- 1Department of Periodontology and Oral Implantology, Maurice H. Kornberg School of Dentistry, Temple University, Philadelphia, PA, United States
- 2Department of Periodontology, Faculty of Dental Medicine, University of Hassan II, Casablanca, Morocco
- 3Oral Microbiome Research Laboratory, Maurice H. Kornberg School of Dentistry, Temple University, Philadelphia, PA, United States
- 4Department of Microbiology, Forsyth Institute, Cambridge, MA, United States
Background: Grade C (previously aggressive) periodontitis (GCP) in adolescents is prevalent in certain parts of Africa where it is associated with JP2 genotype, a highly virulent strain of Aggregatibacter actinomycetemcomitans. The aim of this study was to characterize the subgingival bacteriome in Moroccan subjects with GCP positive to A. actinomycetemcomitans JP2 genotype.
Methods: Subgingival plaque samples were collected from shallow and deep pockets of 8 subjects with GCP (17.2 ± 1.5 years) and from gingival sulci of 13 controls with no periodontitis (14.6 ± 1.1 years). Identification and genotyping of A. actinomycetemcomitans was performed using PCR analysis of the ltx operon, while bacteriome profiling was done by 16S rRNA gene sequencing (V1–V3 region). Groups were compared in terms of microbial diversity, abundances, and dysbiosis.
Results: The shallow and deep pocket sites from GCP cases had a significantly altered microbial composition compared to controls. Species associated with health included Haemophilus parainfluenzae, Lautropia mirabilis, Streptococcus spp., Gemella spp., and Rothia spp. While known periodontal pathogens, including Porphyromonas gingivalis, Tannerella forsythia, Treponema spp. and Fretibacterium spp., were significantly enriched in GCP, non-conventional taxa, including Pseudomonas oral taxon C61 and Enterobacter cloacae were more abundant and showed stronger association with the disease. Less significant differences in abundances of individual taxa were observed between shallow and deep pockets. Overall dysbiosis measured in terms of Subgingival Microbial Dysbiosis Index (SMDI) differentiated between GCP and no-periodontitis with 95% accuracy.
Conclusions: The results suggest that several periodontal pathogens involved in the adult-type periodontitis also play a role in JP2 genotype-associated GCP. The potential role of non-conventional taxa in the pathogenesis of GCP warrants further investigation.
Introduction
Periodontitis encompasses a group of infectious diseases characterized by an inflammatory condition that leads to destruction of the periodontal supporting tissues, and eventual loss of the teeth (1). Grade C (previously aggressive) periodontitis (GCP) (2) is characterized by a disease onset at an early age, and presents as a localized forms that affects especially the molars and incisors, or a generalized form that affects most or all of the teeth (3). The etiology of this disease is not well understood. Present evidence indicates that microorganisms play a primary role in triggering a local inflammatory process which, in susceptible individuals, leads to significant and progressive periodontal tissue loss (4). The host immune response (5), and other factors (6) also play important roles in the development of this disease.
The subgingival bacterium Aggregatibacter actinomycetemcomitans has been widely implicated in the etiology of GCP (7). This microorganism is a fastidious, microaerophilic, gram-negative rod that possesses notable virulence factors, including a leukotoxin (LtxA) which has been shown to kill host leukocytes, induce degranulation in neutrophils and activate secretion of the pro-inflammatory cytokine IL-1β by macrophages (8–10). A. actinomycetemcomitans exhibits 2 different leukotoxic phenotypes: minimally and highly leukotoxic strains (11, 12). For instance, the A. actinomycetemcomitans-JP2 genotype is highly leukotoxic, which is attributed to a 530 bp deletion in the promotor region of the ltx gene operon (12). This genotype is found in subgingival samples of African subjects, and subjects with African ancestry, and is strongly associated with progressive loss of periodontal tissues in GCP (13–15). The genotype has recently been detected in non-African populations as well (16, 17).
Technological advances, including the advent of next generation sequencing technologies, have revolutionized our understanding of the oral bacterial communities (bacteriome) and their role in health and disease (18). Current understanding indicates that in periodontitis there is an imbalance in the composition and function of the subgingival bacteriome (dysbiosis), rather than an infection with specific pathogens (19). This premise seems pertinent also for GCP, where the disease, especially its generalized form, has been associated with a complex subgingival microbial signature that resembles that found in adult periodontitis (7, 20–23).
Our understanding of the bacteriome of GCP is still lacking. Particularly, little is known about the bacteriome associated with JP2 genotype-associated disease. A previous pilot study involving 4 Moroccan GCP patients found that 2 of these were JP2-genotype positive, and reported that the JP2 genotype represented only a minor component of a complex subgingival microbiota (24). Longitudinal studies show that carriage of JP2 genotype is a risk factor of onset and progression of periodontitis (13); suggesting that it plays a role in early stages of the disease as a keystone pathogen, but is then overgrown by a dysbiotic microbiome. Therefore, a more detailed knowledge of the microbial environment associated with the JP2-genotype infection seems pertinent to a better understanding of the pathogenesis of this disease.
The aim of this study was to characterize the subgingival bacteriome in shallow and deep pockets in GCP subjects as compared to subjects without periodontitis (controls) with particular emphasis on subjects positive to A. actinomycetemcomitans-JP2 genotype.
Materials and methods
This study was reviewed and approved by the Institutional Review Board (IRB) at Temple University (project #23242) and by the IRB at the University of Hassan II, Casablanca, Morocco. The children and their parents were provided with sufficient information about the study objectives and methods, and a signed consent form confirming voluntary participation in the study was obtained from the subjects or a legally authorized guardian.
