- Telos-Philosophische Praxis, Buermoos, Austria
In searching for life in extraterrestrial space, it is essential to act based on an unequivocal definition of life. In the twentieth century, life was defined as cells that self-replicate, metabolize, and are open for mutations, without which genetic information would remain unchangeable, and evolution would be impossible. Current definitions of life derive from statistical mechanics, physics, and chemistry of the twentieth century in which life is considered to function machine like, ignoring a central role of communication. Recent observations show that context-dependent meaningful communication and network formation (and control) are central to all life forms. Evolutionary relevant new nucleotide sequences now appear to have originated from social agents such as viruses, their parasitic relatives, and related RNA networks, not from errors. By applying the known features of natural languages and communication, a new twenty-first century definition of life can be reached in which communicative interactions are central to all processes of life. A new definition of life must integrate the current empirical knowledge about interactions between cells, viruses, and RNA networks to provide a better explanatory power than the twentieth century narrative.
Introduction
Scientifically, the first half of the twentieth century was the most successful period for empirically based sciences. Basically, explorations in physics and chemistry paved a path on which science could delimitate validity claims against all other concepts of thoughts such as the broad range of philosophical disciplines, theology, and poetry. Philosophers and physicists such as Wittgenstein, Carnap, Goedel, Russell, and Tarski suggested that exact sciences are strictly based on exact scientific sentences describing empirical facts coherent with observations and measurements in experimental setups (Wittgenstein, 1922; Carnap, 1931, 1939; Gödel, 1931).
The formal language to describe this was mathematical equations that would depict material reality. Information theory and cybernetic systems theory encouraged this progress (Bertalanffy, 1940; Wiener, 1948; Shannon and Weaver, 1949; Turing, 1950; Neumann, 1966). Milestone publication, “Principia Mathematica,” outlined by Bertrand Russel and Alfred North Whitehead was further developed by David Hilberts axiomatic system with error-free logical sentences (Whitehead and Russell, 1910/1912/1913; Hilbert and Bernays, 1934/1939). This exact scientific language was applied to nearly all disciplines of scientific investigations in natural sciences as well as social sciences.
Molecular biology, genetics, and biochemistry started their success stories, which have lasted until today. The role of physicalism in biology was so dominant that biology became a subdiscipline of physics and chemistry (Brenner, 2012). Because biological organisms, cells, tissues, and organs consist of molecules, constructed out of atoms, empirical and measurable features may be described by physics and chemistry. The genetic information storing molecules represent “aperiodic crystal structures” as assumed by Erwin Schroedinger.
The Physicalistic Paradigm in the Biology of the Twentieth Century
“We shall assume the structure of a gene to be that of a huge molecule, capable only of discontinuous change, which consists of a rearrangement of the atoms and leads to an isomeric molecule. The rearrangement may affect only a small region of the gene, and a vast number of different rearrangements may be possible. The energy thresholds, separating the actual configuration from any possible isomeric ones, have to be high enough (compared with the average heat energy of an atom) to make the change-over a rare event. These rare events we shall identify with spontaneous mutations” (Schroedinger, 1944).
These spontaneous mutations occur strictly by chance and are statistically determined, according to natural laws. Evolutionarily relevant changes in molecular structures occur if, in replication processes. The replicated molecular structure is not an identical copy of the former sequence structures but a variation that is not identical with the master blueprint. This variation is a result of a replication error. Erwin Schroedinger introduced the “code-script” to denote that genetic information is a natural code. With the rise of molecular biology, and genetics, he adapted the code-metaphor to describe various features of the genetic code as the result of a molecular ensemble of nucleotides that underlies statistical fluctuations. These are the consequences out of thermodynamics of living systems. Schroedinger's book, “What is Life?” and his suggestion therein, “Life is physics and chemistry,” became one of the most influential works of the twentieth century.
A big step forward in this direction occurred, when Manfred Eigen introduced information theory to molecular biology and adapted information as a molecular property of matter, which may reproduce itself (Eigen, 1971). Eigen's quasispecies theory and its core assumption that mutation caused the diversity of RNA populations dominated paradigmatically nearly half a century (Biebricher and Eigen, 2006). It represents the “exploitation of the formal mathematical analogy of quasispecies dynamics and the statistical mechanics” of quantum theory (Domingo and Schuster, 2016). “Biological selection can be viewed as condensation or localization of sequence distribution in a limited area in sequence space” (Biebricher et al., 1985). That finally should lead to a theory of evolution based upon “biochemical kinetics” (Schuster, 2011).
Eigen follows the basics of information theory as a mathematical theory of communication that is quantifiable and underlies natural laws strictly. And he insisted that the genetic code is a natural language and not a metaphor (Eigen and Winkler, 1983).
All these assumptions are driven by the general agreements of natural scientists at that time: how to discuss matter, natural laws, and biological affairs. To summarize, the physicalistic paradigm in biology that dominated the language of observation as well as the language of theory in biology until today are as follows:
• life is physics and chemistry (and information)
• information is a characteristic of matter
• difference between abiotic matter and biology is gradual
• natural languages and codes are determined by their syntax structure
• syntax structure of natural codes represent the logical structure of matter
• mathematics is the only language that can exactly depict the logical structure of matter
• evolution is variation (mutation) and selection
• mutations are error replications that result out of elementary physical processes and indeterminate due to their quantum-mechanical nature.
What We Know Today About Biological Processes
We may analyze the available parts of a machine to get a functional blueprint for its construction. Then, we may reproduce it trying to optimize the construction. This is an important motif in genetic engineering also. However, if we observe living organisms and their interactional patterns, beginning from single cells to tissues, organs, and complex organisms, we find a variety of non-mechanistic circumstances, features, and capabilities that cannot be part of a mechanistic explanation. These features include common coordination to adapt to new and unexpected environmental circumstances. Machines cannot create new programs out of functional blueprints (Witzany, 1995).
The study of living organisms as machines presupposes biological information as a result of coded content according to principles of biochemical kinetics. Biolinguistics, bioinformatics, information theory, systems theory of (context-free) languages, or similar mathematical theories of language and communication cannot explain the essential features of natural languages and the codes used in communication processes. They investigate quantifiable sets of signs by mathematical procedures as statistical mechanics and, therefore, completely forget the essential agents in real life world being necessary to use languages and codes (Witzany, 1995, 2000, 2010). The formalizable sender–receiver model that was used to describe natural communication processes cannot identify the context dependence of meaning and its deep grammar that may represent different, and even contradictory, meanings to the superficial grammar that is present in identical sign sequences (Austin, 1975; Searle, 1976; Habermas, 1987). The social character of real-life organisms is not within their expertise because social-interacting organisms do not behave like formalizable abiotic elements (Witzany and Baluska, 2012a). Social-interacting living organisms may generate new sign sequences, behavior, and interactional motifs, for which no algorithm is available in principle.
In biology, in contrast to physics, entanglement with language and communication is double-sided. Biological disciplines in the twentieth century were convinced that, if they use the language of physics, they would be accepted as part of the exact science community of natural sciences (Chomsky, 1965). On the other side, the explanatory models to coherently describe genetic information used physics and mathematics to explain the features of DNA sequences as physical properties (Stadler and Schuster, 1992).
Let us remember dominant terms in molecular biology and genetics in the second half of the last century; they include “genetic code,” “code without commas,” “genetic information,” “gene expression,” “transcription,” “translation,” “nucleotide sequences,” “messenger RNA,” “protein-coding sequences,” “cell-to-cell communication,” “open reading frame,” “immune response,” “recognition sites,” and so on. From a philosophy of science perspective, these terms could not be substantiated by the biological disciplines themselves, because they all were defined by physicochemical features, which clearly failed to coherently justify original linguistic terms (Witzany, 1995, 2005).
The search is, thus, for how to combine linguistic terms used in biology with current knowledge about natural languages/codes and communication without entering the “mouse-trap” of physicalism, which leads back to a view that living organisms are mechanistic cause and reaction machines (Witzany, 2017a). If we want to use linguistic terms without their physicalistic paradigm, we must be aware of what natural languages/codes used in communication processes mean, if we study biological processes.
What Do We Know About Natural Languages/Codes and Communication?
If we now consider the basic foundations in the language science and communication theory, we may also find the basic functions and processes in which these terms are appropriate descriptions (Morris, 1946; Wittgenstein, 1953; Austin, 1975; Searle, 1976; Habermas, 1994).
Current empirical facts about communication indicate that communication involves interaction processes between living agents mediated by signs in contrast to interactions in abiotic matter where no signs are present (Witzany, 2011a; Baluška and Witzany, 2014). No signs are present if water freezes to ice.
All living agents that communicate share real-life histories and traditions, and environmental conditions. Experiences and the organization and coordination of everyday life practice dominate communication patterns primarily (Witzany, 2014a, 2015). This everyday life practice is the original source of natural languages.
Communicative interactions need some natural language or code that consists of signs. We may differentiate three kinds of signs used in communicative interactions by competent sign users. Signs may be indices, icons, or symbols. These three kinds of signs found in natural languages or codes are used according to three levels of rules (syntax, semantics, pragmatics). Since Charles Morris, we know that if one level is missing, one cannot speak seriously about a real natural language or code (Morris, 1946). Syntactic rules guarantee the correct generation and combination of signs to sign sequences; semantic rules guarantee the correct combination of signs and signified objects, and pragmatic rules are relevant for the correct combination of signs and the concrete context in which signs are used by a real sign-using agent. Rule-following agents may also fail to follow these rules.
