially different capabilities to modulate host functions by FtxA-expressing strains, as compared to those that do not encode this protein.
Membrane vesicles
A. actinomycetemcomitans outer membrane vesicles (OMVs) can deliver several biologically active virulence factors to host cells, which can modulate the host response (Figure 2). These include Cdt [46], LtxA [47, 48], peptidoglycan-associated lipoprotein (Pal) [49], and the chaperonin GroEL [50]. A. actinomycetemcomitans OMVs carry NOD1- and NOD2-active peptidoglycan, which can be internalized into non-phagocytic human cells including gingival fibroblasts [51]. This supports the role of OMVs as triggers of innate immunity. A. actinomycetemcomitans OMVs can bind to the classical and mannose-binding lectin (MBL) complement inhibitor, C4-binding protein, by means of the outer membrane protein A1 (OmpA1) [52], which is consistent with an ability of the vesicles to mediate serum protection in vitro [53]. The mechanism(s) of how A. actinomycetemcomitans OMVs enter and/or delivers cargo into host cells is not entirely clear. The OMVs appear to enter human cells via clathrin-mediated endocytosis [51, 54], but can also fuse with host cell membranes in a cholesterol-dependent fashion [46]. Toxins delivered via OMVs can act as adhesins in receptor-mediated endocytosis [55], albeit neither LtxA nor Cdt were required for the OMV uptake by host cells [46, 48]. Hence, despite that LtxA has an apparent surface localization on the A. actinomycetemcomitans OMVs, the LtxA receptor LFA-1 is not required for delivering the toxin into the human host cells [39].
Highly purified extracellular vesicles (EVs) released by the F. alocis reference strain ATCC 35896 were recently characterized regarding their proteomic content, using in-gel digestion and liquid chromatography-tandem mass spectrometry (LC-MS/MS) [56]. F. alocis EVs proteomics revealed 28 proteins, including lipoproteins, autolysins, F. alocis complement inhibitor (FACIN), transporter- and metabolism-related proteins, and ribosomal proteins (Figure 1). Interestingly, FtxA, the recently discovered RTX protein family member, [35] according to its GenBank database definition, was identified in the F. alocis EVs proteome [56]. Whether FtxA or any of the other EVs proteins might play a role in the observed immunostimulatory effects of the vesicles on human monocytic and oral keratinocyte cell lines [56], and/or in the EV-mediated inhibition of osteogenesis through TLR2 signaling [57] is not presently clear. However, interestingly, as the osteoclastogenic potency of F. alocis EVs (Figure 1) was reduced upon treatment with lipoprotein lipase, lipoproteins may contribute to the systemic bone loss via TLR2 [58].
Complement
The complement cascade is a process known for its antimicrobial role in bacterial opsonization, which targets the clearance and destruction of the organisms by phagocytes and direct cell lysis by forming the C5b-9 membrane attack complex. However, periodontal pathogens developed effective evasion strategies to counteract complement activation. OMVs appear to play a significant role in the ability of A. actinomycetemcomitans to evade complement attack. These vesicles serve as a decoy that triggers complement activation through lipopolysaccharide (LPS) and takes in complement components [53]. In turn, LPS of some A. actinomycetemcomitans strains (i.e., strain Y4) can bind strongly to C3b, blocking the interaction between complement-derived opsonins with LPS decreasing neutrophils complement-dependent response [59]. Moreover, some of the Omp such as OmpA1 (also known as Omp29 and Omp34), and OmpA2 are important for serum resistance of A. actinomycetemcomitans via binding of C4-binding protein, thereby inhibiting the activation of the classical and MBL complement pathways [52]. In response to H2O2, A. actinomycetemcomitans produces Omp100 [60]. Omp100 captures the alternative complement pathway negative regulator, Factor H, and deposits it at the cell surface, modifying and inactivating C3b [61].
The knowledge of F. alocis' methods to evade the complement cascade are on the rise. Jusko et al. [62] identified the novel complement inhibitory protein FACIN, which is secreted or expressed on the cell surface and binds to C3, blocking all complement pathways. FACIN has dual importance for F. alocis in evading the complement cascade and serving as a cytoplasmic enzyme acetylornithine transaminase involved in arginine catabolism. The authors proposed a mechanism where FACIN binds C3/C3b, yet allows Factor B to bind, then FACIN locks the complex in an inactive state, limiting the C3 convertase as a result (Figure 1).