Study population
The study subjects were identified in a population survey, designed to study the dental health of school children in Morocco (3). In the survey, middle- and high-schools from different regions of Morocco were selected using a probability sampling design. As part of this survey, 984 adolescents 14–18 years old attending schools in the Moroccan cities of Tiznit, Dakhla, and Tangier were interviewed and underwent a comprehensive periodontal examination during the period 2016–1017. The examination consisted of measurement of the probing depth (PD), gingival recession, and bleeding on probing at six sites per tooth on all fully erupted permanent teeth, excluding third molars. The clinical attachment loss (CAL) was then calculated and used to identify subjects with GCP using established criteria (2, 3).
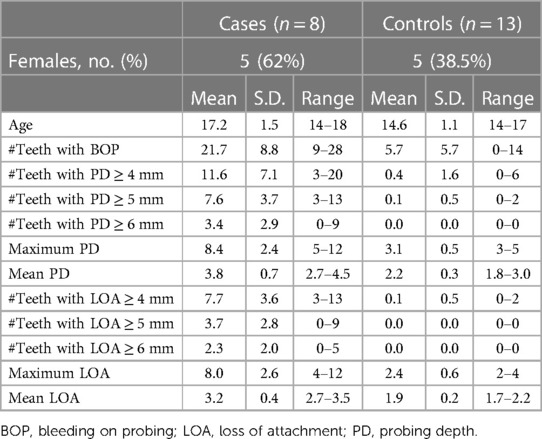
The criteria of inclusion of cases in this study were subjects diagnosed with GCP, tested positive for the A. actinomycetemcomitans JP2 genotype, and who have not had periodontal treatment or used systemic antibiotic during the previous 3 months. In this population, 45 subjects were diagnosed with GCP, and 8 of these fulfilled these criteria and were included in the study. In addition, 13 subjects without periodontitis (controls) were selected randomly from the same population. The clinical characteristics of the two groups are presented in Table 1.
Subgingival plaque sample collection
In the GCP group, subgingival plaque samples were collected from 2 sites with shallow pockets (PD ≤ 4 mm) and 2 sites with deep pockets (PD ≥ 6 mm). In the control group, plaque samples were collected from two sites with PD ≤ 4 mm. The sites were isolated with cotton rolls, and any supragingival plaque was removed; then a sterile paper point was inserted into the depth of the pocket and left in place for 30 s. The paper points from each subject were pooled, separately for shallow and deep pockets in the cases, placed in 1 ml Tris EDTA buffer, and maintained at −20°C until analyzed.
DNA extraction
The samples were vortexed vigorously and pelleted by centrifugation at 13,000 rpm for 10 min. The pellets were each re-suspended in 162 μl PBS with 18 μl Metapolyzme (Sigma, USA), and incubated at 35°C for 4 h. DNA was then extracted using the Invitrogen Purelink Genomic DNA extraction kit (Thermo Fisher Scientific, USA) according to manufacturer’s instructions, quantified using a Qubit 2 Fluorometer (Thermo Fisher Scientific, USA) and stored at −80°C.
Identification and genotyping of A. actinomycetemcomitans
A. actinomycetemcomitans was identified and genotyped by PCR analysis of the ltx operon using the JP2-F3 (5’—TCTATGAAT GGAAACTTGTTCAGA AT−3’) and JP2-R2 (5’—GAA TAA GAT AAC CAA ACC ACA ATA TCC—3’) primers described by Youshida et al (25). While these primers had been originally reported to selectively detect JP2 genotype strains by amplifying a 163-bp fragment, we found they also amplify a 691 bp fragment in non-JP2 genotype strains, which was confirmed by in silico analysis. PCR reactions were performed in Platinum™ II Hot-Start Green PCR Master Mix (Thermo Fisher Scientific, USA), using the following thermal cycling parameters: initial denaturation at 94°C for 2 min, followed by 45 cycles of denaturation at 94°C for 15 s, annealing at 50°C for 15 s, and extension at 68°C for 1 min. Fragment analysis was performed using standard agarose electrophoresis. DNA extracted from A. actinomycetemcomitans non-JP2 strain NCTC 9710 (Public Health England) and JP2 strain ATCC 700685 (American Type Culture Collection, USA) was used as positive control.
16S rRNA sequencing and bioinformatic analysis
Library preparation and sequencing were done at the Australian Centre for Ecogenomics, as described previously (26). Briefly, the degenerate primers 27FYM (27) and 519R (28) were used to prepare indexed libraries of the V1–V3 region of the 16S rRNA gene that were subsequently sequenced on an Illumina Miseq platform using 2*300 bp chemistry.
Paired reads were merged with PEAR (29) and further processed, including quality-filtration, alignment and chimera check, using mothur software package (30) as previously described. The high-quality, non-chimeric sequences were classified to the species-level employing our BLASTN-based algorithm (31). Species represented by less than 100 reads across samples were filtered out. QIIME (Quantitative Insights Into Microbial Ecology) (32) was used for generation of taxonomy plots and calculation of species richness and alpha diversity. Taxonomic read counts were centered log-ratio (CLR) transformed and used for downstream analyses: (1) beta diversity analysis, namely principal component analysis (PCA) and Permutational Multivariate Analysis of Variance (PERMANOVA) using the R packages Vegan, Phyloseq (33) and Microbiome (34); and (2) differential abundance analysis using Microbiome Multivariable Associations with Linear Models (MaAsLin2) (35). Subgingival Microbial Dysbiosis Index (SMDI) as a summary statistic of microbial profiles and measure of dysbiosis was calculated from CLR transformed data as previously described (36). Selected significant results were plotted using ggplot2 package in R.
