- 1Department of Veterinary Pathology, Iowa State University, Ames, IA, United States
- 2Department of Veterinary Microbiology and Preventive Medicine, Iowa State University, Ames, IA, United States
Clostridioides difficile, previously Clostrdium difficile, is a major cause of antibiotic-associated enteric disease in humans in hospital settings. Increased incidence of C. difficile infection (CDI) in community settings raises concerns over an alternative source of CDI for humans. The detection of genetically similar and toxigenic C. difficile isolates in companion animals, including asymptomatic pets, suggests the potential role of household pets as a source of community-associated CDI. The close association between companion animals and humans, in addition to the use of similar antibiotics in both species, could provide a selective advantage for the emergence of new C. difficile strains and thus increase the incidental transmission of CDI to humans. Therefore, screening household pets for C. difficile is becoming increasingly important from a public health standpoint and may become a part of routine testing in the future, for the benefit of susceptible or infected individuals within a household. In this review, we analyze available information on prevalence, pathophysiology, epidemiology, and molecular genetics of C. difficile infection, focusing on companion animals and evaluate the risk of pet-borne transmission of CDI as an emerging public health concern. Molecular epidemiological characterization of companion animal C. difficile strains could provide further insights into the interspecies transmission of CDI. The mosaic nature of C. difficile genomes and their susceptibility to horizontal gene transfer may facilitate the inter-mixing of genetic material, which could increase the possibility of the emergence of new community-associated CDI strains. However, detailed genome-wide characterization and comparative genome analysis are warranted to confirm this hypothesis.
Introduction
Clostridioides difficile is an anaerobic spore-forming bacterium that causes a serious toxin-mediated enteric disease in humans (1). Annually, nearly half a million people in the United States suffer from C. difficile infection (CDI) (2), which incurs ~6.3 billion dollars of treatment and other hospital costs (3). Relapse of CDI usually occurs in ~20% of the individuals within a month after primary treatment (4, 5). Currently, there are no definitive treatment options available for CDI without the possibility of recurrence or relapse (5). A recent study indicated that 1 out of every 11 patients with CDI died within 30 days of diagnosis (2). C. difficile is classically considered a nosocomial pathogen and a major cause of antibiotic-associated diarrhea in hospitalized patients. However, an increase in the number and severity of CDI in humans has been reported outside the hospital environment or in individuals with onset of symptoms 48 h or less after hospital admission, referred to as community-associated infections (6). A paradigm shift has been observed in the CDI epidemiology in recent years and the incidence rate of community-acquired C. difficile infections is over 40% of the total CDI cases reported (2, 7). Moreover, newer reports indicate that the national burden of nosocomial C. difficile infection in the United States has decreased by 36%, whereas community-associated C. difficile infection burden has shown no change in trend (8). Notably, a definitive source of C. difficile in community settings has not been identified so far.
Clostridioides difficile has been isolated repeatedly from the intestinal flora of healthy domesticated animals, including pets, and associated with the sporadic incidence of diarrhea in susceptible animals (9–11). An increase in the isolation of C. difficile from food-animals and animal derived food has been attributed to the increased reports of community-associated human CDI (12). In the past decade, several investigators have isolated and characterized food-animal and meat strains of C. difficile. As an example, a common C. difficile strain isolated in pigs, ribotype (RT) 078, is also a ribotype commonly implicated in human community-associated C. difficile infection (13). However, other studies have questioned the potential foodborne transmission of C. difficile in humans specifically due to the lack of evidence of direct transmission and low prevalence of C. difficile in animal-derived foods (14–16). Therefore, the search for a potential source of C. difficile has recently been focused on companion animals (17). The general public is more intimately associated with pets than food animals, suggesting that C. difficile carriage in pets, especially dogs and cats, poses a relatively high public health risk to humans in household settings.
Reports from various parts of the world suggest household pets are carriers and sources of pathogenic C. difficile to humans. Studies conducted in past years reported an ~4–30 percent prevalence of C. difficile in dogs with several toxigenic isolates, where the toxigenic strains represented nearly 50% in some instances (17–20). Furthermore, C. difficile ribotype RT 106 has now surpassed the hospital-acquired C. difficile RT 027 in becoming the most common ribotype implicated in human CDI in the United States and has been frequently isolated from dogs and cats (19, 21–25). Therefore, screening household pets for C. difficile is becoming increasingly important from a public health point of view and could become routine in the future. In this review, we analyze available information on colonization, pathogenesis, and epidemiology of C. difficile in companion animals, particularly in pets, and examine the potential pet-borne transmission to humans as an emerging public health concern.
C. difficile Colonization in Dogs and Cats
Clostridial species are normal members of the intestinal flora in domestic animal species (26). Several studies indicate varying prevalence of C. difficile in healthy domestic animals with no enteric symptoms (27, 28). Alterations in the enteric microenvironment due to factors like antibiotic treatments, pancreatic exocrine dysfunction, changes in diet, trypsin inhibitors, poor intestinal motility or parasitic infections facilitate overgrowth of C. difficile (26, 29). The stress on the bacteria and overpopulation of the vegetative C. difficile cells triggers sporulation and synchronous secretion of potent exotoxins, toxin A (TcdA) and toxin B (TcdB) (26, 30). The toxins are endocytosed, cleaved, and release the glucosyltransferase domains into the cytosol which inactivate Rho GTPases (30, 31). Inactivation of Rho GTPases causes disruption of the cytoskeleton and intercellular tight junctions, simultaneously stimulating the intestinal epithelial and immune cells to secrete massive amounts of cytokines and chemokines (32, 33), resulting in neutrophilic inflammation and mucosal necrosis (26).
In adult dogs, colonization of toxigenic C. difficile in the gut is predominantly non-clinical and asymptomatic. For example, C. difficile toxins A, B, or combinations of both have been detected in feces of <20% of outpatient and in-patient healthy dogs as well as in-patient diarrheic dogs (27, 34). Conversely, ~90% of puppies had C. difficile isolated from their feces at least once during the first 10 weeks of life, of which more than half of the isolates were toxigenic (35, 36). Carriage of C. difficile in healthy puppies 3 months of age and older is observed to be much lower (35). The carriage rate of C. difficile in cats does not appear to differ from that of dogs (37) although systematic studies on C. difficile cat carriage are limited, in spite of litter boxes thought as a potential additional risk factor for C. difficile transmission within a household.
The pathogenesis and clinical features of CDI in companion animals appear to be strikingly different from that of human CDI. Gut dysbiosis is not a significant feature of CDI in dogs (26, 38), despite being a major factor in the pathogenesis of CDI in humans. Clinical signs such as acute hemorrhagic diarrhea in C. difficile infected dogs do not significantly correlate with the presence of C. difficile in the gut (27, 39). In addition, in the dysbiotic state, dogs tend to show symptoms associated with overgrowth of other cohabitating intestinal bacteria instead of a C. difficile toxin-mediated pathology (40). One case report indicates that cats may present with acute clinical signs of vomiting from CDI (41). Other reported clinical manifestations in cats included gas distension of the small intestines and necrotizing hemorrhagic enterotyphlocolitis (41).
