- 1State Key Laboratory of Agricultural Microbiology, College of Veterinary Medicine, Huazhong Agricultural University, Wuhan, China
- 2Key Laboratory of Development of Veterinary Diagnostic Products, Ministry of Agriculture of the People's Republic of China, Wuhan, China
- 3Key Laboratory of Animal Epidemical Disease and Infectious Zoonoses, Ministry of Agriculture, Huazhong Agricultural University, Wuhan, China
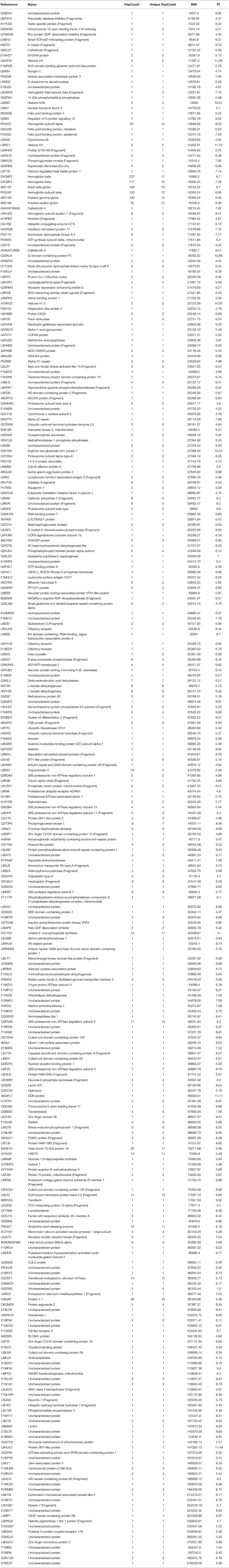
A number of studies have demonstrated that Babesia orientalis (B. orientalis) can only infect water buffalo (Bubalus bubalis) and not dairy cattle (Bos taurus) or beef cattle (Bos taurus), even though all three belong to the tribe Bovini and have close evolutionary relationships. In addition, Babesia species are intracellular protozoans that obligately parasitize in erythrocytes. This may indicate that the infection specificity is due to differences in erythrocyte proteins. Totals of 491, 1,143, and 1,145 proteins were identified from water buffalo, beef cattle, and dairy cattle, respectively, by searching the Uniprot and NCBI databases. The number of proteins identified for water buffalo was far lower than for beef cattle and dairy cattle, particularly in the range from 15 to 25 kDa. Remarkably, 290 identified proteins were unique to water buffalo, of which putative gamma-globin and putative epsilon-globin had a significant possibility of being relevant to the survival of B. orientalis only in water buffalo. A total of 2,222 proteins were annotated in terms of molecular function, biological process, and cellular component according to GO annotation. The number of proteins of water buffalo in oxygen binding was far higher than for beef cattle and dairy cattle. This is the first time that the protein profiles of water buffalo, beef cattle, and dairy cattle have been comparatively analyzed. The uniquely expressed proteins in water buffalo obtained in this study may provide new insights into the mechanism of B. orientalis infection exclusivity in water buffalo and may be a benefit for the development of strategies against B. orientalis.
Introduction
Babesia is a tick-borne apicomplexan parasite that can cause a zoonotic disease known as babesiosis (1–3). A unique characteristic of Babesia is that it is obligate to parasitize and reproduce within erythrocytes. It can infect an extensive range of mammalian and even humans. The main clinical presentations are fever, anemia, hemoglobinuria, jaundice, and even death (2, 4). Babesia gives rise to a massive burden of morbidity, which not only leads to enormous economic losses but also hampers the development of the livestock industry (1, 2, 4, 5). Over 100 species of Babesia have been reported, and they have a worldwide distribution (1). In China, five of these can infect cattle, namely Babesia bovis (B. bovis), Babesia bigemina (B. bigemina), Babesia ovata, Babesia major, and Babesia orientalis (B. orientalis) (4). B. orientalis was first discovered in central and south China in 1984 (4, 6, 7). According to the pathogenicity, morphology, in vitro cultivation characteristics, and phylogenetic analysis of the 18S rRNA gene, it was recognized as a new Babesia species and named B. orientalis in 1997 (4, 8–12). Furthermore, it is only transmitted by Rhipicephalus haemaphysaloides and exclusively parasitizes in the erythrocytes of water buffalo rather than in those of beef cattle or dairy cattle (1, 4, 10, 13–15). In contrast, B. bovis and B. bigemina can infect both water buffalo and cattle through Rhipicephalus and Ixodes (1). Even though substantial efforts have been made in genome sequencing, in vitro cultivation, and with diagnostic methods, the molecular mechanism of the specific invasion of the erythrocytes of water buffalo remains unknown (13, 16, 17). In terms of genome sequencing, only the mitochondrial and apicoplast genomes of B. orientalis and the whole genome of water buffalo have been reported, which provide little information clarifying the mechanisms of invasion specificity (18, 19).