Results
16S sequencing data pre-processing statistics
A total of 1,588,128 paired sequences were obtained (49,629 ± 31472 reads per sample). On average, 88.32% of reads were successfully merged with PEAR and ∼45% were retained after quality filtration and chimera removal steps. Around 90% of the high-quality merged reads were successfully classified to species level. Detailed statistics for preprocessing of each sample is provided in Supplementary File S1. Raw sequences were submitted to Sequence Read Archive (SRA) under project # PRJNA1028864.
A. actinomycetemcomitans infection and genotype status
In the GCP group, 5 subjects (62.5%) were positive for the JP2 genotype but negative for the non-JP2 genotype, while 3 subjects (37.5%) were positive for both the JP2 and non-JP2 genotypes. In the control group, 1 subject (7.7%) was positive for both genotypes; 5 subjects (38.5%) were negative for both strains; 5 subjects (38.5%) were positive for the non-JP2 genotype and negative for the JP2 genotype; and 2 subjects (15.4%) were JP2 genotype positive and non-JP2 genotype negative.
Distinct average subgingival bacteriome profile identified in GCP
A total of 10 bacterial phyla, 109 genera and 308 species were identified in the samples, with a mean of 8.79 ± 1.13 phyla, 60.32 ± 12.96 genera and 152.32 ± 35.13 species per pooled sample. The relative abundances and detection frequencies of the identified phyla, genera and species per sample and per group are provided in Supplementary Files S2–S4. The average relative taxonomic profiles of each of the study groups are presented in Figure 1. While Proteobacteria, Firmicutes, Bacteroidetes and Fusobacteria were overall the most abundant phyla, accounting for >80% of the average bacteriome, their relative abundances substantially varied between the groups with Proteobacteria being the most abundant in GCP (followed by Bacteroidetes and Firmicutes in the deep and shallow pockets, respectively), and Firmicutes followed by Fusobacteria in the controls.
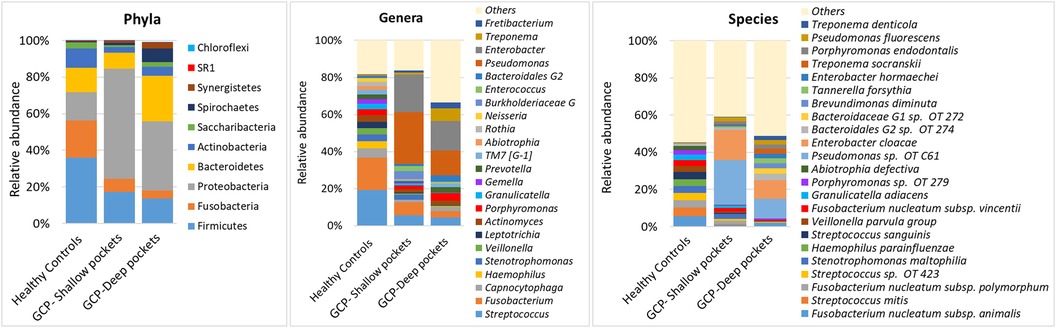
Figure 1. Average microbial profiles. DNA extracted from subgingival plaque samples was sequenced for the V1–V3 region of the 16S rRNA gene using 2 × 300 bp chemistry. The resultant sequences were merged, quality-filtered and classified to the species level using a BLASTn-based algorithm. The stacked bars show the average relative abundances of all phyla, top genera and top species identified in the study groups.
Differences were more evident at the genus and species levels. Streptococcus was the most abundant genus in the controls, followed by Fusobacterium, Capnocytophaga, Haemophilus, Stenotrophomonas, Veillonella, and Leptotrichia, together constituting on average 56% of the microflora. In the GCP group, Enterobacter and Pseudomonas were the most abundant genera accounting alone for 48% and 29% of the average bacteriome in the shallow and deep pockets, respectively, followed by Treponema, Streptococcus, Porphyromonas, Fusobacterium, Bacteroidales genus 2, and Fretibacterium. At the species level, Fusobacterium nucleatum was the most abundant in the controls, followed by S. mitis, Streptococcus oral taxon 423, Stenotrophomonas maltophilia, Heamophilus parainfluenzae, Streptococcus sanguinis, Veilonella parvula group, and Granulicatella adiacens. In GCP, the most abundant species were Pseudomonas oral taxon C61 and Enterobacter cloacae, constituting together about 40% and 20% on average in the shallow and deep pockets, respectively, followed by Bacteroidales oral taxa 274 & 272, Brevundimonas diminuta, Tannerella forsythia, Enterobacter hormaechei, Treponema socranskii, Porphyromonas endodontalis, Pseudomonas fluorescens and Treponema denticola.
GCP associated with altered microbial diversity but not dispersion
There were no differences between the groups in terms of species richness (Figure 2A), but both the shallow and deep pockets showed reduced alpha diversity (Shannon index) compared to the controls, although the difference was only statistically significant for the shallow pockets (Figure 2B). Beta-diversity analysis (PCA and PERMANOVA) revealed significant differences among the groups (Figure 2C), with the controls forming a separate cluster from the shallow and deep pockets (P = 0.001 for both contrasts). There was some separation between the shallow and deep pockets, but the difference did not reach statistical significance (P = 0.09). On the other hand, applying Permutational Multivariate Analysis of Dispersion (PERMDISP) to data identified no significant differences between the groups.
