- Department of Population Health and Pathobiology, College of Veterinary Medicine, North Carolina State University, Raleigh, NC, United States
Nairobi Sheep Disease virus (NSDv) is a zoonotic and tick-borne disease that can cause over 90% mortality in small ruminants. NSDv has historically circulated in East Africa and has recently emerged in the Asian continent. Despite efforts to control the disease, some regions, mostly in warmer climates, persistently report disease outbreaks. Consequently, it is necessary to understand how environmental tolerances and factors that influence transmission may shed light on its possible emergence in other regions. In this study, we quantified the available literature of NSDv from which occurrence data was extracted. In total, 308 locations from Uganda, Kenya, Tanzania, Somalia, India, Sri Lanka and China were coupled with landscape conditions to reconstruct the ecological conditions for NSDv circulation and identify areas of potential disease transmission risk. Our results identified areas suitable for NSDv in Ethiopia, Malawi, Zimbabwe, Southeastern China, Taiwan, and Vietnam. Unsuitable areas included Democratic Republic of Congo, Zambia, and Southern Somalia. In summary, soil moisture, livestock density, and precipitation predispose certain areas to NSDv circulation. It is critical to investigate the epidemiology of NSDv in order to promote better allocation of resources to control its spread in regions that are more at risk. This will help reduce disease impact worldwide as climate change will favor emergence of such vector-borne diseases in areas with dense small ruminant populations.
Introduction
Background
Small ruminant populations have become one of the pillars of socio-economic well-being for developing countries due to their direct contribution to food security, however newly emerging diseases pose a constant threat (1–3). The demand for small ruminant products is growing globally, especially in China, the biggest producer, and importer of ovine meat. Other countries such as the United States, Islamic Republic of Iran, Japan, and Qatar have also increased import demand for these products (4). Some countries such as Somaliland have economies that depend entirely on their livestock industries and employ over 70% of the population (5, 6). Others such as the Kingdom of Saudi Arabia import 5 million live ruminants a year, most of which are sheep and goats (7). However, this economic value is directly dependent on animal health, which has been compromised in regions with high incidence of tick-borne diseases (8–10). The effects of ticks and tick-borne diseases on livestock pose a significant barrier to economic development (11–13), and thus, a better understanding of current emerging disease distributions is important for human socio-economic development and animal welfare.
Some of the most pathogenic diseases of small ruminants include viruses from the Bunyaviridae family, which are spreading to novel areas and currently threaten small ruminant populations worldwide (14–17). There are seven serogroups within the Nairovirus genus, with the most impactful ones being (i) Crimean-Congo hemorrhagic fever group which includes the human pathogen Crimean-Congo hemorrhagic fever virus (CCHFv), and (ii) Nairobi Sheep Disease group that contains Nairobi Sheep Disease virus (NSDv) (18). Due to the similarities between NSDv and CCHFv, studies on NSDv will be useful in furthering our understanding of this important human pathogen (19), and will serve as a good model system to study other nairoviruses (20).
NSDv causes hemorrhagic gastroenteritis, fever, abortion, and high mortality in small ruminants (21) and febrile illness, nausea, vomiting, and headache in humans (22). Mortality rates in susceptible animals exceed 90%, causing significant economic losses for production systems (21). This disease is listed as notifiable to the World Organization for Animal Health (OIE) (23), and it has the potential to impose trade barriers and consequently have a substantial impact on small ruminant producers worldwide (24).
Although the occurrence of NSDv has been reported in numerous countries (15, 25–31), there has been limited understanding of the biogeographic factors shaping its distribution and the potential areas at risk for future epidemics. The available literature on NSDv ranges from 1910 to 2019, with host serology and virus isolation found in 14 of these studies (Table S1). It has been found in environmentally varying areas, leaving an open question regarding the requirements of this virus, and its vectors to efficiently spread infection. For that reason, the aim of the present work was to develop a systematic review and distribution model of NSDv.
NSDv Vectors and Affected Species
Ticks are known to transmit a greater variety of pathogenic microorganisms than any other arthropod vectors, and are among the most important vectors affecting livestock and humans (32–36). NSDv spreads via feeding of competent infected ticks, and its geographic distribution is therefore limited to the areas comprising suitable environment conditions for them (19). The main tick species related to the spread of NSDv are Rhipicephalus appendiculatus in East Africa and Haemaphysalis intermedia in Asia (19). Lewis (37) demonstrated that R. appendiculatus can potentially retain NSDv for long periods (138 to 871 days) depending on the life stage of the tick. NSDv has also been isolated from Amblyomma variegatum (38), Rhipicephalus hemaphysaloides (39), and Haemaphysalis longicornis (15).
NSDv is also known to have the potential to infect humans. The first human infection was recorded in a young boy (40) and some laboratory acquired infections occurred in following years (41, 42). Human sera has been shown to contain antibodies against NSDv in India (40, 43), Uganda (28), Kenya (44), and Sri Lanka (31). Other isolates have also been obtained from different vector species such as Culex vishnui mosquitoes, where the virus was unable to replicate without a tick host (45), Haemaphysalis wellingtoni ticks feeding on red spurfowl (Galloperdix spadicea) (46), and from a pool of Culicoides 23 midges (47) and Culicoides tororensis, although this could be due to blood meal residues in the midges (48). Low titers were found in various wild ruminant species, but this was likely due to an antibody cross-reaction with other viruses (27).
The Emerging Pathway of NSDv
The first reports of NSDv came from Nairobi livestock markets in Kenya in 1910 as the result of an investigation carried out by veterinary pathologist Eustace Montgomery (26). NSDv was first identified as the causative agent in a classic study that showed virus transmission transovarially and transstadially in the Ixodid tick R. appendiculatus (26), findings that were later confirmed by Daubney and Hudson (49). In the following years, it was identified in various other areas of Kenya (27, 38, 50), Uganda (21, 28), Somalia (attributed to Rhipicephalus pulchellus) (29), and Tanzania (51). NSDv is also known as Ganjam virus in Asia. Similarities between NSDv and Ganjam virus were highly debated due to their occurrence on different continents and association with different vectors, but genetic and serologic analyses demonstrated that Ganjam virus is an Asian variant of NSDv, instead of a different virus as previously described (18, 22, 51, 52). NSDv was first isolated on the Asian continent in 1954 from H. intermedia in India (25) and Sri Lanka in 1996 (31). More recently, Gong et al. (15) were the first to discover NSDv present in H. longicornis ticks in China. Small ruminants bred in endemic areas do not appear to be affected by the virus (19), which could be due to the presence of maternal antibodies that provide sufficient protection until the animal's own immunity can be established (19).
In August 2017, H. longicornis was found in the United States for the first time in all three of its life stages, infecting an Icelandic sheep in Hunterdon County (New Jersey) that had not been transported or had any contact with foreign animals (53). The following year, this tick was found in seven other states along the Eastern US and Arkansas (54), and its presence was verified by reexamination of archived historical samples, confirming that H. longicornis was present in West Virginia in 2010 and New Jersey in 2013 (54). Over the past 30 years, the US Department of Agriculture has identified this tick at least six times from imported horses in quarantine (55). H. longicornis is unique for its parthenogenesis, which allows a single female to produce a clonal population without mating with a male (56). H. longicornis is also found in Australia, which may facilitate the appearance of NSDv in the country (57). Due to the potential global distribution of this disease, the Australian Veterinary Association has stated that changes in climate could lead to an increase in disease incidence with devastating consequences (57). H. longicornis is the only exotic tick established in New Zealand that has an economic impact on livestock (58), and NSDv was most recently isolated from this species in China (15). The range of NSDv and its vectors is likely spreading, and it will become even more important as we continue to push for breed improvement and maximizing land use to manage increasing global demands for small ruminant products (20).
Variables Affecting NSDv Spread
Climate change is known to impact the distribution of a great number of diseases (59–63), especially in tropical and subtropical regions (64–66). Our understanding of the environmental preferences facilitating the survival of NSDv vectors and hosts will allow at-risk regions to better prepare for potential incursions of this disease. These tick species mentioned above are known to span large geographical ranges, such as R. appendiculatus, which has a territory that extends from the tropical regions of East Africa to the temperate regions of South Africa (67), as well as India and Pakistan (68), or H. longicornis, which occupies a wide range of climates from equatorial New Guinea and the Pacific Islands, to snow and cool summer conditions in northeast Primorsky Krai, Russia (69, 70). For that reason, a deeper understanding of the environmental conditions facilitating the survival of these ticks is key to predicting novel areas at risk for NSDv spread, which could potentially lessen the impact of this disease on naive populations.
Economic Impact of NSDv
As global trade continues to increase, infectious diseases are posing a greater threat to our economies, food supply, and health (71, 72). NSDv has the potential to have devastating effects on naive sheep and goat populations (21), and therefore, efforts to increase awareness and surveillance should become a priority, especially for countries with economies that rely heavily on small ruminant products. NSDv vectors such as H. longicornis have already adapted to New Zealand and Australia (57, 58), threatening the NSD-free status of these countries. Crossbreeding and the introduction of new breeds of small ruminants have been the target of recent efforts to increase the efficiency of meat production and commerce in some countries. This increases the chances of NSDv outbreaks in populations lacking immunity against the virus (73–75) since the main route for NSDv spread occurs upon movement of susceptible animals into enzootic areas (50). Since small ruminants are vital to people's livelihoods in low-income communities (1), efforts to preserve their health and better understand tick-borne diseases are essential.
Given the critical importance of small ruminant production to global food security and the potential for NSDv spread in new regions, the current information gaps in the epidemiology of this disease have to be explored. This allows us to better predict future outbreaks and potential regions at risk for this disease. This is the first systematic review and predictive model of NSDv, allowing us to assess the potential distribution of the disease and predict regions that are most at risk.
Methods
We divided this study into two main sections: (i) a systematic literature review to identify all susceptible hosts, potential vectors, and the ecological dependencies for virus spread and (ii) an assessment of the geographic and environmental distribution of hosts and vectors using an ecological niche modeling (ENM) approach. Figure 1 illustrates this process.