The meaning of sign sequences is dependent on shared rules of the population members: how to reach a common agreement upon what the signs designate and, most importantly, what the sign user tries to transport or trigger. Without common agreement, no coordination of common behavior can be reached (McCarthy, 1984). Even here, meaning (semantics) is a social function (Mead, 1934). Slight differences in experiences of real-world environments may lead to slightly different sign interpretation, as documented in the variety of dialects, for example, in bee languages as proved by Karl von Frisch (1971). That is important in optimizing energy costs because language/code-using agents do not need new signs for every circumstance but can use limited characters/signs and limited rules to generate unlimited sign sequences. To summarize that:
- Communication depends on natural languages or codes, i.e., signs that can be combined to sign sequences
- No natural language or code speaks itself or codes itself. There must be living agents that use such natural languages or codes
- Inherently, communication is a social interaction
- Correct use of natural languages or codes underlies syntactic (combinatorial), pragmatic (contextual), and semantic (content-specific) rules
- The meaning of information is a social function.
What Remains Today From These Twentieth-Century Narratives?
We now examine the start of the third decade in the twenty-first century: which assumptions of the former narratives are still valid, and which ones must be refuted or revised to better integrate empirical data than the previous ones? The debate in the philosophy of sciences on how to generate correct scientific sentences in observation and theory lasted from 1920 to the 1980's (Witzany, 2010). Some results of these debates were:
1. The concept of an axiomatic system with error-free logical sentences is impossible in principle. The project of exact science and exact scientific language that depicts reality in a 1:1 fashion is a pipe dream. Gödel proved that in every complex system, there is at least one formula or utterance, which can neither be proved nor refuted. If someone thinks about such an undecideable formula in a non-formalizable language, he has the opportunity to determine whether this formula is true or false. For a machine, this is impossible.
2. Universal Turing and von Neuman machines that could reproduce itself have been proposed for more than half a century. No single self-reproducing machine has been built or seen until today because the theoretical construction depends on wrong assumptions.
3. No natural language speaks itself, as no natural code codes itself. Natural languages or codes depend on usage by living agents that generate, arrange, and rearrange sign sequences that are syntactically, pragmatically, and semantically correct. The semiotic features of natural languages or codes exclude randomly derived sequences. No natural language or code emerges as a randomly derived mixture of an alphabet or characters without sign-using agents.
4. In real life, no context-free language exists. The meaning (semantics) of information in natural languages depends on the real-life context (pragmatics) within which linguistic signs are used and not on its syntax.
5. Every natural code or language must embody syntactic (combination), pragmatic (context), and semantic (content) rules, which cannot be reduced to one other. Such semiotic rules are rather conservative but—in contrast to natural laws—under circumstances may change and lead to rearrangements of sequences or their generation de novo.
6. The (1) central dogma of molecular biology (DNA–RNA–protein), (2) “one gene–one protein” thesis, and (3) “non-coding DNA is junk” has been falsified. There are several ways that environmental influences on proteins affect RNAs to react accordingly, which, itself, may be coded into DNA. RNA editing, splicing, and epigenetic imprintings demonstrate that one gene can be transcribed and translated into different proteins with different functions. Non-coding DNA, which is transcribed into non-coding RNAs, leads to an abundance of regulatory agents and networks essential for gene expression.
If we want to use the usual linguistic terms in biology also in the future and cannot use it according to the physicalism of the twentieth century, we should apply current knowledge about natural languages/codes and communication to biological processes (Mattick, 2009; Shapiro, 2009; Witzany, 2014a, 2019).
Life: The Communicative Structure
To investigate communication processes in the living world, we have to identify the various levels of communicative (sign-mediated) interactions. The different levels are intertwined but share level-specific interaction patterns (Witzany, 1993, 2000, 2015).
• Every cell or organism is constantly confronted with abiotic influences such as light, gravity, wind, water, dryness, heat, cold, etc., and has to interpret the relevance for itself and react accordingly to survive. In most cases, these abiotic influences result in experiences that have to be memorized. That facilitates a faster and more appropriate reaction if the same circumstances return.
• Every organism is confronted with organisms that do not relate to the same or similar organisms of its population. These may be hunters and predators and may also be symbiotic partners essential for survival. Sign-mediated interactions between non-related organisms are of a certain quality and help us to understand the rich symbiotic life on this planet.
• The most popular communication processes we can observe and investigate are interorganismic interactions, which mean coordination and organization of common behavior in populations of the same or related organisms.
• Last but not the least, we may observe and investigate communication processes within an organism, i.e., between intercellular and intracellular parts.
All these levels of sign-mediated interactions, i.e., communication processes can be investigated throughout all domains of life as demonstrated within the last decade (Witzany, 2010, 2011b, 2012a, 2014b, 2017b; Witzany and Baluska, 2012b; Witzany and Nowacki, 2016).
I developed biocommunication theory to investigate communication processes within and among cells, tissues, organs, and organisms as sign-mediated interactions. Additionally, biocommunication theory investigates nucleotide sequences as a natural code, that is, language-like text, which follows in parallel three kinds of rules: combinatorial (syntactic), context sensitive (pragmatic), and content specific (semantic) (Witzany, 2015). Natural genome editing from a biocommunicative perspective means competent agent-driven generation and integration of meaningful nucleotide sequences into pre-existing genomic content arrangements and the ability to (re-) combine and re(regulate) them according to context-dependent (i.e., adaptational) purposes of the host organism (Witzany, 2000, 2016a).
Cellular Organisms
Cellular organisms of all domains of life actively compete for environmental resources. They assess their surroundings, estimate how much energy they need for particular goals, and then realize the optimum variant. They take measures to control certain environmental resources. They perceive themselves and can distinguish between self and non-self. These organisms process and evaluate information and then modify their behavior accordingly. All these coordinated activities are the result of communication processes in the four levels mentioned above in Figure 1 (Witzany, 2016a).
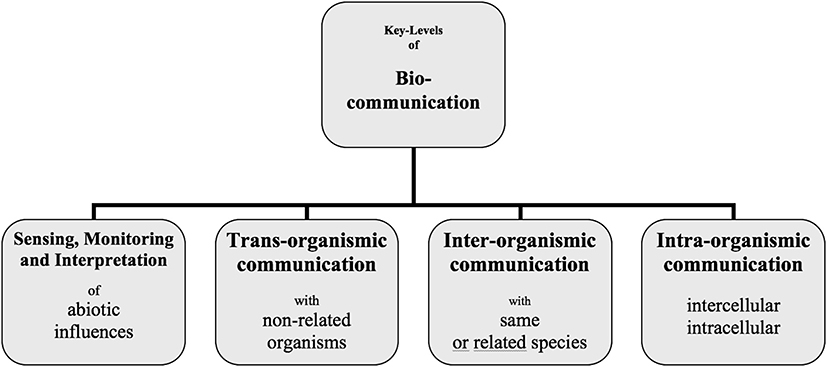
Figure 1. The biocommunication approach identified four levels into which cellular organisms are involved since the start of their life until death.
Akaryotes
Akaryotes, such as bacteria and archaea, have been considered the most primitive organisms. They are also the oldest ones, with which cellular life started approximately 4 billion years ago. For a long time, they have been viewed as determined by a strict input–output reaction pattern. In the last three decades, this picture has changed dramatically. Their capability to organize, e.g., biofilm communities, using several messenger molecules, has been investigated in numerous studies (Witzany, 2011b, 2017b). They can coordinate common behavior like a multicellular organism (interorganismic) (Shapiro, 1998). Quorum sensing is a well-investigated process in akaryotic populations (Bassler, 1999). It is the start of common decision processes important for biofilm formation, bioluminescence, sporulation, or virulence (Kaiser and Losick, 1993; Ben Jacob et al., 2004). Their coordination and organization of common behavior by sign-mediated interactions made them the oldest biological agents that can colonize each ecological niche. Although they may colonize all kingdoms of life (protozoa, animals, fungi, plants) in a parasitic or even neutral way, additionally, they serve as essential symbionts to all higher eukaryotes, without which they could not survive (transorganismic) (Schauder and Bassler, 2001).
They are in a constant struggle with their predators and co-evolutionary partners, that is, bacteriophages, which determine their gene word order (intraorganismic) and provide them with toxin/antitoxin or restriction/modification modules (Villarreal, 2005; Harms et al., 2018). Half of all bacteria in the oceans are killed every day by phages, but they survive because the other half reproduces so fast (Rohwer et al., 2014).
The signaling molecules used in akaryotic communication processes are, for example, acyl-homoserine lactones (AHLs), linear oligopeptides, cyclized oligopeptides, g-butyrolactone, furanosyl diester, cis-11-Methyl-2-dodecanoic acid, 4-hydroxy-2-alkyl quinelines, bacteriocins, cyanobactin, terpene, and palmitic acid methyl ester. Each of these signaling molecules is used for different interactions for coordination purposes (Visick and Fuqua, 2005; Witzany, 2011c; Caetano-Anolles et al., 2017; Wang and Lu, 2017).
Eukaryotes
The emergence of single-celled eukaryotes was surely a key step in biological evolution. What was formerly assumed to be a result of small steps of mutations and their selection was later explained by Lynn Margulis and her serial endosymbiotic theory in a completely contrary way (Bermudes and Margulis, 1989; Margulis, 2004). She proved that the membrane-bound fixation, rather than mutation, of a community of former free-living akaryotes was responsible. The coordination of such a community foundation depends on complex communication processes within the cellular membrane, that are fixed genetically. The crucial difference to akaryotes is the nucleus that assembles chromosomes. There are several indicators that the eukaryotic nucleus stems from a large double-stranded DNA virus that became an essential part of the group identity of the eukaryotic cell (Bell, 2006).
The natural signs in the communication processes of protozoa are hormones and secondary metabolites with which these organisms coordinate their behavioral motifs.
If we focus on protozoa, single-celled eukaryotes like ciliates, we can find various signaling molecules used to coordinate interactions, such as reproduction, mating, feeding, attack, and defense (Luporini et al., 1995, 2006; Jacob et al., 2015). Several classes of hormones and other secretions, such as secondary metabolites, have been identified (Plattner, 2016). Ciliates synthesize and secrete cell-type-specific proteins into their extracellular medium. The proteins are then taken up by species-specific receptors and interpreted by population members to generate appropriate reactions (Witzany, 2016b).
