- 1National Centre for Animal Diseases, Canadian Food Inspection Agency, Lethbridge, AB, Canada
- 2Department of Population Medicine, Ontario Veterinary College, University of Guelph, Guelph, ON, Canada
- 3Department of Pathobiology, Ontario Veterinary College, University of Guelph, Guelph, ON, Canada
- 4National Microbiology Laboratory at Lethbridge, Public Health Agency of Canada, Lethbridge, AB, Canada
- 5Canadian Wildlife Health Cooperative, Ontario Veterinary College, University of Guelph, Guelph, ON, Canada
- 6National Microbiology Laboratory, Public Health Agency of Canada, Winnipeg, MB, Canada
Free-ranging wildlife are increasingly recognized as potential reservoirs of disease-causing Campylobacter species such as C. jejuni and C. coli. Raccoons (Procyon lotor), which live at the interface of rural, urban, and more natural environments, are ideal subjects for exploring the potential role that wildlife play in the epidemiology of campylobacteriosis. We studied the prevalence and genetic diversity of Campylobacter from live-captured raccoons on five swine farms and five conservation areas in southwest Ontario. From 2011 to 2013, we collected fecal swabs (n = 1,096) from raccoons, and (n = 50) manure pit samples from the swine farm environment. We subtyped the resulting Campylobacter isolates (n = 581) using Comparative Genomic Fingerprinting (CGF) and 114 distinct subtypes were observed, including 96 and 18 subtypes among raccoon and manure pit isolates, respectively. Campylobacter prevalence in raccoons was 46.3%, with 98.7% of isolates recovered identified as C. jejuni. Novel raccoon-specific CGF subtypes (n = 40/96) accounted for 24.6% (n = 143/581) of Campylobacter isolates collected in this study. Our results also show that C. jejuni is readily acquired and lost in this wild raccoon population and that a high Campylobacter prevalence is observed despite transient carriage typically lasting 30 days or fewer. Moreover, although raccoons appeared to be colonized by species-adapted subtypes, they also harbored agriculture-associated genotypes that accounted for the majority of isolates observed (66.4%) and that are strongly associated with human infections. This suggests that raccoons may act as vectors in the transmission of clinically-relevant C. jejuni subtypes at the interface of rural, urban, and more natural environments.
Introduction
Campylobacter species are the second most reported bacterial foodborne pathogen in Canada (1), and campylobacteriosis remains one of the most common enteric illnesses worldwide (2). Although human infections are primarily caused by two thermophilic species, C. jejuni and C. coli, several additional species have been reported to cause illness (3). It is generally accepted that the majority of Campylobacter infections are acquired through the handling and ingestion of contaminated poultry products (4–6). While Campylobacter is highly prevalent in poultry and poultry meat (7), it is also commonly found in a wide range of animal hosts that not only include livestock such as cattle and swine, but also household pets and wildlife (6, 8). Given that Campylobacter is actively shed in animal feces, environmental sources such as surface waters may also become contaminated through use by animals or due to surface run-off (9). This may lead to increased environmental transmission and dissemination of Campylobacter across different ecological niches, thereby creating additional routes of exposure to humans or other potential hosts (10).
Campylobacter have been isolated from a diverse array of free-ranging wildlife, including birds such as waterfowl and songbirds and mammals, including rodents, wild boars, and ungulates (8, 11–14). Although the presence of C. jejuni in free-ranging wildlife is not necessarily indicative of a role in the epidemiology of campylobacteriosis, an increasing number of studies is consistent with this possibility. For example, Campylobacter subtypes commonly associated with human illness have been detected in wildlife species (15, 16), and human clinical cases have been linked to wildlife via direct transmission (17–20) or source attribution (21). Furthermore, an increased occurrence of zoonotic pathogens, including Campylobacter and Salmonella, has been observed in free-ranging wildlife living in close proximity to livestock and or human populations (22, 23).
A recent scoping review on bacterial zoonotic pathogens in wild animals reported that the most frequently investigated wildlife groups were birds (47.3%), cervids (15.4%), and rodents (10.5%) (24). Similarly, much Campylobacter research on wildlife has focused on avian species, in large part due to Campylobacter's affinity for avian hosts, but also due to host ecological factors such as increased anthropogenic contact and large habitat ranges. The importance of wild mammals in Campylobacter ecology and epidemiology is less clear. By comparison to avian species, there is a paucity of literature exploring the role of non-avian wildlife on transmission of Campylobacter. There is even less focus on synanthropic species, which live in habitats in close association with humans. In this regard, raccoons (Procyon lotor) represent ideal study subjects because they use a wide variety of habitats (25) and have the potential to move between urban, rural, and forested habitats.
Raccoons are known to carry a number of zoonotic agents (26–29) and they also display distinct social features such as the use of communal latrines (30), which may enhance mechanical transmission of certain microorganisms present in their feces (31). Previous studies have reported Campylobacter prevalence in raccoons ranging from 1% (32) to 41% (33). To our knowledge, there have been few, if any, longitudinal studies examining Campylobacter in a raccoon population over multiple years. The objectives of this study were to: (1) determine the prevalence of Campylobacter in raccoons captured on swine farms and conservation areas; (2) assess the genetic diversity, population structure, and ecology of Campylobacter subtypes observed in raccoons; (3) assess the dynamics of Campylobacter acquisition/loss in individual animals; and finally, (4) compare the subtypes observed in raccoons and human clinical cases to assess the potential role of raccoons in the epidemiology of campylobacteriosis.
Materials and Methods
Animal Trapping and Sample Collection
Procedures for trapping and handling raccoons were approved by the Animal Care Committee at the University of Guelph following the guidelines of the Canadian Committee on Animal Care. For a detailed description and map of the study area as well as trapping and sampling procedures, please refer to Bondo et al. (27) and (29). From May 2011 to November 2013, raccoons were live-trapped on swine farms and conservation areas near the cities of Guelph and Cambridge in southern Ontario, Canada. Individual animals were identified by ear and transponder tags and were sampled only once per monthly trapping week; however, additional samples were collected from the same individual if they were caught in subsequent months. Rectal fecal swabs were collected using Cary-Blair swab applicators [BBL CultureSwab, (BD) Becton, Dickinson and Company, Annapolis, MD, USA]. At each swine farm, one lagoon (i.e., manure pit) sample was collected on the first day of each trapping week as previously described (29). Briefly, to collect lagoon samples, a 24′ Nasco Swing Sampler (Conbar, Monroeville, NJ, USA) was used to collect three sub-samples from three locations around the pit, and up to two depths (i.e., the top 1/3, and mid depth of the storage), for a total of six sub-samples. The samples were then pooled into one sample for analysis. All samples were kept refrigerated or on ice until processing. The median and average number of days between collection and processing was three; the maximum number of days for raccoon swabs was eight whereas for manure pit samples it was 11.