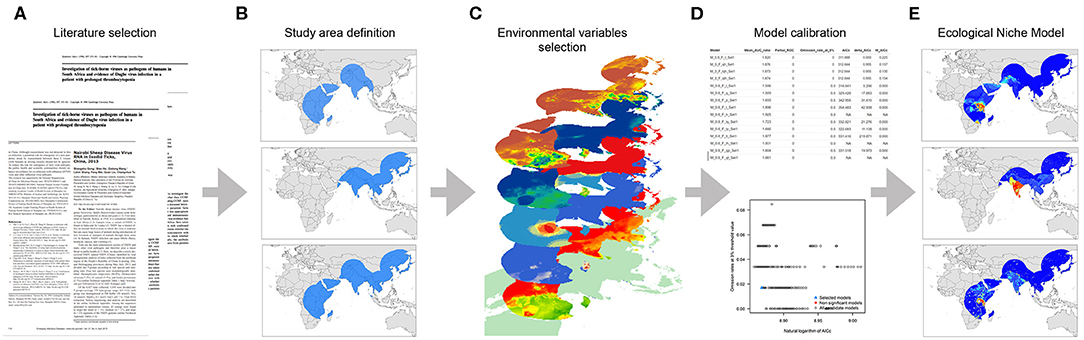
Figure 1. Workflow of the modeling process. (A) Data collection based on literature review, (B) Study area definition, (C) Environmental variables selection, (D) Model calibration, and (E) Final ecological niche model.
Systematic Review
Literature Selection
We followed the steps outlined by O'Connor and Sargeant and others (76–78) (Table S2) to scope the global distribution of NSDv. Our search terms were designed to recover all information and reports under the terms “Nairobi Sheep Disease” and “Ganjam virus.” The study protocol followed the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines (79, 80) (Figure 2).
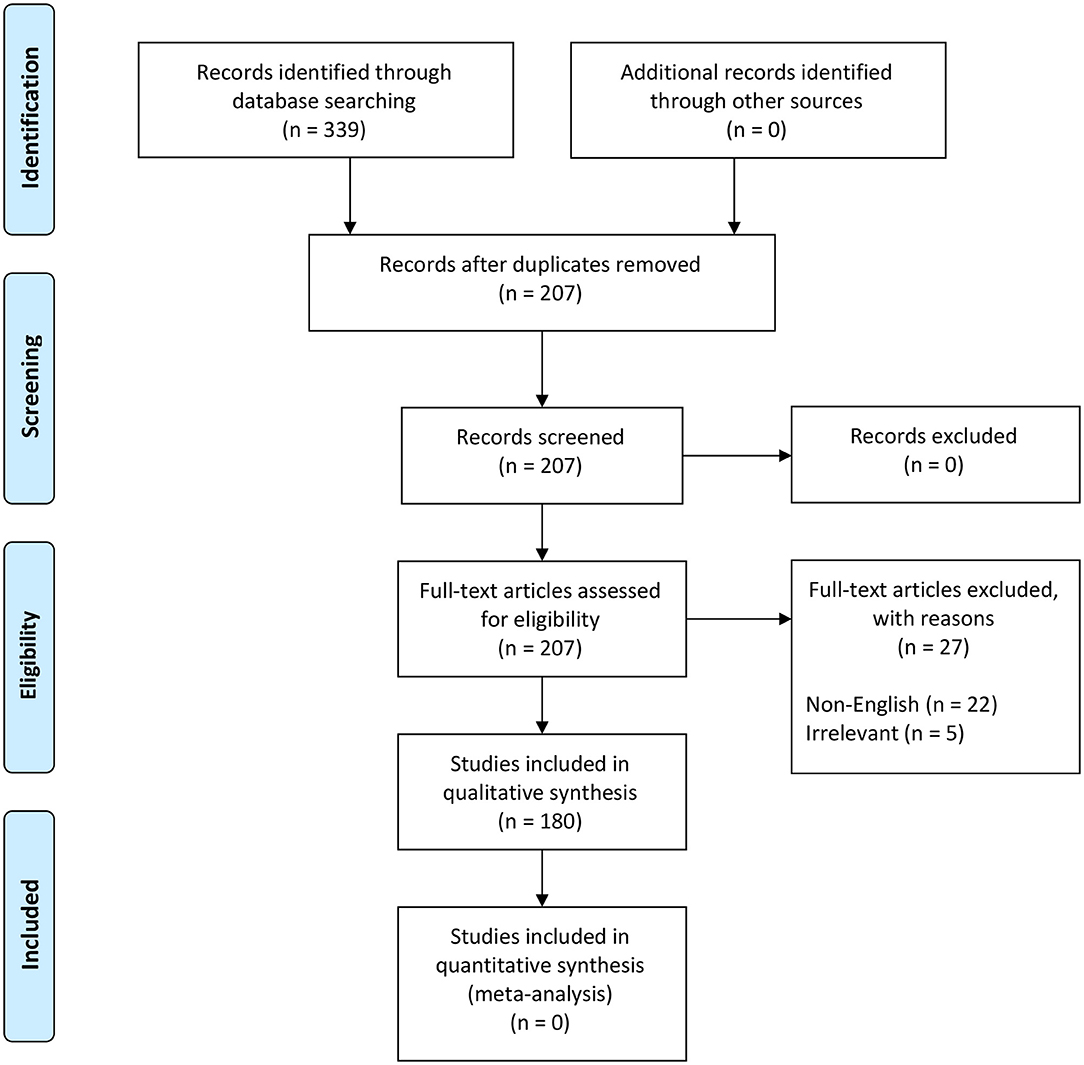
Figure 2. PRISMA flow diagram detailing the literature selection process (79).
These terms were used to search all databases using the EBSCOhost platform, which yielded 339 scientific publications in 11 databases (Table S2). Exclusion criteria included duplicates (132), non-English papers (22), and irrelevant studies (5) which brought the total number of eligible papers for systematic review to 180 (Figure 2). This database search was completed in November 2018.
Ecological Niche Model
Data Collection
From the literature review, we extracted a total of 308 locations from 14 studies. The information retrieved included occurrence data classified as follows: (i) host occurrence- if the virus was identified from sheep, goat, or human serology, (ii) tick occurrence- if the virus was identified directly from tick vectors. Data points were georeferenced to the centroid of the smallest administrative division of each country from where the data was collected. One exception was the use of the locations reported by Gong et al. (15), which were also verified before being considered for analysis. Diagnostics based on post-mortem examinations were not considered since lesions are not specific enough to reliably distinguish NSDv from heartwater, Pasteurella pneumonia, or babesiosis (29). Additional occurrences from the following studies in South Africa (81), Uganda (28), Kenya (38), and India (40) were not included in the study due to non-specific locations from which we were unable to recover the centroid of the smallest administrative boundary.
Model Calibration Area
To define the model calibration area for the circulation of NSDv, we followed the framework proposed by Soberón and Peterson (82), which restricts the ecological niche model to ecological features for the organism in question, the resolution of the environmental variables employed, and the extent of the region where the organisms are able to disperse due to their biogeographic barriers [see M in the BAM framework in Soberón and Peterson (82)] This region is defined as the area accessible to the species that has been sampled, so presence records can exist within a suitable area (83). We measured the maximum distance between all NSDv locations and used this distance as the accessible area for the virus. A buffer around the occurrences was applied to establish the model calibration area.
Environmental Variables Selection
The environmental variables used to estimate the distribution of NSDv were selected based on the described requirements of the hosts and ticks, including their survival in landscapes with suitable temperature and humidity and their amplification by the presence of livestock reservoir species. We used six environmental variables to calibrate all models, and candidate models were selected based on the described requirements of the virus, including its capacity to survive in the landscape based on the presence of susceptible host species. We included the variables shown in Table 1, which will serve as an appropriate approximation of the required biogeographic conditions.
Previous studies have highlighted the importance of temperature for the main host of NSDv, R. appendiculatus (67, 84, 85). Although it is not a critical limiting factor for ticks, it is still important, especially when it approaches the developmental threshold for that species (69). Likewise, variables related to humidity are also important because it has been observed that prolonged drought and dry pastures cause high mortality, especially among engorged H. longicornis (69, 84) and R. appendiculata larvae and eggs (85), and H. intermedia and R. haemaphysaloides numbers which seem to increase after rains (86). For that reason, we used precipitation, runoff, evapotranspiration, and soil moisture as predictors of tick prevalence. Finally, livestock population was represented by the density of sheep and goats, which are most clinically affected by NSDv (52, 87).
Ecological Niche Model
Niche modeling presents a framework based on ecological theory that is widely used to predict, interpret, and understand geographic distributions of species and several other biological phenomena, such as disease transmission (88), impacts of invasive species (89, 90), effects of climate change on species' distributions (91, 92), and reconstruction of ancestral environmental niches (93). For this study, we considered an ecological niche as the set of environmental conditions in a region necessary for a species to persist (83). Information about the presence of susceptible hosts for NSDv were also added to this study through a raster layer that contained sheep and goat densities. We explored three scenarios based on different combinations of occurrences: (i) the occurrences of ticks that tested positive for NSDv; (ii) the occurrences of sheep and goats that tested positive for NSDv (hosts); and (iii) the combination of both positive tick and host occurrences. For these analyses, we considered that the factors explaining the distribution of NSDv and its potential hosts follow the Biotic-Abiotic-Mobility (BAM) framework (82). The analysis was set individually for each occurrence data set for all models in which a hypothesis for each accessible area M was constructed [further details of the importance of this step can be found in Barve et al. (83)]. The study area for positive ticks and host cases was designed based on the union of buffer areas created around each occurrence location. The buffer area was built based on the average distance between the external points and the most central point (centroid). Thus, for positive ticks this buffer was defined as 1,334 km, while for positive hosts, it was 1,555 km. The polygons drawn around each study area were then used to intercept the environmental predictor layers (Table 1).