Most probably, the kingdom of fungi emerged from single-celled eukaryotes, as we can find single-celled as well as multicellular fungi, which are the evolutionary forerunners of animals and plants (Villarreal, 2005; Bonneville et al., 2020). The communication of fungi is rather complex and diverse, with a variety of substances that serve as semiochemicals (Regnier, 1971). Fungi feed on biotic surfaces, degrading them to soluble nutrients especially for plants. Fungi produce, release, and uptake a rich variety of semiochemicals for reproduction purposes, attack and defense activities, as well as developmental processes coordination and virulence (Witzany, 2012a). In single-celled fungi, we find quorum sensing, a similar communication pattern as in akaryotes. Fungi are communicating by semiochemicals such as mitogen-activated protein kinase, cAMPs, RAS, rapamycin, or calcium–calmodulin–calcineurin, to list the most prominent ones, each one used in different contextual needs (Hogan, 2006; Leeder et al., 2011; Potapova, 2012; Soll, 2012).
The evolution of animals started with the emergence of early metazoans and neuronal tissues in jelly fish (Yin et al., 2019). That was the start of electrical signaling in cell-to-cell communication and was unarguably an evolutionary key innovation. The uptake and release of chemical signaling molecules within the whole body such as hormones and secondary metabolites were enriched by the much faster electrical signaling of neuronal networks. The central nervous system enabled electrical communication over far distances within the body (Witzany, 2014b).
Research on neurobiology demonstrated that neuronal communication in animals is the most complex and specialized form of intraorganismic biocommunication on earth (Kandel, 1976). In addition to hormonal and neuronal communication processes, animals communicate interorganismically and transorganismically via auditory, visual, and tactile signs, which lead to an abundance of expression patterns of animal communication. Animal communication is very diverse through genera, families, and species and reaches a complexity peak in human communication that enables human species to act with collective intentionality as a driving force of coordinated and organized division of labor (Tomasello, 2008). The variety of animal communication has been investigated extensively more recently in genera such as chimpanzees, elephants, wolfs, dogs, rodents, mice, rats, spiders, ants, termites, crows, parrots, birds, salamanders, chelonians, cetacean, fish, cephalopods, corals, and nematodes (Witzany, 2014b). Karl von Frisch received a Nobel Prize for deciphering bee languages and their dialects by investigating how moving patterns serve as signals to communicate nutrition sites.
Plants represent the evolutionary youngest organismic kingdom. Up until now, they are regarded as mechanistic growth automatons, maybe because of their sessile lifestyle and rather slow moving patterns in growth and developmental processes in comparism to animals. However, research within the last two decades has changed this perspective dramatically (Baluska et al., 2006; Baluska and Ninkovic, 2010; Perotto and Baluska, 2012; Blande and Glinwood, 2016). The complex communication profiles of plants with non-plants, other plant species, and within plant bodies on the intracellular and intercellular level indicate that plants can communicate at all levels in parallel, in contrast to animals with their centralized nervous system. The interwoven transorganismic communication with bacteria and fungi in the plant root zone and their intraorganismic communication within the plant body has been highly investigated (Bais et al., 2003). Intraorganismic communication in plants coordinates cellular growth, development, shape, and dynamics on the local level and in rather separated parts. Semiochemical communication happens by vesicular trafficking or via plasmodesmata.
Additionally, we can find physical communication by airborne, electric-like, hydraulic, and mechanical signs (Callaway, 2002; Braam, 2005). We may find nucleic acids, oligonucleotides, proteins and peptides, minerals, oxidative signals, gases, mechanical signals, electrical signals, fatty acids, oligosaccharides, growth factors, several amino acids, various secondary metabolite products, and simple sugars as signaling molecules. Today, we know 100,000 different metabolites used in plants (Dunn and Handelsman, 2002; Fleming, 2005).
Virus Communication
Traditionally, viruses have been seen as infective, disease-causing entities with often epidemic consequences that infect all kinds of organisms. Evolutionary, they have been viewed as parasites that escaped out of cells because they cannot reproduce by themselves but need cellular hosts (Villarreal and Witzany, 2010). More recently, this perspective has been corrected (Villarreal, 2005, 2009a; Forterre and Prangishvili, 2013; Moelling and Broecker, 2019). The disease-causing agents are the minority. Most viruses are integrated into the cytoplasm or nucleoplasm of host cells, without harming the host.
The persistent lifestyle is the predominant one, and the symbiotic and coevolutionary lifestyle is omnipresent because, since the start of life, every cell, tissue, organ, and organism on this planet has been persistently infected by multiple viruses (Ryan, 2009; Broecker and Moelling, 2019a; Koonin et al., 2019). Their persistence, in most cases, does not occur as fully functional but as parts of infectious agents that remain as useful tools for cellular needs because such viral parts can be exapted and co-opted by cellular organisms and which we can identify as non-coding RNAs with repetitive sequence syntax (Jurka et al., 2007; Witzany, 2009, 2017c). Defective parts of infectious agents may serve as signals for immune functions against related genetic parasites (Ariza-Mateos and Gómez, 2017).
Viruses and their defective parts play essential roles in genetic content composition and arrangements that help organisms rearrange genetic content for adaptational purposes, such as in immunity systems or in the evolution of new organs, e.g., placenta and the role of syncytin genes (Villarreal, 2009a, 2016a; Perot et al., 2012; Koonin and Krupovic, 2017; Broecker and Moelling, 2019b). We know them as endogenous viruses and defectives, transposons, retrotransposons, long terminal repeats, non-long terminal repeats, long interspersed nuclear elements, short interspersed nuclear elements, alu's, group I introns, group II introns, phages, and plasmids. Because nearly all the remnants of former infectious genetic parasites share a repeat nucleotide syntax, in contrast to the protein-coding non-repeated nucleotide syntax, we now know that, in most cases, they remain as non-coding RNAs (see below) (Witzany, 2011d). In this respect, we may study cellular DNA as the preferred living habitat of an abundance of RNA inhabitants (Brookfield, 2005; Le Rouzic et al., 2007; Vennera et al., 2009; Villarreal and Witzany, 2013a).
We should be reminded that the human genetic content that codes for proteins is 1.5% only, whereas the non-coding but regulation-relevant genetic content is about 98.5% (Boland, 2017). Additionally, we must not forget that viruses represent the most abundant genetic elements on this planet, outnumbering cellular genetic content by 10 times. In this respect, cellular genomes seem like rare islands in an ocean of the global virosphere (Forterre and Prangishvili, 2009; Koonin, 2009; Rohwer et al., 2014).
Many infectious agents have been identified during the last decades as inhabitants of all prokaryotic as well as eukaryotic genomes (Koonin and Dolja, 2014; Koonin et al., 2015). They infect, insert and delete, cut and paste, or copy and paste. Many of them spread within the genome. They can change host genetic identity by insertion, recombination, or epigenetic regulation or re-regulation of genetic content, and co-evolve with the host (Witzany, 2006, 2014c; Catania et al., 2020).
Most interestingly and unknown, the formerly termed RNA virus populations, quasispecies, are now recognized as highly interactive and cooperative agents (Villarreal and Witzany, 2013b). Virus communication demonstrates that quasispecies populations and subpopulations may cooperate and compete in parallel, dependent on the circumstantial context of host life (Villarreal and Witzany, 2019). Social interacting persistent viruses play important roles as host gene regulatory elements—in most cases represented by repetitive sequences—that may react to nearly every unexpected circumstance (Díaz-Muñoz et al., 2017; Sanjuán, 2018).
Viruses are the only biotic agents that can generate code sequences de novo, identify sequence-specific target sites, integrate into pre-existing genetic content, integrate without damage of previous coding regions, recombine according to adaptational purposes, and mark sequence sites to epigenetically fix identity content (Villarreal, 2005, 2009b). The whole range of epigenetic marking, which is so essential for cellular-based organisms to coordinate the variety of developmental stages stems from these infectious agents and has been adapted to cellular needs (Witzany, 2009). Viruses may divide into multipartite genome segments, spread their parts non-randomly throughout host genomes, and reassemble into full functional viral genomes again (Sicard et al., 2016, 2019; Lucía-Sanz and Manrubia, 2017).
Current research demonstrated that viruses communicate to coordinate their behavior: whether it should be lytic or remain in a lysogenic style. The semiochemicals used to communicate at interorganismic levels are peptides (AimP), which reduce the expression of the negative regulator of lysogeny (AimX) by binding to the transcription factor (AimR) promoting lysogeny (Erez et al., 2017; Stokar-Avihail et al., 2019). Interactional motifs in virus communication range from conflict to cooperation in various forms and mimicry, dependent on situational context (Mei and Zhang, 2019; Seligmann, 2019).
Viruses are the only living entities that may exchange genetic sequences as module-like tools between double-stranded DNA, single-stranded DNA, single-stranded RNA, double-stranded RNA, and retroviruses. Most interestingly, they may cooperate and compete as viral clouds in parallel (Koonin et al., 2015; Stedman, 2015, 2018; Berliner et al., 2018).
The most important behavioral motif from both an evolutionary and functional perspective is that viruses can integrate a persistent lifestyle into cellular host organisms by “addiction” modules (Villarreal, 2012, 2016b). This means that former competing viral groups counterbalance each other, together with the host immune system (Koonin et al., 2019). One can find such counter-regulating paired genes of the addiction modules in the restriction/modulation (RM) systems, as well as the toxin/antitoxin (TA) systems (Mruk and Kobayashi, 2014). Insertion/deletion functions represent similar modules as do the RM systems. This infectious technique to colonize host genomes is the key process in generating a new sequence space without error replication (Villarreal, 2009b; Villarreal and Witzany, 2015).