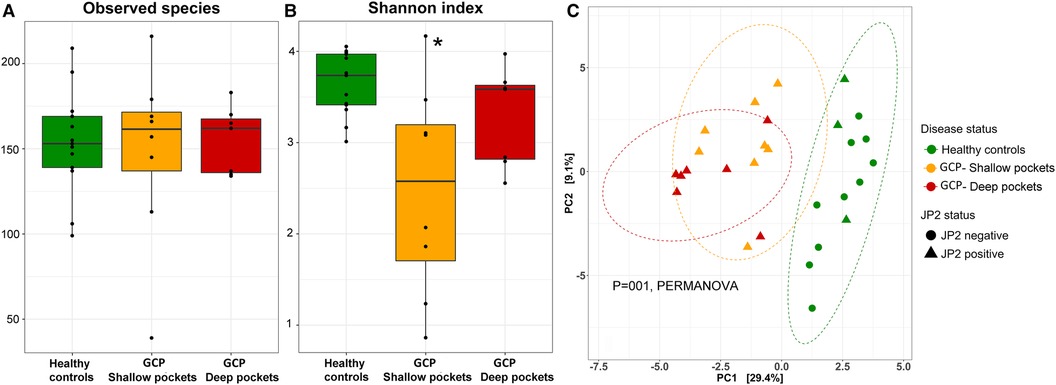
Figure 2. Species richness and diversity. Species count table were rarified and used to calculate observed species richness (A), and alpha diversity [Shannon index, (B)]. The data were then centered log-ratio transformed and used for beta-diversity analysis based on Principle Component Analysis [PCA plot, (C)]. Significance of differences between the groups for (A) and (B) were sought using pairwise Mann-Whitney or Wilcoxon signed rank test as appropriate; *P ≤ 0.05. Significance of differences between the clusters in (C) were assessed with PERMANOVA.
Classical periodontal pathogens and non-conventional taxa enriched in GCP
Analysis with MaAsLin2 identified 46 and 86 differentially abundant genera and species, respectively, between the controls and shallow pockets, and 49 and 127 differentially abundant genera and species, respectively, between the controls and deep pockets at a false discovery rate (FDR) of 0.1 (Supplementary Files S5, S6). The top 20 differentially abundant genera in each contrast are presented in Supplementary Figure S1, and the corresponding top 40 differentially abundant species in each comparison are shown in Figure 3. Compared to both GCP subgroups, the controls had significantly higher abundance of known health-associated bacterial species including H. parainfluenzae, S. mitis, Lautrophia mirabilis, S. sanguinis, V. parvula group, Rothia mucilaginosa, Rothia aeria, Kingella oralis, Streptococcus gordonii and Gemella haemolysans. In contrast, known or putative periodontal pathogens were enriched in both the shallow and deep pockets of GCP compared to the controls including T. forsythia, Treponema socranskii, P. endodontalis, T. denticola, P. gingivalis and Fretibacterium spp. However, several non-conventional species were also abundant, and exclusively found, in the GCP samples including Pseudomonas oral taxon C61, E. Cloacae, Enterobacter cancerogenus and Acinetobacter oral taxon C13. Relative abundances of selected species identified as significantly associated with GCP are depicted in Figure 4.
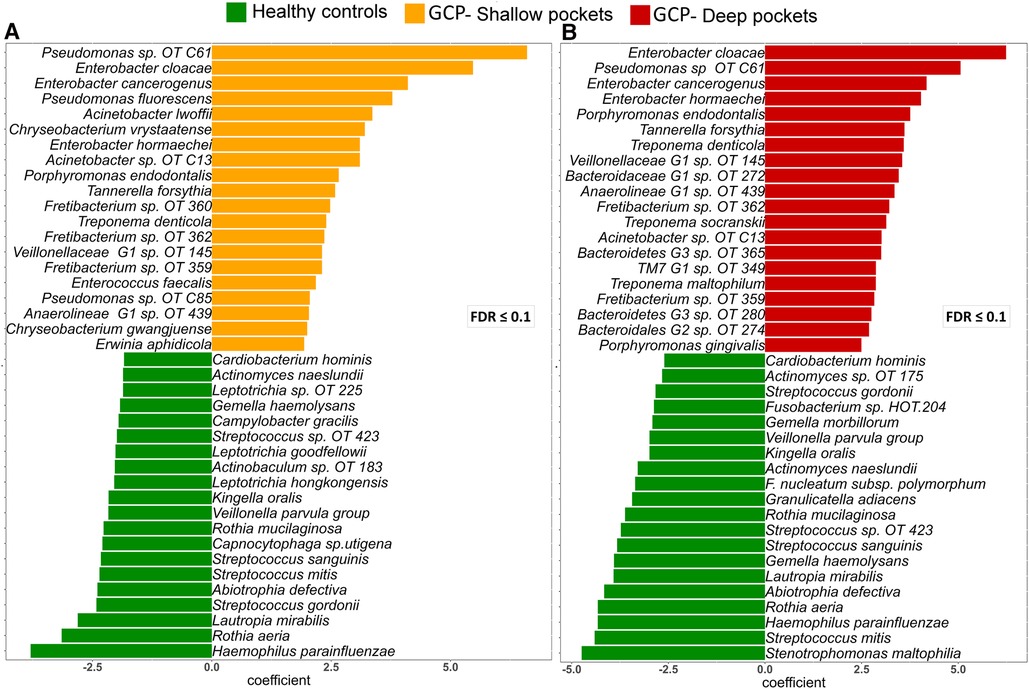
Figure 3. Top differentially abundant species between GCP and health. Centered log ratio (CLR) transformed data were analyzed with MaAsLin2 to identify differentially abundant species between the controls and shallow pockets from the GCP cases (A) and between the controls and deep pockets from the GCP cases (B) Differences with FDR ≤ 0.1 were considered significant. The top 40 species based on magnitude of association (coefficient) are shown in (A) and (B).