Lack of concrete correlation between gut-dysbiosis and CDI in dogs provides insight into the asymptomatic carriage of C. difficile and plausible resistance to clinical CDI in pets. Additionally, the absence of dysbiosis suggests other potential causes or predisposing factors for CDI. Comparative microbiome analysis revealed a marked increase in the abundance of Fusobacteria, Proteobacteria, and Firmicutes, and a decrease in Verrucomicrobia, Bacteroidetes, Euryarchaeota, and Actinobacteria in C. difficile-carrying dogs, whereas, in humans infected with C. difficile, decreases in the abundance of Firmicutes, Actinobacteria, and Euryarchaeota were reported (38). Therefore, the abundance of Firmicutes could be a significant factor potentially associated with a lack of clinical symptoms in C. difficile positive dogs with dysbiosis (38). Notably, Clostridial and Eubacteria species, part of the Firmicutes phylum, possess the ability to convert primary bile acids into secondary bile acids predominantly by 7α-dehydroxylation (42). In humans, 7α-dehydroxylating bacteria increases the level of secondary bile acids, generating an intestinal bile acid profile that is associated with CDI resistance (42). Therefore, such connections should be further explored in dogs and other household pets.
Diet and gut-microbiome play a crucial role in defining the intestinal bile acid profile, thereby directly or indirectly influencing C. difficile colonization and infection in the host gut. In fact, distinct Clostridial species such as Clostridium hiranonis, with demonstrated 7α-dehydroxylating ability, were isolated from the intestines of dogs (38). Clostridial scindens appears to have a beneficial role in mouse models as its abundance correlates with CDI resistance (42, 43). In pet dogs, increases in relative abundance of C. hiranonis have been observed in the gut microbiota of the dogs fed high-intake boiled minced beef compared to dogs fed commercial dry diet (44). This change in microbiome correlated with high levels of secondary bile acids such as deoxycholic acid and ursodeoxycholic acid in the gut (44). Experimentally, C. scindens has previously shown resistance against CDI in an intestinal ex-vivo model when 7α-dehydroxylation is reconstituted to normalize bile acid composition (43). Collectively, these observations suggest a contributory role of commercial pet diet in gut-colonization of C. difficile in dogs. Specifically, dietary changes that promote the growth of 7α-dehydroxylating bacteria in the gut may reduce C. difficile carriage in pets, and thus mitigate potential zoonotic transmission of CDI. A few studies have identified the presence of C. difficile, occasionally toxigenic strains, in raw pet foods, suggesting an increased risk of C. difficile colonization in dogs and cats fed with such diets (45–47). Therefore, further investigations are required to evaluate and address the impact of contaminated pet foods on gut colonization of C. difficile (45).
Although clinical CDI is not well-defined in dogs, antibiotics have been used as a treatment option for enteric clostridial infections in dogs (48). Theoretically, the use of antibiotics against CDI or other disease conditions may cause the emergence of antibiotic-resistant strains of C. difficile within the canine gastrointestinal tract, which could be an added threat in terms of zoonotic transmission of CDI. Although the role of gut-dysbiosis has been described differently in pet CDI pathogenesis, treatments to alleviate dysbiosis have gained favor in efforts to prevent symptoms in pets and humans (49, 50). Since transmission of antibiotic-resistant C. difficile from companion animals appears to be a legitimate concern, antibiotic use in household pets should be revisited to prevent the emergence of antibiotic-resistant C. difficile strains in community settings.
Prevalence and Major Subtypes of C. difficile in Companion Animals
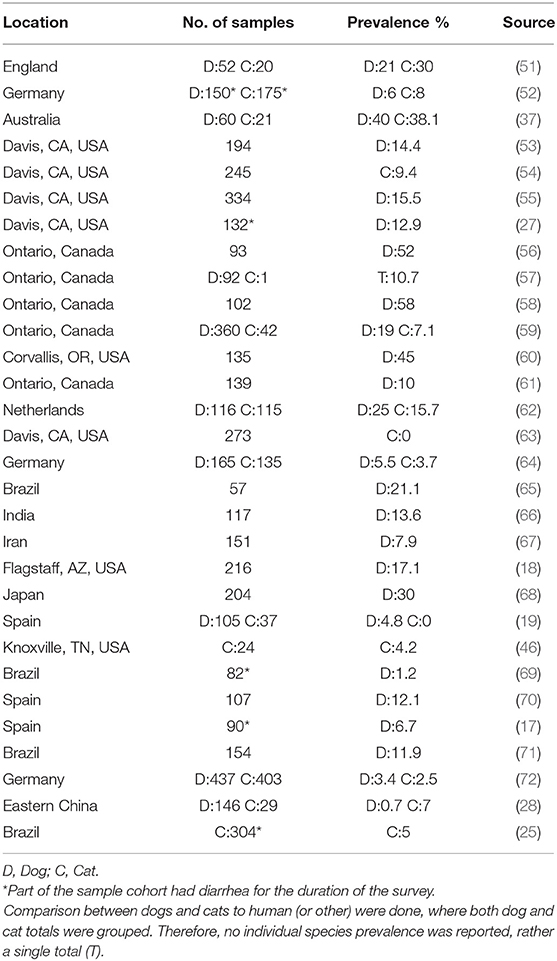
The role of companion animals as a source for human CDI is an emerging public health concern. The lack of association between C. difficile colonization and clinical disease in pets allows for them to be ideal silent reservoirs of toxigenic C. difficile strains. Therefore, prevalence studies on C. difficile carriage rates in household pets are gaining more attention in the public health and medical community. Various studies have isolated toxigenic C. difficile strains at varying prevalence rates in dog and cat feces around the world (Table 1).
C. difficile strains are generally further classified based on the size variation in the 16s and 23s rRNA intergenic spacer region (Ribotype/RT). Most common human C. difficile isolates are RTs 106, 027, 078, 014, 002, and 020 (8, 13, 73–75). Of these, RTs 027 and 078 are generally referred to as hypervirulent strains and are associated with increased toxin production and outbreaks of severe CDI, and carry specific genomic characteristics (76, 77). Specifically, RT 027 is commonly associated with severe human CDI, predominantly in hospital settings (73, 78). This hypervirulent strain emerged and established a significant health problem in the last decade (73). Canadian, Spanish, and German studies identified CDI RTs 027, 078, and 014/0, all known causes of severe humans disease, in dogs (70, 72, 79). Human RT 106, becomes especially important due to its increasing prevalence and noted association with community-associated CDI in the United States and Europe (23, 24). RT 106 is also commonly isolated from dogs and cats (21, 25). Other ribotypes commonly isolated from dogs and cats worldwide include RT 039 in cats; RT 012 in dogs; and RTs 009, 010, and 014/20 overlapping between the two species (62, 64, 75, 80–82). C. difficile isolates from pets are often reported to be resistant to multiple antibiotics, including metronidazole (20, 23, 24, 70, 75, 82, 83). This poses a concern as a metrinidazole antibiotic-resistance adaptation can result in a recurrent CDI (rCDI), as observed in one human case (83). Ribotyping enables clinicians and researchers to quickly identify and predict potentially pathogenic strains of C. difficile that are isolated from clinical or environmental samples. However, C. difficile ribotyping may not be as sensitive as other methods of classification from an evolutionary or phylogenetic point of view, which will be discussed in later sections of this review.