The total occurrences for each set were randomly subdivided into 70% of the data set for model calibration and 30% for model evaluation. This data division allowed for both model calibration and internal testing. For each combination of data, we created 493 candidate models by combining 17 regularization multipliers (0.1–1.0 by the interval of 0.1, from 1 to 10 at the interval of 1), with all 29 possible combinations of the five MaxEnt feature classes (linear=L, quadratic=Q, product=P, threshold=T, and hinge=H). We evaluated and selected the candidate model performances based on significance rates (5%), and model complexity penalizations (AICc). The best models for each dataset were selected according to the following criteria: significant models with omission rates < =5%, and from those selected models, we used delta AICc values of < =2 to determine the final candidates. Both the full model calibration and selection step were performed using the “kuenm” package in the R environment and used Maxent as the modeling algorithm (94).
The final ENM models were performed in Maxent version 3.3.3 k (95). The specific configuration of all models included 10 bootstrap replicates and random seed with logistic outputs. Finally, to identify extrapolation risk in the model transfer steps, we used the mobility-oriented parity (MOP) index for each data set, which is an improved metric proposed by Owens et al. (96). The interpretation of the output models followed Merow et al. (97) as a suitable index to account for the probability of disease risk. To further evaluate model predictions, continuous outputs were converted into binary maps based on a threshold, removing 10% of the calibration occurrences (error = 10%) to reduce the uncertainty of estimations and facilitate model interpretation (98). Finally, the models were transferred to a global scale applying the same predictor layers used for the calibration step (Table 1).
Results
Descriptive Analysis
A descriptive analysis of both the disease and vector distribution is provided in Figure 3, even though the total number of recovered disease occurrences were not used as offset information. A total of 308 data points were extracted from 14 studies included in the literature review, from which 12 data points were isolated from ticks and the rest were obtained from host serology. Data coming from hosts was distributed throughout East Africa, India, and Sri Lanka, while tick data was found only in India, Sri Lanka, and China.
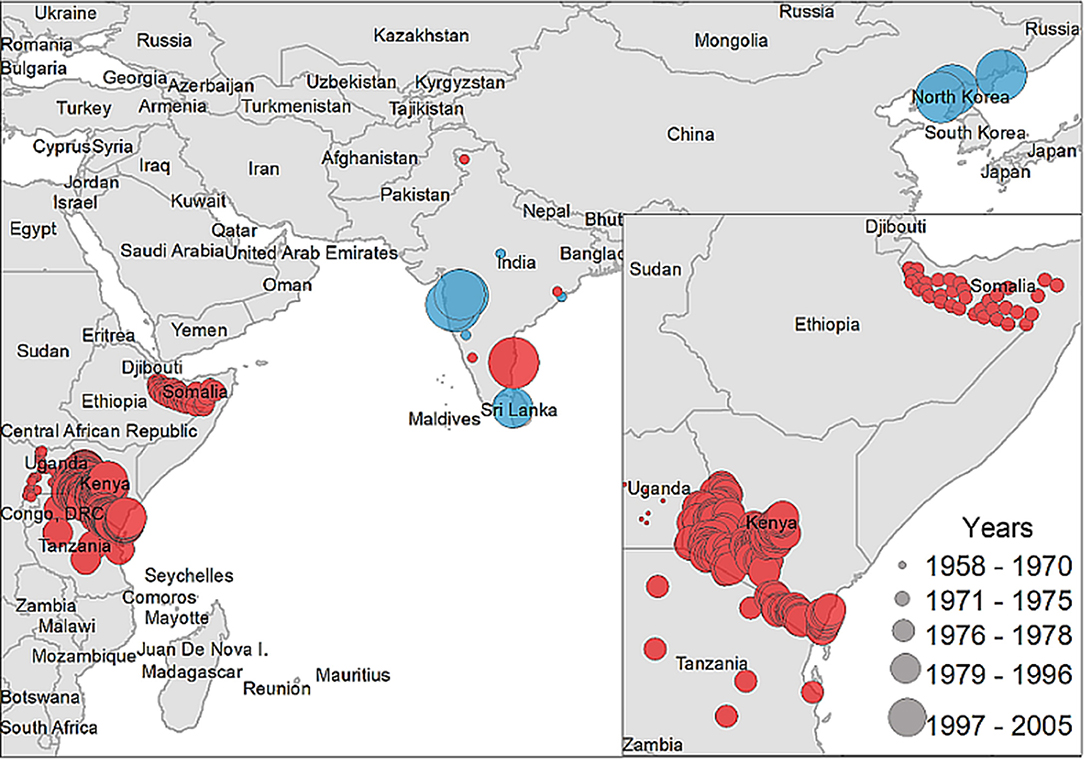
Figure 3. Case distribution of NSDv used for model calibration. Points denote occurrence locations. Color represents positive host (red) and tick (blue) occurrences and size denotes the year of publication.
Ecological Niche Models
NSDv Host Potential Distribution
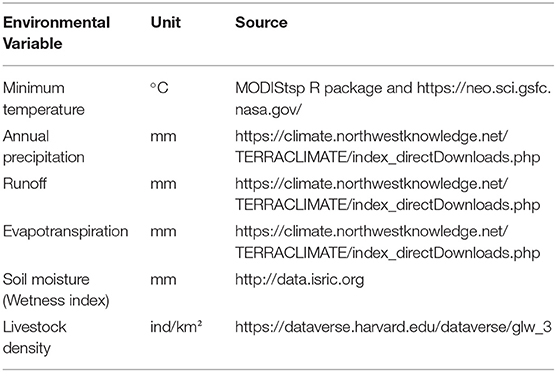
The positive host occurrences of NSDv were used to calibrate 493 models along with the full set of environmental predictors. In total, 490 candidate models were significant from which 157 were significant and met the omission rate criteria. For the global minimum AICc values, 1 model had delta AICc values < =2, and met the full criteria used in the selection step (Figure 4). We emphasize that this selected model met both criteria but did not have lower AICc, which is often utilized as single decision criteria in model selection (99–101) (see the blue triangle which highlights the selected final mode- Figure 4). In order to identify areas of higher risk for NSDv occurrences, the selected model identified the geographic risk areas for small ruminants. This model showed countries presenting low suitability for NSDv spread including Democratic Republic of Congo, Zambia, and southern Somalia. There were clusters of unsuitability throughout the Kingdom of Saudi Arabia, eastern Yemen, Oman, and western China of the study area. High-risk regions were found throughout Eritrea, Ethiopia, Kenya, Uganda, Rwanda, Burundi, Tanzania, Malawi, Zimbabwe, and the southern tip of India (Figure 7). The livestock density variable was the most influential (48.7%) in describing the risk area, followed by runoff (21.8%), evapotranspiration (11.6%), precipitation (10.2%), minimum temperature (4.7%), and soil moisture (3%).
NSDv Tick Potential Distribution
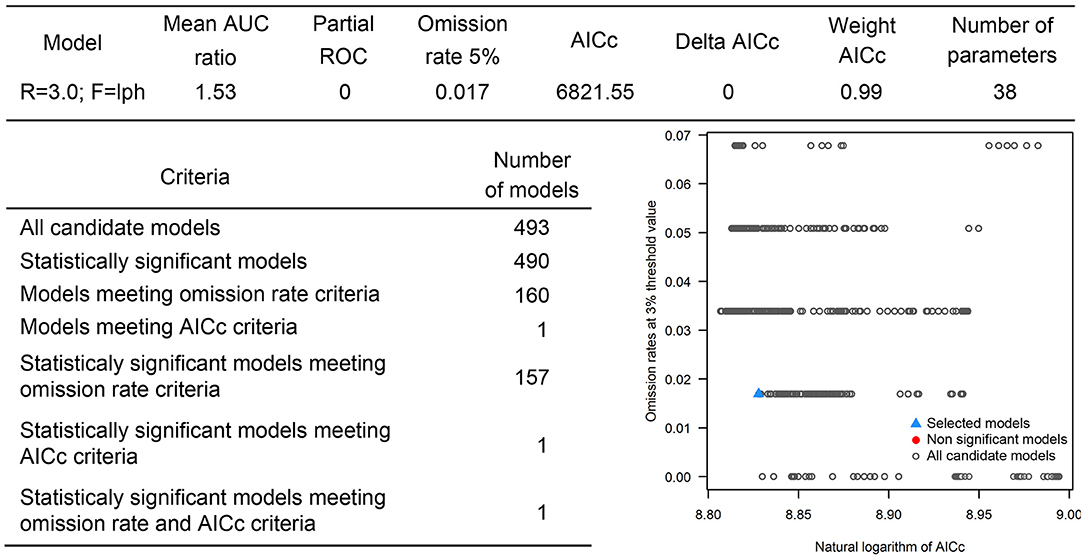
The occurrences of NSDv in vectors was modeled applying the selected parameterizations found in Table 1. Using the locations for tick occurrences, 367 candidate models out of 493 were statistically significant if compared with the assumed null expectations. All 367 of these statistically significant models also met the omission rate criteria. For the global minimum AICc values for ticks, 2 models had delta AICc values < =2 and 1 model met the criteria (Figure 5). The model selected had lower AICc, was significant, and within the omission error cutoff (see the blue triangle which highlights the selected final mode- Figure 5). This model was used to identify areas with a potential circulation of ticks within the calibration study area. It showed unsuitable areas clustered in Sri Lanka, central Vietnam, eastern Taiwan, the western shores of Japan, and southeast China. High-risk regions were found throughout the southern half of India, Sri Lanka, Bangladesh, Myanmar, Laos, Thailand, Cambodia, Taiwan, China's Hainan Island and the southern tip of Guangdong province, and the northern and southern portions of Vietnam (Figure 7). The minimum temperature variable was the most influential (53.5%) in describing the risk areas, followed by livestock (38.4%), soil moisture (4.6%), and runoff (3.5%). Precipitation and evapotranspiration were not influential (0%).