Proteomics, an emerging technology, refers to the protein-expression profiles of a gene, a cell, or a tissue in a particular period (5, 20–22). Unlike the immutable genomics, proteomics is a post-genomic method and is preferentially sensitive to dynamic changes in the parasite and the host; that is, it changes along with the environment. An increasing number of proteomic methods are used to determine the differences between normal and diseased states so as to search for potential drugs and treatment targets (21, 23). For instance, proteomics is used in the analysis of female Rhipicephalus Microplus-stage-specific protein expression of B. bovis and also in finding biomarkers for the diagnosis of Babesia canis (21, 24). In addition, a protein profile of mammalian erythrocyte membranes has been identified by matrix-assisted laser desorption/ionization time-of-flight/mass spectrometer (MALDI-TOF/MS) (22). Proteomics has also been applied to develop vaccines against tick-borne diseases (5). However, no reports have been made of the application of proteomics to B. orientalis, and no comparison has been made of the erythrocyte proteins of water buffalo, beef cattle, and dairy cattle. There are several approaches to the study of proteomics, such as by two-dimensional electrophoresis/mass spectrometer (2-DE/MS), MALDI-TOF/MS, or liquid chromatography mass spectrometer (LC-MS/MS) (25–28). The shotgun method has the advantage of identifying more proteins than other methods of proteomics, including proteins that have extreme isoelectric point (pI) and molecular mass (Mw) values (26, 29, 30).
To clarify the mechanism of infection specificity, three aspects should be considered: the host, the parasite, and both in conjunction. As a result, this article focuses on the host: the erythrocyte of water buffalo. In this study, proteomics were used to find differences among the erythrocytes of water buffalo, beef cattle, and dairy cattle, which may provide new insights into the mechanisms by which B. orientalis exclusively parasitizes the erythrocytes of water buffalo and may be beneficial for devising strategies for inhibiting the survival and replication of B. orientalis in those erythrocytes.
Materials and Methods
Experimental Animals and Blood Collection
A 1-year-old water buffalo, 1-year-old beef cattle and 1-year-old dairy cattle were verified free of B. orientalis by microscopic examination, reverse line blot, and real-time PCR (13, 17). All of the blood samples were withdraw into sterile vacuum tubes containing anticoagulant with EDTA (1.5 mg/ml blood).
Erythrocyte Protein Preparation
The steps taken to purify the red blood cells (RBCs) were essential and indispensable for LC-MS/MS. The procedures used in this article followed the previously reported protocols of Pesciotta et al. (31), Pasini et al. (32), and Bryk and Wisniewski (28) combined. In brief, the small cells, such as platelets and microparticles, were removed by low-speed centrifuge and multiple cold buffer washes. RBCs were centrifuged at 1,500 rpm for 10 min at 4 °C. The supernatant, especially the layer of white blood cells, was removed. The pellet was resuspended in cold phosphate-buffered saline (PBS) to the original volume and then mixed gently. The above steps were repeated three times or more until the supernatant was clean. Even though most of the white blood cells had been discarded by removing the layer of white blood cells, the remaining white blood cells and other cells larger than RBCs needed to be removed by white cell filters (Plasmodipur, Euro-diagnostica, Arnhem, the Netherlands). The purification of the RBC pellet was evaluated by making smears, and the number of non-RBCs was counted by microscopy. Once the ratio of non-RBCs (the number of non-RBCS/total cells) was <0.001, the RBC pellet could be subjected to the next steps (31, 32).
Next, the RBC pellet was lysed with 20 ml of cold red cell lysis buffer (Tiangen Biotech, Beijing, China), standing for 30 min at 4°C. The RBCs were then beaten 15–20 times using a 1-ml syringe. The lysate was then centrifuged at 12,000 rpm for 10 min at 4°C. The pellet was suspended in 30 ml of PBS, and the above step was repeated five more times. Finally, the resuspended protein solution was stored in PBS at −20°C for 1-DE and shotgun analysis.
SDS-PAGE and Silver Staining
One hundred milligrams of proteins from each specimen was denatured in an equal volume of 2 × protein loading buffer (0.2 M DTT, 20% glycerol, 0.1 M Tris-HCl, pH 6.8, 4% SDS, 0.2% bromophnol blue) at 100°C for 12 min. Denatured proteins were separated through 12% SDS-PAGE at 70 V for 30 min and then 100 V for 1 h. The gel was then stained for 30 min in a solution containing 0.07% (wt./vol.) coomassie brilliant blue G250 (CBB) (Invitrogen, Carlsbad, CA, USA). The SDS-PAGE gels were stained with a silver kit (Beyotime Biotechnology, Shanghai, China).
Filter-Aided Sample Preparation (FASP Digestion)
Two hundred micrograms of proteins for each sample was incorporated into 30 μl of SDT buffer (4% SDS, 100 mM DTT, 150 mM Tris-HCl pH 8.0). The detergent, DTT, and other low-molecular-weight components were removed using UA buffer (8 M Urea, 150 mM Tris-HCl pH 8.0) by repeated ultrafiltration (Microcon units, 10 kD). Then, 100 μl of iodoacetamide (100 mM IAA in UA buffer) was added to block reduced cysteine residues, and the samples were incubated for 30 min in darkness. The filters were washed with 100 μl of UA buffer three times and then with 100 μl of 25 mM NH4HCO3 buffer twice. Finally, the protein suspensions were digested with 4 μg of trypsin (Promega, Madison, Wisconsin, US) in 40 μl of 25 mM NH4HCO3 buffer overnight at 37°C, and the resulting peptides were collected as a filtrate. The peptides of each sample were desalted on C18 Cartridges [Empore™ SPE Cartridges C18 (standard density)], bed I.D. 7 mm, volume 3 ml (Sigma), concentrated by vacuum centrifugation and reconstituted in 40 μl of 0.1% (v/v) formic acid. The peptide content was estimated by UV light spectral density at 280 nm using an extinction coefficient of 1.1 of 0.1% (g/l) solution, which was calculated on the basis of the frequency of tryptophan and tyrosine in vertebrate proteins.