Campylobacter Isolation
Isolation of thermophilic Campylobacter species was performed as previously described (34). For samples collected prior to October 2011, the “conventional method” (enrichment followed by direct plating onto selective media) was used; the “membrane method” (enrichment followed by passive membrane filtration onto selective media) was used thereafter. Briefly, fecal swabs were immersed in 20 mL of Bolton broth (BB) containing BB selective supplement (SR0183, Oxoid), (20 mg/L cefoperazone, 20 mg/L vancomycin, 20 mg/L trimethoprim, and 50 mg/L cyclohexamine) and 5% laked horse blood (SR0048) and mixed rapidly for 20 s. Lagoon samples were mixed prior to adding 1 mL of sample to BB with supplement. Bolton broth cultures were incubated at 42°C in microaerobic conditions (10% CO2, 5% O2, 85% N2) for 24 h and subsequently streaked onto modified blood-free charcoal cefoperazone deoxycholate agar supplemented with 32 mg/L cefoperazone and 10 mg/L amphotericin B (mCCDA) or passively filtered for 15 min through 0.65 μM cellulose acetate membrane filters onto the surface of mCCDA. All mCCDA plates were incubated for 48 h under the same microaerobic conditions. Three to five Campylobacter-like colonies were subcultured to blood agar and incubated for 24–48 h. Presumptive Campylobacter spp. colonies were identified on the basis of growth on mCCDA media, colony morphology, and oxidase tests. DNA was extracted from purified cultures using the EZ1 DNA tissue kit (Qiagen) according to the manufacturer's instructions for subsequent PCR speciation of presumptive Campylobacter isolates and subtyping of confirmed Campylobacter isolates.
PCR Confirmation of Campylobacter spp.
Presumptive Campylobacter isolates were confirmed by multiplex PCR targeting a Campylobacter genus-specific region of the 16S rRNA gene, and mapA and ceuE genes for C. jejuni and C. coli identification, respectively (35). Amplicons were visualized using a QIAxcel capillary electrophoresis instrument with the DNA Screening kit. The AM320 separation method was used along with a 15–3,000 bp alignment marker and the QX 100–2.5 kb DNA size marker. Data were analyzed and visualized using the BioCalculator v. 3.0 software.
Comparative Genomic Fingerprinting
Campylobacter spp. isolates were subtyped by Comparative Genomic Fingerprinting (CGF) as previously described (36). Briefly, CGF consists of 8 multiplex PCR reactions that together assess the presence or absence of a set of 40 accessory gene targets found to have variable carriage in the Campylobacter population and that are used to generate a highly discriminatory binary fingerprint. Products from the CGF PCRs were visualized on the QIAxcel as previously described (36) and were scored positive (1) or negative (0) based on presence or absence of each target amplicon using a combination of the BioCalculator software's binary peak calling and confirmed with visual curation. The resulting binary fingerprints were assigned a three-digit CGF subtype (e.g., 0923.002.001) derived from cluster membership in the Canadian Campylobacter CGF database (C3GFdb). Fingerprints identical to those already existing in the database were assigned the appropriate CGF subtype, while novel fingerprints were assigned a CGF subtype based on their similarity to existing fingerprints in the database. Isolates from the same sample with identical subtypes were assumed to be derived from a single clone and only one representative isolate was included in further analyses.
Association of CGF Type With Host Species
At the time of this analysis, the C3GFdb consisted of 4,847 distinct CGF profiles obtained from 19,141 Campylobacter isolates collected from across Canada, primarily from the last decade. These included 23.3% isolates derived from human clinical origin, 32.0% from poultry sources, 20.9% from cattle sources and 14.6% from environmental surface water samples. To compare epidemiologic attributes of subtypes observed among study isolates, we compiled summary statistics based on the composition of host-sources observed in the C3GFdb for each subtype in this study. We classified CGF subtypes observed in this study (n = 114) into four ecological range categories (ERC) based on the composition of sources from which they have been historically observed in the C3GFdb. Subtypes from the present study included: (I) Raccoon Exclusive (RE; n = 44); (II) Raccoon and Environmental Water (REW; n = 16); (III) Swine/Swine Lagoon (SSL; n = 10); and (IV) Mixed Host (MH; n = 44). Phylogenetic and source association analysis was performed using the R language for statistical computing (v.3.13) (37) and the following packages: tidyverse (38) and ggtree (39). A dendrogram was constructed from the 40-gene CGF profile using the “hamming” distance and “average linkage” clustering from the dist and hclust functions, respectively, and visualized using “ggtree.” Clade designations were made based on a tree height “h = 10,” which produced stable, genetically distinct groupings. For each of the CGF subtypes observed in the present study we identified the associated host-sources from the C3GFdb. For associations between CGF subtypes and MLST Clonal Complexes, we used an in-house database created from in silico CGF and MLST predictions generated from publicly available WGS data analyzed using the program Microbial in silico Typer (40); a table of associations is provided as Supplementary Table S1.
Analysis of Strain Dynamics
Forty-three percent (n = 272/628) of the raccoons in this study were captured on multiple occasions, with 18.5% (n = 116/628) captured three or more times, up to maximum of eight captures. These data were analyzed to examine strain dynamics at the level of individual animals as described below.
Statistical Analysis
CGF subtype diversity was assessed using the Simpson's Diversity Index (41) with a 95% confidence interval (42). A chi-square test statistic was used to explore the hypothesis that there was no significant difference in the distribution of isolates from prevalent Campylobacter clades (isolate count n > 9) between each of the location types (e.g., swine farm sites and conservation sites). Chi-square test statistics and post-hoc follow-up calculations for computing adjusted standardized residuals and z-scores for each clade were performed as described by Sharpe (43).
Results
A High Prevalence of Campylobacter Was Found in the Raccoon Population Under Study
From May 2011 to November 2013, we collected 1,096 fecal swabs from 628 raccoons trapped on conservation areas (n = 687) and swine farms (n = 409), and 50 manure pit samples from the environment of the swine farm sites. The prevalence of Campylobacter spp. in raccoon fecal samples was 46.3% (508/1,096) (Table 1). There were no significant differences in Campylobacter prevalence from raccoon fecal swabs with a sample-to-test interval of 1–2 days (46.7%, n = 467) vs. a 3–4 days (46.0%, n = 363), 5–6 days (48.5%, n = 227), or 7–8 days interval (33.3%, n = 39). Similarly, for swine manure pit samples, there were no significant differences in Campylobacter prevalence between sample-to-test intervals of 1–2 days (40.0%, n = 10) vs. 3–4 (38.1%, n = 21), 5–6 (33.3%, n = 9), or 7–11 days (22.2%, n = 9). Among the Campylobacter positive raccoon samples, 502 (98.8%) were positive for C. jejuni, six (1.2%) for Campylobacter spp. (unidentified Campylobacter species), and one for C. coli (Table 1). A single sample was found to harbor mixed Campylobacter species, testing positive for both C. jejuni and an undefined Campylobacter spp. Among swine manure pit samples 34.0% (17/50) were positive for Campylobacter, of which 94.1% (16/17) were positive for C. coli (Table 1). One sample tested positive for Campylobacter spp.
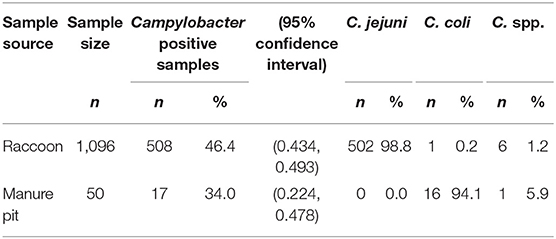
Table 1. Frequency, prevalence and species of Campylobacter in fecal samples obtained from raccoons and swine farm manure pits.