Prevalence of C. difficile in Other Companion Animal Species
The ubiquitous nature of C. difficile spores and their ability to stay in the environment for an extended period render several additional species of animals vulnerable to gut colonization and CDI via the feco-oral route. The organism has been isolated from healthy horses and exotic pets, with some strains more prevalent than others (11, 17, 21, 62, 84–87). Prevalence studies conducted in the Netherlands, Europe, and the Czech Republic demonstrated the presence of toxigenic and non-toxigenic strains of C. difficile in the horse gastrointestinal tract (62, 86, 87). A wide range of prevalence rates and diversity in C. difficile strains have been reported by these investigators. RTs 014 and 078 attracted special attention because they are also associated with human CDI outbreaks (62). Furthermore, multiple antibiotic resistance genes were found to be shared among both human and equine C. difficile isolates (87). As such, the genotypic similarities and overlap between human and equine CDI subtypes raise speculations on the possibility of interspecies transmission or adaptation of different toxigenic C. difficile strains (21, 86, 88).
Due to the limited number of studies conducted in exotic pets, information on toxigenic C. difficile in psittacine birds and small mammals (rabbits, ferrets, and rodents) is sparse (17). Recently, a novel non-toxigenic C. difficile ribotype was isolated from a pet reptile, indicating that exotic pets could carry uncommon C. difficile strains (17). Therefore, further studies are warranted to determine C. difficile prevalence and their zoonotic potential in less common household pets, including reptiles.
Implications of Human-Pet Interactions in CDI Transmission
As asymptomatic carriers, household pets could potentially transmit pathogenic C. difficile strains to susceptible individuals such as the elderly and children, and could further disseminate CDI within a community (51, 89). A British research group investigated C. difficile colonization in infants and observed that a significant proportion of them (30–40%) were colonized with C. difficile, out of which 68% of the isolates were confirmed toxigenic (90). The results from this study pointed out a significant association between the colonization rate and presence of dogs in the household (90). A Canadian study revealed a 26% asymptomatic carriage rate in dogs that are in contact with individuals with CDI in households (91).
In 2006, a pathogenic human strain of C. difficile was identified in a dog that visited patients in a health care facility. Molecular characterization of the C. difficile isolate revealed that this service dog acquired the pathogen most likely from the health care facilities it visited (92). Therefore, an infected human can be considered as a route of initial C. difficile colonization in a susceptible pet. Studies have also demonstrated C. difficile colonization in dogs that participated in animal-assisted care programs in health care settings. Lefebvre et al. (93) observed that dogs visiting the health care facilities had a 2.4 times higher risk of acquiring C. difficile than those involved in other animal-assisted programs. In another study, dogs that had direct human contact, such as licking the patients or receiving treats were found to be at a greater risk of acquiring C. difficile (94). These interactions suggest that CDI may be perpetuated within the community. In a more recent study, spores of toxigenic C. difficile were identified in the nasal secretion of pet dogs adding to the risk of direct transmission of this bacteria to humans in close contact (95). A study conducted in Spain identified toxigenic C. difficile isolates in playground sandboxes that are unprotected from dogs, posing an additional public health risk to a vulnerable young population (96). Additionally, mechanical spread of C. difficile from houses to the community through shoe soles and dog paws have been reported (97).
Recurrence of CDI usually occurs in ~20% of individuals within a month after primary treatment (98). However, a definitive cause of rCDI and a radical method for preventing this recurrence remains unknown. rCDI can be a result of relapse with the same strain or infection with another C. difficile strain (99). Thus, C. difficile transmission between pets and susceptible humans should be considered as one of the possible mechanisms of reinfection in rCDI. As an example, RT 106, commonly found in dogs and cats, has shown to cause a higher recurrence rate in humans as opposed to more virulent strains (24). A possible explanation for this phenomenon could be the reported higher sporulation rate of RT 106, which can increase the chance of reinfection from contaminated surfaces or the retention of spores in the gut (100). However, a higher recurrence rate of this ribotype can also be potentially attributed to the presence of silent carriers of infection, e.g., pets in the household which can harbor, shed, and transmit RT 106 to the patient.
Isolation and molecular typing of C. difficile from rCDI patients are crucial in determining the potential origin of rCDI strains but such data are scanty in the literature. A limited investigation conducted in Minnesota, United States, identified C. difficile-positive humans in homes with pets, where the owner had experienced a previous episode of CDI (101). It was unclear whether the human C. difficile colonization resulted from the previous human CDI or exclusively transmitted from pet and household surfaces. Additionally, the number of households with pets in this study was too small to further examine pets as a valid source (101). As such, owners should be advised to take extra precautions when clostridial diarrhea in their pets, especially in consideration of CDI recurrence.
Although the interspecies transmission of C. difficile between dogs and humans appears to be a legitimate concern, there is a contrasting but beneficial aspect of human-pet interaction for those patients suffering from CDI. Studies have demonstrated that dogs can be trained to detect C. difficile infection at the initial stage of clinical disease and in patients experiencing non-specific symptoms (102–104). A few small scale studies even report a potential protective effect of pet ownership in rCDI (105). However, precautions must still be taken to minimize the risk of further spread of CDI outside of health care facilities through human-pet interactions until the most accurate association is elucidated.
Molecular Epidemiology, Phylogeny, and Potential Interhost Adaptation of Pet C. difficile
Detailed comparative genome-wide characterization of pet C. difficile isolates is required to determine transmission between pets and humans within a household or in a wider environment. Sequence-based genotyping techniques such as Multiple-Locus Variable number tandem repeat Analysis (MLVA), Multilocus Sequence Typing (MLST), Core-genome MLST, or whole-genome Single Nucleotide Polymorphisms (SNP) are based on the changes that occur in conserved parts of the C. difficile genome, which adapts minimally in the course of evolution. Specifically, methods such as maximum likelihood estimations help calculate the length of a branch in a phylogenetic tree and predict the probable evolutionary rates (106). Maximum likelihood analysis conducted on a large database, pubMLST, groups C. difficile isolates diversity into five major distinct clades: clade 1–5 (107). There are three additional cryptic clades, C-I to C-III, which comprise of strains not included in the five major clades (108, 109). Clades are further subcategorized into multiple multilocus Sequence Types (ST) of C. difficile within which different RTs are grouped. Clade 1 has the most diverse STs among all clades, comprised of the most frequent pet associated non-hypervirulent STs. Clade 2 is composed of STs 1, 32, and 67. ST 1 includes the human hypervirulent strain RT 027. A notable member of clade 5 is ST 11, under which the emerging human hypervirulent strain RT 078 is grouped. This RT is widely isolated from food animal species (110). MLST analysis conducted on dog strains isolated in Arizona, United States, demonstrated that several sequence types belong to clade 1 (18). Among these STs, there was a higher frequency of STs 2, 3, 42, and 15. The former three are also observed in equivalent levels in humans (18). Although RTs 027 and 078 are rarely isolated from pets, more general sequence types appear to be shared between dogs and humans, which suggests possible sharing of virulent C. difficile strains.
Although MLVA, MLST, and SNP genotyping techniques are ideal in establishing genetic distance and relatedness, they are less useful in providing information on the unique qualities of individual isolates, such as antibiotic resistance genes, pathogenicity loci, transposons, and mobile elements. Therefore, it is important to study the hypervariable regions of the C. difficile genome from pets, where the acquisition and loss of genetic material can occur, particularly that which may facilitate the rapid adaptation of bacteria in a new environment or host. Such genome-wide characterization can provide this information and other unique features of a given C. difficile isolate and help fill the current large knowledge gap.