To consider the genetic information in cellular genomes of all domains of life without the remnants of persistent viral infections would be as curious as to consider a language text consisting of various characters as molecular bricks without agents that can write coherent linguistic texts and are competent in generating it according to syntactic, pragmatic, and semantic rules (Witzany, 2012b, 2017c).
RNA Communication
The steps from viruses to pure RNA stem-loops can be much easily understood if we consider viroids, that is, short strands of circular, single-stranded RNA virus without a protein coat (Catalán et al., 2019). However, in evolutionary periods, interacting RNA networks most propably predated viruses and cellular life (Root-Bernstein and Dillon, 1997; Witzany, 2011b; Root-Bernstein and Root-Bernstein, 2015; Demongeot and Seligmann, 2019) Here, we focus on the RNA strand level, which clearly shows infective and host-manipulating properties (Diener, 1989; Flores et al., 2012, 2014). This is coherent with the RNA world of RNA stem-loop groups and RNA group identities as result of biotic behavior represented by biological selection processes (Petrov et al., 2014; Ariza-Mateos et al., 2019; Villarreal and Witzany, 2019; Demongeot and Seligmann, 2020).
To understand the social interactions between the proponents of the RNA world means to understand a fascinating sphere of what was formerly assumed to be impossible: that mere sequences of molecules do not behave like physical/chemical entities in an abiotic world but as competent agents on genetic code syntax that cooperate and organize, constitute, and generate sequence structures and groups that depend on group selection (Higgs and Lehman, 2015). As was shown, a single RNA stem-loop behaves like a random assembly of nucleotides without selective forces governed strictly by physical laws (Smit et al., 2006; Vaidya et al., 2012, 2013). Biological selection starts only if they are assembled to groups, consortia (Hayden and Lehman, 2006).
RNA world agents can serve as a template or be active as a catalyst (Gwiazda et al., 2012). This double function of genotype and phenotype is unique in biology, where we can find most organisms divided into genetic coding and protein-based bodies within their real-life interactions. In former times, it was thought that information transfer is a one-way road from genotype to phenotype, but with epigenetics and RNA biology, it became obvious that the reverse direction of information flow—indicating complex sign-mediated interactions (i.e., communication)—plays an important role in evolution, development, and adaptational flexibility (Shapiro, 2009, 2014, 2016; Spadafora, 2016).
RNA stem-loop groups interact with other DNA, RNA, or proteins forming the most important ribo–nucleo–protein complexes (RNP), such as the subgroups of ribosome, spliceosome, and editosome (Mercer and Mattick, 2013). Their active site that leads to group behavior is the single-stranded loops or bulges being essential for self/non-self recognition and group identity.
Additionally, these single-stranded loops are actively prone to integration or rejection of foreign RNA stem-loops. Their highly interaction-prone nucleotide “surface” serves as signs (indices) for competing or cooperating RNA stem-loops, based on complementary base-pairing rules (Schudoma, 2011). This is relevant also in RNA mimicry as demonstrated recently (Ariza-Mateos and Gómez, 2017; Grüll and Massé, 2019).
We should consider such ensembles of RNA stem-loops as RNA populations, which are investigated as varieties of small non-coding RNAs such as snRNA, snoRNAs, piwi RNAs, tRNAs, rRNAs, mRNA, siRNAs, and microRNAs (Bartel, 2004; Carthew and Sontheimer, 2009; Malone and Hannon, 2009; Gebetsberger et al., 2017).
Ensembles of RNA fragments that self-ligate into self-replicating ribozymes may form cooperative networks unexpectedly (Briones et al., 2009; Cheng and Unrau, 2010). It has been shown that three-member networks represent cooperative growth dynamics. When such cooperative networks compete against selfish RNA stem-loop groups, they grow faster. This indicates that RNA populations can evolve higher complexity by cooperative interactions. This also demonstrated that cooperation outcompetes selfishness (Hayden and Lehman, 2006; Vaidya et al., 2012). The primacy of the selfish gene hypothesis, therefore, is outdated on the RNA level.
Non-coding RNAs interact with all relevant key players in the biological world: DNA, RNA, and proteins. They play important roles in nuclear organization, transcription, post-transcriptional, and epigenetic processes and are transcribed in the sense and/or antisense directions (Cech and Steitz, 2014; Long et al., 2017). They may be expressed in different cell types, subcellular compartments, and developmental stages, in every case in a context-dependent manner (Mattick and Gagen, 2001; Mattick, 2003; Clark et al., 2013). Non-coding RNAs can interact cooperatively in a module-like manner (Manrubia and Briones, 2007; Higgs and Lehman, 2015).
Non-coding RNAs may regulate coordinated or independently, autonomously, or functionally interrelated. Such RNAs regulate single genes or bigger genetic networks. They also may control the spatiotemporal coordination of gene expression rather precisely (Witzany, 2009). Additionally, non-coding RNAs can target each other for post-transcriptional regulation, alternative splicing, polyadenylation, and non-templated modifications (Doudna et al., 1989). Another ribonucleoprotein complex, the editosome, plays important roles in transmitting environmental (contextual) information to the epigenome (Cech, 2012). In addition, other ribonucleoprotein complexes may undergo nuclear–cytoplasmic, nuclear–mitochondrial, and axodendritic trafficking. They serve as appropriate RNA–protein consortia that spatiotemporally distribute combinations of ncRNAs, messenger RNAs, and RNA-binding proteins (Atkins et al., 2010; Noller, 2012; Petrov et al., 2015; Villarreal, 2015; Tartaglia, 2016).
Basic tools of such RNA consortia are their complementary composition of base pairing stems and non-base-pairing (single-stranded) loops, which are results of an inherent property of RNA to fold back building stem-loop structures (Villarreal and Witzany, 2013b). The variety of regulations on protein-coding genes, as well as the processing of these regulatory RNAs by a number of defined steps in RNA splicing and RNA editing of RNA transcripts, makes an understanding nearly impossible because of the coordinated complexity (Witzany, 2016c). It is now very clear that non-coding RNAs build those agents that determine the regulation of all steps and substeps of gene regulation in cellular organisms (Mattick, 2009). The interactive connection between the RNA-world agents and cellular organisms is based on the abundance of infectious genetic parasites, viruses, and related agents that transfer and insert all relevant RNA features to the cellular world or, as Frantisek Baluska noted, “without infection, no evolution” (Baluska, 2009; Baluška and Witzany, 2014).
In contrast to DNA viruses, RNA viruses have much smaller genomes on RNA bases without proofreading and repair. “Error-prone” RNA viruses are a key narrative in physicalistic biology. However, it has been formerly assumed as deficit because the “error rate” is very high. The contrary perspective is more effective: to regard it as “innovation rate” because it assembles the property of invention of new sequence contents, de novo, that is, that have not been existent before. This is important for variation as well as infection, immunity, and identity, for both diversified viral and cellular populations and continued interaction between cellular immune systems and infectious genetic parasites throughout the whole history of life (Villarreal, 2015). “Error-threshold” was used to designate the critical state of too many error replications to stabilize newly derived variations (Shah et al., 2019). Replication must be faster than the breakdown rate of the newly derived strands. Because “errors” are not appropriate to describe RNA strand innovation we now may consider these events as “innovation overload.”
Summarizing Current Empirical Data
We have seen that the use of linguistic terms, such as genetic code, code without commas, transcription, translation, reading frame, immune response, etc., in biology, is still possible. However, the physicalistic paradigm that introduced these terms to biology is no longer valid. Physicalism, as well as other mathematical theories of language, cannot substantiate these terms because this is outside their expertise: Natural languages and codes are the result of social interacting agents, according to competent and commonly shared rules of sign use and allow living agents not only to coordinate and organize their behavioral features and capabilities but also to generate completely new sign sequences and behavioral adaptations that cannot be predicted or computed by algorithm-based procedures. Nucleotide sequences of the genetic code are not the result of the self-organization of matter. The genetic code is not the result of statistical mechanics (Table 1).
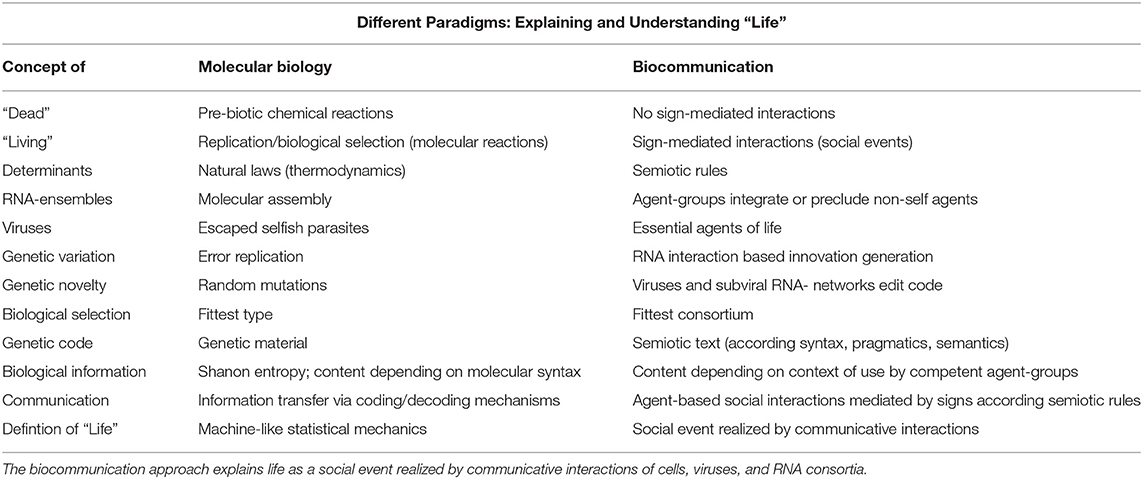
Table 1. Different paradigms investigating living agents and defining life: the molecular biological paradigm explains all life processes primarily by the physical–chemical properties and statistical mechanics.