Campylobacter Circulating in the Raccoon Population Represent a Genetically Diverse Population
A total of 1,555 confirmed Campylobacter isolates derived from Campylobacter-positive raccoon fecal swabs were subtyped by CGF. A further 58 isolates were recovered from swine manure pit samples. After accounting for potentially clonal isolates (i.e., isolates from the same sample sharing the same CGF subtype), 610 isolates remained; this included 581 raccoon isolates and 29 manure pit sample isolates. Among the Campylobacter positive raccoon samples (n = 508), a single CGF subtype was recovered for the majority of the samples (443/508; 87.2%), two CGF types were recovered in 11.2% of samples (57/508), and three subtypes in 1.6% of samples (8/508). We observed a broad genetic diversity in the Campylobacter population, with a total of 114 distinct CGF subtypes identified among study isolates. Subtype clusters ranged in size from 1 to 36 isolates, with 13 subtypes comprising over 50% of isolates in the dataset (n = 329/628) and 51 subtypes detected in single instances. Forty-six subtypes, representing 21.3% of study isolates (n = 130/610), were novel and had not been previously observed in the C3GFdb. There were 96 subtypes observed among raccoon isolates (C. jejuni = 91; Campylobacter spp. = 4; C. coli = 1) and 18 subtypes observed among lagoon isolates (C. coli = 17; Campylobacter spp. = 1); no subtypes were shared between raccoons and manure pit samples. Clustering of the 114 CGF binary profiles revealed 8 major lineages, designated clades A–H (Figure 1). The clades segregated by Campylobacter species: clades A–C, G, and H comprised C. jejuni isolates; clade F comprised C. coli isolates; and clade D comprised non-jejuni/coli Campylobacter species isolates (i.e., undetermined). The majority of raccoon isolates were observed in clade A (52.0%; n = 302), followed by clades B and C, with 30.5% (n = 177) and 14.5% (n = 84) of isolates, respectively. A small number of raccoon isolates (3.1%; n = 18) were found distributed among clades D (n = 5), E (n = 1), F (n = 1), G (n = 2), and H (n = 9). Among swine lagoon isolates, the majority were found in clade F (89.7%; n = 26), with the remaining isolates (10.3%; n = 3) in clade D. Similar levels of diversity were observed between swine farms and conservation areas at the subtype level (Simpson's Index of 0.9648 and 0.9628, respectively). Although overall genotypic richness was similar between swine farms and conservation areas, there was a significant relationship between clade and location type. For clade A, more isolates were identified from swine farm sites compared to conservation sites (p < 0.001). In contrast, for clades B and C, fewer isolates were identified from swine farm sites compared to conservation sites (p < 0.01).
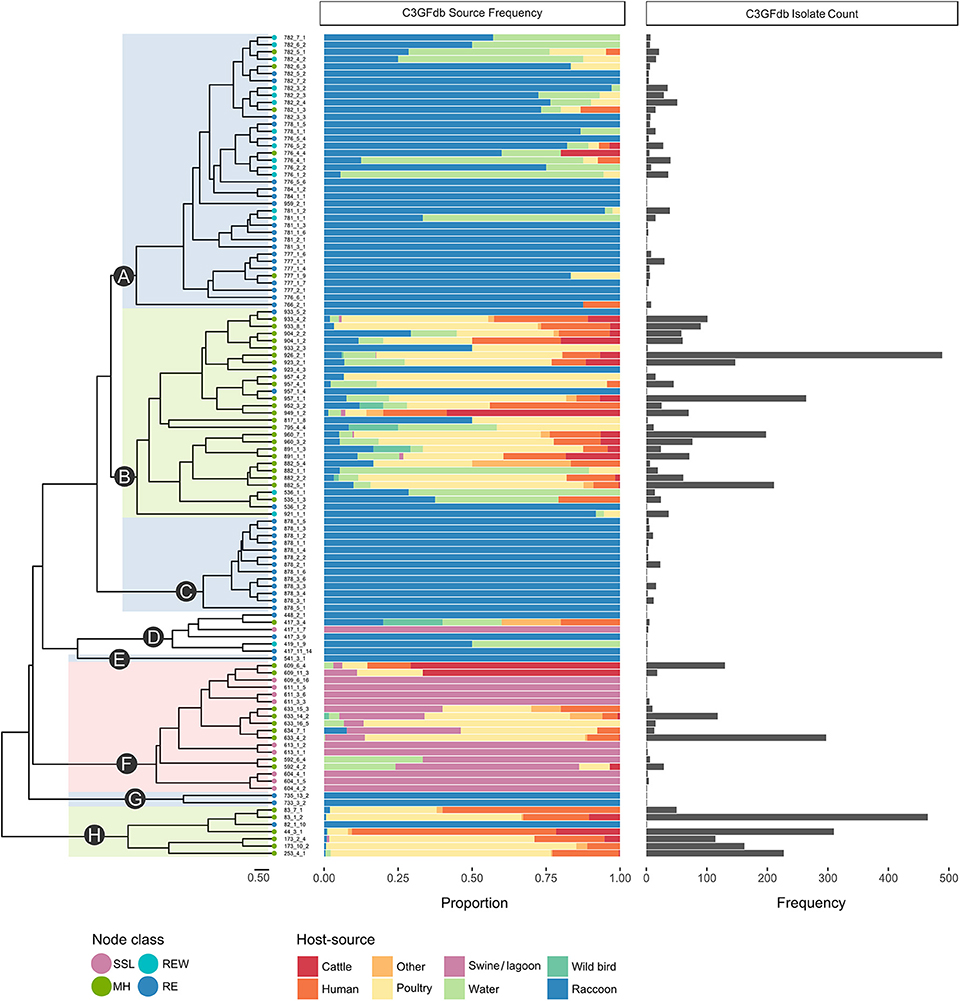
Figure 1. Host-range specificity of Campylobacter subtypes observed in a raccoon population on conservation and swine farm sites in southwest Ontario, Canada. For each of the 114 CGF types in the dendrogram, host-source data from the C3GFdb is used to illustrate relative host-source associations. Terminal nodes of the tree were classified based on source composition of the CGF subtype (MH, mixed host; RE, raccoon exclusive; REW, raccoon and environmental water; SSL, swine and swine lagoon). A large proportion (~97%) of Raccoon isolates from this study are found in clades A, B, and C. Both clade A, which is largely composed of RE and REW subtypes, and clade C, which consists entirely of RE subtypes, appear to be raccoon-associated niche or host-adapted lineages. Clade B is primarily composed of MH subtypes with a large contribution from agriculture and human clinical sources.