NSDv Host and Tick Potential Distribution
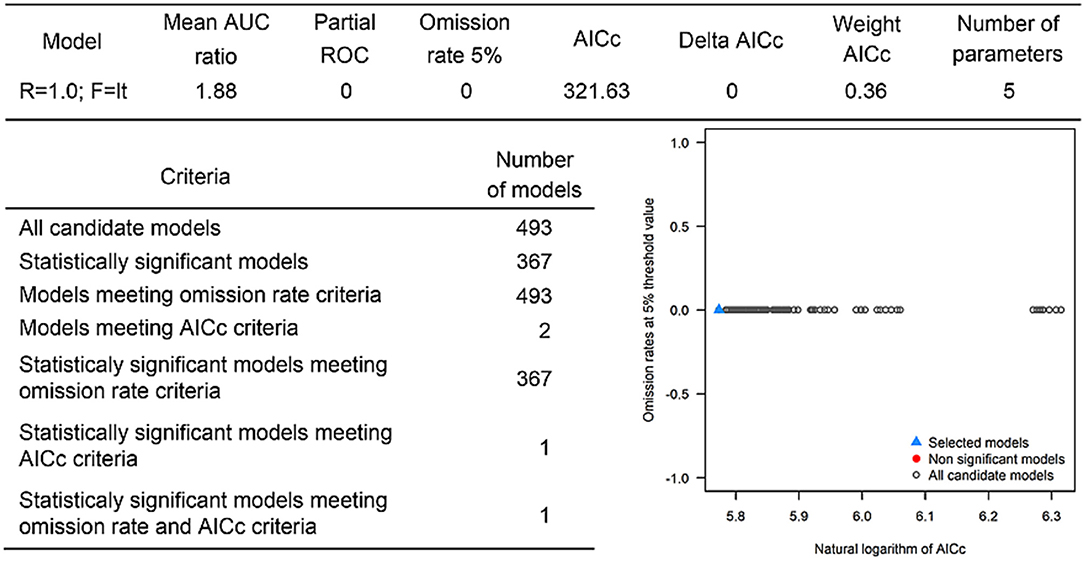
The last model exploration we did included the combination of data associated with infected hosts and the occurrences of potential vector ticks. From 493 candidate models, 209 were significant, from which 109 also met the omission rate criteria. For the global minimum AICc values, 1 model had delta AICc values < =2. Only 1 model met the full criteria used in the model selection step (Figure 6) (see the blue triangle which highlights the selected final model-Figure 6). Suitable areas in the study area included Ethiopia, Uganda, Kenya, Tanzania, the southern tip of India, the southeastern coast of China, and Taiwan (Figure 7). The livestock density variable was the most influential (38.1%) in describing the risk area, followed by evapotranspiration (27.2%), runoff (14.5%), minimum temperature (9.2%), soil moisture (7.8%), and precipitation (3.3%).
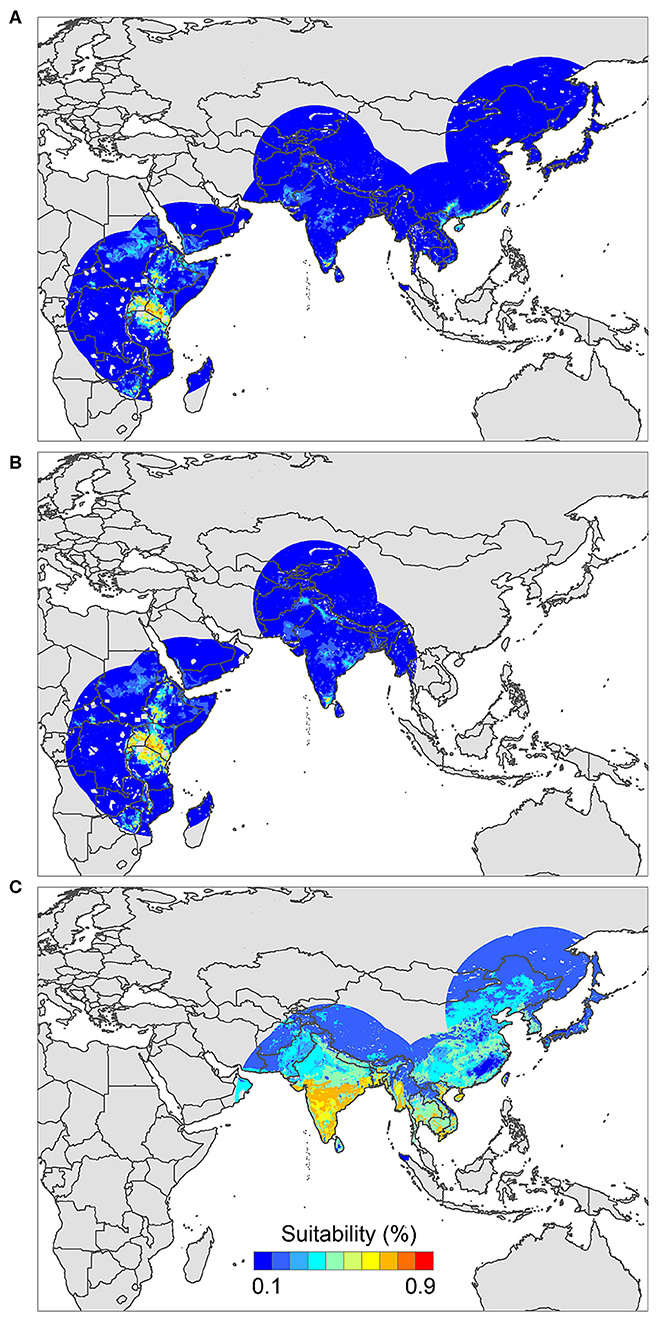
Figure 7. Ecological niche models of (A) hosts and ticks, (B) hosts, and (C) tick circulation. Warmer colors represent areas with higher environmental suitability.
NSDv Environmental Preferences
Overall, our ecological niche models coincided well with the reported geographical locations of the host and tick occurrences identified during the systematic review. These regions are the most environmentally suited for NSDv, and they indicate areas where the disease would most easily become established and continue spreading to naive small ruminant populations. For the host-based model, the most suitable areas occurred with the highest values of evaporation (46.1 mm; IQR 83.0 mm), precipitation (40.3 mm; IQR 63.0 mm), and runoff (2.8 mm; IQR 3.0 mm). In the case of the tick-based model, the most suitable areas presented the highest values of soil moisture (62.2 mm; IQR 588.0 mm) and livestock density (11812.0 lnd/km2; IQR 8953.8 lnd/km2). Finally, the model combining hosts and ticks presented the highest values of minimum temperature (12.86°C; IQR 5.50°C) (see Table 2 for detailed information).
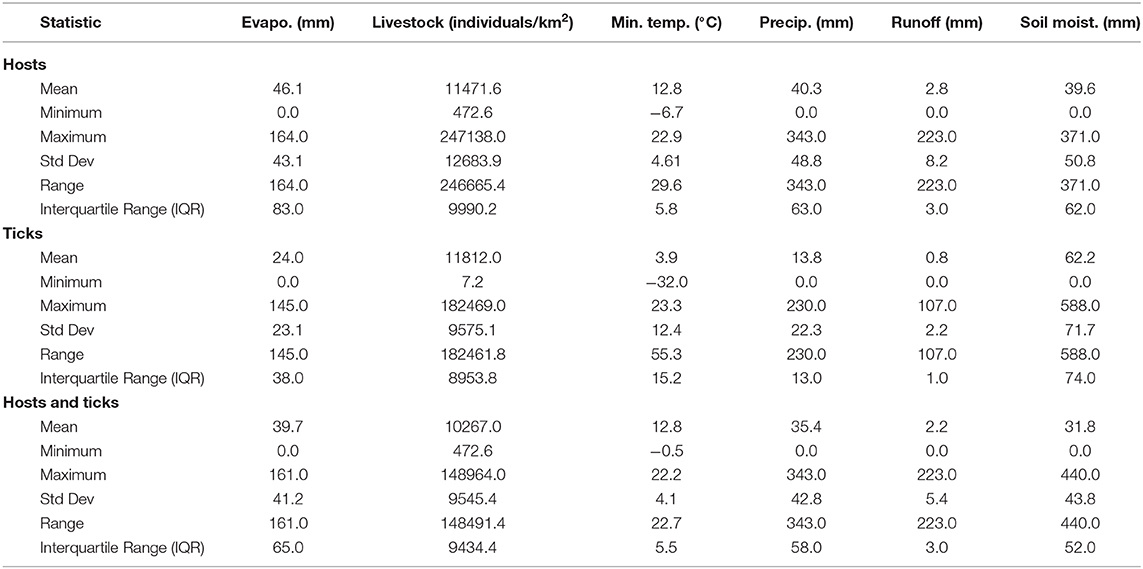
Table 2. Environmental preference of hosts, ticks, and hosts & ticks based on their potential distributions.
Discussion
We identified novel countries at risk for infection with NSDv, including Ethiopia, Malawi, Zimbabwe, Southeastern China, Taiwan, and Vietnam. These findings suggest that NSDv may have a wider range than was previously thought, and therefore, these predictions can help target active surveillance efforts to those regions where NSDv could efficiently spread if introduced in the future.
Livestock diseases can result in devastating trade bans with wide-ranging consequences (6). Specifically, NSDv has the potential to easily spread to naive populations and cause significant economic losses (21). Therefore, it is imperative for at-risk countries to be aware of these risks and take necessary precautions to protect their animals, peoples' livelihoods, local economies, and global trade partners.
To the authors' knowledge, this is the first systematic review of NSDv that has gathered all available geographic locations where this disease has occurred. Systematic reviews are increasingly being used in animal agriculture and veterinary medicine since they offer a replicable evidence-based method to identify, evaluate, and summarize primary research (76). The systematic literature review highlighted knowledge gaps in the epidemiology of NSDv, most notably limitations in surveillance of outbreaks and the identification of major NSDv reservoirs and vectors. It also uncovered the association of multiple tick species as candidate vectors, which have each proven their adaptability to a wide range of environments, making ENM a feasible approach to model the occurrence of the disease. By evaluating environmental drivers behind tick and host distributions, we were able to approximate the impact that climate change might have on their distribution. Additionally, the recent isolation of NSDv in China showed its potential to spread to various parts of the world, and further demonstrated the need for appropriate modeling techniques to help countries target their surveillance efforts.