Today, we know that life is constituted of three levels of interactions: cells that are genetically regulated by RNA networks, which are remnants of former genome-invading agents such as viruses and their relatives. Quantitatively, cellular genes represent rare islands in an ocean of viruses and virus-related infectious genetic parasites (virosphere). The invasion strategy of genomic parasites resulted in persistence within host genomes that now represent evolutionary novel genetic identities than before the invasion. Immune systems in cellular organisms represent communicating networks of persistent genetic parasites that serve as immune function against related parasites. Former competing genetic parasites, together with host immune function, generate a variety of regulatory tools that are counter regulated. The three interaction levels (a) RNA-groups, (b) viruses, and (c) cell-based organisms not only constitute and regulate but also inherently are basically open for generating new, unexpectable, and non-computable interaction profiles. This means genetic de novo sequences, new cooperation pathways, exaptation and new traits of former generated module like parts that evolved for different purposes, re-use of former degraded modules (viral defective minorities), and new behavioral motifs of cellular life forms.
Conclusion
Primarily, life is a process. The main characteristic of this process is the coordinated organization of complex interactions that we see as protein-based organisms of three domains of life, their reproduction, and metabolism all mediated by complex interwoven gene regulation as a result of communication. Living nature is structured and organized by language and communication within and among organisms, viruses, and RNA networks. If communication is damaged or disturbed, coordination and organization may be incomplete, and normal function becomes disregulated, leading to the broad variety of diseases. Without RNA world agents, no cellular gene regulation could take place. Without viruses and related infectious agents, these capabilities of RNA stem-loop group behavior as gene inventors and regulators would not have been integrated into cellular host genomes.
Therefore, we must ask whether mutation (“error replication”) is the correct term to desígnate genetic variation in the future. Error replications, which in most cases means DNA damage without successful repair, is an empirical fact but does not play important roles in genetic innovation. Evolutionary relevant genetic variations are the result of natural genome editing by competent agents such as viruses and RNA networks with their inherent competence to generate and modify nucleotide sequences. This is competent nucleotide sequence editing. In contrast to former convictions, this is an agent-based interaction process, which is far from statistical mechanics and biochemical kinetics. Instead of error replication, we should use now “genetic innovation,” which much better fits to the empirically documented events. Darwinian evolution then could be revised to “innovation and selection.”
If we want a new definition of life for astrobiological research, we must integrate the complementarity of cells, viruses, and RNA networks into a communicative life world. Life as a process depends on these interactional agents. Yes, all living agents are constituted by elements that underlie physics and chemistry. However, in contrast to abiotic planets, life on this planet depends on communication processes involved in all sign-mediated interactions of cells, viruses, and subviral RNA networks. Therefore, we can formulate a new definition of life: life is communicative interaction, which means life is primarily a social event.
Life is a social event. Social events are realized by communicative interactions on three complementary levels in parallel: cell communication, RNA communication, and virus communication.
Data Availability Statement
All datasets generated for this study are included in the article/supplementary material.
Author Contributions
The author confirms being the sole contributor of this work and has approved it for publication.
Conflict of Interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer LV declared a past co-authorship with the author to the handling editor.
References
Ariza-Mateos, A., Briones, C., Perales, C., Domingo, E., and Gómez, J. (2019). The archaeology of coding RNA. Ann. N.Y. Acad. Sci. 1447, 119–134. doi: 10.1111/nyas.14173
Ariza-Mateos, A., and Gómez, J. (2017). Viral tRNA mimicry from a biocommunicative perspective. Front. Microbiol. 8:2395. doi: 10.3389/fmicb.2017.02395
Atkins, J. F., Gesteland, R. F., and Cech, T. (2010). RNA Worlds. From Life's Origins to Diversity in Gene Regulation. New York, NY: Cold Spring Harbor Laboratory Press.
Austin, J. L. (1975). How to Do Things With Words. London: Harvard University Press. doi: 10.1093/acprof:oso/9780198245537.001.0001
Bais, H. P., Park, S. W., Weir, T. L., Callaway, R. M., and Vivanco, J. M. (2003). How plants communicate using the underground information superhighway. Trend. Plant. Sci. 9, 26–32. doi: 10.1016/j.tplants.2003.11.008
Baluska, F. (2009). Cell-cell channels, viruses, and evolution: via infection, parasitism, and symbiosis toward higher levels of biological complexity. Ann. N. Y. Acad. Sci. 1178, 106–119. doi: 10.1111/j.1749-6632.2009.04995.x
Baluska, F., Mancuso, S., and Volkmann, D. (2006). Communication in Plants. Heidelberg: Springer. doi: 10.1007/978-3-540-28516-8
Baluska, F., and Ninkovic, V. (2010). Plant Communication From an Ecological Perspective. Heidelberg: Springer. doi: 10.1007/978-3-642-12162-3
Baluška, F., and Witzany, G. (2014). Life is more than a computer running DNA software. World J. Biol.Chem. 5, 275–278. doi: 10.4331/wjbc.v5.i3.275
Bartel, D. P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297. doi: 10.1016/S0092-8674(04)00045-5
Bassler, B. L. (1999). How bacteria talk to each other: regulation of gene expression by quorumsensing. Curr. Opin. Microbiol. 2, 582–587. doi: 10.1016/S1369-5274(99)00025-9
Bell, P. J. L. (2006). Sex and the eukaryotic cell cycle is consistent with a viral ancestry for the eukaryotic nucleus. J. Theor. Biol. 243, 54–63. doi: 10.1016/j.jtbi.2006.05.015
Ben Jacob, E., Becker, I., Shapira, Y., and Levine, H. (2004). Bacterial linguistic communication and social intelligence. Trends Microbiol. 12, 366–372. doi: 10.1016/j.tim.2004.06.006
Berliner, A. J., Mochizuki, T., and Stedman, K. M. (2018). Astrovirology: viruses at large in the universe. Astrobiol. 18, 207–223. doi: 10.1089/ast.2017.1649
Bermudes, D., and Margulis, L. (1989). Symbiont acquisition as neoseme: origin of species and higher taxa. Symbiosis. 4, 185–198.
Bertalanffy, L. (1940). Der Organismus als physikalisches System betrachtet. Die Naturwissenschaften. 28, 521–531. doi: 10.1007/BF01497764
Biebricher, C. K., and Eigen, M. (2006). What is a quasispecies? Curr. Top. Microbiol. Immunol. 299, 1–31. doi: 10.1007/3-540-26397-7_1
Biebricher, C. K., Eigen, M., and Gardiner, W. C. Jr. (1985). Kinetics of RNA replication: competition and selection among self-replicating RNA species. Biochemistry. 24, 6550–6560. doi: 10.1021/bi00344a037
Blande, J. D., and Glinwood, R. (2016). Deciphering Chemical Language of Plant Communication. Heidelberg: Springer. doi: 10.1007/978-3-319-33498-1
Boland, C. R. (2017). Non-coding RNA: it's not junk. Dig. Dis. Sci. 62, 1107–1109. doi: 10.1007/s10620-017-4506-1
Bonneville, S., Delpomdor, F., Préat, A., Chevalier, C., Araki, T., Kazemian, M., et al. (2020). Molecular identification of fungi microfossils in a Neoproterozoic shale rock. Sci. Adv. 6:eaax7599. doi: 10.1126/sciadv.aax7599
Braam, J. (2005). In touch: plant responses to mechanical stimuli. New Phytol. 165, 373–389. doi: 10.1111/j.1469-8137.2004.01263.x
Briones, C., Stich, M., and Manrubia, S. C. (2009). The dawn of the RNA world: Toward functional complexity through ligation ofrandom RNA oligomers. RNA 15, 743–749. doi: 10.1261/rna.1488609
Broecker, F., and Moelling, K. (2019a). What viruses tell us about evolution and immunity: beyond Darwin? Ann. N. Y. Acad. Sci. 1447, 53–68. doi: 10.1111/nyas.14097
Broecker, F., and Moelling, K. (2019b). Evolution of immune systems from viruses and transposable elements. Front. Microbiol. 10:51. doi: 10.3389/fmicb.2019.00051
Brookfield, J. F. Y. (2005). The ecology of the genome. Mobile DNA elements and their hosts. Nat. Rev. Genet. 6, 128–136. doi: 10.1038/nrg1524
Caetano-Anolles, G., Minhas, B. F., Aziz, F., Mughal, F., Shahzad, K., et al. (2017). “The compressed vocabulary of the proteins of archaea,” in Biocommunication of Archaea, ed G. Witzany (Cham: Springer), 147–174. doi: 10.1007/978-3-319-65536-9_10
Callaway, R. M. (2002). The detection of neighbors by plants. Trend. Ecol. Evol. 1 7, 104–105. doi: 10.1016/S0169-5347(01)02438-7
Carnap, R. (1931). Die physikalische Sprache als Universalsprache der Wissenschaft. Erkenntnis 2, 432–465. doi: 10.1007/BF02028172
Carthew, R. W., and Sontheimer, E. J. (2009). Origins and Mechanisms of miRNAs and siRNAs. Cell 136, 642–655. doi: 10.1016/j.cell.2009.01.035
Catalán, P., Elena, S. F., Cuesta, J. A., and Manrubia, S. (2019). Parsimonious scenario for the emergence of viroid-like replicons de novo. Viruses. 11:E425. doi: 10.3390/v11050425
Catania, S., Dumesic, P. A., Pimentel, H., Nasif, A., Stoddard, C. I., Burke, J. E., et al. (2020). Evolutionary persistence of DNA methylation for millions of years after ancient loss of a de novo methyltransferase. Cell 180, 263–277.e20. doi: 10.1016/j.cell.2019.12.012
Cech, T. R. (2012). The RNA worlds in context. Cold Spring Harb. Perspect. Biol. 4:a006742. doi: 10.1101/cshperspect.a006742
Cech, T. R., and Steitz, J. A. (2014). The non-coding RNA revolution-trashing old rules to forge new ones. Cell 157, 77–94. doi: 10.1016/j.cell.2014.03.008
Cheng, L. K., and Unrau, P. J. (2010). Closing the circle: replicating RNA with RNA. Cold Spring Harb. Perspect. Biol. 2:a002204. doi: 10.1101/cshperspect.a002204
Clark, M. B., Choudhary, A., Smith, M. A., Taft, R. J., and Mattick, J. S. (2013). The dark matter rises: the expanding world of regulatory RNAs. Essays Biochem. 54, 1–16. doi: 10.1042/bse0540001
Demongeot, J., and Seligmann, H. (2019). The uroboros theory of life's origin: 22-nucleotide theoretical minimal RNA rings reflect evolution of genetic code and tRNA-rRNA translation machineries. Acta Biotheor. 67, 273–297. doi: 10.1007/s10441-019-09356-w
Demongeot, J., and Seligmann, H. (2020). Accretion history of large ribosomal subunits deduced from theoretical minimal RNA rings is congruent with histories derived from phylogenetic and structural methods. Gene 3:144436. doi: 10.1016/j.gene.2020.144436
Díaz-Muñoz, S. L., Sanjuán, R., and West, S. (2017). Sociovirology: conflict, cooperation, and communication among viruses. Cell Host Microbe. 22, 437–441. doi: 10.1016/j.chom.2017.09.012
Diener, T. O. (1989). Circular RNAs: relics of precellular evolution? Proc. Natl. Acad. Sci. U. S. A. 86, 9370–9374. doi: 10.1073/pnas.86.23.9370
Domingo, E., and Schuster, P. (2016). Quasispecies: From Theory to Experimental Systems. Cham: Springer. doi: 10.1007/978-3-319-23898-2
Doudna, J. A., Cormack, B. P., and Szostak, J. W. (1989). RNA structure, not sequence, determines the 5'splice-site specicity of a group I intron. Proc. Natl. Acad. Sci. U. S. A. 86, 7402–7406. doi: 10.1073/pnas.86.19.7402
Dunn, A. K., and Handelsman, J. (2002). Toward an understanding of microbial communities through analysisof communication networks. Antonie van Leeuwenhoek 81, 565–574. doi: 10.1023/A:1020565807627
Eigen, M. (1971). Selforganization of matter and the evolution of biological macromolecules. Naturwissenschaften 58, 465–523. doi: 10.1007/BF00623322
Eigen, M., and Winkler, R. (1983). Laws of the Game: How the Principles of Nature Govern Chance. Princeton: Princeton University Press.