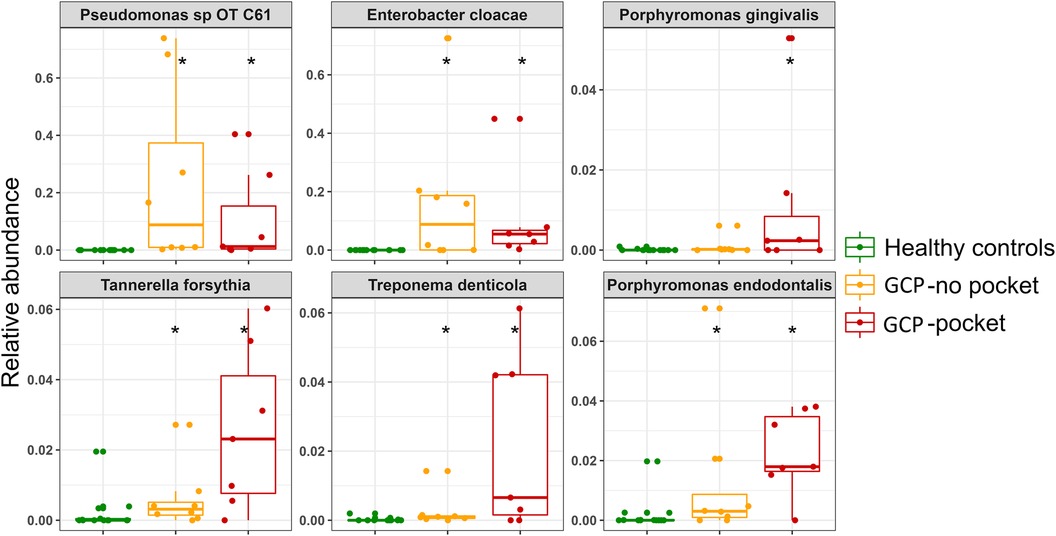
Figure 4. Relative abundances of selected species. Combined box and strip plots depicting the relative abundances of selected known periodontal pathogens as well as non-conventional species that were identified as significantly different between GCP cases and control subjects. *Statistically significant difference based on MaAsLin2 analysis; FDR ≤ 0.1.
No differentially abundant taxa between the shallow and deep pockets were found at an FDR ≤ 0.1. However, using a less stringent cutoff (FDR of 0.25 and P-value of 0.01, for genus and species-level comparisons, respectively) identified a few differences, mainly genus Treponema in association with the deep pockets and genera Gemella, Rothia, and Ganulicatella in addition to S. mitis and the non-conventional species Stenotrophomonas maltophilia in association with the shallow pockets (Supplementary Figure S2).
SMDI discriminates between GCP and health with high accuracy
The level of subgingival microbial dysbiosis, measured in terms of our recently developed SMDI, differed significantly in pairwise comparison between the three groups, being highest in the deep pocket and lowest in the controls (Figure 5). Using an empirical SMDI value of 0 as diagnostic cutoff, discriminated between healthy sites and deep pockets with an accuracy of 95% and between healthy sites and shallow pockets with an accuracy of 81%. Using deep pocket data only, GCP cases were diagnosed with sensitivity of 85.7% and a specificity of 100% (95% accuracy).
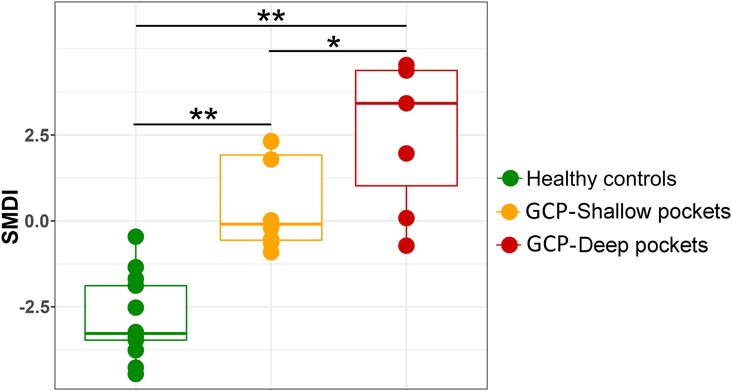
Figure 5. Subgingival microbial dysbiosis in GCP compared to health. Centered log ratio (CLR) transformed data were used to calculate SMDI for each sample as previously described (36) and presented as combined box and strip plot for each group. Significance of differences between the groups were sought using pairwise Mann-Whitney or Wilcoxon signed rank test as appropriate; *P ≤ 0.05; **P ≤ 0.01.
Discussion
Several studies have been conducted to explore the bacteriome of GCP (7, 20–23). However, to the best of our knowledge, the current study is the first attempt to focus on characterizing the bacteriome associated with JP2 genotype-associated GCP. This study included analysis of subgingival plaque samples collected from shallow and deep pockets of 8, previously untreated JP2 genotype-positive GCP cases and from gingival crevices of 13 controls using 16S rRNA gene sequencing. By applying a robust bioinformatic analysis approach to the data, we identified profound and important differences in the microbial community composition between the shallow and deep pockets in the GCP subjects, and between the GCP group and the controls.
Beta-diversity analysis (PCA plot) revealed complete separation between the GCP and controls, which has been previously demonstrated (20, 23). However, there were no differences in beta-dispersion between the 2 groups; i.e., the variation in bacteriome composition among the GCP cases was not different from that among the controls. Therefore, the GCP-associated bacteriome in this study did not follow the Anna Karenina principle (37), which is inconsistent with the findings in a recent study by Altabtbaei et al (20). Whether or not this is specific to JP2 genotype-associated GCP bacteriome entails further investigation. Interestingly, 3 subjects in the control group tested positive for the JP2 genotype, but still clustered with JP2-genotype negative samples. One possible explanation is that these subjects are yet to develop GCP since the average age of the controls was somewhat lower than that of the GCP cases. Alternatively, it may be that bacteriome dysbiosis is a prerequisite for JP2 genotype activity. This is an interesting hypothesis that warrants further exploration.