Identification of human-specific and pet-specific genes could be used as markers of intermixing of C. difficile genetic material to understand host-specific elements that could potentially alter the virulence capacity of C. difficile STs in pets. In 2009, Stabler et al. conducted a study to understand the mechanism of the emergence of human epidemic and hypervirulent C. difficile RT 027 strain. The authors compared the genome of hypervirulent RT 027 to a non-epidemic RT 027 (CD196) identified in very isolated incidents, and C. difficile RT 012 (CD630; the reference genome). The comparative genomic analysis identified a number of recently acquired genetic elements encoding a unique phage island, two-component regulatory systems, and transcription regulators exclusive to the epidemic “hypervirulent” RT 027 strain and the possible cause of its emergence (111). Such an analysis in pet C. difficile, in combination with that of their respective owners, could help predict the possible emergence of C. difficile strains of public health concern.
Understanding genome-wide changes is essential for identifying host-specific adaptation in C. difficile. Within the conserved (core) genome, toxigenic C. difficile encodes for a 19.6-kb Pathogenicity Locus (PaLoc), which constitutes toxins genes (tcdA and tcdB), regulatory genes (tcdC, tcdR), and a holin-like gene (tcdE) responsible for toxin secretion. In contrast, non-toxigenic strains do not exhibit this length of sequence anywhere in their genome (112). Interestingly, non-toxigenic C. difficile strains have acquired toxin production by horizontal gene transfer of the PaLoc (113). Furthermore, a closely related pathogen, C. perfringens, was also found to gain virulence by way of horizontal gene transfer in the gut environment (114). This phenomenon points out the possibility of an alternate mechanism for the emergence of zoonotic C. difficile strains resulting in the intermixing of pet and human C. difficile strains. Furthermore, polymorphisms and deletions exist within the PaLoc that may affect the levels, types, and variants of one or both toxins (115, 116). As the PaLoc is indispensable in CDI pathogenesis, understanding the changes within the PaLoc region of pet and human C. difficile isolates can be useful for predicting the emergence of a hypervirulent and highly toxigenic C. difficile strains.
Conclusion
Clostridioides difficile infection is becoming a significant public health concern as the disease severity, and the proportion of individuals infected in community settings is steadily increasing. Studies from various parts of the world suggest household pets as carriers and potential sources for pathogenic C. difficile to humans. Detection of similar C. difficile isolates from companion animals and humans suggest potential pet-borne transmission of community-associated CDI. However, large scale prevalence studies among pet and owner pairs, with whole-genome characterization of pet and human C. difficile isolates, are necessary to understand host-specific genomic elements, mobile genetic elements, antibiotic resistance genes, and inter- and intra- sequence type variations. Such studies are necessary to predict an already occurring or impending emergence of zoonotic C. difficile strains. Unfortunately, most of the available studies in the literature are conducted on a small scale with limited investigations on genomic details of pet C. difficile isolates. Additionally, systematic studies on C. difficile carriage in cats are limited, even with the potential risks posed by cat litter boxes. Similarly, systematic studies on C. difficile carriage in owner-pet pairs in a household are limited. Therefore, further studies, routine health screening of companion animals and owners for C. difficile carriage, and genomic characterization of pet C. difficile isolates are warranted to address this knowledge gap.
Author Contributions
SM conceptualized the idea. SM and CT designed the project outline. BH, CT, and AV conducted the literature search and analysis. CT, BH, SM, and AV wrote the manuscript. BS reviewed and edited the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Bartlett JG, Chang TW, Gurwith M, Gorbach SL, Onderdonk AB. Antibiotic-associated pseudomembranous colitis due to toxin-producing clostridia. N Engl J Med. (1978) 298:531–4. doi: 10.1056/NEJM197803092981003
2. Lessa FC, Mu Y, Bamberg WM, Beldavs ZG, Dumyati GK, Dunn JR, et al. Burden of Clostridium difficile infection in the United States. N Engl J Med. (2015) 372:825–34. doi: 10.1056/NEJMoa1408913
3. Zhang S, Palazuelos-Munoz S, Balsells EM, Nair H, Chit A, Kyaw MH. Cost of hospital management of Clostridium difficile infection in United States-a meta-analysis and modelling study. BMC Infect Dis. (2016) 16:447–447. doi: 10.1186/s12879-016-1786-6
4. Surawicz CM, McFarland LV, Greenberg RN, Rubin M, Fekety R, Mulligan ME, et al. The search for a better treatment for recurrent clostridium difficile disease: use of high-dose vancomycin combined with saccharomyces boulardii. Clin Infect Dis. (2000) 31:1012–7. doi: 10.1086/318130
5. Dinleyici M, Vandenplas Y. Clostridium difficile colitis prevention and treatment. In: Guandalini S, Indrio F, editors. Probiotics and Child Gastrointestinal Health: Advances in Microbiology, Infectious Diseases and Public Health. Vol. 10. Cham: Springer International Publishing (2019). p. 139–46. doi: 10.1007/5584_2018_322
6. McDonald LC, Coignard B, Dubberke E, Song X, Horan T, Kutty PK. Recommendations for surveillance of Clostridium difficile-associated disease. Infect Control Hosp Epidemiol. (2007) 28:140–5. doi: 10.1086/511798
7. Khanna S, Pardi DS, Aronson SL, Kammer PP, Orenstein R, St Sauver JL, et al. The epidemiology of community-acquired Clostridium difficile infection: a population-based study. Am J Gastroenterol. (2012) 107:89–95. doi: 10.1038/ajg.2011.398
8. Guh AY, Mu Y, Winston LG, Johnston H, Olson D, Farley MM, et al. Trends in U.S. Burden of clostridioides difficile infection and outcomes. N Engl J Med. (2020) 382:1320–30. doi: 10.1056/NEJMoa1910215
9. Songer JG, Uzal FA. Clostridial enteric infections in pigs. J Vet Diagn Invest. (2005) 17:528. doi: 10.1177/104063870501700602