Erez, Z., Steinberger-Levy, I., Shamir, M., Doron, S., Stokar-Avihail, A., Peleg, Y., et al. (2017). Communication between viruses guides lysis-lysogeny decisions. Nature 541, 488–493. doi: 10.1038/nature21049
Fleming, A. J. (2005). Intercellular Communication in Plants. Oxford; Boca Raton, FL: Blackwell Publishing/CRC Press. doi: 10.1093/aob/mcl019
Flores, R., Gago-Zachert, S., Serra, P., Sanjuán, R., and Elena, S. F. (2014). Viroids: survivors from the RNA world? Annu. Rev. Microbiol. 68, 395–414. doi: 10.1146/annurev-micro-091313-103416
Flores, R., Serra, P., Minoia, S., Di Serio, F., and Navarro, B. (2012). Viroids: from genotype to phenotype just relying on RNA sequence and structural motifs. Front. Microbiol. 3:217. doi: 10.3389/fmicb.2012.00217
Forterre, P., and Prangishvili, D. (2009). The great billion-year war between ribosome- and capsid-encoding organisms (cells and viruses) as the major source of evolutionary novelties. Ann. N. Y. Acad. Sci. U. S. A. 1178, 65–77. doi: 10.1111/j.1749-6632.2009.04993.x
Forterre, P., and Prangishvili, D. (2013). The major role of viruses in cellular evolution: facts and hypotheses. Curr. Opin. Virol. 3, 558–565. doi: 10.1016/j.coviro.2013.06.013
Frisch, K. (1971). Bees: Their Vision, Chemical Senses, and Language. Ithaca: Cornell University Press.
Gebetsberger, J., Wyss, L., Mleczko, A. M., Reuther, J., and Polacek, N. (2017). A tRNA-derived fragment competes with mRNA for ribosome binding and regulates translation during stress. RNA Biol. 14, 1364–1373. doi: 10.1080/15476286.2016.1257470
Gödel, K. (1931). Ueber formal unentscheidbare Sätze der Principia Mathematica und verwandter Systeme. Monatsh. Math. Phys. 38, 173–198. doi: 10.1007/BF01700692
Grüll, M. P., and Massé, E. (2019). Mimicry, deception and competition: the life of competing endogenous RNAs. Wiley Interdiscip. Rev. RNA. 10:e1525. doi: 10.1002/wrna.1525
Gwiazda, S., Salomon, K., Appel, B., and Mueller, S. (2012). RNA self-ligation: from oligonucleotides to full length ribozymes. Biochimie 94, 1457–1463. doi: 10.1016/j.biochi.2012.03.015
Habermas, J. (1987). The Theory of Communicative Action. Lifeworld and System: A Critique of Functionalist Reason, Vol. 2. Boston, MA: Beacon Press.
Habermas, J. (1994). Actions, speech acts, linguistically mediated interactions and the lifeworld. Phil. Probl. Tod. 1, 45–74. doi: 10.1007/978-94-017-4522-2_3
Harms, A., Brodersen, D. E., Mitarai, N., and Gerdes, K. (2018). Toxins, targets, and triggers: an overview of toxin-antitoxin biology. Mol. Cell. 70, 768–784. doi: 10.1016/j.molcel.2018.01.003
Hayden, E. J., and Lehman, N. (2006). Self-assembly of a group I intron from inactiveoligonucleotide fragments. Chem. Biol. 13, 909–918. doi: 10.1016/j.chembiol.2006.06.014
Higgs, P. G., and Lehman, N. (2015). The RNA world: molecular cooperation at the origins of life. Nat. Rev. Genet. 16, 7–17. doi: 10.1038/nrg3841
Hilbert, D., and Bernays, P. (1934/1939). Grundlagen der Mathematik. Berlin; New York, NY: Springer.
Hogan, D. A. (2006). Talking to themselves: autoregulation and quorum sensing in fungi. Eukaryot. Cell 584, 613–619. doi: 10.1128/EC.5.4.613-619.2006
Jacob, S., Chaine, A. S., Schtickzelle, N., Huet, M., and Clobert, J. (2015). Social information fromimmigrants: multiple immigrant-based sources of information for dispersal decisions in aciliate. J. Anim. Ecol. 84, 1373–1383. doi: 10.1111/1365–2656.12380
Jurka, J., Kapitonov, V. V., Kohany, O., and Jurka, M. V. (2007). Repetitive sequences in complex genomes: structure and evolution. Annu. Rev. Genomics Hum. Genet. 8, 241–259. doi: 10.1146/annurev.genom.8.080706.092416
Kaiser, D., and Losick, R. (1993). How and why bacteria talk to each other. Cell 73, 873–885. doi: 10.1016/0092-8674(93)90268-U
Kandel, E. (1976). Cellular basis of behavior: An Introduction to Behavioral Neurobiology. New York, NY: Freeman.
Koonin, E. V. (2009). On the origin of cells and viruses: primordial virus world scenario. Ann. N. Y. Acad. Sci. U. S. A. 1178, 47–64. doi: 10.1111/j.1749-6632.2009.04992.x
Koonin, E. V., and Dolja, V. V. (2014). Virus world as an evolutionary network of viruses and capsidless selfish elements. Microbiol. Mol. Biol. Rev. 78, 278–303. doi: 10.1128/MMBR.00049–13
Koonin, E. V., Dolja, V. V., and Krupovic, M. (2015). Origins and evolution of viruses of eukaryotes: the ultimate modularity. Virology 479/480, 2–25. doi: 10.1016/j.virol.2015.02.039
Koonin, E. V., and Krupovic, M. (2017). Polintons, virophages and transpovirons: a tangled web linking viruses, transposons and immunity. Curr. Opin. Virol. 25, 7–15. doi: 10.1016/j.coviro.2017.06.008
Koonin, E. V., Makarova, K. S., Wolf, Y. I., and Krupovic, M. (2019). Evolutionary entanglement of mobile genetic elements and host defence systems: guns for hire. Nat. Rev. Genet. 21, 119–131. doi: 10.1038/s41576-019-0172-9
Le Rouzic, A., Dupas, S., and Capy, P. (2007). Genome ecosystem and transposable elements species. Gene 390, 214–220. doi: 10.1016/j.gene.2006.09.023
Leeder, A. C., Palma-Guerrero, J., and Glass, N. L. (2011). The social network: deciphering fungal language. Nature Rev. Microbiol. 9, 440–451. doi: 10.1038/nrmicro2580
Long, Y., Wang, X., Youmans, D. T., and Cech, T. R. (2017). How do lncRNAs regulate transcription? Sci. Adv. 3:eaao2110. doi: 10.1126/sciadv.aao2110
Lucía-Sanz, A., and Manrubia, S. (2017). Multipartite viruses: adaptive trick or evolutionary treat? N. P. J. Syst. Biol. Appl. 3:34. doi: 10.1038/s41540-017-0035-y
Luporini, P., Vallesi, A., Alimenti, C., and Ortenzi, C. (2006). The cell type-specific signal proteins(pheromones) of protozoan ciliates. Curr. Pharm. Des. 12, 3015–3024. doi: 10.2174/138161206777947452
Luporini, P., Vallesi, A., Miceli, C., and Bradshaw, R. A. (1995). Chemical signaling in ciliates. J. Eukaryot. Microbiol. 42, 208–212. doi: 10.1111/j.1550-7408.1995.tb01567.x
Malone, C. D., and Hannon, G. J. (2009). Small RNAs as guardians of the genome. Cell 136, 656–668. doi: 10.1016/j.cell.2009.01.045
Manrubia, S. C., and Briones, C. (2007). Modular evolution and increase of functional complexity in replicating RNA molecules. RNA 13, 97–107. doi: 10.1261/rna.203006
Margulis, L. (2004). Serial endosymbiotic theory (SET) and composite individuality. Transitionfrom bacterial to eukaryotic genomes. Microbiol. Tod. 31, 173–174.