Based on the reports obtained, we determined the countries which are currently at risk for the introduction of this disease. We performed extensive model fitting from which we selected the best performance-based model, focusing on significance rather than Akaike Information Criterion (AIC) values as our main criteria for model selection. Using significance rather than AIC allows us to choose the single best model which outperforms the rest and will give us the most realistic prediction of occurrence. We found varying results in our models, with notable differences in the environmental preferences needed for the optimal spread of the virus when host and tick-based models were compared. Because our data points included serology from hosts and virus isolation from ticks, these models should be interpreted together rather than separately. Multiple environmental variables play a role in the transmission of NSDv, and they should all be considered holistically. We support that this combination of data would more accurately predict potential regions of outbreak since the main transmission of NSDv will likely occur when both hosts and ticks coincide together in the same environment.
ENMs have been widely applied in epidemiology to assess species introduction into novel areas and disease emergence (99, 102–104), or the effect of climate on disease distribution (91, 105–107). However, previous ENM methods rely solely on the lowest AIC to identify the best fit model, which alone may not lead us to the best option for that set of occurrences (108). Significance should be used as the first criteria when filtering through candidate models, followed by performance and simplicity (94). This approach reduces the time spent on transferring these models, and uses the mobility-oriented parity (MOP) index to prevent outcome over-interpretation and identify extrapolation risks, making it a critical tool when transferring ENMs to current and future scenarios, and resulting in a more accurate prediction of species' ecological niche as the environment changes. These calibrations have become more popular and are being applied throughout epidemiology (94, 109), but our model of NSDv will be the first time that this calibrated approach has been used to model the potential distribution of a virus.
Ticks are described as the main route of NSDv transmission and therefore, they are the most likely route of infection for naive populations given that the virus can survive for such extended periods of time inside its vector (37). International trade is a known pathway for the introduction of exotic ticks into new areas, which have the potential to harbor pathogens with significant consequences for human health and agriculture (110, 111), highlighting the importance of increased surveillance measures, and regulatory framework over animal trade-related interactions. Transhumance pastoralism in the Sudano-Sahel region of Africa results in significant transboundary animal and human movement, and has resulted in the spread of several tick species and livestock diseases (112–115). There have been some efforts to regulate this movement across borders, while also recognizing the importance of this practice to millions of pastoralists and their livestock (116). Other potential routes of disease introduction include migratory birds, which have been shown to spread invasive tick species and vector-borne diseases, including the closely-related CCHFv which affects humans (117, 118).
In the tick-based model, we observed that minimum temperature was the most important variable. This is an important finding considering the constantly rising patterns of air and ocean temperatures over the last century due to global warming, where extreme temperatures are predicted to experience severe changes (119), forcing species to modify their current distribution to new and more-suitable environments (120–122). Previous studies have already documented expansions in the suitable areas of tick-borne diseases (123–125). Likewise, NSDv preferred areas shown to have elevated soil moisture and a denser population of livestock. Although the density of sheep and goats was expected to have a direct influence on the number of infected ticks found in some locations, soil moisture may have facilitated the ability of ticks to survive in the field and continue transmitting the disease to other individuals. In contrast, the model based on host occurrences had elevated mean evaporation, precipitation, and runoff. This may correlate with areas of intense feed and water resources, thus concentrating more animals in one area, which will facilitate the spread of the disease between and within the animal population.
Even though these two models can independently explain and predict areas where NSDv is most likely to occur, the combination and overlap of the most suitable environments for hosts and ticks will represent the areas where hosts and their vectors are more likely to proliferate and coincide, greatly increasing the possibility of disease spread. Thus, we recommend the comparison and combination of these models for future epidemiological assessments in order to get the most accurate prediction of at-risk areas. Given the relatively low surveillance done to control NSDv, there is a possibility that the disease has already reached the predicted area without being yet detected, and thus, we highlight the importance of immediate sampling to ensure the absence of virus in the region.
We encountered some limitations during the development of this study. First, a number of non-English papers (n = 22) were excluded in our systematic review which may have included additional data for our models. We only included data points that had a specific location and excluded those that were labeled as having been collected in general areas or districts, so it is possible that additional occurrences were not recorded. We also used a centroid to approximate the location of occurrences, which entails the possibility of some data points being farther away from the center. Additionally, multiple species of Ixodid ticks were modeled together which may result in overgeneralization of their geographic distribution. Finally, our analyses would have benefited from the availability of detailed information about variables such as international trade or more extensive small ruminant vector-borne disease surveillance, which would have increased the accuracy in the determination of risk areas and transmission routes for NSDv. Besides, our ENMs are mostly based on climatic variables, not considering the physiological tolerances of the organisms in the study, and thus, the presence of the above-mentioned variables would provide a more realistic approximation of their potential distribution (126).
Vector-borne diseases are expected to be the most climate-sensitive subset of infectious diseases (127, 128), and using environmental variables is critical to predicting their distribution. Recent outbreaks of devastating ruminant diseases such as Bluetongue and epizootic hemorrhagic disease, serve as a reminder of the consequences that climate change has on vector-borne diseases (129), and illustrates the importance of using the best modeling techniques we have at our disposal in order to more accurately predict their spread.
Conclusions
The data compiled here will be useful for additional spatial and environmental modeling of NSDv and its vectors, informing governments and policy makers about their prevalence and spread in order to direct surveillance efforts. Further studies would also help the development of case studies for NSDv and other infectious diseases affecting small ruminants, as well as to understand and predict where to best implement surveillance strategies. This can result in improved response time to outbreaks, resulting in decreased economic losses and reduced impact of disease on human and animal health. Further study of the tick species shown to carry and transmit NSDv will be imperative as the distribution of these species changes with environmental factors and increasing international trade. As the ticks' distribution changes, so too should surveillance strategies. Additionally, many countries that could potentially be at risk for NSDv have never had a cross-sectional study done on their livestock or tick species of interest, which would hinder their ability to quickly and effectively respond to a disease outbreak. As an OIE reportable disease, NSDv can cause significant trade implications for countries and could be underreported as a result. Rising global demand for these products and recognition of the socio-economic importance of small ruminants merits further discussion of the diseases that impact them and pose a threat to their populations in other parts of the world and the communities that depend on them.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession numbers can be found in the article/Supplementary Material.
Author Contributions
SK, MJ, and GM contributed to the design of the project. SK collected and curated the data used in both the systematic review and modeling exercise. MJ and GM performed the modeling section. SK and AF-D-D wrote the manuscript. All authors reviewed and edited the manuscript.
Funding
We acknowledge the Department of Population Health and Pathobiology at North Carolina State University for providing startup funds for GM.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2020.00419/full#supplementary-material
References
1. Kosgey IS. Breeding Objectives and Breeding Strategies for Small Ruminants in the Tropics. (2004) Available online at: http://edepot.wur.nl/121527 (accessed March 15, 2019).
2. Umunna MO, Olafadehan OA, Arowona A. Small ruminant production and management systems in urban area of southern guinea Savanna of Nigeria. Asian J Agric Food Sci. (2014) 2:107–14.
3. Adams F. Socio-Economic Analysis of Small Ruminant Livestock Production in Northern Ghana. Socio-Economic Anal Small Rumin Livest Prod North Ghana. (2015). p. 1–200. Available online at: http://ir.knust.edu.gh/xmlui/bitstream/handle/123456789/9264/Faizal_Adams.pdf?sequence=1 (accessed March 15, 2019).
4. FAO. Meat Market Review: 2019 Outlook. (2019). Available online at: http://www.fao.org/3/ca3880en/ca3880en.pdf (accessed April 10, 2020).
5. Godiah LM, Baker D, Elmi II, Costagli R, Gulaid I, Wanyoike F. Enhancing the Provision of Livestock Marketing Information in Somaliland. Nairobi (2014) Available online at: https://cgspace.cgiar.org/bitstream/handle/10568/56737/ilriResearchReport_35.pdf?sequence=8&isAllowed=y (accessed March 17, 2019).
6. Muhumed MM, Yonis AM. The future of somaliland livestock exports: examining the sustainability of livestock trade. Int J Manag Account Econ. (2018) 5. Available online at: https://www.researchgate.net/publication/328319505_The_Future_of_Somaliland_Livestock_Exports_Examining_the_Sustainability_of_Livestock_Trade_International_Journal_of_Management_Accounting_and_Economics_Vol_5_No_8_2018_With_Abdiqadir_M_Yonis (accessed March 17, 2019).
7. Abd El-Rahim IHA, Asghar AH, Mohamed AM, Fat'hi SM. The impact of importation of live ruminants on the epizootiology of foot and mouth disease in Saudi Arabia. Rev Sci Tech. (2016) 35:769–78. doi: 10.20506/rst.35.3.2567
8. Kosgey IS Rowlands GJ van Arendonk JAM Baker RL. Small ruminant production in smallholder and pastoral/extensive farming systems in Kenya. Small Rumin Res. (2008) 77:11–24. doi: 10.1016/j.smallrumres.2008.02.005
9. Yitayew D, Samuel D. Tick borne hemoparasitic diseases of ruminants: a review. Adv Biol Res (Rennes). (2015) 9:210–224. doi: 10.5829/idosi.abr.2015.9.4.9516
10. Mureithi DK, Mukiria EW. An assessment of tick-borne diseases constraints to livestock production in a smallholder livestock production system: a case of Njiru District, Kenya. Int J Res Agric For. (2015) 2:43–49. doi: 10.5455/ijlr.20151222101656
11. Minjauw B, McLeod A. Tick-Borne Diseases And Poverty: The Impact of Ticks and Tick-Borne Diseases on the Livelihoods of Small-Scale and Marginal Livestock Owners in India and Eastern and Southern Africa. Edinburgh: DFID Animal Health Programme, Centre for Tropical Veterinary Medicine. (2003) Available at: https://www.cabdirect.org/cabdirect/abstract/20063155090 (accessed March 17, 2019).