10. Weese JS. Bacterial enteritis in dogs and cats: diagnosis, therapy, and zoonotic potential. Vet Clin North Am Small Anim Pract. (2011) 41:287–309. doi: 10.1016/j.cvsm.2010.12.005
11. Diab SS, Songer G, Uzal FA. Clostridium difficile infection in horses: a review. Vet Microbiol. (2013) 167:42–9. doi: 10.1016/j.vetmic.2013.03.032
12. Rodriguez-Palacios A, Borgmann S, Kline TR, LeJeune JT. Clostridium difficile in foods and animals: history and measures to reduce exposure. Anim Health Res Rev. (2013) 14:11. doi: 10.1017/S1466252312000229
13. Debast SB, van Leengoed LA, Goorhuis A, Harmanus C, Kuijper EJ, Bergwerff AA. Clostridium difficile PCR ribotype 078 toxinotype V found in diarrhoeal pigs identical to isolates from affected humans. Environ Microbiol. (2009) 11:505–11. doi: 10.1111/j.1462-2920.2008.01790.x
14. Limbago B, Thompson AD, Greene SA, MacCannell D, MacGowan CE, Jolbitado B, et al. Development of a consensus method for culture of Clostridium difficile from meat and its use in a survey of U.S. retail meats. Food Microbiol. (2012) 32:448–51. doi: 10.1016/j.fm.2012.08.005
15. Mooyottu S, Flock G, Kollanoor-Johny A, Upadhyaya I, Jayarao B, Venkitanarayanan K. Characterization of a multidrug resistant C. difficile meat isolate Int J Food Microbiol. (2015) 192:111. doi: 10.1016/j.ijfoodmicro.2014.10.002
16. Shaughnessy MK, Snider T, Sepulveda R, Boxrud D, Cebelinski E, Jawahir S, et al. Prevalence and molecular characteristics of clostridium difficile in retail meats, food-producing and companion animals, and humans in minnesota. J Food Prot. (2018) 81:1635–42. doi: 10.4315/0362-028X.JFP-18-104
17. Andres-Lasheras S, Martin-Burriel I, Mainar-Jaime RC, Morales M, Kuijper E, Blanco JL, et al. Preliminary studies on isolates of Clostridium difficile from dogs and exotic pets. BMC Vet Res. (2018) 14:77. doi: 10.1186/s12917-018-1402-7
18. Stone NE, Sidak-Loftis LC, Sahl JW, Vazquez AJ, Wiggins KB, Gillece JD, et al. More than 50% of Clostridium difficile isolates from pet dogs in Flagstaff, USA, carry toxigenic genotypes. PLoS ONE. (2016) 11:e0164504. doi: 10.1371/journal.pone.0164504
19. Álvarez-Pérez S, Blanco JL, Harmanus C, Kuijper EJ, García ME. Prevalence and characteristics of Clostridium perfringens and Clostridium difficile in dogs and cats attended in diverse veterinary clinics from the Madrid region. Anaerobe. (2017) 48:47–55. doi: 10.1016/j.anaerobe.2017.06.023
20. Álvarez-Pérez S, Blanco JL, Peláez T, Lanzarot MP, Harmanus C, Kuijper E, et al. Faecal shedding of antimicrobial-resistant Clostridium difficile strains by dogs. J Small Anim Pract. (2015) 56:190–5. doi: 10.1111/jsap.12311
21. Silva ROS, Rupnik M, Diniz AN, Vilela EG, Lobato FCF. Clostridium difficile ribotypes in humans and animals in Brazil. Mem Inst Oswaldo Cruz. (2015) 110:1062–5. doi: 10.1590/0074-02760150294
22. Rainha K, Fernandes Ferreira R, Trindade CNR, Carneiro LG, Penna B, Endres BT, et al. Characterization of Clostridioides difficile ribotypes in domestic dogs in Rio de Janeiro, Brazil. Anaerobe. (2019) 58:22–9. doi: 10.1016/j.anaerobe.2019.06.007
23. Suarez-Bode L, Barron R, Perez JL, Mena A. Increasing prevalence of the epidemic ribotype 106 in healthcare facility-associated and community-associated Clostridioides difficile infection. Anaerobe. (2019) 55:124–9. doi: 10.1016/j.anaerobe.2018.12.002
24. Carlson TJ, Blasingame D, Gonzales-Luna AJ, Alnezary F, Garey KW. Clostridioides difficile ribotype 106: a systematic review of the antimicrobial susceptibility, genetics, and clinical outcomes of this common worldwide strain. Anaerobe. (2020) 62:102142. doi: 10.1016/j.anaerobe.2019.102142
25. Silva ROS, Ribeiro MG, de Paula CL, Pires IH, Oliveira Junior CA, Diniz AN, et al. Isolation of Clostridium perfringens and Clostridioides difficile in diarrheic and nondiarrheic cats. Anaerobe. (2020) 62:102164. doi: 10.1016/j.anaerobe.2020.102164
26. Uzal FA, Plattner BL, Hostetter JM. Alimentary system: diseases associated with enteric clostridial infections. In: Jubb K, Kennedy P, Palmer N, editors. Pathology of Domestic Animals. Vol. 2. St. Louis: Elsevier (2016). p. 183–94. doi: 10.1016/B978-0-7020-5318-4.00007-3
27. Marks SL, Kather EJ, Kass PH, Melli AC. Genotypic and phenotypic characterization of Clostridium perfringens and Clostridium difficile in diarrheic and healthy dogs. J Vet Intern Med. (2002) 16:533–40. doi: 10.1111/j.1939-1676.2002.tb02383.x
28. Wei Y, Sun M, Zhang Y, Gao J, Kong F, Liu D, et al. Prevalence, genotype and antimicrobial resistance of Clostridium difficile isolates from healthy pets in Eastern China. BMC Infect Dis. (2019) 19:46. doi: 10.1186/s12879-019-3678-z
29. Martirossian G, Sokół-Leszczyńska B, Mierzejewski J, Meisel-Mikołajczyk F. [Occurrence of Clostridium difficile in the digestive system of dogs]. Med Dosw Mikrobiol. (1992) 44:49–54.
30. Voth DE, Ballard JD. Clostridium difficile toxins: mechanism of action and role in disease. Clin Microbiol Rev. (2005) 18:247–63. doi: 10.1128/CMR.18.2.247-263.2005
31. Just I, Wilm M, Selzer J, Rex G, von Eichel-Streiber C, Mann M, et al. The enterotoxin from Clostridium difficile (ToxA) monoglucosylates the Rho proteins. J Biol Chem. (1995) 270:13932–6. doi: 10.1074/jbc.270.23.13932
32. Kelly CP, Pothoulakis C, LaMont JT. Clostridium difficile colitis. N Engl J Med. (1994) 330:257–62. doi: 10.1056/NEJM199401273300406
33. Solomon K. The host immune response to Clostridium difficile infection. Therap Adv Infect Dis. (2013) 1:19–35. doi: 10.1177/2049936112472173
34. Weese JS, Staempfli HR, Prescott JF, Kruth SA, Greenwood SJ, Weese HE. The roles of Clostridium difficile and enterotoxigenic Clostridium perfringens in diarrhea in dogs. J Vet Intern Med. (2001) 15:374–8. doi: 10.1111/j.1939-1676.2001.tb02332.x
35. Perrin J, Buogo C, Gallusser A, Burnens AP, Nicolet J. Intestinal carriage of Clostridium difficile in neonate dogs. Zentralblatt Veterinarmedizin Reihe B. (1993) 40:222–6. doi: 10.1111/j.1439-0450.1993.tb00131.x
36. Buogo C, Burnens AP, Perrin J, Nicolet J. [Presence of Campylobacter spp., Clostridium difficile, C. perfringens and salmonellae in litters of puppies and in adult dogs in a shelter]. Schweiz Arch Tierheilkd. (1995) 137:165–71.