Oxidative stress
The dysregulated inflammation and high abundance of hyperactivated neutrophils contribute to the generation of an oxidative-stress enriched environment in the periodontal pocket [34, 63]. Periodontal pathogens develop different survival strategies to detoxify and resist this toxic environment. Depending on the environmental cues, A. actinomycetemcomitans activates the oxygen resistance transcription regulator (oxyR), which regulates the expression of Omp100 and catalase (KatA) [60] (Figure 2). Catalase aids in the degradation of H2O2 produced by neutrophils and streptococci [64], protecting A. actinomycetemcomitans from oxidative damage. This in turn increases oxygen availability allowing A. actinomycetemcomitans to shift from fermentative to respiratory metabolism.
F. alocis has been reported to possess virulence factors that contribute to the organism's resistance to oxidative stress. Furthermore, in vitro, the growth of F. alocis is stimulated under oxidative stress conditions. F. alocis reference strain ATCC 35896 encodes an antioxidant enzyme, superoxide reductase FA796 (Figure 1), that reduces superoxide radicals into H2O2 [65]. In vitro, FA796 and the hypothetical protein FA519 are involved in resistance to H2O2-induced oxidative stress, protection against superoxide radicals, and air exposure, however, the exact mechanisms are unknown. The FA519 protein might confer F. alocis the ability to resist both H2O2 and nitric oxide-induced oxidative stress [66]. Interestingly, the expression of the FA519 genes was significantly enhanced when F. alocis was co-cultured with P. gingivalis. Gene encoding glutathione peroxidase as well as an alkyl hydroperoxide reductase subunit AhpC are found in the genome of F. alocis and may function in clearing H2O2, however, the partner to the latter mentioned protein (AhpF) is missing in the genome.
Conclusion and future perspective
The observation that the combined presence of A. actinomycetemcomitans and F. alocis in the subgingival plaque increases the risk for progression of attachment loss might be explained by the differences in growth requirements and regional nutrient and atmospheric conditions. While A. actinomycetemcomitans is facultatively anaerobic and able to colonize the gingiva early in the disease process, F. alocis is an obligate anaerobic that will preferentially grow in deeper periodontal pockets. Both species express virulence factors that induce cellular and molecular mechanisms in concordance with the pathogenesis of periodontitis. While the virulence of A. actinomycetemcomitans is strongly linked to the expression of its two exotoxins (LtxA and Cdt, Figure 2), we only recently started to unravel the virulence patterns of F. alocis (Figure 1). The recent report by Miralda et al. [67] with a detailed characterization of F. alocis extending neutrophil lifespan is at odds with the capacity of A. actinomycetemcomitans to kill leukocytes. In this study, F. alocis reference strain ATCC 35896, which expresses the exotoxin, was responsible for extending neutrophil lifespan. These two contradictory properties may be attributed to different toxins (i.e., FtxA and LtxA) of the same toxin superfamily, different expression levels, and differential microbial evasion strategies to overcome neutrophil responses. Here, we reviewed and discussed the virulence factors of A. actinomycetemcomitans and F. alocis and their pathogenic role in periodontitis (Figure 2). Several open questions arise, like the possible role of FtxA in the pathogenesis of periodontitis, which remains to be evaluated, as well as the possible synergies between FtxA and LtxA. Increased knowledge about the virulence of these two bacteria one by one or together might be of importance for improved risk prediction in the future.
Author contributions
HO and IS were involved in drafting some sections of the manuscript, designing Supplementary Table S1, Figure 1 (IS), and Figure 2 (HO). GB, JO, AJ, and SMU were involved in the conceptual idea, writing, and critical revision of the manuscript as well as obtaining funding.