Previous studies have shown that GCP, particularly the generalized form, is associated with a dysbiotic subgingival bacteriome that shares similarities with that found in association with chronic periodontitis. Consistently, we found that several established and emerging periodontal pathogens are significantly more abundant in both shallow and deep probing sites in GCP compared to controls, including members of the “red complex” (P. gingivalis, T. forsythia, and T. denticola), P. endodontalis, Fretibacterium spp. and TM7 spp. Furthermore, applying our recently reported subgingival microbial dysbiosis index (SMDI) to the present data successfully discriminated between the GCP cases and controls with high accuracy (95%). SMDI was developed using a machine learning approach applied to data sets from studies on the bacteriome of chronic periodontitis, providing a diagnostic accuracy of 92%–96%. This striking similarity in the diagnostic accuracy of SMDI between chronic periodontitis and JP2 genotype-associated GCP, suggests that the two diseases share the features of subgingival microbial dysbiosis.
It may be hypothesized that GCP is a consequence of some kind of interactions between the JP2 genotype and subgingival bacteriome dysbiosis. One conceivable mechanism is that the JP2 genotype act as a keystone pathogen that subverts the immune response by its leukotoxicity effect that could disrupts microbial hemostasis, that in turn orchestrates a destructive immune response, consistent with the polymicrobial synergy and dysbiosis hypothesis (38). Indeed, in this study A. actinomycetemcomitans had a very low abundance in the subgingival plaque, which is an important feature of keystone pathogens. Another possibility is that the activity of the JP2 genotype is modulated by the bacteriome: in the presence of microbial dysbiosis, the JP2 genotype becomes more virulent, resulting in rapidly progressive form of periodontitis.
A novel finding in this study is the detection of non-oral gram negative facultatively anaerobic rods, including Pseudomonas, Enterobacter and Acinetobacter spp. in both shallow and deep pockets of the GCP cases. Pseudomonas oral taxon C61 and E. Cloacae together accounted for an average of 30% of the subgingival bacteriome of the GCP subjects. Species belonging to these same genera have been previously isolated from pockets of both chronic and GCP but not in such high abundance as in this material (39–41). It is not likely that detecting these species in the samples was due to, for example contamination during collection or processing of the samples, because these species were not detected in the control samples. However, since this study design is cross-sectional, it is not possible to infer on the role of these species in the pathogenesis of GCP in the present subjects. Pseudomonas oral taxon C61 is closely related to Pseudomonas fluorescens which, as well as E. Cloacae, is a ubiquitous species found in the environment and as a commensal organism in humans, however, in immunocompromised patients, they result in various opportunistic infections (42, 43). It is also notable that enteric rods have been shown previously to correlate with the level of A. actinomycetemcomitans and the severity of periodontal disease (40, 41). Therefore, it is possible that they play an opportunistic role in JP2 genotype-associated GCP.
In conclusion, in JP2 genotype-associated GCP, we identified a dysbiotic subgingival bacteriome that is somewhat similar to the bacteriome associated with chronic periodontitis. SMDI, which was originally developed for chronic periodontitis, characterized GCP cases with high accuracy. The interplay between subgingival dysbiosis and JP2 genotype infection, as well as the potential role of non-oral gram-negative rods in the pathogenesis of GCP warrants further investigation.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA1028864.
Ethics statement
The studies involving humans were approved by IRB at the University of Hassan II, Casablanca, Morocco. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
VM: Conceptualization, Methodology, Writing – original draft, Investigation. JK: Conceptualization, Resources, Writing – review and editing, Investigation. DB: Methodology, Data curation, Formal analysis, Investigation, Project administration, Visualization, Writing – original draft. AG: Investigation, Resources, Writing – review and editing. TC: Data curation, Formal analysis, Resources, Software, Writing – review and editing. NA-H: Conceptualization, Formal analysis, Funding acquisition, Methodology, Project administration, Resources, Supervision, Writing – original draft. JA: Conceptualization, Funding acquisition, Methodology, Resources, Supervision, Writing – review and editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article.