12. Jaime Betancur Hurtado O, Giraldo-Ríos C. Economic and Health Impact of the Ticks in Production Animals. IntechOpen (2019). doi: 10.5772/intechopen.81167
13. Narladkar BW. Projected economic losses due to vector and vector-borne parasitic diseases in livestock of India and its significance in implementing the concept of integrated practices for vector management. Vet World. (2018) 11:151–60. doi: 10.14202/vetworld.2018.151-160
14. Linthicum KJ, Britch SC, Anyamba A. Rift valley fever: an emerging mosquito-borne disease. Annu Rev Entomol. (2016) 61:395–415. doi: 10.1146/annurev-ento-010715-023819
15. Gong S, He B, Wang Z, Shang L, Wei F, Liu Q, et al. Nairobi sheep disease virus RNA in ixodid ticks, China, 2013. Emerg Infect Dis. (2015) 21:718–20. doi: 10.3201/eid2104.141602
16. Armstrong PM, Andreadis TG, Anderson JF. Emergence of a new lineage of Cache Valley virus (Bunyaviridae: Orthobunyavirus) in the northeastern United States. Am J Trop Med Hyg. (2015) 93:11–7. doi: 10.4269/ajtmh.15-0132
17. Elliott RM. Bunyaviruses and climate change. Clin Microbiol Infect. (2009) 15:510–7. doi: 10.1111/j.1469-0691.2009.02849.x
18. Kuhn J, Wiley M, Rodriguez S, Bào Y, Prieto K, Travassos da Rosa A, et al. Genomic characterization of the genus nairovirus (Family Bunyaviridae). Viruses. (2016) 8:164. doi: 10.3390/v8060164
19. Baron MD, Holzer B. Nairobi sheep disease virus/Ganjam virus. Rev Sci Tech. (2015) 34:411–7. doi: 10.20506/rst.34.2.2367
20. Bin Tarif A, Lasecka L, Holzer B, Baron MD. Ganjam virus/Nairobi sheep disease virus induces a pro-inflammatory response in infected sheep. Vet Res. (2012) 43:71. doi: 10.1186/1297-9716-43-71
21. Terpstra C. Nairobi Sheep Disease. Studies on Virus Properties, Epizootiology and Vaccination in Uganda. Nairobi Sheep Dis Stud virus Prop Epizoot Vaccin Uganda. (1969) Available online at: https://www.cabdirect.org/cabdirect/abstract/19702201697 (accessed March 17, 2019).
22. Yadav PD, Vincent MJ, Khristova M, Kale C, Nichol ST, Mishra AC, et al. Genomic analysis reveals Nairobi sheep disease virus to be highly diverse and present in both Africa, and in India in the form of the Ganjam virus variant. Infect Genet Evol. (2011) 11:1111–20. doi: 10.1016/j.meegid.2011.04.001
23. OIE. OIE-Listed Diseases, Infections and Infestations in Force in 2020. (2020) Available online at: https://www.oie.int/animal-health-in-the-world/oie-listed-diseases-2020/ (accessed December 4, 2020).
24. Ibrahim H. Small Ruminant Production Techniques. Nairobi: ILRI (International Livestock Research Institute) (1998). Available online at: https://cgspace.cgiar.org/handle/10568/509 (accessed March 17, 2019).
25. Dandawate CN, Shah KV. Ganjam virus: a new arbovirus isolated from ticks Haemaphysalis intermedia Warburton and Nuttall, 1909 in Orissa, India. Indian J Med Res. (1969) 57:799–804.
26. Montgomery E. On a Tick-borne gastroenteritis of sheep and goats occurring in British East Africa. J Comp Pathol Ther. (1917) 30:28–57. doi: 10.1016/S0368-1742(17)80002-3
27. Davies FG. A survey of Nairobi sheep disease antibody in sheep and goats, wild ruminants and rodents within Kenya. J Hyg (Lond). (1978) 81:251–8. doi: 10.1017/S0022172400025080
28. Weinbren MP, Gourlay RN, Lumsden WHR, Weinbren BM. An epizootic of nairobi sheep disease in Uganda. J Comp Pathol Ther. (1958) 68:174–87. doi: 10.1016/S0368-1742(58)80018-1
29. Edelsten RM. The distribution and prevalence of Nairobi Sheep disease and other tick-borne infections of sheep and goats in northern Somalia. Trop Anim Health Prod. (1975) 7:29–34. doi: 10.1007/BF02383239
30. Jessett DM. Serological evidence of Nairobi sheep disease in Tanzania. Trop Anim Health Prod. (1978) 10:99–100. doi: 10.1007/BF02235317
31. Perera LP, Peiris JSM, Weilgama DJ, Calisher CH, Shope RE. Nairobi sheep disease virus isolated from Haemaphysalis intermedia ticks collected in Sri Lanka. Ann Trop Med Parasitol. (1996) 90:91–3. doi: 10.1080/00034983.1996.11813031
32. Jongejan F, Uilenberg G. The global importance of ticks. Parasitology. (2004) 129:S3–14. doi: 10.1017/S0031182004005967
33. de la Fuente J, Estrada-Pena A, Venzal JM, Kocan KM, Sonenshine DE. Overview: ticks as vectors of pathogens that cause disease in humans and animals. Front Biosci. (2008) 13:6938–46. doi: 10.2741/3200
34. Parola P, Raoult D. Ticks and tickborne bacterial diseases in humans: an emerging infectious threat. Clin Infect Dis. (2001) 32:897–928. doi: 10.1086/319347
35. Laurenson MK, Norman RA, Gilbert L, Reid HW, Hudson PJ. Identifying disease reservoirs in complex systems: mountain hares as reservoirs of ticks and louping-ill virus, pathogens of red grouse. J Anim Ecol. (2003) 72:177–85. doi: 10.1046/j.1365-2656.2003.00688.x
36. Keesing F, Belden LK, Daszak P, Dobson A, Harvell CD, Holt RD, et al. Impacts of biodiversity on the emergence and transmission of infectious diseases. Nature. (2010) 468:647–52. doi: 10.1038/nature09575
37. Lewis EA. Nairobi sheep disease: the survival of the virus in the tick Rhipicephalus appendiculatus. Parasitology. (1946) 37:55–9. doi: 10.1017/S0031182000013159
38. Johnson BK, Chanas AC, Squires EJ, Shockley P, Simpson DIH, Parsons J, et al. Arbovirus isolations from ixodid ticks infesting livestock, Kano Plain, Kenya. Trans R Soc Trop Med Hyg. (1980) 74:732–7. doi: 10.1016/0035-9203(80)90188-1
39. Joshi M V., Geevarghese G, Joshi GD, Ghodke YS, Mourya DT, Mishra AC. Isolation of ganjam virus from ticks collected off domestic animals around Pune, Maharashtra, India. J Med Entomol. (2005) 42:204–6. doi: 10.1093/jmedent/42.2.204
40. Dandawate CN, Work TH, Webb JK, Shah KV. Isolation of ganjam virus from a human case of febrile illness: a report of a laboratory infection and serological survey of human sera from three different states of India. Indian J Med Res. (1969) 57:975–82.
41. Rao CV, Dandawate CN, Rodrigues JJ, Rao GL, Mandke VB, Ghalsasi GR, et al. Laboratory infections with Ganjam virus. Indian J Med Res. (1981) 74:319–24.
42. Banerjee K, Gupta NP, Goverdhan MK. Viral infections in laboratory personnel. Indian J Med Res. (1979) 69:363–73.
43. Joshi M V, Elankumaran S, Joshi GD. A post-epizootic survey of rift valley fever-like illness among sheep at Veerapuram, Chennai, Tamil Nadu. Indian J Virol. (1998) 14:155–7.
44. Morrill JC, Johnson BK, Hyams C, Okoth F, Tukei PM, Mugambi M, et al. Serological evidence of arboviral infections among humans of coastal Kenya. J Trop Med Hyg. (1991) 94:166–8.
45. Dandawate CN, Singh KR, Dhanda V. Experimental infection of certain mosquitoes & ticks with Ganjam virus. Indian J Exp Biol. (1981) 19:1185–6.
46. Rajagopalan PK, Sreenivasan MA, Paul SD. Isolation of Ganjam virus from the bird tick Haemaphysalis wellingtoni Nuttall and Warburton 1907. Indian J Med Res. (1970) 58:1195–6.