HPLC-ESI-MS/MS (Shotgun Analysis)
The peptide mixture (3 ug) was loaded onto a reversed-phase trap column (Thermo Scientific Acclaim PepMap100, 100 μm × 200 mm, nanoViper C18) connected to a C18 reversed-phase analytical column (Thermo Scientific Easy Column, 10 cm long, 75 μm inner diameter, 3 μm resin) in buffer A (0.1% Formic acid) and separated with a linear gradient of buffer B (84% acetonitrile and 0.1% Formic acid) at a flow rate of 300 nl/min controlled by IntelliFlow technology. The linear gradient was determined by the project proposal: 0–35% buffer B for 50 min, 35–100% buffer B for 5 min, then being held in 100% buffer B for 5 min.
LC-MS/MS analysis was performed on a Q Exactive mass spectrometer (Thermo Scientific) that was coupled to an Easy nLC (Thermo Fisher Scientific) for 60 min. The mass spectrometer was operated in positive ion mode. MS data were acquired using a data-dependent top 10 method, dynamically choosing the most abundant precursor ions from the survey scan (300–1,800 m/z) for HCD fragmentation. The automatic gain control (AGC) target was set to 3e6 and the maximum inject time to 10 ms. The dynamic exclusion duration was 40.0 s. Survey scans were acquired at a resolution of 70,000 at m/z 200, the resolution for HCD spectra was set to 17, 500 at m/z 200, and the isolation width was 2 m/z. The normalized collision energy was 30 eV, and the underfill ratio, which specifies the minimum percentage of the target value likely to be reached at maximum fill time, was defined as 0.1%. The instrument was run with peptide recognition mode enabled.
Protein Identification and Annotation
The MS/MS spectra were searched for in the UniProtKB Bovinae and Babesia database (56445 total entries, downloaded 20170807) using the MASCOT engine (Matrix Science, London, UK; version 2.4). For protein identification, the following options were used. Peptide mass tolerance = 20 ppm, MS/MS tolerance = 0.1 Da, enzyme = trypsin, missed cleavage = 2, fixed modification: carbamidomethyl (C), variable modification:Oxidation (M), peptides FDR ≦ 0.01, protein FDR ≦ 0.01.
Gene Ontology (GO) Annotation
The protein sequences of differentially expressed proteins were retrieved in batches from the UniProtKB database (UniProtKB Bovinae database). The retrieved sequences were locally searched for in the SwissProt database (UniProtKB Bovinae database) using NCBI BLAST+ client software to find homolog sequences from which the functional annotation could be transferred to the studied sequences. In this work, the top 10 blast hits with E-values of <1e-3 for each query sequence were retrieved and were loaded into Blast2GO9 (UniProtKB Bovinae database) for GO mapping and annotation. An annotation configuration with an E-value filter of 1e-6, default gradual EC weights, a GO weight of 5, and an annotation cutoff of 75 was chosen. Un-annotated sequences were then re-annotated with more permissive parameters. The sequences without BLAST hits and un-annotated sequences were then selected to go through an InterProScan10 against the EBI database to retrieve functional annotations of protein motifs, and the InterProScan GO terms were merged with the annotation set. The GO annotation results were plotted by R scripts.
Results
SDS-PAGE and Silver Staining
The erythrocyte proteins of water buffalo, beef cattle, and dairy cattle were separated by SDS-PAGE. The SDS-PAGE results were visualized through CBB and silver staining (Figure 1). There were obviously far more protein bands for beef and dairy cattle than for water buffalo, especially in the range from 15 to 25 kDa. The number of proteins detected by the shotgun method was obviously far higher than that through SDS-PAGE.
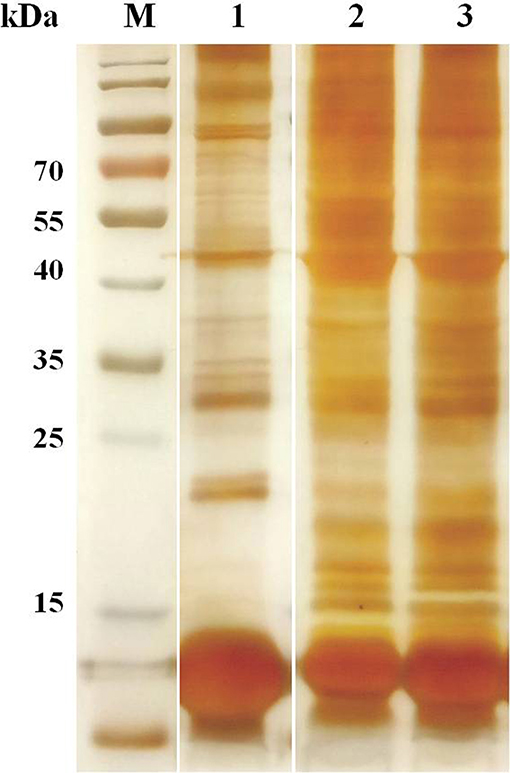
Figure 1. The erythrocyte proteins of water buffalo, beef cattle, and dairy cattle were separated by SDS-PAGE, and then silver staining was performed.
Global Analysis of Erythrocyte Proteomes
Proteins were digested via FASP and were subjected to shotgun LC-ESI-MS/MS analysis. After removing redundant sequences, the identified proteins were searched for in the Uniprot and NCBI databases (Supplementary Table 1). A total of 491, 1,143, and 1,145 proteins (pepcount ≥1) were identified in water buffalo, beef cattle, and dairy cattle, with 4,012 peptides including 1,825 unique peptides, 6,771 peptides including 5,380 unique peptides, and 6,519 peptides including 4,881 unique peptides, respectively.