This work was supported by Graduate Student Research Grant from the Temple University Kornberg School of Dentistry, Philadelphia, USA.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/froh.2023.1288499/full#supplementary-material
References
1. Van Dyke TE, Serhan CN. Resolution of inflammation: a new paradigm for the pathogenesis of periodontal diseases. J Dent Res. (2003) 82(2):82–90. doi: 10.1177/154405910308200202
2. Caton JG, Armitage G, Berglundh T, Chapple ILC, Jepsen S, Kornman KS, et al. A new classification scheme for periodontal and peri-implant diseases and conditions—introduction and key changes from the 1999 classification. J Periodontol. (2018) 89(Suppl 1):S1–8. doi: 10.1111/jcpe.12935
3. Albandar JM. Aggressive periodontitis: case definition and diagnostic criteria. Periodontol 2000. (2014) 65(1):13–26. doi: 10.1111/prd.12014
4. Henderson B, Kaiser F. Bacterial modulators of bone remodeling in the periodontal pocket. Periodontol 2000. (2018) 76(1):97–108. doi: 10.1111/prd.12160
5. Ebersole JL, Dawson D 3rd, Emecen-Huja P, Nagarajan R, Howard K, Grady ME, et al. The periodontal war: microbes and immunity. Periodontol 2000. (2017) 75(1):52–115. doi: 10.1111/prd.12222
6. Albandar JM, Rams TE. Risk factors for periodontitis in children and young persons. Periodontol 2000. (2002) 29:207–22. doi: 10.1034/j.1600-0757.2002.290110.x
7. Könönen E, Müller HP. Microbiology of aggressive periodontitis. Periodontol 2000. (2014) 65(1):46–78. doi: 10.1111/prd.12016
8. Åberg CH, Kelk P, Johansson A. Aggregatibacter actinomycetemcomitans: virulence of its leukotoxin and association with aggressive periodontitis. Virulence. (2015) 6(3):188–95. doi: 10.4161/21505594.2014.982428
9. Herbert BA, Novince CM, Kirkwood KL. Aggregatibacter actinomycetemcomitans, a potent immunoregulator of the periodontal host defense system and alveolar bone homeostasis. Mol Oral Microbiol. (2016) 31(3):207–27. doi: 10.1111/omi.12119
10. Johansson A. Aggregatibacter actinomycetemcomitans leukotoxin: a powerful tool with capacity to cause imbalance in the host inflammatory response. Toxins (Basel). (2011) 3(3):242–59. doi: 10.3390/toxins3030242
11. Kolodrubetz D, Spitznagel J Jr, Wang B, Phillips LH, Jacobs C, Kraig E. cis elements and trans factors are both important in strain-specific regulation of the leukotoxin gene in Actinobacillus actinomycetemcomitans. Infect Immun. (1996) 64(9):3451–60. doi: 10.1128/iai.64.9.3451-3460.1996
12. Brogan JM, Lally ET, Poulsen K, Kilian M, Demuth DR. Regulation of Actinobacillus actinomycetemcomitans leukotoxin expression: analysis of the promoter regions of leukotoxic and minimally leukotoxic strains. Infect Immun. (1994) 62(2):501–8. doi: 10.1128/iai.62.2.501-508.1994
13. Haubek D, Ennibi OK, Poulsen K, Vaeth M, Poulsen S, Kilian M. Risk of aggressive periodontitis in adolescent carriers of the JP2 clone of Aggregatibacter (Actinobacillus) actinomycetemcomitans in Morocco: a prospective longitudinal cohort study. Lancet (London, England). (2008) 371(9608):237–42. doi: 10.1016/S0140-6736(08)60135-X
14. Åberg C H, Haubek D, Kwamin F, Johansson A, Claesson R. Leukotoxic activity of Aggregatibacter actinomycetemcomitans and periodontal attachment loss. PloS One. (2014) 9(8):e104095. doi: 10.1371/journal.pone.0104095
15. Haubek D, Johansson A. Pathogenicity of the highly leukotoxic JP2 clone of Aggregatibacter actinomycetemcomitans and its geographic dissemination and role in aggressive periodontitis. J Oral Microbiol. (2014) 6:jom.v6.23980. doi: 10.3402/jom.v6.23980
16. Stahli A, Sculean A, Eick S. JP2 genotype of Aggregatibacter actinomycetemcomitans in Caucasian patients: a presentation of two cases. Pathogens. (2020) 9(3):178. doi: 10.3390/pathogens9030178
17. Claesson R, Hoglund-Aberg C, Haubek D, Johansson A. Age-related prevalence and characteristics of Aggregatibacter actinomycetemcomitans in periodontitis patients living in Sweden. J Oral Microbiol. (2017) 9(1):1334504. doi: 10.1080/20002297.2017.1334504
18. Kilian M, Chapple IL, Hannig M, Marsh PD, Meuric V, Pedersen AM, et al. The oral microbiome—an update for oral healthcare professionals. Br Dent J. (2016) 221(10):657–66. doi: 10.1038/sj.bdj.2016.865
19. Curtis MA, Diaz PI, Van Dyke TE. The role of the microbiota in periodontal disease. Periodontol 2000. (2020) 83(1):14–25. doi: 10.1111/prd.12296
20. Altabtbaei K, Maney P, Ganesan SM, Dabdoub SM, Nagaraja HN, Kumar PS. Anna karenina and the subgingival microbiome associated with periodontitis. Microbiome. (2021) 9(1):97. doi: 10.1186/s40168-021-01056-3
21. Faveri M, Mayer MP, Feres M, de Figueiredo LC, Dewhirst FE, Paster BJ. Microbiological diversity of generalized aggressive periodontitis by 16S rRNA clonal analysis. Oral Microbiol Immunol. (2008) 23(2):112–8. doi: 10.1111/j.1399-302X.2007.00397.x
22. Li Y, Feng X, Xu L, Zhang L, Lu R, Shi D, et al. Oral microbiome in Chinese patients with aggressive periodontitis and their family members. J Clin Periodontol. (2015) 42(11):1015–23. doi: 10.1111/jcpe.12463