Mattick, J. S. (2003). Challenging the dogma: the hidden layer of non-coding RNAs in complex organisms. BioEssays 25:930. doi: 10.1002/bies.10332
Mattick, J. S. (2009). Deconstructing the dogma: a new view of the evolution and genetic programming of complex organisms. Ann. N. Y. Acad. Sci. U. S. A. 1178, 29–46. doi: 10.1111/j.1749–6632.2009.04991.x
Mattick, J. S., and Gagen, M. J. (2001). The evolution of controlled multitasked gene networks:The role of introns and other non-coding RNAs in the development of complex organisms. Mol. Biol. Evol. 18, 1611–1630. doi: 10.1093/oxfordjournals.molbev.a003951
McCarthy, T. (1984). “Translator's introduction,” in The Theory of Communicative Action 1, ed. J. Habermas (Boston, MA: Beacon Press), vi–xxxix.
Mei, S., and Zhang, K. (2019). In silico unravelling pathogen-host signaling cross-talks via pathogen mimicry and human protein-protein interaction networks. Comput. Struct. Biotechnol. J. 18, 100–113. doi: 10.1016/j.csbj.2019.12.008
Mercer, T. R., and Mattick, J. S. (2013). Structure and function of long non-coding RNAs in epigenetic regulation. Nat. Struct. Mol. Biol. 20, 300–307. doi: 10.1038/nsmb.2480
Moelling, K., and Broecker, F. (2019). Viroids-first - a model for life on earth, mars, and exoplanets. Geosciences. 9:241. doi: 10.3390/geosciences9050241
Morris, C. (1946). Signs, Language, and Behavior. New York, NY: Prentice Hall. doi: 10.1037/14607-000
Mruk, I., and Kobayashi, I. (2014). To be or not to be: regulation of restriction-modification systems and other toxin-antitoxin systems. Nucleic Acids Res. 42, 70–86. doi: 10.1093/nar/gkt711
Neumann, J. (1966). Theory of Self-Reproducing Automata. Urbana/London: University of Illinois Press.
Noller, H. (2012). Evolution of protein synthesis from an RNA world. Cold Spring Harb. Perspect. Biol. 4:a003681. doi: 10.1101/cshperspect.a003681
Perot, P., Bolze, P. A., and Mallett, F. (2012). “From Viruses to Genes: Syncytins,” in Viruses: Essential Agents of Life, ed G. Witzany (Dordrecht: Springer), 325–361. doi: 10.1007/978-94-007-4899-6_17
Perotto, D., and Baluska, F. (2012). Signaling and Communication in Plant Symbiosis. Heidelberg: Springer. doi: 10.1007/978-3-642-20966-6
Petrov, A. S., Bernier, C. R., Hsiao, C., Norris, A. M., Kovacs, N. A., Waterbury, C. C., et al. (2014). Evolution of the ribosome at atomic resolution. Proc. Natl. Acad. Sci. U. S. A. 111:10251–10256. doi: 10.1073/pnas.1407205111
Petrov, A. S., Gulen, B., Norris, A. M., Kovacs, N. A., Bernier, C. R., Lanier, K. A., et al. (2015). History of the ribosome and the origin of translation. Proc. Natl. Acad. Sci. U. S. A. 112:15396–15401. doi: 10.1073/pnas.1509761112
Plattner, H. (2016). “Principles of intracellular signalling in ciliated protozoa – a brief outline,” in Biocomminication of Ciliates, eds G. Witzany and M. Nowacki (Dordrecht: Springer), 13–34. doi: 10.1007/978-3-319-32211-7_2
Potapova, T. (2012). “Cell-to-cell communication in the tip growth of mycelial fungi,” in Biocommunication of Fungi, ed G. Witzany (Dordrecht:Springer), 103–114. doi: 10.1007/978-94-007-4264-2_7
Regnier, F. E. (1971). Semiochemical—structure and function. Biol. Reprod. 4, 309–326. doi: 10.1093/biolreprod/4.3.309
Rohwer, F., Youle, M., Maughan, H., and Hisakawa, N. (2014). Life inOur Phage World. A Centennial Field Guide to the Earth's Most Diverse Inhabitants. San Diego: Wholon.
Root-Bernstein, M., and Root-Bernstein, R. (2015). The ribosome as a missing link in the evolution of life. J. Theor. Biol. 367, 130–158. doi: 10.1016/j.jtbi.2014.11.025
Root-Bernstein, R. S., and Dillon, P. F. (1997). Molecular complementarity I: the complementarity theory of the origin and evolution of life. J. Theor. Biol. 188, 447–479. doi: 10.1006/jtbi.1997.0476
Sanjuán, R. (2018). Collective properties of viral infectivity. Curr. Opin. Virol. 33, 1–6. doi: 10.1016/j.coviro.2018.06.001
Schauder, S., and Bassler, B. L. (2001). The languages of bacteria. Genes Dev. 15, 1468–1480. doi: 10.1101/gad.899601
Schroedinger, E. (1944). What is Life? The Physical Aspect of the Living Cell. London: Cambridge University Press.
Schudoma, C. (2011). It's a loop world—single strands in RNA as structural and functional elements. Biomol. Conc. 2, 171–181. doi: 10.1515/bmc.2011.016
Schuster, P. (2011). Mathematical modeling of evolution. Solved and open problems. Theory Biosci. 130, 71–89. doi: 10.1007/s12064–010-0110-z
Searle, J. (1976). Speech Acts: An Essay in the Philosophy of Language. Cambridge: Cambridge University Press.
Seligmann, H. (2019). Syntenies between cohosted mitochondrial, chloroplast, and phycodnavirus genomes: functional mimicry and/or common ancestry? DNA Cell Biol. 38, 1257–1268. doi: 10.1089/dna.2019.4858
Shah, V., de Bouter, J., Pauli, Q., Tupper, A. S., and Higgs, P. G. (2019). Survival of RNA replicators is much easier in protocells than in surface-based, spatial systems. Life 9:E65. doi: 10.3390/life9030065
Shannon, C. E., and Weaver, W. (1949). The Mathematical Theory of Communication. Urbana: University of Illinois Press.
Shapiro, J. A. (1998). Thinking about bacterial populations as multicellular organisms. Ann. Rev.Microbiol. 52, 81–104. doi: 10.1146/annurev.micro.52.1.81
Shapiro, J. A. (2009). Revisiting the central dogma in the 21st century. Ann. N. Y. Acad. Sci. 1178, 6–28. doi: 10.1111/j.1749-6632.2009.04990.x
Shapiro, J. A. (2014). Epigenetic control of mobile DNA as an interface between experience and genome change. Front. Genet. 5:87. doi: 10.3389/fgene.2014.00087
Shapiro, J. A. (2016). Nothing in evolution makes sense except in the light of genomics: read-write genome evolution as an active biological process. Biology. 5:E27. doi: 10.3390/biology5020027
Sicard, A., Michalakis, Y., Gutiérrez, S., and Blanc, S. (2016). The strange lifestyle of multipartite viruses. PLoS Pathog. 12:e1005819. doi: 10.1371/journal.ppat.1005819
Sicard, A., Pirolles, E., Gallet, R., Vernerey, M. S., Yvon, M., Urbino, C., et al. (2019). A multicellular way of life for a multipartite virus. Elife. 8:e43599. doi: 10.7554/eLife.43599
Smit, S., Yarus, M., and Knight, R. (2006). Natural selection is not required to explain universal compositional patterns in rRNA secondary structure categories. RNA 12, 1–14. doi: 10.1261/rna.2183806
Soll, D. (2012). “Signal transduction pathways regulating switching, mating and biofilm formationin Candida albicans and related species,” in Biocommunication of Fungi, ed G. Witzany (Dordrecht: Springer), 85–102. doi: 10.1007/978-94-007-4264-2_6
Spadafora, C. (2016). Soma to germline inheritance of extrachromosomal genetic information via a LINE-1 reverse transcriptase-based mechanism. Bioessays. 38:726–733. doi: 10.1002/bies.201500197
Stadler, P. F., and Schuster, P. (1992). Mutation in autocatalytic reaction networks. An analysis based on perturbation theory. J. Math. Biol. 30, 597–631. doi: 10.1007/BF00948894
Stedman, K. M. (2015). Deep recombination: RNA and ssDNA virus genes in DNA virus and host genomes. Annu. Rev. Virol. 2, 203–217. doi: 10.1146/annurev-virology-100114–055127
Stedman, K. M. (2018). Viral recombination: ecology, evolution, and pathogenesis. Viruses 10:E358. doi: 10.3390/v10070358
Stokar-Avihail, A., Tal, N., Erez, Z., Lopatina, A., and Sorek, R. (2019). Widespread utilization of peptide communication in phages infecting soil and pathogenic bacteria. Cell Host Mic. 25, 746–755. doi: 10.1016/j.chom.2019.03.017
Tartaglia, G. G. (2016). The grand challenge of characterizing ribonucleoprotein networks. Front. Mol. Biosci. 3:24. doi: 10.3389/fmolb.2016.00024
Tomasello, M. (2008). Origins of Human Communication. Cambridge: MIT Press. doi: 10.7551/mitpress/7551.001.0001
Turing, A. (1950). Computing machinery and intelligence. Mind 59, 433–460. doi: 10.1093/mind/LIX.236.433
Vaidya, N., Manapat, M. L., Chen, I. A., Xulvi-Brunet, R., Hayden, E. J., and Lehman, N. (2012). Spontaneous network formation among cooperative RNA replicators. Nature 491, 72–77. doi: 10.1038/nature11549
Vaidya, N., Walker, S. I., and Lehman, N. (2013). Recycling of informational units leads to selection of replicators in a prebiotic soup. Chem. Biol. 20, 241–252. doi: 10.1016/j.chembiol.2013.01.007
Vennera, S., Feschotte, C., and Biémonta, C. (2009). Transposable elements dynamics: toward a community ecology of the genome. Trends Genet. 25, 317–323. doi: 10.1016/j.tig.2009.05.003
Villarreal, L. P. (2005). Viruses and the Evolution of Life. Washington: ASM Press. doi: 10.1128/9781555817626
Villarreal, L. P. (2009a). The source of self: genetic parasites and the origin of adaptive immunity. Ann. N. Y. Acad. Sci. 1178, 194–232. doi: 10.1111/j.1749–6632.2009.05020.x
Villarreal, L. P. (2009b). Origin of Group Identity: Viruses, Addiction and Cooperation. New York, NY: Springer.