Campylobacter Subtypes in the Raccoon Population Display Different Types of Host Range Specificity
Among the four ecological range categories that we defined for the subtypes in this study (Raccoon Exclusive or RE; Raccoon and Environmental Water or REW; Swine/Swine Lagoon or SSL; and Mixed Host or MH), raccoon isolates were evenly distributed among RE, REW and MH subtypes (RE: 31.3%, n = 182; REW: 34.1%, n = 198; MH: 34.6%, n = 201). Swine lagoon isolates were similarly distributed among SSL and MH subtypes (SSL: 51.7%, n = 15; MH: 48.3%, n = 14). The distribution of ERC subtypes suggests a strong raccoon association for certain clades observed in our analysis (Figure 1). Clade C, which included 14.5% of raccoon isolates in the study, was strictly composed of RE subtypes, and these were novel to the C3GFdb. Clade A, which included the majority of raccoon isolates in the study (52.0%), comprised largely RE and REW subtypes (n = 33/38). Source frequencies from the C3GFdb (Table 2) indicate that a majority of isolates from clade A subtypes (n = 461) include raccoon isolates from this study (65.5%; n = 302) along with historical isolates from environmental water sources (26.2%; n = 121). A small number of isolates (6.9%; n = 32) comprised the remaining isolates in the clade and were derived from cattle, poultry, and human clinical sources (Table 2). Conversely, clades B and H, which included 30.5 and 1.6% of raccoon isolates, respectively, were of MH subtype composition (n = 23/29 and n = 6/7, respectively). Based on C3GFdb source association data, isolates from subtypes in clades B (n = 2,118) and H (n = 1,329) were primarily chicken-associated (clade B = 54.9%; clade H = 55.0%). Importantly, these clades included a large proportion of isolates from human clinical cases (14.4 and 33.3%, respectively), which is in contrast to clades A and C (1.7 and 0.0%, respectively). Unsurprisingly, the C. coli specific-clade F included 26/29 swine manure pit isolates and consisted of 9 SSL subtypes and 9 MH subtypes.
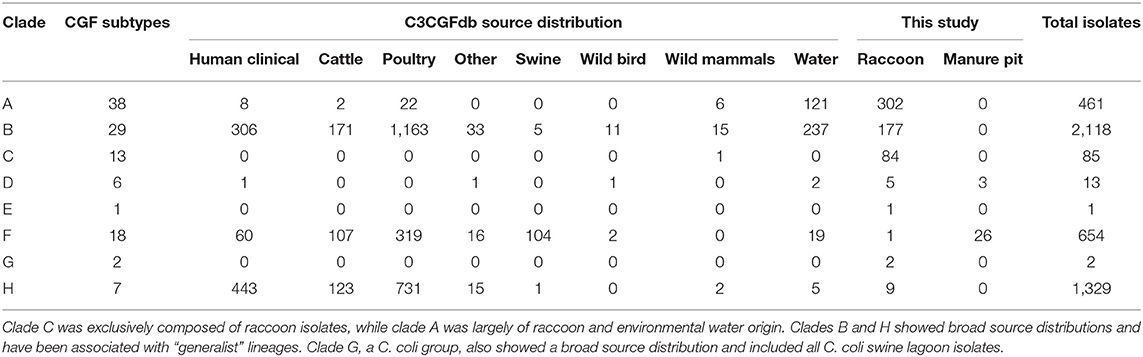
Table 2. Host-source distributions from the C3CGFb for each of the clades observed in raccoon and swine manure pit isolates.
Campylobacter Strain Dynamics in Raccoons Include Both Transient and Longer-Term Carriage Within Individual Animals
A total of 468 samplings (43%; n = 468/1,096) involved consecutive recapture events involving the same animal (Figure 2), with a majority (77%; n = 361/468) occurring within the same sampling year. The positive rate for Campylobacter was 67% (n = 242/361) and 69% (n = 74/107) among recaptures in the same and different sampling years, respectively (Figure 2). Among recaptures within the same sampling year, 58% (n = 209/361) reflected a Campylobacter status change, with either animals that acquired or lost Campylobacter (n = 139), and animals that acquired a different subtype (n = 70) between recaptures. This rate was 67% among recapture events in different sampling years (n = 72/107), including 52 cases of animals acquiring or losing Campylobacter between recaptures and 20 cases of animals acquiring a different subtype. Overall, 60% (n = 281/468) of recapture events reflected a change in Campylobacter status, although the rate rises to 89% (n = 281/316) when excluding recaptures in which the animal tested negative on both occasions (n = 152). There were 125 instances of consecutive positive results, with a median of 35 days between recaptures (minimum = 25 days, maximum = 617 days). Of these, 40.0% (n = 50/125) occurred within a window of 0–30 days (i.e., short), an additional 33.6% (n = 42/125) occurred in a window of 31–60 days (i.e., intermediate), and 26.4% (n = 33/125) occurred in a window of >60 days (i.e., long), which included 22 recaptures that occurred on different sampling years. Overall, 72.0% of consecutive recaptures that were positive on both occasions (n = 90/125) yielded a different subtype on consecutive samplings. Among the 28% of consecutive recaptures yielding the same subtype (n = 35/125), we observed a decreasing proportion of matching subtypes as the time between sampling events increased (Figure 3). Furthermore, when examined in the context of ecological range, we observed that a larger proportion of recaptures in which the same subtype was consecutively isolated involved raccoon-associated subtypes (i.e., categories RE and REW) compared to MH subtypes (Overall: 30/35; 0–30 days: 15/18; 31–60 days: 12/14; and >60 days: 3/3). Among recapture events yielding changes in Campylobacter status on consecutive observations, the ratio of instances of strain acquisition to strain loss was similar across the three major raccoon-containing clades (A: 126/90; B: 66/46; C: 41/24 and across the various ERCs (MH: 73/57; RE: 80/53; REW: 87/56), all of which were similar to the overall ratio of acquisition-to-loss (n = 240/166).
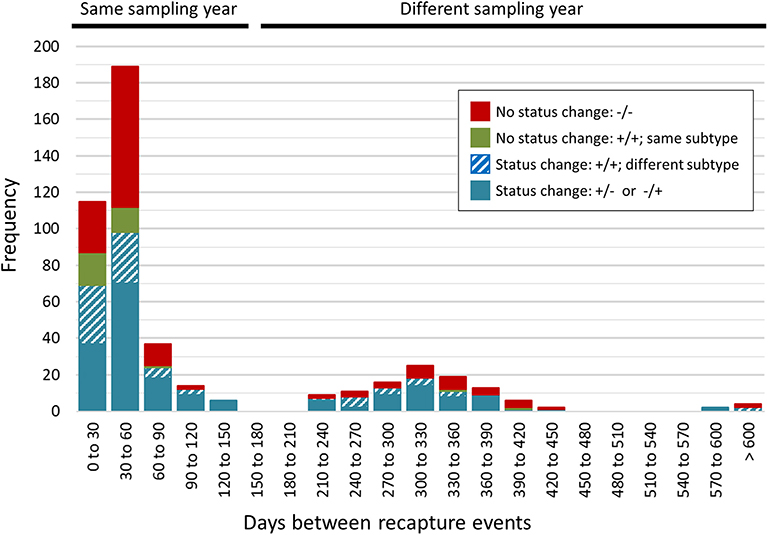
Figure 2. Rapid changes in Campylobacter status in a raccoon population on conservation and swine farm sites in southwest Ontario, Canada. Campylobacter culture status was examined for consecutive recapture events involving the same animal (n = 468). A majority of consecutive recapture events (77%; n = 361) occurred within the same sampling year and 67% (n = 242/361) yielded a Campylobacter positive result. The positive rate among recapture events in different sampling years was 69% (n = 74/107). Overall, 89% of recapture events that involved a Campylobacter positive result (n = 281/316) reflected a Campylobacter status change, with either animals that tested positive after previously testing negative or vice-versa (n = 191), and animals testing positive in both cases but that shed a different subtype (n = 90). These strain dynamics are consistent with the rapid turnover of strains in this raccoon population.