37. Riley TV, Adams JE, O'Neill GL, Bowman RA. Gastrointestinal carriage of Clostridium difficile in cats and dogs attending veterinary clinics. Epidemiol Infect. (1991) 107:659–65. doi: 10.1017/S0950268800049359
38. Stone NE, Nunnally AE, Jimenez V Jr, Cope EK, Sahl JW, Sheridan K, et al. Domestic canines do not display evidence of gut microbial dysbiosis in the presence of Clostridioides (Clostridium) difficile, despite cellular susceptibility to its toxins. Anaerobe. (2019) 58:53–72. doi: 10.1016/j.anaerobe.2019.03.017
39. Allenspach K. Bacteria involved in acute haemorrhagic diarrhoea syndrome in dogs. Vet Rec. (2015) 176:251–2. doi: 10.1136/vr.h986
40. Suchodolski JS, Markel ME, Garcia-Mazcorro JF, Unterer S, Heilmann RM, Dowd SE, et al. The fecal microbiome in dogs with acute diarrhea and idiopathic inflammatory bowel disease. PLoS ONE. (2012) 7:e51907. doi: 10.1371/journal.pone.0051907
41. Walczak R, Paek M, Suran J, Amory JT, Specchi S, Sanchez M. Radiography and ultrasonography of pneumatosis intestinalis in a cat. Vet Radiol Ultrasound. (2018) 61:E26–30. doi: 10.1111/vru.12635
42. Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature. (2015) 517:205–8. doi: 10.1038/nature13828
43. Studer N, Desharnais L, Beutler M, Brugiroux S, Terrazos MA, Menin L, et al. Functional intestinal Bile Acid 7alpha-Dehydroxylation by Clostridium scindens associated with Protection from Clostridium difficile Infection in a gnotobiotic mouse model. Front Cell Infect Microbiol. (2016) 6:191. doi: 10.3389/fcimb.2016.00191
44. Herstad KMV, Ronning HT, Bakke AM, Moe L, Skancke E. Changes in the faecal bile acid profile in dogs fed dry food vs high content of beef: a pilot study. Acta Vet Scand. (2018) 60:29. doi: 10.1186/s13028-018-0383-7
45. Weese JS, Rousseau J, Arroyo L. Bacteriological evaluation of commercial canine and feline raw diets. Can Vet J. (2005) 46:513–6.
46. Hamper BA, Bartges JW, Kirk CA. Evaluation of two raw diets vs a commercial cooked diet on feline growth. J Feline Med Surg. (2017) 19:424–34. doi: 10.1177/1098612X16634388
47. Morelli G, Catellani P, Miotti Scapin R, Bastianello S, Conficoni D, Contiero B, et al. Evaluation of microbial contamination and effects of storage in raw meat-based dog foods purchased online. J Anim Physiol Anim Nutr. (2020) 104:690–7. doi: 10.1111/jpn.13263
48. Silva ROS, de Oliveira Júnior CA, Blanc DS, Pereira ST, de Araujo MCR, Vasconcelos A, et al. Clostridioides difficile infection in dogs with chronic-recurring diarrhea responsive to dietary changes. Anaerobe. (2018) 51:50–3. doi: 10.1016/j.anaerobe.2018.03.011
49. Kaleko M, Bristol JA, Hubert S, Parsley T, Widmer G, Tzipori S, et al. Development of SYN-004, an oral beta-lactamase treatment to protect the gut microbiome from antibiotic-mediated damage and prevent Clostridium difficile infection. Anaerobe. (2016) 41:58–67. doi: 10.1016/j.anaerobe.2016.05.015
50. Kokai-Kun JF, Bristol JA, Setser J, Schlosser M. Nonclinical safety assessment of SYN-004: an Oral β-lactamase for the protection of the gut microbiome from disruption by biliary-excreted, intravenously administered antibiotics. Int J Toxicol. (2016) 35:309–16. doi: 10.1177/1091581815623236
51. Borriello SP, Honour P, Turner T, Barclay F. Household pets as a potential reservoir for Clostridium difficile infection. J Clin Pathol. (1983) 36:84–7. doi: 10.1136/jcp.36.1.84
52. Weber A, Kroth P, Heil G. [The occurrence of Clostridium difficile in fecal samples of dogs and cats]. Zentralblatt Veterinarmedizin Reihe B. (1989) 36:568–76. doi: 10.1111/j.1439-0450.1989.tb00647.x
53. Struble AL, Tang YJ, Kass PH, Gumerlock PH, Madewell BR, Silva JJr. Fecal shedding of Clostridium difficile in dogs: a period prevalence survey in a veterinary medical teaching hospital. J Vet Diagn Invest. (1994) 6:342–7. doi: 10.1177/104063879400600310
54. Madewell BR, Bea JK, Kraegel SA, Winthrop M, Tang YJ, Silva JJr. Clostridium difficile: a survey of fecal carriage in cats in a veterinary medical teaching hospital. J Vet Diagn Invest. (1999) 11:50–4. doi: 10.1177/104063879901100108
55. Cave NJ, Marks SL, Kass PH, Melli AC, Brophy MA. Evaluation of a routine diagnostic fecal panel for dogs with diarrhea. J Am Vet Med Assoc. (2002) 221:52–9. doi: 10.2460/javma.2002.221.52
56. Weese JS, Armstrong J. Outbreak of Clostridium difficile-associated disease in a small animal veterinary teaching hospital. J Vet Intern Med. (2003) 17:813–6. doi: 10.1111/j.1939-1676.2003.tb02519.x
57. Arroyo LG, Kruth SA, Willey BM, Staempfli HR, Low DE, Weese JS. PCR ribotyping of Clostridium difficile isolates originating from human and animal sources. J Med Microbiol. (2005) 54:163–6. doi: 10.1099/jmm.0.45805-0
58. Lefebvre SL, Waltner-Toews D, Peregrine AS, Reid-Smith R, Hodge L, Arroyo LG, et al. Prevalence of zoonotic agents in dogs visiting hospitalized people in Ontario: implications for infection control. J Hosp Infect. (2006) 62:458–66. doi: 10.1016/j.jhin.2005.09.025
59. Clooten J, Kruth S, Arroyo L, Weese JS. Prevalence and risk factors for Clostridium difficile colonization in dogs and cats hospitalized in an intensive care unit. Vet Microbiol. (2008) 129:209–14. doi: 10.1016/j.vetmic.2007.11.013
60. McKenzie E, Riehl J, Banse H, Kass PH, Nelson S, Marks SL. Prevalence of diarrhea and enteropathogens in racing sled dogs. J Vet Intern Med. (2010) 24:97–103. doi: 10.1111/j.1939-1676.2009.0418.x
61. Weese JS, Finley R, Reid-Smith RR, Janecko N, Rousseau J. Evaluation of Clostridium difficile in dogs and the household environment. Epidemiol Infect. (2010) 138:1100–4. doi: 10.1017/S0950268809991312
62. Koene MG, Mevius D, Wagenaar JA, Harmanus C, Hensgens MP, Meetsma AM, et al. Clostridium difficile in Dutch animals: their presence, characteristics and similarities with human isolates. Clin Microbiol Infect. (2012) 18:778–84. doi: 10.1111/j.1469-0691.2011.03651.x
63. Queen EV, Marks SL, Farver TB. Prevalence of selected bacterial and parasitic agents in feces from diarrheic and healthy control cats from Northern California. J Vet Intern Med. (2012) 26:54–60. doi: 10.1111/j.1939-1676.2011.00843.x
64. Schneeberg A, Rupnik M, Neubauer H, Seyboldt C. Prevalence and distribution of Clostridium difficile PCR ribotypes in cats and dogs from animal shelters in Thuringia, Germany. Anaerobe. (2012) 18:484–8. doi: 10.1016/j.anaerobe.2012.08.002
65. Silva RO, Santos RL, Pires PS, Pereira LC, Pereira ST, Duarte MC, et al. Detection of toxins A/B and isolation of Clostridium difficile and Clostridium perfringens from dogs in Minas Gerais, Brazil. Braz J Microbiol. (2013) 44:133–7. doi: 10.1590/S1517-83822013005000008
66. Hussain I, Sharma RK, Borah P, Rajkhowa S, Barkalita LM, Hasin D, et al. Isolation and characterization of Clostridium difficile from pet dogs in Assam, India. Anaerobe. (2015) 36:9–13. doi: 10.1016/j.anaerobe.2015.09.006
67. Ghavidel M, Salari Sedigh H, Razmyar J. Isolation of Clostridium difficile and molecular detection of binary and A/B toxins in faeces of dogs. Iran J Vet Res. (2016) 17:273–6.