The erythrocyte protein profiles of water buffalo, beef cattle, and dairy cattle were analyzed with the Venny 2.1.0 tool (http://bioinfogp.cnb.csic.es/tools/venny/index.html). The resulting Venn diagram is shown in Figure 2. It shows that 67 proteins were common to them all, the majority of which were house-keeping genes including ATP synthase subunits, heat shock proteins (HSP70 and HSP90), actin, tubulin, ribosomal protein, and so on. A total of 63 proteins were common to water buffalo and beef cattle, 71 proteins were common to water buffalo and dairy cattle, and 289 proteins were common to beef cattle and dairy cattle. In obvious contrast to water buffalo, the protein profile of beef cattle was, for the most part, relatively similar to that of dairy cattle. Furthermore, 290 proteins were water buffalo-biased, 718 proteins were dairy cattle-biased, and 724 proteins were beef cattle-biased. The 290 proteins that were only identified in water buffalo are detailed in Table 1; these might be related to the infection specificity.
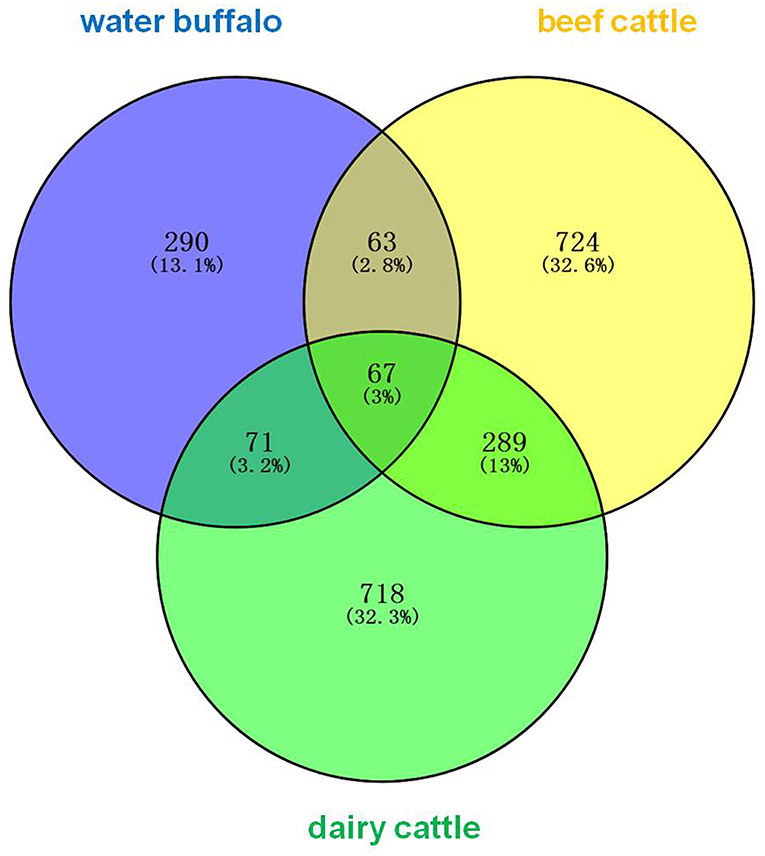
Figure 2. The erythrocyte proteins of water buffalo, beef cattle, and dairy cattle were identified and comparatively analyzed in a Venn diagram. The identified erythrocyte proteins of water buffalo are in blue, those of beef cattle are in yellow, and those of dairy cattle are in green.
Theoretical Two-Dimensional Distribution of the Identified Proteins
The distributions of the Mw and pI values of the identified proteins from water buffalo, dairy cattle, and beef cattle are shown in Figure 3. Mw and pI were calculated by using the compute Mw/pI tool (http://cn.expasy.org/tools/pi_tool.html) according to the predicted amino acid sequences. They both played directive roles in the characterization of the proteins. Most of the identified proteins of water buffalo, dairy cattle, and beef cattle were in the range of 15 to 55 kDa and more than 115 kDa, accounting for 68.4% (336/491), 65.9% (754/1,145), and 67.3% (769/1,143) of their totals, respectively. Analysis of molecular weight revealed that there was a significant difference in the range of 15 to 25 kDa, with the number of proteins being obviously less in water buffalo than in dairy and beef cattle.
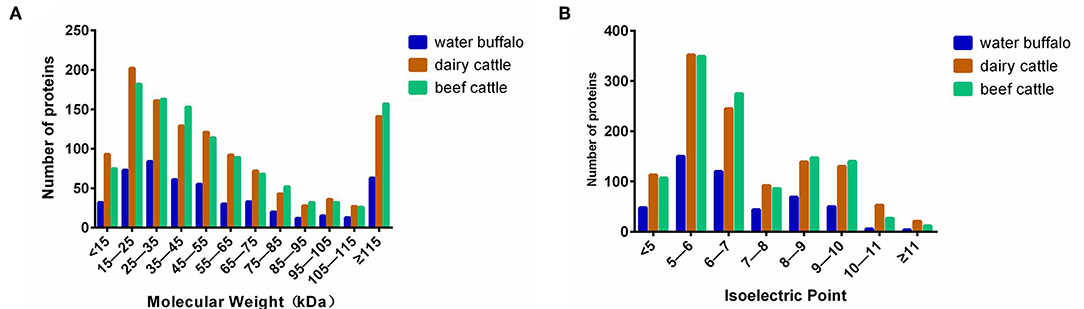
Figure 3. The distributions of the Mw and pI values of the identified erythrocyte proteins of water buffalo, dairy cattle, and beef cattle were comparatively analyzed. That of water buffalo is shown in blue, that of dairy cattle in red, and that of beef cattle in green. (A) The distribution of Mw. (B) The distribution of pI.