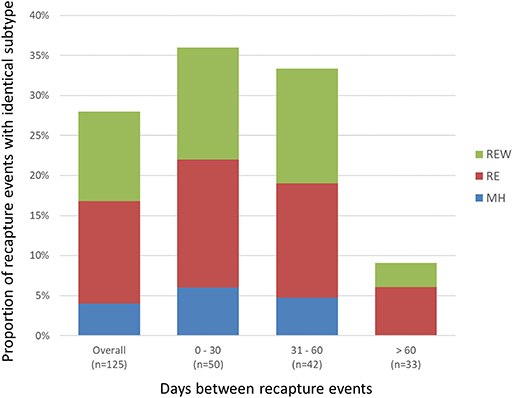
Figure 3. Short- vs. long-term colonization with Campylobacter in a raccoon population on conservation and swine farm sites in southwest Ontario, Canada. Analysis of consecutive Campylobacter positive recapture events yielding an identical subtype indicates that as the duration between captures increased beyond 60 days, there is a large decrease in the number of cases where the same subtype was observed, suggesting that most animals are only transiently colonized. Among the small proportion of recapture events consistent with prolonged colonization (i.e., consecutive and temporally separated Campylobacter-positive recapture events yielding the same subtype), results indicate that these are more likely to involve raccoon-adapted subtypes from RE and REW categories.
Discussion
The prevalence of Campylobacter spp. in raccoon fecal samples in this study was 46%, which is consistent with what was observed in a previous study from the same region of southwestern Ontario (41%) (33). Indeed, given the practical limitations imposed by potential delays in sample processing (3–11 days) due to shipping of fecal swabs from the study site (Guelph, Ontario) to the laboratory where microbiological work was carried out (Lethbridge, Alberta), it is very likely that our prevalence estimates underestimate true prevalence rates in this raccoon population. By contrast, studies from Japan and New York found only 1.3 and 6% of raccoons carried Campylobacter, respectively (32, 44). There are a limited number of studies systematically examining Campylobacter in raccoons or other medium sized mammals. Many studies instead rely on convenience-based sampling methodology, for example, intakes into a wildlife rehabilitation center (20, 45). The Grand River watershed in southern Ontario is heavily impacted by mixed agricultural activities, with farms making up ~75% of the watershed (46).
In this study, we examined raccoons circulating in a range that included several swine farms and more natural environments in order to investigate Campylobacter prevalence, genetic diversity, and strain dynamics in this wild animal population. Significant genetic diversity was observed among raccoon isolates collected in this study, with 96 distinct CGF subtypes and at least eight major lineages observed in this population. Interestingly, there was a distinct lack of overlap in CGF subtypes observed in raccoons trapped at swine farm sites and samples obtained at those farms (i.e., manure pit samples) despite the high rates of Campylobacter recovery in these animals. Swine are known to preferentially harbor C. coli, which was recovered from nearly all manure pit samples analyzed. The near absence of C. coli in raccoons from this study was unexpected, as some level of C. coli exposure would be expected to take place in the swine-farm environment. Nonetheless, our findings are consistent with overall trends in the C3GFdb, in that of nearly 300 subtypes that have been observed in either raccoons or swine, only seven minor subtypes have included isolates recovered from both species. Other studies investigating interactions between wildlife and swine farms have shown little evidence of shared Campylobacter subtypes between farm fecal/manure isolates and small and medium sized wild mammals (such as mice, rats, badger, fox) (11, 47, 48). The swine in this study were housed indoors, which likely prevented raccoons from coming into direct contact with the animals or fresh swine feces. Furthermore, swine fecal wastes are generally managed in a more confined manner (e.g., manure pits) compared to other livestock such as cattle. Lastly, because raccoons have wide-ranging habitats, it is possible that raccoons trapped at swine farm sites may not have routinely used these areas to forage, thereby decreasing their exposure. Thus, there exist possible barriers (i.e., biological, physical, behavioral) that may limit the transmission of C. coli between swine and raccoons.
During the course of this longitudinal study, a significant proportion of samples represented cases in which the same animal was recaptured and these data were used to examine Campylobacter strain dynamics within individual animals, including patterns of short-, intermediate-, and long-term Campylobacter carriage and their shedding. A majority of recapture events (60%) yielded differences in Campylobacter-status, including cases in which the animal appeared to have acquired or lost Campylobacter between recaptures (40%), and cases in which the animal tested positive in both occasions but where we observed a difference in subtype (19%). Overall shifts in Campylobacter status increased to 89% if excluding recaptures in which the animal tested negative in both occasions. Moreover, we observed instances of Campylobacter-positive animals that appeared to revert to negative status on subsequent recaptures in as few as 25 days, with 18 instances occurring within 30 days. This would suggest that although Campylobacter prevalence in this raccoon population is high, carriage and shedding is likely transient, with the majority of raccoons harboring C. jejuni for only short periods of time (≤1 month). We also observed significant strain displacement among animals that tested positive for Campylobacter on consecutive recaptures, with a different subtype observed in 72% of cases. Moreover, although we selected 1,555 isolates for subtyping from among the 508 fecal swabs that tested positive for Campylobacter, a majority of samples (87.2%) yielded a single subtype. Taken together, these data suggest that raccoons readily acquire and lose Campylobacter, with rapid turnover of strains among animals that remained Campylobacter-positive on consecutive samplings. Nonetheless, it is important to note possible confounding factors that could yield data consistent with transient carriage. These include limitations with microbiological and molecular subtyping methods (i.e., limit of detection/recovery of isolates leading to culture-negative samples or limiting the isolation of multiple strains from a single sample).
Our data yielded a small proportion of instances in which consecutive recaptures were Campylobacter positive and yielded the same subtype. Although these tended to be recapture events on consecutive months, a small number of cases included recaptures with significant temporal separation, up to a length of 394 days. It is unclear whether these represent ongoing colonization or re-infection with the same subtype; whole-genome sequence (WGS) analysis of these isolates is likely to shed light on this issue since the much higher discriminatory power of WGS-based subtyping approaches could help differentiate between these two types of events.
Our analysis of host source metadata from the Canadian Campylobacter CGF database (C3GFdb) revealed that the C. jejuni population found in raccoons consisted primarily of subtypes with a strong raccoon association and subtypes with a mixed-host association. These were primarily distributed among three major clades that included the majority of raccoon isolates in this study. Nearly one-third of raccoon isolates had subtypes that were novel to the C3GFdb, with many of these forming a clade that appears to be exclusive to raccoons. Most remaining isolates from raccoon-exclusive subtypes could be found in a clade that was otherwise composed primarily of subtypes that had previously been observed only among isolates from surface water samples. Host-adapted subtypes in wild animals and surface water have been described in previous studies (49–51). Although Stabler et al. (8) have previously described an apparent water and wildlife C. jejuni clade, this is the first report of Campylobacter subtypes that appear to be uniquely host-adapted to raccoons. It is noteworthy that the majority of events in which animals were positive for the same CGF subtype on consecutive recaptures (i.e., consistent with longer-term colonization) involved raccoon-adapted subtypes.