23. Schulz S, Porsch M, Grosse I, Hoffmann K, Schaller HG, Reichert S. Comparison of the oral microbiome of patients with generalized aggressive periodontitis and periodontitis-free subjects. Arch Oral Biol. (2019) 99:169–76. doi: 10.1016/j.archoralbio.2019.01.015
24. Rylev M, Bek-Thomsen M, Reinholdt J, Ennibi OK, Kilian M. Microbiological and immunological characteristics of young Moroccan patients with aggressive periodontitis with and without detectable Aggregatibacter actinomycetemcomitans JP2 infection. Mol Oral Microbiol. (2011) 26(1):35–51. doi: 10.1111/j.2041-1014.2010.00593.x
25. Yoshida A, Ennibi OK, Miyazaki H, Hoshino T, Hayashida H, Nishihara T, et al. Quantitative discrimination of Aggregatibacter actinomycetemcomitans highly leukotoxic JP2 clone from non-JP2 clones in diagnosis of aggressive periodontitis. BMC Infect Dis. (2012) 12:253. doi: 10.1186/1471-2334-12-253
26. Al-Hebshi NN, Nasher AT, Maryoud MY, Homeida HE, Chen T, Idris AM, et al. Inflammatory bacteriome featuring Fusobacterium nucleatum and Pseudomonas aeruginosa identified in association with oral squamous cell carcinoma. Sci Rep. (2017) 7(1):1834. doi: 10.1038/s41598-017-02079-3
27. Frank JA, Reich CI, Sharma S, Weisbaum JS, Wilson BA, Olsen GJ. Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl Environ Microbiol. (2008) 74(8):2461–70. doi: 10.1128/AEM.02272-07
28. Lane DJ, Pace B, Olsen GJ, Stahl DA, Sogin ML, Pace NR. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci U S A. (1985) 82(20):6955–9. doi: 10.1073/pnas.82.20.6955
29. Zhang J, Kobert K, Flouri T, Stamatakis A. PEAR: a fast and accurate illumina paired-End reAd mergeR. Bioinformatics. (2014) 30(5):614–20. doi: 10.1093/bioinformatics/btt593
30. Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. (2009) 75(23):7537–41. doi: 10.1128/AEM.01541-09
31. Al-Hebshi NN, Nasher AT, Idris AM, Chen T. Robust species taxonomy assignment algorithm for 16S rRNA NGS reads: application to oral carcinoma samples. J Oral Microbiol. (2015) 7:28934. doi: 10.3402/jom.v7.28934
32. Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. (2010) 7(5):335–6. doi: 10.1038/nmeth.f.303
33. McMurdie PJ, Holmes S. Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PloS One. (2013) 8(4):e61217. doi: 10.1371/journal.pone.0061217
34. Tools for microbiome analysis in R [Internet]. (2017). Available at: https://github.com/microbiome/microbiome/
35. Mallick H, Rahnavard A, McIver LJ, Ma S, Zhang Y, Nguyen LH, et al. Multivariable association discovery in population-scale meta-omics studies. PLoS Comput Biol. (2021) 17(11):e1009442. doi: 10.1371/journal.pcbi.1009442
36. Chen T, Marsh PD, Al-Hebshi NN. SMDI: an index for measuring subgingival microbial dysbiosis. J Dent Res. (2022) 101(3):331–8. doi: 10.1177/00220345211035775
37. Zaneveld JR, McMinds R, Vega Thurber R. Stress and stability: applying the Anna karenina principle to animal microbiomes. Nat Microbiol. (2017) 2:17121. doi: 10.1038/nmicrobiol.2017.121
38. Hajishengallis G, Lamont RJ. Beyond the red complex and into more complexity: the polymicrobial synergy and dysbiosis (PSD) model of periodontal disease etiology. Mol Oral Microbiol. (2012) 27(6):409–19. doi: 10.1111/j.2041-1014.2012.00663.x
39. Slots J, Feik D, Rams TE. Prevalence and antimicrobial susceptibility of enterobacteriaceae, pseudomonadaceae and acinetobacter in human periodontitis. Oral Microbiol Immunol. (1990) 5(3):149–54. doi: 10.1111/j.1399-302X.1990.tb00413.x
40. da Silva-Boghossian CM, do Souto RM, Luiz RR, Colombo AP. Association of red complex, A. actinomycetemcomitans and non-oral bacteria with periodontal diseases. Arch Oral Biol. (2011) 56(9):899–906. doi: 10.1016/j.archoralbio.2011.02.009
41. Ardila CM, Alzate J, Guzman IC. Relationship between gram negative enteric rods, Aggregatibacter actinomycetemcomitans, and clinical parameters in periodontal disease. J Indian Soc Periodontol. (2012) 16(1):65–9. doi: 10.4103/0972-124X.94607
42. Scales BS, Dickson RP, LiPuma JJ, Huffnagle GB. Microbiology, genomics, and clinical significance of the pseudomonas fluorescens species complex, an unappreciated colonizer of humans. Clin Microbiol Rev. (2014) 27(4):927–48. doi: 10.1128/CMR.00044-14
Keywords: biofilm, dysbiosis, high-throughput nucleotide sequencing, microbiota, periodontitis
Citation: Molli VLP, Kissa J, Baraniya D, Gharibi A, Chen T, Al-Hebshi NN and Albandar JM (2023) Bacteriome analysis of Aggregatibacter actinomycetemcomitans-JP2 genotype-associated Grade C periodontitis in Moroccan adolescents. Front. Oral. Health 4:1288499. doi: 10.3389/froh.2023.1288499
Received: 5 September 2023; Accepted: 23 October 2023;
Published: 14 November 2023.
Edited by:
Sigrun Eick, University of Bern, SwitzerlandReviewed by:
Anders Johansson, Umeå University, SwedenAhmed Hashim, King Faisal University, Saudi Arabia
© 2023 Molli, Kissa, Baraniya, Gharibi, Chen, Al-Hebshi and Albandar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nezar N. Al-Hebshi YWxoZWJzaGlAdGVtcGxlLmVkdQ==
†These authors have contributed equally to this work
 Vijaya Lakshmi Pavani Molli
Vijaya Lakshmi Pavani Molli Jamila Kissa2
Jamila Kissa2 Divyashri Baraniya
Divyashri Baraniya Tsute Chen
Tsute Chen Nezar N. Al-Hebshi
Nezar N. Al-Hebshi