Funding
This work was supported by the NIH-National Institute of Dental and Craniofacial Research (NIDCR) DE024509 and DE014615 (SMU), by Ruth L. Kirschstein National Research Service Award by the NIDCR F31DE027585 (HO), by TUA grants from Region Västerbotten, Sweden [7002667 (AJ) and 7003193 (JO)], and by grants from Insamlingsstiftelsen, Medical Faculty, Umeå University (JO and AJ).
Acknowledgments
The authors acknowledge the omission of citing in this review the work performed by many investigators in the field due to space limitations.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/froh.2022.981343/full#supplementary-material
References
1. Eke PI, Borgnakke WS, Genco RJ. Recent epidemiologic trends in periodontitis in the USA. Periodontol. (2020) 82:257–67. doi: 10.1111/prd.12323
2. Eke PI, Dye BA, Wei L, Slade GD, Thornton-Evans GO, Borgnakke WS, et al. Update on prevalence of periodontitis in adults in the United States: NHANES 2009 to 2012. J Periodontol. (2015) 86:611–22. doi: 10.1902/jop.2015.140520
3. Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, et al. The human oral microbiome. J Bacteriol. (2010) 192:5002–17. doi: 10.1128/JB.00542-10
4. Griffen AL, Beall CJ, Firestone ND, Gross EL, Difranco JM, Hardman JH, et al. CORE: a phylogenetically-curated 16S rDNA database of the core oral microbiome. PLoS ONE. (2011) 6:e19051. doi: 10.1371/journal.pone.0019051
5. Raja M, Ummer F, Dhivakar CP. Aggregatibacter actinomycetemcomitans - a tooth killer? J Clin Diagn Res. (2014) 8:ZE13–16. doi: 10.7860/JCDR/2014/9845.4766
6. Gholizadeh P, Pormohammad A, Eslami H, Shokouhi B, Fakhrzadeh V, Kafil HS. Oral pathogenesis of Aggregatibacter actinomycetemcomitans. Microb Pathog. (2017) 113:303–11. doi: 10.1016/j.micpath.2017.11.001
7. Belibasakis GN, Maula T, Bao K, Lindholm M, Bostanci N, Oscarsson J, et al. Virulence and pathogenicity properties of aggregatibacter actinomycetemcomitans. Pathogens. (2019) 8:222. doi: 10.3390/pathogens8040222
8. Fine DH, Schreiner H, Velusamy SK. Aggregatibacter, a low abundance pathobiont that influences biogeography, microbial dysbiosis, and host defense capabilities in periodontitis: the history of a bug, and localization of disease. Pathogens. (2020) 9:179. doi: 10.3390/pathogens9030179
9. Robertson PB, Lantz M, Marucha PT, Kornman KS, Trummel CL, Holt SC. Collagenolytic activity associated with Bacteroides species and Actinobacillus actinomycetemcomitans. J Periodontal Res. (1982) 17:275–83. doi: 10.1111/j.1600-0765.1982.tb01154.x
10. Brogan JM, Lally ET, Poulsen K, Kilian M, Demuth DR. Regulation of Actinobacillus actinomycetemcomitans leukotoxin expression: analysis of the promoter regions of leukotoxic and minimally leukotoxic strains. Infect Immun. (1994) 62:501–8. doi: 10.1128/iai.62.2.501-508.1994
11. Hritz M, Fisher E, Demuth DR. Differential regulation of the leukotoxin operon in highly leukotoxic and minimally leukotoxic strains of Actinobacillus actinomycetemcomitans. Infect Immun. (1996) 64:2724–9. doi: 10.1128/iai.64.7.2724-2729.1996
12. Jinadasa RN, Bloom SE, Weiss RS, Duhamel GE. Cytolethal distending toxin: a conserved bacterial genotoxin that blocks cell cycle progression, leading to apoptosis of a broad range of mammalian cell lineages. Microbiology (Reading). (2011) 157:1851–75. doi: 10.1099/mic.0.049536-0