In contrast to raccoon-adapted clades, 78% of isolates in clade B, which contained approximately 30% of raccoon isolates in this study, were from subtypes with mixed-host association. Mixed-source genotypes have been well-described and typically include representatives from Clonal Complexes (CCs) described as ‘generalists' including ST-45, ST-21, and ST-48 (52–55). These CCs are highly prevalent worldwide, known for their broad host distribution, including livestock, and high burden of human illness. During the course of validating CGF (36), we established correlations between specific CGF subtypes and corresponding CCs and have subsequently been able to refine these based on in silico subtyping predictions from WGS data (40, 56). Notably, many CGF subtypes within clade B have been characterized as belonging to CC ST-45, including CGF subtype 0926.002.001, which is the third most prevalent genotype in the C3GFdb and ranks sixth in terms of number of human clinical cases associated with it. Although clade H only comprised a small proportion of raccoon isolates in this study (1.6%), it is noteworthy because CGF subtypes from within clade H have been shown to belong to CCs ST-21 and ST-48 (57). Moreover, it includes CGF subtypes 0044.003.001 and 0083.001.002, which are among the most prevalent in Canada and rank second and third in terms of number of associated human clinical cases, respectively. Because of their synanthropic behavior, the potential for acquisition of clinically relevant Campylobacter subtypes by raccoons via the same routes of exposure as humans cannot be ruled out. Our data suggest that the high Campylobacter rates observed in this wild raccoon population are likely due to environmental and ecological factors, including high rates of mixed agriculture activities, which allow for sustained and consistent Campylobacter exposure to agriculture-associated genotypes strongly implicated in human cases of campylobacteriosis.
Conclusions
Prevalence of Campylobacter in the raccoon population under study, which is native to the Grand River Watershed in southwestern Ontario, Canada, was found to be much higher than what has been previously reported for other populations of small to medium-sized wild mammals. This high prevalence was observed despite evidence suggesting that individual animals were only transiently colonized, with a majority of raccoons harboring C. jejuni for only short periods of time (i.e., ≤1 month). Our data show significant genotypic flux within individual animals, which is consistent with the constant acquisition, loss, and replacement of strains, and suggest that this raccoon population is constantly exposed to a wide range of circulating but endemic strains. This may be due to ecological factors within the Grand River Watershed, where agriculture and human activities may give rise to a wide variety of Campylobacter sources. Although raccoons appear to be poor hosts for C. coli strains typically observed in swine, many of the C. jejuni subtypes observed in this raccoon population have been previously associated with agricultural sources (e.g., chickens, cattle) and human illness. Interestingly, raccoons were also found to carry C. jejuni subtypes within two clades that appear to be genetically distinct from genotypes recovered from humans and food animals and that appear to represent raccoon-associated niche- or host-adapted strains; the potential risk to human health from these strains remains unknown. Nevertheless, the high proportion of clinically-relevant generalist Campylobacter subtypes found in raccoon fecal samples suggests that raccoons likely act as transient vectors of Campylobacter and may play a role in the transmission of strains at the interface of rural, urban, and more natural environments.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Ethics Statement
The animal study was reviewed and approved by Animal Care Committee at the University of Guelph.
Author Contributions
SM led all aspects of laboratory and downstream analyses and drafted the manuscript. BH participated in laboratory and downstream analyses and assisted with manuscript preparation. KB participated in study design and fieldwork and assisted with manuscript preparation. VG contributed to study design and manuscript preparation. CJ conceived of the study, participated in its design and coordination, and assisted with manuscript preparation. ET participated in study design and coordination, contributed to downstream analyses, and manuscript preparation.
Funding
Financial support for this work was provided through the Public Health Agency of Canada and the Government of Canada's Genomics Research and Development Initiative (ET), the National Science and Engineering Research Council (CJ). KB received support through the United States Department of Agriculture and an Ontario Graduate Scholarship. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Erin Harkness, Samantha Allen, Samantha Kagan, and Mary Thompson for assisting with field data collection; Bryan Bloomfield for collecting manure pit samples and maintaining relations with landowners; Tami Harvey and Barbara Jefferson for submitting lab samples, and Quentin Papach for assisting with the development of a Microsoft Access database. The authors would also like to thank Morgan Chow, Madeline Tsoi, and Emily Che for technical assistance in microbiological work and the many contributors to the Canadian Campylobacter Comparative Genomic Fingerprinting Database (C3GFdb), without whom this work would not have been possible.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2020.00027/full#supplementary-material
References
1. Thomas MK, Murray R, Flockhart L, Pintar K, Pollari F, Fazil A, et al. Estimates of the burden of foodborne illness in Canada for 30 specified pathogens and unspecified agents, circa 2006. Foodborne Pathog Dis. (2013) 10:639–48. doi: 10.1089/fpd.2012.1389
2. Kaakoush NO, Castaño-Rodríguez N, Mitchell HM, Man SM. Global epidemiology of Campylobacter infection. Clin Microbiol Rev. (2015) 28:687–720. doi: 10.1128/CMR.00006-15
3. Lastovica AJ. Emerging Campylobacter spp.: the tip of the iceberg. Clin Microbiol Newsl. (2006) 28:49–56. doi: 10.1016/j.clinmicnews.2006.03.004
4. Nauta M, Hill A, Rosenquist H, Brynestad S, Fetsch A, van der Logt P, et al. A comparison of risk assessments on Campylobacter in broiler meat. Int J Food Microbiol. (2009) 129:107–23. doi: 10.1016/j.ijfoodmicro.2008.12.001
5. Skarp CPA, Hänninen ML, Rautelin HIK. Campylobacteriosis: the role of poultry meat. Clin Microbiol Infect. (2016) 22:103–9. doi: 10.1016/j.cmi.2015.11.019
6. Pintar KDM, Thomas KM, Christidis T, Otten A, Nesbitt A, Marshall B, et al. A comparative exposure assessment of Campylobacter in Ontario, Canada. Risk Anal. (2017) 37:677–715. doi: 10.1111/risa.12653
7. Suzuki H, Yamamoto S. Campylobacter contamination in retail poultry meats and by-products in the world: a literature survey. J Vet Med Sci. (2009) 71:255–61. doi: 10.1292/jvms.71.255
8. Stabler RA, Larsson JT, Al-Jaberi S, Nielsen EM, Kay E, Tam CC, et al. Characterization of water and wildlife strains as a subgroup of Campylobacter jejuni using DNA microarrays. Environ Microbiol. (2013) 15:2371–83. doi: 10.1111/1462-2920.12111