In terms of pI, the great majority of the identified proteins of water buffalo, dairy cattle, and beef cattle were in the range of 5–7, accounting for 54.9% (270/491), 52.1% (597/1,145), and 54.6% (624/1,143) of their totals, respectively. In the range of 5–6, water buffalo had a significantly lower protein count than do dairy and beef cattle.
Gene Ontology Annotation
A total of 2,222 proteins of water buffalo, dairy cattle, and beef cattle were annotated in terms of molecular function, biological process, and cellular component according to the Gene Ontology Annotation (http://www.ebi.ac.uk/goa/) (Figure 4).
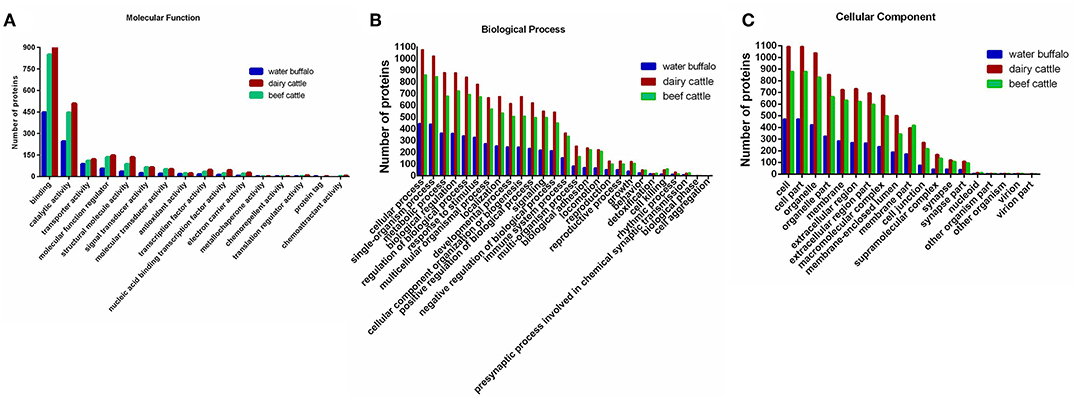
Figure 4. A total of 2,222 identified erythrocyte proteins of water buffalo, dairy cattle, and beef cattle were annotated, respectively, in terms of molecular function, biological process, and cellular component. (A) Molecular function. (B) Biological process. (C) Cellular component. Those of water buffalo are shown in blue, dairy cattle in red, and beef cattle in green.
For the molecular function annotation, the numbers of water buffalo, beef cattle, and dairy cattle in level two were 15, 15, and 16 respectively, of which 14 were common to them all. A large proportion of proteins in level two were assigned to binding (GO:0005488) and catalytic activity (GO:0003824), significantly more than other categories. The majority of proteins in binding categories were assigned to protein binding (GO:0005515), ion binding (GO:0043167), and organic cyclic compound binding (GO:0097159). Remarkably, the number of proteins in oxygen binding (GO:0019825) was higher in water buffalo than in beef cattle and dairy cattle, even though the number was far lower in other subcategories in level three. This may indicate that oxygen binding is more active in water buffalo, which may be a benefit for Babesia survival. Most proteins in catalytic activity were relevant to hydrolase activity (GO:0016787), oxidoreductase activity (GO:0016491), and transferase activity (GO:0016740).
In terms of the biological process categories, most proteins were categorized into metabolic processes (GO:0008152), cellular processes (GO:0009987), and single-organism processes (GO:0044699). Among the GO terms, there was no significant difference in processes between species, even though cell aggregation was exclusive to cattle. It was noteworthy that far fewer proteins were categorized into cellular processes in water buffalo than in dairy cattle and beef cattle, unlike for other processes.
In the cellular component categories, the number of proteins was far higher for dairy cattle than for water buffalo and beef cattle in level two. Most of the proteins were assigned to cell (GO:0005623), cell part (GO:0044464), organelle (GO:0043226), organelle part (GO:0044422), and membrane (GO:0016020). Among these, the numbers of plasma membrane (GO:0005886) components for water buffalo, beef cattle, and dairy cattle were 170, 408, and 403 respectively, of which 92 proteins were unique to water buffalo.
Significant Differences in Water Buffalo, Dairy Cattle, and Beef Cattle
The number of peptides (peptide count) is directly connected with the relative abundance of the proteins in erythrocytes as identified by LC-MS/MS. Therefore, based on the number of peptides, peptide counts of ≥20 of the identified erythrocyte proteins of water buffalo, beef cattle, and dairy cattle were selected and compared with each other. The number of peptide counts ≥20 were 25, 56, and 50 in water buffalo, beef cattle, and dairy cattle, respectively. Even though the number with a peptide count ≥20 in water buffalo was lower than in beef cattle and dairy cattle, the species of those proteins were similar. Most were hemoglobin, skeleton proteins (spectrin, ankyrin, actin), heat shock proteins, anion exchange protein, and so on. Remarkably, putative gamma-globin and putative epsilon-globin were only detected in water buffalo; they were not detected in beef cattle and dairy cattle. Furthermore, the relative abundance of putative gamma-globin and putative epsilon-globin was high in all of the identified proteins, and the peptide counts were 130 and 76, respectively. Therefore, gamma-globin and epsilon-globin may play key roles and are promising explanations for B. orientalis only invading or multiplying in the RBCs of water buffalo.