13. Johansson A. Aggregatibacter actinomycetemcomitans leukotoxin: a powerful tool with capacity to cause imbalance in the host inflammatory response. Toxins (Basel). (2011) 3:242–59. doi: 10.3390/toxins3030242
14. Belibasakis GN, Bostanci N. Inflammatory and bone remodeling responses to the cytolethal distending toxins. Cells. (2014) 3:236–46. doi: 10.3390/cells3020236
15. DiRienzo JM. Breaking the gingival epithelial barrier: role of the aggregatibacter actinomycetemcomitans cytolethal distending toxin in oral infectious disease. Cells. (2014) 3:476–99. doi: 10.3390/cells3020476
16. Periasamy S, Kolenbrander PE. Aggregatibacter actinomycetemcomitans builds mutualistic biofilm communities with Fusobacterium nucleatum and Veillonella species in saliva. Infect Immun. (2009) 77:3542–51. doi: 10.1128/IAI.00345-09
17. Fine DH, Markowitz K, Fairlie K, Tischio-Bereski D, Ferrendiz J, Furgang D, et al. A consortium of Aggregatibacter actinomycetemcomitans, Streptococcus parasanguinis, and Filifactor alocis is present in sites prior to bone loss in a longitudinal study of localized aggressive periodontitis. J Clin Microbiol. (2013) 51:2850–61. doi: 10.1128/JCM.00729-13
18. Nemoto T, Shiba T, Komatsu K, Watanabe T, Shimogishi M, Shibasaki M, et al. Discrimination of bacterial community structures among healthy, gingivitis, and periodontitis statuses through integrated metatranscriptomic and network analyses. mSystems. (2021) 6:e0088621. doi: 10.1128/mSystems.00886-21
19. Könönen E, Gursoy M, Gursoy UK. Periodontitis: a multifaceted disease of tooth-supporting tissues. J Clin Med. (2019) 8:1135. doi: 10.3390/jcm8081135
20. Dahlen G, Basic A, Bylund J. Importance of virulence factors for the persistence of oral bacteria in the inflamed gingival crevice and in the pathogenesis of periodontal disease. J Clin Med. (2019) 8:1339. doi: 10.3390/jcm8091339
21. Henderson B, Kaiser F. Bacterial modulators of bone remodeling in the periodontal pocket. Periodontol. (2018) 76:97–108. doi: 10.1111/prd.12160
22. Van der Velden U, Abbas F, Armand S, Loos BG, Timmerman MF, Van der Weijden GA, et al. Java project on periodontal diseases. The natural development of periodontitis: risk factors, risk predictors and risk determinants. J Clin Periodontol. (2006) 33:540–8. doi: 10.1111/j.1600-051X.2006.00953.x
23. Fine DH, Markowitz K, Furgang D, Fairlie K, Ferrandiz J, Nasri C, et al. Aggregatibacter actinomycetemcomitans and its relationship to initiation of localized aggressive periodontitis: longitudinal cohort study of initially healthy adolescents. J Clin Microbiol. (2007) 45:3859–69. doi: 10.1128/JCM.00653-07
24. Haubek D, Ennibi OK, Poulsen K, Vaeth M, Poulsen S, Kilian M. Risk of aggressive periodontitis in adolescent carriers of the JP2 clone of Aggregatibacter (Actinobacillus) actinomycetemcomitans in Morocco: a prospective longitudinal cohort study. Lancet. (2008) 371:237–42. doi: 10.1016/S0140-6736(08)60135-X
25. Höglund Åberg C, Kwamin F, Claesson R, Dahlen G, Johansson A, Haubek D. Progression of attachment loss is strongly associated with presence of the JP2 genotype of Aggregatibacter actinomycetemcomitans: a prospective cohort study of a young adolescent population. J Clin Periodontol. (2014) 41:232–41. doi: 10.1111/jcpe.12209
26. Rylev M, Kilian M. Prevalence and distribution of principal periodontal pathogens worldwide. J Clin Periodontol. (2008) 35:346–61. doi: 10.1111/j.1600-051X.2008.01280.x