47. Walker AR, Davies FG. A preliminary survey of the epidemiology of bluetongue in Kenya. J Hyg (Lond). (1971) 69:47–60. doi: 10.1017/S0022172400021239
48. Davies FG, Walker AR, Ochieng P, Shaw T. Arboviruses isolated from Culicoides midges in Kenya. J Comp Pathol. (1979) 89:587–95. doi: 10.1016/0021-9975(79)90049-5
49. Daubney R, Hudson JR. Nairobi sheep disease. Parasitology. (1931) 23:507–24. doi: 10.1017/S0031182000013895
50. Davies FG. Nairobi sheep disease in Kenya. The isolation of virus from sheep and goats, ticks and possible maintenance hosts. J Hyg (Lond). (1978) 81:259–65. doi: 10.1017/S0022172400025092
51. Davies FG, Casals J, Jesset DM, Ochieng P. The serological relationships of Nairobi sheep disease virus. J Comp Pathol. (1978) 88:519–23. doi: 10.1016/0021-9975(78)90005-1
52. Marczinke BI, Nichol ST. Nairobi sheep disease virus, an important tick-borne pathogen of sheep and goats in Africa, is also present in Asia. Virology. (2002) 303:146–51. doi: 10.1006/viro.2002.1514
53. Rainey T, Occi JL, Robbins RG, Egizi A. Discovery of Haemaphysalis longicornis (Ixodida: Ixodidae) parasitizing a sheep in New Jersey, United States. J Med Entomol. (2018) 55:757–9. doi: 10.1093/jme/tjy006
54. Beard C Ben, Occi J, Bonilla DL, Egizi AM, Fonseca DM, Mertins JW, et al. Multistate infestation with the exotic disease–vector tick Haemaphysalis longicornis — United States, August 2017–September 2018. MMWR Morb Mortal Wkly Rep. (2018) 67:1310–3. doi: 10.15585/mmwr.mm6747a3
55. Haddow AD. The consequences of medically important invasive arthropods: the longhorned tick, Haemaphysalis longicornis. Clin Infect Dis. (2019) 68:530–1. doi: 10.1093/cid/ciy695
56. Mihara R, Umemiya-Shirafuji R, Abe Y, Matsuo T, Horiuchi N, Kawano S, et al. The development of oocytes in the ovary of a parthenogenetic tick, Haemaphysalis longicornis. Parasitol Int. (2018) 67:465–71. doi: 10.1016/j.parint.2018.04.006
57. Thomson S. Emergency animal diseases bulletin no.119: Nairobi sheep disease. Aust Vet J. (2018) 96:N10.
58. Heath A. Biology, ecology and distribution of the tick, Haemaphysalis longicornis Neumann (Acari: Ixodidae) in New Zealand. N Z Vet J. (2016) 64:10–20. doi: 10.1080/00480169.2015.1035769
59. Dantas-Torres F. Climate change, biodiversity, ticks and tick-borne diseases: the butterfly effect. Int J Parasitol Parasites Wildl. (2015) 4:452–61. doi: 10.1016/j.ijppaw.2015.07.001
60. Purse B V., Mellor PS, Rogers DJ, Samuel AR, Mertens PPC, Baylis M. Climate change and the recent emergence of bluetongue in Europe. Nat Rev Microbiol. (2005) 3:171–81. doi: 10.1038/nrmicro1090
61. Ready PD. Leishmaniasis emergence and climate change. Rev Sci Tech. (2008) 27:399–412. doi: 10.20506/rst.27.2.1803
62. Burge CA, Mark Eakin C, Friedman CS, Froelich B, Hershberger PK, Hofmann EE, et al. Climate change influences on marine infectious diseases: implications for management and society. Ann Rev Mar Sci. (2014) 6:249–77. doi: 10.1146/annurev-marine-010213-135029
63. Wu X, Lu Y, Zhou S, Chen L, Xu B. Impact of climate change on human infectious diseases: empirical evidence and human adaptation. Environ Int. (2016) 86:14–23. doi: 10.1016/j.envint.2015.09.007
64. Lyon B, DeWitt DG. A recent and abrupt decline in the East African long rains. Geophys Res Lett. (2012) 39. doi: 10.1029/2011GL050337
65. McSweeney C, New M, Lizcano G, Lu X, McSweeney C, New M, et al. The UNDP climate change country profiles: improving the accessibility of observed and projected climate information for studies of climate change in developing countries. Bull Am Meteorol Soc. (2010) 91:157–66. doi: 10.1175/2009BAMS2826.1
66. Lott FC, Christidis N, Stott PA. Can the 2011 East African drought be attributed to human-induced climate change? Geophys Res Lett. (2013) 40:1177–81. doi: 10.1002/grl.50235
67. Randolph SE. Abiotic and biotic determinants of the seasonal dynamics of the tick Rhipicephalus appendiculatus in South Africa. Med Vet Entomol. (1997) 11:25–37. doi: 10.1111/j.1365-2915.1997.tb00286.x
68. Ghosh S, Bansal GC, Gupta SC, Ray D, Khan MQ, Irshad H, et al. Status of tick distribution in Bangladesh, India and Pakistan. Parasitol Res. (2007) 101:207–16. doi: 10.1007/s00436-007-0684-7
69. Heath A. Implications for New Zealand of potentially invasive ticks sympatric with Haemaphysalis longicornis Neumann, 1901 (Acari: Ixodidae). Syst Appl Acarol. (2013) 18:1–26. doi: 10.11158/saa.18.1.1
70. Hoogstraal H, Roberts FHS, Kohls GM, Tipton VJ. Review of Haemaphysalis (Kaiseriana) longicornis Neumann (resurrected) of Australia, New Zealand, New Caledonia, Fiji, Japan, Korea, and northeastern China and USSR, and its parthenogenetic and bisexual populations (Ixodoidea, Ixodidae). J Parasitol. (1968) 54:1197–213. doi: 10.2307/3276992
71. Marano N, Arguin PM, Pappaioanou M. Impact of globalization and animal trade on infectious disease ecology. Emerg Infect Dis. (2007) 13:1807–9. doi: 10.3201/eid1312.071276
72. Fèvre EM, Bronsvoort BM de C, Hamilton KA, Cleaveland S. Animal movements and the spread of infectious diseases. Trends Microbiol. (2006) 14:125–31. doi: 10.1016/j.tim.2006.01.004
73. Getachew T, Haile A, Wurzinger M, Rischkowsky B, Gizaw S, Abebe A, et al. Review of sheep crossbreeding based on exotic sires and among indigenous breeds in the tropics: an Ethiopian perspective. African J Agric Res. (2016) 11:901–11. doi: 10.5897/AJAR2013.10626
74. Mbuku SM Okeyo AM Kosgey IS Kahi AK. Optimum crossbreeding systems for goats in low-input livestock production system in Kenya. Small Rumin Res. (2015) 123:55–61. doi: 10.1016/j.smallrumres.2014.10.001
75. Kosgey IS Baker RL Udo HMJ Van Arendonk JAM. Successes and failures of small ruminant breeding programmes in the tropics: a review. Small Rumin Res. (2006) 61:13–28. doi: 10.1016/j.smallrumres.2005.01.003
76. O'Connor AM, Sargeant JM. An introduction to systematic reviews in animal health, animal welfare, and food safety. Anim Heal Res Rev. (2014) 15:3–13. doi: 10.1017/S146625231400005X
77. European Food Safety Authority. Application of systematic review methodology to food and feed safety assessments to support decision making. EFSA J. (2010) 8:1637. doi: 10.2903/j.efsa.2010.1637
78. Grindlay DJC, Brennan ML, Dean RS. Searching the veterinary literature: a comparison of the coverage of veterinary journals by nine bibliographic databases. J Vet Med Educ. (2012) 39:404–12. doi: 10.3138/jvme.1111.109R
79. Moher D, Liberati A, Tetzlaff J, Altman DG, PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. (2009) 6:e1000097. doi: 10.1371/journal.pmed.1000097
80. Liberati A, Altman DG, Tetzlaff J, Mulrow C, Gøtzsche PC, Ioannidis JPA, et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: explanation and elaboration. PLoS Med. (2009) 6:e1000100. doi: 10.1371/journal.pmed.1000100
81. Burt FJ, Spencer DC, Leman PA, Patterson B, Swanepoel R. Investigation of tick-borne viruses as pathogens of humans in South Africa and evidence of Dugbe virus infection in a patient with prolonged thrombocytopenia. Epidemiol Infect. (1996) 116:353–61. doi: 10.1017/S0950268800052687
82. Soberón J, Peterson AT. Interpretation of models of fundamental ecological niches and species' distributional areas. Biodivers Informatics. (2005) 1–10. doi: 10.17161/bi.v2i0.4
83. Barve N, Barve V, Jiménez-Valverde A, Lira-Noriega A, Maher SP, Peterson AT, et al. The crucial role of the accessible area in ecological niche modeling and species distribution modeling. Ecol Modell. (2011) 222:1810–9. doi: 10.1016/j.ecolmodel.2011.02.011
84. Olwoch JM, Reyers B, Engelbrecht FA, Erasmus BFN. Climate change and the tick-borne disease, Theileriosis (East Coast fever) in sub-Saharan Africa. J Arid Environ. (2008) 72:108–20. doi: 10.1016/j.jaridenv.2007.04.003
85. Tukahirwa EM. The effects of temperature and relative humidity on the development of Rhipicephalus appendiculatus Neumann (Acarina, Ixodidae). Bull Entomol Res. (1976) 66:301–12. doi: 10.1017/S0007485300006696
86. Latha BR, Aiyasami SS, Pattabiraman G, Sivaraman T, Rajavelu G. Seasonal activity of ticks on small ruminants in Tamil Nadu State, India. Trop Anim Health Prod. (2004) 36:123–33. doi: 10.1023/B:TROP.0000012108.42526.11
87. Uilenberg G. General review of tick-borne diseases of sheep and goats world-wide. Parassitologia. (1997) 39:161–5.
88. Peterson AT. Ecologic niche modeling and spatial patterns of disease transmission. Emerg Infect Dis. (2006) 12:1822. doi: 10.3201/eid1212.060373
89. Jiménez-Valverde A, Peterson AT, Soberón J, Overton JM, Aragón P, Lobo JM. Use of niche models in invasive species risk assessments. Biol Invasions. (2011) 13:2785–97. doi: 10.1007/s10530-011-9963-4
90. Peterson AT. Predicting the geography of species' invasions via ecological niche modeling. Q Rev Biol. (2003) 78:419–33. doi: 10.1086/378926
91. González C, Wang O, Strutz SE, González-Salazar C, Sánchez-Cordero V, Sarkar S. Climate change and risk of leishmaniasis in North America: predictions from ecological niche models of vector and reservoir species. PLoS Negl Trop Dis. (2010) 4:e585. doi: 10.1371/journal.pntd.0000585
92. Jara M, García-Roa R, Escobar LE, Torres-Carvajal O, Pincheira-Donoso D. Alternative reproductive adaptations predict asymmetric responses to climate change in lizards. Sci Rep. (2019) 9:1–9. doi: 10.1038/s41598-019-41670-8
93. Alvarado-Serrano DF, Knowles LL. Ecological niche models in phylogeographic studies: applications, advances and precautions. Mol Ecol Resour. (2014) 14:233–48. doi: 10.1111/1755-0998.12184
94. Cobos ME, Peterson AT, Barve N, Osorio-Olvera L. kuenm: an R package for detailed development of ecological niche models using Maxent. PeerJ. (2019) 7:e6281. doi: 10.7717/peerj.6281
95. Steven J. Phillips, Miroslav Dudík RES. Maxent Software for Modeling Species Niches and Distributions (Version 3.3.3 k). Available online at: http://biodiversityinformatics.amnh.org/open_source/maxent/ (accessed April 10, 2020).