68. Usui M, Suzuki K, Oka K, Miyamoto K, Takahashi M, Inamatsu T, et al. Distribution and characterization of Clostridium difficile isolated from dogs in Japan. Anaerobe. (2016) 37:58–61. doi: 10.1016/j.anaerobe.2015.10.002
69. Silva ROS, Dorella FA, Figueiredo HCP, Costa EA, Pelicia V, Ribeiro BLD, et al. Clostridium perfringens and C. difficile in parvovirus-positive dogs Anaerobe. (2017) 48:66–9. doi: 10.1016/j.anaerobe.2017.07.001
70. Orden C, Blanco JL, Álvarez-Pérez S, Garcia-Sancho M, Rodriguez-Franco F, Sainz A, et al. Isolation of Clostridium difficile from dogs with digestive disorders, including stable metronidazole-resistant strains. Anaerobe. (2017) 43:78–81. doi: 10.1016/j.anaerobe.2016.12.008
71. Diniz AN, Coura FM, Rupnik M, Adams V, Stent TL, Rood JI, et al. The incidence of Clostridioides difficile. and Clostridium perfringens netF-positive strains in diarrheic dogs. Anaerobe. (2018) 49:58–62. doi: 10.1016/j.anaerobe.2017.12.003
72. Rabold D, Espelage W, Abu Sin M, Eckmanns T, Schneeberg A, Neubauer H, et al. The zoonotic potential of Clostridium difficile from small companion animals and their owners. PLoS ONE. (2018) 13:e0193411. doi: 10.1371/journal.pone.0193411
73. Kuijper EJ, Coignard B, Tull P. Emergence of Clostridium difficile-associated disease in North America and Europe. Clin Microbiol Infect. (2006) 12(Suppl. 6):2–18. doi: 10.1111/j.1469-0691.2006.01580.x
74. Janezic S, Ocepek M, Zidaric V, Rupnik M. Clostridium difficile genotypes other than ribotype 078 that are prevalent among human, animal and environmental isolates. BMC Microbiol. (2012) 12:48–48. doi: 10.1186/1471-2180-12-48
75. Pirš T, Avberšek J, Zdovc I, Krt B, Andlovic A, Lejko-Zupanc T, et al. Antimicrobial susceptibility of animal and human isolates of Clostridium difficile by broth microdilution. J Med Microbiol. (2013) 62:1478–85. doi: 10.1099/jmm.0.058875-0
76. Carlson PE, Walk ST, Bourgis AET, Liu MW, Kopliku F, Lo E, et al. The relationship between phenotype, ribotype, and clinical disease in human Clostridium difficile isolates. Anaerobe. (2013) 24:109–16. doi: 10.1016/j.anaerobe.2013.04.003
77. Kamuju V, Kumar S, Khan WH, Vivekanandan P. Hypervirulent Clostridium difficile ribotypes are CpG depleted. Virulence. (2018) 9:1422–5. doi: 10.1080/21505594.2018.1509669
78. Kuijper EJ, Barbut F, Brazier JS, Kleinkauf N, Eckmanns T, Lambert ML, et al. Update of Clostridium difficile infection due to PCR ribotype 027 in Europe, 2008. Euro Surveill. 13:18942.
79. Murphy CP, Reid-Smith RJ, Boerlin P, Weese JS, Prescott JF, Janecko N, et al. Escherichia coli and selected veterinary and zoonotic pathogens isolated from environmental sites in companion animal veterinary hospitals in southern Ontario. Can Vet J. (2010) 51:963–72.
80. Keel K, Brazier JS, Post KW, Weese S, Songer JG. Prevalence of PCR ribotypes among Clostridium difficile isolates from pigs, calves, and other species. J Clin Microbiol. (2007) 45:1963–4. doi: 10.1128/JCM.00224-07
81. Wetterwik KJ, Trowald-Wigh G, Fernström LL, Krovacek K. Clostridium difficile in faeces from healthy dogs and dogs with diarrhea. Acta Vet Scand. (2013) 55:23. doi: 10.1186/1751-0147-55-23
82. Spigaglia P, Drigo I, Barbanti F, Mastrantonio P, Bano L, Bacchin C, et al. Antibiotic resistance patterns and PCR-ribotyping of Clostridium difficile strains isolated from swine and dogs in Italy. Anaerobe. (2015) 31:42–6. doi: 10.1016/j.anaerobe.2014.10.003
83. Boekhoud IM, Hornung BVH, Sevilla E, Harmanus C, Bos-Sanders IMJG, Terveer EM, et al. Plasmid-mediated metronidazole resistance in Clostridioides difficile. Nat Commun. (2020) 11:598. doi: 10.1038/s41467-020-14382-1
84. Medina-Torres CE, Weese JS, Staempfli HR. Prevalence of Clostridium difficile in horses. Vet Microbiol. (2011) 152:212–5. doi: 10.1016/j.vetmic.2011.04.012
85. Thean S, Elliott B, Riley TV. Clostridium difficile in horses in Australia–a preliminary study. J Med Microbiol. (2011) 60:1188–92. doi: 10.1099/jmm.0.030908-0
86. Rodriguez Diaz C, Seyboldt C, Rupnik M. Non-human difficile Reservoirs, C., and sources: animals, food, environment. Adv Exp Med Biol. (2018) 1050:227–43. doi: 10.1007/978-3-319-72799-8_13
87. Kecerova Z, Cizek A, Nyc O, Krutova M. Clostridium difficile isolates derived from Czech horses are resistant to enrofloxacin; cluster to clades 1 and 5 and ribotype 033 predominates. Anaerobe. (2019) 56:17–21. doi: 10.1016/j.anaerobe.2019.01.005
88. Keessen EC, Gaastra W, Lipman LJ. Clostridium difficile infection in humans and animals, differences and similarities. Vet Microbiol. (2011) 153:205–17. doi: 10.1016/j.vetmic.2011.03.020
89. O'Neill G, Adams JE, Bowman RA, Riley TV. A molecular characterization of Clostridium difficile isolates from humans, animals and their environments. Epidemiol Infect. (1993) 111:257–64. doi: 10.1017/S095026880005696X
90. Stoesser N, Eyre DW, Quan TP, Godwin H, Pill G, Mbuvi E, et al. Epidemiology of Clostridium difficile in infants in Oxfordshire, UK: Risk factors for colonization and carriage, and genetic overlap with regional C. difficile infection strains PLoS ONE. (2017) 12:e0182307. doi: 10.1371/journal.pone.0182307
91. Loo VG, Brassard P, Miller MA. Household transmission of Clostridium difficile to family members and domestic pets. Infect Control Hosp Epidemiol. (2016) 37:1342–8. doi: 10.1017/ice.2016.178
92. Lefebvre SL, Arroyo LG, Weese JS. Epidemic Clostridium difficile strain in hospital visitation dog. Emerging Infect Dis. (2006) 12:1036–7. doi: 10.3201/eid1206.060115
93. Lefebvre SL, Reid-Smith RJ, Waltner-Toews D, Weese JS. Incidence of acquisition of methicillin-resistant Staphylococcus aureus, Clostridium difficile, and other health-care-associated pathogens by dogs that participate in animal-assisted interventions. J Am Vet Med Assoc. (2009) 234:1404–17. doi: 10.2460/javma.234.11.1404