27. Kittichotirat W, Bumgarner RE, Chen C. Evolutionary divergence of Aggregatibacter actinomycetemcomitans. J Dent Res. (2016) 95:94–101. doi: 10.1177/0022034515608163
28. Claesson R, Höglund-Åberg C, Haubek D, Johansson A. Age-related prevalence and characteristics of Aggregatibacter actinomycetemcomitans in periodontitis patients living in Sweden. J Oral Microbiol. (2017) 9:1334504. doi: 10.1080/20002297.2017.1334504
29. Khzam N, Miranda LA, Kujan O, Shearston K, Haubek D. Prevalence of the JP2 genotype of Aggregatibacter actinomycetemcomitans in the world population: a systematic review. Clin Oral Investig. (2022) 26:2317–34. doi: 10.1007/s00784-021-04343-3
30. Miralda I, Vashishta A, Uriarte SM. Neutrophil interaction with emerging oral pathogens: a novel view of the disease paradigm. Adv Exp Med Biol. (2019) 1197:165–78. doi: 10.1007/978-3-030-28524-1_12
31. Aja E, Mangar M, Fletcher HM, Mishra A. Filifactor alocis: recent insights and advances. J Dent Res. (2021) 220345211000656. doi: 10.1177/00220345211000656
32. Dahlen G, Claesson R, Aberg CH, Haubek D, Johansson A, Kwamin F. Subgingival bacteria in Ghanaian adolescents with or without progression of attachment loss. J Oral Microbiol. (2014) 6. doi: 10.3402/jom.v6.23977
33. Hiranmayi KV, Sirisha K, Ramoji Rao MV, Sudhakar P. Novel pathogens in periodontal microbiology. J Pharm Bioallied Sci. (2017) 9:155–63. doi: 10.4103/jpbs.JPBS_288_16
34. Miralda I, Uriarte SM. Periodontal Pathogens' strategies disarm neutrophils to promote dysregulated inflammation. Mol Oral Microbiol. (2021) 36:103–20. doi: 10.1111/omi.12321
35. Oscarsson J, Claesson R, Bao K, Brundin M, Belibasakis GN. Phylogenetic Analysis of Filifactor alocis Strains Isolated from Several Oral Infections Identified a Novel RTX Toxin, FtxA. Toxins (Basel). (2020) 12:687. doi: 10.3390/toxins12110687
36. Bao K, Claesson R, Gehrig P, Grossmann J, Oscarsson J, Belibasakis GN. Proteomic characterization of the oral pathogen filifactor alocis reveals key inter-protein interactions of its RTX toxin: FtxA. Pathogens. (2022) 11. doi: 10.3390/pathogens11050590
37. Johansson A, Dahlén G. “Bacterial virulence factors that contribute to periodontal pathogenesis,” in Bostanci N, Belibasakis G, editor. Pathogenesis of periodontal diseases. (Cham, Switzerland: Springer Nature). (2018) p. 31–49. doi: 10.1007/978-3-319-53737-5_4
38. Li X, Zhou L, Takai H, Sasaki Y, Mezawa M, Li Z, et al. Aggregatibacter actinomycetemcomitans lipopolysaccharide regulates bone sialoprotein gene transcription. J Cell Biochem. (2012) 113:2822–34. doi: 10.1002/jcb.24157
39. Nice JB, Balashova NV, Kachlany SC, Koufos E, Krueger E, Lally ET, et al. Aggregatibacter actinomycetemcomitans leukotoxin is delivered to host cells in an LFA-1-indepdendent manner when associated with outer membrane vesicles. Toxins (Basel). (2018) 10:414. doi: 10.3390/toxins10100414
40. Kelk P, Moghbel NS, Hirschfeld J, Johansson A. Aggregatibacter actinomycetemcomitans leukotoxin activates the NLRP3 inflammasome and cell-to-cell communication. Pathogens. (2022) 11:159. doi: 10.3390/pathogens11020159
41. Sordi MB, Magini RS, Panahipour L, Gruber R. Pyroptosis-mediated periodontal disease. Int J Mol Sci. (2021) 23:372. doi: 10.3390/ijms23010372