96. Owens HL, Campbell LP, Dornak LL, Saupe EE, Barve N, Soberón J, et al. Constraints on interpretation of ecological niche models by limited environmental ranges on calibration areas. Ecol Modell. (2013) 263:10–8. doi: 10.1016/j.ecolmodel.2013.04.011
97. Merow C, Smith MJ, Silander JA. A practical guide to MaxEnt for modeling species' distributions: what it does, and why inputs and settings matter. Ecography (Cop). (2013) 36:1058–69. doi: 10.1111/j.1600-0587.2013.07872.x
98. Peterson AT, Soberón J, Pearson RG, Anderson RP, Martínez-Meyer E, Nakamura M, et al. Ecological Niches and Geographic Distributions (MPB-49). Princeton, NJ: Princeton University Press (2011). doi: 10.23943/princeton/9780691136868.001.0001
99. Machado G, Weiblen C, Escobar LE. Potential distribution of Pythium insidiosum in Rio Grande do Sul, Brazil, and projections to neighbour countries. Transbound Emerg Dis. (2018) 65:1671–9. doi: 10.1111/tbed.12925
100. Magory Cohen T, McKinney M, Kark S, Dor R. Global invasion in progress: modeling the past, current and potential global distribution of the common myna. Biol Invasions. (2019) 21:1295–309. doi: 10.1007/s10530-018-1900-3
101. Sutton GF. Searching for a needle in a haystack: where to survey for climatically-matched biological control agents for two grasses (Sporobolus spp.) invading Australia. Biol Control. (2019) 129:37–44. doi: 10.1016/j.biocontrol.2018.11.012
102. Benedict MQ, Levine RS, Hawley WA, Lounibos LP. Spread of the tiger: global risk of invasion by the mosquito Aedes albopictus. Vector-Borne Zoonotic Dis. (2007) 7:76–85. doi: 10.1089/vbz.2006.0562
103. Peterson AT, Bauer JT, Mills JN. Ecologic and geographic distribution of filovirus disease. Emerg Infect Dis. (2004) 10:40–7. doi: 10.3201/eid1001.030125
104. DeVaney SC, McNyset KM, Williams JB, Peterson AT, Wiley EO. A tale of four “carp”: invasion potential and ecological niche modeling. PLoS ONE. (2009) 4:e5451. doi: 10.1371/journal.pone.0005451
105. Gálvez R, Descalzo MA, Guerrero I, Miró G, Molina R. Mapping the current distribution and predicted spread of the leishmaniosis sand fly vector in the Madrid Region (Spain) based on environmental variables and expected climate change. Vector-Borne Zoonotic Dis. (2011) 11:799–806. doi: 10.1089/vbz.2010.0109
106. Daszak P, Zambrana-Torrelio C, Bogich TL, Fernandez M, Epstein JH, Murray KA, et al. Interdisciplinary approaches to understanding disease emergence: the past, present, and future drivers of Nipah virus emergence. Proc Natl Acad Sci USA. (2013) 110:3681–8. doi: 10.1073/pnas.1201243109
107. Baquero OS, Machado G. Spatiotemporal dynamics and risk factors for human Leptospirosis in Brazil. Sci Rep. (2018) 8:1–4. doi: 10.1038/s41598-018-33381-3
108. Velasco JA, Poe S, González-Salazar C, Flores-Villela O. Solitary ecology as a phenomenon extending beyond insular systems: exaptive evolution in Anolis lizards. Biol Lett. (2019) 15:20190056. doi: 10.1098/rsbl.2019.0056
109. Raghavan RK, Peterson AT, Cobos ME, Ganta R, Foley D. Current and future distribution of the lone star tick, Amblyomma americanum (L.) (Acari: Ixodidae) in North America. PLoS ONE. (2019) 14:e0209082. doi: 10.1371/journal.pone.0209082
110. Burridge MJ. Ticks (Acari: Ixodidae) spread by the international trade in reptiles and their potential roles in dissemination of diseases. Bull Entomol Res. (2001) 91:3–23. doi: 10.1079/BER200071
111. Keirans JE, Durden LA. Invasion: exotic ticks (Acari: Argasidae, Ixodidae) imported into the United States. A review and new records. J Med Entomol. (2001) 38:850–61. doi: 10.1603/0022-2585-38.6.850
112. Adakal H, Biguezoton A, Zoungrana S, Courtin F, De Clercq EM, Madder M. Alarming spread of the Asian cattle tick Rhipicephalus microplus in West Africa-another three countries are affected: Burkina Faso, Mali and Togo.
113. Adinci KJ, Akpo Y, Sessou P, Yessinou RE, Adehan SB, Youssao AKI, et al. Influence of transhumance on the spread of Rhipicephalus microplus (Canestrini, 1888) in Benin. J Adv Vet Anim Res. (2018) 5: doi: 10.5455/javar.2018.e272
114. Byaruhanga C, Collins NE, Knobel D, Kabasa W, Oosthuizen MC. Endemic status of tick-borne infections and tick species diversity among transhumant zebu cattle in Karamoja Region, Uganda: Support for control approaches. Vet Parasitol Reg Stud Reports. (2015) 1–2:21–30. doi: 10.1016/j.vprsr.2015.11.001
115. Macpherson CNL. The effect of transhumance on the epidemiology of animal diseases. Prev Vet Med. (1995) 25:213–24. doi: 10.1016/0167-5877(95)00539-0
116. Apolloni A, Nicolas G, Coste C, EL Mamy AB, Yahya B, EL Arbi AS, et al. Towards the description of livestock mobility in Sahelian Africa: some results from a survey in Mauritania. PLoS ONE. (2018) 13:e0191565. doi: 10.1371/journal.pone.0191565
117. Lindeborg M, Barboutis C, Ehrenborg C, Fransson T, Jaenson TGT, Lindgren PE, et al. Migratory birds, ticks, and crimean- congo hemorrhagic fever virus. Emerg Infect Dis. (2012) 18:2095–7. doi: 10.3201/eid1812.120718
118. Leblebicioglu H, Eroglu C, Erciyas-Yavuz K, Hokelek M, Acici M, Yilmaz H. Role of migratory birds in spreading Crimean-Congo hemorrhagic fever, Turkey. Emerg Infect Dis. (2014) 20:1331–4. doi: 10.3201/eid2008.131547
119. Wolff E, Arnell N, Friedlingstein P, Gregory JM, Haigh J, Haines A, et al. Climate updates: progress since the fifth Assessment Report (AR5) of the IPCC. The Royal Society (2017).
120. Thomas CD, Cameron A, Green RE, Bakkenes M, Beaumont LJ, Collingham YC, et al. Extinction risk from climate change. Nature. (2004) 427:145–8. doi: 10.1038/nature02121
121. Velásquez-Tibatá J, Salaman P, Graham CH. Effects of climate change on species distribution, community structure, and conservation of birds in protected areas in Colombia. Reg Environ Chang. (2013) 13:235–48. doi: 10.1007/s10113-012-0329-y
122. Márquez AL, Real R, Olivero J, Estrada A. Combining climate with other influential factors for modelling the impact of climate change on species distribution. Clim Change. (2011) 108:135–57. doi: 10.1007/s10584-010-0010-8
123. Gray JS, Dautel H, Estrada-Peña A, Kahl O, Lindgren E. Effects of climate change on ticks and tick-borne diseases in Europe. Interdiscip Perspect Infect Dis. (2009) 2009:593232. doi: 10.1155/2009/593232
124. Süss J, Klaus C, Gerstengarbe FW, Werner PC. What makes ticks tick? Climate change, ticks, and tick-borne diseases. J Travel Med. (2008) 15:39–45. doi: 10.1111/j.1708-8305.2007.00176.x
125. Alkishe AA, Peterson AT, Samy AM. Climate change influences on the potential geographic distribution of the disease vector tick Ixodes ricinus. PLoS ONE. (2017) 12:e0189092. doi: 10.1371/journal.pone.0189092
126. Escobar LE, Craft ME. Advances and limitations of disease biogeography using ecological niche modeling. Front Microbiol. (2016) 7:1174. doi: 10.3389/fmicb.2016.01174
127. Caminade C, McIntyre KM, Jones AE. Impact of recent and future climate change on vector-borne diseases. Ann N Y Acad Sci. (2019) 1436:157–73. doi: 10.1111/nyas.13950
128. Tabachnick WJ. Challenges in predicting climate and environmental effects on vector-borne disease episystems in a changing world. J Exp Biol. (2010) 213:946–54. doi: 10.1242/jeb.037564
Keywords: ecological niche model, systematic review, spatial distribution, small ruminants, tick-borne, disease surveillance
Citation: Krasteva S, Jara M, Frias-De-Diego A and Machado G (2020) Nairobi Sheep Disease Virus: A Historical and Epidemiological Perspective. Front. Vet. Sci. 7:419. doi: 10.3389/fvets.2020.00419
Received: 22 April 2020; Accepted: 11 June 2020;
Published: 22 July 2020.
Edited by:
Ferran Jori, Centre de Coopération Internationale en Recherche Agronomique pour le Développement (CIRAD), FranceReviewed by:
Carolina Probst, Friedrich Loeffler Institute, GermanyMohamed Gharbi, National Veterinary School of Sidi Thabet, Tunisia
Copyright © 2020 Krasteva, Jara, Frias-De-Diego and Machado. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gustavo Machado, Z21hY2hhZEBuY3N1LmVkdQ==
 Stephanie Krasteva
Stephanie Krasteva Manuel Jara
Manuel Jara Alba Frias-De-Diego
Alba Frias-De-Diego Gustavo Machado
Gustavo Machado