Discussion
The hemoprotozoan was identified as a novel Babesia species and named B. orientalis in 1997 (13). The only natural host was found to be water buffalo, and not beef cattle and dairy cattle, although all of them belong to the tribe of Bovini (10, 13). In contrast, B. bovis and B. bigemina can infect not only water buffalo but also beef cattle and dairy cattle. To date, no studies or articles have become available regarding this difference. This is because many challenges and difficulties limit the investigation of this problem, including the difficulty of obtaining the parasites, the difficulty of continuous cultivation, the non-applicability of gene-editing techniques (CRISPR), and so on.
Due to the fact that Babesia can only invade and reside in erythrocytes, the parasite will interact with the erythrocyte through ligands and receptors (33). Many studies have focused on this, and several interaction ligands in parasites and receptors in erythrocytes have been characterized in plasmodium (34). However, there was no significant information available on the recognition ligands and receptors in Babesia. Furthermore, most studies pay attention to finding the ligands in the membrane of erythrocytes. However, when parasites reside into the erythrocyte, the contents of the RBC are equally necessary to the parasites. Therefore, this study was from the perspective of the integral erythrocyte proteome including both membrane and cytoplasmic proteins, making it more comprehensive and rigorous. In this study, a comprehensive analysis was performed to compare the erythrocyte proteomes of water buffalo, beef cattle, and dairy cattle. Overall, a total of 491, 1,143, and 1,145 proteins were identified in water buffalo, beef cattle, and dairy cattle, respectively. The number for water buffalo was far less than for beef cattle and dairy cattle, particularly in the range from 15 to 25 kDa, which was also exhibited in the SDS-PAGE results. Furthermore, the erythrocyte protein profile of beef cattle was far more similar to that of dairy cattle, and both were significantly divergent from that of water buffalo. Some significant molecular biases to water buffalo were identified, which may be related to the exclusive survival of B. orientalis in the RBCs of water buffalo. Putative gamma-globin and putative epsilon-globin were not detected, and no information is available for beef cattle and dairy cattle to date. Moreover, all of the identified proteins of water buffalo were relatively rich in putative gamma-globin and putative epsilon-globin. The two proteins were encoded by the hemoglobin subunit beta (HBB) gene and have functions in heme binding, iron ion binding, oxygen binding, and oxygen carrier activity. The number of proteins in oxygen binding (GO:0019825) in water buffalo is far higher than in beef cattle and dairy cattle, which increases the significance of deep investigation of these two proteins. Hemoglobin is vital to hemoprotozoan survival inside the RBC and, to date, it is regarded as the main energy source for most of the hemoprotozoan (35). Moreover, hemoglobin is covalently modified in order to inhibit the intake of amino acids by plasmodium but does not affect the normal functions (36). One article has also reported that malaria can cause an imbalance in the globin expression by using the CD34+ haematopoietic stem cell culture system (37). Therefore, further investigation of whether gamma-globin and epsilon-globin are the main reasons for the water buffalo infection specificity of B. orientalis would be worthwhile.
All in all, this study is the first to characterize and detail the erythrocyte protein profiles of water buffalo, beef cattle, and dairy cattle by using shotgun technology. In combination with bioinformatics analysis, it has clearly represented the differences among the erythrocyte proteomes of water buffalo, beef cattle, and dairy cattle. Even so, there are many challenges and obstacles that must still be faced. This study was the first to try to find some clues and to explain why water buffalo is the only host of B. orientalis and has provided new insights into this question. This study can also act as a guide for the development of vaccines and anti-B. orientalis survival agents.
Conclusion
In conclusion, this study obtained the complete erythrocyte proteomes of water buffalo, beef cattle, and dairy cattle and performed comparative analysis from several aspects including mw, pI, molecular function, biological process, and cellular component. A total of 290 uniquely expressed proteins were identified in water buffalo, which might be related to the infection specificity of B. orientalis to water buffalo. The mechanism for infection specificity is complex, and more work needs to be done to elucidate the reasons for the exclusive survival of B. orientalis in the erythrocytes of water buffalo rather than in beef and dairy cattle.
Data Availability Statement
All data obtained in this study had been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repository with the dataset identifiers PXD011408 and IPX0001371000.
Ethics Statement
The experimental animals were housed and treated in accordance with the stipulated rules for the regulation of the administration of affairs concerning experimental animals of P. R. China. All experiments were performed under the approval of the Laboratory Animals Research Centre of Hubei Province and the Ethics Committee of Huazhong Agricultural University (Permit number: HZAUCA-2016-007).
Author Contributions
JG performed the experiments. JG and YS participated in the data analysis. JG, YS, and YT helped with the diagnostic assays. JG and JZ edited the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by the National Key Research and Development Program of China (Grant No. 2017YFD0500402 and 2017YFD0501201), the National Key Basic Research Program (973 Program) of China (Grant No. 2015CB150302), the National Natural Science Foundation of China (Grant No. 31772729), and the Natural Science Foundation of Hubei Province (Grant No. 2017CFA020).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2019.00346/full#supplementary-material
Supplementary Table 1. The detail results of the mass spectrometry (LC-MS/MS).
Abbreviations
B. orientalis, Babesia orientalis; B. bovis, Babesia bovis; B. bigemina, Babesia bigemina; HSP, Heat shock protein; MALDI-TOF/MS, Matrix-assisted laser desorption/ionization time-of-flight/mass spectrometer; 2-DE/MS, Two-dimensional electrophoresis/mass spectrometer; LC-MS/MS, Liquid chromatography mass spectrometer; RBC, Red blood cell; Ip, Isoelectric point; Mw, Molecular mass; PBS, Phosphate buffered saline; Hp, Haptoglobin; Hb, Hemoglobin; HBB, hemoglobin subunit beta.