42. Ozuna H, Uriarte SM, Demuth DR. The hunger games: aggregatibacter actinomycetemcomitans exploits human neutrophils as an epinephrine source for survival. Front Immunol. (2021) 12:707096. doi: 10.3389/fimmu.2021.707096
43. Belibasakis GN, Johansson A, Wang Y, Chen C, Kalfas S, Lerner UH. The cytolethal distending toxin induces receptor activator of NF-kappaB ligand expression in human gingival fibroblasts and periodontal ligament cells. Infect Immun. (2005) 73:342–51. doi: 10.1128/IAI.73.1.342-351.2005
44. Höglund Åberg C, Antonoglou G, Haubek D, Kwamin F, Claesson R, Johansson A. Cytolethal distending toxin in isolates of Aggregatibacter actinomycetemcomitans from Ghanaian adolescents and association with serotype and disease progression. PLoS ONE. (2013) 8:e65781. doi: 10.1371/journal.pone.0065781
45. Linhartova I, Bumba L, Masin J, Basler M, Osicka R, Kamanova J, et al. RTX proteins: a highly diverse family secreted by a common mechanism. FEMS Microbiol Rev. (2010) 34:1076–112. doi: 10.1111/j.1574-6976.2010.00231.x
46. Rompikuntal PK, Thay B, Khan MK, Alanko J, Penttinen AM, Asikainen S, et al. Perinuclear localization of internalized outer membrane vesicles carrying active cytolethal distending toxin from Aggregatibacter actinomycetemcomitans. Infect Immun. (2012) 80:31–42. doi: 10.1128/IAI.06069-11
47. Kato S, Kowashi Y, Demuth DR. Outer membrane-like vesicles secreted by Actinobacillus actinomycetemcomitans are enriched in leukotoxin. Microb Pathog. (2002) 32:1–13. doi: 10.1006/mpat.2001.0474
48. Demuth DR, James D, Kowashi Y, Kato S. Interaction of Actinobacillus actinomycetemcomitans outer membrane vesicles with HL60 cells does not require leukotoxin. Cell Microbiol. (2003) 5:111–21. doi: 10.1046/j.1462-5822.2003.00259.x
49. Karched M, Ihalin R, Eneslätt K, Zhong D, Oscarsson J, Wai SN, et al. Vesicle-independent extracellular release of a proinflammatory outer membrane lipoprotein in free-soluble form. BMC Microbiol. (2008) 8:18. doi: 10.1186/1471-2180-8-18
50. Goulhen F, Hafezi A, Uitto VJ, Hinode D, Nakamura R, Grenier D, et al. Subcellular localization and cytotoxic activity of the GroEL-like protein isolated from Actinobacillus actinomycetemcomitans. Infect Immun. (1998) 66:5307–13. doi: 10.1128/IAI.66.11.5307-5313.1998
51. Thay B, Damm A, Kufer TA, Wai SN, Oscarsson J. Aggregatibacter actinomycetemcomitans outer membrane vesicles are internalized in human host cells and trigger NOD1- and NOD2-dependent NF-kappaB activation. Infect Immun. (2014) 82:4034–46. doi: 10.1128/IAI.01980-14
52. Lindholm M, Min Aung K, Nyunt Wai S, Oscarsson J. Role of OmpA1 and OmpA2 in Aggregatibacter actinomycetemcomitans and Aggregatibacter aphrophilus serum resistance. J Oral Microbiol. (2019) 11:1536192. doi: 10.1080/20002297.2018.1536192
53. Lindholm M, Metsaniitty M, Granstrom E, Oscarsson J. Outer membrane vesicle-mediated serum protection in Aggregatibacter actinomycetemcomitans. J Oral Microbiol. (2020) 12:1747857. doi: 10.1080/20002297.2020.1747857
54. O'Donoghue EJ, Krachler AM. Mechanisms of outer membrane vesicle entry into host cells. Cell Microbiol. (2016) 18:1508–17. doi: 10.1111/cmi.12655
55. Kesty NC, Mason KM, Reedy M, Miller SE, Kuehn MJ. Enterotoxigenic Escherichia coli vesicles target toxin delivery into mammalian cells. EMBO J. (2004) 23:4538–49. doi: 10.1038/sj.emboj.7600471