9. Jokinen C, Edge TA, Ho S, Koning W, Laing C, Mauro W, et al. Molecular subtypes of Campylobacter spp., Salmonella enterica, and Escherichia coli O157:H7 isolated from faecal and surface water samples in the Oldman River watershed, Alberta, Canada. Water Res. (2011) 45:1247–57. doi: 10.1016/j.watres.2010.10.001
10. Ravel A, Pintar K, Nesbitt A, Pollari F. Non food-related risk factors of campylobacteriosis in Canada: a matched case-control study. BMC Public Health. (2016) 16:1016. doi: 10.1186/s12889-016-3679-4
11. Petersen L, Nielsen EM, Engberg J, On SL, Dietz HH. Comparison of genotypes and serotypes of Campylobacter jejuni isolated from Danish wild mammals and birds and from broiler flocks and humans. Appl Environ Microbiol. (2001) 67:3115–21. doi: 10.1128/AEM.67.7.3115-3121.2001
12. Sippy R, Sandoval-Green CMJ, Sahin O, Plummer P, Fairbanks WS, Zhang Q, et al. Occurrence and molecular analysis of Campylobacter in wildlife on livestock farms. Vet Microbiol. (2012) 157:369–75. doi: 10.1016/j.vetmic.2011.12.026
13. Sasaki Y, Goshima T, Mori T, Murakami M, Haruna M, Ito K, et al. Prevalence and antimicrobial susceptibility of foodborne bacteria in wild boars (Sus scrofa) and wild deer (Cervus nippon) in Japan. Foodborne Pathog Dis. (2013) 10:985–91. doi: 10.1089/fpd.2013.1548
14. Carbonero A, Paniagua J, Torralbo A, Arenas-Montes A, Borge C, García-Bocanegra I. Campylobacter infection in wild artiodactyl species from southern Spain: Occurrence, risk factors and antimicrobial susceptibility. Comp Immunol Microbiol Infect Dis. (2014) 37:115–21. doi: 10.1016/j.cimid.2014.01.001
15. Kwan PSL, Barrigas M, Bolton FJ, French NP, Gowland P, Kemp R, et al. Molecular epidemiology of Campylobacter jejuni populations in dairy cattle, wildlife, and the environment in a farmland area. Appl Environ Microbiol. (2008) 74:5130–8. doi: 10.1128/AEM.02198-07
16. French NP, Midwinter A, Holland B, Collins-Emerson J, Pattison R, Colles F, et al. Molecular epidemiology of Campylobacter jejuni isolates from wild-bird fecal material in children's playgrounds. Appl Environ Microbiol. (2009) 75:779–83. doi: 10.1128/AEM.01979-08
17. Hudson SJ, Lightfoot NF, Coulson JC, Russell K, Sisson PR, Sobo AO. Jackdaws and magpies as vectors of milkborne human Campylobacter infection. Epidemiol Infect. (1991) 107:363–72. doi: 10.1017/S0950268800049001
18. Riordan T, Humphrey TJ, Fowles A. A point source outbreak of Campylobacter infection related to bird-pecked milk. Epidemiol Infect. (1993) 110:261–5. doi: 10.1017/S0950268800068187
19. Gardner TJ, Fitzgerald C, Xavier C, Klein R, Pruckler J, Stroika S, et al. Outbreak of campylobacteriosis associated with consumption of raw peas. Clin Infect Dis. (2011) 53:26–32. doi: 10.1093/cid/cir249
20. Saunders S, Smith K, Schott R, Dobbins G, Scheftel J. Outbreak of campylobacteriosis associated with raccoon contact at a wildlife rehabilitation centre, Minnesota, 2013. Zoonoses Public Health. (2016) 64:222–7. doi: 10.1111/zph.12300
21. Cody AJ, McCarthy ND, Bray JE, Wimalarathna HML, Colles FM, Jansen van Rensburg MJ, et al. Wild bird-associated Campylobacter jejuni isolates are a consistent source of human disease, in Oxfordshire, United Kingdom. Environ Microbiol Rep. (2015) 7:782–8. doi: 10.1111/1758-2229.12314
22. Andrés S, Vico JP, Garrido V, Grilló MJ, Samper S, Gavín P, et al. Epidemiology of subclinical salmonellosis in wild birds from an area of high prevalence of pig salmonellosis: phenotypic and genetic profiles of Salmonella isolates. Zoonoses and Public Health. (2013) 60:355–65. doi: 10.1111/j.1863-2378.2012.01542.x
23. Hald B, Skov MN, Nielsen EM, Rahbek C, Madsen JJ, Wainø M, et al. Campylobacter jejuni and Campylobacter coli in wild birds on Danish livestock farms. Acta Vet Scand. (2016) 58:11. doi: 10.1186/s13028-016-0192-9
24. Greig J, Rajić A, Young I, Mascarenhas M, Waddell L, LeJeune J. A scoping review of the role of wildlife in the transmission of bacterial pathogens and antimicrobial resistance to the food chain. Zoonoses Public Health. (2015) 62:269–84. doi: 10.1111/zph.12147
25. Rosatte R, Ryckman M, Ing K, Proceviat S, Allan M, Bruce L, et al. Density, movements, and survival of raccoons in Ontario, Canada: implications for disease spread and management. J Mammal. (2010) 91:122–35. doi: 10.1644/08-MAMM-A-201R2.1
26. Jardine CM, Janecko N, Allan M, Boerlin P, Chalmers G, Kozak G, et al. Antimicrobial resistance in Escherichia coli isolates from raccoons (Procyon lotor) in southern Ontario, Canada. Appl Environ Microbiol. (2012) 78:3873–9. doi: 10.1128/AEM.00705-12
27. Bondo KJ, Weese JS, Rouseau J, Jardine CM. Longitudinal study of Clostridium difficile shedding in raccoons on swine farms and conservation areas in Ontario, Canada. BMC Vet Res. (2015) 11:254. doi: 10.1186/s12917-015-0563-x
28. Bondo KJ, Pearl DL, Janecko N, Boerlin P, Reid-Smith RJ, Parmley J, et al. Epidemiology of antimicrobial resistance in Escherichia coli isolates from raccoons (Procyon lotor) and the environment on swine farms and conservation areas in Southern Ontario. PLoS ONE. (2016) 11:e0165303. doi: 10.1371/journal.pone.0165303
29. Bondo KJ, Pearl DL, Janecko N, Boerlin P, Reid-Smith RJ, Parmley J, et al. Impact of season, demographic and environmental factors on Salmonella occurrence in raccoons (Procyon lotor) from swine farms and conservation areas in southern Ontario. PLoS ONE. (2016) 11:e0161497. doi: 10.1371/journal.pone.0161497
30. Hirsch BT, Prange S, Hauver SA, Gehrt SD. Patterns of latrine use by raccoons (Procyon lotor) and implication for Baylisascaris procyonis transmission. J Wildl Dis. (2014) 50:243–9. doi: 10.7589/2013-09-251
31. Bondo KJ, Pearl DL, Janecko N, Boerlin P, Reid-Smith RJ, Parmley J, et al. Epidemiology of Salmonella on the paws and in the faeces of free-ranging raccoons (Procyon Lotor) in southern Ontario, Canada. Zoonoses Public Health. (2016) 63:303–10. doi: 10.1111/zph.12232
32. Lee K, Iwata T, Nakadai A, Kato T, Hayama S, Taniguchi T, et al. Prevalence of Salmonella, Yersinia and Campylobacter spp. in feral raccoons (Procyon lotor) and masked palm civets (Paguma larvata) in Japan. Zoonoses and Public Health. (2011) 58:424–31. doi: 10.1111/j.1863-2378.2010.01384.x
33. Viswanathan M, Pearl DL, Taboada EN, Parmley EJ, Mutschall S, Jardine CM. Molecular and statistical analysis of Campylobacter spp. and antimicrobial-resistant Campylobacter carriage in wildlife and livestock from Ontario farms. Zoonoses Public Health. (2016) 64:194–203. doi: 10.1111/zph.12295
34. Jokinen CC, Koot JM, Carrillo CD, Gannon VPJ, Jardine CM, Mutschall SK, et al. An enhanced technique combining pre-enrichment and passive filtration increases the isolation efficiency of Campylobacter jejuni and Campylobacter coli from water and animal fecal samples. J Microbiol Methods. (2012) 91:506–13. doi: 10.1016/j.mimet.2012.09.005
35. Denis M, Soumet C, Rivoal K, Ermel G, Blivet D, Salvat G, et al. Development of a m-PCR assay for simultaneous identification of Campylobacter jejuni and C. coli. Lett Appl Microbiol. (1999) 29:406–10. doi: 10.1046/j.1472-765X.1999.00658.x
36. Taboada EN, Ross SL, Mutschall SK, MacKinnon JM, Roberts MJ, Buchanan CJ, et al. Development and validation of a comparative genomic fingerprinting method for high-resolution genotyping of Campylobacter jejuni. J Clin Microbiol. (2012) 50:788–97. doi: 10.1128/JCM.00669-11
37. R Core Team. R: A Language and Environment for Statistical Computing. (2012). Available online at: http://www.R-project.org (accessed April, 2019).