References
1. Uilenberg G. Babesia–a historical overview. Vet Parasitol. (2006) 138:3–10. doi: 10.1016/j.vetpar.2006.01.035
2. Schnittger L, Rodriguez AE, Florin-Christensen M, Morrison DA. Babesia: a world emerging. Infect Genet Evolut. (2012) 12:1788–809. doi: 10.1016/j.meegid.2012.07.004
3. Gohil S, Herrmann S, Gunther S, Cooke BM. Bovine babesiosis in the 21st century: advances in biology and functional genomics. Int J Parasitol. (2013) 43:125–32. doi: 10.1016/j.ijpara.2012.09.008
4. He L, Liu Q, Yao B, Zhou Y, Hu M, Fang R, et al. A historical overview of research on Babesia orientalis, a protozoan parasite infecting water buffalo. Front Microbiol. (2017) 8:1323. doi: 10.3389/fmicb.2017.01323
5. Marcelino I, de Almeida AM, Ventosa M, Pruneau L, Meyer DF, Martinez D, et al. Tick-borne diseases in cattle: applications of proteomics to develop new generation vaccines. J Proteomics. (2012) 75:4232–50. doi: 10.1016/j.jprot.2012.03.026
6. Liu Q, Zhao JL, Zhou YQ, Liu EY, Yao BA, Fu Y. Study on some molecular characterization of Babesia orientalis. Vet Parasitol. (2005) 130:191–8. doi: 10.1016/j.vetpar.2005.03.021
7. Guo J, Hu J, Sun Y, Yu L, He J, He P, et al. A novel Babesia orientalis 135-kilodalton spherical body protein like: identification of its secretion into cytoplasm of infected erythrocytes. Parasit Vectors. (2018) 11:205. doi: 10.1186/s13071-018-2795-7
8. Liu Z, Zhao J, Ma L, Yao B. Babesia orientalis sp.nov. parasitized in buffalo Bubalus bubalis in China (Piroplasmida: Babeslldae). Acta Vet Zootech Sinica. (1997) 28:84–9.
9. Liu Z, Zhao J, Ma L, Yao B. Studies on buffalo babesiosis in Hubei Province, China. Trop Anim Health Prod. (1997) 29(suppl. 4):33S−6S. doi: 10.1007/BF02632914
10. Yao B, Zhao J, Liu E, Ding S, Shi J, Liu Z. Serological investigations on Babesia orientalis infection. Status of water buffaloes in Hubei Province. Parasitol Res. (2002) 88(suppl. 1):S11–2. doi: 10.1007/s00436-001-0561-8
11. Zhao J, Liu Z, Yao B, Ma L. Culture-derived Babesia orientalis exoantigens used as a vaccine against buffalo babesiosis. Parasitol Res. (2002) 88:S38–40. doi: 10.1007/s00436-001-0569-0
12. He L, Fan L, Hu J, Miao X, Huang Y, Zhou Y, et al. Characterisation of a Babesia orientalis apical membrane antigen, and comparison of its orthologues among selected apicomplexans. Ticks Tick Borne Dis. (2015) 6:290–6. doi: 10.1016/j.ttbdis.2015.01.010
13. Zhao J. The Effects of medium volume, depth and replacing medium interval on the growth of Babesia bovis in vitro. J Huazhong Agric Univ. (1997).
14. He L, Feng HH, Zhang WJ, Zhang QL, Fang R, Wang LX, et al. Occurrence of Theileria and Babesia species in water buffalo (Bubalus babalis, Linnaeus, 1758) in the Hubei province, South China. Vet Parasitol. (2012) 186:490–6. doi: 10.1016/j.vetpar.2011.11.021
15. Vannier E, Krause PJ. Babesiosis in China, an emerging threat. Lancet Infect Dis. (2015) 15:137–9. doi: 10.1016/S1473-3099(14)71062-X
16. He L, Zhou YQ, Oosthuizen MC, Zhao JL. Loop-mediated isothermal amplification (LAMP) detection of Babesia orientalis in water buffalo (Bubalus babalis, Linnaeus, 1758) in China. Vet Parasitol. (2009) 165:36–40. doi: 10.1016/j.vetpar.2009.06.036
17. He L, Feng HH, Zhang QL, Zhang WJ, Khan MK, Hu M, et al. Development and evaluation of real-time PCR assay for the detection of Babesia orientalis in water buffalo (Bubalus bubalis, Linnaeus, 1758). J Parasitol. (2011) 97:1166–9. doi: 10.1645/GE-2819.1
18. He L, Zhang Y, Zhang QL, Zhang WJ, Feng HH, Khan MK, et al. Mitochondrial genome of Babesia orientalis, apicomplexan parasite of water buffalo (Bubalus babalis, Linnaeus, 1758) endemic in China. Parasit Vectors. (2014) 7:82. doi: 10.1186/1756-3305-7-82
19. Huang Y, He L, Hu J, He P, He J, Yu L, et al. Characterization and annotation of Babesia orientalis apicoplast genome. Parasit Vectors. (2015) 8:543. doi: 10.1186/s13071-015-1158-x
20. Resing KA, Meyer-Arendt K, Mendoza AM, Aveline-Wolf LD, Jonscher KR, Pierce KG, et al. Improving reproducibility and sensitivity in identifying human proteins by shotgun proteomics. Anal Chem. (2004) 76:3556–68. doi: 10.1021/ac035229m
21. Adaszek L, Banach T, Bartnicki M, Winiarczyk D, Lyp P, Winiarczyk S. Application the mass spectrometry MALDI-TOF technique for detection of Babesia canis canis infection in dogs. Parasitol Res. (2014) 113:4293–5. doi: 10.1007/s00436-014-4124-1