56. Kim HY, Lim Y, An SJ, Choi BK. Characterization and immunostimulatory activity of extracellular vesicles from Filifactor alocis. Mol Oral Microbiol. (2020) 35:1–9. doi: 10.1111/omi.12272
57. Song MK, Kim HY, Choi BK, Kim HH. Filifactor alocis-derived extracellular vesicles inhibit osteogenesis through TLR2 signaling. Mol Oral Microbiol. (2020) 35:202–10. doi: 10.1111/omi.12307
58. Kim HY, Song MK, Gho YS, Kim HH, Choi BK. Extracellular vesicles derived from the periodontal pathogen Filifactor alocis induce systemic bone loss through Toll-like receptor 2. J Extracell Vesicles. (2021) 10:e12157. doi: 10.1002/jev2.12157
59. Yamaguchi N, Tsuda H, Yamashita Y, Koga T. Binding of the capsule-like serotype-specific polysaccharide antigen and the lipopolysaccharide from Actinobacillus actinomycetemcomitans to human complement-derived opsonins. Oral Microbiol Immunol. (1998) 13:348–54. doi: 10.1111/j.1399-302X.1998.tb00690.x
60. Ramsey MM, Whiteley M. Polymicrobial interactions stimulate resistance to host innate immunity through metabolite perception. Proc Natl Acad Sci U S A. (2009) 106:1578–83. doi: 10.1073/pnas.0809533106
61. Asakawa R, Komatsuzawa H, Kawai T, Yamada S, Goncalves RB, Izumi S, et al. Outer membrane protein 100, a versatile virulence factor of Actinobacillus actinomycetemcomitans. Mol Microbiol. (2003) 50:1125–39. doi: 10.1046/j.1365-2958.2003.03748.x
62. Jusko M, Miedziak B, Ermert D, Magda M, King BC, Bielecka E, et al. FACIN, a Double-edged sword of the emerging periodontal pathogen filifactor alocis: a metabolic enzyme moonlighting as a complement inhibitor. J Immunol. (2016) 197:3245–59. doi: 10.4049/jimmunol.1600739
63. Uriarte SM, Edmisson JS, Jimenez-Flores E. Human neutrophils and oral microbiota: a constant tug-of-war between a harmonious and a discordant coexistence. Immunol Rev. (2016) 273:282–98. doi: 10.1111/imr.12451
64. Stacy A, Everett J, Jorth P, Trivedi U, Rumbaugh KP, Whiteley M. Bacterial fight-and-flight responses enhance virulence in a polymicrobial infection. Proc Natl Acad Sci U S A. (2014) 111:7819–24. doi: 10.1073/pnas.1400586111
65. Mishra A, Aja E, Fletcher HM. Role of superoxide reductase FA796 in oxidative stress resistance in filifactor alocis. Sci Rep. (2020) 10:9178. doi: 10.1038/s41598-020-65806-3
66. Aja E, Mishra A, Dou Y, Fletcher HM. Role of the filifactor alocis hypothetical protein FA519 in oxidative stress resistance. Microbiol Spectr. (2021) 9:e0121221. doi: 10.1128/Spectrum.01212-21
Keywords: periodontitis, Filifactor alocis, Aggregatibacter actinomycetemcomitans, virulence factors, chronic inflammation
Citation: Ozuna H, Snider I, Belibasakis GN, Oscarsson J, Johansson A and Uriarte SM (2022) Aggregatibacter actinomycetemcomitans and Filifactor alocis: Two exotoxin-producing oral pathogens. Front. Oral. Health 3:981343. doi: 10.3389/froh.2022.981343
Received: 29 June 2022; Accepted: 27 July 2022;
Published: 15 August 2022.
Edited by:
Ulvi Kahraman Gürsoy, University of Turku, FinlandReviewed by:
Sarah Anna Kuehne, University of Birmingham, United KingdomCopyright © 2022 Ozuna, Snider, Belibasakis, Oscarsson, Johansson and Uriarte. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Silvia M. Uriarte, c2lsdmlhLnVyaWFydGVAbG91aXN2aWxsZS5lZHU=
 Hazel Ozuna
Hazel Ozuna Ian Snider2
Ian Snider2 Georgios N. Belibasakis
Georgios N. Belibasakis Jan Oscarsson
Jan Oscarsson Anders Johansson
Anders Johansson Silvia M. Uriarte
Silvia M. Uriarte