Villarreal, L. P. (2012). “The addiction module as a social force,” in Viruses: Essential Agents of Life, ed G. Witzany (Dordrecht: Springer Science+Business Media), 107–146. doi: 10.1007/978-94-007-4899-6_6
Villarreal, L. P. (2015). Force for ancient and recent life: viral and stem-loop RNA consortia promote life. Ann. N. Y. Acad. Sci. 1341, 25–34. doi: 10.1111/nyas.12565
Villarreal, L. P. (2016a). Viruses and the placenta: the essential virus first view. APMIS. 124, 20–30. doi: 10.1111/apm.12485
Villarreal, L. P. (2016b). Persistent virus and addiction modules: an engine of symbiosis. Curr. Opin. Microbiol. 31, 70–79. doi: 10.1016/j.mib.2016.03.005
Villarreal, L. P., and Witzany, G. (2010). Viruses are essential agents within the roots and stem of the tree of life. J. Theor. Biol. 262, 698–710. doi: 10.1016/j.jtbi.2009.10.014
Villarreal, L. P., and Witzany, G. (2013a). The DNA habitat and its RNA inhabitants: at the dawn of RNA sociology. Gen. Ins. 6, 1–12. doi: 10.4137/GEI.S11490
Villarreal, L. P., and Witzany, G. (2013b). Rethinking quasispecies theory: from fittesttype to cooperative consortia. World J. Biol. Chem. 4, 79–90. doi: 10.4331/wjbc.v4.i4.79
Villarreal, L. P., and Witzany, G. (2015). When competing viruses unify: evolution, conservation, and plasticity of genetic identities. J. Mol. Evol. 80, 305–318. doi: 10.1007/s00239–015-9683-y
Villarreal, L. P., and Witzany, G. (2019). That is life: communicating RNA networks from viruses and cells in continuous interaction. Ann. N. Y. Acad. Sci. 1447, 5–20. doi: 10.1111/nyas.14040
Visick, K. L., and Fuqua, C. (2005). Decoding microbial chatter: cell–cell communication in bacteria. J. Bacteriol. 187, 5507–5519. doi: 10.1128/JB.187.16.5507-5519.2005
Wang, S., and Lu, Z. (2017). “Secondary Metabolites in Archaea and Extreme Environments,” in Biocommunication of Archaea, ed G. Witzany (Cham: Springer), 235–239. doi: 10.1007/978-3-319-65536-9_14
Whitehead, A. N., and Russell, B. (1910/1912/1913). Principia Mathematica. Cambridge: Cambridge University Press.
Wiener, N. (1948). Cybernetics, or Control and Communication in the Animal and the Machine. New York, NY: Wiley.
Witzany, G. (1993). Natur der Sprache - Sprache der Natur. Sprachpragmatische Philosophie der Biologie. Wuerzburg: K?nigshausen & Neumann.
Witzany, G. (1995). From the “logic of the molecular syntax” to molecular pragmatism. Explanatory deficits in Manfred Eigen's concept of language and communication. Evol. Cogn. 1, 148–168.
Witzany, G. (2005). Natural history of life: history of communication logics and dynamics. S. E. E. D. J. 5, 27–55.
Witzany, G. (2006). Natural genome editing competences of viruses. Acta Biotheor. 54, 235–253. doi: 10.1007/s10441-006-9000-7
Witzany, G. (2009). Non-coding RNAs: persistent viral agents as modular tools for cellular needs. Ann. N. Y. Acad. Sci. 1178, 244–267. doi: 10.1111/j.1749–6632.2009.04989.x
Witzany, G. (2010). Biocommunication and Natural Genome Editing. Dordrecht: Springer. doi: 10.1007/978-90-481-3319-2
Witzany, G. (2011a). Can mathematics explain the evolution of human language? Comm. Integr. Biol. 4/5, 1–5. doi: 10.4161/cib.4.5.16426
Witzany, G. (2011b). Biocommunication in Soil Microorganisms. Heidelberg: Springer. doi: 10.1007/978-3-642-14512-4
Witzany, G. (2011c). “Introduction: key levels of biocommunication of bacteria,” in Biocommunication in Soil Microorganisms, ed G. Witzany (Dortrecht: Springer), 1–34. doi: 10.1007/978-3-642-14512-4_1
Witzany, G. (2011d). The agents of natural genome editing. J. Mol. Cell Biol. 3, 181–189. doi: 10.1093/jmcb/mjr005
Witzany, G. (2012b). “From molecular entities to competent agents: viral infection-derived consortia act asnatural genetic engineers,” in Viruses: Essential Agents of Life, ed G. Witzany (Dordrecht: Springer), 407–419. doi: 10.1007/978-94-007-4899-6_20
Witzany, G. (2014a). “Language and communication as universal requirements for life,” in Astrobiology: An Evolutionary Approach, ed V. Kolb (Boka Raton: CRC Press), 349–370.
Witzany, G. (2014b). Biocommunication of Animals. Dordrecht: Springer. doi: 10.1007/978-94-007-7414-8
Witzany, G. (2014c). Pragmatic turn in biology: from biological molecules to genetic content operators. World J. Biol. Chem. 5, 279–285. doi: 10.4331/wjbc.v5.i3.279
Witzany, G. (2015). Life is physics and chemistry and communication. Ann. N.Y. Acad. Sci. 1341, 1–9. doi: 10.1111/nyas.12570
Witzany, G. (2016a). “Key levels of biocommunication,” in Biocommunication: Sign-Mediated Interactions Between Cells and Organisms, eds R. Gordon and J. Seckbach (Singapore: World Scientific), 37–61. doi: 10.1142/9781786340450_0002
Witzany, G. (2016b). “Introduction: Key Levels of Biocommunication of Ciliates,” in Biocomminication of Ciliates, eds G. Witzany and M. Nowacki (Dordrecht: Springer), 1–12. doi: 10.1007/978-3-319-32211-7_1
Witzany, G. (2016c). Crucial steps to life: from chemical reactions to code using agents. Biosystems. 140, 49–57. doi: 10.1016/j.biosystems.2015.12.007
Witzany, G. (2017a). “Artificial and natural genetic information processing,” in Information Studies and the Quest for Transdisciplinarity, eds M. Burgin and W. Hofkirchner (Singapore: World Scientific), 523–547. doi: 10.1142/9789813109001_0019
Witzany, G. (2017b). Biocommunication of Archaea. Dordrecht: Springer. doi: 10.1007/978-3-319-65536-9
Witzany, G. (2017c). Two genetic codes: repetitive syntax for active non-coding RNAs; non - repetitive syntax for the DNA archives. Comm. Integ. Biol. 10:e1297352. doi: 10.1080/19420889.2017.1297352
Witzany, G. (2019). “Communication as the main characteristic of life,”in Handbook of Astrobiology, ed V. Kolb (Boka Raton: CrC Press), 91–105. doi: 10.1201/b22230-9
Witzany, G., and Baluska, F. (2012a). Life's code script does not code itself. The machine metaphor for living organisms is outdated. EMBO Rep. 13, 1054–1056. doi: 10.1038/embor.2012.166
Witzany, G., and Baluska, F. (2012b). Biocommunication of Plants. Heidelberg: Springer. doi: 10.1007/978-3-642-23524-5
Witzany, G., and Nowacki, M. (2016). Biocommunication of Ciliates. Dordrecht: Springer. doi: 10.1007/978-3-319-32211-7
Keywords: sign mediated interactions, communication, cellular life, viruses, RNAs, evolution, essential agents of life, biocommunication
Citation: Witzany G (2020) What is Life? Front. Astron. Space Sci. 7:7. doi: 10.3389/fspas.2020.00007
Received: 22 January 2020; Accepted: 17 February 2020;
Published: 18 March 2020.
Edited by:
Tetyana Milojevic, University of Vienna, AustriaReviewed by:
Jordi Gómez, Instituto de Parasitología y Biomedicina López-Neyra (IPBLN), SpainLuis Villarreal, University of California, Irvine, United States
Copyright © 2020 Witzany. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guenther Witzany, d2l0emFueUBzYmcuYXQ=
 Guenther Witzany
Guenther Witzany