38. Wickham H. tidyverse. (2017). Available online at: https://cran.r-project.org/web/packages/tidyverse/index.html (accessed April, 2019).
39. Yu G, Smith DK, Zhu H, Guan Y, Lam TTY. ggtree: an r package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol Evol. (2017) 8:28–36. doi: 10.1111/2041-210X.12628
40. Kruczkiewicz P, Mutschall S, Barker D, Thomas JE, Van Domselaar GH, Gannon VP, et al. MIST: a tool for rapid in silico generation of molecular data from bacterial genome sequences. In: Proc 4th Int Conf Bioinforma Models Methods Algorithms. Barcelona (2013). p. 316–23.
42. Grundmann H, Hori S, Tanner G. Determining confidence intervals when measuring genetic diversity and the discriminatory abilities of typing methods for microorganisms. J Clin Microbiol. (2001) 39:4190–2. doi: 10.1128/JCM.39.11.4190-4192.2001
43. Sharpe D. Your Chi-square test is statistically significant: now what? Res Eval. (2015) 20:1–10. doi: 10.12968/prps.2015.Sup170.15
44. Rainwater KL, Marchese K, Slavinski S, Humberg LA, Dubovi EJ, Jarvis JA, et al. Health survey of free-ranging raccoons (Procyon lotor) in central park, New York, New York, USA: implications for human and domestic animal health. J Wildl Dis. (2017) 53:272–84. doi: 10.7589/2016-05-096
45. Hamir AN, Franklin S, Wesley IV, Sonn RJ. Campylobacter jejuni and Arcobacter species associated with intussusception in a raccoon (Procyon lotor). Vet Rec. (2004) 155:338–40. doi: 10.1136/vr.155.11.338
46. GRCA. Grand River Watershed Characterization Report – Executive Summary. (2008). Available online at: https://www.sourcewater.ca/en/source-protection-areas/resources/Documents/Grand/Grand_Reports_Characterization_ES.pdf (accessed November 9, 2019).
47. Jensen AN, Dalsgaard A, Baggesen DL, Nielsen EM. The occurrence and characterization of Campylobacter jejuni and C. coli in organic pigs and their outdoor environment. Vet Microbiol. (2006) 116:96–105. doi: 10.1016/j.vetmic.2006.03.006
48. Meerburg BG, Jacobs-Reitsma WF, Wagenaar JA, Kijlstra A. Presence of Salmonella and Campylobacter spp. in wild small mammals on organic farms. Appl Environ Microbiol. (2006) 72:960–2. doi: 10.1128/AEM.72.1.960-962.2006
49. Sheppard SK, Colles FM, McCarthy ND, Strachan NJC, Ogden ID, Forbes KJ, et al. Niche segregation and genetic structure of Campylobacter jejuni populations from wild and agricultural host species. Mol Ecol. (2011) 20:3484–90. doi: 10.1111/j.1365-294X.2011.05179.x
50. Griekspoor P, Colles FM, McCarthy ND, Hansbro PM, Ashhurst-Smith C, Olsen B, et al. Marked host specificity and lack of phylogeographic population structure of Campylobacter jejuni in wild birds. Mol Ecol. (2013) 22:1463–72. doi: 10.1111/mec.12144
51. Weis AM, Storey DB, Taff CC, Townsend AK, Huang BC, Kong NT, et al. Genomic comparisons and zoonotic potential of Campylobacter between birds, primates, and livestock. Appl Environ Microbiol. (2016) 82:7165–75. doi: 10.1128/AEM.01746-16
52. Gripp E, Hlahla D, Didelot X, Kops F, Maurischat S, Tedin K, et al. Closely related Campylobacter jejuni strains from different sources reveal a generalist rather than a specialist lifestyle. BMC Genom. (2011) 12:584. doi: 10.1186/1471-2164-12-584
53. Sheppard SK, Cheng L, Méric G, de Haan CPA, Llarena AK, Marttinen P, et al. Cryptic ecology among host generalist Campylobacter jejuni in domestic animals. Mol Ecol. (2014) 23:2442–51. doi: 10.1111/mec.12742
54. Dearlove BL, Cody AJ, Pascoe B, Meric G, Wilson DJ, Sheppard SK. Rapid host switching in generalist Campylobacter strains erodes the signal for tracing human infections. ISME J. (2016) 10:721–9. doi: 10.1038/ismej.2015.149
55. Llarena AK, Zhang J, Vehkala M, Välimäki N, Hakkinen M, Hänninen ML, et al. Monomorphic genotypes within a generalist lineage of Campylobacter jejuni show signs of global dispersion. Microb Genom. (2016) 2:e000088. doi: 10.1099/mgen.0.000088
56. Carrillo CD, Kruczkiewicz P, Mutschall S, Tudor A, Clark C, Taboada EN. A framework for assessing the concordance of molecular typing methods and the true strain phylogeny of Campylobacter jejuni and C. coli using draft genome sequence data. Front Cell Infect Microbiol. (2012) 2:57. doi: 10.3389/fcimb.2012.00057
Keywords: Campylobacter, longitudinal surveillance, molecular subtyping, Procyon lotor, raccoon, zoonoses
Citation: Mutschall SK, Hetman BM, Bondo KJ, Gannon VPJ, Jardine CM and Taboada EN (2020) Campylobacter jejuni Strain Dynamics in a Raccoon (Procyon lotor) Population in Southern Ontario, Canada: High Prevalence and Rapid Subtype Turnover. Front. Vet. Sci. 7:27. doi: 10.3389/fvets.2020.00027
Received: 25 November 2019; Accepted: 14 January 2020;
Published: 11 February 2020.
Edited by:
Ozan Gundogdu, University of London, United KingdomReviewed by:
Friederike Hilbert, University of Veterinary Medicine Vienna, AustriaFrances Colles, University of Oxford, United Kingdom
Marja-Liisa Hänninen, University of Helsinki, Finland
Philip Edward Carter, Institute of Environmental Science and Research (ESR), New Zealand
Copyright © 2020 Jardine, Bondo, Hetman, and Her Majesty the Queen in Right of Canada. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Eduardo N. Taboada, ZWR1YXJkby50YWJvYWRhQGNhbmFkYS5jYQ==
†Present address: Benjamin M. Hetman, Canadian Field Epidemiology Program, Center for Emergency Preparedness and Response, Public Health Agency of Canada, Ottawa, ON, Canada
Kristin J. Bondo, Department of Natural Resources Management, Texas Tech University, Lubbock, TX, United States
 Steven K. Mutschall
Steven K. Mutschall Benjamin M. Hetman
Benjamin M. Hetman Kristin J. Bondo
Kristin J. Bondo Victor P. J. Gannon
Victor P. J. Gannon Claire M. Jardine
Claire M. Jardine Eduardo N. Taboada
Eduardo N. Taboada