94. Lefebvre SL, Weese JS. Contamination of pet therapy dogs with MRSA and Clostridium difficile. J Hosp Infect England. (2009) 72:268–9. doi: 10.1016/j.jhin.2009.02.019
95. Rodriguez C, Taminiau B, Bouchafa L, Romijn S, Van Broeck J, Delmee M, et al. Clostridium difficile beyond stools: dog nasal discharge as a possible new vector of bacterial transmission. Heliyon. (2019) 5:e01629. doi: 10.1016/j.heliyon.2019.e01629
96. Orden C, Neila C, Blanco JL, Álvarez-Pérez S, Harmanus C, Kuijper EJ, et al. Recreational sandboxes for children and dogs can be a source of epidemic ribotypes of Clostridium difficile. Zoonoses Public Health. (2018) 65:88–95. doi: 10.1111/zph.12374
97. Janezic S, Mlakar S, Rupnik M. Dissemination of Clostridium difficile spores between environment and households: dog paws and shoes. Zoonoses Public Health. (2018) 65:669–74. doi: 10.1111/zph.12475
98. Kelly CP, LaMont JT. Clostridium difficile—more difficult than ever. N Engl J Med. (2008) 359:1932–40. doi: 10.1056/NEJMra0707500
99. Barbut F, Richard A, Hamadi K, Chomette V, Burghoffer B, Petit JC. Epidemiology of recurrences or reinfections of Clostridium difficile-associated diarrhea. J Clin Microbiol. (2000) 38:2386–8. doi: 10.1128/JCM.38.6.2386-2388.2000
100. Vohra P, Poxton IR. Comparison of toxin and spore production in clinically relevant strains of Clostridium difficile. Microbiology. (2011) 157:1343. doi: 10.1099/mic.0.046243-0
101. Shaughnessy MK, Bobr A, Kuskowski MA, Johnston BD, Sadowsky MJ, Khoruts A, et al. Environmental contamination in households of patients with recurrent clostridium difficile infection. Appl Environ Microbiol. (2016) 82:2686–92. doi: 10.1128/AEM.03888-15
102. Bomers MK, van Agtmael MA, Luik H, van Veen MC, Vandenbroucke-Grauls CM, Smulders YM. Using a dog's superior olfactory sensitivity to identify Clostridium difficile in stools and patients: proof of principle study. BMJ. (2012) 345:e7396. doi: 10.1136/bmj.e7396
103. Bomers MK, van Agtmael MA, Luik H, Vandenbroucke-Grauls CM, Smulders YM. A detection dog to identify patients with Clostridium difficile infection during a hospital outbreak. J Infect. (2014) 69:456–61. doi: 10.1016/j.jinf.2014.05.017
104. Bryce E, Zurberg T, Zurberg M, Shajari S, Roscoe D. Identifying environmental reservoirs of Clostridium difficile with a scent detection dog: preliminary evaluation. J Hosp Infect. (2017) 97:140–5. doi: 10.1016/j.jhin.2017.05.023
105. Redding LE, Kelly BJ, Stefanovski D, Lautenbach JK, Tolomeo P, Cressman L, et al. Pet ownership protects against recurrence of Clostridioides difficile infection. Open Forum Infect Dis. (2020) 7:ofz541. doi: 10.1093/ofid/ofz541
106. Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Bioinformatics. (1997) 13:555–6. doi: 10.1093/bioinformatics/13.5.555
107. Dingle KE, Elliott B, Robinson E, Griffiths D, Eyre DW, Stoesser N, et al. Evolutionary history of the Clostridium difficile pathogenicity locus. Genome Biol Evol. (2014) 6:36–52. doi: 10.1093/gbe/evt204
108. Janezic S, Potocnik M, Zidaric V, Rupnik M. Highly divergent Clostridium difficile strains isolated from the environment. PLoS ONE. (2016) 11:e0167101. doi: 10.1371/journal.pone.0167101
109. Elliott B, Androga GO, Knight DR, Riley TV. Clostridium difficile infection: Evolution, phylogeny and molecular epidemiology. Infect Genet Evol. (2017) 49:1–11. doi: 10.1016/j.meegid.2016.12.018
110. Griffiths D, Fawley W, Kachrimanidou M, Bowden R, Crook DW, Fung R, et al. Multilocus sequence typing of Clostridium difficile. J Clin Microbiol. (2010) 48:770–8. doi: 10.1128/JCM.01796-09
111. Stabler RA, He M, Dawson L, Martin M, Valiente E, Corton C, et al. Comparative genome and phenotypic analysis of Clostridium difficile 027 strains provides insight into the evolution of a hypervirulent bacterium. Genome Biol. (2009) 10:R102. doi: 10.1186/gb-2009-10-9-r102
112. Braun V, Hundsberger T, Leukel P, Sauerborn M, von Eichel-Streiber C. Definition of the single integration site of the pathogenicity locus in Clostridium difficile. Gene. (1996) 181:29–38. doi: 10.1016/S0378-1119(96)00398-8
113. Brouwer MSM, Roberts AP, Hussain H, Williams RJ, Allan E, Mullany P. Horizontal gene transfer converts non-toxigenic Clostridium difficile strains into toxin producers. Nat Commun. (2013) 4:2601. doi: 10.1038/ncomms3601
114. Kobayashi S, Wada A, Shibasaki S, Annaka M, Higuchi H, Adachi K, et al. Spread of a large plasmid carrying the cpe gene and the tcp locus amongst Clostridium perfringens isolates from nosocomial outbreaks and sporadic cases of gastroenteritis in a geriatric hospital. Epidemiol Infect. (2009) 137:108. doi: 10.1017/S0950268808000794
115. Spigaglia P, Mastrantonio P. Molecular analysis of the pathogenicity locus and polymorphism in the putative negative regulator of toxin production (TcdC) among Clostridium difficile clinical isolates. J Clin Microbiol. (2002) 40:3470–5. doi: 10.1128/JCM.40.9.3470-3475.2002
Keywords: companion animals, Clostridioides difficile, prevalence, molecular epidemiology, public health
Citation: Hernandez BG, Vinithakumari AA, Sponseller B, Tangudu C and Mooyottu S (2020) Prevalence, Colonization, Epidemiology, and Public Health Significance of Clostridioides difficile in Companion Animals. Front. Vet. Sci. 7:512551. doi: 10.3389/fvets.2020.512551
Received: 16 November 2019; Accepted: 14 August 2020;
Published: 18 September 2020.
Edited by:
Bernard J. Phiri, Ministry for Primary Industries, New ZealandReviewed by:
Patrizia Spigaglia, National Institute of Health (ISS), ItalyLaurence John Gleeson, LJG Consulting, Australia
Copyright © 2020 Hernandez, Vinithakumari, Sponseller, Tangudu and Mooyottu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shankumar Mooyottu, c2hhYW5AaWFzdGF0ZS5lZHU=; Chandra Tangudu, Y3Rhbmd1ZHVAaWFzdGF0ZS5lZHU=
 Belen G. Hernandez
Belen G. Hernandez Akhil A. Vinithakumari
Akhil A. Vinithakumari Brett Sponseller
Brett Sponseller Chandra Tangudu
Chandra Tangudu Shankumar Mooyottu
Shankumar Mooyottu