22. Sharma S, Punjabi V, Zingde SM, Gokhale SM. A comparative protein profile of mammalian erythrocyte membranes identified by mass spectrometry. J Membr Biol. (2014) 247:1181–9. doi: 10.1007/s00232-014-9718-0
23. Kules J, Mrljak V, Baric Rafaj R, Selanec J, Burchmore R, Eckersall PD. Identification of serum biomarkers in dogs naturally infected with Babesia canis canis using a proteomic approach. BMC Vet Res. (2014) 10:111. doi: 10.1186/1746-6148-10-111
24. Johnson WC, Taus NS, Reif KE, Bohaliga GA, Kappmeyer LS, Ueti MW. Analysis of stage-specific protein expression during Babesia Bovis development within female Rhipicephalus Microplus. J Proteome Res. (2017) 16:1327–38. doi: 10.1021/acs.jproteome.6b00947
25. Matei H, Frentescu L, Benga G. Comparative studies of the protein composition of red blood cell membranes from eight mammalian species. J Cell Mol Med. (2000) 4:270–6. doi: 10.1111/j.1582-4934.2000.tb00126.x
26. Li J, Hosseini Moghaddam SH, Chen X, Chen M, Zhong B. Shotgun strategy-based proteome profiling analysis on the head of silkworm Bombyx mori. Amino Acids. (2010) 39:751–61. doi: 10.1007/s00726-010-0517-3
27. Cheng K, Li J, Yang D, Yang Y, Rao C, Zhang S, et al. 2D-gel based proteomics unravels neurogenesis and energetic metabolism dysfunction of the olfactory bulb in CUMS rat model. Behav Brain Res. (2016) 313:302–9. doi: 10.1016/j.bbr.2016.05.026
28. Bryk AH, Wisniewski JR. Quantitative analysis of human red blood cell proteome. J Proteome Res. (2017) 16:2752–61. doi: 10.1021/acs.jproteome.7b00025
29. Guo D, Zhu Q, Zhang H, Sun D. Proteomic analysis of membrane proteins of vero cells: exploration of potential proteins responsible for virus entry. DNA Cell Biol. (2014) 33:20–8. doi: 10.1089/dna.2013.2193
30. Zhang Z, Xin W, Wang S, Zhang X, Dai H, Sun R, et al. Xylem sap in cotton contains proteins that contribute to environmental stress response and cell wall development. Funct Integr Genomics. (2015) 15:17–26. doi: 10.1007/s10142-014-0395-y
31. Pesciotta EN, Sriswasdi S, Tang HY, Speicher DW, Mason PJ, Bessler M. Dysferlin and other non-red cell proteins accumulate in the red cell membrane of Diamond-Blackfan Anemia patients. PLoS ONE. (2014) 9:e85504. doi: 10.1371/journal.pone.0085504
32. Pasini EM, Kirkegaard M, Mortensen P, Lutz HU, Thomas AW, Mann M. In-depth analysis of the membrane and cytosolic proteome of red blood cells. Blood. (2006) 108:791–801. doi: 10.1182/blood-2005-11-007799
33. Yokoyama N, Okamura M, Igarashi I. Erythrocyte invasion by Babesia parasites: current advances in the elucidation of the molecular interactions between the protozoan ligands and host receptors in the invasion stage. Vet Parasitol. (2006) 138:22–32. doi: 10.1016/j.vetpar.2006.01.037
34. Baum J, Gilberger TW, Frischknecht F, Meissner M. Host-cell invasion by malaria parasites: insights from Plasmodium and Toxoplasma. Trends Parasitol. (2008) 24:557–63. doi: 10.1016/j.pt.2008.08.006
35. Gupta P, Mehrotra S, Sharma A, Chugh M, Pandey R, Kaushik A, et al. Exploring heme and hemoglobin binding regions of plasmodium heme detoxification protein for new antimalarial discovery. J Med Chem. (2017) 60:8298–308. doi: 10.1021/acs.jmedchem.7b00089
36. Geary TG, Delaney EJ, Klotz IM, Jensen JB. Inhibition of the growth of Plasmodium falciparum in vitro by covalent modification of hemoglobin. Mol Biochem Parasitol. (1983) 9:59–72. doi: 10.1016/0166-6851(83)90057-9
Keywords: Babesia orientalis, water buffalo, dairy cattle, beef cattle, erythrocyte, proteome
Citation: Guo J, Sun Y, Tian Y and Zhao J (2019) Comparative Analysis of Erythrocyte Proteomes of Water Buffalo, Dairy Cattle, and Beef Cattle by Shotgun LC-MS/MS. Front. Vet. Sci. 6:346. doi: 10.3389/fvets.2019.00346
Received: 11 July 2019; Accepted: 24 September 2019;
Published: 18 October 2019.
Edited by:
Guiquan Guan, Lanzhou Veterinary Research Institute (CAAS), ChinaReviewed by:
Houshuang Zhang, Shanghai Veterinary Research Institute (CAAS), ChinaRuofeng Yan, Nanjing Agricultural University, China
Copyright © 2019 Guo, Sun, Tian and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Junlong Zhao, emhhb2p1bmxvbmdAbWFpbC5oemF1LmVkdS5jbg==
 Jiaying Guo
Jiaying Guo Yali Sun
Yali Sun Yu Tian
Yu Tian Junlong Zhao
Junlong Zhao